Abstract
Background:
There is a lack of information on CYP2D6, a major metabolizing enzyme, in Africa ethnic nationalities. The objective was to determine CYP2D6 phenotype in Yoruba Nigerians using dextromethorphan (DEX).
Method:
A total of 89 healthy volunteers received 30 mg of DEX orally followed by blood and urine sample collection at 3-hour and over 8 hours post-dose, respectively. DEX and dextrorphan (DOR) concentrations were determined using High Performance Liquid Chromatography (HPLC). The metabolic ratio (MR, DEX/DOR) were plotted for the phenotype determination.
Results:
The log MR that separated poor (PMs) from normal metabolizers (NMs) was 0.28 and 0.75 for urine and plasma, respectively. Two subjects (2.3%) identified as PMs had a mean MR of 17 and 3.2 in plasma and urine, significantly higher than that of NMs (p<0.0001). A positive correlation between urine and plasma MR was noted.
Conclusion:
The prevalence of PMs in the Yoruba Nigerians was similar to that reported among blacks.
Keywords: CYP2D6, dextromethorphan, metabolic ratio, phenotyping, Yoruba Nigerian
1. Introduction
CYP2D6 is responsible for the biotransformation of about 25% of clinically used drugs [1] including antidepressants (e.g. amitriptyline), antiarrhythmics (e.g. flecainide), antipsychotics (e.g. risperidone), β-adrenergic blockers (e.g. metoprolol) and miscellaneous drugs including codeine, tramadol, and tamoxifen[2]. CYP2D6 gene is highly polymorphic, and to date, more than 140 alleles have been discovered (www.cypalleles.ki.se/CYP2D6.htm). The phenotypic expression of these alleles have been grouped as: poor metabolizers (PMs), intermediate metabolizers (IMs), normal metabolizers (NMs), rapid metabolizers (RMs) and ultrarapid metabolizers (UMs) using such probe drugs as debrisoquine, dextromethorphan, sparteine, tramadol and metoprolol [3–5]. Of these probe drugs, dextromethorphan (DEX) appears favoured as it is relatively accessible and well tolerated when compared to the others [6–8]. Liquid chromatography tandem-mass spectrometry (LC-MS/MS) is the most preferred for the analysis of dextromethorphan and its metabolites, but in its absence high performance liquid chromatography (HPLC) with florescence detector is better than ultraviolet (UV) detector.
CYP2D6 polymorphisms may result in up to a 30–40 fold difference in substrate clearance resulting in plasma concentrations that are outside the therapeutic range in patients [9, 10]. The clinical consequence of this may include concentration-related drug toxicity and or therapeutic failure [11, 12]. Therefore, adverse drug reactions resulting from poor drug handling due to enzyme polymorphisms should be preventable if appropriate dosing could be achieved. In this vein, therapeutic failure, and even economic-related losses may also be reduced. It is noteworthy that substantial drug-related morbidity and mortality are recorded yearly. For example, fatal adverse drug reactions were estimated as the 7th leading cause of death in Sweden in 2001 [13].
The prevalence of CYP2D6 phenotypes varies among different populations and races: studies showed 7– 10% PMs among Caucasians [14, 15], 1–2% PMs in the Asian populations [16, 17], 6.9% PMs in the Cuban populations [18], 3.1% PMs in the Ecuadorian populations[19], and 1–4% PMs population in Africa [20–23]. In Nigeria, few available studies among the many ethnic populations indicate a prevalence of PMs of 0–3.5% [24, 25].
The 2006 national census revealed that Nigeria has over 154 million people belonging to major and minor ethnic nationalities [26]. Of this, the Yoruba, a major ethnic nationality, had a population of over 32 million making up over one fifth of the Nigerian population. (www.mapsofworld.com/Nigerian) The Yoruba Nigerians are indigenous to the southwestern Nigeria. This present study, therefore, was aimed at evaluating single 3-hour plasma sample in the determination of CYP2D6 phenotype among the Yoruba Nigerians in order to identify poor metabolizers. Furthermore, the utility of urine samples for phenotype evaluation was explored by determining the metabolic ratio of dextromethorphan in urine compared to that calculated from the plasma samples.
2. Methods
2.1. Setting
Although Nigeria has over 350 ethnic nationalities, the three major ethnic groups; namely Hausa-Fulani, Yoruba, and Ibo constitute over 60% of the entire population. Yorubas are native to the Southwestern and some part of the Northcentral Nigeria, and they constitute about 21% of the total population of Nigeria. The study was conducted at the University College Hospital, Ibadan, Nigeria, a foremost tertiary health facility in the country. Ibadan, is the heartland and political headquarters of the Yoruba ethnic group in Nigeria.
2.2. Study Population and Subject Selection
Subjects’ recruitment was through informal contacts and invitation through social functions and community meetings within and around Ibadan, Oyo State, Nigeria. The primary eligibility criterion was that the participant was, at least, a third generation Yoruba Nigerian with maternal and paternal parents and grandparents being of Yoruba extraction; in good health as confirmed by medical history, physical examination and laboratory investigations. The participants were unrelated. Other inclusion criteria were age between 18 and 50 years, informed consent of the participants, non-smoking, not on any medication including herbal preparations. Exclusion criteria were the chronic use of alcohol or any other recreational drugs, use of oral contraceptives, pregnancy, lactation, known/history suggestive of allergy to dextromethorphan and related drugs.
Eligible participants were interviewed on their medical history; physically examined (including blood pressure, height and weight measurement); blood (5mL) and urine samples were collected for laboratory investigations such as complete blood count, liver and renal function tests, and urinalysis, to further ascertain fitness for the study. The participants were enrolled in batches of 10 or 12 on each of the recruiting days, between 2 September and 30 September 2014 taking full cognizance of their convenience.
The study received ethical approval from the Joint University of Ibadan/University College Hospital (UI/UCH) Ethical Review Committee (UI/EC/12/0186), and was conducted in accordance with the principle of Helsinki declaration. Each participant, prior to the commencement of the study and following a detailed explanation of nature signed an informed consent. Due to the relatively small sample size, the primary objective was modified to identify poor metabolizers.
2.3. Drug administration and sample collection
Following an overnight fast, participants were requested to completely empty their bladder after which 30 mg dextromethorphan hydrobromide, in syrup form, was given orally. Three hours post dextromethorphan administration, 5 mL of blood was collected, immediately centrifuged and the plasma transferred to separate plain plasma tubes. Also urine samples were collected over the first 8-hour period into a container having 2 g of ascorbic acid for acidification. The total urine voided by each participant was recorded before storing 15 mL aliquots in a container with 20 mg ascorbic acid. The plasma and urine samples which were initially stored at −20°C and was later transferred to a −80°C ultralow freezer until analysis. During the nine months’ period of storage, daily charting of the temperature of the freezer was done morning and evening. In addition to the generator, an inverter was in place as an immediate backup in the case of power failure. The samples were shipped in liquid nitrogen from the University College Hospital, Ibadan to the Central Science Laboratory (CSL) at the Obafemi Awolowo University (OAU), Ile-Ife, Nigeria where the analyses were done.
