Abstract
Little is known about the structure-function relationship of membrane-bound lipid desaturases. Using a domain-swapping strategy, we found that the N terminus (comprising the two first transmembrane segments) region of Bacillus cereus DesA desaturase improves Bacillus subtilis Des activity. In addition, the replacement of the first two transmembrane domains from Bacillus licheniformis inactive open reading frame (ORF) BL02692 with the corresponding domain from DesA was sufficient to resurrect this enzyme. Unexpectedly, we were able to restore the activity of ORF BL02692 with a single substitution (Cys40Tyr) of a cysteine localized in the first transmembrane domain close to the lipid–water interface. Substitution of eight residues (Gly90, Trp104, Lys172, His228, Pro257, Leu275, Tyr282, and Leu284) by site-directed mutagenesis produced inactive variants of DesA. Homology modeling of DesA revealed that His228 is part of the metal binding center, together with the canonical His boxes. Trp104 shapes the hydrophobic tunnel, whereas Gly90 and Lys172 are probably involved in substrate binding/recognition. Pro257, Leu275, Tyr282, and Leu284 might be relevant for the structural arrangement of the active site or interaction with electron donors. This study reveals the role of the N-terminal region of Δ5 phospholipid desaturases and the individual residues necessary for the activity of this class of enzymes.
Keywords: Bacillus, fatty acid desaturase, fatty acid biosynthesis, enzyme mechanism, membranes, protein modeling
FA desaturases are proteins universally distributed among living organisms that play a pivotal role in the structure and functioning of biological membranes. These enzymes catalyze the introduction of a double bond into FAs in a reaction that is dependent on a di-iron center, molecular oxygen, and reducing equivalents (1, 2). Desaturases comprise two distinct evolutionary groups: the soluble acyl-acyl carrier protein (acyl-ACP) desaturases, which use acyl-ACP as a substrate, and membrane FA desaturases that display a range of lipid substrate preferences, including phospholipids, galactolipids, acyl-CoAs, and sphingolipids (2, 3). The lack of a structural model has been a central issue in membrane FA desaturase characterization, which has been overcome with the recent publication of the crystal structures of two mammalian stearoyl-CoA desaturases (SCDs). These structures showed the interaction between the lipid substrate and the membrane-bound enzyme (4, 5), providing new insights into the catalytic mechanism of desaturation. However, our understanding of the structure–function relationship of membrane-bound desaturases that are not specific to acyl-CoA remains very limited. These enzymes, like many membrane proteins, have proven difficult to overexpress and purify. However, they are remarkable for their structural similarity and functional diversity (2), and this structural resemblance has provided the basis for the study of structure–function relationships. Thus, site-directed mutagenesis and swapping-domain experiments, with membrane-bound desaturases, have been used by numerous groups to define several regions and amino acid (AA) residues that are critical for the catalytic properties of these enzymes. For instance, based on AA sequence comparison and site-directed mutagenesis, it has been shown that membrane-bound desaturases contain three histidine clusters (His boxes) with the general structure [H(X)3-4H, H(X)2-3HH, and H/Q(X)2-3HH]. These motifs are strongly conserved in all members of this class, and it has been demonstrated that they are essential for enzymatic activity by coordinating the di-iron center of the active site (6, 7).
The Bacillus subtilis Δ5 desaturase (Des), which catalyzes the introduction of cis-double bonds at the Δ5 position of a wide range of saturated FAs (8), is the best studied of the prokaryotic membrane-bound FA desaturases. Earlier studies, using systematic mutagenesis of conserved residues, have clearly demonstrated that the His clusters are essential for Des activity (9). Moreover, based on genetic analysis and in vivo assays, our group has established that Des is specific for phospholipids substrates (10) and utilizes both ferredoxins and flavodoxins as electron donors for the desaturation reaction (11). Recently, we reported the functional characterization of a Δ5 phospholipid desaturase from Bacillus cereus, named DesA, that shares 66.2% identity with Des. Although their primary structures are similar, DesA has a much higher enzyme activity than Des, suggesting that the B. cereus desaturase contain important residues that enhance its activity.
In this work, we took advantage of the high similarity of both Des and DesA with an inactive protein encoded by open reading frame (ORF) BL02692 (coding for a putative desaturase) from Bacillus licheniformis to design domain-swapping and site-directed mutagenesis experiments in order to elucidate the molecular basis of these observed differences. These studies revealed that a key tyrosine at the end of the first transmembrane (TM) helix of DesA, close to the lipid-water interface, is critical for desaturation activity. Additionally, we identified eight new essential residues for desaturase activity, which are completely conserved in a large number of membrane-bound desaturases. To evaluate the impact of these mutations, homology modeling of DesA was carried out based on the human SCD1 crystal structure. These results improve our understanding of the biochemical mechanism of these important lipid-modifying enzymes and could potentially lead to engineering the Bacillus desaturases for biotechnological purposes.
