Abstract
Prior studies have shown that aversive olfactory memory is acquired by dopamine acting on a specific receptor, dDA1, expressed by mushroom body neurons. Active forgetting is mediated by dopamine acting on another receptor, Damb, expressed by the same neurons. Surprisingly, prior studies have shown that both receptors stimulate cAMP accumulation, presenting an enigma of how mushroom body neurons distinguish between acquisition and forgetting signals. Here, we surveyed the spectrum of G protein coupling of dDA1 and Damb and confirmed that both receptors can couple to Gs to stimulate cAMP synthesis. However, the Damb receptor uniquely activates Gq to mobilize Ca2+ signaling with greater efficiency and dopamine sensitivity. The knockdown of Gαq with RNAi in the mushroom bodies inhibits forgetting but has no effect on acquisition. Our findings identify a novel, Damb/Gq signaling pathway that stimulates forgetting, and resolves the opposing effects of dopamine on acquisition and forgetting.
Keywords: dopamine receptors, memory acquisition, memory forgetting, mushroom body, olfactory learning
Introduction
Pioneering neuroscience studies using Drosophila melanogaster have revealed that the brain is constructed with cellular and circuit mechanisms that promote forgetting (Davis and Zhong, 2017). The best-understood mechanism for active forgetting of olfactory memory at the circuit level involves the release of dopamine (DA) from dopaminergic neurons (DAn-forgetting cells) of the PPL1 cluster onto the axons of mushroom body neurons (MBn-engram cells). Forgetting cells exhibit chronic activity (Berry et al., 2012) that slowly promotes forgetting, but are stimulated by sensory information and quieted by sleep (Berry et al., 2015). DA released from the forgetting cells stimulates a specific DA receptor, Damb (DopAmine receptor expressed in Mushroom Bodies), required for normal forgetting (Han et al., 1996; Berry et al., 2012). This receptor mobilizes an intracellular signaling pathway that includes the scaffolding protein, Scribble, and its associated protein components – Rac1 and Cofilin – that may cause forgetting by reversing changes in the neuronal actin cytoskeleton that occur with memory formation (Davis and Zhong, 2017).
Curiously, several studies have shown that DA released from the PPL1 DAn is also required for the acquisition of new, aversive olfactory memories (Schwaerzel et al 2003; Schroll et al., 2006; Claridge-Chang et al., 2009; Aso et al., 2012). The current working model envisions the unconditioned stimulus (US) for olfactory classical conditioning as conveyed by DAn activation of the MBn. How DAn-stimulation of MBn might lead to both acquisition and forgetting of olfactory memories can be partially explained by the existence of a second DA receptor, named dDA1, which, like Damb, is preferentially and uniformly expressed along the axons of the MBn as revealed by light microscopic immunohistochemistry (Han et al., 1996; Kim et al., 2003). Mutants of dDA1 fail to acquire memory (Kim et al., 2007) consistent with a two-receptor model in which the dDA1 receptor mediates the acquisition of olfactory memories by MBn and Damb spurs forgetting (Berry et al., 2012).
However, a two-receptor model presents a mystery of how MBn know whether they are receiving an acquisition or forgetting signal from the PPL1 DAn. The most attractive explanation for this is that the two receptors might have different signaling properties and mobilize distinct signaling cascades. However, prior pharmacological characterization experiments in reconstituted systems classified both receptors as D1-like, stimulating the accumulation of cAMP by activating the heterotrimeric G protein, Gs, and the effector, adenylyl cyclase (Sugamori et al., 1995; Han et al., 1996; Reale et al., 1997). These observations challenge the hypothesis that the two receptors mobilize different intracellular signaling cascades for acquisition and forgetting.
We have revisited the question of whether the two receptors couple to distinct downstream signaling cascades. We show using a real-time BRET (bioluminescence resonance energy transfer) assay for G protein activation by GPCRs (Masuho et al. 2015b) that dDA1 strongly couples to Gαs and increases cAMP accumulation upon stimulation by DA; whereas the Damb receptor couples preferentially to Gαq over Gαs and induces calcium signaling. Consistent with a Damb/Gαq pathway role in forgetting, RNAi knockdown of Gαq in the MBn produces a selective deficit in forgetting. These results reconcile the dilemma of how DA stimulates both acquisition and forgetting, with acquisition occurring through a dDA1/Gαs/cAMP pathway and forgetting through Damb/Gαq/Ca2+.
RESULTS
Damb is a promiscuous GPCR preferentially activating Gq.
We first compared the ability of dDA1 and Damb receptors to modulate changes in cAMP and Ca2+ concentrations in response to DA (Figure 1A). Receptors were reconstituted into HEK293T/17 cells together with firefly luciferase-based GloSensor-22F cAMP sensor (Binkowski et al., 2011) or CalfluxVTN Ca2+ sensor (Yang et al., 2016) that report real time fluctuations in second messenger levels by changes in luminescence and BRET signal, respectively. Consistent with previous reports (Sugamori et al., 1995; Han et al., 1996), we observed that application of DA to cells expressing either dDA1 or Damb stimulated cAMP production (Figure 1B). These responses were driven by the transfected receptors as no significant signal was observed in control experiments where receptor constructs were omitted. The elevation in cAMP levels was significantly greater in cells transfected with dDA1 compared to those transfected with damb. In contrast, the dDA1 receptor promoted a marginal Ca2+ response upon DA application whereas the responses elicited by Damb were robust with fast response kinetics (Figure 1C).
