Abstract
Background
Prostate Imaging-Reporting and Data System version 2 (PI-RADSv2) provides standardized nomenclature for interpretation of prostate multiparametric MRI (mpMRI). Inclusion of additional features for categorization may provide benefit to stratification of disease.
Purpose
To prospectively compare PI-RADSv2 to a qualitative in-house system for detecting prostate cancer on mpMRI.
Study Type
Prospective
Population
338 patients who underwent mpMRI May 2015–May 2016, with subsequent MRI/transrectal ultrasound fusion-guided biopsy.
Field Strength
3T mpMRI [T2W, DWI (ADC map, b-2000 DWI acquisition), and DCE MRI].
Assessment
One genitourinary radiologist prospectively read mpMRIs using both in-house and PI-RADSv2 5-category systems.
Statistical Test
In lesion-based analysis, overall and clinically significant (CS) tumor detection rates (TDR) were calculated for all PI-RADSv2 and in-house categories. Ability of each scoring system to detect cancer was assessed by area under receiver operator characteristic curve (AUC). Within each PI-RADSv2 category, lesions were further stratified by their in-house categories to determine if TDRs can be increased by combining features of both systems.
Results
In 338 patients (median PSA 6.5[0.6–113.6] ng/mL; age 64[44–84] years), 733 lesions were identified (47% tumor-positive). Predictive abilities of both systems were comparable for all (AUC 76–78%) and CS cancers (AUCs 79%). The in-house system had higher overall and CS TDRs than PI-RADSv2 for categories 3 and 4 (p<0.01 for both), with the greatest difference between the scoring systems seen in lesions scored category 4 (CS TDRs: in-house 65%, PI-RADSv2 22.1%). For lesions categorized as PI-RADSv2=4, characterization of suspicious/indeterminate EPE and equivocal findings across all mpMRI sequences contributed to significantly different TDRs for both systems (TDR range 19–75%, p<0.05).
Data Conclusion
PI-RADSv2 behaves similarly to an existing validated system which relies on the number of sequences on which a lesion is seen. This prospective evaluation suggests that sequence positivity and suspicion of EPE can enhance PI-RADSv2 category 4 cancer detection.
Keywords: prostate, cancer, mpMRI, PI-RADSv2, detection
INTRODUCTION
As multiparametric MRI (mpMRI) has become more widespread, there is an increasing demand for standardization of image acquisition and interpretation (1–3). The Prostate Imaging-Reporting and Data System version 2 (PI-RADSv2) utilizes a dominant sequence strategy in each zone, with a final lesion-based category assignment ranging from 1–5 representing low to high likelihood of cancer (5). In the peripheral zone (PZ) diffusion-weighted imaging (DWI) is considered dominant and there is secondary assistance from dynamic contrast-enhanced (DCE) MRI. In the transition zone (TZ) T2-weighted (T2W) imaging is dominant with limited support from DWI characteristics. These recommendations are largely based on expert consensus and data supporting zone-specific dominant sequence differentiation is limited (6, 7). Furthermore, the subjective nature of interpretation within the PI-RADSv2 lexicon may contribute to high inter-reader variability reported in early validation studies (8, 9).
For several years prior to PI-RADSv2 we had successfully used a simple in-house scoring system for prostate MRI which was mostly based on the number of sequences on mpMRI that were positive. Recognizing the benefits of using a standardized nomenclature and diagnostic criteria we rapidly adopted PI-RADSv2 but continued using the in-house scoring system to ease the transition with our referring clinicians who had less familiarity with the new system. Our purpose is to prospectively compare the performance of these two systems in the hopes of finding features in the older system that could be used to improve the performance of PI-RADS v2 as we consider the next version of this scoring system (8, 11–13).
MATERIALS AND METHODS
Study Population
This single-institution prospective Health Insurance Portability and Accountability Act-compliant study was approved by the local institutional review board. All patients gave written informed consent before undergoing mpMRI [T2W, DWI (ADC map, b-2000 DWI acquisition), and DCE MRI] at 3-Tesla (3T). PI-RADSv2 became available in January of 2015. Beginning May 2015 thru May 2016, 963 consecutive patients underwent mpMRI at our institution. Of these patients, 364 underwent MRI/transrectal ultrasound (TRUS) fusion-guided biopsy. Inclusion criteria were having mpMRI at 3T with subsequent MRI/TRUS fusion biopsy. Patients were excluded for not having subsequent MRI/TRUS fusion guided biopsy (n=599), for receiving a prior treatment (n=25) or for missing prospective categorization with the in-house system (n=1); 338 patients were included in the final study population. The patient population in this study is part of a large prospective PI-RADSv2 validation study population. Previous reports on this group include establishing the correlation of PI-RADSv2 scoring system with the International Society of Urological Pathology (ISUP) Prostate Cancer Grade Group System (14). Here we report a prospective comparison of PI-RADSv2 and an in-house method of MRI interpretation.
