RBM3 plays a protective role in embryonic neurogenesis. This study finds that maternal cold stress affects the embryonic brain development via RBM3 and Yap. When RBM3 is knocked down or knocked out under the maternal cold stress, the embryonic neurogenesis was impaired.
Abstract
In mammals, a constant body temperature is an important basis for maintaining life activities. Here, we show that when pregnant mice are subjected to cold stress, the expression of RBM3, a cold-induced protein, is increased in the embryonic brain. When RBM3 is knocked down or knocked out in cold stress, embryonic brain development is more seriously affected, exhibiting abnormal neuronal differentiation. By detecting the change in mRNA expression during maternal cold stress, we demonstrate that Yap and its downstream molecules are altered at the RNA level. By analyzing RNA-binding motif of RBM3, we find that there are seven binding sites in 3′UTR region of Yap1 mRNA. Mechanistically, RBM3 binds to Yap1-3′UTR, regulates its stability, and affects the expression of YAP1. RBM3 and YAP1 overexpression can partially rescue the brain development defect caused by RBM3 knockout in cold stress. Collectively, our data demonstrate that cold temperature affects brain development, and RBM3 acts as a key protective regulator in cold stress.
Introduction
Stress affects neuronal development of the fetus during maternal pregnancy. For example, nutritional status (Steenweg-de Graaff et al., 2017), emotions (Dworkin and Losick, 2001), alcoholism (Tyler and Allan, 2014), and infection (Toyama et al., 2015; Tang et al., 2016) in the progeny have been reported to be associated with the development of the fetal brain.
Among the potential subpopulations vulnerable to temperature changes, pregnant women have received less attention. During the process of pregnancy, the temperature of pregnant mammals remains stable. Compared with exposure to room temperature (24.4°C), weekly exposures during the last 4 wk of pregnancy to extreme cold was found to be associated with a 17.9% increased risk of preterm birth (He et al., 2016).
Cumulative and acute exposures to extremely low temperatures may induce maternal stress during pregnancy (Lin et al., 2017b). There is increasing evidence that temperature plays a role as a trigger of adverse birth outcomes, such as preterm birth, low birth weight, and stillbirth (Ha et al., 2017; Zhang et al., 2017). However, the relationship between maternal temperature and fetal brain development remains unknown. Cold stress is an important stimulus to the mother and fetus during pregnancy (Kali et al., 2016). Maternal cold stress can lead to abnormal fetal development and may even cause miscarriage. However, whether maternal cold stress affects the brain development is largely unclear.
Neocortical development is a spatially and temporally regulated process that is defined by an early expansion of proliferative neural stem cells (NSCs) that reside in the ventricular zone (VZ) of the embryonic cortical epithelium (Fang et al., 2013). In the development process, any external stimuli are likely to affect the fate of NSCs and then affect the structure or function of the brain (Durak et al., 2016).
The biological function of RBM3 during embryonic brain development is little known. RBM3, which was initially defined as an RNA-binding protein (Derry et al., 1995; Dresios et al., 2005), is induced to be expressed at low temperatures (Danno et al., 1997). It is associated with the structural plasticity and protective effects of cooling in neurodegeneration (Peretti et al., 2015). RBM3 is also related to changes in the expression of different RNAs during the circadian rhythm of body temperature (Liu et al., 2013) and regulates the expression of temperature-sensitive miRNAs (Wong et al., 2016). However, whether RBM3 is involved in the temperature-associated regulation of brain development and neural stem cell development is also unknown.
The activity and expression of Yap1 can be rapidly regulated by a variety of life activities (Lin et al., 2015, 2017a). For example, Yap1 has been associated with energy homeostasis (Wang et al., 2015), mechanical pressure (Aragona et al., 2013), G-protein coupled receptor signaling, and oxidative stress (Lehtinen et al., 2006). However, whether YAP1 is able to participate in certain biological processes under a low temperature followed by prolonged cold stress is still unknown.
Here, we examined the in vivo effects of RBM3 disruption on embryonic mammalian brain development under different maternal body temperature environments. We found that knockdown of RBM3 in cold stress, but not in the normal body temperature condition, results in reduced cortical neural progenitor proliferation and altered neurogenesis. In particular, the expression of Yap1 in the sample of the embryonic cerebral cortex in cold stress was increased compared with that at normal temperature. During cold stress, the expression of Yap1 in the sample of the RBM3 knockout embryonic cerebral cortex was decreased compared with that in the littermate control. Furthermore, our results showed that RBM3 regulated the stability of Yap1 mRNA by binding to the 3′UTR region of Yap1 mRNA. We confirmed that knockout of RBM3 led to brain development defects in cold stress.
Together, these observations provide new insights into the roles of cold stress during brain development. Our results reveal the function of RBM3 during neocortical development in prenatal cold stress. The data suggest that RBM3 and Yap1 act as potential cold shock proteins in brain development.
Results
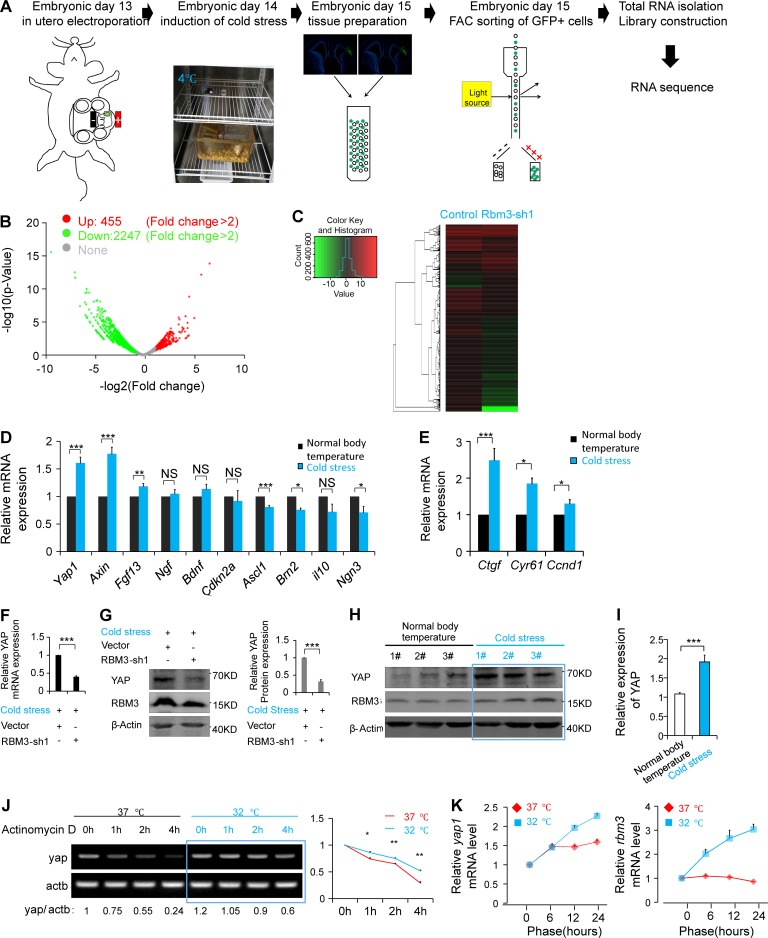
Cold stress affects neurogenesis and induces the expression of RBM3
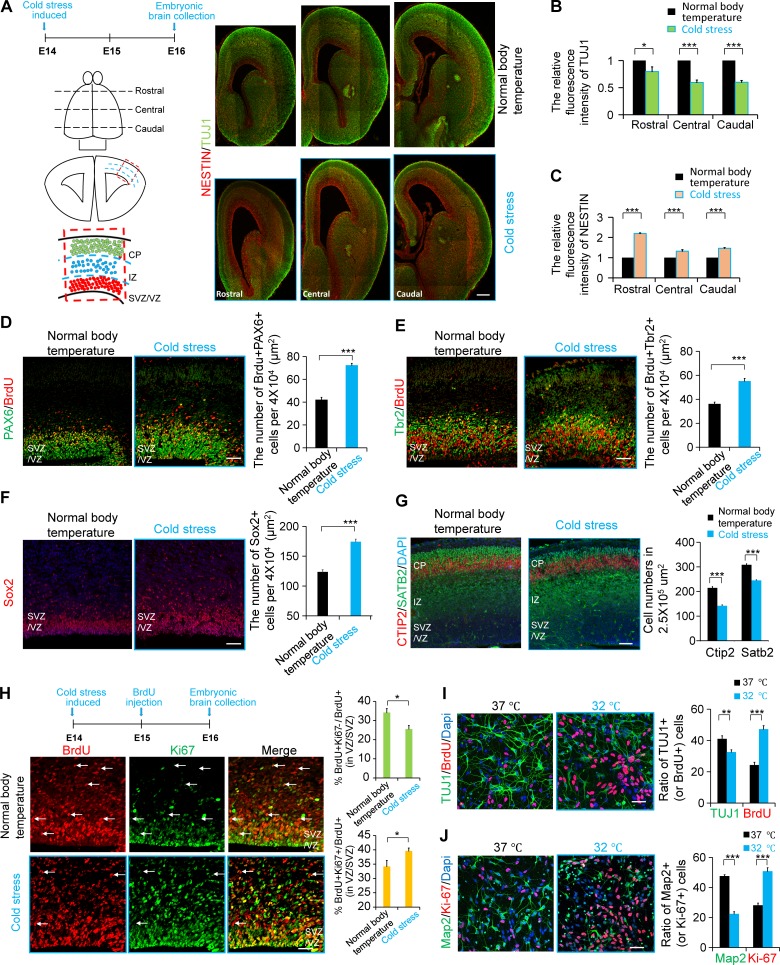
It has been reported that many external environmental factors affect the development of the fetal brain. Whether a change in constant body temperature, an important feature of mammals, affects the development of the cerebral cortex is unknown. To examine the effect of maternal body temperature changes on embryonic brain development, we injected 5′-AMP (0.7 mg/g; Tao et al., 2011; Peretti et al., 2015) into pregnant mice to induce cold stress, and the rectal temperatures were recorded at different time points to determine the reduction in body temperature in these pregnant mice (Fig. S1 A). The immunofluorescence staining results showed that TUJ1+ neuronal cells were decreased after the induction of cold stress in pregnant mice, and the proportion of NESTIN+ NSCs was increased (Fig. 1, A–C). The immunofluorescence staining results also showed that the proportion of PAX6+BrdU+ (or TBR2+BrdU+ or SOX2+) NSCs was increased (Fig. 1, D–F), and SATB2 (or CTIP2)-positive neuronal cells were decreased after the induction of cold stress in pregnant mice (Fig. 1 G). The result of the cell cycle exit experiment showed that, after the induction of maternal cold stress, the proliferation of NSCs was increased, and the percentage of NSCs exiting the cell cycle was decreased (Fig. 1 H). The results of in vitro culture of NSCs also showed that cooling inhibited the differentiation into neurons and promoted the proliferation of NSCs (Fig. 1, I and J). We also found that after induction of cold stress, fetal and neonatal mice had a reduced brain size at embryonic day 16 (E16) and postnatal day 2 (P2) compared with that of the control group (Fig. S1, B–E). Together, these results suggest that maternal body temperature as an important physiological indicator plays an important role in regulating embryonic brain development.
