The identification of voltage-sensing elements in membrane proteins is challenging due to the diversity of voltage-sensing mechanisms. Kasimova et al. present a computational approach to predict the elements involved in voltage sensing, which they validate using voltage-gated ion channels.
Abstract
Voltage-sensitive membrane proteins are united by their ability to transform changes in membrane potential into mechanical work. They are responsible for a spectrum of physiological processes in living organisms, including electrical signaling and cell-cycle progression. Although the mechanism of voltage-sensing has been well characterized for some membrane proteins, including voltage-gated ion channels, even the location of the voltage-sensing elements remains unknown for others. Moreover, the detection of these elements by using experimental techniques is challenging because of the diversity of membrane proteins. Here, we provide a computational approach to predict voltage-sensing elements in any membrane protein, independent of its structure or function. It relies on an estimation of the propensity of a protein to respond to changes in membrane potential. We first show that this property correlates well with voltage sensitivity by applying our approach to a set of voltage-sensitive and voltage-insensitive membrane proteins. We further show that it correctly identifies authentic voltage-sensitive residues in the voltage-sensor domain of voltage-gated ion channels. Finally, we investigate six membrane proteins for which the voltage-sensing elements have not yet been characterized and identify residues and ions that might be involved in the response to voltage. The suggested approach is fast and simple and enables a characterization of voltage sensitivity that goes beyond mere identification of charges. We anticipate that its application before mutagenesis experiments will significantly reduce the number of potential voltage-sensitive elements to be tested.
Introduction
The membrane potential (MP) in living cells results from the uneven distribution of ions between the two sides of the cell membrane (Hille, 2001). It regulates several critical physiological processes, such as the formation and propagation of the action potential in excitable cells (Hille, 2001) and the progression along the cell cycle in nonexcitable ones (Cone, 1969, 1971; Yang and Brackenbury, 2013). Voltage-sensitive membrane proteins are able to detect changes in the MP and in some cases respond directly to it (Bezanilla, 2008). The ability to understand and eventually modulate the function of these proteins provides an interesting strategy to diagnose and treat neurological diseases by changing either the MP or the proteins’ response to it.
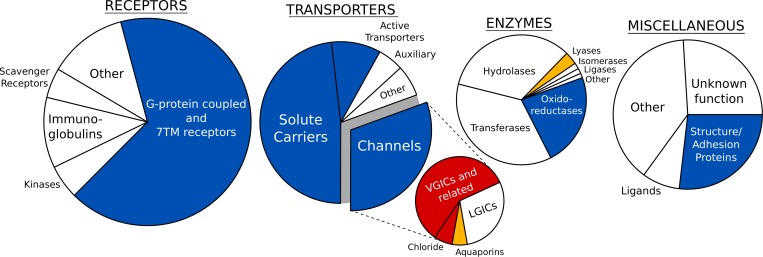
The group of voltage-sensitive membrane proteins (VSMPs) includes representatives from transporters (Nakao and Gadsby, 1986; Catterall, 1988; Weer et al., 1988; Colombini, 1989; Schnetkamp and Szerencsei, 1991; Bernardi, 1992; Parent et al., 1992; Kavanaugh, 1993; Petronilli et al., 1994; Jentsch et al., 1995; Wadiche et al., 1995; Caterina et al., 1997; Halestrap et al., 1997; Scorrano et al., 1997; Kaim and Dimroth, 1998, 1999; Künkele et al., 1998; Lostao et al., 2000; Sugawara et al., 2000; Yao et al., 2000; Zheng et al., 2000; Jutabha et al., 2003, 2011; Mackenzie et al., 2003; Melzer et al., 2003; Bezanilla, 2005, 2008; Anzai et al., 2008; Hub et al., 2010; Zhang et al., 2011; Malhotra et al., 2013; van der Laan et al., 2013; Zander et al., 2013), enzymes (Reddy et al., 1995; Beltrán et al., 1996; Cooper et al., 1998; DeCoursey et al., 2003; Valdez and Boveris, 2007; Rosasco et al., 2015; Vorburger et al., 2016), receptors (Ben-Chaim et al., 2003; Rinne et al., 2013; Vickery et al., 2016b), and proteins playing a role in structure or adhesion (Bargiello et al., 2012; Fig. 1). These proteins are extremely diverse in their structure, but all of them have a common ability to convert the electrical energy into a mechanical response. To achieve this, VSMPs have developed one or more voltage-sensing elements, which detect changes in the MP and alter their conformational state accordingly (Bezanilla, 2008; Swartz, 2008). In voltage-gated potassium and sodium channels, the protein itself plays the role of the voltage sensor: the S4 helix of the S1–S4 helical bundle carries several positively charged residues, which on application of an electric field, move in the direction of this field (Long et al., 2005; DeCaen et al., 2009; Khalili-Araghi et al., 2010; Tao et al., 2010; Wu et al., 2010; Delemotte et al., 2011; Payandeh et al., 2011; Henrion et al., 2012; Jensen et al., 2012; Vargas et al., 2012; Lacroix et al., 2014; Machtens et al., 2017). In most other VSMPs, the voltage-sensing elements are nowhere near as well characterized. Several studies suggest different origins of their voltage sensitivity, including specific protein residues and/or ions trapped inside protein cavities (Nakao and Gadsby, 1986; Weer et al., 1988; Mirzabekov and Ermishkin, 1989; Thomas et al., 1993; Verselis et al., 1994; Pusch et al., 1995; Reddy et al., 1995; Popp et al., 1996; Boukalova et al., 2010; Kwon et al., 2012; Pinto et al., 2016; Vickery et al., 2016a). Ions moving in an electric field have, for instance, been suggested to trigger conformational rearrangements in voltage-gated chloride channels (Pusch et al., 1995; Chen and Miller, 1996; Smith and Lippiat, 2010; Zifarelli et al., 2012; Grieschat and Alekov, 2014; De Jesús-Pérez et al., 2016), solute carriers (Parent et al., 1992), active transporters (Nakao and Gadsby, 1986, 1989; Weer et al., 1988; Holmgren et al., 2000), and G-protein–coupled receptors (Vickery et al., 2016a).
Figure 1.
Membrane protein families, encompassing voltage-sensitive representatives. The families are arranged in four large groups according to Almén et al. (2009); only α-helical membrane proteins are shown. Channels are subdivided in voltage-gated (VGIC), ligand-gated (LGICs), chloride channels and aquaporins. Blue indicates families containing voltage-sensitive membrane proteins, and white indicates families in which voltage-sensitive representatives have not yet been discovered. The families shown in red correspond to membrane proteins for which changes in the MP is the primary stimulus for activation. Finally, the families in orange contain membrane proteins whose voltage sensitivity is controversial. To compose this figure we used data from multiple references (Nakao and Gadsby, 1986; Catterall, 1988; Weer et al., 1988; Colombini, 1989; Schnetkamp and Szerencsei, 1991; Bernardi, 1992; Parent et al., 1992; Kavanaugh, 1993; Petronilli et al., 1994; Jentsch et al., 1995; Reddy et al., 1995; Wadiche et al., 1995; Beltrán et al., 1996; Caterina et al., 1997; Halestrap et al., 1997; Scorrano et al., 1997; Cooper et al., 1998; Kaim and Dimroth, 1998, 1999; Künkele et al., 1998; Lostao et al., 2000; Sugawara et al., 2000; Yao et al., 2000; Zheng et al., 2000; Ben-Chaim et al., 2003; DeCoursey et al., 2003; Jutabha et al., 2003, 2011; Mackenzie et al., 2003; Melzer et al., 2003; Bezanilla, 2005, 2008; Valdez and Boveris, 2007; Anzai et al., 2008; Hub et al., 2010; Zhang et al., 2011; Bargiello et al., 2012; Malhotra et al., 2013; Rinne et al., 2013; van der Laan et al., 2013; Zander et al., 2013; Rosasco et al., 2015; Vorburger et al., 2016).
In this work, we characterize the voltage sensitivity of several selected membrane proteins using a newly developed computational approach. It evaluates the propensity of the molecular system to detect changes in the local electric field and to respond to them. The system’s response propensity is directly connected to the gating charge, a well-known characteristic of voltage sensitivity that corresponds to the amount of charge transferred during protein activation. Our approach also estimates the local electric field response, which reflects how sensitive the local electric field is to the application of an external electric field. We show that, in all tested voltage-sensitive proteins, the system’s response propensity is large compared with voltage-insensitive ones, which suggests this could be an efficient tool to probe voltage sensitivity in other membrane proteins. We also show how the local electric field response can be large in selected regions of voltage-insensitive proteins. However, the lack of charges in these regions results in their inability to respond to changes in the MP. Finally, we investigate six VSMPs for which the voltage sensors have not yet been characterized. For each of them, we depict putative voltage-sensitive residues and/or voltage-sensitive ions trapped in protein cavities.
Our approach only requires knowledge of a protein structure and modest computational resources, which makes it possible to apply before mutagenesis experiments in order to reduce the number of potential voltage-sensitive elements to be tested.
Materials and methods
Systems’ preparation and molecular dynamics simulations
The Charmm-GUI server was used to prepare the systems for molecular dynamics simulations (Jo et al., 2008). Briefly, every protein of interest was embedded into a 1-palmytoyl-2-oleoylphosphatidylcholine bilayer and solvated with 150 mM of either KCl or NaCl solution. The CHARMM36 force field (Mackerell et al., 2004) was used to describe proteins and lipids with TIP3P as a water model (Jorgensen et al., 1983). In the case of NavMs, we also considered a system with a TIP4P water model to test whether the results depend on the water model used. Fig. 2 A shows that our method is indeed robust to changes in the water model. However, the choice of force field parameters, and in particular of partial charge definition, can in principle influence the results. Note that the Na+ ion bound inside the protein cavities of the M2 receptor was not placed there initially; instead, it reached the binding site during the molecular dynamics simulations. For the detailed description of the systems’ composition and properties, see Table 1.
