Abstract
Previously (Glushakova et al. 2017), a cellulose-based cationic (Q) paper derivatized with quaternary ammonium groups was shown to be a convenient platform to collect, preserve, and store nucleic acids (NAs) derived from mosquito vectors infected with pathogens for surveillance. NAs bind electrostatically to Q-paper, but the quantity of NA bound depends on the paper’s binding capacity. To optimize the original technology for mosquito surveillance, factors that affected NA absorbance on Q-paper were evaluated. Sixteen variations of Q-paper were prepared with modifications of the derivatizing reagents and derivatization temperature. The binding capacities of these variations were determined first with 1,3,5-benzenetricarboxylic (BTCA), then viral RNA (purified or in infected mosquito samples) was used for validation. For this, samples with Zika (ZIKV) and chikungunya (CHIKV) RNA or virus-infected Aedes aegypti mosquito bodies were applied to sixteen Q-paper variants. Washing the paper samples with water versus elution with aqueous salt (1 M) gave samples that were analyzed for viral RNA by a PCR-based direct Luminex hybridization assay. The comparison ranked the Q-paper binding capacities from the lowest to the highest. The Q-paper with the highest RNA binding capability was further validated with ZIKV- and CHIKV-infected mosquito saliva.
Keywords: cationic (Q) paper, arboviruses, mosquito
1. Introduction
The emergence of medically important arboviruses such as dengue (DENV), Zika (ZIKV), and chikungunya (CHIKV) are associated with expansion of their geographic range, mosquito vector diversity, and human reservoirs (Benedict et al. 2007, Powell and Tabachnick 2013, Schaffner et al. 2013). In the absence of vaccines and specific antiviral drugs, the most effective strategy to reduce risk of transmission to humans is early pathogen detection in mosquitoes. The yellow fever mosquito Aedes aegypti is the primary vector for these emerging pathogens.
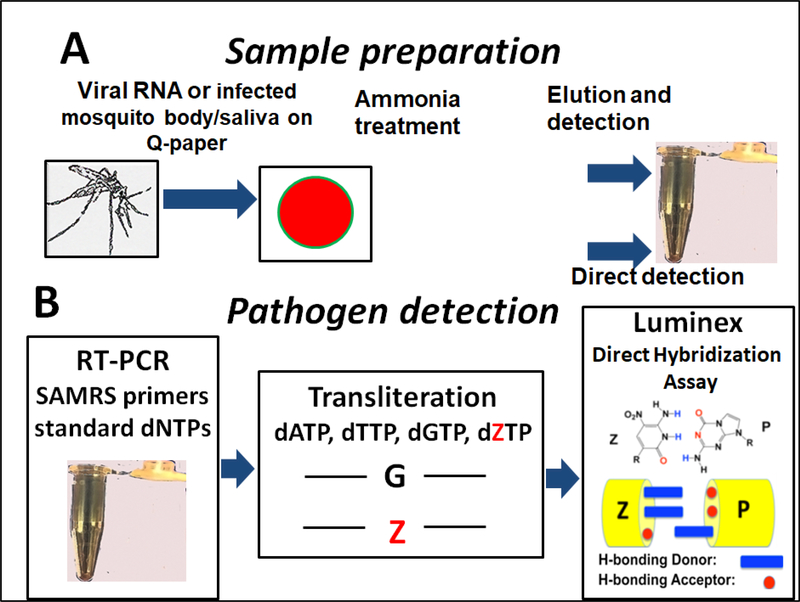
It was shown previously (Glushakova et al. 2017) that a platform that exploited cationic (Q) paper may be used as a component of a simple and inexpensive tool to survey for mosquitoes carrying arboviruses. In a typical workflow, an infected mosquito (or pool of mosquitoes) is squashed onto Q-paper and treated by aqueous ammonia, releasing virus nucleic acids (NA) from mosquito tissues, which is then adsorbed to the surface of the Q-paper via electrostatic interactions (Figure A.1A).
The cationic surface of the Q-paper was created by covalently attaching quaternary ammonium groups (trimethylammonium alkyl) to paper by stepwise treatment with NaOH and then 2,3-epoxypropyltrimethylammonium chloride (Yang et al. 2013). However, the paper produced according to the published protocol had a brittle texture and poor water adsorption. These factors impacted the paper’s adsorbance capacity and, consequently, the detection of pathogens from infected mosquitoes or other biological samples.
Here, we report a study that optimizes the protocol for producing Q-paper by varying the input reagents and temperature regime systematically.
2. Material and Methods
2.1. Preparation of cationic paper (Q-paper) and measuring its binding capacity
Q-paper was originally prepared by modifying the procedure described in (Yang et al. 2013) as previously reported (Glushakova et al. 2017). To determine the combination of factors that impact Q-paper quality and its nucleic acids binding capacity, the input reagents were evaluated (Table 1). For this, each input reagent was tested in the original concentration or a 0.1-fold dilution. All combinations of reagent concentrations and temperatures used for Q-paper production are presented in Table 1.
Table 1.
Q-paper preparation. The binding capacities were determined with 1,3,5benzenetricarboxylic acid (BTCA).
| Q-paper Identity, # | NaOH, % | Mass ratio EPTMAC to paper, MR:1 | Acetic acid, % | Temperature to dry paper, °C | Binding capacity, μmoles/cm2 |
|---|---|---|---|---|---|
| 1 | 18 | 2.8 | 10 | 25 | 0.114 |
| 2 | 18 | 0.28 | 10 | 25 | 0.276 |
| 3 | 18 | 2.8 | 1 | 25 | 0.059 |
| 4 | 18 | 0.28 | 1 | 25 | 0.234 |
| 5 | 1.8 | 2.8 | 10 | 25 | 0.330 |
| 6 | 1.8 | 0.28 | 10 | 25 | 0.090 |
| 7 | 1.8 | 2.8 | 1 | 25 | 0.294 |
| 8 | 1.8 | 0.28 | 1 | 25 | 0.072 |
| 9 | 18 | 2.8 | 10 | 55 | 0.186 |
| 10 | 18 | 0.28 | 10 | 55 | 0.306 |
| 11 | 18 | 2.8 | 1 | 55 | 0.132 |
| 12 | 18 | 0.28 | 1 | 55 | 0.228 |
| 13 | 1.8 | 2.8 | 10 | 55 | 0.324 |
| 14 | 1.8 | 0.28 | 10 | 55 | 0.114 |
| 15 | 1.8 | 2.8 | 1 | 55 | 0.384 |
| 16 | 1.8 | 0.28 | 1 | 55 | 0.102 |
| Filter paper | −0.0002 |
Briefly, one gram of precut filter paper was immersed in 50 mL of NaOH aqueous solution (1.8% or 18%) for 10 min for activation. The treated paper was collected by filtration and immersed in (2,3-epoxypropyl) trimethylammonium chloride, EPTMAC, (Sigma-Aldrich, St. Louis MO) aqueous solution for 24 h at room temperature. The mass ratio of EPTMAC to filter paper was 2.8:1 or 0.28:1. Next, the cationic (Q) paper was collected by vacuum filtration and neutralized with 1% or 10% of acetic acid. The final product was washed three times with ethanol (96%) and dried at 25°C or 55°C in an oven.
