Abstract
Objective
To identify underlying genetic causes in patients with pediatric movement disorders by genetic investigations.
Methods
All patients with a movement disorder seen in a single Pediatric Genetic Movement Disorder Clinic were included in this retrospective cohort study. We reviewed electronic patient charts for clinical, neuroimaging, biochemical, and molecular genetic features. DNA samples were used for targeted direct sequencing, targeted next-generation sequencing, or whole exome sequencing.
Results
There were 51 patients in the Pediatric Genetic Movement Disorder Clinic. Twenty-five patients had dystonia, 27 patients had ataxia, 7 patients had chorea-athetosis, 8 patients had tremor, and 7 patients had hyperkinetic movements. A genetic diagnosis was confirmed in 26 patients, including in 20 patients with ataxia and 6 patients with dystonia. Targeted next-generation sequencing panels confirmed a genetic diagnosis in 9 patients, and whole exome sequencing identified a genetic diagnosis in 14 patients.
Conclusions
We report a genetic diagnosis in 26 (51%) patients with pediatric movement disorders seen in a single Pediatric Genetic Movement Disorder Clinic. A genetic diagnosis provided either disease-specific treatment or effected management in 10 patients with a genetic diagnosis, highlighting the importance of early and specific diagnosis.
Pediatric movement disorders are usually part of complex neurodevelopmental disorders and are associated with global developmental delay, cognitive dysfunction, behavioral disorders, and/or epilepsy. Pediatric movement disorders are classified into 2 categories, including hyperkinetic and hypokinetic movements.1,2 Pediatric hyperkinetic movements, defined as unwanted and excess movements, include dystonia, chorea, athetosis, myoclonus, tremor, tics, and stereotypies based on the definition and classification of The Taskforce on Childhood Movement Disorders.1 Pediatric hypokinetic movements, described as a decrease in the number of movements, is called hypokinetic-rigid syndrome or parkinsonism.2 The underlying etiology of pediatric movement disorders can be acquired or hereditary. Management of movement disorders requires multidisciplinary approach ranging from physiotherapy, pharmacologic treatment, to deep brain stimulation.3–5
Recent advances in molecular genetic investigations, including targeted next-generation sequencing panels for dystonia or parkinsonism and whole exome and genome sequencing research studies, have discovered various novel genes causing pediatric movement disorders as part of neurodevelopmental disorders. Recently, the Movement Disorder Society Task Force for Nomenclature of Genetic Movement Disorders made recommendations for nomenclature of genetic movement disorders.6
We aim to determine prevalence of genetic diagnoses in patients with pediatric movement disorders using genetic investigations in this retrospective cohort study. We also aim to identify disease-specific treatment or management effects based on the patients' genetic diagnoses in pediatric movement disorders.
Methods
This retrospective cohort study was performed in a single Pediatric Genetic Movement Disorder Clinic at an academic pediatric health science center from January 1, 2014 through December, 2016. Inclusion criteria were as follows: (1) one of the pediatric movement disorders; (2) referred or being followed in this Pediatric Genetic Movement Disorder Clinic for investigations of underlying genetic causes (referrals from pediatric neurology, pediatric epilepsy, developmental pediatrics, and genetic clinics).
DNA samples of all patients were used for genetic investigations, depending on their phenotype, including targeted direct sequencing, targeted next-generation sequencing panel of dystonia, of epilepsy, and of cellular energetic, and mitochondrial genome or whole exome sequencing (always performed as trio, patient and both parents) according to various clinical molecular genetics laboratories' methods. All variants reported from the clinical laboratories were annotated using the Alamut database for predictions of pathogenicity, cross-species conservation of nucleotides and amino acid sequences. We used the recommendations from mutation nomenclature (hgvs.org/mutomen) to name variants. American College of Medical Genetics and Genomics guidelines for variant classification for the molecular genetic result were used for interpretation.7
We reviewed electronic patient charts for types of pediatric movement disorders, clinical features accompanying pediatric movement disorders, neuroimaging features, and biochemical investigations. We entered all information into an Excel database. We reported treatment outcome and management implications of a genetic diagnosis in patients with pediatric movement disorders.
Standard protocol approvals, registrations, and patient consents
This study was approved by the Research Ethics Board at The Hospital for Sick Children (Approval#1000054997).
