Multiple ER-phagy receptors have been reported recently, but other mediators or regulators have remained elusive. Liang et al. developed two ER-phagy–specific reporter assays and showed that Atlastins, a family of ER surface GTPases, are positive regulators of ER-phagy that act downstream of an ER-phagy receptor, FAM134B.
Abstract
Specific receptors are required for the autophagic degradation of endoplasmic reticulum (ER), known as ER-phagy. However, little is known about how the ER is remodeled and separated for packaging into autophagosomes. We developed two ER-phagy–specific reporter systems and found that Atlastins are key positive effectors and also targets of ER-phagy. Atlastins are ER-resident GTPases involved in ER membrane morphology, and Atlastin-depleted cells have decreased ER-phagy under starvation conditions. Atlastin’s role in ER-phagy requires a functional GTPase domain and proper ER localization, both of which are also involved in ER architecture. The three Atlastin family members functionally compensate for one another during ER-phagy and may form heteromeric complexes with one another. We further find that Atlastins act downstream of the FAM134B ER-phagy receptor, such that depletion of Atlastins represses ER-autophagy induced by the overexpression of FAM134B. We propose that during ER-phagy, Atlastins remodel ER membrane to separate pieces of FAM134B-marked ER for efficient autophagosomal engulfment.
Introduction
The selective autophagy of organelles (organellophagy) constitutes a major part of cellular proteostasis and homeostasis. Dysregulation in organellophagy particularly impacts differentiated cells, such as neurons. The most notable example is mitophagy, whereby loss-of-function mutations of mitophagy proteins such as PARKIN and PINK1 have been linked to neurodegenerative diseases such as Parkinson’s disease (Pickrell and Youle, 2015).
The ER is a multifunctional organelle that is the major site for protein and lipid synthesis, as well as the quality control of newly synthesized proteins. To prevent the accumulation of toxic protein aggregates, the ER harbors a well-studied quality control pathway known as ER-associated degradation, in which misfolded ER proteins are extracted for destruction by the proteasome (Brodsky, 2012).
Under certain conditions such as starvation, fragments of the ER are engulfed in their entirety by autophagosomes and sent for destruction in acidified lysosomes in a process known as ER-phagy (Mochida et al., 2015; Dikic, 2017). Originally described in yeast, ER-phagy has recently been demonstrated to occur in higher eukaryotic cells (Schuck et al., 2014; Mochida et al., 2015; Nakatogawa, 2015). Several ER surface receptors, including FAM134B, reticulon 3L (RTN3L), Sec62, and CCPG1, have been shown to contain conserved LC3-interacting regions (LIRs) that can act as specific autophagy receptors to allow portions of the larger ER network to be shunted to core autophagy pathways (Khaminets et al., 2015; Fumagalli et al., 2016; Grumati et al., 2017; Smith et al., 2018). ER-phagy is therefore connected to bulk autophagy of the cytoplasm but may have dedicated upstream logic, signals, and mediators that are only beginning to be elucidated. For example, unlike cytoplasm, the ER is composed of a highly interconnected membrane-bound network. It is currently unclear how ER portions targeted for autophagy are isolated from the rest of the ER and packaged into discrete components for delivery to autophagosomes.
The ER network consists of complex connections of ER tubules and sheets that are constantly remodeled during normal homeostasis. This process is fulfilled by a variety of ER membrane surface proteins, such as RTNs and REEPs (involved in ER tubule formation) and CLIMP63 and FAM134B (involved in ER sheet formation; Klopfenstein et al., 1998; Voeltz et al., 2006; Nikonov et al., 2007; Shibata et al., 2008; Sparkes et al., 2010; Khaminets et al., 2015). ER-integral membrane proteins known as Atlastins (ATLs) are also involved in the fusion of ER tubules to form three-way junctions that yield the characteristic weblike network of the ER (Rismanchi et al., 2008; Wang et al., 2016; Zhao et al., 2016).
We hypothesized that the ER should be remodeled before autophagic engulfment and that ER-remodeling proteins might facilitate this process. We adapted several assays previously used to measure general autophagy to instead report on organelle-specific autophagy, with a focus on ER-phagy. With these assays in hand, we used CRISPR transcriptional inhibition (CRISPRi) to show that ATLs are required for ER-phagy in human cells during nutrient starvation. The three human ATL family members are expressed at different levels in various cell types and are functionally redundant during ER-phagy. ATLs contain an N-terminal GTPase domain and two transmembrane (TM) helices close to the C terminus that span the ER membrane, such that both N and C termini face the cytosol (Fig. S1 A). In cells that predominantly express ATL2, we find that ER-phagy requires the N-terminal GTPase domain, proper ER localization through the TM domain, and a C-terminal helical tail that is also required for ER membrane remodeling. Overexpression of FAM134B is sufficient to induce ER-phagy and partial loss of ATL2, suggesting that ATL2 could be a remodeling factor for the same ER subdomain marked by FAM134B. However, removal of ATL2 during FAM134B overexpression abrogates ER-phagy. Our results uncover a new mediator of ER-phagy that we propose is required to remodel and separate the ER components marked for destruction by the FAM134B ER-phagy receptor.
Results and discussion
Quantitative assays for ER-phagy
One of the obstacles in studying ER-phagy is the paucity of robust markers to monitor the process (Khaminets et al., 2015; Mochida et al., 2015; Grumati et al., 2017). Commonly used ER protein markers such as calnexin (CANX), PDI, CLIMP63, TRAF-α, and REEP5 exhibit only a marginal decrease in protein levels during ER-phagy, and it is difficult to use them to establish quantitative measures of ER-phagy. To overcome these limitations, we developed two complementary assays for ER-phagy adapted from previous assays used to measure general autophagy flux.
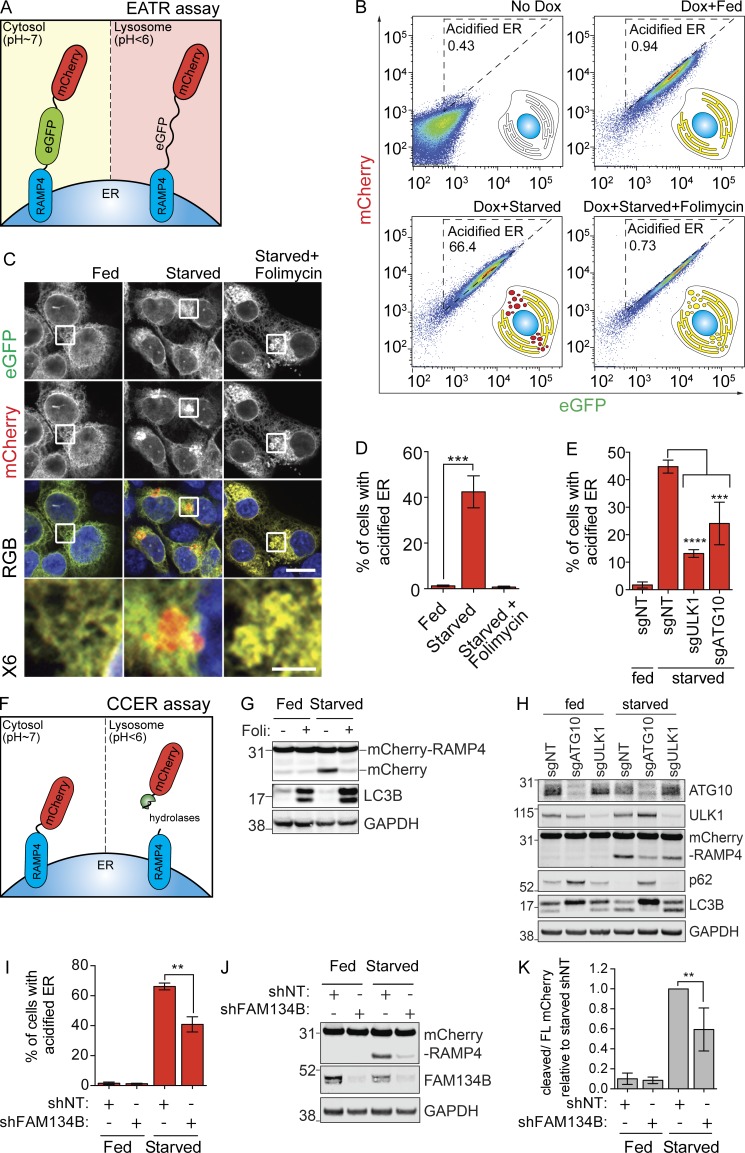
First, we modified a tandem eGFP-mCherry-LC3 reporter often used to measure general autophagy and fused it instead to RAMP4, a subunit of the ER translocon complex, to report specifically on lysosomal degradation of the ER (Kimura et al., 2007; Mizushima et al., 2010). For simplicity, we refer this system as ER autophagy tandem reporter (EATR; Fig. 1 A). EATR is designed such that the fluorescent proteins point toward the cytoplasm so that they are exposed to the lysosome upon engulfment, and takes advantage of the lowered stability of eGFP relative to mCherry when delivered to the acidic environment of the lysosome. Cytosolic ER (pH ∼7) should be fluorescent in both the eGFP and mCherry channels, whereas ER within a lysosomal vesicle (pH <4) would lose eGFP fluorescence but retain mCherry.
Figure 1.