MATERIALS AND METHODS
Bacterial strains and growth conditions
Bacterial strains used in the present study are listed in Table 1. Escherichia coli and B. subtilis strains were routinely grown in Luria Bertani (LB) broth at 37°C. Spizizen salts, supplemented with 0.8% glycerol, 0.01% each tryptophan and phenylalanine, and trace elements were used as the minimal medium (MM) to grown B. subtilis (12). Antibiotics were added to medium at the following concentrations: ampicillin (Amp), 100 µg/ml; kanamycin (Km), 5 µg/ml for B. subtilis experiments and 50 µg/ml for E. coli; and spectinomycin (Sp), 50 µg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strains and plasmids | Relevant characteristics | Source/reference |
| Strains | ||
| Bacillus subtilis | ||
| JH642 | trpC2 pheA1 | Laboratory stock |
| LC5 | JH642 des::Kmr | (8) |
| EV | LC5 amyE::pSG1154 Kmr Spr | (19) |
| DesA | LC5 amyE::Pxil-desA Kmr Spr | (19) |
| Des | LC5 amyE::Pxil-des Kmr Spr | This study |
| BL02692 | LC5 amyE::Pxil-ORFBL02692 Kmr Spr | This study |
| Chimera I | LC5 amyE::Pxil-(desA-des) Kmr Spr | This study |
| Chimera II | LC5 amyE::Pxil-(des-desA) Kmr Spr | This study |
| Chimera III | LC5 amyE::Pxil-(desA-ORFBL02692) Kmr Spr | This study |
| Chimera IV | LC5 amyE::Pxil-(ORFBL02692-desA) Kmr Spr | This study |
| Chimera V | LC5 amyE::Pxil-(desAN1-ORFBL02692(N2+C)) Kmr Spr | This study |
| BL02692 Cys40Tyr | LC5 amyE::Pxil- ORFBL02692 (C40Y) Kmr Spr | This study |
| BL02692 Cys40Ala | LC5 amyE::Pxil- ORFBL02692 (C40A) Kmr Spr | This study |
| DesA Tyr40Cys | LC5 amyE::Pxil-desA (Y40C) Kmr Spr | This study |
| DesA Gly90Arg | LC5 amyE::Pxil-desA (G90R) Kmr Spr | This study |
| DesA Trp104Arg | LC5 amyE::Pxil-desA (W104R) Kmr Spr | This study |
| DesA Arg149Ala | LC5 amyE::Pxil-desA (R149A) Kmr Spr | This study |
| DesA Lys172Ala | LC5 amyE::Pxil-desA (K172A) Kmr Spr | This study |
| DesA Gly201Ala | LC5 amyE::Pxil-desA (G201A) Kmr Spr | This study |
| DesA His228Ala | LC5 amyE::Pxil-desA (H228A) Kmr Spr | This study |
| DesA Pro257Ala | LC5 amyE::Pxil-desA (P257A) Kmr Spr | This study |
| DesA Leu275Arg | LC5 amyE::Pxil-desA (L275R) Kmr Spr | This study |
| DesA Pro277Ala | LC5 amyE::Pxil-desA (P277A) Kmr Spr | This study |
| DesA Tyr282Arg | LC5 amyE::Pxil-desA (Y282R) Kmr Spr | This study |
| DesA Leu284Arg | LC5 amyE::Pxil-desA (L284R) Kmr Spr | This study |
| DesA Tyr26Trp, Ile28Leu | LC5 amyE::Pxil-desA (Y26W-I28L) Kmr Spr | This study |
| DesA Phe51Trp | LC5 amyE::Pxil-desA (F51W) Kmr Spr | This study |
| Escherichia coli DH5α | supE44 thi-1 ΔlacU169 ϕ80lacZ ΔM15 endA1 recA1 hsdR17 gyrA96 relA1 trp6 cysT329::lac inmλpI(209) | Laboratory stock |
| Plasmids | ||
| pCR-Blunt II-Topo | E. coli cloning vector, Kmr | Invitrogen |
| pGEM-T Easy | E. coli cloning vector, Ampr | Promega |
| pSG1154 | Expression vector that integrates at the amyE locus of B. subtilis, Spr | (16) |
General molecular techniques
Chromosomal DNA was isolated using standard techniques (13). In all cases, DNA fragments were obtained by PCR using oligonucleotides described in supplemental Table S3. Oligonucleotides were purchased from Genbiotech SRL (Argentina). PCR products of expected sizes were purified from the gel using the AxyPrep DNA Gel Extraction Kit (Axygen, Bioscience), ligated into a pGEM T-Easy vector or pCR-Blunt II-Topo (Promega, Madison, WI), and transformed in E. coli DH5α (13). Plasmid DNA was prepared using the Wizard DNA purification system (Promega Life Science) and sequenced. Transformation of B. subtilis was carried out by the method of Dubnau and Davidoff-Abelson (14). The amy phenotype was assayed on colonies grown during 48 h in LB starch plates, by flooding the plates with 1% I2-KI solution (15). The amy+ colonies produced a clear halo, whereas amy− colonies gave no halo. All plasmids and primers used in this study are listed in Table 1 and supplemental Table S3, respectively.
Construction of the chimeric enzymes
Chimeric enzymes were constructed using the overlap extension PCR method (16). In the first step, domains N and C of the desaturases were amplified individually from a genomic DNA template with overlapping ends using primers specified in supplemental Table S3. The fusion primers were used to generate PCR fragments corresponding to specific regions of one of the desaturases. Amplifications were carried out with Phusion DNA polymerase High Fidelity (New England Biolabs Inc.). After gel purification, amplified fragments were fused in a subsequent overlap PCR to yield a recombinant molecule, which was amplified using primers comprising the start codon and the stop codon of each desaturase. PCR products were cloned into the pTOPO vector (Invitrogen) and sequenced. Hybrid genes were digested with the corresponding restriction enzymes (XhoI/EcoRI) and cloned into pSG1154 (17) integration vector in-frame with the sequence encoding a C-terminal GFP tag. The constructs were transformed in a B. subtilis LC5 strain (Table 1) that is unable to synthesize unsaturated FAs (UFAs) due to the lack of the Δ5 desaturase.
Site-directed mutagenesis
Single AA substitutions were introduced into desaturases using a QuikChange Site-Directed Mutagenesis Kit (Stratagene). The PCR mixture contained 100 ng of template, primers with each corresponding mutation (10 pmol each), 1 unit of Phusion DNA polymerase (New England Biolabs), 1× Phusion HF reaction buffer, and 0.2 mM dNTPs in a final volume of 50 µl. The thermocycling program included an initial denaturation step at 98°C for 30 s followed by 30 cycles of 98°C for 10 s, 58°C for 30 s, and 72°C (30 s/1 kb). Before transformation, each PCR mixture was digested with 20 units of DpnI (Fermentas) for 3 h at 37°C to remove the original template DNA. Primers used to generate individual codon substitutions are listed in supplemental Table S3. Plasmids resulting from QuikChange® reactions were confirmed by sequencing. Hybrid genes were digested with the corresponding restriction enzymes (XhoI/EcoRI) and cloned into pSG1154 (17) integration vector in-frame with the sequence encoding a C-terminal GFP tag for heterologous expression in B. subtilis LC5 strain.
