Endoplasmic reticulum (ER) morphology is dynamic and key to its function during different cellular processes. Dong et al. now show in convergent studies in human cells and Caenorhabditis elegans neurons that a phosphoinositide phosphatase (INPP5K) is localized on the surface of the ER network and helps control the shape of the ER.
Abstract
INPP5K (SKIP) is an inositol 5-phosphatase that localizes in part to the endoplasmic reticulum (ER). We show that recruitment of INPP5K to the ER is mediated by ARL6IP1, which shares features of ER-shaping proteins. Like ARL6IP1, INPP5K is preferentially localized in ER tubules and enriched, relative to other ER resident proteins (Sec61β, VAPB, and Sac1), in newly formed tubules that grow along microtubule tracks. Depletion of either INPP5K or ARL6IP1 results in the increase of ER sheets. In a convergent but independent study, a screen for mutations affecting the distribution of the ER network in dendrites of the PVD neurons of Caenorhabditis elegans led to the isolation of mutants in CIL-1, which encodes the INPP5K worm orthologue. The mutant phenotype was rescued by expression of wild type, but not of catalytically inactive CIL-1. Our results reveal an unexpected role of an ER localized polyphosphoinositide phosphatase in the fine control of ER network organization.
Introduction
Phosphoinositides are a family of signaling bilayer phospholipids resulting from the reversible phosphorylation of phosphatidylinositol at the 3, 4, and 5 position of the inositol ring. Each of the phosphorylated headgroups recognizes with variable affinity and specificity distinct set of protein motifs and domains, thus helping to recruit and regulate cytosolic proteins at membrane interfaces. Via these interactions, as well as via direct actions on membrane proteins, phosphoinositides play major roles in the control of a variety of physiological processes, including signal transduction, membrane trafficking, cytoskeleton dynamics, and transport of ion and metabolites across bilayers. Key to this function is the heterogeneous distribution of the different phosphoinositides on different membranes, which is achieved and maintained through the subcellular targeting of lipid kinases, lipid phosphatases, and lipid transport proteins (Di Paolo and De Camilli, 2006; Balla, 2013).
Mammalian genomes encode 10 inositol 5-phosphatases. One 5-phosphatase, INPP5A, only acts on soluble inositol polyphosphates, while the other nine have phosphoinositide phosphatase activity (i.e., dephosphorylate the 5 position of lipid-bound inositol polyphosphates), although they can also dephosphorylate soluble inositol polyphosphates (Conduit et al., 2012; Hakim et al., 2012; Pirruccello and De Camilli, 2012). All nine proteins are cytosolic enzymes in which the catalytic module is flanked by domains that mediate their subcellular targeting to membranes where they express their catalytic action. Typically, these 5-phosphatases are targeted to membranes distal to the ER, which include the plasma membrane and membranes of the secretory and endocytic pathways, where the bulk of their substrates are localized (Conduit et al., 2012; Hakim et al., 2012; Pirruccello and De Camilli, 2012). One exception is INPP5K, a 5-phosphatase localized at least in part, on the surface of the ER (Wiradjaja et al., 2001; Gurung et al., 2003). Recombinant full-length INPP5K has 5-phosphatase activity toward PI(4,5)P2 and PI(3,4,5)P3, with marked preference for PI(4,5)P2 (Ijuin et al., 2000; Schmid et al., 2004). However, neither PI(4,5)P2 nor PI(3,4,5)P3 is thought to be concentrated, or even present, in the ER, raising questions about the physiological function of this localization (Di Paolo and De Camilli, 2006; Balla, 2013).
INPP5K, also known as skeletal muscle and kidney-enriched inositol 5-phosphatase (SKIP), is highly expressed in the developing and adult brain, eye, muscle, and kidney (Ijuin et al., 2000). The knockout of INPP5K in mouse results in embryonic lethality (Ijuin et al., 2008). Human biallelic point mutations that impair INPP5K’s phosphatase activity give rise to congenital muscular dystrophy with additional clinical manifestations, including cataracts, intellectual impairments, and short stature (Osborn et al., 2017; Wiessner et al., 2017). Mechanisms of disease, however, remain unclear. Specifically, it is unknown whether the ER localization of INPP5K contributes to the disease, as pools of INPP5K not associated with the ER are present. For example, it was shown that upon growth factor stimulation, a pool of INPP5K can be recruited to the plasma membrane to down-regulate PI(3,4,5)P3 signaling (Gurung et al., 2003).
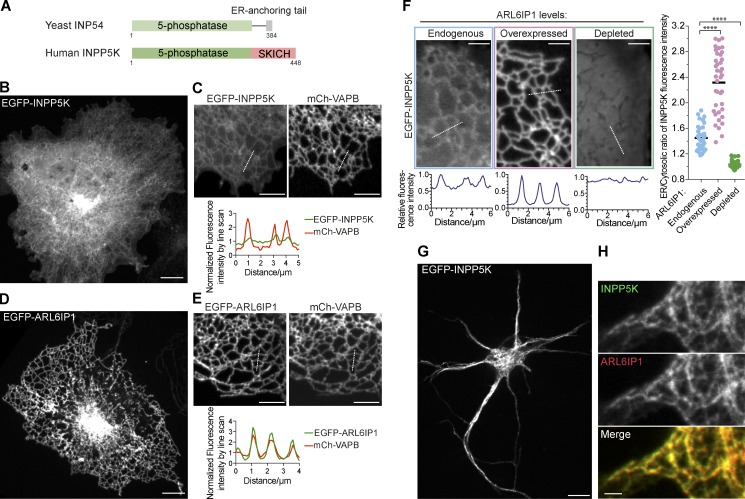
INPP5K has a simple two-domain structure with an N-terminal 5-phosphatase domain followed by a C-terminal SKICH domain, with no transmembrane regions reported. The closest homologue of INPP5K in yeast, the protein INP54, also localizes at the ER surface, suggesting a highly conserved ER-related function of this enzyme. However, INP54, which lacks the SKICH domain, is anchored to the ER via a hydrophobic 13-aa C-terminal sequence that is missing in INPP5K (Fig. 1 A; Wiradjaja et al., 2001). How INPP5K is targeted to the ER remains unknown.
Figure 1.
ER localization of INPP5K and ARL6IP1. (A) Domain organization of yeast INP54 and human INPP5K. Note the presence of an ER-anchoring tail at the C terminus of yeast INP54. Human INPP5K lacks such an anchoring ER and instead contains the SKICH domain. (B and C) Confocal images of COS-7 cells expressing EGFP-INPP5K alone (B) or coexpressing EGFP-INPP5K and the ER marker mCh-VAPB, an intrinsic membrane protein (C), showing the dual localization of EGFP-INPP5K in the cytosol (indicated by the diffuse fluorescence in B) and in the ER (shown by the colocalization with mCh-VAPB on tubular structures and the line scan analysis in C). Representative examples of several cells imaged in at least three independent experiments. All transfected cells exhibited the phenotype shown. Scale bars: 10 µm in B, 5 µm in C. (D and E) Confocal images of COS-7 cells expressing EGFP-ARL6IP1 (C) or coexpressing EGFP-ARL6IP1 and mCh-VAPB (D), demonstrating the localization of EGFP-ARL6IP1 in the ER (shown by the colocalization with mCh-VAPB on tubular structures and the line scan analysis in D). Representative examples of cells imaged in at least three independent experiments. All transfected cells exhibited the phenotype shown. Scale bars: 10 µm in D, 5 µm in E. (F) Representative confocal images and corresponding line-scan analysis of COS-7 cells showing that EGFP-INPP5K has a dual localization in the ER and cytosol in cells only expressing endogenous ARL6IP1, while it is primarily recruited to the ER when coexpressed with mCh-ARL6IP1 and loses its ER localization upon ARL6IP1 knockdown. Scale bars: 5 µm. The ratio of EGFP-INPP5K fluorescence intensity in the ER relative to the adjacent cytosol under the three conditions is plotted in the right panel. Data were acquired by line-scan analyses of multiple individual ER tubules from multiple cells (“Endogenous”: n = 39 ER tubules from 9 cells; “Overexpressed”: n = 39 ER tubules from 11 cells; “Depleted”: n = 33 ER tubules from 10 cells). ****, p < 0.0001 (two-tailed t test). (G and H) Representative confocal image of mouse cortical neurons grown 4 d in vitro and transfected with EGFP-INPP5K and mCh-ARL6IP1, showing their localization in the ER reticular network. Scale bars: 10 µm in G, 2 µm in H.
Here, we have investigated the potential physiological role of an interaction of INPP5K with the ER protein ARL6IP1 reported in a publicly accessible proteomics database (Rual et al., 2005). ARL6IP1 is a protein with reticulon-like features (Yamamoto et al., 2014) whose mutations result in hereditary spastic paraplegia (Novarino et al., 2014; Nizon et al., 2018), a pathology frequently associated with dysfunction of proteins that control the morphology of the ER (Blackstone et al., 2011). We demonstrate that the interaction with ARL6IP1 is responsible for the recruitment of INPP5K to the ER and that, like ARL6IP1, INPP5K is enriched relative to several other ER proteins in newly formed ER tubules. Furthermore, the knockdown of either protein results in the increase of ER sheets. These results converged with those of a forward genetic screen in Caenorhabditis elegans showing that loss-of-function mutations of the worm INPP5K orthologue, cil-1, result in defects of ER morphology in the PVD, a highly branched sensory neuron of this organism.
Results
INPP5K is recruited to the ER by its interaction with ARL6IP1
Inspection of the primary sequence of INPP5K does not predict a transmembrane or C-terminal hydrophobic sequence (Fig. 1 A). Accordingly, when EGFP-tagged human INPP5K was expressed in COS-7 cells (thus resulting in INPP5K overexpression), we observed localization on the ER, as reported previously, but also diffuse cytosolic fluorescence (Fig. 1, B and C).
The search of a high-throughput yeast-two-hybrid interactome (Rual et al., 2005) revealed as one of the top hits ARL6IP1, a multipass ER transmembrane protein (Fig. 1, D and E). Upon overexpression of mCh-ARL6IP1, a robust enhancement of EGFP-INPP5K localization on the ER was detected (Fig. 1 F). Conversely, the ER localization of INPP5K was lost upon ARL6IP1 knockdown (Fig. 1 F). These results support the hypothesis that INPP5K anchoring to the ER membrane is mediated by an interaction with ARL6IP1. Colocalization of cotransfected mCh-ARL6IP1 and EGFP-INPP5K was also observed in neurons, where both proteins were present throughout axons and dendrites (Fig. 1, G and H). As a control, we also coexpressed EGFP-INPP5K together with mCh-MAD2L1BP, a nuclear-enriched protein (Habu et al., 2002) and another potential binding partner of INPP5K revealed by the yeast-two-hybrid interactome (Rual et al., 2005). In cells coexpressing these constructs, both MAD2L1BP and INPP5K were concentrated in the nucleus (Fig. S1 A), validating the interaction but proving that the ER localization of INPP5K in ARL6IP-overexpressing cells is specific. In subsequent experiments, we focused selectively on the ER localization of INPP5K and its interaction with ARL6IP1.