Development of sensitive and quantitative assays to measure ER-phagy. (A) Schematic of the EATR assay. eGFP is quenched as a result of low pH-induced protonation, causing a switch from GFP+/mCherry+ to GFP−/mCherry+ during ER-phagy. (B) HCT116 cells expressing the EATR reporter were starved for 16 h with or without folimycin (100 nM). Cells within the acidified ER gate are undergoing ER-phagy. Reducing lysosomal acidification with folimycin moves cells out of the acidified ER gate. (C) EATR HCT116 cells were starved and fixed to visualize eGFP quenching. Scale bar represents 20 µm. Insets represent sixfold enlargement of the boxed area. Scale bar for sixfold enlarged images represent 5 µm. (D) Flow cytometry measurement of B is quantified as percentage of cells with more acidified ER. Data presented as mean ± SD of three biological replicates. P value indicates two-tailed unpaired t test (***, P < 0.0005). (E) EATR CRISPRi HCT116 cells stably expressing sgRNAs were starved for 16 h. Data presented as mean ± SD of three biological replicates. P value indicates one-way ANOVA with Dunnett’s multiple comparisons test (***, P < 0.001; ****, P < 0.0001). (F) Schematic illustration of the CCER assay. ER targeted for degradation results in the lysosomal cleavage of the mCherry tag from RAMP4, resulting in the formation of a smaller, mCherry-only product that can be resolved by Western blotting. (G) CCER HCT116 cells were starved for 16 h in EBSS ± folimycin before Western blotting. (H) CCER CRISPRi HCT116 cells stably expressing sgRNAs were starved for 16 h before Western blotting. (I) EATR HCT116 cells were transfected with shRNA targeting FAM134B and starved for 16 h before FACS measurement. Data presented as mean ± SD of three biological replicates. P value indicates two-tailed unpaired t test (**, P < 0.001). (J) CCER HCT116 cells were treated as in I, harvested, and subjected to Western blot. (K) Quantification of data in J. Data presented as mean ± SD of three biological replicates. P value indicates two-tailed unpaired t test (**, P < 0.01).
Tandem fluorescent protein systems have previously been adapted to study mitophagy and have even been fused to FAM134B to demonstrate its role as an ER-phagy reporter (Khaminets et al., 2015). We chose RAMP4 as a marker for ER, as it is a small TM ER protein that is ubiquitously expressed in various ER subdomains of the cells. By microscopy, we found that EATR colocalizes with both REEP5 (ER tubules) and CLIMP63 (ER sheets; Fig. S1 B). Expression of EATR does not alter the degradation rate of ER proteins or general autophagy upon starvation compared with parental HCT116 cells (Fig. S1 C). We placed EATR under the control of a doxycycline-inducible promoter to control expression in a defined time period and hence follow a bolus of ER as it transitions from cytoplasmic to lysosomal and is finally degraded (Fig. 1 B).
We created HCT116 cell lines harboring the inducible EATR construct to monitor ER autophagy by fluorescence microscopy. Although EATR is localized to the ER when cells are grown in complete media (“fed”; Fig. 1 C), transitioning cells from fed to Earl’s buffered saline solution (EBSS; “starved”) induces the formation of mCherry-only ER puncta that are recruited to a perinuclear region (Fig. 1 C). This is consistent with studies that lysosomes are perinuclear and that autophagic ER cargoes are trafficked to a perinuclear space (Korolchuk et al., 2011; Khaminets et al., 2015). Treatment with folimycin, an inhibitor of the lysosomal proton pump, rescued GFP fluorescence in perinuclear ER (Fig. 1 C).
We next asked if EATR could be used in a flow cytometry assay to quantify the numbers of cells undergoing various levels of ER-phagy. In this mode, the relative change in eGFP and mCherry fluorescence intensity reports on delivery of the ER to the acidic lysosomal environment. We first validated that doxycycline induction results in expression of mCherry and eGFP, with each cell having roughly equal amounts of mCherry and eGFP fluorescence (Fig. 1 B, top two columns). Upon starvation, we observed a decrease in eGFP signal but not mCherry fluorescence, resulting in a shift of cell population into the “acidified ER” gate (Fig. 1 B, bottom left). Consistent with our microscopy results, folimycin treatment rescued eGFP fluorescence on the ER, presumably because of deacidification of the lysosome (Fig. 1 B, bottom right). Quantifying the number of cells that fall into the acidified ER gate yields a metric of ER-phagy that is reproducible between biological replicates (Fig. 1 D).
To ensure that our EATR assay reports on bona fide autophagy of the ER, we used CRISPRi to knock down ULK1, a core autophagy-activating kinase, and ATG10, the E2-like ligase required for LC3 activation and lipidation (Fig. 1 E). CRISPRi works by stable expression of catalytically inactive dCas9 fused with a KRAB transcriptional repressor domain (dCas9-KRAB) in combination with a single guide RNA (sgRNA) targeting the transcriptional start site of a gene of interest (Gilbert et al., 2014). We chose CRISPRi for gene knockdown because of its high efficacy and low off-target effects compared with siRNA, plus the ability to rapidly assess phenotypes in bulk knockdown cells without the confounding mixed genotypes associated with bulk CRISPR editing (Gilbert et al., 2014; Horlbeck et al., 2016). In EATR CRISPRi HCT116 cells transduced with an sgRNA targeting ULK1 or ATG10, we observed robust inhibition of ER-phagy in both cases, suggesting that EATR reports on ER-phagy that relies on the canonical autophagy pathway for ER degradation (Fig. 1 E). In sum, we find that the EATR system is capable of measuring ER-phagy both visually by microscopy and quantitatively by flow cytometry.
Encouraged by the ER-phagy results with EATR, we adapted the tandem reporter system to study other organellophagies by fusing the tandem fluorescent reporters to different organelle surface proteins, including TOM20 for mitophagy, PMP34 for pexophagy, and LAMP1 for lysophagy (Fig. S1 D). Similar to the EATR construct, the tandem fluorescent reporters were fused to the organelle-specific protein such that they point toward the cytoplasm and allow for the detection of pH change upon lysosomal localization. In all cases, we observed robust and characteristic localization of all tandem fluorescent reporters to their expected organelles upon doxycycline induction (Fig. S1 D). While these reporters are less well characterized than EATR, all the constructs are publicly available on Addgene and could be useful in the future study of other organellophagies.
We desired an orthogonal assay for ER-phagy that does not depend on a change in fluorescence and took inspiration from bulk autophagy assays in yeast that measure lysosomal cleavage of GFP from LC3 as a readout for autophagy (Hosokawa et al., 2007). GFP-LC3 is effective in yeast but not human cells, because of differences in lysosomal acidification (Mizushima et al., 2010). However, we reasoned that the stability of mCherry in the flow assay could be a replacement for GFP to measure tag separation during autophagy. We found that a stably expressed mCherry-RAMP4 fusion (lacking eGFP) is cleaved to free mCherry during starvation, such that Western blotting for mCherry yields two distinct bands (Fig. 1 F and G). Treatment with folimycin prevents release of mCherry, indicating that this separation reports on the internalization of ER by lysosomes (Fig. 1 G). Endogenous staining with LAMP2 (lysosomes) and LC3 (autophagosomes) showed colocalization with mCherry-RAMP4 only during starvation, consistent with the initiation of ER-phagy (Fig. S1, E and F). We further validated that the mCherry reporter does not overflow into other organelles such as the Golgi and mitochondria (Fig. S1 G). For simplicity, we refer to the mCherry-RAMP4 ER-phagy assay as mCherry cleavage from ER (CCER).
CCER allows for more sensitive detection of ER-phagy as opposed to measurement on the loss of endogenous ER protein, since the appearance and accumulation of the mCherry band from the autophagocytosed portion of the ER provides a more obvious change than the marginal loss of endogenous ER proteins relative to the rest of the ER (Fig. S1 H). We found that the time course of ER-phagy measured by CCER is consistent with measurements made using endogenous ER resident proteins (Fig. S1 H). Similar to EATR, knockdown of ULK1 and ATG10 in CCER CRISPRi HCT116 cells also resulted in less mCherry cleavage during starvation, indicating that CCER measurements of ER-phagy depend on the core autophagic pathway (Fig. 1 H).
We next asked how the EATR and CCER systems respond to perturbation of recently described ER-phagy receptors. Both EATR and CCER demonstrated robust and significant inhibition of ER-phagy after knockdown of FAM134B, but modest inhibition after knockdown of RTN3L (Fig. 1, I–K; and Fig. S2, A–C), even though both proteins were depleted >90% as determined by quantitative RT-PCR (qRT-PCR; Fig. S2 D). We found that overexpression of WT FAM134B but not RTN3L strongly induced ER-phagy in both the EATR and CCER systems even in fed conditions (Fig. S2, E and F). As a negative control, overexpression of FAM134B with a mutated LIR (LIRmut) did not induce ER-phagy (Fig. S2, E, G, and H). This is consistent with studies showing that FAM134B overexpression is sufficient to trigger ER-phagy (Khaminets et al., 2015).
RTN3L might not be required for ER-phagy in HCT116 cells, or the EATR and CCER assays might be more sensitive to FAM134B-mediated rather than RTN3-mediated ER-phagy. Considering that only a small fraction of ER is targeted for ER-phagy at a given time, we find that EATR and CCER offer sensitive and quantitative readouts for the progression of ER-phagy via flow cytometry, fluorescence microscopy, and Western blotting (Khaminets et al., 2015; Mochida et al., 2015).
