Autophagosome fusion with vacuoles requires a conserved fusion machinery, though the topology remained unclear. Two papers in this issue, Bas et al. and Gao et al., uncover Ykt6 as the required autophagosomal SNARE.
Abstract
Autophagy is a catabolic pathway that delivers intracellular material to the mammalian lysosomes or the yeast and plant vacuoles. The final step in this process is the fusion of autophagosomes with vacuoles, which requires SNARE proteins, the homotypic vacuole fusion and protein sorting tethering complex, the RAB7-like Ypt7 GTPase, and its guanine nucleotide exchange factor, Mon1-Ccz1. Where these different components are located and function during fusion, however, remains to be fully understood. Here, we present a novel in vitro assay to monitor fusion of intact and functional autophagosomes with vacuoles. This process requires ATP, physiological temperature, and the entire fusion machinery to tether and fuse autophagosomes with vacuoles. Importantly, we uncover Ykt6 as the autophagosomal SNARE. Our assay and findings thus provide the tools to dissect autophagosome completion and fusion in a test tube.
Introduction
Macroautophagy, hereafter referred to as autophagy, is an intracellular degradation and recycling pathway highly conserved among eukaryotes. During autophagy, cellular material such as organelles and other cytosolic components are sequestered by a double-membrane vesicle, the autophagosome, and then exposed to lysosomal luminal content (Lamb et al., 2013b; Ge et al., 2014; Shibutani and Yoshimori, 2014). In yeast, autophagosome diameters typically range from ∼300 to 900 nm (Baba et al., 1994). Flux through the pathway culminates with autophagosome–vacuole fusion, where the external autophagosomal membrane fuses with the vacuole, releasing the inner single-membrane vesicle into the lumen of this organelle. The generated autophagic body is degraded by the resident hydrolases and the resulting metabolites are then exported to the cytoplasm for reuse (Klionsky et al., 2016).
How the autophagosome fuses with the vacuole (or the lysosome in metazoan cells) has not been fully characterized. It has been assumed that their fusion follows a similar pathway as almost all fusion events involved in intracellular trafficking, which includes requirements for a specific Rab GTPase, a tethering factor, and membrane-bound SNARE proteins (Lamb et al., 2013a; Reggiori and Ungermann, 2017).
Rab GTPases are switch-like proteins with a C-terminal prenyl anchor that require a guanine nucleotide exchange factor (GEFs) to localize to a specific membrane. GEFs then trigger exchange of bound GDP for GTP, which enables the Rab to bind effector proteins (Goody et al., 2017). On vacuoles, the RAB7-like Ypt7 binds to the homotypic vacuole fusion and protein sorting (HOPS) tethering complex, a large hexameric complex with two Ypt7-binding sites at opposite ends (Seals et al., 2000; Bröcker et al., 2012). HOPS can tether Ypt7-positive membranes (Hickey and Wickner, 2010; Ho and Stroupe, 2015; Lürick et al., 2017), but it also interacts with SNAREs, promoting their assembly and lipid bilayer mixing of closely opposed membranes (Lobingier and Merz, 2012; Baker et al., 2015; D’Agostino et al., 2017; Orr et al., 2017).
Previous studies demonstrated that the biogenesis and fusion of autophagosomes with the vacuole requires Ypt7, its GEF Mon1-Ccz1; HOPS; the Q-SNAREs Vam3 (Qa), Vti1 (Qb), and Vam7 (Qc); the R-SNARE Ykt6; and the SNARE recycling machinery of Sec18 and Sec17 (Ishihara et al., 2001; Reggiori and Ungermann, 2017; Gao et al., 2018). However, other SNAREs have also been implicated in autophagosome biogenesis and fusion (Nair et al., 2011). Given that essential SNAREs like Ykt6 and Vti1 function in ER and Golgi biogenesis as well (McNew et al., 1997; Fischer von Mollard and Stevens, 1999; Dilcher et al., 2001), in vivo analyses of SNARE mutants could cause indirect defects, which may be misleading in the identification of the autophagosomal SNARE. We recently demonstrated that Mon1-Ccz1 is specifically recruited to autophagosomes by LC3-like Atg8, where it loads Ypt7 onto the autophagosomal membrane (Gao et al., 2018). Ypt7 on autophagosomes could then allow vacuole-bound HOPS to tether and fuse autophagosomes with vacuoles. This process very likely functions in a similar manner in metazoan cells (Hegedüs et al., 2016).
Several assays have been established to follow autophagy in vivo (Noda et al., 1995; Kirisako et al., 1999; Torggler et al., 2017). Although they can point to the involved proteins, these assays fail to dissect the specific requirements of proteins on membranes. One major advantage of an in vitro assay with purified components and organelles is the accessibility to the organelle surface and the possibility to quantitatively alter the amounts of specific proteins, which has been instrumental to unravel, for example, the mechanism of homotypic fusion between vacuoles (Wickner, 2010).
To unveil the mechanism of autophagosome–vacuole fusion and the involved proteins, we have developed a reliable in vitro autophagosome–vacuole fusion assay with purified organelles. This assay allowed us to confirm the direct involvement of Ypt7, HOPS, and Ccz1-Mon1 in this event and identified Ykt6 as the autophagosomal SNARE.
Results
Preparation of an autophagosome-enriched fraction
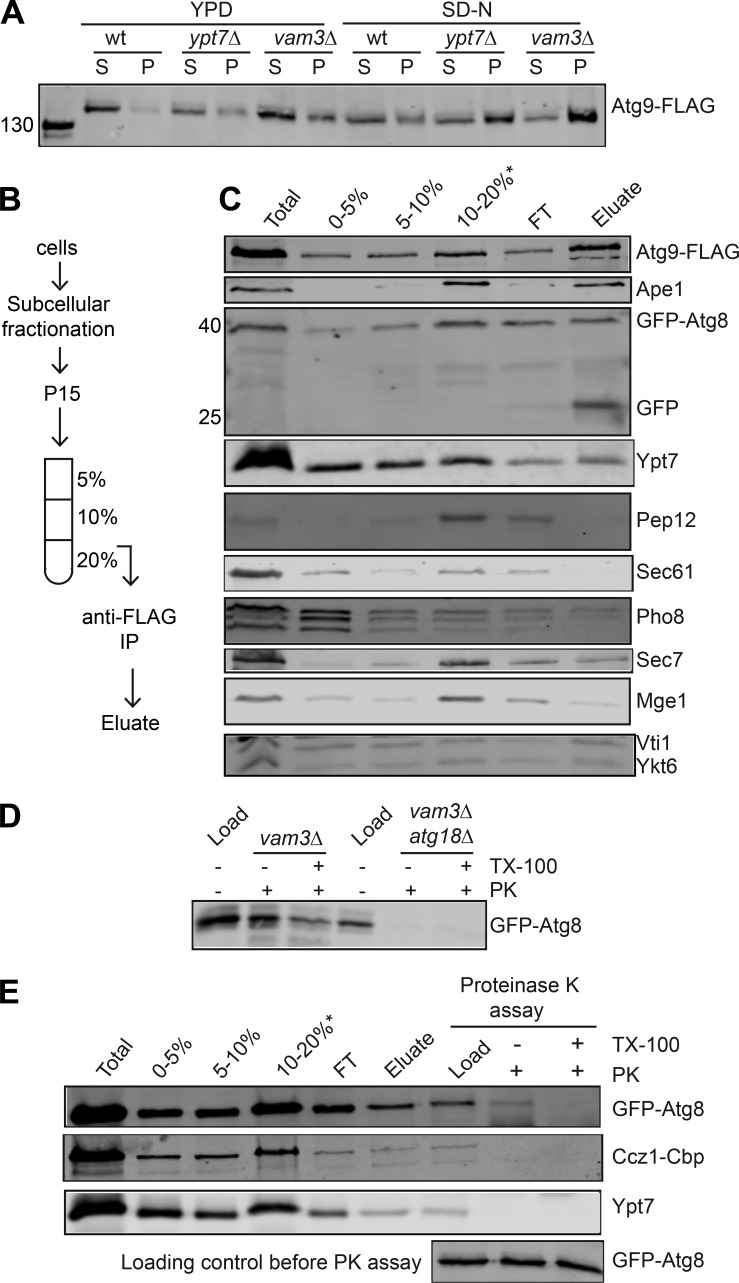
Autophagosomes fail to fuse with the vacuole and accumulate in the cytoplasm upon deletion of VAM3 or YPT7 (Darsow et al., 1997; Wurmser et al., 2000). These autophagosomes carry the transmembrane autophagy protein Atg9 (Cebollero et al., 2012; Yamamoto et al., 2012). To enrich autophagosomes from yeast, we thus tagged Atg9 with 3xFLAG in vam3Δ and ypt7Δ cells, and membranes from starved cells were separated by differential centrifugation at 15,000 g. Atg9 accumulated in this pellet fraction that contained high-density membranes when vam3Δ and ypt7Δ strains, but not wild-type cells, were nitrogen starved (Fig. 1 A), indicating that autophagosomes are enriched in this fraction.
Figure 1.
Purification of an autophagosome-enriched fraction. (A) Detection of autophagosomes by subcellular fractionation assay. The vam3Δ and ypt7Δ mutants expressing 3xFLAG-tagged Atg9 were grown in YPD medium and starved in SD-N medium for 3 h. Cells were opened, and proteins present in the 15,000 g pellet (P15) fraction were analyzed by Western blot (see Materials and methods). (B) Scheme for autophagosome fusion on iodixanol gradients. IP, immunoprecipitation. (C) Purification of autophagosomes. vam3Δ cells expressing 3xFLAG-tagged Atg9 and GFP-tagged Atg8 were grown in YPD medium and then starved in SD-N medium for 3 h. P15 fractions were applied to iodixanol density gradient centrifugation, and autophagosome-enriched fractions were incubated with anti-FLAG beads. Bound autophagosomes were eluted using the FLAG peptide. Eluates were collected, boiled in SDS sample buffer, and analyzed by SDS-PAGE and Western blotting against selected proteins. Atg9, prApe1, and Atg8 are autophagosome marker proteins. The other marker proteins of organelles are endosomes (Pep12), ER (Sec61), vacuoles (Pho8), Golgi (Sec7), and mitochondria (Mge1), Golgi, and endosome/vacuole (Vti1 and Ykt6). FT, flow through. (D and E) Protease protection assay. Autophagosome-enriched fractions were incubated with or without proteinase K (PK) and/or Triton X-100 (TX100). Reactions were stopped by addition of PMSF, boiled in SDS sample buffer, and analyzed by SDS-PAGE and Western blotting. All data are representatives of at least three independent experiments (C–E).