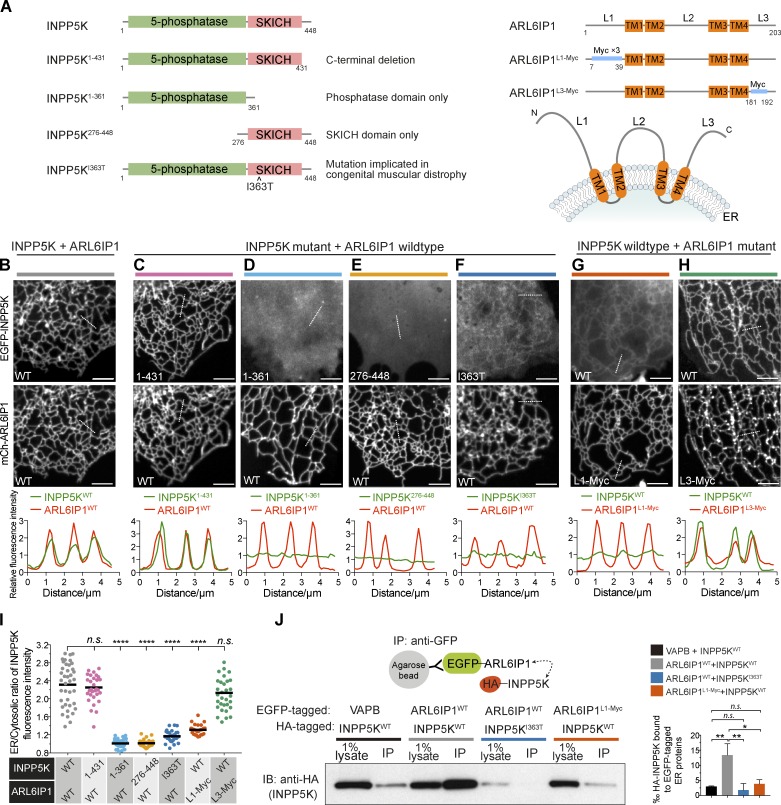
INPP5K comprises an N-terminal 5-phosphatase domain and a C-terminal SKICH domain (Fig. 2 A). Both WT INPP5K and an INPP5K deletion construct lacking the short tail (17 aa) downstream of the SKICH domain localized at the ER when coexpressed with mCh-ARL6IP1 (Fig. 2, B, C, and I). Deletion constructs of INPP5K lacking either the 5-phosphatase domain or the SKICH domain did not localize to the ER with overexpressed ARL6IP1 and were localized exclusively in the cytosol (Fig. 2, D, E, and I). Thus, both domains are required to bind ARL6IP1, possibly reflecting an ARL6IP1-interacting surface comprising both domains. Importantly, an INPP5K construct harboring a single-residue substitution within the SKICH domain that results in congenital muscular dystrophy, I363T (Wiessner et al., 2017), also displayed a significant loss of ER recruitment (Fig. 2, F and I).
Figure 2.
INPP5K is recruited to the ER via the interaction with ARL6IP1. (A) Left: INPP5K domain structure and deletion constructs used for the experiments shown in B and C. Right: ARL6IP1 domain structure and predicted topology. TM, transmembrane regions. L1 to L3, three cytosolically exposed regions. Segments highlighted in blue were replaced with flexible linkers of equivalent length comprising myc tags and these constructs were used for experiments shown in F. (B–H) Representative confocal images and corresponding line-scan analysis of COS-7 cells coexpressing the EGFP-INPP5K and mCh-ARL6IP1 constructs depicted in A. (B) WT INPP5K and WT ARL6IP1 colocalize in the ER. (C) The C-terminal 17-aa segment of INPP5K is dispensable for colocalization with ARL6IP1. (D and E) Neither the 5-phosphatase domain only (INPP5K1–361) nor the SKICH domain only (INPP5K276–448) is sufficient to bind ER-bound ARL6IP1. (F) The INPP5KI363T patient mutation strongly impairs the recruitment to the ER. (G and H) The recruitment of INPP5K to the ER is strongly reduced when the N-terminal region of ARL6IP1 is replaced by another sequence (G) but remains unchanged when the C-terminal region of ARL6IP1 is replaced (H). The replacement of the L1 segment, but not the L3 segment, of ARL6IP1 nearly abolishes the recruitment of WT INPP5K to the ER. Hot spots of ARL6IP1L3-myc possibly reflect misfolded proteins. Note these hot spots are not enriched for INPP5K (see Results). Scale bars: 5 µm. (I) Plot of the ratio of EGFP-INPP5K fluorescence intensity on the ER relative to the adjacent cytosol based on line-scan analysis of individual tubules from multiple cells. Data for cells expressing INPP5KWT and ARL6IP1WT were the same as those shown in Fig. 1 F. (INPP5KWT and ARL6IP1WT: n = 39 ER tubules from nine cells; INPP5K1–361 and ARL6IP1WT: n = 31 ER tubules from four cells; INPP5K276–448 and ARL6IP1WT: n = 35 ER tubules from four cells; INPP5KI363T and ARL6IP1WT: n = 31 ER tubules from four cells; INPP5KWT and ARL6IP1L1-Myc: n = 32 ER tubules from six cells; INPP5KWT and ARL6IP1L3-Myc: n = 32 ER tubules from four cells). ****, p < 0.0001; n.s., not significant (two-tailed t test). (J) Extracts of HeLa cells transfected with HA-INPP5K and the indicated EGFP-tagged constructs were subjected to anti-GFP immunoprecipitation (IP) and then processed for SDS-PAGE and immunoblotting (IB) with anti-HA antibody. Left: Representative blot from three independent experiments. The bar graph on the right shows quantification of HA-INPP5K coprecipitated with EGFP-tagged proteins normalized to input (from densitometric scans of gel bands). Data are presented as mean ± SEM, n = 3; **, p < 0.01; *, p < 0.05; n.s., not significant (two-tailed t test).
ARL6IP1 comprises four predicted transmembrane helices arranged as pairs of hairpins with N and C termini exposed to the cytosol (Fig. 2 A, right). Replacement of the cytoplasmic N terminus of ARL6IP1 with an amino acid flexible linker of equivalent length consisting of myc epitopes (ARL6IP1L1-Myc) resulted in an ER-localized protein that no longer recruited INPP5K (Fig. 2, G and I). Conversely, a similar replacement of the cytosol exposed C-terminal region of ARL6IP1 did not impair the ability of ARL6IP1 to recruit INPP5K, although it also resulted in a clustering of ARL6IP1 into small punctate structures superimposed to the ER network (Fig. 2, H and I). As INPP5K did not cocluster with ARL6IP1 into these structures, they may reflect a pool of misfolded ARL6IP1 due to the amino acid replacement.
To validate the INPP5K-ARL6IP1 interaction biochemically, lysates from cells coexpressing HA-INPP5K and either EGFP-tagged ARL6IP1 or, as a negative control, the ER membrane protein EGFP-VAPB were detergent solubilized and immunoprecipitated using an anti-GFP nanobody (GFP-trap; Fig. 2 J). HA-INPP5KWT robustly coprecipitated with EGFP-ARL6IP1WT, whereas the pool of HA-INPP5KWT that coprecipitated with EGFP-ARL6IP1L1-Myc was much smaller and similar to the pool coprecipitated with EGFP-VAPB (Fig. 2 J), possibly reflecting nonspecific binding. HA-INPP5KI363T mutant, whose expression was nonetheless reduced compared with HA-INPP5KWT in cells transfected with the equal amount of expression vectors, did not robustly coprecipitate with EGFP-ARL6IP1WT (Fig. 2 J).
Collectively, these results implicate the N-terminal region of ARL6IP1 and a surface of INPP5K at the interface of the phosphatase domain and the SKICH domain in the INPP5K-ARL6IP1 interaction.
Mammalian cells also express INPP5J, a paralogue of INPP5K. INPP5J, like INPP5K, contains a 5-phosphatase domain and a SKICH domain, but EGFP-INPP5J did not localize to the ER. Additionally, EGFP-INPP5J did not colocalize with mCh-ARL6IP1 (Fig. S1, B and C). Thus, we did not further investigate this protein.
Enrichment of ARL6IP1 and INPP5K, relative to other ER proteins, in peripheral ER tubules
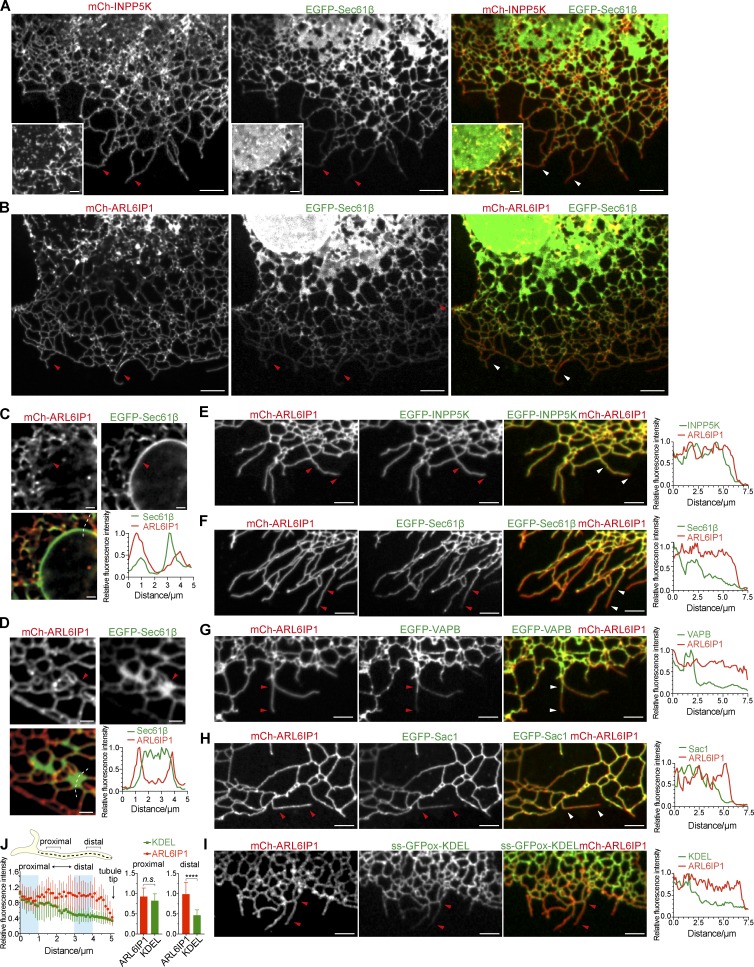
While fluorescently labeled ARL6IP1 and INPP5K always precisely colocalize when expressed in the same cell (Fig. 2 B; see also Fig. 3 E), we observed some discrepancy of localization between ARL6IP1 (and thus, by extension, INPP5K) and other membrane proteins of the ER, such as EGFP-Sec61β, EGFP-VAPB, EGFP-Sac1, and the ER luminal marker ss-GFPox-KDEL. In COS-7 cells transiently expressing mCh-ARL6IP1 together with GFP-Sec61β or ss-GFPox-KDEL, GFP-Sec61β and ss-GFPox-KDEL showed the typical distribution expected for an ER membrane protein (i.e., a localization throughout the ER network), including the nuclear envelope (Fig. 3, A–C; and Fig. S2, A and B), peripheral cisternae (Fig. 3 D; and Fig. S2, A and B) and tubules (Fig. 3, A and E; and Fig. S2, A and B). In contrast, ARL6IP1, as also recently reported (Feng et al., 2017), as well as INPP5K, were present on the entire tubular network and particularly enriched in the peripheral tubular ER (Fig. 3 E), but they were absent from the nuclear envelope (Fig. 3, A and C; and Fig. S2, A and B) and primarily decorated the rims of sheet-like ER elements positive for GFP-Sec61β and ss-GFPox-KDEL (Fig. 3 D; and Fig. S2, A and B).
Figure 3.