ATL knockdown inhibits ER-phagy
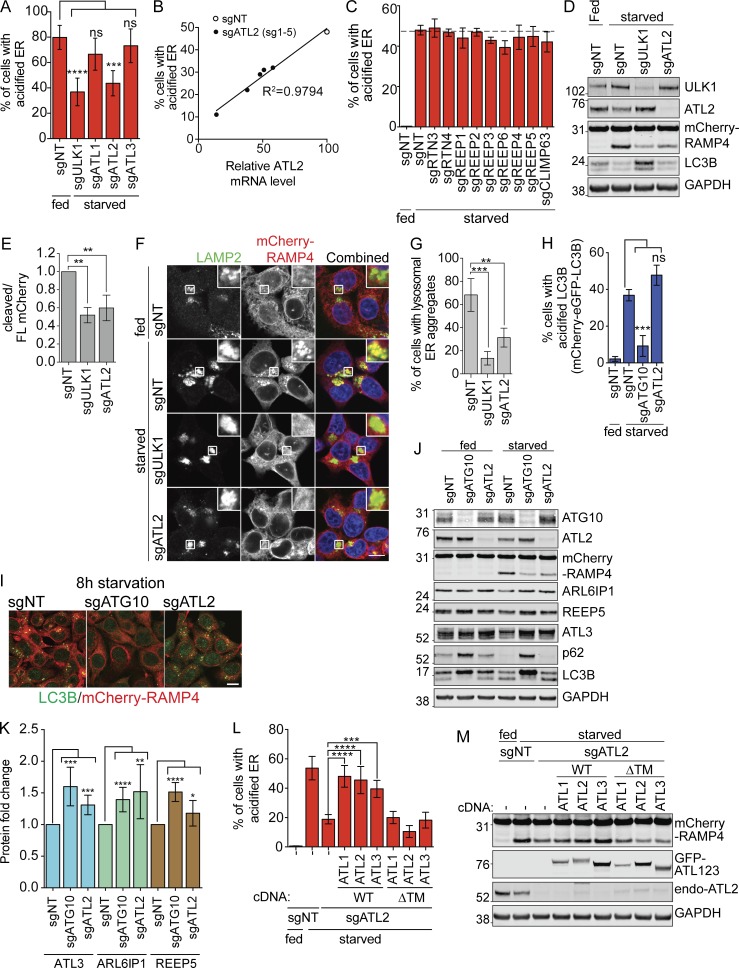
ATLs are ER-resident GTPases involved in ER remodeling at three-way junctions, and so represent promising candidates to separate ER for delivery to autophagosomes. To determine whether ATLs play a role in ER-phagy, we used CRISPRi to stably knock down ATL1, 2, and 3. The three ATLs are paralogs with highly similar protein sequences, but only knockdown of ATL2 strongly inhibited ER-phagy (Fig. 2 A and Fig. S3, A and B). While the knockdown efficiency of sgRNAs for ATL1 and ATL3 did not correlate with the extent of ER-phagy (Fig. S3, A and B), the knockdown efficiency of each ATL2 guide RNA correlated very well with the extent of ER-phagy inhibition (Fig. 2 B).
Figure 2.
EATR and CCER detect changes in ER-phagy induced by manipulation of a known ER-phagy receptor. (A) EATR CRISPRi HCT116 cells expressing the indicated proteins were starved before FACS measurement. Data presented as mean ± SD of three biological replicates. P value indicates one-way ANOVA with Dunnett’s multiple comparisons test (***, P < 0.001; ****, P < 0.0001). (B) The extent of ER-phagy inhibition of five different sgRNAs targeting ATL2 correlates with the knockdown efficiency of each sgRNA as determined by qRT-PCR. (C) EATR CRISPRi HCT116 cells transduced with the indicated sgRNAs were starved for 16 h before FACS measurement. Data presented as mean ± SD of three biological replicates. No statistically significant difference is determined between sgNT and the other sgRNAs based on one-way ANOVA with Dunnett’s multiple comparisons test. (D) CCER CRISPRi HCT116 cells transduced with sgULK1 or sgATL2 were starved, harvested, and Western blotted. (E) Quantification of data from D. Data presented as mean ± SD of three biological replicates. P value indicates one-way ANOVA with Dunnett’s multiple comparisons test (**, P < 0.005). (F) CCER CRISPRi HCT116 cells stably transduced with the indicated sgRNAs were starved and immunostained with LAMP2 antibody. Scale bar represents 20 µm. Inset represents 3× enlargement of boxed area. (G) Quantification of data from F. An average of 100 cells per condition were quantified from three biological replicates. P value indicates one-way ANOVA with Dunnett’s multiple comparisons test (**, P < 0.01; ***, P < 0.001). (H) mCherry-eGFP-LC3B CRISPRi HCT116 cells were starved for 8 h and measured by FACS similar to the EATR assay. Data presented as mean ± SD of three biological replicates. P value indicates one-way ANOVA with Dunnett’s multiple comparisons test (***, P < 0.0005). (I) CCER CRISPRi HCT116 cells expressing sgATG10 or sgATL2 were starved for 8 h, fixed, and immunostained with LC3B antibody. Scale bar represents 20 µm. (J) HCT116 cells expressing sgATG10 or sgATL2 were starved for 16 h, harvested, and subjected to Western blot. (K) Quantification of data from J. Data presented as mean ± SD of n > 4 biological replicates. P value indicates unpaired, two-tailed student t test (*, P < 0.05; **, P < 0.005; ***, P < 0.001; ****, P < 0.0001). (L) EATR CRISPRi HCT116 cells expressing sgATL2 and the indicated ATL constructs were starved for flow cytometry. Data presented as mean ± SD of three biological replicates. P value indicates one-way ANOVA with Dunnett’s multiple comparisons test (***, P < 0.005; ****, P < 0.0001). (M) CCER CRISPRi HCT116 cells expressing sgATL2 and the indicated overexpression constructs were treated the same as L, harvested, and subjected to Western blot.
Using Western blotting, we found that the levels of each ATL vary remarkably in different cellular contexts (Fig. S3 C). Consistent with previous studies, neuroblastoma SH-SY5Y cells have high levels of ATL1 relative to ATL2 and ATL3 (Rismanchi et al., 2008; Muriel et al., 2009). However, in other cell lines, ATL1 can be completely absent. We generally found that there are one or two dominantly expressed ATLs in each line. For example, hTERT-RPE1 cells only express ATL3 but HepG2 cells only express ATL2. In HCT116 cells, ATL2 is the dominant ATL relative to ATL1 and ATL3.
ATLs, REEPs, and RTNs work cooperatively to maintain ER network and morphology, and depletion of all three protein classes has been reported to disrupt ER morphology (Pendin et al., 2011; Wang et al., 2016). Although knockdown of ATL2 inhibits ER-phagy, we did not see an effect when we stably knocked down ER-resident membrane remodeling proteins from the RTN, REEP, and CLIMP63 families (Fig. 2 C and Fig. S3, D and E). Although we tested only a subset of ER remodeling proteins and cannot rule out the involvement of other ER remodelers during ER-phagy, these results indicate that ATL’s role in ER-phagy is not redundant with RTNs and REEPs.
We further validated that ATL2 is required for ER-phagy using an exon-targeting ATL2 siRNA and found by EATR that this also inhibits ER-phagy (Fig. S3 F). Using CCER, we observed less accumulation of cleaved mCherry in both ULK1- and ATL2-depleted cells (both sgRNA and siRNA) compared with the nontargeting (NT) control cells (Fig. 2, D and E; and Fig. S3 G). ULK1 knockdown resulted in the accumulation of more LC3B-I and less conversion into LC3B-II, indicating an expected block in general autophagy (Figs. 2 D and S3 G). In contrast, ATL2 depletion by either sgRNA or siRNA did not affect the degradation of LC3B, indicating that general autophagy was unaffected by loss of ATL2 (Figs. 2 D and S3 G). In CCER-expressing cells, knockdown of ULK1 or ATL2 reduces the formation of mCherry-RAMP4 puncta and its colocalization with the LAMP2 lysosomal marker compared with NT control (Fig. 2, F and G). Importantly, mCherry-RAMP4 still colocalizes with CANX, an endogenous ER marker after ATL2 knockdown, indicating that the lack of ATL2 does not interfere with the insertion or localization of mCherry-RAMP4 (Fig. S3, H and I).
To further validate that ATL2 regulates only ER-phagy and not general autophagy, we used cells stably expressing mCherry-eGFP-LC3B to measure general autophagy flux by flow cytometry. We found that general autophagy is inhibited only in cells lacking ATG10 but not lacking ATL2 (Fig. 2 H). Using confocal microscopy, we observed robust inhibition of endogenous LC3B puncta formation in ATG10- but not ATL2-depleted cells (Fig. 2 I). Western blotting analysis showed robust accumulation of p62 and inhibition of LC3B lipidation upon depletion of ATG10 but not ATL2 (Fig. 2 J).
Because EATR and CCER measure ER-phagy in the context of RAMP4 stable expression, we assessed endogenous ER protein levels to rule out transgenic artifacts. Of the endogenous ER-localized proteins that we probed for, only ARL6IP1, REEP5, and ATL3 showed consistent, albeit modest, accumulation upon ATL2 knockdown. This suggests that ATL2 may regulate the turnover of specific ER domains that these proteins occupy (Fig. 2, J and K). Altogether, our data indicate that the ATL2 ER membrane remodeler specifically mediates ER-phagy but not general autophagy.
ATL1, ATL2, and ATL3 can functionally compensate for one another during ER-phagy
ATL1, ATL2, and ATL3 have highly similar sequences, with 62–65% overall protein homology and especially high homology in their GTPase domains (Fig. S3 J). To find out whether the various ATLs can perform similar functions during ER-phagy, we stably reexpressed cDNAs for ATL1, ATL2, or ATL3 in ATL2-depleted HCT116 cells. During starvation-induced ER-phagy, reexpression of any one of the three ATLs was sufficient to rescue ER-phagy (Fig. 2, L and M; and Fig. S3 K). In contrast, truncated forms of each ATL lacking the ER-localizing TM region (ΔTM) failed to rescue ER-phagy. In addition, immunoprecipitation of ATL2 showed interaction with ATL3 in a manner that is dependent on the GTPase domain but not the TM or C-terminal helix, as shown by the lack of interaction with the dimerization-defective Arg244Gln (R244Q) mutant but stable interaction with other constructs (Fig. S3 L; Rismanchi et al., 2008). This indicates that ATL2 may form a heterodimer with ATL3. Overall, ATL1, ATL2, and ATL3 appear to be functionally interchangeable during ER-phagy, and their ER localization is required for this function.
