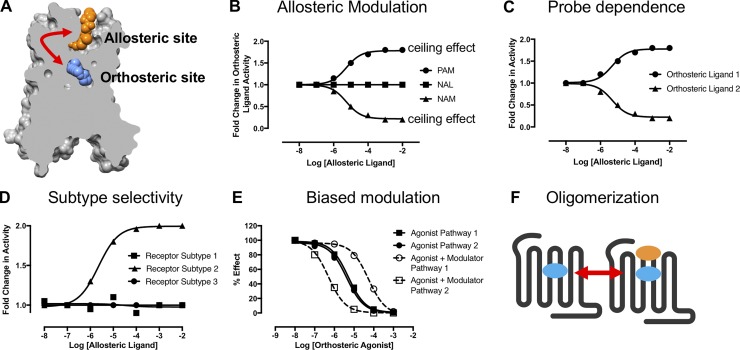
Burger et al. summarize our mechanistic understanding of allostery in the prototypical GPCR, the muscarinic acetylcholine receptor.
Abstract
Recent breakthroughs and developments in structural biology have led to a spate of crystal structures for G protein–coupled receptors (GPCRs). This is the case for the muscarinic acetylcholine receptors (mAChRs) where inactive-state structures for four of the five subtypes and two active-state structures for one subtype are available. These mAChR crystal structures have provided new insights into receptor mechanisms, dynamics, and allosteric modulation. This is highly relevant to the mAChRs given that these receptors are an exemplar model system for the study of GPCR allostery. Allosteric mechanisms of the mAChRs are predominantly consistent with a two-state model, albeit with some notable recent exceptions. Herein, we discuss the mechanisms for positive and negative allosteric modulation at the mAChRs and compare and contrast these to evidence offered by pharmacological, biochemical, and computational approaches. This analysis provides insight into the fundamental pharmacological properties exhibited by GPCR allosteric modulators, such as enhanced subtype selectivity, probe dependence, and biased modulation while highlighting the current challenges that remain. Though complex, enhanced molecular understanding of allosteric mechanisms will have considerable influence on our understanding of GPCR activation and signaling and development of therapeutic interventions.
Introduction
G protein–coupled receptors (GPCRs) are integral membrane proteins composed of seven transmembrane (TM)–spanning helices that are capable of binding to a diverse range of molecules outside of the cell to elicit a chemical response inside the cell (Wacker et al., 2017a). This intracellular response is mediated by G proteins, arrestins, and other proteins that trigger activation of a wide array of signaling pathways and ultimately lead to a physiological response. Because of their central role in determining cellular behavior and responses to external stimuli, GPCRs are recognized as important therapeutic targets. This is reflected in their current prevalence in the drug market, where they are the target of ∼34% of approved drugs (Hauser et al., 2017). Despite this prominence, there are substantial challenges in the generation of drug-like molecules that are specific to a given target GPCR. This is in part because of the similarities of the endogenous (orthosteric) ligand-binding site across closely related receptors. One strategy to circumvent this challenge is to target allosteric sites that are topographically distinct from, but conformationally linked to, orthosteric sites (Fig. 1 A).
Figure 1.
The many facets of GPCR allosteric modulation. (A) Structure of the M2 mAChR highlighting allosteric and orthosteric-binding sites. (B) Allosteric modulators are characterized through three different modes of behavior: PAM, NAM, or NAL. This is represented schematically, where increasing concentrations of different allosteric modulators are titrated against a single concentration of an orthosteric ligand. The saturability of the modulatory effect above a certain concentration of allosteric modulator is indicative of the “ceiling effect,” which is a key molecular characteristic of allosteric drugs. (C) Probe dependence is another unique pharmacological characteristic of allosteric modulators whereby the magnitude and direction of the allosteric effect can change depending on the orthosteric ligand used as a probe of receptor function. Shown is the effect on a fixed concentration of orthosteric ligand 1 or orthosteric ligand 2 at the same receptor in the presence of increasing concentrations of the same allosteric modulator. The binding of orthosteric ligand 1 is increased, whereas the binding of orthosteric ligand 2 is decreased. (D) Allosteric modulators can display selectivity in their ability to only enhance the binding of an orthosteric ligand at one particular subtype relative to other related receptor subtypes. (E) Biased modulation occurs when an allosteric ligand can promote more than one type of active state to the relative exclusion of another, such that the observed effect of the same agonist–modulator pair can vary depending on the signaling pathway that is linked to each receptor conformation. In this example, the modulator increases the potency of an agonist for pathway 1 but decreases the potency of the agonist in pathway 2; the two pathways are thus downstream of two different active receptor states. (F) Allosteric interactions can also arise between binding sites (orthosteric or allosteric) located on individual GPCRs that are arranged in conformationaly linked dimeric or oligomeric arrays.
The turn of the 21st century saw increasing awareness and appreciation for the use of allosteric ligands as therapeutic agents at GPCRs, and it is now well recognized that GPCRs contain allosteric sites (Christopoulos and Kenakin, 2002). Allosteric modulators can exquisitely “fine-tune” orthosteric ligand activity to predetermined levels, akin to the actions of a “dimmer switch,” and thus allosteric modulation represents a completely different paradigm to traditional agonists or antagonists for selectively sculpting GPCR activity, especially in a tissue- or disease-specific manner (Christopoulos et al., 2014). The degree of modulation, or fine-tuning, is indicative of the cooperativity between the orthosteric ligand and the allosteric modulator. Allosteric modulators that enhance the binding or signaling of the orthosteric ligand (positive cooperativity) are referred to as positive allosteric modulators (PAMs), allosteric modulators that reduce the binding or signaling of the orthosteric ligand (negative cooperativity) are referred to as negative allosteric modulators (NAMs), and allosteric ligands that do not influence the binding or signaling of the orthosteric ligand at equilibrium but are able to block the actions of other allosteric modulators acting at the same site are termed neutral allosteric ligands (NALs; Fig. 1 B; May et al., 2007). In addition, the magnitude and direction of the allosteric effect can vary depending on the orthosteric ligand used to interact with the allosteric modulator; this is referred to as probe dependence and has significant implications in the detection and validation of allosteric modulators (Fig. 1 C).
Targeting allosteric binding sites allows for therapeutic benefits that are not attainable when targeting the orthosteric site (Christopoulos et al., 2014). These include a ceiling effect in which pharmacological activity reaches a maximum effect regardless of the administered dose of the allosteric modulator, consequently decreasing the risk of on-target side effects (Fig. 1 B). Moreover, given that the allosteric interaction can only arise in the presence of the endogenous orthosteric ligand, the spatial or temporal signaling profile elicited with the endogenous ligand can be maintained (Kenakin and Christopoulos, 2013). A significant pharmacologic characteristic of allosteric modulators is their ability to selectively modulate one GPCR subtype to the relative exclusion of other subtypes and thus avoid the inherent problem of targeting the conserved orthosteric-binding site that is present on closely related receptors (Fig. 1 D). Some allosteric modulators can also display “biased” modulation; this occurs if the allosteric modulator promotes a distinct subset of receptor conformations to the relative exclusion of others (Kenakin, 2012). This phenomenon can provide enhanced therapeutic targeting through the design of allosteric modulators that, in their interaction with the orthosteric ligand, drive the formation of a receptor conformation linked to the desired therapeutic effect over conformations linked to side effects (Fig. 1 E). Last, a common feature of allosteric proteins is an oligomeric quaternary structure (Changeux and Christopoulos, 2016). Though GPCRs are often cited as a notable exception to this rule, there is experimental evidence to support the potential for allosteric interactions occurring between binding sites on GPCRs within dimeric or oligomeric arrays (Hern et al., 2010; Harikumar et al., 2012; Lane et al., 2014; Redka et al., 2014; Shivnaraine et al., 2016; Fig. 1 F).
Recent developments in structural biology coupled with computational, biochemical, and pharmacological approaches have begun to offer insight into the structural basis of allosteric modulation at many GPCRs. Herein, we discuss our current understanding of the mechanisms underlying allosteric modulation using muscarinic acetylcholine receptors (mAChRs) as an exemplar system that has proven to be generalizable to other GPCRs. Using this mechanistic understanding, we examine how the concepts associated with allosteric modulation are rationalized and compare and contrast the evidence offered from the aforementioned experimental techniques.
mAChRs as tools to understand GPCR allostery
mAChRs are an important subfamily of class A GPCRs and are expressed throughout the central and peripheral nervous systems (Caulfield and Birdsall, 1998). There are five mAChR subtypes (M1–M5), which are not only involved in regulating the vital “rest and digest” functions of the peripheral nervous system (M2, M3) but also implicated in many important neurological disorders, including Alzheimer’s disease (M1), schizophrenia (M1/M4), and drug addiction (M5; Kruse et al., 2014). The five subtypes differ in their expression pattern, physiological function, and G protein coupling. Three subtypes (M1, M3, and M5) signal predominantly through activation of G proteins from the Gq/11 family, whereas M2 and M4 mAChRs primarily signal through the Gi/o family of G proteins.
