Abstract
Blue light affects many aspects of plant growth and development throughout the plant lifecycle. Plant cryptochromes (CRYs) are UV-A/blue light photoreceptors that play pivotal roles in regulating blue light-mediated physiological responses via the regulated expression of more than one thousand genes. Photoactivated CRYs regulate transcription via two distinct mechanisms: indirect promotion of the activity of transcription factors by inactivation of the COP1/SPA E3 ligase complex or direct activation or inactivation of at least two sets of basic helix–loop–helix transcription factor families by physical interaction. Hence, CRYs govern intricate mechanisms that modulate activities of transcription factors to regulate multiple aspects of blue light-responsive photomorphogenesis. Here, we review recent progress in dissecting the pathways of CRY signaling and discuss accumulating evidence that shows how CRYs regulate broad physiological responses to blue light.
INTRODUCTION
The photolyase/cryptochrome (CRY) superfamily is a class of evolutionarily related flavoproteins that share sequence similarity with DNA photolyase (1–4). The CRY/photolyase superfamily consists of five major subgroups, including cyclobutane pyrimidine photolyase, 6–4 photolyase, CRY-DASH, plant CRY and animal CRY. Photolyase and CRY have distinct biochemical and physiological functions. Photolyases catalyze a blue/UV-A light-dependent DNA repair reaction. On the other hand, plant and animal CRYs have lost their DNA repair activity (5–7) and instead acquired regulatory functions during their evolution, although CRY-DASH seems to play a role in the repair of cyclobutane pyrimidine dimmers. CRYs regulate blue light-dependent photomorphogenic growth and developmental processes in plants as well as the circadian clock and magnetoreception in animals (1,2,8,9). Since the first discovery of CRY1 in Arabidopsis in 1993 (1), great progress has been made in understanding the function of CRYs in plants and animals. Specifically, recent works have identified CRYs as more versatile photoreceptors in plants than initially proposed (10). Moreover, CRYs were also found to govern at least three mechanistically independent pathways to regulate gene expression under blue light (11–13). This article focuses on the function and molecular mechanism of CRYs in regulating plant growth and development. Other articles in this special issue describe photolyase and the function of CRYs in animals.
DISCOVERY OF CRY AND ITS PHOTOCHEMICAL PROPERTY
Since Charles Darwin first documented blue light-induced phototropism in 1880 (14), a diverse array of blue light responses has been reported in plants. Currently, plants are known to possess flavoprotein photoreceptors, including CRYs, phototropins and FLAVIN-BINDING KELCH REPEAT F-BOX 1/LOV KELCH PROTEIN 2 (LKP2)/ZEITLUPE (ZTL) proteins, to regulate blue light responses (15). Prior to the discovery of these photoreceptors, the blue light photoreceptors containing UV-A/blue light-absorbing chromophores, such as flavin, pterin and carotenoids, had been recognized (16). Blue light responses exhibit unique action spectra with two peaks, one in the near-UV region and the other in the blue region of the spectrum. However, these blue light photoreceptors long remained unidentified, likely due to the relatively low abundance of these photoreceptors, the lack of an appropriate biochemical assay system for these photoreceptors and contamination by blue light-absorbing free pigments. The term “cryptochrome” was originally proposed as a portmanteau for unidentified blue light receptors because blue light responses were prevalent in cryptogamic plants, but blue light receptors remained cryptic (17).
Since the 1980s, molecular genetic techniques relying on the model species Arabidopsis thaliana (Arabidopsis) have successfully provided great benefits to the field of plant photobiology. For example, the inhibition of hypocotyl elongation under light had been assumed to be controlled by three types of photoreceptors, including blue light receptors, UV-B receptors, red/far-red light-absorbing phytochromes, and screening to identify these photoreceptors on the basis of this phenotype is simple and sophisticated (18–20). In 1980, several Arabidopsis mutants that exhibit long hypocotyls under white light were identified (18). Notably, one of these mutants, the hy4 mutant, features a long hypocotyl specifically in blue light but not in other wavelengths of light, suggesting that this mutant harbors lesions in the blue light photoreceptor or its downstream signaling intermediates (18). Cashmore’s group isolated the HY4 gene (later renamed CRY1) in 1993 (1); the protein encoded by HY4 was labeled as “cryptochrome” because it shares significant homology with DNA photolyases, suggesting that HY4 is a possible blue light photoreceptor that mediates the inhibition of hypocotyl elongation under blue light in Arabidopsis. Intriguingly, DNA photolyase was suspected to be a “cryptochrome” before the HY4 gene was isolated (16,21). On the other hand, it is ironic that a different term, phototropin, was assigned to the blue light photoreceptor responsible for phototropism (22) despite the fact that phototropism was the most prominent blue light response identi-fied for a “cryptochrome” at that time.
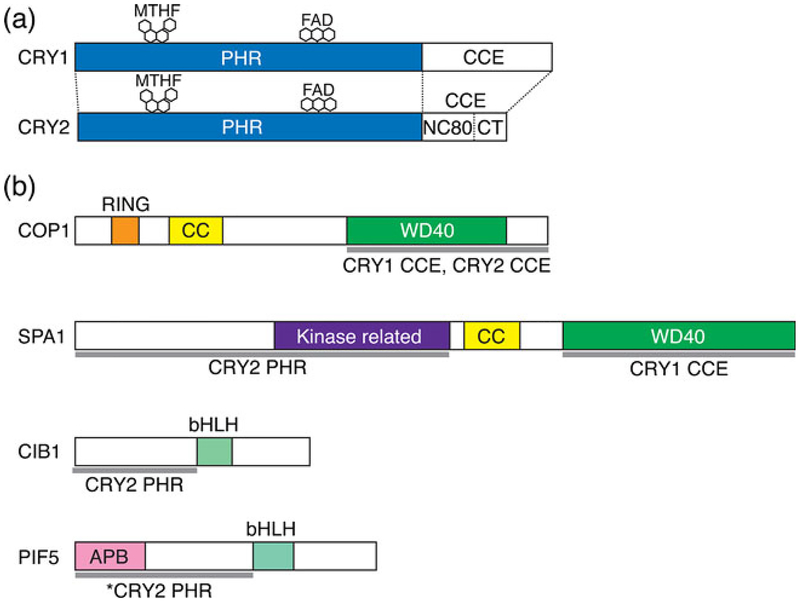
Cashmore’s group and Sancar’s group demonstrated that Arabidopsis CRY1 protein purified from insect cells or Escherichia coli cells stoichiometrically binds to flavin adenine dinucleotide (FAD), which primarily absorbs the blue/UV-A region of light via a noncovalent bond (5,6). In addition, Arabidopsis CRY1 expressed in E. coli cells was found to bind to 5,10-methenyltetrahydrofolate (6). These results suggest that, like photolyase, CRY1 possesses FAD as a primary chromophore and pterin as a secondary chromophore. Interestingly, CRY1 did not exhibit photolyase activity (5,6). Later, CRY-like sequences were identi-fied not only in plants but also in animals, including humans, mice, flies and bacteria (3). Furthermore, Sancar’s group demonstrated that human CRY1 and CRY2, similar to Arabidopsis CRY1, did not exhibit photolyase activity (7). Thus, this class of proteins was proposed to form a new protein family named CRY, although plant CRYs and animal CRYs seem to have evolved independently from divergent photolyase ancestry, with plant CRYs sharing greater sequence similarity with type I microbial photolyases and animal CRYs being more similar to 6–4 photolyases (3). Members of the CRY family of proteins possess an N-terminal photolyase homology region (PHR) domain followed by a C-terminal extension (CCE) domain of varied length (Fig. 1a). CRYs are conserved in all plant species whose genome sequences have been identified. Specifically, the Arabidopsis genome encodes two CRYs, CRY1 and CRY2 (23).
Figure 1.
The domain structure of CRYs and CRY-interacting proteins. (a) Schematic diagram showing domains in CRY1 and CRY2. MTHF, 5-methyltetrahydrofolate; FAD, flavin adenine dinucleotide; PHR, photolyase homology region domain; CCE, C-terminal extension domain; NC80, 80 residues without C-terminal tail in CCE domain; CT, C-terminal tail. (b) Schematic of CRY-interacting proteins showing the functional domains and CRY-binding sites. RING, RING finger domain; CC, coiled-coil domain; WD40, WD40 repeat; kinase-related, protein kinase-like domain; bHLH, basic helix–loop–helix domain, APB, active phytochrome B binding motif. The gray lines at the bottom of each diagram represent the CRY-binding regions. The binding domains of CRYs are also shown underneath each gray line. The asterisk indicates that CRY2 binds PIF5 with a mutation in the APB motif.
Spectroscopic analysis of Arabidopsis CRY1 and CRY2 derived from insect cells demonstrated that CRYs exhibit a photocycle between reduced FAD and oxidized FAD (5,6,24–28). The recombinant CRYs prepared in the dark bind fully oxidized FAD, which primarily absorbs blue light. Upon blue light exposure, a semireduced neutral FAD radical (FADH*) is generated within microseconds. FADH* is unstable and can be readily converted back to the fully oxidized form in a few minutes in the dark. The oxidized FAD, which absorbs blue light, is observed in the resting state of CRYs and is consistent with the observations that all CRY-mediated responses are triggered by blue light. Conversely, green light irradiation partially inhibits the effect of blue light on CRY2 degradation, the CRY-mediated inhibition of hypocotyl elongation and photoperiodic flowering (24–26). These effects were attributed to the shifting of the balance between oxidized FAD and FADH* toward reduced levels of FADH*, which absorbs green light and consequently decreases the amount of activated CRY. This relationship suggests that CRYs contain FADH* in its active state. However, green light irradiation did not inhibit blue light-mediated CRY2 degradation and the expression of the FLOWERING LOCUS T (FT) gene, the floral inducer gene, in another experiment (27). Furthermore, mutations in Tri-Trp, which consists of three tryptophan residues located close to FAD that act as electron donors during the photocycle of FAD, completely disrupt photoreduction in vitro but preserve the effects of blue light in plants (27,28). These results suggest that the photocycle between oxidized FAD and FADH* does not contribute to the photoperception of CRY in plant cells. Moreover, the blocking of photocycle in the other members of the current CRY/photolyase superfamily of proteins such as Drosophila and monarch CRY, and E. coli photolyase does not disrupt their physiological functions (9,29–34). Therefore, the specific mechanism underlying CRY photo-excitation has not yet been identified.
CRY STRUCTURE AND FUNCTION
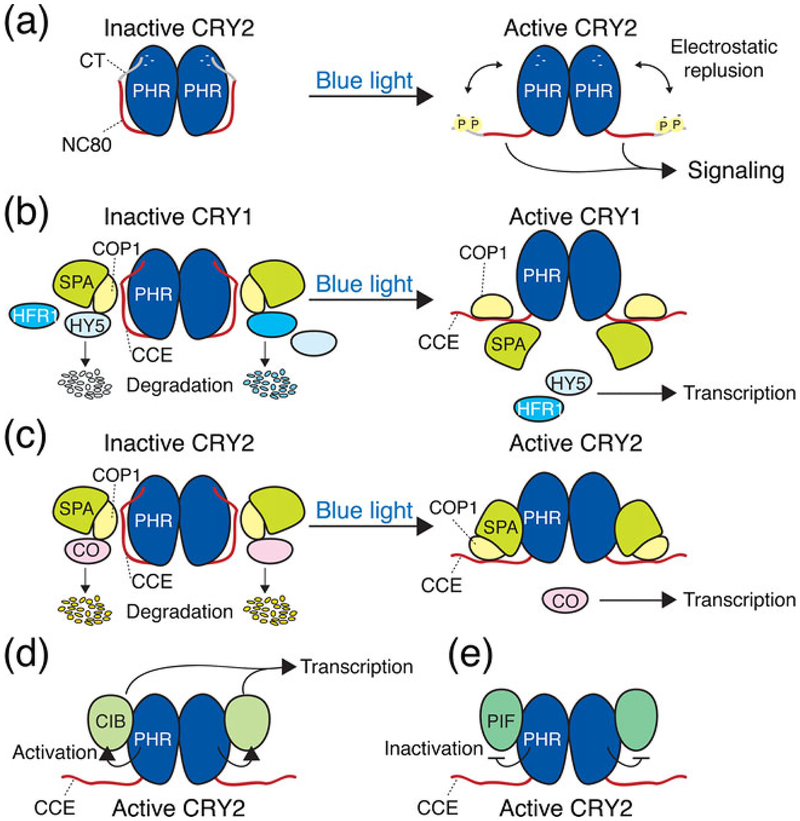
Both Arabidopsis CRY1 and CRY2 exist as dimers in plant cells, and the formation of these dimers is necessary for the function of CRY (35–37). The PHR domain, which contains chromophores, is responsible for not only photoperception but also dimer formation (35,37). The PHR domain is further divided into the N-terminal α/β domain and the C-terminal FAD-binding α domain, which are connected by a long interdomain loop. A crystallographic analysis of the CRY1 PHR domain revealed that similar to photolyases, the protein surface of the PHR domain is predominantly negatively charged and that the α domain consists of a chromophore pocket that harbors FAD in an unusual U-shaped conformation (38). In addition, the PHR domain of CRY1 binds to a nonhydrolysable ATP analog, adenylyl imidodiphosphate (AMP-PNP), near FAD (38). Unlike PHR domains, which are highly conserved among plant CRYs, CCE domains differ considerably in their length and sequence among plant CRYs. For example, the CCE domains of Arabidopsis CRY1 and CRY2 are 180 and 110 amino acid residues in length, respectively (Fig. 1a). Although the CCE domain of CRY1 appears to be intrinsically unstructured (39), CCE domains play an important role in the relay of CRY signaling to downstream components (4). The overexpression of β-glucuronidase fused to the CCE domain of either CRY1 (GUS-CCE1) or CRY2 (GUSCCE2) in Arabidopsis resulted in a strong constitutive photomorphogenic (cop)-like phenotype, and plants with this phenotype exhibit a morphology that mimics that of wild-type plants grown in light, even when grown in the dark (36,40,41). Therefore, the CCE domains of CRY1 and CRY2 exert their functions irrespective of the light condition when they lack the PHR domain. This observation suggested that PHR domains regulate the accessibility or conformation of CCE domains in response to blue light to achieve photomorphogenic responses. Consistent with this intramolecular photoactivation model of CRYs, the CRY1 CCE domain is digested by proteases in a blue light-dependent manner (39). In addition, transient grating spectroscopy revealed a large decrease in the diffusion coefficient in full-length CRY1 in response to blue light, which is indicative of a conformational change, but this decrease was not observed in truncated CRY1, which lacks the CCE domain (42). These observations suggest that blue light triggers dynamic conformational changes in CRYs at the CCE domain to facilitate a CRY signaling relay (Fig. 2a).
