Abstract
In bacteria the ability to remodel membrane underpins basic cell processes such as growth, and more sophisticated adaptations like inter-cell crosstalk, organelle specialisation, and pathogenesis. Here, selected examples of membrane remodelling in bacteria are presented and the diverse mechanisms for inducing membrane fission, fusion, and curvature discussed. Compared to eukaryotes, relatively few curvature-inducing proteins have been characterised so far. Whilst it is likely that many such proteins remain to be discovered, it also reflects the importance of alternative membrane remodelling strategies in bacteria where passive mechanisms for generating curvature are utilised.
Keywords: Membrane, curvature-inducing protein, vesicle, bacteria, remodelling
1. Introduction
Lipid bilayers are indispensable to living systems acting as physical barriers that both separate the inside of the cell from the external milieu, and provide scope for compartmentalisation and specialisation. They must be sufficiently rigid to maintain the integrity of the cell boundary and architecture of organelles, yet plastic to allow for continual remodelling and shape revision. Proteins integrated within the lipid bilayer or attached to its periphery provide a means for exquisite tuning of bilayer consistency and curvature, and ultimately for the evolution of diverse functions. In eukaryotes, classic examples include SNARES (Jahn and Scheller, 2006), dynamin (Praefcke and McMahon, 2004), and BAR-domain containing proteins (Mim and Unger, 2012) all of which have distinct mechanisms for fusing, cutting, and sensing membrane curvature.
Whilst bacteria do not have the same organelle complexity as eukaryotes, they still have extensive membrane reservoirs undergoing continual morphological differentiation, and face many of the same mechanical challenges for membrane remodelling. In comparison to eukaryotes, the number of known bacterial proteins dedicated to this task is surprisingly low. Is this because important families are yet to be discovered or does it reflect a more fundamental difference in the way bacteria approach membrane manipulation? In this review we differentiate between membrane remodelling that requires the active involvement of curvature-inducing proteins (CIPs) often by scaffolding and/or wedging (McMahon and Gallop, 2005), and passive remodelling which occurs in their absence (CIP-free) via mechanisms such as protein crowding (Stachowiak et al., 2012), asymmetric lipid enrichment (McMahon and Gallop, 2005), or membrane blebbing (Schwechheimer and Kuehn, 2015). CIP and passive membrane remodelling processes are not mutually exclusive and will work in concert within a cellular context. In eukaryotes, CIPs are prevalent in many membrane shaping processes, and in this review we assess whether the same phenomenon holds for bacteria.
Sophisticated life cycles, unexpected complex social behaviors, and increasingly diverse morphological specialisations are being discovered in bacteria. As this review describes, these are often critically dependent on membrane remodelling events (Fig. 1) with regions of high local curvature observed at both the cytoplasmic membrane (CM) and outer membrane (OM) (Fig. 2). The mechanics of how such curvature is introduced is relatively well understood in some systems such as cell cytokinesis. However, in many systems almost nothing is known. For example, is Myxococcus xanthus inter-cellular communication and OM exchange a CIP-free membrane remodelling process (Ducret et al., 2013; Pathak et al., 2012)? And might MamY in Magnetospirillum magneticum be a bona fide BAR-domain involved in magnetosome CM curvature (Mim and Unger, 2012)? In such cases are there conserved families of proteins driving these membrane remodelling processes or has each species evolved its own unique toolkit? Are CIPs required? These are the kinds of questions that this review aims to investigate and what makes this emergent field of membrane remodelling in bacteria important.
Fig. 1.
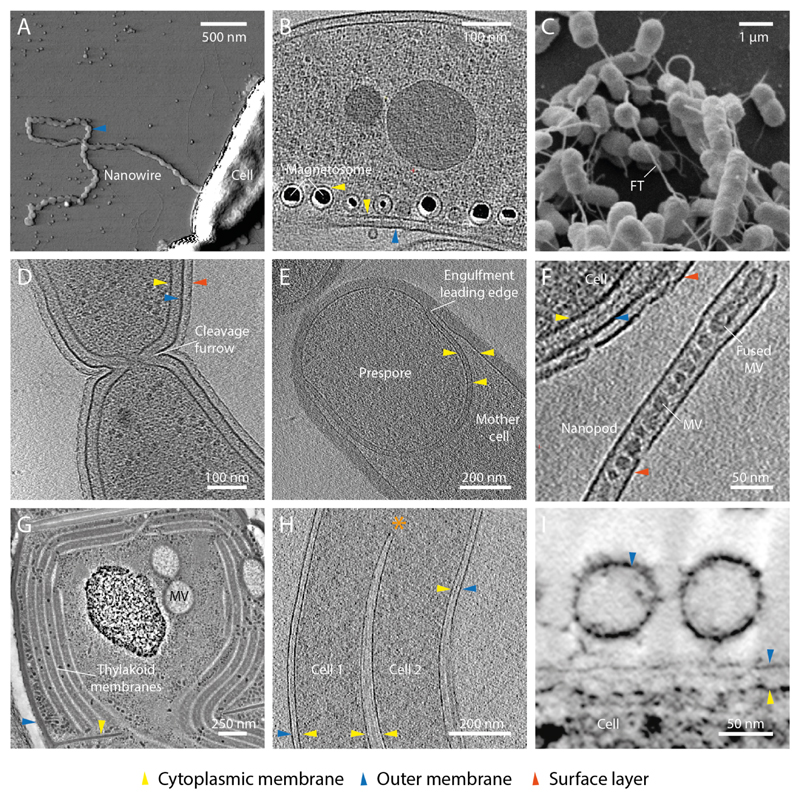
Selected examples of high membrane curvature and remodelling in bacteria. (A) Atomic force microscopy image of a Shewanella oniedensis MR-1 cell with an OM extension that forms a nanowire (Pirbadian et al., 2014). Image courtesy of Sahand Pirbadian and Mohamed El-Naggar. (B) Electron cryo tomogram (ECT) of Magnetospirillum magneticum AMB-1 shows that magnetosomes are invaginations of the CM (Komeili et al., 2006). Image courtesy of Arash Komeili and Grant Jensen. (C) Scanning electron micrograph showing cross-feeding between Escherichia coli and Acinetobacter baylyi connected by nanotubular membrane structures (Pande et al., 2015). FT- feeding tube. Image courtesy of Christian Kost. (D) ECT showing a late stage dividing Caulobacter crescentus cell with invaginated cytokinetic cleavage furrow. Image courtesy of Tanmay Bharat and Jan Löwe. (E) ECT of sporulating Bacillus subtilis. The prespore is in late stage engulfment by the mother cell (Tocheva et al., 2013). Image courtesy of Elitza Tocheva and Grant Jensen. (F) ECT of a Delftia sp. Cs1-4 nanopod (Shetty et al., 2011). MV- membrane vesicle. Image courtesy of Elitza Tocheva, Grant Jensen and William Hickey. (G) Thin-section micrograph of a high-pressure frozen, freeze-substituted Microcoleus sp. cell showing CM vesicles (MV) and extensive thylakoid membrane network (Scheuring et al., 2014). Image courtesy of Dana Charuvi, Reinat Nevo and Ziv Reich. (H) ECT of two Borrelia garinii cells with fused cell envelopes (Kudryashev et al., 2011). Asterisk shows merging of cytoplasmic cylinders and a region of high IM curvature. Image courtesy of Misha Kudryashev and Friedrich Frischknecht. (I) Micrograph of high pressure frozen, freeze substituted Myxococcus xanthus biofilms depicting OM vesicles tethered to cell surface (Palsdottir et al., 2009; Remis et al., 2014). Image courtesy of Manfred Auer.
Fig. 2.
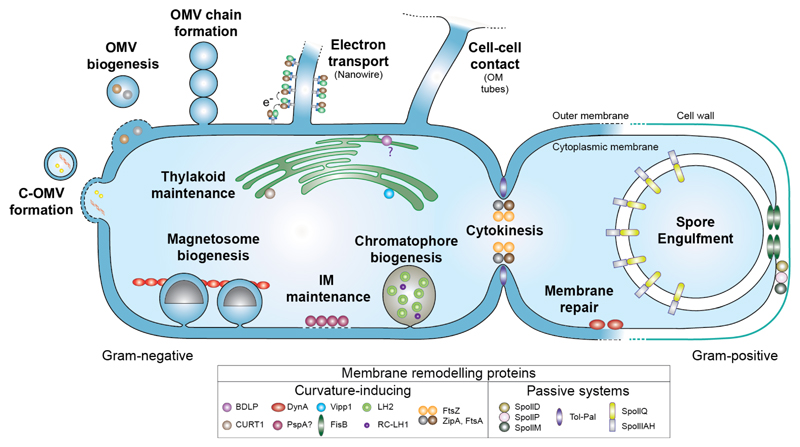
Schematic showing key membrane remodelling events in bacteria. Proteins involved in membrane remodelling are divided into two categories depending on their ability to directly bind membrane and induce its curvature. Note that whilst BDLP1 is known to associate with the cell envelope, its role in thylakoid maintenance is currently only speculative. PspA is included as it senses and binds stored curvature elastic stress in the membrane (McDonald et al., 2015) but direct evidence that it is able to remodel membrane like its homologue Vipp1 is still lacking.
2. CIP-mediated membrane remodelling in bacteria
This section focuses on those examples where active membrane remodelling occurs in bacteria through recruitment of CIPs that bind and directly drive rearrangements in bilayer shape. The aim is to focus specifically on the mechanism by which membrane remodelling is induced and to understand how this feeds through to function.
2.1. Bacterial dynamin-like proteins
2.1.1. The dynamin family and their discovery in bacteria
Dynamin family members (DFMs) are ancient GTPase domain containing CIPs that mediate fission, fusion and active reshaping of membrane. In eukaryotes, they are fundamental in diverse basic cellular processes such as endocytosis, mitochondrial maintenance and viral resistance. The hallmark feature of DFMs is their ability to couple oligomerisation, usually through assembly of a helical scaffold, to induction of high curvature in lipid bilayers (Praefcke and McMahon, 2004). This destabilises the bilayer and promotes membrane fission and fusion probably via a hemi-fission intermediate.