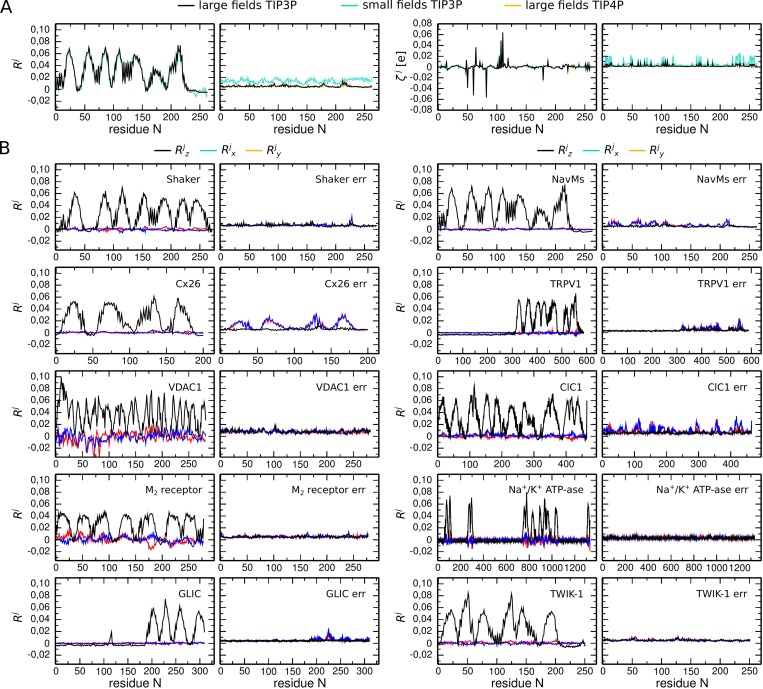
Figure 2.
Per-residue local electric field response Rj values and response propensity ζj values estimated by using different electric fields for a range of systems. (A) and values estimated for the system with NavMs by using large electric fields (between −0.02 and 0.02 V/Å), small electric fields (between −0.005 and 0.005 V/Å), and TIP4P water (large electric fields). The left panels illustrate the average, and the right panels illustrate the standard deviation. Note that in all three cases, the average of and is almost identical. The error is similar for large electric fields and different water models but significantly larger for the small electric fields. (B) and values estimated for Shaker, NavMs, Cx26, TRPV1, VDAC1, ClC1, M2 receptor, Na+/K+ ATPase, GLIC, and TWIK-1. , , and are computed by using the z, x, and y components of the local electric field, respectively (the z axis is the membrane normal, and the direction of the electric field). The left panels illustrate the average, and the right panels illustrate the standard deviation. Based on these data, we chose 0.02 to be the detection threshold for R.
Table 1. Description of the computational systems’ composition and properties.
| Protein | PDB code | Residue patching | System’s dimensions (Å × Å × Å) | Number of atoms | Duration of the equilibration last step |
|---|---|---|---|---|---|
| Shakera | A homology model based on 2R9R (Long et al., 2007); see Yazdi et al. (2016) for the details | None | 130 × 130 × 105 | 191,034 | 1 µs |
| NavMs | 5HVX (Sula et al., 2017) | None | 145 × 145 × 120 | 209,838 | 400 ns |
| GLIC | 4NPQ (Sauguet et al., 2014) | None | 110 × 110 × 145 | 159,083 | 1 µs |
| TWIK-1 | 3UKM (Miller and Long, 2012) | Disulfide bond between C78 and C78 (intersubunit) | 105 × 105 × 125 | 127,219 | 200 ns |
| Cx26 | 2ZW3 (Maeda et al., 2009) | Disulfide bonds between C45 and C156, C50 and C152, C56 and C145 | 130 × 130 × 122 | 189,168 | 200 ns |
| VDAC1 | 3EMN (Ujwal et al., 2008) | None | 85 × 85 × 85 | 54,590 | 200 ns |
| TRPV1b | 3J5R (Cao et al., 2013; Liao et al., 2013; Kasimova et al., 2018) | None | 160 × 140 × 175 | 402,346 | 750 ns |
| ClC1 | 6COY (Park and MacKinnon, 2018) | None | 120 × 120 × 100 | 149,529 | 200 ns |
| M2 receptor | 3UON (Haga et al., 2012) | Disulfide bonds between C96 and C176, C413 and C416 | 90 × 90 × 105 | 76,169 | 200 ns |
| Na+/K+ ATPasec | 3WGU (Kanai et al., 2013) | D195, E358, E779, D804, and E954 are protonated; disulfide bonds between C126 and C149, C159 and C175, C213 and C276 | 115 × 115 × 175 | 250,192 | 100 ns |
The trajectory used for the analysis was taken from Yazdi et al. (2016).
The trajectory for the closed capsaicin-bound state of TRPV1 used for the analysis was taken from Kasimova et al. (2018).
The trajectory for the sodium-bound Na+/K+ ATPase used for the analysis was taken from Razavi et al. (2017).
The molecular dynamics simulations were performed by using GROMACS 2016.1 (Abraham et al., 2015). Each system was equilibrated by following a multistep protocol. During the first 2.5 ns, the protein and lipid headgroups were restrained to their initial positions to allow for the rearrangements of lipid tails and solution. The lipid headgroups were subsequently released, and the simulations continued for 2.5 ns. In the next 35 ns, the restraints applied to the protein backbone and sidechains were gradually decreased from 400 kJ/mol/Å and 200 kJ/mol/Å, respectively, to 0 kJ/mol/Å. Finally, the protein was fully relaxed without restraints until the root mean square deviation with respect to the initial structure reached a plateau value (for the duration of this last step of the equilibration, see Table 1).
A Nosé-Hoover thermostat (Nosé, 1984) and Parrinello-Rahman barostat (Parrinello and Rahman, 1981) were used to keep the temperature (300°K) and pressure (1 atm) constant. A cutoff of 12 Å was applied for short-range electrostatics and Van der Waals interactions. For the latter, a switching function between 10 and 12 Å was applied to smoothly bring the forces to 0 at 12 Å. The particle mesh Ewald summation (Darden et al., 1993) was used for the long-range component of electrostatics. A 1-fs time step was used for the first two steps of the equilibration and 2 fs for the rest of the equilibration and the simulations under an electric field.
Molecular dynamics simulations under an electric field
For each protein of interest, 10 conformational states were extracted from the equilibration trajectory with a stride of 20 ns. For every state, 8 short (2-ns) molecular dynamics simulations were performed with restraints applied to all heavy atoms of the protein and under an electric field; the following values of the field were used: −0.020, −0.015, −0.010, −0.005, 0.005, 0.010, 0.015, and 0.020 V/Å. Note that these values correspond to an MP up to ∼0.5–3.5 V (depending on the system’s size). Smaller electric fields can be used as well but, in such cases, the uncertainty in the estimated variables is larger and the results are more qualitative (Fig. 2 A). Five frames were extracted from each of the trajectories to compute an average local electrostatic potential map by using the PMEpot plugin of VMD (Aksimentiev and Schulten, 2005):
| (1) |
Here, corresponds to a point charge approximated by a spherical Gaussian with an Ewald factor of 0.25. Eq. 1 was solved on a grid with a resolution of 1 Å. R, (per-element local electric field response: ; the average is computed over all atoms of the element j), and were calculated by using an in-house python code (see Results and Discussion for more details). Finally, the average and the standard deviations of and were computed based on all 10 conformational states and all subunits in the case of homomultimers.
The detection threshold was identified based on the calculations of (Fig. 2 B). For every protein of interest, was computed for the two dimensions orthogonal to that of the external electric field (i.e., in the definition of the components of the local field orthogonal to the external field were used). The largest value, 0.02, was further considered as the detection threshold.
Online supplemental material
Video 1 shows the local electric field response estimated for the voltage-gated potassium channel Shaker, projected onto a ribbon representation of the protein backbone. Video 2 shows the local electric field response estimated for the voltage-gated sodium channel NavMs, projected onto a ribbon representation of the protein backbone. Video 3 shows the local electric field response estimated for Cx26, projected onto a ribbon representation of the protein backbone. Video 4 shows the local electric field response estimated for TRPV1, projected onto a ribbon representation of the protein backbone. Video 5 shows the local electric field response estimated for VDAC1, projected onto a ribbon representation of the protein backbone. Video 6 shows the local electric field response estimated for ClC1, projected onto a ribbon representation of the protein backbone. Video 7 shows the local electric field response estimated for the muscarinic acetylcholine receptor M2, projected onto a ribbon representation of the protein backbone. Video 8 shows the local electric field response estimated for the Na+/K+ ATPase, projected onto a ribbon representation of the protein backbone. Video 9 shows the local electric field response estimated for GLIC, projected onto a ribbon representation of the protein backbone. Video 10 shows the local electric field response estimated for the two-pore domain potassium channel TWIK-1, projected onto a ribbon representation of the protein backbone.
Results and discussion
The system’s response propensity and local electric field response R as variables to computationally assess voltage sensitivity of a given membrane protein
Voltage sensitivity occurs in many different membrane proteins with diverse structure and function (Nakao and Gadsby, 1986; Catterall, 1988; Weer et al., 1988; Colombini, 1989; Schnetkamp and Szerencsei, 1991; Bernardi, 1992; Parent et al., 1992; Kavanaugh, 1993; Petronilli et al., 1994; Jentsch et al., 1995; Reddy et al., 1995; Wadiche et al., 1995; Beltrán et al., 1996; Caterina et al., 1997; Halestrap et al., 1997; Scorrano et al., 1997; Cooper et al., 1998; Kaim and Dimroth, 1998, 1999; Künkele et al., 1998; Lostao et al., 2000; Sugawara et al., 2000; Yao et al., 2000; Zheng et al., 2000; Ben-Chaim et al., 2003; DeCoursey et al., 2003; Jutabha et al., 2003, 2011; Mackenzie et al., 2003; Melzer et al., 2003; Bezanilla, 2005, 2008; Valdez and Boveris, 2007; Anzai et al., 2008; Hub et al., 2010; Zhang et al., 2011; Bargiello et al., 2012; Malhotra et al., 2013; Rinne et al., 2013; van der Laan et al., 2013; Zander et al., 2013; Rosasco et al., 2015; Vorburger et al., 2016); the unifying factor is how they are all able to detect changes in the MP and convert these changes into mechanical work (Bezanilla, 2008). The gating charge is a characteristic variable of voltage sensitivity. It was initially proposed to report the amount of charge transferred upon gating in voltage-gated ion channels (Armstrong and Bezanilla, 1973; Aggarwal and MacKinnon, 1996; Noceti et al., 1996) and then generalized to characterize any voltage-sensitive transition between two states of a given membrane protein. Computationally, the gating charge Q can be estimated via the excess free energies caused by the application of the MP (Roux, 2008; Treptow et al., 2009) in Eq. 2:
| (2) |
As such, it measures how the MP perturbs the equilibrium population of the activated versus deactivated states of a protein, referred to as a and d, respectively. It can be further decomposed into individual contributions from different protein elements (residues and ions trapped in cavities; Roux, 2008; Treptow et al., 2009) as , where
| (3) |
In Eq. 3, the summation runs over all charges of the element j; and represent dimensionless coupling of the charge to in the deactivated and activated states (also known as electrical distance). The coupling function can be approximated as the rate of change of the local electrostatic potential with respect to , In practice, when structures of the deactivated and activated states are available, can be computed by using this approximation and Eq. 3. However, for many membrane proteins, the structure of only one state is known, and therefore, direct estimation of is not possible. Instead, one can estimate the fraction of the gating charge, which is transferred by element j upon an infinitesimally small displacement along the direction of the applied electric field. Hereafter, we will consider a scenario in which the applied electric field is aligned with the normal to the membrane, which coincides with the axis. Assuming changes monotonically on , one can obtain:
| (4) |
where corresponds to the local electric field. By using Eq. 4, it is possible to estimate the amount of charge transferred by the element j upon 1 Å displacement along the direction of an applied electric field,
We further show that can be considered as a good predictor for and used for the detection of voltage-sensing elements.