MRI Technique
mpMRIs were obtained at 3-Tesla (Achieva 3.0T-TX, Phillips Healthcare, Best, Netherlands) either using an endorectal coil (BPX-30, Medrad, Pittsburgh, PA, USA) with anterior half of a 32-channel cardiac SENSE coil (InVivo, Gainesville, FL, USA), or using only the 32-channel cardiac SENSE coil. The endorectal coil was filled with 45 mL Fluorinert (3M, Maplewood, MN, USA), and was utilized in the majority of the patients. The mpMRI protocol included T2W (axial, coronal, sagittal), DWI (up to b-2000 mm/sec2 with endorectal coil, up to b-1500 mm/sec2 without endorectal coil) available as an ADC map and high b-value sequence, and DCE MRI and is summarized in Table 1.
Table 1.
Multiparametric MRI acquisition parameters given for prostate imaging at 3-Tesla with and without use of an endorectal coil.
| Multiparametric MR Imaging Sequence Parameters at 3T | ||||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| With endorectal coil | Surface coil only | |||||||
|
| ||||||||
| Parameter | T2 Weighted |
DWI* | High b- Value DWI† |
DCE MR Imaging‡ |
T2 Weighted |
DWI** | High b- Value DWI†† |
DCE MR Imaging‡‡ |
|
| ||||||||
| Field of view (mm) | 140 × 140 | 140 × 140 | 140 × 140 | 262 × 262 | 180 × 180 | 140 × 140 | 140 × 140 | 262 × 262 |
| Acquisition Matrix | 304 × 234 | 112 × 109 | 76 × 78 | 188 × 96 | 320 × 216 | 80 × 79 | 64 × 62 | 176 × 66 |
| Repetition time (msec) | 4434 | 4986 | 6987 | 3.7 | 3686 | 4766 | 7203 | 3.7 |
| Echo time (msec) | 120 | 54 | 52 | 2.3 | 120 | 45 | 47 | 2.3 |
| Flip angle (degrees) | 90 | 90 | 90 | 8.5 | 90 | 90 | 90 | 8.5 |
| Section thickness (mm), no gaps | 3 | 3 | 3 | 3 | 3 | 2.73 | 2.73 | 3 |
| Image reconstruction matrix (pixels) | 512 × 512 | 256 × 256 | 256 × 256 | 256 × 256 | 512 × 512 | 128 × 128 | 128 × 128 | 256 × 256 |
| Reconstruction voxel imaging resolution (mm/pixel) | 0.27 × 0.27 × 3.00 | 0.55 × 0.55 × 2.73 | 0.55 × 0.55 × 2.73 | 1.02 × 1.02 × 3.00 | 0.35 × 0.35 × 3.00 | 1.09 × 1.09 × 2.73 | 1.09 × 1.09 × 2.73 | 1.02 × 1.02 × 3.00 |
| Time for acquisition (min:sec) | 2:48 | 4:54 | 3:50 | 5:16 | 4:48 | 4:54 | 6:07 | 5:16 |
For ADC map calculation. Five evenly-spaced b values (0–750 sec/mm2) were used
b=2000 sec/mm2
DCE Images obtained, before, during, and after a single dose of gadopentetate dimeglumine 0.1mmol/kg at 3 mL/sec. Each sequence obtained at 5.6 s intervals
For ADC map calculation. Three evenly-spaced b values (0–600 sec/mm2) were used
b=1500 sec/mm2
DCE Images obtained, before, during, and after a single dose of gadopentetate dimeglumine 0.1mmol/kg at 3 mL/sec. Each sequence obtained at 5.6 s intervals
MRI Interpretation
All MRI scans were read by one highly experienced genitourinary radiologist (B.T.) (~6000 scans read from 2007–2015), with suspicious lesions identified prospectively as part of the clinical workflow. Suspicious areas were categorized using the 2015 PI-RADSv2 guidelines and simultaneously categorized using the traditional qualitative in-house system in use since 2007. Both systems are 5-category systems. PI-RADSv2 categorization is based on specific descriptions of lesion appearance that are incorporated into a zone-based schema as illustrated in Table 2 (15). The in-house system relies on the number of sequences on which the lesion can be identified plus one point for possible EPE and 2 points for definite EPE (1, 12). The two systems are compared in Table 3.
Table 2.
Guidelines given in PI-RADSv2, published by Weinreb et al. (5). In the peripheral zone (PZ), diffusion weighted imaging (DWI) findings serve as the dominant sequence determinant of final PI-RADSv2 categorization, while in the transition zone (TZ), T2-weighted (T2W) imaging serves as the dominant sequence. Certain other sequence findings can lead to overall upgrade to category 4. In the PZ, this is by dynamic contrast enhanced (DCE) positivity, while in the TZ, this is by a high DWI categorization.
| MRI Findings for DWI Category |
PZ DWI Category |
Other Sequence Findings |
Final PI-RADSv2 Category |
Other Sequence Findings |
TZ T2W Category |
MRI Findings for T2W Category |
|---|---|---|---|---|---|---|
| No abnormality | 1 | 1 | 1 | No abnormality | ||
| Indistinct hypointense on ADC map | 2 | 2 | 2 | Circumscribed hypointense/heterogenous nodules, Benign prostatic hyperplasia | ||
| Focal mildly/moderately hypointense on ADC; isointense/mildly hyperintense on high b-value DWI | 3 | DCE − | 3 | DWI≤4 | 3 | Heterogenous signal intensity, obscured margins, indeterminate |
| DCE + | 4 | DWI =5 | ||||
| Focal markedly hypointense on ADC; markedly hyperintense on high b-value DWI; <1.5 cm | 4 | 4 | Lenticular/non-circumscribed, homogenous, moderately hypointense, <1.5 cm | |||
| Same as 4, but ≥ 1.5 cm or definite extraprostatic extension | 5 | 5 | 5 | Same as 4, but ≥ 1.5 cm or definite extraprostatic extension |
Table 3.