BDLP1 from the cyanobacteria Nostoc punctiforme was the first bacterial DFM characterised and was shown to share the same overall core fold as the canonical human Dynamin 1, excluding its PH domain and proline-rich domain (Fig. 3A) (Bramkamp, 2012; Low and Löwe, 2006; Low and Löwe, 2010). BDLP1 comprises a three-module architecture with an N-terminal GTPase domain, and 4-helix-bundle neck and trunk domains (Low and Löwe, 2006). The trunk tip incorporates a specialised lipid binding region termed the paddle. In bacteria it is not uncommon that this core fold is modified by gene truncation, gene fusion, or addition of domains that include predicted lipid and protein-protein binding motifs (Fig. 3A). In eukaryotic DFMs, domain modification is a mechanism for expanding functional repertoire and the same likely holds for bacteria. Bacterial DFMs are found in many bacterial species, and usually exist in multiple copy number, often as a pair within an operon lying side-by-side or in close proximity. However, some species actually have up to 8 putative homologues in variable genetic arrangements distributed throughout the chromosome (Fig. 3B). In such systems do the DFMs function alone or in hetero-complexes? And might they have more than a single function within a cell? Other than the specialist FtsZ-FtsA cytokinesis motor (Section 2.2), bacterial DFMs are the only identified protein family dedicated to coupling chemical energy release from nucleotide turnover directly to membrane shaping. This makes them a potentially unique cellular resource for heavy duty membrane remodelling in bacteria that can be adapted to different functions depending on the demands of the specific microbial niche. What are the known cellular roles for bacterial DFMs and is there evidence for functional diversification?
Fig. 3.
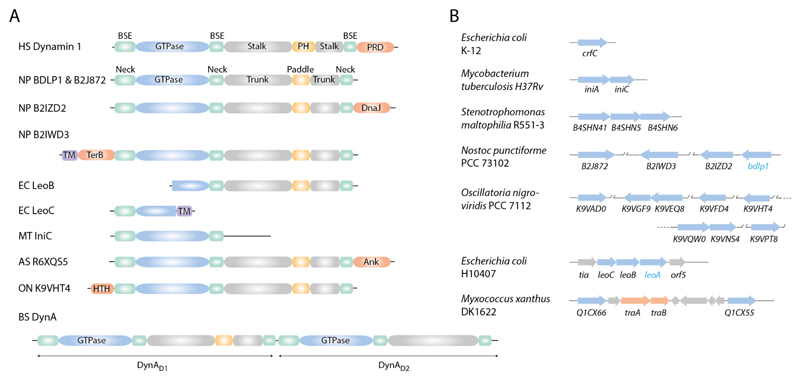
DFM gene structure and gene copy number is variable in bacteria. (A) Schematic showing the conserved modular structure of human Dynamin 1 (Faelber et al., 2011; Ford et al., 2011) and selected bacterial DFMs. Many bacterial DFMs have additional non-canonical domains that likely mediate protein-protein interactions. PRD- proline-rich domain; DnaJ- chaperone protein domain, PF00226; TM- predicted trans-membrane; TerB- tellurite resistance protein domain, PF05099; Ank- ankyrin repeat, PF12796; HTH- helix-turn-helix, PF01381. HS- Homo sapiens; NP- Nostoc punctiforme PCC73102; EC- Escherichia coli H10407; MT- Mycobacterium tuberculosis H37Rv; AS- Alistipes sp. CAG:435; ON- Oscillatoria nigro-viridis PCC 7112. (B) Schematic showing the variable number and genetic arrangement of bacterial DFMs between selected species (blue arrows). Bacterial DFMs usually exist as at least a pair in an operon (Bürmann et al., 2011; Michie et al., 2014). E. coli is an exception with the single DFM crfC (Ozaki et al., 2013). Cyanobacteria such as Nostoc punctiforme and Oscillatoria nigro-viridis usually have between 4-8 DFMs. Crystal structures for BDLP1 (Low and Löwe, 2006) and LeoA (Michie et al., 2014) confirm that their respective genes code for DFMs, whilst all others are assigned by sequence homology. In Myxococcus xanthus, two predicted bacterial DFMs flank the genes traA and traB, which are essential for OM exchange (Ducret et al., 2013; Pathak et al., 2012).
2.1.2. Bacterial DFMs - membrane remodelling machines with multiple functions?
Just a handful of bacterial DFM systems have been described and already there is emerging evidence for the following multiple functions.
Chromosome partitioning: the first functional data assigned to a bacterial DFM was for the Escherichia coli protein CrfC (Ozaki et al., 2013), which has a predicted structural fold that is similar to that of BDLP1 with GTPase, neck and trunk domains (Fig. 3A and Fig. 4A). However, part of the neck domain has been shown to bind DnaN, where DnaN is a DNA binding protein crucial for efficient DNA polymerase III processivity. Through this interaction, CrfC functions in nascent DNA strand colocalisation and chromosome equipartitioning (Ozaki et al., 2013). This is an interesting function as no membrane binding or remodelling behaviour has yet been described. Whether CrfC has some as yet unidentified mechanism for coupling chromosome partitioning to the membrane remains to be determined.
Fig. 4.
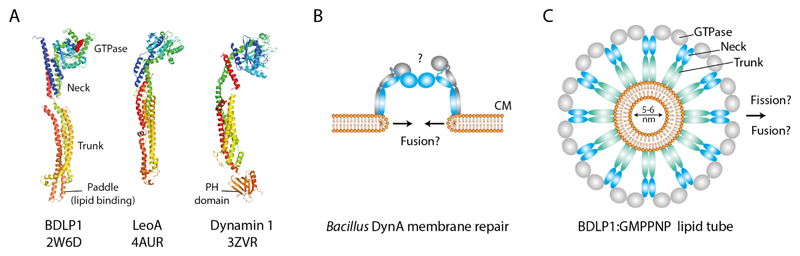
DFM-mediated membrane remodeling mechanisms in bacteria. (A) Bacterial and human DFMs share a conserved structural fold. (B) Bacillus subtilis DynA is implicated in membrane repair through a poorly understood fusion mechanism that appears not to rely on helical self-assembly as for BDLP1. (C) End-on view schematic showing a molecular model of a BDLP1-lipid tube (Low et al., 2009). BDLP1-GMPPNP forms a helical scaffold that forces the bound membrane into a highly curved tube. Through protein crowding and wedging, the paddle at the trunk tip likely displaces much of the outer leaflet.
Membrane stress response: DynA from Bacillus subtilis is distinct as it comprises two DFMs fused together, which probably results from the merging of a side-by-side chromosomal pair (Bürmann et al., 2011) (Fig. 3A). This phenomenon strongly suggests that hetero-oligomerisation of non-fused DFM pairs in other systems will be important for function. In the cell DynA protects against antibiotics that induce membrane pore formation such as nisin, and phage infection. By active membrane remodelling (Section 2.1.3), it likely repairs disordered lipid bilayer and forms a membrane surveillance system responding to specific environmental stress conditions (Sawant et al., 2015) (Fig. 4B). Importantly, this function is consistent with earlier reports of the Mycobacterium bovis DFM IniA, which confers resistance to the antibiotics isoniazid and ethambutol (Colangeli et al., 2005). Whilst these antibiotics target the cell, they also indirectly induce membrane stress due to impaired (un-)decaprenyl phosphate recycling (Grover et al., 2014). Furthermore, iniA exists in an operon side-by-side with another DFM iniC (Fig. 3B), which may form a functional unit equivalent to Bacillus DynA (Bürmann et al., 2011; Colangeli et al., 2005).
Outer membrane vesicle (OMV) biogenesis: enterotoxigenic E. coli H10407 intoxicates target cells by releasing OMVs containing heat labile (LT) enterotoxin from its cell surface. The tia pathogenicity island contributes to LT release as knockout of leoA, one of the five genes within the island, leads to LT being sequestered in the periplasm (Brown and Hardwidge, 2007; Fleckenstein et al., 2000) and an ~50% reduction in LT OMV-mediated secretion (Brown and Hardwidge, 2007; Fleckenstein et al., 2000; Michie et al., 2014). With LeoA recently identified as a bacterial DFM (Fig. 4A) located in the periplasm (Michie et al., 2014), this provides evidence that it may play a direct role in OMV biogenesis. However, that leoA knockout does not fully reduce OMV secretion suggests that other mechanisms for OMV biogenesis are still important and LeoA may only be indirectly involved in vesiculation. Whether LeoA binds nucleotide is unclear (Brown and Hardwidge, 2007; Michie et al., 2014) and certainly it does not readily bind lipid in vitro (Michie et al., 2014). It is speculated that its activation requires the DFMs LeoB and LeoC located beside LeoA within the tia locus (Michie et al., 2014) (Fig. 3). Further studies are therefore required before this exciting idea that DFMs have a conserved role in vesiculation in both eukaryotes and bacteria is proven.
Thylakoid maintenance: cyanobacteria have extensive thylakoid membrane systems with specific shape, ultrastructure, and high membrane curvature. How do cyanobacteria maintain this morphology? In plants, both fission and fusion DFMs play an essential role in thylakoid maintenance and therefore they are obvious candidates for the equivalent function in cyanobacteria, where they are found in particularly high copy number (Fig. 3B). Current evidence for their involvement is only circumstantial and rests on the observation that BDLP1 in the cyanobacteria N. punctiforme is most closely related to FZL, a plant mitofusin-like DFM required for thylakoid integrity, and that they share a similar punctate membrane localization pattern (Gao et al., 2006; Low and Löwe, 2006). More studies are therefore required before a role in thylakoid maintenance can be properly assigned.
2.1.3. Molecular mechanisms for DFM-mediated membrane remodelling in bacteria
From the few studies where the molecular mechanism for bacterial DFM membrane remodelling has been investigated, it is likely that many of the same principles are used to introduce curvature as observed in eukaryotes. This includes oligomerisation, wedging/protein crowding effects mediated by lipid binding domains, and induction of a hemi-fission state. BDLP1 from N. punctiforme remodels membrane by forming a helical scaffold that forces the lipid template, under high curvature, into an ~10 nm diameter tube (Low and Löwe, 2006; Low et al., 2009). Filament assembly requires both lipid and GTP, which induces back-to-back association of subunits and GTPase domain dimerization (Fig. 4C). The paddle lipid-binding motif at the trunk tip wedges into, and probably displaces through protein crowding, much of the membrane outer leaflet, making it amenable to subsequent fission or fusion events. Nucleotide hydrolysis is then coupled to filament collapse and release from the membrane. Although the precise role of BDLP1 is yet to be resolved and it is speculated to be involved in thylakoid maintenance (Low and Löwe, 2006), it is clear that this protein readily oligomerises into supra-molecular assemblies and is a potent tool for inducing membrane curvature.
