Abstract
The inflammatory microenvironment plays an important role in the pathogenesis and progression of tumors and may be associated with somatic genomic alterations. We examined the association of tumor infiltrating T-cell density with clinical-pathologic variables, tumor molecular subtype and oncologic outcomes in surgically-treated primary prostate cancer occurring in patients of European-American or African-American ancestry. We evaluated 312 primary prostate tumors, enriched for patients with African-American ancestry and high grade disease. Tissue microarrays were immunostained for CD3, CD8 and FOXP3 and were previously immunostained for ERG and PTEN using genetically validated protocols. Image analysis for quantification of T-cell density in tissue microarray tumor spots was performed. Automated quantification of T-cell densities in tumor-containing regions of tissue microarray spots and standard histologic sections were correlated (r=0.73, p<0.00001) and there was good agreement between visual and automated T-cell density counts on tissue microarray spots (r=0.93, p<0.00001). There was a significant correlation between CD3+, CD8+ and FOXP3+ T-cell densities (p<0.00001), but these were not associated with most clinical or pathologic variables. Increased T-cell density was significantly associated with ERG positivity (median 309 vs 188 CD3+ T-cells/mm2; p=0.0004) and also with PTEN loss (median 317 vs 192 CD3+ T-cells/mm2; p=0.001) in the combined cohort of matched European-American and African-American ancestry patients. The same association or a similar trend was present in patients of both ancestries when analyzed separately. When the African-American patients from the matched race set were combined with a separate high grade set of African-American cases, there was a weak association of increased FOXP3+ T-cell densities with increased risk of metastasis in multivariable analysis. Though high T-cell density is associated with specific molecular subclasses of prostate cancer, we did not find an association of T-cell density with racial ancestry.
Keywords: Prostatic adenocarcinoma, tumor infiltrating lymphocytes, T-cells, CD3, PTEN, ERG, African-American
Introduction
Elucidating how the immune microenvironment affects cancer development and progression has been one of the most challenging questions in immuno-oncology. We now understand that the immune system likely has dual host-protective and tumor-promoting roles: it can suppress tumor growth by destroying cancer cells, but also promote tumor progression either by selecting for tumor cells that are more fit to survive in an immunocompetent host or by establishing conditions within the tumor microenvironment that facilitate tumor outgrowth (1). Given this complexity, it is not surprising that the association of tumor-infiltrating lymphocytes with clinical outcome and clinical-pathologic parameters varies widely across various types of solid tumors (2–6). For example, in patients with renal cancer, the presence of a pronounced lymphocytic infiltration within the tumor is associated with decreased survival (6, 7). In contrast, lymphocytic infiltration has been associated with improved survival in patients with colorectal cancer (8, 9). In fact, immune scores (IS) appear to be a better prognostic indicator than the TNM system in colorectal cancers (2, 10). The interaction of the immune microenvironment with tumor progression not only depends on the histologic tumor type but is likely further complicated by the underlying molecular subtype of the tumor. In colon cancer, for example, genetic deficiencies in the DNA mismatch repair pathway are relatively common compared to other tumor types, leading to high mutation rates and generation of neo-epitopes that may facilitate immune recognition and destruction of tumor cells (11–13) and our group has demonstrated similar findings for prostate cancer (14).
Aside from these rare cases with mismatch DNA repair defects, prostate cancer is considered to have a relatively immunologically “cold” microenvironment compared to other solid tumor types (15, 16), however the relationship between lymphocytic infiltration and patient outcomes has varied widely in the literature (17–24). A few studies have suggested that low numbers of tumor-infiltrating lymphocytes are associated with a higher risk of disease progression (17, 21), while most other studies have shown the opposite (18–21, 23, 24). Underlying these apparent inconsistencies is the fact that many of these studies have used differing definitions of tumor-infiltrating lymphocytes, variable methodologies to detect tumor-infiltrating lymphocytes, and most have evaluated disparate outcome measures. In addition, few studies have integrated tumor molecular subtyping in the examination of tumor-infiltrating lymphocytes numbers, a variable that may also affect the immune microenvironment in prostate cancer. Finally, to our knowledge, essentially all prior histologic studies of tumor-infiltrating lymphocytes in prostate cancer have used predominantly white or Caucasian cohorts with European ancestry, with limited or no analysis of tumor-infiltrating lymphocytes with respect to oncologic outcomes or molecular parameters in more racially diverse populations. To address these knowledge gaps, we studied T-lymphocyte infiltration in a well-defined clinical cohort with known molecular subtyping information and diverse racial ancestry (25). After validating our method for digital quantification of T-cell density across tissue microarray cores and in relation to whole tissue sections, we assessed the association of the density of different subsets of T-cells (CD3+, CD8+ and FOXP3+) with clinical-pathological variables, PTEN/ERG status, and clinical outcome measures in surgically treated prostate cancer occurring in patients of European-American or African-American descent.
