Abstract
Multicomponent reactions are excellent tools to generate complex structures with broad chemical diversity and fluorescent properties. Herein we describe the adaptation of the fluorescent BODIPY scaffold to multicomponent reaction chemistry with the synthesis of BODIPY adducts with high fluorescence quantum yields and good cell permeability. From this library we identified one BODIPY derivative (PhagoGreen) as a low-pH sensing fluorescent probe that enabled imaging of phagosomal acidification in activated macrophages. The fluorescence emission of PhagoGreen was proportional to the degree of activation of macrophages and could be specifically blocked by bafilomycin A, an inhibitor of phagosomal acidification. PhagoGreen does not impair the normal functions of macrophages and can be used to image phagocytic macrophages in vivo.
Fluorescent probes are chemical entities of enormous importance in biomedical research and medical imaging. In the context of fluorescence live cell imaging, they enable real-time tracking of biomolecules, metabolites and cells under physiological conditions without altering regular cellular functions.1 The 4,4-difluoro-4-bora-3a,4a-diaza-s-indacene (BODIPY) scaffold has played a pivotal role in fluorescent probe development, and it is one of the most exploited fluorophores due to its excellent photophysical properties.2 Amine and carboxylic acid-derivatized BODIPY dyes are readily available, and have been conjugated to numerous biomolecules to develop fluorescent compounds to enable biological interrogation. This approach has rendered a wide variety of BODIPY-based cell imaging probes.3 Combinatorial strategies have recently expanded the chemical diversity of the BODIPY core. These strategies employ efficient and stepwise reactions (e.g., Knoevenagel condensation, ‘click’ chemistry) to implement structural diversification into a presynthesized BODIPY scaffold.4 Subsequent high-throughput screenings of the resulting libraries have significantly accelerated the discovery of new fluorescent probes.5 We envisioned that the use of multicomponent reactions (MCRs)6 for the synthesis of BODIPY fluorescent probes would lead to novel complex structures that are difficult to prepare by conventional synthetic strategies. MCRs can increase the chemical diversity of BODIPY dyes with the formation of unusual C–C bonds and give BODIPY compounds with unexplored chemical connectivity and potentially new features as imaging probes.
Our group and others have described MCRs to prepare complex fluorescent molecules based on 2,6-cyanodianilines,7 isoquinolines,8 naphthalimides,9 benzoazepines10 and imidazoles.11 Balakirev and co-workers recently reported the combinatorial exploitation of three component Ugi MCRs in droplet arrays to successfully discover new fluorophores with drug-like properties.12 Whereas these examples proved the suitability of MCRs to generate de novo fluorescent structures, they employed scaffolds with inherent limitations as fluorophores (e.g., short emission wavelengths, low extinction coefficients, poor quantum yields, compromised cell permeability).
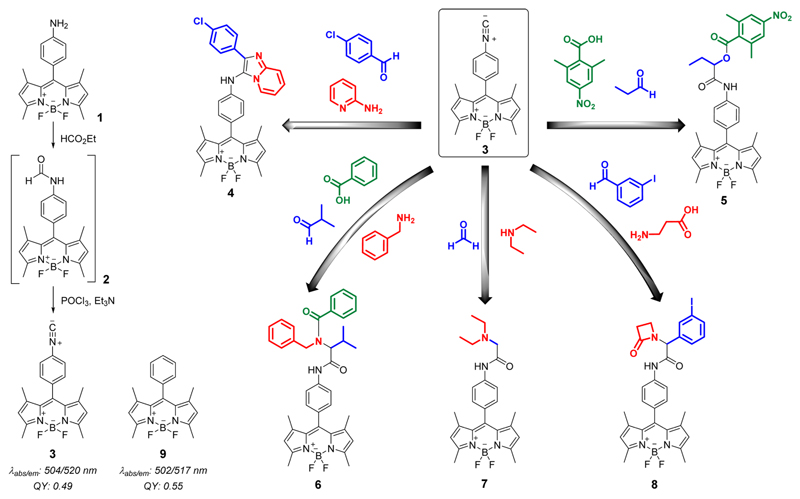
The adaptation of MCRs to the highly fluorescent and cell permeable BODIPY scaffold provides a practical platform to develop novel compounds with unexpected features as cell imaging fluorescent probes. Since the most versatile MCRs are based on isonitrile chemistry,13 we prepared an isonitrile-BODIPY scaffold (3) compatible with several MCRs (Scheme 1). Compound 3 was prepared in a two-step reaction from the BODIPY aniline 1, obtained by reduction of the corresponding nitro compound.14 The BODIPY aniline 1 was formylated with HCO2Et to render the formamide 2, which was subsequently dehydrated under standard conditions with POCl3 to afford the isonitrile 3. Notably, the isonitrile functional group did not affect the fluorescent properties of the BODIPY core (9) (Figure S1 and Table S1 in Supporting Information (SI)). To the best of our knowledge, this is the first report of an isonitrile-functionalized BODIPY dye and its subsequent derivatization using MCRs.
