Abstract
The pathogenicity of Campylobacter concisus, increasingly found in the human gastrointestinal (GI) tract, is unclear. Some studies indicate that its role in GI conditions has been underestimated, whereas others suggest that the organism has a commensal-like phenotype. For the enteropathogen C. jejuni, the lipooligosaccharide (LOS) is a main driver of virulence. We investigated the LOS structure of four C. concisus clinical isolates and correlated the inflammatory potential of each isolate with bacterial virulence. Mass spectrometric analyses of lipid A revealed a novel hexa-acylated diglucosamine moiety with two or three phosphoryl substituents. Molecular and fragment ion analysis indicated that the oligosaccharide portion of the LOS had only a single phosphate and lacked phosphoethanolamine and sialic acid substitution, which are hallmarks of the C. jejuni LOS. Consistent with our structural findings, C. concisus LOS and live bacteria induced less TNF-α secretion in human monocytes than did C. jejuni. Furthermore, the C. concisus bacteria were less virulent than C. jejuni in a Galleria mellonella infection model. The correlation of the novel lipid A structure, decreased phosphorylation, and lack of sialylation along with reduced inflammatory potential and virulence support the significance of the LOS as a determinant in the relative pathogenicity of C. concisus.
Keywords: cytokines, glycolipids, inflammation, lipid A, mass spectrometry, monocytes, phosphorylation, toll-like receptors
Campylobacter concisus is a member of the Campylobacter genus with as-yet-unclear pathogenic potential. Following the initial identification of C. concisus in patients with periodontal disease, recent investigations indicate this non-jejuni and -coli Campylobacter species has a role in acute and chronic human gastrointestinal (GI) conditions (1–5). This notion, however, remains a topic of debate because in general its overall high prevalence in patients and healthy individuals alike suggests a commensal-like nature for C. concisus compared with C. jejuni (6, 7). Nonetheless, increased prevalence of C. concisus DNA in samples of patients with ulcerative colitis, pediatric Crohn’s disease, gastroesophageal reflux, and Barrett’s esophagus disease has been reported (5, 8–12). Furthermore, C. concisus and other non-jejuni and -coli Campylobacter species have been implicated as causal agents in prolonged mild endemic diarrhea in children and in traveler’s diarrhea (11, 13, 14). Recent meta-analyses have also revealed an association of Campylobacter spp., mainly C. concisus and C. showae, with an increased risk of inflammatory bowel disease (15).
C. concisus can attach to and invade intestinal epithelial cells and cause the secretion of proinflammatory cytokines, likely by a toll-like receptor 4 (TLR4)-dependent mechanism (9, 16). Adherence, invasion, and a proinflammatory phenotype may be strain-specific characteristics important in the pathogenesis of Proteobacteria such as C. concisus and some Escherichia coli (17). In general, bacterial lipopolysaccharide (LPS) and lipooligosaccharide (LOS) activate TLR4 and are important virulence factors for Gram-negative bacteria (18–21). In C. jejuni, the hydrophobic lipid A backbone of the LOS is hexa-acylated and is known to be a potent activator of TLR4 (22, 23). The interaction of LOS with TLR4 triggers a downstream signaling cascade by activating the NF-κB transcription factor and subsequent secretion of proinflammatory cytokines such as TNF-α and interleukin 8. Apart from TLR4 engagement, C. jejuni LOS also plays a role in bacterial invasion, colonization, and stress survival (23–25).
The biosynthesis region of C. jejuni LOS is highly variable between species possessing major differences in the content and organization of genes encoding LOS carbohydrate moieties and their linkages. In addition, the LOS gene locus is prone to frequent phase variations, resulting in broad structural heterogeneity among strains (26, 27). Major differences are observed in the outer region of the oligosaccharide (OS) moiety, but variations in lipid A phosphorylation and the number of amide linkages are also present (22). The lipid A disaccharide of C. jejuni comprises either 2-amino-2-deoxy-d-glucose (glucosamine; GlcN) or 2,3-diamino-2,3-dideoxy-d-glucose (GlcN3N); thus, the number of amide linkages to the acyl chains can vary from two (GlcN-GlcN) to four (GlcN3N-GlcN3N). Lipid A is hexa-acylated with either myristic (C14:O) or palmitic (C16:O) fatty acids, four of which are hydroxy fatty acids and are directly linked to the disaccharide residue (22, 28).
There is significant heterogeneity in the position and number of phosphate and phosphoethanolamine (PEA) residues bound to the OS and lipid A of the C. jejuni LOS (28). Amide linkages and lipid A phosphorylation have been shown to affect TLR4 activation (22, 29, 30). Although the core OS of C. jejuni is primarily conserved, significant variations in the outer OS region (31), which can contain N-acetylneuraminic acid (NeuAc, sialic acid) residues that are thought to modulate TLR4 activation, have been found (22, 32, 33).
Although there has been progress in the understanding of C. jejuni LOS and its role in the pathogenesis of human disease (22, 32, 34), the structure and heterogeneity of C. concisus LOS has remained unstudied. Such investigations could be informative regarding the relative pathogenicity and/or commensalism of C. concisus in humans. The aims of this study were to delineate the characteristics of the C. concisus LOS structure and inflammatory potential compared with the characteristics of C. jejuni LOS.
MATERIALS AND METHODS
Bacterial strains and LOS extraction
Clinical C. concisus isolates were isolated from the sigmoid colon biopsy of a newly presenting male pediatric patient diagnosed with Crohn’s disease as part of the BISCUIT study (B38) (35) and from feces of patients with acute gastroenteritis (2010-131105, 2010-347972) (10, 11). The C. concisus NCTC 12408 strain is commercially available and originates from pediatric enteritis. C. jejuni 11168H is a variant of the human diarrhea isolate NCTC 11168 (36). The latter two strains were a kind gifts from David Guilliano (University of East London) and Brendan Wren (London School of Hygiene and Tropical Medicine), respectively. Bacterial strains were grown on blood agar no. 2 containing 0.5% yeast extract (Oxoid, Basingstoke, UK) and 5% defibrinated horse blood (Sigma-Aldrich, Gillingham, UK). All strains were grown at 37°C in a gas jar under microaerobic conditions generated by using a CampyGen sachet (Oxoid). For C. concisus strains the microaerobic atmosphere was supplemented with ∼10% H2 generated with sodium borohydride (Sigma-Aldrich) (37). C. jejuni and C. concisus strains were grown for 24 and 72 h, respectively, harvested, and stored at −80°C prior to use. Bacterial LOS was extracted and purified by a modification of the hot phenol-water method as described previously (22).
SDS-PAGE analysis
LOS samples of 10 µg were subjected to SDS-PAGE using a 12% polyacrylamide gel (PROTEAN II xi cell; Bio-Rad, Hercules, CA) in Tris-glycine running buffer. The gel was fixed for 1 h in 40% methanol and 5% acetic acid, and LOS was visualized by silver staining (38).
Preparation of intact LOS for MALDI-TOF MS
LOS samples were prepared for MS analysis as previously described (18). Briefly, purified LOS (10 mg/ml) was suspended in a methanol-water (1:3) solution containing 5 mM EDTA. An aliquot was desalted with cation exchange beads (Dowex 50WX8-200). The desalted sample solution was mixed with 100 mM dibasic ammonium citrate (9:1 v/v), and 1–2 µl was spotted onto a thin layer of matrix composed of a 4:1 solution of 2,4,6-trihydroxyacetophenone (200 mg/ml in methanol; Sigma-Aldrich) with nitrocellulose (15 mg/ml in acetone-isopropanol [1:1]; Bio-Rad). Samples were left to air dry prior to analysis.
O-deacylation of LOS
Native LOS (∼300 μg) was incubated with 200 μl anhydrous hydrazine (Sigma-Aldrich) at 37°C for 2 h with intermittent vortexing. The reaction was stopped with 1 ml precooled acetone (−20°C), and the O-deacylated LOS samples were pelleted by centrifugation at 10,000 g, washed with chilled acetone, centrifuged again, dissolved in 20 μl water, lyophilized, and stored at −80°C.
HF treatment of LOS
Phosphoesters were partially removed by hydrogen fluoride (HF) treatment. Native LOS (10 mg/ml) was reacted with 48% aqueous HF at 4°C for 16–20 h. Excess HF was removed using a Savant SpeedVac (Thermo Fisher Scientific, Waltham, MA) with an in-line trap.
High-resolution MALDI-TOF and IMS-MS
MALDI-TOF MS and ion-mobility spectrometry (IMS)-MS analyses were performed on a Synapt G2 high-definition MS system (Waters Corporation, Milford, MA) in sensitivity mode. The instrument is equipped with a T-wave ion-mobility cell (TriwaveTM) (39, 40) and was operated in MALDI mode as previously described (41). Spectra were obtained in negative- or positive-ion mode operating a neodymium-doped yttrium aluminum garnet laser at 355 nm and 200 Hz. In general, spectra were acquired for ∼1–2 min with a scan duration of 1.0 s and an overall cycle time of 1.024 s. The instrument was calibrated using the masses of the monoisotopic ions for porcine renin substrate, intact bovine insulin, and B chain.