Guidelines for qualitative in-house system. Baseline sequence-based detection positivity and negativity is established, and cumulative result produces final categorization 1–5.
| Final In-House Category |
T2W | DWI | DCE | Extraprostatic Extension |
|---|---|---|---|---|
| 1 | + | − | − | − |
| − | + | − | − | |
| − | − | + | − | |
| 2 | + | + | − | − |
| + | − | + | − | |
| − | + | + | − | |
| 3 | + | + | + | − |
| 4 | + | + | + | minimal capsular bulge and/or lesion capsule contact length ≥1.5cm |
| 5 | + | + | + | Definite EPE |
Reference Standard
Patients with suspicious lesions identified on mpMRI were recommended for subsequent MRI/transrectal ultrasound (MRI/TRUS) fusion-guided biopsy. All biopsies were performed using the office-based UroNav platform (Invivo Corporation, Gainesville, FL) with prepared lesion targets. Each lesion was sampled with 2 cores: one in the axial plane and the other in the sagittal plane. All procedures were performed by either one urologist (P.A.P.) or by one interventional radiologist (B.J.W.), both of whom had performed >1500 MRI/TRUS fusion guided biopsies at the time of the study.
One genitourinary pathologist (M.J.M.) (>25 years of experience in interpretation of prostate histopathology) analyzed the resultant biopsy cores, with primary and secondary Gleason scores provided. Clinically significant (CS) disease was defined as lesions scored ≥Gleason 3+4.
Statistical Analysis
Tumor detection rate (TDR), defined as the proportion of true positive lesions among all detected lesions, was calculated for each category within the PI-RADSv2 and in-house scoring systems. The standard error (SE) and 95% CI of TDRs were calculated from 2,000 bootstrap samples by a random sampling of patients with replacement. The 95% CIs were obtained from the 2.5% and 97.5% percentiles of the bootstrap resampling distribution. The significance of the difference in TDRs between PI-RADSv2 and in-house systems was evaluated at each category level (1–5) using the Wald test. To partially account for multiple testing a more conservative significance level of 0.01 was used.
Ability of each scoring system to detect cancer was assessed by receiver operator characteristic (ROC) curve analysis, evaluated for all cancers and clinically significant cancers. Area under the curve (AUC) was reported for each system. SE and 95% confidence intervals were calculated using the same bootstrap resampling technique described above.
Within each PI-RADSv2 category, lesions were further stratified by the assigned in-house category for that patient to compare TDRs across concordant and discordant categorization across the two systems. The difference in TDR across consecutive in-house categories within each PI-RADSv2 category was tested using the Wald test based on the bootstrap standard error. All p-values correspond to two-sided tests, with a p-value <0.05 considered to represent a significant difference.
RESULTS
Patient and Lesion Characteristics
The final study population consisted of 338 patients. The median PSA was 6.5 [range 0.6–113.6] ng/mL and median age was 64 [44–84] years. The median time between MRI and MRI/TRUS fusion-guided biopsy was 36 [0–241] days.
A total of 733 lesions were detected and categorized on mpMRI with subsequent MRI/TRUS fusion-guided biopsy, of which 346/733 (47%) were histopathologically proven tumor-positive (n= 110, 120, 45, 54, 17 were Gleason 3+3, 3+4, 4+3, 4+4, 4+5, respectively).
Cancer Detection Performance
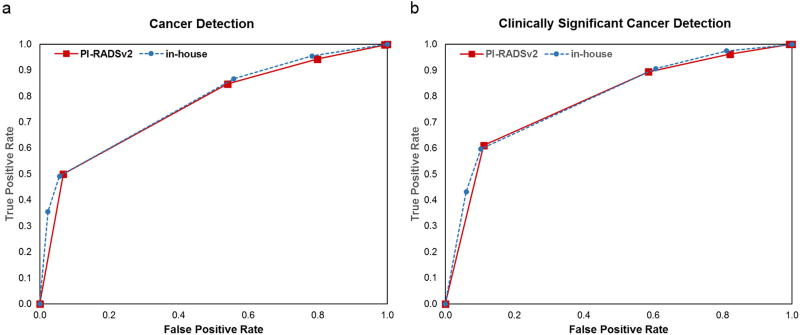
Performance of both systems for detecting cancer is shown for all disease and CS disease in Figure 1. The in-house system showed a non-significant benefit to overall cancer detection compared to PI-RADSv2, with AUC 77.6% and 76.2%, respectively (p=0.20). Both systems demonstrated the ability to locate CS cancer, with AUCs of 0.78 and 0.79 for PI-RADSV2 and the in-house system, respectively (p=0.45).
Figure 1.
Performance of clinically significant cancer detection in PI-RADSv2 (red) vs in-house (blue) systems. ROC curves for (A) all cancer detection with PI-RADSv2 AUC 76% and in-house AUC 78% (p=0.20) and (B) for clinically significant cancer, each with AUC = 79%.