The Q-paper binding capacity was determined with 1,3,5-benzenetricarboxylic acid (BTCA). First, this reagent (2 mM, 5 mL) was neutralized to pH 8 with a NaOH solution (3 mM, ca. 2 mL). The product was diluted to a final concentration of 1 mM, and Q-paper was soaked for 15 min (2 mL of solution for 1 cm2 paper) followed by three consecutive washes with 1 mL dd H2O to remove unbound benzenetricarboxylate reagent. Acetic acid (aqueous, 1 M) was added to release the coulombically bound benzenetricarboxylate from the Q-paper. The total amount of bound benzenetricarboxylate was evaluated by UV measurement.
2.2. Arboviruses in this study
Isolates of CHIKV (Togaviridae/Alphavirus) and ZIKV (Flaviviridae/ Flavivirus) were provided by the Centers for Disease Control and Prevention and University of Texas Medical Branch in Galveston, TX, respectively. All arboviruses were obtained from human clinical samples (ZIKV, Asian lineage, strain PRVABC59; GenBank accession: KU501215.1; CHIKV from British Virgin Islands in 2013, Asian lineage, GenBank accession: KJ451624; Chikungunya virus from La Réunion in 2006, Indian Ocean lineage, GenBank accession: KT449801).
2.3. Propagation of arboviruses.
Virus stocks were produced by propagating viral isolates in cultured African green monkey (Vero) cells. Cells were cultured in M-199 medium supplemented with penicillinstreptomycin (0.2%), antimycotic (0.2%), and fetal bovine serum (10%). Viral titer was determined by plaque assay (Medina et al. 2012, Faye et al. 2013, Kaur et al. 2016).
To obtain fresh virus for infectious blood meals, the viruses were propagated by inoculation of confluent monolayers of Vero cells in tissue culture flasks (T-175 cm2) with diluted viral stocks at a multiplicity of infection of 0.01. After one hour of incubation at 37°C within a carbon dioxide atmosphere (5%), 25 mL media were added to each flask. Following the incubation (two days for CHIKV and seven days for ZIKV), mature viral particles were harvested from cell culture media.
2.4. CHIKV and ZIKV-infected Aedes aegypti mosquitoes
The CHIKV or ZIKV infectious blood meals were used for Aedes aegypti mosquito infection. This procedure was described previously (Glushakova et al. 2017). Freshly harvested media containing mature viral particles was combined with defibrinated bovine blood to produce viral titers similar to viremia in symptomatic patients (Lanciotti et al. 2008, Appassakij et al. 2013).
Mosquitoes were individually dissected to separate the bodies from the legs, which were tested as indicators of arboviral infection. Purified viral RNA was tested for the presence of CHIKV or ZIKV RNA by real time (TaqMan)-PCR according to (Reiskind et al. 2008).
2.5. Saliva from CHIKV- and ZIKV-infected mosquitoes
After ingestion of CHIKV- or ZIKV-infected blood (about seven days), females were individually transferred to plastic tubes (37-mL). The time of saliva collection was estimated based on the highest CHIKV titers expectorated in saliva (Dubrulle et al. 2009, Alto et al. 2017). The procedure for collecting infected saliva was described previously (Glushakova et al. 2018). Briefly, mosquitoes were deprived of sucrose (24 hours) and transferred to tubes fitted with a lid with honey-soaked Q-paper (1 cm diameter). The honey contained a blue colored food dye to indicate that the mosquito ingested honey (Burkett-Cadena et al. 2016). Mosquitoes were examined with a light-emitting diode for blue in their crop after 24 hours. Mosquitoes and Q-paper were stored at −80°C following the transmission assay. Q-paper was tested for the presence of CHIKV and ZIKV RNA only for those mosquitoes that fed on blue honey.
2.6. Saliva collection by capillary tubes
In addition to the collection of mosquito saliva on Q-paper, saliva expectorates were collected from a subset of different mosquitoes using a capillary assay procedure according to methods used in (Alto et al. 2017). Briefly, mosquitoes were cold anesthetized (4°C), legs and wings removed, and proboscis placed in a capillary tube filled with immersion oil heated to 37°C. Mosquito saliva was collected from individual mosquitoes for one hour after which saliva expectorates were expelled into 300 μL of M-199 media. Mosquito body, leg, and saliva samples were stored at −80°C until tested for the presence of viral RNA by conventional reverse transcription PCR and by direct Luminex hybridization assay.
2.7. ZIKV and CHIKV RNA isolation from arboviral stocks
To purify NA from mosquito legs for downstream arboviral infection tests, individual mosquito legs were homogenized in 1.0 mL media using a TissueLyser (Qiagen, Valencia CA) and clarified by centrifugation. Viral RNA was isolated from homogenate (140 μL) with QIAamp viral RNA mini kit (Qiagen, Valencia CA) and eluted in TE buffer (50 μL) (Glushakova et al. 2017). Next, the presence of the CHIKV RNA in isolated NA was determined by quantitative (TaqMan) RT-PCR with Superscript III One-Step qRTPCR kit (Invitrogen, Carlsbad, CA) and the CFX96 Real-time PCR Detection System (Bio-Rad Laboratories, Hercules, CA). Primers and probe (Integrated DNA Technologies, Coralville, IA) had the following sequences:
CHIKV_Forward Primer, 5′-GTACGGAAGGTAAACTGGTATGG-3′
CHIKV_Reverse Primer, 5′-TCCACCTCCCACTCCTTAAT-3′
CHIKV_Probe, 5′-/56-FAM/TGCAGAACCCACCGAAAGGAAACT/3BHQ_1/−3′
The RT-PCR assay consisted of a 30-min RT step at 50°C linked to a 40-cycle PCR (94°C for 10 s and 60°C for 60 s).
