The SAGA coactivator complex and the nuclear pore–associated TREX-2 complex couple transcription with mRNA export. Evangelista et al. identify a novel interplay between TREX-2 and the deubiquitination module of SAGA that is necessary to maintain monoubiquitinated H2B levels required for efficient DNA repair through homologous recombination.
Abstract
DNA repair is critical to maintaining genome integrity, and its dysfunction can cause accumulation of unresolved damage that leads to genomic instability. The Spt–Ada–Gcn5 acetyltransferase (SAGA) coactivator complex and the nuclear pore–associated transcription and export complex 2 (TREX-2) couple transcription with mRNA export. In this study, we identify a novel interplay between human TREX-2 and the deubiquitination module (DUBm) of SAGA required for genome stability. We find that the scaffold subunit of TREX-2, GANP, positively regulates DNA repair through homologous recombination (HR). In contrast, DUBm adaptor subunits ENY2 and ATXNL3 are required to limit unscheduled HR. These opposite roles are achieved through monoubiquitinated histone H2B (H2Bub1). Interestingly, the activity of the DUBm of SAGA on H2Bub1 is dependent on the integrity of the TREX-2 complex. Thus, we describe the existence of a functional interaction between human TREX-2 and SAGA DUBm that is key to maintaining the H2B/HB2ub1 balance needed for efficient repair and HR.
Introduction
Eukaryotic cells are constantly exposed to DNA damage that can arise from exogenous and endogenous sources (Lindahl and Barnes, 2000; Barnes and Lindahl, 2004; Hoeijmakers, 2009). Among the plethora of different lesions that can be generated on the DNA molecules, double-strand breaks (DSBs) are among the most dangerous type of damage because if they are not successfully repaired, they can lead to chromosomal rearrangements (Pfeiffer et al., 2000; Iliakis et al., 2004). The DNA damage response (DDR) is a complex signaling cascade that is activated by DSBs that are exogenously generated by ionizing radiation or by chemicals and programmed DSBs that happen in well-defined locations in the genome, such as in meiosis or at the immunoglobulin class-switch recombination (CSR) locus (Dudley et al., 2005).
The main hallmark of the DDR is the phosphorylated form of the histone variant H2AX on serine 139 (γH2AX) deposited by the activated ATM kinase (Rogakou et al., 1998). Two main DNA repair subpathways exist: nonhomologous end joining (NHEJ) and homologous recombination (HR). Although the former ligates the extremities of the two broken ends and can often leave deletions at the site of the break, HR restores in a faithful way the genetic information at the site and in the vicinity of the break using the homologous chromatid as a template for repair, thus being by definition error free. HR takes place in S and G2 phases of the cell cycle (Orthwein et al., 2015), given the availability of the sister chromatids to be used as a template, whereas NHEJ can be active throughout the cell cycle. For HR to occur, the key step is the production of a 3′ single-stranded DNA (ssDNA) end through the process of resection (Liu and Huang, 2016) that depends on specific cell cycle–regulated proteins (Huertas et al., 2008; Falck et al., 2012; Wang et al., 2013).
The transcription and export complex-2 (TREX-2) complex is involved in mRNA export (Fischer et al., 2002, 2004; Köhler and Hurt, 2007; Faza et al., 2009; Wickramasinghe et al., 2010a,b; Umlauf et al., 2013). The human TREX-2 complex is composed of five subunits, germinal center–associated nuclear protein (GANP), ENY2, PCID2, Centrin2/3, and DSS1 (which in yeast are Sac3, Sus1, Thp1, Cdc31, and Sem1, respectively). Moreover, it has recently been shown that hTREX-2 stably associates with the nuclear pore complex (NPC) and that this interaction is crucial for its role in mRNA export (Umlauf et al., 2013).
The Spt–Ada–Gcn5 acetyltransferase (SAGA) coactivator complex and the TREX-2 complex are evolutionarily conserved among eukaryotes. SAGA is composed of 19 subunits organized in several functional modules with different activities and roles (Koutelou et al., 2010). The histone acetyltransferase module of SAGA is required at gene promoters to induce acetylation of Lysine (K) 9 of histone H3 (H3K9ac) and H3K14. This activity is catalyzed by the GCN5 enzyme (Grant et al., 1997). The deubiquitination module (DUBm) of SAGA is required to remove monoubiquitin from K120 of human H2B (H2Bub1) in gene bodies (Gavin et al., 2002; Sanders et al., 2002; Henry et al., 2003). These activities are required for the transcription of all active genes in both yeast (y) and human (h) cells (Bonnet et al., 2014; Baptista et al., 2017). In human cells, monoubiquitination of histone H2B is deposited by RNF20 and RNF40 (Zhu et al., 2005) and deubiquitinated by SAGA (Gavin et al., 2002; Sanders et al., 2002; Henry et al., 2003). Interestingly, H2B in mammalian cells was found to be monoubiquitinated in response to DNA damage and is necessary for efficient DSB repair (Moyal et al., 2011; Nakamura et al., 2011). Moreover, recent studies have highlighted the importance of the DUB module of SAGA during CSR in mouse B cells (Ramachandran et al., 2016; Li et al., 2018).
TREX-2 and SAGA share one subunit, ySus1/hENY2 (Rodríguez-Navarro et al., 2004). In TREX-2, ySus1/hENY2 binds to the scaffold protein ySac3/hGANP in two copies (Jani et al., 2012). In SAGA, hENY2 binds to ATXN7L3, and together with ATXN7 and the DUB enzyme, USP22, forms the DUBm (Zhang et al., 2008; Lang et al., 2011). Moreover, in human cells, ENY2 and ATXN7L3 form alternative DUBm complexes with the deubiquitinating enzymes USP27X, or USP51, which are both 70–80% identical to USP22. These alternative DUBm members lack ATXN7 and thus do not associate with SAGA. Nevertheless, they can deubiquitinate H2Bub1 in vitro and in vivo (Atanassov et al., 2016). It has been shown that the knockdown (KD) of ENY2 destabilizes hGANP in TREX-2 and ATXN7L3 in the SAGA DUBm (Umlauf et al., 2013; Li et al., 2016). Moreover, the activity of the SAGA DUBm and the above-mentioned alternative DUB modules strictly require the adaptors ENY2 and ATXN7L3 for their activity (Atanassov et al., 2016). For simplicity, hereafter the three SAGA-related DUBm-s will be called DUBm-s.
In yeast, the physical and functional interaction between TREX-2 and SAGA complexes was described and was suggested to connect ongoing transcription with the NPC for immediate RNA export in a process called gene gating (Blobel, 1985; Rodríguez-Navarro et al., 2004; Cabal et al., 2006; Chekanova et al., 2008; Pascual-García et al., 2008; García-Oliver et al., 2012). Destroying this connection at the export level has been shown to induce genome instability as yTREX-2 and yNucleoporins (Nup) mutants but not ySAGA mutants show hyperrecombination phenotype (Gallardo and Aguilera, 2001; Gallardo et al., 2003; Rondón et al., 2003; González-Aguilera et al., 2008).
In mammalian cells, the involvement of Nups and associated proteins in the DDR has not been connected with RNA export or transcription defects (Lemaître et al., 2012; Duheron et al., 2017; Mackay et al., 2017). Moreover, the interplay of mammalian TREX-2 or the SAGA DUBm has not been studied in the context of DNA repair.
In this study, we have investigated the role of TREX-2/GANP and the SAGA DUBm and related DUBm-s in DSB repair. We find that loss of the scaffold subunit of TREX-2 (GANP) results in DNA repair deficiency by HR. This GANP-dependent HR defect is accompanied by decreased resection and is mediated by down-regulation of H2Bub1 levels. In contrast, down-regulation of the DUB activities (by KD of either ENY2 or ATXN7L3) results in up-regulation of HR efficiency and a parallel increase in H2Bub1 levels. Interestingly, concomitant depletion of TREX-2 and the DUB activity restores the HR pathway along with RAD51 recruitment to DNA damage sites, and this results in normal H2Bub1 levels upon DNA damage. These results together demonstrate a functional cross-talk between human TREX-2 and the USP22-related DUBm-s that are important for correct DSB repair during HR.
Results
Depletion of the TREX-2 scaffold protein GANP affects the efficiency of DNA repair
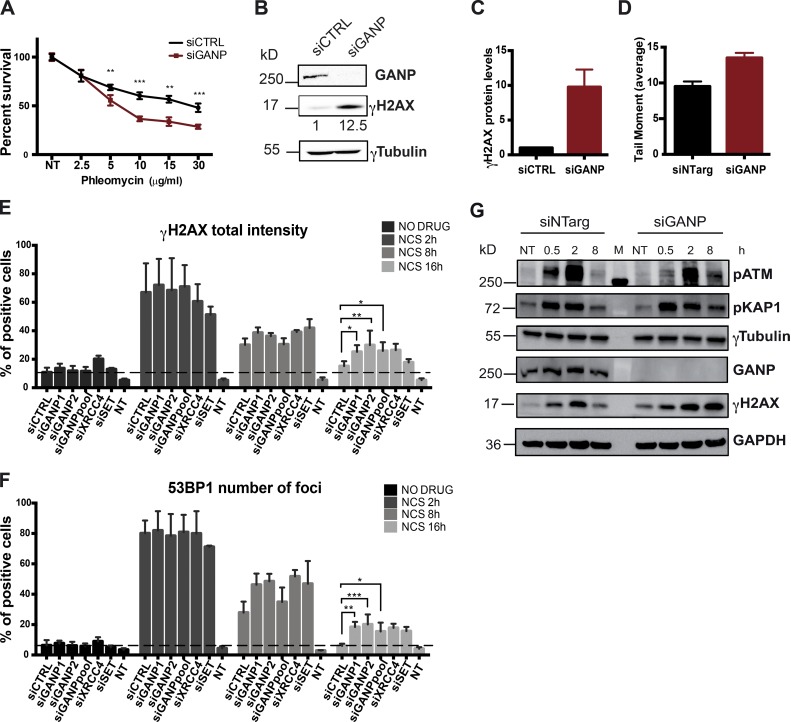
To investigate the role of the TREX-2 complex in genome stability in mammalian cells, we performed clonogenic survival assay in HeLa cells treated with the radiomimetic drug phleomycin in combination with siRNA-mediated KD of GANP. Interestingly, cells depleted of GANP displayed a significant increase in sensitivity to all concentrations of the drug compared with control cells (Fig. 1 A). This result suggests a GANP-dependent defect in either the activation of DDR or repair of DSBs. The efficient GANP depletion by siRNA was monitored by Western blot (Fig. 1 B). Moreover, GANP depletion leads to substantially elevated γH2AX levels (Fig. 1, B and C).
Figure 1.
Depletion of GANP affects DNA repair without affecting the activation of the DDR. (A) Clonogenic survival assay in HeLa cells depleted of GANP and control cells treated with increasing concentration of phleomycin. The graph represents the average of three independent experiments with three technical replicates each, where the number of colonies for each concentration was normalized to the respective untreated condition. Statistical significance was calculated using the Mann–Whitney test (ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001). (B) Western blot analysis of GANP and γH2Ax of RIPA extracts obtained from GANP-depleted cells and control cells. Numbers indicate γH2Ax levels quantified using ImageJ using γTubulin as loading control. (C) Graph represents the average of γH2Ax levels quantified using ImageJ in three independent experiments (n = 3). (D) DNA damage measured with the neutral comet assay of GANP-depleted cells and control cells. The number of breaks are represented in tail moment as the tail length normalized to the percentage of DNA in the tail. Graph represents the average of two independent experiments with n ≥ 100 cells. Error bars represent SEM. (E and F) High-throughput screening performed to investigate the effect of GANP KD in DNA repair in HeLa cells. Graphs represent the percentage of positive cells for γH2AX intensity (E) and 53BP1 foci (F) compared with the nontreated (NT) cells. The x axis shows the corresponding siRNA used. Two different siRNAs for GANP were used (siGANP1 and siGANP2); unless otherwise specified in all the other experiments siGANP2 was used and is labeled siGANP. Black and gray boxes represent untreated cells (NO DRUG), and the time (in hours) of recovery after NCS treatment at 50 ng/ml for 15 min. A positive cell is defined as a cell having γH2AX intensity or number of 53BP1 foci higher than an arbitrary threshold defined on the 5% of nontreated cells. Graphs represent the average of two independent experiments, each of them performed in three technical replicates, with SD. Per each condition, n > 1,000 cells. Statistical significance was calculated using the ANOVA (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001). (G) Western blot analysis of total cell lysate obtained by RIPA extraction from cells treated with GANP siRNA and control cells. Hours represent the time of repair after NCS treatment. For details about statistical analysis, refer to the Statistical analysis section of Materials and methods. siCTRL, control siRNA; siNTarg, nontargeting siRNA (see Table 1).