Importantly, reflects the propensity of the system to respond to changes in the local electric field upon application of . It does not fully correspond to but to a fraction of it, assuming also that the element j is free to displace by 1 Å. In membrane proteins, however, conformational changes triggered by application of voltage can be substantially larger than 1 Å. In voltage-gated ion channels, for instance, S4 is displaced by ∼10 Å between the activated and resting states (Delemotte et al., 2011; Vargas et al., 2012). Such a large displacement is not well described by the first-order derivative (Eq. 4) assumed in our method. Therefore, the identified elements are state-specific; this means that other voltage-sensing elements may remain unidentified if they were not subject to a large local electric field in the examined conformation. Also, the list of identified elements may include false positives if not all of these elements are free to displace along the direction of the applied electric field during a voltage-sensing transition. Because of the potential presence of false-positives, it is important to further validate the predictions made by the suggested approach experimentally.
In addition, we estimate the contribution to caused by the change in the local electric field, This property, which we call the local electric field response R, shows how sensitive the local electric field is to the application of an external electric field, and it is, therefore, useful for detection of protein regions subject to voltage sensitivity.
In voltage-sensitive membrane proteins, charges are located in regions with a large local electric field response
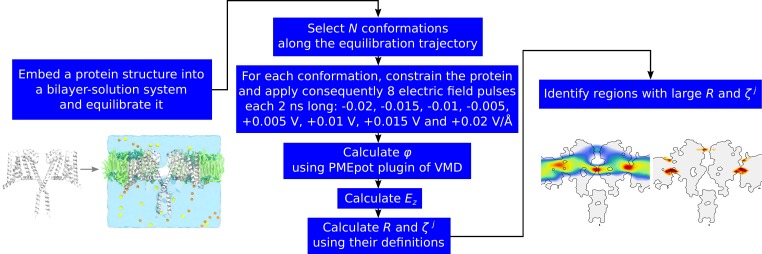
To test our approach, we applied it to a set of voltage-sensitive and voltage-insensitive membrane proteins. In the former case, we considered two voltage-gated ion channels, whose voltage-sensing elements have been well characterized, and six other membrane proteins, for which these elements remain largely unknown (Table 2). For each of these proteins, we calculated the local electric field response R and the system’s response propensity . Fig. 3 shows the workflow of the suggested approach. A structure of the protein is embedded into a bilayer-solution system and equilibrated. Several protein conformations (N in total) are then extracted from the molecular dynamics trajectory, and each of them is further submitted to eight 2-ns-long runs under different values of an applied electric field (Fig. 3). After that, for every produced run, the local electrostatic potential φ is computed by using the PMEpot plugin of VMD (Aksimentiev and Schulten, 2005). R and are finally calculated by using their definitions. Importantly, overall, we performed only N× 2 × 2 ns of molecular dynamics simulations (without considering the equilibration step), which only require modest computational resources.
Table 2. Voltage-sensitive and voltage-insensitive membrane proteins analyzed in the present study.
Figure 3.
Workflow of the suggested approach. The protein is embedded into a bilayer-solution environment and equilibrated. N conformations are extracted from the equilibration trajectory, and for each of them, eight independent runs under different electric fields are performed. For every run, the map of the local electrostatic potential is calculated. Based on this, the local electric field, the local electric field response and the system's response propensity are computed. The system's elements with R and (use greek symbol) above the detection threshold correspond to putative voltage sensors.
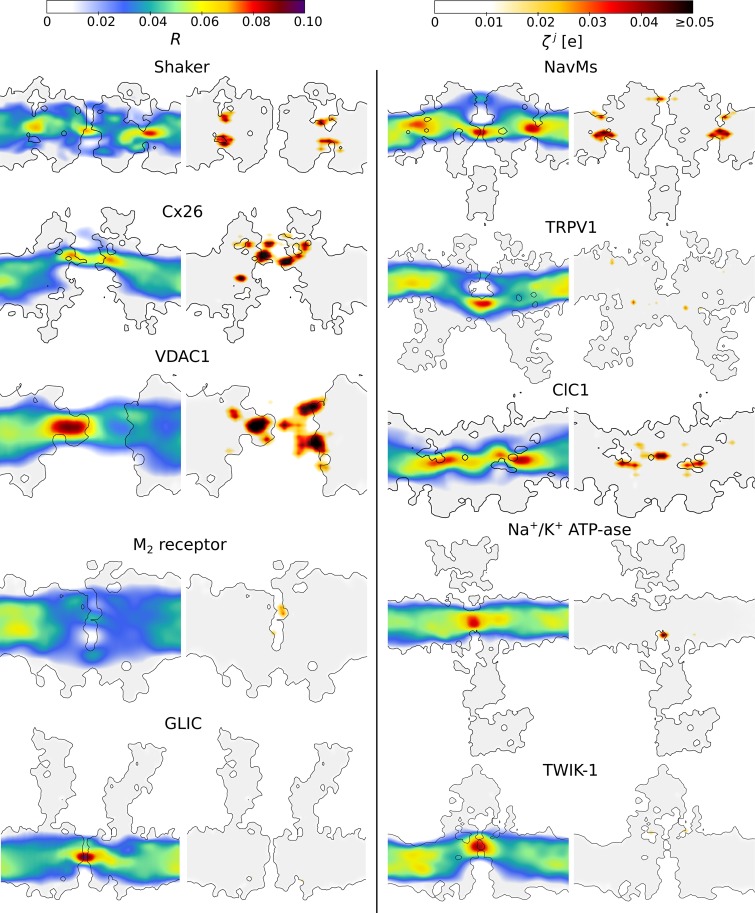
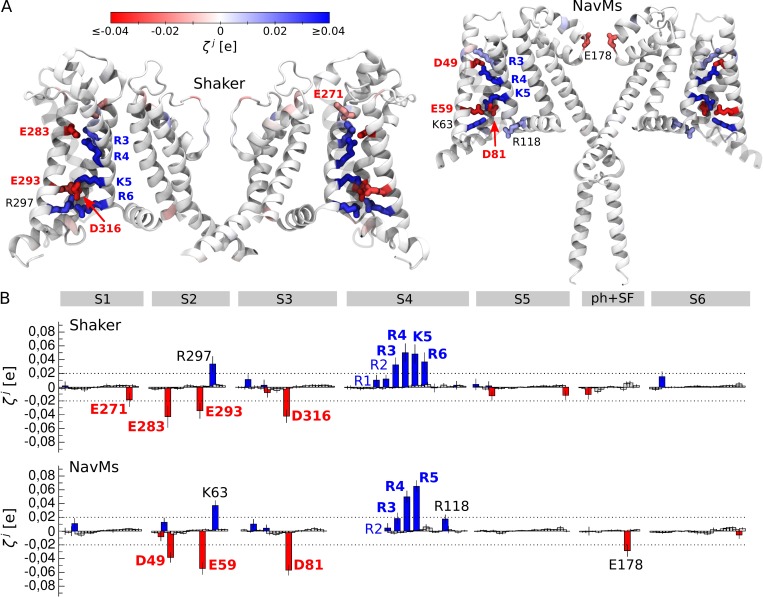
In many voltage-sensitive membrane proteins, we detected regions with a large R value and therefore with a potential to respond to changes in (Fig. 4 and Videos 1–10). The center of the voltage sensors in Shaker and NavMs (Long et al., 2005; DeCaen et al., 2009; Khalili-Araghi et al., 2010; Tao et al., 2010; Wu et al., 2010; Delemotte et al., 2011; Payandeh et al., 2011; Henrion et al., 2012; Jensen et al., 2012; Vargas et al., 2012; Lacroix et al., 2014; Machtens et al., 2017), the N terminus in voltage-dependent anion channel 1 (VDAC1) (Mirzabekov and Ermishkin, 1989; Peng et al., 1992; Thomas et al., 1993; Popp et al., 1996; Song et al., 1998; Teijido et al., 2014; Briones et al., 2016), and the Cl− binding site in ClC1 (Pusch et al., 1995; Chen and Miller, 1996; Smith and Lippiat, 2010; Zifarelli et al., 2012; Grieschat and Alekov, 2014; De Jesús-Pérez et al., 2016) all show R values larger than 0.08. However, in the M2 receptor (Ben-Chaim et al., 2003, 2006; Navarro-Polanco et al., 2011; Rinne et al., 2013; Vickery et al., 2016a,b), which is also known to be voltage-sensitive, R is small and reaches a maximum of only 0.04 in the Na+ binding site. Moreover, in voltage-insensitive membrane proteins, large R values were detected along the conductive pore, with the selectivity filter in the two-pore domain potassium channel (TWIK-1; Schewe et al., 2016) and the gate in the ligand-gated ion channel (GLIC; Bocquet et al., 2007), showing R values as large as 0.08. Therefore, R does not directly correlate with voltage sensitivity and cannot be used to discriminate between voltage-sensitive and voltage-insensitive proteins.
Figure 4.