Biochemical studies focused on B. subtilis DynA (Bürmann et al., 2011) have also been particularly important as this protein results from the genetic fusion of a dynamin pair (DynAD1 and DynAD2) (Fig. 3A) and may represent a paradigm for how other DFM pairs co-depend for function. DynA mixed with liposomes binds and tethers the liposomes together, or actively fuses them in the presence of magnesium. This fusion is independent of nucleotide despite both DynAD1 and DynAD2 being active GTPases. Interestingly, the DynAD1 subunit alone is sufficient for this liposome tethering and fusion (Bürmann et al., 2011). The molecular detail of how DynA induces membrane fusion is still to be dissected, although this phenotype fits plausibly with its role in membrane stress and repair (Section 2.1.2). Certainly, how any eukaryotic or bacterial DFM tethers opposing membranes together before inducing membrane fusion is one of the outstanding questions in the field. DynA does not seem to form a helical polymer and tubulate liposomes like BDLP1, and its membrane association is not nucleotide dependent. These proteins therefore seem to be mechanistically quite diverged, and whether this is a consequence of having different functions remains to be determined.
2.2. Bacterial cytokinesis
Bacteria divide by replicating their chromosome and distributing a single copy to each of the two nascent daughter cells. Localisation of the division site is defined by the Min system (Raskin and de Boer, 1999) and nucleoid occlusion (Bernhardt and de Boer, 2005; Woldringh et al., 1991; Wu and Errington, 2004), which allows for a tightly regulated and complex hierarchical assembly of proteins called the divisome to form exclusively at the mid-cell. The divisome contains the essential machinery for the mechanical invagination and constriction of the CM, de novo peptidoglycan synthesis, and septum formation. This process of cell division has recently been reviewed elsewhere (Egan and Vollmer, 2013; Lutkenhaus et al., 2012; Rico et al., 2013), and so here only those cytokinetic proteins involved in remodelling membrane are focused upon and their mechanisms discussed.
2.2.1. FtsZ, FtsA and ZipA form the membrane constriction complex
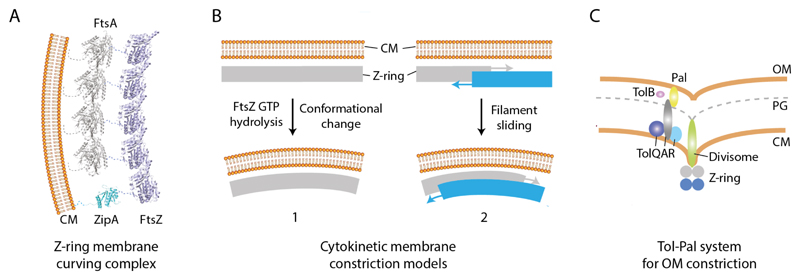
During cell division, the CM is observed to invaginate at the mid-cell so that a localised protrusion of high curvature forms and constricts radially (Bi and Lutkenhaus, 1991; Burdett and Murray, 1974; Judd et al., 2005; Szwedziak et al., 2014). In a broadly conserved mechanism, the proteins FtsZ, FtsA, and ZipA form a core membrane curving complex that drives this CM constriction. The proteins are recruited early to the division site and act as an assembly hub for all downstream divisome components. FtsZ is a tubulin homologue (Löwe and Amos, 1998) that assembles dynamic (Anderson et al., 2004) head-to-tail linear filaments in a GTP-dependent fashion. Recent cryo-electron tomography studies on whole cells offer evidence that FtsZ filaments encircling the constriction site form a continuous, rather than discontinuous (Coltharp et al., 2016; Holden et al., 2014; Li et al., 2007), helical Z-ring (Szwedziak et al., 2014). Anchoring of FtsZ to the membrane is mediated by association with ZipA (Hale and deBoer, 1997) and actin-like FtsA, with the latter the predominant cellular binding partner (Pichoff and Lutkenhaus, 2002; Wang et al., 1997) (Fig. 5A). In B. subtilis, it is estimated that there is a ratio of around 1000 FtsA to 5000 FtsZ molecules in the cell (Feucht et al., 2001), so that FtsZ polymers are likely attached to the membrane by a ring of FtsA polymers interspersed with ZipA molecules.
Fig. 5.
Overview of core cytokinetic CM and OM remodeling machinery. (A) The proteins FtsA, ZipA and FtsZ form the Z-ring, a helical membrane-associated contractile scaffold that localises at the mid-cell. Minimally, FtsA and ZipA tether FtsZ to the membrane. FtsZ is the motor for generating membrane contractile force and curvature. (B) Two prevailing mechanisms for how FtsZ generates membrane curvature. Left, GTP hydrolysis drives FtsZ filament curving which is coupled to membrane bending. Right, Helical FtsZ filaments maximize lateral interactions between neighbouring rungs through filament sliding. This leads to constriction of the helix and the associated membrane. Both mechanisms require iterative cycles of FtsZ assembly and disassembly. (C) Schematic showing the core components of the Tol-Pal complex, which couples the CM to the OM, and is essential for OM constriction. A proton motive force is somehow required for energisation and function of the complex.
Reconstitution experiments in which FtsZ is engineered with a lipid binding motif and incorporated into liposomes show the formation of Z-ring-like contractile girdles inducing membrane invagination (Osawa et al., 2008). More natural reconstitution experiments where FtsA is mixed with native or fluorescently tagged FtsZ also show striking membrane contractile girdles (Osawa and Erickson, 2013; Szwedziak et al., 2014). This reinforces the assertion that these components alone are sufficient to provide the force required to remodel membrane and drive cell division. Such data complements early in vivo data where E.coli FtsZ could be replaced by foreign B. subtilis FtsZ engineered to bind E.coli FtsA and ZipA (Osawa and Erickson, 2006), meaning FtsZ is likely a key driver for membrane constriction.
2.2.2. The mechanics of cytoplasmic membrane constriction
Two principle models predominate that are not necessarily mutually exclusive for how the FtsZ motor drives membrane constriction. The first model has at its core nucleotide driven conformational change that introduces a force generating curvature into FtsZ filaments and consequently the membrane. In vitro FtsZ filaments bound with GTP tend to be linear, but form curved filaments in the presence of GDP (Erickson et al., 1996; Lu et al., 2000). Membrane constriction could occur as straight filamentous scaffolds attached to the membrane bend, and introduce a radial force that pulls inward (Li et al., 2007) (Fig. 5B). Interestingly, the crystal structure of a Mycobacterium FtsZ-GDP protofilament has recently been solved where a curved conformation is observed. Whilst this curvature is observed through a crystal contact, the structure provides a putative mechanism for longitudinal bending between subunits (Li et al., 2013). However, bent filaments do also occur with non-hydrolysable GTP analogues on mica (Horger et al., 2008), and the hydrolysis-deficient FtsZ D212G mutant can form contractile girdles in liposomes (Osawa and Erickson, 2011) and completes cell division in vivo (Trusca et al., 1998). Further work is therefore required to clarify the precise role of nucleotide in driving membrane bending and FtsZ-mediated membrane remodelling.
The second model originates from theoretical studies where FtsZ force generation and membrane curvature is induced by maximization of lateral contacts between filaments through a sliding mechanism (Lan et al., 2009). Complementary modelling work supports the premise that force generation is possible without nucleotide-dependent bending (Ghosh and Sain, 2008; Ghosh and Sain, 2011), and there is growing experimental evidence for this as well. Reconstitution of unmodified native FtsZ and FtsA in liposomes and visualized by cryo-electron tomography represents the most natural in vitro system yet assembled (Szwedziak et al., 2014). Here, constriction sites produced by helical FtsZ-FtsA contractile girdles are likely nucleotide independent and curvature is not obviously increased by the addition of GTP. The observation of spiral domes with tapering filament curvature over a wide range of radii suggests a possible sliding and condensation mechanism (Fig. 5B). Filament sliding could be coupled to increasing membrane constriction if the helical pitch of the Z-ring is constrained, as it would be in vivo. Filament sliding also provides a mechanism for membrane abscission and does not require the addition of still unidentified membrane remodelling machinery to seal the septum (Szwedziak et al., 2014). Note that the Snf7 protein in ESCRT-III has recently been shown to form filamentous spirals that are coupled to membrane deformation albeit through a superficially different mechanism (Chiaruttini et al., 2015). In addition, the Archaea Euryarchaeota and Crenarchaeota phylums use ESCRT-III homologues instead of FtsZ to form a ring-shaped structure that drives membrane constriction during cell division (Lindas et al., 2008; Samson et al., 2008). Central to both the bending and sliding model is the dynamic nature and cycling of FtsZ assembly and disassembly, which is modulated by complex interplay between FtsA, ZipA and other FtsZ associating proteins. The precise role of nucleotide in driving FtsA assembly (Szwedziak et al., 2012) and whether this augments FtsZ-mediated membrane constriction is currently unclear.
There is still a question as to the degree to which the remodelling and synthesis of novel peptidoglycan contributes or even drives CM constriction. Here CM invagination would be driven passively by the ingrowth of nascent peptidoglycan, and the dynamic Z-ring would simply act as a scaffold to spatially organize cell wall remodelling proteins (Loose and Mitchison, 2014). Certainly peptidoglycan synthesis seems a key factor for force generation during forespore engulfment during sporulation in B. subtilis (Meyer et al., 2010) (Section 3.2.1). However, this is hard to reconcile with the observation that Mycoplasma genus bacteria lack a cell wall and likely rely on FtsZ for cell division (Wang and Lutkenhaus, 1996). Similarly, L-form bacteria (Allan et al., 2009), which lack a cell wall, have been of interest as a potential tool for investigating the role of the Z-ring. However, L-form B. subtilis has recently been shown to divide without the normal FtsZ-mediated division apparatus (Leaver et al., 2009). Instead, L-form B. subtilis undergo substantial remodeling of their cell envelope membranes with pseudopodium-like protrusions developing into spherical bodies that are thought to be progeny. The mechanism for these remarkable membrane protrusions is not well understood although it is speculated that active force generation will be required (Leaver et al., 2009).