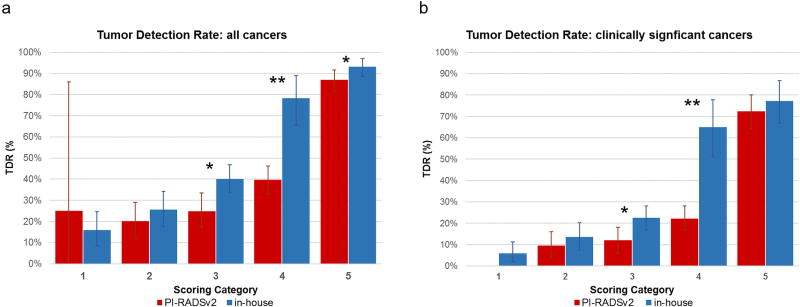
Lesion-based scoring of the two systems is compared in Table 4. Category 5 showed the highest concordance between the two systems with 63.5% (127/199) lesions scored 5 in PI-RADSv2 also scored 5 in the in-house system. Notably, 95.3% of lesions scored as category 5 in both systems were tumor-positive. The distribution of TDRs achieved within scoring categories from each system are shown in Figure 2. The category with the lowest concordance in scoring between the systems (category 4) also demonstrated the greatest difference in tumor detection rates for all cancers (PI-RADSv2 TDR: 39.6%, in-house TDR: 78.3%, p<0.0001) (Figure 2A). This pattern remained consistent in detection rates for CS cancer (PI-RADSv2 CS TDR: 22.1% CS TDR, in-house CS TDR: 65%, p<0.0001) (Figure 2B).
Table 4.
Concordance of lesion categorization by PI-RADSv2 and in-house systems. Parentheses indicate (% true positive for all cancer | % true positive for clinically significant cancer).
| In-house system categorization |
PI-RADSv2 Lesion Categorization | ||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| 1 | 2 (0% | 0%) | 56 (16.1% | 7.1%) | 39 (15.4% | 5.1%) | 3 (33.3% | 0%) | 0 |
| 2 | 2 (50% | 0%) | 29 (24.1% | 13.8%) | 53 (30.2% | 18.9%) | 33 (18.2% | 6.1%) | 0 |
| 3 | 0 | 8 (37.5% | 12.5%) | 37 (24.3% | 8.1%) | 259 (41.3% | 24.0%) | 20 (55% | 35%) |
| 4 | 0 | 0 | 2 (50% | 50%) | 6 (83.3% | 50%) | 52 (78.8% | 67.3%) |
| 5 | 0 | 1 (0% | 0%) | 2 (50% | 0%) | 2 (50% | 0%) | 127 (95.3% | 80.3%) |
Figure 2.
Tumor Detection Rates (TDR) for lesion-based categorization levels 1–5 on PI-RADSv2 (red) vs. in-house (blue) systems, shown for (A) all cancers and (B) clinically significant cancers). *p<0.01, ** p<0.001
Sub-Stratification of PI-RADSv2 Category Assignment
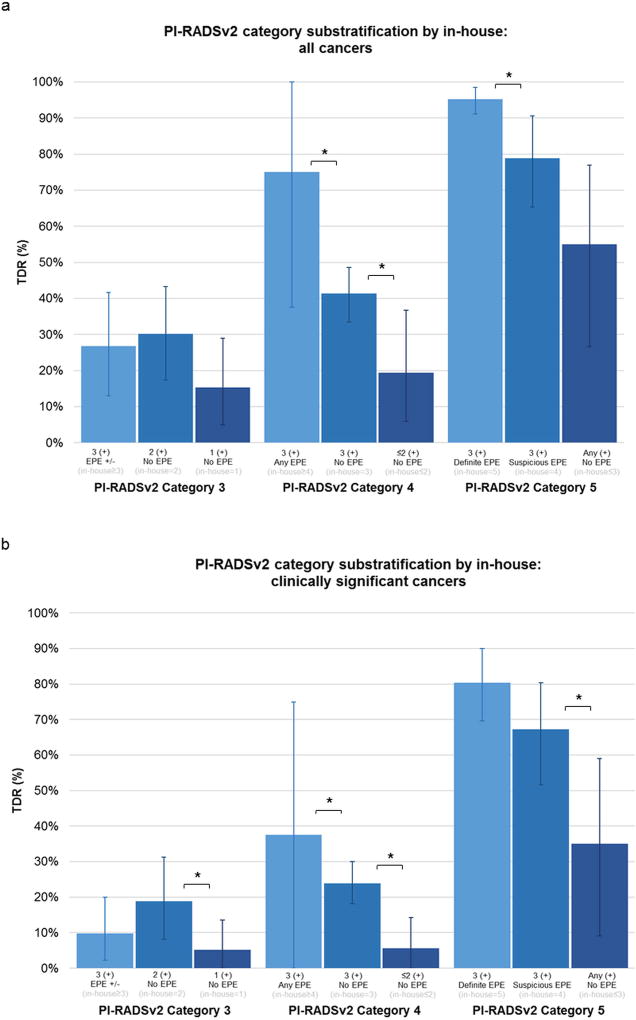
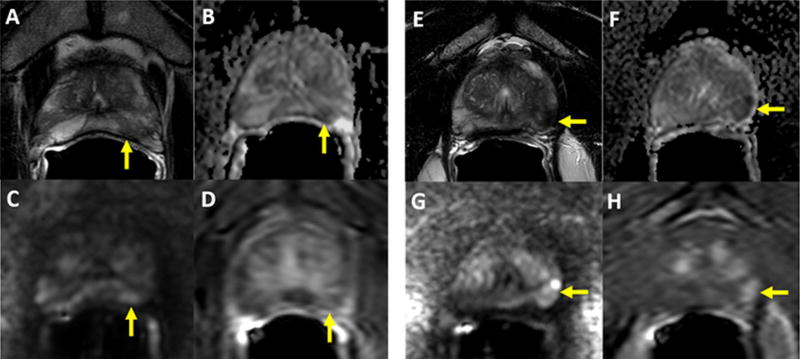
Given the similar overall performance between the two systems we attempted to determine whether PI-RADS v2 scored lesions could be sub-stratified by their in-house score. For instance, lesions assigned PI-RADSv2 = 3, 4, or 5 were sub-stratified by the in-house scoring system (Figure 3). In all cases, lesions with fewer positive findings on mpMRI sequences demonstrated poorer TDR. The greatest differences were seen in lesions classified as PI-RADSv2 category 4, with 75% all cancer TDR in lesions positive on all three sequences and suspicious for EPE (in-house category 4 or higher) compared to 41.3% for all cancer TDR in lesions positive on all three sequences without EPE (in-house category 3) and 19.4% all cancer TDR in lesions positive on only two sequences (in-house category 2), p=0.048 and p=0.014, respectively (Figure 3A). Similar trends were noted for detection of clinically significant disease (Figure 3B), demonstrating CS TDR was lowest in PI-RADSv2 category 4 lesions positive only on two sequences (in-house category 2), p=0.0002 (Figure 3B). Lesions scored PI-RADSv2 category 5 sub-stratified by definite EPE (in-house category 5), suspicious EPE (in-house category 4), and no EPE (in-house category 3) also showed differences in all cancer TDR (95.3%, 7.8%,55.0%, respectively) and CS TDR (80.3%, 67.3%, 35.0%). Examples of two PI-RADSv2 category 4 lesions with different in-house categorization are shown in Figure 4.
Figure 3.