ZIKV RNA was purified from viral stocks in cell culture medium using the Superscript III One-Step qRT-PCR kit. The procedure was performed according to the manufacturer’s protocol (Qiagen, Valencia CA). ZIKV RNA titers in samples were determined by TaqMan RT-PCR with the following oligonucleotides (Integrated DNA Technologies):
ZIKV_Forward Primer, 5′- CTTCTTATCCACAGCCGTCTC-3′
ZIKV_Reverse Primer, 5′- CCAGGCTTCAACGTCGTTAT-3′
ZIKV_Probe, 5′-/56-FAM/AGAAGGAGACGAGATGCGGTACAGG/3BHQ_1/−3′
The program for qRT-PCR consisted of a 30-min step at 50°C linked to a 40-cycle PCR (94°C for 12 s and 58°C for 60 s).
To express the viral titer in mosquito samples and viral stocks, a standard curve was established and compared to cDNA synthesis for a range of stock viral serial dilutions (genomes equivalents). Plaque assays of the same viral dilutions were performed and the titer was expressed as plaque forming unit (PFU) equivalents per mL (Bustin 2000). Presence of arboviral RNA in the legs of mosquitoes indicated a specimen with a disseminated infection (Turell et al. 1984).
2.8. Conventional reverse transcription (RT)-PCR
The PCR primers and Luminex probes were designed as described previously (Glushakova et al. 2015) and are presented in Table 2. CHIKV oligonucleotides were chosen to target non-structural protein NS4B. ZIKV oligonucleotides targeted nonstructural protein NS5. Standard versions of oligonucleotides were ordered from Integrated DNA Technology (IDT, Coralville, USA); primers and probes containing artificial SAMRS (self-avoiding molecular recognition system) and AEGIS (artificially expanded genetic information system) nucleotides were synthesized on an ABI 3900 synthesizer using AEGIS and SAMRS phosphoramides (www.firebirdbio.com).
Table 2.
Standard and self-avoiding molecular recognition system (SAMRS) oligonucleotides used for detection of ZIKV and CHIKV RNA. All reverse primers are 5’-biotinylated. All probes were 5’-amino-C12-modified. P, artificially expanded genetic information system (AEGIS) nucleotide; *, SAMRS nucleotides.
| Oligonucleotides | Forward (F) and Reverse (R) primers’ sequences | Luminex probe sequence |
|---|---|---|
| CHIKV, standard nucleotides | F_CHIKV: CAGATGGCAACGAACAGG R_CHIKV: GGGTCCTCTGAGCTTCTC |
CCTTTGCAAGCTCCAGATC |
| CHIKV, SAMRS nucleotides | F_CHIKV: CAGATGGCAACGAA*C*A*G*G R_CHIKV: GGGTCCTCTGAGCT*T*C*T*C |
CCTTTPCAAPCTCCAPATC |
| ZIKV, standard nucleotides | F_ ZIKV: AGGGACCTCCGACTGATG R_ ZIKV: CCTCAATCCACACTCTGTTC |
GAAAGGGAGAATGGATGACC |
| ZIKV, SAMRS nucleotides | F_ ZIKV: AGGGACCTCCGACT*G*A*T*G R_ ZIKV: CCTCAATCCACACTCT*G*T*T*C |
PAAAPPPAPAATPPATPACC |
Conventional RT-PCRs were executed with SuperScript™ One-Step RT-PCR or SuperScript™ III One-Step RT-PCR System with Platinum™ Taq DNA Polymerase (Life Technologies) according to the manufacturer’s protocol. The full procedure was presented previously (Glushakova et al. 2017). Typically, the final primer concentration was 0.25 – 0.3 μM. For PCR optimization, additional MgSO4 (1.1 – 1.5 mM) was added to the buffer solution (final concentration, 2.7 mM). All reactions were incubated in DNA Engine® Multi-Bay Thermal Cyclers (BioRad, Hercules, CA, USA) or in miniPCR® Thermal Cycler (Carolina Biological Supply Company, Burlington, NC) as follows: One cycle of cDNA synthesis and pre-denaturation (53°C for 30 min and 94°C for 2 min) and 30–35 cycles of PCR (94°C for 15 s, 54°C for 30 s, and 70°C for 30 s) with a final extension at 72°C for 5 min.
The hybridization procedure on Luminex beads was optimized and improved by: (1) preparation of the biotin-tagged amplicon strand in excess, (2) conversion of the dC bases in the resulting amplicon into AEGIS dZ bases (“transliteration”). For these, reverse primer extension reaction (RPER) was executed for each PCR amplicon in the presence of a mixture of dATP, dTTP, dGTP, and dZTP oligonucleotides as described previously (Glushakova et al. 2015, Glushakova et al. 2017). In a pilot experiment, the optimal cycling conditions were established as: 95°C for 1 min and 20–25 cycles (94°C for 20 s, 53°C for 30 s, and 72°C for 30 s). A final incubation at 72°C was run for 3 minutes.
2.9. Probe coupling to Luminex MicroPlex carboxylated micro-spheres
Luminex capture probes were built with standard and dPTP-nucleotides, and each contained an amino-C12 linker at their 5’- ends. These were coupled to Luminex MicroPlex carboxylated microspheres (Luminex, Austin TX) by a carbodiimide-based procedure according to the manufacturer’s protocol, as previously described (Glushakova et al. 2015). Finally, beads were re-suspended in Tris-EDTA (pH 8.0) to a final volume of 100 μL and counted with a light microscope.
2.10. Luminex direct hybridization assay (DHA)
Luminex Direct hybridization assays (DHAs) (Dunbar 2006) were performed in accordance to the “no wash” protocol (http://www.luminexcorp.com/Support/SupportResources/) as presented previously (Glushakova et al. 2015). Each reaction was performed in triplicate, and “no-target” controls were performed in replicates of four or five. The assays were analyzed for internal bead color and R-phycoerythrin reporter fluorescence with a Luminex 200 analyzer (Luminex xMAP Technology) and the xPonent Software solutions. Data were expressed as the median reporter fluorescence intensity (MFI, Luminex unit). The instrument’s gate setting was established before the experiment and maintained throughout the course of the study.
3. Results
3.1. Testing Q-paper binding capacity with Zika virus RNA
To optimize the protocol for producing Q-paper, the reagents and temperature regimes were systematically varied from those described by (Yang et al. 2013). This gave sixteen cationic Q-paper variants (Table 1) that were individually analyzed in three ways: by their texture, by their ability to bind an anionic dye (BTCA), and by their ability to capture RNA that was later recovered.
Regarding texture, simple inspection divided the Q-paper samples into two groups (Appendix D, Table D.1). The first comprised papers with a brittle, hydrophobic texture (#1–4, 9–12); these were produced when the concentration of activating aqueous NaOH was high (18%), the concentration recommended by (Yang et al. 2013). Soft, pliable, and hydrophilic samples of Q-paper were obtained when a lower concentration (1.8%) of aqueous NaOH was used to activate the paper hydroxyl groups for derivatization (#5–8, 13–16).
