Mejlvang et al. show that amino acid starvation of human fibroblasts and a lung cancer cell line induces a rapid and selective degradation of a subset of proteins, including autophagy receptors p62/SQSTM1, NBR1, TAX1BP1, NDP52, and NCOA4, that is independent from mTOR and canonical macroautophagy but dependent on endosomal microautophagy.
Abstract
It is not clear to what extent starvation-induced autophagy affects the proteome on a global scale and whether it is selective. In this study, we report based on quantitative proteomics that cells during the first 4 h of acute starvation elicit lysosomal degradation of up to 2–3% of the proteome. The most significant changes are caused by an immediate autophagic response elicited by shortage of amino acids but executed independently of mechanistic target of rapamycin and macroautophagy. Intriguingly, the autophagy receptors p62/SQSTM1, NBR1, TAX1BP1, NDP52, and NCOA4 are among the most efficiently degraded substrates. Already 1 h after induction of starvation, they are rapidly degraded by a process that selectively delivers autophagy receptors to vesicles inside late endosomes/multivesicular bodies depending on the endosomal sorting complex required for transport III (ESCRT-III). Our data support a model in which amino acid deprivation elicits endocytosis of specific membrane receptors, induction of macroautophagy, and rapid degradation of autophagy receptors by endosomal microautophagy.
Introduction
Starvation is a fundamental type of stress occurring in biological systems. From yeast to humans, starvation-induced degradation by “self-eating” autophagic processes provides metabolic building blocks and energy to sustain core cellular processes (Ohsumi, 2014). Starvation-induced autophagy may also implement adaptations by degrading key regulators of processes disadvantageous for fitness during starvation (Kristensen et al., 2008; Müller et al., 2015).
Short-lived proteins are mainly degraded by the ubiquitin-proteasome system, whereas more long-lived proteins are degraded by the lysosome (Zhang et al., 2016). Extracellular and plasma membrane proteins are degraded by endocytosis, whereas autophagy degrades cytoplasmic and organelle-bound proteins. The autophagic pathways include macroautophagy, chaperone-mediated autophagy, and microautophagy (Mizushima and Komatsu, 2011). In macroautophagy, a double membrane vesicle, the autophagosome, sequesters parts of the cytoplasm. Autophagosomes fuse either directly with lysosomes or first with late endosomes to form amphisomes (Seglen et al., 1991), which subsequently fuse with lysosomes. In the resulting autolysosomes, the contents are degraded and recycled to the cytosol (Lamb et al., 2013). Chaperone-mediated autophagy involves direct uptake of cargo by lysosomes dependent on the chaperone HSC70 and lysosomal membrane protein 2A (LAMP2A; Cuervo and Wong, 2014). In microautophagy, cargo is taken up directly by lysosomes via invagination of their limiting membranes (Marzella et al., 1981). The morphological similarity between microautophagy and generation of intraluminal vesicles during late endosome/multivesicular body (MVB) biogenesis suggests that they are mechanistically related. Studies of microautophagy involving MVBs/late endosomes rather than lysosomes provide evidence for such a relationship (Sahu et al., 2011; Uytterhoeven et al., 2015; Mukherjee et al., 2016). Members of the endosomal sorting complex required for transport (ESCRT), orchestrating inward budding of the endosomal membrane to form intraluminal vesicles (Christ et al., 2017), are required for endosomal microautophagy (Lefebvre et al., 2018).
Macroautophagy can be either nonselective or selective. Selectivity is mediated by autophagy receptors tethering cargo to the growing phagophore (Johansen and Lamark, 2011; Rogov et al., 2014; Stolz et al., 2014; Hamacher-Brady and Brady, 2016). The sequestosome 1–like receptors (SLRs) p62/SQSTM1 (sequestosome 1), NBR1, TAX1BP1, NDP52, and OPTN bind ubiquitylated cargo including protein aggregates, damaged organelles, and intracellular bacterial pathogens (Deretic, 2012; Rogov et al., 2014; Stolz et al., 2014). Some TRIM family ubiquitin E3 ligases may also act as autophagy receptors (Mandell et al., 2014; Chauhan et al., 2016; Kimura et al., 2016). NCOA4 is a specialized cargo receptor for degradation of ferritin to liberate iron (Dowdle et al., 2014; Mancias et al., 2014). There are also organelle-bound receptors for autophagy of mitochondria and ER (Hamacher-Brady and Brady, 2016; Khaminets et al., 2016). Autophagy receptors recruit cargo to phagophores by binding to ATG8 proteins via LC3-interacting region (LIR) motifs (Birgisdottir et al., 2013). Mammals have seven ATG8 isoforms; LC3A, -B, -B2, and -C and GABARAP, GABARAPL1, and GABARAPL2 (Shpilka et al., 2011). ATG8s are covalently conjugated to phosphatidylethanolamine in a reaction dependent on the E3-ubiquitin ligase–like complex ATG12–ATG5–ATG16 (Ichimura et al., 2000). This enables them to bind the phagophore membrane (Kabeya et al., 2004). Turnover of p62, levels of lipidated LC3B, and LC3B puncta formation are commonly used readouts for macroautophagic activity (Klionsky et al., 2016).
Autophagosome formation is positively regulated by the uncoordinated 51-like kinase 1/2 (ULK1/2) complex (also comprising ATG13, ATG101, and FIP200) and requires generation of phosphatidylinositol-3 phosphate (PI3P) on phagophore membranes by the PI3 kinase class 3 (PI3KC3). VPS34 is the catalytic subunit of the PI3KC3 complex 1 also containing Beclin 1, VPS15, and ATG14L (Mizushima et al., 2011). The mechanistic target of rapamycin (mTOR) kinase complex regulates cell growth and metabolism in response to intracellular amino acid levels (Saxton and Sabatini, 2017). In fed cells, mTOR allows cell growth and synthesis of new proteins, actively repressing macroautophagy via phosphorylation of ULK1 (Hosokawa et al., 2009). In starved cells, mTOR is inactivated, and formation of autophagosomes is induced. Basal macroautophagy serves as a quality control system and selectively degrades labeled cargo (Mizushima and Hara, 2006; Johansen and Lamark, 2011; Fimia et al., 2013; Rogov et al., 2014; Stolz et al., 2014; Zhang et al., 2016). We do not know whether starvation-induced macroautophagy retains selectivity or switches to nonselective bulk macroautophagy.
In this study, we show that upon acute amino acid starvation in both the lung cancer cell line A549 and BJ normal diploid fibroblasts, a number of substrates are rapidly degraded by endosomal microautophagy. Among the substrates are the selective autophagy receptors p62/SQSTM1, NBR1, NDP52, TAX1BP1, and NCOA4 and the ATG8 family proteins LC3B and GABARAPL2. Endocytosis of specific membrane proteins and induction of macroautophagy are parts of the response to starvation. However, the rapid degradation of autophagy receptors is mediated by ESCRT-III–dependent delivery of these proteins to vesicles inside late endosomes/MVBs. This immediate autophagic degradation response is induced by the absence of extracellular amino acids and independent of mTORC1.
Results
Identification of starvation-induced changes in protein expression by quantitative proteomics
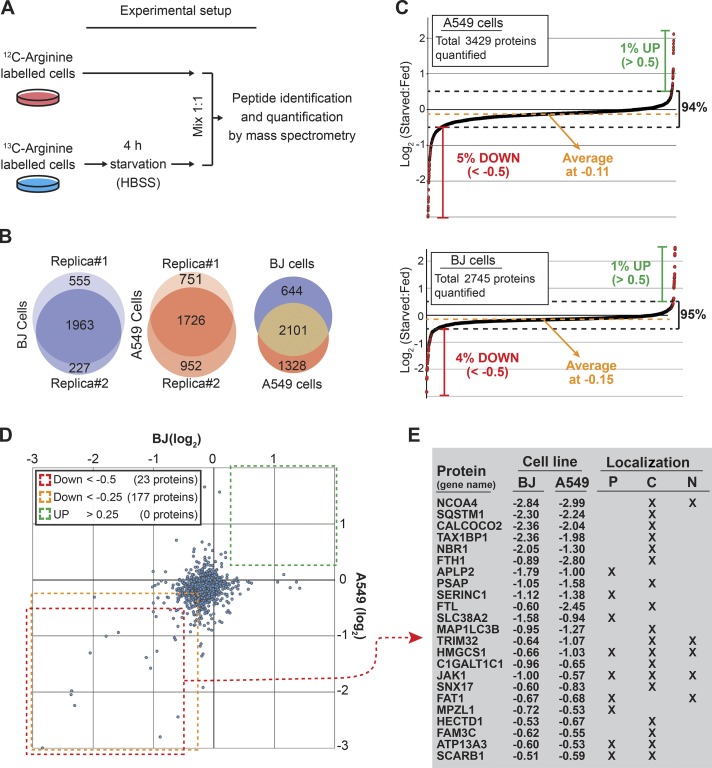
To improve our understanding of how cells respond and adapt to starvation, we aimed to identify which proteins were up- and down-regulated during the initial phase of starvation. Hence, we applied stable isotope labeling by amino acids in cell culture (SILAC) and mass spectrometry to quantify changes in the proteome caused by culturing cells in HBSS for 4 h (see Materials and methods and Fig. 1 A). HBSS is commonly used for starvation studies and contains d-glucose and essential inorganic ions, but it lacks amino acids and serum. We performed the analyses in both epithelial cancer cells (A549) and primary fibroblasts (BJ). Whereas some cell death was observed for the cancer cells during prolonged starvation (2–4 d), shorter periods of starvation did not affect viability of the two cell lines (Fig. S1 A). Proteomic analysis yielded quantifications of 2,745 proteins in BJ and 3,429 in A549 cells compiled in two datasets referred to as the BJ- and A549-starveome (Fig. 1, B and C; and Data S1 and S2). On average, protein expression decreased by 8–10% during the starvation period (−0.15 and −0.11 in log2; Fig. 1 C). This likely reflects reduced growth of cells entering G1 phase during the starvation period (Scott et al., 2000). Consistently, 12.6% of the A549 cells divided during the starvation period. Whereas the majority of detected proteins did not change in expression upon starvation (∼95% were within ±0.5 log2), ∼1% increased and 5% decreased (Fig. 1 C). This clearly supports the well-established hypothesis that starvation induces a cellular response inhibiting anabolic processes and simultaneously stimulates catabolic processes to sustain core metabolic processes needed for survival (Howell and Manning, 2011). We did not detect a single protein that was up-regulated in both cell lines (Fig. 1 D), likely because of the up-regulated proteins being predominantly regulatory, low-abundance proteins like transcription factors. Hence, ATF4 and CHOP, which rapidly increase in expression during starvation (Jiang et al., 2004) were not detected. In contrast, many proteins decreased in both A549 and BJ cells during 4-h starvation. We identified 177 proteins that decreased >15% (−0.25 in log2) and 23 proteins that decreased >30% (−0.5 in log2) in both cell lines (Fig. 1, D and E). Many of these proteins decreased with similar magnitude in the two cell lines (Fig. 1, D and E).
Figure 1.
Identification of starvation-induced changes in protein expression by quantitative proteomics. (A) SILAC combined with mass spectrometry was used to quantify changes in the proteome caused by culturing cells for 4 h in starvation medium (HBSS). Each experiment was based on two biological replicas with reciprocal labeling. Data were normalized to cell number using the mean expression of four highly abundant nuclear proteins linked to nuclear lamina (Lamin B1 and Lamin B2) or chromosomes (Histone H3 and H4). (B) Overlap between identified proteins from two biological replicas (middle and left) and overlap between proteins identified in both replicas from A549 and BJ cells (right). (C) Dot plot showing the effect of starvation on global protein expression in A549 (top) and BJ (bottom). (D) Dot plot showing the effect of starvation on proteins identified in both BJ and A549 cells. (E) List of proteins found to be reduced (less than −0.5 in log2) in both cell lines after 4-h starvation. Numbers indicate the starvation-induced reduction on a log2 scale. Proteins were categorized by their subcellular localization. P, plasma membrane; C, cytoplasm; N, nucleus.