As Vam3 is a bona fide vacuolar SNARE that arrives at vacuoles from the Golgi via the AP-3 pathway (Darsow et al., 1998), we reasoned that autophagosomes from this mutant should have all required fusion factors on their surface. We then enriched autophagosomes as before, applied the corresponding P15 fraction to an iodixanol step gradient, and collected fractions after centrifugation. To biochemically and visually follow autophagosomes, endogenous Atg8 was N-terminally fused with GFP (Kirisako et al., 1999). All makers of autophagosomes (Atg9, Ape1, and Atg8) were enriched in the 10–20% interface of the iodixanol gradient (Fig. 1 C). We collected this fraction and incubated it with anti-FLAG agarose beads to further concentrate autophagosomes by immunoisolation (Fig. 1, B and C). Upon elution with FLAG peptide, we detected the autophagosome marker proteins in the eluates, showing that these fractions indeed contained autophagosomes (Fig. 1 C). Importantly, this preparation also presented significant amounts of Ypt7, whereas organelle protein markers for endosomes (Pep12), ER (Sec61), vacuoles (Pho8), Golgi (Sec7), or mitochondria (Mge1) were depleted. We also observed the SNAREs Vti1 and Ykt6 in the eluate. These data show that autophagosomes can be enriched following our procedure.
To determine whether the purified autophagosomes from vam3Δ cells are intact, we performed a protease protection assay (Fig. 1 D; Klionsky et al., 2016). If autophagosomes are closed, then membrane proteins such as Atg8 are present on the autophagosomal surface and luminal leaflet, and thus only part of it is accessible by externally added protease (Klionsky et al., 2016). We therefore treated autophagosomes with proteinase K in the presence or absence of Triton X-100. In the absence of Triton X-100, approximately half of GFP-tagged Atg8 disappeared and was almost completely turned over in the presence of detergent (Fig. 1 D). In contrast, GFP-Atg8 disappeared completely, regardless of detergent addition, when we analyzed the open autophagosomes obtained from a vam3Δ strain also lacking Atg18, a protein essential for autophagosome formation (Rieter et al., 2013). We thus conclude that our procedure leads to the purification of intact autophagosomes. We noticed that the detected amount of GFP-Atg8 in the load was much less upon deletion of Atg18 than in the vam3Δ strain, showing that knockout of ATG18 results in a block in the generation of autophagosomal structures as expected (Barth et al., 2001; Guan et al., 2001).
We recently reported that Mon1-Ccz1 is specifically recruited to the preautophagosomal structures under starvation conditions (Gao et al., 2018). To confirm that Mon1-Ccz1 and Ypt7 are present on complete autophagosomes, we traced both Ccz1 and Ypt7 during autophagosome purification before conducting a proteinase K protection assay as above. Although GFP-Atg8 was still detectable in preparation exclusively exposed to proteinase K, both Ccz1 and Ypt7 were completely degraded by the same treatment (Fig. 1 E). These data indicate that complete autophagosomes, ready to fuse with vacuoles, present Ypt7 and Mon1-Ccz1 on their surface.
Novel visual assay to monitor autophagosome–vacuole fusion
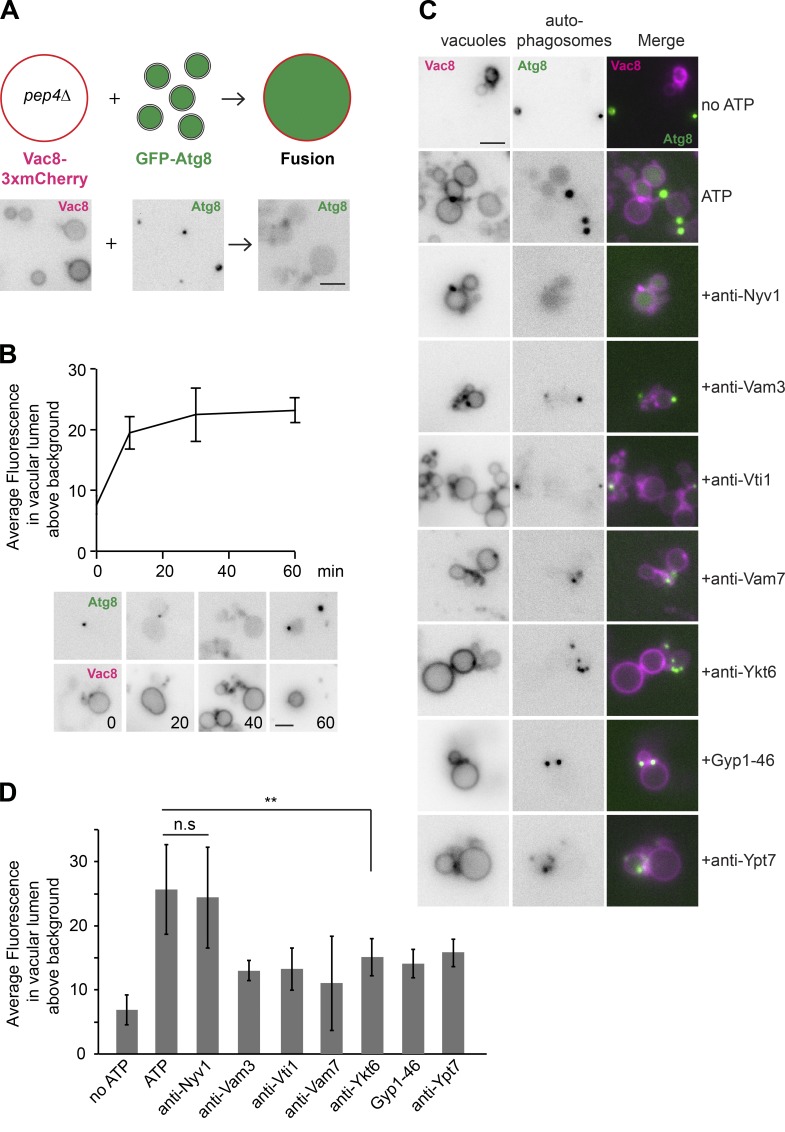
To establish an in vitro autophagosome–vacuole fusion assay, we endogenously tagged the vacuolar membrane protein Vac8 with 3xmCherry in the protease-deficient pep4Δ strain and purified vacuoles following an established protocol (Haas, 1995). Autophagosomes were purified from vam3Δ cells carrying GFP-tagged Atg8 as before. We reasoned that the strongly reduced vacuolar proteolytic activity in the pep4Δ cells should lead to an accumulation of autophagic bodies carrying GFP-Atg8 within the vacuole lumen during autophagosome–vacuole fusion (Fig. 2 A).
Figure 2.
Establishment of in vitro autophagosome–vacuole fusion assay. (A) Scheme of the autophagosome–vacuole fusion (top). Autophagosomes were purified from vam3Δ cells expressing Atg9-3xFLAG, and GFP-Atg8 were starved for 3 h (bottom left). Vacuoles were isolated from pep4Δ cells expressing Vac8-3xmCherry (bottom middle), and then incubated with autophagosomes at 26°C for 1 h with ATP (bottom right). Fusion was analyzed by fluorescence microscopy. Scale bar, 2 µm. (B) Time course of autophagosome–vacuole fusion. Autophagosomes and vacuoles were purified as in A and incubated at 26°C with ATP for the indicated time. Fluorescence intensity of GFP-Atg8 in the vacuolar lumen was quantified by ImageJ using the region of interest manager tool. Separate channels for GFP-Atg8 and Vac8-3xmCherry are shown. Error bars represent standard deviations of three independent experiments. Scale bar, 2 µm. (C) Autophagosomes and vacuoles were purified as in A and incubated at 26°C for 1 h with or without ATP and with the indicated inhibitors. Analysis was performed as in B. (D) Quantification of the time course fusion experiments. Error bars represent standard deviations of three independent experiments. **, P < 0.01 (Student’s t test). All data are representative of at least three independent experiments.
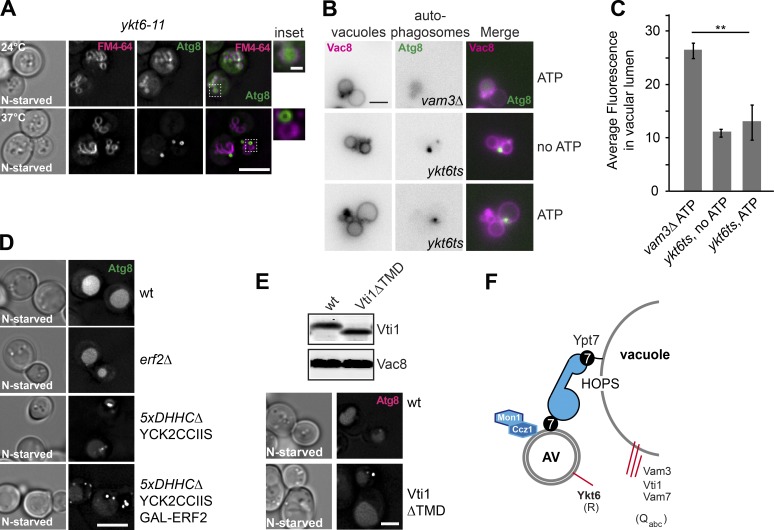
For our fusion assay, we used the established buffer conditions for vacuole–vacuole homotypic fusion (Haas, 1995) and incubated vacuoles with purified autophagosomes at 26°C in the presence of ATP. At the indicated time points, samples were analyzed by fluorescence microscopy to quantify the green fluorescent signal within vacuoles, marked by Vac8-3xmCherry (Fig. 2, A and B). We interpreted the green fluorescence in the vacuole lumen as rapidly moving autophagic bodies. We observed two distinct events: (1) the association of autophagosomes to vacuoles, visible as green puncta proximal to the red vacuoles, likely reflecting the tethering between these two organelles, and (2) the accumulation of green fluorescence within the vacuole lumen. The increase of green fluorescence in the vacuole lumen depended on both temperature and ATP, and it was completed after 60 min, similarly to what was observed for homotypic vacuole fusion (Haas et al., 1994; Fig. 2, B and C). These results strongly suggest that autophagosomes initially tether and then fuse with the vacuole in our in vitro reaction.