Methods
Patients and tissue samples
With Johns Hopkins institutional review board approval, three patient sets were included in this study. The first included a previously described group of 227 patients who underwent radical prostatectomy at Johns Hopkins, designed to evaluate the association of self-identified European-American or African-American racial ancestry with patient outcomes (25). Tissue microarrays were constructed from matched prostate tumors collected from 100 self-identified African-American and 127 self-identified European-American patients who underwent radical prostatectomy from 1995 through 2005 (hereafter referred to as matched race tissue microarray set). An additional set of 85 African-American patients with high grade (Gleason 4+3=7 and higher) disease who underwent radical prostatectomy from 2005–2010 was also studied to enrich for adverse oncologic outcomes (hereafter referred to as high grade African-American set). We had adequate power to examine associations with clinical outcomes only among the combined African-American tumor cohorts (matched and high grade combined together; hereafter referred to as combined African-American set). The clinical outcome measure for this combined cohort was biochemical recurrence or time to metastasis and patients had a median of 5 years of follow-up.
An additional cohort was utilized only for validation of digital quantification of CD3+ T-cell density as measured on tissue microarray versus standard histologic sections (whole slide sections). This group of tumors included a previously published cohort of radical prostatectomies from 2004–2014 with primary Gleason pattern 5 (n=29) (14). This cohort was selected for the validation due to the availability of standard histologic sections from the same paraffin blocks used to make the tissue microarray and the high rate of mismatch repair deficient tumors in this group. We have previously shown that prostate tumors with defects in mismatch repair are associated with a high tumor infiltrating lymphocyte density (14). Thus this cohort uniquely provides samples across a wide dynamic range of tumor-infiltrating lymphocyte densities to evaluate the correlation between CD3+ T-cell density measured on standard histologic tissue sections (whole slide sections) versus tissue microarray spots. All tissue microarrays described above included four individual 0.6 mm punches of the dominant tumor nodule from each case (slightly more than 1 mm2 of tissue for analysis).
Immunohistochemistry
Immunostaining for CD3 and CD8 were performed in a CLIA-accredited laboratory utilizing a polyclonal rabbit antibody for CD3 (A0452, Dako/Agilent, Santa Clara, CA) and a mouse monoclonal for CD8 (clone C8/C8144B, 760–4250; Cell Marque, Rocklin, CA) on the Ventana Benchmark immunostaining system (Ventana/Roche, Tucson, AZ). Immunostaining for FOXP3 utilized a rat monoclonal antibody (FJK-16s, Invitrogen/Thermo Fisher) on the Ventana Discovery Ultra (Ventana/Roche, Tucson, AZ). Immunostaining for ERG and PTEN status utilized previously reported and genetically validated rabbit monoclonal antibodies and staining protocols (EPR3864 from Ventana for ERG and D4.3 from Cell Signaling Technologies for PTEN) on the Ventana Benchmark or Ventana Discovery Ultra (Ventana/Roche) (25). Inter-observer reproducibility for PTEN and ERG scoring is high. For PTEN, we have previously published that among pathologists at the same institution (JHU), there was 96.4% agreement over 2783 cores (κ = 0.905; 95% CI=0.887–0.923) (26). In an additional study of pathologists at one institution (JHU) on an independent cohort, PTEN scores were concordant in 98.4% of cores evaluated by both pathologists (925/940; including a total of 240 with PTEN loss identified by both reviewers), corresponding to a κ=0.984 (27). For ERG, the inter-observer variability is also low, with agreement across 92% of 58 cases in a study of pathologists at one institution (JHU), corresponding to a 3-category κ=0.855 (95%CI=0.738–0.972) (TLL, unpublished data). For CD3, CD8, FOXP3 immunostaining, automated image analysis for number of cells per millimeter squared was performed as below with the Aperio Digital Pathology software (Leica, Wetzlar, Germany) as previously described (14). Visual scoring using a dichotomous and validated scoring system was performed previously for PTEN and ERG (25).