Results
There were 51 patients seen in this Pediatric Genetic Movement Disorder Clinic and included in this study. Demographics, clinical features, and genetic diagnosis of all patients are listed in table 1. Pediatric movement disorders included dystonia in 25 patients (49%) (generalized in 21 patients, focal in 3 patients, and hemidystonia in 1 patient), ataxia in 27 patients (53%), chorea-athetosis in 7 patients (13%), tremor in 8 patients (16%), and hyperkinetic movements in 7 patients (13%). Twenty-three patients (45%) had more than one movement disorder in combination. Global developmental delay or cognitive dysfunction was present in 39 patients (76%). History of developmental regression was reported in 4 patients (8%). Epilepsy was present in 21 patients (41%).
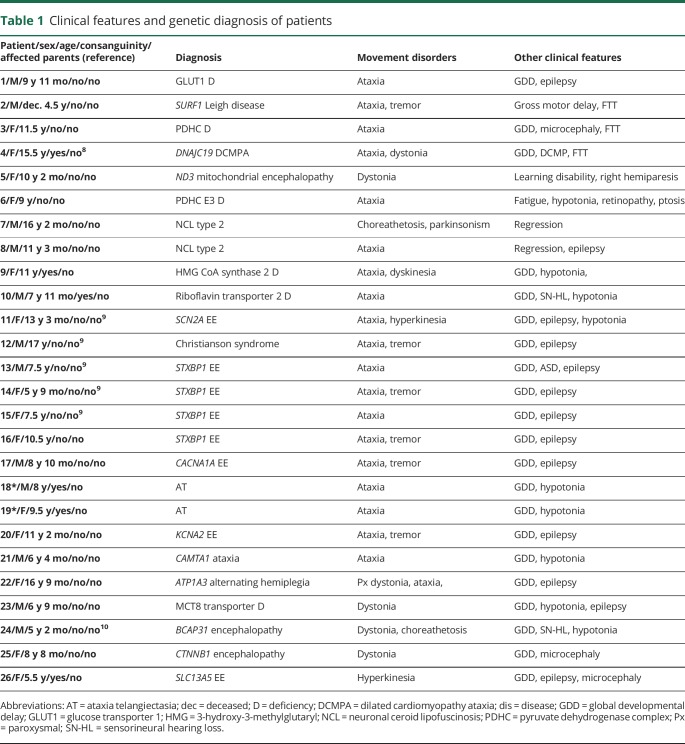
Table 1.
Clinical features and genetic diagnosis of patients
There was an identifiable underlying genetic cause in 26 patients (51%). The most common movement disorder was ataxia (53%) and a genetic diagnosis was confirmed in 74% of those patients. The second most common movement disorder was dystonia (49%), and a genetic diagnosis was confirmed in 24% of those patients. In 2 siblings, based on their phenotype and history of consanguinity in their parents, we suspected ataxia telangiectasia and confirmed the diagnosis by direct sequencing of ATM. In the remaining 24 patients, targeted next-generation sequencing panels (5 patients for epilepsy; 3 patients for cellular energetic; 1 patient for Leigh disease) or whole exome sequencing (n = 14) or mitochondrial genome sequencing (n = 1) confirmed the genetic diagnoses. Ten patients (38%) had autosomal recessive, 11 patients (42%) had autosomal dominant de novo, 4 patients had X-linked inheritance pattern, and 1 patient had mitochondrial inheritance pattern. We identified 26 variants in 21 genes in 26 patients, including 14 novel and 12 known likely pathogenic variants.8–16 None of the novel variants were found in The Single Nucleotide Polymorphism Database (dbSNP) as polymorphisms. The novel variants were moderately or highly conserved across species and reported to be disease causing in Mutation Taster and/or deleterious in Sorting Intolerant From Tolerant prediction programs. In silico analysis results of all variants are listed in supplemental e-table (table e-1, links.lww.com/NXG/A65).