The T-wave device on the Synapt G2 high-definition MS system consists of three cells: a trap cell, IMS cell, and transfer cell. For IMS-MS experiments, the T-wave peak height voltage was 40 V, and the T-wave velocity used was generally a variable wave velocity of 650 to 250 m/s. Typically, the T-wave mobility cell contained nitrogen at a pressure of ∼2 mbar. The trap gas flow was 0.4 ml/min, the helium cell gas flow was 180 ml/min, and the IMS gas flow was 90 ml/min. The trap DC bias was 80 V.
MS/MS with IMS was performed typically by selecting precursor ions with instrument LM and HM resolution settings of 4.7 and 15.0, respectively. In initial TOF MS/MS mode without IMS, fragmentation was achieved by applying collision energy with argon as the collision gas in the trap region of the T-wave ion-mobility cell. Collision energies of 85–110 V were required for optimum fragmentation of intact LOS or prompt fragment ions (pseudo-MS3) in the trap. For IMS-MS/MS experiments, collision energy was also applied with argon as the collision gas in the transfer cell after IMS separation at values ranging from 50 to 90 V, depending on the analyte. For the analysis of OS and lipid A prompt fragments, a T-wave variable wave velocity of 1,100 to 200 m/s was used. 2D IMS spectra were viewed using DriftScope 2.1 software, and selected spectral regions were exported to MassLynx with retention of drift-time information for the generation of mobilograms and subspectra. Chemical structures were generated using the ChemBioDraw Ultra software.
Genomic analysis
Genomic analysis of the C. concisus strains used in our study was carried out to look for the presence or absence of genes involved in PEA transfer and in sialic acid biosynthesis and transfer. The genomes of three of the strains (B38, 2010-131105, and 2010-347972) and 53 additional C. concisus strains were recently whole genome sequenced, and the genomes were assembled (42). In addition, the following analyses were carried out on 36 other publicly available C. concisus strains (2, 43–47).
Gene and protein prediction of genome assemblies was carried out with Prokka (48). The Prokka dependencies BioPerl (49), GNU parallel (50), BLAST+ (51), and Prodigal (52) were utilized along with the recommended and optional tools Aragorn (53), Barrnap (https://github.com/Victorian-Bioinformatics-Consortium/barrnap) (42), HMMER3 (54), Infernal (55), RNAmmer (56), and SignalP (57). Annotation of predicted genes and proteins was carried out with BlastKOALA (58).
PCR validation of LOS genes
PCR validation of the genome analysis was performed using the primer pair eptC-F (CCAGATGAAGCCGGTGAGTT) and eptC-R (T[G/A]CTCCAAGGCT[C/T]TTTGCTT), which amplifies a 578 bp region of the PEA transferase gene eptC, and the primer pair waaC-F (TGGCT[A/C]GTTGATGCCCGTTT) and waaC-R (ATCGCCTCAGCTCTTGC[T/C]TT), which amplifies a 522 bp region of the LOS heptosyl transferase I waaC gene, which is the first within the LOS gene cluster. PCR conditions were as follows: eptC: 94°C for 5 min, 30 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 30 s, followed by 72°C for 10 min; waaC: 94°C for 5 min, 30 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s, followed by 72°C for 10 min.
TNF-α secretion by THP-1 monocytic cells and PBMCs
The human monocytic cell line THP-1 was obtained from the American Type Culture Collection (Manassas, VA) and propagated in RPMI 1640 supplemented with 10% FBS at 37°C in a 5% CO2 atmosphere. The cells were differentiated with 10 ng/ml PMA (Sigma-Aldrich) for 18 h as previously reported (59). The differentiated THP-1 cells were seeded at 1.2 × 105 cells per well in 96-well plates and treated with 100 ng/ml LOS or culture media only for 18 h. The supernatants were collected and stored at −80°C until analysis.
Peripheral blood mononuclear cells (PBMCs) were isolated from venous blood obtained from seven healthy adult volunteers following the manufacturer’s protocol (LymphoprepTM; Axis-Shield, Dundee, UK). PBMCs (2 × 106 cells/ml) were suspended in RPMI 1640 medium with 10% FBS and cocultured with bacteria (multiplicity of infection = 100), and supernatants were collected after 18 h. TNF-α cytokine release was determined using a Ready-Set-Go TNF-α ELISA kit (Affymetrix, San Diego, CA) following the manufacturer’s instructions.
Blood samples were obtained with informed consent and ethical approval from the Institute of Child Health/Great Ormond Street Hospital Research Ethics Committee and in accordance with the Declaration of Helsinki.
Galleria mellonella infection model
G. mellonella larvae (UK Waxworms Ltd., Dinnington, UK) were injected into the second foreleg with a bacterial suspension in 10 µl inocula (n = 15; ∼107 CFU) (60, 61). Larvae were incubated at 37°C, and survival was monitored at 24 h intervals for 5 days. PBS injection served as a control.
Statistical analysis
One-way ANOVA with Tukey posttest was applied for multiple comparison analysis. Survival curves were analyzed by the Mantel-Cox log-rank test. GraphPad Prism 7.00 software was used for analysis.
RESULTS
Heterogeneity between C. concisus LOS structures
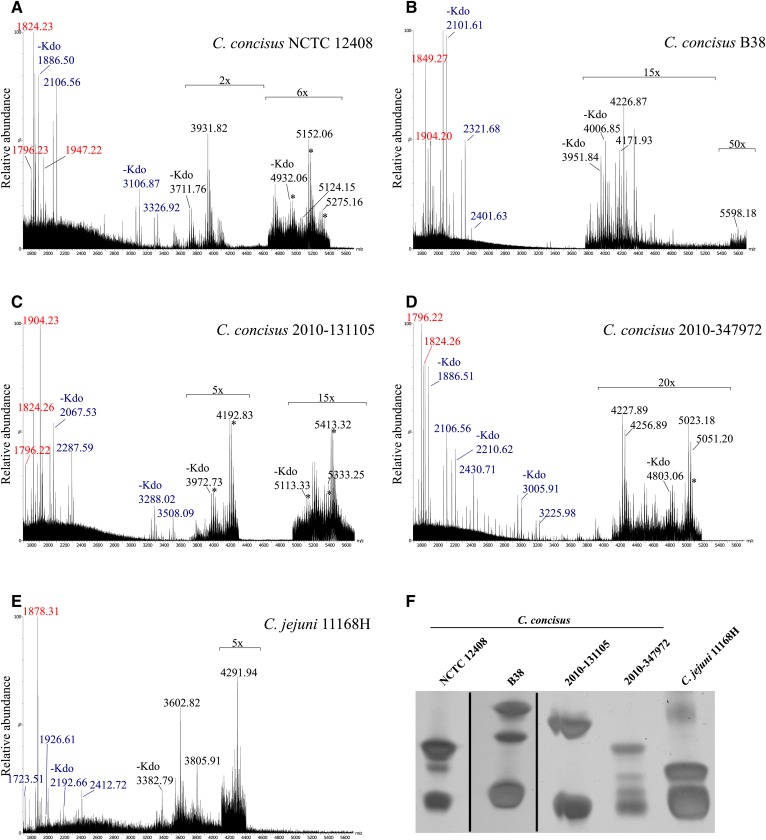
To investigate the structure of C. concisus LOS, we first analyzed intact LOS that was isolated from four C. concisus GI strains by MS and SDS-PAGE. LOS was also extracted from the C. jejuni 11168H strain and used for comparison. Spectra of the intact LOS were obtained in high-resolution reflectron-mode negative-ion MALDI-TOF MS that enabled measuring monoisotopic molecular ions with high mass accuracy (<35 ppm). The spectra provided evidence of varied and multiple LOS molecular ions, with some ions detected above m/z 5,000 in all four C. concisus LOS samples (Fig. 1A–D). Peaks for the molecular ions of the C. jejuni 11168H LOS were of lower mass in the range of m/z 3,600 to 4,300 (Fig. 1E). This is in agreement with previous MS studies of intact C. jejuni LOS from other strains that reported molecular ion peaks of lower mass than those for the LOS of the C. concisus isolates (22, 62, 63).
Fig. 1.
Negative-ion MALDI-TOF spectra and LOS profiles of C. concisus and C. jejuni LOS. A–E: Negative-ion MALDI-TOF MS spectra for the LOS of C. concisus isolates NCTC 12408, B38, 2010-131105, 2010-347972, and C. jejuni 11168H, respectively. The relative abundance of the peaks was magnified where indicated. The m/z values presented are for the monoisotopic peaks observed (red font for lipid A fragments, blue font for OS fragment ions, and black font for molecular ions). Asterisks show peaks corresponding to sodiated molecular or fragment ion peaks. Purified LOS (10 μg per lane) of C. concisus isolates NCTC 12408, B38, 2010-131105, 2010-347972, and C. jejuni 11168H were separated by SDS-PAGE and stained using silver nitrate. F: The gel was cropped where indicated (black lines).
The SDS-PAGE profile of the Campylobacter LOS showed the presence of multiple bands in all samples (Fig. 1F), which was consistent with the heterogeneity in the LOS that was revealed by the MS analyses. In accordance with the MALDI-TOF MS data, the electrophoretic mobility of the slow migrating bands was indicative of larger LOS moieties in the C. concisus isolates compared with the C. jejuni LOS. The C. concisus isolates with bands for the largest LOS moieties were B38 and 2010-131105, and these appear to be in agreement with the molecular ion peaks observed in the negative-ion spectra at m/z 5,598.18 (Fig. 1B) and m/z 5,413.32 (Fig. 1C), respectively.