ATL2 is degraded via the autolysosomal pathway
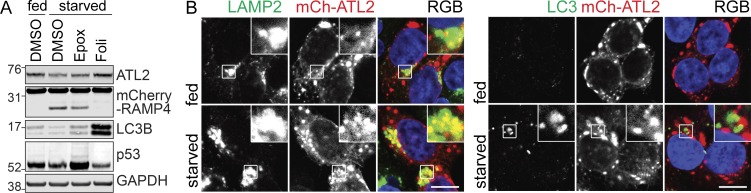
We consistently observed reduced ATL2 protein levels after starvation-induced ER-phagy (Fig. 2, D and J; and Fig. 3 A). In addition, knockdown of the key autophagy genes ULK1 or ATG10 resulted in accumulation of ATL2 during starvation (Fig. 2, D and J). Because ATL2 depletion inhibits ER-phagy, we asked if ATL2 is cotargeted for autophagic degradation. Using epoxomicin and folimycin as inhibitors of the proteasome and lysosome, respectively, we found that the level of ATL2 protein is restored upon lysosome inhibition but not proteasome inhibition (Fig. 3 A). We also observed robust colocalization of mCherry-ATL2 with endogenous staining of the lysosomal marker LAMP2 and proximal association with the autophagy adaptor LC3 (Fig. 3 B).
Figure 3.
ATL2 is degraded via the autolysosomal pathway. (A) CCER HCT116 cells were starved with either epoxomicin or folimycin, harvested, and subjected to Western blot. (B) HCT116 cells stably expressing mCherry-ATL2 were starved, fixed, and immunostained with LAMP2 and LC3B antibodies. Scale bar represents 10 µm. Inset represents threefold enlargement.
We asked if ATL2 is recruited to specific ER domains during localized ER-phagy. We used U2-OS cells stably triple-expressing BFP-RAMP4 (ER marker), GFP-LC3B (autophagy marker), and mCherry-ATL2 (Fig. S4 A). Under fed conditions, mCherry-ATL2 and BFP-RAMP4 colocalize perfectly. After 6 h of starvation, GFP-LC3B forms a ring-like structure surrounding mCherry-ATL2 puncta (Fig. S4, A and B). In certain regions, these mCherry-ATL2 puncta are also devoid of BFP-RAMP4. The number of mCherry-positive, BFP-negative puncta increases over time and is most striking after 16 h of starvation (Fig. S4, A and B). We speculate that these puncta are lysosomal compartments, since BFP and GFP are degraded in an acidic environment, whereas mCherry is resistant to lysosomal degradation. To further interrogate this, we used live-cell imaging in triple-stable cells with BFP-RAMP4 replaced with mLAMP1-BFP. We found that GFP-LC3B is recruited to mCherry-ATL2 before the trafficking of both proteins to BFP-mLAMP1–tagged lysosomes during starvation (Fig. S4 C). Overall, our data suggest that ATL2 is not degraded by the proteasome-mediated ER-associated degradation pathway, and is instead lysosomally degraded during starvation. Because the FAM134B ER-phagy receptor is also lysosomally degraded during ER-phagy, we suggest that membrane proteins specifically located in autolysosome-destined ER may be carried along during ER-phagy.
ATL dimerization and GTPase activity are required for ER-phagy
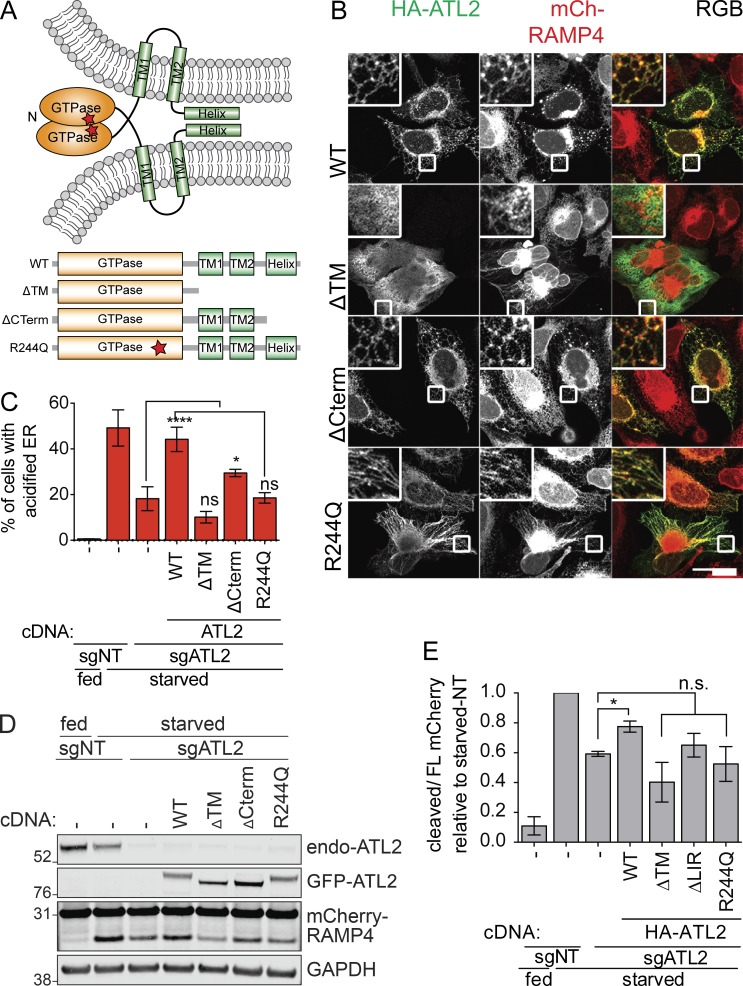
We next asked what molecular features of ATLs are required for ER-autophagy versus ER morphology. ER morphology is maintained by the various domains of ATL2 that perform sequential functions in a GTP-dependent fashion (Fig. 4 A; Moss et al., 2011). GTP loading at the GTPase domain primes ATL2 for dimerization with another activated ATL2, leading to neighboring ER membrane tethering. The C-terminal helix destabilizes ER membrane to promote membrane fusion (Moss et al., 2011). We generated several constructs of ATL2 to compare the importance of each domain for ER morphology and ER-phagy (Fig. 4 A).
Figure 4.
ER-phagy requires a functional ATL GTPase domain and proper ER localization. (A) Schematic of the topology and dimerization of ATLs to facilitate ER membrane fusion. The different truncations and mutations of ATL2 tested are shown underneath. (B) Functional characterization of ATL2 domains required for ER morphology maintenance. U2-OS cells expressing mCherry-RAMP4 were transiently transfected with the indicated ATL2 constructs. Cells were then fixed and immunostained for HA-epitope. Scale bar represents 40 µm. Inset represents 3× enlargement of the boxed areas. (C) EATR CRISPRi HCT116 cells stably expressing sgATL2 and cDNA of ATL2 mutant variants were starved and analyzed by flow cytometry. Data presented as mean ± SD of three biological replicates. P value indicates one-way ANOVA with Dunnett’s multiple comparisons test (*, P < 0.05; ****, P < 0.0001). (D) CCER CRISPRi HCT116 cells with sgATL2 and stably expressing the indicated ATL2 mutants were starved, harvested, and subjected to Western blot. (E) Quantification of data from D. Data presented as mean ± SD of three biological replicates. P value indicates one-way ANOVA with Dunnett’s multiple comparisons test (*, P < 0.05).
Consistent with previous studies, overexpressed WT ATL2 localizes to the ER network, forming a distinctive reticular network with interspersed puncta at the three-way junctions of the ER (Fig. 4 B; Rismanchi et al., 2008). During starvation, stable reexpression of WT ATL2 restored the diminished ER-phagy caused by depletion of ATL2 (Fig. 4, C–E).
In the presence of endogenous ATL2, overexpressed ATL2 lacking the TM helices (ATL-ΔTM) did not localize to a clearly ER structure. Instead, this construct caused mCherry-RAMP4–marked ER to collapse into an aggregated perinuclear cluster with radiating strands (Fig. 4 B). This dominant negative ER morphology phenotype is consistent with reports that free ATL2 GTPase domains serve as dosage-dependent inhibitors of endogenous ATL2 by preventing dimerization between endogenous ATL2 molecules and inhibiting ER membrane fusion (Moss et al., 2011). We found that reexpression of ATL2-ΔTM during ATL2 knockdown did not rescue ER-phagy (Fig. 4, C–E).
We interrogated the requirement of membrane fusion for ER morphology and ER-phagy using a truncated form of ATL2 that retains the TM regions but lacks the C-terminal helical domain (ATL2-ΔCterm). We found that overexpression of ATL2-ΔCterm was not dominant negative and had a similar ER morphology to that of WT ATL2 at basal state (Fig. 4 B). ATL2-ΔCterm expression during ATL2 knockdown partially rescues ER-phagy, indicating that the membrane fusion function of ATL2 is required during ER-phagy (Fig. 4, C–E).
We finally overexpressed an ATL2 mutant that renders the protein incapable of dimerizing (Arg244Gln; Rismanchi et al., 2008). Arg244Gln (R244Q) is equivalent to the human pathogenic Arg217Gln mutation in ATL1 that is implicated in hereditary spastic paraplegia (HSP). Overexpression of R244Q led to ER elongation and lack of reticular connections, but without the collapsed structures observed with the ΔTM construct (Fig. 4 B). The ATL2-R244Q mutant was also unable to rescue the loss of ER-phagy induced by ATL2 knockdown (Fig. 4, C–E). Overall, our data show that the functional GTPase domain and proper ER localization of ATL2 are required for both ER morphology and ER-phagy regulations. While we cannot rule out additional roles of ATL during starvation, our data suggest that ATL’s role during ER-phagy is closely related to its role of ER membrane remodeling.