Starvation triggers a rapid autophagic response
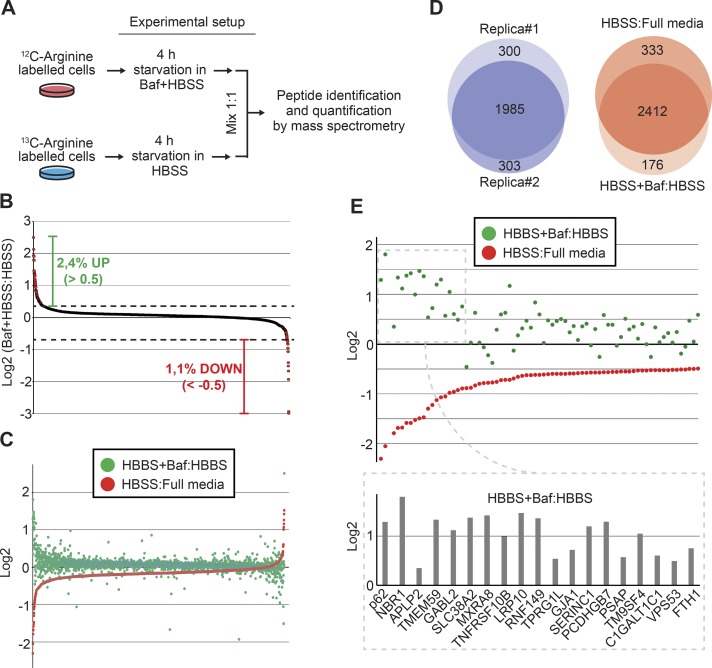
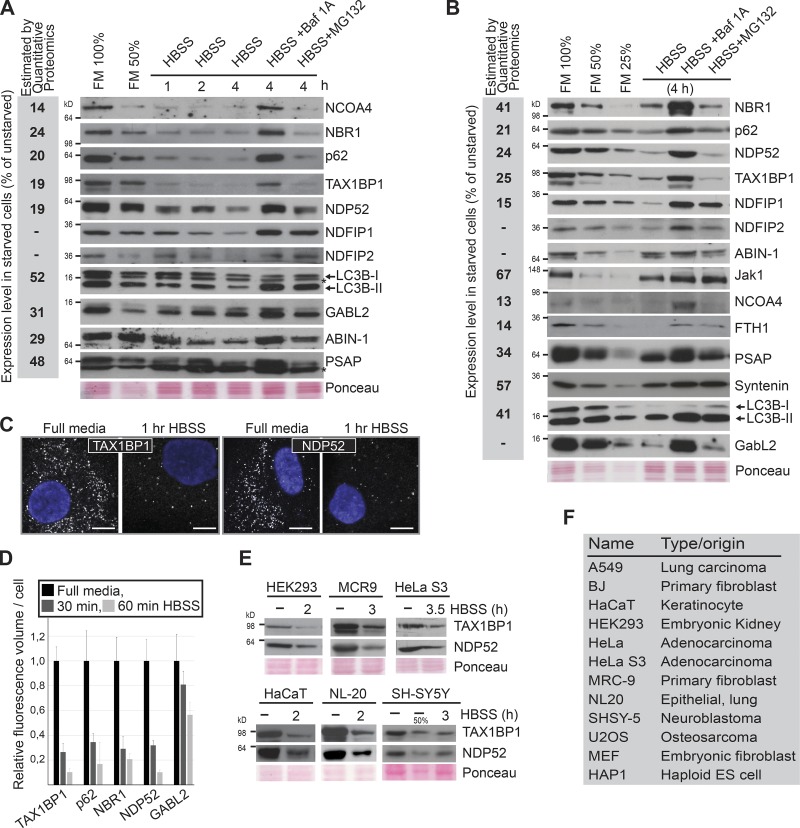
Of the most profoundly degraded proteins, multiple proteins were associated with autophagy. Strikingly, the selective autophagy receptors p62/SQSTM1, NBR1, CALCOCO2/NDP52, and TAX1BP1 (Johansen and Lamark, 2011; Rogov et al., 2014) decreased by as much as 80% (Fig. 1 E), and the autophagosomal marker LC3B decreased by ∼50%. In addition, the expression of the cargo receptor for ferritin, NCOA4 (Dowdle et al., 2014; Mancias et al., 2014), and the two subunits of ferritin (FTL and FTH1) were strongly reduced after the short starvation period. Because these proteins and many others of the most rapidly degraded proteins (Fig. 1 E) are shown to be degraded in the lysosome (Mancias et al., 2014), we tested to what degree the decrease in protein expression caused by 4-h starvation was caused by lysosomal degradation. Hence, we compared protein expression in BJ cells starved for 4 h in the absence/presence of the vacuolar H+-ATPase inhibitor bafilomycin A1, which functionally inhibits lysosomal activity (Fig. 2, A–E; and Data S3). Based on two biological replicas, we quantified 2,588 proteins, of which 2.4% increased in expression (>0.5 in log2) and 1.1% decreased upon lysosomal inhibition (Fig. 2, B and D). The overlap between this dataset and the BJ-starveome allowed us to evaluate whether a protein found to decrease during starvation was degraded by lysosomes (Fig. 2, C and D). Of the 75 most depleted proteins identified, 68% increased (>0.2 in log2) in expression upon lysosomal inhibition (Fig. 2 E). This percentage reached 100% when considering only proteins reduced more than twofold. To validate our measurements of relative protein expression in the quantitative proteomic studies, a diverse panel of proteins found to be down-regulated upon starvation was analyzed by Western blot. These independent experiments mirrored the results obtained by quantitative proteomics with high accuracy (Fig. 3, A and B). Moreover, several down-regulated proteins (e.g., GABARAPL2, ABIN-1, and NDFIP1/2) detected by mass spectrometry only in one of the cell lines because of the low abundance of analyzed peptides, actually turned out to be down-regulated in both when analyzed by Western blotting (Fig. 3, A and B; and Fig. S2). Thus, the true overlap of proteins degraded upon starvation in both cell lines is clearly larger than the proteomic data suggest. In line with the proteomic data, Western blot analysis showed that the degradation of investigated proteins was abrogated upon lysosomal inhibition by either bafilomycin A1 or the lysosomal protease inhibitors E64d and Pepstatin A (Fig. 3, A and B; and Fig. S1 C). In comparison, inhibition of the proteasome by MG132 only affected the starvation-induced reduction of a few proteins (Fig. 3, A and B). The majority of investigated proteins showed a significant decrease in expression within 1 h of starvation (Fig. 3 A). Consistently, quantitative analyses of immunostained cells revealed a significant decrease in intensity for the autophagy receptors TAX1BP1, p62, NBR1, and NDP52 already after 30 min of starvation (Fig. 3, C and D). We additionally observed degradation of NDP52 and TAX1BP1 upon short-term starvation in 10 additional cell lines (Fig. 3, E and F; and Figs. S1 D and S3 D). Taken together, our data suggest that 2–3% of the proteome is rapidly targeted for lysosomal degradation upon acute starvation. Some of these substrates are plasma membrane receptors (e.g., SLC38A2, SERINC1, and TNFRSF10B/D), substantiating that mammalian cells selectively degrade certain receptors by endocytosis in a similar fashion to yeast (Jones et al., 2012; Müller et al., 2015). We could show that GFP-SLC38A2 localized to the plasma membrane and was degraded upon amino acid starvation (Fig. S1 B). Starvation in combination with inhibition of lysosomal degradation (bafilomycin A1) caused a dramatic increase in SLC38A2-positive vesicles within the cytoplasm (Fig. S1 B). However, the many cytoplasmic substrates argue that a general autophagic response selectively targeting specific substrates for degradation is executed within the first 4 h of starvation.
Figure 2.
The rapid degradation triggered by amino acid starvation is dependent on the lysosome. (A) SILAC experiment where bafilomycin A1 (Baf; 200 nM) was used to block lysosomal-mediated protein degradation in BJ cells during 4-h starvation in HBSS. (B) Dot plot of expression changes induced by lysosomal inhibition (Baf) during 4 h in HBSS. (C) Dot plot of expression changes of all proteins identified in both the BJ-starveome and the bafilomycin dataset. (D) Overlap of identified proteins from the two biological replicas (left), and the overlap between the BJ-starveome and bafilomycin dataset (right). (E) Dot plot representation of the 100 most degraded proteins in both the HBSS:Full media and the HBSS+Baf:HBSS experiments in BJ cells. The lower part shows the 20 most degraded proteins.
Figure 3.
Starvation triggers an autophagic response. (A and B) Western blot analyses comparing the expression level of indicated proteins in A549 (A) and BJ (B) cells. The experiments were performed three independent times with similar results. Bafilomycin A1, 200 nM; MG132, 10 µM. The reduction in expression estimated by quantitative proteomics is shown to the left (gray box). (C) Immunofluorescence analysis of TAX1BP1 and NDP52 in BJ cells starved for 1 h. The experiment was performed three independent times with similar results. Bar, 10 µm. (D). Relative quantification of signal intensities of indicated proteins based on immunostained BJ cells (n > 100). Error bars represent SD. (E) Western blot analyses of expression levels of different autophagy receptors in indicated cell lines upon short-time starvation. This experiment was performed three independent times with similar results. (F) Cell lines eliciting rapid degradation of NDP52 and TAX1BP1 upon starvation (see also E).
The rapid autophagic response to starvation is triggered by amino acid deficiency independently of mTOR
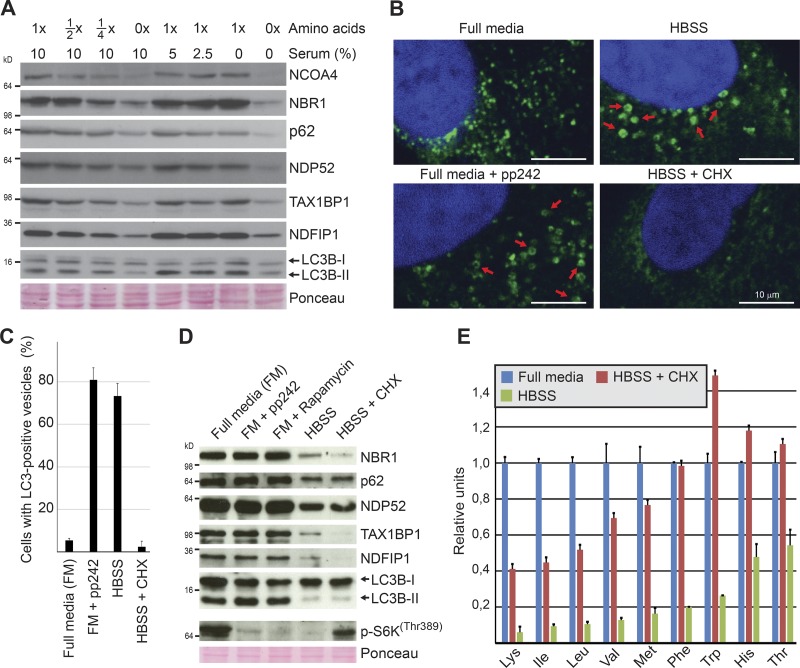
To elucidate how this acute autophagic response was triggered, we tested whether the expression of a panel of substrates was affected by amino acid and/or serum deprivation. 4 h of amino acid deprivation caused a marked degradation of all the substrates, whereas serum removal had little or no effect (Figs. 4 A and S1 E). The degradation was not further enhanced when both cell lines were deprived of both serum and amino acids. Hence, the acute autophagic response caused by starving cells in HBSS is mainly triggered by lack of amino acids. Interestingly, even a 50% reduction of amino acids triggered a degradation response, albeit with a lower magnitude (Figs. 4 A and S1 E). As mTORC1 activity is dependent on intracellular levels of amino acids, we assumed that the degradation was mediated by macroautophagy and elicited by mTORC1 deactivation. However, 4 h of pharmacological inhibition of mTOR by rapamycin or pp242 had only minimal effect on the expression level of our test panel (Figs. 4 D and S1 F). Efficacy of the mTOR inhibitors was confirmed by reduced levels of phospho-S6KThr389 and induction of autophagosome formation revealed by appearance of ring-shaped structures positive for LC3B (Fig. 4, B and C; Kabeya et al., 2004; Mizushima, 2004). As previously noted (Klionsky et al., 2016), the dramatic induction of autophagosomes caused by mTOR inhibition was not accompanied by dramatic changes in the LC3B-I/LC3B-II ratio (Figs. 4 D and S1 F). Blocking protein synthesis by cycloheximide in starving cells partially rescued intracellular levels of amino acids and mTOR activity but did not inhibit the starvation-induced degradation of the test panel (Fig. 4, D and E; and Fig. S1, F and G). Intriguingly, inhibition of mTOR does not play a major role in the rapid autophagic degradation of the investigated substrates occurring within the first 4 h of starvation. The data show that the response is induced by the absence of extracellular rather than intracellular amino acids.