To test whether GANP depletion induces spontaneous DNA damage, we performed a COMET assay on GANP-depleted cells in the absence of drug. Interestingly, GANP depletion leads to increased comet tails that are indicative of increased DNA breaks (Fig. 1 D), further supporting the notion that GANP is key in maintaining genome stability as its absence leads to persistent DNA breaks in the absence of damaging agents.
To further investigate the role of GANP/TREX-2 in repairing DSBs, we monitored the kinetics of repair in HeLa and human telomerase reverse transcriptase (hTERT)-immortalized RPE1 control cells or cells depleted for GANP after DNA damage induction. We treated the cells with the antitumoral antibiotic neocarzinostatin (NCS), which creates ssDNA and dsDNA breaks, and we followed the appearance and disappearance of γH2AX foci and accumulation of the key DDR regulator 53BP1. As positive controls in our assay, we used depletion of XRCC4, involved in the last steps of NHEJ (Li et al., 1995), and depletion of the oncogene SET (Kalousi et al., 2015). As expected, cells treated with control siRNA showed a progressive reduction of both DDR marks (γH2AX and 53BP1) at 8 and 16 h of repair (Fig. 1, E and F). However, KD of GANP delayed the disappearance of both γH2AX and 53BP1 foci after 8 and 16 h of repair in HeLa and RPE1 cells, equivalent to the delay observed upon depletion of XRCC4 and SET (Fig. 1, E and F; and Fig. S1 A).
Next, we analyzed whether the depletion of GANP would affect the DDR activation in NCS-treated cells. Our analyses show that in cells depleted for GANP, the initial recruitment of 53BP1 was not affected (Fig. 1 F), and activation of ATM, monitored by its autophosphorylation on Ser1981 or by the phosphorylation of its substrate KAP1, was comparable with control cells (Fig. 1 G). Interestingly, however, we find that in GANP-depleted cells phospho-ATM and γH2AX were persistent 8 h after NCS treatment, suggesting that GANP is involved in repair of exogenously inflicted DNA breaks (Fig. 1 G). As a consequence, GANP-depleted cells were also more sensitive to NCS, compared with controls (Fig. S1 B).
Because GANP/TREX-2 was shown to be located mainly at the inner side of the NPC (Umlauf et al., 2013), we wondered whether the remaining unrepaired breaks were accumulated at the periphery of the nucleus or were equally distributed between periphery and inner nucleus. Analysis of the raw intensity data for γH2AX showed no shift in the distribution of γH2AX in the two compartments between GANP-depleted cells and control cells or cells depleted of XRCC4, suggesting a more global effect (Fig. S1, C and D). These results suggest that although TREX-2 is predominantly localized at the nuclear pores, it has a rather global role in DSB repair.
Depletion of GANP/TREX-2 affects the efficiency of HR
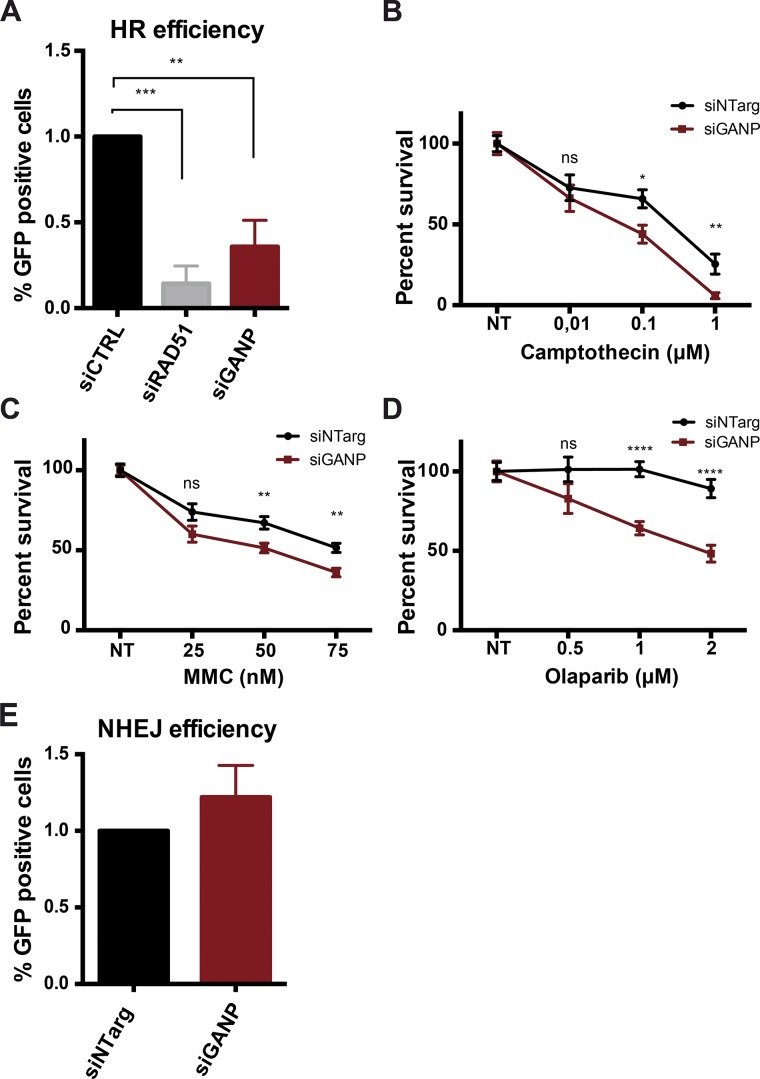
DSBs are repaired by two main repair pathways: NHEJ and HR. We showed that depletion of GANP results in unrepaired persistent breaks throughout the nucleus without affecting the early activation of the DDR, suggesting the involvement of GANP in repair. Thus, we next analyzed the involvement of GANP in the two main repair pathways. To assess the rate of HR, we used a U2OS cell line stably integrating a DR-GFP reporter (Mimori and Hardin, 1986; Weinstock et al., 2006), and to analyze NHEJ, the GCV6 human fibroblast cell line in which the coding frame between a GFP construct and the promoter is restored after the removal of a 34-bp sequence and subsequent rejoining of the two ends by NHEJ (Rass et al., 2009). Interestingly, depletion of GANP reduced the efficiency of HR by 60% (Fig. 2 A). The reduced HR efficiency in GANP-depleted cells was almost as strong as that caused by the depletion of RAD51, a key player in homology-directed repair (Fig. 2 A; Sung et al., 2003). To investigate whether a reduced HR efficiency could also be reflected by increased sensitivity to HR-inducing drugs, we performed clonogenic survival assay using camptothecin and mitomycin C (MMC) that induce DNA lesions in S-phase or lesions that rely in the activation of HR for repair, respectively (Liu et al., 2000). GANP-depleted cells were consistently more sensitive to camptothecin and MMC, confirming the result obtained in the DR-GFP assay (Fig. 2, B and C). HR-deficient cells are generally more sensitive to PARP inhibitors; thus we investigated the response of GANP-depleted cells to the PARP-inhibitor Olaparib. In good agreement, GANP-depleted cells were significantly more sensitive to Olaparib compared with controls (Fig. 2 D). HR is the preferred pathway in S and G2 phases of the cell cycle; thus a possible effect on cell cycle progression could explain the dramatic effect observed on HR efficiency. To verify this possibility, we analyzed cell cycle progression by either propidium iodide incorporation in U2OS cells or EdU incorporation in HeLa cells under GANP KD conditions. We did not observe any significant effect on cell cycle in both cellular systems depleted of GANP (Fig. S1, E and F), and GANP depletion did not affect RAD51 protein levels (Fig. S1 G). In contrast with the role of GANP in HR, we did not detect a significant change in NHEJ after GANP depletion (Fig. 2 E). These experiments together show that GANP is required for promoting HR.
Figure 2.
Depletion of GANP affects the efficiency of HR. (A) HR efficiency in GANP-depleted cells and control cells. The frequency of HR-mediated repair is analyzed by flow cytometry as the percentage of GFP-positive cells (see Materials and methods). Graph represents the merge of three independent experiments with SD (n = 3). RAD51 siRNA was used as a positive control for reduction of HR efficiency. Statistical significance was calculated using ANOVA (**, P ≤ 0.01; ***, P ≤ 0.001). (B–D) Clonogenic survival assay of HeLa cells depleted of GANP and control cells treated with increasing concentration of camptothecin (B), MMC (C), and Olaparib (D). Graphs represent the average of three independent experiments with three technical replicates each, where the number of colonies per each concentration was normalized to the respective untreated condition. Error bars represent SEM. Statistical significance was calculated using the Mann–Whitney test (ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01). (E) NHEJ efficiency in control cells and cells depleted of GANP. The frequency of NHEJ-mediated repair was analyzed by flow cytometry as the percentage of GFP-positive cells (see Materials and methods). Graph represents the mean of four independent experiments with error bars indicating SD. For details about statistical analysis, refer to the Statistical analysis section of Materials and methods. NT, nontreated cells; siCTRL, control siRNA; siNTarg, nontargeting siRNA (see Table 1).
Resection is decreased in the absence of GANP/TREX-2
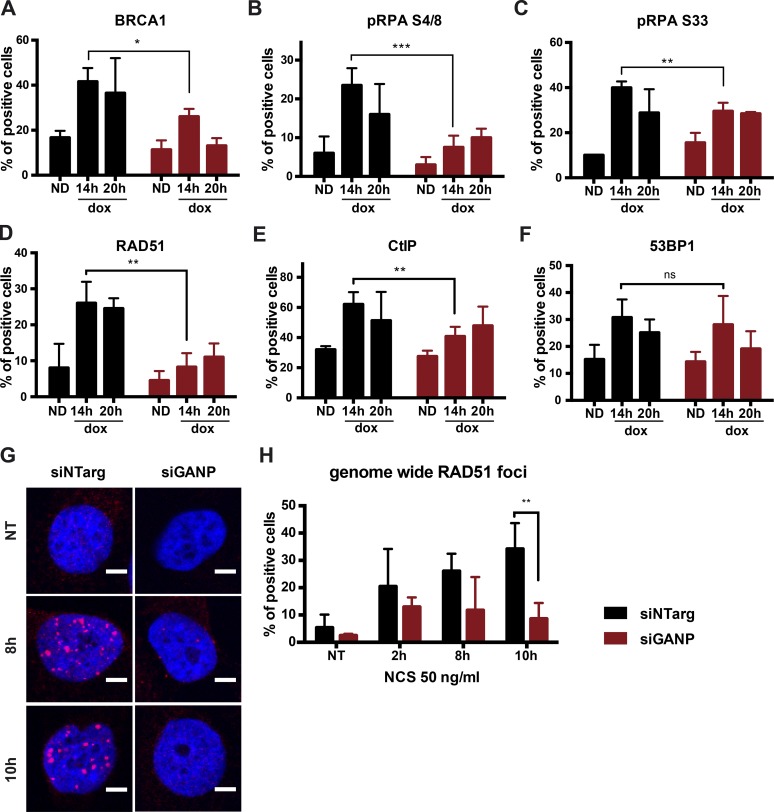
To dissect the steps of HR that are affected by GANP depletion and test whether the above HR defect is accompanied by a decreased end resection, we analyzed the recruitment of proteins involved in this process. To this end, we used U2OS cells stably integrating an ISceI site flanked by the Lac operator (LacO) array and stably expressing the LAC repressor (LacI) fused to GFP that allows visualization of the locus (Lemaître et al., 2014). Addition of doxycycline (dox) enables the expression of the ISceI enzyme and the induction of a single DSB at the lacO locus. Using microscopy, we analyzed the recruitment of a series of factors known to be involved in resection or which serve as markers of resection in human cells such as BRCA1, phospho-RPA, CtBP-interacting protein (CtIP), and RAD51 (Daley et al., 2015). In agreement with our above results, depletion of GANP dramatically reduced recruitment of BRCA1, phosphorylation of RPA at different phosphorylation sites (serine 4/8 and serine 33), and recruitment of RAD51, compared with control cells (Fig. 3, A–D; see Fig. S2 [A–D] for representative images). BRCA1 in association with CtIP and the Mre11–Rad50–Nbs1 complex forms the BRCA1 C complex that is required for the early steps of resection (Yu et al., 1998; Yun and Hiom, 2009). Thus, we also analyzed recruitment of CtIP to further investigate whether HR is affected at the initial stages of resection. Recruitment of CtIP was also affected in GANP-depleted cells, suggesting a defect in resection initiation (Figs. 3 E and S2 E). However, GANP depletion did not affect recruitment of the early DDR factor 53BP1 (Fig. 3 F). Efficient induction of breaks was monitored by γH2AX (Fig. S2, F and G). The defect in resection was also confirmed by the defective formation of RAD51 and RPA foci in HeLa cells treated with NCS and depleted of GANP with two different siRNAs (Fig. 3, G and H; and Fig. S2, H and I). As observed before, there was no difference in 53BP1 recruitment (Fig. S2 L) between GANP-depleted and control cells. All these experiments together indicate that GANP/TREX-2 is required for efficient HR, and that the recruitment of resection factors to DSBs and DNA-end resection is impaired in the absence of GANP.