Preferential localization of ARL6IP1 and INPP5K, relative to other ER proteins, in peripheral ER tubules. (A) COS-7 cell coexpressing the ER membrane marker EGFP-Sec61β, mCh-INPP5K, and Myc-ARL6IP1 imaged by confocal fluorescence microscopy. Both EGFP-Sec61β and mCh-INPP5K fluorescence are present throughout the ER tubular network, but mCh-INPP5K is enriched over EGFP-Sec61β in the peripheral tubular ER (as marked by arrowheads) and is nearly undetectable in ER sheets. Scale bars: 5 µm. Insets at the bottom left corners show the nuclear envelope of a different cell from the same field, demonstrating that mCh-INPP5K fluorescence is absent from the nuclear envelope marked by EGFP-Sec61β fluorescence. Inset scale bars: 2 µm. Representative examples of several cells imaged in at least three independent experiments. (B) COS-7 cell coexpressing EGFP-Sec61β and mCh-ARL6IP1, showing the relative enrichment of mCh-ARL6IP1 over EGFP-Sec61β in the peripheral tubular ER (as marked by arrowheads) and the presence of EGFP-Sec61β, but not mCh-ARL6IP1, on the nuclear envelope. Scale bars: 5 µm. Representative examples of several cells imaged in at least three independent experiments. (C and D) Representative confocal images of COS-7 cells coexpressing mCh-ARL6IP1 and EGFP-Sec61β showing regions including the nuclear envelope (C) or peripheral ER sheets (D), respectively. Graphs show a representative example of the quantification of the normalized fluorescence intensity measured along the dashed lines as delineated in the merged fields. Note the lack of mCh-ARL6IP1 signal from the nuclear envelope (B) and from the peripheral ER sheets, except for their edges (C), while EGFP-Sec61β labels these structures (arrowheads). Scale bars: 2 µm. (E–I) Representative confocal images and respective line-scan analysis of the periphery of COS-7 cells coexpressing mCh-ARL6IP1 and other GFP-tagged proteins as indicated. While ARL6IP1 and INPP5K are homogenously present on all the ER tubules and precisely localized (E), the fluorescence of ER membrane proteins Sec61β, VAPB, and Sac1 and of the luminal ER marker ss-GFPox-KDEL declines toward the ends of the most distal tubules (F–I, arrowheads). Scale bars: 5 µm. Representative examples of several cells imaged in at least three independent experiments. (J) Quantification of normalized fluorescent intensity of mCh-ARL6IP1 and ss-GFPox-KDEL along peripheral ER tubules based on line scans as exemplified on the top. Note that the dashed line elongates beyond the ends of the ER tubules into the background. Data are represented as mean ± SD (n = 13 ER tubules from six cells). The bar graphs at bottom right show the average fluorescence intensity along 1-µm segments at the proximal and distal end, respectively. ****, p < 0.0001; n.s., not significant (two-tailed t test).
Additionally, inspection of ER tubules at the edge of COS-7 cells revealed a continuously homogenous labeling of all tubules by ARL6IP1 and INPP5K (Fig. 3 E), while labeling by EGFP-Sec61β, EGFP-VAPB, EGFP-Sac1, and ss-GFPox-KDEL declined toward the cell periphery, in particular toward the tips of tubules, resulting in a relative enrichment of ARL6IP1 relative to Sec61β, VAPB, Sac1, and ss-GFPox-KDEL at their tips (Fig. 3, F–J).
The strong preference of ARL6IP1 for the tubular (thus high-curvature) ER is consistent with the resemblance of its predicted transmembrane topology with that of the reticulons: two hydrophobic hairpins with a cytosolic N and C-terminal regions (Voeltz et al., 2006). However, while the hairpins of the reticulon are embedded in the bilayer, the bend of the two hairpins of ARL6IP1, like the hairpins of another class of ER-shaping proteins, the atlastins (Liu et al., 2012), are predicted to protrude into the ER lumen, so that the hairpin alone would not be sufficient to generate bilayer asymmetry. Both the reticulons and the atlastins contain conserved amphipathic helices that are thought to interact with the membrane bilayer and help sense/stabilize its curvature (Liu et al., 2012; Brady et al., 2015). Analysis of the secondary structure of ARL6IP1 with several prediction algorithms suggested the presence of a helical amphipathic structure in its conserved cytosolic region bridging TM2 to TM3 (Fig. S2 C). This helix exhibits high hydrophobicity (<H> = 0.341) and high hydrophobic moment (<µH> = 0.438; Drin and Antonny, 2010) and is highly conserved in the ARL6IP1 family (Fig. S2 D). Replacement of the region containing the helical structure with an amino acid flexible linker of equivalent length consisting of myc epitopes (ARL6IP1L2-Myc) or substitution of two bulky hydrophobic residues on the predicted hydrophobic surface of the amphipathic helix with hydrophilic glutamic acids abolished the relative enrichment of ARL6IP1 into the peripheral ER (Fig. S2, E and F). Furthermore, this mutation abolished the preference of this construct for ER tubules relative to ER sheets, as demonstrated by coexpression of WT EGFP-ARL6IP1 with the mCh-ARL6IP1F105E L112E mutant in the same cells (Fig. S2, G and H). Together, these results indicate that bilayer curvature preference of ARL6IP1 is linked to its relative enrichment in peripheral ER tubules.
ER tubules populated by ARL6IP1 and INPP5K undergo rapid motion
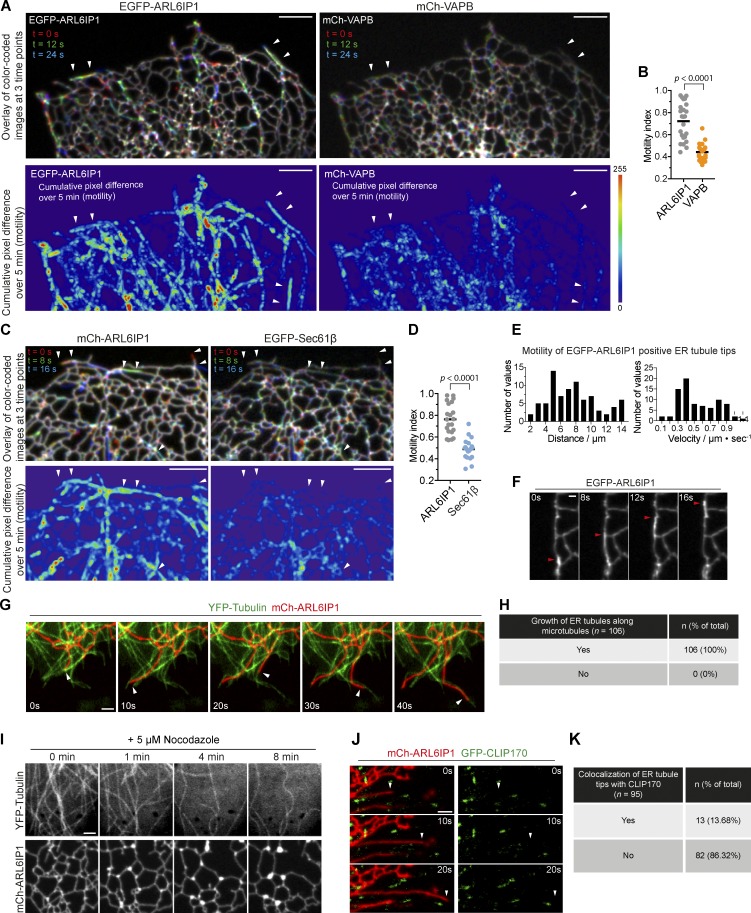
The relative enrichment of INPP5K and ARL6IP1 in the peripheral ER network, and at the distal portions of tubules in particular, raised the possibility that these two proteins populate, preferentially relative to other proteins, newly formed elongating tubules. To address this possibility, we monitored the dynamics of ARL6IP1 and INPP5K relative to other ER markers, such as the ER membrane proteins VAPB (Fig. 4, A and B) and Sec61β (Fig. 4, C and D) and the ER luminal protein ss-GFPox-KDEL (Fig. S3, B–D). Confocal microscopy observation of cell expressing fluorescently tagged Sec61β, VAPB, and KDEL demonstrated an overall rather stable structure of the ER network, with only irregular oscillations over short distances in a direction perpendicular to the tubules, consistent with thermally derived Brownian motion, as described previously (Nixon-Abell et al., 2016). To quantify the difference in motility, the cumulative pixel differences of these ER proteins during a 5-min recording were measured, compared in the heat maps and quantified (see Fig. S3, A and B for methods). The relative motility of EGFP-ARL6IP1 occurs primarily along linear tracks (Fig. 4, A and C; and Fig. S3, B and C) and is higher than that of other ER proteins such as Sec61β and VAPB (a membrane protein) or ss-GFPox-KDEL (a luminal protein; Fig. 4, B and D; and Fig. S3 D).
Figure 4.
ER tubules populated by ARL6IP1 and INPP5K undergo rapid motion. (A–D) Analysis of the motility of ER proteins. (A and C) Representative confocal images of COS-7 cells coexpressing EGFP-ARL6IP1 and mCh-VAPB (top fields of A) or mCh-ARL6IP1 and EGFP-Sec61β (top fields of C). In the images shown, three time points were color-coded and merged, so that the stationary ER elements staying on the same sets of pixels appear white, while the motile ER elements occupying alternate pixels appear in colors. Note the abundance of the colored ER tubules in ARL6IP1 images, relative to the VAPB and Sec61β (arrowheads). Bottom: Graphic display of motility from the same field shown above during a 5-min recording. Differences of fluorescence intensity at each pixel between subsequent time-lapse images were calculated, and these values are added up and pseudocolored (see Fig. S3 for methods). Scale bars: 5 µm. (B and D) Plots of motility index (see Fig. S3 for methods) where cumulative values of motility in several cells, calculated as in A and B, were normalized to the initial fluorescence intensity. n = 21 cells expressing EGFP-ARL6IP1 and mCh-VAPB (C), and n = 24 cells for cells expressing mCh-ARL6IP1 and EGFP-Sec61β (D). Data are represented as scattered dots with the solid black bar as mean (two-tailed t test). (E) Histograms of transport distances and velocity of the tips of motile ER tubules in cells overexpressing EGFP-ARL6IP1 (out of 81 events from five cells). All the events shown represent ER tubule movement that originated and ended during the recorded time. (F) Example of a growing ER tubule sliding along a preexisting ER tubule (arrows point to the tips of a ER tubule). Scale bar: 1 µm. (G) ER tubule extension events were captured during live-cell imaging of cells expressing YFP-α-tubulin and mCh-ARL6IP1 at the times indicated. Arrows depict the movement of ARL6IP1-postive ER tubules along microtubules. Scale bar: 2 µm. (H) Frequency of ARL6IP1-positive ER tubules that grow along microtubules marked by YFP-α-Tubulin (data from 14 cells). (I) Live-cell images of a COS-7 cell expressing YFP-α-Tubulin and mCh-ARL6IP1 upon 5 µM nocodazole treatment. Note the depolymerization of microtubules, the collapse of tubular ER network and the accumulation of bright foci containing mCh-ARL6IP1. Scale bar: 2 µm. Images are representative of three independent experiments. (J) Time-lapse images of cells expressing mCh-ARL6IP1 and GFP-CLIP170, a microtubule plus end-tracking protein. Arrowheads point to the tip of an elongating ARL6IP1-positive ER tubule. Note this tip lacks CLIP170 fluorescence. Scale bar: 2 µm. (K) Frequency of ARL6IP1-positive ER tubules tips adjacent (<1 µm) to CLIP170 puncta (data from 10 cells).