ATL2 colocalizes with and acts downstream of FAM134B during ER-phagy
ATLs are best known for their roles in ER morphology, but their location on the ER membrane raises the possibility that they could play a similar role as FAM134B in recruiting autophagosomes. While we confirmed that HA-FAM134B coimmunoprecipitates with LC3B in an LIR-dependent manner, we found that ATL2 does not bind to LC3B (Fig. 5 A). This indicates that ATLs play a distinct role from FAM134B in promoting ER-phagy.
Figure 5.
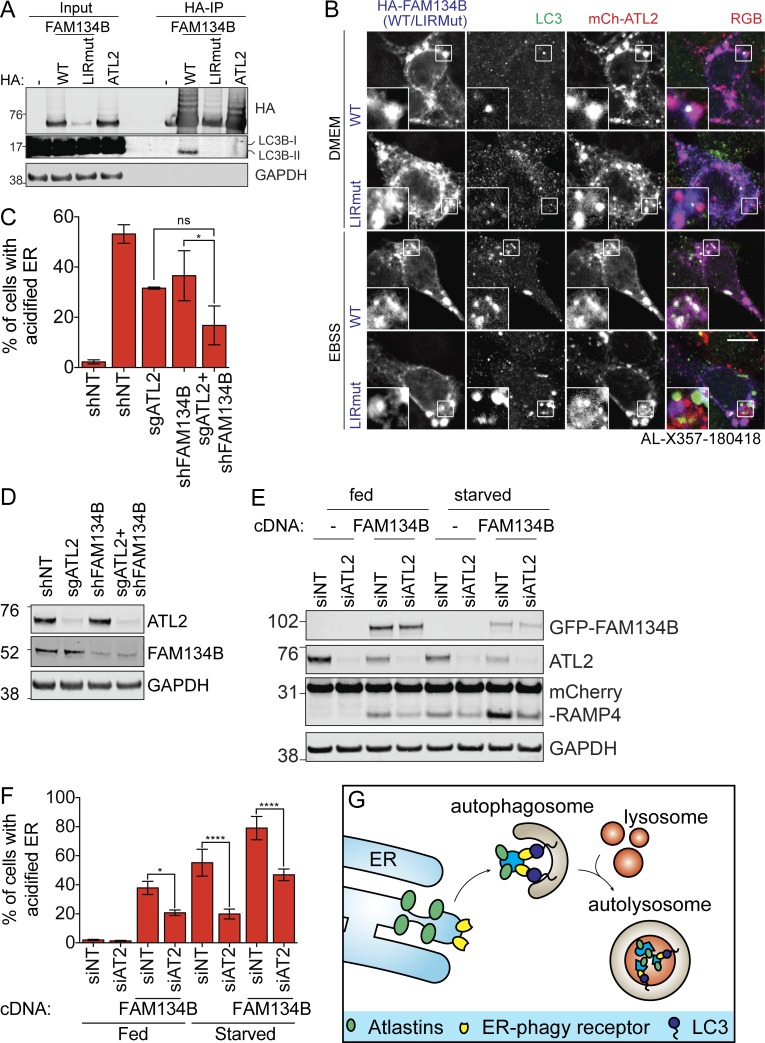
ATLs act downstream of FAM134B-mediated ER-phagy. (A) HEK293T cells transiently expressing the indicated constructs were treated with folimycin for 2 h before HA immunoprecipitation and Western blotting. (B) HCT116 cells expressing mCherry-ATL2 and HA-FAM134B (WT or LIRMut) were starved, fixed, and immunostained with HA and LC3B antibodies. Scale bar represents 10 µm. Inset represents threefold enlargement of boxed area. (C) EATR HCT116 CRISPRi cells stably depleted of the indicated proteins were starved before flow cytometry measurement. Data presented as mean ± SD of three biological replicates. P value indicates one-way ANOVA with Tukey’s multiple comparisons test (*, P < 0.05). (D) EATR HCT116 CRISPRi cells in C were harvested at basal state (fed condition) to assess FAM134B and ATL2 depletions. (E) CCER HCT116 CRISPRi cells stably expressing HA-FAM134B and sgATL2 were starved and Western blotted. (F) EATR CRISPRi HCT116 cells stably expressing GFP-FAM134B and sgATL2 were starved and measured by flow cytometry. Data presented as mean ± SD of three biological replicates. P value indicates one-way ANOVA with Tukey’s multiple comparisons test (*, P < 0.05; ****, P < 0.0001). (G) We propose that FAM134B acts as the ER-phagy receptor that recruits LC3 to the ER, whereas ATLs are involved in ER membrane remodeling to deliver FAM134B-marked ER to the autophagosomes.
We asked whether FAM134B interacts with ATL2 and found that HA-FAM134B weakly coimmunoprecipitates with endogenous ATL2 (Fig. S4 D). However, this interaction requires only the membrane localization of ATL2 or the ability of an isolated ATL2 GTPase domain to dimerize with any membrane-bound ATL2 partner (ATL2-ΔTM, ATL2-R244Q, ATL2-ΔGTPase, and ATL2-ΔCterm, all coimmunoprecipitated). Additionally, ATL2’s apparent interaction with FAM134B is independent of FAM134B’s ability to bind LC3B (Fig. S4 E). Using microscopy, we found that ATL2 and FAM134B colocalize with one another in both fed and starved conditions (Fig. 5 B). The colocalization of FAM13B and ATL2 with starvation-induced LC3B foci was dependent on the LIR region of FAM134B. Hence, it seems that FAM134B and ATL2 occupy the same ER subdomain, causing an apparent interaction during immunoprecipitation. Notably, autophagosomes are recruited to this shared ER subdomain during ER-phagy, and robust ER-phagy requires both FAM134B and ATL2.
FAM134B and ATL2 appear to serve distinct functions during ER-phagy. We therefore asked if they act in series or in parallel. We knocked down ATL2 in either the presence or absence of FAM134B (Fig. 5 C and D). Double knockdown of ATL2 and FAM134B modestly reduced ER-phagy beyond individual knockdown but not statistically significantly, suggesting that the two proteins could act in series. To formally test epistasis between ATL2 and FAM134B, we also knocked down ATL2 in FAM134B-overexpressing cells. FAM134B overexpression induces ER-phagy under fed conditions, and an epistatic interaction would be expected to modulate this effect.
We found that overexpression of FAM134B was sufficient to induce the degradation of ATL2 in fed conditions, and that ATL2 levels are further decreased upon starvation (Fig. 5 E). We also found that FAM134B-induced ER-phagy is diminished by the knockdown of ATL2, and that this is true in both fed and starved conditions (Fig. 5, E and F; and Fig. S4 F). These data indicate epistasis between FAM134B and ATL2: specifically, ATL2 is downstream of FAM134B. We favor an overall model in which FAM134B recruits autophagosomes to the ER and ATL2 helps separate FAM134B-marked ER fragments for autophagosomal engulfment (Fig. 5 G).
Because FAM134B overexpression reduces ATL2 levels, ATLs may be both effectors and cargoes of FAM134B-induced ER-phagy. This could be purposeful lysosomal degradation to reduce levels of ATL2, or ATL2 could be accidentally captured into autophagosomes during the process of engulfment. The latter possibility mirrors reductions in FAM134B observed during ER-autophagy (Fig. 5 G; Khaminets et al., 2015). Future work will hopefully shed light on whether the biophysical role of ATLs in membrane fusion facilitates ER-phagy, since on its face ER fission seems more relevant to dissolve portions of the ER network for degradation. ATL1 has known roles in ER vesiculation during normal homeostasis, and this function could be coopted during ER-phagy (Muriel et al., 2009).
A possible role of ER-autophagy in HSP
Dysregulation in organelle quality control and turnover has detrimental effects on cell fitness and has been linked to multiple diseases. Mounting evidence points toward the importance of organelle quality control for the health of nonregenerative cell types such as neurons. For example, mutations in mitophagy regulators such as PINK1 and Parkin have been reported in patients of familial Parkinsonism (Gegg and Schapira, 2011). The FAM134B ER-phagy receptor has likewise been implicated in hereditary sensory neuropathy, a genetic disorder that results in degeneration of sensory nerves of the extremities. Notably, an ATL1 loss-of-function mutation (SPG3A) has been associated with HSP (Namekawa et al., 2007). HSP is characterized by the loss of axonal cortical motor neurons, resulting in progressive weakness and spasticity of the lower limbs. ATL1 mutations have previously been proposed to lead to HSP through dysregulation of ER morphology (Zhu et al., 2006). Considering the similarities in clinical manifestation between FAM134B and ATL1 mutations, combined with our data demonstrating that ATLs act downstream of FAM134B during ER-phagy, we suggest that an inability to execute ER-phagy might be a contributing factor to HSP caused by ATL1 mutations. It remains to be seen whether HSP is caused by deficits in ER-phagy or dysfunctional ER morphology, and further mechanistic studies will hopefully shed light on the molecular basis of the disease.