Scheme 1. Synthesis of an Isonitrile-BODIPY Scaffold and Its Derivatization Using Different MCRs.
We employed 3 as the starting material for a number of isonitrile-based MCRs, namely Passerini,15 Bienaymé–Blackburn–Groebcke,16 and three variants of the Ugi-MCR17 (Scheme 1). We performed a Bienaymé–Blackburn–Groebcke MCR with 3, α-aminopyridine, and 4-chlorobenzaldehyde to obtain the azaindole 4. A conventional Passerini reaction of 3 with propanal and 2,6-dimethyl-4-nitrobenzoic acid rendered the adduct 5. A four-component Ugi MCR with isobutylaldehyde, benzylamine, and benzoic acid afforded compound 6, whereas the β-amino acid variation led to the β-lactam 8. The adduct 7 was obtained with a variant of the Ugi MCR using formaldehyde and diethylamine. Notably, all adducts (4–8) were isolated in good yields (see details and characterization data in SI) and retained the characteristic fluorescent properties of the BODIPY core (Figure S1 and Table S1 in SI). The synthesis of this collection of BODIPY adducts confirms that the reactivity of the isonitrile group in different MCRs is not hampered by the BODIPY structure. Altogether, the results validate isonitrile-based MCRs as a synthetic platform for the diversification of the BODIPY scaffold toward fluorescent conjugates that might be difficult to prepare by conventional strategies.
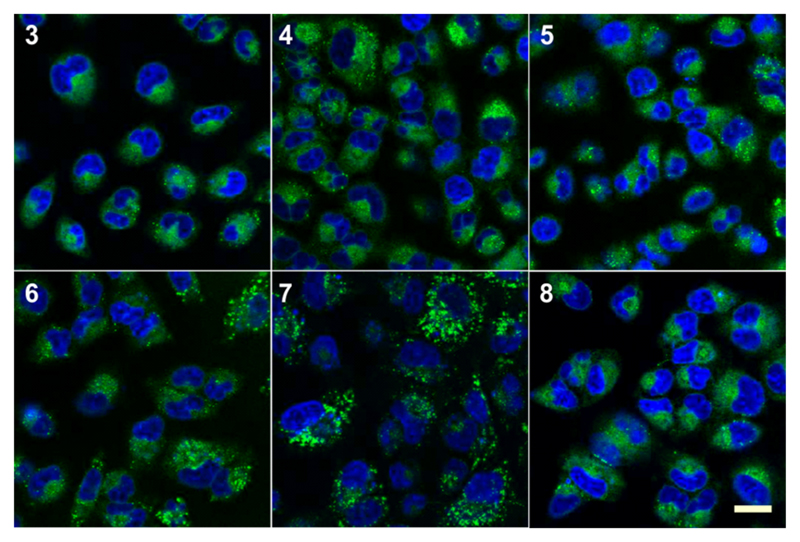
We assessed the cell permeability of compounds 3–8 by imaging their localization in live A549 cells together with different intracellular trackers (Figures S2 and S3 in SI) and observed that all adducts readily entered cells at concentrations in the nanomolar range. While most adducts stained the cytoplasm and some lysosomes, compound 7 exhibited a distinctive, vesicle-like staining pattern (Figure 1). We examined the subcellular localization of 7 in different cell lines by colocalization with LysoTracker Red, a fluorescent dye that labels acidic organelles (Figures S4 and S5 in SI).