Observation of cells expressing fluorescent forms of ARL6IP1 and INPP5K revealed the occurrence of rapidly moving elements positive for these two proteins (speed = 0.54 ± 0.23 µm/s; distance of single moving event = 7.6 ± 3.2 µm; Fig. 4, E and F) superimposed onto the more stable ER network (Video 1). Close inspection of the mobile elements by live-cell nanoscopy imaging (stimulated emission depletion [STED]) revealed that they corresponded to elongating tubules that either grew roughly along preexisting ER tubules (Video 2) or explored new territory (Video 3). As shown by coexpression of either fluorescent ARL6IP1 with YFP-α-tubulin, this elongation occurred along microtubules (Fig. 4, G and H) and stopped upon depolymerization of microtubules with nocodazole (Fig. 4 I). Moreover, it appeared to occur by sliding along the length of existing microtubules, as expected for microtubular motor-based motility (membrane sliding mechanism; Fig. 4 G) and not by attaching to the plus ends of microtubules and extending by a microtubule polymerization-dependent mechanism (tip attachment complex mechanism), as revealed by lack of colocalization with CLIP170 (Fig. 4, J and K; Waterman-Storer and Salmon, 1998; Friedman et al., 2010).
Increased abundance of ER sheets upon the loss of INPP5K or ARL6IP1
The selective enrichment of ARL6IP1/INPP5K on newly formed tubules opens the question of whether presence of these proteins plays a role in ER tubules growth and stability. To address this question, we examined ER morphology upon ARL6IP1 or INPP5K silencing in HeLa cells (Fig. 5, A and B). Typically, ER sheets are primarily confined to the perinuclear region, although peripheral membrane sheets are also seen. In ARL6IP1 or INPP5K knockdown cells, a decrease of ER tubules and expansion of the ER sheets at the cell periphery was observed (Fig. 5 B). This effect was rescued by WT INPP5K, which has PI(4,5)P2 and PI(3,4,5)P3 phosphatase activity as reported (Fig. S4, A and B), but not by mutant INPP5K harboring point mutations that block its phosphatase activity (D192A; Fig. S4, C and D) or impair its recruitment to ARL6IP1 (D361G; Fig. 5 C). Thus, INPP5K and ARL6IP1 play a role in maintaining tubular ER, possibly because their presence is required for efficient growth of new tubules. We did not observe, however, a change in the localization and/or level of a well-established PI(4,5)P2 marker protein (GFP-PHPLCδ1) in INPP5K knockdown cells (Fig. S4, E–G). As in WT cells, this PI(4,5)P2 probe labeled nearly exclusively the plasma membrane. These negative results may indicate that PI(4,5)P2 pools controlled by INPP5K are only very small or that such pools may not be efficiently recognized by the PI(4,5)P2 marker due to the need of a coreceptor (Di Paolo and De Camilli, 2006).
Figure 5.
Increased abundance of ER sheets upon loss of INPP5K or ARL6IP1. (A) Western blots of WT HeLa cells transfected with the indicated siRNAs showing depletion of INPP5K or ARL6IP1 proteins. (B) Top row: Representative confocal images of HeLa cells transfected with the indicated siRNAs and expressing the ER marker EGFP-Sec61β. High magnification of the regions enclosed by dashed boxes in the top row are shown in the bottom row. Scale bars: 5 µm in the top row, 2 µm in the bottom row. Solid red lines denote the edge of the cell. Note the predominant presence of ER tubules in the control cell, but the predominance of ER sheets throughout the cytoplasm of INPP5K or ARL6IP1 siRNA-treated cells. (C) Occupancy of the ER (both tubules and sheets) per unit area of each cell analyzed based on the analysis described in B for cells treated with control or INPP5K siRNA and expressing the indicated EGFP-INPP5K constructs. Automatic thresholding was applied (see Materials and methods for details) to select the total area occupied by the ER network (delineated by solid lines). The occupancy of the ER network per unit cell area serves as a proxy for the abundance of peripheral ER sheets and was calculated by dividing the pixel area occupied by the ER by that of the total cell area. Pooled data from three independent experiments are presented as scattered dots (solid black bars indicating the mean ± SD) and analyzed by two-tailed t-tests. n = 28 cells for control or INPP5K siRNA, 25 cells for INPP5K siRNA + EGFP-INPP5KWT, 25 cells for INPP5K + EGFP-INPP5KD192A, 21 cells for INPP5K + EGFP-INPP5KD361G. ****, p < 0.0001; n.s., p > 0.05 (INPP5K siRNA + EGFP-INPP5KD192A vs. INPP5K siRNA, p = 0.0541; INPP5K siRNA + EGFP-INPP5KD361G vs. INPP5K siRNA, p = 0.057).
Loss of the INPP5K orthologue in C. elegans leads to defects in neuronal ER morphology
The results described above converged with those of an independent and concomitant genetic screen aimed at the identification of genes that control the organization of the ER in the PVD neuron in C. elegans (Fig. 6 A). The morphology of PVD neurons was revealed by the expression of mCherry under the control of a PVD-specific promoter (PVD::mCherry), while the distribution of the ER in these cells was monitored by expression of a fluorescent ER marker derived from the signal peptidase SP12 under the control of a PVD-specific promoter (PVD::GFP::SP12; Rolls et al., 2002). This screen led to the isolation of a cil-1(wy50075) mutant, which showed abnormal ER distribution in PVD dendrites. Based on primary sequence and domain structure similarity (a 5-phosphatase domain followed by SKICH domain), CIL-1 appears to be the orthologue of INPP5K (Fig. S5). As shown in Fig. 6 B, the ER extended into the majority of the dendritic branches of PVD neurons in WT animals. In contrast, although the morphology of PVD neurons in cil-1 mutants remained largely intact, the presence of the ER in the branches of the proximal and posterior regions of cil-1 mutant PVD neurons was more sparse (Fig. 6 C). Additionally, close inspection of the fluorescence of the ER marker (PVD::GFP::SP12) in the cell bodies of PVD neurons shows a difference in the architecture of the ER (Fig. 6, D and E), suggestive of a lower abundance of ER tubules in cil-1 mutants.
Figure 6.
cil-1 functions cell autonomously in PVD to regulate ER morphology. (A) Schematic drawing of PVD dendrites and ER pattern. Note that the ER tubules invade some, but not all, dendritic branches in the PVD. PVD dendrites are divided into three different segments: posterior, proximal, and distal dendrites. Based on the ER branch morphology, ER tubules are divided into three types (I, L, and T) to indicate whether the ER tubules invade just the secondary or tertiary branches. (B) Representative confocal images of transgenic animals expressing a PVD neuron marker ser-2Prom3::mCherry (PVD::mCherry) and an ER resident protein ser-2Prom3::GFP::SP12 (PVD::GFP::SP12) simultaneously. Note that the ER tubules in the branches are reduced in the cil-1 mutant. This defect is restored by reexpresssion of CIL-1 in PVD neurons (PVD::CIL-1). Arrowhead represents an ER branch. Scale bar: 10 µm. (C) Quantification of PVD proximal region ER branches. *, p < 0.05; **, p < 0.01; n.s., not significant by one-way ANOVA. Error bars indicate SEM. n = 40 for each genotype. (D) ER morphology in PVD cell body revealed by SIM imaging. The images are single confocal slices crossing the nuclear envelope at approximately its equatorial region. A roughly concentric circle (red) of 4.6 µm in diameter was drawn around the nucleus. The number of crossings between the red circle and ER tubules was counted. Scale bar: 5 µm. (E) Quantification of the number of crossings between the red circle and ER profiles in cells of the various genotypes. The number of crossings reflects the complexity of the ER. ***, p < 0.001; n.s., not significant by one-way ANOVA. Error bars indicate SEM. n > 48 for each genotype.
Cell-autonomous function of Cil-1 in PVD neurons
We assessed the functional contribution of CIL-1 to ER morphology defects by analyzing ER architecture upon reexpression of cil-1 from a PVD-specific promoter. Overexpression of WT CIL-1 in cil-1 mutants rescued the ER branching defects (Fig. 6, B–E). In contrast, expression of catalytically dead CIL-1 (N175A; Bae et al., 2009) or a SKICH domain–deleted version of CIL-1 failed to rescue the ER morphology phenotype (Fig. 6 D). Importantly, three CIL-1 mutations (I39T, Y261C, and I327T) corresponding to INPP5K mutations found in human patients (Wiessner et al., 2017) also did not rescue the ER morphology phenotype (Fig. 6, D and E).
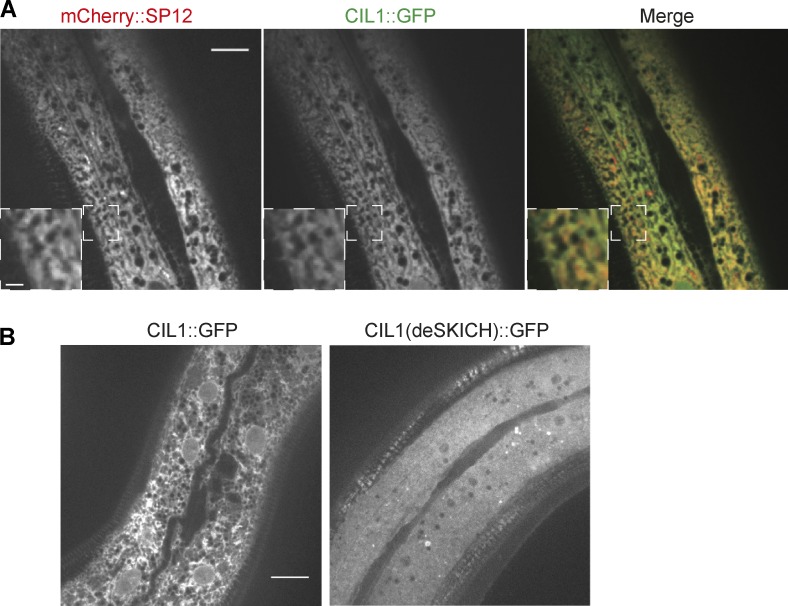
To confirm endogenous expression of CIL-1 in PVD neurons, we created a transgene in which a cil-1 genomic fragment including the endogenous cil-1 promoter and coding regions were fused with GFP. With this transgene, we observed robust expression of GFP in PVD (Fig. S5), suggesting that cil-1 functions cell autonomously in extending the ER into the dendritic branches of PVD cells. To investigate the subcellular localization of CIL-1 in worm cells, we coexpressed CIL-1::GFP and a ER marker (pdpy-7::cil-1::GFP and pdpy-7::mCherry::SP12, respectively) in the hypodermal cells (a large, flat cell where ER networks can be easily visualized). Similar to INPP5K’s localization in mammalian cells, CIL-1::GFP showed reticular distribution and extended colocalization with the ER marker, consistent with a localization of CIL-1 in the ER (Fig. 7 A). Importantly, the SKICH domain deletion construct lost the reticular ER-like localization indicating that CIL-1, like INPP5K, requires the SKICH domain for binding to the ER (Fig. 7 B, right).
Figure 7.
The SKICH domain is essential for CIL-1 ER localization. (A) Representative confocal images of a transgenic worm expressing mCherry::SP12 and CIL-1::GFP driven by the hypodermal cell specific promoter. Scale bar: 10 µm. The pattern of CIL-1::GFP fluorescence is very similar to the pattern of the ER marker mCherry::SP12. Insets showing magnified view of the boxed region (scale bar: 2 µm). (B) Representative confocal images of transgenic worms expressing full-length CIL-1::GFP or a truncated from of CIL-1 lacking the SKICH domain, CIL-1(deSKICH)::GFP. While WT CIL-1 has the reticular distribution in cells expected for an ER protein, the truncated protein has a diffuse cytosolic localization. Scale bar: 10 µm.