2.4. Drug Analysis
Dextromethorphan and dextrorphan concentrations in plasma and urine were analysed using a previously reported methods by Zimova et al [27] and Jurica et al [28] with modifications. Dextromethorphan hydrobromide syrup (TussipectR), (Belgium) 1.5mg/mL was supplied, in part, free by Benjamin Michael Pharmacy, Lagos, Nigeria. The following reference standards, all of HPLC grade, were used including dextromethorphan (drug), dextrorphan (metabolite) and levallorphan (internal standard), β-glucuronidase (Helix Pomata), acetonitrile and methanol, all from the Sigma Aldrich, Steinheim, Germany. Analytical grade of anhydrous sodium carbonate, anhydrous sodium acetate, orthophosphoric acid, potassium dihydrogen orthophosphate (KH2P04), hydrochloric acid and ascorbic acid, all from the British Drug Houses (BDH) Chemical Limited, England. Other chemicals used included the analytical grade of hexane and n-butanol (Scharlau SL Gato Pèrez, Barcelona, Spain) and triethylamine (Burgoyne Burbibges and Co, Mumbai, India). Briefly, the method involved addition of 0.4 mL sodium acetate (0.2 mol per liter) and 4000 units/sample β-glucuronidase to 1 mL of urine and incubated in water bath at 37 °C for 18 hours to enzymatically hydrolyse the sample. The mixture (1 mL) was pipetted into 10-mL screw-capped, tapered glass tubes followed by addition of 20μg/ml of internal standard and 4 mL Na2CO3 (0.5 mol/litre). Samples were extracted with 4 mL of hexane: n-butanol, 9:l(v/v) mixture. After vortexing and centrifuging, the organic layer was separated and kept, but the aqueous phase was reextracted. Both organic phases were subjected to re-extraction into 0.3 mL of 0.1M hydrochloric acid (HCL) (0.1M), and 50 μL of the aqueous phase was injected into the HPLC. Plasma and urine samples were subjected to the same protocol. The high performance liquid chromatographic system used consisted of an Agilent 1260 series HPLC system (Agilent Technologies, Germany) fitted with 1260 quaternary pump VL (model G1311C), an online vacuum degasser (Agilent, Japan), a manual injector valve ((Rheodyne, USA) with a 20 μl sample loop, and sample detector with a variable wavelength (VWD) (190–600nm) standard version (model G1311B). Data acquisition was enabled by an LC3D Chemstation software and window 2007 for system control. Chromatographic separation was achieved with an Eclipse plus C-18 reverse phase HPLC column, (3.5 μm particle size and 100 × 4.6 mm, i.d.), Agilent United States of America (USA). The mobile phase consisted of 30% Acetonitrile: 20% Methanol: 0.06% Triethylamine: 49.94% KH2PO4 (0.01 mol/litre), adjusted to pH of 3.2 by orthophosphoric acid (0.1 mol/litre). The flow rate was 1.5ml/min at the ambient temperature. The wavelength of the Ultraviolet-Visible Spectrophotometry (UV-VIS) detector used was 230nm.
Dextrorphan, levallorphan, and dextromethorphan were distinctly resolved eluting with retention time of 1.3, 1.7 and 3.5 minutes, respectively. The Intra-assay and Inter-assay coefficients of variation for both dextromethorphan and dextrorphan were less than 6% at concentrations of 0.5 and 2μg/mL (n=6). The limit of detection was 0.1 μg/mL while the limits of quantitation was 0.2 μg/mL for both dextromethorphan and dextrorphan in the urine and plasma.
2.5. Data Analysis
The statistical analysis was performed using International Business Machines Corporation-Statistical Package for the Social Sciences (IBM SPSS) Statistics for Windows, version 22 and Microsoft Excel 2013. The socio-demographic variables were summarized using descriptive statistics (frequency and percentage). The age, height, weight and Body Mass Index (BMI) were computed with mean ± standard deviation. The molar concentrations of dextromethorphan and dextrorphan in the plasma and urine samples were measured and summarized as mean, and standard deviation. The metabolic ratio of dextromethorphan/dextrorphan (MRDEX/DOR) was calculated by dividing the molar concentration of dextromethorphan (μg/mL) by the molar concentration of dextrorphan (μg/mL). The log MRDEX/DOR at 3-hour for plasma and 8-hour for urine were calculated and used as the index of CYP2D6 activity. A probit plot of log MR was constructed for plasma and urine, and the anti-mode (cut-off points) that separated the phenotypes (poor and normal metabolizers) were obtained from the graph. The probit plot was constructed with Microsoft Excel 2013, with log MR on the X-axis and probability % on Y-axis on the semi-logarithm graph. The probability % was calculated for each sample (plasma and urine) using the equation: Probability %, P = 100(i-0.5)/T [29], where i = rank of the participant log MR (from 1–89) and T = total number of participants (i.e. 89). Trend lines were added to the probit plot to get the best line of fit. Based on the best line of fit, a polynomial equation of regression was obtained, and intercept at X-axis was considered as the log MR separating poor metabolizers (PMs) from normal metabolizers (NMs). Pearson’s correlation was used to compare the quantitative variables (plasma and urinary metabolic ratios, age, body mass index) while the independent t-test was used to compare mean values in two groups such as male and female. The p-value was set at <0.05 for statistical significance.
3. Results
One hundred and four (110) consented to participate in the study but 11 individuals could not be enrolled for (apparent) inadequate renal function and/or ingestion of medicines or herbal remedies. Of the 99 participants, four samples could either not be found or urine collection discovered to be incomplete, and six samples were removed because the concentrations obtained for dextromethorphan and or dextrorphan were lower than than the limit of quantification (0.2 μg/mL). Of the 89 participants who completed the study per protocol with all relevant samples being available, 58(65.2%) were males, and 31(34.8%) were females. The mean age of the participants was 36.1±9.5 years. The mean weight, height and Body Mass Index (BMI) were 64±13.4 kg, 1.69±0.07 m and 22.4±4.2 kg/m2, respectively. One participant reported mild drowsiness one-hour post-dose which resolved spontaneously without any intervention.
The mean concentrations of the dextromethorphan and dextrorphan in the 8-hour urine were 0.75±0.54μg/ml and 1.01±0.51 μg/mL. respectively. The plasma and urine concentrations of dextromethorphan and dextrorphan are summarized in Table 1.