To test the specificity of this reaction, we focused on the fusion event and therefore used specific antibodies and established inhibitors of vacuole fusion. In vivo analyses revealed that autophagosome fusion with vacuoles requires the SNARE proteins Vti1, Ykt6, Vam3, and Vam7 (Fischer von Mollard and Stevens, 1999; Dilcher et al., 2001; Ishihara et al., 2001; Ohashi and Munro, 2010). However, it had not yet been established whether the essential SNAREs Vti1 and Ykt6 might have already affected earlier steps of autophagosome formation. Importantly, and in agreement with the observed fusion block in vivo, antibodies to Vam3, Vam7, Ykt6, and Vti1 strongly decreased the appearance of intravacuolar green fluorescence, indicating impairment in autophagosome–vacuole fusion (Fig. 2, C and D). This result demonstrates that Vti1 and Ykt6 are indeed directly involved in fusion between autophagosomes and vacuoles. In contrast, antibodies to the vacuolar SNARE Nyv1, which block vacuole homotypic fusion (Ungermann et al., 1999), did not interfere with this event (Fig. 2 D), in agreement with the fact that Nyv1 has been shown to not be involved in autophagy (Fischer von Mollard and Stevens, 1999). To further determine whether the autophagosome–vacuole fusion depends on the Rab Ypt7, we used a promiscuous and catalytic Rab GTPase-activating domain of Gyp1, named Gyp1–46, which blocks fusion of vacuoles by inhibiting Ypt7 (Brett et al., 2008; Zick and Wickner, 2012). This construct also blocked autophagosome–vacuole fusion, as did an antibody to Ypt7 (Fig. 2 D). Altogether, these data suggest that we established a specific in vitro fusion assay that recapitulates the in vivo fusion of autophagosomes with vacuoles.
HOPS and Mon1-Ccz1 drive tethering and fusion
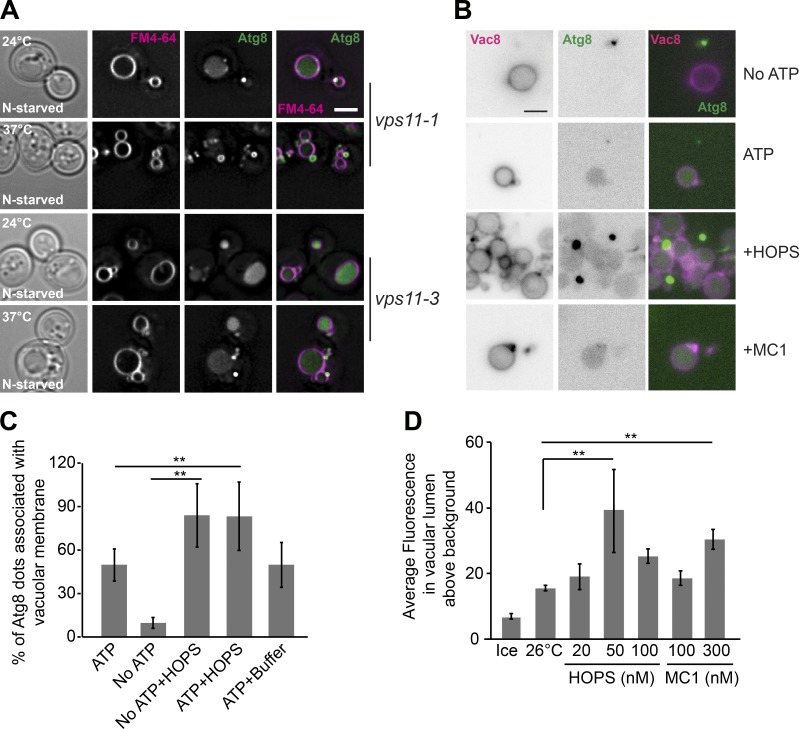
The Ypt7 GTPases requirement implies that the HOPS tethering complex promotes tethering and fusion of autophagosomes with vacuoles. To assess the direct role of HOPS in autophagy, we first analyzed autophagy in vivo. Previously, two temperature-sensitive (ts) alleles in the HOPS subunit Vps11 (i.e., vps11-1 and vps11-3) have been characterized (Peterson and Emr, 2001). Vps11 is also present in the endosomal CORVET (class C core vacuole endosome tethering) complex, which shares four subunits with HOPS (Balderhaar and Ungermann, 2013). Whereas the vps11-1 allele impairs all HOPS functions, the vps11-3 affects specifically the endosomal pathway (Peterson and Emr, 2001), and thus likely CORVET.
To monitor autophagy, we starved vps11 mutant cells expressing GFP-tagged Atg8 at permissive (24°C) or restrictive (37°C) temperature and monitored delivery of GFP-Atg8 to the vacuole lumen. Whereas vps11-3 cells were functional at both temperatures, vps11-1 cells accumulated GFP-Atg8 in large dots proximal to the vacuole if incubated at 37°C (Fig. 3 A). This indicates that HOPS is indeed required for this fusion as implied before (Wurmser et al., 2000; Gao et al., 2018).
Figure 3.
HOPS and Mon1-Ccz1 mediate autophagosome–vacuole fusion. (A) A HOPS-specific mutant blocks in autophagy defects. Cells were transformed with a centromeric plasmid expressing GFP-Atg8 under the control of the CUP1 promoter (Gao et al., 2018), grown at 24°C in synthetic dextran complete medium without uracil (SDC-URA), and then shifted to SD-N (N starved) for 2 h at 24°C or 37°C. Vacuoles were stained with FM4-64 and analyzed relative to GFP-Atg8 by fluorescence microscopy. Scale bar, 2 µm. (B) HOPS and Mon1-Ccz1 stimulates the fusion between autophagosomes and vacuoles. Purified autophagosomes and vacuoles were incubated as in Fig. 2 in the presence or absence of 50 nM HOPS and 300 nM Mon1-Ccz1. Scale bar, 2 µm. (C) Tethering is triggered by HOPS. Autophagosomes and vacuoles were incubated as in B, and Atg8-positive dots proximal to the vacuole were scored (n = 3). **, P < 0.01 (Student’s t test). (D) Titration of HOPS and Mon1-Ccz1 into the fusion assay. Fusion as observed in B was quantified. Error bars represent standard deviation of three independent experiments. **, P < 0.01 (Student’s t test).
To determine whether the HOPS complex promotes tethering and fusion, we added the purified complex to the autophagosome–vacuole fusion assay. In both the presence and absence of ATP, HOPS promoted strong association of GFP-positive autophagosomes with vacuoles (Fig. 3, B and C) and fusion (Fig. 3 D). The optimal concentration of 50 nM was in the range of what was observed for HOPS-stimulated vacuole–vacuole fusion (Lürick et al., 2017), whereas higher concentrations were inhibitory (Fig. 3 D). The Ypt7 GEF Mon1-Ccz1 also stimulated fusion, but to a lesser extent than HOPS (Fig. 3 D), which is likely limited by available Ypt7 and HOPS.
Ykt6 is present on autophagosomes
Autophagosome fusion with vacuole requires an autophagosomal SNARE protein. In higher eukaryotes, this SNARE is Syntaxin17 (Itakura et al., 2012; Takáts et al., 2013). As shown here and implicated by in vivo analyses previously (Darsow et al., 1997; Fischer von Mollard and Stevens, 1999; Ishihara et al., 2001), yeast autophagosome–vacuole fusion requires the SNAREs Vam3, Vam7, Vti1, and Ykt6, though it had not been clarified which SNARE is on autophagosomes. We excluded the Qa-SNARE Vam3, which is delivered to vacuoles via the AP-3 pathway (Darsow et al., 1998), as autophagosomes from vam3Δ cells are fusion competent. Vam7, a Qc-SNARE, was also unlikely because it lacks a transmembrane domain and specifically binds to phosphatidylinositol-3-phosphate on the limiting membrane of vacuoles via its Phox homology domain (Cheever et al., 2001). We considered it likely that yeast autophagosomes receive their SNAREs via ER or Golgi-derived vesicles and thus focused on the promiscuous Qb-SNARE Vti1 and the R-SNARE Ykt6, which have been shown to function in multiple pathways (von Mollard et al., 1997; Fischer von Mollard and Stevens, 1999; Dilcher et al., 2001). Both SNAREs have the necessary membrane anchor (i.e., Vti1 has a transmembrane domain, whereas Ykt6 is bound to membranes via C-terminal prenyl and palmitoyl anchors; von Mollard et al., 1997; Fukasawa et al., 2004; Veit, 2004).