Drugs targeting the mAChRs are currently used in the treatment of chronic obstructive pulmonary disease, motion sickness, overactive bladder, and Sjögren’s syndrome (Kruse et al., 2014). However, the clinical translation of drugs selectivity targeting mAChRs, particularly for the treatment of neurological disorders, has proven elusive. For example, the M1/M4 mAChR-preferring agonist, xanomeline, showed promising results in a phase 3 Alzheimer’s disease trial (Bodick et al., 1997) and a small proof-of-concept trial in schizophrenia (Shekhar et al., 2008) but was ultimately abandoned because of unacceptable side effects, including those mediated by the related M2 and M3 subtypes in the periphery (Kruse et al., 2014). Xanomeline is an orthosteric agonist, binding to the highly conserved acetylcholine-binding site. Because of the difficulty in developing subtype selective drugs that target the acetylcholine-binding site, researchers have increasingly sought ligands that target allosteric sites. As a result, there are a plethora of allosteric ligands at hand for the mAChRs, and these have been extensively used for pharmacological inquiry into allosteric modulation (Bock et al., 2018). The concept of allosteric modulation of GPCRs was first described at this receptor family (Lüllmann et al., 1969; Clark and Mitchelson, 1976), and they are arguably the most well characterized GPCRs in regard to allosteric modulation (Gregory et al., 2007).
Structural features of inactive-state mAChRs
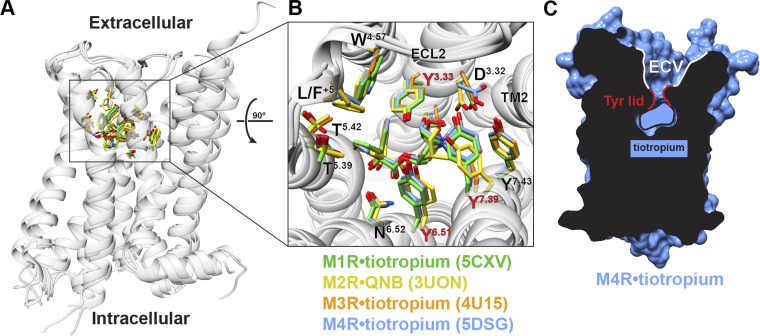
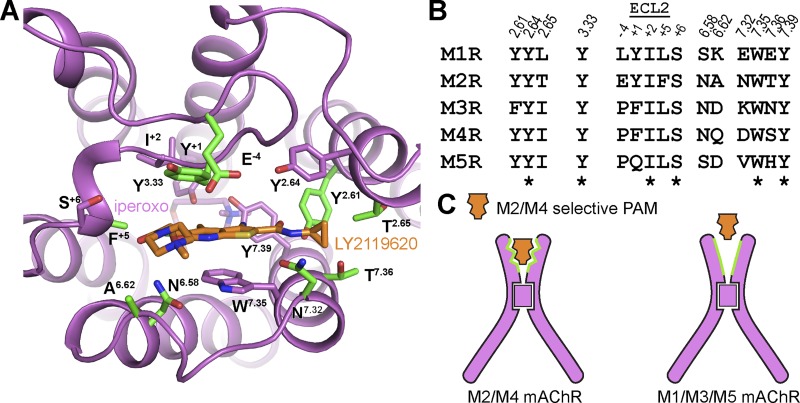
Breakthroughs in GPCR structural biology have led to inactive-state structures for four of the five of the mAChR subtypes (Fig. 2 A and Table 1). Overall, these structures are similar to each other and to those of other biogenic amine GPCRs that have been crystallized over the last decade. The M1, M3, and M4 mAChRs were crystallized with the antagonist tiotropium and the M2 mAChR with 3-quinuclidinyl benzilate (QNB; Table 1). These ligands bind in the orthosteric site, which is located within the extracellular third of the core of the seven-transmembrane domain. Comparison of the orthosteric-binding site across the M1–M4 mAChR structures reveals high similarity in the positions of amino acids that contact tiotropium or QNB, and notably, these residues are highly conserved across all subtypes (Fig. 2 B). This observation provides a molecular basis for the long-standing difficulty associated with the development of highly selective antagonists or agonists that target the mAChR orthosteric-binding site (Thal et al., 2016).
Figure 2.
Inactive-state structures of the M1–M4 mAChRs. (A) Alignment of the inactive-state structures of the M1–M4 mAChRs reveals high similarity in their structure. (B) The orthosteric site has high similarity in the positions and sequence of the amino acid residues that interact with the orthosteric ligand. Highlighted in red are the three tyrosine residues that separate the ECV from the orthosteric-binding site through the formation of a tyrosine lid. (C) Residue L/F+5 in ECL2 is numbered relative to the conserved disulfide bond between ECL2 and TM3. Protein Data Bank codes are indicated in B.
Table 1. Structures of mAChRs.
| Receptor | Ligand | Ligand type | Protein Data Bank accession number | Resolution (Å) | Reference |
|---|---|---|---|---|---|
| M1R-T4L | Tiotropium | Inverse agonist | 5CXV | 2.7 | Thal et al., 2016 |
| M2R-T4L | QNB | Inverse agonist | 3UON | 3.0 | Haga et al., 2012 |
| M2R•Nb9.8 | Iperoxo | Agonist | 4MQS | 3.5 | Kruse et al., 2013 |
| M2R•Nb9.8 | Iperoxo + LY2119620 | Agonist + PAM | 4MQT | 3.7 | Kruse et al., 2013 |
| M3R-T4L | Tiotropium | Inverse agonist | 4DAJ | 3.4 | Kruse et al., 2012 |
| M3R-dsT4L | Tiotropium | Inverse agonist | 4U14 | 3.6 | Thorsen et al., 2014 |
| M3R-mT4L | Tiotropium | Inverse agonist | 4U15 | 2.8 | Thorsen et al., 2014 |
| M3R-mT4L | NMS | Inverse agonist | 4U16 | 3.7 | Thorsen et al., 2014 |
| M4R-mT4L | Tiotropium | Inverse agonist | 5DSG | 2.6 | Thal et al., 2016 |
Notwithstanding the high degree of amino acid sequence conservation within the mAChR orthosteric-binding site, agonists and antagonists with limited subtype selectivity have been identified. This would suggest that selectivity can be achieved through subtle differences in receptor conformation or receptor dynamics. Although comparison of the M1–M4 crystal structures showed that the orthosteric sites are highly similar, some differences were observed. In particular, residues D3.32 and Y7.39 (superscript numbers are from the Ballesteros and Weinstein scheme for conserved class A GPCR residues; Ballesteros and Weinstein, 1995) occupy different rotameric states in the M2 mAChR versus the other subtypes (Fig. 2 B). As a result, the M4 mAChR appears more similar in structure to the M1 mAChR than its more closely related subfamily member, the M2 mAChR. These differences may also rationalize the rank potency order of the clinically used antagonist, pirenzepine (M1>M4>M3>M2). Nonetheless, these subtle conformational differences in the solved structures could also be attributed to the fact that M2 and M1/M4 mAChRs were cocrystallized with different antagonists (Thal et al., 2016).
A relatively unique feature of inactive-state mAChR structures is a large solvent-accessible extracellular vestibule (ECV), which is separated from the orthosteric-binding pocket by three tyrosine residues: Y3.33, Y6.51, and Y7.39 (Fig. 2 C). These tyrosine residues (or “tyrosine lid”; Kruse et al., 2013) are part of a hydrogen bond network that forms a roof over the orthosteric site, thus creating a floor for an allosteric site located in the ECV. Importantly, residues lining the ECV have a lower amino acid sequence conservation than residues in the orthosteric site and have therefore made the ECV an attractive target for the development of subtype-selective allosteric modulators. Indeed, numerous PAMs and NAMs have been discovered that target the ECV, resulting in the ECV being labeled as a “common” allosteric site for mAChRs (Wess, 2005; Stahl and Ellis, 2010; Valant et al., 2012; Abdul-Ridha et al., 2014).
The structural basis of positive allosteric modulation
Recently, crystal structures were determined of an active-state M2 mAChR with the highly potent and efficacious agonist iperoxo in a ternary complex with a G protein mimetic (nanobody) in the presence and absence of the PAM LY2119620 (Table 1; Kruse et al., 2013). When analyzed in the context of previous crystal structures, the M2 mAChR structures provide key insight into the mechanisms behind receptor activation and allosteric modulation.
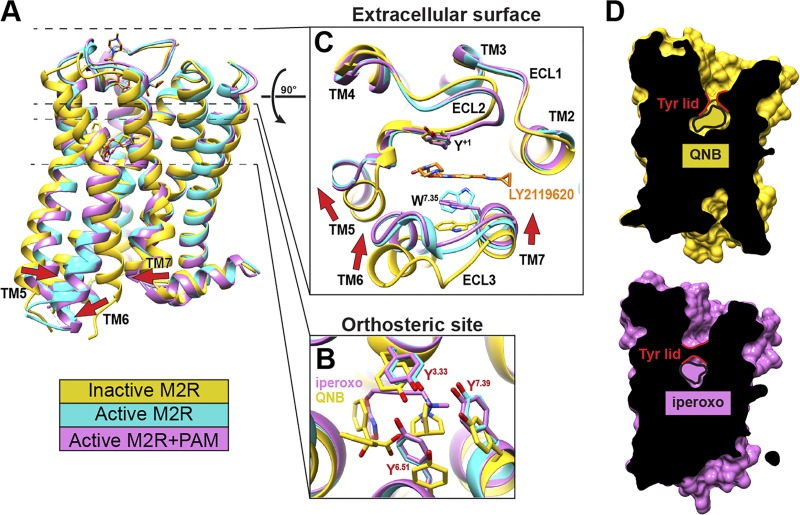
A hallmark feature of GPCR activation that is readily observed in the M2•iperoxo structures is an outward displacement of TM6 at the cytoplasmic end to create a binding site for G proteins (Fig. 3 A; Lebon et al., 2011; Rasmussen et al., 2011b). This displacement is accompanied by additional movements in TM5 and TM7 at the cytoplasmic end and is linked to a striking contraction in the size of the orthosteric-binding site (Fig. 3 B). This is mediated by an inward movement of extracellular ends of TM6 and TM7 at the orthosteric site, which repositions the tyrosine lid into a hydrogen bond network that completely seals the orthosteric site from the ECV (Fig. 3, C and D).
