Myoepithelial cells function collectively as a dynamic barrier to the invasion and dissemination of Twist1+ luminal epithelial cells and both luminal and basal phenotype breast cancer cells. Barrier function depends on myoepithelial abundance and both smooth muscle contractility and intercellular adhesion within the myoepithelium.
Abstract
The mammary epithelium is composed of an inner luminal and surrounding myoepithelial cell layer. The presence of cancer cells beyond the myoepithelium defines invasive breast cancer, yet the role of the myoepithelium during invasion remains unclear. We developed a 3D organotypic culture assay to model this process through lineage-specific expression of the prometastatic transcription factor Twist1. We sought to distinguish the functional role of the myoepithelium in regulating invasion and local dissemination. Myoepithelial-specific Twist1 expression induced cell-autonomous myoepithelial cell escape. Remarkably, luminal-specific Twist1 expression was rarely sufficient for escape. Time-lapse microscopy revealed that myoepithelial cells collectively restrain and reinternalize invading Twist1+ luminal cells. Barrier function correlated with myoepithelial abundance and required the expression of α-smooth muscle actin and P-cadherin. We next demonstrated that myoepithelial cells can restrain and recapture invasive cancer cells. Our data establish the concept of the myoepithelium as a dynamic barrier to luminal dissemination and implicate both smooth muscle contractility and intercellular adhesion in barrier function.
Introduction
The normal mammary gland is organized around a branched ductal network arranged in an epithelial bilayer, with an inner luminal and outer myoepithelial cell layer (Adriance et al., 2005). Myoepithelial cells and the basement membrane separate luminal epithelial cells from the stroma; hence, interactions between luminal cells and the surrounding mesenchyme are largely mediated by the myoepithelium (Deugnier et al., 2002; Faraldo et al., 2005; Gudjonsson et al., 2005). The myoepithelium has been proposed to regulate both normal mammary epithelial development and cancer invasion. However, the cellular and molecular basis of this role remains incompletely understood (Gudjonsson et al., 2005; Hu et al., 2008).
The majority of breast tumors are thought to arise from luminal epithelial cells located inside the myoepithelium (Deugnier et al., 2002; Adriance et al., 2005; Polyak and Hu, 2005; Molyneux et al., 2010; Proia et al., 2011; Keller et al., 2012; Melchor et al., 2014; Tao et al., 2015). The presence of tumor cells outside the myoepithelium distinguishes ductal carcinoma in situ (DCIS) from invasive ductal carcinoma (IDC; Sternlicht et al., 1997; Lerwill, 2004; Man and Sang, 2004). This fundamentally morphological assessment of intercellular spatial relationships remains the most efficient means to distinguish DCIS from IDC despite decades of analysis of mutations, gene expression, and protein localization (Ma et al., 2003; Hu et al., 2008). This clinicopathologic observation suggests that the integrity and extent of coverage of the myoepithelium is of central importance to patient outcomes.
Breast tumors are composed of heterogeneous cancer cell populations, and experiments suggest that a small and specialized subset of cancer cells is responsible for invasion, local dissemination from the primary tumor, systemic spread, and seeding of secondary sites (Sahai, 2007; Kedrin et al., 2008; Cheung et al., 2013). The molecular mechanisms by which epithelial cells acquire the ability to migrate within the tissue and disseminate out of it remain largely unknown, as are the relative roles of different luminal and basal epithelial populations during dissemination. As tumors grow, the cancer cells progressively outnumber the myoepithelial cells, yet it remains unclear how the cancer cells escape (Gusterson et al., 1982). Because of the limitations of tissue architecture modeling in 2D culture, most breast cancer invasion assays do not contain myoepithelial cells in their physiological organization relative to the cancer cells. However, multiple studies have proposed a role for myoepithelial cells as cellular tumor suppressors (Sternlicht et al., 1997) and have identified mechanisms for this function through secretion of protease inhibitors and down-regulation of matrix metalloproteinases (Barsky and Karlin, 2005), which exert antiproliferative effects on cancer cells (Shao et al., 1998) and inhibit angiogenesis (Nguyen et al., 2000). In addition, focal disruptions in the myoepithelial layer are associated with gene expression changes in the tumor cells, a higher rate of proliferation, and leukocyte infiltration (Man and Sang, 2004).
Taken together, data from prior research suggests a critical role for the myoepithelium in maintaining the in situ status of DCIS lesions and imply that breach of myoepithelial integrity is critical for invasion (Adriance et al., 2005). There are, however, two distinct conceptual frameworks for thinking about this barrier. The first derives from histological analysis, in which the myoepithelium is like a wall: once a gap is generated, the cancer cells can rush through without restriction. The second framework derives from real-time analyses of epithelial tissues that reveal dynamic migrations within epithelial populations (Ewald et al., 2008; Neumann et al., 2018) and extensive regulatory interactions between populations (Cerchiari et al., 2015). Accordingly, it was possible that the myoepithelium could respond to and dynamically regulate the migration and invasion of luminal cancer cells.
Observing invasion past the myoepithelium in vivo is challenging because of the limited optical accessibility of the mammary gland and the slow and unpredictable nature of localized invasion events. To overcome these challenges, we adapted a 3D organotypic culture based on normal mouse mammary epithelium to model myoepithelial barrier function. We previously demonstrated that constitutive expression of the prometastatic transcription factor Twist1 (Yang et al., 2004) in primary mouse mammary organoids induces robust local dissemination of cells out of the epithelium into the surrounding ECM (Shamir et al., 2014). In this study, we set out to determine the capacity of myoepithelial cells to limit luminal dissemination and the nature of this regulatory interaction.
Results and discussion
Mouse models used in this study
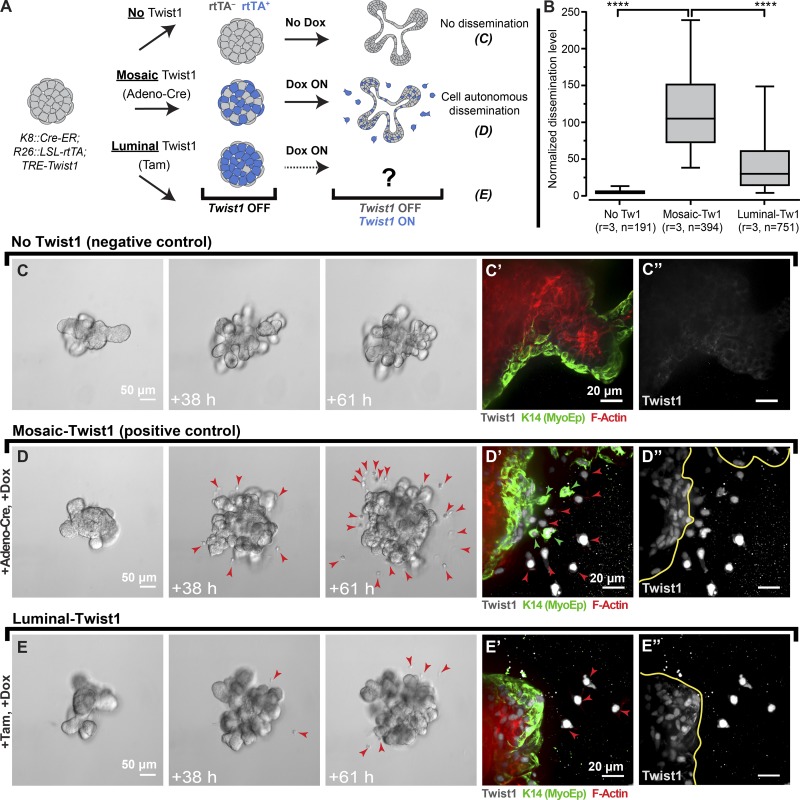
We first sought to test the dissemination potential of different epithelial cell types and visualize interactions between luminal and myoepithelial cells. To accomplish these goals, we developed three inducible and lineage-specific mouse models: Ubiquitous-Twist1, Myoepithelial-Twist1, and Luminal-Twist1. In the Ubiquitous-Twist1 model, we used a cytomegalovirus (CMV) promoter–driven reverse tetracycline transactivator (rtTA), a Tet-responsive Twist1, and a transgenic Keratin 14 (K14) promoter-driven GFP as a myoepithelial-specific reporter allele (Fig. S1 A; CMV::rtTA;TRE-Twist1;K14::Actin-GFP, Ubiquitous-Twist1 model). This combination enabled us to track the interactions of myoepithelial and luminal cell types in real-time while both populations expressed Twist1. We next established a Myoepithelial-Twist1 model in which Twist1 was regulated by a myoepithelial-specific K14 promoter–driven rtTA (K14::rtTA;TRE-Twist1, Myoepithelial-Twist1 model). Finally, we established a Luminal-Twist1 model in which Twist1 was regulated by a luminal-specific Keratin 8 (K8) promoter–regulated rtTA (K8::Cre-ER;R26::LSL-rtTA;TRE-Twist1;mTomato [mT]/mGFP [mG]). Because of its conditional design, the Luminal-Twist1 mouse model can be used to drive Twist1 in a random subset of cells with adenoviral Cre recombinase (Adeno-Cre; designated the Mosaic-Twist1 model) or specifically in luminal cells using tamoxifen (Fig. 1 A; Luminal-Twist1 model). In both settings, the mT/mG Cre reporter serves as an indirect marker of Cre-lox recombination and rtTA expression. These three mouse models together enabled us to isolate the role of the myoepithelium during Twist1-induced dissemination.
Figure 1.