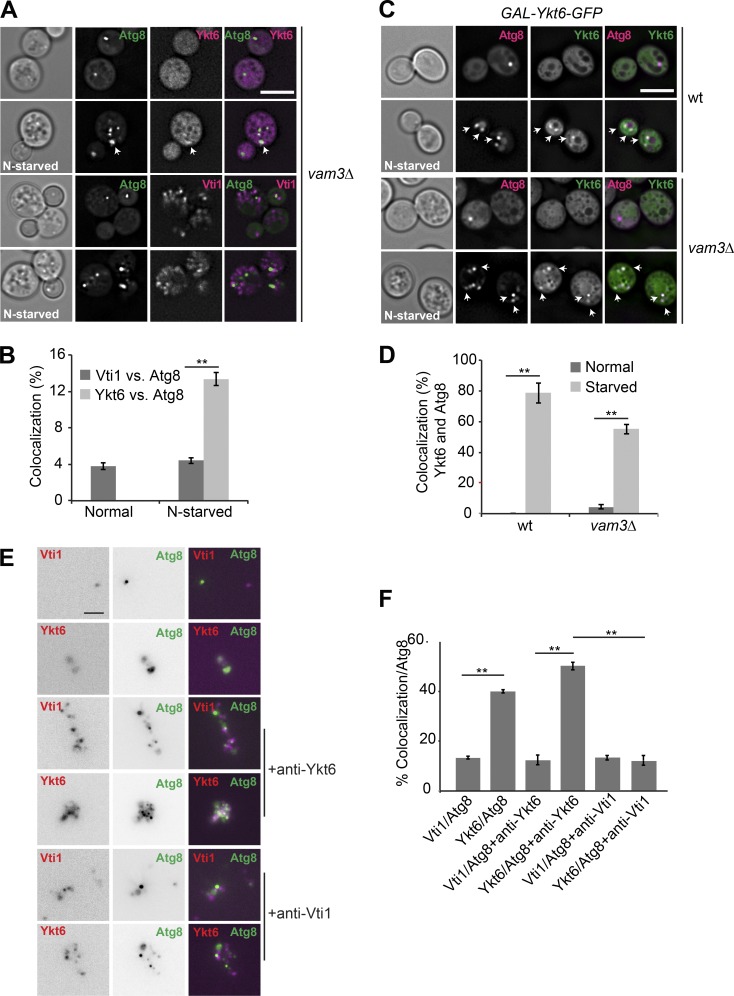
To determine which SNARE is present on autophagosomes, we colocalized mCherry-tagged Ykt6 and Vti1 with GFP-tagged Atg8 in the same vam3Δ cells in vivo. Ykt6 appeared completely cytosolic in nutrient-rich conditions (Fig. 4 A) but localized in weak, yet distinct, puncta upon nitrogen starvation. Most of these puncta were positive for GFP-Atg8 puncta. In contrast, we observed no colocalization between Vti1 and Atg8 puncta before and after starvation (Fig. 4, A and B). These data show that Ykt6, but not Vti1, is recruited to autophagic membranes. We reasoned that better localization could be achieved if Ykt6 is overproduced. We thus used a strain in which GFP-tagged Ykt6 was overproduced from a GAL1 promoter (Meiringer et al., 2008). We indeed observed significant colocalization between GFP-Ykt6 and mCherry-Atg8 puncta under starvation conditions in wild-type cells (Fig. 4, C and D). The same was observed in a strain lacking Vam3 (Fig. 4, C and D). Altogether, our results reveal that Ykt6 localizes to autophagosomes.
Figure 4.
Ykt6 is located on autophagosomes. (A) Localization of Atg8 relative to Ykt6 during normal growth and nitrogen starvation. vam3Δ cells expressing GFP-tagged Atg8 and either mCherry-tagged Ykt6 or Vti1 were grown in YPD or SD-N for 2 h and analyzed by fluorescence microscopy. Single planes of stacks are shown. Scale bar, 5 µm. (B) Percentage of Ykt6 or Vti1 puncta colocalizing with Atg8 under both conditions. The data were quantified from A. Error bars represent standard deviation of three independent experiments. **, P < 0.01 (Student’s t test). (C) Cells expressing GFP-tagged Ykt6 under the control of the GAL1 promoter were transformed with a centromeric plasmid expressing mCherry-Atg8 under the control of the TPI1 promoter. Cells were grown in SGC-LEU or SG-N for 2 h and analyzed by fluorescence microscopy, and are shown as individual slices. Scale bar, 5 µm. (D) Percentage of Ykt6 puncta colocalizing with Atg8 under both conditions. Data were quantified from C. Error bars represent standard deviation of three independent experiments. **, P < 0.01 (Student’s t test). (E) Ykt6 localizes with Atg8 on purified autophagosomes. The vam3Δ mutant expressing 3xFLAG-tagged Atg9, GFP-tagged Atg8, and mCherry-tagged Ykt6 or Vti1 were grown in YPD medium and then starved in SD-N medium for 3 h. Isolated autophagosomes were then incubated with or without an antibody against Ykt6 or Vti1. Scale bar, 2 µm. (F) Quantification of Fig. 4 E. Error bars represent standard deviation of three independent experiments. **, P < 0.01 (Student´s t test).
To confirm that Ykt6 indeed localizes to autophagosomes, we isolated these vesicles from our vam3Δ tester strain expressing either endogenously tagged mCherry-Vti1 or mCherry-Ykt6. When we colocalized these two SNAREs with GFP-Atg8 by fluorescence microscopy, only Ykt6 displayed significant overlap with Atg8, whereas Vti1 was only proximal to autophagosomes and likely on contaminating endosomal and/or Golgi membranes (Fig. 4, E and F). To further substantiate these observations, we added an antibody against either Ykt6 or Vti1 to the same autophagosome preparations. The addition of the Ykt6 antibodies caused a clustering of the GFP-Atg8–positive autophagosomal membranes, which was concomitantly accompanied by a clustering and colocalization of the mCherry-Ykt6 signal. For mCherry-Vti1, we observed just clustering without displaying significant overlap with GFP-Atg8 dots (Fig. 4, E and F). In contrast, the antibody to Vti1 did not lead to a colocalization of the signals, which indicates that Vti1 only clusters other membranes that copurified with autophagosomes. Our combined in vivo and in vitro data thus strongly support the notion that Ykt6 is present on autophagosomes.
Ykt6 is required for the fusion of autophagosomes with vacuoles
The localization of Ykt6 to autophagosomes prompted us to analyze whether Ykt6 is also required for autophagosome–vacuole fusion in vivo. ts mutants of Ykt6 have been characterized previously (Kweon et al., 2003). Among these, the ykt6-11 mutant impairs the transport of CPY from the late Golgi to the vacuole and possibly autophagy. To monitor autophagy, we expressed GFP-tagged Atg8 in ykt6-11 cells and then starved the cells at either the permissive (24°C) or restrictive (37°C) temperature. At permissive temperature, GFP-Atg8 was sorted efficiently to the vacuole lumen. However, at the restrictive temperature of 37°C, GFP-Atg8 accumulated in dots proximal to the vacuole, which sometimes showed a lumen (Fig. 5 A), similar to that observed for the HOPS vps11-1 mutant (Fig. 3 A). These data agree with our model that this mutant accumulates autophagosomes before fusion with the vacuole, indicating that Ykt6 functions in autophagosome–vacuole fusion.
Figure 5.
Ykt6 is required for fusion of autophagosomes with vacuoles. (A) ykt6 mutant cells have impaired autophagosome–vacuole fusion. ykt6-11 ts cells expressing GFP-tagged Atg8 were grown at 24°C in YPD and then shifted to SD-N for 2 h at 24°C or 37°C. Vacuoles were stained with FM4-64 and analyzed by fluorescence microscopy and are shown as individual slices. Scale bar, 5 µm. Scale bar for the inset, 0.5 µm. (B) Purified autophagosomes from ykt6 ts mutant cells cannot fuse with vacuoles in vitro. The vam3Δ or ykt6 ts strain expressing Atg9-3xFLAG and GFP-Atg8 were starved for 3 h before purifying autophagosomes. Vacuoles were isolated from pep4Δ cells expressing Vac8-3xmCherry and then incubated with autophagosomes at 26°C for 1 h with or without ATP. Scale bar, 2 µm. (C) Quantification of Fig. 5 B. Error bars represent standard deviation of three independent experiments. **, P < 0.01 (Student’s t test). (D) Palmitoylation is required for autophagy. Cells were transformed with a centromeric plasmid expressing GFP-Atg8 under the control of the CUP1 promoter (Gao et al., 2018), grown in SDC-URA, and then shifted to SD-N for 2 h. Data were analyzed by fluorescence microscopy, and a single plane from an image stack is shown. (E) Autophagy is functional in cells expressing Vti1 without a transmembrane domain (Vti1ΔTMD). Cells with the wild-type and Vti1ΔTMD, expressing mCherry-tagged Atg8, were grown in rich medium and then shifted to SD-N for 2 h. Top: Western blot of whole-cell lysates of the indicated strains using antibodies against Vti1 and Vac8. Bottom: Fluorescence microscopy images of the indicated cells. Scale bars for D and E, 5 µm. (F) Working model of autophagosome–vacuole (AV) fusion. Red lines indicate the involved R-SNARE Ykt6 and Qabc-SNAREs (Vam3, Vam7, and Vti1). Mon1-Ccz1 and Ypt7 are on the autophagosomes, and Ypt7 is shown to be associated with the HOPS complex.
To further confirm that Ykt6 is the autophagosomal SNARE responsible for fusion with vacuoles in yeast, we purified autophagosomes from either vam3Δ or ykt6 ts strains before performing the fusion assay. We previously observed that some ts mutants display their phenotype already without heat shock due to the purification procedure (Ungermann et al., 1999). In agreement, fusion was blocked when autophagosomes from ykt6 ts mutant cells were used (Fig. 5, B and C). Our data are thus consistent with our interpretation that Ykt6 is located on autophagosomes and is required for fusion with the vacuole.
On membranes, Ykt6 is both prenylated and palmitoylated (Fukasawa et al., 2004). We thus asked whether palmitoylation is important for autophagy, taking into consideration that Ykt6 is so far the only factor among the Atg proteins that may require this modification. Almost all palmitoylation in yeast is mediated by seven polytopic membrane proteins with a central DHHC consensus motif (Roth et al., 2006). To test the role of palmitoylation in autophagy, we monitored GFP-Atg8 sorting by autophagy to the vacuole lumen in several DHHC mutants (Fig. 5 D). Whereas single deletions of the ER-localized Erf2 had no effect on autophagy, deletions of five or six DHHC proteins (Roth et al., 2006) strongly impaired autophagy (Fig. 5 D). Even though indirect, these data agree with a requirement of palmitoylation of proteins (such as Ykt6) for autophagy.