Prompt fragmentation of the labile 2-keto-3-deoxyoctulosonic acid (Kdo)-lipid A bond of the LOS occurring in the MALDI source produced abundant lipid A and OS fragment ions of the LOS. Observed peaks corresponding to the loss of Kdo (220.06 Da) aided in the identification of peaks for OS fragment ions and LOS molecular ions. An attribute of this methodology is that abundant lipid A and OS fragment ions are produced together with molecular ions for the intact LOS that thus enables more certain identification of the latter and provides an opportunity to obtain top-down sequence information. For example, the observed peak at m/z 1,904.23 for the monoisotopic mass of the largest lipid A fragment ion of C. concisus isolate 2010-131105 LOS (Fig. 1C) together with that for the OS fragment ion at m/z 2,287.59 plus a proton gives a value of m/z 4,192.83, corresponding to the monoisotopic mass of the prominent molecular ion peak for intact LOS.
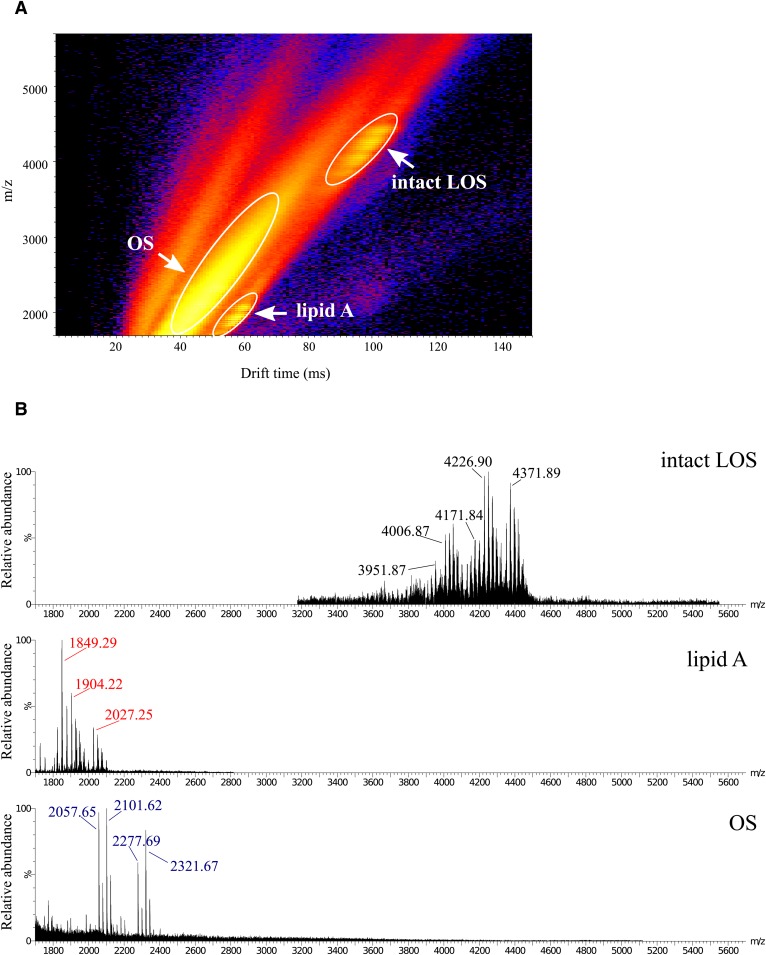
IMS MALDI-TOF MS was used to separate intact LOS and the prompt lipid A and OS fragment ions formed in the source (Fig. 2). Due to the differences in the shape and size of the lipid A and OS fragments and intact LOS, the ions were separated by IMS to provide more evidence of distinct molecular characteristics. In some cases, the ion types occurred in otherwise overlapping regions of the spectra. The IMS enabled the extraction of subspectra of specific ion types, permitting unambiguous classification of ions and mass measurements. The spectra of the LOS from each isolate contained unique OS fragment ions and molecular ions for intact LOS but contained some similar peaks for lipid A fragment ions (Table 1).
Fig. 2.
Analysis of intact LOS from C. concisus B38 by MALDI-TOF MS with IMS. The panel shows a 2D plot of the IMS data set. A: Differences in drift time allow for the separation of low-molecular-weight components of non-LOS origin, OS prompt fragment ions, intact LOS, and lipid A prompt fragment ions. B: Regions of the mass spectra of the (M-H)− molecular ions, lipid A prompt fragments, and OS prompt fragments. Spectra were acquired with the Synapt G2 high-definition mass spectrometer using IMS and analyzed with DriftScope and MassLynx software.
TABLE 1.
Observed (M-H)− peaks for lipid A and OS fragment ions and corresponding intact LOS
| Strain | Lipid A | OS | Intact LOS |
| C. concisus NCTC 12408 | 1,796.23 | 3,326.92 | 5,124.15 |
| 1,824.23 | 3,326.92 | 5,152.06 | |
| 1,947.22 | 3,326.92 | 5,275.16 | |
| C. concisus B38 | 1,849.27 | 2,321.68 | 4,171.93 |
| 1,904.20 | 2,321.68 | 4,226.87 | |
| 5,598.18 | |||
| C. concisus 2010-131105 | 1,904.23 | 2,287.59 | 4,192.83 |
| 1,824.26 | 3,508.09 | 5,333.25 | |
| 1,904.23 | 3,508.09 | 5,413.32 | |
| C. concisus 2010-347972 | 1,796.22 | 2,430.71 | 4,227.89 |
| 1,796.22 | 3,225.98 | 5,023.18 | |
| 1,824.26 | 3,225.98 | 5,051.20 | |
| C. jejuni 11168H | 1,878.31 | 1,723.51 | 3,602.82 |
| 1,878.31 | 1,926.61 | 3,805.91 | |
| 1,878.31 | 2,412.72 | 4,291.94 |
MS/MS analysis of the lipid A moiety
We performed negative-ion MS/MS analysis with IMS to obtain sequence information and structural details regarding the lipid A and OS moieties (supplemental Fig. S1). Differences in the IMS drift time of the lipid A fragment ions compared with ions for OS fragments or non-LOS species of similar m/z values resulted in cleaner MS/MS spectra and enabled more confident interpretations of the MS/MS spectra. This is illustrated by the MS/MS spectrum of the lipid A fragment ions at m/z 1,824.3 (supplemental Fig. S1B, top) from C. concisus LOS strain 2010-131105. Without IMS, we would have detected some of the less abundant but significant non-lipid A fragment ions deriving from the peak at m/z 1,821.60 shown in the MS/MS spectrum (supplemental Fig. S1B, bottom).
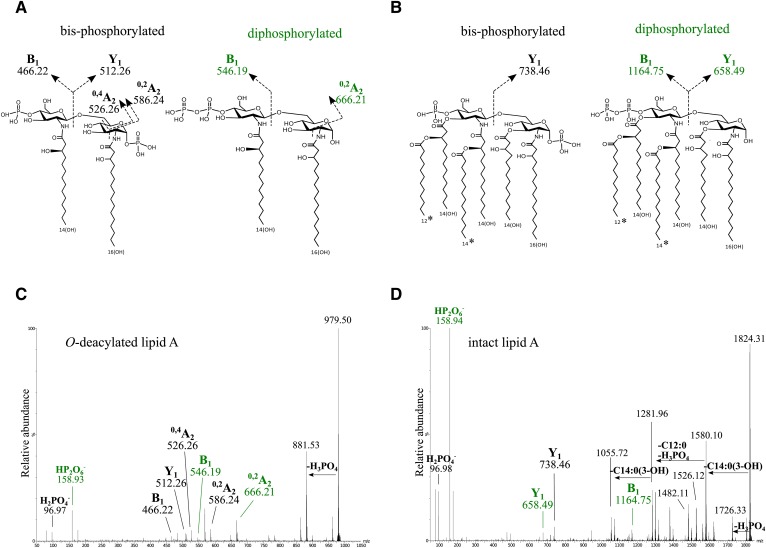
Hydrazine treatment of LOS was performed to hydrolyze the O-linked acyl chains from the lipid A while the N-linked fatty acyl groups remained intact. An IMS-MS/MS negative-ion spectrum (Fig. 3A, C) was obtained of the O-deacylated lipid A fragment ion (m/z 979.5) from the LOS of C. concisus strain 2010-131105. The fragment ion peaks observed were in accordance with the presence of a disaccharide containing two GlcN residues with β-hydroxy myristic acid (C14:0[3-OH]) and β-hydroxy palmitic acid (C16:0[3-OH]) linked to the amino groups on the nonreducing and reducing terminal monosaccharide residues, respectively. Peaks corresponding to B- and Y-type fragment ions as well as cross-ring fragment ions (0,2A and 0,4A) provided evidence for two different lipid A structures (Fig. 3A), one with phosphate on both ends (bisphosphorylated) and one diphosphorylated lipid A with a single pyrophosphoryl group (supplemental Table S1).
Fig. 3.