Luminal-specific Twist1 expression rarely results in dissemination past normal myoepithelium. (A) Mosaic or luminal-specific expression of Twist1 was achieved in K8::Cre-ER;R26::LSL-rtTA;TRE-Twist1 organoids by Adeno-Cre and a tamoxifen (Tam)-inducible Cre-ER, respectively, followed by culture in doxycycline (Dox) to induce Twist1 expression. (B) K8::Cre-ER;R26::LSL-rtTA;TRE-Twist1 organoids from the same mouse were left untreated or treated with Adeno-Cre (Mosaic-Twist1) or tamoxifen (Luminal-Twist1). The number of disseminated cells per organoid was significantly higher in Adeno-Cre–treated organoids compared with tamoxifen-treated organoids (fivefold difference), whereas untreated organoids showed no dissemination across three biological replicates. (C–C′′) The no-Twist1 control showed branching and no dissemination. (D–D′′) Mosaic-Twist1 expression induced branching and dissemination. (E–E′′) Luminal-Twist1 expression rarely led to dissemination. In the Mosaic-Twist1 model, dissemination of both luminal and myoepithelial cells was observed (D′ and D′), whereas in the luminal-Twist1 model, we only observed dissemination of luminal cells (E′ and E′′). n, total number of organoids; r, number of biological replicates. In D and E, red arrowheads mark all disseminated cells. In D′, D″, E′, and E″, red arrowheads mark K14− cells, and green arrowheads mark K14+ cells. Data were analyzed by two-tailed nonparametric Kruskal–Wallis multiple comparisons test; ****, P < 0.0001. Data are presented as box plots, with error bars representing the 5th to 95th percentile.
The myoepithelium appears to regulate dissemination of Twist1+ luminal cells
We first examined the real-time interactions among labeled myoepithelial and luminal cell populations within organoids cultured from Ubiquitous-Twist1 mice using confocal microscopy (Fig. S1 A). Induction of Twist1 led to dissemination of both luminal epithelial and myoepithelial cells, consistent with our previous study (Shamir et al., 2014). Twist1 expression also disrupted the architecture of the organoids such that gaps in basal myoepithelial coverage often emerged. In turn, Twist1+ luminal cells were observed to escape from the organoid through these gaps (Fig. S1 B). Furthermore, we occasionally observed myoepithelial cells wrapping around invasive luminal epithelial cells, giving the appearance that they were restraining the luminal cells from escaping (Fig. S1 C). Based on these observations, we hypothesized that myoepithelial cells actively regulate luminal cell dissemination. However, the expression of Twist1 in both cell populations in the Ubiquitous-Twist1 model limited our analyses. We therefore turned to models in which Twist1 was expressed in specific epithelial compartments and tested the dissemination potential of the various cell types.
Myoepithelial-specific Twist1 expression induces myoepithelial cell dissemination
To study the dissemination potential of myoepithelial cells, we used our Myoepithelial-Twist1 mouse model (Fig. S1 D). We observed branching morphogenesis in both control and Twist1-induced conditions, whereas dissemination was specific to the Twist1-induced organoids (Fig. S1, E–G′). In basal medium without growth factors, Twist1 activation induced high levels of dissemination, which were significantly reduced by the addition of FGF2 (Fig. S1 E′′′). We conclude that the extent of myoepithelial-specific dissemination is regulated by FGF2 signaling. We confirmed by immunofluorescence (IF) that disseminated cells were K14+ (Fig. S1 G′). Conversely, luminal cells in this model were Twist1− and remained within the organoid, demonstrating that myoepithelial-specific Twist1 expression leads to cell-autonomous myoepithelial dissemination. However, it is not sufficient for dissemination of Twist1− luminal epithelial cells.
Luminal-specific Twist1 expression is rarely sufficient for dissemination
We next used the Luminal-Twist1 model to assay the sufficiency of Twist1 to induce dissemination of luminal cells in the presence of a normal myoepithelium (Fig. 1, A and B). As an internal control and to provide a baseline for dissemination levels with this rtTA, we used Adeno-Cre to induce expression of rtTA in a random subset of both luminal and myoepithelial cells (Mosaic-Twist1 model; Fig. 1, A and B). Mosaic-Twist1 organoids were typically branched, with robust levels of dissemination relative to Twist1− control organoids (Fig. 1 D vs. Fig. 1 C). We validated by IF that both luminal and myoepithelial cells disseminated in Mosaic-Twist1 organoids and were Twist1+ (Fig. 1, D′ and D′′). We next induced Twist1 expression specifically in luminal cells (Luminal-Twist1 model) and observed much lower levels of dissemination than in Mosaic-Twist1 organoids even though both were derived from the same mice (Fig. 1, B and E vs. Fig. 1 D). The limited dissemination capacity of Luminal-Twist1 organoids was despite expression and nuclear localization of Twist1 in nondisseminating cells (Fig. 1, E′ and E′′). Quantification across three biologically independent replicates revealed significantly lower dissemination behavior when the myoepithelium was Twist1− (Fig. 1 B). Finally, we confirmed that addition of tamoxifen to organoids did not itself suppress dissemination and that both Mosaic-Twist1 and Luminal-Twist1 organoids expressed comparable levels of Twist1 protein (Fig. S2, A–C). As an additional control, we tested the relative recombination efficiencies when using Adeno-Cre and tamoxifen to induce Twist1 expression and confirmed that induction of mG and doxycycline addition resulted in a large majority of Twist1+ cells in both models, with a high correlation between mG positivity and Twist1 expression (Fig. S2, D–G). Taken together, these data reveal that the presence of an intact myoepithelium correlates with reduced escape of Twist1+ luminal cells. We next sought to determine the nature of the luminal–myoepithelial cell interactions driving this regulation.
Normal myoepithelial cells suppress dissemination of Twist1+ luminal cells
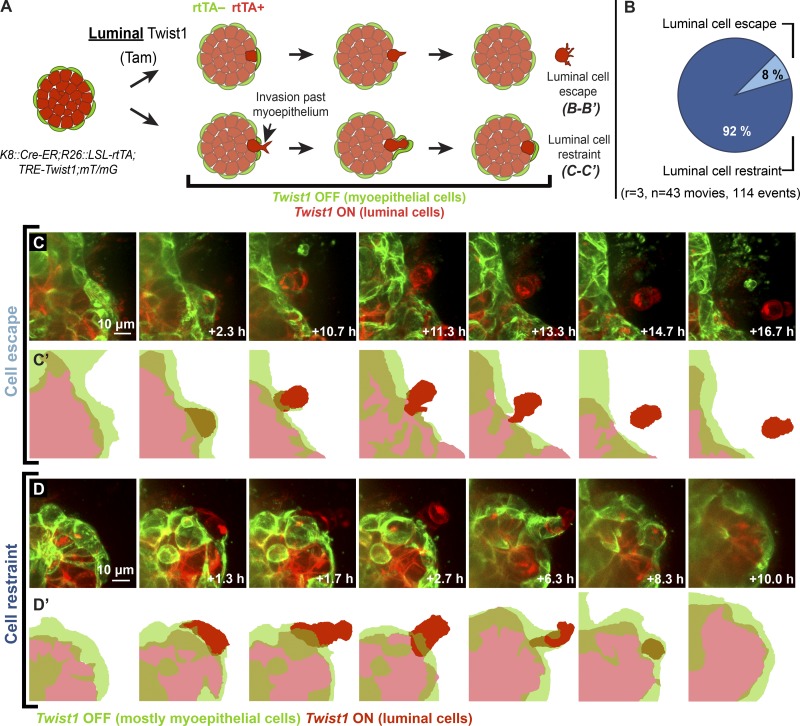
In the Luminal-Twist1 model, Cre-ER activity results in the conversion of a reporter locus from driving mT (Cre−) to mG (Cre+) expression. Because Cre-ER is expressed under the control of the luminal-specific K8 promoter, addition of tamoxifen and doxycycline results in differentially labeled Twist1+ luminal cells and Twist1− myoepithelial cells (Fig. 2 A). For consistency, myoepithelial cells in time-lapse videos are pseudocolored in green throughout the figures. We conducted time-lapse confocal imaging of Twist1-induced organoids from this model and observed that when Twist1+ luminal cells protruded into the ECM, the adjacent Twist1− myoepithelial cells typically covered and contained them, reestablishing a continuous myoepithelial layer (Fig. 2, B and D–D′; and Videos 2, 3, 4, and 5). We observed only rare instances of luminal cells successfully disseminating through a normal myoepithelial layer (Fig. 2, B–C′; and Video 1). When we quantified those events, we found that luminal cell restraint occurred 92% of the time, while luminal cell escape happened only 8% of the time (Fig. 2 B).
Figure 2.
The myoepithelium dynamically restrains protruding Twist1+ luminal cells. (A) Organoids isolated from K8::Cre-ER;R26::LSL-rtTA;TRE-Twist1;mT/mG mice were treated with tamoxifen (Tam) to induce rtTA expression and cultured in doxycycline to induce Twist1 expression specifically in the luminal cell population. (B) Luminal cell restraint by myoepithelial cells quantified across multiple replicates was observed 92% of the time, and luminal cell escape was observed 8% of the time. (C and C′) Luminal cells were very rarely observed to disseminate past the myoepithelium. (D and D′) Luminal cells that protruded past the myoepithelium were typically restrained by myoepithelial cells. Adjacent myoepithelial cells migrated over and appeared to contain protruding luminal cells. n, total number of videos analyzed; r, number of biological replicates. Myoepithelial cells in time-lapse videos are pseudocolored in green throughout the figure.