For OM invagination and constriction, no discrete membrane remodelling machinery has yet been found in Gram-negative bacteria. Instead a poorly understood CIP-free mechanism seems to have evolved where OM invagination is driven predominantly by a coupling to the peptidoglycan layer and CM via the conserved Tol-Pal system that spans the periplasm. Components of this system include the integral CM components TolA, TolR, TolQ, the soluble periplasmic TolB, and OM protein Pal (Sturgis, 2001), and are recruited to the divisome via FtsN (Gerding et al., 2007) (Fig. 5C). Whilst the invagination force likely comes largely from synthesis of nascent peptidoglycan (Gray et al., 2015), the proton motive gradient is also critical for energizing the Tol system (Cascales et al., 2000). Whether Tol components are actively involved in drawing the OM inwards or have more of a signal switch function is still to be resolved (Bonsor et al., 2009; Cascales et al., 2000; Gerding et al., 2007; Germon et al., 2001).
2.3. Thylakoid maintenance in cyanobacteria
2.3.1. Vipp1 can trigger membrane fusion in vitro
Vesicle-inducing protein 1 (Vipp1) is a peripheral membrane protein broadly conserved in cyanobacteria and plants. Vipp1 is involved in thylakoid membrane maintenance and biogenesis, although in vivo functional studies on its exact role are not yet conclusive. For example, the deletion of the vipp1 gene either decreases the formation of CM vesicles and thylakoid membranes (Kroll et al., 2001; Westphal et al., 2001) or has little effect (Fuhrmann et al., 2009a; Nordhues et al., 2012). Two important structural features of Vipp1 are crucial for thylakoid membrane remodelling. Firstly, Vipp1 interacts with the CM of cyanobacteria via its N-terminal amphipathic domain (Otters et al., 2013). Secondly, Vipp1 can form oligomeric structures (Fuhrmann et al., 2009b), which are capable of binding to lipid vesicles in vitro (McDonald et al., 2015). Recent data suggests that the oligomeric ring of Vipp1 can trigger membrane fusion in vitro in the presence of magnesium (Hennig et al., 2015). This is important as cyanobacteria will likely require a fusogenic CIP for maintenance of their thylakoid network. However, whether a membrane fusion role fits with the various proposed functions for Vipp1 in vivo (Kroll et al., 2001; Lo and Theg, 2012; Nordhues et al., 2012; Zhang et al., 2014) remains to be determined. In plants, thylakoid maintenance is mediated by the interplay of fission and fusion DFMs, and whether similar proteins work in concert with Vipp1 in cyanobacteria remains to be determined. Currently there is no protein mediating thylakoid membrane fission, although BDLP1 and its homologues would be obvious candidates (Section 2.1).
Phage shock protein A (PspA) is a broadly conserved homologue of Vipp1 found in both Gram-positive and Gram-negative bacteria. Rather than having a role in thylakoid maintenance, PspA maintains the integrity of the CM by reacting to membrane stress and protecting against loss of the proton motive force (Kobayashi et al., 2007). Despite a lack of in vivo evidence for PspA-induced membrane curvature, its structural similarity to Vipp1, mechanism of membrane binding via its N-terminal amphipathic helix (McDonald et al., 2015), and ability to form oligomeric rings (Hankamer et al., 2004) suggests it may have similar membrane remodelling properties.
2.3.2. CURT1 proteins shape thylakoid membranes
Recently, Curvature Thylakoid 1 (CURT1) proteins were suggested to remodel thylakoids by inducing membrane curvature. The CURT1 family of proteins is highly conserved in plants and cyanobacteria. Deletion of CURT1 genes alters the architecture of thylakoid membranes, and reduces levels of photosynthesis (Armbruster et al., 2013). CURT1A is able to bind to membranes and trigger tubulation of liposomes in vitro. As with Vipp1, CURT1A forms oligomeric structures both in thylakoids and lipid vesicles (Armbruster et al., 2013).
2.4. Cytoplasmic membrane remodelling
2.4.1. Chromatophore biogenesis
Chromatophores of purple phototrophic bacteria are vesicles that originate from the CM and convert light energy into chemical energy. Depending on the stage of development and/or light intensity, chromatophores have been shown to either directly link to the CM, or be detached (Scheuring et al., 2014; Tucker et al., 2010). For full functionality, the chromatophore requires a core complex made up of light-harvesting I (LH1) and reaction centre (RC) complexes, a cytochrome bc1 complex, and a light-harvesting II (LH2) complex.
Studies from the last decade have been directed towards understanding chromatophore biogenesis, with special focus on how membrane curvature is introduced. CM vesiculation and the shape of chromatophores appear to be driven by interplay between RC-LH1 and LH2. Experimental and computational data indicate that both RC-LH1 and LH2 dimers induce membrane curvature by assembling a scaffold (Olsen et al., 2008; Qian et al., 2008). The mechanism behind RC-LH1-induced membrane curvature relies on the ability of RC-LH1, in complex with a small polypeptide called PufX, to bend its conformation after dimerisation (Qian et al., 2008; Siebert et al., 2004). In the absence of LH2, RC-LH1-PufX dimers form tubular chromatophores in Rhodobacter (Chandler et al., 2008; Qian et al., 2008; Siebert et al., 2004). When monomeric, RC-LH1 alone appears to lack membrane-curving properties as observed in species with lamellar chromatophores (Scheuring et al., 2014). Atomic force microscopy studies with LH2 protein complexes show two tilted conformations of LH2 dimer (inwards and outwards) that create a zig-zag lattice on spherical chromatophores (Liu et al., 2008; Qian et al., 2008). In addition, an alternative hexagonal arrangement of LH2 complexes can induce membrane curvature in lamellar chromatophores as observed in Rhodospirillum sp. (Chandler et al., 2008; Chandler et al., 2009; Goncalves et al., 2005). Collectively, the shape of chromatophores depends on the presence and oligomeric state of RC-LH1 and LH2 complexes, which are capable of inducing membrane curvature via protein crowding as well as through self-assembly scaffold mechanisms.
In a more general context, molecular dynamic simulations suggest that crowding and the geometry of membrane protein oligomers can be sufficient to induce membrane curvature. Monte Carlo simulations show that protein complexes with similar curvature properties to RC-LH1 and LH2 may attract and accumulate in the membrane, leading to areas of high local curvature (Frese et al., 2008). Other CM proteins in bacteria may therefore represent an additional source of membrane curvature that is not well understood.
2.4.2. Magnetosome biogenesis
Magnetosomes are innovative systems for detection of magnetic fields developed by magnetotactic bacteria. Electron tomography established that magnetosomes are linearly arranged CM invaginations supported by the actin homologue MamK (Komeili et al., 2006). Interestingly, the recent discovery of MamY, which encodes a putative BAR domain, suggests that there might be a role for CIPs in magnetosome formation. MamY was shown to tubulate liposomes in vitro by direct interaction with phospholipids (Cornejo, 2016; Tanaka et al., 2010). Additionally, membrane invagination during magnetosome biogenesis depends on four genes from the mamAB operon - mamI, mamL, mamB and mamQ (Murat et al., 2010). MamL contains a positively charged C-terminal helix, which is predicted to interact with the CM and trigger membrane curving during magnetosome biogenesis (Komeili, 2012). However, the expression of these four genes and simultaneous knock out of the rest of the mamAB operon is not sufficient to trigger membrane vesiculation, suggesting the involvement of other factors early in magnetosome development (Komeili, 2012; Murat et al., 2010).
3. CIP-free membrane remodelling in bacteria
This section focuses on those examples in bacteria where intensive membrane remodelling is known to occur but appears, given the current state of understanding, not to depend on specific CIPs. This idea of passive membrane remodelling is particularly relevant to the OM in Gram-negative bacteria, which undergoes constant re-shaping and diverse specialisation with no CIP yet conclusively implicated. This may be because one has yet to be found or be fully characterised, with the DFM LeoA (Michie et al., 2014) a putative candidate. Or it may reflect a more fundamental shift in the way bacteria approach membrane remodelling in an environment generally remote from, or with reduced (Mempin et al., 2013), chemical energy sources.
3.1. OM vesicle and tube biogenesis
3.1.1. OMVs - a single tool with multiple functions
OMVs are 20-300 nm in diameter and include periplasmic and surface constituents of Gram-negative bacteria (Beveridge, 1999; Gankema et al., 1980; Kadurugamuwa and Beveridge, 1997; Zhou et al., 1998). They are released individually or conjoined as chains (Remis et al., 2014), and represent a means for bacteria to communicate remotely and modulate the external environment. Their functions are diverse and include the release of virulence factors from pathogens (Ellis and Kuehn, 2010), the promotion of inter-cellular crosstalk within the bacterial community, and nutrient foraging and uptake (Berleman and Auer, 2013). In the current era of rising bacterial antibiotic resistance, some of the most exciting OMV studies examine their ability to enzymatically inactivate antibiotics (Kulkarni and Jagannadham, 2014; Lee et al., 2013), spread antibiotic resistance genes (Jin et al., 2011), and act as decoys by direct binding to antibiotics. Alternatively, the OM may be remodeled into tubes that extend many microns from the host. Such structures are implicated in for example, long-range electron transport in Shewanella (Gorby et al., 2008; Pirbadian et al., 2014) (Fig. 1A), OM exchange in M. xanthus (Ducret et al., 2013), or formation of structured multi-cellular consortia in Chlorochromatium aggregatum (Wanner et al., 2008). OMV and tube formation represent some of the most active membrane remodelling in bacteria and their importance for complex behaviour and niche adaptation should not be understated.