FA extraction and analysis by gas chromatography
To determine the FA composition, cells were grown at 37°C in MM to exponential phase and then shifted to 25°C. The cultures were harvested in stationary phase. Total cellular FAs were prepared by the method of Bligh and Dyer (18). The FA methyl esters (FAMEs) were prepared by transesterification of glycerolipids with 0.5 M sodium methoxide in methanol and then analyzed in a Shimadzu Turbo Mass gas chromatography-mass spectrometer on a capillary column (30 mm × 0.25 mm in diameter) of 100% dimethylpolysiloxane (PE-1, Shimadzu). Helium at 1 ml/min was used as the carrier gas, and the column temperature was programmed to rise by 4°C min−1 from 140°C to 240°C. Branched-chain FAs, straight-chain FAs, and UFAs used as reference compounds were obtained from Sigma Chemical Co. (8).
The amount of UFAs accumulated was quantified by GC/MS (supplemental data) and expressed as a percentage of the total FAs. The positions of the double bonds in UFAs were determined by GC/MS. FAMEs were converted to dimethyl disulfide (DMDS) adducts, as previously described (8), and then separated on a PE-1 column ramped from 140 to 280°C at 4°C min−1. The spectra were recorded in the electron impact mode at 70 eV, using 1 s scans of the m/z 40–400.
Assay of recombinant desaturase activity “in vivo”
B. subtilis LC5 derivative strains carrying the recombinants and WT desaturases were grown overnight at 37°C in MM with the corresponding antibiotics. Cells were resuspended in MM with 0.5% xylose after 4–5 h, having reached OD600 values of 0.4–0.5. At this point, cells were labeled with 1 μCi/ml 1-[14C]palmitate [specific activity 58 mCi/mM] for 3 h at 37°C (10, 19). Following incubation, cells were collected, and lipids were prepared by the method of Bligh and Dyer (18). Labeled FAMEs were applied to 10% silver nitrate-impregnated plates with Silica Gel G (thickness, 0.5 mm; Analtech). Chromatographic separation was achieved with toluene as liquid phase, at −20°C, and the spots of the FAs were detected using a PhosphorImager screen (Typhoon 9200).
Topological prediction
Topology predictions for B. cereus Δ5 desaturase (DesA) were performed using the following online servers: TMpred (https://embnet.vital-it.ch/software/TMPRED_form.html), TMHMMv2.0 (http://www.cbs.dtu.dk/services/TMHMM/), HMMTOP (http://www.enzim.hu/hmmtop/), DAS-TMfilter (http://www.enzim.hu/DAS/DAS.html), Phobius (http://phobius.sbc.su.se/), MEMSAT (PSIPRED) (http://bioinf.cs.ucl.ac.uk/psipred/), SOSUI (http://harrier.nagahama-i-bio.ac.jp/sosui/sosui_submit.html), OCTOPUS (http://octopus.cbr.su.se/), SPLIT 4.0 (http://splitbioinf.pmfst.hr/split/4/), TMMOD (http://liao.cis.udel.edu/website/servers/TMMOD), and TOPCONS (http://single.topcons.net/).
RESULTS
The N-terminal region of Δ5 desaturases is involved in desaturation activity
Integral membrane desaturases have proven to be recalcitrant to overexpression, purification, and characterization; therefore, during the last few years, we have undertaken a genetic and biochemical approach to explore the structure-function relationship in these enzymes using as models several Bacillus spp. In an early report, we showed that B. subtilis has a single acyl-lipid desaturase, encoded by the des gene, which introduces a cis double bond at the Δ5 position of the acyl chain of membrane glycerolipids (8, 20, 21). We also have found that B. cereus strain ATCC 14579 contains two highly active acyl-lipid desaturases, DesA and DesB, which have Δ5 and Δ10 regioselectivities, respectively (19). During the characterization of the UFA biosynthesis in B. licheniformis ATCC 14580, we found that this bacterium synthesizes only Δ5-UFAs (supplemental Fig. S1A, B). This strain contains two ORFs (BL02692 and BL02106), with the encoded proteins sharing 69% of identity. Both ORFs were annotated as Δ5 desaturases, based on sequence similarity with Des from B. subtilis (69% and 91% of identity, respectively; data not shown). When these genes were expressed in the B. subtilis LC5 strain, we determined that, whereas protein encoded by ORF BL02106 showed acyl-lipid Δ5 desaturase activity, the strain expressing ORF BL02692 was found to be devoid of any desaturase activity (supplemental Fig. S1C). Therefore, this inactive ORF, which shares all the characteristic features of membrane-bound acyl lipid desaturases (supplemental Fig. S2), offers a unique opportunity to identify regions of the protein required for enzymatic activity. To this end, we designed chimeric proteins of B. licheniformis ORF BL02692 with the highly active B. cereus Δ5-desaturase DesA and the less active B. subtilis Des, with which it shares 69% of sequence ID (8, 19).
