Abstract
Glutamate is an important metabolite of glutaminolysis, a metabolic pathway employed by many aggressive cancers including triple-negative breast cancer. With the exception of the brain, in vivo detection of glutamate in tissues using 1H magnetic resonance spectroscopy (MRS) is challenging. Compared to MRS, glutamate-weighted chemical exchange saturation transfer MR imaging (GluCEST MRI) offers a more sensitive detection mechanism that is free of glutamine interference. Here we developed a robust, highly repeatable GluCEST MRI protocol in mice bearing human triple-negative breast cancer xenografts and treated with a potent glutaminase inhibitor, CB-839. In paired studies, treatment with CB-839 for 2 days reduced the GluCEST asymmetry value compared to baseline (P < 0.05, n=10). The absolute change of the GluCEST asymmetry value was −2.5 percent points after CB-839 treatment versus +0.3 after vehicle (P < 0.01). Correspondingly, treatment with CB-839 reduced tumor glutamate concentrations by 1.5 mM, consistent with prior calibration between changes of the GluCEST value versus tissue glutamate concentration; CB-839, however, did not change tumor intracellular pH. These results demonstrate in a mouse model of breast cancer the utility of GluCEST MRI to detect the early response to glutaminase inhibition.
Keywords: Chemical Exchange Saturation Transfer, Magnetic Resonance Imaging, Triple Negative Breast Cancer, Glutaminase, Glutaminolysis, Metabolism
Introduction
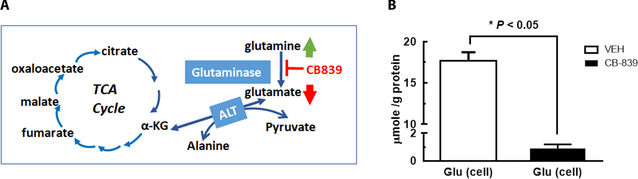
Glutamate plays an important role in cancer metabolism, especially in glutaminolysis pathway, where cellular glutamate level is associated with the catabolism of glutamine by mitochondrial glutaminase, GLS, activity (Figure 1A). Recent discoveries in cancer metabolism have revealed that glutaminolysis is activated in many aggressive forms of human cancers, including the triple negative breast cancers (TNBC). This provides strong rationale to target the cancer-specific metabolic signature. As the first and rate-limiting enzyme of glutaminolysis, inhibition of GLS activity can effectively block glutamine’s utilization for macromolecular synthesis and energy (1,2). Developing GLS-targeted strategy is especially important for TNBC, which has higher recurrence rate after surgical removal of the primary tumor and is not responsive to endocrine (e.g., Tamoxifen) or HER2-targeted therapies. Several small molecule inhibitors of GLS were developed and a highly potent and selective GLS inhibitor, CB-839, is undergoing clinical trials in patients with glutaminolytic cancers including TNBC (2,3). In TNBC tumors which exhibit high GLS and low glutamate synthase activity (3,4), previous studies revealed a reverse relationship between GLS activity and cellular glutamate concentration. Consequently, change in the tumor glutamate level could be a measurable indicator of GLS inhibition.
Figure 1. Metabolic pathway of glutaminolysis in cancer and cellular glutamate level in TNBC cells after vehicle (VEH) or GLS inhibitor (CB-839) treatment.
A: Mitochondrial glutaminase (GLS, kidney type) catalyzes the conversion of glutamine to glutamate, the first and rate-limiting step of glutaminolysis pathway. B: Cellular glutamate concentrations (μmol/g protein) after TNBC cells (HCC1806) were exposed to the vehicle (0.01% DMSO in culture media, n=2) or 1 μM CB-839 (in same amount of DMSO, n=2) for 24 h followed by extraction of aqueous metabolites (Supplemental Methods).