Negative-ion IMS-MS/MS spectra of O-deacylated and intact lipid A of C. concisus 2010-131105. A, C: Proposed structure and fragmentation from MS/MS of O-deacylated lipid A of C. concisus 2010-131105. B, D: Proposed structure and fragmentation from MS/MS of intact lipid A of C. concisus 2010-131105. Fragment ion peaks observed in the MS/MS spectrum of the parent ions at m/z 979.50 of the O-deacylated (C) and parent ions at m/z 1,824.31 in the intact lipid A (D) were consistent with B-, Y-, 0,4A-, and 0,2A-type fragment ions as indicated in the proposed structures. The asterisk indicates potential interchange in the positions of two fatty acids. The green-colored labels are for fragment ions consistent with a lipid A with a single pyrophosphoryl group.
Negative-ion IMS-MS/MS of the m/z 1,824.3 ions for the intact lipid A of strain 2010-131105 produced fragment ion peaks at m/z 1,726.33 corresponding to the loss of H3PO4 (−97.98 Da) at m/z 1,580.10, consistent with the loss of C14:0(3-OH) (−244.20 Da), and at m/z 1281.96, consistent with sequential losses of H3PO4, C14:0(3-OH), and C12:0 (−200.18 Da). The 0,4A-type cross-ring B- and Y-type fragment ions detected in the MS/MS spectrum of the intact lipid A also were in agreement with the expression of di- and bisphosphorylated hexa-acylated lipid A (Fig. 3B, D), confirming our interpretation of the MS/MS spectrum of the O-deacylated lipid A. In addition, low-mass fragment ions were observed in the negative-ion MS/MS spectra at m/z 158.9 for , providing further support for the presence of pyrophosphoryl groups on the diphosphorylated lipid A.
In total, we identified five different lipid A moieties in C. concisus LOS (Table 2). All were in accordance with a disaccharide of two GlcN residues that contained a total of two amide linkages. Fragment ions at m/z 1,824.2 or higher corresponded to the same fatty acid profile as the lower-mass fragment ions at m/z 1,796.2, but with an exchange of a C16:0(3-OH) residue for a C14:0(3-OH). An additional phosphate or PEA moiety accounted for all of the other high-mass lipid A species observed in the spectra of the C. concisus isolates (Table 2).
TABLE 2.
Proposed compositions and observed and calculated negative Y-ion type peaks for lipid A fragment ions of intact C. concisus LOS
| Composition | HexN-HexN | HexN-HexN | HexN-HexN | HexN-HexN | HexN-HexN |
| 4 C14:0(3-OH) C14:0 C12:0 2P |
3 C14:0(3-OH) C16:0(3-OH) C14:0 C12:0 2P |
3 C14:0(3-OH) C16:0(3-OH) C14:0 C12:0 2P PEA(-H3PO4) |
3 C14:0(3-OH) C16:0(3-OH) C14:0 C12:0 3P |
3 C14:0(3-OH) C16:0(3-OH) C14:0 C12:0 2P PEA |
|
| Calculated m/z | 1,796.21 | 1,824.24 | 1,849.27 | 1,904.21 | 1,947.25 |
| Observed m/z (Δppm) | |||||
| NCTC 12408 | 1,796.23 (10.3) | 1,824.23 (−7.1) | 1,947.22 (−16.1) | ||
| B38 | 1,849.27 (−2.4) | 1,904.20 (−4.8) | |||
| 2010-131105 | 1,796.22 (4.7) | 1,824.26 (9.4) | 1,904.23 (10.9) | ||
| 2010-347972 | 1,796.22 (4.7) | 1,824.26 (9.4) | |||
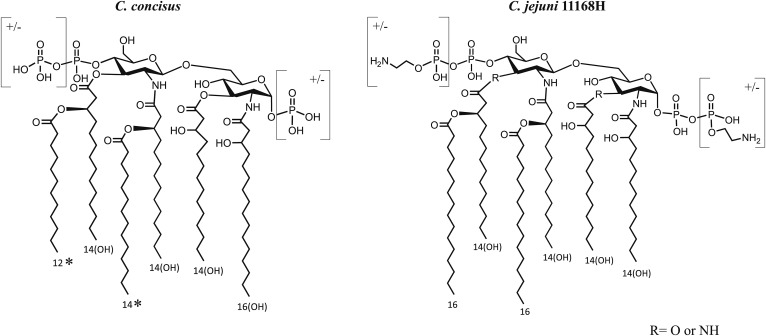
Overall, the data showed that the hexa-acylated C. concisus LOS has some similarity to C. jejuni LOS; however, clear differences were identified (Fig. 4). Both contain four hydroxy fatty acids, but there is a single C16:0(3-OH) in most C. concisus lipid A moieties, in place of one of the C14:0(3-OH) moieties on C. jejuni LOS. There are two secondary fatty acids on the nonreducing terminal glucosamine in lipid A of both species, but these were identified as C14:0 and C12:0 in C. concisus, in contrast to the two C16:0 acyl chains on the nonreducing terminal glucosamine of C. jejuni lipid A (28, 64).
Fig. 4.
Schematic of consensus structures of C. concisus and C. jejuni 11168H lipid A. The asterisk indicates potential interchange in the positions of two fatty acids.
Analyses of the OS moieties
Although the lipid A moieties of the C. concisus strains possessed significant similarities, we identified peaks of unique masses corresponding to OS fragment ions in the spectra of the LOS of each isolate (Fig. 1, Table 1). HF treatment of the LOS was used to remove and aid in establishing the presence of phosphoesters. A comparison of positive-ion MALDI-TOF MS spectra of the HF-treated and untreated LOS of C. concisus NCTC 12408 confirmed that there was a single phosphate substituent (HPO3; 79.97 Da) as part of the inner-core OS observed at m/z 2,108.6 in untreated LOS (supplemental Fig. S2).
The presence of a single phosphate in the core OS was further established by MALDI-TOF IMS MS/MS analyses in the positive-ion mode of the untreated and HF-treated NCTC 12408 LOS. Extensive fragmentation produced losses in accordance with the presence of expected monosaccharide components such as Kdo, heptose (192.06 Da), hexose (162.05 Da), and hexosamine (HexN; 161.07 Da) (supplemental Fig. S3). A loss of 175 Da, possibly corresponding to hexosaminuronic acid (HexNA; 175.05 Da) was detected in some of the spectra and apparently corresponded to low-mass fragment ion peaks observed in positive-ion spectra at m/z 176.06, as observed in supplemental Fig. S3.
Elements of commonality were detected in the spectra of the LOS from C. concisus. Monoisotopic OS fragment ions were observed at m/z 2,106.56 in the negative-ion spectra of intact LOS from both NCTC 12408 and 2010-347972 (Fig. 1A, D), presumably for the core OS. In addition, there was a neutral loss of 1,220.4 Da from both LOS molecular ion and OS fragment ions in spectra of NCTC 12408 (OS peaks at m/z 3,326.92 and 2,106.56) and 2010-131105 (OS peaks at m/z 3,508.09 and m/z 2,287.59; Fig. 1A, C). The OS structures could be related to the semi-rough-type LPS observed in some Gram-negative bacteria that contain one O-antigen repeat unit (65).
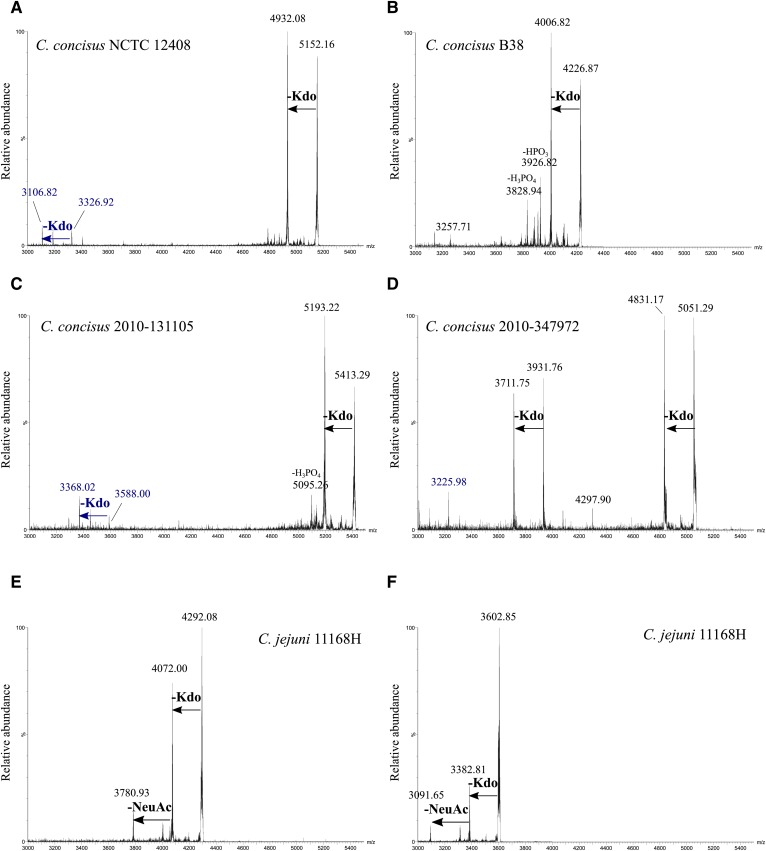
Lack of nonreducing terminal sialic acid (NeuAc) on the C. concisus LOS was established by negative-ion MS/MS of molecular ions of intact LOS (Fig. 5). Collisional activation of the molecular ions of the intact LOS from the four strains of C. concisus (Fig. 5A–D) produced abundant fragment ions for the loss of Kdo (220.06 Da), followed by the loss of HPO3 and H2PO4 (97.98 Da). For example, MS/MS of (M-H)− of intact LOS from NCTC 12408 at m/z 5,152.10 produced a prominent peak at m/z 4,932.02 for the loss of Kdo (Fig. 5A). Similarly, collisional activation of the (M-H)− ions at m/z 4,226.87 for intact LOS from strain B38 produced a prominent peak at m/z 4,006.82 corresponding to a loss of Kdo and a peak at m/z 3,926.82 for the loss of both Kdo and HPO3 (Fig. 5B). Peaks for similar fragments of the LOS molecular ion were also observed for the other two C. concisus strains (Fig. 5C, D).