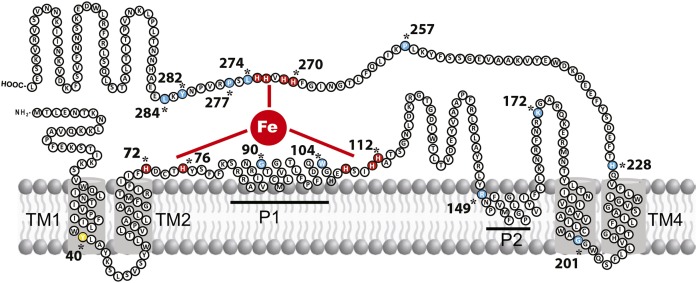
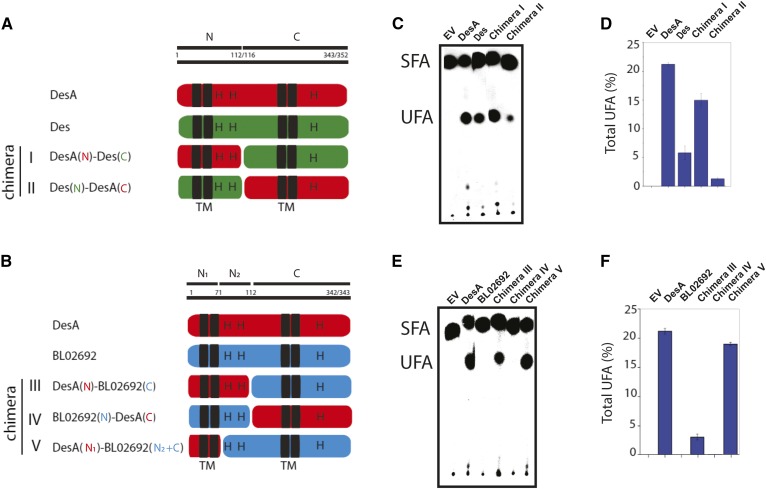
As the first step in chimera constructions, a topological model of DesA was composed using multicomputational analysis of selected full-length protein sequences of desaturases from Firmicutes (supplemental Fig. S2 and Table S1). This analysis, combined with topology prediction servers (OCTOPUS, DAS, and TOPCONS), showed that the model of DesA (Fig. 1) is in close agreement with the predicted and validated topologies of several membrane-bound FA desaturases (1, 6, 22, 23). Indeed, DesA has four TM domains and two peripheral membrane-associated regions (P), with the N and C termini facing the cytoplasm. The hydrophilic regions also contain the three His boxes characteristic of integral membrane desaturases (Fig. 1). Based on this model, B. cereus DesA, B. subtilis Des, and B. licheniformis ORF BL02692 were dissected into two regions (N and C). Region N (112–116 AA length) comprises the cytosolic N-terminal region, two TM helices (TM 1–2), and the first peripheral membrane-associated segment (P1), including the first and second His boxes. Region C (343–352 AA length) is composed of the second peripheral membrane-associated domain (P2), the second set of TM helices (TM 3–4), and the cytosolic C-terminal domain containing the third His box (Figs. 1, 2A). Chimeras constructed combining the N terminus and C terminus of the three Δ5 desaturases are shown in Fig. 2A, B. For functional characterization, these hybrids were expressed in B. subtilis LC5 strain, and the contribution of individual regions to the desaturase activity was assessed as described in Materials and Methods. The results shown in Fig. 2 and supplemental Fig. S3 indicate that, with the exception of chimeric protein IV (BL02692N and DesAC) that completely lost desaturase activity, chimeras I (DesAN and DesC), II (DesN and DesAC), III (DesAN and BL02692C), and V (DesAN1 and BL02692N2+C) were active (Fig. 2C, E). However, these chimeras showed significant variation for UFA production (Fig. 2D, F). Chimera I was more active than Des but less active than DesA, and the activity of chimera II was lower than both Des and DesA WT enzymes (Fig. 2C, D). Because the WT and chimeric desaturases are present at similar levels in membranes of B. subtilis LC5 derivative strains (supplemental Fig. S4), the production of UFAs shown in Fig. 2 is due to different desaturase activities rather than to changes in expression levels or stability of the proteins. These results indicate that the N-terminal region of DesA is important to improve desaturase activity of Des, but is not sufficient to reach the levels observed for DesA. Interestingly, chimera III, carrying region N of DesA and region C of ORF BL02692, produced an active desaturase (Fig. 2E, F), whereas chimera IV, carrying the region N of ORF BL02692 and region C of DesA, did not show any desaturation activity (Fig. 2E, F). These results indicate that B. licheniformis ORF BL02692 possesses a defective N terminus and that this region appears to be critical for desaturase activity.
Fig. 1.
Proposed topological model of the B. cereus acyl-lipid desaturase DesA. This model shows that DesA contains four TM domains (TM1–4), two peripheral helices (P1–2), and both the N and C termini oriented toward the cytosol. The residues colored in red represent the conserved His boxes, which are catalytically essential. Residues colored in light blue represent the AA residues that were mutated. Tyr40 is highlighted in yellow.
Fig. 2.
Construction of chimeras by domain swapping and determination of FA desaturase activities. A, B: Schematic structural diagrams of the WT desaturase and chimeric proteins constructed by domain swapping. C, E: Autoradiogram of the products of [14C]palmitate labeling of B. subtilis LC5 derivative strains expressing the WT and recombinant proteins. EV represents the B. subtilis LC5 strain carrying the empty vector. Cells were grown overnight at 37°C in MM, washed, resuspended in fresh MM, with or without xylose 0.5%, and labeled with 1 μCi/ ml of [14C]palmitate for 3 h at 37°C. FAs were converted to FAMEs and analyzed using 10% silver nitrate-impregnated Silica Gel G plates. The radioactivity on the plates was visualized using a PhosphorImager screen. SFA, saturated fatty acids. D, F: Total UFAs, expressed as proportion (%) of the total FAs, quantified by GC/MS. Data represent means ± SD of three replicate cultures.
To further interrogate the importance of the N region of DesA, we also constructed chimera V containing only the first 70 residues of the N terminus of DesA (N1) and the C-terminal portion plus the N2 region of BL02692, as it is shown in Fig. 2B. The desaturase activity of this chimera was similar to DesA and higher than chimera III (Fig. 2E, F). It should be noted that in this chimera, His clusters belong to the same ORF, suggesting that specific steric and/or folding constraints of these regions optimize the desaturase activity. UFAs synthesized by these chimeras were identified as Δ5 UFAs, and the nature of these isomers was confirmed by unequivocal determination of the double-bond positions through the analysis of the mass spectrum of the DMDS derivative (supplemental Fig. S5). Collectively, these results indicated that the two TM helices of DesA (region N1) were sufficient to restore the function of B. licheniformis ORF BL02692 and that this enzyme exhibited Δ5 desaturase activity.