Non-invasive measurement of tissue glutamate level by in vivo 1H MR spectroscopy (MRS) is established in the brain, which has the highest glutamate concentration (6~12.5 mM) of all tissues (5). In non-neuronal tissues, however, in vivo 1H MRS is not feasible to assess glutamate level even with the sophisticated acquisition and post-processing methods (6) owing to 1) much lower glutamate concentration than the brain, 2) interfering signals from lipids, and 3) overlapping with glutamine resonance peaks. Therefore, a more sensitive glutamate detection method that is free of the above limitations is highly desirable.
Glutamate is among a few endogenous metabolites that exhibit the Chemical Exchange Saturation Transfer (CEST) effect (7), providing an amplification mechanism that allows indirect detection of low concentration solute pool (such as glutamate). Glutamate-weighted CEST (GluCEST) MRI demonstrates greater than 100-fold increase in sensitivity over 1H MRS detection of glutamate in the brain (8). GluCEST has shown great utility in several neurological/psychiatric applications (9–11). However, its ability to assess the pharmacodynamics of drugs targeting cancer glutaminolysis has not been examined.
Radio-labeled glutamate analogs detectable by PET, such as 18F-fluoroglutamate (12) or 18F-FSPG (13), are very sensitive but their signals represent cellular glutamate transporter activity rather than the endogenous glutamate level in the tissue. To our knowledge, GluCEST MRI is the only method that has the sensitivity potentially suitable for non-invasively assessing changes in tumor glutamate level. Based on the mechanistic link between cellular glutamate level and GLS activity in TNBC, we hypothesized that a reduction of overall GluCEST signal would be observed in TNBC tumors responding to GLS inhibitor. To test this hypothesis, we conducted the first feasibility study of GluCEST MRI as a therapeutic marker of GLS inhibitor.
Materials and Methods
Human TNBC cell line, HCC1806 (ATCC, Manassas, Virginia), exhibiting high GLS activity (3), was authenticated using the Short Tandem Repeat DNA profiling and was tested to be free of mycroplasma by agar culture and Hoechst DNA staining. Cells within 50 passages were used. GLS inhibitor, CB-839 (Calithera Biosciences, San Francisco, CA) was obtained via a material transfer agreement.
Tumor inoculation, treatment & imaging schedule, quantification of aqueous metabolites concentration in tumor tissue and cell extracts, in vivo measurement of tumor intracellular pH by 31P MRS are described in the Supplemental Methods. Animal studies were approved by local IACUC.
In vivo GluCEST MR imaging was performed on a 9.4 Tesla horizontal bore MR spectrometer (Agilent, Palo Alto, CA). A custom-built slotted-tube cylindrical resonator (13 mm inner diameter x 16.5 mm height) tuned to 1H was installed underneath a flat plastic platform on which the mouse was lying on its side with the flank tumor fit inside the resonator cylinder. The mouse was anesthetized with 1–2% isoflurane (mixed in air at 1 L/min) while the respiration and temperature were monitored, and the rectal temperature was maintained at 37 ± 0.2 °C by warm air (SA Inc., Stony Brook, NY).
Homogeneity of the static magnetic field (B0) was improved by shimming of the entire tumor using the point-resolved spectroscopy (PRESS) technique. For imaging, a single, 10-mm thick slice was planned to include the entire tumor (<10 mm in any dimension). To minimize the effect of respiration motion, the slice direction was parallel to the diaphragm motion. This slice was used for acquiring the B0 and radiofrequency field (B1) map (14), GluCEST and T2-weighted (T2W) gradient echo images with a matrix size of 128 ×128 over a field of view (FOV) of 25×25 mm2. B0 map was acquired using the Water Saturation Shift Referencing (WASSR) technique (15): a saturation pulse of 200 msec duration was applied with B1 of 0.3 μT and Δω in the range of −1 to +1 ppm with 0.1 ppm increment.
CEST images were acquired using a saturation pulse consisting of 4 square pulses of 250 msec each with a 4 μsec inter-pulse-delay and a 64-segment GRE readout (segment TR/TE = 6.7/3.4 msec). The B1 was optimized to 5.9 μT (250 Hz) to adequately saturate the amine protons on glutamate (9) and was applied with offset frequencies of ±2.5, ±2.75, ±3, ±3.25, ±3.5 ppm in reference to the bulk water. For each offset frequency, two acquisitions were executed with an 8-sec interval to allow T1 recovery. Within-subject variability was assessed by test-retest study, where two GluCEST imaging sessions were performed on the same mice in a single same day and the mice were recovered from the anesthesia and returned to the cage during the interval.