Materials and methods
sgRNA, shRNA, siRNA, and cDNA plasmid cloning procedures
Information for the sgRNA sequences targeting the transcription start site of each gene were obtained from the Weissman CRISPRi-v2 library (Horlbeck et al., 2016). In each case, the top five guides were cloned into pGL1-library vector (Addgene 84832) as previously described (Horlbeck et al., 2016). Knockdown efficiency for each guide was validated by either qRT-PCR or Western blotting. The sgRNA construct and its targeting protospacer sequence for each gene are as follows: pLG1-puro NT sgRNA 1 (5′-GCGCCAAACGTGCCCTGACGG-3′), pLG1-puro NT sgRNA 3 (5′-GGCTCGGTCCCGCGTCGTCG-3′), pLG1-puro-sgULK1-1 (5′-GGCGGCGGCACAGAGACCGT-3′), pLG1-puro-sgULK1-2 (5′-GCCGACTCCGGCTCCAACTA-3′), pLG1-puro-sgATG10-1 (5′-GAGGCCGCGGACCTGACTGA-3′), pLG1-puro-sgATL1-1 (5′-GCAACCTGCGGCCCCGGAGA-3′), pLG1-puro-sgATL1-2 (5′-GGCGCTCGCTGCCTTCTCCG-3′), pLG1-puro-sgATL1-3 (5′-GGGAACCCACAAGTCCCCGG-3′), pLG1-puro-sgATL1-4 (5′-GCGGGGCCGCAGGTTGCTGG-3′), pLG1-puro-sgATL1-5 (5′-GTCCTCCCAACCGATCGCTA-3′), pLG1-puro-sgATL2-1 (5′-GAGGGCAGCAACCGCACCAG-3′), pLG1-puro-sgATL2-2 (5′-GTAGCTGCTGGGAGAACCAG-3′), pLG1-puro-sgATL2-3 (5′-GTCGGTACAAGATGGCGGAG-3′), pLG1-puro-sgATL2-4 (5′-GAAGGTCGGGGCGGACACGG-3′), pLG1-puro-sgATL2-5 (5′-GAGGCCGCCCCCTAAGGTCG-3′), pLG1-puro-sgATL3-1 (5′-GTGGCCCAACGGACAGCCcg-3′), pLG1-puro-sgATL3-2 (5′-GAGCAGGGGTGCAGAGGAGA-3′), pLG1-puro-sgATL3-3 (5′-GCGGCCCTTGGGCCGCCCAG-3′), pLG1-puro-sgATL3-4 (5′-GCAGAGGAGAGGGACGGGTG-3′), pLG1-puro-sgATL3-5 (5′-GGTGCGGGCGGGAACGAACC-3′), pLG1-puro-sgRTN1-1 (5′-GTCGCCGTGGCTCTCCTCGG-3′), pLG1-puro-sgRTN1-2 (5′-GGCTGCGGCTGGGCTCGCAG-3′), pLG1-puro-sgRTN2-1 (5′-GCCACTTCAGTAGAACCCGG-3′), pLG1-puro-sgRTN2-2 (5′-GCATATCGGCTCCGAGTGAG-3′), pLG1-puro-sgRTN3-1 (5′-GCTCGGAGCAGGCGGAGTAA-3′), pLG1-puro-sgRTN3-2 (5′-GCCAAACGCTCGTCCCCGGG-3′), pLG1-puro-sgRTN4-1 (5′-GCGCAGTGCAATCTGTAACT-3′), pLG1-puro-sgRTN4-2 (5′-GTCCACACCCAGAAGACGTC-3′), pLG1-puro-sgREEP1-1 (5′-GGGCGCGCTCAGTCTGGCGG-3′), pLG1-puro-sgREEP1-2 (5′-GAACGCGCGAACGCCGGGCA-3′), pLG1-puro-sgREEP2-1 (5′-GGCTCCTGCGCCCATCCTAG-3′), pLG1-puro-sgREEP2-2 (5′-GTGATGCGGGCTGTGATGGA-3′), pLG1-puro-sgREEP3-1 (5′-GCTGCGGACCAGGCCGCCAA-3′), pLG1-puro-sgREEP3-2 (5′-GCACCGGGCCCGGCTCCTGA-3′), pLG1-puro-sgREEP4-1 (5′-GGTTGCGAACTCCGCCCTGT-3′), pLG1-puro-sgREEP4-2 (5′-GGGCGCGCCCGCTCCCAAGT-3′), pLG1-puro-sgREEP5-1 (5′-GGTCGCCGCTCCAGTCTATC-3′), pLG1-puro-sgREEP5-2 (5′-GAGTGCCGGATAGACTGGAG-3′), pLG1-puro-sgREEP6-1 (5′-GCGAGCTCGCCCGCGGACCA-3′), pLG1-puro-sgREEP6-2 (5′-GGCGCTGGAGGCCAAGACCG-3′), pLG1-puro-sgCLIMP63-1 (5′-GCGGCCCGGGAAACTTGCAG-3′), pLG1-puro-sgCLIMP63-2 (5′-GTCCGCGCCGCCCGACGGGT-3′), and pLG1-hygro-sgATL2-1 (5-GAGGGCAGCAACCGCACCAG-3′).
Short hairpin (sh) NT (5′-CCTAAGGTTAAGTCGCCCTCG-3′), shFAM134B (5′-GAGGTATCCTGGACTGATAAT-3′; Khaminets et al., 2015), and shRTN3 (5′-TATGTTGGCATCGCCCGAGAT-3′; Bastide et al., 2017) were cloned into pLKO.1 puro construct (Addgene 8453) according to the Addgene protocol (http://www.addgene.org/tools/protocols/plko/).
The cDNA of ATL1 was obtained from Harvard PlasmID database (HsCD00326984). cDNAs of ATL2, ATL3, and FAM134B were obtained by PCR amplification of cDNA generated from HCT116 cells. All overexpression constructs were cloned into pLenti-X1-DEST vector (with neomycin-, blastocystin-, or hygromycin-resistant cassette) via Gibson Assembly (Gibson et al., 2009). First, a previously cloned pLenti-X1-pEF-mCherry-eGFP-LC3 construct (by LR clonase reaction) was cut using BamHI and XbaI to liberate the insert. All genes of interest were then Gibson assembled using Gibson Assembly Master Mix (E2611; NEB) together with the desired epitope or fluorescence tag into the pLenti-X1-DEST vector backbone according to the manufacturer’s instructions. All the plasmids used in this study are detailed in Table S1.
All described plasmids are available at Addgene. All GFP-tagged versions of the ATL1, ATL2, ATL3, and FAM134B constructs are available at Addgene (https://www.addgene.org/Jacob_Corn/).
Cell culture
Cell culture was performed at 37°C in a humidified atmosphere containing 5% CO2. HCT116, HEK293T, MCF7, U2-OS, MDA-MB-468, SH-SY5Y, and U2-OS cells were obtained from the Berkeley Cell Culture Facility. All cell lines were cultured in DMEM with high glucose. hTERT-RPE1 cells were cultured in DMEM/F12 with high glucose. All culture media were supplemented with 10% FBS, 100 U/ml penicillin (Gibco), 100 g/ml streptomycin (Gibco), 0.1 mM nonessential amino acids, and 1 mM sodium pyruvate (Gibco). All cell lines were verified mycoplasma free using the MycoAlert Mycoplasma Detection Kit (Lonza).
Lentiviral packaging and transduction
Lentiviral packaging of all constructs was performed in HEK293T cells using TransIT-LT1 Transfection Reagent (Mirus) according to the manufacturer’s instructions. Briefly, plasmids were transfected at a ratio of 1:3 (1 µg plasmid to 3 µl TransIT-LT1 reagent). The plasmid composition follows 50% plasmid with target of interest 40% ΔVPR plasmid and 10% VSVG plasmid. Lentiviruses were harvested at either 48 or 72 h after transfection for transduction. Puromycin treatment was performed at 30 µg/ml, and neomycin (G418) treatment was performed at 2 mg/ml in HCT116 and U2-OS cells. All antibiotic selections were performed for at least two passages to ensure complete selection.
Transient transfection of siRNA and plasmids
Transient transfection of plasmids into HEK293T cells for the purpose of immunoprecipitation was performed using TransIT-LT1, whereas transient transfection of plasmids into U2-OS cells was performed using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions at a ratio of 1:3 (1 µg plasmid to 3 µl Lipofectamine 3000).
siRNA oligonucleotides were transiently transfected into targeted cells using RNAiMAX (Invitrogen) according to the manufacturer’s instructions. For each well of a 12-well plate, 120 pmol siRNA was diluted in 50 µl OptiMEM. 3.6 µl RNAiMAX was diluted in 50 µl OptiMEM. The diluted siRNA was then added to the diluted RNAiMAX mixture and incubated at room temperature for 5 min before adding into cell culture. The siRNAs used in this study are as follows: siNT (ON-TARGETplus NT #1; D-001810-01-05; Dharmacon), siULK1 (siGenome Smartpool; M-005049-00-0005; Dharmacon), siATL2 (5′-GGAGCUAUCCUUAUGAACAUUCAUA-3′; Rismanchi et al., 2008), siATL3 (5′-GGUUAGAGAUUGGAGUUUCCCUUAU-3′; Rismanchi et al., 2008), and siFAM134B (5′-AGGUAUCCUGGACUGAUAAUG-3′; Rivera-Monroy et al., 2016).
Generation of CRISPRi HCT116 cells
HCT116 cells were transduced with lentivirus with an EF1a-dCas9-HA-BFP-KRAB-NLS construct to generate a pool of CRISPRi HCT116 cells. Cells were FACS sorted and single-cell seeded based on BFP expression. Individual clones were validated by probing for HA tag expression via Western blotting. The clones were transduced with a panel of previously validated sgRNAs (SEL1, SYVN, UBE4A, DPH1, and FUT4), and knockdown efficiency of each clone was assessed by qRT-PCR. The selected CRISPRi HCT116 clone was further transfected with either TET-On-mCherry-GFP-RAMP4 (for EATR assay) or mCherry-RAMP4 (for CCER assay), and clonal cell lines were generated for uniform doxycycline response and expression of the ER markers. For simplicity, the two cell lines were named EATR CRISPRi HCT116 and CCER CRISPRi HCT116, respectively.