Tumor Detection Rates (TDR) of PI-RADSv2 Categories based on sub-classification by in-house scoring, shown for (A) all cancers and (B) clinically significant cancers. * p<0.05.
Figure 4.
Panels 4A–4D: 56-year old man with a serum PSA of 3.99ng/ml. Axial T2W MRI shows a heterogeneously hypointense lesion in the left apical PZ (arrow) (A), which shows diffusion restriction on ADC maps (B) and b2000 DW MRI (C) with relatively weak contrast enhancement on DCE MRI (D) (arrow). This lesion was scored as PIRADS 4/5 (T2=3, DWI=4, DCE=negative), whereas its in-house score was 2/5 since it is only positive on T2W and DW MRI. Targeted biopsy revealed benign prostate tissue within this lesion. Panels 4E–4H: 66-year old man with a serum PSA of 13.45ng/ml. Axial T2W MRI shows homogenously hypointense lesion in the left apical PZ with slight capsular bulge (arrow), which shows diffusion restriction on ADC maps (B) and b2000 DW MRI (C) with early contrast enhancement on DCE MRI (D) (arrow). This lesion was scored as PIRADS 4/5 (T2=3, DWI=4, DCE=negative), whereas its inhouse score was 4/5 since it is positive on T2W, DW MRI, DCE MRI with slight capsular bulge. Targeted biopsy revealed Gleason 3+4 prostate adenocarcinoma within this lesion.
DISCUSSION
The ideal categorization system for MRI-detected prostate lesions would rank the likelihood of detecting clinically significant cancer according to each stepwise increase in score, with the highest category achieving the highest detection rates for CS disease. This study demonstrates PI-RADSv2 performed fairly well in this regard, achieving an overall AUC of 0.79 for clinically significant cancer. This compares favorably with another scoring system previously validated at our institution that relied on completely different criteria suggesting that future versions of PI-RADS v2 might benefit from integrating features from other scoring systems.
Prior comparisons of in-house qualitative systems to PI-RADS v1 demonstrated poorer performance of PI-RADSv1 (6). In the present study, PI-RADSv2 performed comparably to an in-house system suggesting that PI-RADSv2 is a marked improvement over the prior version. Our results are in broad agreement with detection AUCs reported for PI-RADSv2 in other studies, which range from 60.2–88% (16, 17). Collectively, these affirm the diagnostic value of PI-RADSv2.
As one might expect, scoring systems tend to perform better for larger and more obvious lesions. For instance, lesions graded Category 5 by either PI-RADSv2 or the in-house system achieved the highest tumor detection rates (>70% for all cancers, >80% for clinically significant cancer). Definite extraprostatic extension (EPE) at mpMRI, a key component of both systems’ categorization to 5, has been linked to detection of clinically significant prostate cancer, higher Gleason scores, and more extensive disease (18–20). However, PI-RADS v2 does not incorporate varying degrees of confidence in the diagnosis of EPE whereas the in-house system assigns one extra point for mild EPE and two points for definite EPE. We found that adding this feature to PI-RADSv2 category 4 lesions allowed for useful stratification of category 4 lesions into lower and higher risk strata. This may indicate that accounting for various degrees of capsular bulge and/or lesion capsule contact length, may improve PI-RADSv2 categorization in some lesions.