Site-directed mutagenesis of B. licheniformis ORF BL02692
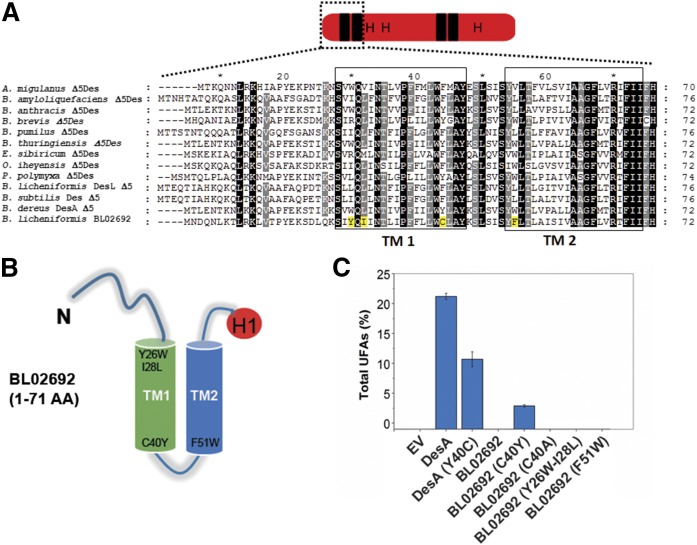
To further identify residues in the N-terminal region of DesA that are responsible for restoring the activity of ORF BL02692, a sequence alignment of several Δ5 desaturases from Firmicutes and this ORF was performed (Fig. 3A). This analysis revealed the presence of four AA residues (Tyr26, Ile28, Cys40, and Phe51), localized in the first two TM domains of ORF BL02692 (Fig. 3B), which have significantly different properties from the corresponding AAs found in the analyzed sequences. To test whether the four divergent AAs were responsible for the lack of activity of ORF BL02692, they were replaced by the corresponding AAs found in active desaturases. Thus, the changes introduced by site-directed mutagenesis in ORF BL02692 were the following: Tyr26Trp–Ile28Leu, Cys40Tyr, and Phe51Trp. These variants were expressed in B. subtilis LC5, and its activities were analyzed by GC/MS and TLC, as described before. Unexpectedly, the results revealed that a single substitution of Cys40 by Tyr, in the first TM domain, was able to restore the desaturase activity of the ORF BL02692 enzyme (Fig. 3C and supplemental Fig. S6), whereas mutants DesA Tyr26Trp–Ile28Leu and DesA Phe51Trp yielded inactive proteins (Fig. 3C and supplemental Fig. S6). To evaluate whether the activity of ORF BL02692 Cys40Tyr was restored by the disruption of a potential intramolecular disulfide bridge, we replaced Cys40 by Ala. This mutation did not have any impact on ORF BL02692 activity (Fig. 3C and supplemental Fig. S6), suggesting that the improved activity observed in ORF BL02692 Cys40Tyr was due to Tyr itself, rather than to the disruption of a disulfide bond. Indeed, when we replaced Tyr40 by Cys in DesA, we found that this mutation decreased the desaturase activity about 50%, mainly toward C17 FAs (supplemental Table S2). Nevertheless, the ORF BL02692 Cys40Tyr has a proportional increase in production of all 16:1 and 17:1 FAs (supplemental Table S2). Together, these results support the functional importance of Tyr40 in desaturase activity and suggest that different specificity-determining residues enable DesA or ORF BL02692 to recognize the length of their acyl chain substrates.
Fig. 3.
Key residues of the first TM domain on the acyl-lipid desaturase activity. A: Partial sequence alignment of Δ5-acyl-lipid desaturases from Firmicutes with B. licheniformis ORF BL02692. The deduced AA sequences of the first 70–76 AA residues were aligned by using ClustalW. Completely conserved residues are shaded in black and partially conserved are shaded in gray. TMI and TM2 are indicated by hollow boxes. The residuesY26, I28, C40, and F51 of ORF BL02692 are highlighted in yellow. B: Scheme of the first two predicted membrane-spanning helices, TM1 and TM2, of ORF BL02692, showing the relative position of the mutated residues. H1 (in red) denotes the first His box. C: Desaturation activity of the WT and mutant desaturases expressed in the B. subtilis LC5 strain. EV represents the B. subtilis LC5 strain carrying the empty vector. Total UFAs are expressed as proportion (%) of the total FAs, quantified by GC/MS.
Mutational analysis of the conserved AA residues of Bacillus acyl-lipid desaturases
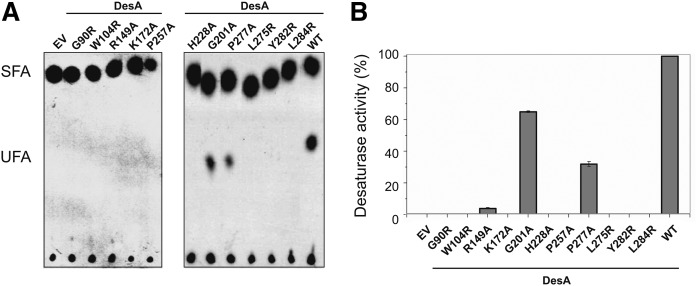
In the search for other critical AA residues for desaturase activity, multiple sequence alignment was performed among known acyl-lipid desaturases from several strains from Firmicutes. The sequence alignment of 14 Δ5-desaturases and 3 divergent Δ10-desaturases allowed us to find 11 completely conserved AA residues, excluding the well-known His boxes (supplemental Fig. S7). These residues included glycine (Gly90 and Gly201), tryptophan (Trp104), arginine (Arg149), lysine (Lys172), leucine (Leu275 and Leu284), proline (Pro257 and Pro277), histidine (His228), and tyrosine (Tyr282). In the predicted topology model of B. cereus DesA (Fig. 1), six of the conserved residues (Gly90, Trp104, Leu275, Pro277, Tyr282, and Leu284) were located in close proximity to the His boxes (supplemental Fig. S7). The importance of each conserved residue in DesA activity was explored by site-directed mutagenesis. The enzymatic activity of mutant desaturases was examined by TLC and GC/MS analysis as described above. According to this analysis, Gly201Ala and Pro277Ala substitution reduced significantly the desaturase activity of DesA, whereas the activity of mutants Arg149Ala, Gly90Arg, Trp104Arg, Lys172Ala, Pro257Ala, His228Ala, Leu275Arg, Tyr282Arg, and Leu284Arg was severely compromised or null (Fig. 4 and supplemental Fig. S8). These results indicate that these conserved residues are important for enzyme activity.