Figure 1. Cell permeability of BODIPY adducts in live cell imaging.
A549 cells were incubated with compounds 3–8 (250 nM) for 30 min and imaged under the confocal fluorescence microscope. DAPI was used for nuclear counterstaining. Scale bar: 20 μm.
The similar staining patterns of 7 and Lysotracker Red indicated that 7 is an acidotropic fluorescent molecule with bright fluorescence emission in subcellular acidic environments. Compound 7 has a pKa of 5.76 ± 0.07 (Figure S6 in SI) and higher sensitivity to pH than LysoTracker Red (Figure S7 in SI).
We envisaged that these remarkable properties of 7 as a cell permeable fluorescent probe for acidic microenvironments may be applied to imaging phagosomal acidification in macrophages. Macrophages are immune cells with key roles in inflammation and tissue homeostasis. Macrophages ingest pathogens and particles by phagocytosis. During the course of phagocytosis the maturation and fusion of endosomes leads to a progressive phagosomal acidification.18 Most currently used probes for activated macrophages target the recognition of enzymes (e.g., cathepsins) or cell surface receptors (e.g., folate, integrins).19 Bogyo and co-workers recently described fluorescent probes to monitor legumain activity in the acidic organelles of activated macrophages.20
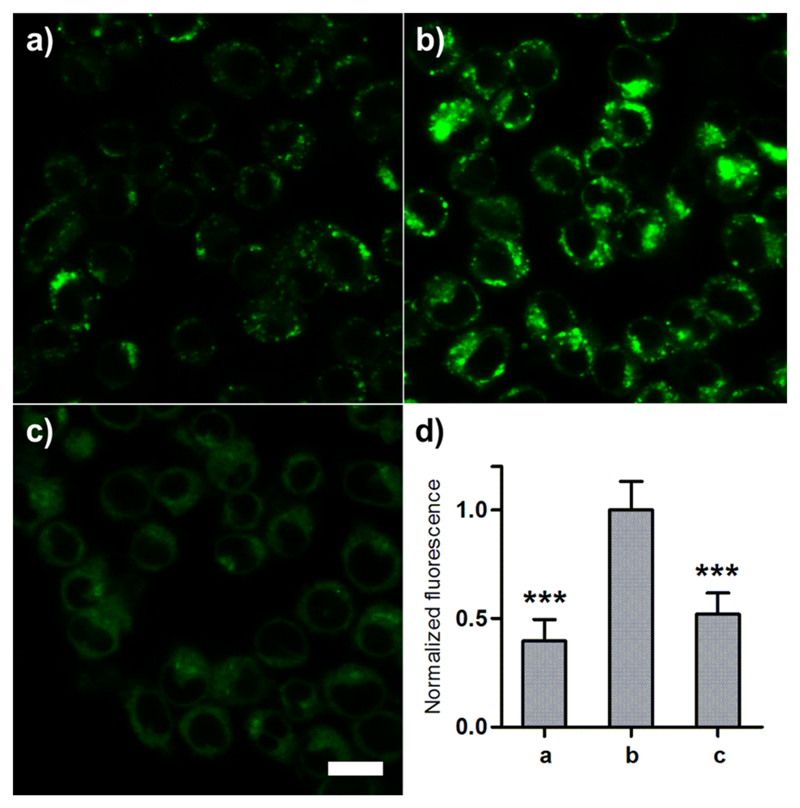
We acquired fluorescent images of RAW264.7 macrophages treated with compound 7 before and after activation with zymosan (Figure 2). Zymosan is a glucan from the yeast cell wall that induces phagosomal acidification in macrophages.21 Zymosan-activated macrophages displayed bright fluorescence in the perinuclear region, where most phagosomes are located (Figure 2b). The fluorescence intensity was significantly brighter in zymosan-activated than in nonactivated macrophages (Figure 2d). The staining of 7 was proportional to the extent of the zymosan treatment, proving that, unlike Lysotracker Red, the emission of 7 correlates with the degree of activation of macrophages (Figures S8 and S9 in SI). We confirmed that the staining of 7 relies on the acidification of the phagosomes. As shown in Figure 2c, the staining of zymosan-activated macrophages decreased upon treatment with bafilomycin A, an inhibitor of the vacuolar ATPase that is required for phagosomal acidification.22
Figure 2. Compound 7 stains macrophages undergoing phagosomal acidification.