Figure 4.
The immediate autophagic response to starvation is triggered by amino acid deficiency independently of mTOR. (A) Western blot analyses assessing the effect of 4-h amino acids versus serum starvation in BJ cells. The experiment was performed three independent times with similar results. (B) Representative immunofluorescence images of endogenous LC3B (in green) in A549 cells treated with the mTOR inhibitor pp242 (250 nM) or starved in HBSS or in HBSS with cycloheximide (CHX; 50 µg/ml) for 4 h. Red arrows depict LC3B-positive vesicles. The experiment was performed more than three independent times with similar results. (C) Quantification of cells with LC3-positive vesicles based on technical triplicate (n > 100). Error bars represent SD. (D) Western blot analyses of expression levels of indicated proteins in BJ cells treated as indicated for 4 h. pp242 (250 nM) and rapamycin (250 nM) were used to inhibit mTOR. Cycloheximide (50 µg/ml) was used to block protein synthesis. The experiment was performed three independent times with similar results. (E) Quantification of free pools of intracellular amino acids by HPLC-MS/MS in BJ cells starved for 1 h with/without cycloheximide (50 µg/ml). Error bars represent SD of a technical triplicate. The experiment was performed three independent times with similar results.
Macroautophagy does not play a key role in the immediate autophagic response to starvation
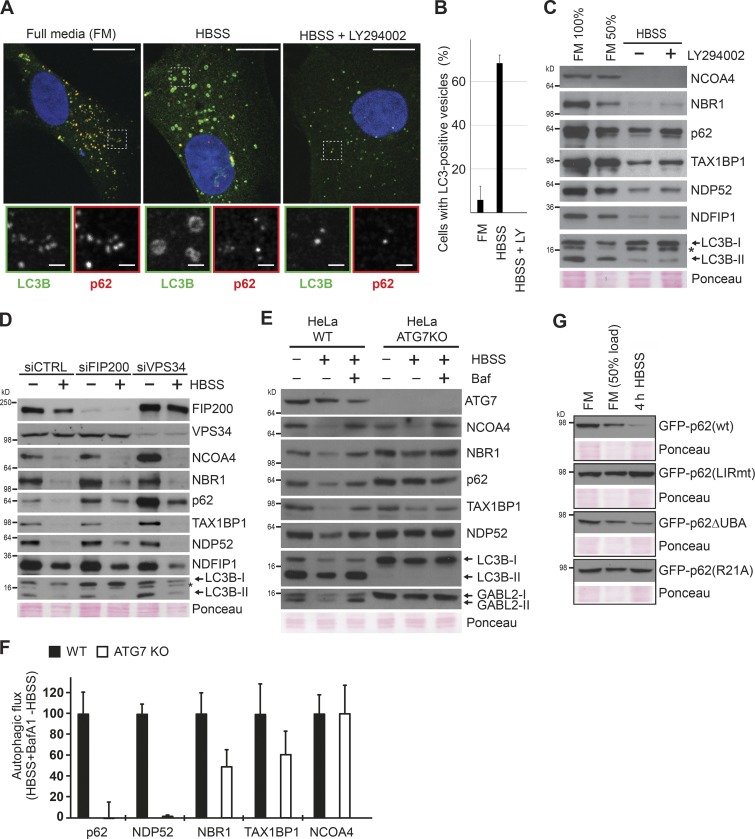
Our finding that pharmacological mTOR inhibition for up to 4 h could not mimic starvation-induced degradation of canonical macroautophagy substrates (e.g., p62) may initially seem surprising. However, although many studies show that 2 h of starvation is sufficient to detect a significant decrease in p62 levels (Wang et al., 2012; McAlpine et al., 2013; Petherick et al., 2013; Dooley et al., 2014; Dowdle et al., 2014; Jiang and Mizushima, 2015; Khaminets et al., 2015), pharmacological mTOR inhibition requires substantially longer treatments (>12 h) to achieve this degradation (Artal-Martinez de Narvajas et al., 2013; Ichimura et al., 2013; Wang et al., 2015; Yun et al., 2016). Furthermore, it has been shown that starvation induces p62 degradation in Torin1-treated cells (Wang et al., 2012). We also observed, as previously reported (Watanabe-Asano et al., 2014), that cells starved in the presence of cycloheximide showed a completely diffuse LC3 staining, indicating lack of macroautophagic activity (Fig. 4, B and C). Taken together, these data could reflect that the proteins degraded most rapidly upon starvation are degraded by a macroautophagy-independent pathway induced in parallel to the slower mTOR-dependent macroautophagy. To test this hypothesis, we assessed to what degree our test panel substrates were subjected to macroautophagic degradation during the first hours of starvation by impairing macroautophagy both genetically and pharmacologically. Autophagosome formation requires de novo phosphorylation of phosphatidylinositol to PI3P to facilitate elongation of the phagophore and recruitment of lipidated LC3. Inhibition of the responsible kinase, VPS34 (PI3KC3), pharmacologically with the PI3K inhibitor LY294002 blocked the starvation-induced formation of LC3B-positive autophagosomes, whereas the staining intensity of the autophagy receptor p62 was strongly reduced upon 1-h starvation (Fig. 5, A and B; and Fig. S3 A). Western blot analyses confirmed efficient starvation-induced degradation of the autophagy receptors p62, NBR1, TAX1BP1, NDP52, and NCOA4 as well as NDFIP1 despite inhibition of macroautophagy with LY294002 (Figs. 5 C and S3 B). Similar results were obtained using the specific VPS34 inhibitors PIK-III or SAR405 (Fig. S3, C–E; Dowdle et al., 2014; Ronan et al., 2014). Consistently, RNAi-mediated depletion of VPS34 inhibited autophagosome formation (Fig. S3 E) without abrogating the starvation-induced degradation of autophagy receptors and NDFIP1 (Fig. 5 D). Previous depletion and knockout (KO) studies of VPS34 have shown that cells form large translucent vacuoles after 4–5 d of VPS34 removal (Johnson et al., 2006; Jaber et al., 2012). This phenotype is associated with accumulation of lipidated LC3B and partial impairment of endocytic transport. Of note, our depletion studies were limited to 72 h, and we observed only a minor increase in lipidated LC3B (Fig. 5 D). It is well established that autophagy receptors are degraded by macroautophagy in full media (FM; Pankiv et al., 2007; Kirkin et al., 2009; Newman et al., 2012). Consistently, basal levels of NCOA4 and p62 increased significantly in VPS34-depleted BJ cells cultured in FM (Fig. 5 D). This increase in basal levels was also seen upon 20-h pharmacological inhibition of VPS34 in DLD1 cells (Dowdle et al., 2014). We consistently found degradation of the autophagy receptors upon either short-term knockdown or short term pharmacological inhibition of VPS34, whereas macroautophagy was inhibited.
Figure 5.
Macroautophagy is not playing a key role in the immediate autophagic response to starvation. (A) Immunofluorescence analysis of indicated proteins in BJ cells starved for 1 h in the presence/absence of the PI3K inhibitor LY294002 (25 µM). Bars: (main images) 20 µm; (insets) 2 μm. The experiment was performed more than three independent times with similar results. (B) Quantification of cells with LC3-positive vesicles based on technical triplicate (n > 100). LY, LY294002 (25 µM). Error bars represent SD. (C) Western blot analyses of expression levels of indicated proteins in BJ cells grown in FM or starved for 4 h in the presence or absence of the inhibitor LY294002 (25 µM). The experiment was performed three independent times with similar results. (D) Western blot analyses assessing the immediate autophagic response to starvation in FIP200- or VPS34-depleted BJ cells. Starvation was initiated 72 h after siRNA transfection. Cells were starved for 4 h in HBSS. The experiment was performed three independent times with similar results. (E) Western blot analyses of expression levels of indicated proteins in WT HeLa and ATG7 KO HeLa cells after 4-h starvation in HBSS. Bafilomycin A1 (Baf) was used (200 nM) to inhibit lysosomal protein degradation. The experiment was performed three independent times with similar results. (F) Autophagic flux in response to HBSS treatment was quantified from Western blots exemplified in E. Levels of the respective proteins in the HBSS-treated samples were subtracted from the levels in HBSS + bafilomycin A1–treated samples. The calculated flux for each protein was normalized to give the value of 100 in the WT cell line. Error bars denote SD from three independent experiments. (G) Western blot analysis of starvation-induced degradation of GFP-tagged p62, p62 LIRmut, PB1 mutant (R21A), and p62ΔUBA. Clones with doxycycline (Dox)-inducible GFP-tagged derivatives of p62 were generated in p62 KO MEFs. Expression was induced overnight by 1 µg/ml doxycycline. This experiment was performed three independent times with similar results.
We next asked whether the ULK1–ATG13–FIP200 complex was required for the immediate autophagic response to starvation by depleting FIP200. Similar to VPS34 depletion, FIP200 depletion blocked starvation-induced macroautophagy as demonstrated by lack of autophagosome formation (Fig. S3 E). However, our test panel substrates were still degraded (Fig. 5 D). Consistent with the independence of FIP200 shown in this study, p62 degradation upon short-term starvation was previously shown to occur in ULK1/2 KO MEFs (McAlpine et al., 2013; Liu et al., 2016).
We then asked whether the rapid starvation-induced degradation of autophagy receptors NBR1, p62, NDP52, TAX1BP1, and NCOA4 also occurred in ATG7 KO cells, where canonical macroautophagy involving lipidated ATG8 proteins is blocked. To this end, we used CRISPR/CAS9 to generate HeLa cells KO for ATG7. Although the levels of the autophagy receptors increased in the KO cells relative to WT cells as a result of loss of the macroautophagic turnover occurring in FM, we observed starvation-induced degradation of NBR1, TAX1BP1, and NCOA4 in ATG7 KO HeLa despite a complete absence of lipidated LC3B (Fig. 5, E and F). However, p62 or NDP52 were not degraded upon starvation in ATG7 KO cells. Consistently, p62 and NDP52 were not degraded upon starvation in human HAP1 cells KO for ATG5, whereas NBR1 and TAX1BP1 were degraded (Fig. S3 F). Both NBR1 and TAX1BP1 were partially affected by ATG7 KO (Fig. 5 F). Hence, their degradation may be mediated by several lysosomal pathways. The degradation of NCOA4, however, was unaffected by ATG7 deficiency. In line with this, Goodwin et al. (2017) recently showed that NCOA4 was degraded via a lysosomal pathway upon iron chelation in ATG7 or ATG3 KO cells. To determine which domains of p62 were required for degradation, we reconstituted p62 KO MEFs with GFP-p62 WT, LIR mutant, R21A PB1 domain mutant, or a UBA deletion mutant. The WT and UBA deletion mutants were degraded, whereas the polymerization defective R21A PB1 domain mutant and the LIR mutant were not (Fig. 5 G). This is consistent with a requirement for interaction with lipidated ATG8s. Taken together, our data reveal that in addition to starvation-induced macroautophagy, starvation induces a hitherto uncharacterized autophagic response that causes rapid degradation of a large pool of specific proteins, including the selective autophagy receptors. This immediate autophagic response is manifested as a reduction in protein expression within 1 h (Fig. 3 A) and possibly initiated within the first 30 min of starvation (Fig. 3 D).