The binding capacity of each cationic paper variant was evaluated using 1,3,5-benzenetricarboxylic acid (BTCA). This dye, anionic at pH 7 and higher, was chosen because it could be quantitated by UV spectroscopy. BTCA adsorption revealed differences in the abilities of Q-paper variants to bind to this small molecule (Table 1). Binding capacities ranged from 0.059 μmoles/cm2 (#3) to 0.384 μmoles/cm2 (#15).
The ability of different Q-papers to adsorb macromolecular RNAs was then compared using full genomic ZIKV and CHIKV RNA. For this comparison, small pieces (3 × 5 mm) of paper were pre-cut from each of the sixteen sheets. Then, aliquots (1 μL) of RNA (about 104 PFU-equivalents of ZIKV and 106 genome equivalents of CHIKV RNA) were applied on top of each piece. The paper samples were air-dried in a biosafety cabinet and then individually placed into centrifuge tubes and kept at −20°C until processed (Figure A.1).
To remove unbound RNA from the surface, the Q-papers were washed with molecular biology grade water (150 μL, 2 hours at 4°C). The tubes were vortexed briefly 2–3 times. After, a sample (75 μL) of washing from each tube was collected for downstream RNA detection.
To recover the RNA adsorbed to the Q-paper, aliquots (75 μL) of aqueous NaCl (2 M) were added to each tube to bring the final salt concentration to 1 M. The tubes were vortexed and samples were stored at 4°C for another two hours. Last, the RNAs eluates in NaCl solutions (1 M) were desalted by gel filtration with Centri-Spin20 columns (Princeton Separations, Adelphia NJ).
The RNA in the samples was then amplified by RT-PCR (35 cycles). The amplicons were quantitated using a Luminex-based direct hybridization assay described previously (Glushakova et al., 2015, 2017). The amplicons were “transliterated” (Glushakova et al., 2017) to give products that had the nonstandard nucleotide Z instead of C (Yang et al., 2013). The transliterated products were hybridized to dPTP-containing probes on a Luminex platform (Figure 1–2).
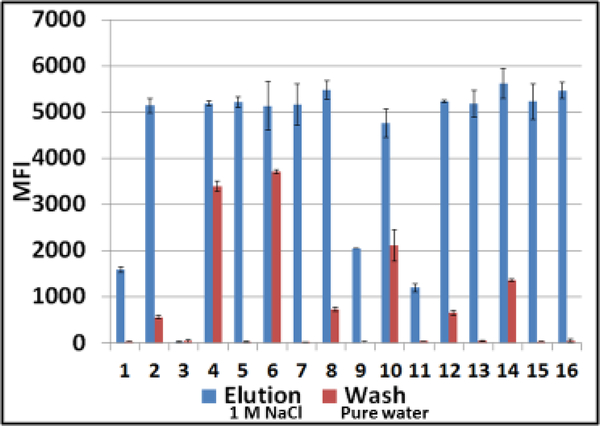
1.
Luminex profiles of samples obtained by washing Q-papers carrying ZIKV RNA with water (red bars) versus Q-paper high salt eluates (blue bars). Samples were analyzed by RT-PCR and downstream hybridization with ZIKV-specific probe on Luminex instrument. MFI, median fluorescent intensity (Luminex unit); #1–16, numbers on the abscissa indicate different Q-paper samples prepared using the protocols #1–16 (Table 1).
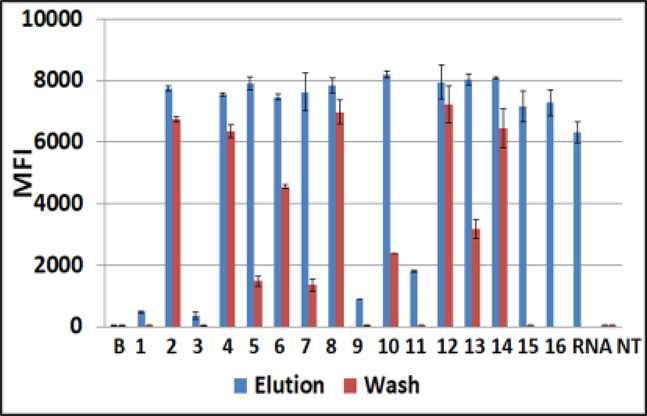
2.
Luminex profiles of water-washes from Q-papers carrying CHIKV RNA (red bars) versus Q-paper high salt eluates (blue bars). Samples were analyzed by RT-PCR followed by downstream hybridization with CHIKV-specific probe on Luminex beads. MFI, median fluorescent intensity (Luminex unit); #1–16, numbers on the abscissa indicate different Q-paper samples prepared using the protocols #1–16 (Table 1). B, background fluorescence, sample buffer added to control wells; RNA, water solution of RNA, 103 genomes; NT, no template RT-PCR negative control; Water-wash sample from Q-paper #16 was not tested.
ZIKV RNA was detected in both water–washes and high salt eluates from Q-papers #2, 4, 6, 8, 10, 12, and 14 (Figure 1). These samples displayed strong Luminex signals in a range of 5,000–5,500 median fluorescence intensity (MFI) units in eluates, but lower signals (with variable values) in water washes. Samples from Q-papers #5, 7, 15 and 16 displayed strong positive signals in the same ranges (high salt eluates). Luminex outcomes for samples from Q-papers #1, 9, and 11 were 2.5-to 5-fold lower, and samples from #3 were at background levels. These observations paralleled those made with BTCA (Table 1), except for samples from Q-papers #6, 8, 14, and 16.
The Q-papers #5, 7 and 15 revealed the highest NA absorbance capacities (Figure 1). The crucial conditions for the preparation of these papers were the concentrations of NOH aqueous solution (1.8%) and EPTMAC (mass ratio EPTMAC to paper was 2.8). Other reagents (acetic acid, 1 or 10%) and drying temperature (25°C or 55°C) seem to be less important.
The data were confirmed with CHIKV RNA on Q-papers #1–16 (Figure 2). These samples were tested using SuperScript™ III (SSIII) One-Step RT-PCR System with Platinum™ Taq High Fidelity DNA Polymerase (SuperScript One-Step RT-PCR System was discontinued by Invitrogen). Due to higher SSIII sensitivity, the CHIKV–specific Luminex fluorescent signals had larger values (7–8,000 MFI), and also, small amounts of CHIKV RNA were detected in water-washes from papers #5, 7, 13 (about 10% of values obtained for eluates from the same samples). In general, these complementary data were in an agreement with the original findings obtained with ZIKV RNA.