Figure 1.
Cooling affects neurogenesis. (A–C) Cold stress was induced at E14, and brains were collected at E16. Embryonic brain sections from rostral to caudal in different temperature conditions were costained with anti-TUJ1 and anti-Nestin antibodies. The quantification shows the relative density of fluorescence. The sketch on the left shows the rostral, central, and caudal sections, and the area selected for the analysis of the phenotype. (n = 3 brains). (D) Cold stress was induced at E14, and brains were collected at E16. BrdU was injected 2 h before brain collection. Embryonic brain sections in different temperature conditions were costained with anti-PAX6 and anti-BrdU antibodies. The bar graph shows the number of BrdU+PAX6+ cells in the VZ/SVZ per 40,000 µm2 (n = 3 brains). (E) Cold stress was induced at E14, and brains were collected at E16. BrdU was injected 2 h before brain collection. Embryonic brain sections in different temperature conditions were costained with anti-Tbr2 and anti-BrdU antibodies. The bar graph shows the number of BrdU+Tbr2+ cells in the VZ/SVZ per 40,000 µm2 (n = 3 brains). (F) Cold stress was induced at E14, and brains were collected at E16. Embryonic brain sections in different temperature conditions were stained with anti-SOX2 antibody. The bar graph shows the number of SOX2 cells in the VZ/SVZ per 40,000 µm2 (n = 3 brains). (G) Cold stress was induced at E14, and brains were collected at E16. Embryonic brain sections in different temperature conditions were costained with anti-SATB2 and anti-CTIP2 antibodies. The bar graph shows the number of SATB2+ (or CTIP2+) cells per 250,000 µm2 (n = 3 brains). (H) Cold stress was induced at E14, and brains were collected at E16. BrdU was injected 24 h before brain collection. Embryonic brain sections in different temperature conditions were costained with anti-Ki67 and anti-BrdU antibodies. The arrows indicate the cells that are BrdU+Ki67−. The bar graph shows the ratio of BrdU+Ki67−/BrdU+ cells (or BrdU+Ki67+/BrdU+ cells) in the VZ/SVZ (n = 3 brains). (I) NSCs were isolated at E12 and then cultured at 37°C or 32°C for 2 d. BrdU was added into the culture medium 1 h before the cells were fixed. In addition, the cells at different temperatures were costained with anti-TUJ1 and anti-BrdU antibodies. The bar graphs represent the ratio of TUJ1+/Dapi+ cells (or BrdU+/Dapi+ cells; n = 3 independent experiments). (J) NSCs were isolated from the E12 brain and then cultured at 37°C or 32°C for 2 d. The cells in different temperature conditions were costained with anti-Ki67 and anti-Map2 antibodies. The bar graphs represent the ratio of Map2+/Dapi+ cells (or Ki67+/Dapi+ cells). (n = 3 independent experiments). Bars: 30 µm (A–C); 20 µm (D, E, and H); 15 µm (F); 50 µm (G); 10 µm (I and J). *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars represent the mean ± SEM.
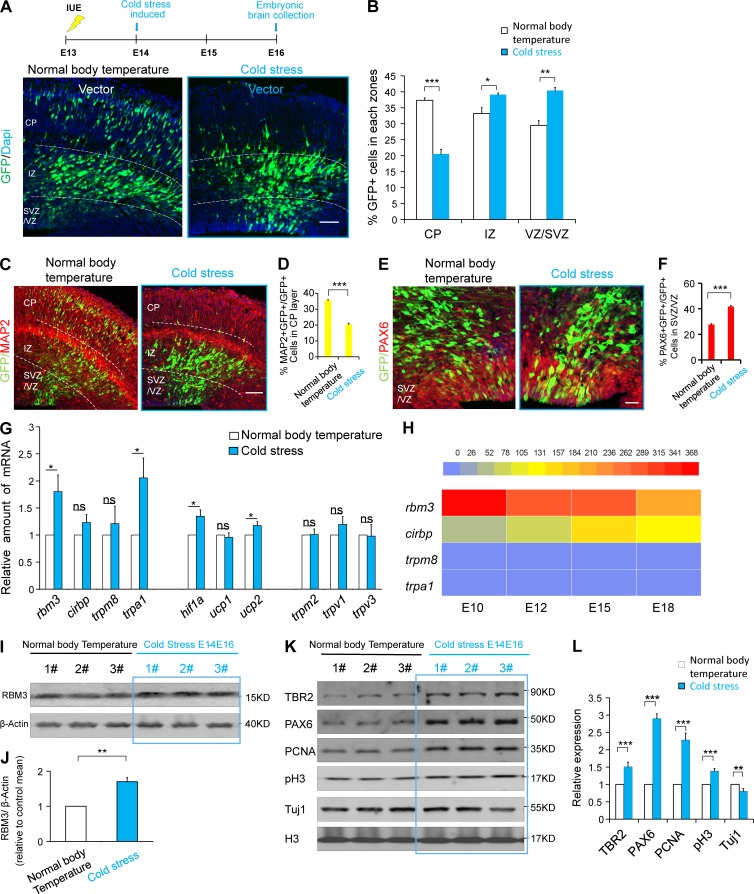
To study the effects of cold stress on embryonic cortical development, in utero electroporation (IUE) was used to introduce a GFP-expression plasmid into the cold stress–induced embryonic cerebral cortex and into the cortex of the normal body temperature group. The results of IUE showed that the induction of maternal cold stress decreased the GFP+ cells in the cortical plate (CP) and increased the GFP+ cells in the VZ/subventricular zone (SVZ) region compare with the control group. (Fig. 2, A and B). The ratio of GFP+MAP2+ neuronal cells in the CP layer decreased, and the ratio of PAX6+GFP+ NSCs increased in the VZ/SVZ region when the maternal cold stress was induced (Fig. 2, C–F). To detect which changes occurred in the gene expression level in the embryonic brain leading to the abnormal development during maternal cold stress, according to the previous studies, the indicated genes clusters were tested, including genes associated with low temperature sensitivity (RBM3 [Danno et al., 1997, Cirbp [Nishiyama et al., 1997], trpm8 [Palkar et al., 2018], and trpa1 [Moore et al., 2018]), metabolic sensitivity (HIF1a [Luo et al., 2018] and Ucp1 and Ucp2 [Margaryan et al., 2017]), and high temperature sensitivity (trpm2, trpv1, and trpv3 [Tan and McNaughton, 2016]; Fig. 2 G). The results showed that the mRNA of the cold-induced protein RBM3 was significantly increased. We also examined the expression of these low temperature sensitive genes (RBM3, Cirbp, trpm8, and trpa1) at different development periods (E10, E12, E15, and E18) and found that RBM3 was the most highly expressed gene in the embryonic brain among the genes analyzed here (Fig. 2 H). Western blotting analysis and immunofluorescence staining of RBM3 showed that the expression of RBM3 (Fig. 2, I and J; and Fig. S1, F and G), NSC markers (Pax6 and Tbr2), and proliferation markers (PCNA and pH3) were increased, but the neuronal marker Tuj1 was decreased (Fig. 2, K and L), when the cold stress was induced. These results indicate that RBM3 is involved in the brain development process under cold stimulation conditions.
Figure 2.
Maternal cold stress can induce the expression of RBM3 and affect the development of the cerebral cortex. (A and B) The results of IUE showed that the induction of maternal cold stress decreased the GFP+ cells in CP and increased the GFP+ cells in the VZ/SVZ region. The GFP-expression plasmid was electroporated into E13 embryonic mouse brains; cold stress was induced at E14, and the phenotype was analyzed at E16. The percentage of GFP-positive cells in each region is shown (n = 3 brains). (C and D) The GFP-expression plasmid was electroporated into E13 embryonic mouse brains, and cold stress was induced at E14; brain sections at the different temperatures were stained by anti-MAP2 antibody at E16. The percentage of GFP and MAP2 double-positive cells in the CP layer is shown (n = 3 brains). (E and F) The GFP-expression plasmid was electroporated into E13 embryonic mouse brains, and cold stress was induced at E14; brain sections at the different temperatures were stained by anti-PAX6 antibody at E16. The percentage of GFP and PAX6 double-positive cells in the VZ/SVZ is shown (n = 3 brains). (G) Cold stress was induced at E14, and the cortex was isolated for mRNA collection at E16. The indicated genes were detected by real-time PCR (n = 3 independent experiments). (H) The indicated cold-related genes’ mRNA levels in the NSCs at different development periods were examined. (I and J) The protein samples from cold stress–induced and normal body temperature embryonic brains were analyzed by Western blotting. The bar graph shows the change in RBM3 under different temperature conditions (n = 6 brains). (K and L) Western-blot analysis showing the neurogenesis-related markers’ changes in embryonic brains from the cold stress–induced and normal body temperature groups. The bar graph shows the changes at the different temperature conditions (n = 6 brains). Bars: 100 µm (A–D); 50 µm (E and F). *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars represent the mean ± SEM.