Fig. 5.
Negative-ion MALDI-TOF MS/MS analysis of intact LOS ions. A–D: Negative-ion MALDI-TOF MS/MS spectra for an LOS molecular ion (M-H)− peak of each C. concisus isolates NCTC 12408, B38, 2010-131105, and 2010-347972. E, F: Negative-ion MALDI-TOF MS/MS spectra for two intact LOS ions of C. jejuni 11168H. The mass of the losses observed are consistent with the fragments of known carbohydrate or phosphoryl moieties as indicated. Labels in blue font indicate fragment ions corresponding with OS fragments as previously observed.
In contrast, the MS/MS spectra of molecular ions of the intact LOS from C. jejuni (Fig. 5E, F) contained fragment ion peaks consistent with the loss of Kdo as well as the loss of both Kdo and NeuAc (291.10 Da). For example, collisional activation of the (M-H)− ions at m/z 4,292.08 (Fig. 5E) from the C. jejuni LOS produced abundant fragment ions at m/z 4,072.00 for the loss of Kdo and a peak at m/z 3,780.93 for the loss of Kdo and NeuAc. Fragment ions from the loss of NeuAc are consistent with nonreducing terminal sialylation that have previously been established as being present on the 11168H LOS (22). However, corresponding fragment ion peaks for the loss of NeuAc were strikingly absent in the MS/MS of intact LOS from the 4 C. concisus strains.
Genomic analyses of LOS biosynthetic genes
We conducted a genomic analysis of three C. concisus strains to look for the presence or absence of genes involved in PEA transfer and in sialic acid biosynthesis and transfer. We looked specifically for the presence of sialic acid synthase (neuB1), sialic acid transferase (cstII), PEA transferase (eptC), and heptosyl transferase (waaC), which is known to be the first gene within the LOS gene cluster (66). An analysis of whole genome sequences indicated that all three strains contained waaC, whereas none of the strains contained neuB1 or cstII, confirming the lack of sialic acid synthesis machinery. The presence of waaC was further confirmed by PCR analysis. This lack of sialic acid synthesis capability was also confirmed in all 89 published C. concisus genomes. When we looked for the presence of the PEA transferase eptC gene within all of the C. concisus genomes, we found that most strains (69%) did not contain the eptC gene; however, all three of the strains used in this study were eptC-positive, which was also confirmed by PCR analysis.
Inflammatory and virulence potential of C. concisus isolates
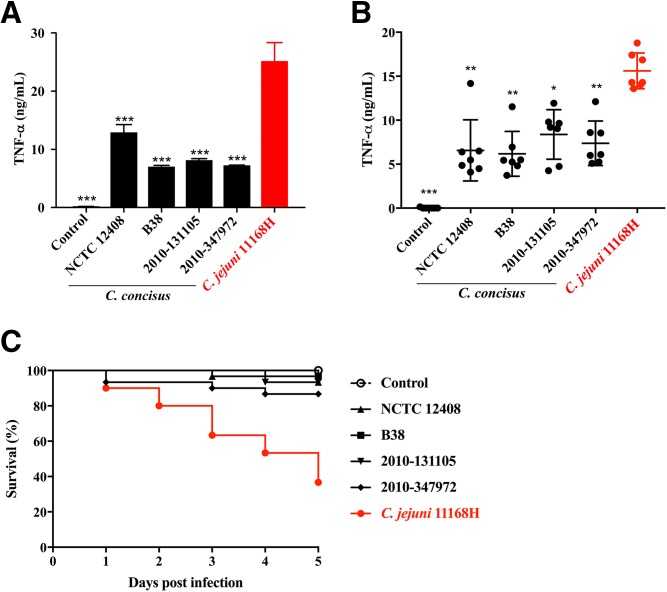
The LOS-induced TLR4 signaling cascade is crucial in triggering monocytic innate immune responses to C. jejuni that result in TNF-α secretion (22). To determine the proinflammatory potential of C. concisus LOS, differentiated THP-1 monocytes were treated with purified LOS from all four C. concisus strains and from the C. jejuni 11168H strain for 18 h prior to quantification of the proinflammatory cytokine TNF-α. Notably, the LOS from all C. concisus strains elicited a significantly reduced TNF-α response compared with the C. jejuni 11168H LOS (Fig. 6A; P < 0.001). To confirm the importance of LOS signaling in this context, PBMCs from seven healthy donors were cocultured with live bacteria of each of the four C. concisus strains and C. jejuni. As seen in the response to purified LOS, the C. concisus live bacteria also induced significantly less TNF-α compared with C. jejuni 11168H (Fig. 6B; P < 0.05).
Fig. 6.
Inflammatory potential and virulence of C. concisus isolates. A: Differentiated THP-1 cells were stimulated with purified (100 ng/ml) LOS for 18 h prior to the quantification of TNF-α secretion by ELISA. Results are expressed as the mean ± SD of eight biological replicates. Stars indicate statistical significant differences compared with C. jejuni as analyzed by ANOVA with Tukey post hoc test. B: Human PBMCs (n = 7) were cocultured with live bacteria at MOI 100 for 18 h prior to the quantification of TNF-α secretion by ELISA. Results are expressed as the mean ± SD of three independent experiments. Stars indicate statistical significant differences compared with C. jejuni as analyzed by ANOVA with a Tukey post hoc test (A, B). C: Survival of G. mellonella larvae exposed to C. concisus isolates and C jejuni 11168H. G. mellonella larvae were injected with a bacterial suspension (∼107 CFU), and their survival was monitored over time. Results represent the mean survival rate (n = 30) pooled from two independent experiments. Survival rates in the C. jejuni group were compared with each of the C. concisus groups (P < 0.001). The Mantel-Cox log-rank test was applied for statistical analysis. *P < 0.05; **P < 0.01; and ***P < 0.001.
To assess bacterial virulence, we used a G. mellonella infection model. Larvae were infected with C. concisus isolates and C. jejuni 11168H, and survival was monitored over 5 days. C. concisus isolates were significantly less virulent (>85% G. mellonella survival) compared with C. jejuni (36% survival) (Fig. 6C; P < 0.001). Thus, our findings show that the C. concisus is less inflammatory and less virulent compared with C. jejuni and, furthermore, that these differences are reflected by the reduced inflammatory potential of the C. concisus LOS compared with the LOS of C. jejuni.
DISCUSSION
C. concisus is being increasingly found in the human GI tract, yet the contribution of this organism to homeostasis in health or to disease states is unclear. In this study, we aimed at defining and correlating the structure of C. concisus LOS with its bioactivity. Four clinical isolates were chosen from adult and pediatric patients presenting with gastroenteritis and Crohn’s disease. The intact LOS of all four C. concisus isolates was larger than that of C. jejuni, which was primarily due to the differential size of the OS moiety. In addition, significant heterogeneity between the OS of the C. concisus isolates was observed. Heterogeneity in the C. jejuni LOS, particularly in the outer region of the OS, has been previously described and was ascribed to differences in the gene content and frequent phase variations in the LOS gene loci (26). Variations primarily were observed in the outer region of the OS, whereas core OS and the general structure of the lipid A were found to be predominantly conserved (31). In line with these findings, we observed structural similarities in the lipid A and in likely monosaccharide components of the core OS in all four C. concisus isolates, whereas there appeared to be greater variability in the outer OS regions.
Hexa-acylation has been found to be a requisite for optimal engagement with TLR4 and its downstream signaling cascade (22). Despite the presence of a hexa-acylated lipid A moiety in the LOS of both C. concisus and C. jejuni, we observed less induction of TNF-α by PBMCs in response to C. concisus strains compared with C. jejuni. This may be due to the recognized relationship between the 3D shape of lipid A and its bioactivity. Specifically, it has been found that lipid A molecules with a more conical wedge-shaped conformation and a hydrophilic backbone that is smaller compared with that of the hydrophobic portion of the molecule as found in C. jejuni induce more inflammatory signaling compared with molecules with a more cylindrical shape, as would be expected for the C. concisus lipid A due to the lack of secondary C16 acyl groups (67). Moreover, although C. jejuni strains can have up to two GlcN3N moieties in the lipid A, our data showed that the lipid A of the four C. concisus isolates contained no GlcN3N moieties, which is thought to affect the recognition of lipid A by the TLR4-MD2 complex due to differences in the flexibility of amide- versus ester-linked acyl chains (22, 30).