Figure 1. G protein selectivity of dDA1 and DAMB.
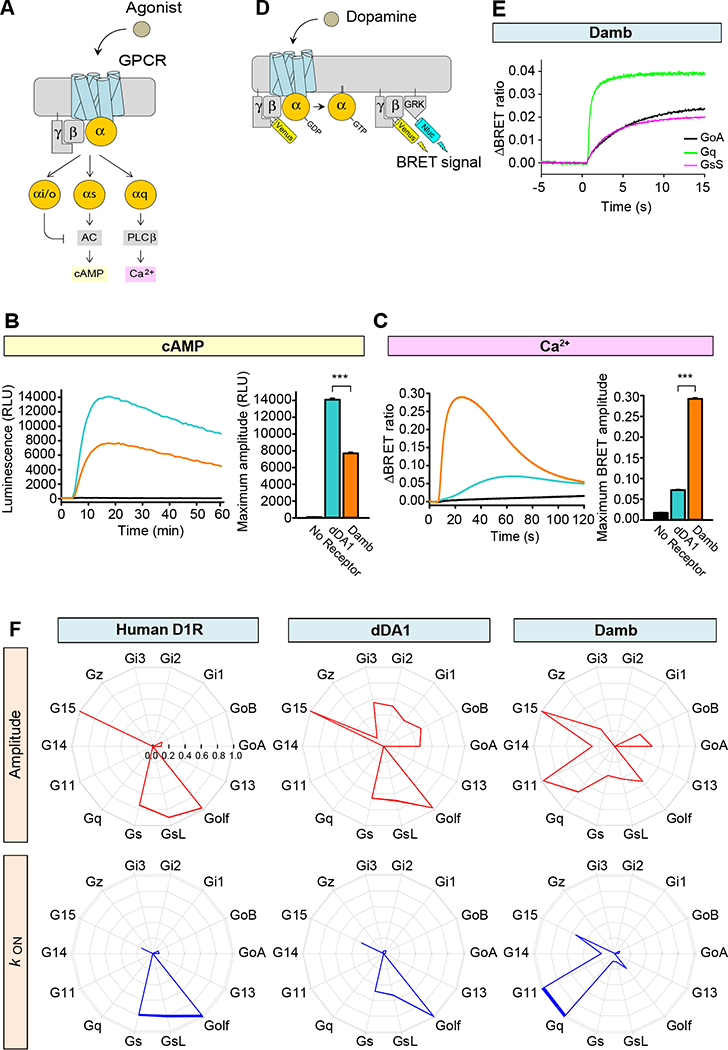
A. Cartoon of GPCR-mediated second messenger regulation. Stimulation with agonist leads to charging of the α subunit with GTP and its dissociation from βγ subunits. The Gαs subunit stimulates adenylyl cyclase (AC) resulting in cyclic AMP (cAMP) generation and Gαq stimulates phospholipase Cβ resulting in calcium release from internal stores.
B. Cyclic AMP assay to examine Gs coupling of the receptors. HEK293T/17 cells were transfected with GloSensor-22F/cAMP without exogenous GPCR (black) or with dDA1 (cyan) or DAMB (orange) expressing plasmids. A saturated concentration of DA (100 μM) was applied at 5 min and luminescence was monitored for 60 min. Each trace represents the mean of 6 replicates. The bar graph at the right quantitates the relative responses. Statistics: Results are expressed as the mean ± SEM. ANOVA with Tukey post hoc, ***p<0.001, n=6 per group.
C. Ca2+ assay to examine Gq coupling. HEK293T/17 cells were transfected with CalfluxVTN without exogenous GPCR or with dDA1 or DAMB expressing cDNAs. 100 μM DA was applied at 5 sec and the BRET ratio was monitored for 2 min. Each trace represents the mean of 6 replicates. The bar graph at the right quantitates the relative responses. Statistics: Results are expressed as the mean ± SEM. ANOVA with Tukey post hoc, ***p<0.001, n=6 per group.
D. Schematic representation of the BRET assay for real-time monitoring of G protein activity. Activation of a GPCR by agonist leads to the dissociation of inactive heterotrimeric G proteins into active GTP-bound Gα and Venus-Gβγ subunits. The released Venus-Gβγ can then interact with the Gβγ effector mimetic masGRK3ct-Nluc to produce the BRET signal. The masGRK3ct-Nluc reporter protein consists of C-terminal fragment of G protein Receptor Kinase, type 3 (GRK3) fused to a myristic acid attachment peptide (mas) and NanoLuc luciferase (Nluc) attached at the C-terminus (ct).
E. Real-time monitoring of G protein activation by the DAMB receptor. HEK293T/17 cells were transfected with the BRET sensor pair (panel D) and DAMB together with GαoA (black), Gαq (green), or GαsS (magenta). DA at 100 μM concentration was applied at time zero and the BRET signal followed across time. This time plot illustrates how the rate and magnitude of G protein coupling were assayed in panel F.