RBM3 regulates neurogenesis during the maternal cold stress
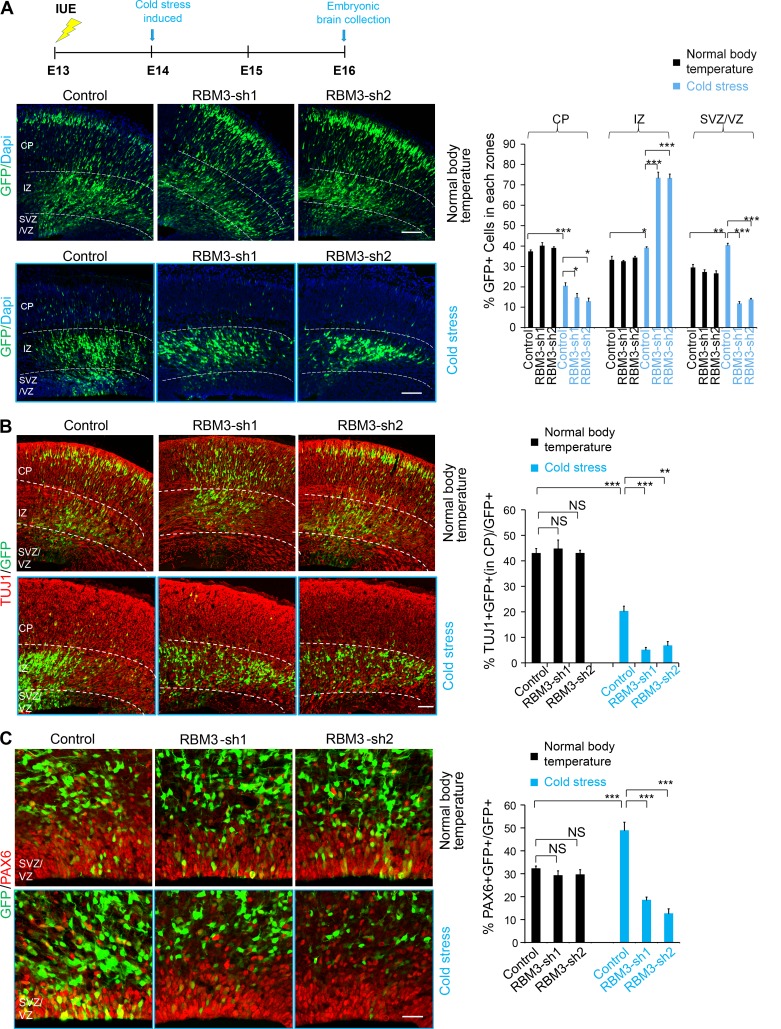
We used IUE to introduce RBM3 shRNA into neural progenitor cells in E13 mouse embryos in pregnant mice under different body temperatures (normal body temperature or induced cold stress). We used immunostaining and Western methods to detect the knockdown efficiency (Fig. S2, A–D). Interestingly, we found that in the induction of maternal cold stress conditions, knockdown of RBM3 led to significant changes in the GFP-positive cell distribution. There was a significant loss of GFP-positive cells in the proliferation VZ/SVZ, and GFP-positive cells in the cortical plate were also reduced (Fig. 3 A). These results indicated that RBM3 was involved in the development of the fetal cerebral cortex in the maternal cold stress state. When the cold stimulation occurred during development, the embryonic cortical development was more seriously affected without RBM3 participation.
Figure 3.
RBM3 regulates NSC proliferation and neuronal differentiation during the maternal cold stress. (A) RBM3 knockdown causes GFP-positive cell positioning changes in the embryonic brain in the cold stress–induced mice. An empty shRNA control or RBM3-KD plasmid was electroporated into E13 embryonic mouse brains, and the phenotype was analyzed at E16. The pregnant mice were cooled using intraperitoneally injected 5′-AMP (0.7 mg/g). The percentage of GFP-positive cells in each region is shown (n = 3 brains). (B) RBM3 knockdown decreases neuronal differentiation. E16 brain sections were stained for TUJ1 after the electroporation of the control or RBM3 knockdown plasmid into the brain at E13; the cold stress was induced at E14, and then the mice were maintained at low temperature or room temperature for 48 h. The percentage of GFP and TUJ1 double-positive cells relative to the total GFP-positive cells is shown as a bar graph (n = 3 brains). Bar: 100 µm. (C) The percentage of GFP and PAX6 double-positive progenitor cells is decreased by RBM3 knockdown in cold stress. The mouse brain was electroporated at E13; the cold stress was induced at E14, and then the mice were maintained at low temperature or room temperature for 48 h. The percentage of GFP+PAX6+ cells relative to the total GFP-positive cells in the VZ/SVZ is shown. (n = 3 brains). Bars: 100 µm (A and B); 50 µm (C). *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars represent the mean ± SEM.
To confirm whether RBM3 knockdown leads to decreased neuronal differentiation when cold stress is induced, we immunostained electroporated brain sections with the neuronal markers TUJ1 and MAP2. The results revealed that knockdown of RBM3 during the maternal cold stress led to a significant decrease in GFP-TUJ1 double-positive cells in CP layer (Fig. 3 B). The quantification of GFP-MAP2 double-positive cells revealed that knockdown of RBM3 led to an obvious decrease in the percentage of MAP2+ neurons (Fig. S2 E). These results demonstrated that RBM3 knockdown in the induced maternal cold stress resulted in a decrease in neuronal differentiation. The result of PAX6 immunofluorescence staining showed that knockdown of RBM3 in cold stress resulted in a decreased proportion of radial glial cells in the VZ/SVZ (Fig. 3 C). To test neural progenitor cell proliferation, pregnant dams were injected with BrdU 2 h before brain collection at E16. The result showed that knockdown of RBM3 during the maternal cold stress caused a reduction of BrdU and GFP double-positive cells versus the control (Fig. S2 F). The result of TBR2 immunofluorescence staining showed that knockdown of RBM3 in cold stress resulted in a decreased proportion of BPs in the VZ/SVZ (Fig. S2 G). IUE under the shorter cold exposure time was preformed to ensure that the abnormal brain development was caused by a primary response to maternal cold stress. Abnormal NSC proliferation and differentiation were also observed with short time cold stress treatment (Fig. S3, A–E).
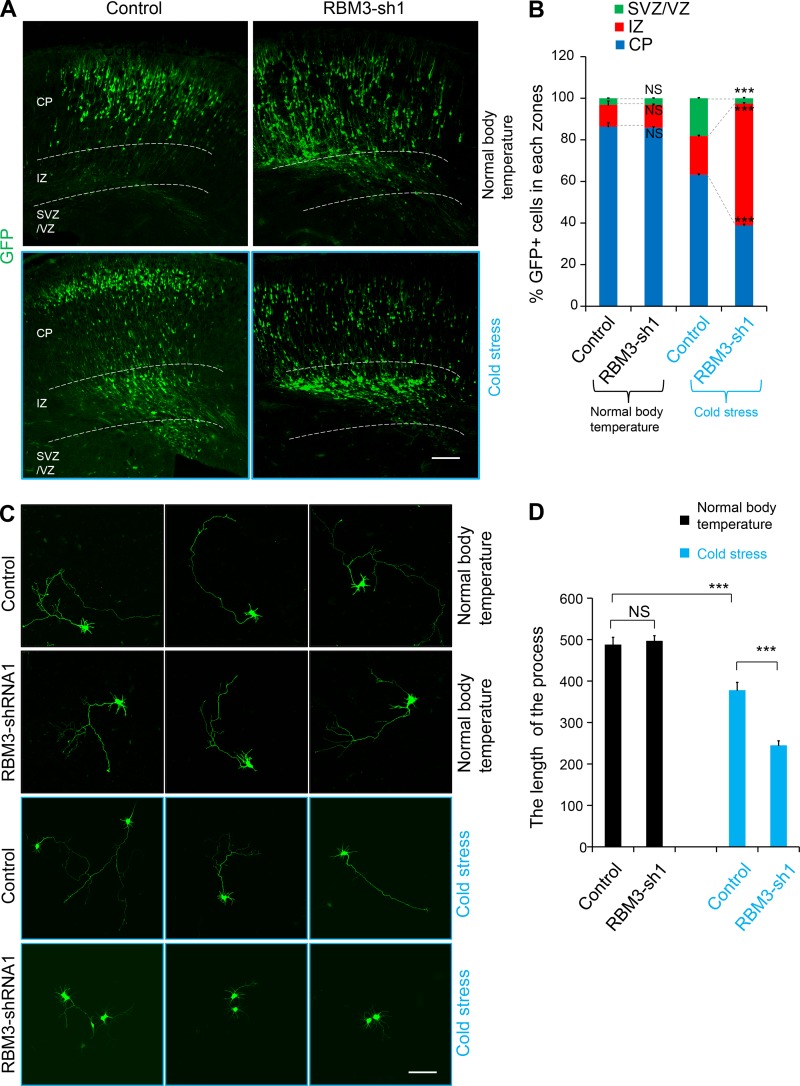
Long-term experimental results (from E13 to P0) showed that the RBM3 knockdown defect phenotype was persistent under the maternal cold stress condition, but there was no obvious phenotype in the normal body temperature group (Fig. 4, A and B). Our results also showed that RBM3 knockdown affected neuronal development in vitro when cells were isolated from the cold stress–induced group, but there was no obvious phenotype for the cells isolated under the normal body temperature condition (Fig. 4, C and D).
Figure 4.
RBM3 knockdown affects the neuronal development in cold stress–induced mice. (A and B) RBM3 knockdown caused GFP-positive cell positioning changes in the cold stress–induced embryonic brain in a long-term experiment. The empty shRNA control or RBM3-KD plasmid was electroporated into E13 embryonic mouse brains, and the phenotype was analyzed at P0. The pregnant mice were cooled at E14. The percentage of GFP-positive cells in each region is shown (n = 3 brains). (C and D) RBM3 knockdown causes abnormal development of GFP-positive cells, which were isolated from the cold stress–induced group. IUE was performed at E13, and then cold stress was induced at E14. Subsequently, the GFP+ cells were isolated from the embryonic brains and cultured for 2 d. The length of the process in each indicated condition is shown (n = 3 independent experiments). Bars: 100 µm. ***, P < 0.001. Error bars represent the mean ± SEM.
Maternal cold stress induces a global change in the transcriptome
To gain a deeper insight into how RBM3 regulates neural progenitor proliferation and differentiation in cold stress, we examined the transcriptome of dividing cells following RBM3 knockdown. We performed IUE using either control or RBM3 shRNA in the cold stress. The GFP+ dividing cells were isolated through fluorescence-activated cell sorting 48 h after the IUE. Following total RNA extraction, transcriptional sequencing was performed to identify differentially expressed genes between control and RBM3-knockdown cells (Fig. 5 A).
Figure 5.