Substrates of the rapid starvation response are associated with the endosomal system
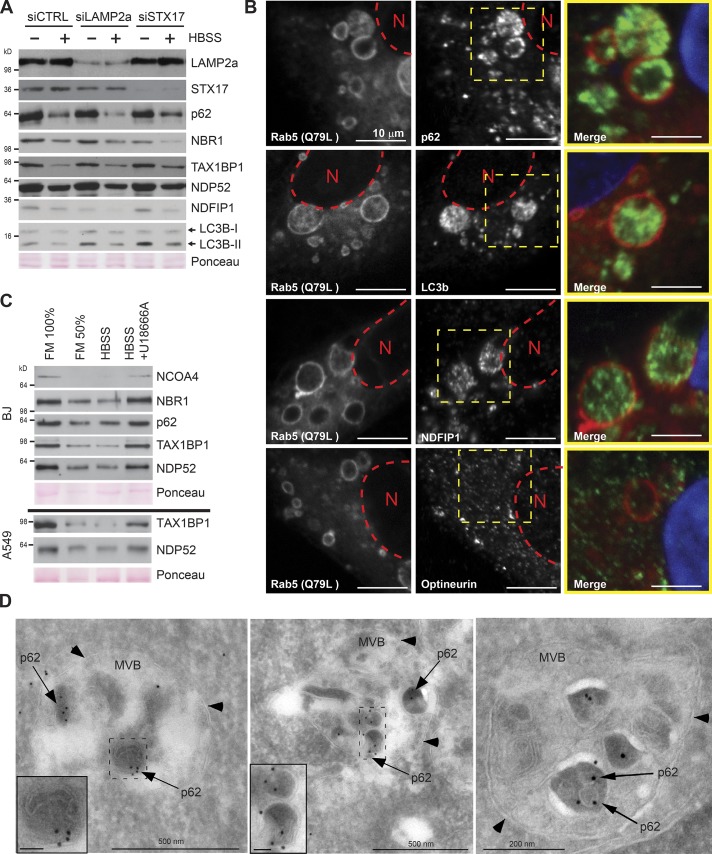
As RNAi-mediated depletion of LAMP2A did not inhibit the degradation caused by 1 h starvation, we excluded chaperone-mediated autophagy from playing a major role in the rapid autophagic response to starvation (Fig. 6 A). We next asked whether the substrates were targeted directly to lysosomes or via earlier structures of the endosomal network. In support of the latter, the endosomal proteins NDFIP1 and NDFIP2 (Mund and Pelham, 2010) are among the most efficiently degraded substrates (Fig. 3, A and B), and the autophagy receptors TAX1BP1 and p62 colocalize with structures positive for the endosomal marker CD63 (Fig. S4 A). Additionally, NDFIP1, p62, TAX1BP1, NDP52, and LC3B accumulated inside giant endosomes generated by overexpression of a hyperactive Rab5 Q79L mutant (Figs. 6 B and S4 B; Stenmark et al., 1994). In comparison, the subcellular localization of Clathrin and the autophagy receptor Optineurin, which are not substrates of the rapid autophagic response to starvation, were unaffected by Rab5 (Q79L) overexpression (Figs. 6 B and S4 B). Immuno-EM verified that p62 resides within late endosomes including MVBs when macroautophagy is blocked by VPS34 inhibition in cells starved for amino acids (Fig. 6 D). This suggests that p62 can enter late endosomes via a macroautophagy-independent mechanism. As p62 is seen as electron-dense bodies surrounded by a single membrane inside MVBs containing smaller intraluminal vesicles, we assume that p62 is taken up by invagination of the limiting membrane of the MVB. The p62-positive vesicles inside MVBs were generally both more electron dense and larger than the typical intraluminal vesicles (Fig. 6 D). The cholesterol transport inhibitor, U18666A, is known to block MVB dynamics and to inhibit endosomal microautophagy (Liscum and Faust, 1989; Sahu et al., 2011). Consistent with degradation via endosomal microautophagy, pharmacological treatment with U18666A abrogated the starvation-induced degradation of the test panel (Fig. 6 C).
Figure 6.
Substrates of the immediate autophagic response to starvation are associated with the endosomal network. (A) Western blots assessing the immediate autophagic response to starvation in LAMP2a- and STX17-depleted BJ cells. Starvation was induced 72 h after siRNA transfection. Cells were starved for 1 h in HBSS. The experiment was performed two independent times with similar results. (B) Immunofluorescence analysis of indicated proteins in A549 cells transiently expressing mCherry-Rab5 (Q79L mutant). Red dotted line indicates the location of nucleus (N). The dotted yellow square represents the enlargement (far right) with merged p62 (green) and Rab5(Q79L; red) signals. The experiment was performed three independent times with similar results. (C) Western blot analysis assessing the influence of cholesterol on immediate autophagic response to 1-h starvation. To deplete cellular membranes for cholesterol before starvation, BJ (top) and A549 (bottom) were treated with U18666A (5 µg/ml) to block MVB dynamics. The experiment was performed two independent times with similar results. (D) Electron micrographs of BJ cells treated with the VPS34 inhibitor PIK-III (5 µM) for 2 h and starved in HBSS with PIK-III for 30 min (left and middle) or 60 min (right). Cryosections were labeled with antibody against p62, followed by 10 nm protein A–gold (black dots depicted by arrows; membranes indicated with arrowheads). Inset bars, 50 nm.
The rapid degradation of selective autophagy receptors upon starvation depends on ESCRT-III and VPS4
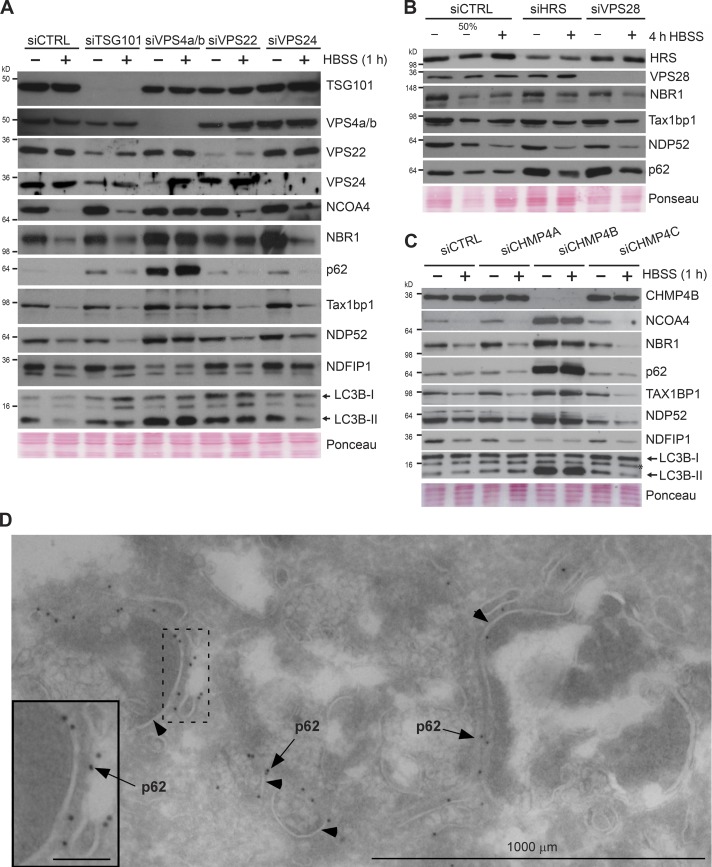
Recent studies suggest that autophagic substrates can enter MVBs via inward budding through a process termed endosomal microautophagy (Sahu et al., 2011; Uytterhoeven et al., 2015; Mukherjee et al., 2016) that consequently leads to their entrapment within intraluminal vesicles. Furthermore, fission yeast Nbr1, a homologue of mammalian autophagy receptor NBR1, can enter the lysosomal vacuole independently of the core autophagy machinery (Liu et al., 2015). As these processes rely on ESCRT-III and its regulatory ATPase VPS4 (Raiborg and Stenmark, 2009), we next asked whether any of these components were essential for the observed immediate autophagic response to starvation. Transient overexpression of an ATPase-defective mutant of VPS4B (EQ) known to inhibit the endosomal pathway (Bishop and Woodman, 2000) caused a clear accumulation of TAX1BP1 and p62 compared with overexpressing WT VPS4 (Fig. S5 A). RNAi-mediated depletion of VPS4A and -B or CHMP4B (ESCRT-III) inhibited starvation-induced degradation of our test panel (Fig. 7, A and C; and Fig. S5, B–F). These results were verified by using alternative siRNAs for CHMP4B (Figs. 7 C and S5 E) and VPS4A and VPS4B (Figs. 7 A and S5, C, D, and F). Interestingly, knockdown of both VPS4A and -B is required to achieve blockade of the rapid starvation-induced degradation (Fig. S5 F). However, we did not find any dependence on ESCRT-I (TSG101 and VPS28) and Hsc70 (Fig. 7, A and B; and Fig. S5 G), which has been reported for endosomal microautophagy of, for example, GAPDH in murine dendritic cells (Sahu et al., 2011). Neither did knockdown of ESCRT-II (VPS22/EAP30) or the ESCRT-III components CHMP3/VPS24, CHMP4A, and CHMP4C have any effect (Fig. 7, A and C). With the limitation that we were unable to achieve knockdown <25% of the normal expression level of Hrs (ESCRT-0); neither did this knockdown affect degradation (Fig. 7 B). The levels of autophagy receptors and lipidated LC3B increased profoundly upon siRNA knockdown of VPS4 and CHMP4B (Fig. 7, A and C). As impairment of VPS4 and ESCRT-III also inhibits macroautophagy by preventing autophagosome-lysosome fusion (Nara et al., 2002; Filimonenko et al., 2007), this likely reflects dual inhibition of the basal levels of endosomal microautophagy and macroautophagy. However, knockdown of the autophagosomal SNARE STX17 (Itakura et al., 2012) did not affect the starvation-induced degradation of autophagy receptors in BJ cells (Fig. 6 A). Knockdown of CHMP4B led to loss of MVB structures, and very interestingly, p62 was enriched at the membrane surfaces (Fig. 7 D). This suggests that p62 has an affinity for these membranes, and this probably enables the rapid internalization and degradation of p62 upon amino acid starvation. Taken together, our data show that starvation-induced endosomal microautophagy plays a key role in mediating the immediate autophagic response to starvation.
Figure 7.
The immediate autophagic response to starvation depends on the ESCRT machinery. (A) Western blot analyses of the immediate autophagic response to starvation in TSG101-, VPS4a/b-, VPS22-, or VPS24-depleted BJ cells. Starvation was initiated 72 h after siRNA transfection. Cells were starved for 1 h in HBSS. The experiments were performed three independent times with similar results. (B) Western blot analyses of the immediate autophagic response to starvation in HRS- or VPS28-depleted BJ cells. Starvation was initiated 72 h after siRNA transfection. Cells were starved for 4 h in HBSS. The experiments were performed three independent times with similar results. (C) Western blot analyses of the immediate autophagic response to starvation in CHMP4A-, CHMP4B-, or CHMP4C-depleted BJ cells. Starvation was initiated 72 h after siRNA transfection. Cells were starved for 1 h in HBSS. The experiments were performed three independent times with similar results. (D) Electron micrograph of BJ cells after siRNA-mediated CHMP4B knockdown and starvation as in C. Cryosections were labeled with antibody against p62, followed by 10 nm protein A-gold (black dots depicted by arrows, membranes indicated with arrowheads). Inset bars, 100 nm.
Discussion
In this study, we present proteomics data from an epithelial lung cancer cell line and normal human fibroblasts showing that mammalian cells respond to amino acid deprivation by eliciting an immediate autophagic response in parallel to the well-described induction of macroautophagy. Starvation-induced macroautophagy is thought of as an unselective and relatively slow bulk degradation process distinct from selective autophagy (Ohsumi, 2014; Szalai et al., 2015; Klionsky et al., 2016; Galluzzi et al., 2017). We describe a starvation-induced, rapid, and selective degradation of a subset of proteins including the autophagy receptors LC3B and GABARAPL2, which is independent of canonical macroautophagy but dependent on endosomal microautophagy, requiring the activities of the ESCRT-III component CHMP4B and the AAA ATPase VPS4. The selective autophagy receptors p62/SQSTM1, NBR1, TAX1BP1, NDP52, and NCOA4 depend on macroautophagy for efficient degradation of their cargo (Pankiv et al., 2007; Kirkin et al., 2009; Thurston et al., 2009; Newman et al., 2012; Dowdle et al., 2014; Mancias et al., 2014). Surprisingly, these proteins along with LC3B and GABARAPL2 were among the most effectively degraded by selective endosomal microautophagy upon amino acid depletion. The significant overlap of substrates identified in both cell lines suggests that the immediate autophagic response to starvation is a basal response targeting a specific subset of proteins. Interestingly, 12 of the 23 most efficiently degraded substrates in both cell lines in our study (Fig. 1 E) were among the most heavily enriched autophagy substrates identified in a recent proteomics study (Mancias et al., 2015). In that SILAC study, performed in FM with two human pancreatic cancer cell lines (PANC-1 and PA-TU-8988T) and the MCF7 breast cancer cell line, autophagic vesicles were enriched by density gradient centrifugation. Hence, in both macroautophagy and the MVB-mediated pathway, a subset of proteins including the mentioned autophagy receptors are the most efficiently degraded substrates. Recent research suggests that a starvation-induced immediate degradation response mediated by the MVB pathway ensures survival in yeast by both implementing adaptations and reinforcing nutrient supplies (Müller et al., 2015). Macroautophagy and starvation-induced endocytosis were simultaneously activated. However, the starvation-induced degradation of membrane proteins peaked much earlier than macroautophagy and was completed within 3 h. These findings suggest an important role for the MVB pathway early during starvation in yeast. Very likely, the immediate response to starvation plays a similar role in mammalian cells, with endocytosis of integrated membrane proteins and endosomal microautophagy as the most rapidly acting degradation pathways. The functional roles of many of the substrates we identified suggest that their degradation could be advantageous for survival upon prolonged starvation. Degradation of glutamine transporters SNAT1 and SNAT2 (SLC38A1 and SLC38A2) may prevent induction of apoptosis in starving cells exposed to glutamine (Bröer et al., 2016; Villar et al., 2017). The degradation of death receptors DR4 and DR5 (TNFRSF10B and D) as well as caspase 3 would attenuate apoptosis triggered by glucose starvation (Iurlaro et al., 2017).