3.2. Testing Q-paper binding capacities with CHIKV-infected Aedes aegypti mosquito bodies
Aedes aegypti mosquitoes were infected by CHIKV, and the infection was confirmed by detecting CHIKV RNA in dissected leg samples (Appendix D, Table D.1). Mosquito bodies positive for infection were squashed onto Q-papers #1–16. Aqueous ammonia (50 μL, 1 M) was added to each sample and allowed to dry (22–24°C) for 20 min before Qpapers were frozen (−80°C) (Glushakova et al. 2017).
Before the downstream analysis, Q-paper samples were thawed and “unbound” NAs were removed by washing with ddH2O. The adsorbed NAs were recovered from the Qpaper surface by elution with aqueous NaCl (1 M final). The eluates were desalted as described above (Section 2.1). All water-washes and desalted eluates were assayed for CHIKV RNA by downstream RT-PCR based Luminex DHAs (Figure 3, Table D.1).
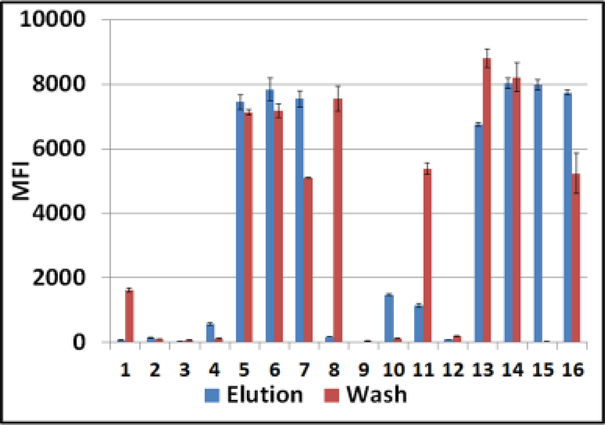
3.
CHIKV-infected Ae. aegypti mosquito bodies on Q-paper. Luminex profiles of CHIKV RNA in water-washes (red bars) versus CHIKV RNA in high salt eluates (blue bars) are presented. All samples were analyzed first by conventional RT-PCR with SAMRS primers then by downstream hybridization with virus-specific AEGIS dP probe on Luminex instrument. MFI, median fluorescent intensity; #1–16, Q-paper variants (Table1).
These assays also showed that different Q-paper samples #1–16 adsorbed viral RNA from mosquito bodies differently (Table D.1 and Figure 3). Hydrophobic Q-papers either did not bind viral RNA (background signals for water-washes and eluates, samples # 2, 3, 9, 12) or bound only with low capacity, giving lower Luminex fluorescent signals (in water-wash #1; in high salt eluates #4 and 10).
The presence of CHIKV RNA in both fractions (water-washes and eluates) was documented for samples # 5–7, 11, 13, 14, and 16. The corresponding Luminex assays displayed strong fluorescence in the 6,000 to 8,500 MFI units range. This group was comprised of papers with a soft hydrophilic texture. However, water-wash from Q-paper #8 (pliable, soft, but produced with less EPTMAC) displayed a strong Luminex output while the fluorescence signal of its high-salt eluate was at background levels.
The highest adsorbance capacity was observed with Q-paper #15. There was no detectable CHIKV RNA in its water-wash, and CHIKV RNA was recovered only by highsalt elution. Q-paper #15 had a soft texture and was prepared with 1.8% NaOH aqueous solution, 2.8:1 EPTMAC:paper mass ratio, 1% acetic acid, and 55°C drying temperature (Table 1). These experiments were repeated with ZIKV-infected Aedes aegypti mosquitoes and general trends were confirmed (Figure 4) with small variations in Luminex profiles, such as absence of ZIKV RNA in water wash for sample #8 and background level of both fractions for sample #11, etc.).
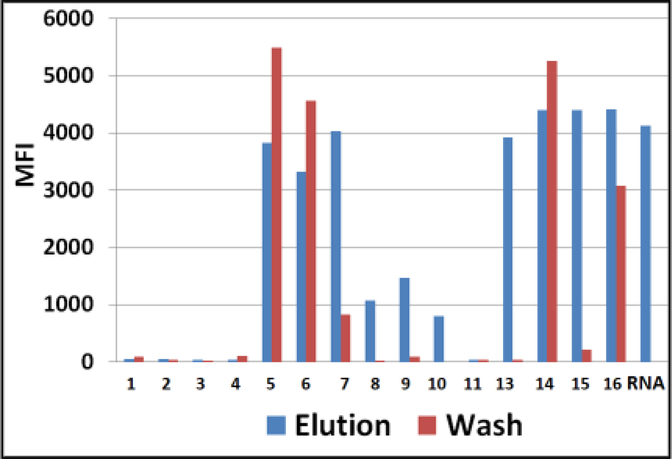
4.
ZIKV-infected Ae. aegypti mosquito bodies on Q-paper. Luminex profiles of ZIKV RNA in water-washes (red bars) versus high salt eluates (blue bars) are presented. The samples were analyzed by conventional RT-PCR followed by downstream hybridization with ZIKV-specific AEGIS dP probe on Luminex beads. MFI, median fluorescent intensity; #1–16, Q-paper variants (Table1). Water-wash of #13 was not tested.
Viral RNA adsorbance profiles obtained for mosquito bodies had more sophisticated patterns than those with pure viral RNA. Mosquito NA and negatively charged proteins might compete with viral RNA and saturate the positive charges on the Q-paper surface. These would impact Q-papers with lesser cationic charges, specifically, those produced with less EPTMAC, which is needed for the introduction of quaternary ammonium groups (trimethylammonium alkyl) (# 2, 4, 8, 10 and 12) (Figure 3–4).
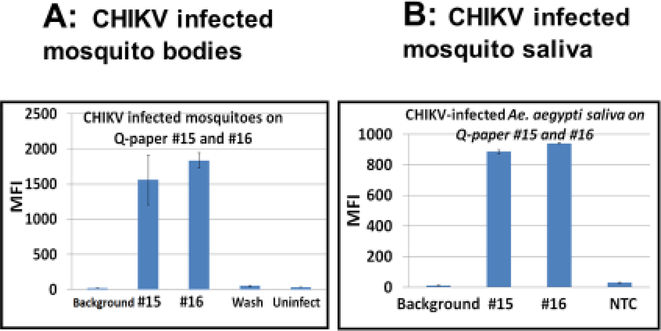
3.3. Detection of CHIKV and ZIKV in infected Ae.aegypti mosquito saliva on Q-papers #15 and #16 by Luminex- based mono- or multiplexed assays (Figures 5–6).
5.