It is generally acknowledged that the CS tumor detection rate for PI-RADSv2 category 4 has been lower than originally expected (5, 9, 25–27). This may be partly due to upgrading of category 3 lesions to 4 based on one of several rules included in PI-RADSv2. Lesions in the PZ with equivocal DWI findings but positive enhancement on DCE MRI are upgraded to category 4, and similarly TZ lesions with equivocal T2W MRI findings but definite positive DWI findings can be upgraded to PI-RADSv2 category 4. It has been documented that the CDR for upgraded (3+1) category 4 lesions is lower than pure category 4 lesions (28) and the addition of 3+1 lesions tends to bring down the overall CDR for category 4. Unlike PI-RADS, the in-house system reserves category 4 status for “absolute positive” findings in all three MR pulse sequences with at least one aggressive feature such as suspicious EPE. Applying additional features of suspicious but not necessarily definite EPE criteria to PI-RADSv2 category 4 lesions appears to sub-stratify lesions categorized as PI-RADSv2 = 4 into a higher risk subset.
Our study utilized a single reader in testing detection rates for the two categorization systems. We have previously published on inter-observer agreement across five readers for both systems and found reasonably comparable agreement for overall suspicion score assigned by the in-house system (kappa = 0.55) versus PI-RADS v2 (kappa = 0.46) (8). Furthermore, in broad agreement with this study, we found that increase in suspicion score correlated with more clinically significant disease. However, the prior study was a retrospective PI-RADS v2 evaluation, and the present study benefits from prospective reading by an expert familiar with both systems. Additionally, the secondary objective within this prospective study was understanding the features that do not overlap between the two scoring paradigms. Utilization of an expert reader provides opportunity to identify differences that can further contribute to stratification of disease.
Additional limitations of the study include that the in-house system has been in use at our institution for mpMRI interpretation for 10 years, while PI-RADSv2-based interpretation began with its release in 2015. We did allow for a 3-month lead-in to gain experience with PI-RADS v2 before beginning accrual to this study. Nonetheless, it is difficult to isolate the advantage that the familiarity with the in-house system provides. It is important to note, however, that our PI-RADSv2 TDRs are in broad agreement with other recent PI-RADSv2 validation data (14). Finally, our study cohort consisted of patients who underwent MRI/TRUS fusion-guided biopsy which is inherently a limited gold standard compared to a radical prostatectomy population. The biopsy system utilized in this study has been proven to have accuracy of within 3 millimeters of lesion targets (35). However, while radical prostatectomy-based histopathology is a more definitive ground truth, the surgical population is biased towards higher-grade disease and not necessarily representative of a typical screening population of at-risk patients undergoing mpMRI for diagnosis for which the scoring systems were designed.
In conclusion, PI-RADSv2 performed comparably to an existing validated in-house scoring system in use for over 10 years. Further analysis of PI-RADSv2 categories 4 and 5 lesions reveals that when features of the in-house system were integrated into PI-RADSv2, sub-stratification into higher and lower risk patients was achieved within each PI-RADSv2 score. Such features include the number of positive MRI sequences and the degree of suspected EPE. These findings should inform future iterations of PI-RADS.
Acknowledgments
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
The National Institutes of Health (NIH) Medical Research Scholars Program is a public-private partnership supported jointly by the NIH and generous contributions to the Foundation for the NIH from the Doris Duke Charitable Foundation, The American Association for Dental Research, the Colgate-Palmolive Company, Genentech and alumni of student research programs and other individual supporters via contributions to the Foundation for the National Institutes of Health.