Figure 3.
Comparison of the inactive and active states of the M2 mAChR. (A) Alignment of the inactive (yellow), active (cyan), and active-state, PAM-bound (pink) M2 mAChR structures reveal reorganization of TMs 5, 6, and 7 in the active state (indicated by arrows). Residue Y+1 in ECL2 is numbered relative to the conserved disulfide bond between ECL2 and TM3. (B and C) Views of the orthosteric- (B) and ECV-binding (C) sites show contraction mediated by the movements of TM5, TM6, ELC3, and TM7. (D) Cross sections of the receptor shown with the interior surface colored black. The tyrosine lid capping the orthosteric site is highlighted in red and completely seals the orthosteric site in the active state. LY2119620 is not shown for clarity.
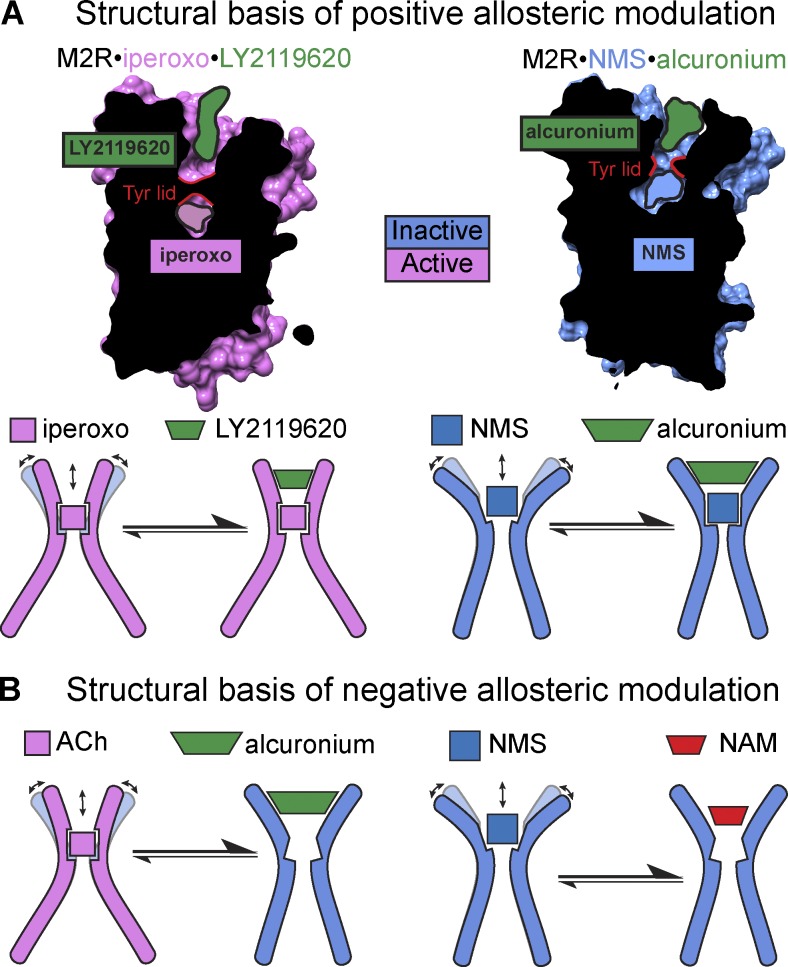
The changes in the orthosteric site that occur upon receptor activation are accompanied by a substantial reduction in size of the ECV and consequently the allosteric site. Interestingly, this occurs regardless of whether or not the PAM LY2119620 is present. Aside from a change in the rotamer of side chain W7.35 (Fig. 3 C), comparison of the two active states with and without the PAM reveal high similarity in the ECV, indicating the presence of a “preformed” allosteric site. Importantly, LY2119620 sits above the closed tyrosine lid interacting with Y7.39, thus further stabilizing closure of the lid. Altogether these observations help explain how mAChR PAMs exhibit positive cooperativity with orthosteric agonists. Through preferentially binding and stabilizing a preexisting closed, active conformation of the ECV, PAMs drive the conformational landscape toward the active conformation and allow for the transmission of positive cooperativity to occur between the two binding sites (Fig. 4 A).
Figure 4.
Structural insights into allosteric modulation of mAChRs. (A) Structural basis of positive allosteric modulation. In the active-state M2R•iperoxo•LY2119620 structure, LY2119620 acts as a small “wedge” by binding to a closed ECV conformation that further stabilizes the active state of the receptor. A similar mechanism can be observed for the inactive state, whereby a NAM of an agonist can behave as a PAM of an inverse agonist. At the M2 mAChR, MD simulations revealed that alcuronium acts a PAM for the inverse agonist NMS by stabilizing the ECV in a more open conformation (relative to not only the active state but also the inactive state in the absence of modulator; Dror et al., 2013). (B) Structural basis of negative allosteric modulation. NAMs of mAChR agonists promote a more open ECV receptor conformation that is detrimental to agonist binding and G protein coupling. In contrast, NAMs of inverse agonists are not large enough to stabilize the open ECV, thus favoring a conformation that precludes inverse agonist binding. Arrows indicate an increase in the dynamics of the ECV that could reflect effects on ligand dissociation.
Recent structural and biophysical studies have explained the long-standing pharmacological observations (De Lean et al., 1980) of G proteins exhibiting an allosteric character (Rasmussen et al., 2011a; DeVree et al., 2016; Staus et al., 2016). Using the M2 mAChR as an example, the binding of a G protein or G protein–mimicking nanobody was shown to substantially enhance the binding of the agonist iperoxo for the receptor, indicating an allosteric interaction between the G protein mimetic and the receptor (DeVree et al., 2016). Additionally, the G protein mimetic promotes a “closed” conformation of the ECV through an ∼2.5-Å inward movement, toward the core of the TM bundle, that is mediated by TM6 and ECL3 (Fig. 3 C; Kruse et al., 2013). At the β2-adrenergic receptor, closure of the ECV was shown to impede the association and dissociation of ligands into and out of the orthosteric site. Similar findings were observed at the M2 mAChR and the µ-opioid receptor, suggesting a common mechanistic rationale for the role of G proteins in the allosteric enhancement of agonist affinity (DeVree et al., 2016). Significantly, the M2 mAChR PAM LY2119620 directly interacts with residues in the closed ECV conformation and coordinates a three-way π–π stacking interaction with Y+1 (numbered relative to the conserved disulfide bond between ECL2 and TM3) and W7.35 (Fig. 3 C; Kruse et al., 2013). Mutation of either of these residues to an alanine at the M4 mAChR completely abolishes the allosteric effect of the highly similar PAM LY2033298 (Nawaratne et al., 2010; Thal et al., 2016). Overall, this indicates that stabilization of the ECV is driven by a concerted and synergistic allosteric interaction from both PAMs and G proteins as the main mechanism through which agonist activity is enhanced, and the extent of such stabilization may be linked to the degree of cooperativity (i.e., high vs. low).
The structural basis of negative allosteric modulation
To date, there are no available crystal structures of mAChRs bound to a small-molecule NAM. Because of a lack of structural data, insight into negative allosteric modulation at mAChRs has been addressed using molecular dynamics (MD) simulations. Dror et al. (2013) used long-timescale MD simulations on the inactive-state M2 mAChR structure to investigate the mechanism of action of a number of mAChR NAMs. The binding modes of these NAMs are quite similar and predicted to involve cation-π interactions with aromatic residues in the common ECV allosteric site, as supported by previous mutagenesis studies. Significantly, the MD simulations revealed that certain NAMs of the inverse agonist, N-methylscopolamine (NMS), such as C7/3-phth and gallamine, occupy a restricted volume of the open ECV NMS-bound conformation (Fig. 4 B), such that cobinding is not favored. In contrast, alcuronium, a PAM of NMS, is bulky and favors a more “open” ECV conformation when the receptor is bound to NMS (Fig. 4 A; Dror et al., 2013). This explains why, pharmacologically, C7/3-phth and gallamine are NAMs of NMS, whereas alcuronium is a PAM of NMS, as it promotes a more open yet still inactive state that can simultaneously accommodate both ligands. However, it should be noted that because in all instances these modulators promote inactive states (albeit to varying extents), they are all NAMs of orthosteric agonists, irrespective of their differential effects on the binding of orthosteric antagonists or inverse agonists (Clark and Mitchelson, 1976; Jakubík et al., 1997; Christopoulos et al., 1999). In addition, although not always appreciated, it should be noted that direct charge–charge interactions between allosteric and orthosteric ligands may be an additional mechanism contributing to the observed allosteric effect over and above discrete changes in receptor conformation (Dror et al., 2013); the generality of this second mechanism of allostery across different classes of GPCRs remains to be determined and thus represents an interesting area for future studies.
Interestingly, structures of NAMs bound to other GPCRs have also been solved, as evidenced, for example, by the NAM-bound crystal structures of the seven-transmembrane domain regions of the class C metabotropic glutamate receptors 1 and 5 (Congreve et al., 2017) and others (Oswald et al., 2016; Zheng et al., 2016; Liu et al., 2017; Song et al., 2017; Thal et al., 2018). However, it should also be noted that none of these (non-mAChR) structures have found the NAM to be in a location similar to the ECV of the mAChRs. Collectively, this suggests that there are multiple structural explanations for explaining GPCR NAM activity based on binding locus, which is not necessarily surprising given the plethora of binding modes and types of ligands that can interact with GPCRs, but, mechanistically, the ability to prevent adoption of an active state likely explains how most such compounds work.
