Abstract
Background
Genome-wide association studies have implicated the transcription factor 7-like 2 (TCF7L2) gene in type 2 diabetes risk, and more recently, in decreased body mass index. Given the contrary direction of genetic effects on these two traits, it has been suggested that the observed association with body mass index may reflect either selection bias or a complex underlying biology at TCF7L2.
Methods
Using 9031 Hispanic/Latino adults (21–76 years) with complete weight history and genetic data from the community-based Hispanic Community Health Study/Study of Latinos (HCHS/SOL, Baseline 2008–2011), we estimated the multivariable association between the additive number of type 2 diabetes increasing-alleles at TCF7L2 (rs7903146-T) and body mass index. We then used structural equation models to simultaneously model the genetic association on changes in body mass index across the life course and estimate the odds of type 2 diabetes per TCF7L2 risk allele.
Results
We observed both significant increases in type 2 diabetes prevalence at examination (independent of body mass index) and decreases in mean body mass index and waist circumference across genotypes at rs7903146. We observed a significant multivariable association between the additive number of type 2 diabetes-risk alleles and lower body mass index at examination. In our structured modeling, we observed non-significant inverse direct associations between rs7903146-T and body mass index at ages 21 and 45 years, and a significant positive association between rs7903146-T and type 2 diabetes onset in both middle and late adulthood.
Conclusions
Herein, we replicated the protective effect of rs7930146-T on body mass index at multiple time points in the life course, and observed that these effects were not explained by past type 2 diabetes status in our structured modeling. The robust replication of the negative effects of TCF7L2 on body mass index in multiple samples, including in our diverse Hispanic/Latino community-based sample, supports a growing body of literature on the complex biologic mechanism underlying the functional consequences of TCF7L2 on obesity and type 2 diabetes across the life course.
Electronic supplementary material
The online version of this article (10.1186/s40608-018-0200-x) contains supplementary material, which is available to authorized users.
Keywords: TCF7L2, Genetics, Obesity, Diabetes, Hispanic/Latinos
Background
Hispanic/Latino adults in the United States (US) are disproportionally affected by obesity and it consequences such as type 2 diabetes (T2D) [1] and this disparity is widening as compared to non-Hispanic Whites [2]. The transcription factor-7 like 2 gene (TCF7L2) was the first locus to be associated with T2D in genome-wide association studies (GWAS) and has been consistently associated with T2D [3, 4], TCF7L2 (previously known as TCF4) encodes a transcription factor that is an effector of the Wnt signaling pathway [5]. Although the underlying biological mechanisms of TCF7L2 remain unclear [6], the consistent association between the TCF7L2 locus and T2D has been generalized to many diverse populations including Hispanic/Latinos [7, 8]. Indeed, the associated risk allele, rs7903146-T, harbored within the fourth intron of TCF7L2 has the largest effect on T2D risk of all GWAS-identified T2D loci reported to date [8]. In Hispanic/Latinos each risk-allele has been associated with a 40% increased odds of T2D [7, 9].
The T2D-increasing allele at TCF7L2 has also been associated with lower body mass index (BMI) [3, 10–12], resulting in a subsequent call for future research [13] given the strong epidemiologic correlation between increasing BMI and risk of T2D [14]. This association has been attributed to a T2D-related ascertainment bias, mainly due to the observation that the strongest and most significant TCF7L2 associations with BMI are seen in T2D cases/controls, as compared to population-based studies [15–17].
There is mounting evidence of a complex biologic story for TCF7L2, explained in part by the bidirectional action of TCF7L2 that may be cell, tissue or metabolically dependent [5]. Functional studies indicate that the rs7903146 variant may act in a cell or tissue-specific manner [18], by influencing alternative splicing of the TCF7L2 [19–21], or by binding affinity of complex transcriptional machinery at an open chromatin region specific to human pancreatic islets [22–25] to modulate pancreatic islet cell insulin production and secretion [17], action in adipose tissue [26], hepatic glucose output [27] or intestinal tissue differentiation [28]. Observational studies indicate that the T2D risk allele at TCF7L2 associates with decreases insulin secretion [29–31]. Thus, we may expect individuals with the T allele have lower BMI values on average, and perhaps a differential pattern of insulin resistance.
Due to the mounting evidence on potential selection bias and the multi-faceted action of TCF7L2 variation on insulin and glucose biology [5, 6, 18], we aimed 1) to replicate the multivariable association between TCF7L2 T2D risk alleles and lower BMI in a population-based study of US Hispanic/Latinos accounting for key covariates, and 2) to model the structured pathways between rs7903146, at TCF7L2, BMI over time, and age of diabetes diagnosis. We performed these analyses in 9031 self-identified Hispanic/Latino adults (21–76 years of age at examination) residing in four US urban centers, who consented to genotyping and provided weight history and T2D diagnosis information the Hispanic Community Health Study/Study of Latinos (HCHS/SOL) baseline examination (2008–2011).
Methods
Study participants
We used data from the HCHS/SOL study, a multi-center, longitudinal, household-based cohort study of 16,415 Hispanic/Latino adults, aged 18–76 years in 2008–2011, who were sampled using a two-stage probability design from four US urban communities (The Bronx, NY; Chicago, IL; Miami, FL; San Diego, CA), as described previously in detail [32, 33]. Briefly, the complex sampling design allowed researchers to 1) over-sample individuals ≥45 years of age who were most likely to experience cardiometabolic disease outcomes either by the baseline examination or during follow up, while 2) capturing the varied socioeconomic and demographic composition of Hispanic/Latino households (as per the 2000 Census block group proportion of residents ≥25 years old with at least a high school education and the proportion Hispanic/Latino residents) and efficiently estimating cardiometabolic disease across the four Hispanic/Latino communities under study. Centrally-trained study personnel conducted the screening and baseline examinations in either English or Spanish based on participant preference.
Body mass index
As part of the HCHS/SOL baseline examination [32], current body weight was self-reported (in whole lb. or kg) and measured (to a tenth of a kg) and height was measured (to whole cm) on participants who were able to stand on both feet. As described previously [34], the accuracy and reliability of the self-reported weights were good (mean differenceself-report–measured = 0.23 kg, r2 = 0.97; inter-rater reliability coefficients, 0.93 and 0.97). Waist circumference was measured in cm at the umbilicus using a tape measure, and body fat percentage estimated by a Tanita Body Composition Analyzer.
Additionally, a weight history questionnaire was used to collect self-reported body weights (in whole lb. or kg, while not pregnant) at 21, 45, and 65 years of age, for individuals 21 years or older at baseline. If participants indicated that they could not remember their exact weight, personnel were instructed to inquire about their best guess. The quality control procedures and data cleaning are described in the Appendix. We converted each weight from the weight histories to kg and rounded each weight to the whole unit, to eliminate measurement error by unit of report (e.g. lb. or kg).
We excluded all weights from women who reported currently being pregnant at baseline or individuals with limb amputations that otherwise did not limit their ability to stand (Additional file 1: Figure S2 and Table S1). Using measured height at baseline, we calculated two baseline BMI measures (kg/m2) and up to three BMIs from the weight histories of individuals at least 21 years of age (corresponding to 21, 45, and 65 years). We further excluded any BMI that was less < 16 or > 70 kg/m2. As measured height is an imperfect proxy of an individual’s height at various times in the past, all models of BMI from the weight histories (at 21, 45, and 65 years) also accounted for the age at baseline as a measure of age at time of recall.
