Targeting of Vps13 to membranes is highly dynamic. Bean et al. identify Ypt35 and Mcp1 as adaptors for Vps13 at endosomes and mitochondria, respectively, and show all known Vps13 adaptors use a related motif to compete for Vps13 membrane recruitment.
Abstract
The regulated expansion of membrane contact sites, which mediate the nonvesicular exchange of lipids between organelles, requires the recruitment of additional contact site proteins. Yeast Vps13 dynamically localizes to membrane contacts that connect the ER, mitochondria, endosomes, and vacuoles and is recruited to the prospore membrane in meiosis, but its targeting mechanism is unclear. In this study, we identify the sorting nexin Ypt35 as a novel adaptor that recruits Vps13 to endosomal and vacuolar membranes. We characterize an interaction motif in the Ypt35 N terminus and identify related motifs in the prospore membrane adaptor Spo71 and the mitochondrial membrane protein Mcp1. We find that Mcp1 is a mitochondrial adaptor for Vps13, and the Vps13–Mcp1 interaction, but not Ypt35, is required when ER-mitochondria contacts are lost. All three adaptors compete for binding to a conserved six-repeat region of Vps13 implicated in human disease. Our results support a competition-based model for regulating Vps13 localization at cellular membranes.
Introduction
Membrane contact sites, regions where two organelles come within close proximity without fusing, are critical regions of lipid and ion transport, lipid biosynthesis, and interorganellar communication (Helle et al., 2013; Eisenberg-Bord et al., 2016; Phillips and Voeltz, 2016). These sites are formed by protein tethers that span the 10–30-nm distance between opposing membranes and link them through transmembrane anchors, lipid-binding domains, and/or protein interactions (Eisenberg-Bord et al., 2016; Gatta and Levine, 2017). The extent of any given contact site is modulated in response to cellular cues and changing metabolic needs, and the relative size of different membrane contacts is tightly coordinated (Elbaz-Alon et al., 2014, 2015; Liu et al., 2016; Murley and Nunnari, 2016; Bian et al., 2018). The expansion or contraction of contact sites requires dynamic changes in the localization of tethers and lipid transfer proteins, but how these changes are coordinated is not well understood.
In Saccharomyces cerevisiae, protein tethers have been identified that create membrane contact sites connecting the ER, mitochondria, and vacuole. The ER–mitochondria encounter structure (ERMES) complex tethers the ER to mitochondria (Kornmann et al., 2009), Vps39 drives formation of the vacuole–mitochondria patch (vCLAMP; Elbaz-Alon et al., 2014; Hönscher et al., 2014), and Nvj1 and Vac8 link the ER to the vacuole to form the nucleus–vacuole junction (NVJ; Pan et al., 2000). These contact sites may functionally overlap as loss of mitochondrial contact sites caused by defects in the ERMES complex causes expansion of vCLAMP contacts, which may provide an alternative pathway for transporting lipids to mitochondria (Elbaz-Alon et al., 2014; Hönscher et al., 2014). Identifying the constituent proteins that regulate contact site assembly and function and understanding how their localization is controlled remain major challenges in the field.
Vps13 is a highly conserved 358-kD protein that is present at multiple cellular locations including vCLAMP and NVJ membrane contact sites, endosomes, and the prospore membrane (Huh et al., 2003; Park et al., 2013, 2016; Lang et al., 2015). Although Vps13 in yeast and other organisms has been implicated in processes including vesicle fusion, autophagy, and actin regulation (Brickner and Fuller, 1997; Föller et al., 2012; Muñoz-Braceras et al., 2015; Dalton et al., 2017; De et al., 2017; Rzepnikowska et al., 2017), its roles at membranes, including membrane contact sites, are not known. Humans contain four VPS13 proteins (VPS13A–D) that when nonfunctional cause different diseases—Chorea-acanthocytosis, Cohen syndrome, Parkinson’s disease, and spastic ataxia, respectively—suggesting they have functionally diverged (Rampoldi et al., 2001; Kolehmainen et al., 2003; Drin et al., 2007; Lesage et al., 2016; Seong et al., 2018). Although mammalian VPS13 proteins have not yet been found at contact sites, dysfunction of membrane contact sites is implicated in neurological and movement disorders (van der Kant and Neefjes, 2014; Paillusson et al., 2016).
The location of Vps13 at contact sites is regulated by nutrient availability. During mitotic growth when glucose is abundant, Vps13 is found primarily at endosomes and is frequently observed near endosome–mitochondria contacts but relocalizes to the NVJ when glucose is limiting (Lang et al., 2015; Park et al., 2016). Vps13 is also reported to localize to vCLAMPs, and this is essential for cell viability when ERMES is disrupted (Lang et al., 2015). Spontaneously occurring dominant mutations in Vps13 suppress ERMES defects and may favor relocalization to vCLAMPs (Lang et al., 2015; Park et al., 2016). This discovery highlights the critical role of Vps13 at membrane contact sites and suggests that uncovering the mechanisms that control Vps13 localization may be key to understanding coordination between contact sites.
Vps13 binds phosphatidic acid and phosphoinositides in vitro (De et al., 2017; Rzepnikowska et al., 2017), but it is unclear whether lipid-binding domains are sufficient for targeting to membranes in vivo. Instead, Vps13 may rely on interactions with membrane-specific adaptor proteins for recruitment. The only known adaptor is the meiosis-specific protein Spo71, which recruits Vps13 to the prospore membrane, where it facilitates prospore membrane expansion and closure (Park and Neiman, 2012; Park et al., 2013). If adaptors regulate Vps13 localization, additional adaptors may exist for recruiting Vps13 to other membranes, including the endosome, vacuole, mitochondria, and/or ER.
In this study, we identify the uncharacterized sorting nexin Ypt35 as the adaptor for Vps13 at endosome and vacuole membranes. We characterize a novel motif on Ypt35 that binds Vps13, and by searching for other proteins that contain a similar sequence, we identify related motifs in the meiosis-specific adaptor Spo71 and the mitochondrial membrane protein Mcp1. Our findings suggest that all three adaptors compete for binding to a highly conserved six-repeat domain in Vps13 to regulate the localization of Vps13 to different membranes.
Results
The sorting nexin Ypt35 recruits Vps13 to endosomes
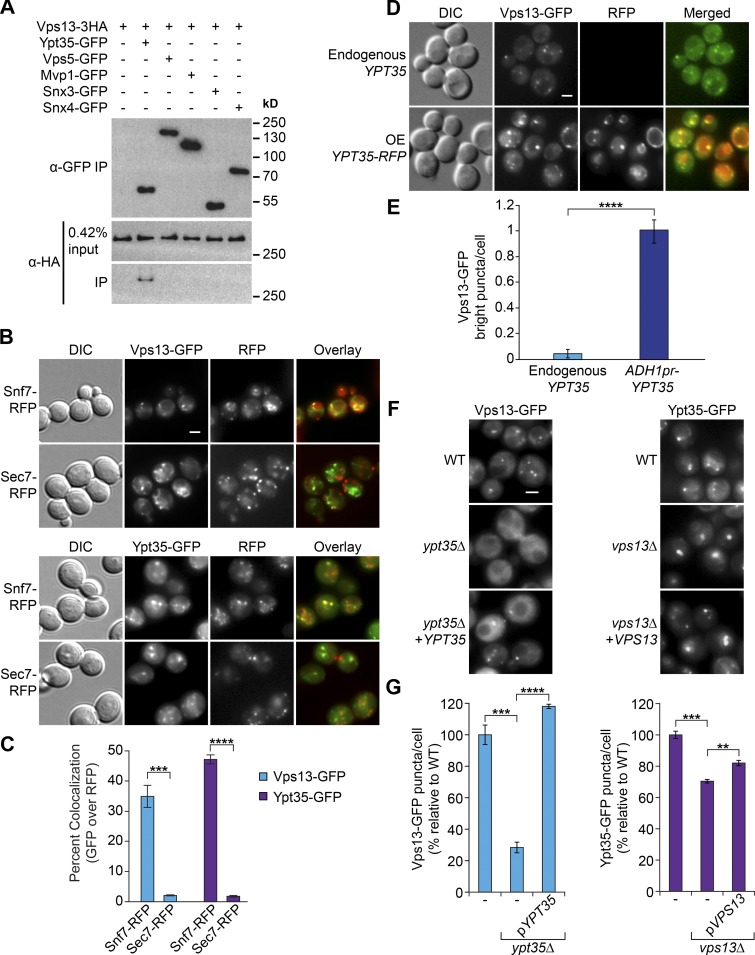
To identify potential Vps13 recruitment factors, we searched available large-scale datasets. An interaction between Vps13 and the endosome-localized PX domain–containing sorting nexin Ypt35 (Huh et al., 2003; Teasdale and Collins, 2012) was identified in a high-throughput proteomics study (Ho et al., 2002), and we found both Ypt35 and Vps13 in a genome-wide endosome recycling screen (Burston et al., 2009; Dalton et al., 2017). Indeed, Vps13-3HA copurified with Ypt35-GFP immunoprecipitated from cells treated with the cross-linker dithiobis(succinimidyl propionate) (DSP; Fig. 1 A). Although only a fraction of Vps13-3HA copurified with Ypt35-GFP, the interaction was specific because no Vps13-3HA was observed after immunoprecipitation of other endosomal sorting nexins (Vps5, Mvp1, Snx3, or Snx4).
Figure 1.
The sorting nexin Ypt35 recruits Vps13 to endosomes. (A) GFP-tagged sorting nexins were immunopurified from lysates cross-linked with 400 µg/ml DSP, and coprecipitating Vps13-HA was detected. All proteins were expressed at endogenous levels. IP, immunoprecipitation. (B) Endogenous Vps13-GFP (top) and Ypt35-GFP (bottom) colocalize with Snf7-RFP–labeled endosomes but not Sec7-RFP labeled Golgi. (C) Quantitation of colocalization. Two-tailed equal-variance t test; n = 3, cells/strain/replicate ≥ 674; ***, P < 0.001; ****, P < 0.0001. (D) Overexpression (OE) of YPT35-RFP from the ADH1 promoter recruits Vps13-GFP to bright colocalizing puncta. DIC, differential interference contrast. (E) Quantitation of bright Vps13-GFP puncta in cells with endogenous or overexpressed YPT35. Two-tailed unequal variance t test; n = 3, cells/strain/replicate ≥ 355; ****, P < 0.0001. (F) Vps13-GFP requires YPT35 for its punctate localization. (G) Quantitation of Vps13-GFP and Ypt35-GFP puncta per cell. Two-tailed equal variance t test; n = 3, cells/strain/replicate ≥ 604; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Bars, 2 µm. Error bars indicate SEM.
Vps13 is reported to act at both the Golgi and endosomes (De et al., 2017). Automated image analysis indicated that the colocalization of Vps13-GFP and Ypt35-GFP with the endosomal marker Snf7-RFP (34.9% and 47.2%, respectively, based on puncta overlap area) was significantly higher than with the late Golgi marker Sec7-RFP (2.1% and 1.8%, respectively; Fig. 1, B and C), suggesting both proteins have substantial endosomal pools at a steady state as reported by others (Huh et al., 2003; Park et al., 2016). We were unable to visualize C-terminally RFP-tagged Ypt35 expressed at endogenous levels. When overexpressed from an ADH1 promoter, Ypt35-RFP localized to the vacuolar rim and to endosomes, where it colocalized with Vps13-GFP (Fig. 1 D). Notably, the intensity of Vps13-GFP puncta was strongly enhanced in these cells (Fig. 1, D and E), suggesting overexpression of Ypt35 drives recruitment of Vps13 to endosomes.