Image analyses
For image analysis of CD3, CD8 and FOXP3 immunostaining, stained standard histologic slides (whole slide sections) and tissue microarray slides were scanned at 20X magnification on Nano Zoomer Digital Pathology scanner (Hamamatsu). The positively stained cells per millimeter squared tissue were quantitatively scored with the Aperio Digital Pathology software (Leica) based on the number of cores evaluable and containing tumor tissue on the tissue microarray for each case. For each immunostained standard slide (whole slide section), all tumor tissue present from the dominant tumor nodule, excluding benign epithelium, areas of artifactual staining or tissue wrinkling and with minimal intervening stromal tissue, was circled using the pen tool in Aperio Image Analysis software and selected for analysis (Supplementary Figure S1). For the tissue microarray sections, all four cores that contained tumor from the dominant tumor nodule were circled and selected for analysis in their entirety; cores were excluded if they did not contain tumor glands (Supplementary Figure S1) and areas of artifactual staining were manually excluded as well (Supplementary Figure S2).
FOXP3+, CD8+ or CD3+ T-cells within the selected tumor area were identified by Aperio software as described previously (14). In brief, the number of positive cells was used by designing a detection mask for brown immunostaining using a modified version of the nuclear analysis parameters in Aperio Image Analysis software. Though CD3, CD8 and FOXP3 are membranous stains, the low cytoplastic volume of T-lymphocytes results in staining of most of the cell’s area, thus nuclear anlaysis works well to quantify the number of positively staining cells. Stain color and scoring theme parameters were set manually in Aperio Image Analysis software, using a pathologist’s visual estimate of which T-cells were positively stained across several representative images. Paramaters used included range of brown intensity, average cell size and cell shape. The software identified count was visually checked by pathologists in at least 50% of tissue microarray spots selected randomly for evaluation and across all standard sections (Supplementary Figure S3). Parameters were set separately for each stain on tissue microarray sections and standard histologic (whole slide) sections but once set, these parameters were uniformly applied across African-American and European-American samples. The cell count in each manually circled area was returned by the Aperio software. The total tumor area analyzed was calculated by Aperio Image Analysis software for standard histologic sections and manually for the tissue microarray spots, assuming that the diameter of each spot was 0.6 mm and multiplying by the total number of spots analyzed for each case. The ratio of positive cells to the total tumor area analyzed was calculated for each case.
Statistical Analysis
To evaluate the correlation between different methods of T-cell density assessments, we calculated Pearson correlations for (1) T-cell density assessed on histologic sections (whole slide sections) and assessed on tissue microarray spots, (2) T-cell density assessed visually and assessed with automated scoring, (3) CD3+ T-cell density assessed on two different tissue microarray sections, and (4) CD8+ T-cell density assessed on two different tissue microarray sections. To evaluate the correlation between lymphocytes of interest assessed for the matched tissue microarray set, we calculated Pearson correlations for (1) CD8+ and CD3+ T-cell densities, (2) FOXP3+ and CD3+ densities, and (3) FOXP3+ and CD8+ T-cell densities.
We calculated proportions (race, stage, grade) and median (age, prostate specific antigen levels) values for demographic and clinico-pathologic factors overall, and compared by race using the chi-square test for proportions and the Kruskal-Wallis test for continuous factors. We calculated median CD3+ T-cell density, CD8+ T-cell density, FOXP3 density and stratified by race. Within race strata, we also calculated median densities by stage, by grade, by ERG presence, and by PTEN loss, by joint categories of ERG presence and PTEN loss. Median densities were compared by race, and within race strata, by stage and by grade using the Kruskal-Wallis test. We calculated a Spearman correlation for each density measure (CD3, CD8, FOXP3) and age and prostate specific antigen levels, overall and within race. For all analyses of African American men, we evaluated measures by tissue microarray set (matched race and high grade African-American sets only) and combined across sets.
We had adequate numbers of adverse oncologic events to examine the association of tumor infiltrating T-cell density with prostate cancer outcomes only in the combined African-American cohort, since it was highly enriched for high Gleason score tumors by design (25). Men were followed for biochemical recurrence, metastasis and cancer specific survival from date of radical prostatectomy until 2017. Biochemical recurrence was defined as a postoperative serum prostate specific antigen level ≥ 0.2 ng/ml which was subsequently confirmed on a second test. Metastatic disease was defined as radiologic evidence of at least one metastatic lesion of non-regional lymph node, bone, or other site (≥M1a).
We categorized men by tertiles of CD3+, CD8+, FOXP3+, and CD8+/FOXP3+ T-cell densities for African-American men. We performed Kaplan-Meier analysis and used Cox proportional hazards regression to estimate the relative hazard (HR) and 95% confidence interval (CI) of biochemical recurrence and metastases for each cell density using men classified in the bottom two tertiles of density as the reference group. All models were adjusted for age, stage, grade, prostate specific antigen levels, and body mass index. All analyses were carried out using SAS 9.4 (Cary, NC). Statistical tests were 2-sided and p<0.05 was considered to be statistically significant.