RAW264.7 cells (preincubated or not with zymosan) were treated with 7 (100 nM) for 15 min and imaged by confocal microscopy. Fluorescence staining of 7 in (a) nonactivated macrophages, (b) zymosan-activated macrophages, and (c) zymosan-activated macrophages treated with bafilomycin A (100 nM); (d) quantification of fluorescence emission represented as means (n = 4) and error bars as SD, *** for p < 0.005 compared to b. Scale bar: 20 μm.
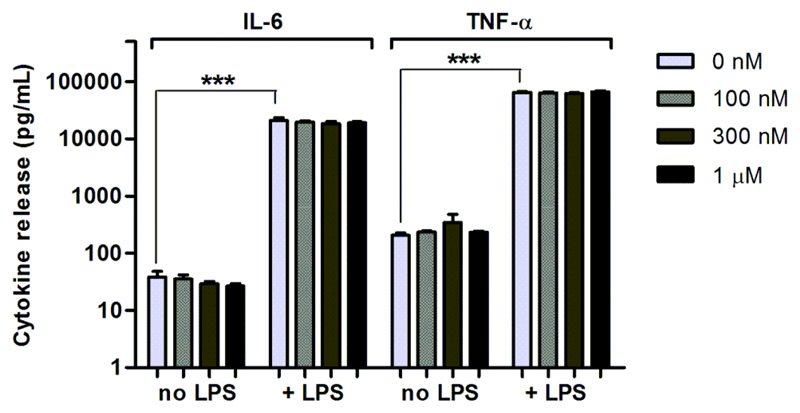
We observed that compound 7 was nontoxic to macrophages at 500 nM, even at long incubation times (Figure S10 in SI). We also studied whether the treatment with compound 7 affected the secretion of TNF-α and IL-6, two major cytokines released by macrophages. As shown in Figure 3, there were no significant differences in the levels of TNF-α and IL-6 secreted by nontreated and 7-treated macrophages before or after stimulation with liposaccharide S (LPS). These results validate 7 as a fluorescent probe to image phagocytic macrophages without impairing their normal function. On account of these observations, compound 7 was named as PhagoGreen.
Figure 3. Cytokine release by macrophages after incubation with PhagoGreen.
RAW264.7 cells (with or without PhagoGreen) were stimulated with LPS (100 ng/mL) for 18 h. The levels of TNF-α and IL-6 in the supernatants were measured by ELISA. Values represented as means (n = 4) and error bars as SD, *** for p < 0.0001.
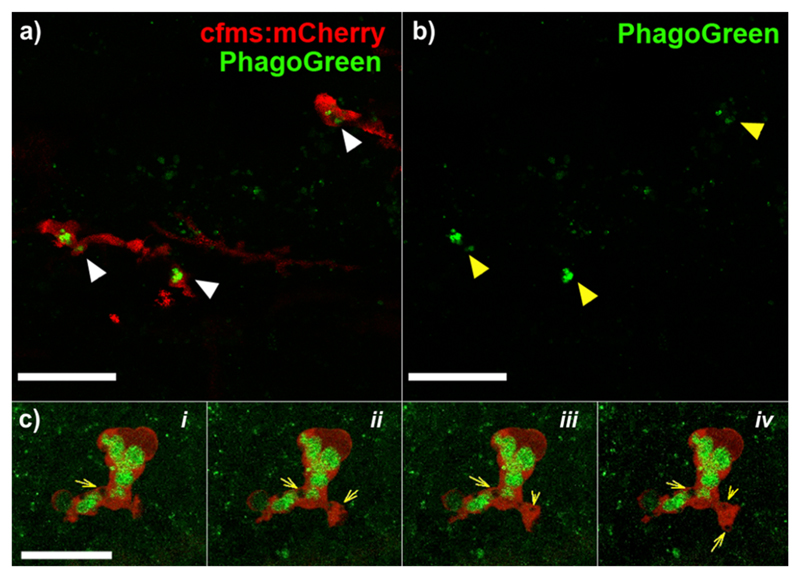
Finally, we examined the application of PhagoGreen to image phagocytic macrophages in vivo. Zebrafish embryos expressing mCherry-labeled phagocytic macrophages23 were imaged after incubation with PhagoGreen (Figure 4).