CEST images were first corrected for B0 and B1 inhomogeneity (8). Based on B0 and B1 map, the tumor region in which the B0 and relative B1 were within [−0.4, 0.4] ppm and [0.8, 1.2], respectively was processed to generate pixel-wise GluCEST asymmetry map using equation [1] (7), where the MTRasymm is normalized to the signal losses by direct water saturation and magnetization transfer contrast at -Δω:
| [1] |
The Msat (± Δω) are images obtained with saturation at a ‘+’ or ‘−’ offset frequency (Δω) to the water resonance. The GluCEST asymmetry value is presented in percent point. In addition to the GluCEST scans, complete MTRasymm analysis was applied on one animal before and after CB-839 treatment. For this, all the acquisition parameters were the same as GluCEST except the Δω was ranged [−5, +5]ppm with the step size of 0.2 ppm. The pre- and post-treatment data was processed using a MATLAB script to generate the z-spectrum and MTRasymm from the tumor region.
Statistics
Data are presented as mean +/− standard deviation (SD) unless specified otherwise. Repeatability was evaluated by Coefficient of variation (CV= mean/SD). Statistical significance was evaluated by two-sided student t-tests with α= 0.05 using GraphPad 6.
Results
To assess the extent of glutamate reduction induced by GLS inhibition (Fig 1A), we first measured the absolute glutamate concentration in TNBC cells using high-resolution 1H MRS of aqueous extracts. We found that CB-839 induced a profound reduction of cellular glutamate concentration: glutamate level in VEH-treated cells was ~19-fold of CB-839 treated cells (Figure 1B), confirming that cellular glutamate level is sensitive to inhibition of GLS activity.
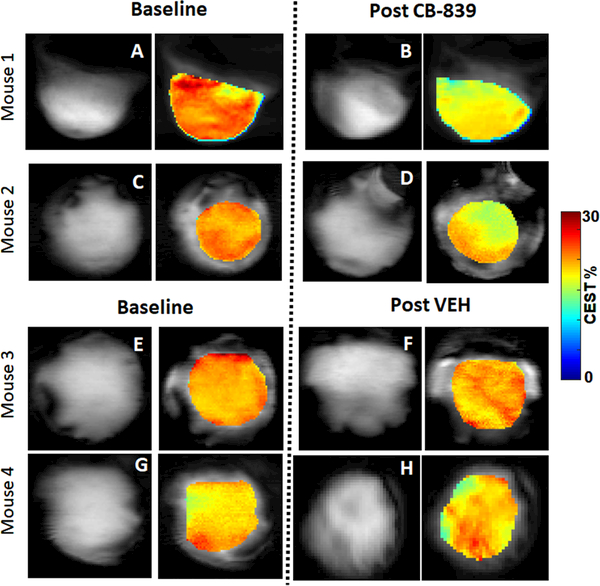
The T2W image and GluCEST map (overlaid on T2W image) from two CB-839 treated mice are shown in Fig 2A-B and C-D, revealing a clear reduction in GluCEST value in response to the GLS inhibition. In contrast, there was no change of average GluCEST value after VEH treatment (Fig 2E-F and G-H). The B0 and B1 maps and their mean values indicate good B0 and B1 homogeneity obtained at baseline and post-treatment scans (supplemental Fig 1A-B). The tumor region that meets both B0 and relative B1 homogeneity cut-off accounted for averagely 85% of the entire tumor volume. Pixel-wise GluCEST was measured from this region to derive GluCEST map. This protocol led to robust test-retest repeatability: the standard deviation (SD) of tumor GluCEST value is 0.3 percent point and CV (SD/mean) is 0.016. Therefore, a measured change of GluCEST value ≥ 1.5 percent points (i.e., 5 × SD) would be highly reliable (not by chance).