F. G protein selectivity diagrams of the human DA D1 receptor (D1R), dDA1, and DAMB. The maximum amplitudes (red) and activation rate constants (blue) from 14 different G proteins were normalized to the largest value and plotted in the wheel diagrams. Line thickness represents the SEM of six technical replicates performed in parallel. Two biological replicates using independent transfections were performed with similar results. Data are shown from one of the experiments.
To examine G protein coupling selectivity of the Drosophila Damb and dDA1 receptors, we employed a recently developed assay system that directly monitors G protein activation by GPCRs by measuring agonist-induced dissociation of mammalian G protein heterotrimers (Masuho et al., 2015b). In this assay, the release of Venus-tagged Gβγ subunits produces BRET responses through interactions with a luciferase-tagged effector mimic, masGRK3ct-Nluc (Figure 1D,E). We used the human DA D1 receptor (D1R) as a reference standard for these experiments. Consistent with its well-established mechanism, this receptor elicited responses with large amplitudes via short and long isoforms of Gαs, the related Gαolf and the promiscuous G protein, Gα15. Minor coupling was observed to GαoA and GαoB (Figure 1F, red wheel diagram). We observed from examining the activation rates, which reflect catalytic efficiency of GPCRs, that the D1R preferentially activates Gs/Golf over all other G protein substrates (Figure 1F, blue wheel diagram). Our analyses of dDA1 showed similar selectivity to D1R in this assay system employing mammalian Gα subunits. It activated all Gi/o subunits, G15, and Gs/olf proteins, but the fastest kinetics were found with Gs and Golf. This indicates that the dDA1 receptor behaves in this assay system like a typical mammalian Gαs-coupled receptor. In contrast, the Damb receptor exhibited a much different coupling profile to human Gα subunits. It activated Go, Gz, Gq, G11, G14, G15 and Gs/olf proteins to differing extents, but exhibited the most rapid activation kinetics for Gq and G11.
Damb and dDA1 couple selectively to Drosophila Gα subunits and differentially rely on the chaperone Ric-8 for function.
We next studied the interaction of Damb and dDA1 receptors with native Drosophila Gα subunits using 2 Gαq proteins, GαqD and GαqG, and 2 Gαs proteins, GαsA and GαsD These proteins are isoforms expressed by the two different genes through alternative splicing and were identified as being expressed in the brain by cDNA cloning (Table S1). The GαsA isoform was determined to be more abundantly expressed than the GαsD isoform based on frequency of clones obtained (Table S1, S2). The GαqG isoform was found as more abundantly expressed than the GαqD transcript through qRT-PCR assays (Table S3). First, we examined their expression in HEK293T/17 cells by monitoring their association with mammalian Gβγ subunits using a masGRK3ct-Nluc competition assay (Figure 2A). Venus-Gβγ and masGRK3ct-Nluc form a complex resulting in a large basal BRET signal in the absence of exogenously added Gα or agonist. Binding of Gα to Gβγ results in a formation of the basal heterotrimeric protein complex displacing the masGRK3ct-Nluc construct reporter and thus reducing the BRET signal. Since the chaperone-like proteins, Ric-8A and Ric-8B, facilitate the expression of many Gα subunits (Chan, et al., 2013; Gabay, et al., 2011; Masuho et al., 2015b), we further studied the effects of their co-expression. We discovered that both GαsA and GαsD were able to form complexes with Gβγ subunits as indicated by the suppression of the basal BRET signal and that this process strictly required the presence of Ric-8B (Figure 2B,C). Similarly, we were able to reconstitute heterotrimeric protein complexes of both GαqD and GαqG with Gβγ subunits. Formation of the heterotrimeric protein complex containing GαqD required the expression of Ric-8A. Complex formation with GαqG occurred without the expression of Ric-8A, but this chaperone further enhanced the formation of the heterotrimer.
Figure 2. Effects of mammalian Ric-8A and Ric-8B on expression of Drosophila Gα subunits.
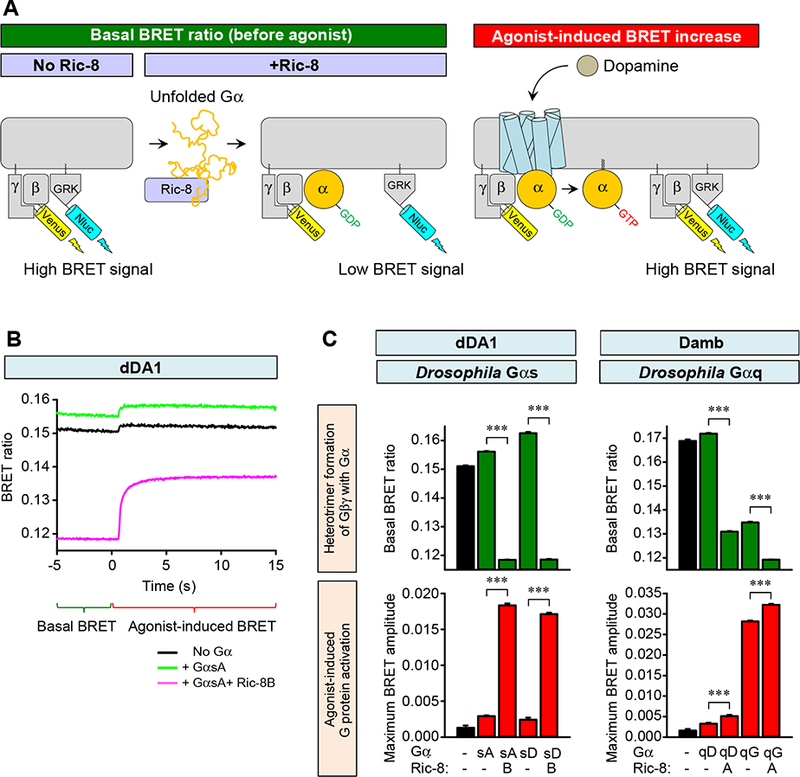
A. Schematic diagram of the Ric-8 effects in BRET assays with and without agonist. Co-transfection of Ric-8 with Gα provided for expression of functional Gα subunits and interaction of Gα with Venus-Gβγ. Functional heterotrimer formation consequently decreased the basal BRET ratio and increased agonist-induced BRET response.