Table 1:
Plasma and urine concentrations of dextromethorphan and dextrorphan in the participants
| Variable(ug/ml) | Mean±SD | Median(range) |
|---|---|---|
| 8-hour urine dextromethorphan | 0.75±0.54 | 0.74(0.2–2.56) |
| 8- hour urine dextrorphan | 1.01±0.51 | 0.95(0.36–3.90) |
| 3-hour plasma dextromethorphan | 3.14±1.78 | 2.5(0.51–9.00) |
| 3-hour plasma Dextrorphan | 1.33±0.79 | 1.03(0.2–4.22) |
The median (range) metabolic ratio and log MR in the 8-hour urine were 0.74 (0.001–4.2) and −0.13 (−2.9–0.6), respectively. There was no statistically significant gender difference in the mean MR urine (t=1.8, p=0.072). Figure 1 provides a histogram of the frequency distribution of the metabolic ratio in the 8-hour urine. As shown in Figure 2, the cut-off (log MR) and the antimode for 8-hour urine separating the poor metabolizers (PMs) from extensive metabolizers(EMs) were 0.28 and 1.91 respectively. Two (2.3%) participants were classified as poor metabolizers (PMs).
Figure 1.
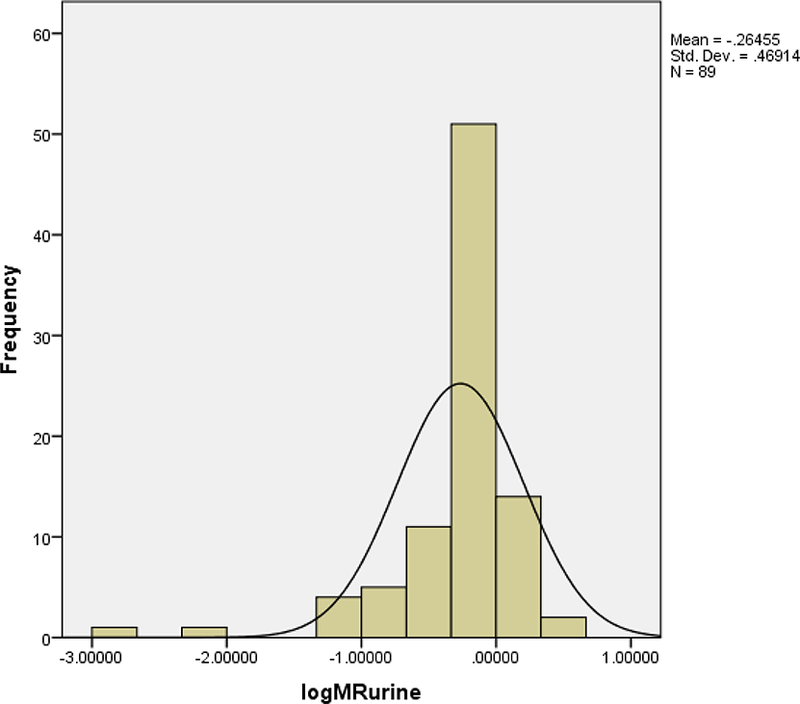
Histogram of the frequency distribution of the log MR in the 8-hour urine.
Figure 2:
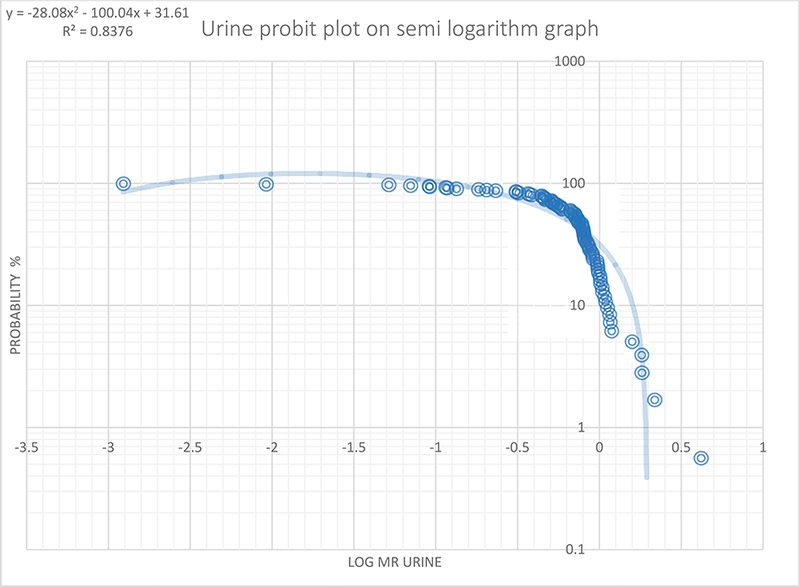
Probit plot of metabolic ratio (n=89) in 8-hour urine samples
The probit plotted using the semi-log graph of probability percent (probability %) against the log MR and the trend line best fit plotted. The log MR (0.28) when the trend line crosses the x-axis was chosen as the anti-mode (1.91) by converting back to MR (i.e. the antilog of log MR) separating the poor metabolizers from the normal metabolizers.
The mean concentration of dextromethorphan in the 3-hour plasma was 3.4±1.8 μg/mL while that of the dextrorphan was 1.33±0.79 μg/mL. The median (range) of the metabolic ratio in the 3-hour plasma was 2.36 (1.4–26.5) while the median log MR was 0.37(0.1–1.4). The MR plasma was not significantly different between genders (p=0.072). Figure 3 provides a histogram of the frequency distribution of the log MR of the 3-hour plasma samples. As shown in probit plot in Figure 4, the log MR that separated the normal metabolizers (NMs) from poor metabolizers (PMs) was 0.75, and the anti-mode was 5.6. Two (2.3%) participants were classified as poor metabolizers (PMs).
Figure 3.
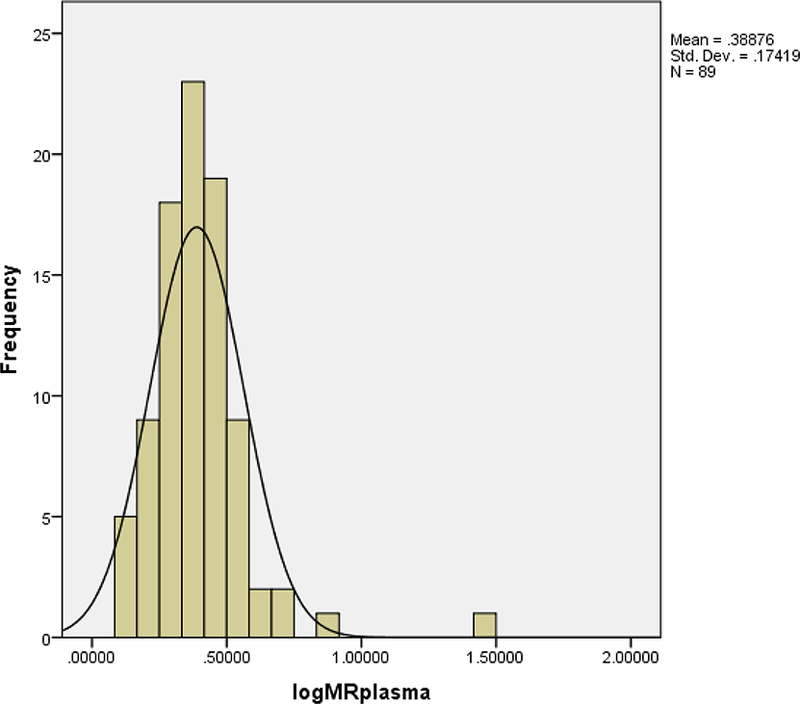
Histogram of the frequency distribution of the log MR in 3-hour plasma.