3.1.2. Is OMV biogenesis a CIP-free process?
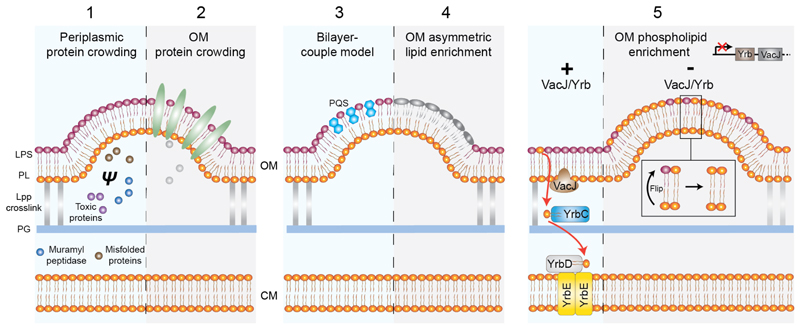
CIPs such as dynamin and clathrin are prevalent in driving membrane vesiculation processes in eukaryotes. However, the same does not appear to hold in primary OMV biogenesis pathways in bacteria, where CIPs have not been readily observed. Current models for OMV biogenesis rely on decoupling the OM from the peptidoglycan, and then promoting its curvature through various CIP-free mechanisms. The OM is connected to the peptidoglycan predominantly through Braun’s lipoprotein (Lpp), which is evenly distributed throughout the cell envelope, and Pal (Fig. 5C), which is located at the cell poles. The mechanisms by which OMV biogenesis then occurs has recently been reviewed (Schwechheimer and Kuehn, 2015) and so only the basic principles for membrane curvature induction are repeated here (Fig. 6). Minimally, loss of peptidoglycan-Lpp linkage appears sufficient to induce membrane vesiculation. Curvature is further augmented by 1) localised concentration of periplasmic components that increases turgor pressure and induces membrane bulging (Hayashi et al., 2002; Zhou et al., 1998); 2) localised crowding of protein cargo destined for export; 3) localised lipid enrichment (Tashiro et al., 2011); 4) the ‘bilayer-coupled model’ which describes small-molecule wedging effects as for Pseudomonas quinolone signal (Schertzer and Whiteley, 2012); 5) phospholipid accumulation in outer leaflet of the OM (Roier et al., 2016). This last model is of particular interest, as it exemplifies how membrane curvature can be achieved in the absence of CIPs and with minimal energetic cost in the relatively energy poor environment of the periplasm. Unlike the CM, the OM is asymmetric with the inner and outer leaflets enriched with phospholipids and lipopolysaccharides, respectively. This important asymmetry is maintained by the VacJ/Yrb transporter system, which shuttles flipped phospholipids from the outer leaflet of the OM back to the CM (Roier et al., 2016). Failure to recover phospholipids from the outer leaflet of the OM has two main consequences that may both promote membrane vesiculation. Firstly, curvature is generated by a bilayer-couple mechanism where accumulation of phospholipid leads to an expansion of the outer leaflet surface area relative to the inner leaflet. Secondly, altered lipid composition in the OM affects membrane fluidity and indirectly contributes to membrane vesiculation.
Fig. 6.
Mechanisms of OMV biogenesis. All models depend on an initial decoupling of the OM from the peptidoglycan layer by breakage of the Lpp crosslink. This decoupling alone is sufficient to induce membrane vesiculation and can be augmented by the following 5 membrane curving mechanisms none of which involve CIPs (Schwechheimer and Kuehn, 2015). 1, Selective local protein crowding in the periplasm increases turgor pressure (ψ) and induces membrane bulging. 2, Selective crowding of cargo proteins such as virulence factors in the OM. 3, A generalised model for OMV formation where molecules such as Pseudomonas quinolone sequence (PQS) increase the surface area of the outer leaflet relative to the inner leaflet by a wedging effect (Schertzer and Whiteley, 2012). 4, Specific enrichment of different fatty acids and LPS subtypes that may be further modified by charge or polysaccharide incorporation. 5, A novel potentially general mechanism for OMV biogenesis based on phospholipid accumulation in the OM outer leaflet. In the absence of the VacJ/YrbABC transport system, phospholipids flip from the OM inner leaflet and are retained in the outer leaflet rather than being recovered to the CM (Roier et al., 2016).
Whilst primary pathways for OMV biogenesis appear to operate in the absence of CIPs, excitingly there may be additional discrete or auxiliary pathways that facilitate vesiculation and where CIPs are important. For example, LeoA in ETEC is a DFM located in the periplasm and implicated in OMV secretion (Section 2.1.2). In addition, we note that in M. xanthus two putative DFMs reside in the same operon with, and just flanking, traA and traB (Fig. 3B), which are essential genes for OM exchange (Pathak et al., 2012) where lipid tubulation and vesiculation appear critical (Ducret et al., 2013). If such DFMs and other CIPs are operating in the periplasm and potentially at the OM interface, how might such systems be energised? LeoA has a GTPase domain, although whether it binds nucleotide is unclear (Brown and Hardwidge, 2007; Michie et al., 2014). Such proteins may be primed by energy transduction from binding partners spanning the CM with exposure to the cytoplasm, or membrane binding energies alone may be sufficient for induction of membrane curvature. Alternatively, indirect energy sources could drive different steps of OMV biogenesis as observed in the Ton system for siderophore and colicin uptake (Kleanthous, 2010). Here proton motive force generated at the CM is coupled to protein conformational changes that are transduced across the periplasm to OM targets.
How OM tubulation occurs is poorly understood although at least in some systems it appears coupled to OMV biogenesis rather than any as yet unidentified bespoke machinery. Uniformly sized OMVs are known to be secreted as extensive chains (Gorby et al., 2008). Atomic force microscopy and TEM studies suggest chain precursors may be modified so that nascent vesicles are merged rather than pinched, and then through an unknown smoothing mechanism form tubes (Pirbadian et al., 2014; Remis et al., 2014).
Recent cryo-electron microscopy studies of Shewanella vesiculosa, and various human pathogens including Neisseria gonorrhoeae and Pseudomonas aeruginosa, indicate that a small proportion of vesicles released are derived from both the CM and OM, and so have a double bilayer structure (C-OMVs) (Perez-Cruz et al., 2015). Interestingly, both double stranded DNA and ATP are found packaged within the C-OMVs. How localised bulging of the CM and OM to generate the vesicle, followed by neck constriction and pinching from the donor membranes occurs is not yet clear. Coordination and remodeling of double bilayer systems is a substantial undertaking as demonstrated by membrane fusion of the mitochondrial inner and outer membrane, which requires complex cross-talk between two DFM pairs located on each membrane as well as support from many other auxiliary proteins (Praefcke and McMahon, 2004). Whether CIPs are involved in this process remains to be seen, although it is interesting to note that in both chromatophore biogenesis (Section 2.4.1) and magnetosome formation (Section 2.4.2) that involve reshaping of the CM, CIPs are either implicated or known to be present.
3.2. Spore morphogenesis in Bacillus subtilis
Gram-positive B. subtilis undergoes a dramatic developmental program during periods of severe resource limitation that results in a durable dormant spore. Initially an asymmetric cell division results in a large mother cell and a future forespore. Once the dividing septum is degraded between these cells, the mother cell membrane migrates around the forespore (Fig. 1E) until the forespore is fully encapsulated and only a thin tubular channel remains. Membrane scission machinery then severs this channel so that the forespore is entirely internalised and detached from the mother cell. Forespore engulfment represents a key paradigm for large-scale membrane remodelling in bacteria where the significant bulk of membrane shaping is seemingly achieved by a CIP-free remodelling mechanism that utilises a minimum of chemical energy.
3.2.1. Rachet-like mechanisms drive membrane remodelling during forespore engulfment
As the mother cell engulfs the forespore, high curvature is induced at the leading edge of the migrating membrane. The force required to reshape the membrane and maintain this movement appears to come from two distinct but synergistic mechanisms.
Firstly, force is generated through the interplay of the outer cell wall synthesis and degradation machinery, and the attached mother cell membrane. SpoIID and SpoIIP are peptidoglycan hydrolases that iteratively bind and degrade peptidoglycan. Complexed with SpoIIM and located within the leading edge of the mother cell membrane (Fig. 2), they function processively to disassemble the cell wall (Gutierrez et al., 2010; Morlot et al., 2010). This function provides an elegant passive mechanism whereby the mother cell membranes are pulled via peptidoglycan retreat around the forespore with no CIP required (Morlot et al., 2010). In addition, light microscopy studies show that peptidoglycan is being actively synthesized along the sporulation septum and that its inhibition halts membrane migration (Meyer et al., 2010). Presumably peptidoglycan synthesis therefore also contributes to membrane migration although the coupling mechanism is still to be resolved.
The second system for membrane engulfment and force generation is revealed when the cell wall is enzymatically removed. Interestingly, engulfment time is markedly reduced and is still completed in the absence of the peptidoglycan remodelling complex SpoIIP, SpoIID and SpoIIM (Broder and Pogliano, 2006). Instead, two membrane proteins, SpoIIQ located on the forespore and SpoIIIAH located on the mother cell (Fig. 2), facilitate engulfment via a putative Brownian ratchet mechanism. Here, thermal fluctuations may shift the leading edge of the mother cell membrane forward whilst SpoIIQ-SpoIIIAH interaction inhibit backward motion so that movement proceeds through a zip-like mechanism (Broder and Pogliano, 2006; Ojkic et al., 2014). Both the above mechanisms will complement each other in intact cells, suggesting what is in effect a dual ratchet mechanism.
3.2.2. Forespore abscission requires the CIP FisB
In the late stages of forespore engulfment the migrating mother cell membranes meet at the cell pole where the final tubular connection between forespore and mother cell is severed. This membrane remodelling event is one of only a few examples in bacteria where a CIP is known to be involved, although the molecular mechanism has not yet been dissected. FisB is a predicted bitopic membrane protein located in the mother cell membrane with a 23 kDa extracellular domain. Mutant studies showed that fisB disruption induces a severely impaired forespore abscission phenotype, whilst FisB fused to GFP or YFP is observed to concentrate at the cell pole during the final stages of engulfment (Doan et al., 2013). Importantly, purified FisB induces liposome mixing when embedded on their surface, and this effect does not require FisB localisation in trans on both opposing membranes. Instead the periplasmic domain preferentially binds cardiolipin, which may be recruited to the positive curvature of the membrane leading edge (Doan et al., 2013). The mechanism by which FisB induces membrane fission, perhaps through oligomerisation, conformational change, or lipid sequestering, is still to be resolved.
4. Conclusion
Membrane remodelling is a fundamental biological process. In bacteria, it is central to morphological differentiation including basic events such as cell division, and more complex behaviours such as OM exchange and inter-cellular communication. Despite many examples of elaborately modelled membrane in bacteria, only a limited number of CIPs have been identified so far. Multiple CIPs may simply be unnecessary in bacteria or considered excessively energetically costly, particularly when operating beyond the cytoplasmic milieu at the OM interface. Bacteria have found elegant mechanistic alternatives such as lipid enrichment, or coupling cell wall remodelling to membrane shaping. CIPs may also be important for viability making them challenging to pick up in mutant screens, and obvious mutant phenotypes may be obscured by an element of redundancy in membrane remodelling systems or by simply having subtle phenotypes.
Alternatively, the relative lack of known CIPs in bacteria compared with eukaryotes is possibly a consequence of having evolved as more specialist tools for complex adaptations that are only just being recognised and investigated. Certainly, the intensive investment in DFMs in so many bacteria supports this idea. DFMs are already implicated in ETEC pathogenesis (Michie et al., 2014) and antibiotic drug tolerance (Sawant et al., 2015) and it is likely that their functional repertoire will expand with future studies.
