Abstract
Mitral valve prolapse (MVP) is a common condition that affects 2%–3% of the general population. MVP is thought to include syndromic forms such as Marfan syndrome and non-syndromic MVP, which is the most frequent form. Myxomatous degeneration and fibroelastic deficiency (FED) are regarded as two different forms of non-syndromic MVP. While FED is still considered a degenerative disease associated with ageing, frequent familial clustering has been demonstrated for myxomatous MVP. Familial and genetic studies led to the recognition of reduced penetrance and large phenotypic variability, and to the identification of prodromal or atypical forms as a part of the complex spectrum of the disease. Whereas autosomal dominant mode is the common inheritance pattern, an X linked form of non-syndromic MVP was recognised initially, related to Filamin-A gene, encoding for a cytoskeleton protein involved in mechanotransduction. This identification allowed a comprehensive description of a new subtype of MVP with a unique association of leaflet prolapse and paradoxical restricted motion in diastole. In autosomal dominant forms, three loci have been mapped to chromosomes 16p11-p12, 11p15.4 and 13q31–32. Although deciphering the underlying genetic defects is still a work in progress, DCHS1 mutations have been identified (11p15.4) in typical myxomatous disease, highlighting new molecular pathways and pathophysiological mechanisms leading to the development of MVP. Finally, a large international genome-wide association study demonstrated the implication of frequent variants in MVP development and opened new directions for future research. Hence, this review focuses on phenotypic, genetic and pathophysiological aspects of MVP.
INTRODUCTION
Mitral valve prolapse (MVP) is a common finding, affecting 2%–3% of the general population, defined by an excess leaflet displacement towards the left atrium (LA) in systole,1 with important adverse prognostic consequences.2–5 Whereas normal mitral leaflet position in the left ventricle (LV) contributes to flow orientation towards LV outflow, leaflet retreat into the LA in MVP creates a pseudoaneurysmal LV chamber.6 MVP is encountered in various conditions. In rare syndromic diseases such as Marfan syndrome MVP is one of multiple manifestations involving several organs. In non-syndromic MVP, the most common form, MVP is generally isolated or associated with benign extra-cardiac manifestations.
Genetic defects identification is a major step to improve our understanding of diseases and refine phenotype as well as to decipher molecular pathways with the ultimate goals of tailoring preventive therapeutics. Numerous genetic defects have been identified so far in syndromic form of MVP whereas genetic analysis of non-syndromic forms remains a work in progress. Hence, this review will focus on the current phenotypic and pathophysiological knowledge in relation to genetic defects leading to syndromic and non-syndromic MVP.
MVP PHENOTYPE
The diagnosis of MVP is classically defined by a displacement of at least 2 mm above the mitral annulus line of one or two leaflets in the parasternal long axis (PSLA) view by echocardiography.1 The PSLA view explores the middle segments of anterior and posterior leaflets (A2-P2) and the medial (A3-P3) and lateral (A1-P1) segments of both leaflets as well as the two commissures by tilting either internally or externally the probe. Additional views such as apical chamber views can facilitate the localisation of prolapse. Specificity of diagnosis, previously strongly limited by M-mode methods and assumptions about annular non-planarity, was strongly improved based on three-dimensional understanding of the saddle shape of the valve, providing a firmer clinical diagnosis and pheno-type for genetic studies.7–9 Whereas normal leaflet length averages 20–23 mm for the anterior leaflet and 12–13 mm for the posterior, leaflets are generally elongated and redundant in MVP. Leaflets are also frequently thickened over the normal range of 1–3 mm, and the mitral annulus is enlarged (normal size 28–30 mm).1,7 Prodromal forms of MVP have been recently evidenced suggesting that MVP encompasses a more complex phenotypic expression than previously thought. Minimal systolic displacement is a displacement <2 mm towards the LA, and the abnormal anterior coaptation is char-acterised by an anterior displacement of the coaptation point (>40%).10 Both MVP and prodromal forms in parents predict the development of MVP in offsprings.11,12 Other atypical manifestations have been highlighted in Filamin-A (FLNA)-MVP such as a restrictive motion in diastole.13 Finally, mitral regurgitation (MR) degree varies from trivial to severe according to the failure of coaptation driven essentially by chordal elongation, chordal rupture or annular enlargement.
