Mechanical forces are generated during epithelial morphogenesis, but how cells maintain adhesion when exposed to these forces is poorly understood. Razzell et al. show that the LIM domain protein Ajuba localizes to adherens junctions under tension in the Drosophila embryo and is required to maintain cell adhesion during epithelial remodeling.
Abstract
The reorganization of cells in response to mechanical forces converts simple epithelial sheets into complex tissues of various shapes and dimensions. Epithelial integrity is maintained throughout tissue remodeling, but the mechanisms that regulate dynamic changes in cell adhesion under tension are not well understood. In Drosophila melanogaster, planar polarized actomyosin forces direct spatially organized cell rearrangements that elongate the body axis. We show that the LIM-domain protein Ajuba is recruited to adherens junctions in a tension-dependent fashion during axis elongation. Ajuba localizes to sites of myosin accumulation at adherens junctions within seconds, and the force-sensitive localization of Ajuba requires its N-terminal domain and two of its three LIM domains. We demonstrate that Ajuba stabilizes adherens junctions in regions of high tension during axis elongation, and that Ajuba activity is required to maintain cell adhesion during cell rearrangement and epithelial closure. These results demonstrate that Ajuba plays an essential role in regulating cell adhesion in response to mechanical forces generated by epithelial morphogenesis.
Introduction
During development, epithelial cells undergo dynamic changes in cell interactions that are necessary for tissue organization, remodeling, and repair. Adherens junction complexes are essential regulators of cell adhesion that are necessary for cell interactions and epithelial integrity. Junctional complexes play critical roles in sustaining and transmitting mechanical forces between cells, but they can also be dynamically assembled and disassembled in response to force (Harris and Tepass, 2010; Lye and Sanson, 2011; Takeichi, 2014). In particular, epithelial cells are exposed to strong mechanical forces during development, when epithelial tissues undergo rapid reorganization in response to developmental signals. Mechanical forces are critical for epithelial morphogenesis in vivo and can influence adherens junction composition, organization, and dynamics in vitro (Gumbiner, 2005; Baum and Georgiou, 2011; Leckband and de Rooij, 2014; Hoffman and Yap, 2015). However, the mechanisms that maintain epithelial integrity under tension, and how these processes are regulated by the physiological forces experienced by cells in vivo, are not well understood.
Mechanical forces can have profound effects on the organization and stability of adherens junction complexes and the nature of the proteins associated with them. In cultured cells, high levels of tension disrupt cell adhesion (Sahai and Marshall, 2002), whereas intermediate levels of tension promote adherens junction clustering (Shewan et al., 2005) and growth (Yamada and Nelson, 2007). Mechanical load produces conformational changes in the core adherens junction protein α-catenin (Yonemura et al., 2010; Yao et al., 2014) and can directly promote the association between α-catenin and F-actin in vitro (Buckley et al., 2014). Many proteins are recruited to sites of increased tension in cells, indicating that a wide range of mechanotransduction mechanisms are activated at adherens junctions (Leerberg and Yap, 2013; Leckband and de Rooij, 2014). One class of proteins that responds to mechanical forces is the zyxin family of LIM domain proteins (Kadrmas and Beckerle, 2004; Schimizzi and Longmore, 2015). Zyxin localizes to sites of increased tension in cells, including adherens junctions, focal adhesions, and actin stress fibers (Yoshigi et al., 2005; Hirata et al., 2008; Sperry et al., 2010; Schiller et al., 2011; Smith et al., 2013; Oldenburg et al., 2015). Other members of the zyxin family, including the Ajuba proteins, localize to adherens junctions and focal adhesions (Marie et al., 2003; Rauskolb et al., 2014; Dutta et al., 2018; Ibar et al., 2018). Ajuba has been implicated in several biological processes, including Hippo signaling (Das Thakur et al., 2010; Reddy and Irvine, 2013; Rauskolb et al., 2014), cell differentiation (Kanungo et al., 2000; Feng et al., 2007), cell migration (Kisseleva et al., 2005; Pratt et al., 2005), and cell proliferation (Kanungo et al., 2000; Hirota et al., 2003). Ajuba binds to the core adherens junction protein α-catenin (Marie et al., 2003) and localizes to adherens junctions in the Drosophila melanogaster wing disc and in cultured mammalian epithelial cells in a tension-dependent fashion (Rauskolb et al., 2014; Ibar et al., 2018). Loss of Ajuba reduces keratinocyte cell aggregation in culture, in part as a result of altered Rac GTPase signaling (Marie et al., 2003; Nola et al., 2011; McCormack et al., 2017). However, despite intriguing links between Ajuba and adherens junctions, it is not known if Ajuba regulates cell adhesion or the cellular response to mechanical forces in vivo.
In contrast to the three Ajuba proteins in mammals—Ajuba, LIMD1, and WTIP—Drosophila has a single Ajuba protein. Drosophila Ajuba regulates Hippo signaling in the larval wing disc (Das Thakur et al., 2010; Reddy and Irvine, 2013; Rauskolb et al., 2014) but has not been shown to affect cell adhesion in this tissue, in which cells are relatively static (Gibson et al., 2006). During convergent extension in the Drosophila embryo, adherens junctions are dynamically remodeled in the plane of the tissue, inducing spatially regulated cell rearrangements that elongate the head-to-tail body axis (Blankenship et al., 2006; Simões et al., 2010; Levayer et al., 2011; Tamada et al., 2012). Planar polarized actomyosin networks associated with adherens junctions generate contractile forces that drive cell rearrangement (Bertet et al., 2004; Zallen and Wieschaus, 2004; Blankenship et al., 2006; Rauzi et al., 2008; Fernandez-Gonzalez et al., 2009). Here we show that Ajuba localizes to adherens junctions in a spatiotemporally regulated fashion during axis elongation. Ajuba localization is modulated by dynamic changes in actomyosin activity, and the tension-sensitive localization of Ajuba requires its N-terminal domain and two of the three LIM domains. We demonstrate that Ajuba stabilizes adherens junction proteins in regions of high tension during axis elongation, and Ajuba activity is required to maintain cell adhesion during cell rearrangement and epithelial closure in the developing embryo. These results demonstrate that Ajuba is a dedicated junctional regulator that is required to maintain cell adhesion in the presence of mechanical forces during epithelial remodeling.
Results
Ajuba localizes to a subset of adherens junctions during convergent extension
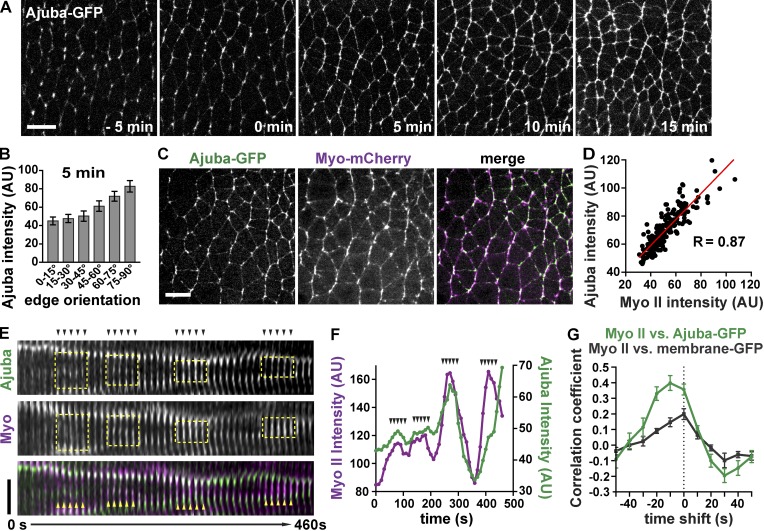
To investigate the role of Ajuba in epithelial remodeling, we first examined Ajuba localization during convergent extension in the Drosophila germband epithelium using a functional Ajuba-GFP fusion (Sabino et al., 2011). In stage 6 embryos before axis elongation, Ajuba-GFP localized primarily to cell vertices (Fig. 1 A, −5 min). During axis elongation in stages 7 and 8, Ajuba-GFP localized to cell–cell contacts in a planar polarized fashion (Fig. 1 A, 0–15 min). Ajuba-GFP was enriched 1.9 ± 0.1-fold (mean ± SEM) at vertical cell edges, which represent interfaces between neighboring cells along the anterior-posterior (AP) axis, compared with horizontal edges, which correspond to interfaces between dorsal and ventral cells (Fig. 1 B). Consistent with findings in other tissues (Marie et al., 2003; Rauskolb et al., 2014), Ajuba junctional localization in the Drosophila embryo requires α-catenin. Reduction of α-catenin levels by RNA interference resulted in a complete loss of Ajuba from cell interfaces, despite the continued presence of β-catenin (Fig. S1, A and B). These results demonstrate that Ajuba localizes to adherens junctions in a dynamic and spatially regulated fashion during convergent extension.
Figure 1.
Ajuba-GFP localization is spatiotemporally regulated and correlates with myosin. (A) Images from a time-lapse video of Ajuba-GFP during convergent extension. Stage 6 (−10 to 0 min), stage 7 (0–10 min), and stage 8 (10–30 min). (B) Ajuba-GFP intensity at cell interfaces of different orientations at t = 5 min). Angles are indicated relative to the AP axis, which is horizontal in all images (n = 6 embryos, 156–372 interfaces analyzed/embryo). (C) Image from a time-lapse video of Ajuba-GFP (green in merge) and Myo-mCherry (magenta in merge). (D) Myo-mCherry and Ajuba-GFP intensity are positively correlated during convergent extension. Each dot shows the mean Myo-mCherry and Ajuba-GFP intensity for a single interface, n = 190 interfaces in one embryo (R values from other embryos: 0.69, 0.72, 0.73, 0.77). (E) Kymograph of Ajuba-GFP (green) and Myo-mCherry (magenta) localization at a single cell interface. Yellow boxes highlight pulses of increased myosin and Ajuba localization. Arrowheads in E correspond to arrowheads in F. Images were acquired at 10-s intervals. (F) Mean Myo-mCherry and Ajuba-GFP intensity at the interface shown in E. (G) Correlation coefficient values between the change in Myo-mCherry interface intensity and the change in Ajuba-GFP (green line) or membrane-GFP (Resille-GFP, gray line). Ajuba-GFP and Resille-GFP data were shifted by the times shown on the x axis (n = 5 embryos/genotype, 10 edges analyzed/embryo). The mean ± SEM between embryos is shown in B and G. Images are anterior left, ventral down. Bars: (A and C) 10 µm; (E) 3 µm. See also Figs. S1 and S2.
Ajuba junctional localization is regulated by actomyosin contractility
We next investigated whether the spatiotemporal pattern of Ajuba localization is controlled by myosin contractility, as Ajuba localization is regulated by myosin activity in other contexts (Rauskolb et al., 2014; Ibar et al., 2018). During convergent extension, myosin II levels and mechanical tension are selectively increased at vertical cell interfaces (Bertet et al., 2004; Zallen and Wieschaus, 2004; Blankenship et al., 2006; Rauzi et al., 2008; Fernandez-Gonzalez et al., 2009; Kasza et al., 2014). In particular, mechanical tension is highest at linked vertical cell edges that form mechanically integrated, supracellular myosin cables within the tissue (Fernandez-Gonzalez et al., 2009). To determine whether Ajuba localization is sensitive to myosin levels, we analyzed the correlation between Ajuba-GFP and Myo-mCherry (a fusion of the myosin II regulatory light chain to mCherry; Martin et al., 2009). Ajuba and myosin levels at cell interfaces were positively correlated; cell interfaces with high myosin levels also had high levels of Ajuba (Fig. 1, C and D). Notably, Ajuba junctional localization was highest at linked vertical interfaces in supracellular cables (Fig. S2, A and B), which have the highest levels of myosin (Fig. S2 C) and are under the highest tension (Fernandez-Gonzalez et al., 2009). Ajuba did not colocalize with other myosin structures, such as myosin pulses at the medial cell cortex (Fig. S1 C), and Ajuba-GFP displayed a punctate localization at vertical cell interfaces that correlated more strongly with E-cadherin than with myosin (Fig. S1, D–F). These results indicate that Ajuba localizes to a subset of adherens junctions that are predicted to be under high tension during convergent extension.