Figure 2.
Hypothetical models depicting a blue light-induced CRY conformational change and CRY signal transduction mechanisms in Arabidopsis. (a) CRYs consist of two domains, the PHR domain and the CCE domain, and they form homodimers via the PHR domains. In the dark, CRYs have a closed conformation. Upon blue light activation, CRYs undergo conformational changes, mostly in CCE domains, to transduce signals to downstream components. Blue light-induced phosphorylation at multiple sites of the C-terminal tail in the CCE domain generates negative charges (–) at these sites, resulting in the electrostatic repulsion of the NC80 domain from the PHR domain, which induces a CRY signaling relay to the downstream components. (b, c) Models depicting how CRY1 and CRY2 regulate the E3 ubiquitin ligase activity of the COP1/SPA complex. In the dark, COP1 and SPA constitute an active complex that promotes the degradation of transcription factors, such as HY5 and CO, which are positive regulators of de-etiolation and flowering, respectively. In blue light, photoactivated CRY1 binds to SPA and dissociates COP1 from SPA1 on the CCE domain. This active CRY1-mediated dissociation inactivates the COP1/SPA complex, and therefore, HY5 accumulates to promote de-etiolation. Unlike CRY1, the blue light-induced interaction between CRY2 and SPA via the PHR domain does not dissociate the COP1/SPA complex but enhances the interaction between CRY2 and COP1. This enhanced connection between CRY2 and COP1 may inactivate the COP1/SPA complex to promote CO accumulation and subsequent FT expression and flowering. (d, e) Model depicting how CRYs regulate the activity of bHLH transcription factors. Blue light-activated CRY2 binds to CIBs via the PHR domain to positively regulate the activity of CIBs, leading to FT expression and subsequent flowering. Photoactivated CRY1 and CRY2 bind to PIFs to inactivate them. The CRY2 PHR domain interacts with PIF5, but the PIF-binding domain of CRY1 has not been mapped yet. The exact mechanism by which CRY binds to CIBs or PIFs to regulate their transcriptional activity remains unknown. CCE, cryptochrome C-terminal extension; CIB, CRY-interacting basic helix–loop–helix; CO, CON-STANS; COP1, CONSTITUTIVE PHOTOMORPHOGENIC 1; CT, C-terminal tail; FT, FLOWERING LOCUS T; HY5, LONG HYPOCOTYL 5; NC80, 80 residues without C-terminal tail in CCE domain; PHR, photolyase-homologous region; PIF, PHYTOCHROME-INTERACTING FACTOR; SPA, SUPPRESSOR OF PHYA-105.
Either CRY1 or CRY2 undergoes blue light-dependent phosphorylation (36,43,44), and the phosphorylation of CRYs closely correlates with the intensity and duration of blue light exposure (43–47). Furthermore, mutations that inhibit the function of CRY1 reduce phosphorylation (44), and mutations of the 13 serine residues of CRY2, including blue light-dependently phosphorylated S598, S599 and S605, reduce the function of CRY2 (47). Thus, the phosphorylation of CRYs is likely critical for their function and regulation. Although GUS-CCE2 proteins are constitutively phosphorylated, GUS fuses to an eighty-amino acid fragment (NC80) that consists of a CCE domain lacking the C-terminal small region (C-terminal tail) (Fig. 1a) exhibited constitutive biological activity without phosphorylation (36). This finding suggested that the CCE domain does not require phosphorylation to mediate the function of CRY2 when it lacks a PHR domain. Importantly, the protein surface of the CRY1 PHR domain is primarily negatively charged (38). Thus, the phosphorylated CCE domain, which becomes negatively charged by the attachment of phosphate groups, is proposed to electrostatically repulse the PHR domain to form an open conformation in blue light. Intriguingly, all three serine residues, which are recognized as blue light-dependent phosphorylation sites, are mapped on the C-terminal tail (47). Therefore, the current model proposes that blue light induces the phosphorylation of the C-terminal tail, thereby triggering the derepression of NC80 from the PHR domain to transduce the signal to the downstream component of the CRY signaling relay (Fig. 2a).
Recombinant CRY1 protein purified from insect cells was phosphorylated in the absence of any kinase in response to blue light in vitro (44,48,49). Whereas CRYs are not similar to the kinase domain, CRY1 stoichiometrically binds to ATP (48,49). In addition, the crystal structure of the PHR domain of CRY1 included AMP-PNP (38). These results suggest that CRY1 is intrinsically competent to catalyze autophosphorylation. CRY2 does not show any detectable autophosphorylation activity (49), but another mechanism has been found to be involved in CRY2 phosphorylation. Specifically, casein kinase 1, CK1.3 and CK1.4, directly binds and phosphorylates CRY2 at Ser-587 and Thr-603 in vitro (46). The loss-of-function and gain-of-function lines of either CK1.3 or CK1.4 showed aberrant CRY2-mediated responses, suggesting that CK1.3 and CK1.4 play a role in CRY2 signal transduction. However, mutations of either CK1.3 or CK1.4 did not significantly affect CRY2 phosphorylation in plant cells. Therefore, other kinases need to be identified to fully understand the phosphorylation process of CRY2.
CRY1 and CRY2 act differently in some ways. For example, CRY2 protein is rapidly degraded by the 26S proteasome system in response to blue light (23,43,45,50–52), whereas CRY1 is stable. CRY2 degradation is mediated in part by phytochrome A (phyA), a major molecular species of phytochrome, and SPA1 (suppressor of PhyA-105), which comprises the E3 ligase complex together with COP1 (constitutive photomorphogenic 1) (52). Due to its rapid degradation under blue light, CRY2 favors the weaker range of blue light intensity to exert its function (23). CRY1 and CRY2 also differ in subcellular localization. Specifically, CRY1 localizes equally to the nucleus and cytoplasm (53,54), but CRY1 in the nucleus regulates all CRY1-mediated responses except for CRY1-mediated cotyledon expansion and root elongation (54). Conversely, CRY2 localizes primarily to the nucleus (36,55). In addition, CRY2 forms the nuclear bodies called as photobodies, in the nucleus in response to blue light (51,56,57). Because photobody formation closely correlates with CRY2 function and degradation (51,55), photobodies have been proposed as sites for CRY2 signal transduction and/or CRY2 degradation. However, little is known regarding the mechanism of CRY2 photobody formation. Notably, CRY2 can also form photobodies in human cells (57), suggesting that CRY2 photo-body formation requires CRY2 itself and cellular components that are common among plants and animals.
CRY-BINDING PROTEINS AND THE SIGNAL TRANSDUCTION MECHANISM OF CRY
CRYs mediate blue light-induced physiological responses primarily by transcriptionally regulating a large number of genes (11,12). For example, CRY1 and CRY2 regulate the transcription of 5–25% of genes in the Arabidopsis genome in response to blue light during seedling development (58–61). The blue light-dependent binding of CRYs to the COP1/SPA complex (11,12) and CRYPTOCHROME-INTERACTING BASIC HELIX–LOOP–HELIX (CIB) proteins (62) was shown to indirectly and directly regulate the transcription of the respective genes. Recently, CRYs have been shown to interact with PHYTO-CHROME-INTERACTING FACTOR (PIF) proteins in a blue light-dependent manner (63,64). Besides these CRY signaling pathways, CRYs are also known to bind to photoreceptors such as phytochromes and ZTL (56,65,66). In addition, we recently identified BLUE-LIGHT INHIBITOR OF CRYPTOCHROMES 1 (BIC1) that interacts with CRYs to modify CRY functions (67). Here, the nature of these interactions and their potential relevance to CRY signal transduction are described in more detail.
COP1/SPA Pathway
COP1 and SPA proteins (SPA1-SPA4 in Arabidopsis) comprise the E3 ubiquitin ligase complex (68). Accordingly, both the cop1 mutant and higher-order combinations of spa1, spa2, spa3 and spa4 mutants displayed strong cop-like phenotypes (69,70), indicating that the COP1 and SPA proteins negatively regulate photomorphogenesis. COP1 consists of an N-terminal RING finger domain, a coiled-coil domain and a C-terminal WD40 repeat (Fig. 1b). SPA proteins also possess a central coiled-coil domain and a C-terminal WD40 repeat, but they contain a protein kinase-like domain in the N-terminal region instead of a RING finger domain (Fig. 1b). COP1 and SPA proteins interact with each other via their central coiled-coil domains to form a complex (71), and the C-terminal WD40 repeat of COP1 recognizes several substrates, such as elongated hypocotyl 5 (HY5) (72, 73) and other transcription factors (68). HY5 is a bZIP transcription factor that promotes photomorphogenesis (74). The COP1/SPA complex recognizes and ubiquitinates HY5 protein in the dark, thereby degrading it via the 26S proteasome. Upon light exposure, CRYs and other photoreceptors inactivate the COP1/SPA complex, and HY5 protein consequently accumulates in the nucleus to induce photomorphogenesis (73).
Consistent with the cop-like phenotype observed in plants overexpressing GUS-CCE1 or GUS-CCE2, the CCE domains of CRY1 and CRY2 bind to COP1 (Fig. 1b) (36,40,41,75). The interaction between CCE domains and COP1 does not depend on blue light (41,75), but CRY1 binds to SPA1 and other SPA proteins in a blue light-dependent manner (76,77). In addition, the binding of the CRY1 CCE domain to the WD40 domain of SPA1 sequesters SPA1 from COP1 on the CRY1 CCE domain (Fig. 2b). Moreover, a tight connection between COP1 and SPA1 is reportedly required for COP1 to exert its E3 ligase activity, and the strong interaction between COP1 and SPA1 is weakened by light exposure (78,79). These observations led to the hypothesis that blue light-dependent CRY1 binding to SPA1 reduces COP1-SPA1 binding, which inactivates COP1, resulting in HY5 accumulation and the consequent transcriptional regulation of a number of genes (Fig. 2b).
Although CRY2 also binds to SPA in a blue light-dependent manner, it regulates COP1/SPA activity in a different manner (80). First, unlike CRY1, CRY2 binds to the N-terminal region of SPA1, including the kinase-like domain, via the PHR domain (Fig. 1b). In addition, the CRY2-SPA1 interaction did not affect the COP1-SPA1 interaction, but it strengthened the CRY2-COP1 interaction. Although the mechanism underlying this strengthening remains unknown, this interaction may reduce the E3 ligase activity of COP1 and thereby lead to the accumulation of CON-STANS (CO), a B-box-type zinc finger transcription factor that binds to the promoter of FT and promotes its expression to accelerate flowering (Fig. 2c).
CIB Pathway
Unlike CRY1, CRY2 binds not only to COP1/SPA and PIFs but also to CIBs in a blue light-dependent manner to promote photoperiodic flowering (62). Five (CIB1–CIB5 in Arabidopsis) of the 18 members of the bHLH transcription factor subfamily 18 have been shown to bind to CRY2 (62,81). Of these transcription factors, CIB1 was first identified as a CRY2-binding protein by yeast two-hybrid screening (62). The PHR domain of CRY2 binds to the N-terminal half of CIB1, which lacks the bHLH domain (Fig. 1b). The overexpression of all CRY2-binding CIBs except for CIB3 resulted in CRY2-dependent early flowering under long day (LD) conditions (81). Conversely, cib1cib5 and cib1cib2cib5 resulted in a late-flowering phenotype under LD conditions, whereas the flowering time of each single mutants was indistinguishable from that of the wild type, indicating the redundant role of CIB proteins in the promotion of photoperiodic flowering. CIB1 forms not only homodimers but also heterodimers with CIB2, CIB4 and CIB5, which bind to the noncanonical E-box found on the FT promoter with high affinity to promote FT expression.