Figure 4. PhagoGreen enables in vivo imaging of phagocytic macrophages.
Transgenic zebrafish with mCherry-labeled macrophages were treated with PhagoGreen (600 nM) for 30 min and imaged by confocal microscopy. (a) Images from a time-lapse movie (Video S1) showing the phagosomal localization of PhagoGreen (white arrowheads) demonstrated by the surrounding red fluorescence of cytoplasmic mCherry; (b) single channel image of (a) showing brighter fluorescence in mature phagosomes (yellow arrowheads); (c) i–iv: sequential images from a time-lapse movie (Video S2) of an actively engulfing macrophage, where the mature phagosome is brightly green fluorescent while the newly formed phagosome is devoid of green fluorescence (yellow arrows). Scale bars (a,b): 20 μm, (c): 15 μm.
As shown in Figure 4, PhagoGreen stained phagosomes in mCherry-labeled phagocytic macrophages. Time-lapse imaging confirmed that PhagoGreen-stained macrophages were actively phagocytic (Video S1). Furthermore, high-resolution time-lapse imaging showed that PhagoGreen did not stain early phagosomes (Figure 4c; Video S2), which corroborates our observation that the mechanism of staining for PhagoGreen relies on phagosomal acidification.
In conclusion, we described the first adaptation of multicomponent reaction chemistry to prepare BODIPY-based imaging probes with broad and unexplored chemical diversity. A novel isonitrile-BODIPY scaffold was successfully used in different MCRs to render highly fluorescent and cell permeable BODIPY adducts. From our library we identified PhagoGreen as a pH-sensitive fluorescent probe to image phagosomal acidification in macrophages. We corroborated the staining mechanism of PhagoGreen in macrophages by different treatments with zymosan, and its specific blocking with bafilomycin A, an inhibitor of phagosomal acidification. PhagoGreen will facilitate the study of the mechanisms underlying macrophage-mediated phagocytosis, as it can image phagocytic macrophages in vivo and in real time without impairing the normal physiology of macrophages.
Supplementary Material
Synthetic procedures, full characterization and spectral data for compounds 1–9, cell viability assays, imaging data including videos. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
This work was supported by DGICYT–Spain (BQU-CTQ2012-30930) and Generalitat de Catalunya (2009SGR 1024). Grupo Ferrer (Barcelona, Spain) is thanked for financial support. M.V. acknowledges the support of the Medical Research Council and the FP7Marie Curie Career Integration Grant. Y.F. is supported by a Wellcome Trust Sir Henry Dale Fellowship (100104/Z/12/Z), and R.J.M. is funded by a Wellcome Trust Intermediate Clinical Fellowship.
Footnotes
Notes
The authors declare no competing financial interest.