Microautophagy occurring by invaginations of the lysosomal membrane is well characterized in yeast. In this study, the process can be both unselective and selective, involving sequestering of specific organelles including mitochondria, peroxisomes, and even portions of the nucleus (Li et al., 2012). Schizosaccharomyces pombe NBR1, a divergent evolutionary ancestor of mammalian NBR1 and p62 (Kraft et al., 2010; Svenning et al., 2011), binds to and transports two cytosolic hydrolases into MVBs. This selective transport occurred independent of macroautophagy components but dependent on ESCRT and ubiquitin (Liu et al., 2015). Hence, there is evolutionary precedence for the ability of p62 and NBR1 to be selectively sorted into MVBs. However, we found p62 and NDP52 to depend on ATG7 and ATG5 and lipidated ATG8 proteins for degradation also via the starvation-induced endosomal pathway whereas NBR1, TAX1BP1, and NCOA4 are still degraded upon starvation in ATG5 or ATG7 KO cells. NCOA4 was recently shown to be degraded also by an alternative lysosomal pathway independent of the ATG8 conjugation system upon iron depletion (Goodwin et al., 2017). In mammalian cells, endosomal microautophagy was reported in a study of murine dendritic cells in which the authors followed the cytosolic proteins GAPDH and aldolase, which were recognized by Hsc70 via their KFERQ-like motifs and sorted into intraluminal vesicles of MVBs in a manner dependent on ESCRT-I, ESCRT-III, and VPS4 (Sahu et al., 2011). We found the same requirement for ESCRT-III and VPS4 and the need for cholesterol trafficking to enable endosomal microautophagy of the selective autophagy receptors upon acute amino acid starvation as reported for GAPDH and aldolase (Sahu et al., 2011). A study of synaptic protein turnover in Drosophila melanogaster also implicated endosomal microautophagy dependent on Hsc70-4 and the KFERQ-like targeting motif (Uytterhoeven et al., 2015). Another study using a biosensor with the KFERQ motif revealed endosomal microautophagy in the larval fat body upon prolonged starvation. The degradation was first seen after 21–22 h of starvation, peaking at 25 h (Mukherjee et al., 2016). However, in the rapid starvation response, we did not find a requirement for ESCRT-0, -I, or -II or for Hsc70. Neither were GAPDH and aldolase degraded selectively during amino acid starvation, suggesting that the degradation process we describe differs both kinetically and mechanistically from the abovementioned studies.
It is unclear exactly how cytoplasmic substrates are targeted to starvation-induced endosomal microautophagy. p62 requires a functional LIR to be degraded by short-term starvation (Fig. 5 G; Itakura and Mizushima, 2011). This is dependent on ATG7 and ATG5 (Fig. 5, E and F; and Fig. S3 F). Hence, it is logical to assume that p62 is targeted to endosomal membranes by lipidated ATG8s in a similar fashion as macroautophagy. This hypothesis is supported by the recent finding that starvation-induced degradation of p62 requires at least one of the ATG8 homologues (Nguyen et al., 2016). Notably, ATG8 proteins are not exclusively confined to autophagosomal membranes but are also found on endolysosomal membranes (Nakatogawa et al., 2012; Jacquin et al., 2017; Fletcher et al., 2018). Consistent with a requirement for ATG8s in the degradation of p62, we did not observe degradation of LC3B or GABARAPL2 in ATG7 KO cells. Thus, at least these two ATG8 homologues require lipidation to be degraded by endosomal microautophagy.
Our data show that amino acid starvation is a composite signal that on one hand involves mTORC1 inactivation with subsequent activation of macroautophagy, and on the other hand leads to induction of endosomal microautophagy independent of mTORC1 inactivation. The two autophagic pathways likely work in concert and may partially compensate for each other. Apparently, this occurs with mitophagy, where mitochondria can be degraded by both macroautophagy and direct uptake into Rab5-positive endosomes (Hammerling et al., 2017). Our results clearly suggest that upon acute depletion of amino acids, it is the extracellular rather than the intracellular levels of amino acids that are sensed. How this sensing is transmitted to initiate selective endosomal microautophagy will be a challenging task for future studies.
There is currently no evidence that p62 acts as a cargo receptor during starvation-induced endosomal microautophagy. No studies show that starvation-induced macroautophagy is selective, and mitochondria and other organelles are not degraded acutely upon starvation (Kristensen et al., 2008; Gomes et al., 2011; Rambold et al., 2011). Likely, the switch from basal macroautophagy to starvation-induced macroautophagy occurs in conjunction with a switch from selective to nonselective macroautophagy. We therefore hypothesize that the rapid degradation of autophagy receptors by starvation-induced endosomal microautophagy could be a precautionary measure to prevent unsolicited selective macroautophagy (Rogov et al., 2014). The rapid degradation of autophagy receptors may be a prerequisite to obtain maximal flux in bulk macroautophagy required upon prolonged starvation.
In conclusion, we show that amino acid starvation induces an immediate autophagic response in addition to induction of bulk macroautophagy. In contrast with bulk macroautophagy, which works constitutively during starvation to recycle cytosolic cargo, the immediate autophagic response mediated by starvation-induced endosomal microautophagy secures rapid degradation of specific substrates to provide resources to sustain (and terminate) ongoing anabolic processes and to implement appropriate adaptations to survive prolonged starvation.
Materials and methods
Cell culture, chemical treatments, and SILAC
SKBR-3 and SHSY5 were cultured in RPMI 1640 supplemented with 10% FBS (S0615; VWR) and 1% streptomycin-penicillin (P4333; Sigma-Aldrich). NL20 was cultured in Opti-MEM (11058021; Thermo Fisher Scientific) supplemented with 5% FBS (S0615; VWR). All other cell lines were cultured in DMEM (D6046; Sigma-Aldrich) supplemented with 10% FBS and 1% streptomycin-penicillin. BJ, A549, and HeLa S3 cells were purchased from ATCC. Cells were treated as indicated with 200 nM bafilomycin A1, 10 µM MG132, 50 µg/ml cycloheximide, 10 µg/ml E64d, 10 µg/ml pepstatin A, 25 µM LY294002, 250 nM pp242, 250 nM rapamycin, 5 µM SAR405, and 2.5 µM PIK-III. 1–24 h pretreatment with U18666A (5 µg/ml) was used to block MVB dynamics. All cell lines were regularly tested for mycoplasma contamination. For SILAC, cells were grown for >7 d in reconstituted medium containing either “light” (12C6 l-lysine and 12C6 l-arginine) or “heavy” (13C6 l-lysine and 13C6 l-arginine) stable isotope–labeled amino acids. SILAC media, serum, and amino acids were from the SILAC Protein Quantitation Kit – DMEM (P#89983; Thermo Fisher Scientific). The additional 13C6 l-arginine was purchased from Thermo Fisher Scientific (P#88210). Additional l-proline was added to the media to prevent conversion of isotope-coded arginine to proline (P0380; Sigma-Aldrich).
Starvation procedure
Cells were grown for a minimum of 24 h to reach ∼60% confluence. Then cells were cultured in fresh media for another 12–24 h before they were briefly washed twice in acclimatized Hanks’ balanced salt solution (sodium bicarbonate buffered HBSS; H8264; Sigma-Aldrich) and finally cultured in acclimatized HBSS for the starvation period. Expression of EGFP-p62 was induced using doxycycline (0.5 µg/ml) for 24 h followed by starvation in HBSS without doxycycline.
Proteomic analysis by mass spectrometry
Cells were directly lysed in 1× Laemmli sample buffer (50 mM Tris, pH 6.8, 2% SDS, and 10% glycerol) and boiled for 10 min, then protein concentration was measured using BCA protein assay kit (23225; Thermo Fisher Scientific). Equal amounts of proteins from starved and unstarved cells were mixed, supplemented with 0.004% bromophenol blue and 10% 2-mercaptoethanol, and boiled for 5 min. The sample was fractionated by SDS-PAGE followed by Coomassie blue staining (P#24615; Thermo Fisher Scientific). Visualized lanes were separated into six fractions, and in-gel trypsin digestion was performed before analysis by high-performance liquid chromatography–tandem mass spectrometry (HPLC-MS/MS). Gel pieces were subjected to in-gel reduction, alkylation, and tryptic digestion using 6 ng/µl trypsin (V511A; Promega; Shevchenko et al., 1996). OMIX C18 tips (Varian) were used for sample cleanup and concentration. Peptide mixtures containing 0.1% formic acid were loaded onto a Thermo Fisher Scientific EASY-nLC1000 system. Samples were injected to a trap column (Acclaim PepMap 75 µm × 2 cm, C18, 3 µm, 100 Å; Thermo Fisher Scientific) for desalting before elution to the separation column (EASY-Spray column, C18, 2 µm, 100 Å, 50 µm, 50 cm; Thermo Fisher Scientific). Peptides were fractionated using a 2–100% acetonitrile gradient in 0.1% formic acid over 180 min at a flow rate of 200 nl/min. The separated peptides was analyzed using a Thermo Fisher Scientific QExactive mass spectrometer. The QExactive was operated in a data-dependent mode with the precursor scan over the range m/z 400–2,000, followed by 10 MS2 scans using parent ions selected from the MS1 scan. The Orbitrap AGC target was set to 3E06, and the MS2 AGC target was 1E05 with maximum injection times of 100 and 50 ms, respectively. For MS/MS, the LTQ isolation width was 2 m/z, and the normalized collision energy was 28%. Raw files from the QExactive MS were analyzed using the quantitative proteomics software MaxQuant (v.1.5.0.30; Cox and Mann, 2008). SILAC pairs were quantitated in MaxQuant, and proteins were identified using the built-in Andromeda search engine using the current human UniprotKB Homo sapiens (human) protein database. Main search peptide tolerance was set to 4.5 ppm, and MS/MS mass tolerance was set to 20 ppm. A false discovery rate of 0.01 was needed for protein identification. At least two peptides had to be quantitated to give a quantitation value. Statistical validation of protein regulation was done with Perseus 1.4.1.3 software.
Antibodies
The following primary antibodies were used in this study. ABIN-1/TNIP1 (37-6100; Invitrogen), ATG7 (8558; Cell Signaling Technology), CHMP4b (Sagona et al., 2010), FIP200 (SAB4200135; Sigma-Aldrich), FTH1 (3998; Cell Signaling Technology), Jak1 (sc-1677; Santa Cruz Biotechnology, Inc.), LAMP2a (ab18528; Abcam), LC3B (L7543; Sigma-Aldrich), NBR1 (sc-130380; Santa Cruz Biotechnology, Inc.), NCOA4 (SAB-1404569; Sigma-Aldrich), NDFIP1 (HPA009682; Sigma-Aldrich), NDFIP2 (HPA009160; Sigma-Aldrich), NDP52 (HPA023195; Sigma-Aldrich), p62/SQSTM1 (GP62-C; Progen, and 610833; BD), p70/S6-kinase (9202; Cell Signaling Technology), p-p70/S6-kinase (9205; Cell Signaling Technology), PSAP (10801-1-AP; Biosite), Syntenin (ab133267; Abcam), TAX1BP1 (HPA024432; Sigma-Aldrich), TSG101 (ab83; Abcam), VPS34 (PA5-34735; Thermo Fisher Scientific), pThr172AMPKα (3537; Cell Signaling Technology), GABARAPL2 (PM038; MBL), PCNA (M0879; Dako), STX-17 (PM076; MBL), VPS4 (SAB4200025; Sigma-Aldrich), VPS4A (Sc-393428; Santa Cruz Biotechnology, Inc.), VPS4B (Sc-377162; Santa Cruz Biotechnology, Inc.), GFP (ab290; Abcam), VPS22 (Malerød et al., 2007), VPS24 (Bache et al., 2006), VPS28 (SC166537; Santa Cruz Biotechnology, Inc.), and HRS (15087; Cell Signaling Technology). The following secondary antibodies were used; HRP-conjugated goat anti-rabbit and anti-mouse (554021 and 554002; BD). Alexa Fluor 555–, 488–, and 647–conjugated antibodies were purchased from Thermo Fisher Scientific and used at 1:500 dilutions.