Type 2 diabetes assessment
HCHS/SOL participants were asked to bring in the medications they were currently taking, during the baseline examination. Individuals were also asked to report if a “doctor ever said that you have diabetes (high sugar in blood or urine)” and the age when this diagnosis was received. Participants were asked to fast overnight (> 8 h) and their glucose was measured in the entire sample, and 2-h post-oral glucose tolerance tests was measured among those who reported never having received a diabetes diagnosis. Impaired fasting glucose among non-diabetics was defined as a fasting glucose 100-125 mg/dL or 140-199 mg/dL after oral glucose challenge. We used the American Diabetes Association definition to identify T2D cases at examination based on fasting glucose (≥126 mg/dL), an oral glucose tolerance test (OGTT, ≥200 mg/dL), percent Glycated Hemoglobin (HbA1C ≥6.5%), or diabetes medications [35]. Controlled diabetes was further defined as % HbA1C < 7%.
Type of diabetes was not reported in HCHS/SOL. Therefore, we used information on age at diabetes diagnosis to create age period-specific T2D diagnosis indicators. If an individual was younger than 45 or 65 years at the baseline examination, then the classification of T2D diagnosis of the incomplete age period was set to missing (e.g. for a 50-year-old, T2D diagnosis for the period of 22–45 years could be yes/no, but would be set to missing for 46–65 years).
Genetic information
Venous blood samples were collected and for all fully consenting participants (i.e. those agreeing to genotyping and sharing of information with HCHS/SOL investigators, those not affiliated with HCHS/SOL, and specialized laboratories) and were analyzed using the MetaboChip (Illumina, Inc., San Diego, CA) (N = 12,209 or 74% of the cohort). The MetaboChip array contains approximately 200,000 single nucleotide polymorphisms (SNPs) at 257 genomic regions previously associated with cardiometabolic traits, including the TCF7L2 region that includes 258 SNPs across over 76,159 bp [36]. HCHS/SOL participants used in this study were genotyped at the Human Genetics Center of the University of Texas-Houston (Houston, TX) and passed person-level quality control filters (< 95% call rate, sex discordance or duplicate).
Based on previous trans-ethnic fine-mapping studies with T2D [37] and BMI [15], we selected rs7903146 as our presumed functional variant of interest at TCF7L2 as it was in strong linkage disequilibrium with several other variants in the area (Additional file 1: Figure S1). In HCHS/SOL, this SNP also had satisfactory quality control measures [38], was in Hardy-Weinberg-Equilibrium (P value = 0.10), and available in the entire sample that passed genetic quality control procedures (n = 12,117). We created an additive score of the number of T2D risk alleles [7, 8] per individual at rs7903146 (e.g. CC = 0, CT = 1, TT = 2). To aid in the interpretability of adjustments for population stratification, we adjusted for continental ancestry proportions, which as reported on previously [39] were designed to represent four a priori-selected ancestral populations using a supervised analysis (K = 4; unrelated 1000 Genomes references representing European: CEU; African: YRI; Northern: MXL; Caribbean/Southern Native American Ancestry: PUR, CLM) in the program ADMIXTURE [40] on a pruned set of more than 45,000 MetaboChip SNPs in low linkage disequilibrium in our sample (r2 < 0.5). Lastly, we also adjusted for the ‘genetic analysis group’ variable from the multidimensional clustering of self-reported Hispanic/Latino background and principal components from genome-wide data on a majority-overlapping sample of 12,803 HCHS/SOL participants (> 99% call rate), as described previously [39].
Statistical analyses
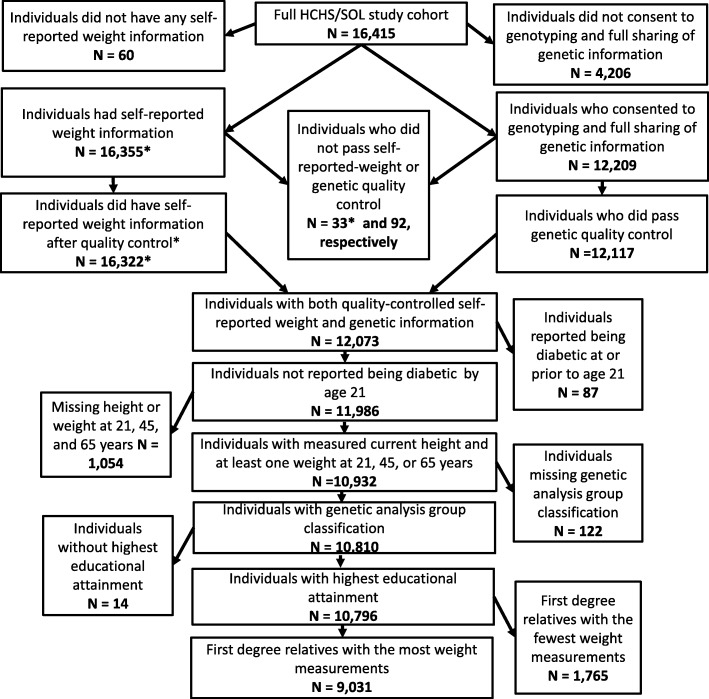
As shown in Fig. 1, of the entire HCHS/SOL baseline cohort of 16,415 participants, 16,322 individuals had self-reported and measured weight values that passed quality control (additional information provided as part of Additional file 1: Figure S2 and Table S1). Of the 12,209 individuals providing their full informed consent for genotyping and data sharing, 12,117 passed genetic quality control, as described above. The union of these two quality controlled data sets included 12,073 individuals (Fig. 1), from which we excluded 87 individuals who reported diabetes diagnosis prior to 22 years of age, to restrict our analysis to those for which a diabetes diagnosis was more likely to be T2D, and 1054 individuals that did not have both a measured current height or at least one self-reported weight at 21, 45 or 65 years and who were therefore unable to contribute to our structural equation modeling. Individuals with missing covariate information, such as missing genetic analysis group (N = 122) or information on their highest education level achieved (categorized as less than or at least a high school diploma or equivalency) (N = 14), were excluded. Lastly, as described previously in HCHS/SOL we used an identity-by-descent analysis in PLINK [41] to identify close relatives (e.g. 0.35 < π < 0.98) [42], and exclude the individual in each pair with the least weight measurements (N = 1765). A total of 9031 individuals remained in the final analytic dataset used for all analyses, and we described their characteristics using descriptive statistics such as means, 95% confidence intervals (CIs), and frequencies.
Fig. 1.
Flowchart of Quality Control and Exclusions Applied to the Hispanic Community Health Study/Study of Latinos (HCHS/SOL) Cohort of 16,415 Individuals (18–76 years), which Resulted in a Final Analytic Sample of 9031 Hispanic/Latino Adults (21–76 years), Which Are Detailed Further in the Additional file 1: Table S1 and Figure S2*
Next, we modeled the association between the additive number of rs7903146-T alleles with multiple BMI measures using multivariable models (e.g. measured and self-reported BMI at baseline, as well as BMI for ages 21, 45 and 65 years), controlling for age at examination, sex, educational attainment, admixture proportions, and genetic analysis group. As an exploratory analysis, we also examined the multivariable associations with the measured BMI stratified by previous diabetes diagnosis, as well as glucose tolerance and diabetic medication at examination.
Using structural equation models, we then examined an a priori-specified set of pathways (Additional file 1: Figure S3) between the additive number of rs7903146-T alleles, BMI (at age 21, 45 and 65), and self-reported T2D status (between ages 22–45 and between ages 46–65). BMI was assumed to be directly associated with T2D status in the period immediately after the BMI measurement (between ages 22–45 or 46–65). Similarly, T2D status during the period of time immediately preceding a given BMI (e.g. T2D between 22 and 45 years and 45-year-old weight) was assumed to be directly associated with the BMI at that time. Direct pathways between rs7903146 to BMIs and T2D measurements were also included. BMI at the previous age was assumed to be directly related to BMI at the following age. Age at baseline examination (age at time of recall), sex, education level, admixture proportions and genetic analysis group, and were included in all pathways to BMI and T2D.