Blue light stabilizes CIB1 protein, but other blue light receptors, ZTL and LKP2, regulate blue light-mediated CIB1 stabilization, suggesting the importance of the proper regulation of CIB1 activity, which is mediated by two different types of blue light receptors (82). Although the CRY2-CIB interaction promotes the activity of CIBs, the exact mechanism underlying CIB activation by CRY2 remains unknown (Fig. 2d).
PIF Pathway
PIFs were identified as proteins that bind to phytochrome in an active phytochrome conformer-specific manner (83). PIFs are also a subset of the bHLH superfamily of transcription factors, but unlike CIBs, they belong to bHLH subfamily 15. To date, PIF1, PIF3, PIF4, PIF5, PIF6, PIF7 and PIF8 have been shown to bind to photoactivated phyB (83), and in addition to the bHLH domain, these PIFs all contain an active phytochrome B binding (APB) motif at the N-terminus, which is necessary and sufficient for phyB binding (Fig. 1b) (84). In addition, PIF1 and PIF3 have been shown to bind to phyA. Upon red light exposure, PIF1, PIF3, PIF4 and PIF5 are phosphorylated and degraded rapidly by the direct binding of phytochrome (85–90). However, phosphorylated PIF7 is stable in red light (91). Furthermore, the pif1pif3pif4pif5 (pifQ) mutant exhibited a weak cop-like phenotype (92,93), suggesting that phytochromes remove the PIF-imposed repression of photo-morphogenesis by rapidly degrading PIF1, PIF3, PIF4 and PIF5 to trigger photomorphogenesis. PIF family member proteins have partly overlapping and distinct functions to regulate the diverse array of phytochrome-mediated physiological responses (83).
Recently, Ma et al. (2016) and Pedmale et al. (2016) demonstrated that CRY1 and CRY2 bind to PIF4 and PIF5 in the presence of light (63,64). Additionally, CRY1 also binds to PIF3 (63). The PHR domain of CRY2 binds to the N-terminal half of PIF5 (Fig. 1b), independently of the APB motif (64), which suggests that CRY2 and phyB recognize the distinct structure of the PIF molecule. These interactions between CRYs and PIFs result in the regulation of low blue light (LBL)-induced shade avoidance response (SAR) and warm temperature-induced cell growth. Unlike the CRY2-CIB interaction, the CRYs-PIF4/5 interaction is proposed to inhibit PIF4/5 activity (Fig. 2e). However, the exact mechanisms underlying CRY-mediated transcriptional inactivation of PIF4/5 are currently unknown.
PIF1 and PIF3 have been shown to rapidly degrade under blue light (94,95). However, blue light-activated phytochromes mediate the rapid degradation of PIF1 and PIF3. This finding is expected because phytochromes absorb not only red/far-red light but also blue light, albeit weakly. Conversely, blue light-induced, phytochrome-mediated PIF1 degradation was accelerated in a cry1cry2 double mutant (94), but CRYs have not been shown to directly bind to PIF1. However, CRYs bind to PIF3, PIF4 and PIF5 (63,64). Therefore, CRY1 and/or CRY2 may directly antagonize phytochrome-induced PIF1 degradation. Alternatively, CRY1 and CRY2 may function indirectly, likely via COP1/SPA activity, because COP1 affects the degradation of PIF3 and likely other PIFs (86). Thus, the mechanisms by which CRYs regulate the stability of PIFs need to be carefully investigated in the future.
HY5 has been shown to physically bind to at least PIF1 and PIF3 to form an antagonizing module (96,97). LONG HYPOCOTYL IN FAR-RED1 (HFR1), a COP1 substrate transcription factor, binds to PIF4 and PIF5 to form nonfunctional heterodimers (98,99). Therefore, the COP1/SPA complex degrades the negative regulators of PIFs. Moreover, HY5 promotes the expression of PIF4 (100,101), and COP1 indirectly regulates the accumulation of PIF3 (86) and possibly other PIFs. Conversely, PIF1 enhances the E3 ligase activity of COP1 in the dark (102). Therefore, the COP1/SPA pathway and PIF pathway seem to interact in a complex manner downstream of the CRY signal transduction chain.
Photoreceptors
CRY1 and CRY2 physically interact with phyA and phytochrome B (phyB), another major molecular species of phytochrome, respectively (56,65). However, an Arabidopsis mutant lacking all five phytochrome species exhibited blue light-induced photomorphogenesis (103), suggesting that the CRY-PHY interaction only modifies rather than determines the CRY signaling relay to the downstream components in the regulation of photo-morphogenesis. Indeed, in vitro CRY1 phosphorylation catalyzed by phyA does not noticeably affect CRY1 phosphorylation in plant (44,65) because CRY1 phosphorylation occurs normally in the phyA mutant and other phytochrome-deficient mutants. In addition to directly binding photoreceptors, CRYs and phytochromes also impinge on the COP1/SPA complex (76,77,80,104,105) and PIFs (63,64,83) to regulate photo-morphogenesis. The binding of CRYs to the COP1/SPA complex or PIFs does not require phytochromes and vice versa. However, whether the CRY-PHY interaction modifies the affinity of CRYs to the COP1/SPA complex or PIFs and whether the affinity of CRYs for these common intermediates modifies phytochrome binding are currently unknown. Nonetheless, these interactions are expected to provide clues to the molecular basis of the cross talk between CRYs and phytochromes observed in a number of physiological responses, such as the inhibition of hypocotyl elongation, anthocyanin accumulation, photoperiodic flowering (106–108). In contrast, phyA has been shown to predominantly degrade SPA2 protein to inactivate the COP1/SPA2 complex under blue light (109), suggesting the selective pathway that relies on common intermediates.
Moreover, other levels of photoreceptor signaling interaction were highlighted by the discovery of the cell type-specific roles of photoreceptors. Specifically, the effect of CRY2 on the regulation of flowering time depends on phyB (106). However, CRY2 promotes flowering in vascular tissue (110), whereas phyB regulates flowering time in mesophyll cells (111), suggesting that CRY2 and phyB interact downstream to regulate flowering time. Therefore, plants integrate photoreceptor-specific functions and multilayered interactions between CRYs and phytochromes, including direct interaction, signaling convergence on common intermediates and intercellular signaling interactions, to respond to fluctuating light in nature.
CRY1 has been also shown to physically bind with ZTL in yeast two-hybrid and in vitro pull-down assays (66). Although both of CRY1 and ZTL have roles in the related physiological responses such as the regulation of circadian clock and photoperiodic flowering, the biological significance of the binding between these photoreceptors remains unknown.
BIC1
BIC1 was identified from a genetic screening of the gain-of-function Arabidopsis mutant pool (67). Three independent dominant mutants (bic1D-1, bic1D-2 and bic1D-3) that exhibit the pheno-types similar to that of cry1cry2 double mutant were obtained. In these mutants, BIC1 gene, which encodes small protein composed of 140 amino acids with no obvious functional motif, is overexpressed. The overexpression of BIC2, a BIC1 homolog in Arabidopsis, also results in blue light-specific long hypocotyl phenotype. On the other hand, bic1bic2 mutant, but not bic1 and bic2 mutants, exhibits CRY-dependent short hypocotyl pheno-type under blue light, suggesting that BIC1 and BIC2 redundantly act to regulate CRY signaling relay. Interestingly, BICs bind with CRYs and negatively regulate CRY dimerization, phosphorylation and CRY binding to SPA1 and CIB1, suggesting that BICs negatively regulate the elementary process of CRY signaling relay (67).
CRY-MEDIATED PHYSIOLOGICAL RESPONSES
The identification of null mutants that are deficient in both CRYs has successfully enabled researchers to elucidate the physiological roles of CRYs in Arabidopsis. Although blue light-mediated de-etiolation and photoperiodic flowering have received the most attention as CRY-mediated functions (1,10,112), CRYs have been shown to regulate more divergent physiological responses, such as stomata opening and development (113–115), the LBL-induced SAR (64), the inhibition of warm temperature-induced growth (63), the light entrainment of the circadian clock (116,117), the temperature compensation of the circadian clock (118), root development (119,120) and programmed cell death (121). Of these responses, the LBL-induced SAR and inhibition of warm temperature-induced cell growth have been recently highlighted by the discovery of the interaction between CRYs and PIFs (63,64). In addition, land plants other than Arabidopsis exhibit CRY-mediated physiological responses that are also observed in Arabidopsis as well as plant-specific responses (10). The molecular mechanism underlying these CRY-mediated physiological responses has been revealed to be more complex than initially proposed because the PIF pathway was found to be a third component of the CRY signal transduction mechanism. The major CRY-mediated physiological responses for which molecular mechanisms have been elucidated are discussed below.
Blue Light-induced De-etiolation
Seeds often germinate in deep soil in nature. Postgerminative seedlings, which germinate in the soil, vigorously elongate their hypocotyls toward the surface of soil with a closed apical hook and cotyledon to protect the apical meristem in the subterranean darkness. When seedlings reach the surface, they stop hypocotyl elongation and simultaneously open the apical hook and cotyledon; then, the cotyledons expand, and the chloroplasts develop. This developmental transition in response to light is called deetiolation. In the de-etiolation process, reciprocal reactions occur in the hypocotyl and cotyledon. Namely, cell growth is inhibited in the hypocotyl but is promoted in the cotyledon.
As described above, the Arabidopsis cry1 mutant exhibits an elongated hypocotyl under blue light but does not show a unique phenotype under red and far-red light (1,18). CRY1 overexpression in Arabidopsis and tobacco increased the sensitivity to blue light during the inhibition of hypocotyl elongation (122,123). These observations indicate that CRY1 predominantly regulates the blue light-induced inhibition of hypocotyl elongation. Conversely, CRY2 has exhibited only limited effects on the de-etiolation process. Whereas a cry2 single mutant does not show an apparent phenotype under high-fluence blue light, the cry1cry2 double mutant exhibits a longer hypocotyl than the cry1 single mutant under this condition (106), suggesting that CRY2 minimally affects the inhibition of hypocotyl elongation under high-fluence blue light. In contrast, the cry2 single mutant exhibited an abnormal phenotype under low to moderate ranges of blue light fluence rate. This observation is consistent with the instability of the CRY2 protein under blue light. CRY1 and CRY2 have also been shown to exert partly overlapping functions; specifically, CRY1 more significantly contributes to other de-etiolation phenotypes, such as cotyledon separation, cotyledon expansion and chlorophyll accumulation (23,123). The hypocotyl length of the phyA mutant is long, albeit much shorter than that of the cry1 mutant under blue light, and the phyAphyB mutant features a longer hypocotyl than the phyA mutant. Conversely, the phyB mutant does not exhibit a long hypocotyl phenotype under this condition (124), indicating that phyA and phyB, in addition to CRYs, regulate the de-etiolation process under blue light.
CRY1 and possibly CRY2 regulate HY5 abundance by inactivating the COP1/SPA complex during CRY-mediated de-etiolation (11,12,76,77). In addition, the nuclear-targeted CRY1 PHR domain has been shown to induce the blue light-mediated inhibition of hypocotyl elongation without the accumulation of HY5 (125). Neither COP1 nor SPA binds to the CRY1 PHR domain. These results suggest that the CRY1 PHR domain can trigger the inhibition of hypocotyl elongation under blue light independently of the COP1/SPA pathway. Nevertheless, the signaling partner protein of the CRY1 PHR domain has not yet been identified. CIBs are not involved in the CRY1 PHR domain-mediated inhibition of hypocotyl elongation because neither loss-of-function nor gain-of-function lines of CIBs exhibited a significant effect on de-etiolation (62,81). On the other hand, PIF3, PIF4 and PIF5, which have been shown to bind to CRY1 and/or CRY2, are known to regulate the de-etiolation process under the control of phytochromes (83,92,93). In fact, the long hypocotyl pheno-type of the cry1 mutant was repressed in pif4 and pifQ mutants under blue light (63), indicating that PIF4 and other PIFs act downstream of CRY1 during hypocotyl growth regulation. In addition, the CRY2 PHR domain, whose sequence is highly similar to that of the CRY1 PHR domain, but not the CCE domain, binds to PIF4 and PIF5 (64), implying that the CRY1 PHR domain may also bind to PIFs. Therefore, the CRY1 PHR domain likely regulates the blue light-induced inhibition of hypocotyl elongation by regulating PIF4 and other PIFs, but this regulation should be experimentally addressed in future work.
The alleles of hfr1 mutants exhibited reduced de-etiolation phenotypes under blue light (126), although they were initially identified as long hypocotyl mutants under far-red light (127) when phyA is exclusively active. A genetic analysis revealed that phyA and hfr1 exert an additive effect, but cry1 is epistatic to hfr1 in the hypocotyl phenotype under blue light. Furthermore, more than 70% of blue light- and CRY1-mediated genes depend on HFR1 (128). These observations indicate that HFR1 acts as a positive regulator of de-etiolation downstream of CRY1, but not phyA, under blue light. Light induces the accumulation of HFR1 by inactivating COP1 (129,130), and HFR1 has been shown to inactivate PIF4 and PIF5 by forming non-DNA-binding heterodimers (98). Therefore, the CRY1-induced reversal of the COP1-mediated degradation of HFR1 may indirectly inhibit the activity of PIFs, at least in part, via nonfunctional HFR1-PIF heterodimer formation.