Figure 4.
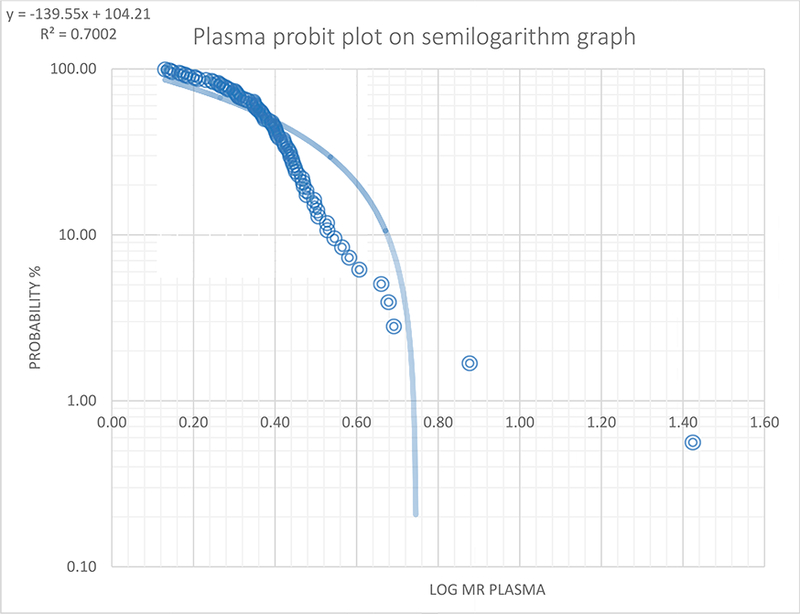
Probit plot of metabolic ratio (n=89) in 3-hour plasma samples
The probit plotted using the semi-log graph of probability percent (probability %) against the plasma log MR and the trend line best fit plotted. The log MR (0.75) corresponding to MR of 5.6 when the trend line crosses the x-axis was chosen as the cut-off separating the poor metabolizers from the normal metabolizers.
Both the (8-hour) urine and the plasma sample obtained 3 hours post-dose identified the two participants (2.3%) as poor metabolizer (PM) phenotype. The two PMs identified were male aged 25 and 27years, respectively. The plasma MRs of the PMs were 7.5 and 26.5, while their urine MRs were 2.2 and 4.2. The NMs has the median (range) plasma and urine MRs of 2.4(1.4–4.9) and 0.7 (0.01–1.8) respectively. There was a strong positive correlation between 8-hour urinary and 3-hour plasma metabolic ratios [r=0.77, p<0.0001, CI (0.22, 0.89)] but no significant correlation between age, body mass index and metabolic ratios as shown in Table 2. Figure 5 shows the scatter plot displaying the linear relationship between the 3- hour plasma and 8-hour urine metabolic ratios.
Table 2:
Correlation of the 8-hour urine MR, 3-hour plasma MRs, age and body mass index
| Variable | Age(years) | BMI(kg/m2) | Urine MR | Plasma MR |
|---|---|---|---|---|
| Urine MR r(p-value) | 0.14(0.188) | −0.09(0.43) | 1 | 0.77(<0.0001)* |
| Plasma MR r(p-value) | 0.09(0.359) | −0.04(0.622) | 0.77(<0.0001)* | 1 |
Statistical significant
Figure 5:
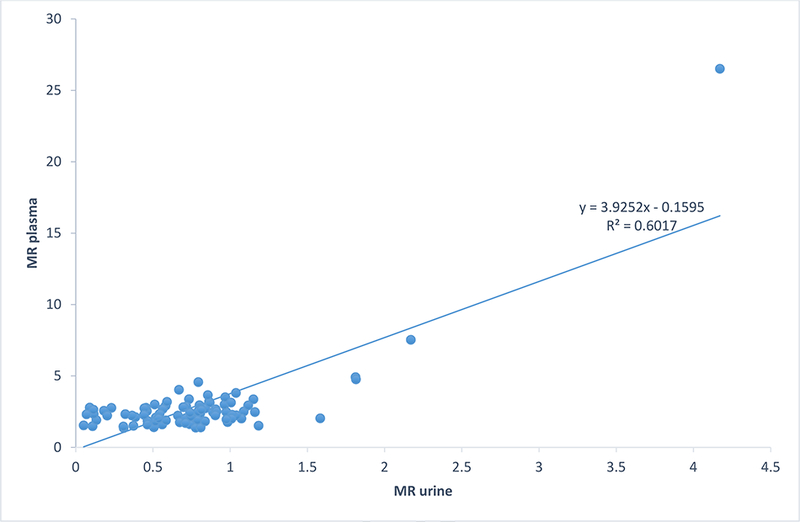
Scatter plot showing the correlation between the Metabolic Ratios(MR) of the 3-hour plasma and 8-hour urine
4. Discussion
A single 3-hour plasma sample was employed in the determination of CYP2D6 among 89 unrelated healthy volunteers of Yoruba ethnic origin in Nigeria. The few existing studies on CYP2D6 phenotyping of specific ethnic nationalities in Nigeria used urine matrix hence the uniqueness of this study [24]. Interestingly, the two (2.3%) PMs identified using 3-hour plasma sample were confirmed by the urine sample, and similar to the documented prevalence of PMs in our area of study. The specific objectives of the study did not include determination of other possible phenotypes such as ultra-rapid and intermediate metabolizers though, it would appear from the various plots that the NMs could be further separated into two sub-groups.