A two-state model of allostery
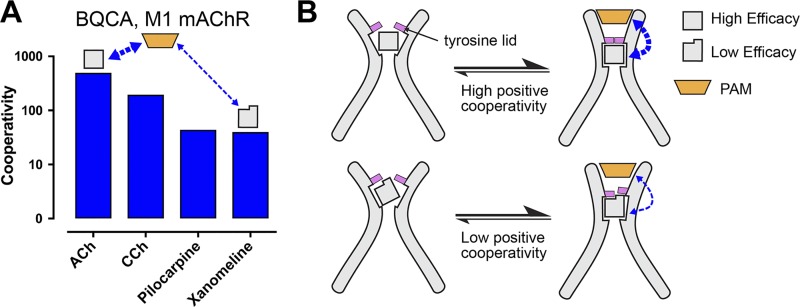
The observations that mAChR allosteric modulators stabilize and select for open and closed conformations of the ECV indicates that the allosteric transition from an inactive to active state generally adheres to a two-state model, as exemplified by the classic Monod–Wyman–Changeux mechanism (Monod et al., 1965). With specific relation to the mAChRs, therefore, one would expect extracellular region–targeting PAMs of agonists to be smaller molecules that select for a closed ECV (i.e., active-state conformation) but concomitantly act as NAMs of antagonists (more correctly, inverse agonists), because the latter orthosteric ligand prefers a more open ECV associated with the inactive-state conformation (Fig. 4). This is best exemplified by pharmacological studies of the M1 mAChR-selective PAM, benzylquinolone carboxylic acid (BQCA), which inhibits the binding of the inverse agonist radioligand, [3H]NMS, while concomitantly enhancing the binding of the agonist carbachol (Canals et al., 2012). This is an instructive example of how BQCA displays a preference for the active-state receptor (via an allosteric site) and thus acts as a NAM for inverse agonists but a PAM for agonists. According to the two-state model, this also implies that the degree of modulation should “track” with the intrinsic efficacy of the orthosteric ligand used as a “probe” against which the actions of the modulator are assessed. Again, this behavior was experimentally verified in signaling assays at the M1 mAChR, where BQCA displayed greater positive cooperativity with full agonists than with partial agonists (Fig. 5 A; Canals et al., 2012). This is an example of “probe dependence” (i.e., a change in the direction and magnitude of allosteric modulation depending on the nature of the orthosteric probe ligand; Christopoulos et al., 2014) that is mechanistically determined by receptor “state dependence” (i.e., the proportion of active vs. inactive states promoted by the orthosteric ligand). Within such a mechanism, a full agonist would be expected to promote a more active receptor conformation than a partial agonist and thus display greater positive allosteric modulation when tested against the same modulator (Fig. 5 B), whereas an inverse agonist would be inhibited depending on how effective it is at promoting an inactive state (i.e., the modulator would be a NAM). An important outcome from these types of findings is that the classification of an allosteric ligand as a PAM, NAM, or even NAL is not absolute but rather context dependent, being additionally determined by the nature of the orthosteric ligand and the resultant receptor conformational state, ultimately manifested by the functional effect on the monitored cellular response (Christopoulos et al., 2014; see below).
Figure 5.
Probe dependence as a consequence of a two-state model of allostery. (A) Cooperativity values for BQCA and different orthosteric agonists at the M1 mAChR from cAMP accumulation assays (y axis values denote the fold increase in agonist potency in the presence of modulator). Acetylcholine (ACh) and carbachol (CCh) are high-efficacy agonists and are potentiated to a greater extent than the low-efficacy agonists pilocarpine and xanomeline. Thus, the degree of cooperativity is correlated with the intrinsic efficacy of each ligand. Data are replotted from Canals et al. (2012). (B) A speculative cartoon for a two-state model of allostery at the M1 mAChR. High efficacy agonists have high cooperativity with PAMs by promoting a fully active conformation, indicated by a fully closed tyrosine lid. Low-efficacy agonists do not promote a fully active conformation, indicated by a partially closed tyrosine lid, and thus have lower cooperativity with BQCA.
The molecular basis of subtype selectivity
The allosteric modulators BQCA and LY2033298 are highly selective PAMs for the M1 and M2/M4 mAChRs, respectively. Two fundamental questions, in the absence of crystallographic data, are where do these ligands bind and how is selectivity between the subtypes mediated? Analysis of residues in the allosteric binding pocket revealed by the M2 mAChR active structure show that many of the residues surrounding LY2119620 are nonconserved residues (Fig. 6, A and B). This suggests that allosteric modulators may display a unique interaction profile at each receptor that is capable of leading to subtype selectivity (Fig. 6 C). Additionally, further selectivity may arise from differences in the conformation of both conserved and nonconserved residues that occur during the allosteric transition between inactive and active states. Through unique interactions of a modulator with nonconserved residues at a given subtype, a more favorable allosteric transition may occur that is not observed at other subtypes, independent of ligand affinity for the unoccupied receptor. Consequently, selectivity can arise through differences in cooperativity, where allosteric modulator binding occurs at all subtypes but only one subtype undergoes an enhanced activation transition that favors agonist interaction. Such is the case for thiochrome, where cooperativity with acetylcholine is the main component for its selective action at the M4 mAChR (Lazareno et al., 2004). However, the mechanistic basis for allosteric effects on activation transition remains speculative and requires further enquiry.
Figure 6.
A potential mechanism of PAM selectivity. (A) The M2/M4 mAChR selective PAM LY2119620 binds in the allosteric pocket, as revealed by the active-state M2 mAChR crystal structure (Protein Data Bank accession no. 4MQT). Residues within 4 Å of LY2119620 (colored orange) are shown as green colored sticks. Residues in ECL2 are numbered relative to the conserved disulfide bond between ECL2 and TM3. (B) Comparison of residues from A across all five human mAChR subtypes. Conserved residues are identified by an asterisk. (C) Nonconserved residues surrounding the LY2119620-binding site may contribute to selectivity.
An allosteric network facilitates conformational selection
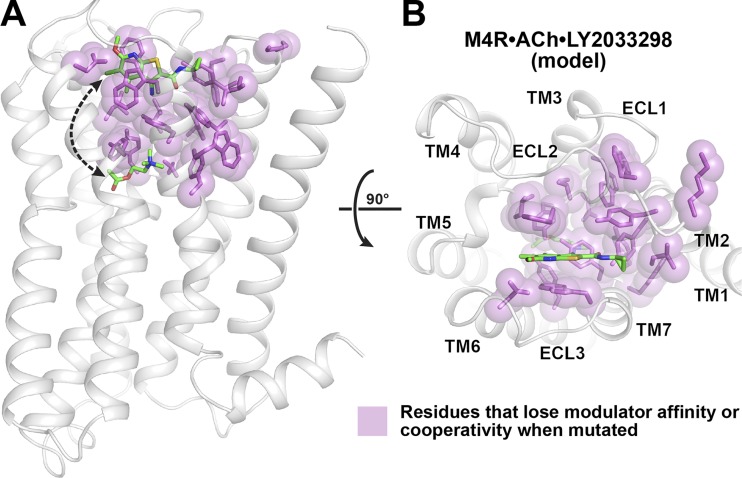
Further evidence for conformational selection as a mechanism for allosteric modulation at the mAChRs comes from recent work on the M1 and M4 mAChRs (Thal et al., 2016). Specifically, by mapping residues that significantly affect PAM cooperativity or binding onto an active-state M4 mAChR model, a “network” was identified that links the allosteric and orthosteric sites (Fig. 7 A). For the M4 receptor, this allosteric network includes residues in TMs 2, 3, 6, and 7 and part of ECL2 (Fig. 7 B). Comparison of the allosteric network residues on the inactive M4 mAChR crystal structure and an active M4 mAChR homology model revealed that residues at the TM2/3/7 interface do not undergo appreciable movement between the inactive and active states (Thal et al., 2016), whereas residues nearer the extracellular surface (highlighted in Fig. 3 C) move significantly between the two states. Given that the TM2/3/7 interface forms part of the hydrophobic core of the receptor, it may act as a “hinge” that facilitates the conformational rearrangement in the ECV that is required for the transition from the inactive to active state. Thus, the binding of an allosteric modulator to the ECV would stabilize the hinge that may otherwise be in a more dynamic state. A corollary to this prediction is that alterations to the TM2/3/7 interface via either mutagenesis or through ligand-induced binding events could alter the nature of the receptor conformation that is stabilized and selected for. This would dictate whether an inactive or active state of the receptor is formed and/or whether positive or negative cooperativity is exerted onto an orthosteric ligand. The “allosteric network” may therefore be a mechanism through which allosteric modulators affect receptor state transitions giving rise to subtype selectivity. Although this structurally linked allosteric network can be envisaged as an energetic “hotwire” linking the orthosteric and allosteric sites and therefore be interpreted as evidence of an induced-fit mechanism (Lockless and Ranganathan, 1999; Süel et al., 2003), it is still consistent with a conformational selection mechanism in that the communication between the hotwire residues changes by virtue of the transition from one state to another.
Figure 7.