Control of Photoperiodic Flowering
Light is one of the most important environmental cues to regulate the transition from the vegetative phase to the reproductive phase, and the day length has an especially profound effect on the determination of flowering time. For example, Arabidopsis is a LD plant because LD conditions favor flowering over short day (SD) conditions. Conversely, rice is an SD plant, which flowers in SD but not LD conditions. In Arabidopsis, CRY2 is a major photoreceptor regulating the photoperiodic control of flowering time. Specifically, cry2 loss-of-function mutant alleles, including fha-1 and fha-2, previously identified photoperiodic pathway-related mutants and displayed significantly late flowering, specifically under LD conditions (112), indicating that CRY2 promotes flowering under LD conditions. A quantitative trait locus analysis identified early day-length insensitive (EDI) as a naturally occurring cry2 allele that is insensitive to changes in day length (131,132). EDI is a gain-of-function allele that is attributed to the C367M mutation on CRY2. Unlike wild-type CRY2, CRY2C367M is stable under SD conditions, resulting in a day-neutral early-flowering phenotype.
CRY1 also plays a role in the photoperiodic control of flowering time. The late-flowering phenotype of the cry1 mutant allele has been observed in some experiments (133,134) but not in others (107,135,136). This discrepancy among experiments is likely due to subtle differences in the conditions of each experiment. For example, the cry1 mutant exhibited a robust late-flowering phenotype at 16°C but not at 23°C (136), indicating that temperature affects the role of CRY1 in the photoperiod control of flowering time. The cry1cry2 double mutant flowers later than either the cry1 or cry2 single mutant under LD or cBL conditions (106). The gain-of-function alleles, which harbor L407F or G389R on CRY1, exhibited a photoperiod-independent early-flowering phenotype (137,138). These observations suggest that CRY1 plays a role in promoting flowering, but the contribution of CRY1 to the photoperiodic control of flowering time appears to be marginal because the effect of the cry1 mutation on the photoperiodic control of flowering could be observed only in certain environmental or genetic conditions. This finding strikingly contrasts the regulation of the de-etiolation process, in which CRY1 exerts a stronger effect than CRY2. Therefore, CRY1 and CRY2 play partly overlapping roles in the regulation of de-etiolation and photoperiodic flowering, but CRY1 and CRY2 predominantly regulate de-etiolation and photoperiodic flowering, respectively.
In Arabidopsis, CRY-mediated photoperiodic flowering is regulated by at least three pathways attributed to the transcriptional regulation of the FT gene (62). The first pathway mediates the circadian regulation of CO gene expression, which results in a CO mRNA abundance peak at dusk under LD conditions. CRY1 and CRY2 likely regulate the entrainment of the circadian clock by regulating COP1-ELF3-mediated GI protein degradation (139). Second, CRY1 and CRY2 regulate the activity of the COP1/SPA complex to stabilize the CO protein under blue light (80,140,141). The peak in CO mRNA abundance and the stabilization of CO protein coincide only under LD conditions and not SD conditions, which results in a CO mRNA abundance peak in the dark (142,143). Therefore, CO protein is stabilized and induces FT expression and subsequent flowering only under LD conditions. The third pathway consists of the CIB-dependent promotion of FT transcription (62). Although this pathway is mechanistically independent of the COP1/SPA pathway, CIB1 activity genetically depends on the function of CO (144). However, how the CIB pathway functionally interacts with CO to regulate floral initiation is presently unclear.
Moreover, whether the PIF pathway is involved in CRY-mediated floral initiation is unknown. Neither pif single mutants nor the pifQ mutant showed an abnormal flowering phenotype, at least at normal temperatures (93). On the other hand, a recent genomewide association study implicated PIF4 in the regulation of flowering time (145). In addition, plants that overexpress PIF4 or PIF5 showed early-flowering phenotypes under LD conditions (146,147). These data imply higher-order functional redundancy among PIFs in the regulation of flowering. Nevertheless, whether the observed PIF-related flowering phenotypes were caused by CRY-PIF or phyB-PIF binding needs to be carefully investigated because the role of CRY2 in the control of photoperiodic flowering strongly depends on phyB function (106).
Blue Light Regulates Stomatal Opening and Development
Stomata regulate gas exchange between the plant and atmosphere to optimize carbon dioxide uptake and the release of oxygen and water vapor in response to environmental and endogenous signals. Very low-fluence blue light, but not red light, is well known to trigger stomata opening (22), suggesting the involvement of blue light photoreceptors in the regulation of stomata opening. Accordingly, phototropins (phot1 and phot2 in Arabidopsis) have been identified as blue light photoreceptors that predominantly promote blue light-mediated stomata opening (148). Moreover, the cry1cry2 mutant showed reduced sensitivity to blue light in terms of stomata opening compared to the wild type (113). Nevertheless, the effect of CRYs is much smaller than that of phototropins and appears only in response to blue light intensities greater than 5 μmol/m2 s, indicating that CRYs are also involved in the regulation of stomata opening at higher blue light intensities. Notably, CRY-deficient and CRY1-overex-pressing plants exhibited enhanced drought tolerance and greater water loss, respectively, largely due to the role of CRYs in the regulation of stomata opening (113).
CRYs and phototropins exert an additive effect on the regulation of stomata opening, suggesting that CRYs and phototropins act independently (113). However, signals from CRYs and phototropins seem to converge on COP1 (113), which negatively regulates stomata opening. The spa1spa2spa3 triple mutant exhibited constitutively opened stomata (115), implying that CRYs regulate COP1 activity via a blue light-dependent CRYSPA interaction to promote stomata opening. Interestingly, constitutive expression of FT in guard cells promotes stomata opening, even in red light (149,150). Conversely, mutations in FT reduce blue light-induced stomata opening. In addition, FT expression in epidermal cells was reduced in the cry1cry2 mutant (149). Therefore, CRY-mediated stomata opening is induced, at least in part, by the expression of FT, as observed for CRY- mediated floral initiation. Stomata opening was reduced in the co mutant, and constitutive CO expression in guard cells resulted in constitutive FT expression and stomata opening (149). Furthermore, CRY1 and CRY2 induce blue light-mediated CO protein stabilization in guard cells (143). Therefore, the photoactivated CRY-induced accumulation of CO is likely mediated by the inactivation of the COP1-SPA complex and appears to regulate FT expression to promote stomata opening. However, whether the CIB pathway is involved in the CRY-regulated expression of FT in epidermal cells is presently unclear. In contrast, except for the observed physical interaction between phot2 and COP1 (151), the mechanistic basis for the regulation of COP1 activity by phototropins has not been demonstrated. Although the overexpression of FT in the guard cells of the phot1phot2 double mutant constitutively opened stomata (150), the expression of FT in the phot1phot2 double mutant was indistinguishable from that of the wild type (149), suggesting that different mechanisms are responsible for phototropin-mediated stomata opening.
In addition to regulating stomata opening, blue light also promotes stomata development. The cry1cry2 double-mutant seedlings exhibit fewer stomata in blue light but not in red light (114). In addition, the overexpression of CRY1 increased the number of stomata, indicating that CRYs promote blue light-mediated stomata formation in cotyledons. Similar to other CRY-mediated responses, CRY-induced stomata formation is negatively regulated by COP1. A genetic analysis revealed that COP1 acts in parallel with the auxin pathway and the receptor-like protein TMM upstream of YDA, a mitogen-activated protein kinase kinase kinase, to regulate the three essential factors SPCH, MUTE and FAMA, which act sequentially to regulate asymmetric cell division and stomatal development. However, the mechanism by which COP1 regulates the activity of YDA is unknown.
As for all other CRY-mediated responses except for LBL-induced shade avoidance and the inhibition of warm temperature-induced cell elongation, little information is available on the possible involvement of PIFs in CRY-mediated stomata opening or stomatal development. The pif3pif4 double mutant exhibits enhanced stomata opening under white light compared to wild type (115). However, the possibility that phyB regulates the observed difference between pif3pif4 and the wild type by binding to PIF3/4 under this condition cannot be excluded because phyB affects stomata opening, albeit weakly (115). PIF4 has been shown to function in phyB-induced stomatal development in mature leaves (152). Specifically, the mature leaves of the phyB mutant and the pif4 mutant exhibit fewer stomata than those of the wild type under high-fluence white light. Under this condition, the pif4phyB double mutant is indistinguishable from the phyB mutant, which has significantly fewer stomata than the pif4 mutant (152), indicating that the promotion of stomata development by PIF4 in mature leaves largely depends on phyB under white light. Thus, the contribution of PIF to CRY-mediated stomatal development is marginal.
LBL-induced Shade Avoidance Response
De-etiolated plants often face unfavorable light conditions, which reinforce the threat to the plant’s survival on the surface of the soil. For example, plants exhibit only limited photosynthetic productivity under a dense canopy. Therefore, plants have evolved a strategy to avoid shade from neighboring plants, which is termed the SAR (153). Specifically, the plant competitively elongates its hypocotyl, stem and petiole in response to shade to expose its photosynthetic organs to favorable light conditions. SAR is induced not only by the simple reduction of light intensity but also by changes in light quality. Because chlorophyll preferentially absorbs blue and red light but transmits green and far-red light, the blue/green and red/far-red light ratios are reduced in the shade. The mechanism by which plants monitor the red/farred ratio has been extensively studied. Specifically, phyB plays a predominant role in sensing the red/far-red ratio via the characteristic photochemical properties of a phytochrome. Phytochromes display photoreversibility between the red light-absorbing Pr form and the far-red-absorbing Pfr form. Red light transforms the Pr form to the Pfr form to activate the phytochrome, which inhibits cell growth in the hypocotyl, stem and petiole. Conversely, far-red light reverses the Pfr form back to the Pr form to inactivate the phytochrome. Therefore, phyB modulates the photoequiliblium between Pr and Pfr to control cell elongation based on the red/far-red ratio under ambient light conditions. Specifically, a low red/far-red ratio reduces the interaction between phyB and PIF7 (154). The reduced interaction results in the dephosphorylation of PIF7, which prompts the binding of PIF7 to the E-box sequences on the promoters of YUCCA genes, which are key auxin biosynthesis genes, to trigger their expression. As a consequence, auxin biosynthesis is enhanced to promote cell elongation (155). PIF4 and PIF5 are also involved in the low red/far-red-induced SAR via the control of their stability by phyB (90), but their effects appear to be limited compared to PIF7 (154).
As observed for the low red/far-red-induced SAR, blue light attenuation also induces the elongation of the hypocotyl, stem and petiole. The cry1 mutant exhibited a constitutive LBL-induced SAR in adult plants, whereas the cry2 mutant remained responsive to LBL (156), indicating that CRY1 is the primary photoreceptor controlling the LBL-induced SAR in adult plants. Conversely, both cry1 and cry2 mutant seedlings respond to LBL in the regulation of hypocotyl elongation, but the responsiveness to LBL is significantly reduced in cry1cry2 double-mutant seedlings, suggesting that both CRY1 and CRY2 regulate the effect of LBL-induced SAR on hypocotyl elongation.
The CRY-PIF pathway is involved in the CRY-mediated SAR. Specifically, a dramatic reduction in LBL-induced cell elongation was observed in the pif4 and pif5 mutants (156), and a genetic analysis revealed that PIF4 and PIF5 act downstream of CRY1 to regulate the LBL-induced SAR. Both CRY1 and CRY2 have been shown to bind to PIF4 and PIF5 (63,64), and both PIF5 protein and CRY2 are stabilized in response to LBL, whereas the levels of PIF4 and CRY1 do not depend on LBL (64). In addition, chromatin immunoprecipitation (ChIP)-sequencing and RNA-sequencing analyses showed that the co-localization of CRY2 and PIF4/5 at PIF4/5-regulated gene promoters to control their gene expression. Compared to the wild type, cry mutants exhibited an enhanced response to LBL, suggesting that CRYs suppress the SAR. Therefore, the accumulation of CRY2 under LBL conditions may negatively regulate the activity of PIF4/5 and thereby prevent excessive elongation (157).
Loss of PIF7 function diminishes the LBL-induced SAR, but the effect of PIF7 is much smaller than that of PIF4/5 because the pif7 mutant remains responsive to LBL, suggesting that PIF4/5 rather than PIF7 predominantly regulates the LBL-induced SAR. This finding strikingly contrasts the low red/far red-induced SAR, for which PIF7 exerts the predominant effect (154). In addition, the LBL-induced SAR and low red/far-red-induced SAR partially differ in their transcriptomic profiles. LBL mainly regulates the expression of genes involved in cell wall modification, whereas low red/far-red light controls the transcription of auxin pathway-related genes. Therefore, despite their similar morphological output, the LBL-induced SAR and low red/far-red light-induced SAR are operated, at least in part, by two independent mechanisms (64,156,158). phyB has also been shown to regulate the LBL-induced SAR. Plants overexpressing phyB did not elongate their hypocotyls in response to LBL, indicating that excess phyB suppresses the LBL-induced SAR (64). Indeed, phyB binds to PIF4/5 under LBL conditions. Conversely, the expression of mutant PIF4 or PIF5, which has mutation on APB motif and does not bind to phyB, did not fully rescue the pif4 or pif5 mutant phenotype under LBL conditions. This observation was interpreted as follows: phyB binds to PIF4/5 via the APB motif to activate PIF4/5 and promote hypocotyl elongation under LBL conditions, which strikingly contrasts the phyB-imposed suppression of PIF activity under red light (83). Notably, CRYs recognize the exterior APB motif of PIF4/5 (Fig. 1b), suggesting that a CRY-PIF4/5-phyB complex may regulate the SAR. In this complex, CRY-PIF4/5 binding may reverse the function of phyB in the regulation of PIF4/5 and vice versa. Therefore, further studies of this apparent disparity in the phyB regulation of PIF4/5 activity might provide insight into the molecular basis of the cross talk between LBL and low red/far-red light signals during the SAR.