Figure 3.
Resection is impaired in GANP-depleted cells. (A–F) Graphs represent time course of the percentage of colocalization of resection factors with the LacO array after dox addition in U2OS 19 ptight 13 cells stably expressing GFP-LacI (Lemaître et al., 2014). Cells were depleted of GANP or treated with control siRNA 48 h before dox addition at a final concentration of 1 µg/ml. Values represent the merge of at least three independent experiments with n ≥ 50 cells per condition. Error bars represent SD. Statistical significance was calculated using t test (ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001). ND, no dox. (G) Representative images of HeLa cells stained with DAPI (in blue) and RAD51 (in red). Hours represent the time of repair after NCS treatment for 15 min at 50 ng/ml. Bars, 5 µm. (H) Graphs represent the percentage of cells showing more than five RAD51 foci of four independent experiments with SD. For each experiment, 10 pictures were acquired using confocal microscope with more than 100 cells per condition. Statistical significance was calculated using the t test (**, P ≤ 0.01). For details about statistical analysis, refer to Statistical analysis paragraph in Materials and methods. NT, nontreated; siNTarg, nontargeting siRNA (see Table 1).
ENY2 and ATXN7L3 are required to avoid unscheduled HR
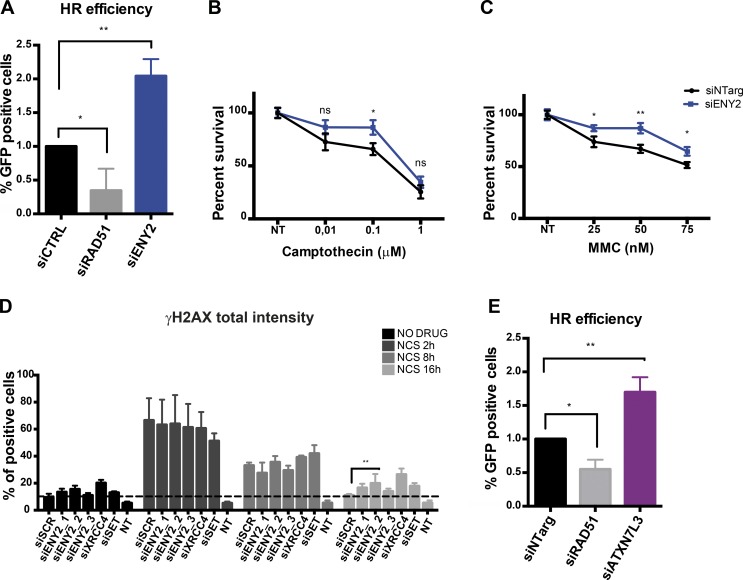
To further investigate the role of the human TREX-2 complex in genome stability, we tested whether depletion of another subunit of the complex would have the same effect on DNA repair. To this aim, we depleted ENY2 that associates with GANP in two copies (Jani et al., 2012). Surprisingly, in contrast with GANP depletion (Fig. 2 A), which decreases HR efficiency by about threefold, siRNA depletion of ENY2 with three different siRNAs increased HR efficiency twofold as monitored by the DR-GFP assay (Figs. 4 A and S2 M). Consistently with the significant increase in the HR efficiency, clonogenic survival assay in HeLa cell performed after ENY2 depletion showed an increased resistance of cells to camptothecin and MMC compared with control cells (Fig. 4, B and C), without affecting cell cycle progression (Fig. S3 A). These experiments suggest that ENY2 plays the opposite role compared with GANP and thus seems to be required to suppress HR.
Figure 4.
ENY2 and ATXN7L3 are required to avoid unscheduled HR. (A and E) Frequency of HR-mediated repair in either ENY2- (A) or ATXN7L3 (E)-depleted cells. Analysis was performed as described in Fig. 2 A. Graphs represent the average of three independent experiments with error bars indicating SD. Statistical significance was calculated using ANOVA (*, P ≤ 0.05; **, P ≤ 0.01). (B and C) Clonogenic survival assay of HeLa cells depleted of ENY2 and control cells treated with increasing concentration of camptothecin (B) and MMC (C). The graph represents the average of three independent experiments with three technical replicates each, where the number of colonies per each concentration was normalized to the respective untreated condition. Error bars represent SEM. Statistical significance was calculated using the Mann–Whitney test (ns, P > 0.05; *, P ≤ 0.05). These experiments were performed in parallel with the experiments in Fig. 2 (B–D); thus, the control is the same. (D) High-throughput screening was performed to investigate the effect of ENY2 KD in DNA repair in HeLa cells (for details, refer to Fig. 1 E). Statistical significance was calculated using ANOVA (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001). Three different siRNAs against ENY2 were used. Error bars represent SEM. Unless otherwise specified, in all other experiments, siENY2_2 was used and is labeled siENY2. For details about statistical analysis, refer to Statistical analysis paragraph in Materials and methods. NT, nontreated; siCTRL, control siRNA; siNTarg, nontargeting siRNA; siSCR, scrambled siRNA (see Table 1).
To understand whether breaks are efficiently repaired in the absence of ENY2, we analyzed the kinetics of repair in asynchronous HeLa and RPE1 cells treated with NCS using γH2AX as a marker. ENY2-depleted cells displayed persistent γH2AX at 16 h after damage induction compared with control cells (Figs. 4 C and S3 B). Moreover, ENY2-depleted cells, although being more resistant to camptothecin and MMC (S and G2 phase–specific damage), were more sensitive to DSBs induced with phleomycin compared with control cells (Fig. S3 C). These results suggest that the down-regulation of ENY2 creates conditions that would favor HR in an unscheduled way being potentially deleterious for the cells.
Interestingly, ENY2 and GANP depletion have opposite effects on DNA repair by HR. ENY2 is an integral component of TREX-2 and several USP22-related DUBm-s (see Introduction). To investigate whether the role of ENY2 in suppressing HR can be ascribed to the DUBm-s or to TREX-2, we performed the DR-GFP assay in cells depleted of another key subunit of the DUBm-s, ATXN7L3. ATXN7L3 depletion had the same effect as depletion of ENY2, suggesting that ENY2 suppresses HR as component of the USP22-related DUB modules (Fig. 4 E; see also Fig. 4 A). These experiments suggest that hTREX-2 and the USP22-related DUB modules play opposite roles in the DNA repair process.
TREX-2 depletion affects the global cellular levels of H2Bub1
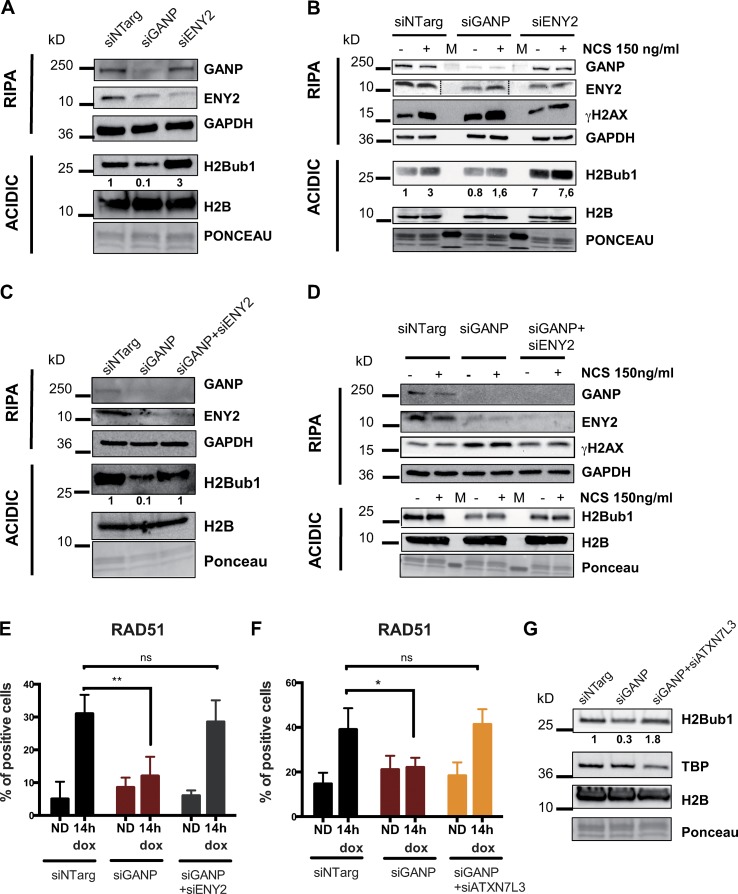
The USP22-related DUB modules are involved in the removal of monoubiquitin from H2Bub1. Although the recruitment of RNF20/40 catalyzing the formation of H2Bub1 after DNA damage has been studied (Moyal et al., 2011; Nakamura et al., 2011; Shiloh et al., 2011), little is known about the importance of H2Bub1 deubiquitination and the balance between histone H2B and H2Bub1 in the response to DNA damage. As formation of H2Bub1 is important for homology-directed repair (Nakamura et al., 2011), an extreme increase in H2Bub1 levels upon DUB depletion could explain unscheduled HR. Therefore, we wondered whether the opposite effects of TREX-2 and DUBm-s in homology-directed repair could be caused by different effects on H2Bub1 levels. We analyzed H2Bub1 levels in HeLa cells depleted of either GANP or ENY2. Interestingly, GANP-depleted cells showed decreased global H2Bub1 (Figs. 5 A and S3 D). However, cells lacking ENY2 showed increased levels of H2Bub1 as a consequence of the disruption of the DUB modules (Figs. 5 A and S3 D; see Introduction), further substantiating the opposite effects of GANP and ENY2 seen in the HR assay. Next, we treated cells with increasing concentrations of NCS. As shown previously (Moyal et al., 2011), the levels of H2Bub1 increased upon DNA damage in a NCS concentration–dependent manner (Fig. S3 E). In GANP-depleted cells, however, there is a minor increase at the level of H2Bub1 upon DNA damage compared with control cells (Fig. 5, B and D; and Fig. S3, F and G). ENY2-depleted cells, however, had elevated H2Bub1 levels even in the absence of DNA damage (Fig. 5, A and B; and Fig. S3 F). These results suggest that the opposite effects observed on the HR/DDR might be caused by the inverse influences of GANP in TREX-2, or ENY2 in the USP22-related DUBm-s on the histone H2Bub1 mark.
Figure 5.
GANP/TREX-2 affects HR through H2Bub1. (A–D and G) Western blot assays of total cell lysate (RIPA) or histone extracts (ACIDIC) in cells treated with indicated siRNAs. Membranes were blotted with depicted antibodies. For specific antibodies used, see Table 2. (E and F) Graphs represent time course of the percentage of colocalization of RAD51 with the LacO array (as described in Fig. 3) in siGANP conditions and codepletion of GANP and ENY2 (E) or GANP and ATXN7L3 (F). Values represent the merge of at least three independent experiments with n ≥ 50 cells per condition. Error bars represent SD. Statistical significance was calculated using ANOVA (ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01). Note that for clearer WB detection, GANP, ENY2, and γH2AX were not always detected on the same membranes due to either very large size differences (GANP and ENY2) or to very small size differences (ENY2 and γH2AX). ND, no dox; siNTarg, nontargeting siRNA (see Table 1).