Myosin localization at the cell cortex during convergent extension is highly dynamic, and pulses of myosin accumulation and dissociation occur on a timescale of seconds (Fig. 1, E and F; Rauzi et al., 2010). Ajuba localization was similarly dynamic: Ajuba levels at adherens junctions rapidly increased after pulses of myosin localization and rapidly diminished as myosin pulses dissipated (Fig. 1, E and F). To investigate the temporal relationship between Ajuba and myosin, we analyzed the relative timing of changes in Ajuba and myosin intensity in time-lapse videos of embryos imaged at 10-s intervals. The strongest correlations between the changes in intensity of Ajuba and myosin were observed when the Ajuba values were shifted back in time by 10 s, suggesting that transient increases in myosin at adherens junctions precede Ajuba localization (Fig. 1 G). Much weaker correlations were detected between myosin and a general membrane marker (Fig. 1 G). These results indicate that Ajuba is rapidly recruited to adherens junctions at the same time as or closely following dynamic increases in myosin localization.
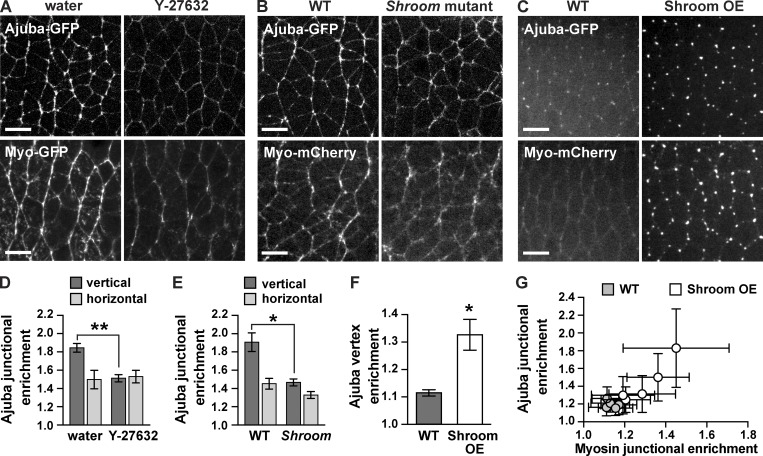
To examine whether the planar polarized localization of Ajuba requires myosin contractility, we used genetic and pharmacological methods to investigate the effects of altering myosin activity. Myosin activity was reduced by perivitelline injection of Y-27632, an inhibitor of Rho-kinase, which is required for myosin cortical localization throughout axis elongation (Bertet et al., 2004; Simões et al., 2010). In addition, we reduced myosin activity partway through elongation by genetically removing the myosin activator Shroom, which is required to maintain myosin cortical localization in stage 8 (Simões et al., 2014). Ajuba-GFP localization to vertical edges was strongly reduced in Y-27632–injected and Shroom mutant embryos, abolishing Ajuba planar polarity (Fig. 2, A, B, D, and E). Conversely, we increased myosin activity by overexpressing the ShroomA isoform of Shroom, which promotes Rho-kinase and myosin localization to adherens junctions (Nishimura and Takeichi, 2008; Bolinger et al., 2010; Simões et al., 2014). ShroomA overexpression enhanced myosin localization at tricellular vertices before axis elongation (Fig. 2, C and F) and at adherens junctions during elongation (Figs. 2 G and S1, D and E). Shroom overexpression recruited Ajuba to both myosin-positive domains (Fig. 2, C, F, and G; and Fig. S1, D and E). Ajuba levels at cell interfaces correlated with myosin levels on an embryo-by-embryo basis in Shroom-overexpressing embryos, suggesting that Ajuba junctional localization is proportional to myosin activity (Fig. 2 G). However, the finer-scale, punctate pattern of Ajuba localization correlated better with E-cadherin than with myosin when only vertical edges were considered (Fig. S1, D–F), consistent with the idea that Ajuba associates with a subset of adherens junctions that are under increased tension. Together, these data demonstrate that myosin promotes Ajuba junctional localization and planar polarity during convergent extension.
Figure 2.
Ajuba localization is sensitive to mechanical tension. (A) Ajuba-GFP and Myo-GFP localization in stage 7/8 embryos injected with water (left) or the Rho-kinase inhibitor Y-27632 (right). (B) Ajuba-GFP and Myo-mCherry in stage 8 WT (left) and Shroom mutant embryos (the progeny of ShroomΔ11/Df males and females; right). The accumulation of Ajuba and myosin at vertical interfaces is lost in Y-27632–injected and Shroom mutant embryos. (C) Ajuba-GFP and Myo-mCherry in stage 6 WT and ShroomA-overexpressing (Shroom OE) embryos. Overexpression of ShroomA recruits Ajuba and myosin to cell vertices. (D) Ajuba-GFP enrichment at vertical edges (oriented at 75–90° relative to the AP axis) and horizontal edges (oriented at 0–15° relative to the AP axis) in stage 7/8 water- or Y-27632–injected embryos (n = 5 embryos/condition, 70–99 edges analyzed/embryo). (E) Ajuba-GFP enrichment at vertical and horizontal edges in stage 8 WT and Shroom mutant embryos (n = 5 embryos/genotype, 106–283 edges analyzed/embryo). Edge intensities were divided by the cytoplasmic intensity to calculate the junctional enrichment. (F) Ajuba-GFP vertex enrichment in stage 6 WT and Shroom OE embryos (n = 5 embryos/genotype, 30 vertices analyzed/embryo). Vertex intensities were divided by the cytoplasmic intensity to calculate the vertex enrichment. (G) Ajuba-GFP enrichment at all edges in stage 7 WT (gray circles) and Shroom OE embryos (white circles; n = 5 embryos/genotype, 75–102 edges analyzed/embryo). The mean ± SEM between embryos is shown in D–F; each dot in G shows the mean ± SD for a single embryo; *, P < 0.01; **, P < 0.001, unpaired Student’s t test. Images are anterior left, ventral down. Bars, 10 µm. See also Fig. S1.
The preLIM domain and LIM domains 1 and 2 are necessary and sufficient for Ajuba tension-sensitive localization
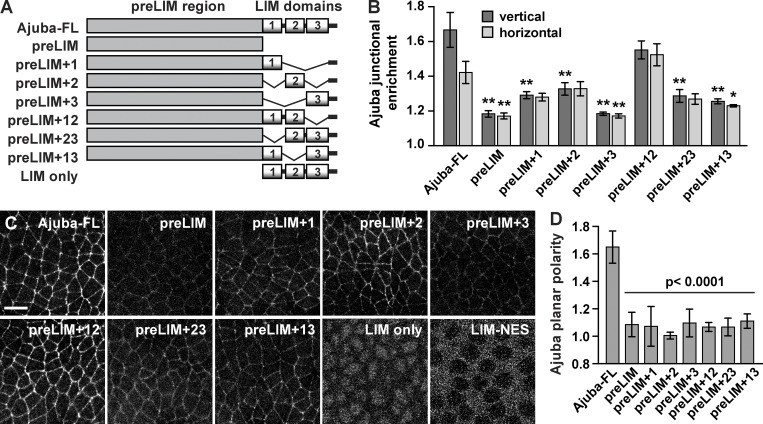
To understand how Ajuba localization is regulated by myosin activity, we first analyzed the domains required for Ajuba junctional localization. Ajuba is a 718-aa protein that contains a 505-aa N-terminal (preLIM) domain and three C-terminal LIM domains, each consisting of two tandem zinc finger motifs (Fig. 3 A; Michelsen et al., 1993; Goyal et al., 1999). The preLIM domain of mammalian Ajuba interacts with Rac GTPase and F-actin, and the LIM domains (and, to a lesser extent, the preLIM domain) interact with α-catenin (Marie et al., 2003; Nola et al., 2011). We tagged Ajuba variants with a C-terminal monomeric superfolder Venus (msVenus) tag and expressed them under the control of a maternal Gal4 driver (Fig. 3 A). Variants that contain the Ajuba preLIM domain were expressed at similar levels, but variants lacking this domain were more weakly expressed (Fig. S3 A). Ajuba-msVenus was enriched at vertical edges in a planar polarized fashion (Fig. 3, B–D), similar to the localization of Ajuba-GFP (Fig. 1 A). In contrast, the Ajuba preLIM domain alone displayed weak cortical localization and was primarily cytoplasmic, and the LIM domains alone localized to the nucleus, failing to associate with the cortex even when fused to a nuclear export sequence that restricted the protein fusion to the cytosol (Fig. 3 C). Adding the preLIM domain to LIM domains 1 or 2, but not LIM domain 3, slightly increased Ajuba junctional localization, and adding both LIM domains 1 and 2 (the preLIM+12 variant) strongly enhanced Ajuba localization to both vertical and horizontal edges compared with the preLIM domain alone (Fig. 3, B and C). Despite the strong junctional localization of the preLIM+12 variant, this variant was not planar polarized (Fig. 3 D). This loss of planar polarity was not caused by a loss of myosin planar polarity, which was unaltered in preLIM+12-expressing embryos (Fig. S3, B and C). Other combinations of two LIM domains were not sufficient to promote strong Ajuba junctional localization (Fig. 3, B and C), and none of the Ajuba deletion variants were planar polarized (Fig. 3 D). These results demonstrate that the LIM domains of Ajuba are functionally distinct, and that the preLIM and all three LIM domains are necessary for Ajuba planar polarity.
Figure 3.
Ajuba junctional localization and planar polarity require the preLIM and LIM domains. (A) Schematic of Ajuba variants. All variants were tagged with C-terminal msVenus and expressed under the control of a maternal Gal4 driver. (B) Junctional enrichment of Ajuba variants at vertical edges (oriented at 75–90° relative to the AP axis) and horizontal edges (oriented at 0–15° relative to the AP axis) in stage 7 embryos. Edge intensities were divided by the cytoplasmic intensity to calculate the junctional enrichment. (C) Confocal images of living stage 7 embryos expressing Ajuba variants tagged with C-terminal msVenus. LIM only and LIM-NES images were acquired at a higher gain and in a more basal plane to show nuclear localization (LIM only) or exclusion (LIM-NES). Images are anterior left, ventral down. Bar, 10 µm. (D) Planar polarized localization of Ajuba variants (the background-subtracted vertical edge intensity divided by the background-subtracted horizontal edge intensity). The mean ± SEM between embryos is shown (n = 5 embryos/variant, 167–416 interfaces analyzed/embryo); *, P < 0.01; **, P < 0.001 compared with full-length Ajuba (Ajuba-FL); one-way ANOVA with Fisher’s least significant difference test. See also Fig. S3.
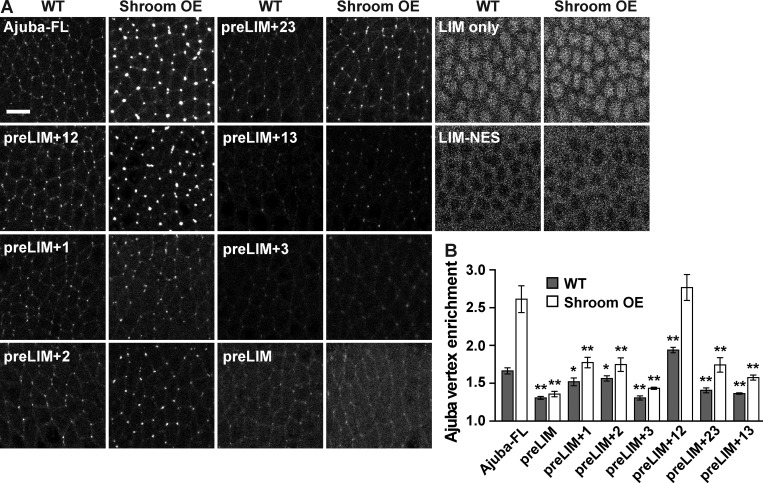
To determine which domains are required for the modulation of Ajuba localization by actomyosin contractility, we analyzed the response of Ajuba variants to increased tension caused by Shroom overexpression. Shroom overexpression in stage 6 embryos strongly enhanced the localization of Ajuba-msVenus to cell vertices (Fig. 4, A and B). In contrast, most of the Ajuba deletion variants responded weakly or not at all to Shroom overexpression (Fig. 4, A and B). The one exception was the Ajuba preLIM+12 variant, which was strongly recruited to vertices in Shroom-overexpressing embryos, to a similar extent as the full-length protein (Fig. 4, A and B). These results indicate that the preLIM domain and LIM domains 1 and 2 are necessary and sufficient for Ajuba to respond to increased tension caused by Shroom overexpression.