MVP is a heterogeneous group of disorders, and is one of the most frequent indications for valvular surgery.1 Clinical, surgical and histological findings10,14 strongly suggest that non-syndromic MVP includes various mitral valve (MV) apparatus phenotypes. Non-syndromic MVP is regarded as a degenerative disease related to either progressive myxomatous degeneration or fibroelastic loss.14–16 The relevance of such stratification remains controversial in view of (1) familial expression variability and (2) the continuum from typical fibroelastic deficiency (FED) disease to typical myxomatous MVP.15 However, due to the absence of gene identification until recently, the pattern of non-syndromic MVP/dystrophy has been poorly refined. On the one hand, comprehensive phenotype assessment of MVP might allow subtypes classification and accelerate genetic discoveries. On the other hand, genetic defects identification will allow a refinement of MVP phenotype, including prodromal lesions.10–12,17
Based on our current knowledge, non-syndromic MVP can be roughly stratified at least in three different major subphenotypes (figure 1).13,15,17–19 First, myxomatous MVP is characterised by bileaflet elongation, thickening and prolapse, mitral annulus dilatation and frequent mitral annular disjunction. MR can occur as a consequence of mitral annular dilatation, leaflet elongation or rupture. Myxomatous degeneration of the spongiosa is the main histological finding. Second, in FED, patients are typically felt to be older; leaflets are thin, translucent, of normal length; and the mitral annulus is mildly dilated. MR occurs as a consequence of chordal rupture, and only the prolapsing or flail segment is thickened and moderately elongated. Histology demonstrates a deficiency in collagen, elastin and proteoglycans. FED is still regarded as a pure degenerative process with remodelling of the prolapsing segment as a consequence of MR. Third, FLNA-MVP or dystrophy, an X linked disease, is characterised by a severe phenotype in male patients and a milder expression in female patients, with frequent polyvalvular involvement, mitral leaflet thickening and elongation. Chordal rupture and mitral annular disjunction have not been described in this subtype of MVP. Myxomatous degeneration has also been reported in FLNA-MVP.13
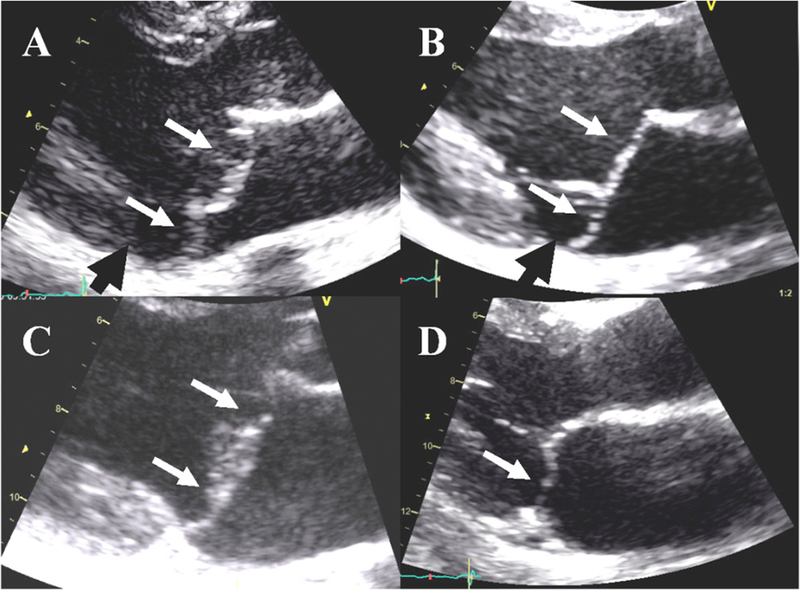
Figure 1.
Mitral valve prolapse (MVP) with no or moderate regurgitation in patients with (A) Marfan syndrome, (B) myxomatous disease, (C) Filamin-A disease and (D) fibroelastic deficiency (FED). Of note, the echocardiographic presentation of MVP is often similar in Marfan syndrome and myxomatous disease, and Filamin-A MVP can be confused with myxomatous MVP. In addition, myxomatous degeneration of mitral leaflets has been described in Marfan syndrome, myxomatous disease and Filamin-A disease, whereas the histological aspect is quite different in FED. White arrows: prolapse/billowing, black head arrows: mitral annular disjunction.
MVP GENETICS
Since MVP is a frequent disease in the general population, the underlying genetic architecture is predicted to follow the common-variant common-disease hypothesis. However, as for most common diseases, a subset of MVP cases cluster within families following a Mendelian inheritance pattern, with rare variants having strong effects leading to disease. In this context, classical linkage analysis has first allowed the mapping of MVP-candidate regions in the genome. More recently, the advent of high-throughput genomics technologies has allowed genome-wide association studies (GWAS) addressing genetic predisposition in the general population. In addition, deep sequencing of all coding sequences, by whole-exome and whole-genome sequencing, is becoming the golden standard for gene discovery and potentially clinical molecular diagnostics. The application of these technologies has facilitated the identification of several key genes associated with MVP including Marfan syndrome.
SYNDROMIC FORMS OF MVP
Syndromic forms of MVP have been linked to genetic defects summarised in table 1.
Table 1.
Genetic anomalies associated with mitral valve prolapse in humans
| Gene or chromosome | Defect localisation/mechanism | |
|---|---|---|
| 1. Rare causal genetic defects | ||
| Syndromic MVP | ||
| Trisomies 18,13,15 | Chr 18, 13, 15 | - |
| Marfan syndrome | FBN1-TGFβR2-TGFβ2 | ECM/TGFβ pathway |
| Loeys-Dietz | TGFβR1-TGFβR2 | TGFβ pathway |
| Juvenile polyposis syndrome | SMAD4-BMPR1A | TGFβ pathway |
| Aneurysm-osteoarthritis syndrome | SMAD3 | TGFβ pathway |
| Ehlers-Danlos syndrome | Collagen types I, III, V, XI and tenascin | ECM |
| Osteogenesis imperfecta | Collagen type I | ECM |
| Williams-Beuren syndrome | Elastin | ECM |
| Pseudoxanthoma elasticum | MRP6 (ABCC6) | ECM |
| BDCS or FTH syndromes | SH3PXD2B | Podosomes/cell migration |
| Larsen-like syndrome | B3GAT3 | ECM/glycosaminoglycans |
| Syndrome with sinus node dysfunction, arrhythmias, LVNC | HCN4 | Ionic channel/heart development |
| Non-syndromic MVP | ||
| Filamin A-MVD | FLNA | Mechanotransduction |
| Myxomatous disease (MMVP2) | DCHS1 | Cell migration and polarity |
| Myxomatous disease (MMVP1–3) | Chr 16–13 | - |
| 2. Frequent variants | ||
| GWAS | LMCD1 | Transcription factor |
| TNS1 | Focal adhesion protein/cytoskeleton organisation | |
BDCS, Borrone dermato-cardio-skeletal; ECM, extracellular matrix; FTH, Frank-Ter Haar; GWAS, genome-wide association study; LVNC, left ventricular non-compaction; MVP, mitral valve prolapse; TGF, transforming growth factor.