Apart from the nature of the acylation of the lipid A, other LOS/LPS modifications have been found to influence engagement with TLR4 and inflammatory signaling (20). Based on an analysis of a panel of livestock and nonlivestock-associated C. jejuni isolates, we previously reported that increased numbers of phosphoryl substituents on the lipid A and the presence of sialic acid in OS contributed to TLR4-mediated inflammatory signaling by THP-1 cells and primary human monocytes, as reflected by TNF-α secretion (22).
C. jejuni produces LOS containing a core OS but lacking the repeating O-antigens, although C. jejuni LPS has been described (68, 69). Indeed, as reported by Karlyshev et al. (70), all C. jejuni strains express LOS, whereas the LPS structure observed in some strains appears to be genetically and structurally unrelated and bears closer resemblance to capsular polysaccharides. LOS of C. jejuni is heavily phosphorylated, containing one or two phosphates and up to three PEA groups on the lipid A and in many strains a PEA on the OS moiety (22). The presence of PEA on the cell surface of C. jejuni is associated with bacterial virulence by various mechanisms. Two recent studies identified the eptC gene encoding a novel transferase conferring PEA expression on flagellar rod proteins, N-linked glycans, and lipid A (71, 72). The addition of PEA to lipid A and the Hep on the core of the OS results in enhanced recognition of TLR4 and resistance to antimicrobial peptides and facilitates the ability of C. jejuni to colonize and survive in avian and murine hosts (29). Variations in PEA and the pyrophosphorylation of lipid A also have been shown to play an important role in TLR4 signaling by the pathogenic species Neisseria meningitidis and N. gonorrheae, whereas PEA and pyrophosphate are absent in the lipid A moiety of most commensal Neisseria strains that rarely cause disease and exhibit lower inflammatory potential (18, 73).
In our analyses of the C. concisus LOS, we found no evidence for PEA in the OS moiety but did detect an 80 Da difference corresponding to phosphate as part of the core of the OS moiety. Overall, most prominent lipid A peaks were consistent with non-PEA containing lipid A, although spectra of the intact LOS from two C. concisus isolates, NCTC 12408 and B38, contained a major peak for a lipid A moiety containing PEA. Interestingly, eptC genes were encoded in all three C. concisus strains subjected to whole genome sequencing, and this was further confirmed by PCR analysis. Despite this, two of the isolates did not express a major peak for PEA containing lipid A, suggesting that PEA expression may undergo genetic regulation. Altogether, no more than three phosphoryl substituents (including both phosphate and PEA) were detected on any of the C. concisus lipid A moieties. In contrast, our previous MS analyses of the C. jejuni LOS revealed that the lipid A contained a total of two to five phosphoryl-containing groups (22). Thus, the extent of phosphorylation and phosphoethanolaminylation of the C. concisus lipid A was less than that of the C. jejuni lipid A, raising the hypothesis that the reduced cytokine and G. mellonella response may at least be partly due to the observed differences in lipid A phosphorylation across this genus.
LOS sialylation is another important virulence factor in C. jejuni that is associated with enhanced inflammation, increased colonization, and increased risk of developing Guillain-Barré syndrome, the last of which arises due to molecular mimicry between the C. jejuni LOS and gangliosides found in peripheral nerves (22, 25, 33, 74). In a previous study we reported the presence of up to two sialic acid residues on the OS of C. jejuni isolates and, importantly, increased sialylation was correlated with enhanced TLR4-mediated TNF-α secretion compared with strains without sialic acid (22). Our MS/MS data indicate the absence of sialic acid in the LOS from the four C. concisus strains analyzed while confirming NeuAc as part of the reference C. jejuni LOS. Furthermore, genomic analyses revealed the lack of sialic acid synthesis capability in all 89 published C. concisus genomes. We previously reported nonsialylated LOS produced by some environmental C. jejuni isolates (nonlivestock-associated), whereas all human isolates tested were sialic acid-positive (22). Given the significance of sialic acid in the virulence of C. jejuni, the absence of sialylation of C. concisus LOS is intriguing because it indicates the lack of an important virulence factor that also contributes to inflammatory signaling. An in-depth analysis of the existence of specific LOS biosynthesis gene loci in C. concisus as well as the presence of inflammation-associated genes involved in sialic acid and PEA biosynthesis and transfer is warranted to further investigate these important structural differences within the Campylobacter genus.
Taken together, our findings show that C. concisus isolates exhibit lipid A moieties and heterogeneous OS with overlapping as well as unique features compared with the well-studied C. jejuni LOS. Despite many similarities in the LOS structures of these two organisms, differences in the lipid A amidation, fatty acid substituents, and phosphorylation as well as the lack of sialic acid substituents in the C. concisus LOS are of significance. The potential association of LOS variations with clinical outcome awaits examination of a larger sample of strains from diverse backgrounds and may give further insight into the remaining question of C. concisus’ pathogenic potential. Furthermore, it is interesting to speculate that these Campylobacter organisms occupy the same ecological niche in the human host as C. jejuni, which may result in interspecies cross-talk and a potential indirect effect on Campylobacter-related pathogenicity. Our findings on LOS composition, proinflammatory potential, and bacterial virulence provide structural and functional insights into the C. concisus LOS that increase our understanding of this underexplored Campylobacter species and its role in human disease.
Supplementary Material
Footnotes
Abbreviations:
- GI
- gastrointestinal
- GlcN
- glucosamine
- GlcN3N
- 2,3-diamino-2,3-dideoxy-d-glucose
- HexN
- hexosamine
- HF
- hydrogen fluoride
- IMS
- ion-mobility spectrometry
- Kdo
- 2-keto-3-deoxyoctulosonic acid
- LOS
- lipooligosaccharide
- LPS
- lipopolysaccharide
- NeuAc
- N-acetylneuraminic acid
- OS
- oligosaccharide
- PBMC
- peripheral blood mononuclear cell
- PEA
- phosphoethanolamine
- TLR4
- toll-like receptor 4
This work was supported in part by Department of Veterans Affairs Merit Review award BX000727 (to G.A.J.). The authors also acknowledge National Institutes of Health National Center for Research Resources Shared Instrumentation Grant S10 RR029446 (to H. E. Witkowska) for acquisition of the Synapt G2 high-definition mass spectrometer, which is located at the University of California, San Francisco Sandler-Moore Mass Spectrometry Core Facility and supported by the Sandler Family Foundation, the Gordon and Betty Moore Foundation, National Institutes of Health/National Cancer Institute Cancer Center Support Grant P30 CA082103, and the Canary Foundation. G.A.J. is a recipient of the Senior Research Career Scientist award from the Department of Veterans Affairs. R.H. is funded by a Career Researcher Fellowship from NHS Research Scotland. The BISCUIT study was funded by a Clinical Academic Training Fellowship from the Chief Scientist Office (CAF/08/01). This is paper number 116 from the Center for Immunochemistry. The contents of this article do not represent the views of the Department of Veterans Affairs or the United States Government. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. K.B. acknowledges funding from the Child Health Research Charitable Incorporated Organisation and the Bogue Fellowship for travel. The authors declare that they have no conflicts of interest with the contents of this article.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Tanner A. C. R., Dzink J. L., Ebersole J. L., and Socransky S. S.. 1987. Wolinella recta, Campylobacter concisus, Bacteroides gracilis, and Eikeneila corrodens from periodontal lesions. J. Periodontal Res. 22: 327–330. [DOI] [PubMed] [Google Scholar]
- 2.Deshpande N. P., Kaakoush N. O., Wilkins M. R., and Mitchell H. M.. 2013. Comparative genomics of Campylobacter concisus isolates reveals genetic diversity and provides insights into disease association. BMC Genomics. 14: 585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaakoush N. O., and Mitchell H. M.. 2012. Campylobacter concisus—a new player in intestinal disease. Front. Cell. Infect. Microbiol. 2: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nielsen H. L., Ejlertsen T., Engberg J., and Nielsen H.. 2013. High incidence of Campylobacter concisus in gastroenteritis in North Jutland, Denmark: a population-based study. Clin. Microbiol. Infect. 19: 445–450. [DOI] [PubMed] [Google Scholar]