The local electric field response R (left) and the system’s response propensity ζj (right) estimated for 10 different membrane proteins. They include voltage-gated potassium channel Shaker (Long et al., 2007; Yazdi et al., 2016), voltage-gated sodium channel NavMs (Sula et al., 2017), Cx26 (Maeda et al., 2009), TRPV1 (Cao et al., 2013; Liao et al., 2013; Kasimova et al., 2018), VDAC1 (Ujwal et al., 2008), ClC1 (Park and MacKinnon, 2018), muscarinic acetylcholine receptor M2 (Haga et al., 2012), Na+/K+ ATPase (Kanai et al., 2013), GLIC (Sauguet et al., 2014), and two-pore domain potassium channel TWIK-1 (Miller and Long, 2012). The slices of the systems along the normal to the membrane are shown. The gray area shows the regions that are not accessible to water (i.e., the proteins and the membrane). To clearly represent , we approximated each point charge of the system element j with a Gaussian distribution (σ = 1.5 Å) and then integrated the signal over 25 slices (each 1 Å wide) parallel to the plane shown in the figure. Only the values above the detection threshold were considered for the integration (see Materials and methods). Shaker, NavMs, Cx26, VDAC1, and ClC1, for which changes in the MP are the primary stimulus for activation, have the largest values, while TRPV1, which is known to be very weakly voltage-sensitive (Caterina et al., 1997; Nilius et al., 2005; Boukalova et al., 2010), has the smallest value among the voltage-sensitive membrane proteins.
, on the other hand, correlates rather well with voltage sensitivity: for all voltage-sensitive membrane proteins, is large, while it is close to or below the detection threshold for all voltage-insensitive ones. Together with the previous observation, this suggests that voltage-sensitive proteins have evolved to place their charges in the regions with a large R value and therefore maximize their potential to respond to changes in . In contrast, in voltage-insensitive proteins, charges are located far from the regions with large R values and thus do not sense changes in the local electric field. This also suggests our approach can be used to probe voltage sensitivity in other membrane proteins in which this property has not yet been tested.
In voltage-gated ion channels, the residues with large values correspond to the true voltage sensors
We further describe the results obtained for the two voltage-gated ion channels, Shaker and NavMs. For voltage-gated ion channels in general, the mechanism of voltage sensitivity has been well characterized based on numerous experimental and computational studies (Long et al., 2005; DeCaen et al., 2009; Khalili-Araghi et al., 2010; Tao et al., 2010; Wu et al., 2010; Delemotte et al., 2011; Payandeh et al., 2011; Henrion et al., 2012; Jensen et al., 2012; Vargas et al., 2012; Lacroix et al., 2014; Machtens et al., 2017). These channels have four-helix bundle domains that sense changes in the MP–voltage sensor domains. One of the four helices (S4) carries several positively charged residues, while the other three (S1–S3) include a few negative countercharges (Long et al., 2005; Payandeh et al., 2011). Upon application of an electric field, the positive charges of S4 are displaced along the direction of this field to trigger conformational rearrangements in the pore domain (DeCaen et al., 2009; Khalili-Araghi et al., 2010; Tao et al., 2010; Wu et al., 2010; Delemotte et al., 2011; Henrion et al., 2012; Jensen et al., 2012; Vargas et al., 2012).
Our approach correctly detects the positively charged residues on S4 and their negative countercharges on S1–S3 as voltage-sensing elements (Fig. 5). Among them, residues located in the center of the voltage sensor domain show large values, while those exposed to the extracellular or cytosolic solutions show values close to the detection threshold (see Materials and methods). This indicates that, in the conformational states of Shaker and NavMs chosen for the analysis, residues such as R4 and K5/R5 contribute most to the gating charge, while the contribution from R1 and R2 is small.
Figure 5.
Detection of the voltage-sensing elements in the two voltage-gated ion channels, Shaker (Long et al., 2007; Yazdi et al., 2016) and NavMs (Sula et al., 2017). (A) Illustrated representation of the two channels. The residues whose is larger than the detection threshold are shown. (B) values estimated for Shaker and NavMs; the average and the standard deviation are shown. The positively and negatively charged residues are shown as blue and red bars, respectively. The dashed lines represent the detection threshold. S1–S6 denotes the transmembrane segments, and ph+SF is the pore helix and the selectivity filter. The residues shown in blue or red and in bold correspond to the known voltage sensors and were detected by our method; those shown in blue and not in bold correspond to the known voltage sensors, for which our method showed below the detection threshold. Finally, residues shown in black were detected by our method but were not yet shown to play a role in voltage sensitivity.
A few other residues show above the detection threshold, including R118 on the S4–S5 linker and E178 in the selectivity filter of NavMs, and a positive residue on S2 in both Shaker and NavMs. To our knowledge, the role of these residues in voltage sensitivity has not yet been assessed, and thus experimental verification will be required to confirm whether they are true- or false-positive signals.
Estimation of the system’s response capacity allows for detection of voltage sensors in uncharacterized voltage-sensitive membrane proteins
The correct identification of voltage sensors in Shaker and NavMs strengthened our confidence in the predictive ability of our approach and suggested applying it to other membrane proteins, whose voltage sensors remain unknown. Overall, we applied it to six membrane proteins, including connexin-26 (Cx26; Maeda et al., 2009), transient receptor potential channel 1 (TRPV1; Cao et al., 2013; Liao et al., 2013; Kasimova et al., 2018), VDAC1 (Ujwal et al., 2008), voltage-gated chloride channel 1 (ClC1; Park and MacKinnon, 2018), muscarinic acetylcholine receptor M2 (Haga et al., 2012), and Na+/K+ ATPase (Kanai et al., 2013; Figs. 6 and 7).
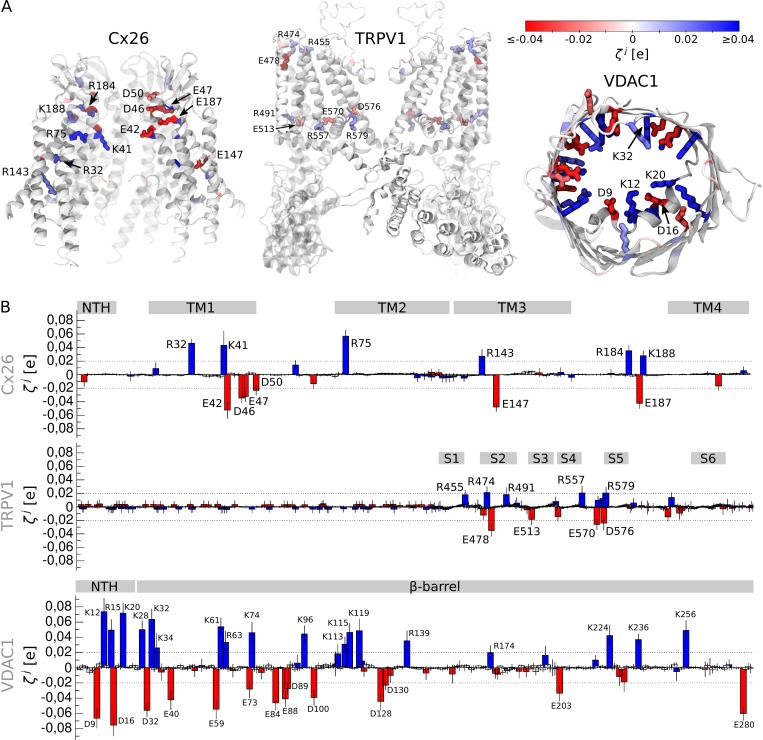
Figure 6.
Detection of the voltage-sensing sensing elements in Cx26 (Maeda et al., 2009), transient receptor potential channel TRPV1 (Cao et al., 2013; Liao et al., 2013; Kasimova et al., 2018), and voltage-dependent anion channel VDAC1 (Ujwal et al., 2008). (A) Illustrated representation of the membrane proteins. The residues whose value is larger than the detection threshold are shown. (B) values estimated for Cx26, TRPV1, and VDAC1; the average and standard deviation are shown. The positively and negatively charged residues are shown as blue and red bars, respectively. The gray rectangles correspond to different regions of the proteins: NTH, the α-helical N terminus in Cx26 and VDAC1; TM1–TM4, transmembrane segments in Cx26; S1–S6, transmembrane segments in TRPV1; and β-barrel, the β-barrel in VDAC1.
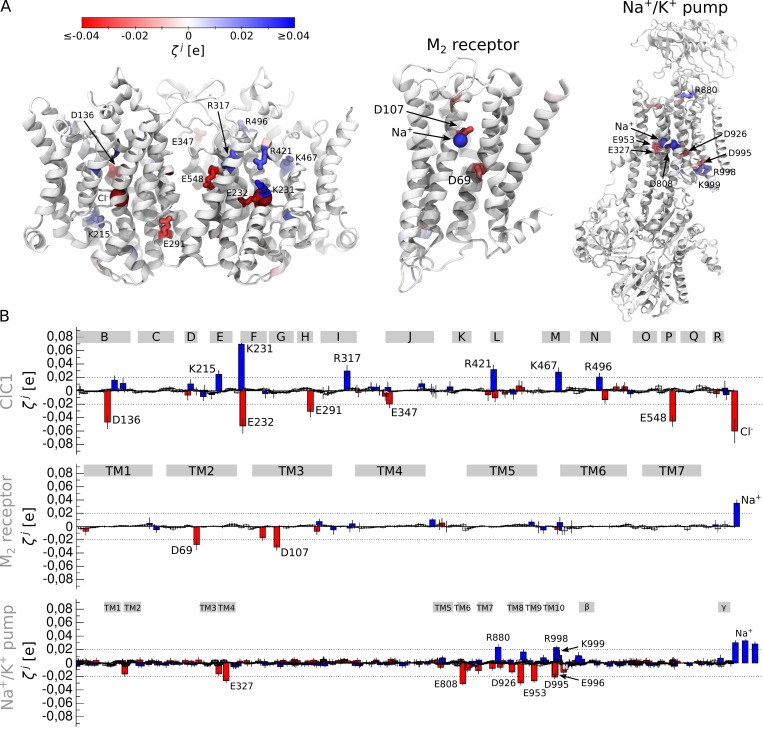
Figure 7.