Whilst it is clear that as a strategy for membrane remodelling bacteria have made effective use of CIP-free mechanisms, CIPs also play an essential part - one that is only just being resolved. This makes the emerging field of membrane remodelling in bacteria a rich ground for future discovery.
Acknowledgements
This work was supported by a Wellcome Trust Career Development Fellowship (097328/Z/11/Z) to HL. We are grateful to Daniel Hansen and Anastasia Chernyatina for manuscript feedback.
References
- Allan EJ, Hoischen C, Gumpert J. Bacterial L-forms. Advances in applied microbiology. 2009;68:1–39. doi: 10.1016/S0065-2164(09)01201-5. [DOI] [PubMed] [Google Scholar]
- Anderson DE, Gueiros-Filho FJ, Erickson HP. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J Bacteriol. 2004;186:5775–5781. doi: 10.1128/JB.186.17.5775-5781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster U, Labs M, Pribil M, Viola S, Xu W, Scharfenberg M, Hertle AP, Rojahn U, Jensen PE, Rappaport F, Joliot P, et al. Arabidopsis CURVATURE THYLAKOID1 proteins modify thylakoid architecture by inducing membrane curvature. The Plant cell. 2013;25:2661–2678. doi: 10.1105/tpc.113.113118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleman J, Auer M. The role of bacterial outer membrane vesicles for intra- and interspecies delivery. Environmental microbiology. 2013;15:347–354. doi: 10.1111/1462-2920.12048. [DOI] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PA. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Molecular cell. 2005;18:555–564. doi: 10.1016/j.molcel.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge TJ. Structures of Gram-negative cell walls and their derived membrane vesicles. J Bacteriol. 1999;181:4725–4733. doi: 10.1128/jb.181.16.4725-4733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Lutkenhaus J. Ftsz ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Bonsor DA, Hecht O, Vankemmelbeke M, Sharma A, Krachler AM, Housden NG, Lilly KJ, James R, Moore GR, Kleanthous C. Allosteric beta-propeller signalling in TolB and its manipulation by translocating colicins. Embo J. 2009;28:2846–2857. doi: 10.1038/emboj.2009.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramkamp M. Structure and function of bacterial dynamin-like proteins. Biological chemistry. 2012;393:1203–1214. doi: 10.1515/hsz-2012-0185. [DOI] [PubMed] [Google Scholar]
- Broder DH, Pogliano K. Forespore engulfment mediated by a ratchet-like mechanism. Cell. 2006;126:917–928. doi: 10.1016/j.cell.2006.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EA, Hardwidge PR. Biochemical characterization of the enterotoxigenic Escherichia coli LeoA protein. Microbiol-Sgm. 2007;153:3776–3784. doi: 10.1099/mic.0.2007/009084-0. [DOI] [PubMed] [Google Scholar]
- Burdett IDJ, Murray RGE. Septum formation in Escherichia coli - characterization of septal structure and effects of antibiotics on cell-division. J Bacteriology. 1974;119:303–324. doi: 10.1128/jb.119.1.303-324.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürmann F, Ebert N, van Baarle S, Bramkamp M. A bacterial dynamin-like protein mediating nucleotide-independent membrane fusion. Mol Microbiol. 2011;79:1294–1304. doi: 10.1111/j.1365-2958.2011.07523.x. [DOI] [PubMed] [Google Scholar]
- Cascales E, Gavioli M, Sturgis JN, Lloubes R. Proton motive force drives the interaction of the inner membrane TolA and outer membrane Pal proteins in Escherichia coli. Mol Microbiol. 2000;38:904–915. doi: 10.1046/j.1365-2958.2000.02190.x. [DOI] [PubMed] [Google Scholar]
- Chandler DE, Hsin J, Harrison CB, Gumbart J, Schulten K. Intrinsic curvature properties of photosynthetic proteins in chromatophores. Biophysical Journal. 2008;95:2822–2836. doi: 10.1529/biophysj.108.132852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler DE, Gumbart J, Stack JD, Chipot C, Schulten K. Membrane curvature induced by aggregates of LH2s and monomeric LH1s. Biophysical Journal. 2009;97:2978–2984. doi: 10.1016/j.bpj.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaruttini N, Redondo-Morata L, Colom A, Humbert F, Lenz M, Scheuring S, Roux A. Relaxation of loaded ESCRT-III spiral springs drives membrane deformation. Cell. 2015;163:866–879. doi: 10.1016/j.cell.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colangeli R, Helb D, Sridharan S, Sun JC, Varma-Basil M, Hazbon MH, Harbacheuski R, Megjugorac NJ, Jacobs WR, Holzenburg A, Sacchettini JC, et al. The Mycobacterium tuberculosis iniA gene is essential for activity of an efflux pump that confers drug tolerance to both isoniazid and ethambutol. Mol Microbiol. 2005;55:1829–1840. doi: 10.1111/j.1365-2958.2005.04510.x. [DOI] [PubMed] [Google Scholar]
- Coltharp C, Buss J, Plumer TM, Xiao J. Defining the rate-limiting processes of bacterial cytokinesis. P Natl Acad Sci USA. 2016;113:E1044–E1053. doi: 10.1073/pnas.1514296113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornejo E, Subramanian P, Li Z, Jensen G, Komeili A. Dynamic remodeling of the magnetosome membrane is triggered by the initiation of biomineralization. mBio. 2016:e01898–01815. doi: 10.1128/mBio.01898-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan T, Coleman J, Marquis KA, Meeske AJ, Burton BM, Karatekin E, Rudner DZ. FisB mediates membrane fission during sporulation in Bacillus subtilis. Gene Dev. 2013;27:322–334. doi: 10.1101/gad.209049.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducret A, Fleuchot B, Bergam P, Mignot T. Direct live imaging of cell-cell protein transfer by transient outer membrane fusion in Myxococcus xanthus. eLife. 2013;2:e00868. doi: 10.7554/eLife.00868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan AJF, Vollmer W. The physiology of bacterial cell division. Ann Ny Acad Sci. 2013;1277:8–28. doi: 10.1111/j.1749-6632.2012.06818.x. [DOI] [PubMed] [Google Scholar]
- Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiology and molecular biology reviews : MMBR. 2010;74:81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP, Taylor DW, Taylor KA, Bramhill D. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. P Natl Acad Sci USA. 1996;93:519–523. doi: 10.1073/pnas.93.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faelber K, Posor Y, Gao S, Held M, Roske Y, Schulze D, Haucke V, Noe F, Daumke O. Crystal structure of nucleotide-free dynamin. Nature. 2011;477:556–560. doi: 10.1038/nature10369. [DOI] [PubMed] [Google Scholar]
- Feucht A, Lucet I, Yudkin MD, Errington J. Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol Microbiol. 2001;40:115–125. doi: 10.1046/j.1365-2958.2001.02356.x. [DOI] [PubMed] [Google Scholar]
- Fleckenstein JM, Lindler LE, Elsinghorst EA, Dale JB. Identification of a gene within a pathogenicity island of enterotoxigenic Escherichia coli H10407 required for maximal secretion of the heat-labile enterotoxin. Infect Immun. 2000;68:2766–2774. doi: 10.1128/iai.68.5.2766-2774.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford MGJ, Jenni S, Nunnari J. The crystal structure of dynamin. Nature. 2011;477:561–566. doi: 10.1038/nature10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frese RN, Pamies JC, Olsen JD, Bahatyrova S, van der Weij-de Wit CD, Aartsma TJ, Otto C, Hunter CN, Frenkel D, van Grondelle R. Protein shape and crowding drive domain formation and curvature in biological membranes. Biophysical Journal. 2008;94:640–647. doi: 10.1529/biophysj.107.116913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann E, Gathmann S, Rupprecht E, Golecki J, Schneider D. Thylakoid membrane reduction affects the photosystem stoichiometry in the cyanobacterium Synechocystis sp. PCC 6803. Plant physiology. 2009a;149:735–744. doi: 10.1104/pp.108.132373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann E, Bultema JB, Kahmann U, Rupprecht E, Boekema EJ, Schneider D. The vesicle-inducing protein 1 from Synechocystis sp. PCC 6803 organizes into diverse higher-ordered ring structures. Molecular biology of the cell. 2009b;20:4620–4628. doi: 10.1091/mbc.E09-04-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gankema H, Wensink J, Guinee PA, Jansen WH, Witholt B. Some characteristics of the outer membrane material released by growing enterotoxigenic Escherichia coli. Infect Immun. 1980;29:704–713. doi: 10.1128/iai.29.2.704-713.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Sage TL, Steryoung KW. FZL, an FZO-like protein in plants, is a determinant of thylakoid and chloroplast morphology. P Natl Acad Sci USA. 2006;103:6759–6764. doi: 10.1073/pnas.0507287103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerding MA, Ogata Y, Pecora ND, Niki H, de Boer PAJ. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol Microbiol. 2007;63:1008–1025. doi: 10.1111/j.1365-2958.2006.05571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germon P, Ray MC, Vianney A, Lazzaroni JC. Energy-dependent conformational change in the TolA protein of Escherichia coli involves its N-terminal domain, TolQ, and TolR. J Bacteriol. 2001;183:4110–4114. doi: 10.1128/JB.183.14.4110-4114.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh B, Sain A. Origin of contractile force during cell division of bacteria. Phys Rev Lett. 2008;101:178101–178104. doi: 10.1103/PhysRevLett.101.178101. [DOI] [PubMed] [Google Scholar]
- Ghosh B, Sain A. Force generation in bacteria without nucleotide-dependent bending of cytoskeletal filaments. Phys Rev E. 2011;83:051924. doi: 10.1103/PhysRevE.83.051924. [DOI] [PubMed] [Google Scholar]
- Goncalves RP, Bernadac A, Sturgis JN, Scheuring S. Architecture of the native photosynthetic apparatus of Phaeospirillum molischianum. Journal of structural biology. 2005;152:221–228. doi: 10.1016/j.jsb.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Gorby Y, McLean J, Korenevsky A, Rosso K, El-Naggar MY, Beveridge TJ. Redox-reactive membrane vesicles produced by Shewanella. Geobiology. 2008;6:232–241. doi: 10.1111/j.1472-4669.2008.00158.x. [DOI] [PubMed] [Google Scholar]