TREX-2 is required to avoid excessive H2Bub1 deubiquitination by the USP22-related DUB modules in response to DNA damage
To investigate whether the observed decrease in H2Bub1 levels upon GANP depletion are the result of an increased DUB activity, we tested whether the concomitant depletion of the three USP22-related DUBm-s through ENY2 KD or ATXN7L3 KD would restore normal H2Bub1 levels. The codepletion of GANP and ENY2 or GANP and ATXN7L3 restored normal H2Bub1 levels compared with H2Bub1 levels in mock-siRNA-treated undamaged or damaged cells (Fig. 5, C, D, and G; and Fig. S3, G and H). These results together suggest that the TREX-2 complex can negatively regulate the deubiquitination of H2Bub1 and that if destabilized through GANP depletion, the activity of the DUB modules is enhanced. To further understand whether the negative effect of TREX-2 on H2Bub1 levels is important in DNA repair by HR, we analyzed the recruitment of RAD51 at the ISceI site in cells either depleted of GANP alone or codepleted of either GANP and ENY2 or GANP and ATXN7L3. Depletion of GANP alone dramatically reduced the number of cells recruiting RAD51 to the LacO array after IsceI cleavage (Figs. 3 D and 5, E and F). However, the additional depletion of ENY2 completely rescued the defect to control levels (Fig. 5 E), as did the depletion of ATXN7L3 (Fig. 5 F). Efficiency of double KD was monitored by RNA levels (Fig. S4, A and B). Because we were able to restore RAD51 recruitment at a single DSB, we wondered whether double depletion of GANP and the DUB module by depletion of either ENY2 or ATXN7L3 would also restore the sensitivity of GANP-depleted cells to HR-inducing drugs. Double KD of GANP and the DUB module subunits ENY2 or ATXN7L3 restored the cells’ sensitivity to both MMC and Olaparib (Fig. S4, C and D). Given the established role of SAGA in transcription elongation, we ruled out that the observed phenotypes would be due to a reduced level of key repair genes by analyzing their mRNAs and protein levels in control cells and cells depleted of GANP or ENY2 (Fig. S4, E and F). These results together indicate that TREX-2 integrity is required to avoid excessive H2Bub1 deubiquitination in response to DNA damage and that the misregulation of H2Bub1 global levels has an impact on HR.
TREX-2-DUB interplay is required to regulate H2Bub1 levels at damaged site
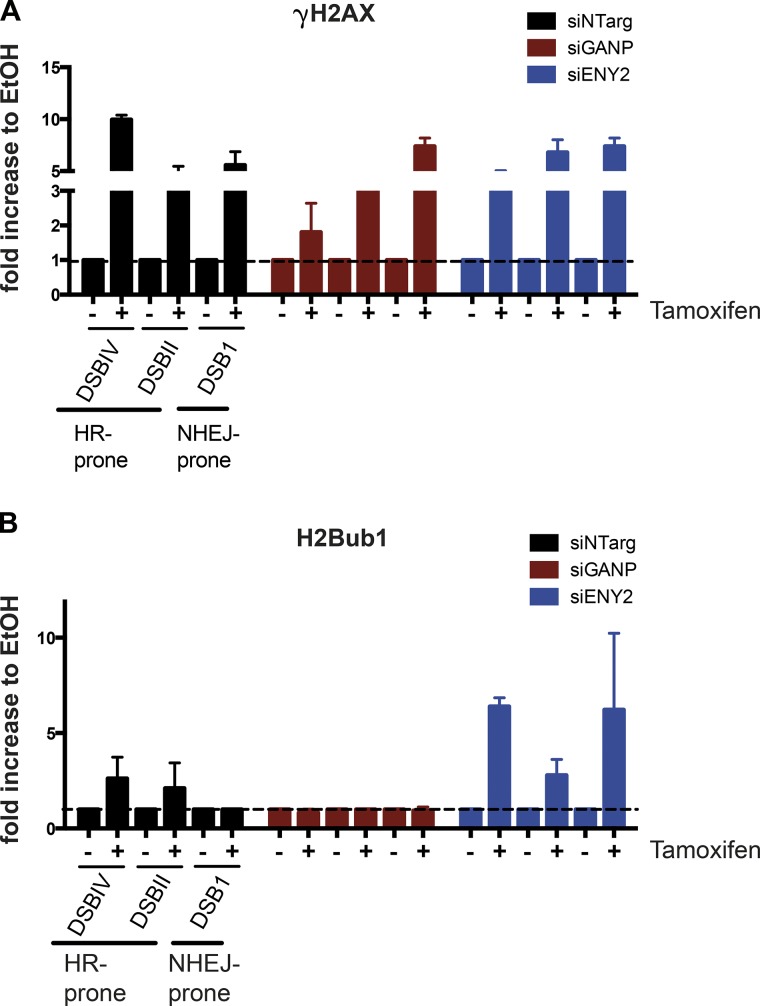
To understand whether the KD of GANP/TREX-2 and the DUBm-s affect H2Bub1 balance at damaged sites, we took advantage of the DSB Inducible via ASISI (DIvA) system in which the ASISI restriction enzyme is integrated in U2OS cells and its nuclear translocation can be induced with 4-OH-tamoxifen (Massip et al., 2010; Caron et al., 2012). This allows the rapid induction of ∼150 DSBs in the human genome at specific locations. Moreover, chromatin immunoprecipitation (IP; ChIP) analyses of HR and NHEJ specific factors showed a difference in the repair pathway for distinct DSBs accordingly to the chromatin landscape, allowing differentiation between HR-prone sites and NHEJ-prone sites (Aymard et al., 2014).
Using this system, we analyzed the presence of H2Bub1 by ChIP followed by quantitative PCR (qPCR) at a subset of damaged sites before and after cut induction. Efficient induction of breaks was monitored by γH2AX ChIP (Fig. 6 A) and by immunofluorescence using γH2AX- and 53BP1-specific antibodies (Fig. S4 G). Our anti-H2Bub1 ChIP-qPCR showed a modest increase of H2Bub1 at damaged sites after ASISI cut in control cells (Fig. 6 B) that was variable, depending on the analyzed sites. Interestingly, we could monitor an increase in H2Bub1 at HR-prone DSBs, whereas we did not detect a significant change in NHEJ-prone sites. However, in GANP-depleted cells, we did not detect any significant H2Bub1 at any of the analyzed sites (Fig. 6 B, red bars) before and after damage. In contrast, in ENY2-depleted cells H2Bub1 induction after damage was increased compared with control cells (Fig. 6 B, blue bars), and it was detected even at NHEJ-prone sites (Fig. 6 B, DSB1). Interestingly, in ENY2-depleted cells, we did not detect increased H2Bub1 levels at these specific ASISI sites before damage induction compared with control cells (not depicted; ChIP values were similar to no-antibody controls), suggesting that the DUBm-s are not operating at these regions before damage. These results suggest that TREX-2 and the SAGA-DUB are required to maintain balanced histone H2Bub1 levels at damaged sites.
Figure 6.
GANP/TREX2 and DUBs are required to maintain regulated H2Bub1 levels at damaged sites. (A and B) ChIP-qPCR analysis of γH2AX (A) and H2Bub1 (B) at ASISI sites in U2OS cells with and without 4-OH tamoxifen addition (see symbols on the x axes). The specific sites analyzed are shown below x axes and are differentiated in HR-prone and NHEJ-prone sites (Aymard et al., 2014). The y axes represent the fold enrichment compared with vehicle-treated cells. Before normalization with vehicle-treated samples the percentage of input was calculated and normalized to the no-antibody control. Graphs represent the average of technical qPCR duplicates of one representative experiments out of two biological replicates (n = 2), which gave comparable results. Error bars represent SD. siNTarg, nontargeting siRNA (see Table 1).
Discussion
In this study, we have described a new functional interaction of the TREX-2 complex with the SAGA-related DUBm-s in DNA repair in human cells. Importantly, our results indicate that the TREX-2 scaffold subunit GANP is required for efficient DSB repair in human cells as GANP-depleted cells accumulate unrepaired breaks as indicated by increased γH2AX signal and comet tail moment, as well as having increased sensitivity to genotoxic agents. In good agreement, a previous study in human cells analyzing the role of PCID2 and DSS1, two other TREX-2 subunits, showed that depletion of PCID2, or DSS1, also causes DNA break accumulation as indicated by an increase in γH2AX staining and comet-tail moment (Bhatia et al., 2014).
In this study, we further show that depletion of GANP/TREX-2 particularly affects the HR pathway. A prerequisite for HR is the production of a 3′ ssDNA overhang in the process known as resection, which will invade the homologous duplex through RAD51. Consistent with a decrease in HR efficiency, GANP-depleted human cells (HeLa and U2OS) are defective in the early stages of resection and subsequently in RAD51 loading both at the IsceI-induced break and genome wide (Fig. 4, D, G, and H). Our data are in accordance with previous research in chicken B cells, where it was shown that GANP stimulates HR (Eid et al., 2014). Note, however, that GANP was also shown to suppress hyperrecombination in mouse cells (Yoshida et al., 2007), suggesting that GANP/TREX-2 might have different roles depending on the origin of the cells studied.
The association of human GANP with the NPC depends on basket nucleoporins NUP153 and TPR (Umlauf et al., 2013). Recent studies described a role for these nucleoporins in DDR, affecting primarily the nuclear import of 53BP1 and its sumoylation (Lemaître et al., 2012; Moudry et al., 2012; Duheron et al., 2017; Mackay et al., 2017). However, we did not detect (a) any 53BP1 import defects in the absence of GANP/TREX-2 (Fig. 1 F), (b) any defects in the recruitment of 53BP1 at the Isce I–induced break (Fig. 3 F), or (c) defects in genome wide formation of 53BP1 foci upon NCS treatment (Fig. S2 L). Thus, the role of GANP/TREX-2 in global DNA repair is likely independent from its interaction with basket nucleoporins. Moreover, we did not detect specific accumulation of unrepaired breaks at the nuclear periphery (Fig. S1, C and D), further suggesting that TREX-2 acts through an NPC-independent mechanism.
Interestingly, the GANP/TREX-2 depletion reduced H2Bub1 levels under either steady-state or damage conditions. The facts that GANP/TREX-2 does not have any known E2/E3 ubiquitin ligase activity and that the DUB module of SAGA is the major DUB responsible for the removal of ubiquitin from H2Bub1 in mouse ES cells (Bonnet et al., 2014) suggest that GANP/TREX-2 might affect histone H2Bub1 levels and consequently DNA repair by affecting the activity of the SAGA DUB module and potentially its related DUB modules with which it shares the ENY2 subunit.
Depletion of the ENY2 alone surprisingly not only does not recapitulate GANP/TREX-2 effect on HR, but it has the opposite effect by stimulating HR efficiency. We ascribed this effect to the functions of the DUBm-s because depletion of ATXN7L3 also increased HR efficiency to the same extent (Fig. 4, A and E). Monoubiquitination of H2B by RNF20/40 E3 ubiquitin ligase happens in response to DNA damage in human cells and is required for recruitment of HR factors (Moyal et al., 2011; Nakamura et al., 2011). Thus, our results are in good accordance with the data showing that depletion of RNF20/40 induces a decrease in HR efficiency and resection (Moyal et al., 2011; Nakamura et al., 2011), resulting in the opposite phenotype than the DUBm-deficient cells.