Detection of the voltage-sensing elements in ClC1 (Park and MacKinnon, 2018), muscarinic acetylcholine receptor M2 (Haga et al., 2012) and Na+/K+ ATPase (Kanai et al., 2013). (A) Illustrated representation of the membrane proteins. The residues whose value is larger than the detection threshold are shown. (B) values estimated for ClC1, M2 receptor, and Na+/K+ ATPase; the average and standard deviation are shown. The positively and negatively charged residues are shown as blue and red bars, respectively. The gray rectangles correspond to different regions of the proteins: B-R, the α-helical transmembrane segments in ClC1; TM1–TM7, the transmembrane segments in the M2 receptor; TM1–TM10, the transmembrane segments in Na+/K+ ATPase; β and γ, auxiliary subunits of Na+/K+ ATPase.
For Cx26, we detected two regions with large values, including the extracellular opening (E42, D46, D50, K41, R75, R184, and K188), and the center of the protein subunit (E147, R32, and R143; Fig. 6). Many of these residues have already been suggested to play crucial roles in voltage sensitivity (Chen et al., 2005; Deng et al., 2006; Kwon et al., 2011, 2012; Pinto et al., 2016). For instance, molecular dynamics simulations (Kwon et al., 2011, 2012) suggested an electrostatic network among E42, D46, R75, R184, E187, and K188 to be a part of the voltage sensor. Experimental evidence indicates that mutagenesis of R75, K41, and E42 significantly alters Cx26 voltage sensitivity (Chen et al., 2005; Deng et al., 2006; Pinto et al., 2016). In particular, K41 neutralization results in an increase of the apparent gating charge, while the double mutant K41E/E42S was shown to be more sensitive to voltage than the wild type (Pinto et al., 2016). Although experimental evidence suggests D2 also contributes to voltage sensitivity (Verselis et al., 1994), this residue did not exhibit any large value in our analysis. We believe this discrepancy might be caused by the low resolution of the N terminus, where D2 is located (Maeda et al., 2009): shortly after the start of molecular dynamics simulations, the N terminus lost its secondary structure and significantly deviated from its initial position, which limits the predictive power in this region.
TRPV1 has a four-helical domain, which is similar to the voltage-sensor domain of Shaker and NavMs (Cao et al., 2013; Liao et al., 2013). However, because this channel does not include several crucial charges on S4 and the corresponding countercharges on S1–S3 (Cao et al., 2013; Liao et al., 2013), it likely responds to changes in the MP through a different mechanism. Our analysis reveals that, in addition to the lack of charges, the four-helical domain also shows small R values, emphasizing that this region is not able to sense changes in the MP (Fig. 4). On the other hand, we found values above the detection threshold for residues of the extra- and intracellular openings of the four-helical domain (R455, R474, E478, R491, and E513) and of the S4–S5 linker (R557, E570, D576, and R579; Fig. 6). Mutagenesis of several of these residues (R557, E570, D576, and R579) has already been shown to significantly affect TRPV1 voltage sensitivity (Boukalova et al., 2010).
In VDAC1, we observed multiple residues spread over the entire protein with large values (Fig. 6). These include 5 residues on the α-helical N terminus and 27 residues on the β-barrel. Experimental and computational evidence also indicates that both the N-terminal α-helix and the β-barrel contribute to voltage sensitivity (Peng et al., 1992; Thomas et al., 1993; Popp et al., 1996; Song et al., 1998; Briones et al., 2016). Together with our data, this suggests a scenario in which the entire protein is involved in the response to changes in the MP rather than a single domain playing the role of a voltage sensor. Among the residues pinpointed by our analysis, mutagenesis of D16, K20, K61, and E84 is known to modulate the steepness of the conductance-voltage relationship (Thomas et al., 1993), while that of D32 and K96 does not significantly affect voltage sensitivity (Thomas et al., 1993). This indicates there are likely a few false-positive signals in our analysis; therefore, it has to be combined with experiments to confirm voltage sensitivity of the detected elements.
Previous experimental studies performed on ClC channels and transporters have revealed two sources of voltage sensitivity: protein residues and chloride ions (Pusch et al., 1995; Chen and Miller, 1996; Smith and Lippiat, 2010; Zifarelli et al., 2012; Grieschat and Alekov, 2014; De Jesús-Pérez et al., 2016). Reducing extracellular Cl− concentration in ClC-0 or a substitution of intracellular Cl− by impermeant anions in ClC-2 shifts their opening to more positive voltages (Pusch et al., 1995; Chen and Miller, 1996; De Jesús-Pérez et al., 2016). Measurements of the gating charge in a permeation-deficient mutant of ClC-5 revealed that Q is reduced upon extracellular ion depletion but does not go to zero, indicating that protein residues contribute to voltage sensitivity along with ions (Smith and Lippiat, 2010; Zifarelli et al., 2012; Grieschat and Alekov, 2014). One of these residues could be the so-called glutamate gate (E232 in ClC1), whose mutagenesis completely abolishes voltage sensitivity (Dutzler et al., 2003; Estévez et al., 2003; Melzer et al., 2003). Our analysis shows large values for both E232 and bound Cl− ions (Fig. 7). It also pinpoints several residues along the conduction path (D136, K215, K231, R421, and K467) and a region at the interface between the two ClC1 subunits (E291, R317, E347, and E548); these residues all show values above the detection threshold, suggesting they are likely significant contributors to the ClC1 gating charge.
In the case of the M2 receptor, the identified voltage-sensing element is complex as well and composed of the protein residues and a Na+ ion bound inside the internal hydrophilic pocket. The role of a Na+ ion in M2 receptor voltage sensitivity was in fact recently suggested based on molecular dynamics simulations (Vickery et al., 2016a). Our data agree with these findings and show large values for a Na+ ion and for D69 and D107 (Fig. 7). In addition, our results support the recent experimental finding that the DRY motif on the TM3 segment does not contribute to voltage sensitivity (Navarro-Polanco et al., 2011): direct measurements of the gating charge revealed that the Q value of the wild-type and the D120N-R121N mutant is similar.
Finally, for the Na+/K+ ATPase, we found three regions where is above the detection threshold, including the ionic binding site and the extracellular and cytosolic openings (Fig. 7). The ionic binding site, which is composed of E327, D808, D926, E953, and three Na+ ions, shows the largest contribution to the gating charge. This agrees with the known data reporting that changes in the extracellular Na+ concentration has a strong effect on voltage dependence of the Na+/K+ ATPase ionic current (Nakao and Gadsby, 1989) and the gating charge (Holmgren et al., 2000).
For the set of membrane proteins investigated here, we were able to rediscover several elements that were previously shown to play a role in voltage sensitivity. It is important to note that in ClC1 the M2 receptor and Na+/K+ ATPase voltage sensitivity was accessed directly by estimating the change in the gating charge upon mutagenesis of residues or upon variation of the ionic concentrations (Nakao and Gadsby, 1989; Holmgren et al., 2000; Ben-Chaim et al., 2006; Smith and Lippiat, 2010; Navarro-Polanco et al., 2011; Zifarelli et al., 2012). In Cx26, VDAC1, and TRPV1, on the other hand, it was accessed indirectly: as a reporter for voltage sensitivity, the effect of a given mutation on the conductance-voltage relationship ( and the voltage-independent component of gating) was used. Such an effect, however, can be possibly caused by structural perturbations of the voltage sensor itself or of other protein domains, and therefore, the experiments indirectly accessing voltage sensitivity must be interpreted with caution.
Traditional mutagenesis tends to introduce perturbations that simultaneously affect electrostatic and steric properties. One can disentangle these effects on protein function by using nonnatural amino acids. For example, citrulline is a neutral and nearly isosteric analogue of arginine that has recently been used to quantify electrostatic contribution of the voltage-sensing residues to gating (Infield et al., 2018). Because estimation of the gating charge is often challenging if at all possible, the usage of nonnatural amino acids can emerge as an alternative strategy to characterize voltage sensitivity.
Conclusions
One of the reasons voltage sensitivity in membrane proteins is hard to detect is that it depends on a combination of two factors: the ability of a protein to focus an electric field in some of its regions and its ability to respond to the electric field based on the presence of charged elements in these regions. We show that it is possible to use a simple and cheap computational tool to distinguish between voltage-sensitive and voltage-insensitive proteins and to detect true voltage-sensitive residues in voltage-gated ion channels. The application of this tool to six membrane proteins without well-characterized voltage sensors leads to the prediction of several candidates for voltage-sensing elements, some of which have already been shown to contribute to voltage sensitivity while others remain to be tested experimentally. The suggested approach is general, does not require extensive computational resources, and can be applied to any other membrane protein regardless of structure or function. This application is important to unravel the variety of ways voltage sensitivity has been manifested in biological molecules, and it should be highly useful both to identify how different factors such as mutations, membrane composition, or ligand binding might alter voltage sensitivity and to engineer this property into voltage-insensitive membrane proteins.
Supplementary Material
Acknowledgments
We acknowledge Kawai Lee, Marie Lycksell, and Rebecca J. Howard for providing the GLIC trajectory, Asghar Razavi for providing the Na+/K+ ATPase trajectory, and Samira Yazdi for providing the Shaker trajectory. The simulations were performed on resources provided by the Swedish National Infrastructure for Computing at PDC Centre for High Performance Computing.
This work was supported by grants from the Vetenskapsrådet (2017-04641) and the Science for Life Laboratory.
The authors declare no competing financial interests.
Author contributions: M.A. Kasimova, E. Lindahl, and L. Delemotte designed the research. M.A. Kasimova performed the simulations, analyzed the data, and wrote the original draft. M.A. Kasimova, E. Lindahl, and L. Delemotte edited the paper.
José D. Faraldo-Gómez served as editor.