- 5.Mukhopadhya I., Thomson J. M., Hansen R., Berry S. H., El-Omar E. M., and Hold G. L.. 2011. Detection of Campylobacter concisus and other Campylobacter species in colonic biopsies from adults with ulcerative colitis. PLoS One. 6: e21490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Etterijck R., Breynaert J., Revets H., Devreker T., Vandenplas Y., Vandamme P., and Lauwers S.. 1996. Isolation of Campylobacter concisus from feces of children with and without diarrhea. J. Clin. Microbiol. 34: 2304–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaakoush N. O., Castaño-Rodríguez N., Mitchell H. M., and Man S. M.. 2015. Global epidemiology of Campylobacter infection. Clin. Microbiol. Rev. 28: 687–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L., Si M. M., Day A. S., Leach S. T., Lemberg D. A., Dutt S., Stormon M., Otley A., O’Loughlin E. V., Magoffin A., et al. . 2009. Detection and isolation of Campylobacter species other than C. jejuni from children with Crohn’s disease. J. Clin. Microbiol. 47: 453–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Man S. M., Kaakoush N. O., Leach S. T., Nahidi L., Lu H. K., Norman J., Day A. S., Zhang L., and Mitchell H. M.. 2010. Host attachment, invasion, and stimulation of proinflammatory cytokines by Campylobacter concisus and other non-Campylobacter jejuni Campylobacter species. J. Infect. Dis. 202: 1855–1865. [DOI] [PubMed] [Google Scholar]
- 10.Nielsen H. L., Engberg J., Ejlertsen T., Bücker R., and Nielsen H.. 2012. Short-term and medium-term clinical outcomes of Campylobacter concisus infection. Clin. Microbiol. Infect. 18: E459–E465. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen H. L., Engberg J., Ejlertsen T., and Nielsen H.. 2013. Clinical manifestations of Campylobacter concisus infection in children. Pediatr. Infect. Dis. J. 32: 1194–1198. [DOI] [PubMed] [Google Scholar]
- 12.Akutko K., and Matusiewicz K.. 2017. Campylobacter concisus as the etiologic agent of gastrointestinal diseases. Adv. Clin. Exp. Med. 26: 149–154. [DOI] [PubMed] [Google Scholar]
- 13.Serichantalergs O., Ruekit S., Pandey P., Anuras S., Mason C., Bodhidatta L., and Swierczewski B.. 2017. Incidence of Campylobacter concisus and C. ureolyticus in traveler’s diarrhea cases and asymptomatic controls in Nepal and Thailand. Gut Pathog. 9: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.François R., Yori P. P., Rouhani S., Siguas Salas M., Paredes Olortegui M., Rengifo Trigoso D., Pisanic N., Burga R., Meza R., Meza Sanchez G., et al. . 2018. The other Campylobacters: not innocent bystanders in endemic diarrhea and dysentery in children in low-income settings. PLoS Negl. Trop. Dis. 12: e0006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castaño-Rodríguez N., Kaakoush N. O., Lee W. S., and Mitchell H. M.. 2017. Dual role of Helicobacter and Campylobacter species in IBD: a systematic review and meta-analysis. Gut. 66: 235–249. [DOI] [PubMed] [Google Scholar]
- 16.Ismail Y., Lee H., Riordan S. M., Grimm M. C., and Zhang L.. 2013. The effects of oral and enteric Campylobacter concisus strains on expression of TLR4, MD-2, TLR2, TLR5 and COX-2 in HT-29 cells. PLoS One. 8: e56888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukhopadhya I., Hansen R., El-Omar E. M., and Hold G. L.. 2012. IBD—what role do Proteobacteria play? Nat. Rev. Gastroenterol. Hepatol. 9: 219–230. [DOI] [PubMed] [Google Scholar]
- 18.Liu M., John C. M., and Jarvis G. A.. 2010. Phosphoryl moieties of lipid A from Neisseria meningitidis and N. gonorrhoeae lipooligosaccharides play an important role in activation of both MyD88- and TRIF-dependent TLR4-MD-2 signaling pathways. J. Immunol. 185: 6974–6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park B. S., Song D. H., Kim H. M., Choi B-S., Lee H., and Lee J-O.. 2009. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 458: 1191–1195. [DOI] [PubMed] [Google Scholar]
- 20.Maeshima N., and Fernandez R. C.. 2013. Recognition of lipid A variants by the TLR4-MD-2 receptor complex. Front. Cell. Infect. Microbiol. 3: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rathinam V. A. K., Appledorn D. M., Hoag K. A., Amalfitano A., and Mansfield L. S.. 2009. Campylobacter jejuni-induced activation of dendritic cells involves cooperative signaling through toll-like receptor 4 (TLR4)-MyD88 and TLR4-TRIF axes. Infect. Immun. 77: 2499–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stephenson H. N., John C. M., Naz N., Gundogdu O., Dorrell N., Wren B. W., Jarvis G. A., and Bajaj-Elliott M.. 2013. Campylobacter jejuni lipooligosaccharide sialylation, phosphorylation, and amide/ester linkage modifications fine-tune human toll-like receptor 4 activation. J. Biol. Chem. 288: 19661–19672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Müller J., Meyer B., Hänel I., and Hotzel H.. 2007. Comparison of lipooligosaccharide biosynthesis genes of Campylobacter jejuni strains with varying abilities to colonize the chicken gut and to invade Caco-2 cells. J. Med. Microbiol. 56: 1589–1594. [DOI] [PubMed] [Google Scholar]
- 24.Naito M., Frirdich E., Fields J. A., Pryjma M., Li J., Cameron A., Gilbert M., Thompson S. A., and Gaynor E. C.. 2010. Effects of sequential Campylobacter jejuni 81–176 lipooligosaccharide core truncations on biofilm formation, stress survival, and pathogenesis. J. Bacteriol. 192: 2182–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Louwen R., Heikema A., Van Belkum A., Ott A., Gilbert M., Ang W., Endtz H. P., Bergman M. P., and Nieuwenhuis E. E.. 2008. The sialylated lipooligosaccharide outer core in Campylobacter jejuni is an important determinant for epithelial cell invasion. Infect. Immun. 76: 4431–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parker C. T., Horn S. T., Gilbert M., Miller W. G., Woodward D. L., and Mandrell R. E.. 2005. Comparison of Campylobacter jejuni lipooligosaccharide biosynthesis loci from a variety of sources. J. Clin. Microbiol. 43: 2771–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker C. T., Gilbert M., Yuki N., Endtz H. P., and Mandrell R. E.. 2008. Characterization of lipooligosaccharide-biosynthetic loci of Campylobacter jejuni reveals new lipooligosaccharide classes: Evidence of mosaic organizations. J. Bacteriol. 190: 5681–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moran A. P., Zahringer U., Seydel U., Scholz D., Stutz P., and Rietschel E. T.. 1991. Structural analysis of the lipid A component of Campylobacter jejuni CCUG 10936 (Serotype O-2) lipopolysaccharide. Description of a lipid A containing a hybrid backbone of 2-amino-2-deoxy-D-glucose and 2,3-diamino-2,3-dideoxy-D-glucose. Eur. J. Biochem. 198: 459–469. [DOI] [PubMed] [Google Scholar]
- 29.Cullen T. W., O’Brien J. P., Hendrixson D. R., Giles D. K., Hobb R. I., Thompson S. A., Brodbelt J. S., and Trent M. S.. 2013. EptC of Campylobacter jejuni mediates phenotypes involved in host interactions and virulence. Infect. Immun. 81: 430–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Mourik A., Steeghs L., Van Laar J., Meiring H. D., Hamstra H. J., Van Putten J. P. M., and Wösten M. M. S. M.. 2010. Altered linkage of hydroxyacyl chains in lipid A of Campylobacter jejuni reduces TLR4 activation and antimicrobial resistance. J. Biol. Chem. 285: 15828–15836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dorrell N., Mangan J. A., Laing K. G., Hinds J., Linton D., Al-Ghusein H., Barrell B. G., Parkhill J., Stoker N. G., Karlyshev A. V., et al. . 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11: 1706–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuijf M. L., Samsom J. N., van Rijs W., Bax M., Huizinga R., Heikema A. P., van Doorn P. A., van Belkum A., van Kooyk Y., Burgers P. C., et al. . 2010. TLR4-mediated sensing of Campylobacter jejuni by dendritic cells is determined by sialylation. J. Immunol. 185: 748–755. [DOI] [PubMed] [Google Scholar]
- 33.Heikema A. P., Koning R. I., Duarte dos Santos Rico S., Rempel H., Jacobs B. C., Endtz H. P., van Wamel W. J. B., and Samsom J. N.. 2013. Enhanced, sialoadhesin-dependent uptake of Guillain-Barré syndrome-associated Campylobacter jejuni strains by human macrophages. Infect. Immun. 81: 2095–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heikema A. P., Bergman M. P., Richards H., Crocker P. R., Gilbert M., Samsom J. N., Van Wamel W. J. B., Endtz H. P., and Van Belkum A.. 2010. Characterization of the specific interaction between sialoadhesin and sialylated Campylobacter jejuni lipooligosaccharides. Infect. Immun. 78: 3237–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen R., Berry S. H., Mukhopadhya I., Thomson J. M., Saunders K. A., Nicholl C. E., Bisset W. M., Loganathan S., Mahdi G., Kastner-Cole D., et al. . 2013. The microaerophilic microbiota of de-novo paediatric inflammatory bowel disease: the BISCUIT study. PLoS One. 8: e58825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karlyshev A. V., Linton D., Gregson N. A., and Wren B. W.. 2002. A novel paralogous gene family involved in phase-variable flagella-mediated motility in Campylobacter jejuni. Microbiology. 148: 473–480. [DOI] [PubMed] [Google Scholar]
- 37.Duffy G., Lynch O. A., and Cagney C.. 2008. Tracking emerging zoonotic pathogens from farm to fork. Meat Sci. 78: 34–42. [DOI] [PubMed] [Google Scholar]
- 38.Apicella M. A. 2008. Isolation and characterization of lipopolysaccharides. Methods Mol. Biol. 431: 3–13. [DOI] [PubMed] [Google Scholar]