Discussion
The results of this study demonstrate that one of the functions of the phosphoinositide phosphatase activity of INPP5K is to participate in the fine control of ER organization. In mammalian cells, INPP5K is recruited to the ER by ARL6IP1, a molecule that share features with ER shaping proteins. Together with ARL6IP1, INPP5K is preferentially and homogeneously localized throughout the tubular ER network relative to sheets, including distal portions of the ER tubules that undergo rapid elongation driven by microtubular motors. In these elongating tubules, both INPP5K and ARL6IP1 are enriched relative to other classical ER marker proteins such as Sec61β, VAPB, and Sac1. Furthermore, their knockdown results in the expansion of ER cisternae at the expense of ER tubules. Expansion of sheets may be the results of decreased formation of new tubules or by their decreased stability once they have formed. These findings raise the possibility that INPP5K may play some role linked to ER tubule formation, although the underlying mechanisms remain unknown. An impact of INPP5K on ER architecture is independently supported by the identification of a requirement for cil-1, the INPP5K orthologue in C. elegans, in the normal extension of ER tubules into the dendritic branches of a model neuron in C. elegans.
Most likely the preference for ER tubules of the INPP5K anchor on the ER, ARL6IP1, reflects properties of ARL6IP1 that are similar to those of ER-shaping proteins such as the reticulons, DP1/Yop1p, and atlastin (Voeltz et al., 2006; Hu et al., 2008; Blackstone et al., 2011). Each of these proteins comprises hydrophobic hairpins that partially or completely span the bilayer and in some cases have an additional amphipathic helix implicated in their curvature-generating/sensing properties (Hu et al., 2008; Shibata et al., 2008; Liu et al., 2012; Brady et al., 2015). As previously suggested, although ARL6IP1 does not share primary sequence homology with any of these ER-shaping proteins, it may adopt a similar topology (Yamamoto et al., 2014). Here, we have also provided evidence for the occurrence of an amphipathic helix on its cytosolic surface between the two hairpins that may contribute to curvature generating/sensing.
The relative enrichment of ARL6IP1, compared with Sec61β, in peripheral ER tubules was also recently reported by endogenously tagging ARL6IP1 with split fluorescent proteins (Feng et al., 2017) and is consistent with the heterogeneity within the ER tubular network that was previously noted for the reticulons (Voeltz et al., 2006). Loss of the reticulons, as the loss of ARL6IP1, results in the expansion of ER sheets (Voeltz et al., 2006). Factors that may contribute to the lag of some ER proteins to populate the newly formed tubules may include (1) lower diffusion rate in the ER, (2) mismatch between curvature of new tubules and curvature preferences of these proteins, and (3) potential selective enrichment of specific lipids in the new tubules, making their bilayer less permissive to invasion by certain ER proteins.
The significance of the presence of a 5-phosphatase throughout the tubular ER, including newly formed tubules, remains unclear. A first major open question is whether INPP5K acts “in cis” on phosphoinositides present the ER membrane or “in trans” at contacts of the ER with other membranes. Several independent studies have reported that the preferred substrates of INPP5K are PI(4,5)P2 and, to a lower extent, PI(3,4,5)P3 (Ijuin et al., 2000; Gurung et al., 2003; Schmid et al., 2004), a finding that we have confirmed. There is no evidence so far for the presence of these phosphoinositides or of other 5-phosphorylated phosphoinositides in the ER (Di Paolo and De Camilli, 2006; Balla and Várnai, 2009; Hammond and Balla, 2015), but one reason for such absence could be precisely the presence of INPP5K (i.e., enzyme that dephosphorylates them). A similar scenario has been proposed to account for the absence of detectable PI4P in the ER, where the inositol 4-phosphatase Sac1 is localized (Nemoto et al., 2000; Zewe et al., 2018). This does not exclude the potential presence of transient pools of phosphorylated phosphoinositides in the ER that are not detectable by available PI(4,5)P2 and PI(3,4,5)P3 probes. Such pools could have three sources: (1) local synthesis by phosphatidylinositol phosphate kinases (but no association of phosphatidylinositol phosphate kinases with the ER have been reported so far), (2) retrograde bulk flow from the Golgi complex to the ER as part of the bilayer of transport vesicles, or (3) transport from other membranes via lipid transport proteins, a mechanism also hypothesized for the delivery of PI4P to the ER (Mesmin et al., 2013). However, even in INPP5K knockdown cells, we did not detect PI(4,5)P2 in the ER by a widely used PI(4,5)P2 probe (GFP-PHPLCδ1).
An alternative scenario is that INPP5K acts in trans on non-ER membranes that contact the ER, for example the plasma membrane, given the abundance of growing ER tubules at the cell periphery. As some proteins that mediate ER–plasma membrane contact site binds PI(4,5)P2 in the plasma membrane, presence of a 5-phosphatase at the tips of ER tubules may play a negative role in the formation of new contacts. Dephosphorylation in trans of plasma membrane phosphoinositides may also help clearing the plasma membrane of the actin cytoskeleton and facilitate dynamics rearrangement of this membrane. On the other hand, we did not find evidence for an increase of PI(4,5)P2 labeling of any membranes after INPP5K knockdown. A similar result was reported in yeast cells upon deletion of the INP54 gene, the closest homologue of INPP5K (Wiradjaja et al., 2007). In yeast, however, the knockout of INP54 in cells harboring a temperature-sensitive mutation in the Sac1 gene resulted in the appearance of PI(4,5)P2 on the vacuole at the restrictive temperature (Wiradjaja et al., 2007). When we knocked down both INPP5K and Sac1 in HeLa cells, no PI(4,5)P2 labeling of intracellular organelles by the PI(4,5)P2 marker GFP-PHPLCδ1 was observed (unpublished data).
Another potential scenario is that presence of INPPK at the ER surface (i.e., next to IP3 receptors; Mignery et al., 1989) may help shut off IP3-mediated signaling, as 5-phosphatases can also act on soluble inositol polyphosphates. Yet another scenario is that ER-bound INPP5K may represent a reserve pool of INPP5K that can be mobilized and translocated to other sites in response to specific stimuli. For example, a pool of INPP5K was shown to be recruited to the plasma membrane to down-regulate PI(3,4,5)P3 signaling in response to growth factor stimulation (Gurung et al., 2003; Ijuin and Takenawa, 2003). However, how these potential actions could relate to our evidence for a role of INPP5K in the control of ER architecture remains unclear.
The identification of CIL-1 in a forward unbiased genetic screen for defects in ER architecture strongly corroborates our findings in mammalian cells. In cil-1 mutant, the ER fails to extend into the majority of small dendritic branches of the PVD neuron, a defect that could be explained by impaired formation/stabilization of new ER tubules. WT Cil-1, but not its catalytically dead mutant, could rescue this defect.
The gene encoding INPP5K was reported to be up-regulated in regenerating mammalian spinal cord axons. The same study showed that overexpression of INPP5K in acutely dissociated mouse cortical neurons enhanced neurite outgrowth and branching during the recovery after in vitro injury (Fink et al., 2017). An interesting possibility is that INPP5K may help coordinate progression of neuronal growth cones with the extension of the ER into its tips. The finding that loss of ARL6IP1 results in degenerative axonal diseases (Blackstone et al., 2011; Novarino et al., 2014; Nizon et al., 2018) and in fragmentation of the smooth ER in axons (Fowler and O’Sullivan, 2016) supports this hypothesis, although ARL61P1 may have independent functions in the control of ER architecture in axons.
It remains unclear whether the clinical manifestations, primarily muscular dystrophy, observed in patients with INPP5K mutations, relate to the impact of INPP5K on ER architecture that we reported here. These patients have additional manifestations, including intellectual disability, consistent with a role of INPP5K in neurons (Fink et al., 2017; Osborn et al., 2017; Wiessner et al., 2017). All these patient mutations likely only result in partial loss of INPP5K function, as complete loss of INPP5K expression in mice results in embryonic lethality (Ijuin et al., 2008). As suggested by the occurrence of INPP5K interactions and localizations that imply non-ER function of INPP5K, including a partnership with MAD2L1BP in the nucleus (Fig. S1 A) and actions at the plasma membrane in response to growth factors (Ijuin et al., 2000; Gurung et al., 2003), this enzyme is expected to have pleiotropic actions that await further investigation.
Materials and methods
Cell culture and transfection
COS-7 cells were cultured in DMEM containing 10% FBS and 1% penicillin/streptomycin at 37°C and 5% CO2. Transfection of plasmids was performed with Lipofectamine 2000 (Life Technologies). For siRNA experiments, HeLa cells were transfected with control or target siRNA oligos by using Lipofectamine RNAi MAX (Life Technologies) and cultured for 72 h before analysis.
Neuron culture and transfection
Primary neurons were cultured from E18 embryos of WT BL6 mice as described previously (Gowrishankar et al., 2017). Briefly, cortices were removed from freshly euthanized E18 embryos in cold HBSS and then minced into small pieces using a sterile scalpel. The tissue was subsequently treated with papain enzyme to digest the extracellular matrix and loosen the cells. After inactivation of the enzyme and double washes with HBSS, the loosened tissue was dissociated by trituration using a pipette to obtain a single-cell suspension. Cells were counted and plated on to poly-d-lysine (20 µg/ml; Sigma-Aldrich)–coated 35-mm MatTek dishes with glass coverslips attached to their bottom (300,000 cells per dish). These neurons were transfected at 7 d in vitro using Lipofectamine 2000 and imaged live at 9 d in vitro on a spinning disk microscope.
DNA plasmids
Sources of plasmids were as follows: YFP-Tubulin, GFP-CLIP170 (D. Toomre, Yale University, New Haven, CT), GFP-PHPLCσ1, EGFP-VAPB, and mCh-VAPB, EGFP-Sac1 (our laboratory). EGFP-Sec61 was obtained from Addgene (Plasmid 62008). ss-GFPox-KDEL (comprising an ER-retention sequence fused to a version of GFP optimized for expression in the ER lumen) was obtained from E.L. Snapp (Albert Einstein College of Medicine, Bronx, NY).
EGFP-INPP5K were generate by PCR amplification from cDNA clone of human INPP5K (clone ID 3354658; Open Biosystems) using the following primers: XhoI_INPP5K_Fw, 5′-GACTCAGATCTCGAGCTTCGATGAGCTCGCGGAAGCTGAGCGGG-3′, EcoRI_Stop_INPP5K_Rv, 5′-CTGCAGAATTCATCTAGATCTGTGGCTGTGCTTCACCCAGTGG-3′. PCR products were ligated between XhoI and EcoRI sites in the pEGFP-C1 vector (Clontech). HA-INPP5K was generated by PCR amplification from the same cDNA clone of human INPP5K using the following primers: AgeI_HA-INPP5K, 5′-GCTACCGGTCATGTACCCATACGATGTTCCAGATTACGCTATGAGCTCGCGGAAGCTG-3′, EcoRI_Stop_INPP5K_Rv, 5′-CTGCAGAATTCATCTAGATCTGTGGCTGTGCTTCACCCAGTGG-3′. PCR products were ligated between AgeI and EcoRI sites in the pEGFP-C1 vector. EGFP-INPP5K276–448 was generated by PCR amplified using the following primers: HindIII_INPP5K_Fw, 5′-AGCTCAAGCTTCGAAGCGGCAGCCCTGTGC-3′, EcoRI_Stop_INPP5K_Rv, 5′-CTGCAGAATTCATCTAGATCTGTGGCTGTGCTTCACCCAGTGG-3′. PCR products were ligated between HindIII and EcoRI sites in the pEGFP-C1 vector.