Fig. 4.
Essential residues of acyl-lipid desaturases from Firmicutes. A: Autoradiogram of the products of [14C]palmitate labeling of B. subtilis strains. Cells were grown overnight at 37°C in MM, washed, resuspended in fresh MM, with or without xylose 0.5%, and labeled with 1 μCi/ml [14C]palmitate for 3 h at 37°C. FAs were converted to FAMEs and analyzed using 10% silver nitrate-impregnated Silica Gel G plates. The radioactivity on the plates was visualized using a PhosphorImager screen. SFA, saturated fatty acids. B: Desaturase activity of DesA and mutants with single AA substitution expressed in B. subtilis LC5 strain. EV represents the B. subtilis LC5 strain carrying the empty vector. FAMEs were prepared from the WT strains and mutants with single AA substitution, and UFAs were quantified by GC/MS. The desaturase activity of DesA was considered as 100%, and UFA production of mutants was expressed as percentage of DesA activity.
Homology modeling of B. cereus DesA
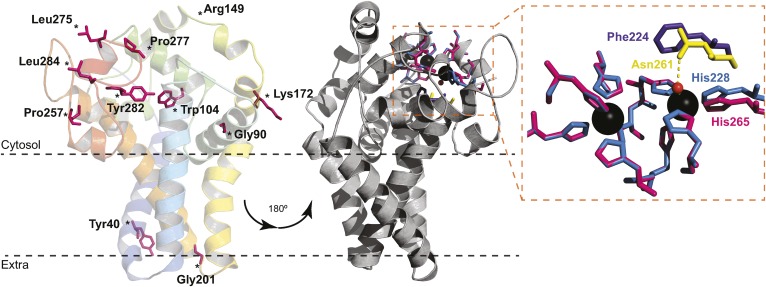
For a better understanding of the structure and function of B. cereus DesA, homology modeling was carried out based on the recently reported crystal structure of human SCD1 (Fig. 5). To this end, residues 18–284 of DesA were modeled (72% of DesA) with 99.9% confidence, and the alignment of the model with the template showed a low root-mean-square deviation of 0.00051Å over 240 residues. In spite of the low sequence identity of these proteins, the tertiary structure of DesA almost completely resembles SCD1. The 3D model is in accordance with the proposed topology (Fig. 1), displaying four TM segments and a cytosolic catalytic domain composed mainly of helical secondary structures. Moreover, the N- and C-terminal ends are located on the cytosolic side. A striking difference emerges from the position of the helix between residues 146 and 160, which locates far from the membrane. The cytosolic catalytic domain is composed mainly of helical secondary structures, with nine His residues forming the metal coordination center. As will be discussed later, this structural model could explain the effects of the AA replacement on DesA activity (see Figs. 4, 5, and supplemental Fig. S8).
Fig. 5.
Structure homology model of B. cereus DesA. The predicted model of acyl-lipid desaturase DesA from B. cereus was performed with the program Phyre2, using the crystal structure of the human SCD1 as a template (Protein Data Bank ID code 4ZYO). The 72% of residues were modeled (18–284 AA) with 99.9% confidence. The model is represented as cartoon and colored in rainbow from N terminus (blue) to C terminus (red). The conserved mutagenized residues are shown as pink sticks. The gray representation is rotated 180°, and the residues involved in metal coordination are represented as sticks (left). Close-up view of the catalytic site pocket (right) shows the nine His residues of SCD1 (pink) and DesA (blue). Phe224 of DesA (purple) cannot bind a water molecule as Asn261 of SCD1 (yellow) to complete the coordination sphere.
DISCUSSION
The FA desaturases from B. subtilis (Des) and B. cereus (DesA) use existing membrane phospholipids as substrates to introduce a cis-double bond at the fifth position of the fatty acyl chain. Because their key role in remodeling the fluidity of the membrane at low growth temperature, these enzymes were extensively characterized by our research group (8, 19, 21). Recently, we demonstrated that DesA has a high desaturation activity, being much more active than Des (21). By taking advantage of high sequence and topological similarity of these two desaturases and one cryptic desaturase coded by the ORF BL02692 from B. licheniformis, we generated sets of chimeric proteins by reciprocal domain swapping of the three desaturases. Expression of these chimeric desaturases in B. subtilis LC5 strain revealed that the two first TM domains of DesA were critical to improve Des activity and to recover the activity of the BL02692 gene product. To identify key residues in the region containing these two TM domains, we carefully examined the AA sequence of several Δ5 desaturases from Firmicutes. Using this analysis combined with site-directed mutagenesis and in vivo assays, we pinpointed a tyrosine residue (Tyr40) in TM1 of DesA that plays a critical role in lipid desaturation.
Membrane proteins need carefully designed anchors to stabilize and integrate with the lipid bilayer (24). The anchoring elements of the protein possess a flexibility necessary for accurately positioning and orientating the protein in the lipid bilayer (25). Tyrosine is an amphipathic and bulky AA with overall hydrophobic characteristics. When it is placed in a TM region, this residue often extends its side chain along the direction perpendicular to the membrane bilayer pointing away from the hydrophobic core (snorkeling effect) (24). Outstandingly, our results indicate that a single substitution of Cys40 by Tyr in the cryptic ORF BL02692 restored its Δ5 desaturase activity. Interestingly, in the homology model of DesA (Fig. 5), Tyr40 is buried into the membrane and fits perfectly with Tyr88 of SCD1, probably anchoring the TM helix to the lipid bilayer (5). Thus, it is tempting to speculate that this conserved Tyr might be snorkeling toward the water phase, pulling TM1 in such a way to allow access of the acyl chain, directly from the lipid bilayer to the substrate tunnel.