Detection of CHIKV in infected Ae. aegypti mosquito bodies (A) and saliva (B) on Qpapers #15 and #16 by monoplexed Luminex-based assay. Representative data are presented. Luminex DHA profiles: A, high salt eluates from Q-paper with CHIKV–infected mosquito bodies; B, high salt eluates from Q-paper with CHIKV–infected mosquito saliva. CHIKV-infected saliva was collected from mosquitoes using a capillary tube (Anderson et al. 2010). MFI, median fluorescent intensity; wash, water-wash from sample #15; NTC, non-infected negative control.
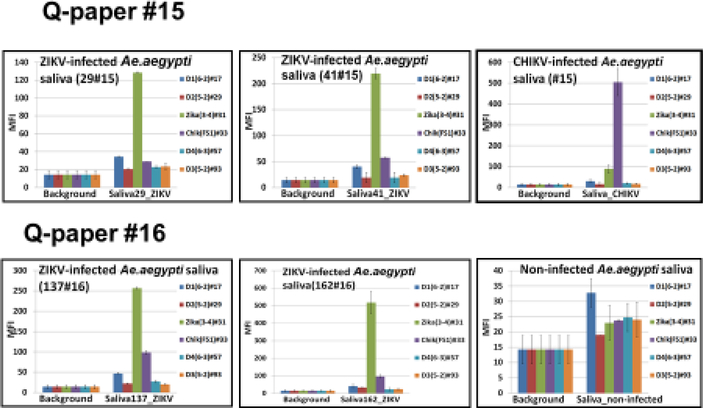
6.
Detection of ZIKV in the infected Ae. aegypti mosquito saliva on Q-papers #15 and #16 by multiplexed Luminex-based assay platform. ZIKV-infected saliva was collected from mosquitoes on Q-paper soaked in honey. Top right panel shows Luminex profiles for CHIKV-infected mosquito saliva (as a comparison). MFI, median fluorescent intensity.
Saliva from infected mosquitoes also contains viruses, and is the principal mode of transmission. Therefore, it was interesting to see whether Q-paper could capture this virus directly from saliva. Results show that it did.
Here, saliva from mosquitoes infected with CHIKV and ZIKV was collected in two ways. First, saliva was collected using capillary tubes (Section 2.6) and then adsorbed on Qpaper (Figure 5). Second, saliva was collected directly onto Q-paper that had been soaked with honey, which induced mosquitos to feed (Section 2.5) (Figure 6). In both cases, the infection status of the mosquitoes was confirmed by quantitative real time RT-PCR of leg samples. Separately, CHIKV-infected mosquito bodies were squashed on top of Q-papers #15 and #16 then analyzed as a reference (representative data shown in Figure 5A). Samples of saliva from five non-infected mosquitoes were used as negative controls. Adsorbed RNA was then eluted from Q-papers as described above and analyzed either by monoplexed Luminex-based assay that targets CHIKV alone using single CHIKV-specific probe (Figure 5B) or that targets a variety of arboviruses (DENV serotypes 1–4, ZIKV and CHIKV) in an assay having by a six-fold multiplexed capability (Figure 6) (Glushakova et al. 2018). All oligonucleotides used in the multiplexed assay are presented in Appendix B (Table B.1).
Seven of ten saliva samples collected in capillaries from CHIKV-infected mosquitoes had positive Luminex outcomes. Samples from non-infected mosquitoes showed only background levels of fluorescence (Figure 5). All eluates from infected mosquito bodies on Q-paper analyzed in this experiment were positive (Luminex profiles of two representative body samples are shown in Figure 5A). Saliva samples generated lower Luminex fluorescent signals (800–1000 MFI units) than body samples (about 2000 MFI units). These observations were consistent with CHIKV loads in mosquito saliva versus mosquito bodies (Dubrulle et al. 2009). Again, different Q-paper variants displayed different binding capacities. For example, #15 displayed the highest binding capacity while paper #16 bound viral RNA more loosely.
Of value for further work, we also found that when saliva was collected directly from ZIKV-infected mosquitoes on honey-treated Q-paper, positive and clear Luminex signals were seen (150 – 250 MFI-units, 5–7 fold higher than background), but averaging 4–5 fold lower then signals seen from saliva collected by capillaries (Figure 5–6). A few samples were only 2–3 times above background (data not shown). These data are consistent with the methods used for saliva collection; mosquitos deliver smaller amounts of saliva to paper soaked with honey than the amounts recovered with capillaries (Alto, unpublished data). Saliva samples from CHIKV-infected mosquitoes were also analyzed (representative data shown in Figure 5, top right panel).
4. Discussion
Since nucleic acids (DNA and RNA) are polyanions, they are expected to be adsorbed onto positively charged surfaces. However, this basic application of Coulombs law is surprisingly underused in molecular biology. Thus, the dominant kits for isolating nucleic acids from biological samples exploit the ability of DNA and RNA to be adsorbed on glass.
Even when cationic surfaces are used to adsorb nucleic acids, they are generally created by tertiary ammonium groups, which lose their positive charges at high pH. This means that alkaline conditions cannot be used for this classical adsorbent. According to the literature, glass and tertiary ammonium groups are used so as to facilitate the elution of the nucleic acids, which are widely regarded as being too tightly bound to a generally cationic surface.
Quaternary ammonium groups do not lose their positive charges at high pH. Accordingly, (Yang et al. 2013) suggested that they would be appropriate to capture anionic carcinogenic dyes from wastewater, including water-soluble anionic Acid Orange 7, Acid Red 18 and Acid Blue 92. Seeking a cheap matrix, Yang et al. (2013) chose cellulosic paper. Seeking a high binding capacity, they further treated that paper with large amounts of a strong base (sodium hydroxide) to deprotonate the hydroxyl groups on the cellulose prior to reacting with an epoxide carrying a trimethylammonium permanent cation.
These authors were, of course, not concerned about the physical properties of the paper, its ability to adsorb macromolecules, or its ability to release the adsorbent. These are all factors that we encountered when we first showed that cellulose-based cationic (Q) paper might be an efficient platform to recover viral and other nucleic acids from biological samples (Glushakova et al. 2017). To our knowledge, this was the first application of cationic paper for this purpose. We were surprised to identify multiple advantages of this strategy, including its ability to support very simple sample preparation. Pathogen nucleic acid was detectable directly off of the Q-paper, without the traditional time-consuming and reagent-consuming steps of RNA purification.
Unfortunately, the paper prepared by the Yang et al. (2013) method was brittle and hydrophobic, and less efficient for NA binding compared to binding of small molecules (as anionic dyes). Therefore, to expand the use of this paper as a matrix to capture saliva from feeding mosquitoes in the wild, which would not need to be physically trapped and killed, we explored improvements in the protocol for preparing Q-paper, hoping to retain its advantages while losing its disadvantages.