Finally, we used an in vivo assay to exclude Vti1 as the autophagosomal SNARE. It has been shown that transmembrane domains of some SNAREs are dispensable, as long as the other SNAREs that they are in complex with are firmly anchored to membranes (Zick and Wickner, 2013; Chen et al., 2016). Cells expressing the essential Vti1 without a transmembrane domain (Vti1ΔTMD) are viable (Chen et al., 2016), and we found that they also sustain autophagy as monitored by starvation-induced migration of GFP-Atg8 to the vacuole lumen (Fig. 5 E). Altogether, these data reveal that Ykt6 is the autophagosomal SNARE in yeast.
Discussion
Autophagosomes need to acquire the machinery for their fusion with lysosomes as they form de novo. Several studies identified the involved machinery using in vivo experiment (Reggiori and Ungermann, 2017), yet only in vitro assays allow us to unravel the precise molecular function of all the participating factors, their topology, and their regulatory crosstalk. Here, we present the development of a novel in vitro assay to study the fusion between of autophagosomes and the lysosome-like vacuoles. The assay relies on a visual inspection of delivery of GFP-Atg8 into the vacuole lumen after fusion, which we detect and quantify by fluorescence microscopy. Our analysis reveals that autophagosome–vacuole fusion requires the Rab Ypt7 GTPase, its GEF Mon1-Ccz1, the HOPS tethering complex, and the SNAREs Vam3, Vam7, Vti1, and Ykt6, in line with previous in vivo analyses (Darsow et al., 1997; Wurmser et al., 2000; Dilcher et al., 2001; Ishihara et al., 2001; Peterson and Emr, 2001; Gao et al., 2018). Visual inspection of the fusion reaction also allowed us to distinguish tethering of autophagosomes to vacuoles, which depends on HOPS (Fig. 3), and thus Ypt7 and its GEF, Mon1-Ccz1 (Nordmann et al., 2010; Bröcker et al., 2012; Langemeyer et al., 2018), and their final fusion, which also requires SNAREs. By combining biochemical and genetic analyses, we demonstrate that Ykt6 is the autophagosomal SNARE. These findings nicely agree with the observations of Bas et al. in this issue on the mechanism of autophagosome–vacuole fusion.
Our observations agree with our working model on autophagosome–vacuole fusion (Fig. 5 F). We recently showed that the Mon1-Ccz1 GEF requires Atg8 for its recruitment onto autophagosomes (Gao et al., 2018), in line with observations in Drosophila melanogaster (Hegedüs et al., 2016). Once localized, Mon1-Ccz1 recruits Ypt7 (or Rab7 in D. melanogaster) to autophagosomes. This overall principle of autophagosome–vacuole tethering is thus very similar to the one occurring between late endosomes and vacuoles. In both cases, Ypt7 is loaded on the incoming organelle and is recognized by the vacuolar HOPS complex (Bröcker et al., 2012; Lürick et al., 2017).
Our in vitro and in vivo data show that Ykt6 is the autophagosomal SNARE. Unlike many SNAREs, Ykt6 does not have a transmembrane domain, but does contain C-terminal prenyl and palmitoyl anchors. It was initially thought that Ykt6 would not be sufficient to promote complete fusion, as a lipid anchor spans just one leaflet of a membrane (McNew et al., 2000). However, even fusion of transmembrane-anchored SNAREs fails if their concentration on membranes is too low, suggesting the need of additional machinery (Zick and Wickner, 2016). Reconstitution of vacuole–vacuole fusion and fusion of synaptic vesicles with the plasma membrane unraveled that SNAREs at their physiological concentrations function together with large tethering complexes and Sec1/Munc18 proteins (Zick and Wickner, 2016; Lai et al., 2017; Wickner and Rizo, 2017). These tethering complexes also provide the essential volume to trigger the transition from hemifusion to fusion and thus have to be considered as an essential component of the fusion machinery, which cooperates with SNAREs (D’Agostino et al., 2017). Lipid-anchored SNAREs are therefore as functional as transmembrane SNAREs (Rohde et al., 2003; Xu et al., 2011). In agreement with this, we observed defective autophagy if palmitoylation is compromised (Fig. 5 D), which could reflect inefficient Ykt6 palmitoylation.
Ykt6 is a highly conserved eukaryotic R-SNARE and functions in SNARE-mediated fusion at the ER, Golgi, endosomes, and vacuole (McNew et al., 1997; Ungermann et al., 1999; Tsui and Banfield, 2000; Dilcher et al., 2001; Hasegawa et al., 2003; Dietrich et al., 2005; Meiringer et al., 2011). As autophagosomes form from multiple membrane sources, Ykt6 likely arrives on autophagosomes via ER- or Golgi-derived vesicles, but a different origin cannot be excluded. Ykt6 requires palmitoylation for function, which likely occurs at the ER or Golgi via the redundantly acting DHHC acyltransferases (Mitchell et al., 2006; Linder and Jennings, 2013). Considering that Ykt6 transport requires an intact endomembrane system, any fusion defect at the Golgi or ER could potentially cause an impairment of autophagy. The same will apply to the only conserved transmembrane protein of autophagy, Atg9. Interestingly, metazoan cells use a different SNARE, Syntaxin17, which is recruited onto autophagosomes after their completion and likely requires a specific machinery for its targeting to autophagosomes (Itakura et al., 2012; Takáts et al., 2014; Arasaki et al., 2015; Kumar et al., 2018). However, two recent studies, which were published while this study was under review, also demonstrate a role of YKT6 in autophagosome–lysosome fusion in metazoan cells (Matsui et al., 2018; Takáts et al., 2018). Both studies find defective fusion in the absence of YKT6 and detect YKT6 on autophagosomes in either D. melanogaster or mammalian cells yet differ in their interpretation of the protein’s function in fusion. While Takáts et al. (2018) suggest a regulatory role of YKT6 in Syntaxin17-dependent fusion, Matsui et al. (2018) have evidence for a direct role of YKT6 as an alternative SNARE in fusion. Our results also suggest that Ykt6 is the SNARE of autophagosomes in yeast, which could be the more ancient role of Ykt6 in autophagy. This does not exclude an additional function of YKT6 as a supporting SNARE in Syntaxin17-mediated fusion (Takáts et al., 2018), which deserves further exploration.
In vitro fusion assays have been of fundamental importance to dissect the involved machinery in homotypic vacuole fusion (Wickner and Rizo, 2017) and recently in endosome–vacuole fusion (Karim and Brett, 2018; Karim et al., 2018). While enzymatic assays facilitate manipulation, visual assays allow the direct inspection of tethering and fusion. Our work, together with the study of Bas et al. (2018), provides a novel assay to address fundamental questions regarding the mechanism and regulation of autophagosome–vacuole fusion and may even be extended to the analysis of autophagosome completion before fusion.
Materials and methods
Yeast strains, molecular biology, and antibodies
Strains used in this study are listed in Table 1. Deletions and tagging of genes were done by PCR-based homologous recombination with appropriate primers (Puig et al., 1998; Janke et al., 2004). pRS416-pCuGFP-ATG8 and pRS415-pmCherry-V5-ATG8 were generated in the Reggiori laboratory.
Table 1. Strains used in this study.
| Strains | Genotype | Source |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Euroscarf Library |
| SEY6210 | MATα leu2-3 leu2-112 ura3-52 his3-Δ200 trp1-Δ101 lys2-801 suc2-Δ9 GAL | F. Reggiori |
| BJ3505 | MATa pep4Δ::HIS3 prb1-Δ1.6R lys2-208 trp1Δ101 ura3-52 gal2 | B. Jones, Carnegie Mellon University, Pittsburgh, PA |
| CUY10047 | BY4741; ATG9-3xFLAG::hphNT1 | This study |
| CUY10048 | BY4741; vam3Δ::kanMX ATG9-3xFLAG::hphNT1 | This study |
| CUY10049 | BY4741; ypt7Δ::kanMX ATG9-3xFLAG::hphNT1 | This study |
| CUY10050 | BY4741; ATG9-3xFLAG::hphNT1 pRS416-pCuGFP-ATG8::URA | This study |
| CUY10051 | BY4741; vam3Δ::kanMX ATG9-3xFLAG::hphNT1 pRS416-pCuGFP-ATG8::URA | This study |
| CUY10052 | BY4741; ypt7Δ::kanMX ATG9-3xFLAG::hphNT1 pRS416-pCuGFP-ATG8::URA | This study |
| CUY10171 | BY4741; vam3Δ::kanMX ATG9-3xFLAG::hphNT1 GFP-ATG8::natNT2 | This study |
| CUY10447 | BJ3505; VAC8-3xmCherry::natNT2 | This study |
| CUY10922 | BJ3505; VAC8-3xmCherry::natNT2 atg15Δ::TRP | This study |
| CUY10923 | BY4741; vam3Δ::kanMX ATG9-3xFLAG::hphNT1 GFP-ATG8::natNT2 atg15Δ::HIS | This study |
| CUY13332 | SEY6210; mel-ykt6-12 ATG9-3xFLAG::hphNT1 | This study |
| CUY11466 | SEY6210; mel-ykt6-11 pRS416-pCuGFP-ATG8::URA | This study |
| CUY11467 | BY4741; ykt6Δ::MET pRS403-GAL1pr-YKT6-GFP::HIS vam3Δ::natNT2 | This study |
| CUY11468 | BY4741; ykt6Δ::MET pRS403-GAL1pr-YKT6-GFP::HIS vam3Δ::natNT2 pRS415-pmCherry-V5-ATG8::LEU | This study |
| CUY11469 | BY4741; ykt6Δ::MET pRS403-GAL1pr-YKT6-GFP::HIS pRS415-pmCherry-V5-ATG8::LEU | This study |
| CUY11470 | MATα his3Δ leu2Δ ura3Δ pRS416-pCuGFP-ATG8::URA | This study |
| CUY11471 | MATα his3Δ leu2Δ ura3Δ erf2Δ pRS416-pCuGFP-ATG8::URA | This study |
| CUY11472 | MATα his3Δ leu2Δ ura3Δ akr1Δ ark2Δ pfa5Δ::G418R YCK2(CCIIS)::Lys2 pfa3Δ::LEU pfa4Δ pRS416-pCuGFP-ATG8::URA | This study |
| CUY11473 | MATα his3Δ leu2Δ ura3Δ akr1Δ ark2Δ pfa5Δ::G418R YCK2(CCIIS)::Lys2 pfa3Δ pfa4Δ GAL-ERF2 pRS416-pCuGFP-ATG8::URA | This study |
Antibodies against Vam7, Vti1, Ykt6, Vam3, Atg8, and Ape1 have been described (Ungermann and Wickner, 1998; Ungermann et al., 1998, 1999; Mari and Reggiori, 2010). Antibodies against Ypt7 and Sec61 were gifts from William Wickner (Dartmouth Medical School, Hanover, NH), against Mge1 from Walter Neupert (University of Munich, Munich, Germany), against Pep12 from Scott Emr (Cornell University, Ithaca, NY), and against Sec7 from Chris Fromme (Cornell University, Ithaca, NY). Antibodies against GFP were obtained from Roche.