Aneuploidy
Aneuploidy syndromes such as trisomies 18, 13 and 15 comprise multiple cardiac malformations including valve alterations that can share some features with Marfan disease and non-syndromic MVP. For instance, polyvalvular disease is frequent in trisomy-18, affects both atrioventricular and semilunar valves, and is characterised by redundant and thick myxomatous leaflets, long chordae tendineae, and hypoplastic or absence of papillary muscles.20
Marfan syndrome
Fibrilline-1 (FBN1) mutations21 lead to type 1 Marfan syndrome, its most common form. In Marfan syndrome, MVP is frequent and can be the first finding leading to the diagnosis. Penetrance and prevalence of MVP increase over time from 43% at 30 years to 77% at 60 years.22 Surgery for MR only is uncommon, carried out in 13% of patients with Marfan syndrome at 60 years.22 Some mutations of transforming growth factor-β2 (TGFβ2) and TGFβ receptor-2 (TGFβR2) genes have also been associated with Marfan syndrome.
Loeys-Dietz syndrome
Mutations in TGFβR1 and TGFβR2 are involved in Loeys-Dietz syndrome.23 In a recent study, MV involvement was less frequent in TGFβR2 compared with FBN1 patients: MVP was found in 21% vs 45% of patients (P=0.001), MR in 35% vs 56% (P<0.0001) and referral to MV surgery was rare in TGFβR2 patients.24
Juvenile polyposis syndrome
Mutations in SMAD-4 (mothers against decapentaplegic homo-logue 4 or SMAD family member-4) and BMPR1A (bone morphogenetic protein receptor, type 1A) genes involved in the TGFβ pathway elicit juvenile polyposis syndrome. MVP has been reported in a family harbouring an SMAD-4 gene mutation.25
Aneurysm-osteoarthritis syndrome
Mutations in SMAD-3 (SMAD family member-3) gene have been identified as the leading cause of aneurysm-osteoarthritis syndrome. In a small series of 44 patients, up to 50% of mutated patients expressed MV alterations (prolapse 14%, billowing 14%, moderate or severe regurgitation 14%), suggesting that MVP is a part of this syndrome.26 FBN1, TGFβR1, TGFβR 2 and SMAD-4 genes are all involved in the TGFβ signalling network.
Williams-Beuren syndrome
Williams-Beuren syndrome has been related to elastine gene (ELN) mutations. In a series of 150 patients with Williams-Beuren syndrome, MVP was found in 6.2% but frequently associated with supravalvular aortic stenosis or pulmonary stenosis.27
Ehlers-Danlos syndrome
Ehlers-Danlos syndrome is related to mutations in collagen type I (COL-1), III, V, XI and tenascin genes. Mild MVP was found in up to 6% of 252 children but mild to moderate MVP in only 0.4% leaving doubt about the strength of the genetic link between MVP and Ehlers-Danlos syndrome.28
Pseudoxanthoma elasticum
Pseudoxanthoma elasticum, a disorder associated with MRP6 (multidrug resistance-associated protein 6, also known as the ABCC6) mutations, is classically considered a cause of MVP. However, evidence is limited for a genetic association, and a recent report found only 3 (4.5%) MVP out of 67 patients.29
Osteogenesis imperfecta
MVP has been occasionally reported in osteogenesis imperfecta, heterogeneous group of genetic diseases characterised mainly by recurrent bone fractures related to impaired collagen synthesis. However, the link between both manifestations has not been clearly established and remains uncertain.
Larsen-like syndrome
In this syndrome, heart defects including MVP are associated with joint dislocations resulting from altered initiation of proteoglycan synthesis.30
Borrone dermato-cardio-skeletal and Frank-Ter Haar syndromes
Both syndromes are autosomal-recessive disorders characterised by cutaneous, cardiovascular (with MVP), skeletal and other abnormalities owing to homozygote mutations in the SH3PXD2B gene (encoding the protein SH3 and PX domains 2B). Interestingly, knockout mice recapitulate the human pheno-type (tables 1 and 2).
Table 2.