We next tested whether Ypt35 is required for recruiting Vps13 to endosomes. Deletion of YPT35 strongly reduced the number of clearly defined Vps13-GFP puncta to near-background levels (P < 0.001), and these puncta were restored by introducing a plasmid encoding YPT35 (P < 0.0001; Fig. 1, F and G). In contrast, loss of VPS13 did not cause an obvious change in Ypt35-GFP localization (Fig. 1 F), although a slight decrease in the number of Ypt35-GFP puncta and a corresponding increase in average puncta area were detected by automated analysis (P < 0.01; Fig. S1 A), suggesting some consolidation of Ypt35-GFP puncta in the absence of Vps13.
Although the C-terminal GFP tag does not interfere with Vps13 function in prospore membrane formation or CPY sorting, it causes synthetic lethality with ERMES mutants and may perturb a putative C-terminal PH domain (Park and Neiman, 2012; Lang et al., 2015; Fidler et al., 2016; Park et al., 2016). We therefore tested whether the endosomal localization of Vps13 with an internal GFP tag after residue 499 (Vps13^GFP; Lang et al., 2015), which does not disrupt known Vps13 functions, was similarly dependent on Ypt35. Because Vps13^GFPpuncta are only slightly brighter than the background mitochondrial autofluorescence, we used automated image analysis to quantitate the overlap with the endosomal marker Vps17-RFP (Fig. S1, B and C). Deletion of YPT35 resulted in a strong decrease in the localization of Vps13^GFP to endosomes (39% of WT levels; P < 0.01), which was rescued by introducing a YPT35 plasmid (120% of WT; P < 0.0001).
Redistribution of Vps13^GFP to other organelles was not detected above the background mitochondrial fluorescence. Collectively, these results indicate that Ypt35 is an adaptor that localizes Vps13 to endosomal membranes.
Vps13 and Ypt35 are interdependent for recruitment to the NVJ
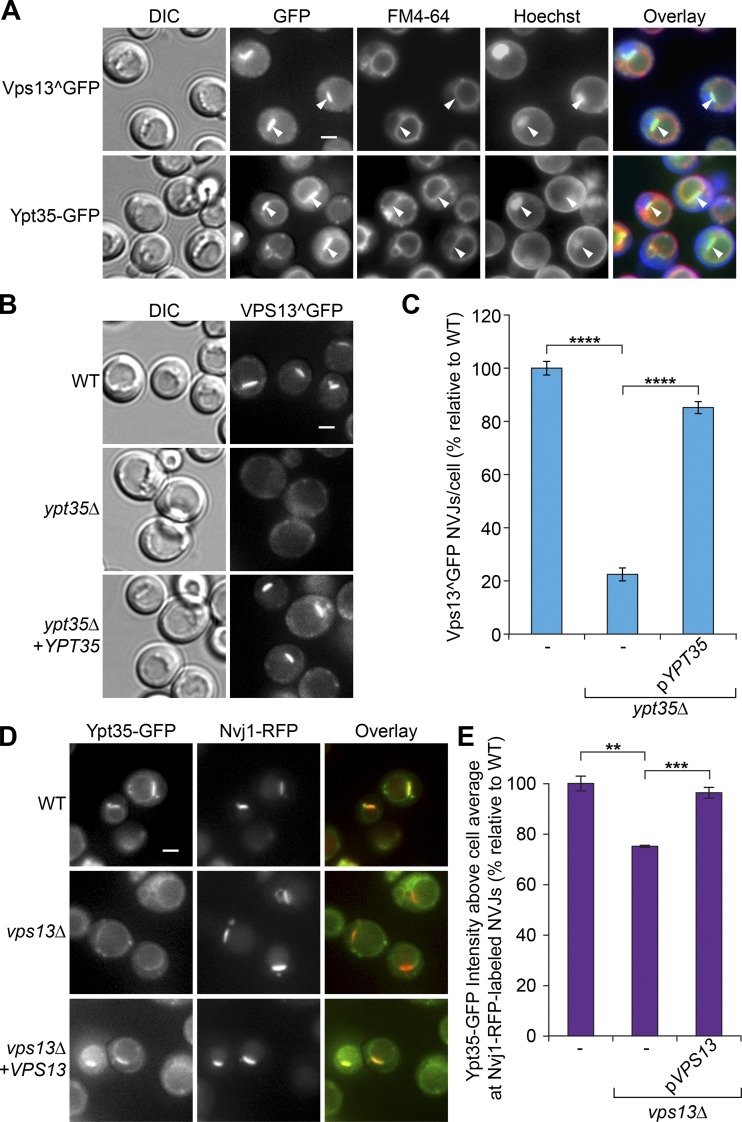
Upon glucose starvation, Vps13 shifts from endosomes to the NVJ (Lang et al., 2015). Given that Ypt35 binds Vps13 and can localize to the vacuolar rim, we tested whether Ypt35 is also present at NVJs after growth on a nonfermentable carbon source. Although low levels of Ypt35-GFP remained at the vacuolar rim, most Ypt35-GFP relocalized to NVJs in acetate-based media, indicating Ypt35 is a novel NVJ-associated protein (Fig. 2 A). Furthermore, Vps13^GFPand Ypt35-RFP colocalized at the NVJ (Fig. S1 D).
Figure 2.
Vps13 and Ypt35 are interdependent for recruitment to the NVJ. (A) Growth in acetate-based media shifts Vps13^GFP and Ypt35-GFP to the NVJ at the interface of the vacuole (FM4-64; red) and nucleus (Hoechst; blue). White arrowheads indicate NVJs. (B) Localization of Vps13^GFP to the NVJ in acetate-based media is Ypt35 dependent. (C) Quantitation of Vps13^GFP-labeled NVJs. Two-tailed equal variance t test; n = 3, cells/strain/replicate ≥ 1,149; ****, P < 0.0001. DIC, differential interference contrast. (D) Ypt35 is dependent on Vps13 for localization to the NVJ, labeled in this figure with Nvj1-RFP, in acetate-based media. (E) Quantitation of the average Ypt35-GFP intensity at Nvj1-RFP–marked NVJs relative to the cell background. Two-tailed equal variance t test; n = 3, cells/strain/replicate ≥ 1,672; **, P < 0.01; ***, P < 0.001. Bars, 2 µm. Error bars indicate SEM.
Deletion of YPT35 abolished Vps13 recruitment to the NVJ (P < 0.0001; Fig. 2, B and C), indicating that Ypt35 localizes Vps13 to the vacuole membrane. Strikingly, VPS13 deletion strongly reduced the NVJ enrichment of Ypt35-GFP, which instead was redistributed to the vacuolar rim and puncta (Fig. 2 D). Quantification of Ypt35-GFP intensity at regions of the vacuole membrane labeled with the NVJ marker Nvj1-RFP showed a significant reduction of NVJ enrichment in vps13 mutants (P < 0.01) that was restored by a VPS13 plasmid (P < 0.001; Fig. 2 E). These data suggest Ypt35 and Vps13 are interdependent for localization to NVJs and support a model in which Ypt35 recruits Vps13 to the vacuole membrane and is subsequently concentrated at the NVJ through the interaction of Vps13 with a component of the ER or NVJ.
Ypt35 contains a novel Vps13 recruitment motif
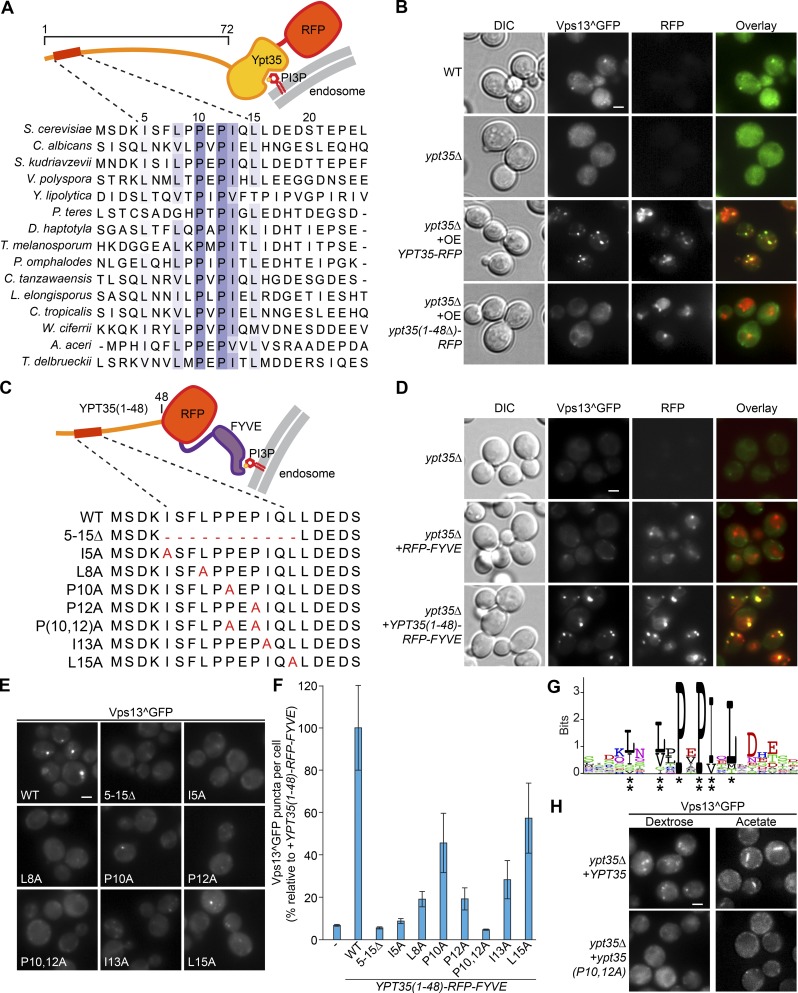
We next mapped the region of Ypt35 responsible for recruiting Vps13 to endosomal and vacuolar membranes. Ypt35 consists of a C-terminal phosphatidylinositol 3-phosphate binding PX domain (Vollert and Uetz, 2004) and an unstructured 72-residue N-terminal extension (Fig. 3 A). Alignment of Ypt35 with Saccharomycetales-level fungal homologues revealed two invariant prolines and several highly conserved hydrophobic amino acids between residues 5 and 20, roughly adhering to the consensus ΦxxΦxPxPΦxΦ, where Φ is a hydrophobic amino acid (Fig. 3 A). Expression of Ypt35 with a truncated N terminus, Ypt35(1–48Δ)-RFP, could not recruit Vps13^GFP to puncta, indicating that residues 1–48, containing the conserved region, are necessary for the Ypt35 adaptor function (Fig. 3 B).
Figure 3.