All analyses accounted for the HCHS/SOL complex sampling design, including primary sampling unit, strata and sampling weights, yielding valid estimates of the disease distribution in the source population. Descriptive statistics were estimated using SAS 9.3 (Research Triangle Park, NC). All multivariable and structural equation models were estimated using Mplus 7.11 software [43], using full-information maximum likelihood methods to account for missing outcome data. Additionally, we identified our analysis subpopulation (N = 9031), or the subpopulation of interest in any stratified models, and used the complex sampling information on the entire cohort in the variance calculations to ensure valid estimates for the source population of HCHS/SOL.
Results
Our weighted sample included women (50%) and men, an average age of 44 years at baseline examination (Table 1). Five percent of those who were at least 45 years old (unweighted n = 5605) received a T2D diagnosis by age 45 (Table 1). In the subsample of participants who were at least 65 years of age (n = 729), 23% reported received a diabetes diagnosis by age 65, with 3% being diagnosed by age 45 and 20% diagnosed between ages 46–65 years. Average BMIs increased across age of recalled weight (24 kg/m2 at 21 years to 29 kg/m2 at 65 years).
Table 1.
Weighted Descriptive Statistics of Hispanic Community Health Study/Study of Latinos Baseline (2008–2011) Analytic Sample of 9031 Adults (21–76 years) from Four Urban United States Centers: The Bronx, NY; Chicago, IL; Miami, FL; San Diego, CA
| Unweighted Analytic Sample Size and Weighted Frequency or Means (95% Confidence Interval) | ||
| Female (%) | n = 5187 | 50% |
| Mean (95% CI) Age at Baseline Examination | n = 9031 | 43.67 (43.12, 44.21) |
| Has High School Diploma or Equivalency (%) | n = 5768 | 69% |
| Genetic Analysis Group (%) | South American (n = 676) | 6% |
| Central American (n = 1069) | 8% | |
| Cuban (n = 1764) | 27% | |
| Dominican (n = 797) | 9% | |
| Mexican (n = 3169) | 33% | |
| Puerto Rican (n = 1556) | 17% | |
| rs7903146 Genotype Frequency (%) | CC (n = 5040) | 55% |
| CT (n = 3373) | 38% | |
| TT (n = 618) | 7% | |
| Mean (95% CI) BMI (kg/m2) | At 21 Years (n = 8759) | 23.77 (23.61, 23.93) |
| At 45 Years (n = 5605) | 27.48 (27.30, 27.66) | |
| At 65 Years (n = 729) | 28.99 (28.46, 29.53) | |
| % with Self-Reported Diabetes Diagnosis | Between the Ages of 22 and 45 Years | 5% |
| Between the Ages of 46 and 65 Years | 23% | |
Abbreviations: BMI body mass index, 95% CI 95% confidence interval
The number of T2D-risk alleles at rs7903146 associated with an increase in T2D prevalence by 7% (P value = 0.0002) and decreased obesity prevalence by 3–5%, based on either the use of measured or self-reported weights (P value < 0.04, Table 2). Mean BMI and waist circumference at examination showed similar quantitative decreases by 0.5–0.6 kg/m2 and 1.1 cm as the number of T2D-risk allele increased (P values< 0.1). Additionally, among the subsample without a past diagnosis of T2D, at examination mean OGTT glucose levels increased by (4 mg/dL difference; P value = 0.06) and HOMA Index of Beta Cell function decreased (12 point difference; P value = 0.07). Other T2D-related measures, such as fasting glucose, insulin, and HbA1C exhibited similar trends across genotypes, but these trends were not statistically significant (P values ≥ 0.1). Further stratification of BMIs by T2D status/age at examination suggested that both increased age and T2D status corresponded to higher average BMIs, regardless of the timing of T2D diagnosis (Table 3). The subset of participants > 65 years at examination self-reported weights corresponding to a mean BMI increase of 2.8 kg/m2 between 45 and 65 years of age among those without T2D at baseline, and of up to 3.4 kg/m2 among those that were diagnosed with T2D after age 65.
Table 2.
Anthropometric Measures (Body Mass Index; Weight; Height; Waist Circumference; Overall and Abdominal Obesity; Percentage Body Fat), Fasting Insulin and Glucose, Post-Oral Glucose Tolerance Test Response, and Diabetic Control Characteristics Weighted Means (Standard Deviations) or Frequencies across rs7903146 Genotypes (n = 9031) at the Hispanic Community Health Study/Study of Latinos Baseline Examination
| CC | CT | TT | P-value | |
|---|---|---|---|---|
| Unweighted total analytic sample size | n = 5040 | n = 3373 | n = 618 | |
| Measured BMIa (kg/m2) | 29.74 (0.17) | 29.26 (0.13) | 29.21 (0.28) | 0.06 |
| Self-Reported BMIa (kg/m2) | 29.81 (0.17) | 29.34 (0.13) | 29.23 (0.27) | 0.05 |
| Measured Weight (kg) | 80.09 (0.52) | 78.71 (0.39) | 78.58 (0.79) | 0.1 |
| Self-Reported Weight (kg) | 80.29 (0.53) | 78.94 (0.39) | 78.69 (0.76) | 0.1 |
| Measured Height (cm) | 163.93 (0.18) | 163.83 (0.19) | 163.94 (0.32) | 0.9 |
| % Overall Measured Obesity (≥30 kg/m2 Measured BMI) | 42.43% | 37.89% | 38.55% | 0.03 |
| % Overall Self-Reported Obesity (≥30 kg/m2 Self-Reported BMI) | 43.29% | 38.85% | 37.66% | 0.03 |
| Waist Circumference (cm) | 98.41 (0.40) | 97.37 (0.31) | 97.34 (0.65) | 0.09 |
| % Abdominal Measured Obesity (≥120 cm for men; ≥88 cm for women) | 56.1% | 55.2% | 57.1% | 0.7 |
| % Body Fat | 33.58 (0.21) | 32.96 (0.19) | 33.18 (0.4) | 0.08 |
| % Identified as Diabetic Before or at Baseline Examinationb | 14.77% | 18.24% | 21.01% | 0.0002 |
| Unweighted non-diabetic subsample | n = 4107 | n = 2586 | n = 454 | |
| Fasting Insulin (mU/L) | 11.98 (0.2) | 11.96 (0.19) | 11.53 (0.47) | 0.7 |
| Fasting Glucose (mg/dL) | 93.78 (0.18) | 94.12 (0.23) | 94.67 (0.51) | 0.2 |
| HOMA Index of Beta Cell Function | 145.17 (2.59) | 140.81 (2.33) | 133.28 (4.81) | 0.07 |
| HOMA Index of Insulin Resistance | 2.82 (0.05) | 2.84 (0.05) | 2.75 (0.12) | 0.8 |
| post OGTT Insulin (mU/L)c | 78.06 (1.81) | 81.09 (1.84) | 75.39 (3.2) | 0.2 |
| post OGTT, Glucose (mg/dL)c | 112.25 (0.78) | 114.23 (0.85) | 115.85 (1.6) | 0.06 |
| Glycated Hemoglobin (mmol/mol) | 35.77 (0.1) | 35.99 (0.1) | 35.72 (0.22) | 0.2 |
| % Glycated Hemoglobin | 5.42 (0.01) | 5.44 (0.01) | 5.41 (0.02) | 0.2 |
| Unweighted diabetic b subsample | n = 933 | n = 787 | n = 164 | |
| Glycated Hemoglobin (mmol/mol) | 57.55 (1.25) | 57.79 (0.94) | 60.06 (2.06) | 0.6 |
| % Glycated Hemoglobin | 7.42 (0.11) | 7.44 (0.09) | 7.64 (0.19) | 0.6 |
| % Controlled Diabetes (< 7% Glycated Hemoglobin) | 26.1% | 31.2% | 35.3% | 0.1 |
All weighted means and standard deviations (or percentages) for anthropometric measures (weight, height, body mass index, waist circumference, fat percentage, overall and abdominal obesity) were estimated from regression models, which accounted for the complex sampling design and age, sex, and ancestry proportions. Additionally, all other weighted means and standard deviations (or percentages) were adjusted for body mass index (BMI) at examination. rs7903146 genotypes were modeled dis-jointly (i.e. no additive model was assumed)
aMeasured and self-reported BMI values at baseline were based off of measured weight and height, and self-reported weight and measured height, respectively
bThe diabetes subsample included individuals reporting having received a previous diabetes diagnosis at baseline examination, or being identified as diabetic at the baseline examination
c2-h Oral Glucose Tolerance Test (OGTT) was conducted in only individuals who did not report having had a previous diabetes diagnosis
Table 3.