Moreover, the COP1/SPA complex has been shown to positively regulate the SAR (159). Shade condition with the attenuation of both of blue light and the red/far-red ratio was recently shown to decrease the stability of the HFR1 protein (160). However, it increased the abundance of the HY5 protein, which likely antagonizes the function of PIFs to avoid excess elongation. In addition, shade-induced HFR1 protein degradation depends on COP1. Specifically, a genetic analysis revealed that the diminished SAR in the cop1 mutant largely depends on the effect of HFR1, and the enhanced SAR in the hfr1 mutant was impaired in a pif4pif5 mutant background. Given the formation of nonfunctional heterodimers between HFR1 and PIF4 or PIF5 (98,99), the COP1/SPA complex likely degrades HFR1 to abolish the HFR1-mediated repression of PIF4/5 function and to elongate the hypocotyl in the shade (160). However, how COP1 differentially recognizes HY5 and HFR1 under this condition and the extent to which the CRY-COP1/SPA interaction impacts this COP1-mediated regulation of HFR1 are currently unknown.
Blue Light Inhibition of Warm Temperature-induced Hypocotyl Elongation
Temperature is another important environmental cue that affects plant growth and development. Warm temperatures that do not cause heat shock accelerate plant growth, including hypocotyl elongation, petiole elongation and leaf expansion. Light signals are closely related to temperature signals. For example, the plant response to red light depends on the temperature signal. Whereas red light inhibits hypocotyl elongation in Arabidopsis at low temperatures (16°C) and normal temperatures (21–23°C), which favor Arabidopsis growth, it promotes hypocotyl elongation at 27°C (161). The effect of blue light on hypocotyl growth differs from that of red light. In the dark, wild-type Arabidopsis exhibits an elongated hypocotyl at 28°C compared to 22°C. This warm temperature-induced hypocotyl growth is inhibited by blue light. However, the cry1 mutant exhibits exacerbated warm temperature-induced hypocotyl elongation under blue light (63), suggesting that blue light inhibits warm temperature-mediated hypocotyl growth by activating CRY1.
The CRY-PIF pathway plays important roles in the CRY1-mediated inhibition of warm temperature-induced hypocotyl elongation. Warm temperature-induced hypocotyl elongation in the cry1 mutant under blue light is partially and significantly repressed in the pif4 and pifQ mutants (63), suggesting that PIF4 and other PIFs act downstream of CRY1 to promote warm temperature-induced hypocotyl growth. Consistently, CRY1 binds to PIF4 and other PIFs, including PIF3 and PIF5, under light conditions (63). A gene expression analysis and ChIP-PCR revealed that CRY1 represses warm temperature-induced auxin biosynthesis by downregulating YUCCA gene expression via the inactivation of PIF4, which binds to the YUCCA gene promoter (63).
In addition to PIFs, COP1 plays important roles in regulating the ambient warm temperature response because the cop1 mutant is partially insensitive to elevated temperatures, even in the dark (63). The loss-of-function mutant of HFR1, whose protein is targeted by COP1 for degradation in the dark, exhibits longer hypocotyls than the wild type under LD conditions at 28°C but not 22°C. HFR1 binds to PIF4 in the nucleus and inhibits PIF4 to activate the YUCCA8 promoter. These observations suggest that CRY1-COP1 binding regulates PIF4 activity via HFR1. Additionally, COP1 likely regulates the transcription of PIF4 probably via HY5. Therefore, the COP1-mediated ambient temperature response is at least partially mediated by the indirect regulation of PIF4 activity (63). However, whether COP1 also regulates the ambient temperature response independently of PIF4 is an important topic of future research.
CRY-regulated Responses in Other Plants
CRY-mediated physiological responses have been analyzed mostly in Arabidopsis, but they have also been investigated in many other major land plants, not including gymnosperms (10). Not only primitive land plants, such as moss and fern, but also monocot and even dicot plants exhibit CRY-mediated physiological responses that have not been observed in Arabidopsis. These physiological responses are briefly described here.
In Physcomitrella patens, a moss, PpCRY1a and PpCRY1b redundantly regulate several developmental processes under blue light, including the induction of side branching in protonema and gametophore growth and differentiation (162). In Adiantum capillus-veneris, a fern, five CRY genes have been identified to date. Of the corresponding gene products, AcCRY3 and AcCRY4 localize in the nucleus during the gametophyte phase (163). Earlier studies with the microbeam irradiation showed that the blue light photoreceptors involved in the spore germination processes are localized in or close to the nucleus (164,165). Thus, the function of AcCRY3 and AcCRY4 is implicated in the regulation of spore germination (163).
In monocots, the functions of CRY genes from rice and barley have been analyzed. The rice genome encodes three CRY genes, OsCRY1a, OsCRY1b and OsCRY2 (166). Of these genes, OsCRY1a and OsCRY1b regulate the blue light-mediated growth inhibition of coleoptiles and leaves (166,167). Moreover, OsCRY2 promotes flowering under both LD and SD conditions (166). In barley, HvCRY1a, HvCRY1b and HvCRY2 have been identified (168), and HvCRY1a and HvCRY1b were found to induce grain dormancy via the expression of 9-cis-epoxycarotenoid dioxygenase, an abscisic acid biosynthesis gene (169). Intriguingly, seed germination is predominantly induced by phyB under blue light in Arabidopsis (170). Hence, the involvement of HvCRY1a and HvCRY1b in the regulation of grain dormancy and germination is a barley-specific response.
In dicots, the functions of CRY have been analyzed in several species, such as rapeseed, tomato, pea and soybean. Generally speaking, CRY genes regulate de-etiolation, stem elongation, leaf expansion and photoperiodic flowering in these species, as observed in Arabidopsis. However, several minor differences exist among the species. For example, GmCRY1a, but not GmCRY2a, plays a predominant role in the regulation of photoperiodic flowering in soybean (171) because GmCRY1a strongly promoted flowering in Arabidopsis, and GmCRY1a, but not GmCRY2a, exhibited photoperiod-dependent circadian rhythmic protein expression. In addition, the photoperiod-dependent expression of GmCRY1a was closely related to photoperiodic flowering and the latitudinal distribution of different soybean accessions.
In addition, species-specific CRY-mediated responses, such as CRY-mediated lycopene accumulation in tomato fruit, have also been identified (172). The most prominently depicted CRY-mediated physiological response in this category is GmCRY2a-mediated leaf senescence in soybean (173). Specifically, the overexpression of GmCRY2a in soybean delays leaf senescence, whereas decreased GmCRY2a expression accelerates this response, suggesting that GmCRY2a negatively regulates leaf senescence in soybean. Conversely, CRYs have not been reported to affect leaf senescence in Arabidopsis or other species. Moreover, the overexpression of GmCIB1 also accelerates leaf senescence. Although genetic evidence showing the interaction between GmCRY2a and GmCIB1 is lacking, GmCRY2a binds to GmCIB1 in a blue light-dependent manner. In addition, the transient expression of GmCRY2a and/or GmCIB1 in tobacco together with an electrophoresis mobility shift assay revealed that GmCRY2a-GmCIB1 binding suppresses the binding of GmCIB1 to DNA, thereby repressing the expression of GmWRKY53b, whose Arabidopsis homolog, AtWRKY53, is known to promote leaf senescence (174). These results suggest that GmCRY2a represses the expression of GmWRKY53b to suppress leaf senescence by sequestering GmCIB1 from the promoter sequence of GmWRKY53b via blue light-dependent direct binding to GmCIB1 (173). Nevertheless, the function of GmWRKY53b needs to be confirmed in soybean. Notably, CRY2-CIB binding does not regulate leaf senescence in Arabidopsis. Furthermore, GmCRY2a-GmCIB1 binding suppresses the activity of GmCIB1 in soybean (173), whereas CRY2-CIB1 binding promotes the transcriptional activity of CIB1 to induce FT expression in Arabidopsis (62). Thus, these results suggest that the role of CRY2-CIBs binding depends on the surrounding environment in nature during the evolution and/or domestication process of dicots.
As described above, CRYs are responsible for several blue light responses in land plants other than Arabidopsis (10). However, except for the mechanisms contributing to GmCRY2a-GmCIB1-regulated leaf senescence in soybean, the molecular mechanisms underlying these responses have not been elucidated. COP1-, SPA-, CIB- and PIF-like sequences have been identified in all land plants whose genome sequences are known. For example, a COP1-like sequence from Physcomitrella patens can partially rescue the cop1 mutant phenotype in Arabidopsis (175). A PIF-like sequence from Marchantia polymorpha, a liverwort, acts in the phytochrome regulation of signal transduction in M. polymorpha (176). These observations imply that COP1/SPA, CIB and PIF may function, at least in part, in some CRY-mediated physiological responses in land plants. Further investigation of these factors in primitive plants might provide clues to understand the evolution of CRY functions and adaptive mechanisms to ambient light environments. In addition, the functional analyses of CRYs and their signaling components in crops might not only provide insight into the CRY signaling mechanism and its ecological and evolutionary significance but also contribute to the improvement of important agronomic traits in crops, including plant height, flowering time, drought tolerance and leaf senescence, and solutions to global warming.
CONCLUDING REMARKS
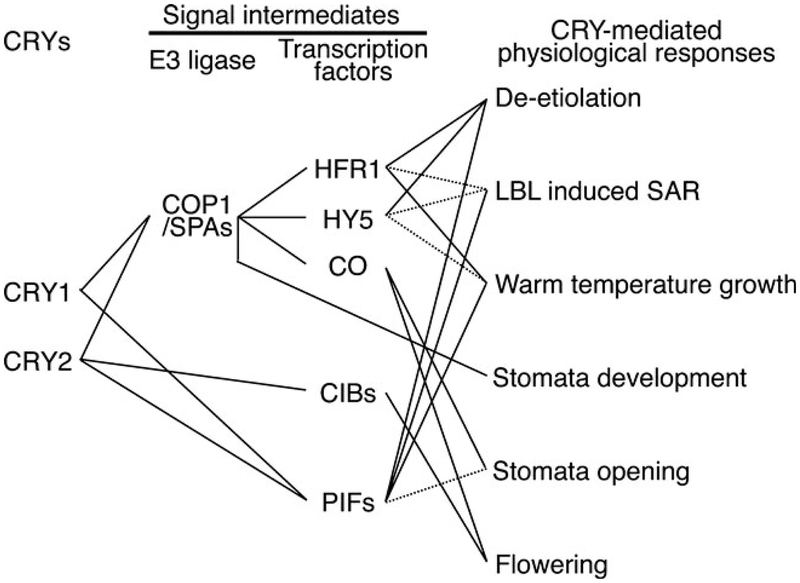
Since CRY1 was first discovered in Arabidopsis in 1993 (1), the mechanisms of how plants response to blue light irradiation have been largely elucidated at the molecular level (12). CRYs exhibit a diverse array of physiological functions throughout plant development as shown by Arabidopsis mutants deficient in CRY1 and/or CRY2 (10). Furthermore, a number of physiological functions have been identified for CRYs in other land plants. These observations clearly indicate the functional importance of CRYs in plant adaptation to ambient environments and consequently in the evolution of plants. In addition, CRYs have been found to directly and indirectly regulate transcription factors in response to blue light. Specifically, CRYs bind to the COP1/SPA complex to regulate the abundance of transcription factors (11,76,77,80), and they directly interact with two sets of bHLH transcription factor family proteins, CIBs and PIFs (62–64). In addition, target transcription factors of the COP1/SPA complex interact with PIFs (96–99). These findings depict the complexity of the CRY signaling relay, in which CRYs play important roles as conductors by orchestrating a complex transcription factor network to regulate gene expression (Fig. 3). This complex CRY-regulated network provides the molecular basis by which plants fine-tune their growth and development to respond to fluctuating blue light environments.
Figure 3.
CRYs directly and indirectly influence transcription factors, which act redundantly, antagonistically or independently. Schematic summary of the network of examined (solid line) or possible (dashed line) pathways downstream of the CRY signaling relay.