References
- 1.Siddiqui MM, Rais-Bahrami S, Turkbey B, George AK, Rothwax J, Shakir N, et al. Comparison of MR/ultrasound fusion-guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. Jama. 2015;313(4):390–7. doi: 10.1001/jama.2014.17942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rhudd A, McDonald J, Emberton M, Kasivisvanathan V. The role of the multiparametric MRI in the diagnosis of prostate cancer in biopsy-naive men. Curr Opin Urol. 2017 doi: 10.1097/MOU.0000000000000415. [DOI] [PubMed] [Google Scholar]
- 3.Bratan F, Niaf E, Melodelima C, Chesnais AL, Souchon R, Mege-Lechevallier F, et al. Influence of imaging and histological factors on prostate cancer detection and localisation on multiparametric MRI: a prospective study. Eur Radiol. 2013;23(7):2019–29. doi: 10.1007/s00330-013-2795-0. [DOI] [PubMed] [Google Scholar]
- 4.Barentsz JO, Richenberg J, Clements R, Choyke P, Verma S, Villeirs G, et al. ESUR prostate MR guidelines 2012. Eur Radiol. 2012;22(4):746–57. doi: 10.1007/s00330-011-2377-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinreb JC, Barentsz JO, Choyke PL, Cornud F, Haider MA, Macura KJ, et al. PI-RADS Prostate Imaging - Reporting and Data System: 2015, Version 2. Eur Urol. 2016;69(1):16–40. doi: 10.1016/j.eururo.2015.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vache T, Bratan F, Mege-Lechevallier F, Roche S, Rabilloud M, Rouviere O. Characterization of prostate lesions as benign or malignant at multiparametric MR imaging: comparison of three scoring systems in patients treated with radical prostatectomy. Radiology. 2014;272(2):446–55. doi: 10.1148/radiol.14131584. [DOI] [PubMed] [Google Scholar]
- 7.Baur AD, Maxeiner A, Franiel T, Kilic E, Huppertz A, Schwenke C, et al. Evaluation of the prostate imaging reporting and data system for the detection of prostate cancer by the results of targeted biopsy of the prostate. Invest Radiol. 2014;49(6):411–20. doi: 10.1097/RLI.0000000000000030. [DOI] [PubMed] [Google Scholar]
- 8.Muller BG, Shih JH, Sankineni S, Marko J, Rais-Bahrami S, George AK, et al. Prostate Cancer: Interobserver Agreement and Accuracy with the Revised Prostate Imaging Reporting and Data System at Multiparametric MR Imaging. Radiology. 2015;277(3):741–50. doi: 10.1148/radiol.2015142818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenkrantz AB, Oto A, Turkbey B, Westphalen AC. Prostate Imaging Reporting and Data System (PI-RADS), Version 2: A Critical Look. AJR Am J Roentgenol. 2016;206(6):1179–83. doi: 10.2214/AJR.15.15765. [DOI] [PubMed] [Google Scholar]
- 10.Rosenkrantz AB, Babb JS, Taneja SS, Ream JM. Proposed Adjustments to PI-RADS Version 2 Decision Rules: Impact on Prostate Cancer Detection. Radiology. 2017;283(1):119–29. doi: 10.1148/radiol.2016161124. [DOI] [PubMed] [Google Scholar]
- 11.Turkbey B, Mani H, Aras O, Ho J, Hoang A, Rastinehad AR, et al. Prostate cancer: can multiparametric MR imaging help identify patients who are candidates for active surveillance? Radiology. 2013;268(1):144–52. doi: 10.1148/radiol.13121325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rais-Bahrami S, Siddiqui MM, Turkbey B, Stamatakis L, Logan J, Hoang AN, et al. Utility of multiparametric magnetic resonance imaging suspicion levels for detecting prostate cancer. The Journal of urology. 2013;190(5):1721–7. doi: 10.1016/j.juro.2013.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rastinehad AR, Turkbey B, Salami SS, Yaskiv O, George AK, Fakhoury M, et al. Improving detection of clinically significant prostate cancer: magnetic resonance imaging/transrectal ultrasound fusion guided prostate biopsy. J Urol. 2014;191(6):1749–54. doi: 10.1016/j.juro.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehralivand S, Bednarova S, Shih JH, Mertan FV, Gaur S, Merino MJ, et al. Prospective Evaluation of Prostate Imaging-Reporting and Data System Version 2 Using the International Society of Urological Pathology Prostate Cancer Grade Group System. J Urol. 2017 doi: 10.1016/j.juro.2017.03.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barentsz JO, Weinreb JC, Verma S, Thoeny HC, Tempany CM, Shtern F, et al. Synopsis of the PI-RADS v2 Guidelines for Multiparametric Prostate Magnetic Resonance Imaging and Recommendations for Use. European urology. 2016;69(1):41–9. doi: 10.1016/j.eururo.2015.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seo JW, Shin SJ, Taik Oh Y, Jung DC, Cho NH, Choi YD, et al. PI-RADS Version 2: Detection of Clinically Significant Cancer in Patients With Biopsy Gleason Score 6 Prostate Cancer. AJR Am J Roentgenol. 2017;209(1):W1–W9. doi: 10.2214/AJR.16.16981. [DOI] [PubMed] [Google Scholar]
- 17.Purysko AS, Bittencourt LK, Bullen JA, Mostardeiro TR, Herts BR, Klein EA. Accuracy and Interobserver Agreement for Prostate Imaging Reporting and Data System, Version 2, for the Characterization of Lesions Identified on Multiparametric MRI of the Prostate. AJR Am J Roentgenol. 2017:1–7. doi: 10.2214/AJR.16.17289. [DOI] [PubMed] [Google Scholar]