Two recent studies also demonstrated that deubiquitination of histone H2Bub1 impacts the early stages of the DDR and is required for DNA repair (Ramachandran et al., 2016; Li et al., 2018). Interestingly, these studies described that the SAGA DUB module is required for optimal irradiation-induced γH2AX formation in CSR in mouse B cells by promoting NHEJ. In our study, we did not detect defects in global γH2AX formation at radiomimetic drug-induced DSBs after depletion of the DUB module subunit ENY2. These contrasting results may be explained by a scenario in which the H2Bub1 histone mark regulates repair differently in distinct species and/or in different genomic loci such as immunoglobulin switch regions. Note that we did not investigate the role of the USP22 KD given the presence of at least two alternative DUB enzymes in human cells (see Introduction; Atanassov et al., 2016). However, depletion of the DUBm adaptors ENY2 and/or ATXN7L3 would eliminate the activity of all the USP22-related DUBm-s (independently of the cellular expression levels of the three related USPs). Thus, ENY2 and/or ATXN7L3 depletion will affect all histone H2Bub1 deubiquitination activities (Atanassov et al., 2016).
We hypothesize that GANP/TREX-2 regulates HR through controlling the DUB module activity because we were able to rescue H2Bub1 global levels, RAD51 recruitment at the ISceI-induced break, and the sensitivity to HR-inducing drugs of GANP-deficient cells by further abolishing the DUBm functions through additional ENY2 or ATXN7L3 depletion (Figs. 5 and S4, C and D). Moreover, although KD of the DUB module generates a global H2Bub1 increase that is damage independent, it is also important to mention that at specific chromatin locations (i.e., ASISI sites) we detected an excess of H2Bub1 levels in ENY2-depleted cells only after damage induction. This suggests different roles for the DUB module; a transcription-related role that reflects changes in transcription of all active genes (Bonnet et al., 2014), and a repair-related role demonstrated by a specific increase in H2Bub1 at damaged sites in ENY2-depleted cells. Nevertheless, our results confirm that H2B/H2Bub1 balance is important for HR-mediated repair and show that H2B/H2Bub1 balance is controlled by a regulated interplay between TREX-2 and SAGA DUB subunits ENY2 and ATXN7L3 (Fig. 7). How the H2Bub1 mark regulates repair is not clear. It was speculated that the HR impairment in RNF20-depleted cells was dependent on a defect in chromatin relaxation ascribed to the H2Bub1-dependent methylation of H3K4 (Nakamura et al., 2011). However, in a second study, H3K4 methylation was maintained unaltered in RNF20 KD, and accumulation of H3K4me was not observed at damaged sites (Moyal et al., 2011). Although a defect in chromatin relaxation was speculated in both studies, it cannot be excluded that H2Bub1 mark would be necessary for the direct recruitment of specific DNA repair factors to break sites.
Figure 7.
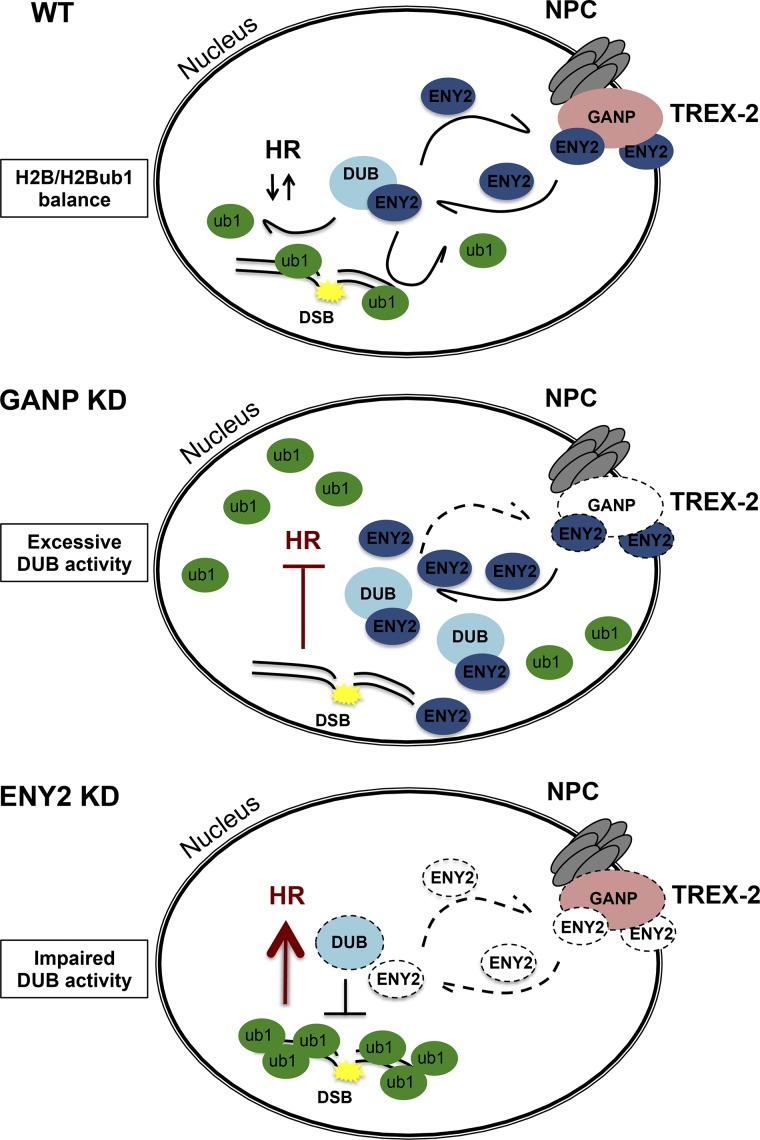
Model. In steady-state conditions (WT), TREX-2 and SAGA DUB share the ENY2 subunit, and H2B/H2Bub1 balance is maintained. Under GANP KD conditions, newly synthesized ENY2 is redistributed to the DUB, and this contributes to increase removal of ubiquitin from H2Bub1, and consequently HR is impaired. Under ENY2 KD conditions, the DUB module is destabilized, and this contributes to increase H2Bub1 levels, and consequently unscheduled HR.
In yeast, physical interactions between TREX-2 and SAGA have been described (Rodríguez-Navarro et al., 2004; Köhler et al., 2008); nevertheless, such interactions have not been detected in Drosophila melanogaster (Weake et al., 2011) and mammalian cells (Umlauf et al., 2013). It is conceivable, however, that in mammalian cells TREX-2 and SAGA DUB also interact in a more dynamic and less stable manner that could be more difficult to detect in different metazoan cell extracts. It seems that the interaction between TREX-2 and the DUB modules can fine-tune DUB activity. It has been shown that the DUB module of SAGA exists in metazoan cells without incorporating in SAGA (Nagy et al., 2009; Li et al., 2017) and that additional related DUBm-s exist, all able to deubiquitinate H2Bub1 (Atanassov et al., 2016). Moreover, the SAGA DUB module was suggested to have SAGA-independent functions (Li et al., 2017). Thus, the regulatory interplay between TREX-2 and the different DUB modules may not necessarily involve the SAGA complex. The fact that the NPC-associated TREX-2 complex regulates the activity of the nucleoplasmic DUB modules might also suggest that a nucleoplasmic pool of TREX-2 exists. Nevertheless, future investigations will be needed to understand the exact mechanism through which TREX-2 can control the DUB activity and consequently DNA repair.
A similar regulatory interplay has been already described, showing that cytoplasmic ATXN7L3B, which is encoded by a pseudogene that arose from the retrotransposition of ATXN7L3, may regulate the nuclear function of SAGA DUB module and H2Bub1 levels through competition for ENY2 binding (Li et al., 2016). Thus, it seems that the different ENY2-containing complexes—TREX-2, the SAGA DUB, the different SAGA-independent DUB modules, and the cytoplasmic ATXN7L3B/ENY2 association—may all have important regulatory functions to achieve well-balanced H2B/H2Bub1 levels that are required for efficient DNA repair and HR. These different complexes may compete with each other for ENY2 and thus, depending on the availability of ENY2 for TREX-2 or the DUB modules, regulate H2Bub1 levels. How exactly neosynthesized ENY2 is distributed in the different complexes by distinct assembly pathways need to be further investigated.
Materials and methods
Cell lines, NCS treatment, and transfections
HeLa cells (human cervix adenocarcinoma) and U2OS cells (human bone osteosarcoma) were cultured in 1 g/liter DMEM supplemented with 5% and 10% FCS, respectively. hTERT-RPE1 cells (epithelial cells immortalized with hTERT) were cultured in DMEM/F12 supplemented with 10% FCS. These cells were purchased from ATCC. U2OS 19 ptight 13 GFP-LacI (Lemaître et al., 2014) were cultured in DMEM 4.5 g/liter phenol-red free supplemented with 10% tet-free FCS. While in culture, cells were maintained in medium containing 2 mM IPTG to avoid GFP-LacI binding to the array. During the experiment, before siRNA treatment, cells were plated in medium without IPTG. U2OS DR-GFP Cherry-IsceI-GR cells were cultured in 1 g/liter phenol red–free DMEM. GCV6 cells were cultured in 1 g/liter DMEM supplemented with 10% FCS. U2OS DIvA (Massip et al., 2010; Caron et al., 2012) were cultured in 1 g/liter phenol red–free DMEM supplemented with 5% tet-free FCS. For NCS treatments, cells were treated with 50 ng/ml (for immunofluorescence) or 150–250 ng/ml (for Western blot analysis) NCS (N9162; Sigma-Aldrich) for 15 min. Medium was refreshed, and cells were allowed to repair for indicated times before fixation or protein extraction. For high-throughput screening in HeLa and hTERT RPE1 cells, siRNAs were transfected in 96-well plates with 25 nM siRNA per well (4,000 cells plated per well) using 5 µl Interferin (Polyplus-transfection) diluted 1/20. For all other experiments and cell lines, indicated siRNAs were transfected using Lipofectamine 2000 (Thermo Fisher Scientific) following the manufacturer’s instructions. For high-throughput screening, two different siRNAs for GANP (individually and pool of the two) and three different siRNAs from ENY2 (individually and pool of the three) were used to transfect cells in 96-well plates (5 pmol siRNA was transfected in each well). For all the other experiments, siGANP2 was used (referred as siGANP) and siENY2_2 was used (referred as siENY2). For siRNAs sequences and references, see Table 1.
Table 1. List of siRNAs used.
| siRNA | Sequence (5′–3′) | Reference |
|---|---|---|
| siCTRL | GAGAGGUCCAAAGUCAAUA | siGANPc; Wickramasinghe et al., 2010a |
| siGANP1 | GAGAGGACCUAAGUCAAUA | Wickramasinghe et al., 2010a |
| siGANP2 | AGCUUGCAGUGGUACAUUU | Wickramasinghe et al., 2010a |
| siSCR | GUUAACGAUAAUUAGAUAA | 5′-GUUAACGAUAAUUAGAUAA-3′ purchased from Sigma-Aldrich |
| siENY2_1 | AGAGCAGCGAUUAACCAAA | Ambion (s32449) |
| siENY2_2 | GUAAAGAGGUAAUUAAAGA | Ambion (s32447) |
| siENY2_3 | AGAGCUAAAUUAAUUGAAU | Ambion (s226899) |
| siXRCC4 | GAAUCCACCUUGUUUCUGA | GE Healthcare, siGenome, Smart Pool (M-004494-02-0020) |
| GAGAAUCAGCUUCAAGAAA | ||
| UGACCGAGAUCCAGUCUAU | ||
| GAACCCAGUAUAACUCAUU | ||
| siSET | NA | GE Healthcare, on-TARGETplus (L-019586-00-0005) |
| siNTarg | UGGUUUACAUGUCGACUAA | GE Healthcare, ON-TARGET plus, (D-001810-10-20) |
| UGGUUUACAUGUUGUGUGA | ||
| UGGUUUACAUGUUUUCUGA | ||
| UGGUUUACAUGUUUUCCUA | ||
| siRAD51 | CAGGGUAAUCACUAAUCA | Ambion (s11735) |
| siATXN7L3 | NA | Thermo Fisher Scientific (271960) |
NA, not available.
Clonogenic survival assay
HeLa cells were transfected with indicated siRNAs, and 48 h after transfection, cells were counted and seeded in triplicates in six-well plates (500 cells per well). The day after, cells were treated with indicated concentration of the following drugs: phleomycin (Sigma-Aldrich; 1-h treatment), camptothecin (C9911; Euromedex; 1-h treatment), MMC (Sigma-Aldrich; EMD Millipore; 16-h treatment), Olaparib (SC-302017; Santa Cruz Biotechnology), and NCS (N9162; Sigma-Aldrich; 15-min treatment). For phleomycin, camptothecin, and MMC, medium was refreshed and cells were then cultured for 10 d. Treatment with Olaparib was maintained for the whole experiment. Colonies were stained with 0.1% crystal violet and counted using Fiji (ImageJ; National Institutes of Health).