Maternal cold stress induces a global change in the transcriptome and promotes high expression of Yap1 in NSCs. (A) Schematic explaining the steps of the RNA-Seq experiment. (B and C) The volcano plot and heat map analysis showed the gene profiling expression. Gene analysis revealed that the transcript levels of 2,702 genes were up-regulated (455 genes) or down-regulated (2,247 genes) in the RBM3-knockdown and cold stress–induced group. (D) The indicated genes associated with neurogenesis were detected by real-time PCR in the cold stress–induced and normal body temperature groups. The cold stress was induced at E14, and then total mRNA from the embryonic brain was collected for quantitative analysis at E16 (n = 5 independent experiments). (E) The expression of Yap1 downstream genes, including ctgf, cyr61, and ccnd1, were increased in the cold stress–induced brains (n = 5 independent experiments). (F and G) IUE was performed at E13, and then cold stress was induced at E14. Next, GFP+ cells were isolated from the embryonic brains. Real-time PCR (F) and Western blotting (G) were used to detect the expression of YAP (n = 3 independent experiments). (H and I) The protein level of Yap1 was increased when cold stress was induced. Cold stress was induced at E14, and the embryonic cortex was collected at E16 for Western blotting (n = 6 brain samples). (J) Yap1 mRNA was degraded faster in NSCs cultured at 37°C versus 32°C, as measured by an mRNA semiquantitative experiment. Actinomycin D (5 µg/ml) was added into the culture medium to inhibit the transcription. The same amount of mRNA in the indicated time phase was reverse transcribed, and PCR was performed (n = 3 independent experiments). (K) Under the 32°C culture conditions, Yap increased faster at the mRNA level in the NSCs. The NSCs isolated from the E12 brains were cultured in the proliferation medium at 37°C for 24 h and then placed at the indicated temperature for analysis at different time phases (n = 3 independent experiments). *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars represent the mean ± SEM.
Our results revealed that 2,702 genes (including 2,247 down-regulated genes and 455 up-regulated genes; fold change >2) in the RBM3 knockdown samples were differentially expressed compared with control samples when cold stress was induced (Fig. 5, B and C). Together, these data suggest that RBM3 has an essential role in regulating the prenatal genes necessary for temperature-related embryonic development. In addition, cold stress did not change the expression of certain important neuronal migration-related genes, and further study revealed that the abnormal phenotype induced by RBM3 knockdown was also not caused by neuronal migration (Fig. S3, F and G).
Maternal cold stress promotes high expression of Yap1 in NSCs
To determine which genes were changed in the induction of maternal cold stress that was associated with the NSC proliferation and differentiation in our experiment, we detected some of the NSC proliferation- and differentiation-related genes. The results showed that Yap1 and its downstream genes (ctgf, cyr61, and ccnd1) were significantly up-regulated when cold stress was induced (Fig. 5, D and E). After checking the RNA-Seq data, we found that there was 60% decrease for Yap1 expression in RBM3 knockdown sample under the cold stress–induced condition. Real-time PCR and Western blotting were used for further confirmation of YAP reduction (Fig. 5, F and G).
Western blot analysis showed that the expression of YAP1 was increased in the cold stress embryonic cortex (Fig. 5, H and I). To investigate whether low temperature stress affects the stability of Yap1 mRNA and thus affects the expression of YAP1, we checked the half-life of Yap1 mRNA at 37°C and 32°C in NSCs by adding actinomycin D, which is an intercalating transcription inhibitor. The result of the mRNA semiquantitative experiment showed that the half-life of Yap1 mRNA was extended at the 32°C condition relative to the 37°C condition (Fig. 5 J). In addition, under the 32°C culture condition, Yap1 increased faster at the mRNA levels in the NSCs (Fig. 5 K). These results indicate that Yap1 may respond to cold stimuli and act as a potential cold-induced protein.
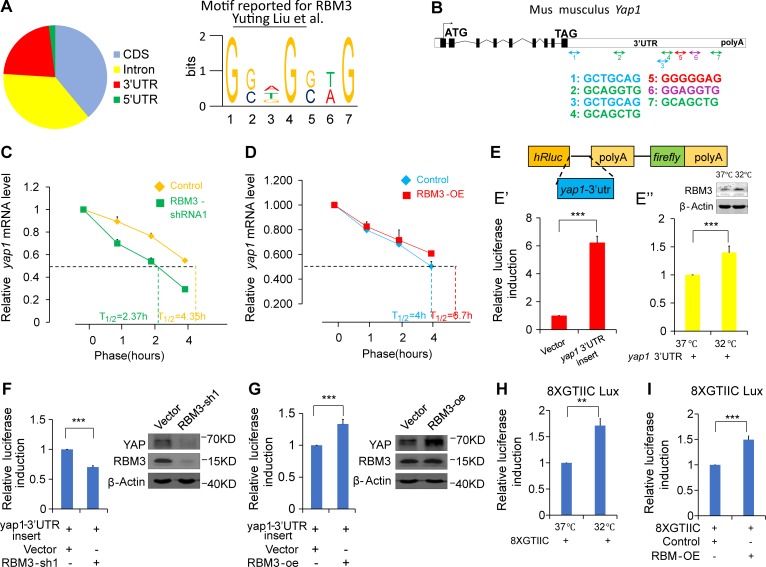
RBM3 promotes YAP expression by binding to the Yap1 mRNA 3′UTR and increasing its stability
Knowing that the expression of Yap1 and RBM3 may have a certain correlation at low temperatures, we wanted to determine the intrinsic molecular mechanism. The RBM3 binding motif has been reported in the previous study (Liu et al., 2013; Fig. 6 A), and we found that RBM3 could bind to the 3′UTR to increase the stability of some mRNAs. After analyzing the 3′UTR region of the mouse Yap1 mRNA, we found that there are seven RBM3 binding sequences in the 3′UTR of Yap1 (Fig. 6 B). When RBM3 was knocked down in the NSCs, the half-life of Yap1 mRNA was reduced (Fig. 6 C), but the half-life was extended when the RBM3 was overexpressed (Fig. 6 D) in the NSCs maintained at 32°C. To further verify that RBM3 stabilizes the Yap1 mRNA though binding to the Yap1-3′UTR region, we constructed a luciferase plasmid containing the full-length Yap1-3′UTR region. At 32°C, there was a sixfold increase in the translation of the message containing the Yap1-3′UTR, compared with the empty vector control (Fig. 6 E’). Further, the expression of RBM3 was increased by culturing cells at 32°C, which resulted in a 1.4-fold increase in translation of the message compared with that of cells that were cultured at 37°C (Fig. 6 E”). To further prove that RBM3 mediates the stability of the Yap1′ mRNA, knockdown of RBM3 was performed in N2A cells, which were cultured at 32°C. The result showed that the activity of the Yap1-3′UTR–containing luciferase was decreased (Fig. 6 F). RBM3 overexpression increased the activity of the Yap1-3′UTR–containing luciferase (Fig. 6 G). To show the specificity of the YAP pathway activation, a cell model of cooling using an 8×GTIIC-luciferase reporter was used to show specific YAP activity. The result showed that the relative activity of the 8×GTIIC-luciferase was increased in cells that were cultured at 32°C versus the cells cultured at 37°C (Fig. 6 H). The overexpression of RBM3 induced the increase in the relative activity of the 8×GTIIC-luciferase (Fig. 6 I). These results suggest that the 3′UTR region of the Yap1 mRNA and the low temperature–induced high expression of RBM3 improved the stability of the RNA. Collectively, the data support a role for RBM3 in controlling the Yap1 expression through binding to its 3′UTR, and Yap1 acts downstream of RBM3 in regulating embryonic cortical development.
Figure 6.
RBM3 promotes the stability of Yap1 mRNA by binding to the 3′UTR of Yap1 mRNA. (A) Schematic showing the motif that has been published for the mouse RBM3 mRNA-recognition motif. (B) Schematic showing there were seven sequences located in the Yap1-3′UTR region matching the mouse RBM3 recognition motif by sequence alignment. (C and D) The half-life of Yap1 mRNA was prolonged by RBM3. The results showed that when RBM3 was knocked down in the NSCs, the half-life of Yap1 mRNA was shortened (C). After overexpression of RBM3, the Yap1 half-life was extended (D). The quantification shows the relative Yap1 mRNA at each time point, compared with the level of Yap1 mRNA at time zero (taken as 1). In addition, the half-life of Yap1 mRNA was calculated and is shown as the t1/2 value (n = 3 independent experiments). (E) The full-length Yap1-3′UTR was cloned into a pisCheck2-vector. HEK293 cells were transfected with a plasmid containing the Yap1-3′UTR or the control empty vector and then cultured at 32°C. The relative luciferase activity was detected 48 h after the transfection and is presented as a bar graph (E’). HEK293 cells were transfected with a plasmid containing the Yap1-3′UTR or the control vector and then were cultured at 32°C or 37°C (E”). The relative luciferase activity was detected 48 h after the transfection and is presented as a bar graph (n = 3 independent experiments). (F) N2A cells were transfected with vector and Yap1-3′UTR luciferase plasmids or (RBM3-sh1 and Yap1-3′UTR luciferase plasmids), and cultured at 32°C. The relative luciferase activity was detected 48 h after the transfection and is presented as a bar graph (n = 3). (G) N2A cells were transfected with vector and Yap1-3′UTR luciferase plasmids (or RBM3-OE and Yap1-3′UTR luciferase plasmids) and then cultured at 32°C. The relative luciferase activity was detected 48 h after the transfection and is presented as a bar graph (n = 3). (H) HEK293 cells were transfected with a plasmid containing the 8XGTIIC luciferase reporter and then cultured at 32°C or 37°C. The relative luciferase activity was detected 48 h after the transfection and is presented as a bar graph (n = 4). (I) HEK293 cells were transfected with the indicated plasmid combinations. The relative luciferase activity was detected 48 h after the transfection and is presented as a bar graph (n = 4). **, P < 0.01; ***, P < 0.001. Error bars represent the mean ± SEM.