References
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., and Lindahl E.. 2015. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 1–2:19–25. 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- Aggarwal S.K., and MacKinnon R.. 1996. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 16:1169–1177. 10.1016/S0896-6273(00)80143-9 [DOI] [PubMed] [Google Scholar]
- Aksimentiev A., and Schulten K.. 2005. Imaging alpha-hemolysin with molecular dynamics: ionic conductance, osmotic permeability, and the electrostatic potential map. Biophys. J. 88:3745–3761. 10.1529/biophysj.104.058727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almén M.S., Nordström K.J.V., Fredriksson R., and Schiöth H.B.. 2009. Mapping the human membrane proteome: a majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC Biol. 7:50 10.1186/1741-7007-7-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzai N., Ichida K., Jutabha P., Kimura T., Babu E., Jin C.J., Srivastava S., Kitamura K., Hisatome I., Endou H., and Sakurai H.. 2008. Plasma urate level is directly regulated by a voltage-driven urate efflux transporter URATv1 (SLC2A9) in humans. J. Biol. Chem. 283:26834–26838. 10.1074/jbc.C800156200 [DOI] [PubMed] [Google Scholar]
- Armstrong C.M., and Bezanilla F.. 1973. Currents related to movement of the gating particles of the sodium channels. Nature. 242:459–461. 10.1038/242459a0 [DOI] [PubMed] [Google Scholar]
- Bargiello T.A., Tang Q., Oh S., and Kwon T.. 2012. Voltage-dependent conformational changes in connexin channels. Biochim. Biophys. Acta. 1818:1807–1822. 10.1016/j.bbamem.2011.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrán C., Zapata O., and Darszon A.. 1996. Membrane potential regulates sea urchin sperm adenylylcyclase. Biochemistry. 35:7591–7598. 10.1021/bi952806v [DOI] [PubMed] [Google Scholar]
- Ben-Chaim Y., Tour O., Dascal N., Parnas I., and Parnas H.. 2003. The M2 muscarinic G-protein-coupled receptor is voltage-sensitive. J. Biol. Chem. 278:22482–22491. 10.1074/jbc.M301146200 [DOI] [PubMed] [Google Scholar]
- Ben-Chaim Y., Chanda B., Dascal N., Bezanilla F., Parnas I., and Parnas H.. 2006. Movement of ‘gating charge’ is coupled to ligand binding in a G-protein-coupled receptor. Nature. 444:106–109. 10.1038/nature05259 [DOI] [PubMed] [Google Scholar]
- Bernardi P. 1992. Modulation of the mitochondrial cyclosporin A-sensitive permeability transition pore by the proton electrochemical gradient. Evidence that the pore can be opened by membrane depolarization. J. Biol. Chem. 267:8834–8839. [PubMed] [Google Scholar]
- Bezanilla F. 2005. Voltage-gated ion channels. IEEE Trans. Nanobioscience. 4:34–48. 10.1109/TNB.2004.842463 [DOI] [PubMed] [Google Scholar]
- Bezanilla F. 2008. How membrane proteins sense voltage. Nat. Rev. Mol. Cell Biol. 9:323–332. 10.1038/nrm2376 [DOI] [PubMed] [Google Scholar]
- Bocquet N., Prado de Carvalho L., Cartaud J., Neyton J., Le Poupon C., Taly A., Grutter T., Changeux J.-P., and Corringer P.-J.. 2007. A prokaryotic proton-gated ion channel from the nicotinic acetylcholine receptor family. Nature. 445:116–119. 10.1038/nature05371 [DOI] [PubMed] [Google Scholar]
- Boukalova S., Marsakova L., Teisinger J., and Vlachova V.. 2010. Conserved residues within the putative S4-S5 region serve distinct functions among thermosensitive vanilloid transient receptor potential (TRPV) channels. J. Biol. Chem. 285:41455–41462. 10.1074/jbc.M110.145466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briones R., Weichbrodt C., Paltrinieri L., Mey I., Villinger S., Giller K., Lange A., Zweckstetter M., Griesinger C., Becker S., et al. 2016. Voltage Dependence of Conformational Dynamics and Subconducting States of VDAC-1. Biophys. J. 111:1223–1234. 10.1016/j.bpj.2016.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao E., Liao M., Cheng Y., and Julius D.. 2013. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature. 504:113–118. 10.1038/nature12823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caterina M.J., Schumacher M.A., Tominaga M., Rosen T.A., Levine J.D., and Julius D.. 1997. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 389:816–824. 10.1038/39807 [DOI] [PubMed] [Google Scholar]
- Catterall W.A. 1988. Structure and function of voltage-sensitive ion channels. Science. 242:50–61. 10.1126/science.2459775 [DOI] [PubMed] [Google Scholar]
- Chen T.Y., and Miller C.. 1996. Nonequilibrium gating and voltage dependence of the ClC-0 Cl- channel. J. Gen. Physiol. 108:237–250. 10.1085/jgp.108.4.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Deng Y., Bao X., Reuss L., and Altenberg G.A.. 2005. Mechanism of the defect in gap-junctional communication by expression of a connexin 26 mutant associated with dominant deafness. FASEB J. 19:1516–1518. 10.1096/fj.04-3491fje [DOI] [PubMed] [Google Scholar]
- Colombini M. 1989. Voltage gating in the mitochondrial channel, VDAC. J. Membr. Biol. 111:103–111. 10.1007/BF01871775 [DOI] [PubMed] [Google Scholar]
- Cone C.D. 1969. Section of biological and medical sciences: electroosmotic interactions accompanying mitosis initiation in sarcoma cells in vitro. Trans. N. Y. Acad. Sci. 31:404–427. 10.1111/j.2164-0947.1969.tb02926.x [DOI] [PubMed] [Google Scholar]
- Cone C.D., Jr 1971. Unified theory on the basic mechanism of normal mitotic control and oncogenesis. J. Theor. Biol. 30:151–181. 10.1016/0022-5193(71)90042-7 [DOI] [PubMed] [Google Scholar]
- Cooper D.M.F., Schell M.J., Thorn P., and Irvine R.F.. 1998. Regulation of adenylyl cyclase by membrane potential. J. Biol. Chem. 273:27703–27707. 10.1074/jbc.273.42.27703 [DOI] [PubMed] [Google Scholar]
- Darden T., York D., and Pedersen L.. 1993. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089–10092. 10.1063/1.464397 [DOI] [Google Scholar]
- DeCaen P.G., Yarov-Yarovoy V., Sharp E.M., Scheuer T., and Catterall W.A.. 2009. Sequential formation of ion pairs during activation of a sodium channel voltage sensor. Proc. Natl. Acad. Sci. USA. 106:22498–22503. 10.1073/pnas.0912307106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey T.E., Morgan D., and Cherny V.V.. 2003. The voltage dependence of NADPH oxidase reveals why phagocytes need proton channels. Nature. 422:531–534. 10.1038/nature01523 [DOI] [PubMed] [Google Scholar]
- De Jesús-Pérez J.J., Castro-Chong A., Shieh R.-C., Hernández-Carballo C.Y., De Santiago-Castillo J.A., and Arreola J.. 2016. Gating the glutamate gate of CLC-2 chloride channel by pore occupancy. J. Gen. Physiol. 147:25–37. 10.1085/jgp.201511424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delemotte L., Tarek M., Klein M.L., Amaral C., and Treptow W.. 2011. Intermediate states of the Kv1.2 voltage sensor from atomistic molecular dynamics simulations. Proc. Natl. Acad. Sci. USA. 108:6109–6114. 10.1073/pnas.1102724108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Chen Y., Reuss L., and Altenberg G.A.. 2006. Mutations of connexin 26 at position 75 and dominant deafness: essential role of arginine for the generation of functional gap-junctional channels. Hear. Res. 220:87–94. 10.1016/j.heares.2006.07.004 [DOI] [PubMed] [Google Scholar]
- Dutzler R., Campbell E.B., and MacKinnon R.. 2003. Gating the selectivity filter in ClC chloride channels. Science. 300:108–112. 10.1126/science.1082708 [DOI] [PubMed] [Google Scholar]
- Estévez R., Schroeder B.C., Accardi A., Jentsch T.J., and Pusch M.. 2003. Conservation of chloride channel structure revealed by an inhibitor binding site in ClC-1. Neuron. 38:47–59. 10.1016/S0896-6273(03)00168-5 [DOI] [PubMed] [Google Scholar]
- Grieschat M., and Alekov A.K.. 2014. Multiple discrete transitions underlie voltage-dependent activation in CLC Cl(-)/H(+) antiporters. Biophys. J. 107:L13–L15. 10.1016/j.bpj.2014.07.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga K., Kruse A.C., Asada H., Yurugi-Kobayashi T., Shiroishi M., Zhang C., Weis W.I., Okada T., Kobilka B.K., Haga T., and Kobayashi T.. 2012. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 482:547–551. 10.1038/nature10753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halestrap A.P., Woodfield K.Y., and Connern C.P.. 1997. Oxidative stress, thiol reagents, and membrane potential modulate the mitochondrial permeability transition by affecting nucleotide binding to the adenine nucleotide translocase. J. Biol. Chem. 272:3346–3354. 10.1074/jbc.272.6.3346 [DOI] [PubMed] [Google Scholar]
- Henrion U., Renhorn J., Börjesson S.I., Nelson E.M., Schwaiger C.S., Bjelkmar P., Wallner B., Lindahl E., and Elinder F.. 2012. Tracking a complete voltage-sensor cycle with metal-ion bridges. Proc. Natl. Acad. Sci. USA. 109:8552–8557. 10.1073/pnas.1116938109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B. 2001. Ion Channels of Excitable Membranes. Third edition Sinauer Associates, Inc.,, Sunderland, MA: 814 pp. [Google Scholar]
- Holmgren M., Wagg J., Bezanilla F., Rakowski R.F., De Weer P., and Gadsby D.C.. 2000. Three distinct and sequential steps in the release of sodium ions by the Na+/K+-ATPase. Nature. 403:898–901. 10.1038/35002599 [DOI] [PubMed] [Google Scholar]
- Hub J.S., Aponte-Santamaría C., Grubmüller H., and de Groot B.L.. 2010. Voltage-regulated water flux through aquaporin channels in silico. Biophys. J. 99:L97–L99. 10.1016/j.bpj.2010.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infield D.T., Lee E.E.L., Galpin J.D., Galles G.D., Bezanilla F., and Ahern C.A.. 2018. Replacing voltage sensor arginines with citrulline provides mechanistic insight into charge versus shape. J. Gen. Physiol. 150:1017–1024. 10.1085/jgp.201812075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M.Ø., Jogini V., Borhani D.W., Leffler A.E., Dror R.O., and Shaw D.E.. 2012. Mechanism of voltage gating in potassium channels. Science. 336:229–233. 10.1126/science.1216533 [DOI] [PubMed] [Google Scholar]