A motif in the N terminus of Ypt35 is necessary and sufficient for Vps13 recruitment. (A) An alignment of Ypt35 with fungal species identifies conserved residues in the N terminus. (B) Deletion of the N-terminal 48 residues of Ypt35 blocks Ypt35-RFP–induced recruitment of Vps13^GFP to puncta. (C) Schematic of the Ypt35(1–48)-RFP-FYVE construct used for Vps13^GFP localization experiments and mutant versions. (D) Ypt35(1–48)-RFP-FYVE recruits Vps13^GFP to puncta. DIC, differential interference contrast. (E) Vps13^GFP recruitment to puncta by Ypt35(1–48)-RFP-FYVE in a ypt35 strain was reduced by mutation of conserved N-terminal Ypt35 residues. (F) Quantitation of Vps13^GFPrecruitment to puncta in cells expressing WT and mutant forms of Ypt35(1–48)-RFP-FYVE. n = 3, cells/strain/replicate ≥ 913. (G) WebLogo (Crooks et al., 2004) of the Ypt35 PxP motif. Residues that when mutated to alanine caused a >40% loss of puncta (*) and >60% loss of puncta (**) are indicated. (H) Ypt35(P10,12A) is unable to recruit Vps13^GFP to puncta or NVJs in dextrose or acetate-based media, respectively. Bars, 2 µm. Error bars indicate SEM.
To test whether the Ypt35 N terminus is responsible for Vps13 recruitment, we fused this sequence to the PI3P-binding FYVE domain from EEA1 (Fig. 3 C). The resulting Ypt35(1–48)-RFP-FYVE chimera localized to puncta, similar to the RFP-FYVE control, but only the chimera induced formation of Vps13^GFP puncta (Fig. 3 D), indicating the first 48 residues of Ypt35 contain a Vps13 recruitment signal. Furthermore, fusing these residues to a tandem repeat of the plasma membrane–targeting PH domains from PLCδ (Stefan et al., 2002) recruited Vps13 to the plasma membrane, indicating the Ypt35(1–48) region is sufficient for Vps13 localization (Fig. S1, E and F). To map the signal, we mutated conserved residues in this region (Fig. 3 C). All mutant chimeras were stably expressed but were substantially less effective at inducing Vps13^GFP puncta (Fig. 3, E and F; and Fig. S1 G). Quantitation of Vps13^GFP puncta indicated the I5A, L8A, P12A, and I13A mutations had the largest effects as single mutants, whereas the P10,12A double mutant reduced Vps13^GFP puncta to background levels (Fig. 3, F and G). Mutation of these invariant prolines in the context of full-length Ypt35, creating Ypt35(P10,12A), blocked recruitment of Vps13^GFP to endosomal puncta in glucose media or to NVJs in acetate-based media, suggesting a loss of adaptor function (Fig. 3 H). Our results indicate that the conserved residues in the N terminus of Ypt35 form a Vps13 interaction motif, which we refer to as a PxP motif, that is necessary and sufficient for Vps13 recruitment.
The Vps13 DUF1162 domain binds the Ypt35 PxP motif
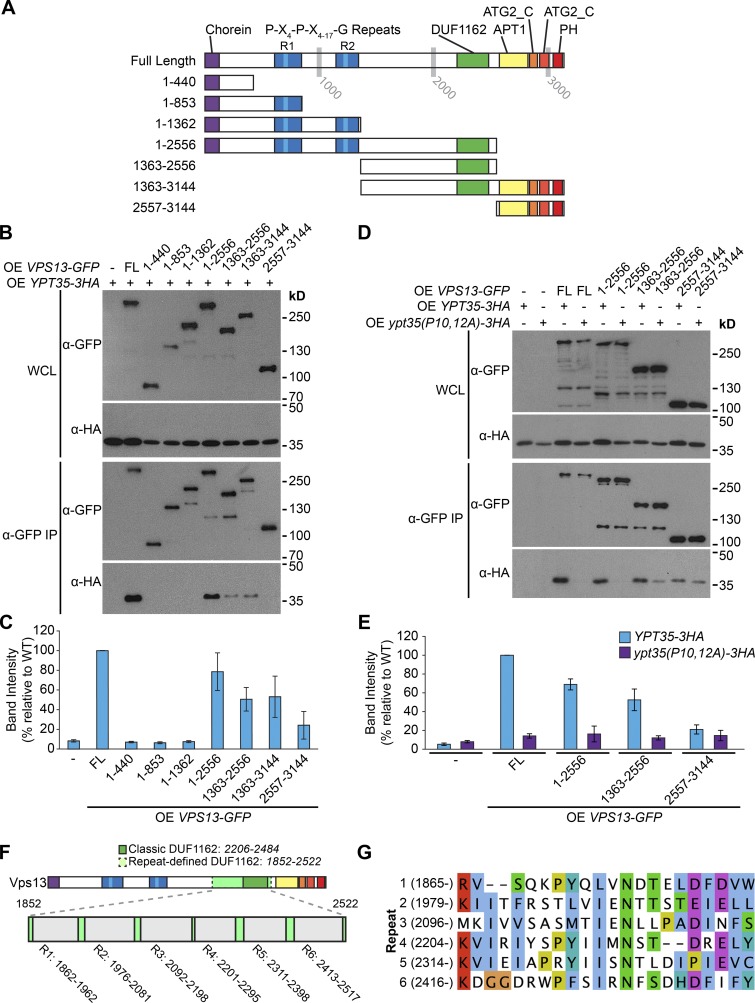
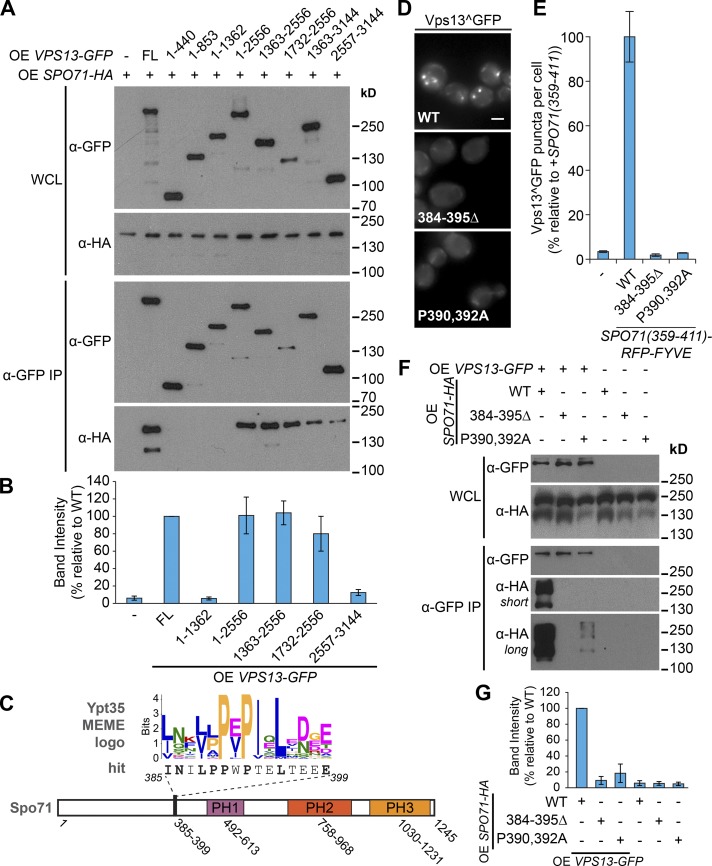
Vps13 contains several conserved domains. These include an N-terminal Chorein domain, a repeat region, a central domain of unknown function (DUF1162), an APT1 domain, two ATG2 C-terminal domains, and a putative lipid-binding pleckstrin homology (PH) domain (Velayos-Baeza et al., 2004; Fidler et al., 2016; Rzepnikowska et al., 2017). To identify the part of Vps13 that interacts with Ypt35, we created a series of Vps13-GFP truncations that separate these Vps13 domains (Fig. 4 A). To detect the interaction in the absence of cross-linkers, the Vps13 fragments and Ypt35 were overexpressed from the ADH1 promoter. All truncated proteins were expressed, but only truncations containing the DUF1162 domain or to a lesser extent the C terminus were able to copurify Ypt35-3HA (Fig. 4, B and C). Although the interaction with the C terminus was reproducible, it was relatively weak, and a fragment containing both the DUF1162 domain and the C terminus did not display enhanced binding. In contrast, interactions with DUF1162-containing fragments were more robust. A smaller DUF1162-containing fragment, Vps13(1,732–2,556), was unstable but able to copurify Ypt35-3HA (Fig. S2 A), suggesting the PxP motif binds Vps13, directly or indirectly, at or near the DUF1162 domain.
Figure 4.
A central region of Vps13 binds Ypt35 in a PxP motif–dependent manner. (A) Schematic of Vps13 truncations. (B) Coimmunoprecipitation of Ypt35-3HA with full-length and truncated forms of Vps13-GFP. Both proteins were overexpressed (OE) from the ADH1 promoter. (C) Densitometry of B; n = 3. (D) Coimmunoprecipitation of WT or mutant Ypt35-3HA with Vps13-GFP fragments indicates that interactions with DUF1162-containing regions are proline dependent. IP, immunoprecipitation; WCL, whole-cell lysate. (E) Densitometry of D; n ≥ 2. (F) Schematic of S. cerevisiae Vps13, highlighting an extended DUF1162 domain defined by six repeats identified by using HHrepID (Biegert and Söding, 2008). (G) An alignment of the most conserved section of the S. cerevisiae DUF1162 repeats. Error bars indicate SEM.
To test the importance of the PxP motif for these interactions, we tested binding of a subset of the Vps13 fragments to the Ypt35(P10,12A)-3HA mutant (Fig. 4, D and E). Full-length Vps13-GFP or DUF1162-containing Vps13-GFP truncations were dependent on the conserved prolines for interactions with Ypt35. In contrast, binding to the C-terminal fragment was largely PxP independent. These results strongly suggest that the Ypt35 PxP motif binds Vps13 either through the DUF1162 domain or an adjacent region.
The Vps13 DUF1162 domain is part of a six-repeat region
Ypt35 is the second DUF1162-interacting protein to be identified. Previously, yeast two-hybrid and split YFP experiments in Arabidopsis thaliana indicated that the Vps13 homologue SHRUBBY interacts with the SHORT-ROOT (SHR) transcription factor through the DUF1162 domain, now commonly referred to as a SHR binding domain (SHR_BD; Koizumi and Gallagher, 2013). Analysis of the smallest Ypt35-interacting Vps13 fragment (1,732–2,556) using HHrepID (Biegert and Söding, 2008) identified six high-probability repeats spanning residues 1,852–2,522, roughly twice the length of the classic DUF1162 domain (2,206–2,284; Fig. 4 F). The first ∼25 residues of each ∼100–amino acid repeat were the most highly conserved and contained an invariant asparagine residue (Figs. 4 G and S3 A).
The DUF1162 repeats are also present in human VPS13A/B/C/D genes (Fig. S3 B). Similar to yeast Vps13, the N-terminal region of each repeat is the most conserved and contains a nearly invariant asparagine residue (Fig. S3, C and D). All human VPS13 genes encode six repeats, but two VPS13B repeats do not contain the highly conserved asparagine residue. We suggest that this extended repeat structure represents a complete DUF1162 domain.
Spo71 binds the Vps13 DUF1162 domain through a PxP motif
Having determined that Ypt35 interacts with the DUF1162 domain-containing region of Vps13, we wondered whether Spo71, an established Vps13 prospore membrane adaptor, binds Vps13 in a similar manner. Using the same panel of overexpressed Vps13-GFP truncations (Fig. 4 A), we found that Spo71 also binds DUF1162 domain-containing fragments and, to a lesser extent, the C-terminal domain (Fig. 5, A and B). To identify candidate PxP-based binding motifs in Spo71, we generated a Multiple Expectation Maximization Algorithms for Motif Elucidation (MEME) motif (Bailey and Elkan, 1994) from Saccharomycetales-level Ypt35 homologues and used it in a Find Individual Motif Occurrences (FIMO; Grant et al., 2011) search of the S. cerevisiae proteome. Among the hits, only one was identified within Spo71 at residues 385–399 (Fig. 5 C). This hit matched the Ypt35 PxP motif consensus with the exception of a threonine in one hydrophobic position. Manual inspection of Spo71 revealed multiple proline-rich tracts and five additional sequences with fewer than four mismatches to the Ypt35 consensus (Fig. S1 H).