An allosteric network at the M4 mAChR. (A) Previous studies on the M4 mAChR have identified residues that either disrupt the binding of LY2033298 or alter the cooperativity between the allosteric and orthosteric site. These residues are shown in pink, modeled on to an active-state M4 mAChR homology model (white) based on the M2•iperoxo•LY2119620 crystal structure (Protein Data Bank accession no. 4MQT; Nawaratne et al., 2010; Leach et al., 2011; Thal et al., 2016). Views from the side (A) and extracellular surface (B) are shown.
Biased allosteric modulation
Despite the evidence that a two-state model of allostery can operate at the mAChRs, interesting pharmacological observations have been reported that are not compatible with the existence of a single active or inactive state. This implies that allosteric modulators are capable of stabilizing multiple active or inactive receptor conformations independent from the orthosteric ligand efficacy for a given pathway (i.e., state dependence) and therefore leading to a different manifestation of probe dependence, both with respect to magnitude and direction, a phenomenon commonly known as biased modulation. As mentioned to above, biased modulation refers to the ability of an allosteric ligand to stabilize more than one type of active or inactive receptor conformation over others, each linked to distinct cellular signaling outcomes (Kenakin and Christopoulos, 2013; Edelstein and Changeux, 2016; Fig. 1 E). Numerous examples of biased modulation have been described (Gentry et al., 2015), including the M1 mAChR allosteric modulator VU0029767 that shows pathway bias by acting as a PAM of acetylcholine for intracellular Ca2+ mobilization while acting as a NAL of acetylcholine for phospholipase D activation (Marlo et al., 2009). Another intriguing example is observed with the allosteric ligand LY2033298 at the M2 mAChR, where biased modulation manifests as a function of the orthosteric ligand used as a probe, as well as the cellular mechanism being measured. For example, in ERK1/2 phosphorylation assays, LY2033298 acts as a PAM for acetylcholine but as a NAM for the agonists pilocarpine and xanomeline (Fig. 8; Valant et al., 2012). However, the modulator is a PAM of all agonists when monitored for its effects on agonist-binding affinity. Thus, the combination of both an agonist and LY2033298 must stabilize different subsets of states that are not normally of high abundance when either ligand is bound alone. Because the phenomenon is both pathway and ligand dependent, this is an example of probe dependence that arises as a consequence of biased modulation, in contrast to the simpler mechanism of probe dependence within a two-state formalism outlined above. In the broadest sense, the existence of biased modulation requires the ability of some ligands to select between multiple active conformations, which is consistent with the notion that GPCRs can explore multiple conformational ensembles. However, atomistic level information on how such biased conformational selection occurs is largely lacking. Because of the differences in the agonists studied, particularly in terms of their size and functional groups, the size and shape of the orthosteric-binding pocket may vary at the microscopic level. For example, bulkier agonists could sterically restrict closure of the orthosteric site, causing incomplete “closure” of the key tyrosine lid associated with mAChR activation (Fig. 5 B). As a result, the allosteric interaction with LY2033298 may differ, resulting in the promotion of distinct conformational states that drive the observed bias. Alternatively, it is possible that LY2033298 is influenced by conformational changes associated outside of the binding pocket, albeit linked to it, via the aforementioned allosteric network to mediate its effects and drive bias (Fig. 7). In fact, residues near allosteric networks involving ECL2, ECL3, and TM2/6/7 have been implicated in mediating biased signaling of orthosteric ligands at several other GPCRs (Steen et al., 2013; Wacker et al., 2013, 2017b; Weston et al., 2016; Wootten et al., 2016; Dal Maso et al., 2018; Lei et al., 2018), perhaps suggesting that alteration of an allosteric network may be an important trigger in the propagation of biased signaling irrespective of whether the ligand is orthosteric or allosteric. At this stage, a more thorough interpretation of how allosteric modulators modify the conformational landscape will likely require complementary approaches, such as NMR and MD, that can better capture conformational dynamics and discrete receptor states, as recently shown for the β2-adrenergic receptor and M2 mAChR (Nygaard et al., 2013; Basith et al., 2018; Jiménez-Rosés et al., 2018).
Figure 8.
Probe dependence as a consequence of biased modulation at the M2 mAChR. Cooperativity values of LY2033298 for different orthosteric agonists at the M2 mAChR from ERK1/2 phosphorylation assays. Note that the magnitude and direction of the allosteric effect are not related to the degree of intrinsic efficacy of the orthosteric agonist. Data represent mean ± SEM and are replotted from Valant et al. (2012).
Oligomerization of mAChRs
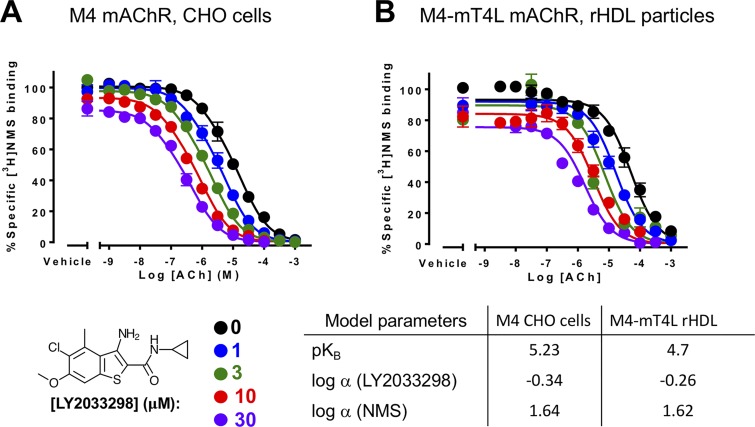
The oligomerization of protein molecules is a central feature of allosteric modulation for many classes of signaling proteins (Changeux and Christopoulos, 2016). However, the dimerization or oligomerization of GPCRs has remained a controversial topic for the last few decades. Although it is clear that dimerization is a key feature for the function of class C GPCRs (Pin et al., 2009), the role of oligomerization for other GPCRs remains less clear, as numerous biochemical and biophysical studies have indicated that monomers are sufficient to act as the minimal functional unit, at least for G protein activation (Gurevich and Gurevich, 2018). Nevertheless, GPCR oligomerization remains a heavily studied and important area of research, and as a model system, mAChRs have played an important role in many of these studies, as exemplified by a recent and extensive review by Marsango et al. (2017). Notably, key findings from previous research supporting oligomerization of mAChRs include multiphasic competition binding curves between an orthosteric ligand and a prototypical mAChR allosteric modulator that are suggestive of a tetrameric assembly between mAChR molecules. In those studies, the authors conclude that cooperativity occurs between orthosteric sites of the tetramer (Shivnaraine et al., 2012). Such results have led to speculation that the observed cooperativity between orthosteric ligands and allosteric modulators may be mediated by receptor oligomers. To begin to address this issue, we have recently introduced purified M4 mAChRs into reconstituted high-density lipoprotein (rHDL) particles, or nanodiscs, to probe the binding properties of monomeric receptor, as was previously done for the β2-adrenergic receptor and the glucagon-like peptide-1 receptor (Whorton et al., 2007; Cai et al., 2017). As shown in Fig. 9, the pharmacological properties of the PAM LY2033298 in radioligand competition binding assays, for both M4 mAChRs expressed in CHO membranes and M4 mAChRs purified into rHDL particles, are nearly identical. This includes the degree of positive modulation with the agonist acetylcholine, the degree of negative modulation with the radiolabeled inverse agonist [3H]NMS, and the binding affinity of LY2033298. Although preliminary, these findings indicate that, at least for the M4 mAChR with ACh and LY2033298, a monomer is the minimal functional unit for observing positive cooperativity between the orthosteric and allosteric sites. Likewise, similar observations were seen at the glucagon-like peptide-1 receptor, where small-molecule PAM effects on orthosteric peptide signaling were maintained in dimer-deficient receptors (Harikumar et al., 2012). A possible explanation for some of the differences between studies that conclude allostery requires an obligatory oligomeric mechanism relative to those consistent with allostery within a receptor monomer may be related to differences in the assay conditions, such as kinetic (Avlani et al., 2004; Lane et al., 2017) or desensitization (Insel et al., 1983; Toews et al., 1983; Toews and Perkins, 1984) mechanisms that contribute to complex binding or concentration–response curves.
Figure 9.
Positive allosteric modulation of monomeric M4 mAChRs. (A and B) [3H]NMS interaction binding studies between the PAM, LY2033298, and acetylcholine at the M4 mAChR expressed in either CHO cells (A; replotted from Leach et al., 2010) or purified from Sf9 cells (B; M4R-mT4L; for methods, see Thal et al., 2016) and reconstituted into rHDL particles, as was previously done for rhodopsin (Whorton et al., 2007). Nearly identical levels of cooperativity were observed, indicating that LY2033298 is able to influence the binding of acetylcholine in a monomeric mAChR and is not dependent on oligomerization status of the receptor. Data from B represent the mean ± SEM of n = 3 experiments performed in duplicate.
Interestingly, a recent study has shown that orthosteric antagonists can differentially alter the ratio of receptors from a monomeric to oligomeric state at the M1 mAChRs (Pediani et al., 2016). For example, the M1 selective antagonists pirenzepine and telenzepine promoted oligomerization of the M1 mAChR, whereas neither atropine nor NMS produced an effect. Furthermore, this differential oligomerization appeared to be receptor specific, as pirenzepine and telenzepine did not affect M3 mAChR oligomerization when tested at concentrations appropriate for the lower-affinity binding (Pediani et al., 2016). Overall, these results suggest that ligand-mediated regulation of oligomerization is “probe dependent” and may be receptor selective. It is intriguing to speculate that this oligomeric probe dependence may be linked to the observed probe dependence between orthosteric and allosteric sites and potentially even receptor selectivity. Certainly, more studies will be required to understand the molecular basis of oligomerization and underpin its role in pharmacology and physiology at mAChRs and more broadly at other GPCRs.