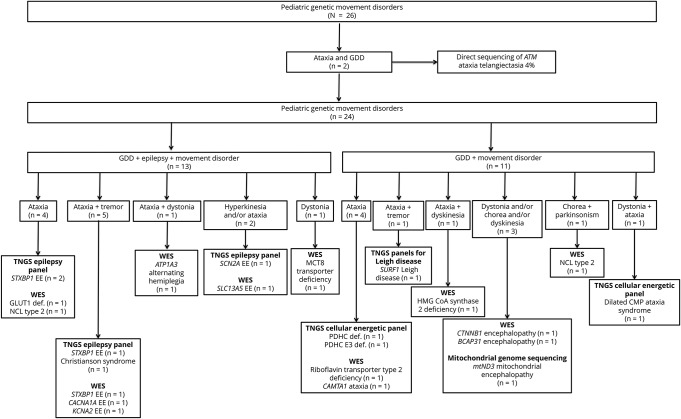
We summarized all genetic causes of pediatric movement disorders and the type of their confirmatory genetic test result in figure 1. Ten patients had one of the inherited metabolic disorders, including glucose transporter 1 (GLUT1) deficiency (n = 1), neuronal ceroid lipofuscinosis type 2 (n = 2), SURF1-associated Leigh disease (n = 1), X-linked pyruvate dehydrogenase complex (PDHC) deficiency (n = 1), dilated cardiomyopathy ataxia syndrome (3-methylglutaconic aciduria) due to homozygous DNAJC19 likely pathogenic variant (n = 1),8 mitochondrial encephalopathy due to mitochondrial ND3 likely pathogenic variant (n = 1), riboflavin transporter type 2 deficiency by homozygous likely pathogenic variant in SLC52A2 (n = 1), mitochondrial 3-hydroxy-3-methylglutaryl (HMG) CoA synthase 2 deficiency (n = 1), PDHC E3 deficiency (n = 1). In the remaining 16 patients, one of the neurogenetic disorders were identified, including STXBP1-associated epileptic encephalopathy (n = 4) (3 patients were reported previously9), SCN2A-associated epileptic encephalopathy (n = 1),9 SLC9A6-associated intellectual disability (Christianson syndrome) (n = 1),9 CACNA1A-associated epileptic encephalopathy (n = 1), ataxia telangiectasia (n = 2), KCNA2-associated epileptic encephalopathy (n = 1), CAMTA1-associated cerebellar ataxia (n = 1), ATP1A3-associated alternating hemiplegia and dystonia (n = 1), Allan-Herndon-Dudley syndrome (MCT8-specific thyroid hormone cell membrane transporter deficiency; n = 1), BCAP31-associated encephalopathy, deafness, dystonia, and cerebral hypomyelination syndrome (n = 1),10 CTNNB1-associated encephalopathy (n = 1), and SLC13A5-associated epileptic encephalopathy (n = 1).
Figure 1. All patients with pediatric genetic movement disorders, their genetic diagnoses, and type of genetic investigations.
Abbreviations: CMP = cardiomyopathy; EE = epileptic encephalopathy; GDD = global developmental delay; NCL = neuronal ceroid lipofuscinosis; TNGS = targeted next generation sequencing; WES = whole exome sequencing.
In one patient with GLUT1 deficiency, despite seizure onset in the first year of life, whole exome sequencing confirmed the diagnosis at the age of 7 years 8 months. One patient with neuronal ceroid lipofuscinosis type 2 presented with seizures and slowly progressive developmental regression from the age of 5 years. His seizures were well controlled with levetiracetam, which was discontinued 3 years later with no seizure recurrence. He was diagnosed by whole exome sequencing at the age of 11 years. In one patient with STXBP1-associated epileptic encephalopathy and in one patient with CACNA1A-associated epileptic encephalopathy, whole exome sequencing was requested due to ataxia and tremor in addition to seizures and global developmental delay and confirmed the diagnosis at the age of 9.5 and 8 years, respectively. One patient with SLC13A5-associated epileptic encephalopathy underwent targeted next-generation sequencing panel for epilepsy, which did not have this gene included at the time of the genetic test. Currently, some of the commercially available targeted next-generation sequencing panels for epilepsy include all those 5 genes and would have been identified those genetic disorders without application of whole exome sequencing.
All patients underwent metabolic investigations, including plasma amino acids, acylcarnitine profile, total and free carnitine, homocysteine, urine organic acids, or urine guanidinoacetate and creatine to creatinine ratio (only in patients with global developmental delay or cognitive dysfunction with no brain magnetic resonance spectroscopy). Eighteen patients (35% of all patients) underwent CSF metabolite measurements. There was no suspected diagnosis in any of those patients. In two patients with neuronal ceroid lipofuscinosis type 2, blood dot spot tripeptidyl peptidase 1 activity, a noninvasive biochemical investigation, was measured after the genetic diagnosis of neuronal ceroid lipofuscinosis type 2.