References
- (1).(a) Kobayashi H, Ogawa M, Alford R, Choyke PL, Urano Y. Chem Rev. 2010;110:2620. doi: 10.1021/cr900263j. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Chan J, Dodani SC, Chang CJ. Nat Chem. 2012;4:973. doi: 10.1038/nchem.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Vendrell M, Zhai D, Er JC, Chang YT. Chem Rev. 2012;112:4391. doi: 10.1021/cr200355j. [DOI] [PubMed] [Google Scholar]; (d) Yuan L, Lin W, Zheng K, He L, Huang W. Chem Soc Rev. 2013;42:622. doi: 10.1039/c2cs35313j. [DOI] [PubMed] [Google Scholar]; (e) Wysocki LM, Lavis LD. Curr Opin Chem Biol. 2011;15:752. doi: 10.1016/j.cbpa.2011.10.013. [DOI] [PubMed] [Google Scholar]
- (2).(a) Loudet A, Burgess K. Chem Rev. 2007;107:4891. doi: 10.1021/cr078381n. [DOI] [PubMed] [Google Scholar]; (b) Ulrich G, Ziessel R, Harriman A. Angew Chem, Int Ed. 2008;47:1184. doi: 10.1002/anie.200702070. [DOI] [PubMed] [Google Scholar]; (c) Boens N, Leen V, Dehaen W. Chem Soc Rev. 2012;41:1130. doi: 10.1039/c1cs15132k. [DOI] [PubMed] [Google Scholar]
- (3).(a) Myochin T, Hanaoka K, Komatsu T, Terai T, Nagano T. J Am Chem Soc. 2012;134:13730. doi: 10.1021/ja303931b. [DOI] [PubMed] [Google Scholar]; (b) Michel BW, Lippert AR, Chang CJ. J Am Chem Soc. 2012;134:15668. doi: 10.1021/ja307017b. [DOI] [PubMed] [Google Scholar]; (c) Dodani SC, Leary SC, Cobine PA, Winge DR, Chang CJ. J Am Chem Soc. 2011;133:8606. doi: 10.1021/ja2004158. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Isik M, Ozdemir T, Turan IS, Kolemen S, Akkaya EU. Org Lett. 2013;15:216. doi: 10.1021/ol303306s. [DOI] [PubMed] [Google Scholar]; (e) Gavande N, Kim H-L, Doddareddy MR, Johnston GAR, Chebib M, Hanrahan JR. ACS Med Chem Lett. 2013;4:402. doi: 10.1021/ml300476v. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Komatsu T, Urano Y, Fujikawa Y, Kobayashi T, Kojima H, Terai T, Hanaoka K, Nagano T. Chem Commun. 2009:7015. doi: 10.1039/b917209b. [DOI] [PubMed] [Google Scholar]
- (4).(a) Lee JS, Kang NY, Kim YK, Samanta A, Feng S, Kim HK, Vendrell M, Park JH, Chang YT. J Am Chem Soc. 2009;131:10077. doi: 10.1021/ja9011657. [DOI] [PubMed] [Google Scholar]; (b) Vendrell M, Krishna GG, Ghosh KK, Zhai D, Lee JS, Zhu Q, Yau YH, Shochat SG, Kim H, Chung J, Chang YT. Chem Commun. 2011;47:8424. doi: 10.1039/c1cc11774b. [DOI] [PubMed] [Google Scholar]; (c) Zhai D, Lee SC, Vendrell M, Leong LP, Chang YT. ACS Comb Sci. 2012;14:81. doi: 10.1021/co200136b. [DOI] [PubMed] [Google Scholar]; (d) Er JC, Tang MK, Chia CG, Liew H, Vendrell M, Chang YT. Chem Sci. 2013;4:2168. [Google Scholar]
- (5).(a) Ahn YH, Lee JS, Chang YT. J Am Chem Soc. 2007;129:4510. doi: 10.1021/ja068230m. [DOI] [PubMed] [Google Scholar]; (b) Lee JS, Kim HK, Feng S, Vendrell M, Chang YT. Chem Commun. 2011;47:2339. doi: 10.1039/c0cc04495d. [DOI] [PubMed] [Google Scholar]
- (6).For an overview, see: Zhu J, Bienaymé H, editors. Multicomponent reactions. Wiley-VCH; Weinheim: 2005. ; Domling A, Wang W, Wang K. Chem Rev. 2012;112:3083. doi: 10.1021/cr100233r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Cui SL, Lin XF, Wang YG. J Org Chem. 2005;70:2866. doi: 10.1021/jo047823h. [DOI] [PubMed] [Google Scholar]
- (8).Kielland N, Vendrell M, Lavilla R, Chang YT. Chem Commun. 2012;48:7401. doi: 10.1039/c2cc32292g. [DOI] [PubMed] [Google Scholar]
- (9).Rotstein BH, Mourtada R, Kelley SO, Yudin AK. Chem — Eur J. 2011;17:12257. doi: 10.1002/chem.201102096. [DOI] [PubMed] [Google Scholar]