Future perspectives and conclusions
A range of crystal structures are now available for the mAChRs, including inactive, active, and allosteric modulator-bound structures. Nevertheless, a number of key structures remain unsolved that would add much-needed insight into the structural basis of allostery. For example, NAM-bound inactive-state structures with and without an orthosteric ligand would show whether or not negative allosteric modulation at mAChRs occurs through a mechanism similar to that evidenced by cobound structures of other GPCRs. Although there is a PAM-bound M2 mAChR structure, given the vast range of pharmacological behaviors displayed by different PAMs across the five mAChR subtypes, additional PAM-bound structures would be of significant interest. For instance, given the state dependence displayed by BQCA, one could envisage that atomistic level differences may be directly observed in GPCR structures solved with the PAM cobound with a full agonist relative to structures solved with the same PAM cobound with a partial agonist. Though a repertoire of structural information exists for orthosteric full and partial agonist active states at other receptors, such as the adenosine A2A receptor and the β2 adrenergic receptor (Lebon et al., 2011; Rasmussen et al., 2011b; Rosenbaum et al., 2011), the effects of allosteric ligands on these conformational states remain largely unknown. Even more strikingly, as evidence by the biased modulation exhibited by LY2033298 at the M2 mAChR, it would be of substantial interest to see how this PAM actually promotes distinct receptor conformations depending on the cobound orthosteric ligand. This structural information could then be used to explain how different receptor conformations drive biased modulation.
Despite the sequence and structural breadth within the GPCR superfamily, intracellular signaling is achieved through the recruitment of only a handful of transducers. Most GPCRs display selectivity in the G proteins that they couple to, yet others have broader G protein–coupling profiles. How this is achieved and what drives the selective coupling of a specific G protein remains largely unknown. Though recent studies have pointed toward the affinity of the G protein for the receptor and the existence of universally conserved patterns of amino acids as the mechanism through which selective coupling is achieved (Flock et al., 2017; Ilyaskina et al., 2018), this area remains worthy of further exploration with evidence for ligand-dependent effects on the rate of G protein activation that are distinct from efficiency of recruitment (Furness et al., 2016). Furthermore, though the role of the G protein in driving extracellular changes has been somewhat explored at the M2 mAChR (DeVree et al., 2016), a broader understanding of how transducer proteins drive changes in the ECV remains absent. This includes the impact of arrestin engagement; little is known about its potential role in causing changes in the ECV that would affect allosterism. Given that G proteins and arrestins are, by their very nature, endogenous allosteric modulator partners of GPCRs, there is potential for transducer proteins to select for specific conformational states in coordination with orthosteric and/or allosteric ligands. In turn, this could manifest into biased modulation and probe dependence driven by the availability of effector proteins, a concept that is poorly explored in the context of allosteric modulation at GPCRs.
Structural information at the mAChRs has revealed why it is difficult to obtain selective orthosteric ligands while providing insights into the mechanism of how some allosteric modulators achieve selectivity. Unraveling the role of potential allosteric networks that link orthosteric and modulator binding sites in driving selectivity would provide further structural and mechanistic rationales for how allosteric ligands can achieve receptor subtype selectivity through cooperativity. Such understanding would be broadly relevant for other GPCR families. We expect that addressing these remaining questions will expand our knowledge of the structural basis of allosteric modulation and receptor dynamics and facilitate the development and therapeutic application of allosteric modulators.
Acknowledgments
This work was supported by the National Health and Medical Research Council of Australia project grant APP1138448. P.M. Sexton and A. Christopoulos are National Health and Medical Research Council Senior Principal Research Fellows, and D.M. Thal is an Australian Research Council Discovery Early Career Research Fellow.
The authors declare no competing financial interests.
Author contributions: W.A.C. Burger, P.M. Sexton, A. Christopoulos, and D.M. Thal wrote the manuscript.
Lesley C. Anson served as editor.
References
- Abdul-Ridha A., López L., Keov P., Thal D.M., Mistry S.N., Sexton P.M., Lane J.R., Canals M., and Christopoulos A.. 2014. Molecular determinants of allosteric modulation at the M1 muscarinic acetylcholine receptor. J. Biol. Chem. 289:6067–6079. 10.1074/jbc.M113.539080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avlani V., May L.T., Sexton P.M., and Christopoulos A.. 2004. Application of a kinetic model to the apparently complex behavior of negative and positive allosteric modulators of muscarinic acetylcholine receptors. J. Pharmacol. Exp. Ther. 308:1062–1072. 10.1124/jpet.103.059840 [DOI] [PubMed] [Google Scholar]
- Ballesteros J.A., and Weinstein H.. 1995. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods in Neurosciences. 366–428. 10.1016/S1043-9471(05)80049-7 [DOI] [Google Scholar]
- Basith S., Lee Y., and Choi S.. 2018. Understanding G protein-coupled receptor allostery via molecular dynamics simulations: Implications for drug discovery. Methods Mol. Biol. 1762:455–472. 10.1007/978-1-4939-7756-7_23 [DOI] [PubMed] [Google Scholar]
- Bock A., Schrage R., and Mohr K.. 2018. Allosteric modulators targeting CNS muscarinic receptors. Neuropharmacology. 136:427–437. 10.1016/j.neuropharm.2017.09.024 [DOI] [PubMed] [Google Scholar]
- Bodick N.C., Offen W.W., Levey A.I., Cutler N.R., Gauthier S.G., Satlin A., Shannon H.E., Tollefson G.D., Rasmussen K., Bymaster F.P., et al. . 1997. Effects of xanomeline, a selective muscarinic receptor agonist, on cognitive function and behavioral symptoms in Alzheimer disease. Arch. Neurol. 54:465–473. 10.1001/archneur.1997.00550160091022 [DOI] [PubMed] [Google Scholar]
- Cai Y., Liu Y., Culhane K.J., DeVree B.T., Yang Y., Sunahara R.K., and Yan E.C.Y.. 2017. Purification of family B G protein-coupled receptors using nanodiscs: Application to human glucagon-like peptide-1 receptor. PLoS One. 12:e0179568 10.1371/journal.pone.0179568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canals M., Lane J.R., Wen A., Scammells P.J., Sexton P.M., and Christopoulos A.. 2012. A Monod-Wyman-Changeux mechanism can explain G protein-coupled receptor (GPCR) allosteric modulation. J. Biol. Chem. 287:650–659. 10.1074/jbc.M111.314278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caulfield M.P., and Birdsall N.J.M.. 1998. International Union of Pharmacology. XVII. Classification of muscarinic acetylcholine receptors. Pharmacol. Rev. 50:279–290. [PubMed] [Google Scholar]
- Changeux J.P., and Christopoulos A.. 2016. Allosteric modulation as a unifying mechanism for receptor function and regulation. Cell. 166:1084–1102. 10.1016/j.cell.2016.08.015 [DOI] [PubMed] [Google Scholar]
- Christopoulos A., and Kenakin T.. 2002. G protein-coupled receptor allosterism and complexing. Pharmacol. Rev. 54:323–374. 10.1124/pr.54.2.323 [DOI] [PubMed] [Google Scholar]
- Christopoulos A., Sorman J.L., Mitchelson F., and El-Fakahany E.E.. 1999. Characterization of the subtype selectivity of the allosteric modulator heptane-1,7-bis-(dimethyl-3′-phthalimidopropyl) ammonium bromide (C7/3-phth) at cloned muscarinic acetylcholine receptors. Biochem. Pharmacol. 57:171–179. 10.1016/S0006-2952(98)00277-9 [DOI] [PubMed] [Google Scholar]
- Christopoulos A., Changeux J.P., Catterall W.A., Fabbro D., Burris T.P., Cidlowski J.A., Olsen R.W., Peters J.A., Neubig R.R., Pin J.P., et al. . 2014. International Union of Basic and Clinical Pharmacology. XC. multisite pharmacology: recommendations for the nomenclature of receptor allosterism and allosteric ligands. Pharmacol. Rev. 66:918–947. 10.1124/pr.114.008862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A.L., and Mitchelson F.. 1976. The inhibitory effect of gallamine on muscarinic receptors. Br. J. Pharmacol. 58:323–331. 10.1111/j.1476-5381.1976.tb07708.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congreve M., Oswald C., and Marshall F.H.. 2017. Applying structure-based drug design approaches to allosteric modulators of GPCRs. Trends Pharmacol. Sci. 38:837–847. 10.1016/j.tips.2017.05.010 [DOI] [PubMed] [Google Scholar]