- Gray AN, Egan AJ, Van't Veer IL, Verheul J, Colavin A, Koumoutsi A, Biboy J, Altelaar AF, Damen MJ, Huang KC, Simorre JP, et al. Coordination of peptidoglycan synthesis and outer membrane constriction during Escherichia coli cell division. eLife. 2015;4:e07118. doi: 10.7554/eLife.07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover S, Alderwick LJ, Mishra AK, Krumbach K, Marienhagen J, Eggeling L, Bhatt A, Besra GS. Benzothiazinones mediate killing of Corynebacterineae by blocking decaprenyl phosphate recycling involved in cell wall biosynthesis. J Biol Chem. 2014;289:6177–6187. doi: 10.1074/jbc.M113.522623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez J, Smith R, Pogliano K. SpoIID-mediated peptidoglycan degradation is required throughout engulfment during Bacillus subtilis sporulation. J Bacteriol. 2010;192:3174–3186. doi: 10.1128/JB.00127-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CA, deBoer PAJ. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in Escherichia coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- Hankamer BD, Elderkin SL, Buck M, Nield J. Organization of the AAA(+) adaptor protein PspA is an oligomeric ring. J Biol Chem. 2004;279:8862–8866. doi: 10.1074/jbc.M307889200. [DOI] [PubMed] [Google Scholar]
- Hayashi J, Hamada N, Kuramitsu HK. The autolysin of Porphyromonas gingivalis is involved in outer membrane vesicle release. FEMS microbiology letters. 2002;216:217–222. doi: 10.1111/j.1574-6968.2002.tb11438.x. [DOI] [PubMed] [Google Scholar]
- Hennig R, Heidrich J, Saur M, Schmüser L, Roeters SJ, Hellmann N, Woutersen S, Bonn M, Weidner T, Markl J. IM30 triggers membrane fusion in cyanobacteria and chloroplasts. Nature communications. 2015;6 doi: 10.1038/ncomms8018. [DOI] [PubMed] [Google Scholar]
- Holden SJ, Pengo T, Meibom KL, Fernandez CF, Collier J, Manley S. High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. P Natl Acad Sci USA. 2014;111:4566–4571. doi: 10.1073/pnas.1313368111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horger I, Velasco E, Mingorance J, Rivas G, Tarazona P, Velez M. Langevin computer simulations of bacterial protein filaments and the force-generating mechanism during cell division. Phys Rev E. 2008;77:011902. doi: 10.1103/PhysRevE.77.011902. [DOI] [PubMed] [Google Scholar]
- Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nature reviews. Molecular cell biology. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Jin JS, Kwon SO, Moon DC, Gurung M, Lee JH, Kim SI, Lee JC. Acinetobacter baumannii secretes cytotoxic outer membrane protein A via outer membrane vesicles. PloS one. 2011;6:e17027. doi: 10.1371/journal.pone.0017027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd EM, Comolli LR, Chen JC, Downing KH, Moerner WE, McAdams HH. Distinct constrictive processes, separated in time and space, divide Caulobacter inner and outer membranes. J Bacteriol. 2005;187:6874–6882. doi: 10.1128/JB.187.20.6874-6882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadurugamuwa JL, Beveridge TJ. Natural release of virulence factors in membrane vesicles by Pseudomonas aeruginosa and the effect of aminoglycoside antibiotics on their release. The Journal of antimicrobial chemotherapy. 1997;40:615–621. doi: 10.1093/jac/40.5.615. [DOI] [PubMed] [Google Scholar]
- Kleanthous C. Swimming against the tide: progress and challenges in our understanding of colicin translocation. Nat Rev Microbiol. 2010;8:843–848. doi: 10.1038/nrmicro2454. [DOI] [PubMed] [Google Scholar]
- Kobayashi R, Suzuki T, Yoshida M. Escherichia coli phage-shock protein A (PspA) binds to membrane phospholipids and repairs proton leakage of the damaged membranes. Mol Microbiol. 2007;66:100–109. doi: 10.1111/j.1365-2958.2007.05893.x. [DOI] [PubMed] [Google Scholar]
- Komeili A. Molecular mechanisms of compartmentalization and biomineralization in magnetotactic bacteria. FEMS microbiology reviews. 2012;36:232–255. doi: 10.1111/j.1574-6976.2011.00315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A, Li Z, Newman DK, Jensen GJ. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311:242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- Kroll D, Meierhoff K, Bechtold N, Kinoshita M, Westphal S, Vothknecht UC, Soll J, Westhoff P. VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc Natl Acad Sci USA. 2001;98:4238–4242. doi: 10.1073/pnas.061500998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudryashev M, Cyrklaff M, Alex B, Lemgruber L, Baumeister W, Wallich R, Frischknecht F. Evidence of direct cell-cell fusion in Borrelia by cryogenic electron tomography. Cell Microbiol. 2011;13:731–741. doi: 10.1111/j.1462-5822.2011.01571.x. [DOI] [PubMed] [Google Scholar]
- Kulkarni HM, Jagannadham MV. Biogenesis and multifaceted roles of outer membrane vesicles from Gram-negative bacteria. Microbiology. 2014;160:2109–2121. doi: 10.1099/mic.0.079400-0. [DOI] [PubMed] [Google Scholar]
- Lan GH, Daniels BR, Dobrowsky TM, Wirtz D, Sun SX. Condensation of FtsZ filaments can drive bacterial cell division. P Natl Acad Sci USA. 2009;106:121–126. doi: 10.1073/pnas.0807963106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver M, Dominguez-Cuevas P, Coxhead JM, Daniel RA, Errington J. Life without a wall or division machine in Bacillus subtilis. Nature. 2009;457:849–853. doi: 10.1038/nature07742. [DOI] [PubMed] [Google Scholar]
- Lee J, Lee EY, Kim SH, Kim DK, Park KS, Kim KP, Kim YK, Roh TY, Gho YS. Staphylococcus aureus extracellular vesicles carry biologically active beta-lactamase. Antimicrobial agents and chemotherapy. 2013;57:2589–2595. doi: 10.1128/AAC.00522-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hsin J, Zhao LY, Cheng YW, Shang WN, Huang KC, Wang HW, Ye S. FtsZ protofilaments use a hinge-opening mechanism for constrictive force generation. Science. 2013;341:392–395. doi: 10.1126/science.1239248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Trimble MJ, Brun YV, Jensen GJ. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. Embo J. 2007;26:4694–4708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindas AC, Karlsson EA, Lindgren MT, Ettema TJG, Bernander R. A unique cell division machinery in the Archaea. P Natl Acad Sci USA. 2008;105:18942–18946. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LN, Aartsma TJ, Frese RN. Dimers of light-harvesting complex 2 from Rhodobacter sphaeroides characterized in reconstituted 2D crystals with atomic force microscopy. The FEBS Journal. 2008;275:3157–3166. doi: 10.1111/j.1742-4658.2008.06469.x. [DOI] [PubMed] [Google Scholar]
- Lo SM, Theg SM. Role of vesicle-inducing protein in plastids 1 in cpTat transport at the thylakoid. The Plant journal : for cell and molecular biology. 2012;71:656–668. doi: 10.1111/j.1365-313X.2012.05020.x. [DOI] [PubMed] [Google Scholar]
- Loose M, Mitchison TJ. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nature cell biology. 2014;16:38–46. doi: 10.1038/ncb2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low HH, Löwe J. A bacterial dynamin-like protein. Nature. 2006;444:766–769. doi: 10.1038/nature05312. [DOI] [PubMed] [Google Scholar]
- Low HH, Löwe J. Dynamin architecture - from monomer to polymer. Curr Opin Struc Biol. 2010;20:791–798. doi: 10.1016/j.sbi.2010.09.011. [DOI] [PubMed] [Google Scholar]
- Low HH, Sachse C, Amos LA, Löwe J. Structure of a bacterial dynamin-like protein lipid tube provides a mechanism for assembly and membrane curving. Cell. 2009;139:1342–1352. doi: 10.1016/j.cell.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löwe J, Amos LA. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- Lu CL, Reedy M, Erickson HP. Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J Bacteriol. 2000;182:164–170. doi: 10.1128/jb.182.1.164-170.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J, Pichoff S, Du SS. Bacterial cytokinesis: From Z ring to divisome. Cytoskeleton. 2012;69:778–790. doi: 10.1002/cm.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald C, Jovanovic G, Ces O, Buck M. Membrane stored curvature elastic stress modulates recruitment of maintenance proteins PspA and Vipp1. mBio. 2015;6:e01188–01115. doi: 10.1128/mBio.01188-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- Mempin R, Tran H, Chen CN, Gong H, Ho KK, Lu SW. Release of extracellular ATP by bacteria during growth. Bmc Microbiol. 2013;13 doi: 10.1186/1471-2180-13-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P, Gutierrez J, Pogliano K, Dworkin J. Cell wall synthesis is necessary for membrane dynamics during sporulation of Bacillus subtilis. Mol Microbiol. 2010;76:956–970. doi: 10.1111/j.1365-2958.2010.07155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michie KA, Boysen A, Low HH, Moller-Jensen J, Löwe J. LeoA, B and C from enterotoxigenic Escherichia coli (ETEC) are bacterial dynamins. PloS one. 2014;9:e107211. doi: 10.1371/journal.pone.0107211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mim C, Unger VM. Membrane curvature and its generation by BAR proteins. Trends in biochemical sciences. 2012;37:526–533. doi: 10.1016/j.tibs.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlot C, Uehara T, Marquis KA, Bernhardt TG, Rudner DZ. A highly coordinated cell wall degradation machine governs spore morphogenesis in Bacillus subtilis. Gene Dev. 2010;24:411–422. doi: 10.1101/gad.1878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat D, Quinlan A, Vali H, Komeili A. Comprehensive genetic dissection of the magnetosome gene island reveals the step-wise assembly of a prokaryotic organelle. Proc Natl Acad Sci U S A. 2010;107:5593–5598. doi: 10.1073/pnas.0914439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordhues A, Schottler MA, Unger AK, Geimer S, Schonfelder S, Schmollinger S, Rutgers M, Finazzi G, Soppa B, Sommer F, Muhlhaus T, et al. Evidence for a role of VIPP1 in the structural organization of the photosynthetic apparatus in Chlamydomonas. The Plant cell. 2012;24:637–659. doi: 10.1105/tpc.111.092692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojkic N, Lopez-Garrido J, Pogliano K, Endres RG. Bistable forespore engulfment in Bacillus subtilis by a zipper mechanism in absence of the cell wall. PloS Comput Biol. 2014;10:e1003912. doi: 10.1371/journal.pcbi.1003912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JD, Tucker JD, Timney JA, Qian P, Vassilev C, Hunter CN. The organization of LH2 complexes in membranes from Rhodobacter sphaeroides. J Biol Chem. 2008;283:30772–30779. doi: 10.1074/jbc.M804824200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Erickson HP. FtsZ from divergent foreign bacteria can function for cell division in Escherichia coli. J Bacteriol. 2006;188:7132–7140. doi: 10.1128/JB.00647-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Erickson HP. Inside-out Z rings - constriction with and without GTP hydrolysis. Mol Microbiol. 2011;81:571–579. doi: 10.1111/j.1365-2958.2011.07716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Erickson HP. Liposome division by a simple bacterial division machinery. P Natl Acad Sci USA. 2013;110:11000–11004. doi: 10.1073/pnas.1222254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Anderson DE, Erickson HP. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320:792–794. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otters S, Braun P, Hubner J, Wanner G, Vothknecht UC, Chigri F. The first alpha-helical domain of the vesicle-inducing protein in plastids 1 promotes oligomerization and lipid binding. Planta. 2013;237:529–540. doi: 10.1007/s00425-012-1772-1. [DOI] [PubMed] [Google Scholar]