B. Effect of Ric-8B, GαsA and DA on signaling through the dDA1 receptor. HEK293T/17 cells were transfected with the BRET sensor pair and dDA1 without exogenous Gα (black), with GαsA (green) or GαsA and Ric-8B (magenta). DA was added at time zero at a concentration of 100 μM. Functional GαsA (with Ric-8A) decreased the basal BRET ratio indicating heterotrimer formation. DA agonist produced a robust and quantifiable response.
C. Effects of DA, Ric-8A or −8B, and GαsA, GαsG, GαqD or GαqG on signaling through the dDA1 and Damb receptors. The cellular BRET assay for the dDA1 receptor was performed with GαsA and GαsG (left), with and without Ric-8B; and for the Damb receptor with GαqD and GαqG (right), with and without Ric-8A. The basal BRET ratio and agonist-induced maximum response amplitude were plotted as bar graphs. Statistics: Results are expressed as the mean ± SEM. ANOVA with Tukey post hoc, ***p<0.001, n=6 per group.
When stimulated with DA, the dDA1 receptor supported robust generation of the BRET signal with both GαsA and GαsD only in the presence of Ric-8B, indicating the ability of this protein combination to fully support signaling upon activation of the receptor (Figure 2B,C). In contrast, only GαqG supported an increase in BRET signal upon activation of Damb by DA and this effect was only slightly potentiated by Ric-8A. Along with results indicating much higher expression of GαqG mRNA in the Drosophila brain over GαqD, these results indicate that Damb likely couples primarily to GαqG.
Damb-GαqG is a high sensitivity dopamine detector.
We then compared the responses of Damb and dDA1 to DA with combinations of native Drosophila Gα subunits. To enable quantitative comparisons, we first titrated the Gα subunit expression to achieve stoichiometric heterotrimer formation (Figure S1). Under these optimal conditions, the dDA1 receptor supported rapid activation kinetics using GαsA but not GαqG (Figure 3). The Damb receptor supported signaling with both Drosophila GαqG and GαsA, similar to its signaling with mammalian G proteins (Figure 1), but with very different potencies. The rate of activation was 5-fold faster for GαqG than for GαsA (2.71 ± 0.04 s−1 and 0.46 ± 0.01 s−1, respectively), and the amplitude of the response 6-fold higher (Figure 3). We further explored the sensitivity of dDA1 and Damb to DA in dose-response experiments. Strikingly, the results revealed that Damb activates GαqG at a 100-fold lower concentration of DA than GαsA (EC50=7.37 ± 0.69 μM for GαsA and 56.7 ± 7.7 nM for GαqG). Furthermore, the potency of Damb-GαqG coupling was an order of magnitude higher than that of dDA1-GαsA (EC50=0.61 ± 0.07 μM). Interestingly, at saturating concentrations of DA, Damb and dDA1 elicited responses using GαsA of similar magnitude, consistent with the substantial cAMP production driven by Damb observed above (Figure 1B). Control experiments measuring the proteolytically stable heterotrimer in transfected cells indicated that differences in expression levels of Drosophila receptor/G protein combinations could not account for the differential G protein coupling described above (Figure S1C, Figure 3). Together, these findings establish Damb as a dual specificity GPCR signaling primarily through GqG at low levels of DA and additionally through GsA at higher levels.
Figure 3. Biochemical characterization of dDA1 and DAMB mediated signaling.