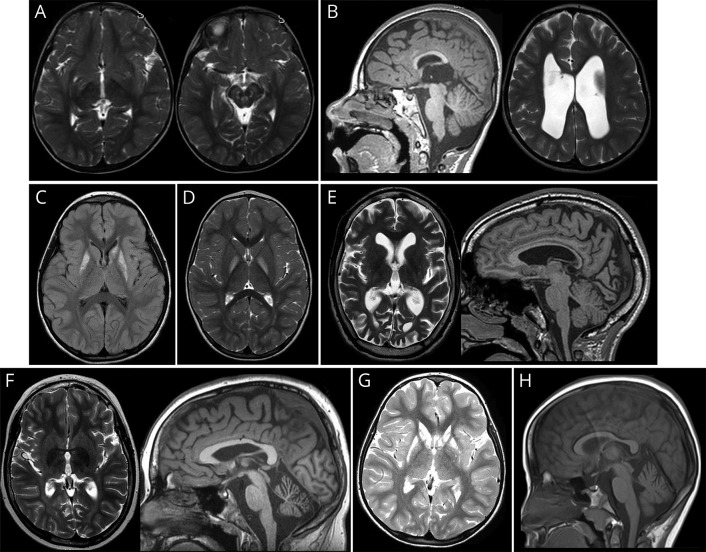
Brain MRI was abnormal in 20 of 24 patients with a genetic diagnosis (2 patients had no brain MRI). Abnormal brain MRI of 8 patients is demonstrated in figure 2. Abnormal brain MRI of 2 patients (patient 48 and patient 2410) were reported previously.
Figure 2. Specific brain MRI findings of 8 patients.
(A) Brain MRI of patient 2 with Leigh disease shows increased T2 signal in the subthalamic nuclei and brain stem in axial image at the age of 3 years. (B) Brain MRI of patient 3 with PDHC deficiency shows dysgenesis of corpus callosum and in T1 sagittal image and dilated ventricles in T2 axial image at the age of 4 years. (C) Brain MRI of patient 5 with mitochondrial encephalopathy shows increased fluid-attenuated inversion recovery signal in bilateral putamen and left caudate head and body at the age of 6.5 years. (D) Brain MRI of patient 6 with PDHC E3 deficiency shows increased T2 signal in bilateral globus pallidi in axial image at the age of 3.5 years. (E) Brain MRI of patient 7 with neuronal ceroid lipofuscinosis type 2 shows increased T2 signal in cerebral white matter, cerebral atrophy and small thalami in axial image, and thin corpus callosum in T1 sagittal image at the age of 13 years. (F) Brain MRI of patient 8 with neuronal ceroid lipofuscinosis type 2 shows increased T2 white matter signal intensity and decreased T2 thalami signal intensity in axial image and cerebellar atrophy in T1 sagittal image at the age of 11 years. (G) Brain MRI of patient 9 with HMG CoA synthase 2 deficiency shows symmetrical increased signal intensity in putamen and caudate nucleus in T2 axial image at the age of 5 years. (H) Brain MRI of patient 20 with KCNA2-associated epileptic encephalopathy shows cerebellar atrophy in T1 sagittal image at the age of 10 years. Abbreviations: HMG = 3-hydroxy-3-methylglutaryl; PDHC = pyruvate dehydrogenase complex.
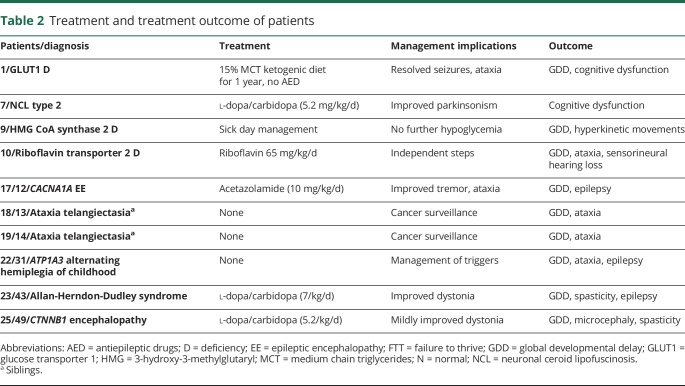
Treatment outcome of patients with genetic pediatric movement disorders are listed in table 2. There were 10 patients (38% of patients with a genetic diagnosis and 20% of all patients) with disease-specific or symptomatic treatment due to their confirmed genetic diagnosis. Because of late diagnosis and/or compliance problems, patients with GLUT1 (n = 1) and riboflavin transporter type 2 (n = 1) deficiencies did not achieve normal neurodevelopmental outcome at the time of this study. A genetic diagnosis guided symptomatic medical treatment in 4 patients (2 with neuronal ceroid lipofuscinosis type 2; 1 with CACNA1A-associated epileptic encephalopathy, and 1 with Allan-Herndon-Dudley syndrome [MCT8-specific thyroid hormone cell membrane transporter deficiency]). A genetic diagnosis guided preventative or sick day management or cancer screening for early detection in 4 patients (in HMG CoA synthase 2 deficiency, ATP1A3-associated alternating hemiplegia of childhood and ataxia telangiectasia).