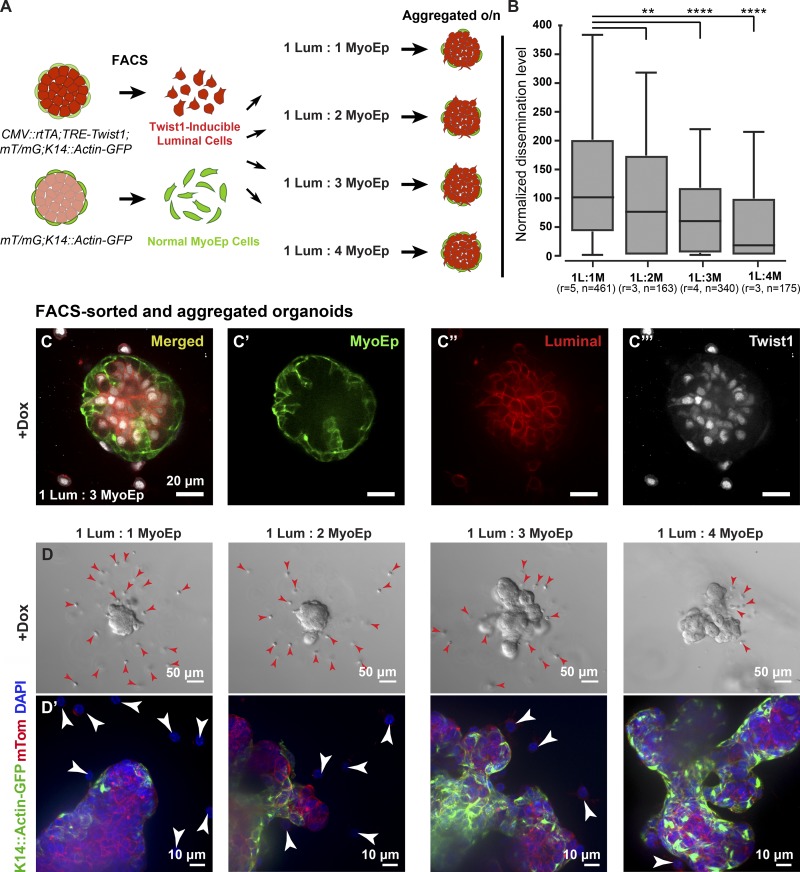
To test directly whether myoepithelial cells are regulating luminal cell dissemination, we combined clustered Twist1+ luminal cells with varying ratios of normal (Twist1−) myoepithelial cells. To achieve this goal, we isolated differentially labeled cell populations via FACS: for Twist1-inducible luminal cells, we used the Ubiquitous-Twist1 mouse (CMV::rtTA;TRE-Twist1;K14::Actin-GFP;mT/mG), and for normal myoepithelial cells, we used a WT mouse that expresses a GFP under the K14 promoter (K14::Actin-GFP;mT/mG). The two cell populations, luminal (GFP−mT+) and myoepithelial (GFP+mT+), were aggregated overnight at different ratios and embedded in Matrigel the next day (Fig. 3 A). Myoepithelial cells localized to the outside of aggregated organoids surrounding the luminal cells. Upon addition of doxycycline, luminal cells expressed Twist1 while myoepithelial cells remained Twist1− (Fig. 3, C–C′′′). The number of disseminated cells per aggregated organoid decreased as the number of normal myoepithelial cells increased (Fig. 3, B and D). In addition, we observed that myoepithelial coverage was more complete with an increased number of myoepithelial cells (Fig. 3 D′), suggesting that the completeness of the myoepithelial layer had a direct impact on the likelihood of cell escape. Collectively, our data demonstrate that myoepithelial cells regulate dissemination of Twist1+ luminal cells and establish the concept of the myoepithelium as a dynamic barrier to cell escape. We next sought insight into the molecular mechanism underlying this barrier function.
Figure 3.
Myoepithelial cell abundance regulates the extent of Twist1+ luminal cell dissemination. (A) Twist1-inducible luminal cells were isolated from CMV::rtTA;TRE-Twist1;mT/mG;K14::Actin-GFP mice, and normal K14+ (mostly myoepithelial) cells were isolated from mT/mG;K14::Actin-GFP mice via FACS using cell type–specific fluorescent reporters. Twist1-inducible luminal cells (red) were aggregated with normal myoepithelial cells (green) at different ratios to form clusters, and aggregated clusters were cultured in 3D Matrigel. (B) The number of disseminated Twist1+ cells per organoid significantly decreased with an increasing number of normal myoepithelial cells. (C–C′′′) Addition of doxycycline (dox) to aggregated organoids induced luminal-specific Twist1 expression. (D and D′) Myoepithelial coverage increased and dissemination decreased with the addition of more myoepithelial cells. Arrowheads indicate disseminated cells. n, total number of organoids; r, number of biological replicates. Data were analyzed by two-tailed nonparametric Kruskal–Wallis multiple comparisons test: **, P < 0.01; ****, P < 0.0001. Data are presented as box plots, with error bars representing the 5th to 95th percentile.
Depletion of α-smooth muscle actin (SMA) or P-cadherin compromises the myoepithelial barrier
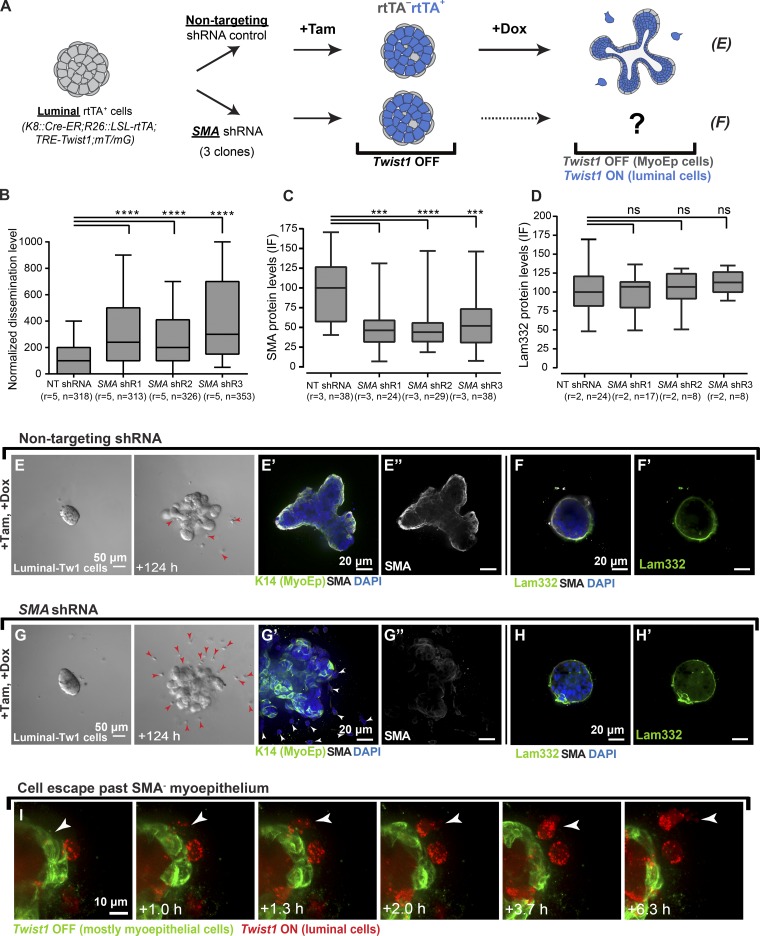
Myoepithelial cells express a smooth muscle contractile molecular program, and it is critical to their role in milk ejection (Haaksma et al., 2011; Weymouth et al., 2012). Markers for the smooth muscle program are also commonly used in pathology to assay the integrity of the myoepithelium and distinguish between in situ and invasive breast carcinoma (Werling et al., 2003; Hilson et al., 2009). Because SMA has a critical role in both development and cancer diagnosis, we sought to test its requirement for myoepithelial regulation of dissemination. To achieve this goal, we knocked down SMA in luminal-Twist1 organoids (K8::Cre-ER;R26::LSL-rtTA;TRE-Twist1;mT/mG) using lentivirally delivered SMA shRNA (Knott et al., 2014) with nontargeting shRNA used as a control, and we monitored for differences in dissemination after Twist1 induction (Fig. 4 A). Control organoids with Twist1+ luminal cells and Twist1− myoepithelial cells had rare disseminated cells (Fig. 4, E–E′′; similar to Fig. 1, E–E′′). In contrast, SMA knockdown organoids displayed a significant increase in dissemination (Fig. 4, G–G′′). Specifically, organoids with depleted SMA showed an approximately threefold increase in the number of disseminated cells as compared with control across three different shRNA clones, each of which significantly reduced SMA protein levels (Fig. 4, B and C). In contrast, there was no difference in the level of the basement membrane protein Laminin-332 in control versus SMA knockdown organoids (Fig. 4, D, F, F′, H, H′). This finding suggests that SMA is not required for establishing the basement membrane and that the presence of basement membrane is not sufficient for limiting cell dissemination. We next used confocal microscopy to monitor cell–cell interactions after SMA knockdown. Upon Twist1 induction, we observed luminal Twist1+ cells protruding and disseminating into the ECM without evident restraint of migration by the surrounding myoepithelial cells (Fig. 4 I and Video 6). Knockdown of the myoepithelial-specific classical cadherin P-cadherin also significantly increased dissemination of Twist1+ cells (Fig. S2, H–K′′). Taken together, these data implicate both SMA-dependent contractility and intercellular adhesion in myoepithelial barrier function.
Figure 4.
Depletion of α-SMA leads to an increase in cell dissemination. (A) Organoids isolated from K8::Cre-ER;R26::LSL-rtTA;TRE-Twist1;mT/mG mice were treated with lentiviral shRNA against a nontargeting (NT) control and against three different clones of α-SMA (myoepithelial smooth muscle gene). Organoids were treated with tamoxifen (Tam) to induce rtTA expression and cultured in doxycycline (Dox) to induce Twist1 expression in the luminal cell compartment. After Twist1 expression, organoids were monitored for dissemination. (B) Organoids were divided into nontargeting and SMA knockdown treatment groups. The number of disseminated cells per organoid was significantly higher in SMA knockdown organoids compared with the nontargeting control (two- to threefold difference) across five biological replicates. (C) SMA protein levels quantified from IF images confirmed a decrease in SMA expression in SMA knockdown organoids. (D) Laminin-332 protein levels quantified from IF images did not significantly differ between SMA knockdown organoids and controls. (E–F′) IF indicated retention of K14, SMA and Laminin-332 in control organoids. (G–H′) IF indicated retention of K14, loss of SMA, and retention of Laminin-332 in SMA knockdown organoids. Red and white arrowheads indicate disseminated cells. (I) Protruding Twist1+ luminal cells were observed to invade past the SMA-depleted myoepithelium and were able to disseminate (arrowheads). n, total number of organoids; r, number of biological replicates. Data were analyzed by two-tailed nonparametric Kruskal–Wallis multiple comparisons test: ***, P < 0.001; ****, P < 0.0001. Data are presented as box plots, with error bars representing the 5th to 95th percentile. Myoepithelial cells in time-lapse videos are pseudocolored in green in I.