Weighted Mean Body Mass Indices at 45 and 65 years by Categories of Baseline Examination and Diabetes Diagnosis Ages in the Hispanic Community Health Study/Study of Latinos among Participants > 45 Years of Age (n = 5643)
| Unweighted Sample Size | BMI (95% CI) at Age 45 (kg/m2) | BMI (95% CI) at Age 65 (kg/m2) | |
|---|---|---|---|
| Between 46 and 65 Years of Age ( n = 4,914) | |||
| No Diabetes Diagnosis Reporting at Baseline Examinationa | n = 4094 | 27.49 (27.29, 27.69) | – |
| Diagnosed Between 22 and 45 | n = 318 | 30.36 (29.53, 31.19) | – |
| Diagnosed Between 46 and Age at Examination | n = 502 | 29.30 (28.68, 29.93) | – |
| Between 66 and 76 Years of Age ( n = 729) | |||
| No Diabetes Diagnosis Reported at Baseline Examinationa | n = 502 | 25.67 (25.16, 26.18) | 28.43 (27.72, 29.14) |
| Diagnosed Between 22 and 45 | n = 26 | 26.51 (25.03, 27.99) | 29.71 (27.56, 31.86) |
| Diagnosed Between 46 and 65 | n = 153 | 27.75 (27.06, 28.45) | 30.65 (29.53, 31.77) |
| Diagnosed Between 66 and Age at Examination | n = 48 | 25.89 (24.71, 27.07) | 29.32 (27.58, 31.07) |
Abbreviations: BMI body mass index, 95% CI 95% confidence interval
aThis categorization includes individuals who were first diagnosed at the baseline clinic visit
Multivariable association analyses
We observed an association between the rs7903146 T2D-increasing allele and lower BMI, after adjustment for age, sex, education level, admixture proportions, and genetic analysis group. Specifically, we found that each T allele associated with lower BMI at examination (21–76 years), based on either measured or self-reported weight (Table 4). As described previously [15], we also observed significant inverse associations between each rs7903146-T allele and BMI (− 0.37 kg/m2, 95% CI: -0.69, − 0.06). Additionally, we also observed non-significant multivariable associations between rs7903146-T and lower BMIs at 21, and 45 years of age, and non-significant increases in BMI at 65 years of age.
Table 4.
Adjusted Parameter Estimatesa between rs7903146-T and Body Mass Indicesb at the Hispanic Community Health Study/Study of Latinos Baseline Examination Representing 9031 Individuals (21–76 Years of Age), and at Several Ages Across the Lifecourse
| Sample Sizec | Estimated Change in kg/m2 (95% CI) per T2D risk allele | P-value | |
|---|---|---|---|
| Measured BMI at Examination | n = 9012 | −0.37 (− 0.69, − 0.06) | 0.019 |
| Self-reported BMI at Examination | n = 8921 | −0.38 (− 0.69, − 0.08) | 0.015 |
| Self-reported BMI at 21 Years | n = 8759 | −0.20 (− 0.45, 0.04) | 0.109 |
| Self-reported BMI at 45 Years | n = 5605 | − 0.18 (− 0.43, 0.06) | 0.134 |
| Self-reported BMI at 65 Years | n = 729 | 0.01 (− 0.82, 0.84) | 0.980 |
Abbreviations: BMI Body mass index, T2D Type 2 diabetes, 95% CI 95% confidence interval
aCorresponding to the additive number of T2D risk alleles at TCF7L2 (rs7903146-T) and then adjusted for age, sex, education level, genetic ancestry group and ancestry proportions
bBody mass index (BMI) was based on self-reported or measured weight in kg, and divided by squared measured height at examination in meters (kg/m2)
cUnweighted sample size differences were a result of individuals missing certain weight measurements. For example, only individuals that were had reached 21, 45 or 65 years of age were asked to provide their weight for that particular age
Using data from the baseline examination, we also ran these BMI models stratified by previous diabetes diagnosis, glucose tolerance and medication status at the baseline examination (Additional file 1: Table S2). Weaker effects per allele on BMI were estimated among participants who reported having diabetes at examination as compared to those without diabetes, regardless of the use of measured or self-reported BMI at examination (− 0.30 to − 0.17 versus − 0.45 to − 0.48 kg/m2 per allele). Compared to our significant protective effect on BMI among all individuals without a prior diabetes diagnosis (− 0.45 kg/m2 per allele), the subset of individuals who had impaired fasting glucose or undiagnosed T2D at the examination (N = 4284) appeared to have an even stronger protective estimated effect on BMI (− 0.66 and − 0.85 kg/m2, respectively). No significant effects were seen for individuals taking diabetes medication at examination.
Structured association analyses
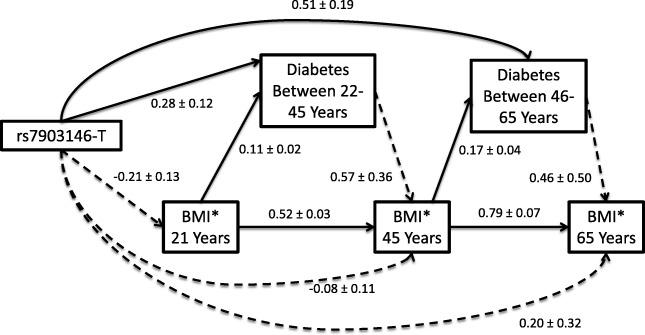
In a structural equation model, we noted that each T allele at rs7903146 was directly associated with a 1.32 (95% CI: 1.05, 1.67) higher odds of T2D diagnosis between the ages of 22 and 45 years, and a 1.67 (95% CI: 1.15, 2.42) higher odds of T2D diagnosis between 22 and 65 years of age. We did not find any significant direct associations between the rs7903146-T and BMI at any age; however, the direction of estimated effect was inverse on BMI at 21 and 45 years (Fig. 2). Furthermore, we found no evidence of indirect associations between rs7903146 and either BMI or T2D at any time point (Additional file 1: Table S3). Similarly, the indirect association between rs7903146 and BMI at 45 and 65 years, as mediated through a previous T2D diagnosis, was negative but non-significant (Additional file 1: Table S4).
Fig. 2.
Illustration of structured pathways (effect estimates and standard errors) between the additive number of rs7903146-T alleles, diabetes, and Body Mass Index (BMI), showing paths with P values < 0.05 in solid lines
Discussion
In this study we successfully replicated the previously-reported association between T2D risk alleles at TCF7L2 (rs7903146-T) and decreased BMI [3, 10–12], within a population-based cohort of US Hispanic/Latino adults of multiple background groups living in four urban communities (21–76 years of age at examination). We also observed consistently protective, albeit non-significant, associations on BMI at 21 and 45 years. In contrast, among the subset of individuals 65 years or older, the non-significant association between T2D-risk variants and BMI at 65 years of age was positive.