Genetically engineered mice model presenting mitral valve defects
| Gene | Protein | Murin valve phenotype |
|---|---|---|
| ADAM17 | ADAM metallopeptidase domain 17 | Abnormal mitral valve morphology |
| ADAM19 | ADAM metallopeptidase domain 19 | Abnormal mitral valve morphology |
| ARSB | Arylsulfatase B | Abnormal mitral valve morphology |
| Mitral valve regurgitation | ||
| BMPR1A | Bone morphogenetic protein receptor type-1 A | Mitral valve regurgitation |
| Abnormal mitral valve morphology | ||
| CREBbp | CREB-binding protein | Abnormal mitral valve morphology |
| CXCR4 | Chemokine (C-X-C motif) receptor 4 | Thickened mitral valve |
| CYR61 | Cysteine-rich, angiogenic inducer, 61 | Abnormal mitral valve morphology |
| DNAH11 | Dynein, axonemal, heavy chain 11 | Abnormal mitral valve morphology |
| D0CK1 | Dedicator of cytokinesis 1 | Thickened mitral valve |
| EFNA1 | Ephrine A | Thickened mitral valve |
| EFNB2 | Ephrine-2B | Thickened mitral valve |
| EHMT1 | Euchromatic histone-lysine N-methyltransferase 1 | Abnormal mitral valve morphology |
| GJA5 | Connexine 40 | Mitral valve stenosis |
| HBEGf | Heparin-binding EGF-like growth factor | Abnormal mitral valve morphology |
| HEY2 | Hairy/enhancer-of-split related with YRPW motif protein 2 | Abnormal mitral valve morphology |
| Mitral valve regurgitation | ||
| Thickened mitral valve | ||
| IDUA | Iduronidase alpha L | Abnormal mitral valve morphology |
| Mitral valve regurgitation | ||
| MEGF8 | Multiple EGF-like-domains 8 | Abnormal mitral valve morphology |
| NF1 | Neurofibromin | Abnormal mitral valve morphology |
| NFATC1 | Nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent 1 | Abnormal mitral valve morphology |
| NTF3 | Neurotrophine 3 | Thickened mitral valve |
| NTRK3 | Neurotrophic tyrosine kinase, receptor, type 3 | Abnormal mitral valve morphology |
| PDLIM7 | PDZ and LIM domain 7 | Abnormal mitral valve morphology |
| Abnormal mitral valve annulus | ||
| RXRA | Retinoid X receptor, alpha | Abnormal mitral valve morphology |
| Mitral valve stenosis | ||
| Mitral valve atresia | ||
| SH3PXD2B | SH3 and PX domains 2B (SH3PXD2B) | Abnormal mitral valve morphology |
| Mitral valve prolapse | ||
| SMAD6 | SMAD family member 6 | Thickened mitral valve |
| SOX9 | SRY (sex determining region Y)-box 9 | Abnormal mitral valve morphology |
| Calcified mitral valve | ||
| TLL1 | Tolloid-like 1 | Abnormal mitral valve morphology |
HCN4 mutation
By contrast to the above syndromes, the link between mutations of HCN4 (hyperpolarisation-activated cyclic nucleotide channel 4) and MVP, recently reported, is more intriguing. The pathology associates sinus node dysfunction/bradycardia, ventricular and atrial arrhythmias, LV non-compaction and MVP.31,32 It is noteworthy that HCN4 is involved in early embryonic heart development, helping to form myocardium and the conduction system. It has been hypothesised that the HCN4 loss of function might interfere with molecular mechanisms leading to MVP.
GENETICS OF NON-SYNDROMIC MVP
Apart from syndromic forms of MVP, familial clustering of MVP with an X linked inheritance pattern was first suggested in 1969 by Monteleone and Fagan.33 A large family from the western part of France was eventually identified with the same mode of transmission.17,34 In the early 1980s, familial studies demonstrated that MVP was inherited in >60% of cases, irrespective of clinical presentation in the proband. Other important information was the age-dependent and sex-dependent expression, the reduced penetrance and the high phenotypic variability within the same family.35 The transmission mode was autosomal dominant and authors concluded that MVP was the most common Mendelian cardiovascular abnormality in humans.35 Later, linkage analysis in large MVP families identified three loci in chromosomes 16p11.2-p12.1 (MMVP1), 11p15.4 (MMVP2) and 13q31.3–31.2 (MMVP3).10,36,37
In 1998, the X linked MVP inheritance17,34 was genetically mapped to chromosome Xq28 (XMVD). A standard positional cloning approach identified a P637Q mutation in the FLNA gene in all affected members. The causal relation of FLNA to XMVD was demonstrated by the identification of three other, smaller, unrelated families affected by FLNA-MVP. The penetrance of the disease was complete in men and incomplete in women. Additional FLNA mutations have recently been identified in families with an X linked form of MVP/dystrophy.13 FLNA-MVP pheno-type is now clearly characterised and comprises both congenital and degenerative alterations of the MV apparatus and frequent polyvalvular involvement in men. Furthermore, FLNA-MVP associates myxomatous-type MVP and a paradoxical restricted motion in diastole, a unique feature in MVP diseases.
In 2015, a targeted resequencing approach allowed Durst et al to identify the MMVP2 disease-causing gene. They identified a loss-of-function mutation in the DCHS1 (Dachsous1) gene (R2513H), which encodes a member of the cadherin superfamily and is important for planar cell polarity (PCP), in the initial MVP pedigree.38 Further genetic studies identified two additional deleterious DCHS1 mutations (R2513H and R2330C). DCHS1 mutations reduce protein stability. Dchs1(+/−) mice develop prolapse of thickened mitral leaflets, which could be traced back to developmental errors in valve morphogenesis, with disorganised MV interstitial cells (MVIC). DCHS1 deficiency in MVICs of patients with MVP or mouse causes altered migration and cellular patterning, supporting these processes as aetiological underpinnings for the disease.