- Jentsch T.J., Günther W., Pusch M., and Schwappach B.. 1995. Properties of voltage-gated chloride channels of the ClC gene family. J. Physiol. 482(suppl):19S–25S. 10.1113/jphysiol.1995.sp020560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S., Kim T., Iyer V.G., and Im W.. 2008. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29:1859–1865. 10.1002/jcc.20945 [DOI] [PubMed] [Google Scholar]
- Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., and Klein M.L.. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926–935. 10.1063/1.445869 [DOI] [Google Scholar]
- Jutabha P., Kanai Y., Hosoyamada M., Chairoungdua A., Kim D.K., Iribe Y., Babu E., Kim J.Y., Anzai N., Chatsudthipong V., and Endou H.. 2003. Identification of a novel voltage-driven organic anion transporter present at apical membrane of renal proximal tubule. J. Biol. Chem. 278:27930–27938. 10.1074/jbc.M303210200 [DOI] [PubMed] [Google Scholar]
- Jutabha P., Anzai N., Wempe M.F., Wakui S., Endou H., and Sakurai H.. 2011. Apical voltage-driven urate efflux transporter NPT4 in renal proximal tubule. Nucleosides Nucleotides Nucleic Acids. 30:1302–1311. 10.1080/15257770.2011.616564 [DOI] [PubMed] [Google Scholar]
- Kaim G., and Dimroth P.. 1998. Voltage-generated torque drives the motor of the ATP synthase. EMBO J. 17:5887–5895. 10.1093/emboj/17.20.5887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaim G., and Dimroth P.. 1999. ATP synthesis by F-type ATP synthase is obligatorily dependent on the transmembrane voltage. EMBO J. 18:4118–4127. 10.1093/emboj/18.15.4118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai R., Ogawa H., Vilsen B., Cornelius F., and Toyoshima C.. 2013. Crystal structure of a Na+-bound Na+,K+-ATPase preceding the E1P state. Nature. 502:201–206. 10.1038/nature12578 [DOI] [PubMed] [Google Scholar]
- Kasimova M.A., Yazici A., Yudin Y., Granata D., Klein M.L., Rohacs T., and Carnevale V.. 2018. Ion Channel Sensing: Are Fluctuations the Crux of the Matter? J. Phys. Chem. Lett. 9:1260–1264. 10.1021/acs.jpclett.7b03396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanaugh M.P. 1993. Voltage dependence of facilitated arginine flux mediated by the system y+ basic amino acid transporter. Biochemistry. 32:5781–5785. 10.1021/bi00073a009 [DOI] [PubMed] [Google Scholar]
- Khalili-Araghi F., Jogini V., Yarov-Yarovoy V., Tajkhorshid E., Roux B., and Schulten K.. 2010. Calculation of the gating charge for the Kv1.2 voltage-activated potassium channel. Biophys. J. 98:2189–2198. 10.1016/j.bpj.2010.02.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künkele K.-P., Juin P., Pompa C., Nargang F.E., Henry J.-P., Neupert W., Lill R., and Thieffry M.. 1998. The isolated complex of the translocase of the outer membrane of mitochondria. Characterization of the cation-selective and voltage-gated preprotein-conducting pore. J. Biol. Chem. 273:31032–31039. 10.1074/jbc.273.47.31032 [DOI] [PubMed] [Google Scholar]
- Kwon T., Harris A.L., Rossi A., and Bargiello T.A.. 2011. Molecular dynamics simulations of the Cx26 hemichannel: evaluation of structural models with Brownian dynamics. J. Gen. Physiol. 138:475–493. 10.1085/jgp.201110679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon T., Roux B., Jo S., Klauda J.B., Harris A.L., and Bargiello T.A.. 2012. Molecular dynamics simulations of the Cx26 hemichannel: insights into voltage-dependent loop-gating. Biophys. J. 102:1341–1351. 10.1016/j.bpj.2012.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix J.J., Hyde H.C., Campos F.V., and Bezanilla F.. 2014. Moving gating charges through the gating pore in a Kv channel voltage sensor. Proc. Natl. Acad. Sci. USA. 111:E1950–E1959. 10.1073/pnas.1406161111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Cao E., Julius D., and Cheng Y.. 2013. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 504:107–112. 10.1038/nature12822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S.B., Campbell E.B., and Mackinnon R.. 2005. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 309:897–903. 10.1126/science.1116269 [DOI] [PubMed] [Google Scholar]
- Long S.B., Tao X., Campbell E.B., and MacKinnon R.. 2007. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 450:376–382. 10.1038/nature06265 [DOI] [PubMed] [Google Scholar]
- Lostao M.P., Mata J.F., Larrayoz I.M., Inzillo S.M., Casado F.J., and Pastor-Anglada M.. 2000. Electrogenic uptake of nucleosides and nucleoside-derived drugs by the human nucleoside transporter 1 (hCNT1) expressed in Xenopus laevis oocytes. FEBS Lett. 481:137–140. 10.1016/S0014-5793(00)01983-9 [DOI] [PubMed] [Google Scholar]
- Machtens J.-P., Briones R., Alleva C., de Groot B.L., and Fahlke C.. 2017. Gating Charge Calculations by Computational Electrophysiology Simulations. Biophys. J. 112:1396–1405. 10.1016/j.bpj.2017.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie B., Schäfer M.K.-H., Erickson J.D., Hediger M.A., Weihe E., and Varoqui H.. 2003. Functional properties and cellular distribution of the system A glutamine transporter SNAT1 support specialized roles in central neurons. J. Biol. Chem. 278:23720–23730. 10.1074/jbc.M212718200 [DOI] [PubMed] [Google Scholar]
- Mackerell A.D. Jr., Feig M., and Brooks C.L. III. 2004. Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 25:1400–1415. 10.1002/jcc.20065 [DOI] [PubMed] [Google Scholar]
- Maeda S., Nakagawa S., Suga M., Yamashita E., Oshima A., Fujiyoshi Y., and Tsukihara T.. 2009. Structure of the connexin 26 gap junction channel at 3.5 A resolution. Nature. 458:597–602. 10.1038/nature07869 [DOI] [PubMed] [Google Scholar]
- Malhotra K., Sathappa M., Landin J.S., Johnson A.E., and Alder N.N.. 2013. Structural changes in the mitochondrial Tim23 channel are coupled to the proton-motive force. Nat. Struct. Mol. Biol. 20:965–972. 10.1038/nsmb.2613 [DOI] [PubMed] [Google Scholar]
- Melzer N., Biela A., and Fahlke C.. 2003. Glutamate modifies ion conduction and voltage-dependent gating of excitatory amino acid transporter-associated anion channels. J. Biol. Chem. 278:50112–50119. 10.1074/jbc.M307990200 [DOI] [PubMed] [Google Scholar]
- Miller A.N., and Long S.B.. 2012. Crystal structure of the human two-pore domain potassium channel K2P1. Science. 335:432–436. 10.1126/science.1213274 [DOI] [PubMed] [Google Scholar]
- Mirzabekov T.A., and Ermishkin L.N.. 1989. The gate of mitochondrial porin channel is controlled by a number of negative and positive charges. FEBS Lett. 249:375–378. 10.1016/0014-5793(89)80662-3 [DOI] [PubMed] [Google Scholar]
- Morth J.P., Pedersen B.P., Toustrup-Jensen M.S., Sørensen T.L.-M., Petersen J., Andersen J.P., Vilsen B., and Nissen P.. 2007. Crystal structure of the sodium-potassium pump. Nature. 450:1043–1049. 10.1038/nature06419 [DOI] [PubMed] [Google Scholar]
- Nakao M., and Gadsby D.C.. 1986. Voltage dependence of Na translocation by the Na/K pump. Nature. 323:628–630. 10.1038/323628a0 [DOI] [PubMed] [Google Scholar]
- Nakao M., and Gadsby D.C.. 1989. [Na] and [K] dependence of the Na/K pump current-voltage relationship in guinea pig ventricular myocytes. J. Gen. Physiol. 94:539–565. 10.1085/jgp.94.3.539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Polanco R.A., Moreno Galindo E.G., Ferrer-Villada T., Arias M., Rigby J.R., Sánchez-Chapula J.A., and Tristani-Firouzi M.. 2011. Conformational changes in the M2 muscarinic receptor induced by membrane voltage and agonist binding. J. Physiol. 589:1741–1753. 10.1113/jphysiol.2010.204107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilius B., Talavera K., Owsianik G., Prenen J., Droogmans G., and Voets T.. 2005. Gating of TRP channels: a voltage connection? J. Physiol. 567:35–44. 10.1113/jphysiol.2005.088377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noceti F., Baldelli P., Wei X., Qin N., Toro L., Birnbaumer L., and Stefani E.. 1996. Effective gating charges per channel in voltage-dependent K+ and Ca2+ channels. J. Gen. Physiol. 108:143–155. 10.1085/jgp.108.3.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosé S. 1984. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 81:511–519. 10.1063/1.447334 [DOI] [Google Scholar]
- Parent L., Supplisson S., Loo D.D.F., and Wright E.M.. 1992. Electrogenic properties of the cloned Na+/glucose cotransporter: II. A transport model under nonrapid equilibrium conditions. J. Membr. Biol. 125:63–79. 10.1007/BF00235798 [DOI] [PubMed] [Google Scholar]
- Park E., and MacKinnon R.. 2018. Structure of the CLC-1 chloride channel from Homo sapiens. eLife. 7:e36629 10.7554/eLife.36629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello M., and Rahman A.. 1981. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 52:7182–7190. 10.1063/1.328693 [DOI] [Google Scholar]
- Payandeh J., Scheuer T., Zheng N., and Catterall W.A.. 2011. The crystal structure of a voltage-gated sodium channel. Nature. 475:353–358. 10.1038/nature10238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S., Blachly-Dyson E., Forte M., and Colombini M.. 1992. Large scale rearrangement of protein domains is associated with voltage gating of the VDAC channel. Biophys. J. 62:123–131. 10.1016/S0006-3495(92)81799-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronilli V., Nicolli A., Costantini P., Colonna R., and Bernardi P.. 1994. Regulation of the permeability transition pore, a voltage-dependent mitochondrial channel inhibited by cyclosporin A. Biochim. Biophys. Acta. 1187:255–259. 10.1016/0005-2728(94)90122-8 [DOI] [PubMed] [Google Scholar]