I363T, D192A, and D361G mutations in INPP5K were generated using a mutagenesis kit (QuikChange II XL; Agilent Technologies), from WT INPP5K as template, with the following primers, respectively (targeted nucleotides are shown in lowercase): INPP5K_I363T_Fw, 5′-GCAGCCCGTGGGACTGGAcTGGACTGTACAAGGTGGGGCTG-3′, INPP5K_I363T_Rv, 5′-CAGCCCCACCTTGTACAGTCCAgTCCAGTCCCACGGGCTGC-3′; INPP5K_D192A_Fw, 5′-CATTATCTGGTTTGGAGcCATGAACTTTCGGATC-3′, INPP5K_D192A_Rv, 5′-GATCCGAAAGTTCATGgCTCCAAACCAGATAATG-3′; INPP5K_D361G_Fw, 5′-CCAGCAGCCCGTGGGgCTGGATTGGACTGTACAAG-3′, INPP5K_D36G_Rv, 5′-CTTGTACAGTCCAATCCAGcCCCACGGGCTGCTGG-3′. INPP5K1-361 and INPP5K1-431 truncations were generated by introducing stop codons at indicated location into the WT INPP5K template, with the following primers for mutagenesis PCR reactions, respectively (targeted nucleotides are shown in lowercase and stop codons in bold): INPP5K-1-361_Stop_Fw, 5′-CAGCAGCCCGTGGGACTGaATTGGACTGTACAAGG-3′, INPP5K-1-361_Stop_Rv, 5′-CCTTGTACAGTCCAATtCAGTCCCACGGGCTGCTG-3′; INPP5K-1-431_Fw, 5′-CAGACCCTTCCAGATCtaGCCTGGCTCCTTGAGGGAGG-3′, INPP5K-1-431_Rv, 5′-CCTCCCTCAAGGAGCCAGGCtaGATCTGGAAGGGTCTG-3′.
To clone fluorescence-tagged ARL6IP1 constructs, cDNA of ARL6IP1 was PCR amplified from a total human brain cDNA library and sequence validated to match the GenBank entry for ARL6IP1 (NM_015161.2). The following primers were used: HindIII_ARL6IP1_Fw, 5′-GCTCAAGCTTCGATGGCGGAGGGAGATAATCG-3′, BamHI_Stop_ARL6IP1_Rv, 5′-CCGGTGGATCCTCATTCGTTTTTCTTTTCTTTTTG-3′. PCR products were ligated between HindIII and BamHI in the pEGFP-C1 or pmCherry-C1 vectors (Clontech), to generate EGFP-ARL6IP1 and mCh-ARL6IP1, respectively. F105E and L112E double mutations were introduced into the WT ARL6IP1 template via two sequential mutagenesis, with the following primers, respectively (targeted nucleotides are shown in lowercase): ARL6IP1_F105E_Fw, 5′-CACTGAACAACAGCAAAGAgaaCATGAAATTTGCAGCAATC-3′, ARL6IP1_F105E_Rv, 5′-GATTGCTGCAAATTTCATGttcTCTTTGCTGTTGTTCAGTG-3′; ARL6IP1_L112E_Fw, 5′-CATGAAATTTGCAGCAATgaAGTAAAAACTCGACGCAG-3′, ARL6IP1_L112E_Rv, 5′-CTGCGTCGAGTTTTTACTtcATTGCTGCAAATTTCATG-3′.
ARL6IP1L1-Myc and ARL6IP1L3-Myc were cloned by synthesizing gBlocks (Integrated DNA Technologies) containing ARL6IP1 cDNAs whose segments on L1 or L3 were replaced by flexible linkers of equivalent length consisting of myc tags; sequences of myc tags were codon optimized to avoid repetitive and GC-rich sequences, which pose challenge for gBlock synthesis. Sequences of the gBlock are as follows (with the replaced nucleotide in lowercase):
ARL6IP1L1-Myc, 5′-ATGGCGGAGGGAGATAATatggaacagaagctcatcagcgaagaagaccttatggaacagaagctcatttccgaggaggatcttatggagcaaaagctgataagcgaggaggacctgGAAAGAGCCTGGTTTCCACCTGCCATCATGGGTGTGGTTTCTTTGGTGTTTCTGATTATCTACTATCTAGATCCATCTGTTCTGTCCGGCGTTTCCTGTTTTGTTATGTTTTTGTGCTTGGCTGACTACCTTGTTCCCATTCTAGCGCCTAGAATTTTTGGCTCCAATAAATGGACCACTGAACAACAGCAAAGATTCCATGAAATTTGCAGCAATCTAGTAAAAACTCGACGCAGAGCTGTGGGTTGGTGGAAACGCCTCTTCACACTAAAGGAAGAAAAACCTAAGATGTACTTCATGACCATGATCGTTTCCCTTGCTGCGGTTGCTTGGGTGGGACAACAAGTCCACAACCTGCTTCTCACCTACCTGATAGTGACTTCCTTACTATTGCTTCCTGGACTAAACCAACATGGAATCATTTTGAAGTACATTGGAATGGCCAAGAGGGAGATAAACAAACTTCTCAAACAAAAAGAAAAGAAAAACGAATGA-3′; and
ARL6IP1L3-Myc, 5′-ATGGCGGAGGGAGATAATCGCAGCACCAACCTGCTGGCTGCAGAGACTGCAAGTCTGGAAGAACAGCTGCAAGGATGGGGAGAAGTGATGCTGATGGCTGATAAAGTCCTCCGATGGGAAAGAGCCTGGTTTCCACCTGCCATCATGGGTGTGGTTTCTTTGGTGTTTCTGATTATCTACTATCTAGATCCATCTGTTCTGTCCGGCGTTTCCTGTTTTGTTATGTTTTTGTGCTTGGCTGACTACCTTGTTCCCATTCTAGCGCCTAGAATTTTTGGCTCCAATAAATGGACCACTGAACAACAGCAAAGATTCCATGAAATTTGCAGCAATCTAGTAAAAACTCGACGCAGAGCTGTGGGTTGGTGGAAACGCCTCTTCACACTAAAGGAAGAAAAACCTAAGATGTACTTCATGACCATGATCGTTTCCCTTGCTGCGGTTGCTTGGGTGGGACAACAAGTCCACAACCTGCTTCTCACCTACCTGATAGTGACTTCCTTACTATTGCTTCCTGGACTAAACCAACATGGAagtggtggcatggaacagaagctcatcagcgaagaagaccttagcggaggcAAACAAAAAGAAAAGAAAAACGAATGA-3′.
These gBlocks were used as template for PCR amplification with the primers HindIII_ARL6IP1_Fw and BamHI_Stop_ARL6IP1_Rv. PCR products were ligated between HindIII and BamHI in the pmCherry-C1 vector to generate mCh-ARL6IP1L1-Myc and mCh-ARL6IP1L3-Myc, respectively.
To clone mCherry-tagged MAD2L1BP construct, cDNA of MAD2L1BP was PCR amplified from a total human brain cDNA library and sequence validated to match the GenBank entry for ARL6IP1 (NM_001003690.1). The following primers were used: HindIII_MAD2L1BP_Fw, 5′-GCTCAAGCTTCGATGGCCCGCGTGCCGCTGGG-3′, BamHI_stop_MAD2L1BP_Rv, 5′-CGGTGGATCCCGTCACTCGCGGAAGCCTTTAAATGTC-3′. PCR products were ligated between HindIII and BamHI in the pmCherry-C1 vector to generate mCh-MAD2L1BP.
EGFP-INPP5J was generate by PCR amplification from cDNA clone of human INPP5J (clone ID 40034071; Open Biosystems) using the following primers: XhoI_INPP5J_Fw, 5′-CAGATCTCGAGCTATGGCCCTCCCAAGGCTTGGCACAC-3′, HindIII_Stop_INPP5J_Rv, 5′-AATTCGAAGCTTGTCAGGGCCCCAGGCCCCCTTC-3′. This cDNA matches the GenBank entry of INPP5J (NM_001284289.1). Compared with the longest isoform, this variant differs in its 5′ UTR, lacks a portion of the 5′ coding region, and thus encodes isoform lacking the N-terminal proline-rich domain but retains the intact 5-phosphatase domain and the SKICH domain. PCR products were ligated between XhoI and EcoRI sites in the pEGFP-C1 vector (Clontech) to generate EGFP-INPP5J.
siRNAs
Double-stranded siRNAs were purchased from Integrated DNA Technologies with the following references and duplex sequences: INPP5K (hs.Ri.INPP5K.13.1), 5′-rGrArCrArUrGrArUrGrGrUrCrArGrCrUrArCrUrCrUrUrCAA-3′, 5′-rUrUrGrArArGrArGrUrArGrCrUrGrArCrCrArUrCrArUrGrUrCrArU-3′; ARL6IP1 (hs.Ri.ARL6IP1.13.2), 5′-rGrUrArCrArGrUrUrCrArArGrUrGrArArUrCrUrGrGrArUAA-3′, 5′-rUrUrArUrCrCrArGrArUrUrCrArCrUrUrGrArArCrUrGrUrArCrUrA-3′; and negative control (NC1), 5′-rCrGrUrUrArArUrCrGrCrGrUrArUrArArUrArCrGrCrGrUAT-3′, 5′-rArUrArCrGrCrGrUrArUrUrUrArCrGrCrGrArUrUrArArCrGrArC-3′.
Antibodies and chemicals
Primary antibodies were as follows (with sources and references codes in parentheses): rabbit monoclonal anti-HA (C29F4; Cell Signaling), rabbit polyclonal anti-INPP5K (GTX32681; GeneTex), rabbit polyclonal anti-ARL6IP1 (GTX85516; GeneTex). Nocodazole was purchased from Sigma-Aldrich (product number M1404).
Fluorescence microscopy
Live cells
Cells were plated on 35-mm glass-bottom dishes (MatTek Corp) at low density allowing attachment overnight and then transfected and imaged with a spinning disc confocal microscope 16–20 h after transfection. Before imaging, cells were transferred to imaging buffer containing 125 mM NaCl, 5 mM KCl, 1.3 mM CaCl2, 1.2 mM MgCl2, 25 mM Hepes, and 3 mM d-glucose with pH adjusted to 7.4 with NaOH.
Imaging
Spinning disc confocal microscopy was performed using the Improvision UltraVIEW VoX system (Perkin-Elmer) built around a Nikon Ti-E inverted microscope and a Hamamatsu C9100-50 camera, equipped with PlanApo objectives (60×/1.49 NA) and controlled by Volocity (Improvision) software.
Image quantification
Fluorescence signals was quantified using Fiji. Data were processed with Excel (Microsoft) and plotted with Prism7 (GraphPad).
Quantification of the occupancy of the ER network in the cell periphery (Fig. 5, B and C)
Cells were plated on MatTek dishes coated with fibronectin so that a single layer of ER tubular network could be resolved. Confocal images of cells expressing EGFP-Sec61b were first converted from 16 to 8 bits, where the minimum intensity of the 16-bit image was 0 in the 8-bit image and the maximum intensity of the 16-bit image was 255. Cytoplasmic regions were manually selected, beginning at the nuclear envelope and ending at the cell periphery. Automatic threshold segmentation was then done to convert a grayscale image to a binary image for the total ER network based on a global pixel intensity thresholding above the background. Area in pixels of the total ER binary image and cytoplasmic region was respectively measured. Dividing the number of ER pixels by the number of cytoplasmic pixels gives the occupancy of the ER per unit cytoplasmic area.
Analysis of the motility of fluorescent-tagged ER proteins
COS-7 cells coexpressing EGFP-ARL6IP1 and mCherry (mCh)-VAPB were imaged by confocal live-cell microscopy. EGFP and mCh signals were separated into two stacks of sequential images, and differences of fluorescence intensity at each pixel between two subsequent images were calculated (“Δ Intensity”). The differences at each pixel were added up to generate a cumulative intensity difference (“∑ Δ intensity”) and pseudocolored using the Physics look-up table in Fiji (see Fig. S3 A). Motility of ER proteins was quantified with a motility index, which reflects normalized cumulative fluorescence intensity changes as an ER protein moves over time. The index is defined as a ratio between the cumulative intensity difference (“∑ Δ intensity”) and the fluorescence intensity at initial time point (“Intensity [Imaget = 0]”).