The Yoruba Nigerians are considered a homogenous population and of common ancestry. Considerable efforts were made to ensure that only participants with sufficiently long Yoruba ancestry were enrolled. In addition, inclusion criteria also took into cognizance other factors that are known to influence the rate and extent of drug metabolism, and therefore potential confounders. For example, within the age range of the participants, there was a lack of correlation between age and the metabolic ratios of the participants. It is also noteworthy that the two poor metabolizers that were identified were aged 25 and 27 years. The age range in this study was similar to previous studies investigating CYP2D6 phenotype [24, 30, 31]. The established tolerability profile of dextromethorphan was further demonstrated by the fact that only one (~1%) participant in our study had mild and transient drowsiness [24, 28, 31–33]. In CYP2D6 phenotyping, the requisite cutoff(s) may be obtained by subjecting data to a probit analysis, Receiver Operating Characteristics (ROC) or other similar statistical analysis [28, 33, 34]. On the other hand, some studies have simply adopted cut-off [35, 36]. The former option was chosen in this case as there is no immediate record of an identical study, that is, determination of CYP2D6 phenotype in the Yoruba Nigerians using a single plasma sample. Further, some researchers have suggested against the utilization of a “universal” cutoff as it may be misleading [28, 37]. The histogram reveals the bimodal distribution and the same was reinforced by the probit plots similar to the previous findings [33, 37]. Studies have reported different MR cutoffs separating the PMs from NMs, while some adopted a universal MR of 0.3 obtained among Caucasians in both 8-hour urine [32, 35], and 3-hour plasma[38, 39]. Some studies reported MR cutoffs for 8-hour urine were 0.3 in Swiss[40], Cuba[18], and Gabon[41], 0.24 in Spain (Spaniards) [42], and 0.32 by Ebeshi et al. in Nigeria [24]. For the 3-hour plasma, some reported log MR cutoff was 0.742 by Jurica et al. in Czech Republic [28], 0.8 by Dorado et al. in Ecuador[43] and 0.142 by Gogtay et al. reported in India [33].
This study explored the comparability of the well-established non-invasive but time-consuming urine versus the relatively invasive but less cumbersome single plasma collection methods. Evaluation of drug metabolism phenotypes has evolved. Though hitherto, a 24-hour urine collection was required, recent studies have used 4 and 8-hour urine sample collection [44, 45]. In addition, a single 3-hour plasma sample has been found to be an equally useful alternative [28, 46]. For emphasis, previous studies have found plasma matrix for phenotyping with dextromethorphan accurate, convenient and more rapid than the standard urine approach [28, 46–48]. Besides, the use of single plasma sample appears to assist in avoiding potential error that preferential accumulation of metabolites due to impaired renal function may introduce in the determination of metabolic ratio.
The frequency of CYP2D6 poor metabolizers obtained in this study is similar to previous studies in Nigeria and other African countries. Ebeshi et al. obtained a prevalence of 3.5% poor metabolizers with urinary dextromethorphan/dextrorphan metabolic ratio among heterogeneous Nigerians[24]. Other studies in Africa also showed a poor metabolizer prevalence of 1.2% in South Africa [31] and 1–4% in other African countries [20, 21, 49]. These findings are similar to that of Asian countries with a poor metabolizer prevalence of 1–4% [16, 17, 33]. The prevalence of poor metabolizer phenotype among the African-Americans appears to be slightly higher at between 5.3% and 7.7%, [50, 51] and even higher still among the Caucasians, 7–10% [28, 36]. The clinical implications of poor metabolizers include slower drug metabolism with potential for adverse drug interactions. There may also be the slower conversion of a prodrug to active metabolites with potential lower efficacy [52, 53].
There were some challenges with the assay of DEX and DOR, particularly for resource-poor countries where sophisticated analytic facilities may be lacking. Although, HPLC with UV detection used in this study was not best suited for determination of dextromethorphan and dextrorphan when compared to LC-MS/MS or HPLC florescent detector with superior sensitivity, this method described by Zimova and Jurica with modifications appeared to be adequate particularly with the assumption that the two major metabolites would have been affected to similar extent.
An accurate determination of CYP2D6-genotype predicted phenotype is required in the clinical settings for precision medicine. However, CYP2D6 genotype-phenotype matching alone may not produce a desirable result, as studies have reported discordance in genotype-predicted PMs [54–56], and recent studies also suggested that CYP2D6 genotype was not a good predictor of ultra rapid metabolism[18, 19, 57]. Gaedigk et al.[58], and Zineh et al.[59], have proposed an Activity Score (AS) that are calculated from the CYP2D6 gene and use to determine the CYP2D6 metabolic activities of an individual. Vanwong et al. using risperidone as a probe [60], Puangpetch et al. [61], in Thailand, and Dodgen et al. in South Africa [31], reported a good correlation between AS, CYP2D6 genotypes and measured phenotypes.
We recognized that a genotype-phenotype pairing and prediction of CYP2D6 enzyme activities with AS would have enhanced a study like this but the genotype study could not be performed due to resource constraint. Although limited by the small sample size and rather conformational findings, the relevance of the present study could be justified by lmited information of CYP2D6 phenotypes available from only two completed studies in Yoruba Nigerians who constitute 40 million people accounting for ~20% Nigerian population. Additionally, dextromethorphan as a common probe drug for CYP2D6 phenotyping might not be optimal for estimating in vivo enzyme activity due to its limitation such as no linear correlation existing between enzyme activity and urinary ratios and its dependence on renal and hepatic transport. Moreover, CYP3A4/5 polymorphisms also contribute to the variability in the clearance of dextromethorphan. Nevertheless, the use of dextromethorphan in this study was largely owing to its accessibility in lower and mid-income settings. Despite other limitations including the relatively poor distinction between NM and PM or ultra rapid from EM phenotypes based on the urinary metabolic ratio, we were able to identify PM using this approach. It is also noted that this was the very first report where DEX in plasma was used to phenotype CYP2D6 in Nigerian populations. This study revealed a significant positive correlation between 8-hour urine and 3-hour plasma metabolic ratios and indeed demonstrated that a single 3-hour plasma sample following administration of DEX to Yoruba Nigerians appears adequate in segregating NMs and PMs on the CYP2D6 metabolized drugs.
5. Conclusions
Single 3-hour plasma sample following administration of DEX revealed similar PMs to 8-hour urine sample among the 89 Yoruba ethnic group of Nigerians studied.
6. Key issues.
CY2D6 is highly polymorphic and responsible for the metabolism of about 25% of all clinically used drugs.
Dextromethorphan has been widely utilized as a probe for CYP2D6 phenotype.
There is a dearth of information on CYP2D6 phenotype regarding Africa ethnic nationalities including the Yoruba Nigerian.
The metabolic ratios of dextromethorphan in urine and plasma could be used to separate poor CYP2D6 metabolizers from extensive metabolizers.
A significant positive correlation between urine and plasma metabolic ratios was noted.
Approximately 2–3% of the Yoruba Nigerians identified as poor metabolizers can be expected to benefit from CYP2D6 phenotype evaluation to avoid drug-related adverse effects.