The first improvement was seen by lowering the amount of sodium hydroxide. In principle, for high capacity adsorbance, one might expect to see a correlation where more base leads to more molecules adsorbed. In fact, this was generally the opposite of what was seen. A milder, more dilute, sodium hydroxide treatment was entirely compatible with the need to activate the paper for derivatization. Moreover, a milder sodium hydroxide treatment gave paper that was physically easier to handle. Further, it appeared to be less “hydrophobic”, easier to wet and therefore easier to interact with typical biological samples.
The second concern was the ability to release RNA from the Q-paper surface after it had been adsorbed. Here, the initially tight binding could be overcome by relatively modest concentrations of salt in the aqueous equate (1 M). Upon dilution, this was entirely compatible with downstream amplification of the adsorbed nucleic acids using RT-PCR. Indeed, the system is entirely compatibly with previously developed technologies from our laboratories that use the latest tools of synthetic biology to get sensitive Luminex-based pathogen detection. Nevertheless, these advantages are expected to be general for any downstream analysis.
Most promising, however, was the observation that virus obtained from mosquitoes feeding on Q-paper that had been treated with honey could also be recovered. This approach avoids yet another sample preparation step: the capture and squishing of captured mosquito onto the paper, as described previously. It seems sufficient to simply allow infected mosquitoes to feed. We are presently exploring the possibility of using honey-treated Q-paper as a capture surface and as a part of a mechanical trap.
Highlights.
A cellulose-based cationic (Q) paper derivatized with quaternary ammonium groups is a convenient platform to collect, preserve, and store nucleic acids (NA).
Factors that affected arboviral RNA adsorbance on Q-paper were evaluated.
Q-paper produced by optimized technology is an efficient tool for downstream infected mosquito surveillance.
Viral RNA could be recovered from infected mosquito saliva follow feeding on Q-paper treated with honey.
Acknowledgments
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIH) (R21AI128188), Florida Department of Health (FDOH) (7ZK15), and the Florida Department of Agriculture and Consumer Services (FDACS) (contract 024376). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH, FDOH or FDACS.
Appendix A.
Figure A.1.
Overview of nucleic acids detection on cationic (Q) paper ((adaptation from (Glushakova et al. 2017) with permission)). A, Sample preparation; B, Luminex direct hybridization assay (DHA) platform. SAMRS primers are presented in Table B.1. tructure of SAMRS nucleotides are presented in Figure C.1.
Appendix B.
Table B.1.
PCR primers and Luminex probes in the six-fold multiplexed assays. These oligonucleotides were also used in (Glushakova et al., 2018, submitted to the J Virol Methods) and published here with permission. All reverse primers are 5’- biotinylated. All probes have 5’-amino-C12-modified.
| Oligonucleotide identity | Sequences | Position, nucleotid es bases | GenBank accession number | Targeting region |
|---|---|---|---|---|
| DENV1 Forward primer |
GGCCRGATTAAGCC*A*T*A*G | 10267–10349 | KY926849.1 | UTR |
| DENV1 Probe |
APAPCTATPCTPCCTPT | |||
| DENV1 Reverse primer |
GCTTTCGGCCTGA*C*T*T*C | |||
| DENV2 Forward primer |
CGTGTCRACTGTRCA*A*C*A*G | 17–136 | KY461768.1 | Capsid/premembrane protein gene |
| DENV2 Probe |
ATTCTCACTTPPAATPCTPC | |||
| DENV2 Reverse primer |
ARTATCCCTGCTGTT*G*G*T*G | |||
| DENV3 Forward primer |
AACACTCTGGGAAGGAT*C*A*C*C | 7405–7507 | KT726361.1 | Non-structural Protein NS4B |
| DENV3 Probe |
TTPPAACACCACPATAPCT | |||
| DENV3 Reverse primer |
AGCAAGCCCAGCT*C*C*T*G | |||
| DENV4 Forward primer |
GCAGGCAAAAGCCA*C*A*A*G | 3289–3430 | AH0119363.2 | Non-structural protein NS2A |
| DENV4 Probe |
APTPPACPPPATAACAPT | |||
| DENV4 Reverse primer |
CATGACCTGCCCTA*A*T*T*G | |||
| CHIKV Forward primer |
CAGATGGCAACGAA*C*A*G*G | 6083–6273 | KY575571.1 | Non-structural protein NS4B |
| CHIKV FS-1 Probe |
CCTTTPCAAPCTCCAPATC | |||
| CHIKV Reverse primer |
GGGTCCTCTGAGCT*T*C*T*C | |||
| ZIKV Forward primer | AGGGACCTCCGACT*G*A*T*G | 9981–10112 | KY415991.1 | Non-structural protein NS5 |
| ZIKV Probe | PAAAPPPAPAATPPATPACC | |||
| ZIKV Reverse primer | CCTCAATCCACACTCT*G*T*T*C |
P, AEGIS nucleotide.*, SAMRS nucleotides. R, mixed A and G bases. Position of primers and probes refer to the arbitrary chosen sequence of the viral strains.
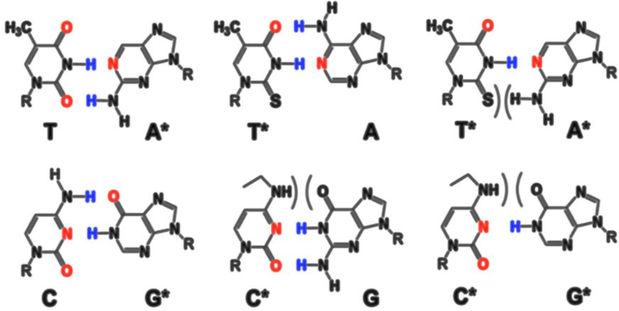
Appendix C.
Figure C.1.
(adapted from (Glushakova et al., 2015) with permission): Self-avoiding molecular recognition systems (SAMRS) were developed by the strategic removal of hydrogen bonding groups. PCR-primers are built with standard and artificial SAMRS nucleotides (3’-ends). *, SAMRS nucleotides.
Appendix D.
Table D.1.