GENETICS OF ISOLATED MVP: COMMON RISK ALLELES FOR MVP
In 2015, Dina et al conducted a GWAS including 1412 isolated MVP cases and 2439 controls.39 Three loci showed genome-wide significant associations with MVP, respectively, at 2q35, 17p13 and 22q12. By replicating association signals among 2864 cases and 9218 controls, three additional loci significantly associated with MVP were identified. Among the genes located within the associated haplotypes, they identified LMCD1 (LIM and cysteine domain transcription factor), which encodes a transcription factor involved in atrioventricular valve defect with regurgitation, as shown by morpholino knock-down in zebrafish. A similar phenotype was obtained in zebrafish when knocking down the TNS1 (Tensin1) gene encoding the focal adhesion protein tensin1. TNS1 and LMCD1 are both implicated in cell proliferation and migration and contribute to MVP, possibly during valve development. This study identified the first risk loci for MVP and suggests new mechanisms involved in MVP regurgitation development.
ANIMAL MODELS AND MVP
Canine myxomatous MVP is the most common heart disease in dogs, accounting for approximately 75% of all dogs with heart disease and an important cause of heart failure and mortality. The disease has many similarities with and is considered a potential model for human MVP. Myxomatous MV disease is most prevalent in small to medium-sized dog breeds. In King Charles cavalier spaniels, a GWAS identified loci associated with development of MVP in a 1.58 Mb region on CFA13 and a 1.68 Mb region on CFA14.40 More recently, a similar association study on whippet dogs identified new severity loci for MV degeneration on CFA15 and secondary loci of suggestive association on CFA2. Positional candidate genes were identified within the primary and secondary loci, including follistatin-related protein-5 precursor (FSTL5) and Rho GTPase-activating protein 26 (ARHGAP26). These results support the hypothesis that MVP in whippets has a genetic basis.41
Genetically engineered mice mimicking syndromic forms of MVP have been reported for FBN1,42 PTPN11, ADAMTS12, TGβR1, TGβR2 and SMAD-4, and also for non-syndromic MVP such as FLNA,43,44 DCHS1 and TNS1.39 Furthermore, structural or functional abnormalities of the MV have been described in a series of genetically engineered mice models (http://www.mouse-mine.org/mousemine/begin.do) (table 2).
PATHOPHYSIOLOGY AND MOLECULAR PATHWAYS
Although it is well established that the degenerative process of leaflets typically involves accumulation of myxoid extracellular matrix (ECM), and fragmentation of collagen and elastin associated with ‘activated’ myofibroblastic interstitial cells, the specific signalling pathways affected and the molecular players involved remain elusive.
The early identification of mutations in the gene encoding the structural ECM protein fibrillin1, eliciting Marfan syndrome, pointed to the critical role of TGF-β signalling pathway in MVP. Fibrillin1 sequesters large latent TGF-β binding complex which finely controls the level of TGF-β activity in the ECM.45 Elegant studies from Dietz’s lab revealed that mice expressing mutant Fibrillin1 have longer and thicker valves than wild-type animals. Importantly, these valvular defects were prevented by TGF-β neutralising antibodies.42 Increased TGF-β signalling was also demonstrated in MVP associated with Loeys-Dietz syndrome despite the loss of function mutations in TGF-β receptors (TGFβR1 and TGFβR2) identified in those patients.23 The mechanisms underlying this paradoxical effect remain unknown. The intense haemodynamic and mechanical stresses the valves endure could modulate TGF-β signalling and potentially the mutation effect.
Besides the genes involved in syndromic forms of MVP, mutations in FLNA and DCHS1 genes were identified in familial non-syndromic MVP. Here again, the impact of haemodynamic and mechanical stresses on the function of mutant FLNA and Dachsous proteins may constitute the unifying feature explaining why patients specifically develop MVP despite the widespread expression of the mutant proteins in other organs. Furthermore, FLNA is known to stabilise the cellular F-actin network and to constitute a scaffolding hub that orchestrates many signalling pathways including mechanotransduction (figure 2).46 Interestingly, although other FLNA mutations were associated with a wide spectrum of congenital anomalies, MVP-causing mutations are all clustered in a specific structural module located in the N-terminal region of the protein. These mutations alter the balance between the activities of two master pieces of actomyosin complex regulation and mechanotransduction pathway, the small GTPases RhoA and Rac1, and destabilise FLNA interactions with the tyrosine phosphatase PTPN12 (PTPPEST). Importantly, PTPN12 substrates include mechanotransduction and integrin signalling pathway actors such as Src kinase, p130Cas and paxillin, as well as small GTPase regulators.46–49
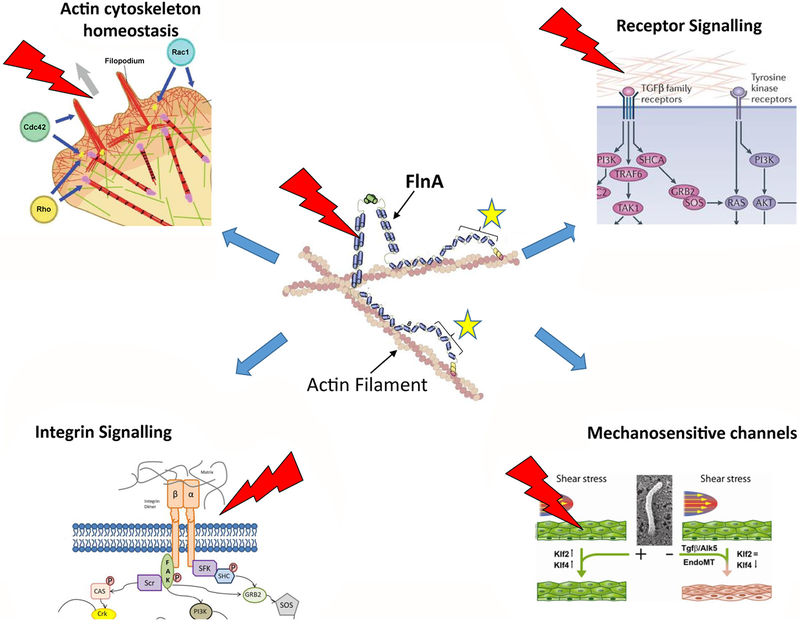
Figure 2.