- Ozaki S, Matsuda Y, Keyamura K, Kawakami H, Noguchi Y, Kasho K, Nagata K, Masuda T, Sakiyama Y, Katayama T. A replicase clamp-binding dynamin-like protein promotes colocalization of nascent DNA strands and equipartitioning of chromosomes in E. coli. Cell Rep. 2013;4:985–995. doi: 10.1016/j.celrep.2013.07.040. [DOI] [PubMed] [Google Scholar]
- Palsdottir H, Remis JP, Schaudinn C, O'Toole E, Lux R, Shi WY, McDonald KL, Costerton JW, Auer M. Three-dimensional macromolecular organization of cryofixed Myxococcus xanthus biofilms as revealed by electron microscopic tomography. J Bacteriol. 2009;191:2077–2082. doi: 10.1128/JB.01333-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande S, Shitut S, Freund L, Westermann M, Bertels F, Colesie C, Bischofs IB, Kost C. Metabolic cross-feeding via intercellular nanotubes among bacteria. Nature Communications. 2015;6 doi: 10.1038/ncomms7238. [DOI] [PubMed] [Google Scholar]
- Pathak DT, Wei X, Bucuvalas A, Haft DH, Gerloff DL, Wall D. Cell contact-dependent outer membrane exchange in myxobacteria: genetic determinants and mechanism. PLoS genetics. 2012;8:e1002626. doi: 10.1371/journal.pgen.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Cruz C, Delgado L, Lopez-Iglesias C, Mercade E. Outer-inner membrane vesicles naturally secreted by gram-negative pathogenic bacteria. PloS one. 2015;10:e0116896. doi: 10.1371/journal.pone.0116896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. Embo J. 2002;21:685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirbadian S, Barchinger SE, Leung KM, Byun HS, Jangir Y, Bouhenni RA, Reed SB, Romine MF, Saffarini DA, Shi L, Gorby YA, et al. Shewanella oneidensis MR-1 nanowires are outer membrane and periplasmic extensions of the extracellular electron transport components. P Natl Acad Sci USA. 2014;111:12883–12888. doi: 10.1073/pnas.1410551111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke GJ, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nature reviews. Molecular cell biology. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- Qian P, Bullough PA, Hunter CN. Three-dimensional reconstruction of a membrane-bending complex: the RC-LH1-PufX core dimer of Rhodobacter sphaeroides. J Biol Chem. 2008;283:14002–14011. doi: 10.1074/jbc.M800625200. [DOI] [PubMed] [Google Scholar]
- Raskin DM, de Boer PA. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remis JP, Wei D, Gorur A, Zemla M, Haraga J, Allen S, Witkowska HE, Costerton JW, Berleman JE, Auer M. Bacterial social networks: structure and composition of Myxococcus xanthus outer membrane vesicle chains. Environmental microbiology. 2014;16:598–610. doi: 10.1111/1462-2920.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico AI, Krupka M, Vicente M. In the beginning, Escherichia coli assembled the proto-ring: an initial phase of division. J Biol Chem. 2013;288:20830–20836. doi: 10.1074/jbc.R113.479519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roier S, Zingl FG, Cakar F, Durakovic S, Kohl P, Eichmann TO, Klug L, Gadermaier B, Weinzerl K, Prassl R, Lass A, et al. A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nature communications. 2016;7:10515. doi: 10.1038/ncomms10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in archaea. Science. 2008;322:1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawant P, Eissenberger K, Karier L, Mascher T, Bramkamp M. A dynamin-like protein involved in bacterial cell membrane surveillance under environmental stress. Environmental microbiology. 2015 doi: 10.1111/1462-2920.13110. [DOI] [PubMed] [Google Scholar]
- Schertzer JW, Whiteley M. A bilayer-couple model of bacterial outer membrane vesicle biogenesis. mBio. 2012;3:e00297. doi: 10.1128/mBio.00297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuring S, Nevo R, Liu LN, Mangenot S, Charuvi D, Boudier T, Prima V, Hubert P, Sturgis JN, Reich Z. The architecture of Rhodobacter sphaeroides chromatophores. Biochim Biophys Acta. 2014;1837:1263–1270. doi: 10.1016/j.bbabio.2014.03.011. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Kuehn MJ. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol. 2015;13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty A, Chen S, Tocheva EI, Jensen GJ, Hickey WJ. Nanopods: a new bacterial structure and mechanism for deployment of outer membrane vesicles. PloS one. 2011;6:e20725. doi: 10.1371/journal.pone.0020725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert CA, Qian P, Fotiadis D, Engel A, Hunter CN, Bullough PA. Molecular architecture of photosynthetic membranes in Rhodobacter sphaeroides: the role of PufX. Embo J. 2004;23:690–700. doi: 10.1038/sj.emboj.7600092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak JC, Schmid EM, Ryan CJ, Ann HS, Sasaki DY, Sherman MB, Geissler PL, Fletcher DA, Hayden CC. Membrane bending by protein-protein crowding. Nature cell biology. 2012;14:944–949. doi: 10.1038/ncb2561. [DOI] [PubMed] [Google Scholar]
- Sturgis JN. Organisation and evolution of the Tol-Pal gene cluster. J Mol Microb Biotech. 2001;3:113–122. [PubMed] [Google Scholar]
- Szwedziak P, Wang Q, Freund SMV, Löwe J. FtsA forms actin-like protofilaments. Embo J. 2012;31:2249–2260. doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwedziak P, Wang Q, Bharat TAM, Tsim M, Löwe J. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. eLife. 2014;3:e04601. doi: 10.7554/eLife.04601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Arakaki A, Matsunaga T. Identification and functional characterization of liposome tubulation protein from magnetotactic bacteria. Mol Microbiol. 2010;76:480–488. doi: 10.1111/j.1365-2958.2010.07117.x. [DOI] [PubMed] [Google Scholar]
- Tashiro Y, Inagaki A, Shimizu M, Ichikawa S, Takaya N, Nakajima-Kambe T, Uchiyama H, Nomura N. Characterization of phospholipids in membrane vesicles derived from Pseudomonas aeruginosa. Bioscience, biotechnology, and biochemistry. 2011;75:605–607. doi: 10.1271/bbb.100754. [DOI] [PubMed] [Google Scholar]
- Tocheva EI, Lopez-Garrido J, Hughes HV, Fredlund J, Kuru E, VanNieuwenhze MS, Brun YV, Pogliano K, Jensen GJ. Peptidoglycan transformations during Bacillus subtilis sporulation. Mol Microbiol. 2013;88:673–686. doi: 10.1111/mmi.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusca D, Scott S, Thompson C, Bramhill D. Bacterial SOS checkpoint protein SulA inhibits polymerization of purified FtsZ cell division protein. J Bacteriol. 1998;180:3946–3953. doi: 10.1128/jb.180.15.3946-3953.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker JD, Siebert CA, Escalante M, Adams PG, Olsen JD, Otto C, Stokes DL, Hunter CN. Membrane invagination in Rhodobacter sphaeroides is initiated at curved regions of the cytoplasmic membrane, then forms both budded and fully detached spherical vesicles. Mol Microbiol. 2010;76:833–847. doi: 10.1111/j.1365-2958.2010.07153.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Lutkenhaus J. Characterization of the ftsZ gene from Mycoplasma pulmonis, an organism lacking a cell wall. J Bacteriol. 1996;178:2314–2319. doi: 10.1128/jb.178.8.2314-2319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XD, Huang JA, Mukherjee A, Cao C, Lutkenhaus J. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J Bacteriol. 1997;179:5551–5559. doi: 10.1128/jb.179.17.5551-5559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner G, Vogl K, Overmann J. Ultrastructural characterization of the prokaryotic symbiosis in "Chlorochromatium aggregatum". J Bacteriol. 2008;190:3721–3730. doi: 10.1128/JB.00027-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal S, Heins L, Soll J, Vothknecht UC. Vipp1 deletion mutant of Synechocystis: a connection between bacterial phage shock and thylakoid biogenesis? Proc Natl Acad Sci USA. 2001;98:4243–4248. doi: 10.1073/pnas.061501198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldringh CL, Mulder E, Huls PG, Vischer N. Toporegulation of bacterial division according to the nucleoid occlusion model. Res Microbiol. 1991;142:309–320. doi: 10.1016/0923-2508(91)90046-d. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell. 2004;117:915–925. doi: 10.1016/j.cell.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Zhang SY, Shen GZ, Li ZK, Golbeck JH, Bryant DA. Vipp1 is essential for the biogenesis of Photosystem I but not thylakoid membranes in Synechococcus sp PCC 7002. J Biol Chem. 2014;289:15904–15914. doi: 10.1074/jbc.M114.555631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Srisatjaluk R, Justus DE, Doyle RJ. On the origin of membrane vesicles in gram-negative bacteria. FEMS microbiology letters. 1998;163:223–228. doi: 10.1111/j.1574-6968.1998.tb13049.x. [DOI] [PubMed] [Google Scholar]