RBM3 knockout affects embryonic neurogenesis under cold stress
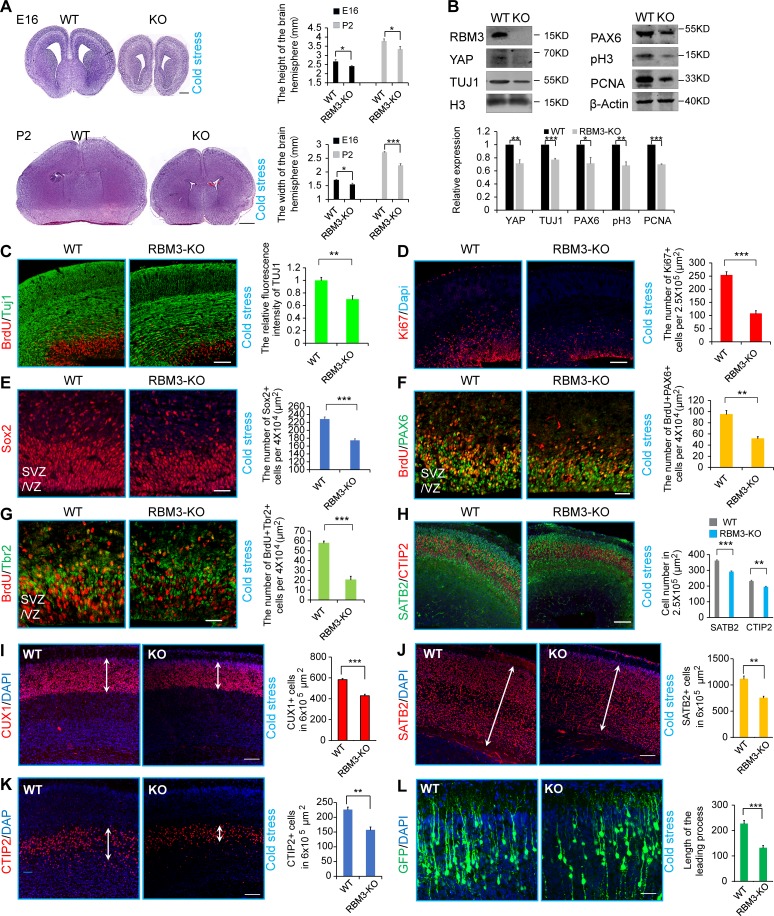
To confirm the phenotype of the RBM3 knockdown, we constructed RBM3 knockout mice through a CRISPR-Cas9 system. No expression of RBM3 was detected in knockout mice (Fig. S4, A–D). In the cold stress–induced embryonic mice cortex, histological analysis showed the changes between the RBM3 knockout and the WT (Fig. 7 A). Further Western blot analysis showed that some neurogenesis associated markers changed between the RBM3 knockout and the WT when cold stress was induced (Fig. 7 B).
Figure 7.
RBM3 knockout decreases the differentiation and proliferation of NSCs under maternal cold stress. (A) H&E staining showed the difference between the RBM3 knockout mouse brain and the WT mouse brain at E16 and P2. Maternal cold stress was induced at E14, and then the brain was collected at E16 or P2 for H&E staining. The bar graph shows the height or width of the indicated brains at different stages (n = 3 brains). (B) Western blot analysis showed that the indicated proliferation and differentiation markers were reduced in cold stress–induced RBM3 knockout embryos. Maternal cold stress was induced at E14 and then maintained at the low temperature until E16 for Western analysis (n = 3 experiments). (C) The ratio of Tuj1-positive cells was reduced in the RBM3 knockout in cold stress. Maternal cold stress was induced at E14, and then the embryonic brain was collected at E16. The bar graph shows the fluorescence intensity of Tuj1 (n = 3 brains). (D) The number of Ki67-positive NSCs was reduced in the cold stress–induced and RBM3 knockout brain sections. The bar graph shows the number of Ki67+ cells in the VZ/SVZ per 250,000 µm2 (n = 3 brains). (E) The number of SOX2-positive NSCs was reduced in the cold stress–induced and RBM3 knockout brain sections. The bar graph shows the number of SOX2+ cells in the VZ/SVZ per 40,000 µm2 (n = 3 brains). (F) The number of BrdU and PAX6 double-positive intermediate progenitors was decreased in cold stress–induced RBM3 knockout brains. The cold stress was induced at E14 and then maintained at the cold temperature until E16. The bar graph shows the number of BrdU+PAX6+ cells in the VZ/SVZ per 40,000 µm2 (n = 3 brains). (G) The number of BrdU and TBR2 double-positive intermediate progenitors was decreased in the cold stress–induced RBM3 knockout brains. The cold stress was induced at E14 and then maintained at the cold temperature until E16. The bar graph shows the number of BrdU+TBR2+ cells in the VZ/SVZ per 40,000 µm2 (n = 3 brains). (H) The number of SATB2- (or CTIP2-) positive neurons was reduced in the cold stress–induced and RBM3 knockout brain sections. The bar graph shows the number of SATB2- (or CTIP2-) positive cells in the VZ/SVZ per 250,000 µm2 (n = 3 brains). (I–K) The number of CUX1-, SATB2-, and CTIP2-positive cells was reduced in the RBM3 knockout in cold stress at P2. The maternal cold stress was induced at E14, and then the brain was collected at P2. The bar graph shows the number of CUX1-, SATB2-, and CTIP2-positive cells (n = 3 brains). (L) RBM3 knockout caused abnormal development of GFP-positive cells at P2 when maternal cold stress was induced. IUE was performed at E13, and then cold stress was induced at E14. After that, the brains were collected at P2. The length of the leading process in each indicated condition is shown (n = 3 independent experiments). Bars: 500 µm (A, E16); 750 µm (A, P2); 50 µm (C, H, and L) 20 µm (D–G); 100 µm (I–K). *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars represent the mean ± SEM.
Our results showed that the TUJ1-positive cells in the RBM3 knockout mice at E16 was decreased compared with the WT mice (Fig. 7 C). The immunostaining results showed that the proliferation markers of Ki67, SOX2, PAX6, and TBR2 were decreased (Fig. 7, D–G), and neuronal markers of STAB2 and CTIP2 were also decreased (Fig. 7 H). In addition, apoptosis was not significantly affected by RBM3 knockout (Fig. S4 E).
In the cold stress–induced RBM3 knockout mice, we also found that the neuronal differentiation was decreased at P2 (Fig. 7, I–K); RBM3 knockout affected the neuronal development in vivo when maternal cold stress was induced (Fig. 7 L). However, there was no significant difference between WT and knockout in the normal body temperature group (Fig. S4, F–H).
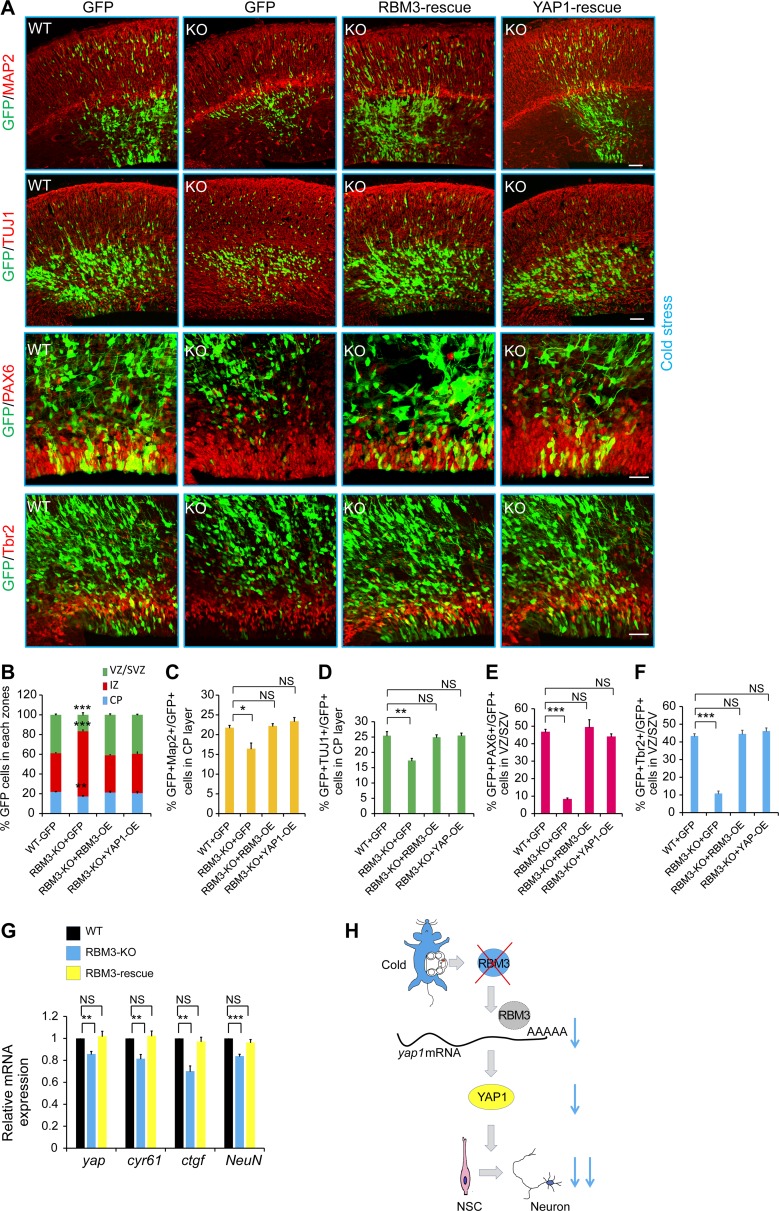
To confirm the phenotype, IUE was used in the E13 knockout embryo, and the phenotype was analyzed 72 h later. At normal body temperature, the RBM3 knockout mice showed no difference in GFP distribution compared with the WT mice. However, the RBM3 knockout phenotype was similar to that of the RBM3 knockdown in the cold stress–induced condition. In the cold stress–induced RBM3 knockout mice, the number of GFP cells was decreased in the VZ/SVZ and CP. Analysis of neuronal differentiation showed that the MAP2+ (or TUJ1+) cells in the CP of the RBM3 knockout embryonic mice were also decreased in cold stress. The PAX6 (or Tbr2) immunofluorescence results showed that the proliferation of the NSCs was decreased in the RBM3 knockout mice during cold stress. When RBM3-OE or YAP1-OE was electroporated into the RBM3 knockout mice, the phenotype of the GFP+ cell distribution and the ratio of GFP+MAP2+ (or GFP+TUJ1+, GFP+PAX6+, and GFP+Tbr2+) cells could be partly rescued (Fig. 8, A–F). To gain a deeper insight into how RBM3 regulates neural progenitor proliferation and differentiation in cold stress, we examined the transcriptome of the embryonic cortex following RBM3 knockout in the maternal cold stress–induced condition. These data also suggested that RBM3 had an essential role in regulating gene expression during embryonic development (Fig. S4, I and J). RBM3 overexpression could rescue the expression of Yap and its downstream molecules (cyr61 and ctgf) in the RBM3 knockout NSCs, which were isolated at E13 and cultured at 32°C (Fig. 8 G). The number of TUJ1+ (or MAP2+) neurons in CP layer were also decreased at P30 in the RBM3 knockout and maternal cold stress–induced mice cortex (Fig. S5, A and B). In addition, YAP1 knockdown in the maternal cold stress–induced mice had the same phenotype as the RBM3 knockout (Fig. S5, C and D). These results further confirm that RBM3 is involved in the regulation of the brain development process when maternal cold stress was induced (Fig. 8 H).