Figure 5.
The Vps13 DUF1162 domain recognizes the meiosis-specific adaptor Spo71 through a PxP motif. (A) Coimmunoprecipitation of TEF1pr-driven Spo71-HA with ADH1pr-driven Vps13-GFP fragments indicates that fragments containing the DUF1162 domain are sufficient for the interaction. (B) Densitometry of selected strains from A; n = 3. (C) A schematic of Spo71 highlighting a putative Vps13 interaction motif identified by a FIMO search by using a MEME motif generated from Ypt35 homologues. (D) Vps13^GFP puncta in ypt35 strains expressing the indicated WT or mutant Spo71(359–411)-RFP-FYVE constructs. (E) Quantitation of Vps13^GFP puncta per cell; n = 3, cells/strain/replicate ≥ 1,473. (F) ADH1pr-driven Vps13-GFP was immunopurified and coprecipitating WT or mutant TEF1pr-expressed Spo71-HA was detected. IP, immunoprecipitation; WCL, whole-cell lysate. (G) Densitometry of F; n = 3. Bars, 2 µm. Error bars indicate SEM.
To test whether any of these putative motifs could interact with Vps13, we made Spo71-RFP-FYVE chimeras using segments of Spo71 (Fig. S1 H). Of the constructs tested, only those containing the FIMO-predicted motif induced Vps13^GFP puncta (Fig. 5, C and D). Vps13^GFP recruitment by the Spo71(359–411) chimera was abolished by mutations that disrupt this motif (P390,392A; 384–395Δ), suggesting that the Spo71 PxP motif identified by the FIMO search is necessary and sufficient for Vps13 interaction (Fig. 5, D and E). The presence of a polar threonine at a conserved position in the motif indicates there is greater amino acid variability than our initial analysis suggested (Fig. 3, A, E, and F).
Although a chimera containing the Spo71 PxP motif recruited Vps13 to membranes, full-length Spo71 may contain redundant signals. We used coimmunoprecipitation to determine whether the motif at residues 385–399 is necessary for Vps13–Spo71 interaction. Although WT Spo71-HA efficiently copurified with Vps13^GFP, the interaction was severely disrupted by the P390,392A mutation and abolished by deletion of the motif (Fig. 5, F and G). A low level of binding to the Spo71(P390,392A) mutant indicates that other residues such as the conserved hydrophobic amino acids may be capable of residual binding in the absence of the invariant prolines. Our results suggest Spo71 contains a PxP motif that is necessary and sufficient for Vps13 binding, indicating that the mechanism for Vps13 binding is conserved between proteins.
Mcp1 is a mitochondrial PxP motif–containing Vps13 adaptor
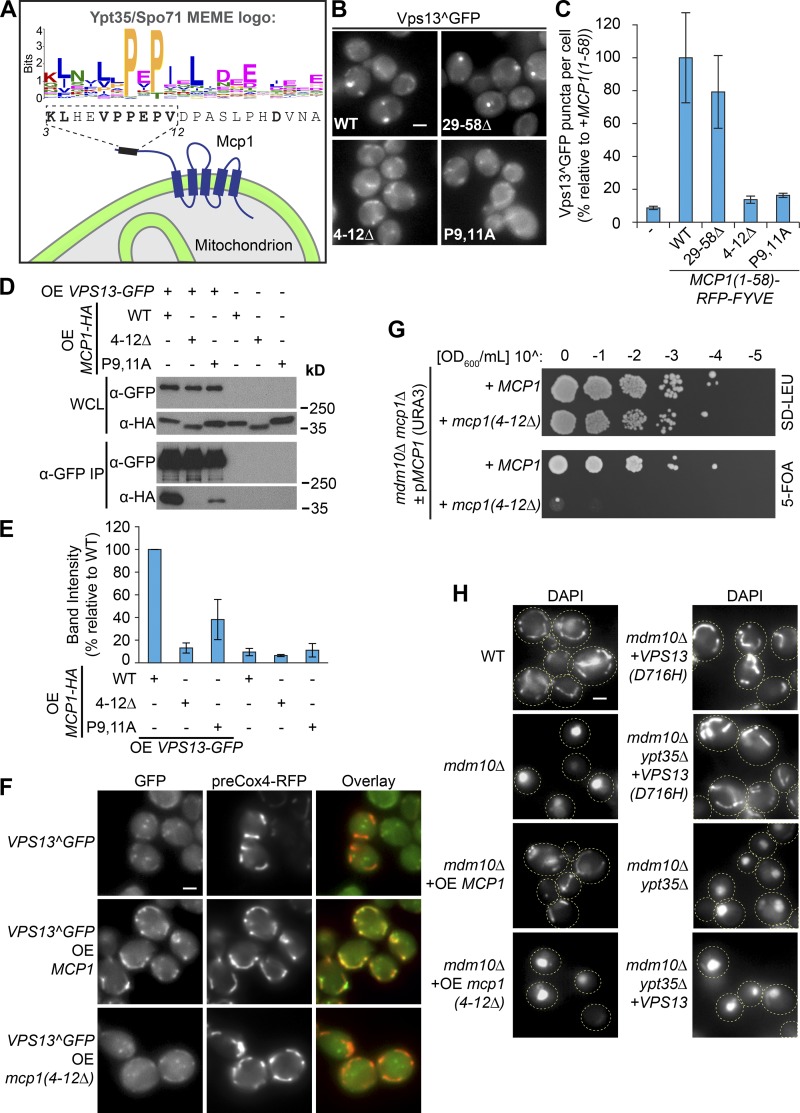
We reasoned that a search of the yeast proteome for sequences similar to the PxP motif might identify additional Vps13 adaptors. We pooled Saccharomycetales-level motif alignments from Ypt35 and Spo71 to generate a new MEME motif (Fig. 6 A). A FIMO search of the S. cerevisiae proteome using this motif identified 296 hits at a P < 10−4 threshold (Table S1), which included a putative PxP motif in the outer mitochondrial membrane protein Mcp1 (P < 3.7 × 10−5). Mcp1 was a strong adaptor candidate because dominant Vps13 alleles suppress ERMES mutants only when Mcp1 is present, and high levels of Mcp1 also suppress ERMES defects (Tan et al., 2013; Lang et al., 2015).
Figure 6.
Mcp1 is a mitochondrial PxP motif–containing Vps13 adaptor that interacts with Vps13 to suppress ERMES mutants. (A) Schematic of Mcp1 highlighting a putative PxP interaction motif identified by FIMO by using a MEME motif generated from Ypt35 and Spo71 homologues. (B) Mutation of the motif (4–12Δ, P9,11A) blocked the ability of the Mcp1(1–58)-RFP-FYVE chimera to induce Vps13^GFP puncta in a ypt35 strain. (C) Quantitation of Vps13^GFP puncta per cell; n = 3, cells/strain/replicate ≥ 1,410. (D) The motif is required for coprecipitation of Vps13 and Mcp1. Vps13-GFP and WT and mutant forms of Mcp1-HA were overexpressed (OE) from the ADH1 promoter. IP, immunoprecipitation; WCL, whole-cell lysate. (E) Densitometry of D; n = 3. (F) The interaction between ADH1pr-driven overexpression of WT Mcp1 but not Mcp1 lacking the PxP motif recruits Vps13^GFP to mitochondria marked by preCox4-RFP (red). (G) Loss of the Mcp1 PxP motif is synthetic lethal with mdm10Δ. (H) Overexpression of WT but not mutant Mcp1suppresses the mitochondrial morphology defect of a strain lacking the ERMES subunit Mdm10. In contrast, loss of YPT35 does not rescue the mdm10Δ mitochondrial morphology defect with or without expression of VPS13 from two copies of the gene, nor does it block rescue by the dominant suppressor Vps13(D716H). DAPI was used as a mitochondrial marker. Bars, 2 µm. Error bars indicate SEM.
The predicted PxP motif in Mcp1, K3LHEVPPEPV12, is conserved in other fungal species and resides in a predicted 58 residue N-terminal cytosolic domain that is followed by five predicted transmembrane helices (Fig. 6 A). To test whether this motif interacts with Vps13, we made a series of Mcp1-RFP-FYVE chimeras. Chimeras with fragments of the N-terminal domain containing the putative PxP motif localized to puncta and recruited Vps13^GFP, but chimeras in which the putative motif was deleted or mutated (4–12Δ; P9,11A) failed to recruit Vps13^GFP (Fig. 6, B and C). This demonstrates that the FIMO-predicted motif is sufficient for Vps13^GFP recruitment.
We next tested whether we could observe a physical interaction between Vps13 and full-length Mcp1. Mcp1-3HA copurified with Vps13-GFP (Fig. 6, D and E), and this interaction was disrupted when either the putative motif was deleted or the core prolines were mutated to alanines (Fig. 6, D and E). Mcp1-3HA showed a weak PxP-dependent binding to the Vps13(1,363–2,556)-GFP DUF1162 domain-containing fragment, whereas the low level of binding to the Vps13 C-terminal fragment was largely unaffected by mutation of the PxP motif (Fig. S2 B). Our results indicate that Mcp1 contains a bona fide PxP motif that is necessary and sufficient for interaction with Vps13.
We hypothesized that if Mcp1 acts as a Vps13 adaptor, high levels of Mcp1 should recruit Vps13 to mitochondria. When Mcp1 was overexpressed from the ADH1 promoter, Vps13^GFP was recruited to mitochondria labeled with preCox4-RFP (Fig. 6 F; Veatch et al., 2009). In contrast, when Mcp1 lacking the PxP motif was overexpressed, this enhanced mitochondrial fluorescence was not observed, and only background mitochondrial autofluorescence remained. Together, these results show that Mcp1 recruits Vps13 to mitochondria through a PxP motif.
The Vps13–Mcp1 interaction, but not Ypt35, is required to suppress ERMES defects
Previous work suggested that Vps13 compensates for loss of ERMES at ER–mitochondria contact sites by relocating to vCLAMPs, creating a bypass for lipid transport (Lang et al., 2015). If Ypt35 and Mcp1 are the respective adaptors for Vps13 at vacuole and mitochondrial membranes, the interaction of each adaptor with Vps13 should be required for rescuing the growth and mitochondrial morphology defects of ERMES mutants.
We used two assays to test the importance of the Mcp1–Vps13 interaction. First, loss of the ERMES subunit Mdm10 is lethal when combined with loss of either vps13 or mcp1 (Tan et al., 2013; Lang et al., 2015). Using a plasmid shuffle assay, we found that plasmids expressing WT MCP1 but not those expressing the PxP motif deletion mutant mcp1(4–12Δ) support the growth of an mcp1 mdm10 strain, indicating that the motif is important for viability when ERMES is defective (Fig. 6 G). Second, high levels of Mcp1 suppress the mitochondrial morphology defects of ERMES mutants (Tan et al., 2013). Mcp1 overexpressed from an ADH1 promoter restored the elongated mitochondrial morphology of an mdm10 strain, whereas deletion of the Mcp1 PxP motif blocked the rescue (Fig. 6 H). Together, these results suggest that Mcp1 must recruit Vps13 to mitochondrial membranes to compensate for ERMES defects.