DR-GFP assays
For HR efficiencies, U2OS DR-GFP Cherry-IsceI-GR cells (Shahar et al., 2012) were transfected with the indicated siRNA, and 24 h later, triamcinolone acetonide was added to allow translocation of Cherry-IsceI-GR into the nucleus for 48 h. For NHEJ efficiency, GCV6 cell line (Rass et al., 2009) bearing the GFP-based substrates was used. Cells were first transfected with the indicated siRNAs, and 48 h later, they were transfected with HA-I-SceI expression vector (pCBASce) using jetPei (Polyplus) following manufacturer’s instructions. GFP intensities were measured by FACS and analyzed using FlowJo software (TreeStar).
Immunofluorescence
Genome-wide recruitment of RPA, RAD51, and 53BP1 was assessed in HeLa cells plated in 24-well plates on round cover glasses (VWR). Recruitment of DNA repair factors at a single IsceI cut site was assessed in U2OS 19 ptight 13 GFP-LacI cells. Immunofluorescence was performed equally in both cell lines. At the decided time point, cells were washed carefully with PBS 1×. Cells stained for RAD51 and RPA were treated with preextraction buffer (0.5% Triton X-100, 50 mM Hepes, pH 7, 150 mM NaCl, 10 mM EGTA, and 2 mM MgCl2) for 10 s before fixation with 4% PFA (Electron Microscopy Science) for 10 min at RT. Cells stained for other factors were fixed with PFA immediately after washing. After fixation, cells were washed with 1× PBS, permeabilized with sterile 0.3% Triton X-100 in PBS for 10 min at RT, and blocked with sterile 5% BSA for 1 h. Primary antibodies were added for 1 h in 1× 0.01% BSA/PBS at RT, and cells were washed and stained with secondary fluorescent antibodies. For primary antibodies, see Table 2. All the fluorescent secondary antibodies were Alexa Fluor antibodies from Invitrogen and were added at a final concentration of 1:1,000 for determination of RAD51, RPA, and 53BP1 foci formation after NCS treatment. In the LacO array colocalization experiments, secondary fluorescent antibodies were added at a final concentration of 1:250 for 1 h. DAPI (Sigma-Aldrich; D9542) was added at 0.3 mM final concentration for 2 min, and coverslips were mounted using ProLong Gold Antifade Mountant (Thermo Fisher Scientific; P36934).
Table 2. List of antibodies used.
| Antibody | Reference | Working dilution |
|---|---|---|
| Mouse anti-γH2AX | Abcam (ab22551) | Western blot and IF (1:1,000) |
| Rabbit anti-GANP | Abcam (113295) | Western blot 1:500 |
| Rabbit anti 53BP1 | Novus Biologicals (NB100-304) | IF (1:1,000) |
| Rabbit anti-Phospho RPA32 (S4/8) | Bethyl (A300-245A) | IF (1:500) |
| Rabbit anti-Phospho RPA 32 (S33) | Bethyl (A300-246A) | IF (1:1,000) |
| Rabbit anti-RAD51 | EMD Millipore (PC130) | IF (1:500) Western blot (1:1,000) |
| Mouse anti-ATM pS1981 | Rockland (200-301-400) | Western blot (1:2,000) |
| Rabbit anti-ENY2 | Santa Cruz Biotechnology (sc-87712) | Western blot (1:200) |
| Mouse anti-γTUBULIN | Sigma-Aldrich (T6557) | Western blot (1:1,000) |
| Rabbit anti-phospho KAP1 | Bethyl (A300-767A) | Western blot (1:500) |
| Mouse anti-GAPDH | EMD Millipore (MAB374) | Western blot (1:1,000) |
| Rabbit anti-BRCA1 | Santa Cruz Biotechnology(C-20 SC642) | IF (1:100) |
| Rabbit anti-CtIP | Bethyl (A300-488A) | IF (1:100) |
| Rabbit anti-H2Bub1 | Cell Signaling Technology (CS55465) | Western blot (1:10,000) |
| Mouse anti-H2B | H2-2A4 (Bonnet et al., 2010) | Western blot (1:10,000) |
| Mouse anti-TBP | 3G3 (Brou et al., 1993) | Western blot (1:1,000) |
| Mouse anti-αTUBULIN | Sigma-Aldrich (T9026) | Western blot (1:1,000) |
| Mouse anti-γH2AX | EMD Millipore (05-636) | ChIP (2 µg/50 µg of chromatin) |
| Mouse anti-H2Bub1 | Medimab | ChIP (4 µg/50 µg of chromatin) |
| Mouse anti-RPA | Genetex (GTX22175) | IF (1:500) |
IF, immunofluorescence.
Microscopy and image acquisition
Representative images of LacO-array colocalization with repair factors and of RAD51, RPA, and 53BP1 foci after NCS treatment were acquired with the Leica Microsystems TCS SP5 inverted confocal microscope equipped with a 458/476/488/496/514-nm argon laser, a 561-nm diode-pumped solid-state laser, a 594-nm HeNe laser, a 633-nm HeNe laser, and a 405-nm laser diode. Cells were visualized using the objective HCX Plan Apochromat 63×/1.40–0.60 oil Lbd Bl with 63× magnification (NA 1.4), and images were taken using the hybrid detector photocounting mode. Images were acquired with LAS AF acquisition software (Leica Microsystems). Secondary fluorescent antibodies were goat anti-rabbit or goat anti-mouse Alexa Fluor 488 or 568 from Invitrogen. For colocalization analyses of DNA repair factors with the LacO array, colocalization was counted at the fluorescence microscope Leica Microsystems DM 4000 B in n ≥ 50 cells per condition.
Cellular extracts and Western blotting
Total protein lysate was obtained by adding cold radioimmunoprecipitation assay (RIPA) buffer (Cold Spring Harbor Protocols) on the cell pellet. Histone proteins extract was obtained by adding acidic buffer (10 mM Hepes, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT. and 0.2 M HCl) on cell pellet. 1× Proteinase Inhibitor Cocktail (cOmplete Mini EDTA free, Proteinase Inhibitor Cocktail; 11836170001; Roche), Phosphatase Inhibitor Cocktail (PhosSTOP; PHOSS-RO; Roche), and 10 mM N-ethylmaleimide (NEM E3876; Sigma-Aldrich) were added in all buffers. Samples were run on 4–20% Precast gels (Mini-Protean TGX Stain-Free Protein Gels; Bio-Rad), and membranes were blotted with indicated antibodies. Western blots were revealed using peroxidase-conjugated secondary antibodies from Jackson ImmunoResearch Laboratories. Images were acquired using Chemidoc Touch Imaging system (Bio-Rad). Protein levels were quantified by Fiji.
Cell cycle analysis
For propidium iodide staining, after treatment with the indicated siRNAs, U2OS cells were fixed in 70% EtOH overnight at −20°C, treated with RNase A (100 µg/ml), and stained with propidium iodide (40 µg/ml). For S-phase analysis, after treatment with the indicated siRNAs, HeLa cells were collected and stained with EdU using the Click-It kit (C10632; Thermo Fisher Scientific) following the manufacturer’s instructions. In both cases, the acquisition was performed on a FACSCalibur (BD). Results were analyzed using FlowJo software. For propidium iodide staining, the cell cycle phases were assigned manually.
Comet assay
For the comet assay, HeLa cells were treated with siRNAs for 48 h before collection. Neutral COMET assay was performed using CometAssay Kit Trevigen (4205-050-K) following manufacturer’s instructions. Cells were analyzed using OpenCOMET plugin on ImageJ.
High-throughput screening
Cells were treated with specific siRNA in 96-well plates in three technical replicates (XRCC4 and SET siRNAs were used as positive controls). After 72 h, cells were treated with 50 ng/ml NCS for 15 min. Medium was refreshed and cells were allowed to repair for 2, 8, and 16 h. Cells were fixed and stained with DAPI-, γH2AX-, and 53BP1-specific antibodies. Image acquisition was done using Cell Insight and a 20× objective. γH2AX intensity and 53BP1 foci number were analyzed with HCS studio. For γH2AX intensity distribution among inner nuclear environment and nuclear periphery, a mask of 2.5 µm (6 pixels) on the DAPI staining was applied to define the nuclear periphery. For each cell, the intensity of the γH2AX signal in the periphery and in the whole nucleus was measured. The peripheral signal per each cell is represented by the ratio between the two values. The inner signal per each cell is represented by the ratio between the inner signal and the total signal.
RNA extracts and qPCR
RNA was extracted using NucleoSpin RNA Kit (Macherey-Nagel), and cDNA was obtained with Superscript II Reverse transcription (Invitrogen) following manufacturer’s instructions. qPCR was performed using LightCycler 480 SYBR Green I Master (Roche) following manufacturer’s instructions. For primer sequence, see Table 3. All the primers were purchased from Sigma-Aldrich.
Table 3. List of primers used.
| Primer name | Sequence (5′–3′) |
|---|---|
| GANP_fw | CACGAGCCAGCAGCAGAAGTTC |
| GANP_rev | CATCCTGTATCGTCCGACCA |
| ENY2_fw | GGAGAAAGAGAACGCCTCAAA |
| ENY2_rev | AGTGATTTCAGCCACCAAGTCA |
| ATXN7L3_fw | CTGGGAATGGGTCGGAACAG |
| ATXN7L3_rev | CCGAGCCATAGGACCAGTCG |
| GAPDH_fw | TCGACAGTCAGCCGCATCTTCTTT |
| GAPDH_rev | ACCAAATCCGTTGACTCCGACCTT |
| BRCA1_fw | CTGAAGACTGCTCAGGGCTATC |
| BRCA1_rev | AGGGTAGCTGTTAGAAGGCTGG |
| RPA_fw | GAGCACCTATCAGCAATCCAGG |
| RPA_rev | CCTTCAGGTCTTGGACAAGCCT |
| 53BP1_fw | AAGCCAGGCAAGAGAATGAGGC |
| 53BP1_rev | GGCTGTTGACTCTGCCTGATTG |
| ATM_fw | TGTTCCAGGACACGAAGGGAGA |
| ATM_rev | CAGGGTTCTCAGCACTATGGGGA |
| CtIP_fw | TGGCAGACAGTTTCTCCCAAGC |
| CtIP_rev | GGCTCCACAAACGCTTTCTGGCT |
| ChIP primer DSB1_fw | GGAGAAGTGGCAGGACAATG |
| ChIP primer DSB1_rev | CAAGGCAAATTTGGGGACTA |
| ChIP primer DSBIV_fw | GAGGAACCATTCGGACAAGA |
| ChIP primer DSBIV_rev | CTGACCAAGGAAGCCTCAAG |
| ChIP primer DSBII_fw | GGGTATGGAGCTGCCTCTAA |
| ChIP primer DSBII_rev | GACAAAGATGGCTGGAGGAG |
ChIP followed by qPCR in U2OS DIvA
Cells were treated with specific siRNAs for 72 h, and then 4-OH tamoxifen was added at a final concentration of 300 nM for 4 h. Cells were cross-linked with formaldehyde 4% for 10 min. Chromatin was sonicated using sonication buffer (0.1% SDS, 10 mM EDTA, and 50 mM Tris-HCl) in the Covaris E220 for 7 min per samples. Approximately 80 µg chromatin was used for IP with the specific antibody and for the no-antibody control using protein G Sepharose beads (Sigma-Aldrich). IP was performed overnight at 4°C, and the beads were added next morning at a 1:10 dilution for 2 h. For specific antibodies, see Table 2. Washing steps were performed with wash buffer (0.1% SDS, 0.5% NP-40, 2 mM EDTA, 150 mM NaCl, and 20 mM Tris-HCl, pH 8), final wash buffer (0.1% SDS, 0.5% NP-40, 2 mM EDTA, 500 mM NaCl, and 20 mM Tris-HCl, pH 8), and LiCl buffer (50 mM Tris-HCl, pH 8, 2 mM EDTA, 0.1% NP-40, and 10% glycerol). Protease inhibitors (cOmplete Mini EDTA-free Protease Inhibitor Cocktail; Sigma-Aldrich) were added in all buffers. Elution was obtained with elution buffer (1% SDS and 0.1 M NaHCO3) at 65°C for 5 min. DNA was purified with phenol:chloroform:isoamyl alcohol 25:24:1 (Sigma-Aldrich). qPCR was performed with LightCycler 480 SYBR Green I Master (Roche) in two technical replicates. For primer sequences, see Table 3.