- 18.Rouviere O, Dagonneau T, Cros F, Bratan F, Roche L, Mege-Lechevallier F, et al. Diagnostic value and relative weight of sequence-specific magnetic resonance features in characterizing clinically significant prostate cancers. PLoS One. 2017;12(6):e0178901. doi: 10.1371/journal.pone.0178901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krishna S, Lim CS, McInnes MD, Flood TA, Shabana WM, Lim RS, et al. Evaluation of MRI for diagnosis of extraprostatic extension in prostate cancer. J Magn Reson Imaging. 2017 doi: 10.1002/jmri.25729. [DOI] [PubMed] [Google Scholar]
- 20.Kongnyuy M, Sidana A, George AK, Muthigi A, Iyer A, Ho R, et al. Tumor contact with prostate capsule on magnetic resonance imaging: A potential biomarker for staging and prognosis. Urol Oncol. 2017;35(1):30, e1–e8. doi: 10.1016/j.urolonc.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim CS, McInnes MDF, Lim RS, Breau RH, Flood TA, Krishna S, et al. Prognostic value of Prostate Imaging and Data Reporting System (PI-RADS) v. 2 assessment categories 4 and 5 compared to histopathological outcomes after radical prostatectomy. J Magn Reson Imaging. 2017;46(1):257–66. doi: 10.1002/jmri.25539. [DOI] [PubMed] [Google Scholar]
- 22.Turkbey B, Mani H, Aras O, Rastinehad AR, Shah V, Bernardo M, et al. Correlation of magnetic resonance imaging tumor volume with histopathology. J Urol. 2012;188(4):1157–63. doi: 10.1016/j.juro.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim KH, Lim SK, Shin TY, Kang DR, Han WK, Chung BH, et al. Tumor volume adds prognostic value in patients with organ-confined prostate cancer. Ann Surg Oncol. 2013;20(9):3133–9. doi: 10.1245/s10434-013-3016-4. [DOI] [PubMed] [Google Scholar]
- 24.Knoedler JJ, Karnes RJ, Thompson RH, Rangel LJ, Bergstralh EJ, Boorjian SA. The association of tumor volume with mortality following radical prostatectomy. Prostate Cancer Prostatic Dis. 2014;17(2):144–8. doi: 10.1038/pcan.2013.61. [DOI] [PubMed] [Google Scholar]
- 25.Mertan FV, Greer MD, Shih JH, George AK, Kongnyuy M, Muthigi A, et al. Prospective Evaluation of the Prostate Imaging Reporting and Data System Version 2 for Prostate Cancer Detection. The Journal of urology. 2016;196(3):690–6. doi: 10.1016/j.juro.2016.04.057. [DOI] [PubMed] [Google Scholar]
- 26.Woo S, Kim SY, Lee J, Kim SH, Cho JY. PI-RADS version 2 for prediction of pathological downgrading after radical prostatectomy: a preliminary study in patients with biopsy-proven Gleason Score 7 (3+4) prostate cancer. Eur Radiol. 2016;26(10):3580–7. doi: 10.1007/s00330-016-4230-9. [DOI] [PubMed] [Google Scholar]
- 27.Greer MD, Brown AM, Shih JH, Summers RM, Marko J, Law YM, et al. Accuracy and agreement of PIRADSv2 for prostate cancer mpMRI: A multireader study. J Magn Reson Imaging. 2016 doi: 10.1002/jmri.25372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greer MD, Shih JH, Lay N, Barrett T, Kayat Bittencourt L, Borofsky S, et al. Validation of the Dominant Sequence Paradigm and Role of Dynamic Contrast-enhanced Imaging in PI-RADS Version 2. Radiology. 2017;285(3):859–69. doi: 10.1148/radiol.2017161316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akin O, Sala E, Moskowitz CS, Kuroiwa K, Ishill NM, Pucar D, et al. Transition zone prostate cancers: features, detection, localization, and staging at endorectal MR imaging. Radiology. 2006;239(3):784–92. doi: 10.1148/radiol.2392050949. [DOI] [PubMed] [Google Scholar]
- 30.Hoeks CM, Hambrock T, Yakar D, Hulsbergen-van de Kaa CA, Feuth T, Witjes JA, et al. Transition zone prostate cancer: detection and localization with 3-T multiparametric MR imaging. Radiology. 2013;266(1):207–17. doi: 10.1148/radiol.12120281. [DOI] [PubMed] [Google Scholar]
- 31.Verma S, Rajesh A, Morales H, Lemen L, Bills G, Delworth M, et al. Assessment of aggressiveness of prostate cancer: correlation of apparent diffusion coefficient with histologic grade after radical prostatectomy. AJR Am J Roentgenol. 2011;196(2):374–81. doi: 10.2214/AJR.10.4441. [DOI] [PubMed] [Google Scholar]
- 32.Tan CH, Wang J, Kundra V. Diffusion weighted imaging in prostate cancer. Eur Radiol. 2011;21(3):593–603. doi: 10.1007/s00330-010-1960-y. [DOI] [PubMed] [Google Scholar]
- 33.Polanec S, Helbich TH, Bickel H, Pinker-Domenig K, Georg D, Shariat SF, et al. Head-to-head comparison of PI-RADS v2 and PI-RADS v1. Eur J Radiol. 2016;85(6):1125–31. doi: 10.1016/j.ejrad.2016.03.025. [DOI] [PubMed] [Google Scholar]
- 34.Rosenkrantz AB, Ayoola A, Hoffman D, Khasgiwala A, Prabhu V, Smereka P, et al. The Learning Curve in Prostate MRI Interpretation: Self-Directed Learning Versus Continual Reader Feedback. AJR Am J Roentgenol. 2016:W1–W9. doi: 10.2214/AJR.16.16876. [DOI] [PubMed] [Google Scholar]
- 35.Xu S, Kruecker J, Turkbey B, Glossop N, Singh AK, Choyke P, et al. Real-time MRI-TRUS fusion for guidance of targeted prostate biopsies. Computer aided surgery : official journal of the International Society for Computer Aided Surgery. 2008;13(5):255–64. doi: 10.1080/10929080802364645. [DOI] [PMC free article] [PubMed] [Google Scholar]