We next tested whether ERMES bypass suppression requires Ypt35 to anchor Vps13 to the vacuole membrane at vCLAMPs. We found YPT35 was not required for the viability of an mdm10 strain (Fig. S2 C), nor was it needed for suppression of the mdm10 mitochondrial morphology defect by the dominant Vps13(D716H) allele (Fig. 6 H). These results suggest that either there is a redundant adaptor that directs Vps13 to the vacuole at vCLAMPs or that Vps13 does not need to be associated with the vacuole membrane to suppress ERMES mutants.
Vps13(D716H) and other dominant Vps13 ERMES suppressors show reduced localization to the NVJ, which was suggested to enhance their availability at mitochondria and/or vCLAMPs (Lang et al., 2015). Because Ypt35 is required for Vps13 localization to the NVJ, Ypt35 could oppose suppression by sequestering Vps13 at the NVJ or in another endosomal or vacuolar pool. However, we did not observe any rescue of mdm10 mitochondrial morphology defects when YPT35 was deleted, even when levels of Vps13 were increased (Fig. 6 H).
Vps13 adaptors compete to recruit Vps13 to different membranes
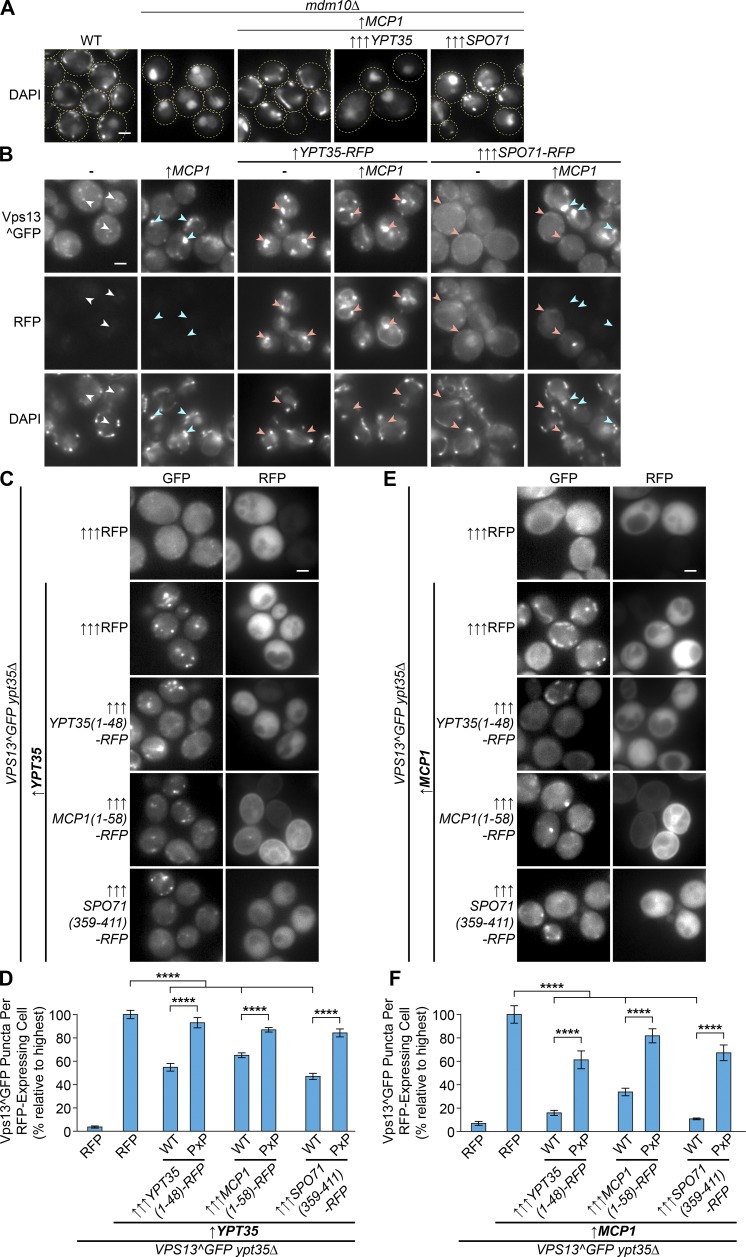
The observation that Mcp1 and Ypt35 use the same motif to recruit Vps13 to mitochondrial and vacuole membranes suggests these adaptors may not bind simultaneously to anchor Vps13 at vCLAMPs. We hypothesized that high levels of Ypt35 or Spo71 would compete with Mcp1 for binding to Vps13 and block ERMES suppression. Consistent with this hypothesis, overexpression of Spo71 or Ypt35 reduced the ability of overexpressed Mcp1 to rescue the mitochondrial morphology defects of an mdm10 strain (Fig. 7 A).
Figure 7.
Vps13 adaptors compete to recruit Vps13 to different membranes. (A) Suppression of the mdm10Δ mitochondrial morphology defect by moderately overexpressed Mcp1 (driven by the ADH1 promoter; ↑) is partially blocked by strong overexpression of Spo71 (driven by the TEF1 promoter; ↑↑↑) and completely blocked by strong overexpression of Ypt35 (driven by the GPD1 promoter; ↑↑↑). (B) High levels of Spo71-RFP or Ypt35-RFP block the Mcp1-dependent mitochondrial targeting of Vps13^GFP, which is instead relocalized to the plasma membrane or to endosomes, respectively. DAPI (blue) marks mitochondria. Arrowheads indicate Vps13^GFP colocalization with RFP (red), mitochondria (blue), or neither (white). (C) Vps13^GFP recruitment to puncta by YPT35 (ADH1pr; ↑) is reduced by highly expressed (TEF1pr; ↑↑↑) soluble RFP chimeras with the PxP motif–containing regions of Ypt35, Mcp1, or Spo71. (D) Quantitation of Ypt35-induced Vps13^GFP puncta in RFP-expressing cells shows a significant PxP-dependent reduction of puncta when the soluble chimeras are expressed. Unpaired one-way ANOVA: n = 5, ≥890 cells/strain/replicate; P < 0.0001 overall; Holm-Sidak's multiple comparisons test, ****, P < 0.0001. (E) Vps13^GFP recruitment to puncta by moderately overexpressed MCP1 (ADH1pr; ↑) is similarly reduced by strong overexpression (TEF1pr; ↑↑↑) of soluble RFP chimeras with the PxP motif–containing regions of Ypt35, Mcp1, or Spo71. (F) Quantitation of the Mcp1-induced Vps13^GFP puncta in RFP-expressing cells shows a significant PxP-dependent reduction of puncta when the soluble chimeras are expressed. Unpaired one-way ANOVA: n = 5, ≥719 cells/strain/replicate; P < 0.0001 overall; Holm-Sidak's multiple comparisons test, ****, P < 0.0001. Error bars indicate SEM. Bars, 2 µm.
If Spo71 and Ypt35 successfully compete for a limited Vps13 pool, cells with high levels of Spo71 or Ypt35 should have an altered Vps13^GFP localization. We have shown Ypt35 recruits Vps13 to endosomes (Fig. 1, B and F), and Park et al. (2013) demonstrated that ectopic expression of Spo71 recruits Vps13 to the plasma membrane in a subset of cells. In line with these observations, concurrent overexpression of Ypt35-RFP or Spo71-RFP with Mcp1 relocalized Vps13^GFP to internal puncta or the plasma membrane, respectively (Fig. 7 B).
The spatial segregation of Vps13 by high levels of Ypt35 and Spo71 could prevent it from encountering Mcp1 at mitochondria. To determine whether adaptors compete for Vps13 in the cytosol or if they bind Vps13 only at membranes, we appended the PxP motif–containing regions of Ypt35, Mcp1, and Spo71 to RFP. We first tested whether overexpression of the soluble RFP fusion proteins from the strong TEF1 promoter blocks the ability of Ypt35, expressed from the moderate ADH1 promoter, to recruit Vps13^GFP to endosomes. Cells expressing high levels of each RFP fusion had fewer Vps13^GFP puncta, suggesting these fusions compete with Ypt35 for Vps13^GFP (Fig. 7 C). Using automated methods to quantify GFP puncta in cells whose RFP signal exceeds a minimum threshold, we found that all three soluble RFP fusions reduced the localization of Vps13^GFP to endosomes by 35–53% (P < 0.0001) and that this competition depended on the integrity of each PxP motif (Fig. 7 D).
A parallel experiment testing the ability of each RFP fusion to disrupt the Mcp1-induced recruitment of Vps13^GFP to mitochondria showed an even stronger PxP-dependent effect (Fig. 7, E and F). In this experiment, all three RFP fusions reduced Vps13^GFP localization by 66–89%. That the RFP fusions were more effective at out-competing Mcp1 suggests Mcp1 has a lower affinity for Vps13 compared with Ypt35. Collectively, our results strongly suggest that the Vps13 adaptors compete for the same binding site or sites on Vps13. Thus, changes in adaptor expression, or PxP motif exposure, are plausible mechanisms for regulating the localization of Vps13.
Discussion
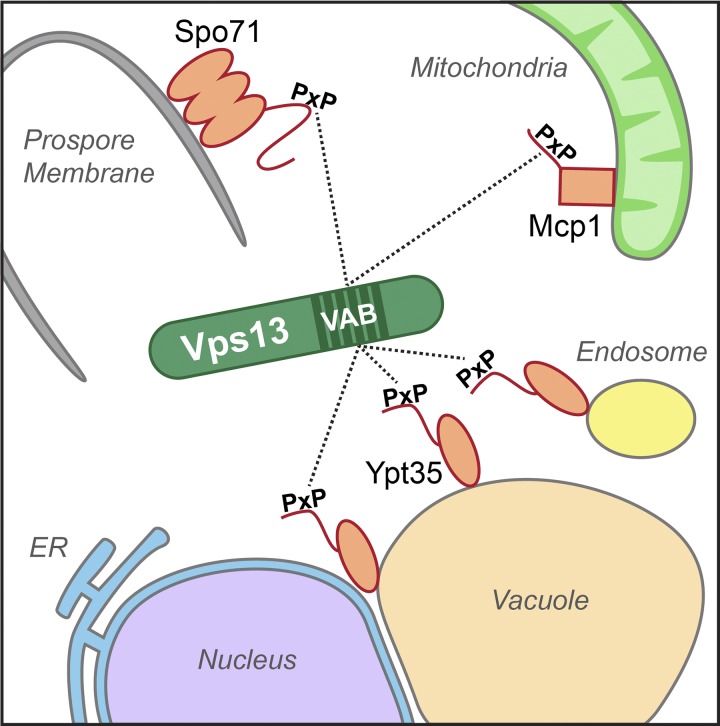
We have identified Ypt35 as the adaptor protein that recruits Vps13 to endosomal and vacuolar membranes and Mcp1 as the adaptor for Vps13 at mitochondria. Together with the prospore membrane adaptor Spo71, these proteins direct Vps13 to distinct cellular locations (Fig. 8). Remarkably, all three adaptors use a related motif to bind a conserved six-repeat region that includes the DUF1162 domain. We recommend this region be renamed the VPS13 adaptor binding (VAB) domain. Our results suggest that Vps13 localization may be determined largely by competition between adaptors. Changes in the level of a particular adaptor or in the exposure of its recruitment motif could drive the localization of Vps13 to membrane contact sites in response to environmental stimuli or altered lipid homeostasis.