Statistical analyses
All statistical analyses were performed with GraphPad Prism. For all survival assays, the Mann–Whitney t test (unpaired nonparametric test) was used to identify statistical significance among the different concentrations and siRNAs. The confidence interval was set at 95% (definition of statistical significance: P < 0.05). For cell cycle analyses, multiple t test was used correcting for multiple comparisons using the Holm–Sidak method. The confidence interval was set at 95% (definition of statistical significance: P < 0.05). For colocalization of the LacO with repair factors and for statistical analysis of RAD51 recruitment after NCS treatment (Fig. 3), statistical significance between siNTarg and siGANP at 14 h dox was calculated using the unpaired t test (definition of statistical significance: P < 0.05). To compare more than two groups (colocalization of LacO with RAD51 in the double KD [Fig. 5, E and F] and DR-GFP assays), the one-way ANOVA test was used to compare the three groups. Multiple comparisons were done using Dunnett’s correction, and all samples were compared with the control siRNA (siNTarg or siCtrl). Quantification of H2Bub1 Western blot depicted in graphs (Fig. S3, D and H) was analyzed with unpaired t test comparing separately each siRNA with the control (siNTarg). Number of experiments and error bars in graphic representation of pooled data are described in all figure legends.
Online supplemental material
Fig. S1 shows an additional high-throughput screening in RPE1 cells (A), clonogenic survival assay with NCS (B), the distribution of intensity for γH2AX in the nucleus upon GANP KD and control cells after NCS treatment (C and D), and the effects of GANP KD on cell cycle and on RAD51 protein levels (E–G). Fig. S2 shows representative images and graphs for the colocalization of specific HR factors with the LacO array after DSB induction (A–G) and formation of RPA (H), RAD51 (I), and 53BP1 (L) foci after NCS treatment in GANP-depleted cells. It shows also DR-GFP assay with different siRNAs for GANP and ENY2 (M). Fig. S3 shows the effects of ENY2 KD on cell cycle progression (A), γH2AX resolution after NCS treatment (B), and phleomycin sensitivity (C). It also shows quantitative graphs for H2Bub1 levels in control cells and cells depleted of GANP and ENY2 in the absence (D and H) and presence (F and G) of DNA damage. Fig. S4 shows efficiency of double KD (A and B), rescue of sensitivity to HR-inducing drugs in double KD (C and D), the effects of ENY2 and GANP KD on the levels of key repair genes (E and F), and efficient induction of breaks with 4-OH tamoxifen in DIvA cells (G).
Supplementary Material
Acknowledgments
We thank G. Legube for the U2OS DIvA cells and helpful discussions and K. Tsouroula and A. Kalousi for thoughtful discussions and experimental suggestions throughout the course of the work. We are grateful to the Institut de Génétique et de Biologie Moléculare et Cellulaire cell culture facility for cells and media. We thank the Mediaprep service of the Institut de Génétique et de Biologie Moléculare et Cellulaire for buffer preparations. We are grateful to the E. Guiot and the Imaging and Microscopy platform of the Institut de Génétique et de Biologie Moléculare et Cellulaire for assistance in confocal microscopy.
This work was supported by funds from Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale, and University of Strasbourg. This study was also supported by the European Research Council Advanced Grant (ERC-2013-340551, Birtoaction to L. Tora) and Agence Nationale de la Recherche grant ANR-10-LABX-0030-INRT, a French state fund managed by the Agence Nationale de la Recherche under the frame program Investissements d’Avenir (ANR-10-IDEX-0002-02). E. Soutoglou acknowledges support from Institut National du Cancer (PLBio-15-199).
The authors declare no competing financial interests.
Author contributions: F.M. Evangelista performed most of the experiments. A. Maglott-Roth and L. Brino performed the high-throughput screening experiments. M. Stierle contributed to the execution of molecular laboratory work. F.M. Evangelista, E. Soutoglou, and L. Tora designed the study, analyzed data, and wrote the paper. All authors contributed text and figure panels to the manuscript. All authors gave final approval for publication.
References
- Atanassov B.S., Mohan R.D., Lan X., Kuang X., Lu Y., Lin K., McIvor E., Li W., Zhang Y., Florens L., et al. 2016. ATXN7L3 and ENY2 coordinate activity of multiple H2B deubiquitinases important for cellular proliferation and tumor growth. Mol. Cell. 62:558–571. 10.1016/j.molcel.2016.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymard F., Bugler B., Schmidt C.K., Guillou E., Caron P., Briois S., Iacovoni J.S., Daburon V., Miller K.M., Jackson S.P., and Legube G.. 2014. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 21:366–374. 10.1038/nsmb.2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista T., Grünberg S., Minoungou N., Koster M.J.E., Timmers H.T.M., Hahn S., Devys D., and Tora L.. 2017. SAGA is a general cofactor for RNA polymerase II transcription. Mol. Cell. 68:130–143.e5. 10.1016/j.molcel.2017.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes D.E., and Lindahl T.. 2004. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 38:445–476. 10.1146/annurev.genet.38.072902.092448 [DOI] [PubMed] [Google Scholar]
- Bhatia V., Barroso S.I., García-Rubio M.L., Tumini E., Herrera-Moyano E., and Aguilera A.. 2014. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 511:362–365. 10.1038/nature13374 [DOI] [PubMed] [Google Scholar]
- Blobel G. 1985. Gene gating: a hypothesis. Proc. Natl. Acad. Sci. USA. 82:8527–8529. 10.1073/pnas.82.24.8527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet J., Wang Y.H., Spedale G., Atkinson R.A., Romier C., Hamiche A., Pijnappel W.W., Timmers H.T., Tora L., Devys D., and Kieffer B.. 2010. The structural plasticity of SCA7 domains defines their differential nucleosome-binding properties. EMBO Rep. 11:612–618. 10.1038/embor.2010.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet J., Wang C.Y., Baptista T., Vincent S.D., Hsiao W.C., Stierle M., Kao C.F., Tora L., and Devys D.. 2014. The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev. 28:1999–2012. 10.1101/gad.250225.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brou C., Chaudhary S., Davidson I., Lutz Y., Wu J., Egly J.M., Tora L., and Chambon P.. 1993. Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J. 12:489–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabal G.G., Genovesio A., Rodriguez-Navarro S., Zimmer C., Gadal O., Lesne A., Buc H., Feuerbach-Fournier F., Olivo-Marin J.C., Hurt E.C., and Nehrbass U.. 2006. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 441:770–773. 10.1038/nature04752 [DOI] [PubMed] [Google Scholar]
- Caron P., Aymard F., Iacovoni J.S., Briois S., Canitrot Y., Bugler B., Massip L., Losada A., and Legube G.. 2012. Cohesin protects genes against γH2AX induced by DNA double-strand breaks. PLoS Genet. 8:e1002460 10.1371/journal.pgen.1002460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova J.A., Abruzzi K.C., Rosbash M., and Belostotsky D.A.. 2008. Sus1, Sac3, and Thp1 mediate post-transcriptional tethering of active genes to the nuclear rim as well as to non-nascent mRNP. RNA. 14:66–77. 10.1261/rna.764108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley J.M., Niu H., Miller A.S., and Sung P.. 2015. Biochemical mechanism of DSB end resection and its regulation. DNA Repair (Amst.). 32:66–74. 10.1016/j.dnarep.2015.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley D.D., Chaudhuri J., Bassing C.H., and Alt F.W.. 2005. Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. Adv. Immunol. 86:43–112. 10.1016/S0065-2776(04)86002-4 [DOI] [PubMed] [Google Scholar]
- Duheron V., Nilles N., Pecenko S., Martinelli V., and Fahrenkrog B.. 2017. Localisation of Nup153 and SENP1 to nuclear pore complexes is required for 53BP1-mediated DNA double-strand break repair. J. Cell Sci. 130:2306–2316. 10.1242/jcs.198390 [DOI] [PubMed] [Google Scholar]
- Eid M.M., Maeda K., Almofty S.A., Singh S.K., Shimoda M., and Sakaguchi N.. 2014. GANP regulates the choice of DNA repair pathway by DNA-PKcs interaction in AID-dependent IgV region diversification. J. Immunol. 192:5529–5539. 10.4049/jimmunol.1400021 [DOI] [PubMed] [Google Scholar]
- Falck J., Forment J.V., Coates J., Mistrik M., Lukas J., Bartek J., and Jackson S.P.. 2012. CDK targeting of NBS1 promotes DNA-end resection, replication restart and homologous recombination. EMBO Rep. 13:561–568. 10.1038/embor.2012.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faza M.B., Kemmler S., Jimeno S., González-Aguilera C., Aguilera A., Hurt E., and Panse V.G.. 2009. Sem1 is a functional component of the nuclear pore complex-associated messenger RNA export machinery. J. Cell Biol. 184:833–846. 10.1083/jcb.200810059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T., Strässer K., Rácz A., Rodriguez-Navarro S., Oppizzi M., Ihrig P., Lechner J., and Hurt E.. 2002. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 21:5843–5852. 10.1093/emboj/cdf590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T., Rodríguez-Navarro S., Pereira G., Rácz A., Schiebel E., and Hurt E.. 2004. Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nat. Cell Biol. 6:840–848. 10.1038/ncb1163 [DOI] [PubMed] [Google Scholar]
- Gallardo M., and Aguilera A.. 2001. A new hyperrecombination mutation identifies a novel yeast gene, THP1, connecting transcription elongation with mitotic recombination. Genetics. 157:79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo M., Luna R., Erdjument-Bromage H., Tempst P., and Aguilera A.. 2003. Nab2p and the Thp1p-Sac3p complex functionally interact at the interface between transcription and mRNA metabolism. J. Biol. Chem. 278:24225–24232. 10.1074/jbc.M302900200 [DOI] [PubMed] [Google Scholar]
- García-Oliver E., García-Molinero V., and Rodríguez-Navarro S.. 2012. mRNA export and gene expression: the SAGA-TREX-2 connection. Biochim. Biophys. Acta. 1819:555–565. 10.1016/j.bbagrm.2011.11.011 [DOI] [PubMed] [Google Scholar]
- Gavin A.C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 415:141–147. 10.1038/415141a [DOI] [PubMed] [Google Scholar]
- González-Aguilera C., Tous C., Gómez-González B., Huertas P., Luna R., and Aguilera A.. 2008. The THP1-SAC3-SUS1-CDC31 complex works in transcription elongation-mRNA export preventing RNA-mediated genome instability. Mol. Biol. Cell. 19:4310–4318. 10.1091/mbc.e08-04-0355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P.A., Duggan L., Côté J., Roberts S.M., Brownell J.E., Candau R., Ohba R., Owen-Hughes T., Allis C.D., Winston F., et al. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640–1650. 10.1101/gad.11.13.1640 [DOI] [PubMed] [Google Scholar]
- Henry K.W., Wyce A., Lo W.S., Duggan L.J., Emre N.C., Kao C.F., Pillus L., Shilatifard A., Osley M.A., and Berger S.L.. 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17:2648–2663. 10.1101/gad.1144003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers J.H. 2009. DNA damage, aging, and cancer. N. Engl. J. Med. 361:1475–1485. 10.1056/NEJMra0804615 [DOI] [PubMed] [Google Scholar]
- Huertas P., Cortés-Ledesma F., Sartori A.A., Aguilera A., and Jackson S.P.. 2008. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 455:689–692. 10.1038/nature07215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliakis G., Wang H., Perrault A.R., Boecker W., Rosidi B., Windhofer F., Wu W., Guan J., Terzoudi G., and Pantelias G.. 2004. Mechanisms of DNA double strand break repair and chromosome aberration formation. Cytogenet. Genome Res. 104:14–20. 10.1159/000077461 [DOI] [PubMed] [Google Scholar]