Acknowledgements
We acknowledge the assistance provided by Mr. M. Adegoke of the Central Science Laboratory, Obafemi Awolowo University (OAU), Ile-Ife, Nigeria, and Dr. Jan Jurica, Clinical Pharmacology Department, Faculty of Medicine, Brno, the Czech Republic in the development of methods. Mr. Nathaniel K. Afolabi, Department of Pediatrics, University College Hospital, Ibadan, Nigeria, provided support in the sample storage. We thank Drs Tunde Adedokun and Bidemi Yusuf of the Epidemiology and Medical Statistics Department, the University of Ibadan for their assistance in the statistical analysis of this work.
Funding
The paper has been funded by University of Ibadan in Nigeria.
Footnotes
Declaration of Interest
Q Ma is currently supported in part by NIH K08MH098794. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
Papers of special note have been highlighted as * = of interest, ** = of considerable interest
- 1.Ingelman-Sundberg M, Pharmacogenetics of cytochrome P450 and its applications in drug therapy: the past, present and future. Trends in pharmacological sciences 2004, 25 (4), 193–200. [DOI] [PubMed] [Google Scholar]
- 2.Brunton LL; Lazo JS; Parker KL, Goodman & Gilman’s the pharmacological basis of therapeutics. McGrawHill, New York: 2006, 11, 737–8. [Google Scholar]
- 3.Zanger UM; Raimundo S; Eichelbaum M, Cytochrome P450 2D6: overview and update on pharmacology, genetics, biochemistry. Naunyn-Schmiedeberg’s archives of pharmacology 2004, 369 (1), 23–37. [DOI] [PubMed] [Google Scholar]
- 4.Zhou S-F, Polymorphism of human cytochrome P450 2D6 and its clinical significance. Clinical pharmacokinetics 2009, 48 (12), 761–804. [DOI] [PubMed] [Google Scholar]
- 5.Caudle KE; Dunnenberger HM; Freimuth RR; Peterson JF; Burlison JD; Whirl-Carrillo M; Scott SA; Rehm HL; Williams MS; Klein TE; Relling MV; Hoffman JM, Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genetics in medicine : official journal of the American College of Medical Genetics 2017, 19 (2), 215–223. * The Clinical Pharmacogenetics Implementation Consortium (CPIC) in this original research proposed a standardized pharmacogenomics terms to ensure maintainance of consistent numenclature. For example, proposed five phenotypes for CYP2D6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Özdemir M; Crewe KH; Tucker GT; Rostami-Hodjegan A, Assessment of in vivo CYP2D6 activity: differential sensitivity of commonly used probes to urine pH. The Journal of Clinical Pharmacology 2004, 44 (12), 1398–1404. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka E; Kurata N; Yasuhara H, How useful is the ‘cocktail approach’for evaluating human hepatic drug metabolizing capacity using cytochrome P450 phenotyping probes in vivo? Journal of clinical pharmacy and therapeutics 2003, 28 (3), 157–165. [DOI] [PubMed] [Google Scholar]
- 8.Frank D; Jaehde U; Fuhr U, Evaluation of probe drugs and pharmacokinetic metrics for CYP2D6 phenotyping. European journal of clinical pharmacology 2007, 63 (4), 321–333. [DOI] [PubMed] [Google Scholar]
- 9.Gasche Y; Daali Y; Fathi M; Chiappe A; Cottini S; Dayer P; Desmeules J, Codeine intoxication associated with ultrarapid CYP2D6 metabolism. New England Journal of Medicine 2004, 351 (27), 2827–2831. [DOI] [PubMed] [Google Scholar]
- 10.Kirchheiner J; Heesch C; Bauer S; Meisel C; Seringer A; Goldammer M; Tzvetkov M; Meineke I; Roots I; Brockmoller J, Impact of the ultrarapid metabolizer genotype of cytochrome P450 2D6 on metoprolol pharmacokinetics and pharmacodynamics. Clinical Pharmacology & Therapeutics 2004, 76 (4), 302–312. [DOI] [PubMed] [Google Scholar]
- 11.Rau T; Wohlleben G; Wuttke H; Thuerauf N; Lunkenheimer J; Lanczik M; Eschenhagen T, CYP2D6 genotype: impact on adverse effects and nonresponse during treatment with antidepressants-a pilot study. Clin Pharmacol Ther 2004, 75 (5), 386–93. [DOI] [PubMed] [Google Scholar]
- 12.Breil F; Verstuyft C; Orostegui L; Buhl C; Alvarez J-C; Chouinard G; Becquemont L; Corruble E, Non-response to consecutive antidepressant therapy caused by CYP2D6 ultrarapid metabolizer phenotype. International Journal of Neuropsychopharmacology 2008, 11 (5), 727–728. [DOI] [PubMed] [Google Scholar]
- 13.Wester K; Jonsson AK; Spigset O; Druid H; Hägg S, Incidence of fatal adverse drug reactions: a population based study. British journal of clinical pharmacology 2008, 65 (4), 573–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fux R; Mörike K; Pröhmer AM; Delabar U; Schwab M; Schaeffeler E; Lorenz G; Gleiter CH; Eichelbaum M; Kivisto KT, Impact of CYP2D6 genotype on adverse effects during treatment with metoprolol: a prospective clinical study. Clinical Pharmacology & Therapeutics 2005, 78 (4), 378–387. [DOI] [PubMed] [Google Scholar]
- 15.Bertilsson L; Lou YQ; Du YL; Liu Y; Kuang TY; Liao XM; Wang KY; Reviriego J; Iselius L; Sjoqvist F, Pronounced differences between native Chinese and Swedish populations in the polymorphic hydroxylations of debrisoquin and S-mephenytoin. Clinical pharmacology and therapeutics 1992, 51 (4), 388–97. [DOI] [PubMed] [Google Scholar]
- 16.Kim K; Johnson JA; Derendorf H, Differences in drug pharmacokinetics between East Asians and Caucasians and the role of genetic polymorphisms. The Journal of Clinical Pharmacology 2004, 44 (10), 1083–1105. [DOI] [PubMed] [Google Scholar]
- 17.Xie H-G; Kim RB; Wood AJ; Stein CM, Molecular basis of ethnic differences in drug disposition and response. Annual review of pharmacology and toxicology 2001, 41 (1), 815–850. [DOI] [PubMed] [Google Scholar]
- 18.Dorado P; Gonzalez I; Naranjo ME; de Andres F; Penas-Lledo EM; Calzadilla LR; A LL, Lessons from Cuba for Global Precision Medicine: CYP2D6 Genotype Is Not a Robust Predictor of CYP2D6 Ultrarapid Metabolism. Omics : a journal of integrative biology 2017, 21 (1), 17–26. [DOI] [PubMed] [Google Scholar]