Figure 8.
Model of Vps13 recruitment by adaptor proteins with PxP motifs. Ypt35, Spo71, and Mcp1 all contain PxP motifs that compete to recruit Vps13 to endosomes/vacuole/NVJ, the prospore membrane, and mitochondria respectively. Each adaptor shows PxP-dependent binding to a central repeat region of Vps13 that overlaps the DUF1162 domain; we propose this domain be named the VPS13 adaptor binding (VAB) domain.
Ypt35 recruits Vps13 to the endosome and vacuole
Recruitment of Vps13 to endosomes and to ER–vacuole contact sites is the first identified function for Ypt35. Ypt35 is not the only PX domain-containing protein that localizes to endolysosomal contact sites. The mammalian PX-BAR sorting nexin Snx2 functions with retromer to form recycling tubules at endosomes and interacts with the ER protein VAP to create membrane contact sites that promote tubule scission (Dong et al., 2016). Although many sorting nexins contain curvature-sensing PX-BAR domains and are involved in receptor trafficking, Ypt35 lacks a BAR domain and is not required for sorting the CPY receptor Vps10, nor does it appear to sort other cargo (Bean et al., 2017). Recruiting Vps13 to endosome and vacuole membranes may thus be its primary role.
The ER protein Mdm1/Snx13 uses its PX domain to bind to the vacuole in trans, which anchors it to both NVJs and to vacuolar non-NVJ cytoplasmic ER contact sites (Henne et al., 2015; Murley et al., 2015; Wong and Levine, 2016). The Mdm1 and Ypt35 PX domains bind selectively and with high affinity to phosphatidylinositol 3-phosphate, a lipid found at endosomes and vacuoles (Gillooly et al., 2000; Yu and Lemmon, 2001; Di Paolo and De Camilli, 2006). Whereas Mdm1 is a constitutive component of the NVJ, Ypt35 is primarily at endosomes when nutrients are abundant and moves to the NVJ when glucose is limiting. Thus, other factors beyond the PX domain are important for the NVJ localization of Vps13.
Vps13 adaptors compete for Vps13 binding
We identified a short motif within the unstructured N terminus of all three Vps13 adaptors that bind directly or indirectly to a conserved region of Vps13 containing the DUF1162 domain. The core motif present in all three adaptors consists of prolines flanked by hydrophobic residues that conform to a ΦxxΦxPxP(Φ/T) consensus. In each adaptor, the motif is located in an unstructured region ∼50–100 amino acids N-terminal to lipid binding or transmembrane domains, which may help position Vps13 relative to the membrane.
The similarity between recruitment motifs suggests that adaptors compete for binding to a single site on Vps13. Indeed, we found that soluble fusion proteins containing these motifs can displace Vps13 from endosomes or mitochondria. Our copurification and competition experiments suggest Spo71 has the strongest affinity for Vps13, whereas Mcp1 has the weakest. Slight differences between motifs, such as the presence of hydrophobic or acidic residues after the PxP sequence, may explain these different signal strengths. The relatively high affinity of Spo71 for Vps13 could explain how Vps13 is efficiently relocalized to the prospore membrane during meiosis, when Spo71 expression is induced (Park et al., 2013). At steady state, the relative affinities of Mcp1 and Ypt35 motifs may dictate Vps13 distribution between endosomes and mitochondria. Furthermore, this distribution could be fine-tuned by changes in the relative abundance of Ypt35 and Mcp1 or posttranslational modifications that modulate the conformation or affinity of their binding motifs.
Are there other adaptors? Vps13 can localize to peroxisomes (John Peter et al., 2017), and although we found little evidence of Golgi targeting in nutrient-rich conditions, Vps13 is required for homotypic fusion of Golgi membranes and for Golgi/endosomal trafficking (Brickner and Fuller, 1997; Dalton et al., 2017; De et al., 2017). These functions may not involve membrane contact sites (Dalton et al., 2017; De et al., 2017) but likely require association of Vps13 with membranes. A search of the yeast proteome successfully identified recruitment signals in Mcp1 and Spo71, suggesting this approach may identify new adaptors that use a PxP motif to recruit Vps13 to other membranes.
How does Vps13 suppress ERMES defects?
A previous study suggested that Vps13 compensates for ERMES defects by acting at vCLAMPs, which provides a bypass pathway for lipid transfer (Lang et al., 2015). While our work was in progress, John Peter et al. (2017) identified Mcp1 as the mitochondrial adaptor for Vps13. Our results extended these findings by identifying a PxP motif within Mcp1 that mediates the interaction with the conserved DUF1162 domain in Vps13. Mcp1 requires this motif to bypass ERMES defects, demonstrating that Vps13 recruitment to mitochondria is essential for suppression. However, because Mcp1 and Ypt35 compete for binding Vps13, it is unlikely these mitochondrial and vacuolar adaptors bind simultaneously to Vps13 to tether it at the vCLAMP, and we found that YPT35 is not required for ERMES suppression.
How can we reconcile these findings? A second vacuolar adaptor could recruit Vps13 to vCLAMPs. An alternative hypothesis is that Vps13 does not act at vCLAMPs but instead tethers a different organelle to the mitochondrial membrane, creating a three-way contact site that includes the vCLAMP. A similar model has been advanced to explain how the ER-localized GRAM domain protein Lam6 regulates the expansion of vCLAMPs when ERMES is lost (Wong and Levine, 2016). High levels of Lam6, which are sufficient for vCLAMP expansion, cause proliferation of ER tubules that invade vCLAMPs (Elbaz-Alon et al., 2015), which may create a three-way ER–mitochondrial–vacuole junction.
Our data indicate that Vps13 binds directly or indirectly to ER membranes. Under starvation conditions, Vps13 pulls Ypt35 to the NVJ, suggesting Vps13 engages in simultaneous interactions with a PxP-containing adaptor and an ER component to bridge two membranes. Indeed, PxP motifs are found in unstructured linkers that could stretch up to 16–36 nm to extend across a contact site, similar to the positioning of StARkin lipid transfer domains 16–21 nm from transmembrane domains (Gatta et al., 2015; Wong and Levine, 2016; Gatta and Levine, 2017). By bridging ER and mitochondria membranes, Vps13 could provide a secondary tether at mitochondrial–ER contact sites that compensates for loss of ERMES.
Although this is an appealing model, many of the dominant Vps13 alleles that suppress ERMES defects have reduced localization at the NVJ (Lang et al., 2015; Park et al., 2016). This suggests NVJ sequestration of Vps13 blocks its action at mitochondria, yet other mutations that release Vps13 from the NVJ, including loss of Nvj1 (Lang et al., 2015) or Ypt35 (this study), do not suppress ERMES. Defining the full set of Vps13 binding partners may clarify the mechanism of ERMES suppression.
Does Vps13 need to bridge different organelle membranes?
It remains to be determined whether Vps13 acts as a tether, a scaffold, and/or a lipid-transfer protein. Vps13 does not act as a primary tether at NVJs because loss of Vps13 does not block NVJ formation. Vps13 may have a redundant tethering role at the NVJ and could also contribute to the ER–vacuolar non-NVJ cytoplasmic ER sites that persist in nvj1 mutants (Henne et al., 2015; Murley et al., 2015; Wong and Levine, 2016). Vps13 may not need to bridge two membranes. Instead, it could provide a scaffold to enrich lipid transfer proteins at membrane contact sites. Vps13 contains several domains that bind lipids (De et al., 2017; Rzepnikowska et al., 2017). Although no direct role in lipid transfer has been demonstrated, it is interesting that John Peter et al. (2017) found a function for Mcp1 in ERMES suppression beyond Vps13 recruitment and speculated that Vps13 shuttles lipids to Mcp1.
Conserved lipid-binding domains in Vps13 could work together with PxP sequences for membrane recruitment. The APT1 domain may bind PI3P to enhance endosomal targeting, whereas the C-terminal PH domain (Fidler et al., 2016) may contribute to Vps13 localization at mitochondria. Because all three adaptors displayed a weak PxP-independent binding to the Vps13 C terminus, these domains may make other contacts with adaptors or recruit cofactors. For example, the C terminus of VPS13B binds Rab6 for recruitment to the Golgi in human cells (Seifert et al., 2015), suggesting combinatorial interactions with Rab GTPases or lipids may contribute to Vps13 localization.
The adaptor-binding domain of Vps13 is a conserved six-repeat region
We found the VAB domain is composed of six repeats spanning a region roughly twice the size of the previously defined DUF1162/SHR-BD domain (Rzepnikowska et al., 2017). All six repeats, each featuring an invariant asparagine residue, are present in most Vps13 family members including human VPS13A, VPS13C, and VPS13D. Although VPS13B contains six identifiable repeats, two lack the invariant asparagine. Mutation of the key asparagine in the most conserved repeat (N2993S) of VPS13B causes Cohen syndrome (Kolehmainen et al., 2003), whereas mutation of the invariant asparagine in the last repeat of VPS13D (N3521S) is associated with a childhood-onset movement disorder (Gauthier et al., 2018; Seong et al., 2018). This suggests these repeats have a critical functional role, although candidate binding partners have yet to be identified.
Human VPS13 proteins are ubiquitously expressed and localize to a variety of organelles including Golgi, lipid droplets, and mitochondria (Seifert et al., 2015; Yang et al., 2016; Ramseyer et al., 2018). Although they have not yet been demonstrated to be at contact sites, a recent study found VPS13C on a subdomain of lipid droplets proximal to mitochondria (Ramseyer et al., 2018). Moreover, VPS13A/C contain predicted FFAT motifs, suggesting they interact with VAPA/B at the ER (Mikitova and Levine, 2012; Huttlin et al., 2015). The functional divergence of human VPS13 family members may allow them to fulfill specialized roles at different organelles involving unique adaptor repertoires, explaining how dysfunction of these related proteins can cause diverse diseases. Further studies in yeast promise to shed more light on this family of proteins.
Materials and methods
Yeast strains and plasmids
Strains and plasmids are listed in Table S2 (A and B). Yeast strains were made by standard PCR-based homologous recombination (Longtine et al., 1998; Janke et al., 2004). Gene deletions were confirmed by colony PCR. Plasmids were made via homologous recombination in yeast by cotransforming linearized plasmids with PCR products generated from the primers listed in Table S2 C. The plasmids were recovered in Escherichia coli and sequenced.
Fluorescence microscopy
Log phase yeast were grown in synthetic dextrose-based media unless otherwise mentioned, transferred to concanavalin A–treated glass bottom MatriPlates (Brooks) and imaged on a DMi8 microscope (Leica Microsystems) with a high-contrast Plan Apochromat 63×/1.30 Glyc CORR CS objective (Leica Microsystems), an ORCA-Flash4.0 digital camera (Hamamatsu Photonics), and MetaMorph 7.8 software (MDS Analytical Technologies). Images were acquired at room temperature. For Fig. 1 (E and F) and Fig. S1 A, a high-contrast Plan Apochromat 100×/1.40 Oil STED white objective (Leica Microsystems) was used instead. Alternatively, for Fig. 1 D, log-phase yeast were imaged with a Plan Apochromat 100×/1.40 NA oil immersion objective lens on an Axioplan 2 fluorescence microscope (ZEISS) with a CoolSNAP camera (Roper Scientific) and MetaMorph 7.7 software. All measurements were done on raw images; for presentation, images were processed by using MetaMorph, and cropping and adjustment of brightness and contrast was performed in Photoshop CS5 (Adobe).