Figure 4.
Ajuba variants are differentially regulated by tension. (A) Images of Ajuba-msVenus variants in stage 6 WT and ShroomA-overexpressing (Shroom OE) embryos. LIM only and LIM-NES images were acquired at a higher gain and in a more basal plane to show nuclear localization (LIM only) or exclusion (LIM-NES). Images are anterior left, ventral down. Bar, 10 µm. (B) Vertex enrichment of Ajuba variants in WT (gray) and Shroom OE (white) embryos. Vertex intensities were divided by the cytoplasmic intensity to calculate the vertex enrichment. The mean ± SEM between embryos is shown (n = 8 embryos/variant, 30 vertices analyzed/embryo); *, P < 0.05; **, P < 0.001 compared with full-length Ajuba (Ajuba-FL); one-way ANOVA with Fisher’s least significant difference test. See also Fig. S3.
Ajuba regulates dynamic cell rearrangements during convergent extension
Thus far, we have shown that Ajuba localizes to adherens junctions in a tension-dependent fashion during convergent extension, during which planar polarized contractile forces drive oriented cell rearrangements within the tissue (Bertet et al., 2004; Zallen and Wieschaus, 2004; Blankenship et al., 2006; Rauzi et al., 2008; Fernandez-Gonzalez et al., 2009). These results raise the possibility that Ajuba could play a role in regulating cell adhesion in response to actomyosin contractility. To test this idea, we performed time-lapse imaging of convergent extension in embryos expressing β-catenin-GFP (McCartney et al., 2001; Videos 1 and 2). Embryos that lack maternal Ajuba expression were generated using two null alleles: AjubaII, which removes Ajuba and the neighboring gene (AMP-deam; Das Thakur et al., 2010), and Ajuba54, a targeted deletion of the Ajuba ORF that we generated using CRISPR mutagenesis (Fig. S4, A and B; Port et al., 2014). The complete loss of maternal and zygotic Ajuba function caused 100% penetrant lethality by the pupal stage, with some lethality at earlier stages (Fig. S4, C and D). The effects on convergent extension were similar in the presence or absence of zygotic Ajuba activity (Fig. S5, A, B, and D–G), and these embryos were combined for analysis (Figs. 5, 6, and 7).
Figure 5.
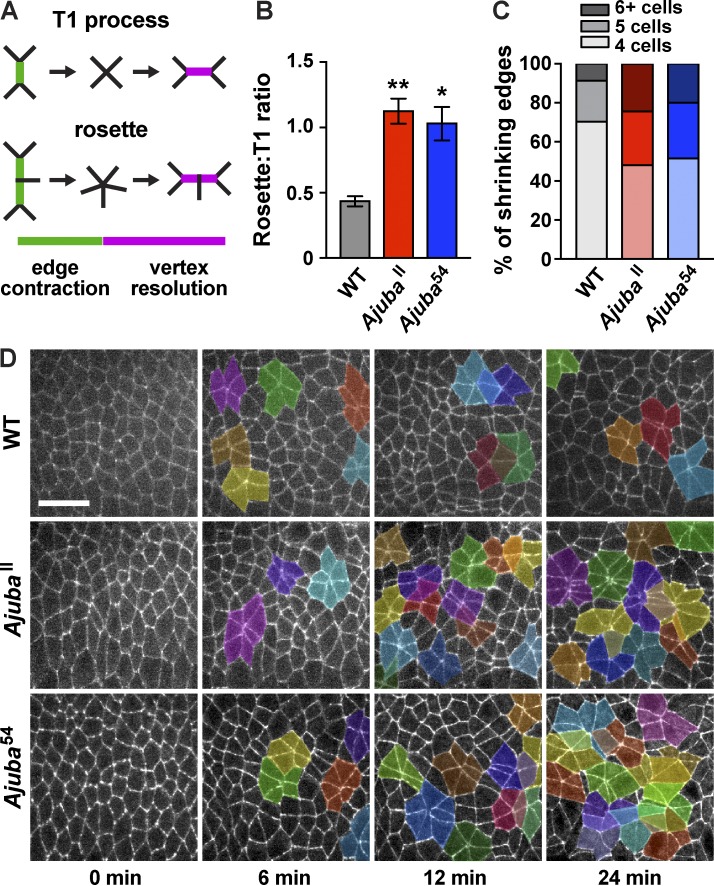
Loss of Ajuba activity enhances higher-order rosette behaviors. (A) Schematics of T1 and rosette processes. (B) Rosette:T1 ratio, calculated as the ratio of the number of shrinking edges that joined rosettes or T1 processes during axis elongation in WT, AjubaII, and Ajuba54 mutant embryos. The mean ± SEM between embryos is shown; *, P < 0.001; **, P < 0.0001; one-way ANOVA with Fisher’s least significant difference test. (C) Percentage of shrinking edges that formed four-cell vertices (T1 processes, light shading), five-cell rosettes (medium shading), or rosettes with six or more cells (dark shading; n = 12 embryos/genotype, 33–49 shrinking edges analyzed/embryo in B and C). (D) Stills from videos of WT and Ajuba mutant embryos with rosettes of five or more cells highlighted. Anterior left, ventral down. Bar, 20 µm. See also Figs. S4 and S5 and Videos 1 and 2.
Figure 6.
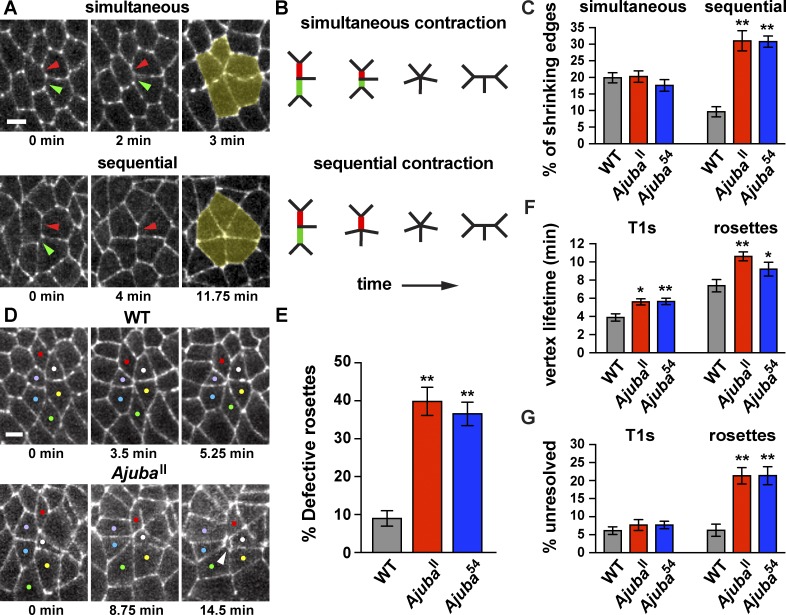
Ajuba mutants display defective cell adhesion in rosettes. (A and B) Images (A) and schematics (B) of rosette formation through simultaneous or sequential contraction. Shrinking edges indicated by red and green arrowheads; rosettes highlighted in yellow. (C) Percentage of shrinking edges that formed rosettes through simultaneous or sequential contraction. (D) Images of rosettes from videos of WT and AjubaII mutant embryos. Neighboring cells are closely apposed during rosette formation in WT, but gaps appear between cells in Ajuba mutants (white arrowhead). Colored dots indicate cells in rosettes. (E) Percentage of rosettes that displayed gaps at rosette vertices. (F) Lifetime of high-order vertices in T1 processes (T1s) or rosettes. (G) Percentage of shrinking edges that formed vertices that did not resolve. The mean ± SEM between embryos is shown (n = 12 embryos/genotype, 33–49 shrinking edges analyzed/embryo in C, F, and G; 7–21 rosettes analyzed/embryo in E); *, P < 0.05; **, P < 0.001; one-way ANOVA with Fisher’s least significant difference test. Cells are labeled with β-catenin-GFP. Images are anterior left, ventral down. Bar, 5 µm. See also Figs. S4 and S5 and Videos 3 and 4.
Figure 7.
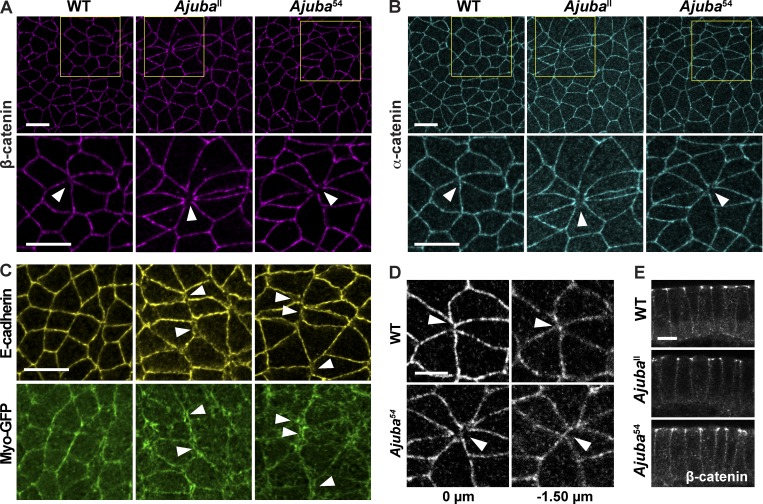
Ajuba mutants have defects in junctional protein localization at sites of multicellular adhesion. (A and B) WT and Ajuba mutant embryos immunostained for β-catenin (A) or α-catenin (B). Gaps devoid of β-catenin and α-catenin occur at rosette vertices in Ajuba mutants (arrowheads). Yellow boxes (top) show the location of cells in the high-magnification images (bottom). (C) Embryos immunostained for E-cadherin and Myo-GFP. In WT embryos, E-cadherin and myosin in neighboring cells are closely apposed at cell interfaces. In Ajuba mutants, E-cadherin signal is disrupted at vertical edges in forming rosettes, and myosin separates into two parallel bands (arrowheads). (D) Z slices of rosettes from confocal images in B of WT and Ajuba54 mutant embryos immunostained for α-catenin. (E) Cross-sections of WT and Ajuba mutant embryos immunostained for β-catenin. Images are anterior left, ventral down in A–D, and apical up in E. Bars: (A–C) 10 µm; (D and E) 5 µm. All embryos are shown at stage 8. See also Fig. S4.
Cell intercalation during convergent extension in Drosophila is driven by two types of cell rearrangement: rearrangements among four cells, in which a single vertical edge contracts to form a four-cell vertex, also known as a T1 process (Weaire and Rivier, 1984; Bertet et al., 2004), and rosette rearrangements in which multiple linked edges contract to bring five or more cells together at a single point (Blankenship et al., 2006; Fig. 5 A). In both cases, the formation of new contacts between cells that were previously separated promotes vertex resolution, which completes the cell rearrangement. As Ajuba preferentially localizes to linked shrinking edges in forming rosettes (Fig. S2, A and B), we analyzed rosette behaviors in Ajuba mutants. In WT embryos, T1 processes and rosettes occur in an ∼2:1 ratio (Fig. 5 B; Farrell et al., 2017). In contrast, more shrinking edges joined rosettes in Ajuba mutants, resulting in an increase in the number of rosette structures and nearly equal frequencies of rosettes and T1 processes (Fig. 5, B–D). This phenotype was rescued by expressing full-length Ajuba, demonstrating that the cell rearrangement defects in Ajuba mutants are caused by the loss of Ajuba activity (Fig. S3, D and E). In addition, the increased rosette formation in Ajuba mutants was rescued by the Ajuba preLIM+12 variant, but not the preLIM+3 variant (Fig. S3, D and E). These results indicate that the localization of Ajuba to adherens junctions, but not necessarily its planar polarized distribution, is important for its functions in cell rearrangement.