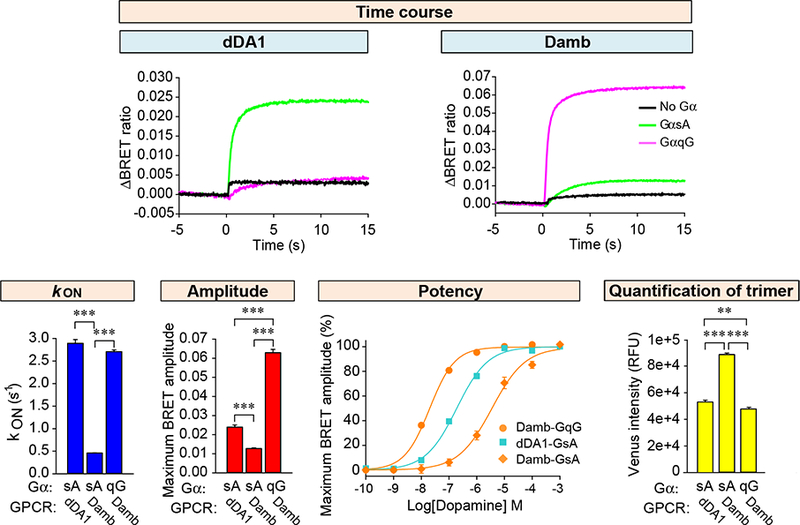
Time course of G protein activation mediated by the dDA1 and DAMB receptors shown at the top of the figure. DA at 100 μM was applied at time zero. Activation rate constants and agonist-induced maximum BRET amplitudes plotted as bar graphs. Dose response analyses were performed for dDA1 with the GαsA combination, and DAMB with GαsA and GαqG combinations. Statistics: Results are expressed as the mean ± SEM. ANOVA with Tukey post hoc, ***p<0.001, n=6 per group. Quantification of trimeric G protein in transfected cells was performed as described in Figure S1C. The intensity of Venus in cells transfected without Gα was subtracted from the intensity in cells transfected with Gα. Statistics: Results are expressed as the mean ± SEM. ANOVA with Tukey post hoc, **p<0.01, ***p<0.001, n=8 per group.
Knockdown of Gαq leads to memory enhancement.
We subsequently tested the hypothesis that Gαq mediates the intrinsic forgetting function of the Damb receptor on olfactory memories with Gal4-uas RNAi knockdown experiments (Figure 4A). Our initial experiments employed Gal4 lines expressed pan-neuronally or in the MBn, but we found using Nsyb-gal4 (all neurons), R13-gal4 (MBn), and c772-gal4 (MBn) that adult flies failed to eclose or that they had impaired olfactory learning, consistent with previously reported neurodevelopmental roles for Gαq (Ratnaparkhi et al., 2002). To avoid these developmental complications, we controlled expression of RNAi transgenes only during adulthood using tub-gal80ts (Figure 4B; McGuire et al., 2003). Experimental flies expressing Gαq RNAi2 in adult neurons showed an increase in memory of ~30% compared to control flies when tested at 3 hr after conditioning (Figure 4B). We tested two additional Gαq RNAi lines targeting different sequences of Gαq (Figure 4A; S2A) to confirm these behavioral results and to test the role of knockdown in the MBn. Similar to the results obtained with adult, pan-neuronal expression of RNAi2, knockdown with these RNAi’s in adult MBn produced an increase in the Performance Index, mapping the forgetting deficit to the MBn and showing that the phenotype observed with RNAi2 was not due to off-target effects (Figure 4C).
Figure 4. RNAi knock down of Gαq impairs forgetting.
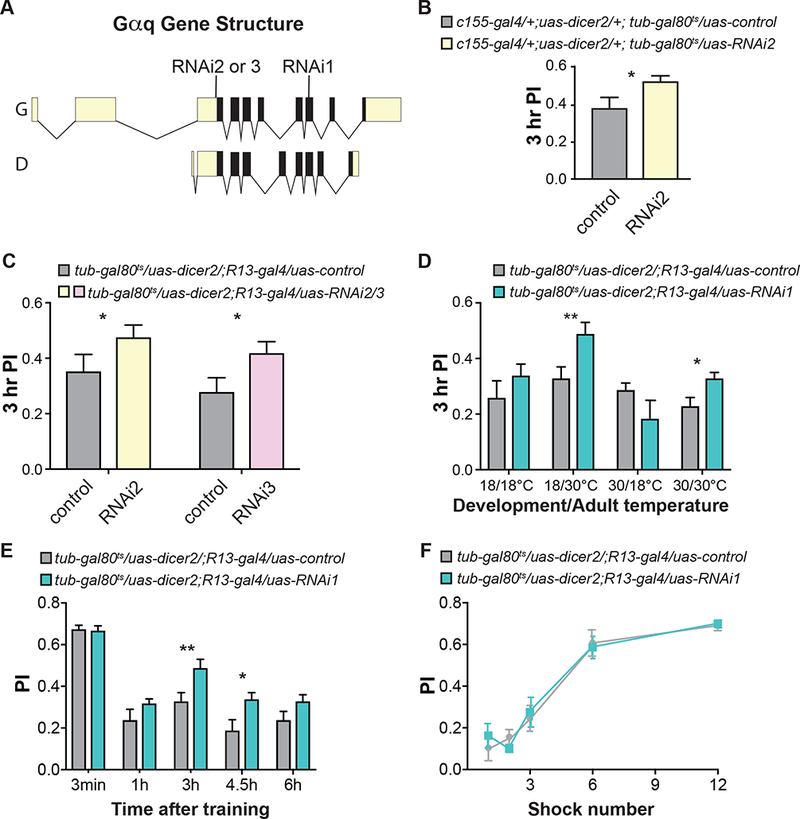
A. The graphic shows the structure of the Gαq gene with two annotated isoforms, G and D. Two different transcriptional units and alternative splicing encode these forms. The open reading frame is illustrated as solid black with 5’ and 3’ untranslated regions colored light yellow. Three different RNAi lines were used in this study. All three Gαq RNAi transgenes encode short hairpin RNAi sequences complementary to 24 nucleotides of two annotated isoforms, G and D, with RNAi2 and 3 targeting the 5’ UTR in exon 4 and Gαq RNAi1 targeting sequences in exon 10. Supplementary Figure 3 illustrates an expanded exon 4 and 10 and the specific sequences targeted by the three RNAi lines.