In a recent breakthrough, CRYs were found to not only bind to the COP1/SPA complex and CIBs but also to PIFs to regulate gene expression (63,64). This finding answered many questions related to CRY signal transduction, but it also gave rise to a number of new questions. The function of CRY-PIF binding has only been reported for the LBL-induced SAR and the inhibition of warm temperature-induced hypocotyl elongation (63,64). However, considering the versatility of PIFs (83), CRY-PIF binding can be expected to play many other roles in CRY-mediated responses. The exact mechanism by which the direct binding to bHLH transcription factors regulates the activity of PIFs and CIBs should also be elucidated in the future. This binding is evolutionarily interesting because animal CRYs also bind to bHLH transcription factors to constitute the circadian clock. In addition, additional efforts are necessary to address how three mechanistically independent pathways can converge to delineate CRY signal transduction. Continued exploration to answer these and other unresolved questions regarding CRY will undoubtedly provide more invaluable insights into not only the CRY signal transduction mechanism and plant photomorphogenesis but also the evolution of CRY family proteins.
Acknowledgements—
This work was supported by Fujian-Taiwan Joint Innovative Center for Germplasm Resources and Cultivation of Crop (FJ 2011 Program, No. 2015–75, China), the Program for New Century Excellent Talents in Fujian Province University and the School Special Development program of Fujian Agriculture and Forestry University (6112C035001).
Biographies

Zhaohe Yang is a Ph.D. candidate at Basic Forestry and Proteomics Research Center, Fujian Agriculture and Forestry University (FAFU), under the direction of Dr. Chen-tao Lin and Dr. Yoshito Oka. He obtained M.S. degree from FAFU. Currently, his research focuses on the cryptochrome regulation of plant blue light responses.

Bobin Liu is a lecturer at College of Forestry, FAFU. He obtained Ph.D. in biochemistry and molecular biology from Nanjing Forestry University in 2014. His present research interest is the light regulation of plant regeneration.

Jun Su received Ph.D. in forest conservation from FAFU. He joined Basic Forestry and Proteomics Research Center, FAFU, as a postdoctoral researcher in 2015. Currently, his research focuses on the blue light-dependent transcriptional regulation.

Jiakai Liao is currently a Ph.D. student in genetics at the College of Life Science, FAFU. He obtained M.S. degree from the Agricultural College of the Nanjing Agriculture University in 2014. His present research focuses on the light-dependent mechanism of transcriptional regulation.

Chentao Lin is currently a professor at the Department of Molecular, Cell & Developmental Biology, University of California, Los Angeles. He got his B.S. degree in agronomy at the South China College of Tropical Crops and obtained M.S. degree in Iowa State University. After receiving his Ph.D. in genetics at Michigan State University, he started the research on cryptochrome in Dr. Anthony Cashmore’s laboratory at the University of Pennsylvania. Since then, he has devoted himself in the research on the mechanism of how cryptochromes regulate blue light responses in plants.

Yoshito Oka is currently a professor at Basic Forestry and Protomics Research Center, FAFU. He obtained B.S. and M.S. degrees from the Tokyo University of Science and he received his Ph.D. in science from the Kyoto University in 2005. After he had worked as a postdoctoral researcher at the Kyoto University, University of California, Berkeley, and RIKEN, he moved to FAFU in 2014. During his research career, he has mostly focused on the phytochrome and cryptochrome regulation of plant light responses. Now he is working on the activation and inactivation mechanisms of plant cryptochrome.
REFERENCES
- 1.Ahmad M and Cashmore AR (1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366, 162–166. [DOI] [PubMed] [Google Scholar]
- 2.Sancar A (2003) Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev 103, 2203–2237. [DOI] [PubMed] [Google Scholar]
- 3.Lin C and Todo T (2005) The cryptochromes. Genome Biol. 6, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Partch C and Sancar A (2005) Photochemistry and photobiology of cryptochrome blue-light photopigments: The search for a photo-cycle. Photochem. Photobiol 81, 1291–1304. [DOI] [PubMed] [Google Scholar]
- 5.Lin C, Robertson DE, Ahmad M, Raibekas AA, Jorns MS, Dutton PL and Cashmore AR (1995) Association of flavin ade-nine dinucleotide with the Arabidopsis blue light receptor CRY1. Science 269, 968–970. [DOI] [PubMed] [Google Scholar]
- 6.Malhotra K, Kim ST, Batschauer A, Dawut L and Sancar A (1995) Putative blue-light photoreceptors from Arabidopsis thaliana and Sinapis alba with a high degree of sequence homology to DNA photolyase contain the two photolyase cofactors but lack DNA repair activity. Biochemistry 34, 6892–6889. [DOI] [PubMed] [Google Scholar]
- 7.Hsu DS, Zhao X, Zhao S, Kazantsev A, Wang RP, Todo T, Wei YF and Sancar A (1996) Putative human blue-light photoreceptors hCRY1 and hCRY2 are flavoproteins. Biochemistry 35, 13871–13877. [DOI] [PubMed] [Google Scholar]
- 8.Lin C and Shalitin D (2003) Cryptochrome structure and signal transduction. Annu. Rev. Plant Biol 54, 469–496. [DOI] [PubMed] [Google Scholar]
- 9.Gegear RJ, Foley LE, Casselman A and Reppert SM (2010) Animal cryptochromes mediate magnetoreception by an unconventional photochemical mechanism. Nature 463, 804–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Wang Q, Nguyen P and Lin C (2014) Cryptochrome-mediated light responses in plants. Enzymes 35, 167–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu H, Liu B, Zhao C, Pepper M and Lin C (2011) The action mechanisms of plant cryptochromes. Trends Plant Sci. 16, 684–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu B, Yang Z, Gomez A, Liu B, Lin C and Oka Y (2016) Signaling mechanisms of plant cryptochromes in Arabidopsis thaliana. J. Plant. Res 129, 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu PB, Lian HL, Wang WX, Xu F and Yang HQ (2016) Pivotal roles of the phytochrome-interacting factors in cryptochrome signaling. Mol. Plant 9, 496–497. [DOI] [PubMed] [Google Scholar]
- 14.Darwin C and Darwin F (1880) The Power of Movement in Plants. John Murray, London, England. [Google Scholar]
- 15.Kami C, Lorrain S, Hornitschek P and Fankhauser C (2010) Light-regulated plant growth and development. Curr. Top. Dev. Biol 91, 29–66. [DOI] [PubMed] [Google Scholar]
- 16.Galland P and Senger H (1991) Flavins as possible blue light photoreceptors In Photoreceptor Evolution and Function (Edited by Holmes MG), pp. 65–124. Academic Press, London, England. [Google Scholar]
- 17.Gressel J (1979) Blue Light Photoreception. Photochem. Photo-biol 30, 749–754. [Google Scholar]
- 18.Koornneef M, Rolff E and Spruit C (1980) Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z. Pflanzenphysiol 100, 147–160. [Google Scholar]
- 19.Chory J, Peto CA, Ashbaugh M, Saganich R, Pratt L and Ausubel F (1989) Different roles for phytochrome in etiolated and green plants deduced from characterization of Arabidopsis thaliana mutants. Plant Cell 1, 867–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koornneef M and Kendrick RE (1994) Photomorphogenic mutants of higher plants In Photomorphogenesis in Plants (Edited by Kendrick RE and Kronenberg GHM), pp. 601–628. Kluwer Academic, Dordrecht. [Google Scholar]
- 21.Lipson ED and Horwitz BA (1991) Photosensory reception and transduction In Sensory Receptors and Signal Transduction (Edited by u JL and Satir BH), pp. 1–64. Wiley-Liss, New York. [Google Scholar]
- 22.Briggs WR and Huala E (1999) Blue-light photoreceptors in higher plants. Annu. Rev. Cell Dev. Biol 15, 33–62. [DOI] [PubMed] [Google Scholar]
- 23.Lin C, Yang H, Guo H, Mockler T, Chen J and Cash-more AR (1998) Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc. Natl Acad. Sci. USA 95, 2686–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banerjee R, Schleicher E, Meier S, Viana RM, Pokorny R, Ahmad M, Bittl R and Batschauer A (2007) The signaling state of Arabidopsis cryptochrome 2 contains flavin semiquinone. J. Biol. Chem 282, 14916–14922. [DOI] [PubMed] [Google Scholar]
- 25.Zeugner A, Byrdin M, Bouly JP, Bakrim N, Giovani B, Brettel K and Ahmad M (2005) Light-induced electron transfer in Arabidopsis cryptochrome-1 correlates with in vivo function. J. Biol. Chem 280, 19437–19440. [DOI] [PubMed] [Google Scholar]
- 26.Bouly JP, Schleicher E, Dionisio-Sese M, Vandenbussche F, Van Der Straeten D, Bakrim N, Meier S, Batschauer A, Gal-land P, Bittl R and Ahmad M (2007) Cryptochrome blue light photoreceptors are activated through interconversion of flavin redox states. J. Biol. Chem 282, 9383–9391. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Wang Q, Yu X, Liu H, Yang H, Zhao C, Liu X, Tan C, Klejnot J, Zhong D and Lin C (2011) Arabidopsis cryptochrome 2 (CRY2) functions by the photoactivation mechanism distinct from the tryptophan (trp) triad-dependent photoreduction. Proc. Natl Acad. Sci. USA 108, 20844–20849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao J, Wang X, Zhang M, Bian M, Deng W, Zuo Z, Yang Z, Zhong D and Lin C (2015) Trp triad-dependent rapid photoreduction is not required for the function of Arabidopsis CRY1. Proc. Natl Acad. Sci. USA 112, 9135–9140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li YF and Sancar A (1990) Active site of Escherichia coli DNA photolyase: Mutations at Trp277 alter the selectivity of the enzyme without affecting the quantum yield of photorepair. Biochemistry 29, 5698–5706. [DOI] [PubMed] [Google Scholar]
- 30.Li YF, Heelis PF and Sancar A (1991) Active site of DNA photolyase: Tryptophan-306 is the intrinsic hydrogen atom donor essential for flavin radical photoreduction and DNA repair in vitro. Biochemistry 30, 6322–6329. [DOI] [PubMed] [Google Scholar]
- 31.Kavakli IH and Sancar A (2004) Analysis of the role of intraprotein electron transfer in photoreactivation by DNA photolyase in vivo. Biochemistry 43, 15103–15110. [DOI] [PubMed] [Google Scholar]
- 32.Song SH, Ozturk N, Denaro TR, Arat NO, Kao YT, Zhu H, Zhong D, Reppert SM and Sancar A (2007) Formation and function of flavin anion radical in cryptochrome 1 blue-light photoreceptor of monarch butterfly. J. Biol. Chem 282, 17608–17612. [DOI] [PubMed] [Google Scholar]
- 33.Ozturk N, Song SH, Selby CP and Sancar A (2008) Animal type 1 cryptochromes. Analysis of the redox state of the flavin cofactor by site-directed mutagenesis. J. Biol. Chem 283, 3256–3263. [DOI] [PubMed] [Google Scholar]
- 34.Ozturk N, Selby CP, Annayev Y, Zhong D and Sancar A (2011) Reaction mechanism of Drosophila cryptochrome. Proc. Natl Acad. Sci. USA 108, 516–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sang Y, Li QH, Rubio V, Zhang YC, Mao J, Deng XW and Yang HQ (2005) N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTO-CHROME 1. Plant Cell 17, 1569–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu X, Shalitin D, Liu X, Maymon M, Klejnot J, Yang H, Lopez J, Zhao X, Bendehakkalu KT and Lin C (2007) Derepression of the NC80 motif is critical for the photoactivation of Arabidopsis CRY2. Proc. Natl Acad. Sci. USA 104, 7289–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenfeldt G, Viana RM, Mootz HD, von Arnim AG and Batschauer A (2008) Chemically induced and light-independent cryptochrome photoreceptor activation. Mol. Plant 1, 4–14. [DOI] [PubMed] [Google Scholar]
- 38.Brautigam CA, Smith BS, Ma Z, Palnitkar M, Tom-chick DR, Machius M and Deisenhofer J (2004) Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 101, 12142–12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Partch CL, Clarkson MW, Ozgür S, Lee AL and Sancar A (2005) Role of structural plasticity in signal transduction by the cryptochrome blue-light photoreceptor. Biochemistry 44, 3795–3805. [DOI] [PubMed] [Google Scholar]
- 40.Yang HQ, Wu YJ, Tang RH, Liu D, Liu Y and Cashmore AR (2000) The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103, 815–827. [DOI] [PubMed] [Google Scholar]
- 41.Yang HQ, Tang RH and Cashmore AR (2001) The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13, 2573–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kondoh M, Shiraishi C, Müller P, Ahmad M, Hitomi K, Getzoff ED and Terazima M (2011) Light-induced conformational changes in full-length Arabidopsis thaliana cryptochrome. J. Mol. Biol 413, 128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shalitin D, Yang H, Mockler TC, Maymon M, Guo H, Whitelam GC and Lin C (2002) Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417, 763–767. [DOI] [PubMed] [Google Scholar]
- 44.Shalitin D, Yu X, Maymon M, Mockler T and Lin C (2003) Blue light-dependent in vivo and in vitro phosphorylation of Arabidopsis cryptochrome 1. Plant Cell 15, 2421–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu X, Klejnot J, Zhao X, Shalitin D, Maymon M, Yang H, Lee J, Liu X, Lopez J and Lin C (2007) Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell 19, 3146–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan ST, Dai C, Liu HT and Xue HW (2013) Arabidopsis casein kinase1 proteins CK1.3 and CK1.4 phosphorylate cryptochrome 2 to regulate blue light signaling. Plant Cell 25, 2618–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Q, Barshop WD, Bian M, Vashisht AA, He R, Yu X, Liu B, Nguyen P, Liu X, Zhao X, Wohlschlegel JA and Lin C (2015) The blue light-dependent phosphorylation of the CCE domain determines the photosensitivity of Arabidopsis CRY2. Mol. Plant 8, 631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bouly JP, Giovani B, Djamei A, Mueller M, Zeugner A, Dudkin EA, Batschauer A and Ahmad M (2003) Novel ATP-binding and autophosphorylation activity associated with Arabidopsis and human cryptochrome-1. Eur. J. Biochem 270, 2921–2928. [DOI] [PubMed] [Google Scholar]