Myoepithelial cells appear to suppress invasion of luminal and basal phenotype tumor organoids
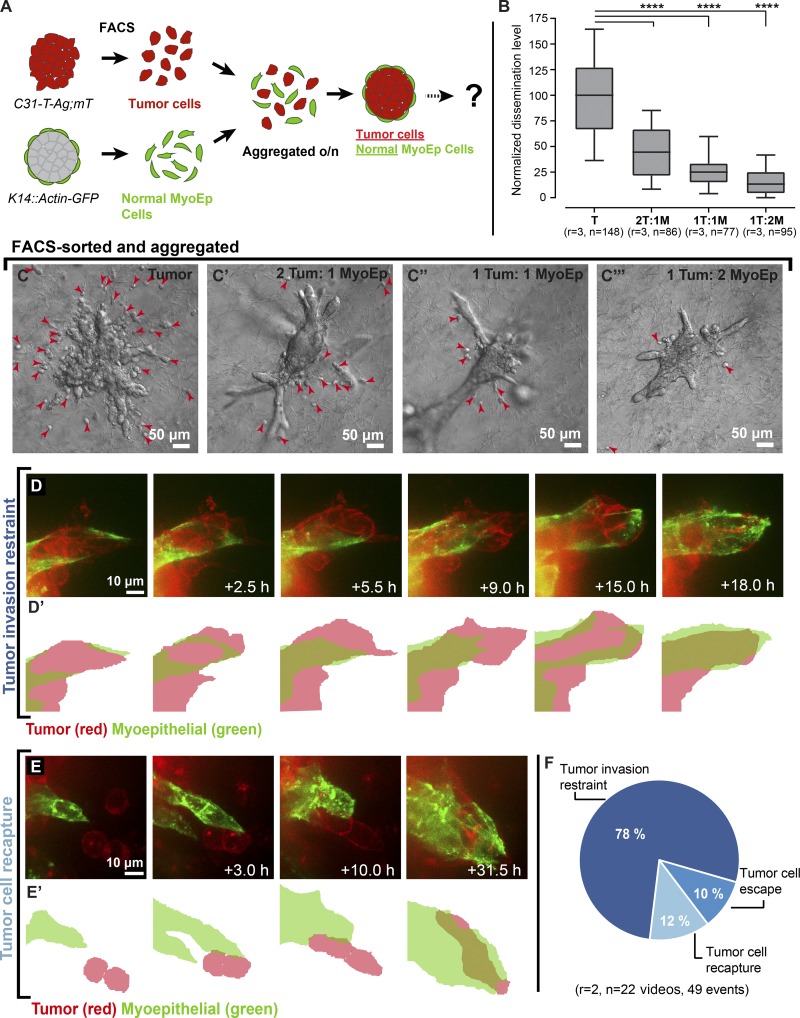
Our data thus far revealed that myoepithelial cells regulate dissemination in our Twist1+ model system. We next sought to test their capacity to restrain tumor invasion using tumor organoids isolated from a mouse model of basal breast cancer (C31-T-Ag;mT/mG). We combined varying ratios of GFP+ myoepithelial cells isolated from WT mice (K14::Actin-GFP) with mT+ tumor cells and cultured the aggregated organoids in collagen I gels, which model the stromal microenvironment around invasive carcinomas (Fig. 5, A and B). Myoepithelial cells spontaneously localized to the outside of the tumor aggregates. Organoids consisting only of tumor cells showed robust dissemination (Fig. 5 C). In contrast, as we increased the number of normal myoepithelial cells, the number of disseminated cells per tumor organoid decreased markedly (Fig. 5, B and C′–C′′′). We next used real-time confocal microscopy to study cell–cell interactions between myoepithelial and tumor cells during this process. Myoepithelial cells were observed to wrap around tumor cells as they protruded into collagen I and to limit the extent of their invasion (Fig. 5, D–D′′; and Video 7). In addition, we observed myoepithelial cells capture already disseminated tumor cells and reestablish their connection to the main tumor mass (Fig. 5, E–E′′; and Video 8). Quantification revealed the relative frequency of cell restraint (78%), recapture (12%), and escape past the myoepithelium (10%; Fig. 5 F). Similar apparent restraint of invasion and recapture of disseminated cells was observed in tumor organoids derived from a mouse model of luminal breast cancer (MMTV::PyMT;βactin-CFP; Fig. S3, A–D). We conclude that the myoepithelium can actively regulate and suppress tumor cell invasion and dissemination through cell migrations and rearrangements that dynamically maintain overall tissue architecture.
Figure 5.
Normal myoepithelial cells suppress invasion and dissemination of C3-1-T tumor cells. (A) Tumor cells (mT+; red) were isolated from C3-1-Tag;mT/mG mice and normal myoepithelial cells (GFP+; green) were isolated from K14::Actin-GFP mice via FACS. Tumor cells were aggregated with normal myoepithelial cells at different ratios to form clusters, and aggregated clusters were cultured in 3D collagen I. (B–C′′′) The clusters were divided into four groups and monitored for dissemination. The number of disseminated cells per cluster decreased with an increasing number of normal myoepithelial cells. Arrowheads indicate disseminated cells. (D and D′) Myoepithelial cells appeared to contain invasion past the myoepithelial layer and reestablish a continuous myoepithelial layer. (E and E′) Myoepithelial cells were also observed to extend into the collagen and capture disseminated tumor cells. (F) Tumor cell restraint, recapture, and dissemination quantified from real-time confocal videos across multiple replicates. n, total number of organoids; r, number of biological replicates. Data were analyzed by two-tailed nonparametric Kruskal–Wallis multiple comparisons test: ****, P < 0.0001. Data are presented as box plots, with error bars representing the 5th to 95th percentile. Myoepithelial cells in time-lapse videos are pseudocolored in green.
Our focus in this study was on testing the role of the myoepithelium in restraining epithelial invasion and local dissemination in organoid models. Prior studies had proposed that myoepithelial cells could act as cellular tumor suppressors (Sternlicht et al., 1997; Deugnier et al., 2002; Adriance et al., 2005; Gudjonsson et al., 2005; Polyak and Hu, 2005; Hu et al., 2008; Polyak, 2010). However, the nature of this interaction was unclear. We developed lineage-specific expression models to enable the induction of Twist1 in either luminal or myoepithelial cells. Myoepithelial cells directly contact the ECM and were observed to disseminate in a cell-autonomous fashion in response to Twist1 expression. In contrast, luminal epithelial cells are surrounded by contractile myoepithelial cells and so must negotiate this layer before they can access the ECM. Expression of Twist1 specifically in luminal cells rarely resulted in dissemination in these 3D cultures; we instead typically observed active migration of myoepithelial cells to enclose and restrain invasive luminal epithelial cells. Barrier function depended on both the abundance of myoepithelial cells and on their molecular properties. Expression of Twist1 or knockdown of either of the myoepithelial-specific genes SMA or P-cadherin was sufficient to compromise the myoepithelial barrier. We then demonstrated that normal myoepithelial cells could also restrain invasive cancer cells in 3D organoid culture. Our data establish the concept of the myoepithelium as a dynamic barrier to epithelial invasion and local dissemination through active cell migration and intercellular interactions.
We speculate that our conclusions have implications for understanding the role of the myoepithelium during breast cancer progression in patients. The integrity of the myoepithelium is used to distinguish DCIS from IDC, and even microscopic gaps in the myoepithelium correlate with worse patient outcomes (Sternlicht et al., 1997; Man et al., 2003; Man and Sang, 2004). As such, myoepithelial cells are quite literally positioned to regulate invasion. However, an invasion-suppressing role could be accomplished by a passive mechanical barrier, similar to a stone wall. Once breached, it would not further resist invasion. Instead, in our organoid cultures, we observed a dynamic myoepithelial barrier capable of restraining invasion and even recapturing disseminated cells. We speculate that pathological evidence of microinvasion past an otherwise intact myoepithelium could be a snapshot in time of a myoepithelium that was still actively resisting invasion and dissemination in that patient (Man and Sang, 2004). Furthermore, a recent single-cell sequencing study revealed that most mutations and copy-number changes are shared between DCIS and IDC in the same patient (Casasent et al., 2018), lending plausibility to the idea that initiation of invasive behavior may relate more to the overcoming of local barriers such as the myoepithelium than the acquisition of novel proinvasive mutations. We also note that the cancer cells that lead collective invasion express K14 but lack expression of smooth muscle markers such as SMA (Cheung et al., 2013). There are therefore distinct invasion suppressing (myoepithelial; K14+SMA+) and invasion promoting (K14+SMA−) epithelial populations expressing K14 within breast tumors.
We demonstrated that the myoepithelium is a dynamic barrier to luminal epithelial dissemination and relies upon both contractility and adhesion for this function. This concept is supported by recent papers that have elucidated self-organizing principles governing tissue architecture. Briefly, the organization of cell populations within tissues can be regulated by their relative cell–cell and cell–matrix adhesion (Cerchiari et al., 2015), mutual adhesion and cortical tension (Maître et al., 2012), interfacial tensions (Neumann et al., 2018), and proteolytically driven differences in motility (Mori et al., 2009). Accordingly, it will be important to deepen our understanding of how molecular differences among cells drive cell behavior and ultimately tissue and organ structure and function. Based on past work, we speculate that the highly cohesive nature of the myoepithelium may underlie its ability to capture invasive luminal cells and restore normal tissue architecture (Cerchiari et al., 2015).
Further studies are now needed to identify the sensing mechanisms used by the myoepithelium to detect and respond to invasive luminal cells, particularly once they have already disseminated. It will also be important to elucidate the mechanisms by which the myoepithelial barrier eventually fails and tumor cells succeed in metastasizing to distant sites. A better understanding of myoepithelial function may also help inform the diagnosis or management of breast cancer patients.