Next, we employed a structural equation model to examine the direct and indirect pathways between rs7903146, T2D and BMI, which revealed that this suggestive protective effect between T2D-risk variants and BMI at 21 and 45 years of age remained even after controlling for earlier BMI. These results collectively suggest that there may be a persistent independent protective effect of TCF7L2 T2D risk alleles on BMI across most of adulthood. In contrast to a previous cross-sectional study of 1235 Hispanic/Latinos, which estimated a larger effects of T2D-risk alleles at TCF7L2 on BMI by adjusting for concurrent T2D status (− 0.3 to − 1.1 kg/m2 for rs12255372-T) [12], our large and diverse study of US Hispanic/Latinos estimated more modest effects of T2D-risk alleles on BMI (− 0.4 kg/m2 for rs7903146-T; unadjusted for T2D status) and leveraged information on weight and T2D histories collected during the HCHS/SOL baseline examination to further decompose the complex relationships between prior BMI and T2D (Effect of each T2D-risk allele on BMI ranged from − 0.2 to 0.2 kg/m2 at 21 and 65 years of age, respectively).
Our findings shed light on the two predominant hypotheses put forth to explain the inverse direction of association between T2D and BMI at TCF7L2, as captured by variation in rs7903146. First, it has been suggested that case ascertainment bias [17] may drive the association of TCF7L2 T2D risk alleles and lower BMI, as previous GWAS have shown attenuated effects of TCF7L2 on BMI among population-based samples as compared to the effect sizes in samples of T2D cases [15]. Specifically, collider stratification may bias the TCF7L2-BMI association downwards when the ratio T2D cases to controls has been distorted to over-represent cases, or cases with more favorable insulin resistance profiles [44]. The active HCHS/SOL community engagement, household sampling, and location of clinic sites in the local community all served to minimize selection bias.
The consistent negative association between T2D risk alleles and BMI in early and mid-adulthood seen in this and previous work [3, 10, 11] may point to another explanation. A growing body of literature implicates pleiotropy at TCF7L2 in both T2D and BMI [5]. We observed protective associations on BMI at 21 and 45 years of age, which were not explained by accounting for indirect pathways through T2D or earlier BMI in our structured modeling. This work leverages detailed weight history data to provide further evidence for a complex mechanism underlying TCF7L2 action across the life course that may explain its associations with both T2D and BMI [3, 10–12], or the apparent statistical interaction between TCF7L2 genotype and adiposity on T2D related traits seen in previous cross-sectional studies of US Hispanic/Latinos [31]. Yet, clearly future functional or longitudinal analyses in population-based samples are required to substantiate our study’s findings.
Herein, we were also able to explore for the first time to our knowledge, what might be the direct effect of T2D diagnosis on subsequent BMI in the context of TCF7L2 genetic effects. The receipt of a T2D diagnosis between 22 and 45 years of age was significantly associated with an average increase in BMI at 45 years, as compared to those that never received a diagnosis during this time (Fig. 2). We did observe a similar, but non-significant association of T2D diagnosis between 46 and 65 years on BMI at 65 years. This indicates that the possible impact of pre-diagnosis metabolic dysfunction, T2D-related lifestyle counseling, or medical intervention also does not fully explain the apparent negative association between the TCF7L2 T2D risk allele and BMI [15]. This was further supported by our non-significant TCF7L2 associations on BMI at the examination among T2D individuals concurrently taking medications (Additional file 1: Table S2).
This current analysis is additionally strengthened by its focus on adults of varied Hispanic/Latino backgrounds [45]. Our sampling weights accounted for non-response and our statistical modeling approach also allowed us to account for missing data under the assumption of non-informative missingness and to base our variance calculations on information on the full population-based sample. In our dataset, missingness for age-specific BMIs was primarily determined by one’s age (BMI at 45 and 65 years of age would be missing for a 35-year-old participant).
Even though a previous study, which did not genotype rs7903146 directly, has posited that their best marker SNP at TCF7L2 (rs12255372, r2 = 0.7 in AMR with rs7903146) may capture a secondary BMI signal in Hispanic/Latinos [12], subsequent trans-ethnic fine-mapping studies of BMI and T2D including diverse Hispanic/Latino samples [8, 46] and large Hispanic/Latino studies have not supported the presence of multiple signals [47]. This gives us confidence that rs7903146, the lead variant for the single T2D signal observed in HCHS/SOL [48], is the best available SNP marker to simultaneously investigate allelic effects on BMI and T2D diagnosis within a structural equation modeling framework. Nonetheless, we do acknowledge that our current results do not capture all possible sources of pleiotropy at the TCF7L2 locus, which warrants further study.
Our structural equation results are also limited by our reliance on self-reported age of diabetes diagnosis, instead of repeated quantitative measures of T2D or its successful control. Among Hispanic/Latinos 15–19 year old, less than two thirds of diabetes cases may be Type 1, but the type distribution of cases steadily trends towards more T2D cases into early adulthood [49]—a period captured in HCHS/SOL. For this reason, we excluded a small number of individuals reporting early onset (< 22 years, N = 87) of diabetes. In HCHS/SOL, an additional N = 344 individuals reported a diabetes diagnosed between 22 and 45 years of age, only 30% of which were taking insulin by the baseline examination. Without medical or medication histories, we were unable to validate if these were T2D, or Latent Autoimmune Diabetes in Adults cases who would be expected to be leaner on average [50]. Nonetheless we take confidence in the observation that the association of TCF7L2 T2D-risk alleles and BMI was stronger among those without previous T2D diagnosis. In fact, individuals with impaired glucose tolerance and undiagnosed diabetes at examination had the greatest protective effect on BMI of T2D-risk alleles at TCF7L2. Forthcoming HCHS/SOL, or other prospective cohort follow up data will allow future investigators to explore the contemporaneous and interacting relationships between TCF7L2, BMI and T2D status across the adult life course.
Similarly, our structural equation modeling was notably limited by its reliance on self-reported weight histories, and height measured at the baseline examination to approximate the BMIs at 21, 45 and 65 years of age. Nonetheless this study cohort self-reported their current weight with good accuracy and reliability at baseline [34], and we robustly replicated our unstructured TCF7L2 associations with BMI at examination (21–76 years) using both measured and self-reported current weights. Lastly, we cannot rule out the role of birth cohort or healthy immigrant effects in shaping the characteristics of our sample of predominantly foreign-born middle-aged adults, especially among the subset of older adults in HCHS/SOL (e.g. ≥65 years of age) who were healthy enough to be community-dwelling at the time of recruitment, and willing to participate in the extensive baseline examination. Our structured modeling sheds light on this survival bias, as T2D-risk alleles were non-significantly associated with an increased BMI at 65 years of age, independent of earlier BMIs and T2D statuses.
Conclusions
Our significant population-based associations between T2D risk alleles at TCF7L2 (rs7903146) and lower BMI do not support selection bias as the sole explanation of the TCF7L2-BMI association. This work contributes to a mounting body of literature reporting consistent protective effects of T2D risk alleles at TCF7L2 and BMI, which points to a complex mechanistic structure underlying the functional consequences of TCF7L2 on both T2D and BMI. Yet, future functional work is needed to describe the specific cell or tissue types that are most relevant to the observed TCF7L2 action. Observational analyses may be particularly useful for estimating causal effects at this genetic locus and pinpointing windows of susceptibility for future public health interventions in populations, like US Hispanic/Latinos, which carry disproportionate burdens of both T2D and obesity.