Purification of proteins
HOPS and Mon1-Ccz1 were purified from an overexpression strain via the tandem affinity tag as described before (Lürick et al., 2017; Gao et al., 2018). Yeast cells were grown in YPG (1% yeast extract, 1% peptone, and 2% galactose) at 30°C to an OD600 of 6, harvested, and lysed in lysis buffer (300 mM NaCl, 50 mM Hepes-NaOH, pH 7.4, 1.5 mM MgCl2, 1×FY protease inhibitor mix [Serva], 0.5 mM PMSF, and 1 mM DTT). After centrifugation of lysates for 1 h at 100,000 g, the supernatant was incubated with IgG Sepharose beads (GE Healthcare) for 2 h at 4°C with in lysis buffer containing 10% glycerol. Collected beads were washed with ice-cold 15 ml lysis buffer containing 10% glycerol and 0.5 mM DTT. Bound proteins were eluted by tobacco etch virus protease cleavage overnight at 4°C (Mon1-Ccz1) or 1 h at 16°C (HOPS). Purified proteins were analyzed on SDS-PAGE. Purification of His-Sec18 was done as described previously (Lürick et al., 2017).
Light microscopy and image analysis
Yeast cells were cultured in YPD (1% yeast extract, 2% peptone, and 2% glucose) medium to log phase before to be transferred into synthetic minimal medium lacking nitrogen (SD-N; 0.17% yeast nitrogen base without amino acids and ammonium sulfate and 2% glucose) for the indicated times to induce autophagy. Cells or purified organelles were imaged on a Deltavision Elite imaging system based on an inverted microscopy, equipped with 100× NA 1.49 and 60× NA 1.40 objectives, a sCMOS camera (PCO), an Insight SSI (TM) illumination system, and SoftWoRx software (Applied Precision). Stacks of six or eight images with 0.2-µm spacing were collected. For purified autophagosmes and vacuoles, no Z stacks were recorded to avoid the bleaching of the fluorescence signal.
Isolation of autophagosomes
Yeast cells (i.e., the vam3Δ strain expressing GFP-Atg8 and Atg9-3xFLAG) were first grown to an OD600 of ∼1.0 in 1 liter of YPD medium and then nitrogen starved in SD-N medium for 3 h to induce autophagy. Cells were harvested by centrifugation. The resulting pellets were resuspended in the 0.1 M Tris/HCl, pH 9.4, buffer containing 10 mM DTT and incubated for 15 min at 30°C. Cells were centrifuged again and incubated with the spheroplasting buffer (0.16× SD-N, 0.6 M sorbitol, and 50 mM KPi, pH 7.4) and 0.3 mg/ml lyticase for 30 min at 30°C. Spheroplasts were collected by centrifugation at 1,000 g for 3 min and resuspended in lysis buffer (0.2 M sorbitol, 50 mM KOAc, 2 mM EDTA, and 20 mM Hepes/KOH, pH 6.8) containing a protease inhibitor cocktail (0.1 mg/ml of leupeptin, 1 mM o-phenanthroline, 0.5 mg/ml of pepstatin A, and 0.1 mM Pefabloc), 1 mM PMSF, and 1 mM DTT. The resuspended cells were incubated with 0.04 mg/ml DEAE Dextran (Sigma-Aldrich) on ice for 5 min and then heat shocked for 2 min at 30°C. These suspensions were then centrifuged at 400 g for 10 min at 4°C. After centrifugation, the supernatant was centrifuged again at 15,000 g for 15 min at 4°C, and then the pellets were resuspended in 1 ml lysis buffer. These suspensions were layered onto discontinuous iodixanol gradients (1.5 ml of 20%, 6 ml of 10%, and 4 ml of 5%) in an SW40 tube (Seton). Loaded gradients were spun at 100,000 g for 60 min at 4°C. Fractions at the 10–20% interface (1 ml) were collected and incubated with anti-FLAG beads (Sigma-Aldrich) overnight. Beads were washed with 2 ml ice-cold lysis buffer. Bound autophagosomes were eluted with 150 µl lysis buffer containing 0.25 µg/µl FLAG-peptide, and the suspension containing the eluate was collected by centrifugation at 30 g for 2 min at 4°C. The concentration of autophagosomes was determined by the method of Bradford with RotiQuant-solution (Roth) using bovine serum albumin as the reference standard.
Proteinase K protection assay
5 mg of purified autophagosomes were first treated with and without 1% Triton X-100 and then immediately incubated with 0.1 mg/ml proteinase K on ice for 20 min (total volume, 30 µl). The reactions were terminated by addition of 2 mM PMSF. Samples were dissolved in SDS-PAGE sample buffer, separated on SDS-PAGE gels, and subjected to immunoblot analysis.
Isolation of vacuoles
Yeast cells (i.e., the pep4Δ strain expressing Vac8-3xmCherry) were grown to an OD600 of ∼0.9 to 1.0 in 1 liter of YPD medium. After harvesting cells by centrifugation (5 min, 2,000 g), pellets were resuspended in buffer containing 0.1 M Tris/HCl, pH 9.4, and 10 mM DTT, and incubated for 10 min at 30°C. Cells were centrifuged as before and resuspended in the 25-ml spheroplasting buffer containing 0.3 mg/ml lyticase and then incubated for 25 min at 30°C. Spheroplasts were collected by centrifugation at 1,000 g for 3 min at 4°C and resuspended in 2.5 ml 15% Ficoll (15% wt/vol Ficoll in 0.2 M sorbitol and 10 mM Pipes/KOH, pH 6.8 [GE Healthcare]). After slow resuspension, cells were incubated with 0.02 mg/ml DEAE Dextran (Sigma-Aldrich) on ice for 5 min and then heat shocked for 2 min at 30°C. These suspensions were transferred to SW40 tube (Seton), and sequential layers of Ficoll solution in the aforementioned buffer (0.2 M sorbitol and 10 mM Pipes/KOH, pH 6.8; 3 ml of 8%, 3 ml of 4%, fill with 0%) were slowly added on top of the suspensions. Gradients were centrifuged at 100,000 g for 90 min at 4°C in an SW40 rotor. Vacuoles were collected at the 0–4% interface. The concentration of vacuoles was measured in the same way as described for purified autophagosomes.
Autophagosome–vacuole fusion assay
Yeast cells used for vacuole preparation (from the pep4Δ strain carrying Vac8-3xmCherry) were first grown to OD600 = ∼1.0 in 1 liter YPD medium before being nitrogen starved in SD-N medium for 3 h to induce autophagy. Vacuoles were purified via Ficoll gradient centrifugation as previously described (Haas, 1995). Vacuoles and autophagosomes were then diluted to 0.3 mg/ml and 1.5 mg/ml, respectively, with 0% Ficoll buffer (10 mM Pipes/KOH, pH 6.8, 200 mM sorbitol, and 0.1× protease inhibitor cocktail). A standard fusion reaction contained 3 µg of vacuoles and 15 µg of autophagosomes. Reactions were incubated in fusion buffer (125 mM KCl, 5 mM MgCl2, 20 mM sorbitol, and 1 mM Pipes/KOH, pH 6.8) containing 10 µM CoA, 0.01 µg of 6xHis-Sec18, and an ATP-regenerating system (5 mM ATP, 1 mg/ml creatine kinase, 400 mM creatine phosphatase, 10 mM Pipes/KOH, pH 6.8, and 0.2 M sorbitol) at 26°C for 30 min. Fusion efficiency was finally assessed by fluorescence microscopy.
Acknowledgments
We thank Johannes Numrich and Mareike Nolte for preliminary experiments and support.
F. Reggiori was supported by ALW Open Program (822.02.014), Deutsche Forschungsgemeinschaft/Netherlands Organisation for Scientific Research cooperation (DN82-303), Swiss National Science Foundation Sinergia (CRSII3_154421), Marie Skłodowska-Curie Cofund (713660), and ZonMW Vici (016.130.606) grants. C. Ungermann was supported by a Deutsche Forschungsgemeinschaft grant (UN111/7-3) and the Sonderforschungsbereich 944 (Project P11).
The authors declare no competing financial interests.
Author contributions: J. Gao, F. Reggiori, and C. Ungermann conceived and designed experiments. J. Gao performed all experiments. All authors analyzed the results. J. Gao, F. Reggiori, and C. Ungermann wrote the manuscript.