To identify the mechanisms that lead to increased rosette formation in Ajuba mutants, we analyzed cell rearrangements in more detail. Rosettes could form in one of two ways. In one mechanism, two or more connected edges could contract simultaneously (Fig. 6, A and B, top). Alternatively, edges could contract sequentially, first producing a four-cell vertex that remains stable for an extended period of time (defined here as ≥3 min) before joining a rosette (Fig. 6, A and B, bottom). Simultaneous contraction is the predominant mechanism of rosette formation in WT (Fig. 6 C). In addition, the percentage of shrinking edges that formed rosettes through simultaneous contraction was unchanged in Ajuba mutants. In contrast, Ajuba mutants displayed a more than threefold increase in the percentage of shrinking edges that formed rosettes through sequential contraction (Figs. 6 C and S5 E), accompanied by a decrease in the frequency of T1 processes (Figs. 5 C and S5 B). Therefore, the loss of Ajuba results in an increase in rosette formation through the conversion of T1 processes into rosettes through sequential contraction, a mechanism that rarely leads to rosette formation in WT.
The higher number of rosettes in Ajuba mutants could be caused by an increase in actomyosin contractility, which is required for rosette formation, or a defect in cell adhesion, which is dynamically regulated during rosette formation and resolution. To distinguish between these possibilities, we used laser ablation, a measure of the relative forces acting at cell edges, to investigate whether myosin contractility is altered in Ajuba mutants. The initial retraction velocity in response to ablation is predicted to be proportional to the force acting on that edge before ablation, assuming that the viscoelastic properties of the tissue are constant (Hutson et al., 2003). Retraction velocities after ablation are higher at vertical edges than at horizontal edges in WT embryos, demonstrating that mechanical forces in this tissue are planar polarized (Rauzi et al., 2008; Fernandez-Gonzalez et al., 2009). We observed a similar spatial pattern of retraction velocities in Ajuba mutants, indicating that mechanical forces are correctly generated and organized in the absence of Ajuba activity (Fig. S5, H and I). Consistent with these results, the total number of shrinking edges was not altered in Ajuba mutants. In WT embryos, 87 ± 1% of edges that were vertical at the onset of elongation contracted to a vertex, compared with 82 ± 2% in AjubaII and 87 ± 2% in Ajuba54 embryos (mean ± SEM, n = 12 embryos, 50 edges tracked per embryo). These results indicate that Ajuba does not affect the level or distribution of myosin contractility. To test the alternative possibility that Ajuba mutants have defects in cell adhesion, we analyzed the formation of new contacts between cells. We found that high-order vertices were slower to resolve for both T1 processes and rosettes in Ajuba mutants (Figs. 6 F and S5 F). In addition, there was a significant increase in the percentage of rosettes that did not resolve by the end of elongation (Figs. 6 G and S5 G). Despite these defects, a majority of rosette structures completed resolution, and the extent of tissue elongation was not affected in Ajuba mutants (Fig. S5 C). These results indicate that Ajuba regulates the nature and dynamics of cell rearrangement during convergent extension.
Ajuba is required to maintain cell adhesion in regions of high tension
The defective rosette behaviors that occur despite normal levels of myosin contractility in Ajuba mutants are suggestive of a role for Ajuba in regulating cell adhesion. However, it is not immediately clear how defects in cell adhesion could lead to increased rosette formation and reduced rosette resolution. In one model, cell adhesion at multicellular vertices could be increased in Ajuba mutants, allowing cells to remain in higher-order configurations for a longer time. Alternatively, cell adhesion could be reduced in Ajuba mutants, producing gaps between cells that impede the formation of new cell contacts required for vertex resolution. To distinguish between these possibilities, we analyzed cell behaviors in time-lapse videos of Ajuba mutant embryos. In WT embryos, adherens junction proteins are tightly apposed at interfaces between adjacent cells (Fig. 6 D and Video 3). This distribution indicates that cell adhesion is normally maintained throughout junctional remodeling. In contrast, Ajuba mutants often displayed aberrant gaps between cells that appeared to represent breaks in adhesion (Fig. 6 D and Video 4). These defects were most pronounced in rosettes: 40 ± 4% of rosettes in AjubaII mutants and 37 ± 3% of rosettes in Ajuba54 mutants displayed a visible gap between cell interfaces at late stages of rosette formation, compared with 9 ± 2% of rosettes in WT embryos (Figs. 6 E and S5 D). Transient gaps between cells in Ajuba mutants could account for the delay in the resolution of high-order vertices (Fig. 6, F and G). These results indicate that Ajuba is required to maintain cell adhesion at late stages of rosette formation, when cells are predicted to be under the highest level of tension within the tissue.
We next asked if the cell adhesion defects in Ajuba mutants are caused by a mislocalization of adherens junction proteins. The apical distributions of the core adherens junction proteins α-catenin and β-catenin were not obviously affected in Ajuba mutants (Fig. 7, A, B, and E). However, unlike WT embryos, α-catenin and β-catenin were often absent from rosette vertices in Ajuba mutants (Fig. 7, A and B). In addition, E-cadherin localization at shrinking edges was often faint and diffuse, and cortical myosin structures in neighboring cells often appeared as two parallel lines in Ajuba mutants, in contrast to a single line in WT (Fig. 7 C). Defects in myosin localization at shrinking edges were not associated with a loss of myosin planar polarity or with obvious gaps between cells until late stages of rosette formation. These results suggest that subtle defects in protein localization at shrinking edges in Ajuba mutants precede a more pronounced separation of cells at or near rosette vertices. Although α-catenin localization was visibly disrupted at the apical cell surface, cell contacts appeared intact in more basal planes (Fig. 7 D). These results indicate that Ajuba is required to stabilize cell adhesion and apical adherens junction localization during convergent extension.
Dorsal closure defects in Ajuba mutants are enhanced by reducing E-cadherin
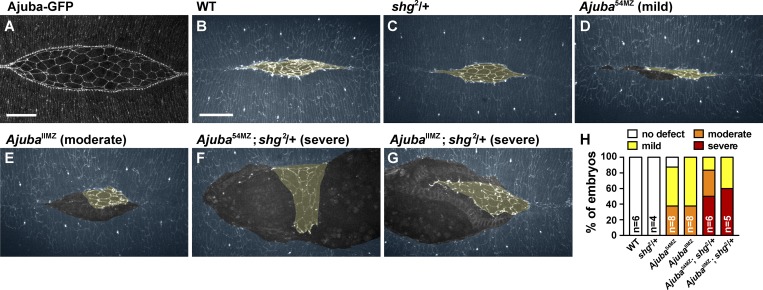
We next sought to determine whether the roles of Ajuba during convergent extension reflect a broader requirement for Ajuba activity in stabilizing cell adhesion under tension. In particular, Ajuba function is expected to be particularly important for tissues in which the level of tension is high or the level of adhesion is low. To investigate this possibility, we analyzed epithelial remodeling in another context in which cells are exposed to strong mechanical forces during development. Dorsal closure is a morphogenetic process driven by actomyosin contractility in the late Drosophila embryo (Kiehart et al., 2000; Hutson et al., 2003). During this process, lateral epidermal sheets on both sides of the embryo come together over the amnioserosa and fuse at the dorsal midline (Fig. 8 B and Video 5; Harris, 2017). High levels of tension are generated at the interface between the lateral epidermis and the amnioserosa, but, despite this tension, these tissues remain tightly apposed throughout closure. Mutants defective for adherens junction proteins disrupt this attachment, resulting in the separation of the amnioserosa from the lateral epidermis (Gorfinkiel and Martinez-Arias, 2007). We found that Ajuba-GFP localizes to the leading edge of the lateral epidermis in a punctate fashion (Fig. 8 A), coinciding with adherens junctions that are associated with a tensile actomyosin cable (Kiehart et al., 2000; Gorfinkiel and Martinez-Arias, 2007). Time-lapse imaging of dorsal closure in stage 14 embryos revealed the formation of small gaps between the lateral epithelium and the amnioserosa during the final zippering phase of dorsal closure in Ajuba mutants (mild defect, Fig. 8 D and Video 7). In addition, a subset of embryos displayed a larger separation of the epidermis from the amnioserosa at earlier stages of closure (moderate defect, Fig. 8 E and Video 8). These results indicate that Ajuba is required to maintain cell adhesion under tension during dorsal closure. If this is the case, then reducing cell adhesion is predicted to enhance the defects in Ajuba mutants. To test this, we introduced one mutant allele of E-cadherin (Drosophila shotgun (shg)) into Ajuba maternal and zygotic mutants to reduce zygotic E-cadherin levels. Reducing zygotic E-cadherin expression by half did not disrupt dorsal closure on its own (Fig. 8 C and Video 6). However, this significantly enhanced the dorsal closure defects in Ajuba mutants, resulting in a catastrophic separation of the epidermis from the amnioserosa early in dorsal closure (severe defect, Fig. 8, F–H; and Videos 9 and 10). These results demonstrate that Ajuba is required to stabilize cell adhesion during multiple epithelial remodeling events in the Drosophila embryo.
Figure 8.
Loss of Ajuba enhances the dorsal closure defects in cell adhesion mutants. (A) Ajuba-GFP localization in a living embryo during dorsal closure. (B–G) Stills from time-lapse videos of stage 14 embryos expressing β-catenin-GFP. WT (B), E-cadherin heterozygote (shg2/+) (C), Ajuba54MZ maternal/zygotic mutant (D), AjubaIIMZ maternal/zygotic mutant (E), Ajuba54MZ maternal/zygotic mutant that is also heterozygous for E-cadherin (F; Ajuba54MZ; shg2/+), and AjubaIIMZ maternal/zygotic mutant that is also heterozygous for E-cadherin (G; AjubaIIMZ; shg2/+). The lateral epidermis (annotated in blue) is connected to the amnioserosa (annotated in yellow) in WT but becomes separated in Ajuba mutants. (H) Percentage of embryos showing no defect or mild, moderate, or severe dorsal closure phenotypes. Images are dorsal views, anterior left. Bars, 30 µm. See also Fig. S4 and Videos 5–10.
Discussion
The conserved LIM domain protein Ajuba has been shown to localize to adherens junctions in a tension-sensitive fashion, but has not previously been demonstrated to regulate cell adhesion in vivo. Here we show that Ajuba is required to stabilize adherens junctions under tension during multiple epithelial remodeling events in the Drosophila embryo. Ajuba localizes to adherens junctions in a dynamic and planar polarized fashion that is spatially and temporally regulated by actomyosin contractility. The Ajuba preLIM domain and LIM domains 1 and 2 are necessary for Ajuba to localize strongly to adherens junctions and respond to changes in myosin activity. Ajuba mutants display defects in the localization of adherens junction proteins in multicellular rosette structures, altering the nature and dynamics of cell rearrangement during convergent extension. In addition, the loss of Ajuba activity results in the formation of large gaps between tissues during dorsal closure in embryos with reduced levels of E-cadherin. These results demonstrate that Ajuba is required to maintain cell adhesion at sites of dynamic changes in cell interactions induced by mechanical forces during epithelial remodeling.