Filamin A: a hub protein involved in mechanotransduction pathways. Dimers of Filamin A protein organise the filamentous actin network and participate, through their interactions with >80 binding partners, to many signalling pathways including: transforming growth factor (TGF)-β and tyrosine kinase receptors, integrin macromolecular complex, mechanosensitive channels (Piezzo) and actin/actomyosin homeostasis. All these signalling pathways are modulated by mechanical stress symbolised by red marks. The location of mitral valve prolapse (MVP)-associated Filamin-A mutations is indicated by yellow stars.
Recently, the second MVP gene (DCHS1) was identified. Dachsous is a key player of the PCP pathway, which coordinates through cytoskeletal rearrangements and cell migration, the polarisation of cells in the tissue plane, and has a fundamental role in morphogenesis. Notably, Dachsous polarises the myosin Dachs in cell-cell junctions, which in turn promotes the anisotropy of junction tension which shapes the epithelia drosophila.50 MVP-associated mutations were shown to decrease the stability of this protocadherin and alter cell migration while DCHS1-deficient mice (Dchs1+/−) exhibited thickened mitral leaflets which could be traced back to defaults during embryonic valvulogenesis.38
Although MVP is generally recognised as a degenerative disease worsening with ageing, developmental defects were also identified in Filamin-A-deficient (FlnA−/−) mice and in zebrafish knocked down for DCHS1 as well as the TNS1 and LMCD1 genes.38,39,44 Knock-in animal and/or in vitro patient- induced pluripotent stem cells (IPS)-derived valvular cellular models are thus now required to assure these observations do not result from pleiotropic effects due to the complete absence of the proteins and to identify the specific molecular mechanisms that link TGF-β and ECM homeostasis signalling pathways with mechanotransduction and organogenesis dysfunctions involved in MV diseases.
PERSPECTIVES
Genetic studies are just starting in non-syndromic MVP but have already changed the general understanding of the disease. Although the few genes identified explain a small proportion of non-syndromic myxomatous MVP, this disease is now recognised as a genetic, inherited condition, but with various phenotypic expressions. Thanks to genetics, new phenotypes are emerging such as FLNA-MVP, and new pathophysiological hypotheses based on specific molecular pathways such as mechanotransduction, TGF-β or PCP alterations are highlighted. Although the ultimate goal is tailoring preventive therapeutics, deciphering underlying genetic and molecular defects of MVP is still a work in progress. Genetic and phenotypic knowledge progress would allow a personalised management of patient with MVP in a near future. In addition, targeting the above-mentioned molecular pathways by a specific drug or a genetic approach, directing progenitor cells towards valvular leaflets or developing tissue-engineered heart valves are potential future therapeutics of MVP.
Acknowledgments
Funding Leducq Foundation Transatlantic Network of Excellence in Mitral Valve Disease (2008–2013, Paris, France); Fondation GenaVie (TLT, 2010, Nantes, France); Fédération Française de Cardiologie (TLT, 2011 and 2014, Paris, France); Fondation Coeur & Recherche (TLT, 2013, Paris, France); French Ministry of Health ‘PHRC National 2007’ (VP, HLM, no 20 17); and ‘PHRC I 2012’ (TLT, API12/N/019, Paris, France). NIH (RAL): HL109506, HL128099 and R01 HL141917. Ellison Foundation (RAL, Boston, MA). Inserm Translational Research Grant (2012–2016, Paris, France) to TLT.
Footnotes
Competing interests None declared.
Provenance and peer review Commissioned; externally peer reviewed.