B. RNAi knock down of Gαq using the pan-neuronal c155-gal4 driver and tub-gal80ts to control expression only during adulthood with a temperature shift from 18°C to 30°C after eclosion. The tub-gal80ts transgene expresses temperature-sensitive Gal80ts protein that inhibits Gal4 activity at 18°C but not at 30°C. The tubulin (tub) promoter drives expression of gal80ts in all cells. Knockdown of Gαq with RNAi2 increased memory expression relative to the “uas-control”. The “uas-control” is a chromosome containing the attP40 docking site used for the insertion of the RNAi constructs. Statistics: Results are expressed as the mean ± SEM. Two-tailed, two-sample Student’s t-test, *p<0.05, n=13 per group.
C. RNAi knock down of Gαq expression in the MBn using the R13-gal4 driver only during adulthood increased memory expression. Statistics: Results are expressed as the mean ± SEM. Two-tailed, two-sample Student’s t-test, *p<0.05, **p<0.001, n=12–16 per group.
D. RNAi knock down of Gαq expression only during adulthood with two additional RNAi transgenes increased memory performance. Statistics: Results are expressed as the mean ± SEM. Two-tailed, two-sample Student’s t-test, *p<0.05, n=12 per group.
E. Memory retention was enhanced with Gαq-RNAi expression in the MBn only at intermediate time – 3 and 4.5 hr – after conditioning. Statistics: Results are expressed as the mean ± SEM. Two-tailed, two-sample Student’s t-test, *p<0.05, **p<0.01, n=8–12 per group.
F. RNAi knock down of Gαq expression in adult MBn did not alter the memory acquisition curve with flies trained with an increasing number of electric shock pulses. Statistics: Results are expressed as the mean ± SEM. Two factor ANOVA, n.s., n=6 per group.
We also tested flies expressing the RNAi transgenes in the adult MBn for possible defects in odor and electric shock avoidance, the two sensory channels for delivering the CS and US stimuli in olfactory classical conditioning. No significant difference was found in avoidance responses between flies expressing the RNAi transgenes in the MBn compared to the control genotype relative to either type of stimulus (Figure S2B–D), precluding the possibility that the behavioral effects observed were due to altered sensory perception.
Ascribing the forgetting phenotype to insulting Gαq expression in adult MBn does not eliminate the possibility of an adult behavioral role from developmental expression. We explored this possibility by shifting the incubation temperature of flies to conditionally express Gαq RNAi in MBn (Figure 4D). While memory was improved with expression of Gαq RNAi in adult MBn, expression during development produced a non-significant trend towards impairment (Figure 4D). Thus, the deficit in forgetting ensues from an adult, physiological role for Gαq. We also measured memory expression at multiple times after conditioning to obtain a better understanding of the temporal forms of memory altered by Gαq blockade (Figure 4E). The results indicated that RNAi knockdown had no effect on memory measured immediately or at 1 hr after conditioning. We again observed impaired forgetting at 3 hr after training and at 4.5 hr. These results indicate that forgetting was impaired across a time window typically considered as intermediate-term memory (Davis, 2011). The magnitude of the observed deficit was more modest than that measured in flies carrying a damb mutation (Berry et al., 2012), but this quantitative difference can be accounted for by the greater strength of a genomic null mutation compared to RNAi knockdown.
To test whether the Gαq RNAi knockdown leads to increased acquisition that could produce enhanced memory, we performed acquisition experiments by training flies with an increasing number of shock pulses (Figure 4F). The experimental flies performed equivalently to the controls in this experiment, indicating that acquisition of memory was comparable between the two groups and arguing against increased acquisition as a possible explanation for the enhanced memory observed with Gαq RNAi knockdown.
Discussion
Here we provide biochemical and behavioral evidence that the Drosophila DA receptor, Damb, couples preferentially to Gαq to mediate signaling by Damb for active forgetting. This conclusion offers an interesting contrast to the role of the dDA1 receptor in MBn for acquisition, and resolves the issue of how MBn distinguish DA-mediated instructions to acquire memory vs those to forget. Prior studies (Sugamori et al., 1995; Han et al., 1996; Reale et al., 1997) had classified both dDA1 and Damb as cAMP stimulating receptors, similar to mammalian D1/D5 DA receptors that work through Gαs/olf. Our results extend prior studies of dDA1 by examining coupling of this receptor with multiple heterotrimeric G proteins to show that the receptor strongly and preferentially couples to Gs proteins. This affirms the receptor’s role in the acquisition of memory (Kim et al., 2007) consistent with the tight link between acquisition and cAMP signaling (Davis, 2005; Tomchik and Davis, 2009). We found that the Damb receptor can weakly couple to Gs proteins but preferentially engages Gq to trigger Ca2+ signaling pathway, a feature not displayed by dDA1. Comparing the two Gαq paralogs of Drosophila (G and D) with a human ortholog shows that Drosophila GαqG and human Gαq share a conserved C-terminus, essential for selective coupling to GPCRs, but quite distinct in sequence to the GαqD C-terminus (Figure S3). Since GαqD is a photoreceptor selective G protein that couples with rhodopsin (Lee et al., 1990; Lee et al 1994), we propose that GαqG is the isoform that relays Damb’s signals to spur forgetting.