- Jani D., Lutz S., Hurt E., Laskey R.A., Stewart M., and Wickramasinghe V.O.. 2012. Functional and structural characterization of the mammalian TREX-2 complex that links transcription with nuclear messenger RNA export. Nucleic Acids Res. 40:4562–4573. 10.1093/nar/gks059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalousi A., Hoffbeck A.S., Selemenakis P.N., Pinder J., Savage K.I., Khanna K.K., Brino L., Dellaire G., Gorgoulis V.G., and Soutoglou E.. 2015. The nuclear oncogene SET controls DNA repair by KAP1 and HP1 retention to chromatin. Cell Reports. 11:149–163. 10.1016/j.celrep.2015.03.005 [DOI] [PubMed] [Google Scholar]
- Köhler A., and Hurt E.. 2007. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8:761–773. 10.1038/nrm2255 [DOI] [PubMed] [Google Scholar]
- Köhler A., Schneider M., Cabal G.G., Nehrbass U., and Hurt E.. 2008. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat. Cell Biol. 10:707–715. 10.1038/ncb1733 [DOI] [PubMed] [Google Scholar]
- Koutelou E., Hirsch C.L., and Dent S.Y.. 2010. Multiple faces of the SAGA complex. Curr. Opin. Cell Biol. 22:374–382. 10.1016/j.ceb.2010.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G., Bonnet J., Umlauf D., Karmodiya K., Koffler J., Stierle M., Devys D., and Tora L.. 2011. The tightly controlled deubiquitination activity of the human SAGA complex differentially modifies distinct gene regulatory elements. Mol. Cell. Biol. 31:3734–3744. 10.1128/MCB.05231-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaître C., Fischer B., Kalousi A., Hoffbeck A.S., Guirouilh-Barbat J., Shahar O.D., Genet D., Goldberg M., Betrand P., Lopez B., et al. 2012. The nucleoporin 153, a novel factor in double-strand break repair and DNA damage response. Oncogene. 31:4803–4809. 10.1038/onc.2011.638 [DOI] [PubMed] [Google Scholar]
- Lemaître C., Grabarz A., Tsouroula K., Andronov L., Furst A., Pankotai T., Heyer V., Rogier M., Attwood K.M., Kessler P., et al. 2014. Nuclear position dictates DNA repair pathway choice. Genes Dev. 28:2450–2463. 10.1101/gad.248369.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Irrazabal T., So C.C., Berru M., Du L., Lam E., Ling A.K., Gommerman J.L., Pan-Hammarström Q., and Martin A.. 2018. The H2B deubiquitinase Usp22 promotes antibody class switch recombination by facilitating non-homologous end joining. Nat. Commun. 9:1006 10.1038/s41467-018-03455-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Atanassov B.S., Lan X., Mohan R.D., Swanson S.K., Farria A.T., Florens L., Washburn M.P., Workman J.L., and Dent S.Y.. 2016. Cytoplasmic ATXN7L3B interferes with nuclear functions of the SAGA deubiquitinase module. Mol. Cell. Biol. 36:2855–2866. 10.1128/MCB.00193-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Seidel C.W., Szerszen L.T., Lange J.J., Workman J.L., and Abmayr S.M.. 2017. Enzymatic modules of the SAGA chromatin-modifying complex play distinct roles in Drosophila gene expression and development. Genes Dev. 31:1588–1600. 10.1101/gad.300988.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Otevrel T., Gao Y., Cheng H.L., Seed B., Stamato T.D., Taccioli G.E., and Alt F.W.. 1995. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell. 83:1079–1089. 10.1016/0092-8674(95)90135-3 [DOI] [PubMed] [Google Scholar]
- Lindahl T., and Barnes D.E.. 2000. Repair of endogenous DNA damage. Cold Spring Harb. Symp. Quant. Biol. 65:127–133. 10.1101/sqb.2000.65.127 [DOI] [PubMed] [Google Scholar]
- Liu T., and Huang J.. 2016. DNA end resection: facts and mechanisms. Genomics Proteomics Bioinformatics. 14:126–130. 10.1016/j.gpb.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.F., Desai S.D., Li T.K., Mao Y., Sun M., and Sim S.P.. 2000. Mechanism of action of camptothecin. Ann. N. Y. Acad. Sci. 922:1–10. 10.1111/j.1749-6632.2000.tb07020.x [DOI] [PubMed] [Google Scholar]
- Mackay D.R., Howa A.C., Werner T.L., and Ullman K.S.. 2017. Nup153 and Nup50 promote recruitment of 53BP1 to DNA repair foci by antagonizing BRCA1-dependent events. J. Cell Sci. 130:3347–3359. [DOI] [PubMed] [Google Scholar]
- Massip L., Caron P., Iacovoni J.S., Trouche D., and Legube G.. 2010. Deciphering the chromatin landscape induced around DNA double strand breaks. Cell Cycle. 9:2963–2972. 10.4161/cc.9.15.12412 [DOI] [PubMed] [Google Scholar]
- Mimori T., and Hardin J.A.. 1986. Mechanism of interaction between Ku protein and DNA. J. Biol. Chem. 261:10375–10379. [PubMed] [Google Scholar]
- Moudry P., Lukas C., Macurek L., Neumann B., Heriche J.K., Pepperkok R., Ellenberg J., Hodny Z., Lukas J., and Bartek J.. 2012. Nucleoporin NUP153 guards genome integrity by promoting nuclear import of 53BP1. Cell Death Differ. 19:798–807. 10.1038/cdd.2011.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyal L., Lerenthal Y., Gana-Weisz M., Mass G., So S., Wang S.Y., Eppink B., Chung Y.M., Shalev G., Shema E., et al. 2011. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol. Cell. 41:529–542. 10.1016/j.molcel.2011.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Z., Riss A., Romier C., le Guezennec X., Dongre A.R., Orpinell M., Han J., Stunnenberg H., and Tora L.. 2009. The human SPT20-containing SAGA complex plays a direct role in the regulation of endoplasmic reticulum stress-induced genes. Mol. Cell. Biol. 29:1649–1660. 10.1128/MCB.01076-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K., Kato A., Kobayashi J., Yanagihara H., Sakamoto S., Oliveira D.V., Shimada M., Tauchi H., Suzuki H., Tashiro S., et al. 2011. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol. Cell. 41:515–528. 10.1016/j.molcel.2011.02.002 [DOI] [PubMed] [Google Scholar]
- Orthwein A., Noordermeer S.M., Wilson M.D., Landry S., Enchev R.I., Sherker A., Munro M., Pinder J., Salsman J., Dellaire G., et al. 2015. A mechanism for the suppression of homologous recombination in G1 cells. Nature. 528:422–426. 10.1038/nature16142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual-García P., Govind C.K., Queralt E., Cuenca-Bono B., Llopis A., Chavez S., Hinnebusch A.G., and Rodríguez-Navarro S.. 2008. Sus1 is recruited to coding regions and functions during transcription elongation in association with SAGA and TREX2. Genes Dev. 22:2811–2822. 10.1101/gad.483308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer P., Goedecke W., and Obe G.. 2000. Mechanisms of DNA double-strand break repair and their potential to induce chromosomal aberrations. Mutagenesis. 15:289–302. 10.1093/mutage/15.4.289 [DOI] [PubMed] [Google Scholar]
- Ramachandran S., Haddad D., Li C., Le M.X., Ling A.K., So C.C., Nepal R.M., Gommerman J.L., Yu K., Ketela T., et al. 2016. The SAGA deubiquitination module promotes DNA repair and class switch recombination through ATM and DNAPK-mediated γH2AX formation. Cell Reports. 15:1554–1565. 10.1016/j.celrep.2016.04.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass E., Grabarz A., Plo I., Gautier J., Bertrand P., and Lopez B.S.. 2009. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 16:819–824. 10.1038/nsmb.1641 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Navarro S., Fischer T., Luo M.J., Antúnez O., Brettschneider S., Lechner J., Pérez-Ortín J.E., Reed R., and Hurt E.. 2004. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 116:75–86. 10.1016/S0092-8674(03)01025-0 [DOI] [PubMed] [Google Scholar]
- Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., and Bonner W.M.. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858–5868. 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- Rondón A.G., García-Rubio M., González-Barrera S., and Aguilera A.. 2003. Molecular evidence for a positive role of Spt4 in transcription elongation. EMBO J. 22:612–620. 10.1093/emboj/cdg047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders S.L., Jennings J., Canutescu A., Link A.J., and Weil P.A.. 2002. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 22:4723–4738. 10.1128/MCB.22.13.4723-4738.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahar O.D., Raghu Ram E.V., Shimshoni E., Hareli S., Meshorer E., and Goldberg M.. 2012. Live imaging of induced and controlled DNA double-strand break formation reveals extremely low repair by homologous recombination in human cells. Oncogene. 31:3495–3504. 10.1038/onc.2011.516 [DOI] [PubMed] [Google Scholar]
- Shiloh Y., Shema E., Moyal L., and Oren M.. 2011. RNF20-RNF40: a ubiquitin-driven link between gene expression and the DNA damage response. FEBS Lett. 585:2795–2802. 10.1016/j.febslet.2011.07.034 [DOI] [PubMed] [Google Scholar]
- Sung P., Krejci L., Van Komen S., and Sehorn M.G.. 2003. Rad51 recombinase and recombination mediators. J. Biol. Chem. 278:42729–42732. 10.1074/jbc.R300027200 [DOI] [PubMed] [Google Scholar]
- Umlauf D., Bonnet J., Waharte F., Fournier M., Stierle M., Fischer B., Brino L., Devys D., and Tora L.. 2013. The human TREX-2 complex is stably associated with the nuclear pore basket. J. Cell Sci. 126:2656–2667. 10.1242/jcs.118000 [DOI] [PubMed] [Google Scholar]
- Wang H., Shi L.Z., Wong C.C., Han X., Hwang P.Y., Truong L.N., Zhu Q., Shao Z., Chen D.J., Berns M.W., et al. 2013. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS Genet. 9:e1003277 10.1371/journal.pgen.1003277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake V.M., Dyer J.O., Seidel C., Box A., Swanson S.K., Peak A., Florens L., Washburn M.P., Abmayr S.M., and Workman J.L.. 2011. Post-transcription initiation function of the ubiquitous SAGA complex in tissue-specific gene activation. Genes Dev. 25:1499–1509. 10.1101/gad.2046211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock D.M., Nakanishi K., Helgadottir H.R., and Jasin M.. 2006. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 409:524–540. 10.1016/S0076-6879(05)09031-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe V.O., McMurtrie P.I., Mills A.D., Takei Y., Penrhyn-Lowe S., Amagase Y., Main S., Marr J., Stewart M., and Laskey R.A.. 2010a mRNA export from mammalian cell nuclei is dependent on GANP. Curr. Biol. 20:25–31. 10.1016/j.cub.2009.10.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe V.O., Stewart M., and Laskey R.A.. 2010b GANP enhances the efficiency of mRNA nuclear export in mammalian cells. Nucleus. 1:393–396. 10.4161/nucl.1.5.12351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., Kuwahara K., Shimasaki T., Nakagata N., Matsuoka M., and Sakaguchi N.. 2007. GANP suppresses DNA recombination, measured by direct-repeat beta-galactosidase gene construct, but does not suppress the type of recombination applying to immunoglobulin genes in mammalian cells. Genes Cells. 12:1205–1213. 10.1111/j.1365-2443.2007.01119.x [DOI] [PubMed] [Google Scholar]
- Yu X., Wu L.C., Bowcock A.M., Aronheim A., and Baer R.. 1998. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J. Biol. Chem. 273:25388–25392. 10.1074/jbc.273.39.25388 [DOI] [PubMed] [Google Scholar]
- Yun M.H., and Hiom K.. 2009. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 459:460–463. 10.1038/nature07955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.Y., Varthi M., Sykes S.M., Phillips C., Warzecha C., Zhu W., Wyce A., Thorne A.W., Berger S.L., and McMahon S.B.. 2008. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol. Cell. 29:102–111. 10.1016/j.molcel.2007.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Zheng Y., Pham A.D., Mandal S.S., Erdjument-Bromage H., Tempst P., and Reinberg D.. 2005. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol. Cell. 20:601–611. 10.1016/j.molcel.2005.09.025 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.