Cell treatments
For both EATR and CCER assays, cells were seeded and grown for 48 h before starvation. In the EATR assay, doxycycline was added at 4 µg/ml 24 h before starvation to induce the expression of eGFP-mCherry-RAMP4. Cells were starved using EBSS with calcium, magnesium, and phenol red (Invitrogen 24010043) to induce ER-phagy. As opposed to starved condition, fed condition indicates incubation in normal DMEM with the usual supplements. Unless stated otherwise, co-treatment with folimycin (Millipore) was performed at 100-nM final concentration. All starvations and/or drug treatments were performed for 16 h unless otherwise indicated.
qRT-PCR
Cultured cells were harvested for RNA extraction using Direct-zol RNA miniprep kit (Zymogen) according to the manufacturer’s instructions. 1 µg of total RNA was used for reverse transcription using Superscript III First Strand Synthesis SuperMix for qRT-PCR (Invitrogen) according to the manufacturer’s instructions. qRT-PCR was performed in triplicate using Fast SYBR Green Master Mix (Applied Biosystems) and StepOne Plus Real-Time PCR System (Applied Biosystems). Samples were analyzed with two-step amplification, and melt curves were analyzed after 40 cycles. The Ct value for test genes was normalized to β-actin, and the expression of each gene was represented as 2-[ΔΔCt] relative to the NT control. All primers used are as follows: ULK1 (forward: 5′-GTCGCCGTCAAGTGCATTAACA-3′; reverse: 5′-CGTACAGGGCCACGATGTTTTC-3′), ATL1 (forward: 5′-CAGCACCTCCAGCTTTTCACTG-3′; reverse: 5′-CACCACCATCGGCTCCATATGA-3′), ATL2 (forward: 5′-TCCTTTTGCCACATCCTGGTCT-3′; reverse: 5′-GCAAGCAGCAATGGAACCAGAT-3′), ATL3 (forward: 5′-AAGATCTGCCTCACCCCAAGTC-3′; reverse: 5′-CTCCCCCACAAACCTCTTCCAT-3′), FAM134B (forward: 5′-CCAGATGAAAGACCCAGGCTCA-3′; reverse: 5′-TGCACACACTACAGACCAGGAG-3′), RTN3* (forward: 5′-CTTTCCCTGGCAGCTTTCAGTG-3′; reverse: 5′-GTTTCAGGGCCCTGTTGATGTG-3′), β-actin (forward: 5′-GGGTCAGAAGGATTCCTATG-3′; reverse: 5′-GGTCTCAAACATGATCTGGG-3′), REEP1 (forward: 5′-CATTTGTAGCCTGGCTGCTGTC-3′; reverse: 5′-CGGTCTTTTGCTTGGACCAGAC-3′), REEP2 (forward: 5′-TGCTCATCTTTGGCACCCTGTA-3′; reverse: 5′-CCGTGGTGAAGAAGGCAAAGAC-3′), REEP3 (forward: 5′-AGCAGTAAAGAGCCAAGGAGCA-3′; reverse: 5′-TCACTGGGGGCTGGTTTACTTT-3′), REEP4 (forward: 5′-TTTGCACTCTTCATGGCAGCAG-3′; reverse: 5′-CAGCACGAAGGCCATCTTGATC-3′), REEP5 (forward: 5′-GTTCCTGCACGAGAAGAACTGC-3′; reverse: 5′-ACCAGTCCGATGACACCAAGAG-3′), REEP6 (forward: 5′-TATCTGCTGTTCGGCTACGGAG-3′; reverse: 5′-TGCTTGGGCTCTCGATAGCTTT-3′), RTN1 (forward: 5′-AGGAACTGAGGAGGCTCTTCCT-3′; reverse: 5′-AACGTAGGTCAGGAGCCACATC-3′), RTN2 (forward: 5′-ATTCACCATCCCCCTGCTGTAC-3′; reverse: 5′-AACTGATTGGTCACCAACCCCA-3′), RTN3-all isoforms (forward: 5′-GGAACTCAGGCGCCTCTTCTTA-3′; reverse: 5′-TCTGTGCCTGATGCCGTTCATA-3′), and CLIMP63 (forward: 5′-CGAAGTGCTGCAGAAACTCCAG-3′; reverse: 5′-ATGGATTTGGTGAGCTCCGTCA-3′).
Western blotting
After treatment, cells were lysed using RIPA buffer (Millipore) supplemented with Halt protease inhibitor cocktail (Thermo Fisher) and Halt phosphatase inhibitor cocktail (Thermo Fisher). Protein concentration was determined by Bradford assay (VWR), and NuPAGE LDS sample buffer (Invitrogen) was added to the concentration-normalized protein samples. Samples were resolved on NU-PAGE Noves Bis-Tris 4–12% gels using 2-(N-morpholino)ethanesulfonic acid buffer at 200 V for 40 min and transferred to 0.4-µm nitrocellulose membranes at 1.3 A and 25 V for 15 min using semidry transfer apparatus (Bio-Rad). After protein transfer, membranes were blocked in 5% nonfat dry milk in Tris-buffered saline with 1% Tween-20 (TBS-T) for 30 min. Primary antibodies were diluted in 5% BSA in TBS-T, and Western blots were incubated in primary antibody for either 1 h at room temperature or overnight at 4°C on a rocker. Western blots were then washed twice in TBS-T for 5 min each time, followed by secondary antibody incubation using Li-Cor near-infrared fluorescence secondary antibodies. All blots were scanned using Li-Cor’s Near-InfraRed fluorescence Odyssey CLx Imaging System with the exception of any goat primary antibody, which was visualized by chemiluminescence using Bio-Rad’s Clarity Western ECL Substrate kit and Chemidoc XRS+ system. Protein densitometry measurement was performed using Li-Cor’s in-house ImageStudio software.
Immunofluorescence
Cells were plated on glass coverslips and grown for 48 h before any starvation or drug treatment. After treatment, cells were fixed with 4% (wt/vol) paraformaldehyde (Electron Microscopy Sciences) for 15 min. Cells were permeabilized using 0.1% Triton X-100 in PBS for all endogenous protein staining except when LC3 staining was required. For endogenous LC3 staining, cells were first fixed in 4% paraformaldehyde followed by 100% methanol for 10 min. Cells were blocked in 1% BSA in PBS for 20 min. Primary antibody staining was performed for 1 h in 1% BSA in PBS, followed by two PBS washes for 5 min each. Secondary antibody staining was performed for 30 min using Alexa Fluor Dyes (Molecular Probes) in 1% BSA in PBS. Coverslips were mounted onto glass slides using ProLong Gold antifade reagent with or without DAPI for nuclear staining. Images were taken using Zeiss LSM 710 Axio Observer confocal microscope with 63× objective lens. Image postprocessing was performed using Adobe Photoshop for inset enlargement and image alignment. Live-cell montage was first generated in ImageJ before further separation of RGB channels in Adobe Photoshop.
Primary and secondary antibodies for Western blotting and immunofluorescence
All primary antibodies used for Western blotting in this study are detailed in Table S2. The secondary antibodies used for Western blotting were obtained from Li-Cor and are as follows: donkey anti-mouse IRDye 680CW (926-32222), donkey anti-mouse IRDye 800CW (926-32212), donkey anti-rabbit IRDye 680CW (926-32223), and donkey anti-rabbit IRDye 800CW (926-32213). We also used donkey anti-goat IgG HRP (SC-2020; Santa Cruz) for the case of a goat primary antibody (ATL2; SC-109213; Santa Cruz; used in Fig. S3 C only).
The secondary antibodies used for Immunofluorescence were from Invitrogen and are as follows: goat anti-mouse AF405 (A31553), donkey anti-mouse AF488 (A21202), donkey anti-mouse AF594 (A21203), goat anti-rabbit AF405 (A31556), donkey anti-rabbit AF488 (A21206), and donkey anti-rabbit AF594 (A21207).
Live-cell imaging
U2-OS cells were transduced with lentivirus to stably express mLAMP1-BFP, eGFP-LC3B, and mCherry-ATL2. Cells that expressed all three fluorescence markers were sorted by FACS. Cells were preseeded a day before experiment onto a 35-mm imaging dish (81156; Ibidi). Cells were starved for either 6 or 16 h before imaging using the aforementioned confocal microscope in an incubator at 37°C and 5% CO2.
Flow cytometry analysis of EATR cells
After treatment, cells were trypsinized and resuspended in complete medium without fixation for immediate flow cytometry analysis. All EATR ER-phagy experiments were analyzed with live cells, because we found that cell permeabilization and fixation reverses eGFP quenching owing to loss of lysosomal acidification. On average, 5,000 mCherry- and eGFP-positive cells were analyzed per sample. Data were analyzed using FlowJo software. Single cells were gated based on eGFP and mCherry fluorescence intensity at fed condition to determine the acidified ER gate. Cells that undergo ER-phagy are defined and quantified based on the shift of cell population into the acidified ER gate due to loss of eGFP fluorescence.
Immunoprecipitation
Coimmunoprecipitation of LC3 with HA-FAM134B or HA-ATL2 was performed in HEK293T cells by transient transfection with 10 µg plasmid for 24 h. Coimmunoprecipitation of ATL3 with HA-ATL2 constructs was performed in HCT116 cells stably expressing the different HA-ATL2 mutant constructs. Immunoprecipitation of all HA-cDNA was performed using Pierce Anti-HA Magnetic Beads Kit (Thermo Fisher) according to the manufacturer’s instructions. Briefly, cells were lysed using the IP-lysis buffer supplemented with Halt protease inhibitor cocktail and Halt phosphatase inhibitor cocktail. Equal amounts of lysates (∼5 mg) with 100 µl HA-magnetic bead slurry were used to perform each immunoprecipitation. Immunoprecipitation was performed at 4°C for 2 h. The beads were washed twice using IP lysis buffer supplemented with 500 mM NaCl, with 5-min incubation on a rotor. The final wash was performed using regular IP lysis buffer. Immunoprecipitated proteins were eluted by boiling the samples at 98°C in 1× NuPAGE LDS sample buffer (Thermo Fisher) for 5 min supplemented with NU-PAGE sample-reducing agent at final 1× concentration.
Statistical analysis
Unless otherwise stated, all statistical analyses were performed using data from three or more independent experiments. Depending on the context of the experiment, statistical scores were derived using either unpaired Student’s t test or ANOVA (with Tukey’s or Dunnett’s multiple comparison test) as stated in each figure legend. Data distribution was assumed to be normal, but this was not formally tested.