We envision that memory acquisition triggered by strong DA release from electric shock pulses used for aversive conditioning drives both cAMP and Ca2+ signaling through dDA1 and Damb receptors in the MBn (Figure S4). Forgetting occurs from weaker DA release after acquisition through restricted Damb/Gαq/Ca2+ signaling in the MBn. The coupling of Damb to Gs at high DA concentrations also explains why Damb mutants have a slight acquisition defect after training with a large number of shocks (Berry et al., 2012). Although the model allows the assignment of acquisition and forgetting to two distinct intracellular signaling pathways, it does not preclude the possibility that other differences in signaling distinguish acquisition from forgetting. These include the possible use of different presynaptic signals such as a co-neurotransmitter released only during acquisition or forgetting.
Experimental Procedures
Cell culture and transfections.
HEK293T/17 cells were seeded and grown for 4 hr prior to transfection with the appropriate constructs. For the cAMP assays, Drosophila DA receptor, pGloSensor-22F cAMP, and PTX-S1 constructs were used. Transfected cells were detached and transferred to microtitre plates containing the GloSensor cAMP Reagent prepared according to the manufacturer’s directions. After 2 hr at room temperature, luminescence was monitored continuously with a microplate reader. DA in solvent was then applied to cells. For the Ca2+ assays, Drosophila DA receptor, CalfluxVTN, and PTX-S1 constructs were used. PTX-S1, was co-transfected to inhibit the possible coupling of endogenous Gi/o to DA receptors. This ensures that all signal recorded in these assays is generated exclusively by the activation of Gs or Gq. For the assay, transfected cells were harvested and resuspended in PBS with MgCl2 and glucose. The cells were distributed in microplates and the Nluc substrate, furimazine, was added. BRET measurements were made using a microplate reader with the signal calculated by measuring the ratio of the light emitted by the Venus reporter and the Nluc reporter. The average baseline value (basal BRET ratio) recorded prior to agonist stimulation was subtracted from the experimental BRET signal values to obtain the ΔBRET ratio. The largest agonist-induced ΔBRET ratio was plotted as the maximum BRET amplitude. Cellular measurements of BRET between Venus-Gβγ and masGRK3ct-Nluc were performed (Masuho et al., 2015a; Masuho et al., 2015b) with minor modifications to monitor G protein activity in real-time with BRET sensors.
Fly Behavioral Experiments.
We used 6 day-old flies for all behavioral experiments. For conditioning, ~60 flies in a conditioning tube received 30 sec of fresh air, 1 min of an odor paired with electric shock pulses (CS+), 30 sec of air, 1 min of a second odor with no electric shock pulses (CS-), and finally 30 sec of air. 3-octanol (OCT) and benzaldehyde (BEN) as standard odorants in our experiments. Flies were tested in T-mazes in which they were allowed to choose between the CS+ and CS- presented in two different arms of the maze. Memory was quantified by calculating the Performance Index (PI) as the (number of flies in the CS- arm)-(number of flies in the CS+ arm)/(number of flies in the CS- arm)+(number of flies in the CS+ arm). Shock and odor avoidance tests were performed on naïve flies to control for any possible change in odor/shock perception and avoidance. The TARGET system (McGuire et al., 2003) was used to control uas-transgene expression with temperature. This employs constitutive expression of a temperature-sensitive Gal4 inhibitor, Gal80ts. The 30°C temperature destabilizes Gal80ts and allows Gal4 to drive the expression of the uas-transgene present.
Statistical Analyses.
Data were analyzed using Student’s t test, or ANOVA for multiple group comparisons as specified in the Figure legends.
Supplementary Material
Acknowledgements
Research the Davis laboratory was supported by grants 4R37NS019904, 5R01NS052351 and 1R35NS097224 from the NINDS. Research in the Martemyanov laboratory was supported by grants DA036596, DA026405 and MH105482. We thank TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) and Janelia Farms for providing transgenic fly stocks used in this study.