Table 2.
Treatment and treatment outcome of patients
In the remaining 25 patients with no genetic diagnosis, 18 patients had targeted next-generation sequencing panel for dystonia, 1 patient had mitochondrial genome sequencing, and 10 patients had whole exome sequencing. Eight patients underwent both dystonia panel and whole exome sequencing. There was no genetic diagnosis in any of those patients.
Discussion
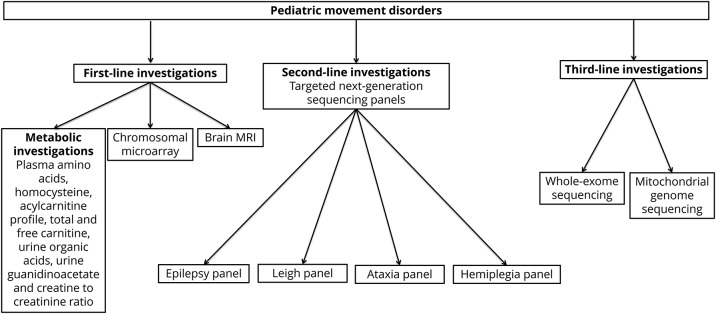
Our retrospective cohort study reports 51% prevalence of genetic diagnoses in patients with pediatric movement disorders from a single Pediatric Genetic Movement Disorder Clinic. There were 21 different genetic disorders combining inherited metabolic disorders (20% of all patients) and neurogenetic disorders (31% of all patients) as underlying genetic causes of pediatric movement disorders. Only 4% of the patients were diagnosed by targeted direct sequencing based on a clinical suspicion of ataxia telangiectasia. Targeted next-generation sequencing panels, including epilepsy, cellular energetic, and Leigh disease, confirmed a genetic diagnosis in 18% of the patients (9/51). Whole exome sequencing identified a genetic diagnosis in 27% of the patients (14/51). Mitochondrial genome sequencing confirmed a genetic diagnosis in 2% of the patients (1/51). Biochemical investigations did not guide diagnoses in any of the patients. The most common inheritance pattern was autosomal dominant or X-linked de novo inheritance. In pediatric movement disorders, we would recommend investigations in a stepwise approach for diagnostic workup and depicted this approach in figure 3. Whole exome sequencing does not capture some of the intragenic rearrangements, whereas targeted next-generation sequencing panels are designed with additional Sanger sequencing fill-ins or deletion or duplication tests to confirm genetic diagnosis as reported recently.17 A genetic diagnosis provided either disease-specific or symptomatic treatment in 38% of the patients with pediatric movement disorders. Our study highlights the importance of genetic investigations and effects of genetic diagnosis on the treatment and management of patients with pediatric movement disorders.
Figure 3. Algorithm for diagnostic workup in pediatric movement disorders.
To the best of our knowledge, there are 2 studies using targeted next-generation or whole exome sequencing to investigate underlying genetic causes of movement disorders in combined pediatric and adult patient populations.18,19 In the first study, 61 patients with childhood and adult-onset dystonia (age range, 1–73 years) were investigated for underlying genetic causes using targeted next-generation sequencing dystonia panel of 94 genes. In that study, 9 of 61 patients (14.8%) had a genetic diagnosis, including PRKRA-associated dystonia, tyrosine hydroxylase deficiency, glutaric aciduria type I, Niemann-Pick type C, PRRT2-associated paroxysmal kinesigenic dyskinesia (three patients), and Rett syndrome (two patients).18 In the second study, 50 patients with movement disorders underwent whole exome sequencing, which targeted 151 genes, and the diagnostic yield was 20%. The type of movement disorders included hereditary spastic paraplegia (58.8%), cerebellar ataxia (23.5%), and dystonia (17.6%). Eighty percent of the patients with a genetic diagnosis had hereditary spastic paraplegia.19 The diagnostic yield of genetic investigations was higher in our retrospective cohort study compared with the previous studies despite similar number of patients and genetic investigations.