- (10).Pan HR, Wang XR, Yan CX, Sun ZX, Cheng Y. Org Biomol Chem. 2011;9:2166. doi: 10.1039/c0ob00746c. [DOI] [PubMed] [Google Scholar]
- (11).(a) Bienayme H, Bouzid K. Angew Chem, Int Ed. 1998;37:2234. doi: 10.1002/(SICI)1521-3773(19980904)37:16<2234::AID-ANIE2234>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]; (b) Wang W, Cao H, Wolf S, Camacho-Horvitz MS, Holak TA, Domling A. Bioorg Med Chem. 2013;21:3982. doi: 10.1016/j.bmc.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Burchak ON, Mugherli L, Ostuni M, Lacapere JJ, Balakirev MY. J Am Chem Soc. 2011;133:10058. doi: 10.1021/ja204016e. [DOI] [PubMed] [Google Scholar]
- (13).For an overview, see: Nenadjenko VG, editor. Isocyanide Chemistry. Wiley-VCH; Weinheim: 2012. ; (b) Domling A. Chem Rev. 2006;106:17. doi: 10.1021/cr0505728. [DOI] [PubMed] [Google Scholar]; (c) Domling A, Ugi II. Angew Chem, Int Ed. 2000;39:3168. doi: 10.1002/1521-3773(20000915)39:18<3168::aid-anie3168>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- (14).Imahori H, Norieda H, Yamada H, Nishimura Y, Yamazaki I, Sakata Y, Fukuzumi S. J Am Chem Soc. 2001;123:100. doi: 10.1021/ja002154k. [DOI] [PubMed] [Google Scholar]
- (15).Banfi L, Riva R. Org React. 2005;65:1. [Google Scholar]
- (16).(a) Bienaymé H, Bouzid K. Angew Chem, Int Ed. 1998;37:2234. doi: 10.1002/(SICI)1521-3773(19980904)37:16<2234::AID-ANIE2234>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]; (b) Blackburn C, Guan B, Fleming P, Shiosaki K, Tsai S. Tetrahedron Lett. 1998;39:3635. [Google Scholar]; (c) Groebcke K, Weber L, Mehlin F. Synlett. 1998:661. [Google Scholar]
- (17).Ugi I, Steinbruckner C. Angew Chem. 1960;72:267. [Google Scholar]
- (18).Jiang L, Salao K, Li H, Rybicka JM, Yates RM, Luo XW, Shi XX, Kuffner T, Tsai VWW, Husaini Y, Wu L, et al. J Cell Sci. 2012;125:5479. doi: 10.1242/jcs.110072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).(a) Verdoes M, Edgington LE, Scheeren FA, Leyva M, Blum G, Weiskopf K, Bachmann MH, Ellman JA, Bogyo M. Chem Biol. 2012;19:619. doi: 10.1016/j.chembiol.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Chen WT, Khazaie K, Zhang G, Weissleder R, Tung CH. Mol Imaging. 2005;4:67. doi: 10.1162/15353500200504199. [DOI] [PubMed] [Google Scholar]; (c) Waldeck J, Hager F, Holtke C, Lanckohr C, von Wallbrunn A, Torsello G, Heindel W, Theilmeier G, Schafers M, Bremer C. J Nucl Med. 2008;49:1845. doi: 10.2967/jnumed.108.052514. [DOI] [PubMed] [Google Scholar]; (d) Saxena A, Kessinger CW, Thompson B, McCarthy JR, Iwamoto Y, Lin CP, Jaffer FA. Mol Imaging Biol. 2013;15:282. doi: 10.1007/s11307-012-0599-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).(a) Lee J, Bogyo M. ACS Chem Biol. 2010;5:233. doi: 10.1021/cb900232a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Edgington LE, Verdoes M, Ortega A, Withana NP, Lee J, Syed S, Bachmann MH, Blum G, Bogyo M. J Am Chem Soc. 2013;135:174. doi: 10.1021/ja307083b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Haggie PM, Verkman AS. J Biol Chem. 2007;282:31422. doi: 10.1074/jbc.M705296200. [DOI] [PubMed] [Google Scholar]
- (22).Lukacs GL, Rotstein OD, Grinstein S. J Biol Chem. 1990;265:21099. [PubMed] [Google Scholar]
- (23).Gray C, Loynes CA, Whyte MK, Crossman DC, Renshaw SA, Chico TJ. Thromb Haemost. 2011;105:811. doi: 10.1160/TH10-08-0525. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.