- Dal Maso E., Zhu Y., Pham V., Reynolds C.A., Deganutti G., Hick C.A., Yang D., Christopoulos A., Hay D.L., Wang M.W., et al. . 2018. Extracellular loops 2 and 3 of the calcitonin receptor selectively modify agonist binding and efficacy. Biochem. Pharmacol. 150:214–244. 10.1016/j.bcp.2018.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lean A., Stadel J.M., and Lefkowitz R.J.. 1980. A ternary complex model explains the agonist-specific binding properties of the adenylate cyclase-coupled beta-adrenergic receptor. J. Biol. Chem. 255:7108–7117. [PubMed] [Google Scholar]
- DeVree B.T., Mahoney J.P., Vélez-Ruiz G.A., Rasmussen S.G.F., Kuszak A.J., Edwald E., Fung J.-J., Manglik A., Masureel M., Du Y., et al. . 2016. Allosteric coupling from G protein to the agonist-binding pocket in GPCRs. Nature. 535:182–186. 10.1038/nature18324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror R.O., Green H.F., Valant C., Borhani D.W., Valcourt J.R., Pan A.C., Arlow D.H., Canals M., Lane J.R., Rahmani R., et al. . 2013. Structural basis for modulation of a G-protein-coupled receptor by allosteric drugs. Nature. 503:295–299. 10.1038/nature12595 [DOI] [PubMed] [Google Scholar]
- Edelstein S.J., and Changeux J.P.. 2016. Biased Allostery. Biophys. J. 111:902–908. 10.1016/j.bpj.2016.07.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flock T., Hauser A.S., Lund N., Gloriam D.E., Balaji S., and Babu M.M.. 2017. Selectivity determinants of GPCR-G-protein binding. Nature. 545:317–322. 10.1038/nature22070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furness S.G.B., Liang Y.L., Nowell C.J., Halls M.L., Wookey P.J., Dal Maso E., Inoue A., Christopoulos A., Wootten D., and Sexton P.M.. 2016. Ligand-dependent modulation of G protein conformation alters drug efficacy. Cell. 167:739–749.e11. 10.1016/j.cell.2016.09.021 [DOI] [PubMed] [Google Scholar]
- Gentry P.R., Sexton P.M., and Christopoulos A.. 2015. Novel allosteric modulators of G protein-coupled receptors. J. Biol. Chem. 290:19478–19488. 10.1074/jbc.R115.662759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory K.J., Sexton P.M., and Christopoulos A.. 2007. Allosteric modulation of muscarinic acetylcholine receptors. Curr. Neuropharmacol. 5:157–167. 10.2174/157015907781695946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich V.V., and Gurevich E.V.. 2018. GPCRs and Signal Transducers: Interaction Stoichiometry. Trends Pharmacol. Sci. 39:672–684. 10.1016/j.tips.2018.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga K., Kruse A.C., Asada H., Yurugi-Kobayashi T., Shiroishi M., Zhang C., Weis W.I., Okada T., Kobilka B.K., Haga T., and Kobayashi T.. 2012. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 482:547–551. 10.1038/nature10753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harikumar K.G., Wootten D., Pinon D.I., Koole C., Ball A.M., Furness S.G., Graham B., Dong M., Christopoulos A., Miller L.J., and Sexton P.M.. 2012. Glucagon-like peptide-1 receptor dimerization differentially regulates agonist signaling but does not affect small molecule allostery. Proc. Natl. Acad. Sci. USA. 109:18607–18612. 10.1073/pnas.1205227109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser A.S., Attwood M.M., Rask-Andersen M., Schiöth H.B., and Gloriam D.E.. 2017. Trends in GPCR drug discovery: new agents, targets and indications. Nat. Rev. Drug Discov. 16:829–842. 10.1038/nrd.2017.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hern J.A., Baig A.H., Mashanov G.I., Birdsall B., Corrie J.E., Lazareno S., Molloy J.E., and Birdsall N.J.. 2010. Formation and dissociation of M1 muscarinic receptor dimers seen by total internal reflection fluorescence imaging of single molecules. Proc. Natl. Acad. Sci. USA. 107:2693–2698. 10.1073/pnas.0907915107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyaskina O.S., Lemoine H., and Bünemann M.. 2018. Lifetime of muscarinic receptor-G-protein complexes determines coupling efficiency and G-protein subtype selectivity. Proc. Natl. Acad. Sci. USA. 115:5016–5021. 10.1073/pnas.1715751115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel P.A., Mahan L.C., Motulsky H.J., Stoolman L.M., and Koachman A.M.. 1983. Time-dependent decreases in binding affinity of agonists for beta-adrenergic receptors of intact S49 lymphoma cells. A mechanism of desensitization. J. Biol. Chem. 258:13597–13605. [PubMed] [Google Scholar]
- Jakubík J., Bacáková L., El-Fakahany E.E., and Tucek S.. 1997. Positive cooperativity of acetylcholine and other agonists with allosteric ligands on muscarinic acetylcholine receptors. Mol. Pharmacol. 52:172–179. 10.1124/mol.52.1.172 [DOI] [PubMed] [Google Scholar]
- Jiménez-Rosés M., Matsoukas M.-T., Caltabiano G., and Cordomí A.. 2018. Ligand-triggered structural changes in the M2 muscarinic acetylcholine receptor. J. Chem. Inf. Model. 58:1074–1082. 10.1021/acs.jcim.8b00108 [DOI] [PubMed] [Google Scholar]
- Kenakin T.P. 2012. Biased signalling and allosteric machines: new vistas and challenges for drug discovery. Br. J. Pharmacol. 165:1659–1669. 10.1111/j.1476-5381.2011.01749.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenakin T., and Christopoulos A.. 2013. Signalling bias in new drug discovery: detection, quantification and therapeutic impact. Nat. Rev. Drug Discov. 12:205–216. 10.1038/nrd3954 [DOI] [PubMed] [Google Scholar]
- Kruse A.C., Hu J., Pan A.C., Arlow D.H., Rosenbaum D.M., Rosemond E., Green H.F., Liu T., Chae P.S., Dror R.O., et al. . 2012. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 482:552–556. 10.1038/nature10867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse A.C., Ring A.M., Manglik A., Hu J., Hu K., Eitel K., Hübner H., Pardon E., Valant C., Sexton P.M., et al. . 2013. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature. 504:101–106. 10.1038/nature12735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse A.C., Kobilka B.K., Gautam D., Sexton P.M., Christopoulos A., and Wess J.. 2014. Muscarinic acetylcholine receptors: novel opportunities for drug development. Nat. Rev. Drug Discov. 13:549–560. 10.1038/nrd4295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane J.R., Donthamsetti P., Shonberg J., Draper-Joyce C.J., Dentry S., Michino M., Shi L., López L., Scammells P.J., Capuano B., et al. . 2014. A new mechanism of allostery in a G protein-coupled receptor dimer. Nat. Chem. Biol. 10:745–752. 10.1038/nchembio.1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane J.R., May L.T., Parton R.G., Sexton P.M., and Christopoulos A.. 2017. A kinetic view of GPCR allostery and biased agonism. Nat. Chem. Biol. 13:929–937. 10.1038/nchembio.2431 [DOI] [PubMed] [Google Scholar]
- Lazareno S., Dolezal V., Popham A., and Birdsall N.J.. 2004. Thiochrome enhances acetylcholine affinity at muscarinic M4 receptors: receptor subtype selectivity via cooperativity rather than affinity. Mol. Pharmacol. 65:257–266. 10.1124/mol.65.1.257 [DOI] [PubMed] [Google Scholar]
- Leach K., Loiacono R.E., Felder C.C., McKinzie D.L., Mogg A., Shaw D.B., Sexton P.M., and Christopoulos A.. 2010. Molecular mechanisms of action and in vivo validation of an M4 muscarinic acetylcholine receptor allosteric modulator with potential antipsychotic properties. Neuropsychopharmacology. 35:855–869. 10.1038/npp.2009.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach K., Davey A.E., Felder C.C., Sexton P.M., and Christopoulos A.. 2011. The role of transmembrane domain 3 in the actions of orthosteric, allosteric, and atypical agonists of the M4 muscarinic acetylcholine receptor. Mol. Pharmacol. 79:855–865. 10.1124/mol.111.070938 [DOI] [PubMed] [Google Scholar]
- Lebon G., Warne T., Edwards P.C., Bennett K., Langmead C.J., Leslie A.G.W., and Tate C.G.. 2011. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature. 474:521–525. 10.1038/nature10136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei S., Clydesdale L., Dai A., Cai X., Feng Y., Yang D., Liang Y.L., Koole C., Zhao P., Coudrat T., et al. . 2018. Two distinct domains of the glucagon-like peptide-1 receptor control peptide-mediated biased agonism. J. Biol. Chem. 293:9370–9387. 10.1074/jbc.RA118.003278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Ahn S., Kahsai A.W., Meng K.C., Latorraca N.R., Pani B., Venkatakrishnan A.J., Masoudi A., Weis W.I., Dror R.O., et al. . 2017. Mechanism of intracellular allosteric β2AR antagonist revealed by X-ray crystal structure. Nature. 548:480–484. 10.1038/nature23652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockless S.W., and Ranganathan R.. 1999. Evolutionarily conserved pathways of energetic connectivity in protein families. Science. 286:295–299. 10.1126/science.286.5438.295 [DOI] [PubMed] [Google Scholar]
- Lüllmann H., Ohnesorge F.K., Schauwecker G.C., and Wassermann O.. 1969. Inhibition of the actions of carbachol and DFP on guinea pig isolated atria by alkane-bis-ammonium compounds. Eur. J. Pharmacol. 6:241–247. 10.1016/0014-2999(69)90181-2 [DOI] [PubMed] [Google Scholar]