Materials and methods
Mouse strains
Mice used in this study were backcrossed to FVB/n background and maintained in accord with a protocol approved by the Johns Hopkins Institutional Animal Care and Use Committee. The Twist1-tetO7-luc (TRE-Twist1) transgenic line contains the mouse Twist1 cDNA under the control of a bidirectional tetracycline responsive element (tetO7) and which regulates the luciferase gene as previously described (Tran et al., 2012). The CMV::rtTA transgenic line was a gift from F. Cong and H. Varmus (National Cancer Institute, Bethesda, MD). These mice carry the transgene encoding the reverse tetracycline-controlled transactivator protein (rtTA) under the control of a human CMV early enhancer. Crossing of TRE-Twist1 and CMV::rtTA transgenic lines resulted in a transgenic line that contained both the rtTA activator and the tetracycline inducible Twist1-tetO7 transgene. Addition of doxycycline in these mice triggers a conformational change that enables tetO7 binding, activation, and Twist1 and luc transcription. The K14::Actin-GFP transgenic line was a gift from E. Fuchs (The Rockefeller University, New York, NY). In this line, the K14-GFP-actin transgene construct containing the human K14 promoter, rabbit β-globin intron, and EGFP-actin cDNA was randomly inserted, and as a result, the mice express EGFP-tagged murine β-actin under the control of the K14 promoter as described previously (Vaezi et al., 2002). The K14::rtTA line was derived by using a construct containing rtTA2S-M2-VP16 (rtTA) preceded by a β-globin intron and under the control of the K14 promoter as described previously (Nguyen et al., 2006). Crossing the K14::rtTA line to theTRE-Twist1 line leads to expression of Twist1 protein in the basal cell lineage upon administration of doxycycline. The K8::Cre-ER line was generated by randomly inserting the K8-CreER fragment containing the CreERT2 fragment preceded by the β-globin intron and followed by a simian virus 40 (SV40) polyA signal. This resulted in mice expressing CreER in the luminal lineage using the K8 promoter as previously described (Van Keymeulen et al., 2011). The mT/mG mouse line was derived by inserting the mT/mG construct containing the β-actin core promoter with a CMV enhancer driving a loxP-flanked coding sequence of membrane-targeted tandem dimer Tomato (tdTomato) into the murine ROSA26 locus as described previously (Muzumdar et al., 2007). These mice ubiquitously expressed tdTomato with membrane localization. After breeding mT/mG mice with the K8::Cre-ER line, and upon tamoxifen administration, there is Cre-mediated excision of the mT sequence, allowing expression of the membrane-localized EGFP (mG). The R26::Lox-Stop-Lox-rtTA-IRES-EGFP (R26::LSL-rtTA) mouse line was generated by a targeted insertion of a conditional rtTA-IRES-EGFP transgene between the first and second exons of the murine ROSA26 locus as previously described (Belteki et al., 2005). In these mice, the upstream sequence is coding for rtTA, and the downstream sequence is coding for EGFP. These two coding sequences were preceded by a loxP site–flanked selectable marker. Prior to Cre-mediated excision of the loxP sequence, the downstream coding sequences are not expressed. Crossing the R26::LSL-rtTA line to the K8::Cre-ER and CMV::rtTA;TRE-Twist1 lines leads to expression of Twist1 protein in the luminal cell lineage upon administration of tamoxifen and doxycycline. To derive the FVB-Tg(C3-1-TAg)cJeg/JegJ line, the rat prostatic steroid-binding protein (C3-1) gene 5′ regulatory region and the simian virus 40 (SV40) large tumor antigen (TAg) were combined to create a C3-1-TAg fusion sequence, which was inserted into mouse embryos as described previously (Maroulakou et al., 1994). The C3-1 regulatory region was used to direct expression of SV40 TAg to the prostate and mammary glands, resulting in a transgenic model for studying tumorigenesis in these tissues. For generation of the MMTV::PyMT line, cDNA encoding the mouse mammary tumor virus (MMTV) and the polyoma virus middle T antigen (PyVT) was inserted into the expression vector pMMTV-SV40 as previously described (Guy et al., 1992). MMTV::PyMT mice express the PyVT under the direction of the MMTV promoter and develop mammary tumors that metastasize to the lung. The βactin-CFP line was derived by inserting a transgenic construct containing an enhanced cyan fluorescent protein gene under the control of the chicken β-actin promoter coupled with the CMV enhancer into embryonic stem cells. These mice express CFP in all cell types. K14::rtTA, K8::Cre-ER, mT/mG, R26::LSL-rtTA, C3-1-Tag, MMTV::PyMT, and βactin-CFP mouse lines were obtained from The Jackson Laboratory. For confocal time-lapse experiments and FACS experiments, different transgenic lines were crossed with βactin-CFP, mT/mG and/or K14::Actin-GFP mice.
Isolation of primary murine mammary organoids
Epithelial fragments termed organoids were isolated from murine mammary glands and mammary tumors using previously described techniques (Ewald et al., 2008; Nguyen-Ngoc et al., 2015). Normal mammary glands were harvested from 8- to 12-wk-old mice, MMTV-PyMT tumors were harvested from mice at 12–16 wk of age, and C3-1-Tag tumors were harvested from mice at 20–24 wk of age. Briefly, we surgically removed no. 3 and no. 4 mammary glands from normal mice and the largest tumors from tumor mice. Dissected tissue was then subjected to a combination of mechanical disruption, collagenase-trypsin digestion, and DNase treatment to separate epithelial tissue from fat and stromal cells. The epithelial fragments were separated from single cells through differential centrifugation. The final pellet contained thousands of organoids, each composed of a couple hundred cells.
3D culture of primary murine mammary organoids
The isolated organoids were suspended in Matrigel (354230; BD) or rat-tail collagen I (354236; Corning) at a concentration of 1–2 organoids/µl and plated as 100–120 µl suspensions in 24-well glass bottom plates (655892; Greiner Bio One) over a 37°C heating block. Acid-solubilized rat-tail collagen I gels (3 mg/ml collagen I, pH 7–7.5) were prepared as described previously (Ewald et al., 2008; Nguyen-Ngoc et al., 2015). Matrigel and collagen I gels were polymerized at 37°C for 45 min before addition of organoid medium: DMEM (D6546; Sigma-Aldrich) with 1% insulin-transferrin-selenium (51500–056; Gibco) and 1% penicillin-streptomycin (P4333; Sigma-Aldrich).
Cell type–specific Twist1 activation in 3D culture
In organoids isolated from CMV::rtTA;TRE-Twist1;mT/mG;K14::Actin-GFP and K14::rtTA;TRE-Twist1 mice, Twist1 expression was induced by supplementing 3D Matrigel with 2.5 nM FGF2 and 5 µg/ml doxycycline organoid medium (Shanghai RenYoung Pharmaceutical Co., Ltd) on day 1 in culture. The organoid medium was not supplemented with 2.5 nM FGF2 in experiments where FGF2 effects were tested on cell dissemination in K14::rtTA;TRE-Twist1 organoids. Organoid medium was changed every 48 h for all doxycycline induction experiments. In organoids isolated from K8::Cre-ER;R26::LSL-rtTA;TRE-Twist1;mT/mG mice, mosaic rtTA expression was induced by incubating organoids with Adeno-Cre (1045; Vector Laboratories) at a ratio of 107 PFU per 1,000 organoids for 1–2 h at 37°C before suspension in Matrigel, resulting in recombination of 50–75% cells. To induce rtTA expression in luminal cells, organoids embedded in 3D Matrigel were cultured continuously with 50 nM 4-hydroxy tamoxifen (H7904; Sigma-Aldrich), which activated Cre-ER in cytokeratin 8 (K8)–positive cells. The next day, for both mosaic and luminal rtTA expression, the organoids were supplemented with 5 µg/ml doxycycline and 2.5 nM FGF2 organoid medium.
Differential interference contrast (DIC) microscopy
Live imaging of organoids was conducted using an LD Plan-Neofluar 20×/0.4 Korr Ph2 objective lens and a Zeiss Cell Observer system with a Zeiss AxioObserver Z1 and an AxioCam MRM camera. For time-lapse imaging, images were acquired at 20-min intervals for 5–7 d, with 100–200 videos collected in parallel. For quantification of dissemination and branching, images were acquired on day 1 and days 5–9 in culture. Temperature was maintained at 37°C and CO2 was held at 5%. AxioVision64 software (Zeiss) was used to analyze time-lapse videos and export individual TIFF files. ImageJ (National Institutes of Health) was used to count disseminated cells, crop and calibrate images, and place scale bars.
Confocal microscopy
Confocal imaging was conducted on a Solamere Technology Group spinning-disk confocal microscope with an iXon3 885 electron-multiplying charge-coupled device camera (Andor). An LD C-Apochromat 40×/1.1 W Korr objective lens (Zeiss) was used for single and time-lapse image acquisition using water for still images and Immersol (Zeiss) for time lapse as the imaging medium. For time-lapse imaging, images were acquired at 20-min time intervals with 15–20 videos collected in parallel using one to three channels (excitation at 405 nm, 488 nm, and 561 nm). For z stacks, 2-µm spacing was used. Acquisition of both time-lapse and still images was performed using µManager (Stuurman et al., 2010) and Piper (Stanford Photonics, Inc.). Temperature was maintained at 37°C and CO2 at 5% for live imaging. Imaris 8 (Bitplane) was used to analyze videos, export individual TIFFs, and adjust brightness and contrast of the images in each channel to maximize the clarity. Adjustments were always made across entire images. A minimal 0.137-µm Gaussian image filter was applied across images in Fig. 1 (C and D) to maximize image clarity. ImageJ was used to crop and calibrate images and place scale bars.