Additional file
Figure S1. LDlink plot showing the regional r2 patterning between 1000 Genomes AMR and EUR reference populations at TCF7L2 around rs7903146 (shown in blue) and rs12255372 (alternative marker of the rs7903146 signal), with the bubble size representing the frequency of each SNP and the support for each SNP’s regulatory potential shown numerically (strong to weak: 1–7 RegulomeDB Scores). Table S1 Staged data cleaning and outlier identification on total sample of 40,525 self-reported weights from 16,355 adult participants (18–76 years) in the Hispanic Community Health Study/Study of Latinos (HCHS/SOL). Figure S2. Flow chart of staged quality control on 16,355 adult Hispanic/Latino participants (18–76 years) with at least one self-reported weight, as part of the anthropometric exam or weight history questionnaire, at the baseline examination (2008–2011) of the Hispanic Community Health Study/Study of Latinos (HCHS/SOL), resulting in 54 self-reported weights recoded due to unit confusion, 541 individuals excluded, and a final analytic sample of 16,322 participants. Table S2. Parameter Estimates of the Effect per Type 2 Diabetes Risk Allele (rs7903146-T) in a Multivariable Model Using Measured and Self-Reported Weight Measurements in the Analytic Sample Stratified by Diabetes Status at Baseline Examination and Medication. Figure S3. Illustration of all possible pathways in Structural Equation Model. Table S3. Parameter Estimates from Pathway Model Results. Table S4. Parameter Estimates for Select Indeirect Pathway Model Results. (DOCX 787 kb)
Acknowledgements
The authors thank the staff and participants of HCHS/SOL for their important contributions.
Funding
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (Grant 5R01DK101855). LFR was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (T32-HD007168) and the Carolina Population Center (P2C-HD050924).
Additionally, the Hispanic Community Health Study/Study of Latinos was carried out as a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute to the University of North Carolina (Grant N01-HC65233), University of Miami (Grant N01-HC65234), Albert Einstein College of Medicine (Grant N01-HC65235), Northwestern University (Grant N01-HC65236), and San Diego State University (Grant N01-HC65237). The following Institutes/Centers/Offices contributed to the HCHS/SOL through a transfer of funds to the NHLBI: National Center on Minority Health and Health Disparities, the National Institute of Deafness and Other Communications Disorders, the National Institute of Dental and Craniofacial Research, the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Neurological Disorders and Stroke, and the Office of Dietary Supplements.
Availability of data and materials
The HCHS/SOL data is available following an approved manuscript proposal. Further information can be found at investigators website at https://sites.cscc.unc.edu/hchs/publications-pub.
Abbreviations
- BMI
Body Mass Index
- GWAS
Genome-wide Association Study
- HCHS/SOL
Hispanic Community Health Study/Study of Latinos
- OGTT
Oral Glucose Tolerance Test
- HbA1C
Glycated Hemoglobin
- SNP
Single Nucleotide Polymorphism
- T2D
Type 2 Diabetes
- TCF7L2
Transcription Factor 7-like 2
- US
United States
Appendix
Between 2008 and 2011, up to 15 participants per site were invited to repeat the Hispanic Community Health Study/Study of Latinos (HCHS/SOL) examination, including the weight history questionnaire, one to three months after their initial baseline examination without additional reimbursement of their time (N = 56). A total of 52 participants reported body weights a median of 40 days after their initial baseline examination (range: 0 to 107 days). The mean differences of the 21 and 45-year old self-reported weight were − 0.9 kg (− 2.7, 1.0) and − 1.3 kg (− 3.8, 1.1) resulting in good reliabilities and low coefficients of variation (21-year recall: ICC = 0.89, CV = 7.7% n = 52; 45-year recall: 0.86, 6.7%, n = 29). Only two participants, ≥65 years of age, recalled their 65-year old weight twice, which precluded reliability calculations for 65-year old weights.
HCHS/SOL personnel (requiring a minimum of 5 practice subjects with 0.5 kg weight and 0.5 cm height agreement between a trainee and expert) measured height (cm) and weight (kg) during the anthropometric examination of the full HCHS/SOL cohort using a fixed (wall mounted) stadiometer (inspected daily) and a digital scale (scales zero balanced daily and calibrated weekly) on all participants that were able to stand on both feet while wearing a scrub suit or examination gown and no shoes [34]. Currently pregnant women were rescheduled for the HCHS/SOL baseline examination approximately 3 months after their delivery. The validity of self-reported weight, and inter-rater reliability of self-reported weight, measured weight and height have been reported to be good for the HCHS/SOL baseline examination [34].
HCHS/SOL study personnel were centrally trained and periodically observed. In addition we applied a data quality control protocol, as described previously for self-reported current weight collected as part of the anthropometric examination [34] to 1) minimize potential instances of unit confusion in the self-report (lb versus kg) or 2) exclude self-reported weights during pregnancy reported during the baseline examination, self-reported or measured weights made by individuals with past limb amputation or when scaled by measured baseline height that correspond to extreme underweight (< 16 kg/m2) or obesity (> 70 kg/m2). However, herein, we extended this protocol to also include recalled weights at 21, 45, or 65 years of age. We first flagged entire weight histories with either a ≥ 15 kg difference between current self-reported and measured weight, or at least two ≥15 kg pairwise fluctuations in weight (Additional file 1: Figure S1 and Table S1). Of the entire set of 16,355 individuals with at least one self-reported weight in adulthood (40,525 observations) and based on our staged quality control protocol, we recoded a total of 54 observations, and excluded an additional 541 observations (Additional file 1: Table S1). Only six self-reported weights from the anthropometric examination (age range 27–64 years) were recoded and retained in our final analytic sample.
Authors’ contributions
The coauthors of this work have collectively contributed to the manuscript the following ways: LFR conceived of the study’s aims, conducted all data quality control measures, and oversaw the statistical analyses, which were conducted by AGH using genetic variables generated by MG and CCL. LFR and AGH then drafted the manuscript jointly. CRI, HMH, KLY, ES, JEB, QQ, AEJ, SFG, CH, RJFL, and KEN have contributed to the content of the final manuscript. CRI, RCK, and GP participated in the design of the HCHS/SOL. SFG, CH, RJFL, and KEN each contributed substantially to the background and discussion of our study findings. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All HCHS/SOL participants provided their written informed consent, following Institutional Review Board approval from all participating academic centers, such as the approval received from the University of North Carolina at Chapel Hill Biomedical Institutional Review Board by the HCHS/SOL Coordinating Center located at the Collaborative Studies Coordinating Center at the University of North Carolina at Chapel Hill.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services.
Lindsay Fernández-Rhodes and Annie Green Howard contributed equally to this work.
Contributor Information
Lindsay Fernández-Rhodes, Phone: 919-260-4855, Email: fernandez-rhodes@unc.edu.
Annie Green Howard, aghoward@email.unc.edu.
Mariaelisa Graff, migraff@email.unc.edu.
Carmen R. Isasi, Email: carmen.isasi@einstein.yu.edu
Heather M. Highland, Email: heather.highland@unc.edu
Kristin L. Young, Email: kristin.young@unc.edu
Esteban Parra, Email: esteban.parra@utoronto.ca.
Jennifer E. Below, Email: piper.below@gmail.com
Qibin Qi, Email: qibin.qi@einstein.yu.edu.
Robert C. Kaplan, Email: robert.kaplan@einstein.yu.edu
Anne E. Justice, Email: aejustice1@geisinger.edu
George Papanicolaou, Email: gjp@nhlbi.nih.gov.
Cathy C. Laurie, Email: cclaurie@u.washington.edu
Struan F. A. Grant, grants@email.chop.edu
Christopher Haiman, Email: Christopher.Haiman@med.usc.edu.