- Marlo J.E., Niswender C.M., Days E.L., Bridges T.M., Xiang Y., Rodriguez A.L., Shirey J.K., Brady A.E., Nalywajko T., Luo Q., et al. . 2009. Discovery and characterization of novel allosteric potentiators of M1 muscarinic receptors reveals multiple modes of activity. Mol. Pharmacol. 75:577–588. 10.1124/mol.108.052886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsango S., Ward R.J., Alvarez-Curto E., and Milligan G.. 2017. Muscarinic receptor oligomerization. Neuropharmacology. 136:401–410. 10.1016/j.neuropharm.2017.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May L.T., Leach K., Sexton P.M., and Christopoulos A.. 2007. Allosteric modulation of G protein-coupled receptors. Annu. Rev. Pharmacol. Toxicol. 47:1–51. 10.1146/annurev.pharmtox.47.120505.105159 [DOI] [PubMed] [Google Scholar]
- Monod J., Wyman J., and Changeux J.-P.. 1965. On the nature of allosteric transitions: A plausible model. J. Mol. Biol. 12:88–118. 10.1016/S0022-2836(65)80285-6 [DOI] [PubMed] [Google Scholar]
- Nawaratne V., Leach K., Felder C.C., Sexton P.M., and Christopoulos A.. 2010. Structural determinants of allosteric agonism and modulation at the M4 muscarinic acetylcholine receptor: identification of ligand-specific and global activation mechanisms. J. Biol. Chem. 285:19012–19021. 10.1074/jbc.M110.125096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygaard R., Zou Y., Dror R.O., Mildorf T.J., Arlow D.H., Manglik A., Pan A.C., Liu C.W., Fung J.J., Bokoch M.P., et al. . 2013. The dynamic process of β(2)-adrenergic receptor activation. Cell. 152:532–542. 10.1016/j.cell.2013.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald C., Rappas M., Kean J., Doré A.S., Errey J.C., Bennett K., Deflorian F., Christopher J.A., Jazayeri A., Mason J.S., et al. . 2016. Intracellular allosteric antagonism of the CCR9 receptor. Nature. 540:462–465. 10.1038/nature20606 [DOI] [PubMed] [Google Scholar]
- Pediani J.D., Ward R.J., Godin A.G., Marsango S., and Milligan G.. 2016. Dynamic regulation of quaternary organization of the M1 muscarinic receptor by subtype-selective antagonist drugs. J. Biol. Chem. 291:13132–13146. 10.1074/jbc.M115.712562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin J.P., Comps-Agrar L., Maurel D., Monnier C., Rives M.L., Trinquet E., Kniazeff J., Rondard P., and Prézeau L.. 2009. G-protein-coupled receptor oligomers: two or more for what? Lessons from mGlu and GABAB receptors. J. Physiol. 587:5337–5344. 10.1113/jphysiol.2009.179978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S.G., Choi H.J., Fung J.J., Pardon E., Casarosa P., Chae P.S., Devree B.T., Rosenbaum D.M., Thian F.S., Kobilka T.S., et al. . 2011a Structure of a nanobody-stabilized active state of the β(2) adrenoceptor. Nature. 469:175–180. 10.1038/nature09648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S.G.F., DeVree B.T., Zou Y., Kruse A.C., Chung K.Y., Kobilka T.S., Thian F.S., Chae P.S., Pardon E., Calinski D., et al. . 2011b Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 477:549–555. 10.1038/nature10361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redka D.S., Morizumi T., Elmslie G., Paranthaman P., Shivnaraine R.V., Ellis J., Ernst O.P., and Wells J.W.. 2014. Coupling of g proteins to reconstituted monomers and tetramers of the M2 muscarinic receptor. J. Biol. Chem. 289:24347–24365. 10.1074/jbc.M114.559294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum D.M., Zhang C., Lyons J.A., Holl R., Aragao D., Arlow D.H., Rasmussen S.G.F., Choi H.J., Devree B.T., Sunahara R.K., et al. . 2011. Structure and function of an irreversible agonist-β(2) adrenoceptor complex. Nature. 469:236–240. 10.1038/nature09665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar A., Potter W.Z., Lightfoot J., Lienemann J., Dubé S., Mallinckrodt C., Bymaster F.P., McKinzie D.L., and Felder C.C.. 2008. Selective muscarinic receptor agonist xanomeline as a novel treatment approach for schizophrenia. Am. J. Psychiatry. 165:1033–1039. 10.1176/appi.ajp.2008.06091591 [DOI] [PubMed] [Google Scholar]
- Shivnaraine R.V., Huang X.P., Seidenberg M., Ellis J., and Wells J.W.. 2012. Heterotropic cooperativity within and between protomers of an oligomeric M(2) muscarinic receptor. Biochemistry. 51:4518–4540. 10.1021/bi3000287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivnaraine R.V., Kelly B., Sankar K.S., Redka D.S., Han Y.R., Huang F., Elmslie G., Pinto D., Li Y., Rocheleau J.V., et al. . 2016. Allosteric modulation in monomers and oligomers of a G protein-coupled receptor. eLife. 5:e11685 10.7554/eLife.11685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G., Yang D., Wang Y., de Graaf C., Zhou Q., Jiang S., Liu K., Cai X., Dai A., Lin G., et al. . 2017. Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators. Nature. 546:312–315. 10.1038/nature22378 [DOI] [PubMed] [Google Scholar]
- Stahl E., and Ellis J.. 2010. Novel allosteric effects of amiodarone at the muscarinic M5 receptor. J. Pharmacol. Exp. Ther. 334:214–222. 10.1124/jpet.109.165316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staus D.P., Strachan R.T., Manglik A., Pani B., Kahsai A.W., Kim T.H., Wingler L.M., Ahn S., Chatterjee A., Masoudi A., et al. . 2016. Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation. Nature. 535:448–452. 10.1038/nature18636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen A., Thiele S., Guo D., Hansen L.S., Frimurer T.M., and Rosenkilde M.M.. 2013. Biased and constitutive signaling in the CC-chemokine receptor CCR5 by manipulating the interface between transmembrane helices 6 and 7. J. Biol. Chem. 288:12511–12521. 10.1074/jbc.M112.449587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Süel G.M., Lockless S.W., Wall M.A., and Ranganathan R.. 2003. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat. Struct. Biol. 10:59–69. 10.1038/nsb881 [DOI] [PubMed] [Google Scholar]
- Thal D.M., Sun B., Feng D., Nawaratne V., Leach K., Felder C.C., Bures M.G., Evans D.A., Weis W.I., Bachhawat P., et al. . 2016. Crystal structures of the M1 and M4 muscarinic acetylcholine receptors. Nature. 531:335–340. 10.1038/nature17188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thal D.M., Glukhova A., Sexton P.M., and Christopoulos A.. 2018. Structural insights into G-protein-coupled receptor allostery. Nature. 559:45–53. 10.1038/s41586-018-0259-z [DOI] [PubMed] [Google Scholar]
- Thorsen T.S., Matt R., Weis W.I., and Kobilka B.K.. 2014. Modified T4 lysozyme fusion proteins facilitate G protein-coupled receptor crystallogenesis. Structure. 22:1657–1664. 10.1016/j.str.2014.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toews M.L., and Perkins J.P.. 1984. Agonist-induced changes in beta-adrenergic receptors on intact cells. J. Biol. Chem. 259:2227–2235. [PubMed] [Google Scholar]
- Toews M.L., Harden T.K., and Perkins J.P.. 1983. High-affinity binding of agonists to beta-adrenergic receptors on intact cells. Proc. Natl. Acad. Sci. USA. 80:3553–3557. 10.1073/pnas.80.12.3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valant C., Felder C.C., Sexton P.M., and Christopoulos A.. 2012. Probe dependence in the allosteric modulation of a G protein-coupled receptor: implications for detection and validation of allosteric ligand effects. Mol. Pharmacol. 81:41–52. 10.1124/mol.111.074872 [DOI] [PubMed] [Google Scholar]
- Wacker D., Wang C., Katritch V., Han G.W., Huang X.P., Vardy E., McCorvy J.D., Jiang Y., Chu M., Siu F.Y., et al. . 2013. Structural features for functional selectivity at serotonin receptors. Science. 340:615–619. 10.1126/science.1232808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker D., Stevens R.C., and Roth B.L.. 2017a How ligands illuminate GPCR molecular pharmacology. Cell. 170:414–427. 10.1016/j.cell.2017.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker D., Wang S., McCorvy J.D., Betz R.M., Venkatakrishnan A.J., Levit A., Lansu K., Schools Z.L., Che T., Nichols D.E., et al. . 2017b Crystal structure of an LSD-bound human serotonin receptor. Cell. 168:377–389.e12. 10.1016/j.cell.2016.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wess J. 2005. Allosteric binding sites on muscarinic acetylcholine receptors. Mol. Pharmacol. 68:1506–1509. [DOI] [PubMed] [Google Scholar]
- Weston C., Winfield I., Harris M., Hodgson R., Shah A., Dowell S.J., Mobarec J.C., Woodcock D.A., Reynolds C.A., Poyner D.R., et al. . 2016. Receptor activity-modifying protein-directed G protein signaling specificity for the calcitonin gene-related peptide family of receptors. J. Biol. Chem. 291:25763 10.1074/jbc.A116.751362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whorton M.R., Bokoch M.P., Rasmussen S.G., Huang B., Zare R.N., Kobilka B., and Sunahara R.K.. 2007. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc. Natl. Acad. Sci. USA. 104:7682–7687. 10.1073/pnas.0611448104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootten D., Reynolds C.A., Smith K.J., Mobarec J.C., Koole C., Savage E.E., Pabreja K., Simms J., Sridhar R., Furness S.G.B., et al. . 2016. The extracellular surface of the GLP-1 receptor is a molecular trigger for biased agonism. Cell. 165:1632–1643. 10.1016/j.cell.2016.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Qin L., Zacarías N.V., de Vries H., Han G.W., Gustavsson M., Dabros M., Zhao C., Cherney R.J., Carter P., et al. . 2016. Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists. Nature. 540:458–461. 10.1038/nature20605 [DOI] [PMC free article] [PubMed] [Google Scholar]