In our retrospective cohort study, 41% of the patients had epilepsy in addition to pediatric movement disorder. We confirmed a genetic diagnosis in 62% of these patients (13/21). In a study, 400 patients with early-onset seizure disorders and/or severe developmental delay were investigated by targeted next-generation sequencing of 46 epilepsy genes. Seventy-one patients (18%) had a confirmed genetic diagnosis. Movement disorders were reported in 4 patients (6%) with SCN2A-, SCN8A-, and FOXG1-associated epilepsy.15 We think that we will see increasing number of reports for overlapping phenotypes of pediatric movement disorders and epilepsy in the future because of application of genetic investigations and identification of variable phenotypes for the same genotype.
Diagnostic yield of chromosomal microarray was reported in 25 patients with pediatric movement disorders, including dystonia (n = 10), paroxysmal kinesigenic dyskinesia (n = 5), tremor (n = 4), chorea (n = 3), myoclonus (n = 2), and paroxysmal non-kinesigenic dyskinesia (n = 1). Seven of 25 patients were diagnosed with a microdeletion (size ranged 160 kb to 3.27 Mb) as the potential cause of movement disorders. In 3 patients, microdeletion was inherited from an affected parent.20 Of interest, chromosomal microarray was applied to 67% of the patients in our study cohort, and none of the patients had any likely pathogenic copy number variants. Despite the higher number of patients who underwent chromosomal microarray in our study compared with previous study, none of the patients with likely pathogenic copy number variants in chromosomal microarray raises the question of whether chromosomal microarray should be used as a first-line investigation in all patients with pediatric movement disorders. We would recommend applying chromosomal microarray to patients with dysmorphic features, developmental disability, epilepsy, and movement disorders as complex neurodevelopmental disorders.
Of interest, we report 2 new patients with attenuated neuronal ceroid lipofuscinosis type 2, who presented with childhood-onset developmental regression and movement disorder with or without seizures. Both patients were diagnosed more than 5 years after their initial presentation by whole exome sequencing. So far, less than 20 patients have been reported in the literature presenting with attenuated or juvenile onset neuronal ceroid lipofuscinosis type 2.21–25 Pediatric movement disorders varied from isolated childhood onset progressive ataxia22 to dystonia-parkinsonism.23 We think that history of developmental regression and progressive movement disorder with or without seizures should prompt direct sequencing of TPP1 or measurement of tripeptidyl peptidase 1 activity. Recently, intracerebroventricular enzyme replacement therapy has been approved for the treatment of neuronal ceroid lipofuscinosis type 2.25 Early diagnosis and initiation of this treatment will likely improve neurodevelopmental outcome of patients with neuronal ceroid lipofuscinosis type 2.
In our retrospective cohort study, basal ganglia or gray matter abnormalities with or without white matter changes were present in 67% of the patients (6/9) with inherited metabolic disorders. Whereas, 40% of the patients with neurogenetic disorders had nonspecific white matter changes or delayed myelination. We think that, in patients with basal ganglia or gray matter changes in brain MRI, inherited metabolic disorders should be in the differential diagnosis.
Despite high prevalence of genetic diagnosis in our retrospective cohort study, we have some limitations, including: (1) our clinic is not a referral clinic for patients with isolated dystonia, chorea, myoclonus, tics, tremor, stereotypic movements, or transient movement disorders; (2) whole exome sequencing was not a first-line genetic testing because of limited resources, despite the majority of the patients underwent whole exome sequencing; (3) we will not be able to know, if the diagnostic yield would have been higher, if all patients would have undergone whole exome sequencing as first-line genetic testing; (4) we will not be able to know, if the diagnostic yield would have been lower, if all patients with various movement disorders would have been investigated for genetic causes. Despite all these limitations, we think that the number of patients and genetic diagnosis confirmed in our study population is still valuable to draw conclusions regarding the diagnostic yield of targeted next-generation sequencing or whole exome sequencing in pediatric movement disorders.