Figure 8.
The abnormal neurogenesis process caused by RBM3 knockout in the maternal cold stress group could be rescued. (A–F) RBM3 knockout caused GFP-positive cell positioning changes in the cold stress–induced embryonic brain, and the RBM3 or Yap1 overexpression could partially rescue this phenotype. The indicated plasmid was electroporated at E13, and the phenotype was analyzed at E16. The pregnant mice were cooled by using 5′-AMP injection. The percentage of GFP-positive cells in each region is shown (B), and the GFP+Map2+ cells in the CP (C), GFP+TUJ1+ cells in the CP (D), GFP+PAX6+ neural progenitors in VZ/SVZ (E), or GFP+Tbr2+ neural progenitors in VZ/SVZ (F) were calculated (n = 3 brains). Bars: 50 µm (A). (G) The expression of Yap and its downstream molecules (cyr61 and ctgf) could be rescued by RBM3 overexpression in the RBM3 knockout NSCs. The WT and RBM3 knockout NSCs were isolated at E13, and the RBM3-overexpression virus was used to rescue the level of RBM3. The indicated gene expression was detected by real-time PCR (n = 3 independent experiments). (H) Schematic diagram: cold stress affects brain development, and RBM3 deficiency reduces neuronal differentiation. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars represent the mean ± SEM.
Discussion
Cold stress is a common physiological phenomenon (Byard and Bright, 2017; Moler et al., 2017), and it has been reported that deep cold stress causes neurological dysfunction (Gatti et al., 2017). However, its roles in pregnancy are little studied. Our results show that induction of maternal cold stress can lead to abnormal development of embryonic mice, especially with respect to NSC development. Here, we report that RBM3 is one of molecules that mediates the effects of cold stress on brain development. Our results show that cold stress affects the expression of RBM3 in the embryonic cerebral cortex.
During the cold stress, RBM3 is induced. RBM3 has been reported to be associated with neuroprotective functions, and RBM3 regulates protein expression by selectively binding to different poly(A)-tail sites during the process of circadian rhythms (Liu et al., 2013). However, the role of RBM3 in embryonic brain neurogenesis was unclear. Our results showed that RBM3 in the embryo may regulate the proliferation and differentiation of NSCs during maternal cold stress by binding to the Yap1-3′UTR, promoting Yap1 expression by stabilizing its mRNA. A maternal hypothermic environment will lead to fetal cerebral cortex development abnormalities. In addition, when RBM3 was knocked down in the cold stress–induced embryonic cortex, the neuronal differentiation and proliferation of NSCs were reduced. We obtained the same phenotype in the RBM3 knockout mice. These results confirm that RBM3 is involved in the development of the cerebral cortex at cold stress.
To explain the intrinsic causes of these results, we screened molecules and found that Yap1 may be the downstream target of RBM3 under cold stress conditions. We observed that the half-life of the Yap1 mRNA in NSCs was longer at 32°C versus 37°C. When RBM3 was knocked down in cultured NSCs at 32°C, the half-life of the Yap1 mRNA was shortened. In contrast, when RBM3 was overexpressed in the NSCs cultured at 32°C, the half-life of the Yap1 mRNA was longer. Our results showed that a low temperature induced a high expression of RBM3, which could enhance the expression of Yap1 by stabilizing its 3′UTR region. In addition, the results of our RNA sequencing analysis showed that the induction of cold stress can affect the development of the fetal cerebral cortex. Interestingly, our results show that Yap1 may also be a potential cold shock protein, and the abundance of Yap1 mRNA and protein was changed at low temperature. These results may enhance our understanding of Yap1.
However, RBM3 knockdown or knockout did not affect the neurogenesis process when the body temperature was normal. These differences may be caused by cooling-induced transcriptome reprogramming. Under the condition of cold stimulation, RBM3 was highly expressed, which may have increased the stability of intracellular protein transcription. It may be explained that RBM3, as an RNA-binding protein, could bind to different regions of mRNAs, which may affect the normal development process, including the development of the cerebral cortex.
RBM3-overexpression and YAP-overexpression partially rescue the RBM3 knockout phenotype when the maternal cold stress was induced. The RBM3 pathway is likely to be one of the effector pathways, which are important for responses to cold stress. It is possible that there are other pathways, which plays function in maternal cold stress condition.
In the IUE experiment, the neurons are still stuck in the intermediate zone and do not properly differentiate or migrate in the absence of RBM3 when the maternal cold stress was induced. We detected some migration related genes (lis1, rac1, dcx, cdh2, and cdk5) expression and did not find significant expression change of these genes. However, we cannot exclude the possibility that cold stress affects migration.
In conclusion, we report that, following the induction of maternal cold stress, there are changes in the expression of specific cold-inducible proteins, including the reported cold shock protein, RBM3, and the never-reported protein Yap1. Interestingly, we show that Yap1 is a downstream target of RBM3. Additionally, our results suggest that RBM3 and Yap1 are involved in the brain development under cold stress.
Materials and methods
Mice
ICR pregnant mice were purchased from Vital River. The mice were maintained in standard conditions (Mice were housed in a 12 h:12 h light:dark cycle with lights on at 7 a.m.). All mice were taken care according to the Guide for the Care and Use of Laboratory Animals. All animal experiments were approved by the Animal Committee of the Institute of Zoology, Chinese Academy of Sciences. The RBM3 knockout mice were generated by using CRISPR-Cas9 system.
RBM3 knockout mice generation
RBM3 knockout mice were generated using the CRISPR-Cas9 system. In brief, upstream-gRNA1: 5′-CGGGCGTGCTGCATCGGAACGGG-3′, upstream-gRNA2: 5′-5GCGGGCGTGCTGCATCGGAACGG-3′; downstream-gRNA1: 5′-GTATTCTTGAGATTGGTACATGG-3′; and downstream-gRNA2: 5′-ATGTGGCTTGGATATATCAAAGG-3′ were cloned into pUC57-kan-T7-gRNA vector. After transcription and purification, the gRNAs and Cas9 expressing vector were microinjected into fertilized one-cell embryos. Then the injected zygotes were implanted into the host dams. When the offspring were born, the genotyping PCR experiments were done, and the PCR production were sequenced to identify the mutations. Following genetic testing, mice with 1,980-bp deletion were screened and bred for the sequential experiments. The genotyping primers used for RBM3 knockout mice were RBM3-269F: 5′-CTCGTTCCCCACCTCACTTC-3′ and RBM3-2575R: 5′-GAGTAGCGGTCATAGCCACC-3′. The PCR products were ∼2,000 bp for WT and 200 bp for RBM3 knockout mice.
Plasmid constructs
The sequences for small hairpin RNA are as follows: RBM3-shRNA1, 5′-GTTGATCATGCAGGAAAGTCT-3′; RBM3-shRNA2, 5′-GTCGTCCTGGAGGATATGGAT-3′; YAP-shRNA1, 5′-CGGTTGAAACAACAGGAATTA-3′.
The shRNAs were subcloned into the pSicoR-GFP vector. RBM3 and YAP cDNA was amplifed by PCR and subcloned into pCDH-CMV-GFP vector.
Induction of cold stress
The pregnant mice were cooled using 5′-AMP as described. Freshly prepared 5′-AMP (Sigma) was injected intraperitoneally (0.7 mg/g). Control mice were injected with saline. Mice were maintained at room temperature 1 h until core body temperature decreased to 33°C. Subsequently, mice were transferred to a refrigerator (5° C) and core body temperature lowered to ∼30°C (Peretti et al., 2015), and then the pregnant mice were housed 24 h at 10°C and 24 h at 5°C (Fischer et al., 2017). For short-term cold experiment, after the maternal body temperature dropped to 30°C, the pregnant mice were housed 4 h at 10°C, and then they were housed in the room temperature. During the experiment we observe the physiological status of pregnant mice every hour to ensure the normal survival of pregnant mice.
IUE
The pregnant mice were anesthetized by given an injection of pentobarbital sodium, and the uterine horns were exposed. Recombinant plasmid mixed with Venus-GFP at a 3:1 molar ratio, and fast green (2 mg/ml; Sigma) was microinjected into the fetal brain ventricles. And the mixed plasmid was electroporated into the brain ventricle cells. After electroporation, the pregnant mice were sacrificed at different time point for phenotype analysis. The fetal brains were fixed in 4% PFA overnight and then dehydrated in 30% sucrose at 4°C. For knockdown tissue RNA-sequence, IUE of dividing cells with either control or RBM3 shRNA was used. GFP-containing tissue from three to five embryos were collected under the microscope, followed by FACS sorting of GFP-positive cells. The total RNA of these cells was collected by using trace RNA extraction kit for RNA-seq.
Cell culture
293FT (or N2A) cells were cultured in DMEM that contained 10% FBS, non-essential amino acid, and penicillin/streptomycin.
Lentiviral package DNA and the core DNA was transfected into HEK293FT by GenEscort (Wisegen). The supernatant containing the virus was collected at 24, 48, and 72 h after transfection, and the cell debris was removed by centrifugation at 3,000 rpm for 5 min.
NSCs were isolated from E12 cortex. In brief, E12 brain cortex tissue around the ventricle was dissected out and digested in papain (20 U/mg; Worthington) for 5 min at 37°C to acquire cells suspension. Cells suspension was washed three times by NSCs culture medium (NeuroBasal/DMEM/F12 with penicillin-streptomycin, GlutaMAX, nonessential amino acids, B27 supplement [2%], bFGF [5 ng/ml], and EGF [5 ng/ml]) to remove the papain. Cell suspensions were then filtered through a 40-µm strainer. Acquired cells were seeded in plates coated with Poly-d-Lysine and Laminin before cell culture. 1.5 million cells per well were for a 6-well plate, while 5,000 cells per well were for a 24-well plate. Cells were infected with lentivirus by the addition of 2 µg/ml polybrene to improve the efficacy of infection. 8 h later, the proliferation medium was changed into a differentiation medium, which consisted of low glucose DMEM (Gibco), 2% B27, and 1% FBS (Invitrogen).