These studies demonstrate a novel role for Ajuba in regulating dynamic cell behaviors in actively remodeling epithelial tissues. Cell adhesion is not generally disrupted in Ajuba mutants. Instead, we demonstrate that the ability to rapidly resolve higher-order interactions among cells requires Ajuba activity. In particular, Ajuba is specifically required for dynamic changes in adhesion in multicellular rosette structures, which are under high levels of tension during convergent extension. Although increased actomyosin contractility can enhance rosette formation (Kasza et al., 2014), myosin activity was not affected in Ajuba mutants. Instead, we show that Ajuba mutants have defects in cell adhesion and adherens junction localization at high-order vertices, which could impede the formation of new contacts between cells and delay the resolution of T1 processes, resulting in the conversion of these structures into rosettes. Rosette behaviors, originally discovered in Drosophila (Blankenship et al., 2006), are commonly observed during convergent extension in vertebrates, including the mouse and Xenopus laevis kidney (Lienkamp et al., 2012), the mouse cochlea and limb bud (Chacon-Heszele et al., 2012; Lau et al., 2015), the chick primitive streak (Rozbicki et al., 2015), and the chick and mouse neural plate (Nishimura and Takeichi, 2008; Nishimura et al., 2012; Williams et al., 2014). Rosettes also occur in other processes of epithelial remodeling, such as in the zebrafish lateral line (Lecaudey et al., 2008), during wound healing (Razzell et al., 2014), and during ommatidial rotation (Mirkovic et al., 2011). These structures likely represent specialized sites of increased tension within tissues, which may place unique demands on adherens junctions and necessitate distinct mechanisms of junctional regulation. Notably, the loss of Ajuba causes striking defects in embryos with reduced cell adhesion, resulting in the large-scale separation of tissues during dorsal closure. This process is also characterized by strong mechanical forces (Kiehart et al., 2000; Hutson et al., 2003). We propose that Ajuba activity may be generally required to reinforce cell adhesion in situations when cell adhesion is low or myosin contractility is high, which are common characteristics of actively remodeling epithelia.
Ajuba localization is highly dynamic and correlates with changes in junctional myosin levels, following peaks of myosin localization within seconds. How Ajuba responds to tension on such a rapid timescale is unclear. In one model, Ajuba could act as a mechanosensor, undergoing a conformational change in response to mechanical force that stabilizes its association with adherens junctions. Alternatively, Ajuba could be recruited to adherens junctions by force-dependent changes in other proteins. A likely candidate for recruiting Ajuba to adherens junctions is α-catenin, which binds to Ajuba and is required for Ajuba junctional localization in vitro (Marie et al., 2003) and in vivo (Rauskolb et al., 2014; this study). Mechanical stretching of α-catenin exposes a central domain in α-catenin that allows it to bind to vinculin (Ishiyama et al., 2013; Yao et al., 2014). In addition, tension stabilizes the interaction between α-catenin and F-actin through a catch bond mechanism (Buckley et al., 2014). Other known binding partners of Ajuba family proteins include Shroom (Chu et al., 2018) and the actin cytoskeleton (Marie et al., 2003; Nola et al., 2011). As Ajuba localization in the Drosophila embryo requires both α-catenin and actomyosin contractility, these results raise the possibility that both actomyosin structures and junctional components contribute to Ajuba recruitment to adherens junctions under tension. Consistent with this possibility, we show that Ajuba localization, function, and response to mechanical forces requires the preLIM domain, which binds to F-actin, as well as the first two LIM domains, which are part of a region that interacts with α-catenin. These two structural requirements for force-dependent Ajuba localization indicate that multiple binding sites are required to recruit Ajuba to adherens junctions under tension.
Interactions between adherens junctions and the actin cytoskeleton are essential for cell adhesion and tissue integrity (Yonemura et al., 2010; Desai et al., 2013). In particular, the interaction between α-catenin and F-actin has been proposed to function as a catch bond that is stabilized in the presence of force (Buckley et al., 2014). This raises the question of why additional proteins such as Ajuba are required to stabilize adhesion complexes, if conformational changes in α-catenin alone are sufficient to strengthen the association between adherens junctions and F-actin. One possibility is that the interaction between α-catenin and F-actin is not stable enough to withstand the strong mechanical forces required for complex junctional remodeling events involving multiple cells. Ajuba could act as an additional, stabilizing link between adherens junctions and the cortical actomyosin network in high-stress regions, reminiscent of the role of vinculin, which stabilizes the interaction between α-catenin and F-actin (le Duc et al., 2010; Huveneers et al., 2012; Twiss et al., 2012). Alternatively, Ajuba could recruit other proteins to adherens junctions that modulate junctional organization or dynamics. Further studies are needed to define the mechanisms that promote the dynamic remodeling of adhesion under tension. Ajuba has been shown to function in a wide range of processes, including cell differentiation, cell proliferation, cell migration, and Hippo signaling. It will be interesting to determine how the effects of Ajuba on dynamic transitions in cell adhesion during tissue remodeling are related to its diverse roles in epithelial development.
Materials and methods
Fly stocks and genetics
Stocks used for live or fixed imaging were the Ajuba-GFP BAC (Sabino et al., 2011), Myo-mCherry (sqh-mCherry, the myosin regulatory light chain fused to mCherry expressed from the sqh promoter; Martin et al., 2009), Resille-GFP (gift of A. Debec, Institut Jacques Monod, Paris, France), Myo-GFP (sqh-GFP expressed from the sqh promoter; Royou et al., 2004), E-cadherin-mTomato (shg-mTomato expressed from the endogenous promoter; Huang et al., 2009), β-catenin-GFP (arm-GFP expressed from the endogenous promoter; McCartney et al., 2001), gap43-mCherry (expressed from the sqh promoter; Martin et al., 2010), ShroomΔ11 (Simões et al., 2014), Df(2R)Exel7131 (Parks et al., 2004), UAST-ShroomA (Bolinger et al., 2010), UASp-Ajuba-msVenus variants (this study), AjubaII (Das Thakur et al., 2010), Ajuba54 (this study), and shg2 (E-cadherin; Nüsslein-Volhard et al., 1984). Shroom mutants were the progeny of ShroomΔ11/Df(2R)Exel7131 females and males that were heterozygous for Ajuba-GFP BAC or sqh-mCherry (III). Shroom-overexpressing embryos were the F2 progeny of UAST-ShroomA; sqh-mCherry; Ajuba-GFP BAC or UAST-ShroomA; E-cadherin-mTomato; Ajuba-GFP BAC males × matαtub67;15 Gal4 females (gift of D. St Johnston, University of Cambridge, Cambridge, UK). The localization of Ajuba-msVenus variants was analyzed in the F2 progeny of UASp-Ajuba-msVenus variant males × matαtub67;15 Gal4 females. The localization of Ajuba-msVenus variants in Shroom-overexpressing embryos was analyzed in the F2 progeny of UAST-ShroomA; UASp-Ajuba-msVenus variant males × matαtub67;15 Gal4 females. The localization of sqh-mCherry in Ajuba-msVenus variant–expressing embryos was analyzed in the F2 progeny of sqh-mCherry; UASp-Ajuba-msVenus variant males × matαtub15 Gal4 females. The α-catenin knockdown (KD) embryos were the F2 progeny of matαtub67 Gal4 females × P{TRiP.HMS00317}attP2 males (generated by the Transgenic RNAi Project at Harvard Medical School; Perkins et al., 2015). Crosses and embryo collections for imaging Ajuba-GFP, Ajuba-msVenus variants, and Shroom mutants were performed at 25°C. Shroom-overexpressing embryos and controls were collected at 18°C, and Ajuba mutant embryos and controls were collected at 20°C. All embryos were imaged live at room temperature except for the fixed embryos in Figs. 7, S1 B, S2 (C and D), and S3 (D and E).
AjubaII and Ajuba54 (simplified to Ajuba* here and in Fig. S4) germline clone embryos were made by heat-shocking flies of the following genotypes as larvae:
Ajuba*, FRT19A/ovoD2, FRT19A; arm-GFP, hs-flp/+
Ajuba*, FRT19A/ovoD2, FRT19A; sqh-GFP, hs-flp/+
Ajuba*, FRT19A/ovoD2, FRT19A; hs-flp/+; matαtub15/Ajuba-msVenus variants
These females were crossed to FM7a, Dfd-YFP/Y males, and progeny were imaged live or fixed for immunostaining. For live imaging, embryos that were maternally mutant for Ajuba (genotyped by the presence of Dfd-YFP signal) and embryos that were maternally and zygotically mutant for Ajuba (genotyped by the absence of Dfd-YFP signal) were analyzed separately (Fig. 8 and Fig. S5, A, B, and D–G) or combined for analysis (Figs. 5, 6, 7, and S5, C, H, and I). For dorsal closure videos, Ajuba*, FRT19A/ovoD2, FRT19A; arm-GFP, hs-flp/+ females were heat-shocked as larvae and crossed to FM7a, Dfd-YFP/Y or FM7a, Dfd-YFP/Y; shg2/CyO, twi-Gal4, UAS-GFP males. Embryos that were maternally and zygotically mutant for Ajuba and heterozygous for shg2 were identified by the absence of GFP signal from the FM7a, Dfd-YFP and CyO, twi-Gal4, UAS-GFP balancers. For the lethality analysis (Fig. S4, C and D), Ajuba maternal mutants were selected as stage 14–17 embryos or as L1 larvae and genotyped based on the presence or absence of the FM7a, Dfd-YFP balancer and allowed to develop before calculating the percentage of crawling larvae (for embryos) or pupal cases and adult flies (for larvae).
Ajuba deletion allele
The Ajuba54 allele was generated using CRISPR mutagenesis (Fig. S4, A and B; Port et al., 2014). Two gRNAs were ubiquitously expressed in transgenic embryos using the pCFD4 plasmid backbone. One gRNA targeted the 5′ end of the Ajuba ORF, just after the start codon, and the other gRNA targeted the Ajuba 3′ UTR. The pCFD4-Ajuba-gRNA plasmid was cloned by two-part Gibson assembly of BbsI-digested pCFD4 with a gel-purified PCR fragment amplified from pCFD4 using Ajuba gRNA primers 5′-TATATAGGAAAGATATCCGGGTGAACTTCGAATGGCGCCCGGTGAGCGCGTTTTAGAGCTAGAAATAGCAAG-3′ and 5′-ATTTTAACTTGCTATTTCTAGCTCTAAAACCTCAGTCGAAGGTCGGAAGGCGACGTTAAATTGAAAATAGGTC-3′ with NEBuilder HiFi 2× Master Mix (New England Biolabs) according to the protocol on http://www.crisprflydesign.org. The pCFD4-Ajuba-gRNA plasmid was inserted into the attP2 landing site on chromosome III. Transgenic males bearing pCFD4-Ajuba-gRNA were crossed to vasa:Cas9 females, and the female F1 progeny were crossed to FM7a, Dfd:YFP/Y males. Individual F2 flies were balanced with FM7a, Dfd:YFP. Deletion of the Ajuba ORF was detected by PCR with primers flanking the gRNA target sites (5′-GGAGCAGGATCTGGTGGATA-3′ and 5′-TGCCAACTGTCGGAGATTTT-3′) and confirmed by sequencing the deletion breakpoints (a 3.4-kb deletion from just inside the first exon to the beginning of the 3′ UTR; Fig. S4 A). The Ajuba54 mutation was recombined with FRT19A for use in making germline clones, and the presence of FRT19A was confirmed by PCR using published primers (Collins et al., 2012).
Transgenic lines
To generate the UASp-Ajuba-msVenus constructs, the full-length Ajuba ORF (2,154 nucleotides) was PCR-amplified and cloned into pEntr/D-TOPO (Invitrogen) using the following primers: 5′-CACCATGACCACCCAGCGGACGCA-3′ and 5′-TCCCATATACTGGTACGAAG-3′. The full-length Ajuba coding sequence was PCR amplified from pEntr-Ajuba and cloned into a UASp C-terminal msVenus backbone digested with BamH1 by Gibson assembly (NEBuilder HiFi 2x Master Mix; New England Biolabs). The monomeric superfolder Venus (msVenus) tag was a gift of B. Glick (University of Chicago, Chicago, IL). Ajuba deletion variants were cloned as two- or three-part Gibson assembly reactions using the same backbone with a combination of PCR fragments amplified from UASp-Ajuba-msVenus and gBlock (IDT) sequences for the deletion variants. Transgenes were inserted into the attP2 site on chromosome III. The following C-terminally msVenus-tagged constructs were generated in this study: Ajuba-FL (aa 1–718), Ajuba preLIM (aa 1–505), Ajuba LIM only (aa 506–718), Ajuba LIM-NES (aa 506–718 plus the NES sequence from PKI, LALKLAGLDI [Wen et al., 1995], fused at the C terminus after msVenus), Ajuba preLIM+1 (aa 1–559 and 693–718), Ajuba preLIM+2 (aa 1–505, 571–623, and 693–718), Ajuba preLIM+3 (aa 1–505 and 631–718), Ajuba preLIM+12 (aa 1–623 and 693–718), Ajuba preLIM+23 (aa 1–505, 571–718), and Ajuba preLIM+13 (aa 1–570 and 631–718).