Our retrospective cohort study reports high prevalence of genetic diagnoses and 21 different genetic disorders showing genetic landscape of pediatric movement disorders. Targeted next-generation sequencing panels and whole exome sequencing increased diagnostic yield to more than 40% in our study. A genetic diagnosis provided either disease-specific or symptomatic therapy in 38% of the patients with a genetic diagnosis, highlighting the importance of genetic investigations to confirm underlying genetic cause in patients with pediatric movement disorders.
Acknowledgment
The authors would like to thank Dr. Michal Inbar-Feigenberg, Dr. Julian Raiman, Dr. Robert Munn, Dr. Sandra Farrell, and Dr. Cynthia Forster-Gibson for referring their patients. The authors would like to thank all staff physicians, including general pediatrics, neurology, and clinical genetics at The Hospital for Sick Children who referred their patients for diagnostic workup to our clinic. The authors would like to thank Stacy Hewson for genetic counseling for the genetic test results. The authors would like to thank Ashley Wilson for her work on the Research Ethics Board application for this study. The authors would like to thank Khadine Wiltshire for formatting this manuscript according to journal's requirements.
Glossary
- GLUT1
glucose transporter 1
- HMG
3-hydroxy-3-methylglutaryl
- MCT
medium chain triglyceride
- PDHC
pyruvate dehydrogenase complex
Author contributions
D. Cordeiro: data analysis, arrangement of clinical genetic tests, drafting the manuscript, and critique of the final version of the manuscript. G. Bullivant: application of in silico tools to all variants in various genes using database, review and critique of the final version of the manuscript. K. Siriwardena, A. Evans, J. Kobayashi, R. D. Cohn: review and critique of the final version of the manuscript. S. Mercimek-Andrews: design, execution, organization and conceptualization of the study, analysis and interpretation of the data, and drafting and finalizing the manuscript for intellectual content.
Study funding
No targeted funding reported.
Disclosure
K. Siriwardena has received funding for travel/speaker honoraria from Biomarin; has received research support from Biomarin. R. D. Cohn has received research funding from the NIH and the Muscular Dystrophy Association. S. Mercimek-Andrews has served on scientific advisory boards of The Hospital for Sick Children, Biomarin, Cost Effectiveness of ERT, and the Homocysteine and Betaine Treatment for Recordati Rare Diseases Canada; has received funding for travel/speaker honoraria from Biomarin; serves on the editorial board for the Journal of Pediatric Genetics; has received industry funding for work on the prevalence of mucopolysaccharidosis in rheumatology and nerve conduction velocity study patients. D. Cordeiro, G. Bullivant, A. Evans, and J. Kobayashi report no disclosures. Full disclosure form information provided by the authors is available with the full text of this article at Neurology.org/NG.
References
- 1.Sanger TD, Chen D, Fehlings DL, et al. . Definition and classification of hyperkinetic movements in childhood. Mov Disord 2010;25:1538–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia-Cazorla A, Duarte ST. Parkinsonism and inborn errors of metabolism. J Inherit Metab Dis 2014;37:627–642. [DOI] [PubMed] [Google Scholar]
- 3.Kurian MA, Dale RC. Movement disorders presenting in childhood. Continuum (Minneap Minn) 2016;22:1159–1185. [DOI] [PubMed] [Google Scholar]
- 4.Koy A, Lin JP, Sanger TD, Marks WA, Mink JW, Timmermann L. Advances in management of movement disorders in children. Lancet Neurol 2016;15:719–735. [DOI] [PubMed] [Google Scholar]
- 5.Lumsden DE, Kaminska M, Ashkan K, Selway R, Lin JP. Deep brain stimulation for childhood dystonia: is “where” as important as in “whom”? Eur J Paediatr Neurol 2017;21:176–184. [DOI] [PubMed] [Google Scholar]
- 6.Marras C, Lang A, van de Warrenburg BP, et al. . Nomenclature of genetic movement disorders: recommendations of the international Parkinson and movement disorder society task force. Mov Disord 2016;31:436–457. [DOI] [PubMed] [Google Scholar]