Analysis of cell dissemination, branching, and cellular protrusions
DIC images were used to analyze branching and dissemination of organoids. Branching was scored by counting the number of organoids with three or more distinct elongated buds. Dissemination was scored by counting the number of adjacent single cells in the ECM separated from the organoid they originated from. An organoid was categorized as disseminated if it had at least two adjacent cells. Both dissemination and branching were quantified for organoids at 7–9 d in culture. Cell counting was performed using ImageJ software. For each experiment, dissemination of the treatment conditions was normalized to the median dissemination of the control, which was set to have a value of 100. Normalized data from multiple independent biological replicates were pooled, and the statistical analysis was performed using Prism (GraphPad Software). The P value was determined by a nonparametric Mann-Whitney-Wilcoxon test (comparing two groups) or a Kruskal-Willis test (comparing multiple groups). Dissemination of cell types with different fluorescent markers (GFP and mT) was scored by analyzing confocal videos using Imaris. Myoepithelial restraint of luminal cell dissemination in K8::Cre-ER;R26::LSL-rtTA;TRE-Twist1;mT/mG organoids was scored by counting the number of events of luminal cells protruding past the myoepithelial layer and being recaptured by myoepithelial cells. Myoepithelial restraint of tumor cell invasion, tumor cell recapture, and tumor cell dissemination in clusters aggregated from cells from mT/mG;K14::Actin-GFP, C31-T-Ag;mT/mG and MMTV::PyMT; βactin-CFP mice was quantified by scoring the number of these events observed in the real-time confocal videos. Restraint of tumor cells was defined as myoepithelial cells wrapping around tumor cells that protruded past the myoepithelial layer and pulling them back, tumor cell recapture was defined as myoepithelial cells reaching out and pulling back fully disseminated tumor cells, and tumor cell dissemination was defined as cells protruding past the myoepithelial layer, fully detaching, and migrating away.
SMA and P-cadherin knockdown experiments
After isolation of K8::Cre-ER;R26::LSL-rtTA;TRE-Twist1(mT/mG) organoids, 0.5–1 ml of a 10-ml organoid suspension was incubated with a cell dissociation reagent TrypLE Express Enzyme (1×; 12604013; Thermo Fisher Scientific) for 5–10 min to dissociate organoids into single cells. Trypan Blue Solution (152250061; Thermo Fisher Scientific) was added to the dissociated cell solution, and a hematocytometer was used to count the number of cells. The resulting number was used to calculate an average organoid size and the number of cells in our original organoid suspension. 1,000–1,200 organoids were plated in a nonadherent 96-well plate (655970; Greiner Bio-One), and organoids were allowed to settle for 1–1.5 h at 37°C. Three different clones of lentiviral SMA shRNA and a nontargeting control shRNA were selected from the transOMIC technologies shERWOOD-UltramiR shRNA library (at >108 TU/ml): ULTRA-3452234 Acta2, 5′-TGCTGTTGACAGTGAGCGATGACGCTGAAGTATCCGATAATAGTGAAGCCACAGATGTATTATCGGATACTTCAGCGTCAGTGCCTACTGCCTCGGA-3′, ULTRA-3201946 Acta2, 5′-TGCTGTTGACAGTGAGCGCCCTCATGAAGATCCTGACTGATAGTGAAGCCACAGATGTATCAGTCAGGATCTTCATGAGGTTGCCTACTGCCTCGGA-3′, ULTRA-3201947 Acta2, 5′-TGCTGTTGACAGTGAGCGCGACTCTCTTCCAGCCATCTTATAGTGAAGCCACAGATGTATAAGATGGCTGGAAGAGAGTCTTGCCTACTGCCTCGGA-3′, and TLNVU4420 nontargeting control shRNA, 5′-TGCTGTTGACAGTGAGCGAAGGCAGAAGTATGCAAAGCATTAGTGAAGCCACAGATGTAATGCTTTGCATACTTCTGCCTTGCCTACTGCCTCGGA-3′. Two different clones of lentiviral P-cadherin shRNA and Luciferase control shRNA were selected from the Sigma-Aldrich MISSION pLKO.1-puro library (at >106 TU/ml): Cdh3 shRNA 1 (TRCN0000430304), Cdh3 shRNA 2 (TRCN0000094414), and Luciferase shRNA (SHC007V). The oligonucleotide sequence used for Cdh3 shRNA#1 was 5′-CCGGCACGTATGACTTGCATCTTTCCTCGAGGAAAGATGCAAGTCATACGTGTTTTTTG-3′ and for Cdh3 shRNA 2 was 5′-CCGGTGAACTGGAGGTGAAGATTCGCTCGAGCGAATCTTCACCTCCAGTTCATTTTTTG-3′. The lentiviral particles were thawed on ice, mixed with 3 µl of ViroMag R/L nanoparticles (RL40200; OZ Biosciences), and incubated at room temperature for 30 min. Most of the media was removed from the 96-well plate with the organoids, and the ViroMag/lentivirus mix was added. The organoids were transfected with lentiviral particles with an MOI of 7. Suspensions were well mixed to disperse the organoids and minimize aggregation. One well with organoids was reserved as a puromycin control to evaluate killing efficiency. The 96-well plate was incubated on top of a magnetic plate (MF10000OZ; OZ Biosciences) for 1.5 h at 37°C, and then the plate was taken off the magnetic plate and incubated overnight at 37°C. On day 2, 200 µl prewarmed organoid medium with 2.5 nM FGF2 was added to all the wells, the plate was allowed to settle for 1 h, ∼200 µl media was removed, another 200 µl fresh FGF2 organoid medium was added, and the suspensions were mixed to redisperse the organoids. On day 4, 200 µl media was removed from all wells, and 200 µl organoid medium with 2.5 nM FGF2 and 2 µg/ml puromycin was added to each well. The efficiency of puromycin selection was evaluated by adding 2 µg/ml puromycin to noninfected organoids. On day 7, the organoids surviving puromycin selection were harvested for 3D culture.
FACS to purify mammary gland cell populations
For each experiment, organoids were isolated from CMV::rtTA;TRE-Twist1;mT/mG;K14::Actin-GFP, mT/mG;K14::Actin-GFP, C31-T-Ag;mT/mG, and MMTV::PyMT;βactin-CFP mice. For CMV::rtTA;TRE-Twist1;mT/mG;K14::Actin-GFP and mT/mG;K14::Actin-GFP mouse lines, three to five mice were used per experiment, and the epithelial tissue was pooled together. For the C31-T-Ag;mT/mG and MMTV::PyMT;βactin-CFP mouse lines, a single mouse was used for each experiment. Isolated organoids were incubated with 5 ml PBS at room temperature for 5 min followed by a 5-min centrifugation at 1,500 rpm. Organoid pellets were resuspended in a cell dissociation reagent TrypLE Express Enzyme (1×; 12604013; Thermo Fisher Scientific) and gently pipetted for 5–10 min at room temperature until dissociated into single cells. The dissociation reagent was neutralized by addition of 10 ml of 5% FBS organoid medium. The cells were centrifuged for 10 min at 1,500 rpm, and the pellets were resuspended in 2.5 nM FGF2 organoid medium. The resulting cell suspensions were filtered through 100-µm cell strainers (352235; Corning) and sorted on a Beckman Coulter MoFlo Cytometer. The cellular distributions were gated based on their endogenous fluorescence reporters (GFP+, GFP+/mT+, and mT+). Dead cells were excluded by propidium iodide fluorescence. The sorted cells were resuspended in 2.5 nM FGF2 organoid medium and plated in a nonadherent U-bottom 96-well plate (655970; Greiner Bio One) or a nonadherent flat bottom 96-well plate (655970; Greiner Bio-One). The number of luminal cells was held constant between wells within an experiment but varied between experiments based on FACS yield. A typical experiment would have 25,000 luminal cells and varying ratios of myoepithelial cells, resulting in a total cell number per well of between 50,000 and 100,000 cells. Cells were allowed to aggregate overnight at 37°C. The next day, the clusters formed in each well were collected and cultured in Matrigel or collagen I as described above. The clusters were imaged by confocal microscopy to confirm the presence of the corresponding fluorescence reporters as well as to examine the architecture of the overall cluster. The clusters were imaged by DIC microscopy on the first and last day of culture, and the number of disseminated cells per cluster was scored using ImageJ software.
Protein isolation
To dissolve Matrigel and isolate protein from organoids, gels were placed into BSA-coated tubes and incubated in 1× Protease-Phosphatase Inhibitor stock at 4°C for 30 min. 1× of Protease-Phosphatase Inhibitor stock had final concentrations of 10% of a Roche Complete Mini tablet (11836153001) and 10% of a Roche PhosSTOP tablet (4906837001). The tubes containing dissolved gels were centrifuged at 400 rcf for 5 min at 4°C. The pellets were resuspended and washed twice with cold 1× PBS to remove remaining medium and Matrigel and centrifuged at 400 rcf for 5 min at 4°C in between washes. The lysis buffer for protein isolation was prepared as following: 5% glycerol (G5516; Sigma-Aldrich), 1× RIPA buffer (20–188; EMD Millipore), 0.1% SDS, and 1× of Protease-Phosphatase Inhibitor stock. Lysis buffer was added to pellets and left on ice for 20 min. The samples were centrifuged for 10 min at 18,000 rcf at 4°C. The lysates were transferred to prechilled tubes and stored at −80°C.
Western blotting
Protein lysates were thawed on ice along with the BSA standard solutions (0–2,000 µm/ml). Protein was quantified using a bicinchoninic assay (23227; Thermo Fisher Scientific). Standards were loaded in triplicate and samples in duplicate on a 96-well plate (3595; Corning). The plate was incubated for 30 min at 37°C, and the absorbance was measured at 562 nm using a Biotek Synergy Mx microplate reader. A linear best-fit line was used to calculate the protein concentrations of samples. Samples resuspended in SDS protein loading buffer (LB0100; Morganville Scientific) and β-mercaptoethanol (M3148; Sigma-Aldrich) were heated at 95°C for 5 min and loaded onto 4–15% Mini-PROTEAN TGX gels (456-1084; Bio-Rad Laboratories) along with a Chameleon Duo prestained ladder (928–6000; Li-Cor). The gels were run in 1× TGS running buffer (161-0772; Bio-Rad Laboratories) at 120 V for ∼1.5 h. Gels were transferred onto hydrophobic polyvinylidene difluoride membranes (IPFL07810; EMD Millipore) in cold 1× TG-MeOH (161–0771; Bio-Rad Laboratories) at 100 V for 1 h at 4°C. Membranes were blocked in Odyssey Blocking Buffer (TBS) for 1–1.5 h. Primary antibodies were prepared in 50% Odyssey Blocking Buffer (TBS)/50% TBS-T and incubated at 4°C overnight on an orbital shaker. Primary antibodies used were mouse anti-Twist1 (sc-81417; 1:100; Santa Cruz Biotechnology, Inc.) and rabbit anti-actin (A2103; 1:1,000; Sigma-Aldrich). The next day, the membranes were washed three times for 5 min each in TBS-T. Secondary antibodies were incubated in 50% Odyssey Blocking Buffer (TBS)/50% TBS-T for 1 h at room temperature on an orbital shaker. Secondary antibodies used were IRDye 800 CW and 680RD (1:10,000; Li-Cor). The blots were washed 2 times for 5 min each in TBS-T and one time for 10 min in TBS and scanned on a Li-Cor Odyssey CLx.