BrdU labeling
For NPC proliferation analysis, BrdU (50 mg/kg) was administered to pregnant female mice for 2 h before harvesting the embryonic brains. For the birth dating experiment, BrdU (50 mg/kg) was administered to pregnant mice 24 h after electroporation, and the embryonic brains were collected 3 d later for phenotype analysis. For the cell-cycle exit experiment, BrdU (50 mg/kg) was administered to pregnant mice for 24 h before the mouse brains were separated and processed. Then, the brains were costained with anti-Ki67 and anti-BrdU antibodies for analysis.
Western blotting
Cells were lysed with radioimmunoprecipitation assay in combination protease inhibitor on ice for 5 min, and the cell debris was eliminate by centrifuged at 4°C for 5 min. The BCA method was used for measuring the concentration of protein. The protein samples were loaded onto SDS-PAGE gel for electrophoresis, and the bands were transferred to nitrocellulose or polyvinylidene fluoride membranes. The membranes were blocked in 5% nonfat milk in PBST (PBS with 0.1% Tween-20) for 1 h at room temperature, and incubated with primary antibody at 4°C. The different secondary antibodies were used to visualize the bands.
Immunostaining
Immunostaining for cultured cells or brain slices was performed according to the following procedure: the samples were washed with PBST (1% Triton X-100 in 1 M PBS), fixed in 4% PFA, blocked by 5% BSA (in 1% PBST) for 1 h, incubated with primary antibodies overnight at 4°C, and visualized using fluorescence-labeling secondary antibodies.
RNA half-life test
The freshly prepared Actinomycin D (5 µg/ml, Sigma; Jiang et al., 2016) was added in cultured cells to stop transcription. After, cells were collect at different time point (0, 1, 2, and 4 h). RNA was extracted using TRIzol reagent (Invitrogen Life Technologies) according to the manufacturer's instructions. Total mRNA was reversed into cDNA using RNA reverse-transcription kit. Real-time reverse transcription (RT) PCR was then performed as previously described; mouse Yap1 mRNA levels were determined by qRT-PCR and normalized by β-Actin. The expression of the Yap mRNA at the starting point was taken as 1. t1/2 was calculated from RNA level trend plot.
Antibodies
Antibodies used were as follows: anti-β III Tubulin (MAB1637; Millipore and T2200; Sigma), anti-PCNA (SC7907; Santa Cruz), anti-PAX6 (AB2237; Millipore), anti-TBR2(AB23345; Abcam), anti-Histone H3 (4499; Cell Signaling Technology), anti–β-ACTIN (20536-1-Ap; Proteintech), anti-NESTIN (MAB353; Millipore), anti-Flag (F1804; Sigma), anti–phospho-Histone H3(Ser10) (3377; Cell Signaling Technology), anti-BrdU (AB6326; Abcam), anti-RBM3 (14363-1-AP; Proteintech), anti-Map2 (MAB3418; Millipore), anti-Ki67 (ab15580; Abcam), anti-TUJ1(MAB1637; Millipore), anti-YAP (4912; Cell Signaling Technology), anti-SOX2 (3728; Cell Signaling Technology), anti-CTIP2 (ab18465; abcam), and anti-SATB2 (ab51502; Abcam).
Real-time PCR
The total RNA was extracted by using the TRIzol (Invitrogen) according to the instructions. FastQuant RT kit (with DNase [TIANGEN]) was used to get first-strand cDNA. Real-time PCR was performed on the real-time PCR machine (ABI).
Confocal imaging and statistical analysis
All confocal images were acquired with the Zeiss LSM 780 microscope in room temperature and analyzed with Photoshop CS4 (Adobe); Detector is PMT; Plan-Apochromat 10/0.45 or Plan-Apochromat 20/0.8 was used; 50% Glycerin was applied as imaging medium; The software for image acquisition and processing was ZEN 2010.
The fluorescence density is calculated using the ImageJ and Zeiss ZEN2012 blue software; for fluorescence density detecting, we use the same concentration of antibodies for different samples, the same incubation time, the same confocal microscope fluorescence excitation light intensity, and the fluorescence density of the control group is taken as one.
Statistical analyses were performed using Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). All bar graphs in the figures are shown as the means ± SEM.
For transcriptome analysis, the IUE was used to introduce the RBM3 knockdown GFP plasmid or control GFP plasmids into the E13 embryonic cortex. Then, GFP-positive NSCs were isolated from E15 mice by FACS. RNA used for global transcriptome analysis were extracted from these NSC and sequenced by Hiseq 2500. Significantly differentially expressed genes were identified when we compared normalized reads count between groups with P < 0.05 and log2 fold change > 1.
Primers for real-time PCR
Primers for RT-PCR are listed in Table 1.
Table 1. Primers used for RT-PCR.
| Primer | Forward or reverse | Sequence (5′–3′) |
|---|---|---|
| RBM3 | Forward | ATGCAGGAAAGTCTGCCAGG |
| Reverse | TCTCTAGACCGCCCATACCC | |
| CIRBP | Forward | TCTCCGAAGTGGTGGTGGTA |
| Reverse | CATCATGGCGTCCTTAGCGT | |
| trpm8 | Forward | TATGAGACCCGAGCAGTGGA |
| Reverse | GGCTGAGCGATGAAATGCTG | |
| trpa1 | Forward | GGCAATGTGGAGCAATAGCG |
| Reverse | TGAGGCCAAAAGCCAGTAGG | |
| Hif1a | Forward | AGGATGAGTTCTGAACGTCGAAA |
| Reverse | GGGGAAGTGGCAACTGATGA | |
| ucp1 | Forward | CGTCCCCTGCCATTTACTGT |
| Reverse | CCCTTTGAAAAAGGCCGTCG | |
| ucp2 | Forward | GGCCTCTGGAAAGGGACTTC |
| Reverse | GACCACATCAACAGGGGAGG | |
| trpv1 | Forward | TCACCGTCAGCTCTGTTGTC |
| Reverse | GATCATAGAGCCTTGGGGGC | |
| trpv3 | Forward | GACCCATAAGACGGACAGCA |
| Reverse | ACCGACGTTTCTGGGAATTCAT | |
| trpm2 | Forward | CTGACTGTCACCCTGCTCTG |
| Reverse | TCTGTGTGTTCCTGCACCTC | |
| Yap1 | Forward | TTCGGCAGGCAATACGGAAT |
| Reverse | GCATTCGGAGTCCCTCCATC | |
| ctgf | Forward | AAGGACCGCACAGCAGTTG |
| Reverse | GGCTCGCATCATAGTTGGGT | |
| cyr61 | Forward | CTTTTCAACCCTCTGCACGC |
| Reverse | CTCGTGTGGAGATGCCAGTT | |
| il10 | Forward | CCAGCTGGACAACATACTGC |
| Reverse | TTCTGGGCCATGCTTCTCTG | |
| ngn3 | Forward | CTCACCATCCAAGTGTCCCC |
| Reverse | AGTCACCCACTTCTGCTTCG | |
| Ascl1 | Forward | GGAACTGATGCGCTGCAAAC |
| Reverse | GTGGCAAAACCCAGGTTGAC | |
| Brn2 | Forward | GTTTGCTCTATTCGCAGCCG |
| Reverse | TCTGCATGGTGTGGCTCATC | |
| Bdnf | Forward | CCGGTATCCAAAGGCCAACT |
| Reverse | CTGCAGCCTTCCTTGGTGTA | |
| Ngf | Forward | ACAGCCACAGACATCAAGGG |
| Reverse | TGACGAAGGTGTGAGTCGTG | |
| Fgf13 | Forward | CGCGAGAAATCCAATGCCTG |
| Reverse | TTCTGCGCCTCTTCTTGGAG | |
| Axin | Forward | CCCGGAGCTATTCCGAGAAC |
| Reverse | TCTCAGCGTCCTCTGTGGTA | |
| actb | Forward | GCAAGTGCTTCTAGGCGGAC |
| Reverse | AAGAAAGGGTGTAAAACGCAGC | |
| ccnd1 | Forward | CCCTGGAGCCCTTGAAGAAG |
| Reverse | AGATGCACAACTTCTCGGCA | |
| cdkn2a | Forward | GTCGCAGGTTCTTGGTCACT |
| Reverse | CATGTTCACGAAAGCCAGAGC |
Luciferase assays
Luciferase assays were performed in HEK293 or N2A with the Yap1-3′UTR insert psi-Check2 luciferase reporter in different conditions as previously reported (Jiang et al., 2016).
8×GTIIC-lux Luciferase assays were performed in HEK293 cells with the YAP/TAZ-responsive reporter 8×GTIIC-Lux according to previous study (Dupont et al., 2011).
The indicated gene expression vectors were transfected into the cells, and the sample was collected 48 h later; the Dual-Luciferase Reporter Assay System (Promega) was used to detect the expression of the luciferase.
The GEO accession numbers
The GEO accession nos. for the cold stress–induced control and RBM3-knockdown transcriptome sequencing data reported in this paper is GSE104300. The GEO accession nos. for the cold stress–induced WT and RBM3 knockout transcriptome sequencing data reported in this paper is GSE109362.
Online supplemental material
Fig. S1 shows that induction of maternal cold stress can affect the development of offspring. Fig. S2 shows that RBM3 regulates NSC proliferation and neuronal differentiation during the maternal cold stress. Fig. S3 shows that RBM3 regulates NSC proliferation and neuronal differentiation during short-term cold stress (4-h exposure). Fig. S4 shows that RBM3 knockout does not affect the neuronal differentiation in the normal body temperature group. Fig. S5 shows that RBM3 knockout decreases the differentiation under maternal cold stress conditions at P30.
Supplementary Material
Acknowledgments
This work was supported by grants from Chinese Academy of Science Strategic Priority Research Program (XDA16020602), the National Natural Science Foundation of China (31730033 and 31621004), the National Key Basic Research Program of China (2015CB964501 and 2014CB964903), and the K.C. Wong Education Foundation.
The authors declare no competing financial interests.
Author contributions: W. Xia and J. Jiao designed the research; W. Xia and L. Su performed the research, analyzed the data, and wrote the manuscript; J. Jiao supervised the project and obtained funding support.