Quantification of the plasma membrane localization of the PI(4,5)P2 probe via line scans
The GFP-PHPLCσ1 probe was used to label PI(4,5)P2 in HeLa cells transfected with control or INPP5K siRNA. A line of 5 µm in length was manually drawn perpendicular to the plasma membrane (see dashed lines in Fig. S4 E). Fluorescence pixel intensity along the line was calculated and normalized by dividing the fluorescence of each pixel by the average fluorescence intensity of the line in the cytosolic region.
Immunoprecipitation and immunoblotting
HeLa cells expressing the indicated constructs were washed in cold PBS and lysed on ice in lysis buffer (50 mM Tris, 150 mM NaCl, 1% NP-40, 0.5 mM EDTA, 10% glycerol, pH 7.4, and protease inhibitor cocktail; Roche). Cell lysates were then centrifuged at 21,000 g for 20 min at 4°C. For anti-GFP immunoprecipitation, supernatants were incubated with Chromotek GFP-trap agarose beads (Allele Biotech) for 2 h at 4°C under rotation. Subsequently, beads were washed in lysis buffer containing 1% NP-40 three times. Afterward, immunoprecipitated proteins bound to the beads were eluted by incubation in PAGE sample loading buffer (containing 2% SDS) at 60°C for 20 min and 70°C for 10 min. Immunoprecipitates were processed for SDS-PAGE and transferred to nitrocellulose membranes with a iBlot2 Gel Transfer Device (Invitrogen). Membrane were blocked with 5% skim milk in TBS and 0.1% Tween-20 and probed with anti-HA primary antibody overnight at 4°C followed by IRDye 800CW goat anti-rabbit secondary antibody (LI-COR Biosciences) for 1 h at room temperature. Blots were visualized using Odyssey Fc Imager (LI-COR Biosciences) at 800 nm channel and quantified using the gel analysis commands in Fiji.
Live-cell nanoscopy imaging
COS-7 were seeded at a concentration of 10–15 × 104 cells/cm2 per dish on glass-bottomed dishes (MatTek), treated with 1 M KOH, and subsequently coated with 0.005 mg/ml human plasma fibronectin (EMD Millipore) at 37°C for 30 min. Cells were transfected after 24 h with Lipofectamine 2000, following the manufacturer’s instructions, with cDNAs encoding SNAP-ARL6IP1 or Halo-ARL6IP1 in OptiMEM-I (Thermo Fisher Scientific). The two vectors were generated replacing EGFP, in the EGFP-ARL6IP1 vector cut at the AgeI and BsrGI sites, with SNAP tag or Halo tag. PCR amplification of the fragments and subsequent ligation was performed using the In-Fusion Cloning Kit and online tools (BD Clontech, Takara Bio). 24 h after transfection, cells expressing ARL6IP1 SNAP or Halo tag were labeled immediately before imaging with JF646-SNAP-tag ligand or JF646-HaloTag ligand, respectively (gift from L. Lavis, Janelia Research Campus, Howard Hughes Medical Institute, Ashburn, VA) at a concentration of 1 µM for 1 h at 37°C. After labeling, cells were rinsed three times with fresh growth media and then incubated in growth media for 1 h at 37°C before imaging. Live-cell imaging was perfomed at 37°C and with 5% CO2 using a stage incubator and objective heater. Cells were imaged in Live Cell Imaging Solution (Thermo Fisher Scientific). Images were acquired using a Leica SP8 STED 3X equipped with a SuperK Extreme EXW-12 (NKT Photonics) pulsed white-light laser as an excitation source and a Onefive Katana-08HP pulsed laser as a depletion light source (775 nm wavelength). Images were acquired using a HC PL APO 100×/1.40 NA oil CS2 objective, with a pixel size of 22.73 nm (1,024 × 1,024 pixels), a line scan speed of 8,000 Hz, and a 16-line average. JF646 was imaged with 633-nm excitation and 775-nm depletion wavelengths, and fluorescence emission light was collected with a HyD hybrid detector between 650 and 750 nm. The detection gate was set to 1.53–6 ns during STED imaging. The pinhole size was set to 1 Airy unit. Images were acquired with a frame rate of 2.63 s. The time-lapse acquired were subsequently smoothed with a 0.5-pixel sigma Gaussian blur, and minimum and maximum brightness values were adjusted linearly for visualization using ImageJ and bleach-corrected using Huygens Essential X11 software.
C. elegans strains
C. elegans strains used in this study were cultured at 20°C on nematode growth medium (NGM) plates seeded with the Escherichia coli OP50 using standard methods. The N2 is the standard strain and all the mutants were isolated from N2. Worm strains used in this study are listed in Table S1.
C. elegans constructs and transgenes
PCR and molecular cloning techniques were used to construct plasmids. Plasmids used for generating transgenic worms in this study are listed in Table S2. Transgenic worms were generated by injection using standard methods. Podr-1::GFP and Podr-1::RFP were used as the coinjection marker.
C. elegans fluorescent imaging
Young adult animals were mounted to a drop of M9 containing 1 mg/ml levamisole on a 3% agar pad. The confocal images were taken using micro-Manger and processed by ImageJ. 3D structured illumination microscopy (SIM) images of worms were acquired on the DeltaVision OMX V3 imaging system (GE Healthcare) with a 100×/1.40 NA oil objective (UPlanSApo; Olympus), solid-state multimode lasers (488 and 561 nm), and electron-multiplying charge-coupled device cameras (Evolve 256×256; Photometrics). Serial Z-stack sectioning was done at 125-nm intervals for SIM mode. To obtain optimal images, immersion oils with refractive indices of 1.520 were used for worms on glass coverslips. The microscope is routinely calibrated with 100-nm fluorescent spheres to calculate both the lateral and axial limits of image resolution. SIM image stacks were reconstructed using softWoRx 6.1.1 (GE Healthcare) with the following settings: pixel size, 39.5 nm; channel-specific optical transfer functions; Wiener filter constant, 0.0010; discard negative intensities background; drift correction with respect to first angle; and custom K0 guess angles for camera positions. The reconstructed images were further processed for maximum-intensity projections with softWoRx 6.1.1.Pixel registration was corrected to be less than 1 pixel for all channels using 100-nm Tetraspeck beads.
Quantification of C. elegans fluorescent images
For Fig. 6, B and C, the ER branches were counted from the whole animal with ZEISS compound fluorescent microscope. The worm is divided to three parts (Fig. 6 A), and the ER branches are divided to three types (I, L, and T) according to its shape.
For Fig. 6, D and E, the pictures of ER morphology of PVD cell body region were taken using SIM. The clearest slice of the cell nucleus was used to quantify tubular ER. A red circle of 4.6 µm in diameter is arbitrarily drawn around the nucleus, which was used to quantify the number of tubular ERs crossing with this circle. All worms were counted at the young adult stage.
Cloning, overexpression, and purification of INPP5K, and INPP5K activity assay
Coding sequences of mouse INPP5K was cloned into a modified pCMV6-AN-His vector (OriGene), which has a GFP tag inserted just after His tag. Active site mutation D210A was constructed by site directed mutagenesis. The plasmid was transfected into Expi293 cells (Thermo Fisher Scientific) for protein expression. After three days of transfection, cells were collected and lysed by three cycles of freeze and thaw (liquid N2 and 37°C water bath). His-tagged GFP-INPP5K was first purified by His60 Nickel Resin (Clontech) and then further purified by gel filtration on a Superdex200 column (GE Healthcare). The gel filtration buffer contained 20 mM Tris-HCl, pH 8.0, 300 mM NaCl, and 0.5 mM TCEP.
One-point activity assay was done by incubating 50 nM INPP5K WT or D210A mutant with 40 µg/ml diC8 PtdInsPs at 37°C for 1 h. The reaction buffer contained 50 mM Hepes, pH 7.4, 100 mM KCl, 2 mM MgCl2, and 1 mM EGTA. The time-course activity assay mixture contained 50 nM INPP5K and 40 µM diC8 PI(4,5)P2 or diC8 PI(3,4,5)P3. The reaction was incubated at room temperature for 20 min. Phosphate release was measured by absorbance at OD620 after addition of malachite green reagent (Maehama et al., 2000).
Online supplemental material
Fig. S1 shows the recruitment of INPP5K to the nucleus by MAD2L1BP and the lack of ER localization of INPP5J. Fig. S2 shows the domain organization of ARL6IP1 and the characterization of the ARL6IP1–INPP5K interaction. Fig. S3 demonstrates the quantitative analysis of the motility of EGFP-ARL6IP1 and mCh-VAPB in the ER. Fig. S4 shows the phosphatase activity of INPP5K by in vitro assay and the lack of an obvious effect of its knockdown on PI(4,5)P2 distribution. Fig. S5 shows the domain organization and phylogenetics of CIL-1 and the endogenous expression of CIL-1 in PVD neurons. Video 1 shows live-cell confocal imaging of a COS-7 cell coexpressing EGFP-ARL6IP1 and mCh-VAPB, highlighting the motility of ARL6IP1 relative to VAPB in the ER. Video 2 shows live-cell nanoscopy imaging of Snap-ARL6IP1 in elongating tubules that grow along preexisting ER tubules. Video 3 shows live-cell nanoscopy imaging of Halo-ARL6IP1 enriched in ER tubules that explore new territory at the periphery of the cell. Table S1 lists C. elegans strains used in this study. Table S2 lists the plasmids used in this study to generate transgenic worms.
Supplementary Material
Acknowledgments
We thank Karin Reinisch for discussion. We would also like to thank Joerg Bewersdorf for assistance with STED microscopy and the Center for Biological Imaging at the Institute of Biophysics, Chinese Academy of Science for assistance with SIM work.
This work was supported in part by the National Institutes of Health (grants NS036251, DK45735, and DK082700 to P. De Camilli and grant NS082208 to K. Shen), the Kavli Foundation (P. De Camilli), the Howard Hughes Medical Institute (P. De Camilli and K. Shen), a China Scholarship Council–Yale World Scholars Program scholarship (R. Dong), a BrightFocus Foundation postdoctoral fellowship (S. Gowrishankar), a grant from the National Natural Science Foundation of China (grant 31571061 to X. Wang), the Chinese Academy of Sciences/State Administration of Foreign Experts Affairs International Partnership Program for Creative Research Teams (K. Shen), and Youth Innovation Promotion Association of the Chinese Academy of Sciences funding (X. Wang).
The authors declare no competing financial interests.
Author contributions: Conceptualization, R. Dong, P. De Camilli, and K. Shen. Methodology, R. Dong, P. De Camilli, and K. Shen. Validation, R. Dong, P. De Camilli, K. Shen, and X. Wang. Formal analysis (application of statistical, mathematical, computational, or other techniques to analyze or synthesize data) R. Dong and X. Wang. Investigation (conducting the research and investigation process, specifically performing the experiments or data collection), R. Dong, L. Benedetti, S. Gowrishankar, T. Zhu, and H. Deng. Writing – Original Draft, R. Dong, P. De Camilli, and K. Shen. Writing – Review & Editing, R. Dong, P. De Camilli, and K. Shen. Visualization, R. Dong and X. Wang. Supervision, P. De Camilli and K. Shen. Project Administration, P. De Camilli and K. Shen). Funding Acquisition: P. De Camilli and K. Shen.