- 19.De Andres F; Teran S; Hernandez F; Teran E; A LL, To Genotype or Phenotype for Personalized Medicine? CYP450 Drug Metabolizing Enzyme Genotype-Phenotype Concordance and Discordance in the Ecuadorian Population. Omics : a journal of integrative biology 2016, 20 (12), 699–710. [DOI] [PubMed] [Google Scholar]
- 20.Dandara C; Masimirembwa CM; Magimba A; Sayi J; Kaaya S; Sommers DK; Snyman J; Hasler JA, Genetic polymorphism of CYP2D6 and CYP2C19 in east-and southern African populations including psychiatric patients. European journal of clinical pharmacology 2001, 57 (1), 11–17. [DOI] [PubMed] [Google Scholar]
- 21.Wennerholm A; Johansson I; Hidestrand M; Bertilsson L; Gustafsson LL; Ingelman- Sundberg M, Characterization of the CYP2D6* 29 allele commonly present in a black Tanzanian population causing reduced catalytic activity. Pharmacogenetics and Genomics 2001, 11 (5), 417–427. [DOI] [PubMed] [Google Scholar]
- 22.Dandara C; Swart M; Mpeta B; Wonkam A; Masimirembwa C, Cytochrome P450 pharmacogenetics in African populations: implications for public health. Expert Opin Drug Metab Toxicol 2014, 10 (6), 769–85. * The article provides a table that show CYP allele frequencies in different population. It also highlights the heterogenous and genetically diversed nature of Africa [DOI] [PubMed] [Google Scholar]
- 23.Aklillu E; Persson I; Bertilsson L; Johansson I; Rodrigues F; Ingelman-Sundberg M, Frequent distribution of ultrarapid metabolizers of debrisoquine in an ethiopian population carrying duplicated and multiduplicated functional CYP2D6 alleles. J Pharmacol Exp Ther 1996, 278 (1), 441–6. [PubMed] [Google Scholar]
- 24.Ebeshi BU; Bolaji OO; Masimirembwa C, Cytochrome P450 2D6 (CYP2D6) genotype and phenotype determination in the Nigerian population. Asian J Pharm Hea Sci 2011, 1, 47–54. **Study CYP2D6 polymorphism among Nigerian using dextromethorphan as a probe with 8-hour urine collection [Google Scholar]
- 25.Lennard MS; Iyun AO; Jackson PR; Tucker GT; Woods HF, Evidence for a dissociation in the control of sparteine, debrisoquine and metoprolol metabolism in Nigerians. Pharmacogenetics 1992, 2 (2), 89–92. * Provides early evidence of CYP2D6 polymorphism among Nigerias [DOI] [PubMed] [Google Scholar]
- 26.Commission NP, National population census. In Abuja, Nigeria: National Population Commission, 2006. [Google Scholar]
- 27.Zimova G; Chladek J; Martinkova J; Beranek M, HPLC determination of dextromethorphan and its metabolites in urine. Chemicke Listy 2000, 94, 4 ** The method together that of Jurica et al. was adapted for the extraction and drug analysis [Google Scholar]
- 28.Jurica J; Bartecek R; Zourkova A; Pindurová E; Sulcova A; Kasparek T; Zendulka O, Serum dextromethorphan/dextrorphan metabolic ratio for CYP2D6 phenotyping in clinical practice. Journal of clinical pharmacy and therapeutics 2012, 37 (4), 486–490. ** The method together that of Zimova et al. was adapted for the extraction and drug analysis [DOI] [PubMed] [Google Scholar]
- 29.Burgess C, Valid analytical methods and procedures. Royal Society of Chemistry: 2000. *Described the calculation and drawing of probit plots [Google Scholar]
- 30.Afshar M; Rouini M; Ala S, Dextromethorphan metabolic phenotyping in an Iranian population. Eur J Clin Pharmacol 2005, 60 (12), 849–54. [DOI] [PubMed] [Google Scholar]
- 31.Dodgen TM; Labuschagne CJ; van Schalkwyk A; Steffens FE; Gaedigk A; Cromarty AD; Alessandrini M; Pepper MS, Pharmacogenetic comparison of CYP2D6 predictive and measured phenotypes in a South African cohort. Pharmacogenomics Journal 2016, 16(6):566–572. *An African study with evidence of correlation bewtween CYP2D6 genotypes, measured phenotypes and predicted phenotypes with AS [DOI] [PubMed] [Google Scholar]
- 32.Chladek J; Zimova G; Beranek M; Martinkova J, In-vivo indices of CYP2D6 activity: comparison of dextromethorphan metabolic ratios in 4-h urine and 3-h plasma. European journal of clinical pharmacology 2000, 56 (9–10), 651–657. [DOI] [PubMed] [Google Scholar]
- 33.Gogtay NJ; Mali NB; Iyer K; Kadam PP; Sridharan K; Shrimal D; Thatte UM, Evaluation of cytochrome P450 2D6 phenotyping in healthy adult Western Indians. Indian journal of pharmacology 2014, 46 (3), 266–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson P; Tucker G; Woods H, Testing for bimodality in frequency distributions of data suggesting polymorphisms of drug metabolism-histograms and probit plots. British journal of clinical pharmacology 1989, 28 (6), 647–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ito T; Kato M; Chiba K; Okazaki O; Sugiyama Y, Estimation of the interindividual variability of cytochrome 2D6 activity from urinary metabolic ratios in the literature. Drug metabolism and pharmacokinetics 2010, 25 (3), 243–253. [DOI] [PubMed] [Google Scholar]
- 36.Wojtczak A; Rychlik-Sych M; Krochmalska-Ulacha E; Skretkowicz J, CYP2D6 phenotyping with dextromethorphan. Pharmacological reports : PR 2007, 59 (6), 734–8. [PubMed] [Google Scholar]
- 37.Othman A, Kadi HO; Thabit AAM, Prevalence of CYP2D6 phenotypes in a Yemeni population. World journal of pharmaceutical research 2014, 3(4), 84–91 [Google Scholar]
- 38.Hartter S; Baier D; Dingemanse J; Ziegler G; Hiemke C, Automated determination of dextromethorphan and its main metabolites in human plasma by high-performance liquid chromatography and column switching. Therapeutic drug monitoring 1996, 18 (3), 297–303. [DOI] [PubMed] [Google Scholar]
- 39.Afshar M; Rouini M; Ala S, Dextromethorphan metabolic phenotyping in an Iranian population. European journal of clinical pharmacology 2005, 60 (12), 849–854. [DOI] [PubMed] [Google Scholar]
- 40.Schmid B; Bircher J; Preisig R; Küpfer A, Polymorphic dextromethorphan metabolism: Co-segregation of oxidative O-demethylation with debrisoquin hydroxylation. Clinical Pharmacology & Therapeutics 1985, 38 (6), 618–624. [DOI] [PubMed] [Google Scholar]