RNAi
All siRNAs were purchased from Sigma-Aldrich as custom-made RNA oligonucleotides. The following sequences were used as targets: CHMP4a, 5′-AAGUAUGGGACCAAGAAUA-3′; CHMP4b 1, 5′-CGAUAAAGUUGAUGAGUUA-3′; CHMP4b 2 5′-AGAAGAGUUUGACGAGGAU-3′; CHMP4c, 5′-UGGCAGAACUUGAAGAAUU-3′; TSG101, 5′-CGAUGGCAGUUCCAGGGAA-3′ (Doyotte et al., 2005); VPS4a 1, 5′-CUGUGGUUUGCAUGUCGGA-3′; VPS4a 2, 5′-CCGAGAAGCUGAAGGAUUA-3′; VPS4b 1, 5′-CCAAAGAAGCACUGAAAGA-3′; VPS4b 2, 5′-GGAUGUCCCUGGAGAUAAA-3′; FIP200, 5′-ACGCAAAUCAGUUGAUGAUUA-3′ (Hirota et al., 2015); VPS34, 5′-CAGAUGGAUCAGAACCCACAA-3′ (Dall’Armi et al., 2010); LAMP2a, 5′-GACUGCAGUGCAGAUGAAG-3′ (Massey et al., 2006); STX17, 5′-CCGAAAGGAUGACCUAGUA-3′ (Guo et al., 2014); VPS28, 5′-GGCUCAGAAAUCAGCUCUA-3′; and HRS, 5′-CGACAAGAACCCACACGU-3′ (Bache et al., 2003).
Western blotting
Samples for Western blotting were harvested in 1× SDS buffer (50 mM Tris, pH 6.8, 2% SDS, and 10% glycerol), boiled for 5 min and protein concentrations measured using BCA Protein Assay Kit (23227; Pierce) and calibrated. Bromophenol blue and DTT were added to final concentrations of 0.1%, and 100 mM, respectively, before samples were run on SDS-PAGE gels at 120/160V for 90 min.
For blotting, semidry transfer was used, 100 mA per blot for 1 h using a buffer containing 14 mM glycine, 48 mM Tris, 0.03% SDS, and 0.15% ethanol. Membranes were stained with Ponceau S before blocking in 5% dry milk in 1× PBS-T for 30 min. Incubation with primary antibody was overnight at 4°C. Membranes were washed six times with 1× PBS-T before incubating with secondary antibody for 1 h at RT. Membranes were washed six times using 1× PBS-T.
Membranes were developed using SuperSignal West Femto Chemiluminescent Substrate (34095; Pierce) and SuperSignal West Pico PLUS Chemiluminescent Substrate (34078; Pierce) on Hyperfilm ECL (18 × 24; GE28-9068-37; GE Healthcare) in a Curix 60 (AGFA) or by chemiluminescence detection using ImageQuant LAS 4000 (GE Healthcare).
Quantification of amino acids by HPLC-MS/MS
850 µl buffer was evaporated and resolubilized in 20 µl methanol before derivatization with Accq-TAG (Waters Corporation) according to the manufacturer’s protocol. α-Amino butyric acid was used as internal standard. Amino acids were separated by a reverse phase X-Bridge column by a linear gradient of acetonitrile and detected by a Waters TSQ tandem quadruple instrument operated in multiple reaction mode with optimized cone voltage and collision energies for each derivatized amino acid.
Immunocytochemistry and microscopy
Cells grown on #1 or #1.5 round 12-mm coverslips were fixed for 10 min in cold (−20°C) methanol and rinsed twice with cold PBS, then incubated in 3% normal goat serum in PBS for 60 min at RT to block unspecific binding. Subsequent incubations with indicated primary antibodies and fluorescence-conjugated secondary antibodies were performed in PBS (containing either 1% normal goat serum or BSA) at RT for 60 and 30 min, respectively. Coverslips were rinsed 6× for 2 min with PBS after both incubations. Coverslips were mounted using Duolink DAPI (DUO82040; Sigma-Aldrich) and imaged by widefield or confocal microscopy.
For widefield microscopy, we used either a DMI6000B inverted microscope (Leica Microsystems) equipped with a 40× NA0.6 Plan Fluotar objective and a DFC320 charge-coupled device camera operated in monochrome mode, or an AxioObserver Z.1 inverted microscope (ZEISS) equipped with a 63× NA1.4 Plan Apochromat objective and a Rolera EM-C2 electron-multiplying charge-coupled device camera (QImaging). Image channels were acquired sequentially using appropriate filter sets for DAPI, EGFP, mCherry, and Alexa Fluor 555, 488, and 647 and combined in the corresponding microscope software.
For confocal microscopy, we used either a TCS SP5 (Leica Microsystems) or an LSM780 (ZEISS), both equipped with 63× NA1.4 Plan Apochromat objectives. For high-resolution images, z stacks were acquired with a pixel size of 50 nm and z step size of 100 nm and then iteratively deconvolved in Huygens Essential v.14.10 (Scientific Volume Imaging) using automatic theoretical point spread functions for each fluorochrome. Otherwise, pixel size was adjusted to satisfy Nyquist sampling. All imaging was performed at RT.
Image channels were acquired simultaneously or sequentially using appropriate lasers and detector settings for the above fluorochromes. Fluorescence intensity was quantified in Volocity v.6.3 (PerkinElmer) on z stacks acquired in at least 20 randomly chosen coordinates on each coverslip. Stacks were autofocused using the DAPI channel and acquired with identical (nonsaturating) scan settings for each experiment. Colocalization of fluorescent signals was visualized using the positive product of the difference from the mean (PDM) channel in Volocity. In all experiments, images shown in individual panels were acquired using identical exposure times or scan settings and adjusted identically for brightness and contrast using Photoshop CS5 (Adobe).
Cells for immuno-EM were fixed and embedded as previously described (Peters et al., 1991) with the exception that cells were fixed in 0.1 M phosphate buffer containing 4% formaldehyde and 0.15% glutaraldehyde. Small blocks were cut and infused with 2.3 M sucrose for 1 h, mounted on silver pins, and frozen in liquid nitrogen. Ultrathin cryosections were cut at −110°C on an ultramicrotome (Ultracut; Leica Biosystems) and collected with a 1:1 mixture of 2% methylcellulose and 2.3 M sucrose. Sections were transferred to formvar/carbon-coated grids and labeled with primary antibodies followed by 10 nm protein A–gold conjugates essentially as described previously (Slot et al., 1991). After embedding in 2% methyl cellulose/0.4% uranyl acetate, we observed sections at 80 kV in an electron microscope (JEOL-JEM 1230).
Rab5 overexpression and stably expressing GFP p62 MEFs
For the Rab5 overexpression studies, subconfluent cells were transfected with mCherry-labeled Rab5 plasmids (Pankiv et al., 2010) using TransIT-LT1 (MIR2300; Mirus) following the supplier’s instructions. After 24 h, fresh medium was added, and after 48 h, cells were fixed in ice-cold methanol for 10 min before immunolabeling. Cell lines stably expressing the different GFP-p62 constructs as well as GFP-SLC38A2 were made in p62 KO MEFs (gift from M. Komatsu, Niigata University, Niigata, Japan) by transfection of Platinum Retroviral Packaging cell line (RV-101; Cell Biolabs) with doxycycline-inducible pDest-LTR plasmids (Skytte Rasmussen et al., 2017) containing GFP-p62 constructs or GFP-SLC38A2 (pDONR221-SLC38A2; DNASU Plasmid Repository ID: HsCD00043884). Transfer of pENTR constructs to destination vectors was performed using the Gateway system (11791020; Thermo Fisher Scientific).
Generation of ATG7 KO HeLa cells
HeLa ATG7 KO cells were generated using CRISPR-Cas9 technology as described previously (Ran et al., 2013). Guide RNA (gRNA) targeting exon 4 of human ATG7 gene (5′-ATCCAAGGCACTACTAAAAG-3′) was annealed and ligated into the Cas9 vector (62988; Addgene) carrying both the CRISPR-associated protein 9 from Streptococcus pyogenes and the puromycin-resistance gene. HeLa cells transfected with gRNA-containing Cas9 vector were selected with 1 µg/ml puromycin for 36 h. Single cells were then sorted and plated into a 96-well plate. Single colonies were grown and KO validated by immunoblotting. Confirmed KO clones were further screened by genomic sequencing to identify indels.
Online supplemental material
Fig. S1 shows that amino acid starvation triggers a lysosomal degradation response independently of mTOR. Fig. S2 shows the top 50 most degraded proteins in BJ and A459 cells upon 4-h amino acid starvation based on SILAC experiments. Fig. S3 shows how macroautophagy does not play a major role in the immediate autophagic response to amino acid starvation. Fig. S4 shows how substrates of the immediate autophagic response to starvation are associated with the endosomal network. Fig. S5 shows that the immediate autophagic response to amino acid starvation depends on VPS4 but not HSPA8. Data S1 includes the relative quantification of proteins in BJ cells grown in full media versus starved for 4 h. Data S2 shows the relative quantification of proteins in A549 cells grown in full media versus starved for 4 h. Data S3 shows the relative quantification of proteins in BJ cells starved for 4 h versus in absence or presence of bafilomycin A1.
Supplementary Material
Acknowledgments
We thank F. Berditchevski (University of Birmingham, Birmingham, UK) for reagents and discussions. The Tromsø University core facilities for proteomics and advanced bioimaging are acknowledged for help with mass spectrometry analyses and microscopy. We thank all members of the Molecular Cancer Research Group for general assistance.
J. Mejlvang holds a career development grant from the Norwegian Cancer Society. T. Johansen was supported by Norwegian Research Council grant 196898 and Norwegian Cancer Society grant 71043-PR-2006-0320. H. Stenmark was supported by a European Research Council Advanced Grant.
The authors declare no competing financial interests.
Author contributions: J. Mejlvang, H. Olsvik, and T. Johansen wrote the manuscript. T. Hansen performed the measurements of amino acids. J.-A. Bruun performed the proteomic data analysis. T.E. Hansen generated stable cell lines expressing various GFP-p62 constructs in p62 KO MEFs. K.B. Larsen assisted with microscopy. J. Mejlvang, S. Svenning, Y.P. Abudu, and H. Brenne performed the experiments. H. Stenmark assisted in the experimental planning and result interpretations. A. Brech performed EM. All authors edited the manuscript.