Figure 2. In vivo GluCEST MRI of the Tumor.
Representative T2W anatomic image and GluCEST map (overlaid on T2W image) at baseline and post-treatment from two CB-839 treated mice (A-B and C-D) and two VEH treated mice (E-F and G-H). The GluCEST map (in color scale) includes the region that meets both B0 and B1 homogeneity criteria (see text), thus it is slightly smaller than the tumor delineated on the T2W image (in grayscale). Note that the single slice includes the entire tumor, although the tumor appears slightly different in the baseline and post-treatment image due to the positioning of the mice.
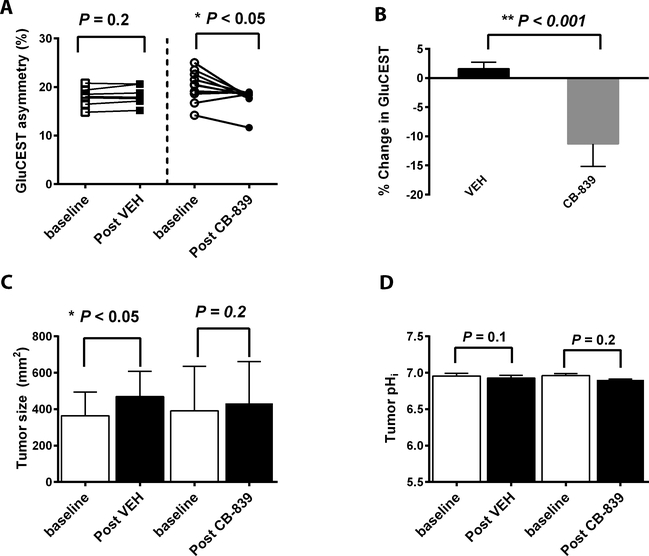
Paired GluCEST values were obtained at baseline and post-treatment (Fig 3A): CB-839 induced a reduction of GluCEST in 9 out 10 mice (P <0.05 comparing baseline versus post), whereas little change detected in VEH-group (n=7). On average, CB-839 induced an absolute 2.5 percent points decrease of GluCEST value while VEH led to 0.28 percent point increase. Compared to the baseline, GluCEST was decreased 11.3% after CB-839 but increased 1.6% after VEH treatment (P < 0.001, Fig 3B). Analyses of z-spectra obtained before and after CB-839 treatment were consistent with the extent of MTRasymm reduction at ~ 3ppm (supplemental Fig 2). Tumor size did not change significantly after the short course of CB-839 treatment whereas it was increased significantly in the VEH group (Fig 3C). Importantly, the two-day exposure to the GLS inhibitor had little effect on tumor pHi estimated by in vivo 31P MR spectroscopy: pHi was 6.96 at baseline, 6.90 after CB-839 and 6.93 after VEH treatment (Fig 3D).
Figure 3. The GluCEST MRI value, tumors size and intracellular pH in response to glutaminase inhibitor treatment.
A: Paired GluCEST MRI values pre and after VEH (n = 7) or CB-839 (n = 10) treatment. B: Percent change of the GluCEST value (= post/pre-1). Sample size is the same as in A. C: Tumor size pre and after treatment (the same cohorts of mice as in A). D: In a separate cohort, intracellular pH of tumor pre and post treatment (n=3 for each treatment arm).