Live imaging
Embryos were dechorionated for 2 min in 50% bleach and mounted in a 1:1 mixture of halocarbon oils 27 and 700 (Sigma) or halocarbon oil 27 alone on a gas-permeable membrane (YSI). For imaging the Ajuba-GFP BAC in WT, Shroom mutant, and Y-27632–injected embryos and the Ajuba-msVenus variants, z-stacks were acquired on a Zeiss LSM 700 confocal microscope with a PlanNeo 40×/1.3-NA oil-immersion objective (1.5 µm optical section and 0.76 µm z-steps) using Zen software (Zeiss). For imaging the Ajuba-GFP BAC with Myo-mCherry or E-cadherin-mTomato in WT and Shroom-overexpressing embryos, z-stacks were acquired at 0.5-µm z-steps on a PerkinElmer Ultraview Vox spinning disk confocal with a Zeiss Plan Neofluor 63×/1.4-NA oil-immersion objective and a Hamamatsu Orca-R2 camera using Volocity software. For time-lapse imaging of axis elongation in WT and Ajuba mutant embryos, z-stacks of embryos expressing β-catenin–GFP were acquired at 15-s intervals and 0.5-µm z-steps on an Ultraview Vox spinning disk confocal with a Zeiss Plan Neofluor 40×/1.3-NA oil-immersion objective using Volocity software. For time-lapse imaging of dorsal closure, z-stacks were acquired at 1-min intervals and 0.5-µm z-steps on a PerkinElmer Ultraview RS5 spinning disk confocal with a Zeiss Plan Neofluor 40×/1.3-NA oil-immersion objective and a Hamamatsu Orca-ER camera using Volocity software. A maximum-intensity projection of the apical-most slices was used for analysis. Pupae were imaged on a Zeiss stereo microscope with a Canon EOS T6i camera using EOS Utility 3 software (Canon).
Laser ablations
Embryos expressing β-catenin–GFP were ablated using an N2 Micropoint laser (Photonics Instruments) tuned to 365 nm. An Ultraview RS5 spinning disk confocal with a Zeiss Plan Neofluor 63×/1.4-NA oil-immersion objective was used to focus the ablation laser and acquire images before and after ablation. Z-stacks at 0.5-µm steps in the region of the adherens junctions were acquired every 2.7 s. The retraction velocity of the vertices after ablation was measured in ImageJ (National Institutes of Health). The retraction velocity was calculated as the velocity at which the two vertices of the ablated interface move apart in the first 2.7 s after ablation.
Immunohistochemistry
Fixed embryos were analyzed in Figs. 7, S1 B, S2 (C and D), and S3 (D and E). All other images shown in this study are of living embryos. To visualize α-catenin and armadillo/β-catenin, embryos were boiled for 10 s in 0.03% Triton X-100/0.4% NaCl and devitellinized in heptane/methanol. To visualize Myo-GFP and E-cadherin, embryos were fixed for 10 min in a 1:1 mixture of 37% FA/0.1 M phosphate buffer, pH 7.2, and hand devitellinized in 0.1 M phosphate buffer. Antibodies used were anti–α-catenin (1:50; Developmental Studies Hybridoma Bank), rabbit anti–armadillo/β-catenin (1:100, made by J.A. Zallen as described by Riggleman et al., 1990), rat anti–E-cadherin (1:50; Developmental Studies Hybridoma Bank), and rabbit anti-GFP (1:150; Torrey Pines). Secondary antibodies conjugated to Alexa Fluor 488, 546, or 647 fluorophores (Molecular Probes) were used at a concentration of 1:500. Embryos were mounted in ProLong Gold (Invitrogen) between two coverslips for imaging. Images were acquired on a Zeiss LSM 700 confocal microscope with a PlanNeo 40×/1.3-NA oil-immersion objective (1.1-µm optical section and 0.56-µm z-steps).
Drug injections
For injections of Rho-kinase inhibitor (Y-27632), stage 6 embryos were dechorionated for 2 min in 100% bleach, attached to coverslips with heptane glue, and desiccated for 7–9 min in Drierite. Embryos were covered in a 1:1 mixture of halocarbon oils 27 and 700 (Sigma) and microinjected laterally into the perivitelline space with 1 mM Y-27632 (Millipore) diluted in water, or water alone as a control. The Y-27632 concentration is predicted to be diluted 50-fold in the embryo. Embryos were imaged live 2–8 min after injection.
Immunoblotting
For Western blot analysis, 40 stage 6–8 embryos were hand-selected, crushed with a glass needle, and boiled in 20 µl of 1.5× SDS buffer with 2% β-mercaptoethanol. Samples were run on Bis-Tris 4–12% protein gels (NuPage; Invitrogen) and transferred onto polyvinylidene fluoride membrane (Immobilon-P; Millipore). Antibodies used were mouse anti-GFP (1:2,000, Roche) and mouse anti–armadillo/β-catenin (1:250; Developmental Studies Hybridoma Bank). Protein bands were detected using chemiluminescence with goat anti–mouse IgG HRP-conjugated secondary antibodies (Jackson Laboratory) and Amersham ECL prime reagent (GE Healthcare). Immunoblots were imaged using a Fujifilm LAS-3000 imager.
Edge and vertex intensity measurements
Protein localization was analyzed in maximum-intensity projections of 1.5–4.5 µm in the region of the apical adherens junctions using SIESTA software (Fernandez-Gonzalez and Zallen, 2011) or ImageJ. Lines on cell edges were drawn by the user in the anterior and central regions of the germband, and the mean pixel intensity and orientation were measured for each edge. Intensities were averaged for all edges in a 0–15° angular range (horizontal edges) and 75–90° angular range (vertical edges) relative to the AP axis. Vertex and edge intensities were divided by the cytoplasmic intensity to calculate the relative enrichment. Ajuba-GFP BAC vertex enrichment (Fig. 2 F) was lower than for UAS-Ajuba FL-msVenus (Fig. 4 B) because different transgenes were used. The cytoplasmic pixel intensity was calculated in SIESTA as the mean intensity of all pixels not included in the edge measurements (Fig. 1 B and Fig. 2, D and E) or in ImageJ by averaging the mean intensity of a circular cytoplasmic region for ≥10 cells (Fig. 2, F and G; Fig. 3, B and D; Fig. 4 B; and Fig. S3 C). Planar polarity was calculated as (mean pixel intensity at vertical edges – mean cytoplasmic pixel intensity)/(mean pixel intensity at horizontal edges – mean cytoplasmic pixel intensity) (Fig. 3 D). In Fig. S2, lines on cell edges were manually drawn on single frames from a time-lapse video (Fig. S2 B) or on images of immunostained embryos (Fig. S2, C and D). Vertex intensities were measured in ImageJ using circular regions of interest of 2.4 µm (Fig. 2 F) or 1.0 µm (Fig. 4 B) in diameter. Only tricellular vertices were analyzed.
Correlation analysis
To measure the spatial correlation between Ajuba-GFP and Myo-mCherry (Fig. 1 D), edge intensities were measured using SIESTA (Fernandez-Gonzalez and Zallen, 2011), and the R value for the linear best fit line was calculated in Prism (GraphPad). To measure the temporal correlation between Ajuba-GFP and Myo-mCherry, or Resille-GFP and Myo-mCherry (Fig. 1 G), we used previously described methods (Martin et al., 2009). Time-lapse videos were made of five embryos each. Z-stacks were acquired at 0.5-µm z-steps and 10-s intervals on an Ultraview Vox spinning disk confocal with a Zeiss Plan Neofluor 63×/1.4-NA oil-immersion objective. A maximum-intensity projection of a z-stack encompassing 4–7 µm in the region of the adherens junctions, which includes the majority of Ajuba-GFP and Myo-mCherry signal, was used for analysis. The mean intensity at each edge was measured in ImageJ. Edge intensity was averaged over a window of three consecutive time points and assigned to the central frame. The changes in Ajuba-GFP, Resille-GFP, and Myo-mCherry smoothed intensities were plotted as a scatter plot, and the R value for the linear best fit line was calculated in Prism. The Ajuba-GFP or Resille-GFP datasets were shifted backward or forward in time, and new R values from these plots were calculated in the same way. To measure the spatial correlation between Ajuba and myosin or E-cadherin (Fig. S1 F), lines were drawn on 50 near-vertical edges per image in ImageJ, the intensity of all pixels under these lines was plotted as an individual scatter plot for each embryo, and the R value was calculated using linear regression analysis.
Edge tracking and analysis
To analyze cell behaviors in WT and Ajuba mutant embryos (Figs. 5, 6, and S5), 50 vertical cell interfaces that were present at the beginning of germband extension in stage 7 (t = 0) were tracked for up to 30 min in each embryo (n = 12 embryos/genotype). Edges were assigned to T1 processes if they contracted into a four-cell vertex that went on to resolve. Edges were assigned to simultaneous rosettes if they contracted into a four-cell vertex that joined a vertex of five or more cells within 3 min, or if they contracted into a vertex of five or more cells that had not previously been a four-cell vertex for more than 3 min. Edges were assigned to sequential rosettes if they contracted into a four-cell vertex that joined a vertex of five or more cells after more than 3 min, or if they contracted into a vertex of five or more cells that had previously been a four-cell vertex for more than 3 min. The rosette:T1 ratio in Figs. 5 B and S5 A was the number of shrinking edges that formed rosettes divided by the number of shrinking edges that formed T1 processes. Rosettes were scored as having an adhesion defect if there was a visible gap in the β-catenin-GFP signal in time-lapse videos. These defects were observed only at late stages of rosette formation. Vertex lifetime and the percentage of unresolved vertices were scored for each pair of cells that shared a shrinking edge. Vertex lifetime was the time from when that edge contracted to a vertex to the time when the two cells no longer shared a vertex. A vertex was scored as unresolved if the two cells still shared a vertex at t = 30 min. The rosette:T1 ratio in Fig. S3 E was calculated at a single time point in fixed embryos immunostained for E-cadherin and was the total number of rosette intermediates (groups of five or more cells that were connected by a common vertex or by short edges that were <1.2 µm long) divided by the total number of T1 intermediates (groups of four cells that were connected by a common vertex or by a short edge that was <1.2 µm). To analyze tissue elongation, two cells that were located at least 30–40 cells apart along the AP axis and 1–2 cell diameters away from the ventral furrow at end of the fast phase of germband extension in late stage 8 were manually tracked back to the beginning of germband extension in stage 7. Tissue elongation was calculated as the fold-change in distance between these cells along the AP axis.
Online supplemental material
Fig. S1 shows the localization of Ajuba in WT, α-catenin KD, and Shroom-overexpressing embryos. Fig. S2 provides additional analysis of Ajuba-GFP localization to different subsets of junctions. Fig. S3 shows the expression levels of Ajuba-msVenus variants and their effects on myosin localization and rosette formation. Fig. S4 describes the genetic crosses used to generate the Ajuba54 allele and the lethality of Ajuba mutants. Fig. S5 shows analysis of cell behavior in Ajuba maternal mutants separated by zygotic genotype. Videos show cell dynamics during convergent extension (Videos 1 and 2), rosette behaviors (Videos 3 and 4), and dorsal closure (Videos 5–10) in WT and Ajuba mutant embryos.
Supplementary Material
Acknowledgments
We are grateful to Chris Fincher for preliminary experiments with Drosophila Ajuba, Ben Glick for the msVenus tag, Matt Laurie for cloning assistance, Masako Tamada for preliminary experiments demonstrating the effects of ShroomA overexpression on myosin and adherens junction localization, and Huapeng Yu for helpful discussions and advice on Y-27632 injections. We thank Eric Brooks, Marissa Gredler, Karl Palmquist, Adam Paré, and Germán Sabio for comments on the manuscript. Stocks obtained from the Bloomington Drosophila Stock Center (National Institutes of Health P4OODO18537) were used in this study.