Detection of CHIKV RNA in infected Ae. aegypti mosquitoes on Q-paper. Fluorescent signal: (+), low, <1000 median fluorescence intensity (MFI); (+++), high >5,000 MFI; (−) not detectable. A note: lower viral titers are expected in the legs than in the bodies, as shown previously for DENV and CHIKV infection (Richards et al. 2010, Alto and Bettinardi 2013).
| Q-paper Identity, # (Table1) | CHIKV titers in mosquito legs by real time-PCR, genome equivalents/mL | Q-paper’s texture | CHIKV RNA detection by Luminex DHA | |
|---|---|---|---|---|
| Water-wash | Eluted by 1M NaCl solution | |||
| 1 | 6.86 ×104 | brittle | − | + |
| 2 | 9.14 ×104 | brittle | − | − |
| 3 | 1.2 ×105 | brittle | − | − |
| 4 | 1.63 ×104 | brittle | + | − |
| 5 | 3.72 ×104 | pliable | +++ | +++ |
| 6 | 3.17 ×104 | pliable | +++ | +++ |
| 7 | 5.79 ×104 | pliable | ++ | +++ |
| 8 | 6.86 ×104 | pliable | +++ | − |
| 9 | 9.14 ×104 | brittle | − | − |
| 10 | 1.2 ×105 | brittle | − | + |
| 11 | 1.63 ×104 | brittle | +++ | + |
| 12 | 3.72 ×104 | brittle | − | − |
| 13 | 3.17 ×104 | pliable | +++ | +++ |
| 14 | 5.79 ×104 | pliable | +++ | +++ |
| 15 | 6.86 ×104 | pliable | − | +++ |
| 16 | 9.14 ×104 | pliable | ++ | +++ |
Footnotes
Competing interests
Some authors of this paper and their institutions own intellectual properties associated with the assay, including SAMRS, AEGIS, and the conversion/capture technologies. Several of the items mentioned here are sold by Firebird Biomolecular Sciences LLC, which employs the indicated authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Alto BW, and Bettinardi D. 2013. Temperature and dengue virus infection in mosquitoes: independent effects on the immature and adult stages. Am J Trop Med Hyg 88: 497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alto BW, Wiggins K, Eastmond B, Velez D, Lounibos LP, and Lord CC. 2017. Transmission risk of two chikungunya lineages by invasive mosquito vectors from Florida and the Dominican Republic. PLoS Negl Trop Dis 11: e0005724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SL, Richards SL, and Smartt CT. 2010. A simple method for determining arbovirus transmission in mosquitoes. J Am Mosq Control Assoc. 26(1):108–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appassakij H, Khuntikij P, Kemapunmanus M, Wutthanarungsan R, and Silpapojakul K. 2013. Viremic profiles in asymptomatic and symptomatic chikungunya fever: a blood transfusion threat? Transfusion 53: 2567–2574. [DOI] [PubMed] [Google Scholar]
- Benedict MQ, Levine RS, Hawley WA, and Lounibos LP. 2007. Spread of the tiger: global risk of invasion by the mosquito Aedes albopictus. Vector Borne Zoonotic Dis 7: 76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkett-Cadena ND, Gibson J, Lauth M, Stenn T, Acevedo C, Xue RD, McNelly J, Northey E, Hassan HK, Fulcher A, Bingham AM, van Olphen J, van Olphen A, and Unnasch TR. 2016. Evaluation of the honey-card technique for detection of transmission of arboviruses in Florida and comparison with sentinel chicken seroconversion. J Med Entomol 53: 1449–1457. [DOI] [PubMed] [Google Scholar]
- Bustin SA 2000. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25: 169–193. [DOI] [PubMed] [Google Scholar]
- Dubrulle M, Mousson L, Moutailler S, Vazeille M, and Failloux AB. 2009. Chikungunya virus and Aedes mosquitoes: saliva is infectious as soon as two days after oral infection. PLoS One 4: e5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar SA 2006. Applications of Luminex xMAP technology for rapid, highthroughput multiplexed nucleic acid detection. Clin Chim Acta 363: 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye O, Faye O, Diallo D, Diallo M, Weidmann M, and Sall AA. 2013. Quantitative real-time PCR detection of Zika virus and evaluation with field-caught mosquitoes. Virol J 10: 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glushakova LG, Alto BW, Kim MS, Bradley A, Yaren O, and Benner SA. 2017. Detection of chikungunya viral RNA in mosquito bodies on cationic (Q) paper based on innovations in synthetic biology. J Virol Methods 246: 104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glushakova LG, Alto BW, Kim MS, Hutter D, Bradley A, Bradley K, Burkett-Cadena N, and Benner SA 2018. Multiplexed kit based on Luminex technology and achievements in synthetic biology discriminates Zika, chikungunya, and four serotypes of dengue viruses in mosquitoes. J Virol Methods (submitted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glushakova LG, Bradley A, Bradley KM, Alto BW, Hoshika S, Hutter D, Sharma N, Yang Z, Kim MJ, and Benner SA. 2015. High-throughput multiplexed xMAP Luminex array panel for detection of twenty two medically important mosquito-borne arboviruses based on innovations in synthetic biology. J Virol Methods 214: 60–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur P, Lee RC, and Chu JJ. 2016. Infectious viral auantification of Chikungunya virus-virus plaque assay. Methods Mol Biol 1426: 93–103. [DOI] [PubMed] [Google Scholar]
- Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, Stanfield SM, and Duffy MR. 2008. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis 14: 1232–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina F, Medina JF, Colon C, Vergne E, Santiago GA, and Munoz-Jordan JL. 2012. Dengue virus: isolation, propagation, quantification, and storage. Curr Protoc Microbiol Chapter 15: Unit 15D 12. [DOI] [PubMed] [Google Scholar]
- Powell JR, and Tabachnick WJ. 2013. History of domestication and spread of Aedes aegypti--a review. Mem Inst Oswaldo Cruz 108 Suppl 1: 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiskind MH, Pesko K, Westbrook CJ, and Mores CN. 2008. Susceptibility of Florida mosquitoes to infection with chikungunya virus. Am J Trop Med Hyg 78: 422–425. [PMC free article] [PubMed] [Google Scholar]
- Richards SL, Anderson SL, and Smartt CT. 2010. Vector competence of Florida mosquitoes for chikungunya virus. J Vector Ecol 35: 439–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner F, Bellini R, Petric D, Scholte EJ, Zeller H, and Rakotoarivony LM. 2013. Development of guidelines for the surveillance of invasive mosquitoes in Europe. Parasit Vectors 6: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turell MJ, Gargan, TP 2nd, and Bailey CL. 1984. Replication and dissemination of Rift Valley fever virus in Culex pipiens. Am J Trop Med Hyg 33: 176–181. [DOI] [PubMed] [Google Scholar]
- Yang F, Song X, and Yan L. 2013. Preparation of cationic waste paper and its application in poisonous dye removal. Water Sci Technol 67: 2560–2567. [DOI] [PubMed] [Google Scholar]
- Yang Z, Durante M, Glushakova LG, Sharma N, Leal NA, Bradley KM, Chen F, Benner SA.2013. Conversion strategy using an expanded genetic alphabet to assay nucleic acids. Anal Chem. 85: 4705–12. [DOI] [PMC free article] [PubMed] [Google Scholar]