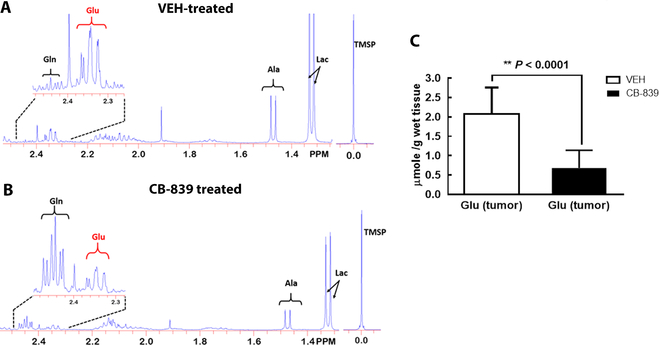
To assess the potential contribution of tumor glutamate and other metabolites to the GluCEST MRI signal, tumor tissues from mice treated by CB-839 or VEH (the same dose/duration as the imaging study) were harvested by freeze-clamping and extracted for 1H MRS (16) (detailed in SI). Representative 1H MR spectra of the tumor extract are shown for VEH (Fig 4A) and CB-839 treated mouse (Fig 4B). Compared to the VEH controls, CB-839 treated group had a 3-fold lower glutamate concentration (P= 5.4 × 10−5, Fig 4C). Besides Glu, other 1H MRS quantifiable major metabolites were listed in Supplemental Table-1, which revealed a moderate decrease of tumor alanine (9.7%) in response to CB-839 treatment but no significant change in choline or lactate level. This suggests that commonly used breast cancer markers (choline and lactate) detectable by in vivo 1H MRS are not sensitive for early detection of GLS inhibition.
Figure 4. Metabolites in perchloric acid (PCA) extracted tumor samples.
Representative 1H MR spectrum of the PCA extract of the tumor from VEH (A) and CB-839 (B) treated mouse. Both spectra were scaled to the reference trimethylsilyl propanoic acid (TMSP) peak at 0 ppm. Insets: the H4 resonances of glutamate and glutamine. C: The tumor glutamate concentration (μmol/g of wet tissue) after 4 doses of VEH or CB-839 treatment. Glu = glutamate, Gln = glutamine, Ala = alanine, Lac = lactate.
Discussion
GLS inhibitors represent a new category of drugs, which are directed at specific metabolic signatures of the tumor, and unlike chemotherapy, they do not induce rapid cell death or tumor shrinkage (3). Hence their therapeutic effect, especially early responses of the tumor cannot be sufficiently evaluated by the tumor size. Meanwhile, other established markers for breast cancer therapy such as FDG-PET, lactate and/or choline detected by in vivo MRS are not sensitive to GLS inhibition (Supplemental Table-1). In this study, we demonstrated the feasibility of GluCEST MRI to detect early therapeutic responses of tumor to GLS inhibition. CEST MRI of endogenous metabolites including APT (17) and GluCEST has a great potential to reach the clinic because it does not need any exogenous contrast media and can be performed on the existing 3T (for APT) and high field 7T (for GluCEST) MRI scanners (18).
Glutamate-weighted nature of the GluCEST value is supported by our experimental condition and results. First, the amide proton transfer (APT) and glutamine have negligible contribution because the B1 saturation required for GluCEST detection is not suitable for observing APT effect (19). Minimal contribution of glutamine to the GluCEST signal was demonstrated at 7T MRI scanner using phantoms containing 2 mM (8) and 10 mM glutamine (both phantoms pH ~7) (Supplemental Fig 3), respectively. Under our study conditions with tumor glutamine at 0.5–1.5 mM range and pH ~7, glutamine contribution to GluCEST signal would be negligible. Second, owing to short treatment regimen (2-day) and specificity of CB-839 (3), which blocks conversion of glutamine to glutamate, the mobile protein concentration may remain unchanged while the change of glutamate concentration would have major contribution to the change of GluCEST asymmetry with modest contribution from Ala (Supplemental Table 1). Third, CB-839 lowered the tumor glutamate concentration by ~1.4 mM corresponding to 2.5 percent points decrease in tumor GluCEST value, suggesting that each mM change in glutamate concentration corresponds to 1.7 percent points change of the GluCEST value, consistent with the GluCEST sensitivity estimated in our previous study (20) plus modest contribution of Ala.