Ruth J. F. Loos, Email: ruth.loos@mssm.edu
Kari E. North, Email: kari_north@unc.edu
References
- 1.Davidson JA, Kannel WB, Lopez-Candales A, Morales L, Moreno PR, Ovalle F, Rodriguez CJ, Rodbard HW, Rosenson RS, Stern M. Avoiding the looming Latino/Hispanic cardiovascular health crisis: a call to action. J Cardiometab Syndr. 2007;2(4):238–243. doi: 10.1111/j.1559-4564.2007.07534.x. [DOI] [PubMed] [Google Scholar]
- 2.Bhupathiraju SN, Hu FB. Epidemiology of obesity and diabetes and their cardiovascular complications. Circ Res. 2016;118(11):1723–1735. doi: 10.1161/CIRCRESAHA.115.306825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38(3):320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 4.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445(7130):881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 5.Jin T. Current understanding on role of the Wnt signaling pathway effector TCF7L2 in glucose homeostasis. Endocr Rev. 2016:er20151146. [DOI] [PubMed]
- 6.McCarthy MI, Rorsman P, Gloyn AL. TCF7L2 and diabetes: a tale of two tissues, and of two species. Cell Metab. 2013;17(2):157–159. doi: 10.1016/j.cmet.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 7.Williams AL, Jacobs SB, Moreno-Macias H, Huerta-Chagoya A, Churchhouse C, Marquez-Luna C, Garcia-Ortiz H, Gomez-Vazquez MJ, Burtt NP, Aguilar-Salinas CA, et al. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature. 2014;506(7486):97–101. doi: 10.1038/nature12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahajan A, Go MJ, Zhang W, Below JE, Gaulton KJ, Ferreira T, Horikoshi M, Johnson AD, Ng MC, Prokopenko I, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46(3):234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parra EJ, Cameron E, Simmonds L, Valladares A, McKeigue P, Shriver M, Wacher N, Kumate J, Kittles R, Cruz M. Association of TCF7L2 polymorphisms with type 2 diabetes in Mexico City. Clin Genet. 2007;71(4):359–366. doi: 10.1111/j.1399-0004.2007.00780.x. [DOI] [PubMed] [Google Scholar]
- 10.Cauchi S, Meyre D, Dina C, Choquet H, Samson C, Gallina S, Balkau B, Charpentier G, Pattou F, Stetsyuk V, et al. Transcription factor TCF7L2 genetic study in the French population: expression in human beta-cells and adipose tissue and strong association with type 2 diabetes. Diabetes. 2006;55(10):2903–2908. doi: 10.2337/db06-0474. [DOI] [PubMed] [Google Scholar]
- 11.Florez JC, Jablonski KA, Bayley N, Pollin TI, de Bakker PI, Shuldiner AR, Knowler WC, Nathan DM, Altshuler D. Diabetes prevention program research G: TCF7L2 polymorphisms and progression to diabetes in the diabetes prevention program. N Engl J Med. 2006;355(3):241–250. doi: 10.1056/NEJMoa062418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salinas YD, Wang L, DeWan AT. Multiethnic genome-wide association study identifies ethnic-specific associations with body mass index in Hispanics and African Americans. BMC Genet. 2016;17(1):78. doi: 10.1186/s12863-016-0387-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeggini E, McCarthy MI. TCF7L2: the biggest story in diabetes genetics since HLA? Diabetologia. 2007, 50(1):1–4. [DOI] [PMC free article] [PubMed]
- 14.Gustafson B, Hedjazifar S, Gogg S, Hammarstedt A, Smith U. Insulin resistance and impaired adipogenesis. Trends Endocrinol Metab. 2015;26(4):193–200. doi: 10.1016/j.tem.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich M, Consortium G. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helgason A, Palsson S, Thorleifsson G, Grant SF, Emilsson V, Gunnarsdottir S, Adeyemo A, Chen Y, Chen G, Reynisdottir I, et al. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet. 2007;39(2):218–225. doi: 10.1038/ng1960. [DOI] [PubMed] [Google Scholar]
- 17.Timpson NJ, Lindgren CM, Weedon MN, Randall J, Ouwehand WH, Strachan DP, Rayner NW, Walker M, Hitman GA, Doney AS, et al. Adiposity-related heterogeneity in patterns of type 2 diabetes susceptibility observed in genome-wide association data. Diabetes. 2009;58(2):505–510. doi: 10.2337/db08-0906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey KA, Savic D, Zielinski M, Park SY, Wang LJ, Witkowski P, Brady M, Hara M, Bell GI, Nobrega MA. Evidence of non-pancreatic beta cell-dependent roles of Tcf7l2 in the regulation of glucose metabolism in mice. Hum Mol Genet. 2015;24(6):1646–1654. doi: 10.1093/hmg/ddu577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prokunina-Olsson L, Welch C, Hansson O, Adhikari N, Scott LJ, Usher N, Tong M, Sprau A, Swift A, Bonnycastle LL, et al. Tissue-specific alternative splicing of TCF7L2. Hum Mol Genet. 2009;18(20):3795–3804. doi: 10.1093/hmg/ddp321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mondal AK, Das SK, Baldini G, Chu WS, Sharma NK, Hackney OG, Zhao J, Grant SF, Elbein SC. Genotype and tissue-specific effects on alternative splicing of the transcription factor 7-like 2 gene in humans. J Clin Endocrinol Metab. 2010;95(3):1450–1457. doi: 10.1210/jc.2009-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pradas-Juni M, Nicod N, Fernandez-Rebollo E, Gomis R. Differential transcriptional and posttranslational transcription factor 7-like regulation among nondiabetic individuals and type 2 diabetic patients. Mol Endocrinol. 2014;28(9):1558–1570. doi: 10.1210/me.2014-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia Q, Deliard S, Yuan CX, Johnson ME, Grant SF. Characterization of the transcriptional machinery bound across the widely presumed type 2 diabetes causal variant, rs7903146, within TCF7L2. Eur J Hum Genet. 2015;23(1):103–109. doi: 10.1038/ejhg.2014.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaulton KJ, Nammo T, Pasquali L, Simon JM, Giresi PG, Fogarty MP, Panhuis TM, Mieczkowski P, Secchi A, Bosco D, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42(3):255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaulton KJ, Ferreira T, Lee Y, Raimondo A, Magi R, Reschen ME, Mahajan A, Locke A, Rayner NW, Robertson N, et al. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat Genet. 2015;47(12):1415–1425. doi: 10.1038/ng.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Y, Oskolkov N, Shcherbina L, Ratti J, Kock KH, Su J, Martin B, Oskolkova MZ, Goransson O, Bacon J, et al. HMGB1 binds to the rs7903146 locus in TCF7L2 in human pancreatic islets. Mol Cell Endocrinol. 2016;430:138–145. doi: 10.1016/j.mce.2016.01.027. [DOI] [PubMed] [Google Scholar]
- 26.Kaminska D, Kuulasmaa T, Venesmaa S, Kakela P, Vaittinen M, Pulkkinen L, Paakkonen M, Gylling H, Laakso M, Pihlajamaki J. Adipose tissue TCF7L2 splicing is regulated by weight loss and associates with glucose and fatty acid metabolism. Diabetes. 2012;61(11):2807–2813. doi: 10.2337/db12-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyssenko V, Lupi R, Marchetti P, Del Guerra S, Orho-Melander M, Almgren P, Sjogren M, Ling C, Eriksson KF, Lethagen AL, et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Invest. 2007;117(8):2155–2163. doi: 10.1172/JCI30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19(4):379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 29.Loos RJ, Franks PW, Francis RW, Barroso I, Gribble FM, Savage DB, Ong KK, O'Rahilly S, Wareham NJ. TCF7L2 polymorphisms modulate proinsulin levels and beta-cell function in a British Europid population. Diabetes. 2007;56(7):1943–1947. doi: 10.2337/db07-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palmer ND, Lehtinen AB, Langefeld CD, Campbell JK, Haffner SM, Norris JM, Bergman RN, Goodarzi MO, Rotter JI, Bowden DW. Association of TCF7L2 gene polymorphisms with reduced acute insulin response in Hispanic Americans. J Clin Endocrinol Metab. 