- 49.Ozgur S and Sancar A (2006) Analysis of autophosphorylating kinase activities of Arabidopsis and human cryptochromes. Biochemistry 45, 13369–13374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahmad M, Jarillo JA and Cashmore AR (1998) Chimeric proteins between cry1 and cry2 Arabidopsis blue light photoreceptors indicate overlapping functions and varying protein stability. Plant Cell 10, 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu X, Sayegh R, Maymon M, Warpeha K, Klejnot J, Yang H, Huang J, Lee J, Kaufman L and Lin C (2009) Formation of nuclear bodies of Arabidopsis CRY2 in response to blue light is associated with its blue light-dependent degradation. Plant Cell 21, 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weidler G, Zur Oven-Krockhaus S, Heunemann M, Orth C, Schleifenbaum F, Harter K, Hoecker U and Batschauer A (2012) Degradation of Arabidopsis CRY2 is regulated by SPA proteins and phytochrome A. Plant Cell 24, 2610–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cashmore AR, Jarillo JA, Wu YJ and Liu D (1999) Cryptochromes: Blue light receptors for plants and animals. Science 284, 760–765. [DOI] [PubMed] [Google Scholar]
- 54.Wu G and Spalding EP (2007) Separate functions for nuclear and cytoplasmic cryptochrome 1 during photomorphogenesis of Arabidopsis seedlings. Proc. Natl Acad. Sci. USA 104, 18813–18818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zuo ZC, Meng YY, Yu XH, Zhang ZL, Feng DS, Sun SF, Liu B and Lin CT (2012) A study of the blue-light-dependent phosphorylation, degradation, and photobody formation of Arabidopsis CRY2. Mol. Plant 5, 726–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.M as P, Devlin PF, Panda S and Kay SA (2000) Functional interaction of phytochrome B and cryptochrome 2. Nature 408, 207–211. [DOI] [PubMed] [Google Scholar]
- 57.Ozkandagliyan I, Chiou YY, Ye R, Hassan BH, Ozturk N and Sancar A (2013) Formation of Arabidopsis Cryptochrome 2 photobodies in mammalian nuclei: Application as an optogenetic DNA damage checkpoint switch. J. Biol. Chem 288, 23244–23251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Folta KM, Pontin MA, Karlin Neumann G, Bottini R and Spalding EP (2003) Genomic and physiological studies of early cryptochrome 1 action demonstrate roles for auxin and gibberellin in the control of hypocotyl growth by blue light. Plant J. 36, 203–214. [DOI] [PubMed] [Google Scholar]
- 59.Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H and Deng XW (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13, 2589–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ohgishi M, Saji K, Okada K and Sakai T (2004) Functional analysis of each blue light receptor, cry1, cry2, phot1, and phot2, by using combinatorial multiple mutants in Arabidopsis. Proc. Natl Acad. Sci. USA 101, 2223–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sellaro R, Hoecker U, Yanovsky M, Chory J and Casal JJ (2009) Synergism of red and blue light in the control of Arabidopsis gene expression and development. Curr. Biol 19, 1216–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D and Lin C (2008) Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322, 1535–1539. [DOI] [PubMed] [Google Scholar]
- 63.Ma D, Li X, Guo Y, Chu J, Fang S, Yan C, Noel JP and Liu H (2016) Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl Acad. Sci. USA 113, 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pedmale UV, Huang SS, Zander M, Cole BJ, Hetzel J, Ljung K, Reis PA, Sridevi P, Nito K, Nery JR, Ecker JR and Chory J (2016) Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164, 233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ahmad M, Jarillo JA, Smirnova O and Cashmore AR (1998) The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol. Cell 1, 939–948. [DOI] [PubMed] [Google Scholar]
- 66.Jarillo JA, Capel J, Tang RH, Yang HQ, Alonso JM, Ecker JR and Cashmore AR (2001) An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature 410, 487–490. [DOI] [PubMed] [Google Scholar]
- 67.Wang Q, Zuo Z, Wang X, Gu L, Yoshizumi T, Yang Z, Yang L, Liu Q, Liu W, Han Y-J, Kim J-I, Liu B, Wohlschlegel JA, Matsui M, Oka Y and Lin C (2016) Photoactivation and inactivation of Arabidopsis cryptochrome 2. Science, 354, 343–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lau OS and Deng XW (2012) The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17, 584–593. [DOI] [PubMed] [Google Scholar]
- 69.Laubinger S, Fittinghoff K and Hoecker U (2004) The SPA quartet: A family of WD-repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. Plant Cell 16, 2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Laubinger S, Marchal V, Le Gourrierec J, Wenkel S, Adrian J, Jang S, Kulajta C, Braun H, Coupland G and Hoecker U (2006) Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133, 3213–3222. [DOI] [PubMed] [Google Scholar]
- 71.Yang J and Wang H (2006) The central coiled-coil domain and carboxyl-terminal WD-repeat domain of Arabidopsis SPA1 are responsible for mediating repression of light signaling. Plant J. 47, 564–576. [DOI] [PubMed] [Google Scholar]
- 72.Holm M, Hardtke CS, Gaudet R and Deng XW (2001) Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 47, 233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Osterlund MT, Hardtke CS, Wei N and Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466. [DOI] [PubMed] [Google Scholar]
- 74.Yi C and Deng XW (2005) COP1-from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 15, 618–625. [DOI] [PubMed] [Google Scholar]
- 75.Wang H, Ma LG, Li JM, Zhao HY and Deng XW (2001) Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294, 154–158. [DOI] [PubMed] [Google Scholar]
- 76.Lian HL, He SB, Zhang YC, Zhu DM, Zhang JY, Jia KP, Sun SX, Li L and Yang HQ (2011) Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 25, 1023–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu B, Zuo Z, Liu H, Liu X and Lin C (2011) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 25, 1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U and Deng XW (2003) The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 17, 2642–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML and Chua NH (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423, 995–999. [DOI] [PubMed] [Google Scholar]
- 80.Zuo Z, Liu H, Liu B, Liu X and Lin C (2011) Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr. Biol 21, 841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu Y, Li X, Li K, Liu H and Lin C (2013) Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet. 9, e1003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu H, Wang Q, Liu Y, Zhao X, Imaizumi T, Somers DE, Tobin EM and Lin C (2013) Arabidopsis CRY2 and ZTL mediate blue-light regulation of the transcription factor CIB1 by distinct mechanisms. Proc. Natl Acad. Sci. USA 110, 17582–17587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Leivar P and Quail PH (2011) PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 16, 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Khanna R, Huq EKikis EA, Al-Sady B, Lanzatella C and Quail PH (2004) A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 16, 3033–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Park E, Kim J, Lee Y, Shin J, Oh E, Chung WI, Liu JR and Choi G (2004) Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 45, 968–975. [DOI] [PubMed] [Google Scholar]
- 86.Bauer D, Viczián A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KC, Ádám É, Fejes E and Schäfer E (2004) Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16, 1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Al-Sady B, Ni W, Kircher S, Schäfer E and Quail PH (2006) Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23, 439–446. [DOI] [PubMed] [Google Scholar]
- 88.Shen H, Zhu L, Castillon A, Majee M, Downie B and Huq E (2008) Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR 1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20, 1586–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shen Y, Khanna R, Carle CM and Quail PH (2007) Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 145, 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lorrain S, Allen T, Duek PD, Whitelam GC and Fankhauser C (2008) Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53, 312–323. [DOI] [PubMed] [Google Scholar]
- 91.Leivar P, Monte E, Al-Sady B, Carle C, Storer A, Alonso JM, Ecker JR and Quail PH (2008) The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20, 337–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E and Quail PH (2008) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol 18, 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D and Choi G (2009) Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl Acad. Sci. USA 106, 7660–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Castillon A and Shen HE (2009) Blue light induces degradation of the negative regulator Phytochrome Interacting Factor 1 to promote photomorphogenic development of Arabidopsis seedlings. Genetics 182, 161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bu Q, Castillon A, Chen F, Zhu L and Huq E (2011) Dimerization and blue light regulation of PIF1 interacting bHLH proteins in Arabidopsis. Plant Mol. Biol 77, 501–511. [DOI] [PubMed] [Google Scholar]
- 96.Chen D, Xu G, Tang W, Jing Y, Ji Q, Fei Z and Lin R (2013) Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 25, 1657–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Toledo-Ortiz G, Johansson H, Lee KP, Bou-Torrent J, Stewart K, Steel G, Rodr ıguez-Concepci on M and Halliday KJ (2014) The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet. 10, e1004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hornitschek P, Lorrain S, Zoete V, Michielin O and Fankhauser C (2009) Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 28, 3893–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Galstyan A, Cifuentes-Esquivel N, Bou-Torrent J and Martinez-Garcia JF (2011) The shade avoidance syndrome in Arabidopsis: A fundamental role for atypical basic helix-loop-helix proteins as transcriptional cofactors. Plant J. 66, 258–267. [DOI] [PubMed] [Google Scholar]
- 100.Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I and Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19, 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Delker C, Sonntag L, James GV, Janitza P, Ibañez C, Zier-mann H, Peterson T, Denk K, Mull S and Ziegler J (2014) The DET1-COP1-HY5 pathway constitutes a multipurpose signaling module regulating plant photomorphogenesis and thermomorpho-genesis. Cell Rep. 9, 1983–1989. [DOI] [PubMed] [Google Scholar]
- 102.Xu X, Paik I, Zhu L, Bu Q, Huang X, Deng XW and Huq E (2014) PHYTOCHROME INTERACTING FACTOR1 enhances the E3 ligase activity of CONSTITUTIVE PHOTOMORPHOGENIC1 to synergistically repress photomorphogenesis in Arabidopsis. Plant Cell 26, 1992–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Strasser B, S anchez-Lamas M, Yanovsky MJ, Casal JJ and Cerd an PD (2010) Arabidopsis thaliana life without phytochromes. Proc. Natl Acad. Sci. USA 107, 4776–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lu XD, Zhou CM, Xu PB, Luo Q, Lian HL and Yang HQ (2015) Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol. Plant 8, 467–478. [DOI] [PubMed] [Google Scholar]
- 105.Sheerin DJ, Menon C, zur Oven Krockhaus S, Enderle B, Zhu L, Johnen P, Schleifenbaum F, Stierhof YD, Huq E and Hiltbrunner A (2015) Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27, 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mockler TC, Guo H, Yang H, Duong H and Lin C (1999) Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126, 2073–2082. [DOI] [PubMed] [Google Scholar]
- 107.Mockler T, Yang H, Yu X, Parikh D, Cheng YC, Dolan S and Lin C (2003) Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc. Natl Acad. Sci. USA 100, 2140–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Casal JJ (2000) Phytochromes, cryptochromes, phototropin: Photoreceptor interactions in plants. Photochem. Photobiol 71, 1–11. [DOI] [PubMed] [Google Scholar]
- 109.Chen S, Lory N, Stauber J and Hoecker U (2015) Photoreceptor specificity in the light-induced and COP1-mediated rapid degradation of the repressor of photomorphogenesis SPA2 in Arabidopsis. PLoS Genet. 11, e1005516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Endo M, Mochizuki N, Suzuki T and Nagatani A (2007) CRYPTOCHROME 2 in vascular bundles regulates flowering in Arabidopsis. Plant Cell 19, 84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Endo M, Nakamura S, Araki T, Mochizuki N and Nagatani A (2005) Phytochrome B in the mesophyll delays flowering by suppressing FLOWERING LOCUS T expression in Arabidopsis vascular bundles. Plant Cell 17, 1941–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Guo H, Yang H, Mockler TC and Lin C (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279, 1360–1363. [DOI] [PubMed] [Google Scholar]