Mitochondrial DNA was labeled with 1 µg/ml DAPI (Sigma-Aldrich) for 20–30 min at 30°C, nuclei were labeled with 5 µg/ml Hoechst 33342 (Sigma-Aldrich) for 15 min at 30°C, and the vacuolar rim was labeled with 4 µM FM4-64 (Thermo Fisher Scientific) for 30 min at 30°C. Cells were washed before imaging. For FM4-64 labeling, washed cells were incubated an additional 60 min at 30°C before imaging.
For NVJ localization (Fig. 2), 2 OD/ml of log phase yeast were incubated at 30°C for 18 h in synthetic complete media with 2% acetate as a carbon source. Yeast were imaged in MatriPlates as above except that washed cells were maintained in nutrient-depleted synthetic acetate media during imaging.
Quantitation of microscopy images
Images were analyzed by using custom MetaMorph 7.8 journals. Briefly, live cells were identified by using several iterations of the Count Nuclei function. Puncta number and size were identified with the Granularity function. Puncta attributed to dead cells and extracellular noise were masked by using the LogicalAND function, and colocalization was measured with the built in Colocalization application. NVJs were identified by either locating bright objects with TopHat (Nvj1-RFP) or using TopHat and Integrated Morphometry Analysis to identify weaker signals (Vps13^GFP) based on size and shape. Significance was determined by Student’s t test or ANOVA followed by Holm-Sidak’s multiple comparison test. Data distribution was assumed to be normal, but this was not formally tested.
Coimmunoprecipitation
To perform the cross-linking coimmunoprecipitation, 20 OD600 of log phase yeast grown in synthetic dextrose media was spheroplasted with zymolyase (MJS BioLynx). Samples were cross-linked for 30 min with 400 µg/ml DSP in cross-linking buffer (20 mM Hepes, 0.7 M sorbitol, and 100 mM NaCl, pH 7.4), and after quenching with 100 mM Tris buffer, an equal volume of 2× lysis buffer (100 mM Hepes, 0.2% NP-40, 100 mM NaCl, 2 mM EDTA, 200 mM PMSF, and 2× HALT protease inhibitor cocktail, pH 7.4) was added. The lysates were incubated with rabbit anti–HA (sc-805; Santa Cruz Biotechnology) followed by protein A–Sepharose beads (GE Healthcare) at 4°C. Washed beads were heated in sample buffer at 70°C for 5 min. Samples were run on SDS-PAGE gels, transferred to nitrocellulose membranes, and blotted with mouse anti–GFP (11–814-460-001; Roche) or mouse anti–HA (MMS-101R; Covance) followed by goat anti–mouse conjugated to HRP (115–035-146; Jackson ImmunoResearch Laboratories, Inc.). West Pico (Pierce) chemiluminescent reagent was added to the blot and used to expose Amersham Hyperfilm (GE Healthcare).
For coimmunoprecipitations that did not involve cross-linking, 40 OD600 of yeast spheroplasts was lysed in lysis buffer (50 mM Hepes, 0.1% Tween-20, 50 mM NaCl, 1 mM EDTA, 100 mM PMSF, and 1× HALT protease inhibitor cocktail, pH 7.4). The lysates were incubated with rabbit anti–GFP (EU2; Eusera) and processed as above. Films were scanned, and densitometry was performed in Fiji (Schindelin et al., 2012).
Plasmid shuffle growth assay
mdm10 mcp1 strains with a MCP1 (URA3) vector and either an MCP1 (LEU2) or mcp1(4–12Δ) (LEU2) vector were grown to saturation in selective media, reinoculated at 0.4 OD600/ml in media without uracil selection, and grown 24 h at 30°C. 4 µl of serially diluted cultures was spotted on plates containing 0.1% 5-fluoroorotic acid to select against the URA3 vector, or on plates lacking leucine as a control, and incubated at 30°C for 3 d.
Tetrad dissection
Yeast diploids were sporulated by incubating 1–2 OD600/ml cultures in sporulation media (0.3% potassium acetate and 0.2% raffinose) at 25°C for 5 d. 50 µl of this culture was digested with 50 µl of 1 M sorbitol containing 1.5 mg/ml zymolyase at 25°C for 8–9 min before spores were separated by using a dissection microscope.
MEME and FIMO-based motif search
Saccharomycetales-level homologues of Ypt35 and Spo71 were identified by OrthoDB (Zdobnov et al., 2017) and aligned with MUSCLE (Edgar, 2004). Sequences that contained three or more residues consistent with the Ypt35/Spo71 consensus and that were not identical to either Ypt35 or Spo71 were selected. A region of 15–29 residues within the alignment centered on the core prolines was analyzed using MEME (Bailey and Elkan, 1994). In each case we selected the most significant MEME motif as an input for a FIMO search of the S. cerevisiae proteome with a threshold of P < 0.0001 (Grant et al., 2011).
Online supplemental material
Fig. S1 shows additional metrics for Vps13-GFP, Ypt35-GFP, and Vps13^GFP localization, provides additional RFP-FYVE and RFP-2×PH chimera controls and constructs, and shows the distribution of FYVE-constructs in the RFP channel. Fig. S2 contains additional coimmunoprecipitations between Vps13-GFP and Ypt35-3HA or Mcp1-HA and shows an mdm10× ypt35 tetrad dissection. Fig. S3 displays the complete alignments of yeast and human DUF1162 repeats. Table S1 lists hits for the MEME/FIMO Vps13 adaptor motif search. Table S2, A, B, and C, respectively, list the yeast strains, plasmids, and primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Benoît Kornmann (ETH Zürich, Zürich, Switzerland), Dr. Aaron M. Neiman (Stony Brook University, Stony Brook, NY), and Dr. Mike Henne (University of Texas Southwestern Medical School, Dallas, TX) for reagents; Dr. Luc Berthiaume (University of Alberta, Edmonton, Canada) for his generous gift of rabbit anti–GFP serum; and Rory Long for technical assistance.
This work was supported by funding from Canadian Institutes of Health Research grants 247169 and 365914 and the Canada Foundation for Innovation (Leading Edge Fund 30636).
The authors declare no competing financial interests.
Author contributions: conceptualization, E. Conibear; methodology, E. Conibear and B.D.M. Bean; investigation, B.D.M. Bean, S.K. Dziurdzik, K.L. Kolehmainen, C.M.S. Fowler, W.K. Kwong, L.I. Grad, M. Davey, and C. Schluter; writing - original draft, B.D.M. Bean; review and editing, E. Conibear, B.D.M. Bean, S.K. Dziurdzik, K.L. Kolehmainen, C.M.S. Fowler, W.K. Kwong, L.I. Grad, M. Davey, and C. Schluter; funding acquisition, E. Conibear; supervision, E. Conibear and B.D.M. Bean.
References
- Bailey T.L., and Elkan C.. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28–36. [PubMed] [Google Scholar]
- Bean B.D., Davey M., and Conibear E.. 2017. Cargo selectivity of yeast sorting nexins. Traffic. 18:110–122. 10.1111/tra.12459 [DOI] [PubMed] [Google Scholar]
- Bian X., Saheki Y., and De Camilli P.. 2018. Ca2+ releases E-Syt1 autoinhibition to couple ER-plasma membrane tethering with lipid transport. EMBO J. 37:219–234. 10.15252/embj.201797359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biegert A., and Söding J.. 2008. De novo identification of highly diverged protein repeats by probabilistic consistency. Bioinformatics. 24:807–814. 10.1093/bioinformatics/btn039 [DOI] [PubMed] [Google Scholar]
- Brickner J.H., and Fuller R.S.. 1997. SOI1 encodes a novel, conserved protein that promotes TGN-endosomal cycling of Kex2p and other membrane proteins by modulating the function of two TGN localization signals. J. Cell Biol. 139:23–36. 10.1083/jcb.139.1.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burston H.E., Maldonado-Báez L., Davey M., Montpetit B., Schluter C., Wendland B., and Conibear E.. 2009. Regulators of yeast endocytosis identified by systematic quantitative analysis. J. Cell Biol. 185:1097–1110. 10.1083/jcb.200811116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., and Brenner S.E.. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton L.E., Bean B.D.M., Davey M., and Conibear E.. 2017. Quantitative high-content imaging identifies novel regulators of Neo1 trafficking at endosomes. Mol. Biol. Cell. 28:1539–1550. 10.1091/mbc.e16-11-0772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De M., Oleskie A.N., Ayyash M., Dutta S., Mancour L., Abazeed M.E., Brace E.J., Skiniotis G., and Fuller R.S.. 2017. The Vps13p-Cdc31p complex is directly required for TGN late endosome transport and TGN homotypic fusion. J. Cell Biol. 216:425–439. 10.1083/jcb.201606078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G., and De Camilli P.. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature. 443:651–657. 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- Dong R., Saheki Y., Swarup S., Lucast L., Harper J.W., and De Camilli P.. 2016. Endosome-ER contacts control actin nucleation and retromer function through VAP-dependent regulation of PI4P. Cell. 166:408–423. 10.1016/j.cell.2016.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G., Casella J.F., Gautier R., Boehmer T., Schwartz T.U., and Antonny B.. 2007. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat. Struct. Mol. Biol. 14:138–146. 10.1038/nsmb1194 [DOI] [PubMed] [Google Scholar]
- Edgar R.C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg-Bord M., Shai N., Schuldiner M., and Bohnert M.. 2016. A tether is a tether is a tether: Tethering at membrane contact sites. Dev. Cell. 39:395–409. 10.1016/j.devcel.2016.10.022 [DOI] [PubMed] [Google Scholar]
- Elbaz-Alon Y., Rosenfeld-Gur E., Shinder V., Futerman A.H., Geiger T., and Schuldiner M.. 2014. A dynamic interface between vacuoles and mitochondria in yeast. Dev. Cell. 30:95–102. 10.1016/j.devcel.2014.06.007 [DOI] [PubMed] [Google Scholar]
- Elbaz-Alon Y., Eisenberg-Bord M., Shinder V., Stiller S.B., Shimoni E., Wiedemann N., Geiger T., and Schuldiner M.. 2015. Lam6 regulates the extent of contacts between organelles. Cell Reports. 12:7–14. 10.1016/j.celrep.2015.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidler D.R., Murphy S.E., Courtis K., Antonoudiou P., El-Tohamy R., Ient J., and Levine T.P.. 2016. Using HHsearch to tackle proteins of unknown function: A pilot study with PH domains. Traffic. 17:1214–1226. 10.1111/tra.12432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Föller M., Hermann A., Gu S., Alesutan I., Qadri S.M., Borst O., Schmidt E.M., Schiele F., vom Hagen J.M., Saft C., et al. . 2012. Chorein-sensitive polymerization of cortical actin and suicidal cell death in chorea-acanthocytosis. FASEB J. 26:1526–1534. 10.1096/fj.11-198317 [DOI] [PubMed] [Google Scholar]
- Gatta A.T., and Levine T.P.. 2017. Piecing together the patchwork of contact sites. Trends Cell Biol. 27:214–229. 10.1016/j.tcb.2016.08.010 [DOI] [PubMed] [Google Scholar]
- Gatta A.T., Wong L.H., Sere Y.Y., Calderón-Noreña D.M., Cockcroft S., Menon A.K., and Levine T.P.. 2015. A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. eLife. 4:1–21. 10.7554/eLife.07253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier J., Meijer I.A., Lessel D., Mencacci N.E., Krainc D., Hempel M., Tsiakas K., Prokisch H., Rossignol E., Helm M.H., et al. . 2018. Recessive mutations in >VPS13D cause childhood onset movement disorders. Ann. Neurol. 10.1002/ana.25204 [DOI] [PubMed] [Google Scholar]