- 7.Richards S, Aziz N, Bale S, et al. . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of medical genetics and genomics and the association for molecular pathology. Genet Med 2015;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al Teneiji A, Siriwardena K, George K, Mital S, Mercimek-Mahmutoglu S. Progressive cerebellar atrophy and a novel homozygous pathogenic DNAJC19 variant as a cause of dilated cardiomyopathy ataxia syndrome. Pediatr Neurol 2016;62:58–61. [DOI] [PubMed] [Google Scholar]
- 9.Mercimek-Mahmutoglu S, Patel J, Cordeiro D, et al. . Diagnostic yield of genetic testing in epileptic encephalopathy in childhood. Epilepsia 2015;56:707–716. [DOI] [PubMed] [Google Scholar]
- 10.Albanyan S, Al Teneiji A, Monfared N, Mercimek-Mahmutoglu S. BCAP31-associated encephalopathy and complex movement disorder mimicking mitochondrial encephalopathy. Am J Med Genet A 2017;173:1640–1643. [DOI] [PubMed] [Google Scholar]
- 11.Lee IC, El-Hattab AW, Wang J, Li FY, et al. . SURF1-associated Leigh syndrome: a case series and novel mutations. Hum Mutat 2012;33:1192–1200. [DOI] [PubMed] [Google Scholar]
- 12.Imbard A, Boutron A, Vequaud C, et al. . Molecular characterization of 82 patients with pyruvate dehydrogenase complex deficiency. Structural implications of novel amino acid substitutions in E1 protein. Mol Genet Metab 2011;104:507–516. [DOI] [PubMed] [Google Scholar]
- 13.Taylor RW, Singh-Kler R, Hayes CM, Smith PE, Turnbull DM. Progressive mitochondrial disease resulting from a novel missense mutation in the mitochondrial DNA ND3 gene. Ann Neurol 2001;50:104–107. [DOI] [PubMed] [Google Scholar]
- 14.Shaag A, Saada A, Berger I, et al. . Molecular basis of lipoamide dehydrogenase deficiency in Ashkenazi Jews. Am J Med Genet 1999;82:177–182. [PubMed] [Google Scholar]
- 15.Trump N, McTague A, Brittain H, et al. . Improving diagnosis and broadening the phenotypes in early-onset seizure and severe developmental delay disorders through gene panel analysis. J Med Genet 2016;53:310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Syrbe S, Hedrich UBS, Riesch E, et al. . De novo loss- or gain-of-function mutations in KCNA2 cause epileptic encephalopathy. Nat Genet 2015;47:393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breen DP, Mercimek-Andrews S, Lang AE. Infantile-onset hand dystonia with intellectual disability: clues to ARX mutations. Neurology 2018;90:333–335. [DOI] [PubMed] [Google Scholar]
- 18.van Egmond ME, Lugtenberg CHA, Brouwer OF, et al. . A post hoc study on gene panel analysis for the diagnosis of dystonia. Mov Disord 2017;32:569–575. [DOI] [PubMed] [Google Scholar]
- 19.Neveling K, Feenstra I, Gilissen C, et al. . A post-hoc comparison of the utility of sanger sequencing and exome sequencing for the diagnosis of heterogeneous diseases. Hum Mutat 2013;34:1721–1726. [DOI] [PubMed] [Google Scholar]
- 20.Dale RC, Grattan-Smith P, Nicholson M, Peters GB. Microdeletions detected using chromosome microarray in children with suspected genetic movement disorders: a single-centre study. Dev Med Child Neurol 2012;54:618–623. [DOI] [PubMed] [Google Scholar]
- 21.Saini AG, Sankhyan N, Singhi P. Chorea in late-infantile neuronal ceroid lipofuscinosis: an Atypical presentation. Pediatr Neurol 2016;60:75–78. [DOI] [PubMed] [Google Scholar]
- 22.Dy ME, Sims KB, Friedman J. TPP1 deficiency: rare cause of isolated childhood-onset progressive ataxia. Neurology 2015;85:1259–1261. [DOI] [PubMed] [Google Scholar]
- 23.Di Giacopo R, Cianetti L, Caputo V, et al. . Protracted late infantile ceroid lipofuscinosis due to TPP1 mutations: clinical, molecular and biochemical characterization in three sibs. J Neurol Sci 2015;356:65–71. [DOI] [PubMed] [Google Scholar]
- 24.Kousi M, Lehesjoki AE, Mole SE. Update of the mutation spectrum and clinical correlations of over 360 mutations in eight genes that underlie the neuronal ceroid lipofuscinoses. Hum Mutat 2012;33:42–63. [DOI] [PubMed] [Google Scholar]
- 25.Elleder M, Dvoráková L, Stolnaja L, et al. . Atypical CLN2 with later onset and prolonged course: a neuropathologic study showing different sensitivity of neuronal subpopulations to TPP1 deficiency. Acta Neuropathol 2008;116:119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]