The crystal structure of SCD1, used as template for the B. cereus DesA model, shows the acyl-CoA substrate bound to the cytosolic side of the protein through the CoA moiety, whereas the acyl chain is observed within the hydrophobic tunnel (4, 5). As expected from the dissimilar substrate specificities of DesA and SCD1, the region of DesA modeled with less confidence corresponds to the CoA binding site of SCD1, i.e., the cytosolic domain of the protein located between TM2 and TM3. Based on these observations, we speculate that such a cytosolic domain might differ somehow in DesA in order to bind phospholipids embedded into the membrane. Moreover, the modeled hydrophobic tunnel of DesA fits very well with the template, and, therefore, the desaturation mechanism of DesA might be analogous to that of SCD1, with a pronounced kink providing the structural constraint to position C5–C6 atoms of the substrate acyl chain close to the catalytic site.
Earlier studies have shown that the eight His residues of the three histidine-rich domain, identified in most desaturases, were essential for the catalytic activity of B. subtilis Des (6, 9). Evidence presented here demonstrated that a new residue, His228, is critical in B. cereus DesA activity. According to the model shown in Fig. 5, nine histidine residues form the metal coordination center, with eight histidine residues located in the conserved boxes. The ninth histidine, His228, is placed near the cytoplasmic water–lipid interface, where TM4 exits the membrane resembling His265 in SCD1 (5). With such architecture, the inhibition of desaturase activity by His228Ala mutation is explained by the alteration of the metal binding site. It should be noted that ,even though the nine His AAs fit almost perfectly with the template, Phe224 cannot imitate Asn261 of SCD1 to accommodate a water molecule near the metal ions, implying that DesA folding differs somehow in order to complete the coordination sphere.
In addition, residues flanking the His boxes are critical for the enzymatic activity, as it was demonstrated for several membrane-bound desaturases (5, 26–31). In this work, we were able to identify 11 highly conserved AA residues among Δ5 and Δ10 acyl-lipid desaturases (besides the conserved His), which may be important for desaturase catalytic activity. We replaced these 11 residues in B. cereus DesA with (Ala or Arg) and found that the enzyme activity of eight mutants (Gly90, Trp104, Lys172, His228, Pro257, Leu275, Tyr282, and Leu284) was completely abolished. A closer inspection of the B. cereus DesA model (Fig. 5) suggests that mutation Trp104Arg produces an inactive variant altering the hydrophobic tunnel where the acyl chain is enclosed. In addition, mutations Gly90Arg, Arg149Ala, and Lys172Ala affect desaturase activity, possibly modifying substrate recognition and/or entrance into the hydrophobic interior of the tunnel (Fig. 5). The moderate decrease in desaturase activity of the Gly201Ala mutant might be due to a partially conservative replacement of a residue located in the extracellular membrane–water interface of TM3. This substitution presumably produces a slight conformational change in the protein, affecting either the hydrophobic tunnel or substrate recognition and binding. The loss of desaturase activity caused by the substitutions Pro257Ala, Leu275Arg, Pro277Ala, Tyr282Arg, and Leu284Arg, which are located close to the di-iron center, could result either from disruption of the metal coordination sphere or by weakening the intermolecular interactions of DesA with the ferredoxin/flavodoxin proteins, which transport the electrons needed for the desaturation reaction.
The results presented in this work are a step forward in our understanding of the structure–function relationship of phospholipid desaturases and will aid in fully unlocking the remarkable enzymatic plasticity displayed by this class of enzymes. MUFAs are the best components for biodiesel when considering the low temperature fluidity and oxidative stability (32). Therefore, the rational design of membrane-bound desaturases with increased activity guided by the structure–function principle elucidated in this paper would offer a powerful tool to engineer FAs of industrial importance.
Supplementary Material
Acknowledgments
The authors thank Guillermo Marcuzzi for technical assistance.
Footnotes
Abbreviations:
- AA
- amino acid
- Des
- Bacillus subtilis Δ5 desaturase
- DesA
- Δ5 phospholipid desaturase from Bacillus cereus
- FAME
- FA methyl ester
- Km
- kanamycin
- MM
- minimal medium
- ORF
- open reading frame
- SCD
- stearoyl-CoA desaturase
- Sp
- spectinomycin
- TM
- transmembrane
- UFA
- unsaturated FA
This work was supported by Fondo para la Investigación Científica y Tecnológica and Agencia Nacional de Promoción Científica y Tecnológica Grant PICT 2008 N 1258 and by Consejo Nacional de Investigaciones Científicas y Técnicas Grant P-UE 2016-IBR. D.E.S. had a Fondo para la Investigación Científica y Tecnológica postdoctoral fellowship. E.S. has a Consejo Nacional de Investigaciones Científicas y Técnicas postdoctoral fellowship. S.G.A., A.D.U., and D.d.M. are members of the Carrera del Investigador Científico, Consejo Nacional de Investigaciones Científicas y Técnicas, Argentina.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Shanklin J., and Cahoon E. B.. 1998. Desaturation and related modifications of fatty acids1. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 611–641. [DOI] [PubMed] [Google Scholar]
- 2.Sperling P., Ternes P., Zank T. K., and Heinz E.. 2003. The evolution of desaturases. Prostaglandins Leukot. Essent. Fatty Acids. 68: 73–95. [DOI] [PubMed] [Google Scholar]
- 3.Li D., Moorman R., Vanhercke T., Petrie J., Singh S., and Jackson C. J.. 2016. Classification and substrate head-group specificity of membrane fatty acid desaturases. Comput. Struct. Biotechnol. J. 14: 341–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H., Klein M. G., Zou H., Lane W., Snell G., Levin I., Li K., and Sang B. C.. 2015. Crystal structure of human stearoyl-coenzyme A desaturase in complex with substrate. Nat. Struct. Mol. Biol. 22: 581–585. [DOI] [PubMed] [Google Scholar]