- Gillooly D.J., Morrow I.C., Lindsay M., Gould R., Bryant N.J., Gaullier J.M., Parton R.G., and Stenmark H.. 2000. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19:4577–4588. 10.1093/emboj/19.17.4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant C.E., Bailey T.L., and Noble W.S.. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics. 27:1017–1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helle S.C.J., Kanfer G., Kolar K., Lang A., Michel A.H., and Kornmann B.. 2013. Organization and function of membrane contact sites. Biochim. Biophys. Acta. 1833:2526–2541. 10.1016/j.bbamcr.2013.01.028 [DOI] [PubMed] [Google Scholar]
- Henne W.M., Zhu L., Balogi Z., Stefan C., Pleiss J.A., and Emr S.D.. 2015. Mdm1/Snx13 is a novel ER-endolysosomal interorganelle tethering protein. J. Cell Biol. 210:541–551. 10.1083/jcb.201503088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y., Gruhler A., Heilbut A., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., et al. . 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 415:180–183. 10.1038/415180a [DOI] [PubMed] [Google Scholar]
- Hönscher C., Mari M., Auffarth K., Bohnert M., Griffith J., Geerts W., van der Laan M., Cabrera M., Reggiori F., and Ungermann C.. 2014. Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev. Cell. 30:86–94. 10.1016/j.devcel.2014.06.006 [DOI] [PubMed] [Google Scholar]
- Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., and O’Shea E.K.. 2003. Global analysis of protein localization in budding yeast. Nature. 425:686–691. 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Huttlin E.L., Ting L., Bruckner R.J., Gebreab F., Gygi M.P., Szpyt J., Tam S., Zarraga G., Colby G., Baltier K., et al. . 2015. The BioPlex network: A systematic exploration of the human interactome. Cell. 162:425–440. 10.1016/j.cell.2015.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., and Knop M.. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962. 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- John Peter A.T., Herrmann B., Antunes D., Rapaport D., Dimmer K.S., and Kornmann B.. 2017. Vps13-Mcp1 interact at vacuole-mitochondria interfaces and bypass ER-mitochondria contact sites. J. Cell Biol. 216:3219–3229. 10.1083/jcb.201610055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi K., and Gallagher K.L.. 2013. Identification of SHRUBBY, a SHORT-ROOT and SCARECROW interacting protein that controls root growth and radial patterning. Development. 140:1292–1300. 10.1242/dev.090761 [DOI] [PubMed] [Google Scholar]
- Kolehmainen J., Black G.C.M., Saarinen A., Chandler K., Clayton-Smith J., Träskelin A.L., Perveen R., Kivitie-Kallio S., Norio R., Warburg M., et al. . 2003. Cohen syndrome is caused by mutations in a novel gene, COH1, encoding a transmembrane protein with a presumed role in vesicle-mediated sorting and intracellular protein transport. Am. J. Hum. Genet. 72:1359–1369. 10.1086/375454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B., Currie E., Collins S.R., Schuldiner M., Nunnari J., Weissman J.S., and Walter P.. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 325:477–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang A.B., John Peter A.T., Walter P., and Kornmann B.. 2015. ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J. Cell Biol. 210:883–890. 10.1083/jcb.201502105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage S., Drouet V., Majounie E., Deramecourt V., Jacoupy M., Nicolas A., Cormier-Dequaire F., Hassoun S.M., Pujol C., Ciura S., et al. International Parkinson’s Disease Genomics Consortium (IPDGC) . 2016. Loss of VPS13C function in autosomal-recessive Parkinsonism causes mitochondrial dysfunction and increases PINK1/Parkin-Dependent mitophagy. Am. J. Hum. Genet. 98:500–513. 10.1016/j.ajhg.2016.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.K., Choudhary V., Toulmay A., and Prinz W.A.. 2016. An inducible ER-Golgi tether facilitates ceramide transport to alleviate lipotoxicity. J. Cell Biol. 216:131–147. 10.1083/jcb.201606059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., and Pringle J.R.. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Mikitova V., and Levine T.P.. 2012. Analysis of the key elements of FFAT-like motifs identifies new proteins that potentially bind VAP on the ER, including two AKAPs and FAPP2. PLoS One. 7:e30455 10.1371/journal.pone.0030455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Braceras S., Calvo R., and Escalante R.. 2015. TipC and the chorea-acanthocytosis protein VPS13A regulate autophagy in Dictyostelium and human HeLa cells. Autophagy. 11:918–927. 10.1080/15548627.2015.1034413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murley A., and Nunnari J.. 2016. The emerging network of mitochondria-organelle contacts. Mol. Cell. 61:648–653. 10.1016/j.molcel.2016.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murley A., Sarsam R.D., Toulmay A., Yamada J., Prinz W.A., and Nunnari J.. 2015. Ltc1 is an ER-localized sterol transporter and a component of ER-mitochondria and ER-vacuole contacts. J. Cell Biol. 209:539–548. 10.1083/jcb.201502033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillusson S., Stoica R., Gomez-Suaga P., Lau D.H.W., Mueller S., Miller T., and Miller C.C.J.. 2016. There’s something wrong with my MAM; the ER-mitochondria axis and neurodegenerative diseases. Trends Neurosci. 39:146–157. 10.1016/j.tins.2016.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Roberts P., Chen Y., Kvam E., Shulga N., Huang K., Lemmon S., and Goldfarb D.S.. 2000. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol. Biol. Cell. 11:2445–2457. 10.1091/mbc.11.7.2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.S., and Neiman A.M.. 2012. VPS13 regulates membrane morphogenesis during sporulation in Saccharomyces cerevisiae. J. Cell Sci. 125:3004–3011. 10.1242/jcs.105114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-S., Okumura Y., Tachikawa H., and Neiman A.M.. 2013. SPO71 encodes a developmental stage-specific partner for Vps13 in Saccharomyces cerevisiae. Eukaryot. Cell. 12:1530–1537. 10.1128/EC.00239-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.S., Thorsness M.K., Policastro R., McGoldrick L.L., Hollingsworth N.M., Thorsness P.E., and Neiman A.M.. 2016. Yeast Vps13 promotes mitochondrial function and is localized at membrane contact sites. Mol. Biol. Cell. 27:2435–2449. 10.1091/mbc.e16-02-0112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M.J., and Voeltz G.K.. 2016. Structure and function of ER membrane contact sites with other organelles. Nat. Rev. Mol. Cell Biol. 17:69–82. 10.1038/nrm.2015.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampoldi L., Dobson-Stone C., Rubio J.P., Danek A., Chalmers R.M., Wood N.W., Verellen C., Ferrer X., Malandrini A., Fabrizi G.M., et al. . 2001. A conserved sorting-associated protein is mutant in chorea-acanthocytosis. Nat. Genet. 28:119–120. 10.1038/88821 [DOI] [PubMed] [Google Scholar]
- Ramseyer V.D., Kimler V.A., and Granneman J.G.. 2018. Vacuolar protein sorting 13C is a novel lipid droplet protein that inhibits lipolysis in brown adipocytes. Mol. Metab. 7:57–70. 10.1016/j.molmet.2017.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzepnikowska W., Flis K., Kaminska J., Grynberg M., Urbanek A., Ayscough K.R., and Zoladek T.. 2017. Amino acid substitution equivalent to human chorea-acanthocytosis I2771R in yeast Vps13 protein affects its binding to phosphatidylinositol 3-phosphate. Hum. Mol. Genet. 26:1497–1510. 10.1093/hmg/ddx054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. . 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert W., Kühnisch J., Maritzen T., Lommatzsch S., Hennies H.C., Bachmann S., Horn D., and Haucke V.. 2015. Cohen syndrome-associated protein COH1 physically and functionally interacts with the small GTPase RAB6 at the Golgi complex and directs neurite outgrowth. J. Biol. Chem. 290:3349–3358. 10.1074/jbc.M114.608174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong E., Insolera R., Dulovic M., Kamsteeg E.J., Trinh J., Brüggemann N., Sandford E., Li S., Ozel A.B., Li J.Z., et al. . 2018. Mutations in VPS13D lead to a new recessive ataxia with spasticity and mitochondrial defects. Ann. Neurol. 10.1002/ana.25220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan C.J., Audhya A., and Emr S.D.. 2002. The yeast synaptojanin-like proteins control the cellular distribution of phosphatidylinositol (4,5)-bisphosphate. Mol. Biol. Cell. 13:542–557. 10.1091/mbc.01-10-0476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan T., Ozbalci C., Brügger B., Rapaport D., and Dimmer K.S.. 2013. Mcp1 and Mcp2, two novel proteins involved in mitochondrial lipid homeostasis. J. Cell Sci. 126:3563–3574. 10.1242/jcs.121244 [DOI] [PubMed] [Google Scholar]
- Teasdale R.D., and Collins B.M.. 2012. Insights into the PX (phox-homology) domain and SNX (sorting nexin) protein families: structures, functions and roles in disease. Biochem. J. 441:39–59. 10.1042/BJ20111226 [DOI] [PubMed] [Google Scholar]
- van der Kant R., and Neefjes J.. 2014. Small regulators, major consequences - Ca2+ and cholesterol at the endosome-ER interface. J. Cell Sci. 127:929–938. 10.1242/jcs.137539 [DOI] [PubMed] [Google Scholar]
- Veatch J.R., Mcmurray M.A., Nelson Z.W., and Gottschling D.E.. 2009. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell. 137:1247–1258. 10.1016/j.cell.2009.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velayos-Baeza A., Vettori A., Copley R.R., Dobson-Stone C., and Monaco A.P.. 2004. Analysis of the human VPS13 gene family. Genomics. 84:536–549. 10.1016/j.ygeno.2004.04.012 [DOI] [PubMed] [Google Scholar]
- Vollert C.S., and Uetz P.. 2004. The phox homology (PX) domain protein interaction network in yeast. Mol. Cell. Proteomics. 3:1053–1064. 10.1074/mcp.M400081-MCP200 [DOI] [PubMed] [Google Scholar]
- Wong L.H., and Levine T.P.. 2016. Lipid transfer proteins do their thing anchored at membrane contact sites… but what is their thing? Biochem. Soc. Trans. 44:517–527. 10.1042/BST20150275 [DOI] [PubMed] [Google Scholar]
- Yang R.Y., Xue H., Yu L., Velayos-Baeza A., Monaco A.P., and Liu F.T.. 2016. Identification of VPS13C as a galectin-12-binding protein that regulates galectin-12 protein stability and adipogenesis. PLoS One. 11:e0153534 10.1371/journal.pone.0153534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.W., and Lemmon M.A.. 2001. All phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol 3-phosphate. J. Biol. Chem. 276:44179–44184. 10.1074/jbc.M108811200 [DOI] [PubMed] [Google Scholar]
- Zdobnov E.M., Tegenfeldt F., Kuznetsov D., Waterhouse R.M., Simão F.A., Ioannidis P., Seppey M., Loetscher A., and Kriventseva E.V.. 2017. OrthoDB v9.1: cataloging evolutionary and functional annotations for animal, fungal, plant, archaeal, bacterial and viral orthologs. Nucleic Acids Res. 45(D1):D744–D749. 10.1093/nar/gkw1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.