REFERENCES
- 1.Levine RA, Hagége AA, Judge DP, et al. Mitral valve disease-morphology and mechanisms. Nat Rev Cardiol 2015;12:689–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tribouilloy C, Rusinaru D, Grigioni F, et al. Long-term mortality associated with left ventricular dysfunction in mitral regurgitation due to flail leaflets: a multicenter analysis. Circ Cardiovasc Imaging 2014;7:363–70. [DOI] [PubMed] [Google Scholar]
- 3.Le Tourneau T, Richardson M, Juthier F, et al. Echocardiography predictors and prognostic value of pulmonary artery systolic pressure in chronic organic mitral regurgitation. Heart 2010;96:1311–7. [DOI] [PubMed] [Google Scholar]
- 4.Le Tourneau T, Messika-Zeitoun D, Russo A, et al. Impact of left atrial volume on clinical outcome in organic mitral regurgitation. J Am Coll Cardiol 2010;56:570–8. [DOI] [PubMed] [Google Scholar]
- 5.Enriquez-Sarano M, Avierinos JF, Messika-Zeitoun D, et al. Quantitative determinants of the outcome of asymptomatic mitral regurgitation. N Engl J Med 2005;352:875–83. [DOI] [PubMed] [Google Scholar]
- 6.Barlow JB, Bosman CK. Aneurysmal protrusion of the posterior leaflet of the mitral valve. An auscultatory-electrocardiographic syndrome. Am Heart J 1966;71:166–78. [DOI] [PubMed] [Google Scholar]
- 7.Freed LA, Levy D, Levine RA, et al. Prevalence and clinical outcome of mitral-valve prolapse. N Engl J Med 1999;341:1–7. [DOI] [PubMed] [Google Scholar]
- 8.Levine RA, Handschumacher MD, Sanfilippo AJ, et al. Three-dimensional echocardiographic reconstruction of the mitral valve, with implications for the diagnosis of mitral valve prolapse. Circulation 1989;80:589–98. [DOI] [PubMed] [Google Scholar]
- 9.Levine RA, Triulzi MO, Harrigan P, et al. The relationship of mitral annular shape to the diagnosis of mitral valve prolapse. Circulation 1987;75:756–67. [DOI] [PubMed] [Google Scholar]
- 10.Nesta F, Leyne M, Yosefy C, et al. New locus for autosomal dominant mitral valve prolapse on chromosome 13: clinical insights from genetic studies. Circulation 2005;112:2022–30. [DOI] [PubMed] [Google Scholar]
- 11.Delling FN, Rong J, Larson MG, et al. Familial clustering of mitral valve prolapse in the community. Circulation 2015;131:263–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delling FN, Rong J, Larson MG, et al. Evolution of mitral valve prolapse: insights from the framingham heart study. Circulation 2016;133:1688–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Tourneau T, Le Scouarnec S, Cueff C, et al. New insights into mitral valve dystrophy: a Filamin-A genotype-phenotype and outcome study. Eur Heart J 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fornes P, Heudes D, Fuzellier JF, et al. Correlation between clinical and histologic patterns of degenerative mitral valve insufficiency: a histomorphometric study of 130 excised segments. Cardiovasc Pathol 1999;8:81–92. [DOI] [PubMed] [Google Scholar]
- 15.Adams DH, Rosenhek R, Falk V. Degenerative mitral valve regurgitation: best practice revolution. Eur Heart J 2010;31:1958–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anyanwu AC, Adams DH. Etiologic classification of degenerative mitral valve disease: Barlow’s disease and fibroelastic deficiency. Semin Thorac Cardiovasc Surg 2007;19:90–6. [DOI] [PubMed] [Google Scholar]
- 17.Trochu JN, Kyndt F, Schott JJ, et al. Clinical characteristics of a familial inherited myxomatous valvular dystrophy mapped to Xq28. J Am Coll Cardiol 2000;35:1890–7. [DOI] [PubMed] [Google Scholar]
- 18.Kyndt F, Gueffet JP, Probst V, et al. Mutations in the gene encoding filamin A as a cause for familial cardiac valvular dystrophy. Circulation 2007;115:40–9. [DOI] [PubMed] [Google Scholar]
- 19.Lardeux A, Kyndt F, Lecointe S, et al. Filamin-a-related myxomatous mitral valve dystrophy: genetic, echocardiographic and functional aspects. J Cardiovasc Transl Res 2011;4:748–56. [DOI] [PubMed] [Google Scholar]
- 20.Van Praagh S, Truman T, Firpo A, et al. Cardiac malformations in trisomy-18: a study of 41 postmortem cases. J Am Coll Cardiol 1989;13:1586–97. [DOI] [PubMed] [Google Scholar]
- 21.Lee B, Godfrey M, Vitale E, et al. Linkage of Marfan syndrome and a phenotypically related disorder to two different fibrillin genes. Nature 1991;352:330–4. [DOI] [PubMed] [Google Scholar]
- 22.Détaint D, Faivre L, Collod-Beroud G, et al. Cardiovascular manifestations in men and women carrying a FBN1 mutation. Eur Heart J 2010;31:2223–9. [DOI] [PubMed] [Google Scholar]
- 23.Loeys BL, Chen J, Neptune ER, et al. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFβR1 or TGFβR2. Nat Genet 2005;37:275–81. [DOI] [PubMed] [Google Scholar]
- 24.Attias D, Stheneur C, Roy C, et al. Comparison of clinical presentations and outcomes between patients with TGFβR2 and FBN1 mutations in Marfan syndrome and related disorders. Circulation 2009;120:2541–9. [DOI] [PubMed] [Google Scholar]
- 25.Andrabi S, Bekheirnia MR, Robbins-Furman P, et al. SMAD4 mutation segregating in a family with juvenile polyposis, aortopathy, and mitral valve dysfunction. Am J Med Genet A 2011;155A:1165–9. [DOI] [PubMed] [Google Scholar]