- Pinto B.I., García I.E., Pupo A., Retamal M.A., Martínez A.D., Latorre R., and González C.. 2016. Charged Residues at the First Transmembrane Region Contribute to the Voltage Dependence of the Slow Gate of Connexins. J. Biol. Chem. 291:15740–15752. 10.1074/jbc.M115.709402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp B., Court D.A., Benz R., Neupert W., and Lill R.. 1996. The role of the N and C termini of recombinant Neurospora mitochondrial porin in channel formation and voltage-dependent gating. J. Biol. Chem. 271:13593–13599. 10.1074/jbc.271.23.13593 [DOI] [PubMed] [Google Scholar]
- Pusch M., Ludewig U., Rehfeldt A., and Jentsch T.J.. 1995. Gating of the voltage-dependent chloride channel CIC-0 by the permeant anion. Nature. 373:527–531. 10.1038/373527a0 [DOI] [PubMed] [Google Scholar]
- Razavi A.M., Delemotte L., Berlin J.R., Carnevale V., and Voelz V.A.. 2017. Molecular simulations and free-energy calculations suggest conformation-dependent anion binding to a cytoplasmic site as a mechanism for Na+/K+-ATPase ion selectivity. J. Biol. Chem. 292:12412–12423. 10.1074/jbc.M117.779090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy R., Smith D., Wayman G., Wu Z., Villacres E.C., and Storm D.R.. 1995. Voltage-sensitive adenylyl cyclase activity in cultured neurons. A calcium-independent phenomenon. J. Biol. Chem. 270:14340–14346. 10.1074/jbc.270.24.14340 [DOI] [PubMed] [Google Scholar]
- Rinne A., Birk A., and Bünemann M.. 2013. Voltage regulates adrenergic receptor function. Proc. Natl. Acad. Sci. USA. 110:1536–1541. 10.1073/pnas.1212656110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosasco M.G., Gordon S.E., and Bajjalieh S.M.. 2015. Characterization of the Functional Domains of a Mammalian Voltage-Sensitive Phosphatase. Biophys. J. 109:2480–2491. 10.1016/j.bpj.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B. 2008. The membrane potential and its representation by a constant electric field in computer simulations. Biophys. J. 95:4205–4216. 10.1529/biophysj.108.136499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauguet L., Shahsavar A., Poitevin F., Huon C., Menny A., Nemecz À., Haouz A., Changeux J.-P., Corringer P.-J., and Delarue M.. 2014. Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation. Proc. Natl. Acad. Sci. USA. 111:966–971. 10.1073/pnas.1314997111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schewe M., Nematian-Ardestani E., Sun H., Musinszki M., Cordeiro S., Bucci G., de Groot B.L., Tucker S.J., Rapedius M., and Baukrowitz T.. 2016. A Non-canonical Voltage-Sensing Mechanism Controls Gating in K2P K(+) Channels. Cell. 164:937–949. 10.1016/j.cell.2016.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnetkamp P.P., and Szerencsei R.T.. 1991. Effect of potassium ions and membrane potential on the Na-Ca-K exchanger in isolated intact bovine rod outer segments. J. Biol. Chem. 266:189–197. [PubMed] [Google Scholar]
- Scorrano L., Petronilli V., and Bernardi P.. 1997. On the voltage dependence of the mitochondrial permeability transition pore. A critical appraisal. J. Biol. Chem. 272:12295–12299. 10.1074/jbc.272.19.12295 [DOI] [PubMed] [Google Scholar]
- Smith A.J., and Lippiat J.D.. 2010. Voltage-dependent charge movement associated with activation of the CLC-5 2Cl-/1H+ exchanger. FASEB J. 24:3696–3705. 10.1096/fj.09-150649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Midson C., Blachly-Dyson E., Forte M., and Colombini M.. 1998. The sensor regions of VDAC are translocated from within the membrane to the surface during the gating processes. Biophys. J. 74:2926–2944. 10.1016/S0006-3495(98)78000-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara M., Nakanishi T., Fei Y.J., Martindale R.G., Ganapathy M.E., Leibach F.H., and Ganapathy V.. 2000. Structure and function of ATA3, a new subtype of amino acid transport system A, primarily expressed in the liver and skeletal muscle. Biochim. Biophys. Acta. 1509:7–13. 10.1016/S0005-2736(00)00349-7 [DOI] [PubMed] [Google Scholar]
- Sula A., Booker J., Ng L.C.T., Naylor C.E., DeCaen P.G., and Wallace B.A.. 2017. The complete structure of an activated open sodium channel. Nat. Commun. 8:14205 10.1038/ncomms14205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz K.J. 2008. Sensing voltage across lipid membranes. Nature. 456:891–897. 10.1038/nature07620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X., Lee A., Limapichat W., Dougherty D.A., and MacKinnon R.. 2010. A gating charge transfer center in voltage sensors. Science. 328:67–73. 10.1126/science.1185954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijido O., Rappaport S.M., Chamberlin A., Noskov S.Y., Aguilella V.M., Rostovtseva T.K., and Bezrukov S.M.. 2014. Acidification asymmetrically affects voltage-dependent anion channel implicating the involvement of salt bridges. J. Biol. Chem. 289:23670–23682. 10.1074/jbc.M114.576314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas L., Blachly-Dyson E., Colombini M., and Forte M.. 1993. Mapping of residues forming the voltage sensor of the voltage-dependent anion-selective channel. Proc. Natl. Acad. Sci. USA. 90:5446–5449. 10.1073/pnas.90.12.5446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treptow W., Tarek M., and Klein M.L.. 2009. Initial response of the potassium channel voltage sensor to a transmembrane potential. J. Am. Chem. Soc. 131:2107–2109. 10.1021/ja807330g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujwal R., Cascio D., Colletier J.-P., Faham S., Zhang J., Toro L., Ping P., and Abramson J.. 2008. The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating. Proc. Natl. Acad. Sci. USA. 105:17742–17747. 10.1073/pnas.0809634105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez L.B., and Boveris A.. 2007. Mitochondrial nitric oxide synthase, a voltage-dependent enzyme, is responsible for nitric oxide diffusion to cytosol. Front. Biosci. 12:1210–1219. 10.2741/2139 [DOI] [PubMed] [Google Scholar]
- van der Laan M., Schrempp S.G., and Pfanner N.. 2013. Voltage-coupled conformational dynamics of mitochondrial protein-import channel. Nat. Struct. Mol. Biol. 20:915–917. 10.1038/nsmb.2643 [DOI] [PubMed] [Google Scholar]
- Vargas E., Yarov-Yarovoy V., Khalili-Araghi F., Catterall W.A., Klein M.L., Tarek M., Lindahl E., Schulten K., Perozo E., Bezanilla F., and Roux B.. 2012. An emerging consensus on voltage-dependent gating from computational modeling and molecular dynamics simulations. J. Gen. Physiol. 140:587–594. 10.1085/jgp.201210873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verselis V.K., Ginter C.S., and Bargiello T.A.. 1994. Opposite voltage gating polarities of two closely related connexins. Nature. 368:348–351. 10.1038/368348a0 [DOI] [PubMed] [Google Scholar]
- Vickery O.N., Machtens J.-P., Tamburrino G., Seeliger D., and Zachariae U.. 2016a Structural Mechanisms of Voltage Sensing in G Protein-Coupled Receptors. Structure. 24:997–1007. 10.1016/j.str.2016.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickery O.N., Machtens J.-P., and Zachariae U.. 2016b Membrane potentials regulating GPCRs: insights from experiments and molecular dynamics simulations. Curr. Opin. Pharmacol. 30:44–50. 10.1016/j.coph.2016.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorburger T., Nedielkov R., Brosig A., Bok E., Schunke E., Steffen W., Mayer S., Götz F., Möller H.M., and Steuber J.. 2016. Role of the Na(+)-translocating NADH:quinone oxidoreductase in voltage generation and Na(+) extrusion in Vibrio cholerae. Biochim. Biophys. Acta. 1857:473–482. 10.1016/j.bbabio.2015.12.010 [DOI] [PubMed] [Google Scholar]
- Wadiche J.I., Arriza J.L., Amara S.G., and Kavanaugh M.P.. 1995. Kinetics of a human glutamate transporter. Neuron. 14:1019–1027. 10.1016/0896-6273(95)90340-2 [DOI] [PubMed] [Google Scholar]
- Weer P.D., Gadsby D.C., and Rakowski R.F.. 1988. Voltage dependence of the Na-K pump. Annu. Rev. Physiol. 50:225–241. 10.1146/annurev.ph.50.030188.001301 [DOI] [PubMed] [Google Scholar]
- Wu D., Delaloye K., Zaydman M.A., Nekouzadeh A., Rudy Y., and Cui J.. 2010. State-dependent electrostatic interactions of S4 arginines with E1 in S2 during Kv7.1 activation. J. Gen. Physiol. 135:595–606. 10.1085/jgp.201010408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., and Brackenbury W.J.. 2013. Membrane potential and cancer progression. Front. Physiol. 4:185 10.3389/fphys.2013.00185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao D., Mackenzie B., Ming H., Varoqui H., Zhu H., Hediger M.A., and Erickson J.D.. 2000. A novel system A isoform mediating Na+/neutral amino acid cotransport. J. Biol. Chem. 275:22790–22797. 10.1074/jbc.M002965200 [DOI] [PubMed] [Google Scholar]
- Yazdi S., Stein M., Elinder F., Andersson M., and Lindahl E.. 2016. The Molecular Basis of Polyunsaturated Fatty Acid Interactions with the Shaker Voltage-Gated Potassium Channel. PLOS Comput. Biol. 12:e1004704 10.1371/journal.pcbi.1004704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander C.B., Albers T., and Grewer C.. 2013. Voltage-dependent processes in the electroneutral amino acid exchanger ASCT2. J. Gen. Physiol. 141:659–672. 10.1085/jgp.201210948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zander C.B., and Grewer C.. 2011. The C-terminal domain of the neutral amino acid transporter SNAT2 regulates transport activity through voltage-dependent processes. Biochem. J. 434:287–296. 10.1042/BJ20100507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Shen W., He D.Z.Z., Long K.B., Madison L.D., and Dallos P.. 2000. Prestin is the motor protein of cochlear outer hair cells. Nature. 405:149–155. 10.1038/35012009 [DOI] [PubMed] [Google Scholar]
- Zifarelli G., De Stefano S., Zanardi I., and Pusch M.. 2012. On the mechanism of gating charge movement of ClC-5, a human Cl(-)/H(+) antiporter. Biophys. J. 102:2060–2069. 10.1016/j.bpj.2012.03.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.