Online supplemental material
Fig. S1 shows control experiments to demonstrate the specificity of EATR and CCER assays as ER-phagy reporters. Fig. S2 shows that EATR and CCER are able to detect ER-phagy induced by overexpression. Fig. S3 demonstrates ATL2 as a specific, positive regulator of ER-phagy and that the ATLs are functionally interchangeable during ER-phagy. Fig. S4 shows that ATL2 is a target substrate for autolysosomal degradation and that ATLs occupy the same ER domain as FAM134B. Table S1 lists all overexpression constructs used in this study. Table S2 lists all primary antibodies used in this study.
Supplementary Material
Acknowledgments
We thank members of the Corn laboratories for suggestions and technical assistance. We thank Holly Aaron and Feather Ives for their microscopy training and assistance.
J.E. Corn is a recipient of the National Institutes of Health New Innovator Grant (DP2-HL-141006). Confocal live and fixed cell imaging experiments were conducted at the Cancer Research Laboratory-Molecular Imaging Center, supported by the Gordon and Betty Moore Foundation.
The authors declare no competing financial interests.
Author contributions: J.R. Liang and E. Lingeman designed and optimized conditions for the EATR and CCER assays. J.R. Liang and S. Ahmed contributed to the generation of stable cell lines and Western blotting for ATL2 experiments. J.R. Liang, E. Lingeman, and J.E. Corn contributed to the design and analysis of the experiments. J.R. Liang and J.E. Corn contributed to the writing of the manuscript.
References
- Bastide A., Peretti D., Knight J.R.P., Grosso S., Spriggs R.V., Pichon X., Sbarrato T., Roobol A., Roobol J., Vito D., et al. . 2017. RTN3 Is a Novel Cold-Induced Protein and Mediates Neuroprotective Effects of RBM3. Curr. Biol. 27:638–650. 10.1016/j.cub.2017.01.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky J.L. 2012. Cleaning up: ER-associated degradation to the rescue. Cell. 151:1163–1167. 10.1016/j.cell.2012.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I. 2017. Proteasomal and Autophagic Degradation Systems. Annu. Rev. Biochem. 86:193–224. 10.1146/annurev-biochem-061516-044908 [DOI] [PubMed] [Google Scholar]
- Fumagalli F., Noack J., Bergmann T.J., Cebollero E., Pisoni G.B., Fasana E., Fregno I., Galli C., Loi M., Soldà T., et al. . 2016. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat. Cell Biol. 18:1173–1184. 10.1038/ncb3423 [DOI] [PubMed] [Google Scholar]
- Gegg M.E., and Schapira A.H.. 2011. PINK1-parkin-dependent mitophagy involves ubiquitination of mitofusins 1 and 2: Implications for Parkinson disease pathogenesis. Autophagy. 7:243–245. 10.4161/auto.7.2.14332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A. III, and Smith H.O.. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 6:343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Gilbert L.A., Horlbeck M.A., Adamson B., Villalta J.E., Chen Y., Whitehead E.H., Guimaraes C., Panning B., Ploegh H.L., Bassik M.C., et al. . 2014. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 159:647–661. 10.1016/j.cell.2014.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumati P., Morozzi G., Hölper S., Mari M., Harwardt M.I., Yan R., Müller S., Reggiori F., Heilemann M., and Dikic I.. 2017. Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. eLife. 6:911 10.7554/eLife.25555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlbeck M.A., Gilbert L.A., Villalta J.E., Adamson B., Pak R.A., Chen Y., Fields A.P., Park C.Y., Corn J.E., Kampmann M., and Weissman J.S.. 2016. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife. 5:914 10.7554/eLife.19760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Hara Y., and Mizushima N.. 2007. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 581:2623–2629. 10.1016/j.febslet.2007.05.061 [DOI] [PubMed] [Google Scholar]
- Khaminets A., Heinrich T., Mari M., Grumati P., Huebner A.K., Akutsu M., Liebmann L., Stolz A., Nietzsche S., Koch N., et al. . 2015. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 522:354–358. 10.1038/nature14498 [DOI] [PubMed] [Google Scholar]
- Kimura S., Noda T., and Yoshimori T.. 2007. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 3:452–460. 10.4161/auto.4451 [DOI] [PubMed] [Google Scholar]
- Klopfenstein D.R.C., Kappeler F., and Hauri H.-P.. 1998. A novel direct interaction of endoplasmic reticulum with microtubules. EMBO J. 17:6168–6177. 10.1093/emboj/17.21.6168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk V.I., Saiki S., Lichtenberg M., Siddiqi F.H., Roberts E.A., Imarisio S., Jahreiss L., Sarkar S., Futter M., Menzies F.M., et al. . 2011. Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 13:453–460. 10.1038/ncb2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., and Levine B.. 2010. Methods in mammalian autophagy research. Cell. 140:313–326. 10.1016/j.cell.2010.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K., Oikawa Y., Kimura Y., Kirisako H., Hirano H., Ohsumi Y., and Nakatogawa H.. 2015. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 522:359–362. 10.1038/nature14506 [DOI] [PubMed] [Google Scholar]
- Moss T.J., Andreazza C., Verma A., Daga A., and McNew J.A.. 2011. Membrane fusion by the GTPase atlastin requires a conserved C-terminal cytoplasmic tail and dimerization through the middle domain. Proc. Natl. Acad. Sci. USA. 108:11133–11138. 10.1073/pnas.1105056108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriel M.-P., Dauphin A., Namekawa M., Gervais A., Brice A., and Ruberg M.. 2009. Atlastin-1, the dynamin-like GTPase responsible for spastic paraplegia SPG3A, remodels lipid membranes and may form tubules and vesicles in the endoplasmic reticulum. J. Neurochem. 110:1607–1616. 10.1111/j.1471-4159.2009.06258.x [DOI] [PubMed] [Google Scholar]
- Nakatogawa H. 2015. Eating the ER and the nucleus for survival under starvation conditions. Mol. Cell. Oncol. 3:e1073416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namekawa M., Muriel M.-P., Janer A., Latouche M., Dauphin A., Debeir T., Martin E., Duyckaerts C., Prigent A., Depienne C., et al. . 2007. Mutations in the SPG3A gene encoding the GTPase atlastin interfere with vesicle trafficking in the ER/Golgi interface and Golgi morphogenesis. Mol. Cell. Neurosci. 35:1–13. 10.1016/j.mcn.2007.01.012 [DOI] [PubMed] [Google Scholar]
- Nikonov A.V., Hauri H.-P., Lauring B., and Kreibich G.. 2007. Climp-63-mediated binding of microtubules to the ER affects the lateral mobility of translocon complexes. J. Cell Sci. 120:2248–2258. 10.1242/jcs.008979 [DOI] [PubMed] [Google Scholar]
- Pendin D., McNew J.A., and Daga A.. 2011. Balancing ER dynamics: shaping, bending, severing, and mending membranes. Curr. Opin. Cell Biol. 23:435–442. 10.1016/j.ceb.2011.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell A.M., and Youle R.J.. 2015. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 85:257–273. 10.1016/j.neuron.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rismanchi N., Soderblom C., and Stadler J.. 2008. Atlastin GTPases are required for Golgi apparatus and ER morphogenesis. Hum. Mol. Genet. 17:1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Monroy J., Musiol L., Unthan-Fechner K., Farkas Á., Clancy A., Coy-Vergara J., Weill U., Gockel S., Lin S.-Y., Corey D.P., et al. . 2016. Mice lacking WRB reveal differential biogenesis requirements of tail-anchored proteins in vivo. Sci. Rep. 6:39464 10.1038/srep39464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S., Gallagher C.M., and Walter P.. 2014. ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J. Cell Sci. 127:4078–4088. 10.1242/jcs.154716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y., Voss C., Rist J.M., Hu J., Rapoport T.A., Prinz W.A., and Voeltz G.K.. 2008. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J. Biol. Chem. 283:18892–18904. 10.1074/jbc.M800986200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.D., Harley M.E., Kemp A.J., Wills J., Lee M., Arends M., von Kriegsheim A., Behrends C., and Wilkinson S.. 2018. CCPG1 Is a Non-canonical Autophagy Cargo Receptor Essential for ER-Phagy and Pancreatic ER Proteostasis. Dev. Cell. 44:217–232.e11. 10.1016/j.devcel.2017.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I., Tolley N., Aller I., Svozil J., Osterrieder A., Botchway S., Mueller C., Frigerio L., and Hawes C.. 2010. Five Arabidopsis reticulon isoforms share endoplasmic reticulum location, topology, and membrane-shaping properties. Plant Cell. 22:1333–1343. 10.1105/tpc.110.074385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz G.K., Prinz W.A., Shibata Y., Rist J.M., and Rapoport T.A.. 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 124:573–586. 10.1016/j.cell.2005.11.047 [DOI] [PubMed] [Google Scholar]
- Wang S., Tukachinsky H., Romano F.B., and Rapoport T.A.. 2016. Cooperation of the ER-shaping proteins atlastin, lunapark, and reticulons to generate a tubular membrane network. eLife. 5:209 10.7554/eLife.18605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G., Zhu P.-P., Renvoisé B., Maldonado-Báez L., Park S.H., and Blackstone C.. 2016. Mammalian knock out cells reveal prominent roles for atlastin GTPases in ER network morphology. Exp. Cell Res. 349:32–44. 10.1016/j.yexcr.2016.09.015 [DOI] [PubMed] [Google Scholar]
- Zhu P.-P., Soderblom C., Tao-Cheng J.-H., Stadler J., and Blackstone C.. 2006. SPG3A protein atlastin-1 is enriched in growth cones and promotes axon elongation during neuronal development. Hum. Mol. Genet. 15:1343–1353. 10.1093/hmg/ddl054 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.