We have established a protocol that was insensitive to respiration motion and has robust test-retest repeatability (CV= 0.016), and as such, the 2.3 percent points reduction in GluCEST value after CB-839 treatment is reliable and significant (Fig 3A). This absolute change represents 11.3% reduction from the baseline value (P < 0.001 compared to VEH controls). The minimal decrease of tumor pHi (<1%, Fig 3D) induced by CB-839 was not likely to impact the GluCEST value, but even it had, it would cause an increase of post-treatment GluCEST value, leading to an underestimation of the treatment effect. Meanwhile, a modest decrease (9.7%) in tumor Ala level (Supplemental Table-1) could have contributed to the overall reduction of GluCEST value mediated by GLS inhibition. Since Ala also exhibits GluCEST effect and the cellular glutamate and Ala pool are coupled via the alanine transferase (ALT) reaction (Fig 1A), a decrease of Ala concentration accompanying the decrease of glutamate concentration would serendipitously increase the GluCEST sensitivity to GLS inhibition.
The change in the glutamate level in response to GLS inhibitor is much larger in cells (Fig 1B) than in tumors (Fig 4C), likely due to high drug concentration maintained in the culture media compared to mice in which rapid clearance and poor perfusion would certainly reduce tumor exposure to the drug. Our data revealed a relatively high baseline tumor GluCEST value of 18 ± 2 % corresponding to a relatively low glutamate concentration in the tumor (2.1 ± 0.7 mM), suggesting contributions of other amine protons. Compared to normal tissues such as the brain, proliferating tumors have a higher level of mobile proteins hence the associated amine protons could have contributed to the absolute GluCEST value. Therefore, it is not the absolute GluCEST value but its decrease that serves as a marker of GLS inhibition.
GLS inhibition results in a decrease of steady-state cellular glutamate concentration and an increase of glutamine concentration. Our earlier work has shown that PET imaging of [18F](2S,4R)4-Fluoroglutamine, a glutamine analog, can detect changes of the tumor glutamine level induced by CB-839 (16). Since the glutamate-to-glutamine-ratio would better represent the degree of GLS inhibition and more sensitive than glutamate or glutamine alone, GluCEST MRI and Fluoroglutamine PET could be combined to optimally evaluate GLS inhibiting drugs in future studies.
Our study has limitations. We examined a TNBC tumor model with relatively high GLS activity (3). Future studies should include a panel of TNBC lines with a range of GLS activities to further test the GluCEST utility. An intrinsic limitation of the GluCEST MRI is the requirement of high magnetic field (≥ 7 T) due to the intermediate exchange rate of the glutamate amine proton (9). Recent FDA approval of 7T MRI scanner will boost the applications of GluCEST, which has been successfully implemented to investigate human neurological/psychological diseases (11). While challenges such as the presence of tumor microcalcification are expected, the clinical application of GluCEST may be motivated by its unique ability to assess cancer glutaminolysis and therapies targeting this metabolic signature.
Supplementary Material
Acknowledgment
We thank Calithera for providing CB-839. We are grateful to Drs. Suzanne Wehrli, Susan Demo and Hoon Choi and Jianbo Cao for technical assistance and helpful discussions. Technical support from the SAIF (Small Animal Imaging Facility) of Radiology Department is acknowledged. This study is supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health Grant P41-EB015893 (Reddy), R01NS087516 (Reddy), Abramson Cancer Center Breast Cancer Pilot Grant, BCPG2016 (Zhou), R21CA198563 (Zhou), R01CA211337 (Mankoff and Zhou) and Komen SAC130060 (Mankoff).
Footnotes
Conflict of interest statement
The authors disclose no potential conflicts of interest.