References
- Aso Y, Herb A, Ogueta M, Siwanowicz I, Templier T, Friedrich AB, Ito K, Scholz H and Tanimoto H (2012). Three dopamine pathways induce aversive odor memories with different stability. PLoS Genet. 8, e1002768. doi: 10.1371/journal.pgen.1002768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry JA, Cervantes-Sandoval I, Chakraborty M, and Davis RL (2015). Sleep facilitates memory by blocking dopamine neuron-mediated forgetting. Cell 161, 1656–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry JA, Cervantes-Sandoval I, Nicholas EP, and Davis RL (2012). Dopamine is required for learning and forgetting in Drosophila. Neuron 74, 530–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binkowski BF, Butler BL, Stecha PF, Eggers CT, Otto P, Zimmerman K, Vidugiris G, Wood MG, Encell LP, Fan F, et al. (2011) A luminescent biosensor with increased dynamic range for intracellular cAMP. ACS Chem. Biol 6, 1193–1197. [DOI] [PubMed] [Google Scholar]
- Chan P, Thomas CJ, Sprang SR, and Tall GG (2013). Molecular chaperoning function of Ric-8 is to fold nascent heterotrimeric G protein α subunits. Proc. Natl. Acad. Sci. U S A. 110, 3794–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claridge-Chang A, Roorda RD, Vrontou E, Sjulson L, Li H, Hirsh J, and Miesenbock G (2009). Writing memories with light-addressable reinforcement circuitry. Cell 139, 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL (2005). Olfactory memory formation in Drosophila: From molecular to systems neuroscience. Ann Rev Neurosci 28, 275–302. [DOI] [PubMed] [Google Scholar]
- Davis RL (2011). Traces of Drosophila memory. Neuron 70, 8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL and Zhong Y (2017). The biology of forgetting. Neuron 95, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay M, Pinter ME, Wright FA, Chan P, Murphy AJ, Valenzuela DM, Yancopoulos GD, Tall GG (2011). Ric-8 proteins are molecular chaperones that direct nascent G protein α subunit membrane association. Sci. Signal. 4(200):ra7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han KA, Millar NS, Grotewiel MS, and Davis RL (1996). DAMB, a novel dopamine receptor expressed specifically in Drosophila mushroom bodies. Neuron 6, 1127–1135. [DOI] [PubMed] [Google Scholar]
- Kim YC, Lee H-G, Seong C-S, and Han K-A (2003). Expression of a D1 dopamine receptor dDA1/DmDop1 in the central nervous system of Drosophila melanogaster. Gene Exp Patterns 3, 237–245. [DOI] [PubMed] [Google Scholar]
- Kim YC, Lee HG, and Han KA (2007). D1 dopamine receptor dDA1 is required in the mushroom body neurons for aversive and appetitive learning in Drosophila. J Neurosci. 27, 7640–7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Shah S, Suzuki E, Zars T, ODay PM, and Hyde DR (1994). The Drosophila dgq gene encodes a G alpha protein that mediates phototransduction. Neuron 13, 1143–1157. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, and Davis RL (2003). Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302, 1765–1768. [DOI] [PubMed] [Google Scholar]
- Masuho I, Martemyanov KA, and Lambert NA (2015a). Monitoring G Protein Activation in Cells with BRET. Methods in molecular biology 1335, 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuho I, Ostrovskaya O, Kramer GM, Jones CD, Xie K, and Martemyanov KA (2015b). Distinct profiles of functional discrimination among G proteins determine the actions of G protein-coupled receptors. Science signaling 8, ra123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Liu LP, Binari R, Hardy R, Shims HS, Cavallaro A, Booker M, Pfeiffer BD, Markstein M, Wang H et al. (2009). A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics 182, 1089–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnaparkhi A, Banerjee S, Hasan G. (2002). Altered levels of Gq activity modulate axonal pathfinding in Drosophila. J Neurosci. 22, 4499–4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reale V, Hannan F, Hall LM, Evans PD. (1997). Agonist-specific coupling of a cloned Drosophila melanogaster D1-like dopamine receptor to multiple second messenger pathways by synthetic agonists. J. Neurosci 17, 6545–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroll C, Riemensperger T, Bucher D, Ehmer J, Völler T, Erbguth K, Gerber B, Hendel T, Nagel G, Buchner E and Fiala A (2006). Light-induced activation of distinct modulatory neurons triggers appetitive or aversive learning in Drosophila larvae. Curr. Biol 16, 1741–1747. [DOI] [PubMed] [Google Scholar]
- Schwaerzel M, Monastirioti M, Scholz H, Friggi-Grelin F, Birman S, and Heisenberg M (2003). Dopamine and octopamine differentiate between aversive and appetitive olfactory memories in Drosophila. J Neurosci 23, 10495–10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugamori KS, Demchyshyn LL, McConkey F, Forte MA, and Niznik HB (1995). A primordial dopamine D1-like adenylyl cyclase-linked receptor from Drosophila melanogaster displaying poor affinity for benzazepines. FEBS Lett 362, 131–138. [DOI] [PubMed] [Google Scholar]
- Tomchik SM and Davis RL (2009). Dynamics of learning-related cAMP signaling and stimulus integration in the Drosophila olfactory pathway. Neuron 64, 510–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Dannecker LE, Mercadante AF, and Malnic B (2006). Ric-8B promotes functional expression of odorant receptors. Proc. Natl. Acad. Sci. U S A 103, 9310–9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Cumberbatch D, Centanni S, Shi SQ, Winder D, Webb D, and Johnson CH (2016). Coupling optogenetic stimulation with NanoLuc-based luminescence (BRET) Ca++ sensing. Nat. Comm 7, 13268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