IF staining
Organoid-containing Matrigel and collagen I gels were fixed in 4% paraformaldehyde for 10 min at 37°C. The fixed gels were either stored for further staining in PBS at 4°C or embedded in optimal cutting temperature compound (OCT; Sakura) and frozen at −80°C. OCT blocks were cut into 50-µm sections on a cryostat at −20°C. For antibody staining, sections were thawed and rinsed twice in 1× PBS for 10 min to remove OCT. Both fixed gels and sections were permeabilized with 0.5% Triton X-100/PBS for 1 h, rinsed three times with PBS, blocked for 1–2 h with 10% FBS/1% BSA/PBS, incubated with primary antibodies in 1% FBS/1% BSA/PBS overnight at 4°C at the dilutions listed below, and rinsed three times with PBS for 15 min. Secondary antibodies diluted in 1% FBS/1% BSA/PBS were incubated for 4–6 h at room temperature or overnight at 4°C. Gels were rinsed three times with PBS for 15 min and were ready for imaging. The slides were rinsed three times in PBS for 15 min, mounted with Fluoromount (F4680; Sigma-Aldrich), and sealed with coverslips.
Primaries antibodies used and their dilutions were as follows: rabbit anti–cytokeratin 14 (PRB-155P; 1:500; Covance), rat anti–cytokeratin 8 (TROMA-I; 1:100; Developmental Studies Hybridoma Bank), mouse anti–αSMA (A5228; 1:250; Sigma-Aldrich; and ab7817, 1:250; Abcam), mouse anti-Twist1 (sc-81417, 1:50; Santa Cruz Biotechnology, Inc.), rat anti–P-cadherin (clone PCD-1, 1:50; a gift from M. Takeichi, RIKEN Center for Developmental Biology, Kobe, Japan), and rabbit anti–Laminin-332 (1:500; gifts from P. Marinkovich, Stanford University, Stanford, CA, and M. Aumailley, University of Cologne, Cologne, Germany). Secondary antibodies used were coupled to Alexa Fluor 405, 488, 568, or 647 (1:150; Invitrogen). F-actin–positive cell membranes were stained with Alexa Fluor phalloidin (1:100; Thermo Fisher Scientific), and nuclei were stained with DAPI (D3571; 1:1,000; Thermo Fisher Scientific).
Quantification of SMA and P-cadherin protein levels in SMA and P-cadherin knockdown organoids
SMA and P-cadherin knockdown and control organoids cultured in Matrigel were fixed, stained with anti–αSMA, anti–P-cadherin, and anti–cytokeratin 14 antibodies, and imaged on the confocal microscope. Imaris was used to adjust brightness of the images in each channel, and to export individual TIFF files. The mean intensity of IF staining for SMA, P-cadherin, and K14 antibodies was measured in ImageJ. K14 protein levels were used as a control for SMA and P-cadherin protein levels in control and knockdown organoids. Prism was used to plot all the final values and perform the statistical analysis.
Quantification of Laminin-332 protein levels in SMA knockdown organoids
SMA knockdown and control organoids cultured in Matrigel were fixed, stained with anti–αSMA and anti–Laminin-332, and imaged on the confocal microscope. Imaris was used to adjust brightness of the images in each channel, and to export individual TIFF files. The mean intensity of IF staining for Laminin-332 antibody was measured in ImageJ. Prism was used to plot all the final values and perform the statistical analysis.
Online supplemental material
Fig. S1 shows the myoepithelium restraining a luminal Twist1+ cell from escaping in the Ubiquitous-Twist1 model and cell-autonomous Twist1 dissemination in the Myoepithelial-Twist1 model. Fig. S2 demonstrates that luminal-specific Twist1 expression leads to low dissemination and is increased by P-cadherin knockdown. Fig. S3 demonstrates how myoepithelial cells suppress invasion and dissemination of PyMT tumor cells. Videos are 3D confocal time-lapse videos. Videos 1 and 2 show luminal cell escape and cell restraint, respectively, and correspond with the still images in Fig. 2 (C and D). Videos 3, 4, and 5 represent additional examples of the myoepithelial dynamic barrier function described in Fig. 2. Video 6 shows luminal cell dissemination past SMA-depleted myoepithelium and corresponds with still images in Fig. 4 I. Video 7 shows myoepithelial cells limiting invasion of basal tumor cells and corresponds with still images in Fig. 5 D. Video 8 shows myoepithelial cells recapturing disseminated basal tumor cells and corresponds with still images in Fig. 5 E.
Supplementary Material
Acknowledgments
We thank members of the Ewald laboratory for helpful discussions. We thank Hao Zhang of The Johns Hopkins School of Public Health Flow Cytometry Core Facility for assistance with FACS and Dr. Masatoshi Takeichi for the gift of P-cadherin antisera.
A.J. Ewald received support for this project through grants from the Breast Cancer Research Foundation/Pink Agenda (BCRF-16-048 and BCRF-17-048), the Metastatic Breast Cancer Network, the National Institutes of Health/National Cancer Institute (U01CA217846 and U54CA2101732), and the American Cancer Society (RSG-12-141-01-CSM).
The authors declare no competing financial interests.
Author contributions: Conceptualization: O.K. Sirka, E.R. Shamir, A.J. Ewald; Methodology: O.K. Sirka, E.R. Shamir, and A.J. Ewald; Validation: O.K. Sirka, E.R. Shamir, and A.J. Ewald; Formal analysis: O.K. Sirka, E.R. Shamir, and A.J. Ewald; Investigation: O.K. Sirka and E.R. Shamir; Writing: O.K. Sirka, E.R. Shamir, and A.J. Ewald; Visualization: O.K. Sirka and E.R. Shamir; Supervision: A.J. Ewald; Funding acquisition: A.J. Ewald.
References
- Adriance M.C., Inman J.L., Petersen O.W., and Bissell M.J.. 2005. Myoepithelial cells: good fences make good neighbors. Breast Cancer Res. 7:190–197. 10.1186/bcr1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsky S.H., and Karlin N.J.. 2005. Myoepithelial cells: autocrine and paracrine suppressors of breast cancer progression. J. Mammary Gland Biol. Neoplasia. 10:249–260. 10.1007/s10911-005-9585-5 [DOI] [PubMed] [Google Scholar]
- Belteki G., Haigh J., Kabacs N., Haigh K., Sison K., Costantini F., Whitsett J., Quaggin S.E., and Nagy A.. 2005. Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res. 33:e51 10.1093/nar/gni051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casasent A.K., Schalck A., Gao R., Sei E., Long A., Pangburn W., Casasent T., Meric-Bernstam F., Edgerton M.E., and Navin N.E.. 2018. Multiclonal Invasion in Breast Tumors Identified by Topographic Single Cell Sequencing. Cell. 172:205–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerchiari A.E., Garbe J.C., Jee N.Y., Todhunter M.E., Broaders K.E., Peehl D.M., Desai T.A., LaBarge M.A., Thomson M., and Gartner Z.J.. 2015. A strategy for tissue self-organization that is robust to cellular heterogeneity and plasticity. Proc. Natl. Acad. Sci. USA. 112:2287–2292. 10.1073/pnas.1410776112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.J., Gabrielson E., Werb Z., and Ewald A.J.. 2013. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell. 155:1639–1651. 10.1016/j.cell.2013.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deugnier M.A., Teulière J., Faraldo M.M., Thiery J.P., and Glukhova M.A.. 2002. The importance of being a myoepithelial cell. Breast Cancer Res. 4:224–230. 10.1186/bcr459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald A.J., Brenot A., Duong M., Chan B.S., and Werb Z.. 2008. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev. Cell. 14:570–581. 10.1016/j.devcel.2008.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraldo M.M., Teulière J., Deugnier M.-A., Taddei-De La Hosseraye I., Thiery J.P., and Glukhova M.A.. 2005. Myoepithelial cells in the control of mammary development and tumorigenesis: data from genetically modified mice. J. Mammary Gland Biol. Neoplasia. 10:211–219. 10.1007/s10911-005-9582-8 [DOI] [PubMed] [Google Scholar]
- Gudjonsson T., Adriance M.C., Sternlicht M.D., Petersen O.W., and Bissell M.J.. 2005. Myoepithelial cells: their origin and function in breast morphogenesis and neoplasia. J. Mammary Gland Biol. Neoplasia. 10:261–272. 10.1007/s10911-005-9586-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusterson B.A., Warburton M.J., Mitchell D., Ellison M., Neville A.M., and Rudland P.S.. 1982. Distribution of myoepithelial cells and basement membrane proteins in the normal breast and in benign and malignant breast diseases. Cancer Res. 42:4763–4770. [PubMed] [Google Scholar]
- Guy C.T., Cardiff R.D., and Muller W.J.. 1992. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell. Biol. 12:954–961. 10.1128/MCB.12.3.954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaksma C.J., Schwartz R.J., and Tomasek J.J.. 2011. Myoepithelial cell contraction and milk ejection are impaired in mammary glands of mice lacking smooth muscle alpha-actin. Biol. Reprod. 85:13–21. 10.1095/biolreprod.110.090639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilson J.B., Schnitt S.J., and Collins L.C.. 2009. Phenotypic alterations in ductal carcinoma in situ-associated myoepithelial cells: biologic and diagnostic implications. Am. J. Surg. Pathol. 33:227–232. 10.1097/PAS.0b013e318180431d [DOI] [PubMed] [Google Scholar]