- 39.Pringle S. D., Giles K., Wildgoose J. L., Williams J. P., Slade S. E., Thalassinos K., Bateman R. H., Bowers M. T., and Scrivens J. H.. 2007. An investigation of the mobility separation of some peptide and protein ions using a new hybrid quadrupole / travelling wave IMS / oa-ToF instrument. Int. J. Mass Spectrom. 261: 1–12. [Google Scholar]
- 40.Giles K., Pringle S. D., Worthington K. R., Little D., Wildgoose J. L., and Bateman R. H.. 2004. Applications of a travelling wave-based radio-frequency-only stacked ring ion guide. Rapid Commun. Mass Spectrom. 18: 2401–2414. [DOI] [PubMed] [Google Scholar]
- 41.Phillips N. J., John C. M., and Jarvis G. A.. 2016. Analysis of bacterial lipooligosaccharides by MALDI-TOF MS with traveling wave ion mobility. J. Am. Soc. Mass Spectrom. 27: 1263–1276. [DOI] [PubMed] [Google Scholar]
- 42.Gemmell M. R., Berry S., Mukhopadhya I., Hansen R., Nielsen H. L., Bajaj-Elliott M., Nielsen H., and Hold G. L.. 2018. Comparative genomics of Campylobacter concisus: analysis of clinical strains reveals genome diversity and pathogenic potential. Emerg. Microbes Infect. 7: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung H. K. L., Tay A., Octavia S., Chen J., Liu F., Ma R., Lan R., Riordan S. M., Grimm M. C., and Zhang L.. 2016. Genome analysis of Campylobacter concisus strains from patients with inflammatory bowel disease and gastroenteritis provides new insights into pathogenicity. Sci. Rep. 6: 38442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L., Budiman V., Day A. S., Mitchell H., Lemberg D. A., Riordan S. M., Grimm M., Leach S. T., Ismail Y.. 2010. Isolation and detection of Campylobacter concisus from saliva of healthy individuals and patients with inflammatory bowel disease. J. Clin. Microbiol. 48: 2965–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mahendran V., Riordan S. M., Grimm M. C., Tran T. A. T., Major J., Kaakoush N. O., Mitchell H., and Zhang L.. 2011. Prevalence of Campylobacter species in adult Crohn’s disease and the preferential colonization sites of Campylobacter species in the human intestine. PLoS One. 6: e25417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ismail Y., Mahendran V., Octavia S., Day A. S., Riordan S. M., Grimm M. C., Lan R., Lemberg D., Tran T. A. T., and Zhang L.. 2012. Investigation of the enteric pathogenic potential of oral Campylobacter concisus strains isolated from patients with inflammatory bowel disease. PLoS One. 7: e38217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahendran V., Tan Y. S., Riordan S. M., Grimm M. C., Day A. S., Lemberg D. A., Octavia S., Lan R., and Zhang L.. 2013. The prevalence and polymorphisms of zonula occluden toxin gene in multiple Campylobacter concisus strains isolated from saliva of patients with inflammatory bowel disease and controls. PLoS One. 8: 375525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 30: 2068–2069. [DOI] [PubMed] [Google Scholar]
- 49.Stajich J. E., Block D., Boulez K., Brenner S. E., Chervitz S. A., Dagdigian C., Fuellen G., Gilbert J. G. R., Korf I., Lapp H., et al. . 2002. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 12: 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tange O. 2011. GNU parallel: the command-line power tool. ;login. 36: 42–47. [Google Scholar]
- 51.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., and Madden T. L.. 2009. BLAST+: architecture and applications. BMC Bioinformatics. 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hyatt D., Chen G. L., LoCascio P. F., Land M. L., Larimer F. W., and Hauser L. J.. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 11: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laslett D., and Canback B.. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32: 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Finn R. D., Clements J., and Eddy S. R.. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39: W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kolbe D. L., and Eddy S. R.. 2011. Fast filtering for RNA homology search. Bioinformatics. 27: 3102–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lagesen K., Hallin P., Rødland E. A., Stærfeldt H. H., Rognes T., and Ussery D. W.. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35: 3100–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petersen T. N., Brunak S., Von Heijne G., and Nielsen H.. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 8: 785–786. [DOI] [PubMed] [Google Scholar]
- 58.Kanehisa M., Sato Y., and Morishima K.. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428: 726–731. [DOI] [PubMed] [Google Scholar]
- 59.John C. M., Liu M., and Jarvis G. A.. 2009. Natural phosphoryl and acyl variants of lipid A from Neisseria meningitidis strain 89I differentially induce tumor necrosis factor-α in human monocytes. J. Biol. Chem. 284: 21515–21525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harding C. R., Schroeder G. N., Collins J. W., and Frankel G.. 2013. Use of Galleria mellonella as a model organism to study Legionella pneumophila infection. J. Vis. Exp. e50964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Champion O. L., Karlyshev A. V., Senior N. J., Woodward M., La Ragione R., Howard S. L., Wren B. W., and Titball R. W.. 2010. Insect infection model for Campylobacter jejuni reveals that O-methyl phosphoramidate has insecticidal activity. J. Infect. Dis. 201: 776–782. [DOI] [PubMed] [Google Scholar]
- 62.Dzieciatkowska M., Brochu D., Van Belkum A., Heikema A. P., Yuki N., Houliston R. S., Richards J. C., Gilbert M., and Li J.. 2007. Mass spectrometric analysis of intact lipooligosaccharide: direct evidence for O-acetylated sialic acids and discovery of O-linked glycine expressed by Campylobacter jejuni. Biochemistry. 46: 14704–14714. [DOI] [PubMed] [Google Scholar]
- 63.Dzieciatkowska M., Liu X., Heikema A. P., Houliston R. S., Van Belkum A., Schweda E. K. H., Gilbert M., Richards J. C., and Li J.. 2008. Rapid method for sensitive screening of oligosaccharide epitopes in the lipooligosaccharide from Campylobacter jejuni strains isolated from Guillain-Barré syndrome and Miller Fisher syndrome patients. J. Clin. Microbiol. 46: 3429–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Szymanski C. M., St. Michael F., Jarrell H. C., Li J., Gilbert M., Larocque S., Vinogradov E., and Brisson J. R.. 2003. Detection of conserved N-linked glycans and phase-variable lipooligosaccharides and capsules from Campylobacter cells by mass spectrometry and high resolution magic angle spinning NMR spectroscopy. J. Biol. Chem. 278: 24509–24520. [DOI] [PubMed] [Google Scholar]
- 65.Caroff M., and Karibian D.. 2003. Structure of bacterial lipopolysaccharides. Carbohydr. Res. 338: 2431–2447. [DOI] [PubMed] [Google Scholar]
- 66.Richards V. P., Lefébure T., Pavinski Bitar P. D., and Stanhope M. J.. 2013. Comparative characterization of the virulence gene clusters (lipooligosaccharide [LOS] and capsular polysaccharide [CPS]) for Campylobacter coli, Campylobacter jejuni subsp. jejuni and related Campylobacter species. Infect. Genet. Evol. 14: 200–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brandenburg K., and Seydel U.. 2010. Conformation and supramolecular structure of lipid A. Adv. Exp. Med. Biol. 667: 25–38. [DOI] [PubMed] [Google Scholar]
- 68.Aspinall G. O., McDonald A. G., and Pang H.. 1992. Structures of the O chains from lipopolysaccharides of Campylobacter jejuni serotypes O:23 and O:36. Carbohydr. Res. 231: 13–30. [DOI] [PubMed] [Google Scholar]
- 69.Penner J. L., and Aspinall G. O.. 1997. Diversity of lipopolysaccharide structures in Campylobacter jejuni. J. Infect. Dis. 176(Suppl 2): S135–S138. [DOI] [PubMed] [Google Scholar]
- 70.Karlyshev A. V., Linton D., Gregson N. A., Lastovica A. J., and Wren B. W.. 2000. Genetic and biochemical evidence of a Campylobacter jejuni capsular polysaccharide that accounts for Penner serotype specificity. Mol. Microbiol. 35: 529–541. [DOI] [PubMed] [Google Scholar]
- 71.Scott N. E., Nothaft H., Edwards A. V. G., Labbate M., Djordjevic S. P., Larsen M. R., Szymanski C. M., and Cordwell S. J.. 2012. Modification of the Campylobacter jejuni N-linked glycan by EptC protein-mediated addition of phosphoethanolamine. J. Biol. Chem. 287: 29384–29396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cullen T. W., and Trent M. S.. 2010. A link between the assembly of flagella and lipooligosaccharide of the Gram-negative bacterium Campylobacter jejuni. Proc. Natl. Acad. Sci. USA. 107: 5160–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.John C. M., Liu M., Phillips N. J., Yang Z., Funk C. R., Zimmerman L. I., Griffiss M., Stein D. C., and Jarvis G. A.. 2012. Lack of lipid A pyrophosphorylation and functional lptA reduces inflammation by Neisseria commensals. Infect. Immun. 80: 4014–4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mortensen N. P., Kuijf M. L., Ang C. W., Schiellerup P., Krogfelt K. A., Jacobs B. C., van Belkum A., Endtz H. P., and Bergman M. P.. 2009. Sialylation of Campylobacter jejuni lipo-oligosaccharides is associated with severe gastro-enteritis and reactive arthritis. Microbes Infect. 11: 988–994. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.