References
- Arasaki K., Shimizu H., Mogari H., Nishida N., Hirota N., Furuno A., Kudo Y., Baba M., Baba N., Cheng J., et al. . 2015. A role for the ancient SNARE syntaxin 17 in regulating mitochondrial division. Dev. Cell. 32:304–317. 10.1016/j.devcel.2014.12.011 [DOI] [PubMed] [Google Scholar]
- Baba M., Takeshige K., Baba N., and Ohsumi Y.. 1994. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J. Cell Biol. 124:903–913. 10.1083/jcb.124.6.903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker R.W., Jeffrey P.D., Zick M., Phillips B.P., Wickner W.T., and Hughson F.M.. 2015. A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science. 349:1111–1114. 10.1126/science.aac7906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderhaar H.J.K., and Ungermann C.. 2013. CORVET and HOPS tethering complexes - coordinators of endosome and lysosome fusion. J. Cell Sci. 126:1307–1316. 10.1242/jcs.107805 [DOI] [PubMed] [Google Scholar]
- Barth H., Meiling-Wesse K., Epple U.D., and Thumm M.. 2001. Autophagy and the cytoplasm to vacuole targeting pathway both require Aut10p. FEBS Lett. 508:23–28. 10.1016/S0014-5793(01)03016-2 [DOI] [PubMed] [Google Scholar]
- Bas L., Papinski D., Licheva M., Torggler R., Rohringer S., Schuschnig M., and Kraft C.. 2018. Reconstitution reveals Ykt6 as the autophagosomal SNARE in autophagosome–vacuole fusion. J. Cell Biol. 10.1083/jcb.201804028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett C.L., Plemel R.L., Lobingier B.T., Vignali M., Fields S., and Merz A.J.. 2008. Efficient termination of vacuolar Rab GTPase signaling requires coordinated action by a GAP and a protein kinase. J. Cell Biol. 182:1141–1151. 10.1083/jcb.200801001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröcker C., Kuhlee A., Gatsogiannis C., Balderhaar H.J., Hönscher C., Engelbrecht-Vandré S., Ungermann C., and Raunser S.. 2012. Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proc. Natl. Acad. Sci. USA. 109:1991–1996. 10.1073/pnas.1117797109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebollero E., van der Vaart A., Zhao M., Rieter E., Klionsky D.J., Helms J.B., and Reggiori F.. 2012. Phosphatidylinositol-3-phosphate clearance plays a key role in autophagosome completion. Curr. Biol. 22:1545–1553. 10.1016/j.cub.2012.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheever M.L., Sato T.K., de Beer T., Kutateladze T.G., Emr S.D., and Overduin M.. 2001. Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat. Cell Biol. 3:613–618. 10.1038/35083000 [DOI] [PubMed] [Google Scholar]
- Chen L., Lau M.S.Y., and Banfield D.K.. 2016. Multiple ER-Golgi SNARE transmembrane domains are dispensable for trafficking but required for SNARE recycling. Mol. Biol. Cell. 27:2633–2641. 10.1091/mbc.e16-05-0277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agostino M., Risselada H.J., Lürick A., Ungermann C., and Mayer A.. 2017. A tethering complex drives the terminal stage of SNARE-dependent membrane fusion. Nature. 551:634–638. [DOI] [PubMed] [Google Scholar]
- Darsow T., Rieder S.E., and Emr S.D.. 1997. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J. Cell Biol. 138:517–529. 10.1083/jcb.138.3.517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsow T., Burd C.G., and Emr S.D.. 1998. Acidic di-leucine motif essential for AP-3-dependent sorting and restriction of the functional specificity of the Vam3p vacuolar t-SNARE. J. Cell Biol. 142:913–922. 10.1083/jcb.142.4.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich L.E.P., LaGrassa T.J., Rohde J., Cristodero M., Meiringer C.T.A., and Ungermann C.. 2005. ATP-independent control of Vac8 palmitoylation by a SNARE subcomplex on yeast vacuoles. J. Biol. Chem. 280:15348–15355. 10.1074/jbc.M410582200 [DOI] [PubMed] [Google Scholar]
- Dilcher M., Köhler B., and von Mollard G.F.. 2001. Genetic interactions with the yeast Q-SNARE VTI1 reveal novel functions for the R-SNARE YKT6. J. Biol. Chem. 276:34537–34544. 10.1074/jbc.M101551200 [DOI] [PubMed] [Google Scholar]
- Fischer von Mollard G., and Stevens T.H.. 1999. The Saccharomyces cerevisiae v-SNARE Vti1p is required for multiple membrane transport pathways to the vacuole. Mol. Biol. Cell. 10:1719–1732. 10.1091/mbc.10.6.1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa M., Varlamov O., Eng W.S., Söllner T.H., and Rothman J.E.. 2004. Localization and activity of the SNARE Ykt6 determined by its regulatory domain and palmitoylation. Proc. Natl. Acad. Sci. USA. 101:4815–4820. 10.1073/pnas.0401183101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Langemeyer L., Kümmel D., Reggiori F., and Ungermann C.. 2018. Molecular mechanism to target the endosomal Mon1-Ccz1 GEF complex to the pre-autophagosomal structure. eLife. 7:765 10.7554/eLife.31145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L., Baskaran S., Schekman R., and Hurley J.H.. 2014. The protein-vesicle network of autophagy. Curr. Opin. Cell Biol. 29:18–24. 10.1016/j.ceb.2014.02.005 [DOI] [PubMed] [Google Scholar]
- Goody R.S., Müller M.P., and Wu Y.-W.. 2017. Mechanisms of action of Rab proteins, key regulators of intracellular vesicular transport. Biol. Chem. 398:565–575. 10.1515/hsz-2016-0274 [DOI] [PubMed] [Google Scholar]
- Guan J., Stromhaug P.E., George M.D., Habibzadegah-Tari P., Bevan A., Dunn W.A. Jr., and Klionsky D.J.. 2001. Cvt18/Gsa12 is required for cytoplasm-to-vacuole transport, pexophagy, and autophagy in Saccharomyces cerevisiae and Pichia pastoris. Mol. Biol. Cell. 12:3821–3838. 10.1091/mbc.12.12.3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A. 1995. A quantitative assay to measure homotypic vacuole fusion in vitro. Methods Cell Sci. 17:283–294. 10.1007/BF00986234 [DOI] [Google Scholar]
- Haas A., Conradt B., and Wickner W.. 1994. G-protein ligands inhibit in vitro reactions of vacuole inheritance. J. Cell Biol. 126:87–97. 10.1083/jcb.126.1.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa H., Zinsser S., Rhee Y., Vik-Mo E.O., Davanger S., and Hay J.C.. 2003. Mammalian ykt6 is a neuronal SNARE targeted to a specialized compartment by its profilin-like amino terminal domain. Mol. Biol. Cell. 14:698–720. 10.1091/mbc.e02-09-0556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedüs K., Takats S., Boda A., Jipa A., Nagy P., Varga K., Kovacs A.L., and Juhasz G.. 2016. The Ccz1-Mon1-Rab7 module and Rab5 control distinct steps of autophagy. Mol. Biol. Cell. 27:3132–3142. 10.1091/mbc.e16-03-0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey C.M., and Wickner W.. 2010. HOPS initiates vacuole docking by tethering membranes before trans-SNARE complex assembly. Mol. Biol. Cell. 21:2297–2305. 10.1091/mbc.e10-01-0044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho R., and Stroupe C.. 2015. The HOPS/class C Vps complex tethers membranes by binding to one Rab GTPase in each apposed membrane. Mol. Biol. Cell. 26:2655–2663. 10.1091/mbc.e14-04-0922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N., Hamasaki M., Yokota S., Suzuki K., Kamada Y., Kihara A., Yoshimori T., Noda T., and Ohsumi Y.. 2001. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell. 12:3690–3702. 10.1091/mbc.12.11.3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi-Itakura C., and Mizushima N.. 2012. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 151:1256–1269. 10.1016/j.cell.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., and Knop M.. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962. 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Karim M.A., and Brett C.L.. 2018. The Na+(K+)/H+ exchanger Nhx1 controls multivesicular body-vacuolar lysosome fusion. Mol. Biol. Cell. 29:317–325. 10.1091/mbc.e17-08-0496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim M.A., Samyn D.R., Mattie S., and Brett C.L.. 2018. Distinct features of multivesicular body-lysosome fusion revealed by a new cell-free content-mixing assay. Traffic. 19:138–149. 10.1111/tra.12543 [DOI] [PubMed] [Google Scholar]
- Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., and Ohsumi Y.. 1999. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147:435–446. 10.1083/jcb.147.2.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Abdelmohsen K., Abe A., Abedin M.J., Abeliovich H., Acevedo-Arozena A., Adachi H., Adams C.M., Adams P.D., Adeli K., et al. . 2016. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 12:1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Jain A., Farzam F., Jia J., Gu Y., Choi S.W., Mudd M.H., Claude-Taupin A., Wester M.J., Lidke K.A., et al. . 2018. Mechanism of Stx17 recruitment to autophagosomes via IRGM and mammalian Atg8 proteins. J. Cell Biol. 217:997–1013. 10.1083/jcb.201708039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweon Y., Rothe A., Conibear E., and Stevens T.H.. 2003. Ykt6p is a multifunctional yeast R-SNARE that is required for multiple membrane transport pathways to the vacuole. Mol. Biol. Cell. 14:1868–1881. 10.1091/mbc.e02-10-0687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y., Choi U.B., Leitz J., Rhee H.J., Lee C., Altas B., Zhao M., Pfuetzner R.A., Wang A.L., Brose N., et al. . 2017. Molecular Mechanisms of Synaptic Vesicle Priming by Munc13 and Munc18. Neuron. 95:591–607.e10. 10.1016/j.neuron.2017.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C.A., Dooley H.C., and Tooze S.A.. 2013a Endocytosis and autophagy: Shared machinery for degradation. BioEssays. 35:34–45. 10.1002/bies.201200130 [DOI] [PubMed] [Google Scholar]
- Lamb C.A., Yoshimori T., and Tooze S.A.. 2013b The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 14:759–774. 10.1038/nrm3696 [DOI] [PubMed] [Google Scholar]
- Langemeyer L., Perz A., Kümmel D., and Ungermann C.. 2018. A guanine nucleotide exchange factor (GEF) limits Rab GTPase-driven membrane fusion. J. Biol. Chem. 293:731–739. 10.1074/jbc.M117.812941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder M.E., and Jennings B.C.. 2013. Mechanism and function of DHHC S-acyltransferases. Biochem. Soc. Trans. 41:29–34. 10.1042/BST20120328 [DOI] [PubMed] [Google Scholar]