- 41.Panserat S; Sica L; Gerard N; Mathieu H; Jacqz-Aigrain E; Krishnamoorthy R, CYP2D6 polymorphism in a Gabonese population: contribution of the CYP2D6*2 and CYP2D6*17 alleles to the high prevalence of the intermediate metabolic phenotype. Br J Clin Pharmacol 1999, 47 (1), 121–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henthorn TK; Benitez J; Avram MJ; Martinez C; Llerena A; Cobaleda J; Krejcie TC; Gibbons RD, Assessment of the debrisoquin and dextromethorphan phenotyping tests by gaussian mixture distributions analysis. Clinical pharmacology and therapeutics 1989, 45 (3), 328–33. [DOI] [PubMed] [Google Scholar]
- 43.Dorado P; Heras N; Machin E; Hernandez F; Teran E; Llerena A, CYP2D6 genotype and dextromethorphan hydroxylation phenotype in an Ecuadorian population. Eur J Clin Pharmacol 2012, 68 (5), 637–44. [DOI] [PubMed] [Google Scholar]
- 44.Chladek J; Zimova G; Martinkova J; Tůma I, Intra-individual variability and influence of urine collection period on dextromethorphan metabolic ratios in healthy subjects. Fundamental & clinical pharmacology 1999, 13 (4), 508–515. [DOI] [PubMed] [Google Scholar]
- 45.Droll K; Bruce-Mensah K; Otton SV; Gaedigk A; Sellers EM; Tyndale RF, Comparison of three CYP2D6 probe substrates and genotype in Ghanaians, Chinese and Caucasians. Pharmacogenetics 1998, 8 (4), 325–33. [DOI] [PubMed] [Google Scholar]
- 46.Shiran M; Chowdry J; Rostami-Hodjegan A; Ellis S; Lennard M; Iqbal M; Lagundoye O; Seivewright N; Tucker G, A discordance between cytochrome P450 2D6 genotype and phenotype in patients undergoing methadone maintenance treatment. British journal of clinical pharmacology 2003, 56 (2), 220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borges S; Li L; Hamman MA; Jones DR; Hall SD; Gorski JC, Dextromethorphan to dextrorphan urinary metabolic ratio does not reflect dextromethorphan oral clearance. Drug metabolism and disposition 2005, 33 (7), 1052–1055. [DOI] [PubMed] [Google Scholar]
- 48.Kohler D; Hartter S; Fuchs K; Sieghart W; Hiemke C, CYP2D6 genotype and phenotyping by determination of dextromethorphan and metabolites in serum of healthy controls and of patients under psychotropic medication. Pharmacogenetics 1997, 7 (6), 453–61. [DOI] [PubMed] [Google Scholar]
- 49.Griese E; Asante-Poku S; Ofori-Adjel D; Mikus G; Eichelbaum M, Analysis of the CYP2D6 gene mutations and their consequences for enzyme function in a West African population. Pharmacogenetics and Genomics 1999, 9 (6), 715–724. [PubMed] [Google Scholar]
- 50.Yee MM; Josephson C; Hill CE; Harrington R; Castillejo MI; Ramjit R; Osunkwo I, Cytochrome P450 2D6 polymorphisms and predicted opioid metabolism in African American children with sickle cell disease. Journal of pediatric hematology/oncology 2013, 35 (7), e301–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gaedigk A; Bradford LD; Marcucci KA; Leeder JS, Unique CYP2D6 activity distribution and genotype-phenotype discordance in black Americans. Clinical pharmacology and therapeutics 2002, 72 (1), 76–89. [DOI] [PubMed] [Google Scholar]
- 52.Bernard S; Neville KA; Nguyen AT; Flockhart DA , Interethnic differences in genetic polymorphisms of CYP2D6 in the US population: clinical implications. The oncologist 2006, 11 (2), 126–135. [DOI] [PubMed] [Google Scholar]
- 53.Rau T; Wohlleben G; Wuttke H; Thuerauf N; Lunkenheimer J; Lanczik M; Eschenhagen T, CYP2D6 genotype: impact on adverse effects and nonresponse during treatment with antidepressants— a pilot study. Clinical Pharmacology & Therapeutics 2004, 75 (5), 386–393. [DOI] [PubMed] [Google Scholar]
- 54.Shiran MR; Chowdry J; Rostami-Hodjegan A; Ellis SW; Lennard MS; Iqbal MZ; Lagundoye O; Seivewright N; Tucker GT, A discordance between cytochrome P450 2D6 genotype and phenotype in patients undergoing methadone maintenance treatment. Br J Clin Pharmacol 2003, 56 (2), 220–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bailey B; Daneman R; Daneman N; Mayer JM; Koren G, Discrepancy between CYP2D6 phenotype and genotype derived from post-mortem dextromethorphan blood level. Forensic science international 2000, 110 (1), 61–70. [DOI] [PubMed] [Google Scholar]
- 56.Leathart JB; London SJ; Steward A; Adams JD; Idle JR; Daly AK, CYP2D6 phenotype-genotype relationships in African-Americans and Caucasians in Los Angeles. Pharmacogenetics 1998, 8 (6), 529–41. [DOI] [PubMed] [Google Scholar]
- 57.Naranjo M; de Andrés F; Delgado A; Cobaleda J; Peñas-Lledó E; LLerena A, High frequency of CYP2D6 ultrarapid metabolizers in Spain: controversy about their misclassification in worldwide population studies. The pharmacogenomics journal 2016, 16 (5), 485–490. [DOI] [PubMed] [Google Scholar]
- 58.Gaedigk A; Simon SD; Pearce RE; Bradford LD; Kennedy MJ; Leeder JS, The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clinical pharmacology and therapeutics 2008, 83 (2), 234–42. *Describe the Activity Score(AS) for predicting CYP2D6 phenotypes [DOI] [PubMed] [Google Scholar]
- 59.Zineh I; Beitelshees AL; Gaedigk A; Walker JR; Pauly DF; Eberst K; Leeder JS; Phillips MS; Gelfand CA; Johnson JA, Pharmacokinetics and CYP2D6 genotypes do not predict metoprolol adverse events or efficacy in hypertension. Clinical pharmacology and therapeutics 2004, 76 (6), 536–44. *Describe the Activity Score(AS) for predicting CYP2D6 phenotypes [DOI] [PubMed] [Google Scholar]
- 60.Vanwong N; Ngamsamut N; Hongkaew Y; Nuntamool N; Puangpetch A; Chamnanphon M; Sinrachatanant A; Limsila P; Sukasem C, Detection of CYP2D6 polymorphism using Luminex xTA technology in autism spectrum disorder: CYP2D6 activity score and its association with risperidone levels. Drug Metab Pharmacokinet 2016, 31 (2), 156–62. [DOI] [PubMed] [Google Scholar]
- 61.Puangpetch A; Vanwong N; Nuntamool N; Hongkaew Y; Chamnanphon M; Sukasem C, CYP2D6 polymorphisms and their influence on risperidone treatment. Pharmacogenomics and personalized medicine 2016, 9, 131–147. [DOI] [PMC free article] [PubMed] [Google Scholar]