- 5.Bai Y., McCoy J. G., Levin E. J., Sobrado P., Rajashankar K. R., Fox B. G., and Zhou M.. 2015. X-ray structure of a mammalian stearoyl-CoA desaturase. Nature. 524: 252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shanklin J., Whittle E., and Fox B. G.. 1994. Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry. 33: 12787–12794. [DOI] [PubMed] [Google Scholar]
- 7.Sayanova O., Beaudoin F., Libisch B., Castel A., Shewry P. R., and Napier J. A.. 2001. Mutagenesis and heterologous expression in yeast of a plant Delta6-fatty acid desaturase. J. Exp. Bot. 52: 1581–1585. [DOI] [PubMed] [Google Scholar]
- 8.Altabe S. G., Aguilar P., Caballero G. M., and de Mendoza D.. 2003. The Bacillus subtilis acyl lipid desaturase is a delta5 desaturase. J. Bacteriol. 185: 3228–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diaz A. R., Mansilla M. C., Vila A. J., and de Mendoza D.. 2002. Membrane topology of the acyl-lipid desaturase from Bacillus subtilis. J. Biol. Chem. 277: 48099–48106. [DOI] [PubMed] [Google Scholar]
- 10.Grau R., and de Mendoza D.. 1993. Regulation of the synthesis of unsaturated fatty acids by growth temperature in Bacillus subtilis. Mol. Microbiol. 8: 535–542. [DOI] [PubMed] [Google Scholar]
- 11.Chazarreta-Cifre L., Martiarena L., de Mendoza D., and Altabe S. G.. 2011. Role of ferredoxin and flavodoxins in Bacillus subtilis fatty acid desaturation. J. Bacteriol. 193: 4043–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spizizen J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. USA. 44: 1072–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook J. 1989. Molecular Cloning: A Laboratory Manual. 2nd edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 14.Dubnau D., and Davidoff-Abelson R.. 1971. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J. Mol. Biol. 56: 209–221. [DOI] [PubMed] [Google Scholar]
- 15.Sekiguchi J., Takada N., and Okada H.. 1975. Genes affecting the productivity of alpha-amylase in Bacillus subtilis Marburg. J. Bacteriol. 121: 688–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horton R. M., Cai Z. L., Ho S. N., and Pease L. R.. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 8: 528–535. [PubMed] [Google Scholar]
- 17.Lewis P. J., and Marston A. L.. 1999. GFP vectors for controlled expression and dual labelling of protein fusions in Bacillus subtilis. Gene. 227: 101–110. [DOI] [PubMed] [Google Scholar]
- 18.Bligh E. G., and Dyer W. J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 19.Chazarreta Cifré L., Alemany M., de Mendoza D., and Altabe S.. 2013. Exploring the biosynthesis of unsaturated fatty acids in Bacillus cereus ATCC 14579 and functional characterization of novel acyl-lipid desaturases. Appl. Environ. Microbiol. 79: 6271–6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aguilar P. S., Cronan J. E. Jr., and de Mendoza D.. 1998. A Bacillus subtilis gene induced by cold shock encodes a membrane phospholipid desaturase. J. Bacteriol. 180: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altabe S. 2013. Remodeling of membrane phospholipids by bacterial desaturases. In Stearoyl-CoA Desaturase Genes in Lipid Metabolism. Springer, New York. 209–231. [Google Scholar]
- 22.Los D. A., and Murata N.. 1998. Structure and expression of fatty acid desaturases. Biochim. Biophys. Acta. 1394: 3–15. [DOI] [PubMed] [Google Scholar]
- 23.Stukey J. E., McDonough V. M., and Martin C. E.. 1990. The OLE1 gene of Saccharomyces cerevisiae encodes the delta 9 fatty acid desaturase and can be functionally replaced by the rat stearoyl-CoA desaturase gene. J. Biol. Chem. 265: 20144–20149. [PubMed] [Google Scholar]
- 24.Liang J., Adamian L., and Jackups R. Jr.. 2005. The membrane-water interface region of membrane proteins: structural bias and the anti-snorkeling effect. Trends Biochem. Sci. 30: 355–357. [DOI] [PubMed] [Google Scholar]
- 25.Stopar D., Spruijt R. B., and Hemminga M. A.. 2006. Anchoring mechanisms of membrane-associated M13 major coat protein. Chem. Phys. Lipids. 141: 83–93. [DOI] [PubMed] [Google Scholar]
- 26.Shanklin J., Guy J. E., Mishra G., and Lindqvist Y.. 2009. Desaturases: emerging models for understanding functional diversification of diiron-containing enzymes. J. Biol. Chem. 284: 18559–18563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hongsthong A., Subudhi S., Sirijuntarat M., and Cheevadhanarak S.. 2004. Mutation study of conserved amino acid residues of Spirulina delta 6-acyl-lipid desaturase showing involvement of histidine 313 in the regioselectivity of the enzyme. Appl. Microbiol. Biotechnol. 66: 74–84. [DOI] [PubMed] [Google Scholar]
- 28.Liu J., Li D., Yin Y., Wang H., Li M., and Yu L.. 2011. Delta6-Desaturase from Mortierella alpina: cDNA cloning, expression, and phylogenetic analysis. Biotechnol. Lett. 33: 1985–1991. [DOI] [PubMed] [Google Scholar]
- 29.Broadwater J. A., Whittle E., and Shanklin J.. 2002. Desaturation and hydroxylation. Residues 148 and 324 of Arabidopsis FAD2, in addition to substrate chain length, exert a major influence in partitioning of catalytic specificity. J. Biol. Chem. 277: 15613–15620. [DOI] [PubMed] [Google Scholar]
- 30.Gagné S. J., Reed D. W., Gray G. R., and Covello P. S.. 2009. Structural control of chemoselectivity, stereoselectivity, and substrate specificity in membrane-bound fatty acid acetylenases and desaturases. Biochemistry. 48: 12298–12304. [DOI] [PubMed] [Google Scholar]
- 31.Vanhercke T., Shrestha P., Green A. G., and Singh S. P.. 2011. Mechanistic and structural insights into the regioselectivity of an acyl-CoA fatty acid desaturase via directed molecular evolution. J. Biol. Chem. 286: 12860–12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao Y., Liu W., Xu X., Zhang H., Wang J., and Xian M.. 2014. Production of free monounsaturated fatty acids by metabolically engineered Escherichia coli. Biotechnol. Biofuels. 7: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.