- Lobingier B.T., and Merz A.J.. 2012. Sec1/Munc18 protein Vps33 binds to SNARE domains and the quaternary SNARE complex. Mol. Biol. Cell. 23:4611–4622. 10.1091/mbc.e12-05-0343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lürick A., Gao J., Kuhlee A., Yavavli E., Langemeyer L., Perz A., Raunser S., and Ungermann C.. 2017. Multivalent Rab interactions determine tether-mediated membrane fusion. Mol. Biol. Cell. 28:322–332. 10.1091/mbc.e16-11-0764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari M., and Reggiori F.. 2010. Atg9 reservoirs, a new organelle of the yeast endomembrane system? Autophagy. 6:1221–1223. 10.4161/auto.6.8.13792 [DOI] [PubMed] [Google Scholar]
- Matsui T., Jiang P., Nakano S., Sakamaki Y., Yamamoto H., and Mizushima N.. 2018. Autophagosomal YKT6 is required for fusion with lysosomes independently of syntaxin 17. J. Cell Biol. 217:2633–2645. 10.1083/jcb.201712058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNew J.A., Sogaard M., Lampen N.M., Machida S., Ye R.R., Lacomis L., Tempst P., Rothman J.E., and Söllner T.H.. 1997. Ykt6p, a prenylated SNARE essential for endoplasmic reticulum-Golgi transport. J. Biol. Chem. 272:17776–17783. 10.1074/jbc.272.28.17776 [DOI] [PubMed] [Google Scholar]
- McNew J.A., Parlati F., Fukuda R., Johnston R.J., Paz K., Paumet F., Söllner T.H., and Rothman J.E.. 2000. Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature. 407:153–159. 10.1038/35025000 [DOI] [PubMed] [Google Scholar]
- Meiringer C.T., Auffarth K., Hou H., and Ungermann C.. 2008. Depalmitoylation of Ykt6 prevents its entry into the multivesicular body pathway. Traffic. 9:1510–1521. 10.1111/j.1600-0854.2008.00778.x [DOI] [PubMed] [Google Scholar]
- Meiringer C.T.A., Rethmeier R., Auffarth K., Wilson J., Perz A., Barlowe C., Schmitt H.D., and Ungermann C.. 2011. The Dsl1 protein tethering complex is a resident endoplasmic reticulum complex, which interacts with five soluble NSF (N-ethylmaleimide-sensitive factor) attachment protein receptors (SNAREs): implications for fusion and fusion regulation. J. Biol. Chem. 286:25039–25046. 10.1074/jbc.M110.215327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D.A., Vasudevan A., Linder M.E., and Deschenes R.J.. 2006. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J. Lipid Res. 47:1118–1127. 10.1194/jlr.R600007-JLR200 [DOI] [PubMed] [Google Scholar]
- Nair U., Jotwani A., Geng J., Gammoh N., Richerson D., Yen W.-L., Griffith J., Nag S., Wang K., Moss T., et al. . 2011. SNARE proteins are required for macroautophagy. Cell. 146:290–302. 10.1016/j.cell.2011.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T., Matsuura A., Wada Y., and Ohsumi Y.. 1995. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 210:126–132. 10.1006/bbrc.1995.1636 [DOI] [PubMed] [Google Scholar]
- Nordmann M., Cabrera M., Perz A., Bröcker C., Ostrowicz C., Engelbrecht-Vandré S., and Ungermann C.. 2010. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr. Biol. 20:1654–1659. 10.1016/j.cub.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Ohashi Y., and Munro S.. 2010. Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol. Biol. Cell. 21:3998–4008. 10.1091/mbc.e10-05-0457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr A., Song H., Rusin S.F., Kettenbach A.N., and Wickner W.. 2017. HOPS catalyzes the interdependent assembly of each vacuolar SNARE into a SNARE complex. Mol. Biol. Cell. 28:975–983. 10.1091/mbc.e16-10-0743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M.R., and Emr S.D.. 2001. The class C Vps complex functions at multiple stages of the vacuolar transport pathway. Traffic. 2:476–486. 10.1034/j.1600-0854.2001.20705.x [DOI] [PubMed] [Google Scholar]
- Puig O., Rutz B., Luukkonen B.G., Kandels-Lewis S., Bragado-Nilsson E., and Séraphin B.. 1998. New constructs and strategies for efficient PCR-based gene manipulations in yeast. Yeast. 14:1139–1146. [DOI] [PubMed] [Google Scholar]
- Reggiori F., and Ungermann C.. 2017. Autophagosome Maturation and Fusion. J. Mol. Biol. 429:486–496. 10.1016/j.jmb.2017.01.002 [DOI] [PubMed] [Google Scholar]
- Rieter E., Vinke F., Bakula D., Cebollero E., Ungermann C., Proikas-Cezanne T., and Reggiori F.. 2013. Atg18 function in autophagy is regulated by specific sites within its β-propeller. J. Cell Sci. 126:593–604. 10.1242/jcs.115725 [DOI] [PubMed] [Google Scholar]
- Rohde J., Dietrich L., Langosch D., and Ungermann C.. 2003. The transmembrane domain of Vam3 affects the composition of cis- and trans-SNARE complexes to promote homotypic vacuole fusion. J. Biol. Chem. 278:1656–1662. 10.1074/jbc.M209522200 [DOI] [PubMed] [Google Scholar]
- Roth A.F., Wan J., Bailey A.O., Sun B., Kuchar J.A., Green W.N., Phinney B.S., Yates J.R. III, and Davis N.G.. 2006. Global analysis of protein palmitoylation in yeast. Cell. 125:1003–1013. 10.1016/j.cell.2006.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seals D.F., Eitzen G., Margolis N., Wickner W.T., and Price A.. 2000. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc. Natl. Acad. Sci. USA. 97:9402–9407. 10.1073/pnas.97.17.9402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibutani S.T., and Yoshimori T.. 2014. A current perspective of autophagosome biogenesis. Cell Res. 24:58–68. 10.1038/cr.2013.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takáts S., Nagy P., Varga Á., Pircs K., Kárpáti M., Varga K., Kovács A.L., Hegedűs K., and Juhász G.. 2013. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J. Cell Biol. 201:531–539. 10.1083/jcb.201211160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takáts S., Pircs K., Nagy P., Varga Á., Kárpáti M., Hegedűs K., Kramer H., Kovács A.L., Sass M., and Juhász G.. 2014. Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila. Mol. Biol. Cell. 25:1338–1354. 10.1091/mbc.e13-08-0449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takáts S., Glatz G., Szenci G., Boda A., Horváth G.V., Hegedűs K., Kovács A.L., and Juhász G.. 2018. Non-canonical role of the SNARE protein Ykt6 in autophagosome-lysosome fusion. PLoS Genet. 14:e1007359 10.1371/journal.pgen.1007359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torggler R., Papinski D., and Kraft C.. 2017. Assays to Monitor Autophagy in Saccharomyces cerevisiae. Cells. 6:23 10.3390/cells6030023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui M.M., and Banfield D.K.. 2000. Yeast Golgi SNARE interactions are promiscuous. J. Cell Sci. 113:145–152. [DOI] [PubMed] [Google Scholar]
- Ungermann C., and Wickner W.. 1998. Vam7p, a vacuolar SNAP-25 homolog, is required for SNARE complex integrity and vacuole docking and fusion. EMBO J. 17:3269–3276. 10.1093/emboj/17.12.3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungermann C., Nichols B.J., Pelham H.R., and Wickner W.. 1998. A vacuolar v-t-SNARE complex, the predominant form in vivo and on isolated vacuoles, is disassembled and activated for docking and fusion. J. Cell Biol. 140:61–69. 10.1083/jcb.140.1.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungermann C., von Mollard G.F., Jensen O.N., Margolis N., Stevens T.H., and Wickner W.. 1999. Three v-SNAREs and two t-SNAREs, present in a pentameric cis-SNARE complex on isolated vacuoles, are essential for homotypic fusion. J. Cell Biol. 145:1435–1442. 10.1083/jcb.145.7.1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit M. 2004. The human SNARE protein Ykt6 mediates its own palmitoylation at C-terminal cysteine residues. Biochem. J. 384:233–237. 10.1042/BJ20041474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mollard G., Nothwehr S., and Stevens T.. 1997. The yeast v-SNARE Vti1p mediates two vesicle transport pathways through interactions with the t-SNAREs Sed5p and Pep12p. J. Cell Biol. 137:1511–1524. 10.1083/jcb.137.7.1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W. 2010. Membrane fusion: five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu. Rev. Cell Dev. Biol. 26:115–136. 10.1146/annurev-cellbio-100109-104131 [DOI] [PubMed] [Google Scholar]
- Wickner W., and Rizo J.. 2017. A cascade of multiple proteins and lipids catalyzes membrane fusion. Mol. Biol. Cell. 28:707–711. 10.1091/mbc.e16-07-0517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmser A.E., Sato T.K., and Emr S.D.. 2000. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J. Cell Biol. 151:551–562. 10.1083/jcb.151.3.551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Zick M., Wickner W.T., and Jun Y.. 2011. A lipid-anchored SNARE supports membrane fusion. Proc. Natl. Acad. Sci. USA. 108:17325–17330. 10.1073/pnas.1113888108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H., Kakuta S., Watanabe T.M., Kitamura A., Sekito T., Kondo-Kakuta C., Ichikawa R., Kinjo M., and Ohsumi Y.. 2012. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J. Cell Biol. 198:219–233. 10.1083/jcb.201202061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zick M., and Wickner W.. 2012. Phosphorylation of the effector complex HOPS by the vacuolar kinase Yck3p confers Rab nucleotide specificity for vacuole docking and fusion. Mol. Biol. Cell. 23:3429–3437. 10.1091/mbc.e12-04-0279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zick M., and Wickner W.. 2013. The tethering complex HOPS catalyzes assembly of the soluble SNARE Vam7 into fusogenic trans-SNARE complexes. Mol. Biol. Cell. 24:3746–3753. 10.1091/mbc.e13-07-0419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zick M., and Wickner W.. 2016. Improved reconstitution of yeast vacuole fusion with physiological SNARE concentrations reveals an asymmetric Rab(GTP) requirement. Mol. Biol. Cell. 27:2590–2597. 10.1091/mbc.e16-04-0230 [DOI] [PMC free article] [PubMed] [Google Scholar]