- Hu M., Yao J., Carroll D.K., Weremowicz S., Chen H., Carrasco D., Richardson A., Violette S., Nikolskaya T., Nikolsky Y., et al. 2008. Regulation of in situ to invasive breast carcinoma transition. Cancer Cell. 13:394–406. 10.1016/j.ccr.2008.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedrin D., Gligorijevic B., Wyckoff J., Verkhusha V.V., Condeelis J., Segall J.E., and van Rheenen J.. 2008. Intravital imaging of metastatic behavior through a mammary imaging window. Nat. Methods. 5:1019–1021. 10.1038/nmeth.1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller P.J., Arendt L.M., Skibinski A., Logvinenko T., Klebba I., Dong S., Smith A.E., Prat A., Perou C.M., Gilmore H., et al. 2012. Defining the cellular precursors to human breast cancer. Proc. Natl. Acad. Sci. USA. 109:2772–2777. 10.1073/pnas.1017626108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott S.R.V., Maceli A., Erard N., Chang K., Marran K., Zhou X., Gordon A., Demerdash O.E., Wagenblast E., Kim S., et al. 2014. A computational algorithm to predict shRNA potency. Mol. Cell. 56:796–807. 10.1016/j.molcel.2014.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerwill M.F. 2004. Current practical applications of diagnostic immunohistochemistry in breast pathology. Am. J. Surg. Pathol. 28:1076–1091. 10.1097/01.pas.0000126780.10029.f0 [DOI] [PubMed] [Google Scholar]
- Ma X.-J., Salunga R., Tuggle J.T., Gaudet J., Enright E., McQuary P., Payette T., Pistone M., Stecker K., Zhang B.M., et al. 2003. Gene expression profiles of human breast cancer progression. Proc. Natl. Acad. Sci. USA. 100:5974–5979. 10.1073/pnas.0931261100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maître J.L., Berthoumieux H., Krens S.F., Salbreux G., Jülicher F., Paluch E., and Heisenberg C.P.. 2012. Adhesion functions in cell sorting by mechanically coupling the cortices of adhering cells. Science. 338:253–256. 10.1126/science.1225399 [DOI] [PubMed] [Google Scholar]
- Man Y.-G., and Sang Q.-X.A.. 2004. The significance of focal myoepithelial cell layer disruptions in human breast tumor invasion: a paradigm shift from the “protease-centered” hypothesis. Exp. Cell Res. 301:103–118. 10.1016/j.yexcr.2004.08.037 [DOI] [PubMed] [Google Scholar]
- Man Y.G., Tai L., Barner R., Vang R., Saenger J.S., Shekitka K.M., Bratthauer G.L., Wheeler D.T., Liang C.Y., Vinh T.N., and Strauss B.L.. 2003. Cell clusters overlying focally disrupted mammary myoepithelial cell layers and adjacent cells within the same duct display different immunohistochemical and genetic features: implications for tumor progression and invasion. Breast Cancer Res. 5:R231–R241. 10.1186/bcr653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroulakou I.G., Anver M., Garrett L., and Green J.E.. 1994. Prostate and mammary adenocarcinoma in transgenic mice carrying a rat C3(1) simian virus 40 large tumor antigen fusion gene. Proc. Natl. Acad. Sci. USA. 91:11236–11240. 10.1073/pnas.91.23.11236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchor L., Molyneux G., Mackay A., Magnay F.A., Atienza M., Kendrick H., Nava-Rodrigues D., López-García M.A., Milanezi F., Greenow K., et al. 2014. Identification of cellular and genetic drivers of breast cancer heterogeneity in genetically engineered mouse tumour models. J. Pathol. 233:124–137. 10.1002/path.4345 [DOI] [PubMed] [Google Scholar]
- Molyneux G., Geyer F.C., Magnay F.A., McCarthy A., Kendrick H., Natrajan R., Mackay A., Grigoriadis A., Tutt A., Ashworth A., et al. 2010. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 7:403–417. 10.1016/j.stem.2010.07.010 [DOI] [PubMed] [Google Scholar]
- Mori H., Gjorevski N., Inman J.L., Bissell M.J., and Nelson C.M.. 2009. Self-organization of engineered epithelial tubules by differential cellular motility. Proc. Natl. Acad. Sci. USA. 106:14890–14895. 10.1073/pnas.0901269106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar M.D., Tasic B., Miyamichi K., Li L., and Luo L.. 2007. A global double-fluorescent Cre reporter mouse. Genesis. 45:593–605. [DOI] [PubMed] [Google Scholar]
- Neumann N.M., Perrone M.C., Veldhuis J.H., Huebner R.J., Zhan H., Devreotes P.N., Brodland G.W., and Ewald A.J.. 2018. Coordination of Receptor Tyrosine Kinase Signaling and Interfacial Tension Dynamics Drives Radial Intercalation and Tube Elongation. Dev. Cell. 45:67–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H., Rendl M., and Fuchs E.. 2006. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell. 127:171–183. 10.1016/j.cell.2006.07.036 [DOI] [PubMed] [Google Scholar]
- Nguyen M., Lee M.C., Wang J.L., Tomlinson J.S., Shao Z.M., Alpaugh M.L., and Barsky S.H.. 2000. The human myoepithelial cell displays a multifaceted anti-angiogenic phenotype. Oncogene. 19:3449–3459. 10.1038/sj.onc.1203677 [DOI] [PubMed] [Google Scholar]
- Nguyen-Ngoc K.-V., Shamir E.R., Huebner R.J., Beck J.N., Cheung K.J., and Ewald A.J.. 2015. 3D culture assays of murine mammary branching morphogenesis and epithelial invasion. Methods Mol. Biol. 1189:135–162. 10.1007/978-1-4939-1164-6_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K. 2010. Molecular markers for the diagnosis and management of ductal carcinoma in situ. J. Natl. Cancer Inst. Monogr. 2010:210–213. 10.1093/jncimonographs/lgq019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K., and Hu M.. 2005. Do myoepithelial cells hold the key for breast tumor progression? J. Mammary Gland Biol. Neoplasia. 10:231–247. 10.1007/s10911-005-9584-6 [DOI] [PubMed] [Google Scholar]
- Proia T.A., Keller P.J., Gupta P.B., Klebba I., Jones A.D., Sedic M., Gilmore H., Tung N., Naber S.P., Schnitt S., et al. 2011. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell. 8:149–163. 10.1016/j.stem.2010.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E. 2007. Illuminating the metastatic process. Nat. Rev. Cancer. 7:737–749. 10.1038/nrc2229 [DOI] [PubMed] [Google Scholar]
- Shamir E.R., Pappalardo E., Jorgens D.M., Coutinho K., Tsai W.-T., Aziz K., Auer M., Tran P.T., Bader J.S., and Ewald A.J.. 2014. Twist1-induced dissemination preserves epithelial identity and requires E-cadherin. J. Cell Biol. 204:839–856. 10.1083/jcb.201306088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z.-M., Nguyen M., Alpaugh M.L., O’Connell J.T., and Barsky S.H.. 1998. The human myoepithelial cell exerts antiproliferative effects on breast carcinoma cells characterized by p21WAF1/CIP1 induction, G2/M arrest, and apoptosis. Exp. Cell Res. 241:394–403. 10.1006/excr.1998.4066 [DOI] [PubMed] [Google Scholar]
- Sternlicht M.D., Kedeshian P., Shao Z.M., Safarians S., and Barsky S.H.. 1997. The human myoepithelial cell is a natural tumor suppressor. Clin. Cancer Res. 3:1949–1958. [PubMed] [Google Scholar]
- Stuurman N., Edelstein A.D., Amodaj N., Hoover K.H., and Vale R.D.. 2010. Computer Control of Microscopes using μManager. Curr. Protoc. Mol. Biol. 14:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L., van Bragt M.P., and Li Z.. 2015. A Long-Lived Luminal Subpopulation Enriched with Alveolar Progenitors Serves as Cellular Origin of Heterogeneous Mammary Tumors. Stem Cell Reports. 5:60–74. 10.1016/j.stemcr.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran P.T., Shroff E.H., Burns T.F., Thiyagarajan S., Das S.T., Zabuawala T., Chen J., Cho Y.-J., Luong R., Tamayo P., et al. 2012. Twist1 suppresses senescence programs and thereby accelerates and maintains mutant Kras-induced lung tumorigenesis. PLoS Genet. 8:e1002650 10.1371/journal.pgen.1002650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaezi A., Bauer C., Vasioukhin V., and Fuchs E.. 2002. Actin cable dynamics and Rho/Rock orchestrate a polarized cytoskeletal architecture in the early steps of assembling a stratified epithelium. Dev. Cell. 3:367–381. 10.1016/S1534-5807(02)00259-9 [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A., Rocha A.S., Ousset M., Beck B., Bouvencourt G., Rock J., Sharma N., Dekoninck S., and Blanpain C.. 2011. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 479:189–193. 10.1038/nature10573 [DOI] [PubMed] [Google Scholar]
- Werling R.W., Hwang H., Yaziji H., and Gown A.M.. 2003. Immunohistochemical distinction of invasive from noninvasive breast lesions: a comparative study of p63 versus calponin and smooth muscle myosin heavy chain. Am. J. Surg. Pathol. 27:82–90. 10.1097/00000478-200301000-00009 [DOI] [PubMed] [Google Scholar]
- Weymouth N., Shi Z., and Rockey D.C.. 2012. Actin isoform specificity is required for the maintenance of lactation. Dev. Biol. 363:1–14. 10.1016/j.ydbio.2011.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Mani S.A., Donaher J.L., Ramaswamy S., Itzykson R.A., Come C., Savagner P., Gitelman I., Richardson A., and Weinberg R.A.. 2004. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 117:927–939. 10.1016/j.cell.2004.06.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.