- 26.van der Linde D, van de Laar IM, Bertoli-Avella AM, et al. Aggressive cardiovascular phenotype of aneurysms-osteoarthritis syndrome caused by pathogenic SMAD3 variants. J Am Coll Cardiol 2012;60:397–403. [DOI] [PubMed] [Google Scholar]
- 27.Del Pasqua A, Rinelli G, Toscano A, et al. New findings concerning cardiovascular manifestations emerging from long-term follow-up of 150 patients with the Williams-Beuren-Beuren syndrome. Cardiol Young 2009;19:563–7. [DOI] [PubMed] [Google Scholar]
- 28.Atzinger CL, Meyer RA, Khoury PR, et al. Cross-sectional and longitudinal assessment of aortic root dilation and valvular anomalies in hypermobile and classic Ehlers-Danlos syndrome. J Pediatr 2011;158:826–30. [DOI] [PubMed] [Google Scholar]
- 29.Prunier F, Terrien G, Le Corre Y, et al. Pseudoxanthoma elasticum: cardiac findings in patients and Abcc6-deficient mouse model. PLoS One 2013;8:e68700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baasanjav S, Al-Gazali L, Hashiguchi T, et al. Faulty initiation of proteoglycan synthesis causes cardiac and joint defects. Am J Hum Genet 2011;89:15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schweizer PA, Schröter J, Greiner S, et al. The symptom complex of familial sinus node dysfunction and myocardial noncompaction is associated with mutations in the HCN4 channel. J Am Coll Cardiol 2014;64:757–67. [DOI] [PubMed] [Google Scholar]
- 32.Milano A, Vermeer AM, Lodder EM, et al. HCN4 mutations in multiple families with bradycardia and left ventricular noncompaction cardiomyopathy. J Am Coll Cardiol 2014;64:745–56. [DOI] [PubMed] [Google Scholar]
- 33.Monteleone PL, Fagan LF. Possible X-linked congenital heart disease. Circulation 1969;39:611–4. [DOI] [PubMed] [Google Scholar]
- 34.Kyndt F, Schott JJ, Trochu JN, et al. Mapping of X-linked myxomatous valvular dystrophy to chromosome Xq28. Am J Hum Genet 1998;62:627–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Devereux RB, Brown WT, Kramer-Fox R, et al. Inheritance of mitral valve prolapse: effect of age and sex on gene expression. Ann Intern Med 1982;97:826–32. [DOI] [PubMed] [Google Scholar]
- 36.Freed LA, Acierno JS, Dai D, et al. A locus for autosomal dominant mitral valve prolapse on chromosome 11p15.4. Am J Hum Genet 2003;72:1551–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Disse S, Abergel E, Berrebi A, et al. Mapping of a first locus for autosomal dominant myxomatous mitral-valve prolapse to chromosome 16p11.2-p12.1. Am J Hum Genet 1999;65:1242–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Durst R, Sauls K, Peal DS, et al. Mutations in DCHS1 cause mitral valve prolapse. Nature 2015;525:109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dina C, Bouatia-Naji N, Tucker N, et al. Genetic association analyseshighlight biological pathways underlying mitral valve prolapse. Nat Genet 2015;47:1206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madsen MB, Olsen LH, Häggström J, et al. Identification of 2 loci associated with development of myxomatous mitral valve disease in Cavalier King Charles Spaniels. J Hered 2011;102(Suppl 1):S62–7. [DOI] [PubMed] [Google Scholar]
- 41.Stern JA, Hsue W, Song KH, et al. Severity of mitral valve degeneration is associated with chromosome 15 loci in whippet dogs. PLoS One 2015;10:e0141234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ng CM, Cheng A, Myers LA, et al. TGF-beta-dependent pathogenesis of mitral valve prolapse in a mouse model of Marfan syndrome. J Clin Invest 2004;114:1586–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norris RA, Moreno-Rodriguez R, Wessels A, et al. Expression of the familial cardiac valvular dystrophy gene, filamin-A, during heart morphogenesis. Dev Dyn 2010;239:2118–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sauls K, de Vlaming A, Harris BS, et al. Developmental basis for filamin-A-associated myxomatous mitral valve disease. Cardiovasc Res 2012;96:109–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Annes JP, Munger JS, Rifkin DB. Making sense of latent TGFbeta activation. J Cell Sci 2003;116:217–24. [DOI] [PubMed] [Google Scholar]
- 46.Ehrlicher AJ, Nakamura F, Hartwig JH, et al. Mechanical strain in actin networks regulates FilGAP and integrin binding to filamin A. Nature 2011;478:260–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohta Y, Hartwig JH, Stossel TP. FilGAP, a Rho- and ROCK-regulated GAP for Rac binds filamin A to control actin remodelling. Nat Cell Biol 2006;8:803–14. [DOI] [PubMed] [Google Scholar]
- 48.Duval D, Labbé P, Bureau L, et al. MVP-Associated Filamin A Mutations Affect FlnAPTPN12 (PTP-PEST) Interactions. J Cardiovasc Dev Dis 2015;2:233–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duval D, Lardeux A, Le Tourneau T, et al. Valvular dystrophy associated filamin A mutations reveal a new role of its first repeats in small-GTPase regulation. Biochim Biophys Acta 2014;1843:234–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bosveld F, Bonnet I, Guirao B, et al. Mechanical control of morphogenesis by Fat/Dachsous/Four-jointed planar cell polarity pathway. Science 2012;336:724–7. [DOI] [PubMed] [Google Scholar]