References
- Aragona M., Panciera T., Manfrin A., Giulitti S., Michielin F., Elvassore N., Dupont S., and Piccolo S.. 2013. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 154:1047–1059. 10.1016/j.cell.2013.07.042 [DOI] [PubMed] [Google Scholar]
- Byard R.W., and Bright F.M.. 2017. Lethal hypothermia - a sometimes elusive diagnosis. Forensic Sci. Med. Pathol. 10.1007/s12024-017-9916-z [DOI] [PubMed] [Google Scholar]
- Danno S., Nishiyama H., Higashitsuji H., Yokoi H., Xue J.H., Itoh K., Matsuda T., and Fujita J.. 1997. Increased transcript level of RBM3, a member of the glycine-rich RNA-binding protein family, in human cells in response to cold stress. Biochem. Biophys. Res. Commun. 236:804–807. 10.1006/bbrc.1997.7059 [DOI] [PubMed] [Google Scholar]
- Derry J.M., Kerns J.A., and Francke U.. 1995. RBM3, a novel human gene in Xp11.23 with a putative RNA-binding domain. Hum. Mol. Genet. 4:2307–2311. 10.1093/hmg/4.12.2307 [DOI] [PubMed] [Google Scholar]
- Dresios J., Aschrafi A., Owens G.C., Vanderklish P.W., Edelman G.M., and Mauro V.P.. 2005. Cold stress-induced protein Rbm3 binds 60S ribosomal subunits, alters microRNA levels, and enhances global protein synthesis. Proc. Natl. Acad. Sci. USA. 102:1865–1870. 10.1073/pnas.0409764102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S., et al. 2011. Role of YAP/TAZ in mechanotransduction. Nature. 474:179–183. 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- Durak O., Gao F., Kaeser-Woo Y.J., Rueda R., Martorell A.J., Nott A., Liu C.Y., Watson L.A., and Tsai L.H.. 2016. Chd8 mediates cortical neurogenesis via transcriptional regulation of cell cycle and Wnt signaling. Nat. Neurosci. 19:1477–1488. 10.1038/nn.4400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin J., and Losick R.. 2001. Linking nutritional status to gene activation and development. Genes Dev. 15:1051–1054. 10.1101/gad.892801 [DOI] [PubMed] [Google Scholar]
- Fang W.Q., Chen W.W., Fu A.K., and Ip N.Y.. 2013. Axin directs the amplification and differentiation of intermediate progenitors in the developing cerebral cortex. Neuron. 79:665–679. 10.1016/j.neuron.2013.06.017 [DOI] [PubMed] [Google Scholar]
- Fischer K., Ruiz H.H., Jhun K., Finan B., Oberlin D.J., van der Heide V., Kalinovich A.V., Petrovic N., Wolf Y., Clemmensen C., et al. 2017. Alternatively activated macrophages do not synthesize catecholamines or contribute to adipose tissue adaptive thermogenesis. Nat. Med. 23:623–630. 10.1038/nm.4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti G., Benussi B., Currò P., Forti G., Rauber E., Minati A., Gabrielli M., Tognolli U., Sinagra G., and Pappalardo A.. 2017. The Risk of Neurological Dysfunctions after Deep Hypothermic Circulatory Arrest with Retrograde Cerebral Perfusion. J. Stroke Cerebrovasc. Dis. 26:3009–3019. 10.1016/j.jstrokecerebrovasdis.2017.07.034 [DOI] [PubMed] [Google Scholar]
- Ha S., Zhu Y., Liu D., Sherman S., and Mendola P.. 2017. Ambient temperature and air quality in relation to small for gestational age and term low birthweight. Environ. Res. 155:394–400. 10.1016/j.envres.2017.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.R., Liu Y., Xia X.Y., Ma W.J., Lin H.L., Kan H.D., Lu J.H., Feng Q., Mo W.J., Wang P., et al. 2016. Ambient Temperature and the Risk of Preterm Birth in Guangzhou, China (2001-2011). Environ. Health Perspect. 124:1100–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Lv X., Lei X., Yang Y., Yang X., and Jiao J.. 2016. Immune Regulator MCPIP1 Modulates TET Expression during Early Neocortical Development. Stem Cell Reports. 7:439–453. 10.1016/j.stemcr.2016.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kali G.T., Martinez-Biarge M., Van Zyl J., Smith J., and Rutherford M.. 2016. Therapeutic hypothermia for neonatal hypoxic-ischaemic encephalopathy had favourable outcomes at a referral hospital in a middle-income country. Acta Paediatr. 105:806–815. 10.1111/apa.13392 [DOI] [PubMed] [Google Scholar]
- Lehtinen M.K., Yuan Z., Boag P.R., Yang Y., Villén J., Becker E.B., DiBacco S., de la Iglesia N., Gygi S., Blackwell T.K., and Bonni A.. 2006. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 125:987–1001. 10.1016/j.cell.2006.03.046 [DOI] [PubMed] [Google Scholar]
- Lin K.C., Moroishi T., Meng Z., Jeong H.S., Plouffe S.W., Sekido Y., Han J., Park H.W., and Guan K.L.. 2017a Regulation of Hippo pathway transcription factor TEAD by p38 MAPK-induced cytoplasmic translocation. Nat. Cell Biol. 19:996–1002. 10.1038/ncb3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Sabnis A.J., Chan E., Olivas V., Cade L., Pazarentzos E., Asthana S., Neel D., Yan J.J., Lu X., et al. 2015. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat. Genet. 47:250–256. 10.1038/ng.3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Hu W., Xu J., Luo Z., Ye X., Yan C., Liu Z., and Tong S.. 2017b Association between temperature and maternal stress during pregnancy. Environ. Res. 158:421–430. 10.1016/j.envres.2017.06.034 [DOI] [PubMed] [Google Scholar]
- Liu Y., Hu W., Murakawa Y., Yin J., Wang G., Landthaler M., and Yan J.. 2013. Cold-induced RNA-binding proteins regulate circadian gene expression by controlling alternative polyadenylation. Sci. Rep. 3:2054 10.1038/srep02054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B., Xiang D., Wu D., Liu C., Fang Y., Chen P., and Hu Y.P.. 2018. Hepatic PHD2/HIF-1α axis is involved in postexercise systemic energy homeostasis. FASEB J.:fj201701139R 10.1096/fj.201701139R [DOI] [PubMed] [Google Scholar]
- Margaryan S., Witkowicz A., Partyka A., Yepiskoposyan L., Manukyan G., and Karabon L.. 2017. The mRNA expression levels of uncoupling proteins 1 and 2 in mononuclear cells from patients with metabolic disorders: obesity and type 2 diabetes mellitus. Postepy Hig. Med. Dosw. 71:895–900. 10.5604/01.3001.0010.5386 [DOI] [PubMed] [Google Scholar]
- Moler F.W., Silverstein F.S., Holubkov R., Slomine B.S., Christensen J.R., Nadkarni V.M., Meert K.L., Browning B., Pemberton V.L., Page K., et al. THAPCA Trial Investigators . 2017. Therapeutic Hypothermia after In-Hospital Cardiac Arrest in Children. N. Engl. J. Med. 376:318–329. 10.1056/NEJMoa1610493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C., Gupta R., Jordt S.E., Chen Y., and Liedtke W.B.. 2018. Regulation of Pain and Itch by TRP Channels. Neurosci. Bull. 34:120–142. 10.1007/s12264-017-0200-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama H., Itoh K., Kaneko Y., Kishishita M., Yoshida O., and Fujita J.. 1997. A glycine-rich RNA-binding protein mediating cold-inducible suppression of mammalian cell growth. J. Cell Biol. 137:899–908. 10.1083/jcb.137.4.899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palkar R., Ongun S., Catich E., Li N., Borad N., Sarkisian A., and McKemy D.D.. 2018. Cooling Relief of Acute and Chronic Itch Requires TRPM8 Channels and Neurons. J. Invest. Dermatol. 138:1391–1399. 10.1016/j.jid.2017.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretti D., Bastide A., Radford H., Verity N., Molloy C., Martin M.G., Moreno J.A., Steinert J.R., Smith T., Dinsdale D., et al. 2015. RBM3 mediates structural plasticity and protective effects of cooling in neurodegeneration. Nature. 518:236–239. 10.1038/nature14142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenweg-de Graaff J., Roza S.J., Walstra A.N., El Marroun H., Steegers E.A.P., Jaddoe V.W.V., Hofman A., Verhulst F.C., Tiemeier H., and White T.. 2017. Associations of maternal folic acid supplementation and folate concentrations during pregnancy with foetal and child head growth: the Generation R Study. Eur. J. Nutr. 56:65–75. 10.1007/s00394-015-1058-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.H., and McNaughton P.A.. 2016. The TRPM2 ion channel is required for sensitivity to warmth. Nature. 536:460–463. 10.1038/nature19074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Hammack C., Ogden S.C., Wen Z., Qian X., Li Y., Yao B., Shin J., Zhang F., Lee E.M., et al. 2016. Zika Virus Infects Human Cortical Neural Progenitors and Attenuates Their Growth. Cell Stem Cell. 18:587–590. 10.1016/j.stem.2016.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z., Zhao Z., and Lee C.C.. 2011. 5′- Adenosine monophosphate induced hypothermia reduces early stage myocardial ischemia/reperfusion injury in a mouse model. Am. J. Transl. Res. 3:351–361. [PMC free article] [PubMed] [Google Scholar]
- Toyama R.P., Xikota J.C., Schwarzbold M.L., Frode T.S., Buss Z.S., Nunes J.C., Funchal G.D., Nunes F.C., Walz R., and Pires M.M.. 2015. Dose-dependent sickness behavior, abortion and inflammation induced by systemic LPS injection in pregnant mice. J. Matern. Fetal Neonatal Med. 28:426–430. 10.3109/14767058.2014.918600 [DOI] [PubMed] [Google Scholar]
- Tyler C.R., and Allan A.M.. 2014. Prenatal alcohol exposure alters expression of neurogenesis-related genes in an ex vivo cell culture model. Alcohol. 48:483–492. 10.1016/j.alcohol.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xiao Z.D., Li X., Aziz K.E., Gan B., Johnson R.L., and Chen J.. 2015. AMPK modulates Hippo pathway activity to regulate energy homeostasis. Nat. Cell Biol. 17:490–499. 10.1038/ncb3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J.J., Au A.Y., Gao D., Pinello N., Kwok C.T., Thoeng A., Lau K.A., Gordon J.E., Schmitz U., Feng Y., et al. 2016. RBM3 regulates temperature sensitive miR-142-5p and miR-143 (thermomiRs), which target immune genes and control fever. Nucleic Acids Res. 44:2888–2897. 10.1093/nar/gkw041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yu C., and Wang L.. 2017. Temperature exposure during pregnancy and birth outcomes: An updated systematic review of epidemiological evidence. Environ. Pollut. 225:700–712. 10.1016/j.envpol.2017.02.066 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.