References
- Artal-Martinez de Narvajas A., Gomez T.S., Zhang J.S., Mann A.O., Taoda Y., Gorman J.A., Herreros-Villanueva M., Gress T.M., Ellenrieder V., Bujanda L., et al. 2013. Epigenetic regulation of autophagy by the methyltransferase G9a. Mol. Cell. Biol. 33:3983–3993. 10.1128/MCB.00813-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bache K.G., Raiborg C., Mehlum A., and Stenmark H.. 2003. STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J. Biol. Chem. 278:12513–12521. 10.1074/jbc.M210843200 [DOI] [PubMed] [Google Scholar]
- Bache K.G., Stuffers S., Malerød L., Slagsvold T., Raiborg C., Lechardeur D., Wälchli S., Lukacs G.L., Brech A., and Stenmark H.. 2006. The ESCRT-III subunit hVps24 is required for degradation but not silencing of the epidermal growth factor receptor. Mol. Biol. Cell. 17:2513–2523. 10.1091/mbc.e05-10-0915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgisdottir A.B., Lamark T., and Johansen T.. 2013. The LIR motif—Crucial for selective autophagy. J. Cell Sci. 126:3237–3247. [DOI] [PubMed] [Google Scholar]
- Bishop N., and Woodman P.. 2000. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol. Biol. Cell. 11:227–239. 10.1091/mbc.11.1.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröer A., Rahimi F., and Bröer S.. 2016. Deletion of amino acid transporter ASCT2 (SLC1A5) reveals an essential role for transporters SNAT1 (SLC38A1) and SNAT2 (SLC38A2) to sustain glutaminolysis in cancer cells. J. Biol. Chem. 291:13194–13205. 10.1074/jbc.M115.700534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S., Kumar S., Jain A., Ponpuak M., Mudd M.H., Kimura T., Choi S.W., Peters R., Mandell M., Bruun J.A., et al. 2016. TRIMs and galectins globally cooperate and TRIM16 and galectin-3 co-direct autophagy in endomembrane damage homeostasis. Dev. Cell. 39:13–27. 10.1016/j.devcel.2016.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ L., Raiborg C., Wenzel E.M., Campsteijn C., and Stenmark H.. 2017. Cellular functions and molecular mechanisms of the ESCRT membrane-scission machinery. Trends Biochem. Sci. 42:42–56. 10.1016/j.tibs.2016.08.016 [DOI] [PubMed] [Google Scholar]
- Cox J., and Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cuervo A.M., and Wong E.. 2014. Chaperone-mediated autophagy: Roles in disease and aging. Cell Res. 24:92–104. 10.1038/cr.2013.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Armi C., Hurtado-Lorenzo A., Tian H., Morel E., Nezu A., Chan R.B., Yu W.H., Robinson K.S., Yeku O., Small S.A., et al. 2010. The phospholipase D1 pathway modulates macroautophagy. Nat. Commun. 1:142 10.1038/ncomms1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V. 2012. Autophagy as an innate immunity paradigm: Expanding the scope and repertoire of pattern recognition receptors. Curr. Opin. Immunol. 24:21–31. 10.1016/j.coi.2011.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley H.C., Razi M., Polson H.E., Girardin S.E., Wilson M.I., and Tooze S.A.. 2014. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol. Cell. 55:238–252. 10.1016/j.molcel.2014.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle W.E., Nyfeler B., Nagel J., Elling R.A., Liu S., Triantafellow E., Menon S., Wang Z., Honda A., Pardee G., et al. 2014. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat. Cell Biol. 16:1069–1079. 10.1038/ncb3053 [DOI] [PubMed] [Google Scholar]
- Doyotte A., Russell M.R., Hopkins C.R., and Woodman P.G.. 2005. Depletion of TSG101 forms a mammalian “Class E” compartment: A multicisternal early endosome with multiple sorting defects. J. Cell Sci. 118:3003–3017. 10.1242/jcs.02421 [DOI] [PubMed] [Google Scholar]
- Filimonenko M., Stuffers S., Raiborg C., Yamamoto A., Malerød L., Fisher E.M., Isaacs A., Brech A., Stenmark H., and Simonsen A.. 2007. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 179:485–500. 10.1083/jcb.200702115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimia G.M., Kroemer G., and Piacentini M.. 2013. Molecular mechanisms of selective autophagy. Cell Death Differ. 20:1–2. 10.1038/cdd.2012.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher K., Ulferts R., Jacquin E., Veith T., Gammoh N., Arasteh J.M., Mayer U., Carding S.R., Wileman T., Beale R., and Florey O.. 2018. The WD40 domain of ATG16L1 is required for its non-canonical role in lipidation of LC3 at single membranes. EMBO J. 37:e97840 10.15252/embj.201797840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L., Baehrecke E.H., Ballabio A., Boya P., Bravo-San Pedro J.M., Cecconi F., Choi A.M., Chu C.T., Codogno P., Colombo M.I., et al. 2017. Molecular definitions of autophagy and related processes. EMBO J. 36:1811–1836. 10.15252/embj.201796697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes L.C., Di Benedetto G., and Scorrano L.. 2011. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 13:589–598. 10.1038/ncb2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin J.M., Dowdle W.E., DeJesus R., Wang Z., Bergman P., Kobylarz M., Lindeman A., Xavier R.J., McAllister G., Nyfeler B., et al. 2017. Autophagy-independent lysosomal targeting regulated by ULK1/2-FIP200 and ATG9. Cell Reports. 20:2341–2356. 10.1016/j.celrep.2017.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., Liang Q., Li L., Hu Z., Wu F., Zhang P., Ma Y., Zhao B., Kovács A.L., Zhang Z., et al. 2014. O-GlcNAc-modification of SNAP-29 regulates autophagosome maturation. Nat. Cell Biol. 16:1215–1226. 10.1038/ncb3066 [DOI] [PubMed] [Google Scholar]
- Hamacher-Brady A., and Brady N.R.. 2016. Mitophagy programs: Mechanisms and physiological implications of mitochondrial targeting by autophagy. Cell. Mol. Life Sci. 73:775–795. 10.1007/s00018-015-2087-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerling B.C., Najor R.H., Cortez M.Q., Shires S.E., Leon L.J., Gonzalez E.R., Boassa D., Phan S., Thor A., Jimenez R.E., et al. 2017. A Rab5 endosomal pathway mediates Parkin-dependent mitochondrial clearance. Nat. Commun. 8:14050 10.1038/ncomms14050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota Y., Yamashita S., Kurihara Y., Jin X., Aihara M., Saigusa T., Kang D., and Kanki T.. 2015. Mitophagy is primarily due to alternative autophagy and requires the MAPK1 and MAPK14 signaling pathways. Autophagy. 11:332–343. 10.1080/15548627.2015.1023047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S., Natsume T., Takehana K., Yamada N., et al. 2009. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 20:1981–1991. 10.1091/mbc.e08-12-1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell J.J., and Manning B.D.. 2011. mTOR couples cellular nutrient sensing to organismal metabolic homeostasis. Trends Endocrinol. Metab. 22:94–102. 10.1016/j.tem.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., et al. 2000. A ubiquitin-like system mediates protein lipidation. Nature. 408:488–492. 10.1038/35044114 [DOI] [PubMed] [Google Scholar]
- Ichimura Y., Waguri S., Sou Y.S., Kageyama S., Hasegawa J., Ishimura R., Saito T., Yang Y., Kouno T., Fukutomi T., et al. 2013. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol. Cell. 51:618–631. 10.1016/j.molcel.2013.08.003 [DOI] [PubMed] [Google Scholar]
- Itakura E., and Mizushima N.. 2011. p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J. Cell Biol. 192:17–27. 10.1083/jcb.201009067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi-Itakura C., and Mizushima N.. 2012. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 151:1256–1269. 10.1016/j.cell.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Iurlaro R., Püschel F., León-Annicchiarico C.L., O’Connor H., Martin S.J., Palou-Gramón D., Lucendo E., and Muñoz-Pinedo C.. 2017. Glucose deprivation induces ATF4-mediated apoptosis through TRAIL death receptors. Mol. Cell. Biol. 37:e00479–16. 10.1128/MCB.00479-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaber N., Dou Z., Chen J.S., Catanzaro J., Jiang Y.P., Ballou L.M., Selinger E., Ouyang X., Lin R.Z., Zhang J., and Zong W.X.. 2012. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc. Natl. Acad. Sci. USA. 109:2003–2008. 10.1073/pnas.1112848109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquin E., Leclerc-Mercier S., Judon C., Blanchard E., Fraitag S., and Florey O.. 2017. Pharmacological modulators of autophagy activate a parallel noncanonical pathway driving unconventional LC3 lipidation. Autophagy. 13:854–867. 10.1080/15548627.2017.1287653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P., and Mizushima N.. 2015. LC3- and p62-based biochemical methods for the analysis of autophagy progression in mammalian cells. Methods. 75:13–18. 10.1016/j.ymeth.2014.11.021 [DOI] [PubMed] [Google Scholar]
- Jiang H.Y., Wek S.A., McGrath B.C., Lu D., Hai T., Harding H.P., Wang X., Ron D., Cavener D.R., and Wek R.C.. 2004. Activating transcription factor 3 is integral to the eukaryotic initiation factor 2 kinase stress response. Mol. Cell. Biol. 24:1365–1377. 10.1128/MCB.24.3.1365-1377.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T., and Lamark T.. 2011. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 7:279–296. 10.4161/auto.7.3.14487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.E., Overmeyer J.H., Gunning W.T., and Maltese W.A.. 2006. Gene silencing reveals a specific function of hVps34 phosphatidylinositol 3-kinase in late versus early endosomes. J. Cell Sci. 119:1219–1232. 10.1242/jcs.02833 [DOI] [PubMed] [Google Scholar]
- Jones C.B., Ott E.M., Keener J.M., Curtiss M., Sandrin V., and Babst M.. 2012. Regulation of membrane protein degradation by starvation-response pathways. Traffic. 13:468–482. 10.1111/j.1600-0854.2011.01314.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., and Yoshimori T.. 2004. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117:2805–2812. 10.1242/jcs.01131 [DOI] [PubMed] [Google Scholar]
- Khaminets A., Heinrich T., Mari M., Grumati P., Huebner A.K., Akutsu M., Liebmann L., Stolz A., Nietzsche S., Koch N., et al. 2015. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 522:354–358. 10.1038/nature14498 [DOI] [PubMed] [Google Scholar]
- Khaminets A., Behl C., and Dikic I.. 2016. Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. 26:6–16. 10.1016/j.tcb.2015.08.010 [DOI] [PubMed] [Google Scholar]
- Kimura T., Mandell M., and Deretic V.. 2016. Precision autophagy directed by receptor regulators—Emerging examples within the TRIM family. J. Cell Sci. 129:881–891. 10.1242/jcs.163758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V., Lamark T., Sou Y.S., Bjørkøy G., Nunn J.L., Bruun J.A., Shvets E., McEwan D.G., Clausen T.H., Wild P., et al. 2009. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 33:505–516. 10.1016/j.molcel.2009.01.020 [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., Abdelmohsen K., Abe A., Abedin M.J., Abeliovich H., Acevedo Arozena A., Adachi H., Adams C.M., Adams P.D., Adeli K., et al. 2016. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 12:1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C., Peter M., and Hofmann K.. 2010. Selective autophagy: Ubiquitin-mediated recognition and beyond. Nat. Cell Biol. 12:836–841. 10.1038/ncb0910-836 [DOI] [PubMed] [Google Scholar]
- Kristensen A.R., Schandorff S., Høyer-Hansen M., Nielsen M.O., Jäättelä M., Dengjel J., and Andersen J.S.. 2008. Ordered organelle degradation during starvation-induced autophagy. Mol. Cell. Proteomics. 7:2419–2428. 10.1074/mcp.M800184-MCP200 [DOI] [PubMed] [Google Scholar]
- Lamb C.A., Yoshimori T., and Tooze S.A.. 2013. The autophagosome: Origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 14:759–774. 10.1038/nrm3696 [DOI] [PubMed] [Google Scholar]
- Lefebvre C., Legouis R., and Culetto E.. 2018. ESCRT and autophagies: Endosomal functions and beyond. Semin. Cell Dev. Biol. 74:21–28. 10.1016/j.semcdb.2017.08.014 [DOI] [PubMed] [Google Scholar]
- Li W.W., Li J., and Bao J.K.. 2012. Microautophagy: Lesser-known self-eating. Cell. Mol. Life Sci. 69:1125–1136. 10.1007/s00018-011-0865-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum L., and Faust J.R.. 1989. The intracellular transport of low density lipoprotein-derived cholesterol is inhibited in Chinese hamster ovary cells cultured with 3-beta-[2-(diethylamino)ethoxy]androst-5-en-17-one. J. Biol. Chem. 264:11796–11806. [PubMed] [Google Scholar]