References
- 1.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 2008;7:11–20 [DOI] [PubMed] [Google Scholar]
- 2.Wang JB, Erickson JW, Fuji R, Ramachandran S, Gao P, Dinavahi R, et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell 2010;18:207–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gross MI, Demo SD, Dennison JB, Chen L, Chernov-Rogan T, Goyal B, et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol Cancer Ther 2014;13:890–901 [DOI] [PubMed] [Google Scholar]
- 4.Kung HN, Marks JR, Chi JT. Glutamine synthetase is a genetic determinant of cell type-specific glutamine independence in breast epithelia. PLoS genetics 2011;7:e1002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Graaf RA. In Vivo NMR Spectroscopy – 2nd Edition: Principles and Techniques. John Wiley & Sons, Ltd; 2007. [Google Scholar]
- 6.Begley JK, Redpath TW, Bolan PJ, Gilbert FJ. In vivo proton magnetic resonance spectroscopy of breast cancer: a review of the literature. Breast Cancer Research 2012;14:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu G, Song X, Chan KW, McMahon MT. Nuts and bolts of chemical exchange saturation transfer MRI. NMR Biomed 2013;26:810–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai K, Haris M, Singh A, Kogan F, Greenberg JH, Hariharan H, et al. Magnetic resonance imaging of glutamate. Nat Med 2012;18:302–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crescenzi R, DeBrosse C, Nanga RP, Reddy S, Haris M, Hariharan H, et al. In vivo measurement of glutamate loss is associated with synapse loss in a mouse model of tauopathy. Neuroimage 2014;101:185–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pepin J, Francelle L, Carrillo-de Sauvage MA, de Longprez L, Gipchtein P, Cambon K, et al. In vivo imaging of brain glutamate defects in a knock-in mouse model of Huntington’s disease. Neuroimage 2016;139:53–64 [DOI] [PubMed] [Google Scholar]
- 11.Davis KA, Nanga RP, Das S, Chen SH, Hadar PN, Pollard JR, et al. Glutamate imaging (GluCEST) lateralizes epileptic foci in nonlesional temporal lobe epilepsy. Sci Transl Med 2015;7:309ra161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ploessl K, Wang L, Lieberman BP, Qu W, Kung HF. Comparative evaluation of 18F-Labeled glutamic acid and glutamine as tumor metabolic imaging agents. Journal of Nuclear Medicine 2012;53:1616–24 [DOI] [PubMed] [Google Scholar]
- 13.Baek S, Choi C-M, Ahn SH, Lee JW, Gong G, Ryu J-S, et al. Exploratory Clinical Trial of (4S)-4-(3-[18F]fluoropropyl)-l-glutamate for Imaging xC−Transporter Using Positron Emission Tomography in Patients with Non–Small Cell Lung or Breast Cancer. Clinical Cancer Research 2012;18:5427–37 [DOI] [PubMed] [Google Scholar]
- 14.Singh A, Cai K, Haris M, Hariharan H, Reddy R. On B1 inhomogeneity correction of in vivo human brain glutamate chemical exchange saturation transfer contrast at 7T. Magn Reson Med 2013;69:818–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim M, Gillen J, Landman BA, Zhou J, van Zijl PC. Water saturation shift referencing (WASSR) for chemical exchange saturation transfer (CEST) experiments. Magn Reson Med 2009;61:1441–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou R, Pantel AR, Li S, Lieberman BP, Ploessl K, Choi H, et al. [18F](2S,4R)4-Fluoroglutamine PET Detects Glutamine Pool Size Changes in Triple-Negative Breast Cancer in Response to Glutaminase Inhibition. Cancer Res 2017;77:1476–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones CK, Schlosser MJ, van Zijl PC, Pomper MG, Golay X, Zhou J. Amide proton transfer imaging of human brain tumors at 3T. Magn Reson Med 2006;56:585–92 [DOI] [PubMed] [Google Scholar]
- 18.Vinogradov E, Sherry AD, Lenkinski RE. CEST: from basic principles to applications, challenges and opportunities. J Magn Reson 2013;229:155–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin T, Wang P, Zong X, Kim SG. MR imaging of the amide-proton transfer effect and the pH-insensitive nuclear overhauser effect at 9.4 T. Magn Reson Med 2013;69:760–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bagga P, Pickup S, Crescenzi R, Martinez D, Borthakur A, D’Aquilla K, et al. In vivo GluCEST MRI: Reproducibility, background contribution and source of glutamate changes in the MPTP model of Parkinson’s disease. Scientific reports 2018;8:2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.