2008;93(1):304–309. doi: 10.1210/jc.2007-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe RM, Allayee H, Xiang AH, Trigo E, Hartiala J, Lawrence JM, Buchanan TA. Transcription factor 7-like 2 (TCF7L2) is associated with gestational diabetes mellitus and interacts with adiposity to alter insulin secretion in Mexican Americans. Diabetes. 2007;56(5):1481–1485. doi: 10.2337/db06-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sorlie PD, Avilés-Santa LM, Wassertheil-Smoller S, Kaplan RC, Daviglus ML, Giachello AL, Schneiderman N, Raij L, Talavera G, Allison M, et al. Design and implementation of the Hispanic community health study/study of Latinos. Ann Epidemiol. 2010;20(8):629–641. doi: 10.1016/j.annepidem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LaVange LM, Kalsbeek WD, Sorlie PD, Avilés-Santa LM, Kaplan RC, Barnhart J, Liu K, Giachello A, Lee DJ, Ryan J, et al. Sample design and cohort selection in the Hispanic community health study/study of Latinos. Ann Epidemiol. 2010;20(8):642–649. doi: 10.1016/j.annepidem.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernandez-Rhodes L, Robinson WR, Sotres-Alvarez D, Franceschini N, Castaneda SF, Buelna C, Moncrieft A, Llabre M, Daviglus ML, Qi Q, et al. Accuracy of self-reported weight in Hispanic/Latino adults of the Hispanic community health study/study of Latinos. Epidemiology. 2017;28(6):847–853. doi: 10.1097/EDE.0000000000000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.American Diabetes A. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2010;33(Suppl 1):S62–S69. doi: 10.2337/dc10-S062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voight BF, Kang HM, Ding J, Palmer CD, Sidore C, Chines PS, Burtt NP, Fuchsberger C, Li Y, Erdmann J, et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8(8):e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maller JB, McVean G, Byrnes J, Vukcevic D, Palin K, Su Z, Howson JM, Auton A, Myers S, Morris A, et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet. 2012;44(12):1294–1301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buyske S, Wu Y, Carty CL, Cheng I, Assimes TL, Dumitrescu L, Hindorff LA, Mitchell S, Ambite JL, Boerwinkle E, et al. Evaluation of the metabochip genotyping array in African Americans and implications for fine mapping of GWAS-identified loci: the PAGE study. PLoS One. 2012;7(4):e35651. doi: 10.1371/journal.pone.0035651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conomos MP, Laurie CA, Stilp AM, Gogarten SM, McHugh CP, Nelson SC, Sofer T, Fernandez-Rhodes L, Justice AE, Graff M, et al. Genetic diversity and association studies in US Hispanic/Latino populations: applications in the Hispanic community health study/study of Latinos. Am J Hum Genet. 2016;98(1):165–184. doi: 10.1016/j.ajhg.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin DY, Tao R, Kalsbeek WD, Zeng D, Gonzalez F, 2nd, Fernandez-Rhodes L, Graff M, Koch GG, North KE, Heiss G. Genetic association analysis under complex survey sampling: the Hispanic community health study/study of Latinos. Am J Hum Genet. 2014;95(6):675–688. doi: 10.1016/j.ajhg.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muthen B, Muthen L. Mplus User’s Guide. Seventh ed. Los Angeles, CA: Muthén & Muthén; 1998-2012.
- 44.Yaghootkar H, Bancks MP, Jones SE, McDaid A, Beaumont R, Donnelly L, Wood AR, Campbell A, Tyrrell J, Hocking LJ, et al. Quantifying the extent to which index event biases influence large genetic association studies. Hum Mol Genet. 2017;26(5):1018–1030. doi: 10.1093/hmg/ddw433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daviglus ML, Talavera GA, Aviles-Santa ML, Allison M, Cai J, Criqui MH, Gellman M, Giachello AL, Gouskova N, Kaplan RC, et al. Prevalence of major cardiovascular risk factors and cardiovascular diseases among Hispanic/Latino individuals of diverse backgrounds in the United States. JAMA. 2012;308(17):1775–1784. doi: 10.1001/jama.2012.14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernandez-Rhodes L, Gong J, Haessler J, Franceschini N, Graff M, Nishimura KK, Wang Y, Highland HM, Yoneyama S, Bush WS, et al. Trans-ethnic fine-mapping of genetic loci for body mass index in the diverse ancestral populations of the population architecture using genomics and epidemiology (PAGE) study reveals evidence for multiple signals at established loci. Hum Genet. 2017;136(6):771–800. doi: 10.1007/s00439-017-1787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Acosta JL, Hernandez-Mondragon AC, Correa-Acosta LC, Cazanas-Padilla SN, Chavez-Florencio B, Ramirez-Vega EY, Monge-Cazares T, Aguilar-Salinas CA, Tusie-Luna T, Del Bosque-Plata L. Rare intronic variants of TCF7L2 arising by selective sweeps in an indigenous population from Mexico. BMC Genet. 2016;17(1):68. doi: 10.1186/s12863-016-0372-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qi Q, Stilp AM, Sofer T, Moon JY, Hidalgo B, Szpiro AA, Wang T, Ng MCY, Guo X, ME-aotDiAA C, et al. Genetics of type 2 diabetes in U.S. Hispanic/Latino individuals: results from the Hispanic community health study/study of Latinos (HCHS/SOL) Diabetes. 2017;66(5):1419–1425. doi: 10.2337/db16-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pettitt DJ, Talton J, Dabelea D, Divers J, Imperatore G, Lawrence JM, Liese AD, Linder B, Mayer-Davis EJ, Pihoker C, et al. Prevalence of diabetes in U.S. youth in 2009: the SEARCH for diabetes in youth study. Diabetes Care. 2014;37(2):402–408. doi: 10.2337/dc13-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cervin C, Lyssenko V, Bakhtadze E, Lindholm E, Nilsson P, Tuomi T, Cilio CM, Groop L. Genetic similarities between latent autoimmune diabetes in adults, type 1 diabetes, and type 2 diabetes. Diabetes. 2008;57(5):1433–1437. doi: 10.2337/db07-0299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. LDlink plot showing the regional r2 patterning between 1000 Genomes AMR and EUR reference populations at TCF7L2 around rs7903146 (shown in blue) and rs12255372 (alternative marker of the rs7903146 signal), with the bubble size representing the frequency of each SNP and the support for each SNP’s regulatory potential shown numerically (strong to weak: 1–7 RegulomeDB Scores). Table S1 Staged data cleaning and outlier identification on total sample of 40,525 self-reported weights from 16,355 adult participants (18–76 years) in the Hispanic Community Health Study/Study of Latinos (HCHS/SOL). Figure S2. Flow chart of staged quality control on 16,355 adult Hispanic/Latino participants (18–76 years) with at least one self-reported weight, as part of the anthropometric exam or weight history questionnaire, at the baseline examination (2008–2011) of the Hispanic Community Health Study/Study of Latinos (HCHS/SOL), resulting in 54 self-reported weights recoded due to unit confusion, 541 individuals excluded, and a final analytic sample of 16,322 participants. Table S2. Parameter Estimates of the Effect per Type 2 Diabetes Risk Allele (rs7903146-T) in a Multivariable Model Using Measured and Self-Reported Weight Measurements in the Analytic Sample Stratified by Diabetes Status at Baseline Examination and Medication. Figure S3. Illustration of all possible pathways in Structural Equation Model. Table S3. Parameter Estimates from Pathway Model Results. Table S4. Parameter Estimates for Select Indeirect Pathway Model Results. (DOCX 787 kb)
Data Availability Statement
The HCHS/SOL data is available following an approved manuscript proposal. Further information can be found at investigators website at https://sites.cscc.unc.edu/hchs/publications-pub.