- 113.Mao J, Zhang YC, Sang Y, Li QH and Yang HQ (2005) A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc. Natl Acad. Sci. USA 102, 12270–12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kang CY, Lian HL, Wang FF, Huang JR and Yang HQ (2009) Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 21, 2624–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang FF, Lian HL, Kang CY and Yang HQ (2010) Phytochrome B is involved in mediating red light-induced stomatal opening in Arabidopsis thaliana. Mol. Plant 3, 246–259. [DOI] [PubMed] [Google Scholar]
- 116.Somers DE, Devlin PF and Kay SA (1998) Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282, 1488–1490. [DOI] [PubMed] [Google Scholar]
- 117.Devlin PF and Kay SA (2000) Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell 12, 2499–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gould PD, Ugarte N, Domijan M, Costa M, Foreman J, Macgregor D, Rose K, Griffiths J, Millar AJ, Finkenstadt B, Penfield S, Rand DA, Halliday KJ and Hall AJ (2013) Network balance via CRY signalling controls the Arabidopsis circadian clock over ambient temperatures. Mol. Syst. Biol 9, 650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Canamero RC, Bakrim N, Bouly J-P, Garay A, Dudkin EE, Habricot Y and Ahmad M (2006) Cryptochrome photoreceptors cry1 and cry2 antagonistically regulate primary root elongation in Arabidopsis thaliana. Planta 224, 995–1003. [DOI] [PubMed] [Google Scholar]
- 120.Zeng J, Wang Q, Lin J, Deng K, Zhao X, Tang D and Liu X (2010) Arabidopsis cryptochrome-1 restrains lateral roots growth by inhibiting auxin transport. J. Plant Physiol 167, 670–673. [DOI] [PubMed] [Google Scholar]
- 121.Danon A, Coll NS and Apel K (2006) Cryptochrome-1-dependent execution of programmed cell death induced by singlet oxygen in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 103, 17036–17041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lin C, Ahmad M, Gordon D and Cashmore AR (1995) Expression of an Arabidopsis cryptochrome gene in transgenic tobacco results in hypersensitivity to blue, UV-A, and green light. Proc. Natl Acad. Sci. USA 92, 8423–8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lin C, Ahmad M and Cashmore AR (1996) Arabidopsis cryptochrome 1 is a soluble protein mediating blue light-dependent regulation of plant growth and development. Plant J. 10, 893–902. [DOI] [PubMed] [Google Scholar]
- 124.Neff MM and Chory J (1998) Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 118, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.He SB, Wang WX, Zhang JY, Xu F, Lian HL, Li L and Yang HQ (2015) The CNT1 domain of Arabidopsis CRY1 alone is sufficient to mediate blue light inhibition of hypocotyl elongation. Mol. Plant 8, 822–825. [DOI] [PubMed] [Google Scholar]
- 126.Duek PD and Fankhauser C (2003) HFR1, a putative bHLH transcription factor, mediates both phytochrome A and cryptochrome signalling. Plant J. 34, 827–836. [DOI] [PubMed] [Google Scholar]
- 127.Fairchild CD, Schumaker MA and Quail PH (2000) HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 14, 2377–2391. [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang X, Wu Y, Tobias JW, Brunk BP, Deitzer GF and Liu D (2008) HFR1 is crucial for transcriptome regulation in the cryptochrome 1-mediated early response to blue light in Arabidopsis thaliana. PLoS ONE 3, e3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Duek PD, Elmer MV, van Oosten VR and Fankhauser C (2004) The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr. Biol 14, 2296–2301. [DOI] [PubMed] [Google Scholar]
- 130.Yang J, Lin R, Sullivan J, Hoecker U, Liu B, Xu L, Deng XW and Wang H (2005) Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17, 804–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.El-Assal SE-D, Alonso-Blanco C, Peeters AJ, Raz V and Koornneef M (2001) A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat. Genet 29, 435–440. [DOI] [PubMed] [Google Scholar]
- 132.El-Assal SE-D, Alonso-Blanco C, Peeters AJ, Wagemaker C, Weller JL and Koornneef M (2003) The role of cryptochrome 2 in flowering in Arabidopsis. Plant Physiol. 133, 1504–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mozley D and Thomas B (1995) Developmental and photobiological factors affecting photoperiodic induction in Arabidopsis thaliana Heynh, Landsberg erecta. J. Exp. Bot 46, 173–179. [Google Scholar]
- 134.Bagnall DJ, King RW and Hangarter RP (1996) Blue-light promotion of flowering is absent in hy4 mutants of Arabidopsis. Planta 200, 278–280. [DOI] [PubMed] [Google Scholar]
- 135.Goto N, Kumagai T and Koornneef M (1991) Flowering responses to light-breaks in photomorphogenic mutants of Arabidopsis thaliana, a long-day plant. Physiol. Plant 83, 209–215. [Google Scholar]
- 136.Blazquez MA, Ahn JH and Weigel D (2003) A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat. Genet 33, 168–171. [DOI] [PubMed] [Google Scholar]
- 137.Exner V, Alexandre C, Rosenfeldt G, Alfarano P, Nater M, Caflisch A, Gruissem W, Batschauer A and Hennig L (2010) A gain-of-function mutation of Arabidopsis cryptochrome 1 promotes flowering. Plant Physiol. 154, 1633–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gu NN, Zhang YC and Yang HQ (2012) Substitution of a conserved glycine in the PHR domain of Arabidopsis cryptochrome 1 confers a constitutive light response. Mol. Plant 5, 85–97. [DOI] [PubMed] [Google Scholar]
- 139.Yu JW, Rubio V, Lee NY, Bai S, Lee SY, Kim SS, Liu L, Zhang Y, Irigoyen ML and Sullivan JA (2008) COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell 32, 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F and Coupland G (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27, 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L and Yang HQ (2008) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20, 292–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Searle I and Coupland G (2004) Induction of flowering by seasonal changes in photoperiod. EMBO J. 23, 1217–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A and Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003–1006. [DOI] [PubMed] [Google Scholar]
- 144.Yu X, Liu H, Klejnot J and Lin C (2010) The cryptochrome blue light receptors In Arabidopsis Book, pp. e0135 American Society of Plant Biologists, Rockville. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Brock MT, Maloof JN and Weinig C (2010) Genes underlying quantitative variation in ecologically important traits: PIF4 (phytochrome interacting factor 4) is associated with variation in intern-ode length, flowering time, and fruit set in Arabidopsis thaliana. Mol. Ecol 19, 1187–1199. [DOI] [PubMed] [Google Scholar]
- 146.Nozue K, Harmer SL and Maloof JN (2011) Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis. Plant Physiol. 156, 357–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kumar SV, Lucyshyn D, Jaeger KE, Alos E, Alvey E, Harberd NP and Wigge PA (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484, 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kinoshita T, Doi M, Suetsugu N, Kagawa T, Wada M and Shimazaki K (2001) Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414, 656–660. [DOI] [PubMed] [Google Scholar]
- 149.Ando E, Ohnishi M, Wang Y, Matsushita T, Watanabe A, Hayashi Y, Fujii M, Ma JF, Inoue S and Kinoshita T (2013) TWIN SISTER OF FT, GIGANTEA, and CONSTANS have a positive but indirect effect on blue light-induced stomatal opening in Arabidopsis. Plant Physiol. 162, 1529–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kinoshita T, Ono N, Hayashi Y, Morimoto S, Nakamura S, Soda M, Kato Y, Ohnishi M, Nakano T, Inoue S and Shimazaki K (2011) FLOWERING LOCUS T regulates stomatal opening. Curr. Biol 21, 1232–1238. [DOI] [PubMed] [Google Scholar]
- 151.Jeong RD, Chandra-Shekara AC, Barman SR, Navarre D, Klessig DF, Kachroo A and Kachroo P (2010) Cryptochrome 2 and phototropin 2 regulate resistance protein-mediated viral defense by negatively regulating an E3 ubiquitin ligase. Proc. Natl Acad. Sci. USA 107, 13538–13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Casson SA, Franklin KA, Gray JE, Grierson CS, Whitelam GC and Hetherington AM (2009) Phytochrome B and PIF4 regulate stomatal development in response to light quantity. Curr. Biol 19, 229–234. [DOI] [PubMed] [Google Scholar]
- 153.Casal JJ (2012) Shade avoidance The Arabidopsis Book. pp. e0157 American Society of Plant Biologists, Rockville. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, Cole BJ, Ivans LJ, Pedmale UV, Jung HS, Ecker JR, Kay SA and Chory J (2012) Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26, 785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, Cheng Y, Lim J, Zhao Y, Ballare CL, Sandberg G, Noel JP and Chory J (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Keller MM, Jaillais Y, Pedmale UV, Moreno JE, Chory J and Ballare CL (2011) Cryptochrome 1 and phytochrome B control shade-avoidance responses in Arabidopsis via partially independent hormonal cascades. Plant J. 67, 195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fraser DP, Hayes S and Franklin KA (2016) Photoreceptor crosstalk in shade avoidance. Curr. Opin. Plant Biol 33, 1–7. [DOI] [PubMed] [Google Scholar]
- 158.Keuskamp DH, Sasidharan R, Vos I, Peeters AJ, Voesenek LA and Pierik R (2011) Blue-light-mediated shade avoidance requires combined auxin and brassinosteroid action in Arabidopsis seedlings. Plant J. 67, 208–217. [DOI] [PubMed] [Google Scholar]
- 159.Rolauffs S, Fackendahl P, Sahm J, Fiene G and Hoecker U (2012) Arabidopsis COP1 and SPA genes are essential for plant elongation but not for acceleration of flowering time in response to a low red light to far-red light ratio. Plant Physiol. 160, 2015–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pacin M, Semmoloni M, Legris M, Finlayson SA and Casal JJ (2016) Convergence of CONSTITUTIVE PHOTOMORPHO-GENESIS 1 and PHYTOCHROME INTERACTING FACTOR signalling during shade avoidance. New Phytol., 211, 967–979. [DOI] [PubMed] [Google Scholar]
- 161.Johansson H, Jones HJ, Foreman J, Hemsted JR, Stewart K, Grima R and Halliday KJ (2014) Arabidopsis cell expansion is controlled by a photothermal switch. Nat. Commun 5, 4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Imaizumi T, Kadota A, Hasebe M and Wada M (2002) Cryptochrome light signals control development to suppress auxin sensitivity in the moss Physcomitrella patens. Plant Cell 14, 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Imaizumi T, Kanegae T and Wada M (2000) Cryptochrome nucleocytoplasmic distribution and gene expression are regulated by light quality in the fern Adiantum capillus-veneris. Plant Cell 12, 81–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wada M and Furuya M (1978) Effects of narrow-beam irradiations with blue and far-red light on the timing of cell division in Adiantum gametophytes. Planta 138, 85–90. [DOI] [PubMed] [Google Scholar]
- 165.Furuya M, Kanno M, Okamoto H, Fukuda S and Wada M (1997) Control of mitosis by phytochrome and a blue-light receptor in fern spores. Plant Physiol. 113, 677–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hirose F, Shinomura T, Tanabata T, Shimada H and Takano M (2006) Involvement of rice cryptochromes in de-etiolation responses and flowering. Plant Cell Physiol. 47, 915–925. [DOI] [PubMed] [Google Scholar]
- 167.Zhang YC, Gong SF, Li QH, Sang Y and Yang HQ (2006) Functional and signaling mechanism analysis of rice CRYPTOCHROME 1. Plant J. 46, 971–983. [DOI] [PubMed] [Google Scholar]
- 168.Perrotta G, Yahoubyan G, Nebuloso E, Renzi L and Giuliano G (2001) Tomato and barley contain duplicated copies of cryptochrome 1. Plant, Cell Environ. 24, 991–998. [Google Scholar]
- 169.Barrero JM, Downie AB, Xu Q and Gubler F (2014) A role for barley CRYPTOCHROME 1 in light regulation of grain dormancy and germination. Plant Cell 26, 1094–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Poppe C, Sweere U, Drumm Herrel H and Schäfer E (1998) The blue light receptor cryptochrome 1 can act independently of phytochrome A and B in Arabidopsis thaliana. Plant J. 16, 465–471. [DOI] [PubMed] [Google Scholar]
- 171.Zhang Q, Li H, Li R, Hu R, Fan C, Chen F, Wang Z, Liu X, Fu Y and Lin C (2008) Association of the circadian rhythmic expression of GmCRY1a with a latitudinal cline in photoperiodic flowering of soybean. Proc. Natl Acad. Sci. USA 105, 21028–21033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Giliberto L, Perrotta G, Pallara P, Weller JL, Fraser PD, Bramley PM, Fiore A, Tavazza M and Giuliano G (2005) Manipulation of the blue light photoreceptor cryptochrome 2 in tomato affects vegetative development, flowering time, and fruit antioxidant content. Plant Physiol. 137, 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Meng Y, Meng Y, Li H, Wang Q, Liu B and Lin C (2013) Blue light-dependent Interaction between cryptochrome 2 and CIB1 regulates transcription and leaf senescence in soybean. Plant Cell 25, 4405–4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zentgraf U, Laun T and Miao Y (2010) The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur. J. Cell Biol 89, 133–137. [DOI] [PubMed] [Google Scholar]
- 175.Ranjan A, Dickopf S, Ullrich KK, Rensing SA and Hoecker U (2014) Functional analysis of COP1 and SPA orthologs from Physcomitrella and rice during photomorphogenesis of transgenic Arabidopsis reveals distinct evolutionary conservation. BMC Plant Biol. 14, 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Inoue K, Nishihama R, Kataoka H, Hosaka M, Manabe R, Nomoto M, Tada Y, Ishizaki K and Kohchi T (2016) Phytochrome signaling is mediated by PHYTOCHROME INTERACTING FACTOR in the liverwort Marchantia polymorpha. Plant Cell 28, 1406–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]