References
- Bae Y.-K., Kim E., L’hernault S.W., and Barr M.M.. 2009. The CIL-1 PI 5-phosphatase localizes TRP Polycystins to cilia and activates sperm in C. elegans. Curr. Biol. 19:1599–1607. 10.1016/j.cub.2009.08.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T. 2013. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93:1019–1137. 10.1152/physrev.00028.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T., and Várnai P.. 2009. Visualization of cellular phosphoinositide pools with GFP-fused protein-domains. Curr. Protoc. Cell Biol. Chapter 24:Unit 24.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackstone C., O’Kane C.J., and Reid E.. 2011. Hereditary spastic paraplegias: membrane traffic and the motor pathway. Nat. Rev. Neurosci. 12:31–42. 10.1038/nrn2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady J.P., Claridge J.K., Smith P.G., and Schnell J.R.. 2015. A conserved amphipathic helix is required for membrane tubule formation by Yop1p. Proc. Natl. Acad. Sci. USA. 112:E639–E648. 10.1073/pnas.1415882112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit S.E., Dyson J.M., and Mitchell C.A.. 2012. Inositol polyphosphate 5-phosphatases; new players in the regulation of cilia and ciliopathies. FEBS Lett. 586:2846–2857. 10.1016/j.febslet.2012.07.037 [DOI] [PubMed] [Google Scholar]
- Di Paolo G., and De Camilli P.. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature. 443:651–657. 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- Drin G., and Antonny B.. 2010. Amphipathic helices and membrane curvature. FEBS Lett. 584:1840–1847. 10.1016/j.febslet.2009.10.022 [DOI] [PubMed] [Google Scholar]
- Feng S., Sekine S., Pessino V., Li H., Leonetti M.D., and Huang B.. 2017. Improved split fluorescent proteins for endogenous protein labeling. Nat. Commun. 8:370 10.1038/s41467-017-00494-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink K.L., López-Giráldez F., Kim I.-J., Strittmatter S.M., and Cafferty W.B.J.. 2017. Identification of Intrinsic Axon Growth Modulators for Intact CNS Neurons after Injury. Cell Reports. 18:2687–2701. 10.1016/j.celrep.2017.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler P.C., and O’Sullivan N.C.. 2016. ER-shaping proteins are required for ER and mitochondrial network organization in motor neurons. Hum. Mol. Genet. 25:2827–2837. 10.1093/hmg/ddw139 [DOI] [PubMed] [Google Scholar]
- Friedman J.R., Webster B.M., Mastronarde D.N., Verhey K.J., and Voeltz G.K.. 2010. ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J. Cell Biol. 190:363–375. 10.1083/jcb.200911024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar S., Wu Y., and Ferguson S.M.. 2017. Impaired JIP3-dependent axonal lysosome transport promotes amyloid plaque pathology. J. Cell Biol. 216:3291–3305. 10.1083/jcb.201612148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung R., Tan A., Ooms L.M., McGrath M.J., Huysmans R.D., Munday A.D., Prescott M., Whisstock J.C., and Mitchell C.A.. 2003. Identification of a novel domain in two mammalian inositol-polyphosphate 5-phosphatases that mediates membrane ruffle localization. The inositol 5-phosphatase skip localizes to the endoplasmic reticulum and translocates to membrane ruffles following epidermal growth factor stimulation. J. Biol. Chem. 278:11376–11385. 10.1074/jbc.M209991200 [DOI] [PubMed] [Google Scholar]
- Habu T., Kim S.H., Weinstein J., and Matsumoto T.. 2002. Identification of a MAD2-binding protein, CMT2, and its role in mitosis. EMBO J. 21:6419–6428. 10.1093/emboj/cdf659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim S., Bertucci M.C., Conduit S.E., Vuong D.L., and Mitchell C.A.. 2012. Inositol polyphosphate phosphatases in human disease. Curr. Top. Microbiol. Immunol. 362:247–314. 10.1007/978-94-007-5025-8_12 [DOI] [PubMed] [Google Scholar]
- Hammond G.R.V., and Balla T.. 2015. Polyphosphoinositide binding domains: Key to inositol lipid biology. Biochim. Biophys. Acta. 1851:746–758. 10.1016/j.bbalip.2015.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Shibata Y., Voss C., Shemesh T., Li Z., Coughlin M., Kozlov M.M., Rapoport T.A., and Prinz W.A.. 2008. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 319:1247–1250. 10.1126/science.1153634 [DOI] [PubMed] [Google Scholar]
- Ijuin T., and Takenawa T.. 2003. SKIP negatively regulates insulin-induced GLUT4 translocation and membrane ruffle formation. Mol. Cell. Biol. 23:1209–1220. 10.1128/MCB.23.4.1209-1220.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijuin T., Mochizuki Y., Fukami K., Funaki M., Asano T., and Takenawa T.. 2000. Identification and characterization of a novel inositol polyphosphate 5-phosphatase. J. Biol. Chem. 275:10870–10875. 10.1074/jbc.275.15.10870 [DOI] [PubMed] [Google Scholar]
- Ijuin T., Yu Y.E., Mizutani K., Pao A., Tateya S., Tamori Y., Bradley A., and Takenawa T.. 2008. Increased insulin action in SKIP heterozygous knockout mice. Mol. Cell. Biol. 28:5184–5195. 10.1128/MCB.01990-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T.Y., Bian X., Sun S., Hu X., Klemm R.W., Prinz W.A., Rapoport T.A., and Hu J.. 2012. Lipid interaction of the C terminus and association of the transmembrane segments facilitate atlastin-mediated homotypic endoplasmic reticulum fusion. Proc. Natl. Acad. Sci. USA. 109:E2146–E2154. 10.1073/pnas.1208385109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehama T., Taylor G.S., Slama J.T., and Dixon J.E.. 2000. A sensitive assay for phosphoinositide phosphatases. Anal. Biochem. 279:248–250. 10.1006/abio.2000.4497 [DOI] [PubMed] [Google Scholar]
- Mesmin B., Bigay J., Moser von Filseck J., Lacas-Gervais S., Drin G., and Antonny B.. 2013. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell. 155:830–843. 10.1016/j.cell.2013.09.056 [DOI] [PubMed] [Google Scholar]
- Mignery G.A., Südhof T.C., Takei K., and De Camilli P.. 1989. Putative receptor for inositol 1,4,5-trisphosphate similar to ryanodine receptor. Nature. 342:192–195. 10.1038/342192a0 [DOI] [PubMed] [Google Scholar]
- Nemoto Y., Kearns B.G., Wenk M.R., Chen H., Mori K., Alb J.G. Jr., De Camilli P., and Bankaitis V.A.. 2000. Functional characterization of a mammalian Sac1 and mutants exhibiting substrate-specific defects in phosphoinositide phosphatase activity. J. Biol. Chem. 275:34293–34305. 10.1074/jbc.M003923200 [DOI] [PubMed] [Google Scholar]
- Nixon-Abell J., Obara C.J., Weigel A.V., Li D., Legant W.R., Xu C.S., Pasolli H.A., Harvey K., Hess H.F., Betzig E., et al. . 2016. Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER. Science. 354:aaf3928 10.1126/science.aaf3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizon M., Küry S., Péréon Y., Besnard T., Quinquis D., Boisseau P., Marsaud T., Magot A., Mussini J.-M., Mayrargue E., et al. . 2018. ARL6IP1 mutation causes congenital insensitivity to pain, acromutilation and spastic paraplegia. Clin. Genet. 93:169–172. 10.1111/cge.13048 [DOI] [PubMed] [Google Scholar]
- Novarino G., Fenstermaker A.G., Zaki M.S., Hofree M., Silhavy J.L., Heiberg A.D., Abdellateef M., Rosti B., Scott E., Mansour L., et al. . 2014. Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science. 343:506–511. 10.1126/science.1247363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn D.P.S., Pond H.L., Mazaheri N., Dejardin J., Munn C.J., Mushref K., Cauley E.S., Moroni I., Pasanisi M.B., Sellars E.A., et al. . 2017. Mutations in INPP5K Cause a Form of Congenital Muscular Dystrophy Overlapping Marinesco-Sjögren Syndrome and Dystroglycanopathy. Am. J. Hum. Genet. 100:537–545. 10.1016/j.ajhg.2017.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirruccello M., and De Camilli P.. 2012. Inositol 5-phosphatases: insights from the Lowe syndrome protein OCRL. Trends Biochem. Sci. 37:134–143. 10.1016/j.tibs.2012.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls M.M., Hall D.H., Victor M., Stelzer E.H.K., and Rapoport T.A.. 2002. Targeting of rough endoplasmic reticulum membrane proteins and ribosomes in invertebrate neurons. Mol. Biol. Cell. 13:1778–1791. 10.1091/mbc.01-10-0514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual J.F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G.F., Gibbons F.D., Dreze M., Ayivi-Guedehoussou N., et al. . 2005. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 437:1173–1178. 10.1038/nature04209 [DOI] [PubMed] [Google Scholar]
- Schmid A.C., Wise H.M., Mitchell C.A., Nussbaum R., and Woscholski R.. 2004. Type II phosphoinositide 5-phosphatases have unique sensitivities towards fatty acid composition and head group phosphorylation. FEBS Lett. 576:9–13. 10.1016/j.febslet.2004.08.052 [DOI] [PubMed] [Google Scholar]
- Shibata Y., Voss C., Rist J.M., Hu J., Rapoport T.A., Prinz W.A., and Voeltz G.K.. 2008. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J. Biol. Chem. 283:18892–18904. 10.1074/jbc.M800986200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz G.K., Prinz W.A., Shibata Y., Rist J.M., and Rapoport T.A.. 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 124:573–586. 10.1016/j.cell.2005.11.047 [DOI] [PubMed] [Google Scholar]
- Waterman-Storer C.M., and Salmon E.D.. 1998. Endoplasmic reticulum membrane tubules are distributed by microtubules in living cells using three distinct mechanisms. Curr. Biol. 8:798–806. 10.1016/S0960-9822(98)70321-5 [DOI] [PubMed] [Google Scholar]
- Wiessner M., Roos A., Munn C.J., Viswanathan R., Whyte T., Cox D., Schoser B., Sewry C., Roper H., Phadke R., et al. . 2017. Mutations in INPP5K, Encoding a Phosphoinositide 5-Phosphatase, Cause Congenital Muscular Dystrophy with Cataracts and Mild Cognitive Impairment. Am. J. Hum. Genet. 100:523–536. 10.1016/j.ajhg.2017.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiradjaja F., Ooms L.M., Whisstock J.C., McColl B., Helfenbaum L., Sambrook J.F., Gething M.J., and Mitchell C.A.. 2001. The yeast inositol polyphosphate 5-phosphatase Inp54p localizes to the endoplasmic reticulum via a C-terminal hydrophobic anchoring tail: regulation of secretion from the endoplasmic reticulum. J. Biol. Chem. 276:7643–7653. 10.1074/jbc.M010471200 [DOI] [PubMed] [Google Scholar]
- Wiradjaja F., Ooms L.M., Tahirovic S., Kuhne E., Devenish R.J., Munn A.L., Piper R.C., Mayinger P., and Mitchell C.A.. 2007. Inactivation of the phosphoinositide phosphatases Sac1p and Inp54p leads to accumulation of phosphatidylinositol 4,5-bisphosphate on vacuole membranes and vacuolar fusion defects. J. Biol. Chem. 282:16295–16307. 10.1074/jbc.M701038200 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., Yoshida A., Miyazaki N., Iwasaki K., and Sakisaka T.. 2014. Arl6IP1 has the ability to shape the mammalian ER membrane in a reticulon-like fashion. Biochem. J. 458:69–79. 10.1042/BJ20131186 [DOI] [PubMed] [Google Scholar]
- Zewe J.P., Wills R.C., Sangappa S., Goulden B.D., and Hammond G.R.V.. 2018. SAC1 degrades its lipid substrate PtdIns4P in the endoplasmic reticulum to maintain a steep chemical gradient with donor membranes. eLife. 7:e35588 10.7554/eLife.35588 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.