This work was supported by National Institutes of Health/National Institute of General Medical Sciences grant R01 GM102803 to J.A. Zallen and National Institutes of Health T32 training grant (T32 HD060600) to M.E. Bustillo. W. Razzell is a Howard Hughes Medical Institute Fellow of the Damon Runyon Cancer Research Foundation (DRG-2226-15). J.A. Zallen is an Investigator of the Howard Hughes Medical Institute.
The authors declare no competing financial interests.
W. Razzell and J.A. Zallen designed the study. W. Razzell and M.E. Bustillo performed the experiments. W. Razzell, M.E. Bustillo, and J.A. Zallen analyzed the data. W. Razzell and J.A. Zallen wrote the paper, and all authors participated in producing the final version of the paper.
References
- Baum B., and Georgiou M.. 2011. Dynamics of adherens junctions in epithelial establishment, maintenance, and remodeling. J. Cell Biol. 192:907–917. 10.1083/jcb.201009141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertet C., Sulak L., and Lecuit T.. 2004. Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature. 429:667–671. 10.1038/nature02590 [DOI] [PubMed] [Google Scholar]
- Blankenship J.T., Backovic S.T., Sanny J.S.S.P., Weitz O., and Zallen J.A.. 2006. Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev. Cell. 11:459–470. 10.1016/j.devcel.2006.09.007 [DOI] [PubMed] [Google Scholar]
- Bolinger C., Zasadil L., Rizaldy R., and Hildebrand J.D.. 2010. Specific isoforms of drosophila shroom define spatial requirements for the induction of apical constriction. Dev. Dyn. 239:2078–2093. 10.1002/dvdy.22326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley C.D., Tan J., Anderson K.L., Hanein D., Volkmann N., Weis W.I., Nelson W.J., and Dunn A.R.. 2014. The minimal cadherin-catenin complex binds to actin filaments under force. Science. 346:1254211 10.1126/science.1254211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacon-Heszele M.F., Ren D., Reynolds A.B., Chi F., and Chen P.. 2012. Regulation of cochlear convergent extension by the vertebrate planar cell polarity pathway is dependent on p120-catenin. Development. 139:968–978. 10.1242/dev.065326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C.W., Xiang B., Ossipova O., Ioannou A., and Sokol S.Y.. 2018. The Ajuba family protein Wtip regulates actomyosin contractility during vertebrate neural tube closure. J. Cell Sci. 131:jcs213884 10.1242/jcs.213884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K.A., Callicoat J.G., Lake C.M., McClurken C.M., Kohl K.P., and Hawley R.S.. 2012. A germline clone screen on the X chromosome reveals novel meiotic mutants in Drosophila melanogaster. G3 (Bethesda). 2:1369–1377. 10.1534/g3.112.003723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Thakur M., Feng Y., Jagannathan R., Seppa M.J., Skeath J.B., and Longmore G.D.. 2010. Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr. Biol. 20:657–662. 10.1016/j.cub.2010.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai R., Sarpal R., Ishiyama N., Pellikka M., Ikura M., and Tepass U.. 2013. Monomeric α-catenin links cadherin to the actin cytoskeleton. Nat. Cell Biol. 15:261–273. 10.1038/ncb2685 [DOI] [PubMed] [Google Scholar]
- Dutta S., Mana-Capelli S., Paramasivam M., Dasgupta I., Cirka H., Billiar K., and McCollum D.. 2018. TRIP6 inhibits Hippo signaling in response to tension at adherens junctions. EMBO Rep. 19:337–350. 10.15252/embr.201744777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell D.L., Weitz O., Magnasco M.O., and Zallen J.A.. 2017. SEGGA: a toolset for rapid automated analysis of epithelial cell polarity and dynamics. Development. 144:1725–1734. 10.1242/dev.146837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Zhao H., Luderer H.F., Epple H., Faccio R., Ross F.P., Teitelbaum S.L., and Longmore G.D.. 2007. The LIM protein, Limd1, regulates AP-1 activation through an interaction with Traf6 to influence osteoclast development. J. Biol. Chem. 282:39–48. 10.1074/jbc.M607399200 [DOI] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R., and Zallen J.A.. 2011. Oscillatory behaviors and hierarchical assembly of contractile structures in intercalating cells. Phys. Biol. 8:045005 10.1088/1478-3975/8/4/045005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R., Simoes S.M., Röper J.C., Eaton S., and Zallen J.A.. 2009. Myosin II dynamics are regulated by tension in intercalating cells. Dev. Cell. 17:736–743. 10.1016/j.devcel.2009.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson M.C., Patel A.B., Nagpal R., and Perrimon N.. 2006. The emergence of geometric order in proliferating metazoan epithelia. Nature. 442:1038–1041. 10.1038/nature05014 [DOI] [PubMed] [Google Scholar]
- Gorfinkiel N., and Martinez-Arias A.. 2007. Requirements for adherens junction components in the interaction between epithelial tissues during dorsal closure in Drosophila. J. Cell Sci. 120:3289–3298. 10.1242/jcs.010850 [DOI] [PubMed] [Google Scholar]
- Goyal R.K., Lin P., Kanungo J., Payne A.S., Muslin A.J., and Longmore G.D.. 1999. Ajuba, a novel LIM protein, interacts with Grb2, augments mitogen-activated protein kinase activity in fibroblasts, and promotes meiotic maturation of Xenopus oocytes in a Grb2- and Ras-dependent manner. Mol. Cell. Biol. 19:4379–4389. 10.1128/MCB.19.6.4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner B.M. 2005. Regulation of cadherin-mediated adhesion in morphogenesis. Nat. Rev. Mol. Cell Biol. 6:622–634. 10.1038/nrm1699 [DOI] [PubMed] [Google Scholar]
- Harris T.J.C. 2017. Sculpting epithelia with planar polarized actomyosin networks: Principles from Drosophila. Semin. Cell Dev. Biol.:S1084-9521(16)30342-1 10.1016/j.semcdb.2017.07.042 [DOI] [PubMed] [Google Scholar]
- Harris T.J.C., and Tepass U.. 2010. Adherens junctions: from molecules to morphogenesis. Nat. Rev. Mol. Cell Biol. 11:502–514. 10.1038/nrm2927 [DOI] [PubMed] [Google Scholar]
- Hirata H., Tatsumi H., and Sokabe M.. 2008. Mechanical forces facilitate actin polymerization at focal adhesions in a zyxin-dependent manner. J. Cell Sci. 121:2795–2804. 10.1242/jcs.030320 [DOI] [PubMed] [Google Scholar]
- Hirota T., Kunitoku N., Sasayama T., Marumoto T., Zhang D., Nitta M., Hatakeyama K., and Saya H.. 2003. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell. 114:585–598. 10.1016/S0092-8674(03)00642-1 [DOI] [PubMed] [Google Scholar]
- Hoffman B.D., and Yap A.S.. 2015. Towards a Dynamic Understanding of Cadherin-Based Mechanobiology. Trends Cell Biol. 25:803–814. 10.1016/j.tcb.2015.09.008 [DOI] [PubMed] [Google Scholar]
- Huang J., Zhou W., Dong W., Watson A.M., and Hong Y.. 2009. From the Cover: Directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc. Natl. Acad. Sci. USA. 106:8284–8289. 10.1073/pnas.0900641106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson M.S., Tokutake Y., Chang M.-S., Bloor J.W., Venakides S., Kiehart D.P., and Edwards G.S.. 2003. Forces for morphogenesis investigated with laser microsurgery and quantitative modeling. Science. 300:145–149. 10.1126/science.1079552 [DOI] [PubMed] [Google Scholar]
- Huveneers S., Oldenburg J., Spanjaard E., van der Krogt G., Grigoriev I., Akhmanova A., Rehmann H., and de Rooij J.. 2012. Vinculin associates with endothelial VE-cadherin junctions to control force-dependent remodeling. J. Cell Biol. 196:641–652. 10.1083/jcb.201108120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibar C., Kirichenko E., Keepers B., Enners E., Fleisch K., and Irvine K.D.. 2018. Tension-dependent regulation of mammalian Hippo signaling through LIMD1. J. Cell Sci. 131:jcs214700 10.1242/jcs.214700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiyama N., Tanaka N., Abe K., Yang Y.J., Abbas Y.M., Umitsu M., Nagar B., Bueler S.A., Rubinstein J.L., Takeichi M., and Ikura M.. 2013. An autoinhibited structure of α-catenin and its implications for vinculin recruitment to adherens junctions. J. Biol. Chem. 288:15913–15925. 10.1074/jbc.M113.453928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadrmas J.L., and Beckerle M.C.. 2004. The LIM domain: from the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 5:920–931. 10.1038/nrm1499 [DOI] [PubMed] [Google Scholar]
- Kanungo J., Pratt S.J., Marie H., and Longmore G.D.. 2000. Ajuba, a cytosolic LIM protein, shuttles into the nucleus and affects embryonal cell proliferation and fate decisions. Mol. Biol. Cell. 11:3299–3313. 10.1091/mbc.11.10.3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasza K.E., Farrell D.L., and Zallen J.A.. 2014. Spatiotemporal control of epithelial remodeling by regulated myosin phosphorylation. Proc. Natl. Acad. Sci. USA. 111:11732–11737. 10.1073/pnas.1400520111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehart D.P., Galbraith C.G., Edwards K.A., Rickoll W.L., and Montague R.A.. 2000. Multiple forces contribute to cell sheet morphogenesis for dorsal closure in Drosophila. J. Cell Biol. 149:471–490. 10.1083/jcb.149.2.471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisseleva M., Feng Y., Ward M., Song C., Anderson R.A., and Longmore G.D.. 2005. The LIM protein Ajuba regulates phosphatidylinositol 4,5-bisphosphate levels in migrating cells through an interaction with and activation of PIPKI alpha. Mol. Cell. Biol. 25:3956–3966. 10.1128/MCB.25.10.3956-3966.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau K., Tao H., Liu H., Wen J., Sturgeon K., Sorfazlian N., Lazic S., Burrows J.T.A., Wong M.D., Li D., et al. 2015. Anisotropic stress orients remodelling of mammalian limb bud ectoderm. Nat. Cell Biol. 17:569–579. 10.1038/ncb3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecaudey V., Cakan-Akdogan G., Norton W.H.J., and Gilmour D.. 2008. Dynamic Fgf signaling couples morphogenesis and migration in the zebrafish lateral line primordium. Development. 135:2695–2705. 10.1242/dev.025981 [DOI] [PubMed] [Google Scholar]
- Leckband D.E., and de Rooij J.. 2014. Cadherin adhesion and mechanotransduction. Annu. Rev. Cell Dev. Biol. 30:291–315. 10.1146/annurev-cellbio-100913-013212 [DOI] [PubMed] [Google Scholar]
- le Duc Q., Shi Q., Blonk I., Sonnenberg A., Wang N., Leckband D., and de Rooij J.. 2010. Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner. J. Cell Biol. 189:1107–1115. 10.1083/jcb.201001149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leerberg J.M., and Yap A.S.. 2013. Vinculin, cadherin mechanotransduction and homeostasis of cell-cell junctions. Protoplasma. 250:817–829. 10.1007/s00709-012-0475-6 [DOI] [PubMed] [Google Scholar]
- Levayer R., Pelissier-Monier A., and Lecuit T.. 2011. Spatial regulation of Dia and Myosin-II by RhoGEF2 controls initiation of E-cadherin endocytosis during epithelial morphogenesis. Nat. Cell Biol. 13:529–540. 10.1038/ncb2224 [DOI] [PubMed] [Google Scholar]
- Lienkamp S.S., Liu K., Karner C.M., Carroll T.J., Ronneberger O., Wallingford J.B., and Walz G.. 2012. Vertebrate kidney tubules elongate using a planar cell polarity-dependent, rosette-based mechanism of convergent extension. Nat. Genet. 44:1382–1387. 10.1038/ng.2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lye C.M., and Sanson B.. 2011. Tension and epithelial morphogenesis in Drosophila early embryos. Curr. Top. Dev. Biol. 95:145–187. 10.1016/B978-0-12-385065-2.00005-0 [DOI] [PubMed] [Google Scholar]