- Liu C.C., Lin Y.C., Chen Y.H., Chen C.M., Pang L.Y., Chen H.A., Wu P.R., Lin M.Y., Jiang S.T., Tsai T.F., and Chen R.H.. 2016. Cul3-KLHL20 ubiquitin ligase governs the turnover of ULK1 and VPS34 complexes to control autophagy termination. Mol. Cell. 61:84–97. 10.1016/j.molcel.2015.11.001 [DOI] [PubMed] [Google Scholar]
- Liu X.M., Sun L.L., Hu W., Ding Y.H., Dong M.Q., and Du L.L.. 2015. ESCRTs cooperate with a selective autophagy receptor to mediate vacuolar targeting of soluble cargos. Mol. Cell. 59:1035–1042. 10.1016/j.molcel.2015.07.034 [DOI] [PubMed] [Google Scholar]
- Malerød L., Stuffers S., Brech A., and Stenmark H.. 2007. Vps22/EAP30 in ESCRT-II mediates endosomal sorting of growth factor and chemokine receptors destined for lysosomal degradation. Traffic. 8:1617–1629. 10.1111/j.1600-0854.2007.00630.x [DOI] [PubMed] [Google Scholar]
- Mancias J.D., Wang X., Gygi S.P., Harper J.W., and Kimmelman A.C.. 2014. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 509:105–109. 10.1038/nature13148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancias J.D., Pontano Vaites L., Nissim S., Biancur D.E., Kim A.J., Wang X., Liu Y., Goessling W., Kimmelman A.C., and Harper J.W.. 2015. Ferritinophagy via NCOA4 is required for erythropoiesis and is regulated by iron dependent HERC2-mediated proteolysis. eLife. 4:e10308 10.7554/eLife.10308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell M.A., Jain A., Arko-Mensah J., Chauhan S., Kimura T., Dinkins C., Silvestri G., Münch J., Kirchhoff F., Simonsen A., et al. 2014. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev. Cell. 30:394–409. 10.1016/j.devcel.2014.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzella L., Ahlberg J., and Glaumann H.. 1981. Autophagy, heterophagy, microautophagy and crinophagy as the means for intracellular degradation. Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 36:219–234. 10.1007/BF02912068 [DOI] [PubMed] [Google Scholar]
- Massey A.C., Kaushik S., Sovak G., Kiffin R., and Cuervo A.M.. 2006. Consequences of the selective blockage of chaperone-mediated autophagy. Proc. Natl. Acad. Sci. USA. 103:5805–5810. 10.1073/pnas.0507436103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlpine F., Williamson L.E., Tooze S.A., and Chan E.Y.. 2013. Regulation of nutrient-sensitive autophagy by uncoordinated 51-like kinases 1 and 2. Autophagy. 9:361–373. 10.4161/auto.23066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. 2004. Methods for monitoring autophagy. Int. J. Biochem. Cell Biol. 36:2491–2502. 10.1016/j.biocel.2004.02.005 [DOI] [PubMed] [Google Scholar]
- Mizushima N., and Hara T.. 2006. Intracellular quality control by autophagy: How does autophagy prevent neurodegeneration? Autophagy. 2:302–304. 10.4161/auto.2945 [DOI] [PubMed] [Google Scholar]
- Mizushima N., and Komatsu M.. 2011. Autophagy: Renovation of cells and tissues. Cell. 147:728–741. 10.1016/j.cell.2011.10.026 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., and Ohsumi Y.. 2011. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27:107–132. 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- Mukherjee A., Patel B., Koga H., Cuervo A.M., and Jenny A.. 2016. Selective endosomal microautophagy is starvation-inducible in Drosophila. Autophagy. 12:1984–1999. 10.1080/15548627.2016.1208887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M., Schmidt O., Angelova M., Faserl K., Weys S., Kremser L., Pfaffenwimmer T., Dalik T., Kraft C., Trajanoski Z., et al. 2015. The coordinated action of the MVB pathway and autophagy ensures cell survival during starvation. eLife. 4:e07736 10.7554/eLife.07736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mund T., and Pelham H.R.. 2010. Regulation of PTEN/Akt and MAP kinase signaling pathways by the ubiquitin ligase activators Ndfip1 and Ndfip2. Proc. Natl. Acad. Sci. USA. 107:11429–11434. 10.1073/pnas.0911714107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H., Ishii J., Asai E., and Ohsumi Y.. 2012. Atg4 recycles inappropriately lipidated Atg8 to promote autophagosome biogenesis. Autophagy. 8:177–186. 10.4161/auto.8.2.18373 [DOI] [PubMed] [Google Scholar]
- Nara A., Mizushima N., Yamamoto A., Kabeya Y., Ohsumi Y., and Yoshimori T.. 2002. SKD1 AAA ATPase-dependent endosomal transport is involved in autolysosome formation. Cell Struct. Funct. 27:29–37. 10.1247/csf.27.29 [DOI] [PubMed] [Google Scholar]
- Newman A.C., Scholefield C.L., Kemp A.J., Newman M., McIver E.G., Kamal A., and Wilkinson S.. 2012. TBK1 kinase addiction in lung cancer cells is mediated via autophagy of Tax1bp1/Ndp52 and non-canonical NF-κB signalling. PLoS One. 7:e50672 10.1371/journal.pone.0050672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.N., Padman B.S., Usher J., Oorschot V., Ramm G., and Lazarou M.. 2016. Atg8 family LC3/GABARAP proteins are crucial for autophagosome–lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J. Cell Biol. 215:857–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y. 2014. Historical landmarks of autophagy research. Cell Res. 24:9–23. 10.1038/cr.2013.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., and Johansen T.. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145. 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Pankiv S., Alemu E.A., Brech A., Bruun J.A., Lamark T., Overvatn A., Bjørkøy G., and Johansen T.. 2010. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end–directed vesicle transport. J. Cell Biol. 188:253–269. 10.1083/jcb.200907015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters P.J., Neefjes J.J., Oorschot V., Ploegh H.L., and Geuze H.J.. 1991. Segregation of MHC class II molecules from MHC class I molecules in the Golgi complex for transport to lysosomal compartments. Nature. 349:669–676. 10.1038/349669a0 [DOI] [PubMed] [Google Scholar]
- Petherick K.J., Williams A.C., Lane J.D., Ordóñez-Morán P., Huelsken J., Collard T.J., Smartt H.J., Batson J., Malik K., Paraskeva C., and Greenhough A.. 2013. Autolysosomal β-catenin degradation regulates Wnt-autophagy-p62 crosstalk. EMBO J. 32:1903–1916. 10.1038/emboj.2013.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C., and Stenmark H.. 2009. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 458:445–452. 10.1038/nature07961 [DOI] [PubMed] [Google Scholar]
- Rambold A.S., Kostelecky B., Elia N., and Lippincott-Schwartz J.. 2011. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. USA. 108:10190–10195. 10.1073/pnas.1107402108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov V., Dötsch V., Johansen T., and Kirkin V.. 2014. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell. 53:167–178. 10.1016/j.molcel.2013.12.014 [DOI] [PubMed] [Google Scholar]
- Ronan B., Flamand O., Vescovi L., Dureuil C., Durand L., Fassy F., Bachelot M.F., Lamberton A., Mathieu M., Bertrand T., et al. 2014. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat. Chem. Biol. 10:1013–1019. 10.1038/nchembio.1681 [DOI] [PubMed] [Google Scholar]
- Sagona A.P., Nezis I.P., Pedersen N.M., Liestøl K., Poulton J., Rusten T.E., Skotheim R.I., Raiborg C., and Stenmark H.. 2010. PtdIns(3)P controls cytokinesis through KIF13A-mediated recruitment of FYVE-CENT to the midbody. Nat. Cell Biol. 12:362–371. 10.1038/ncb2036 [DOI] [PubMed] [Google Scholar]
- Sahu R., Kaushik S., Clement C.C., Cannizzo E.S., Scharf B., Follenzi A., Potolicchio I., Nieves E., Cuervo A.M., and Santambrogio L.. 2011. Microautophagy of cytosolic proteins by late endosomes. Dev. Cell. 20:131–139. 10.1016/j.devcel.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton R.A., and Sabatini D.M.. 2017. mTOR signaling in growth, metabolism, and disease. Cell. 169:361–371. 10.1016/j.cell.2017.03.035 [DOI] [PubMed] [Google Scholar]
- Scott L., Lamb J., Smith S., and Wheatley D.N.. 2000. Single amino acid (arginine) deprivation: Rapid and selective death of cultured transformed and malignant cells. Br. J. Cancer. 83:800–810. 10.1054/bjoc.2000.1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seglen P.O., Gordon P.B., Holen I., and Høyvik H.. 1991. Hepatocytic autophagy. Biomed. Biochim. Acta. 50:373–381. [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., and Mann M.. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850–858. 10.1021/ac950914h [DOI] [PubMed] [Google Scholar]
- Shpilka T., Weidberg H., Pietrokovski S., and Elazar Z.. 2011. Atg8: An autophagy-related ubiquitin-like protein family. Genome Biol. 12:226 10.1186/gb-2011-12-7-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skytte Rasmussen M., Mouilleron S., Kumar Shrestha B., Wirth M., Lee R., Bowitz Larsen K., Abudu Princely Y., O’Reilly N., Sjøttem E., Tooze S.A., et al. 2017. ATG4B contains a C-terminal LIR motif important for binding and efficient cleavage of mammalian orthologs of yeast Atg8. Autophagy. 13:834–853. 10.1080/15548627.2017.1287651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot J.W., Geuze H.J., Gigengack S., Lienhard G.E., and James D.E.. 1991. Immuno-localization of the insulin regulatable glucose transporter in brown adipose tissue of the rat. J. Cell Biol. 113:123–135. 10.1083/jcb.113.1.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H., Parton R.G., Steele-Mortimer O., Lütcke A., Gruenberg J., and Zerial M.. 1994. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 13:1287–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A., Ernst A., and Dikic I.. 2014. Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol. 16:495–501. 10.1038/ncb2979 [DOI] [PubMed] [Google Scholar]
- Svenning S., Lamark T., Krause K., and Johansen T.. 2011. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy. 7:993–1010. 10.4161/auto.7.9.16389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalai P., Hagen L.K., Sætre F., Luhr M., Sponheim M., Øverbye A., Mills I.G., Seglen P.O., and Engedal N.. 2015. Autophagic bulk sequestration of cytosolic cargo is independent of LC3, but requires GABARAPs. Exp. Cell Res. 333:21–38. 10.1016/j.yexcr.2015.02.003 [DOI] [PubMed] [Google Scholar]
- Thurston T.L., Ryzhakov G., Bloor S., von Muhlinen N., and Randow F.. 2009. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 10:1215–1221. 10.1038/ni.1800 [DOI] [PubMed] [Google Scholar]
- Uytterhoeven V., Lauwers E., Maes I., Miskiewicz K., Melo M.N., Swerts J., Kuenen S., Wittocx R., Corthout N., Marrink S.J., et al. 2015. Hsc70-4 deforms membranes to promote synaptic protein turnover by endosomal microautophagy. Neuron. 88:735–748. 10.1016/j.neuron.2015.10.012 [DOI] [PubMed] [Google Scholar]
- Villar V.H., Nguyen T.L., Delcroix V., Terés S., Bouchecareilh M., Salin B., Bodineau C., Vacher P., Priault M., Soubeyran P., and Durán R.V.. 2017. mTORC1 inhibition in cancer cells protects from glutaminolysis-mediated apoptosis during nutrient limitation. Nat. Commun. 8:14124 10.1038/ncomms14124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Wang X., Su Z., Fei H., Liu X., and Pan Q.. 2015. The novel mTOR inhibitor Torin-2 induces autophagy and downregulates the expression of UHRF1 to suppress hepatocarcinoma cell growth. Oncol. Rep. 34:1708–1716. 10.3892/or.2015.4146 [DOI] [PubMed] [Google Scholar]
- Wang R.C., Wei Y., An Z., Zou Z., Xiao G., Bhagat G., White M., Reichelt J., and Levine B.. 2012. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science. 338:956–959. 10.1126/science.1225967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe-Asano T., Kuma A., and Mizushima N.. 2014. Cycloheximide inhibits starvation-induced autophagy through mTORC1 activation. Biochem. Biophys. Res. Commun. 445:334–339. 10.1016/j.bbrc.2014.01.180 [DOI] [PubMed] [Google Scholar]
- Yun W.J., Kim E.Y., Park J.E., Jo S.Y., Bang S.H., Chang E.J., and Chang S.E.. 2016. Microtubule-associated protein light chain 3 is involved in melanogenesis via regulation of MITF expression in melanocytes. Sci. Rep. 6:19914 10.1038/srep19914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Shen S., Qu J., and Ghaemmaghami S.. 2016. Global analysis of cellular protein flux quantifies the selectivity of basal autophagy. Cell Reports. 14:2426–2439. 10.1016/j.celrep.2016.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.