- Marie H., Pratt S.J., Betson M., Epple H., Kittler J.T., Meek L., Moss S.J., Troyanovsky S., Attwell D., Longmore G.D., and Braga V.M.M.. 2003. The LIM protein Ajuba is recruited to cadherin-dependent cell junctions through an association with α-catenin. J. Biol. Chem. 278:1220–1228. 10.1074/jbc.M205391200 [DOI] [PubMed] [Google Scholar]
- Martin A.C., Kaschube M., and Wieschaus E.F.. 2009. Pulsed contractions of an actin-myosin network drive apical constriction. Nature. 457:495–499. 10.1038/nature07522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A.C., Gelbart M., Fernandez-Gonzalez R., Kaschube M., and Wieschaus E.F.. 2010. Integration of contractile forces during tissue invagination. J. Cell Biol. 188:735–749. 10.1083/jcb.200910099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney B.M., McEwen D.G., Grevengoed E., Maddox P., Bejsovec A., and Peifer M.. 2001. Drosophila APC2 and Armadillo participate in tethering mitotic spindles to cortical actin. Nat. Cell Biol. 3:933–938. 10.1038/ncb1001-933 [DOI] [PubMed] [Google Scholar]
- McCormack J.J., Bruche S., Ouadda A.B.D., Ishii H., Lu H., Garcia-Cattaneo A., Chávez-Olórtegui C., Lamarche-Vane N., and Braga V.M.M.. 2017. The scaffold protein Ajuba suppresses CdGAP activity in epithelia to maintain stable cell-cell contacts. Sci. Rep. 7:9249 10.1038/s41598-017-09024-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelsen J.W., Schmeichel K.L., Beckerle M.C., and Winge D.R.. 1993. The LIM motif defines a specific zinc-binding protein domain. Proc. Natl. Acad. Sci. USA. 90:4404–4408. 10.1073/pnas.90.10.4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkovic I., Gault W.J., Rahnama M., Jenny A., Gaengel K., Bessette D., Gottardi C.J., Verheyen E.M., and Mlodzik M.. 2011. Nemo kinase phosphorylates β-catenin to promote ommatidial rotation and connects core PCP factors to E-cadherin-β-catenin. Nat. Struct. Mol. Biol. 18:665–672. 10.1038/nsmb.2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T., and Takeichi M.. 2008. Shroom3-mediated recruitment of Rho kinases to the apical cell junctions regulates epithelial and neuroepithelial planar remodeling. Development. 135:1493–1502. 10.1242/dev.019646 [DOI] [PubMed] [Google Scholar]
- Nishimura T., Honda H., and Takeichi M.. 2012. Planar cell polarity links axes of spatial dynamics in neural-tube closure. Cell. 149:1084–1097. 10.1016/j.cell.2012.04.021 [DOI] [PubMed] [Google Scholar]
- Nola S., Daigaku R., Smolarczyk K., Carstens M., Martin-Martin B., Longmore G., Bailly M., and Braga V.M.M.. 2011. Ajuba is required for Rac activation and maintenance of E-cadherin adhesion. J. Cell Biol. 195:855–871. 10.1083/jcb.201107162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Wieschaus E., and Kluding H.. 1984. Mutations affecting the pattern of the larval cuticle inDrosophila melanogaster : I. Zygotic loci on the second chromosome. Wilehm Roux Arch Dev Biol. 193:267–282. 10.1007/BF00848156 [DOI] [PubMed] [Google Scholar]
- Oldenburg J., van der Krogt G., Twiss F., Bongaarts A., Habani Y., Slotman J.A., Houtsmuller A., Huveneers S., and de Rooij J.. 2015. VASP, zyxin and TES are tension-dependent members of Focal Adherens Junctions independent of the α-catenin-vinculin module. Sci. Rep. 5:17225 10.1038/srep17225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks A.L., Cook K.R., Belvin M., Dompe N.A., Fawcett R., Huppert K., Tan L.R., Winter C.G., Bogart K.P., Deal J.E., et al. 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36:288–292. 10.1038/ng1312 [DOI] [PubMed] [Google Scholar]
- Perkins L.A., Holderbaum L., Tao R., Hu Y., Sopko R., McCall K., Yang-Zhou D., Flockhart I., Binari R., Shim H.S., et al. 2015. The transgenic RNAi project at Harvard medical school: Resources and validation. Genetics. 201:843–852. 10.1534/genetics.115.180208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F., Chen H.-M., Lee T., and Bullock S.L.. 2014. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA. 111:E2967–E2976. 10.1073/pnas.1405500111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt S.J., Epple H., Ward M., Feng Y., Braga V.M., and Longmore G.D.. 2005. The LIM protein Ajuba influences p130Cas localization and Rac1 activity during cell migration. J. Cell Biol. 168:813–824. 10.1083/jcb.200406083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb C., Sun S., Sun G., Pan Y., and Irvine K.D.. 2014. Cytoskeletal tension inhibits Hippo signaling through an Ajuba-Warts complex. Cell. 158:143–156. 10.1016/j.cell.2014.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauzi M., Verant P., Lecuit T., and Lenne P.-F.. 2008. Nature and anisotropy of cortical forces orienting Drosophila tissue morphogenesis. Nat. Cell Biol. 10:1401–1410. 10.1038/ncb1798 [DOI] [PubMed] [Google Scholar]
- Rauzi M., Lenne P.-F., and Lecuit T.. 2010. Planar polarized actomyosin contractile flows control epithelial junction remodelling. Nature. 468:1110–1114. 10.1038/nature09566 [DOI] [PubMed] [Google Scholar]
- Razzell W., Wood W., and Martin P.. 2014. Recapitulation of morphogenetic cell shape changes enables wound re-epithelialisation. Development. 141:1814–1820. 10.1242/dev.107045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy B.V.V.G., and Irvine K.D.. 2013. Regulation of Hippo signaling by EGFR-MAPK signaling through Ajuba family proteins. Dev. Cell. 24:459–471. 10.1016/j.devcel.2013.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggleman B., Schedl P., and Wieschaus E.. 1990. Spatial expression of the Drosophila segment polarity gene armadillo is posttranscriptionally regulated by wingless. Cell. 63:549–560. 10.1016/0092-8674(90)90451-J [DOI] [PubMed] [Google Scholar]
- Royou A., Field C., Sisson J.C., Sullivan W., and Karess R.. 2004. Reassessing the role and dynamics of nonmuscle myosin II during furrow formation in early Drosophila embryos. Mol. Biol. Cell. 15:838–850. 10.1091/mbc.e03-06-0440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozbicki E., Chuai M., Karjalainen A.I., Song F., Sang H.M., Martin R., Knölker H.J., MacDonald M.P., and Weijer C.J.. 2015. Myosin-II-mediated cell shape changes and cell intercalation contribute to primitive streak formation. Nat. Cell Biol. 17:397–408. 10.1038/ncb3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabino D., Brown N.H., and Basto R.. 2011. Drosophila Ajuba is not an Aurora-A activator but is required to maintain Aurora-A at the centrosome. J. Cell Sci. 124:1156–1166. 10.1242/jcs.076711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E., and Marshall C.J.. 2002. ROCK and Dia have opposing effects on adherens junctions downstream of Rho. Nat. Cell Biol. 4:408–415. 10.1038/ncb796 [DOI] [PubMed] [Google Scholar]
- Schiller H.B., Friedel C.C., Boulegue C., and Fässler R.. 2011. Quantitative proteomics of the integrin adhesome show a myosin II-dependent recruitment of LIM domain proteins. EMBO Rep. 12:259–266. 10.1038/embor.2011.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimizzi G.V., and Longmore G.D.. 2015. Ajuba proteins. Curr. Biol. 25:R445–R446. 10.1016/j.cub.2015.02.034 [DOI] [PubMed] [Google Scholar]
- Shewan A.M., Maddugoda M., Kraemer A., Stehbens S.J., Verma S., Kovacs E.M., and Yap A.S.. 2005. Myosin 2 is a key Rho kinase target necessary for the local concentration of E-cadherin at cell-cell contacts. Mol. Biol. Cell. 16:4531–4542. 10.1091/mbc.e05-04-0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões S. de M., Blankenship J.T., Weitz O., Farrell D.L., Tamada M., Fernandez-Gonzalez R., and Zallen J.A.. 2010. Rho-kinase directs Bazooka/Par-3 planar polarity during Drosophila axis elongation. Dev. Cell. 19:377–388. 10.1016/j.devcel.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões S.M., Mainieri A., and Zallen J.A.. 2014. Rho GTPase and Shroom direct planar polarized actomyosin contractility during convergent extension. J. Cell Biol. 204:575–589. 10.1083/jcb.201307070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.A., Blankman E., Deakin N.O., Hoffman L.M., Jensen C.C., Turner C.E., and Beckerle M.C.. 2013. LIM domains target actin regulators paxillin and zyxin to sites of stress fiber strain. PLoS One. 8:e69378 10.1371/journal.pone.0069378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperry R.B., Bishop N.H., Bramwell J.J., Brodeur M.N., Carter M.J., Fowler B.T., Lewis Z.B., Maxfield S.D., Staley D.M., Vellinga R.M., and Hansen M.D.H.. 2010. Zyxin controls migration in epithelial-mesenchymal transition by mediating actin-membrane linkages at cell-cell junctions. J. Cell. Physiol. 222:612–624. 10.1002/jcp.21977 [DOI] [PubMed] [Google Scholar]
- Takeichi M. 2014. Dynamic contacts: rearranging adherens junctions to drive epithelial remodelling. Nat. Rev. Mol. Cell Biol. 15:397–410. 10.1038/nrm3802 [DOI] [PubMed] [Google Scholar]
- Tamada M., Farrell D.L., and Zallen J.A.. 2012. Abl regulates planar polarized junctional dynamics through β-catenin tyrosine phosphorylation. Dev. Cell. 22:309–319. 10.1016/j.devcel.2011.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twiss F., Le Duc Q., Van Der Horst S., Tabdili H., Van Der Krogt G., Wang N., Rehmann H., Huveneers S., Leckband D.E., and De Rooij J.. 2012. Vinculin-dependent Cadherin mechanosensing regulates efficient epithelial barrier formation. Biol. Open. 1:1128–1140. 10.1242/bio.20122428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaire D., and Rivier N.. 1984. Soap, cells and statistics—random patterns in two dimensions. Contemp. Phys. 25:59–99. 10.1080/00107518408210979 [DOI] [Google Scholar]
- Wen W., Meinkoth J.L., Tsien R.Y., and Taylor S.S.. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell. 82:463–473. 10.1016/0092-8674(95)90435-2 [DOI] [PubMed] [Google Scholar]
- Williams M., Yen W., Lu X., and Sutherland A.. 2014. Distinct apical and basolateral mechanisms drive planar cell polarity-dependent convergent extension of the mouse neural plate. Dev. Cell. 29:34–46. 10.1016/j.devcel.2014.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S., and Nelson W.J.. 2007. Localized zones of Rho and Rac activities drive initiation and expansion of epithelial cell-cell adhesion. J. Cell Biol. 178:517–527. 10.1083/jcb.200701058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M., Qiu W., Liu R., Efremov A.K., Cong P., Seddiki R., Payre M., Lim C.T., Ladoux B., Mège R.M., and Yan J.. 2014. Force-dependent conformational switch of α-catenin controls vinculin binding. Nat. Commun. 5:4525 10.1038/ncomms5525 [DOI] [PubMed] [Google Scholar]
- Yonemura S., Wada Y., Watanabe T., Nagafuchi A., and Shibata M.. 2010. α-Catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 12:533–542. 10.1038/ncb2055 [DOI] [PubMed] [Google Scholar]
- Yoshigi M., Hoffman L.M., Jensen C.C., Yost H.J., and Beckerle M.C.. 2005. Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskeletal reinforcement. J. Cell Biol. 171:209–215. 10.1083/jcb.200505018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen J.A., and Wieschaus E.. 2004. Patterned gene expression directs bipolar planar polarity in Drosophila. Dev. Cell. 6:343–355. 10.1016/S1534-5807(04)00060-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.