Results
Validation of automated T-cell quantification on tissue microarrays
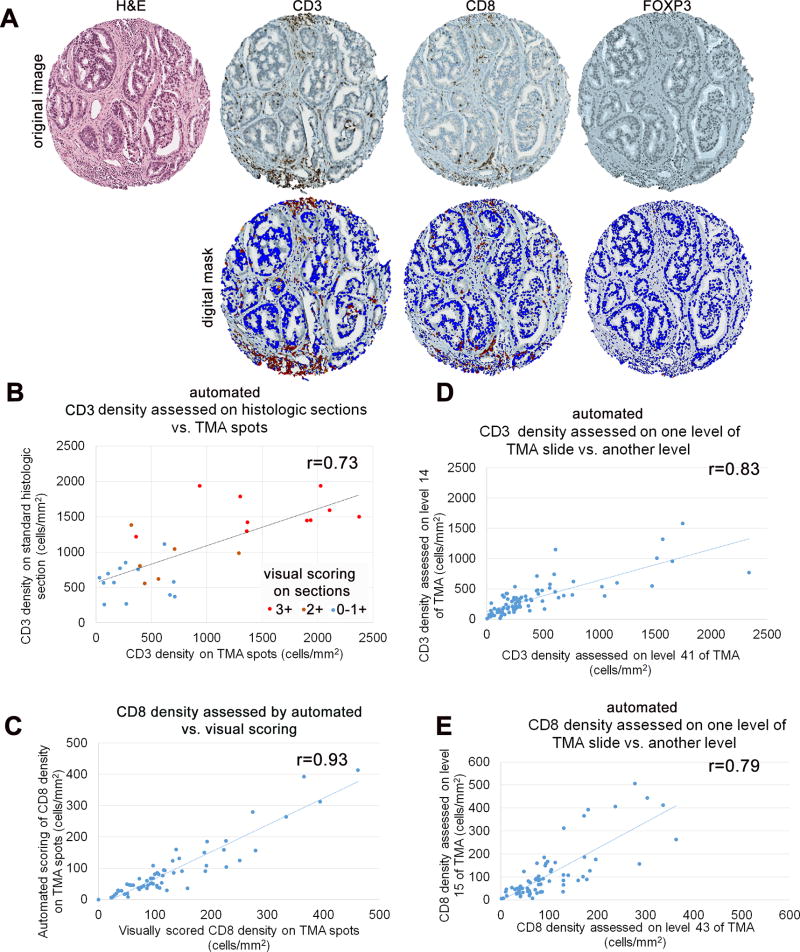
To initially validate the use of tissue microarray spots as a surrogate for quantification of T-cell density in standard histologic sections (whole slide sections) of the dominant tumor nodule, we used a cohort of 29 prostate tumors with primary Gleason pattern 5 (Supplementary Figure S1). We chose to use this tissue microarray set for this purpose because we have previously shown that this subset of prostate tumors demonstrates a wide dynamic range in tumor infiltrating T-cell densities, likely due to the high rate of mismatch repair genomic defects in these cases (14). We compared automated CD3+ T-cell density assessed in 4 tissue microarray spots from each dominant tumor nodule (Figure 1A, Supplementary Figure S2) with automated CD3+ T-cell density assessed in a single standard histologic section of the dominant tumor nodule from the same tissue block that was punched to make the tissue microarray. For the histologic sections, we also compared automated CD3+ T-cell density with a visual counting of CD3+ T-cell density. Overall, there was good correlation between automated CD3+ T-cell density assessed in tissue microarray spots and automated CD3+ T-cell density assessed on standard histologic sections of the dominant tumor nodule (r=0.73, p<0.00001) and qualitative correlation between the visual scoring system and the automated density quantification on the standard sections (Figure 1B).
Figure 1. Validation of automated T-cell density measurements on tissue microarray.
A) Representative H&E staining and lymphocyte immunostaining in prostate tumor on tissue microarray cores. Immunostaining for CD3, CD8 and FOXP3 identifies respective specific subsets of tumor infiltrating lymphocytes in prostate tumor (top). Aperio image software identifies CD3+ T-cells, CD8+ T-cells and FOXP3+ T-cells (red) in selected tumor regions and surrounding tumor and stromal nuclei (blue; bottom). B) CD3+ T-cell density (cells/mm2) on standard histologic sections correlates significantly with CD3+ T-cell density (cells/mm2) on tissue microarray (TMA) spots, when assessed by automated digital quantification. (Pearson’s correlation coefficient r= 0.73, p<0.00001). C) Automated digitally scored T-cell densities (cells/mm2) correlate significantly with manual visually scored T-cell densities (cells/mm2) on tissue microarray spots, thus validating the methodology used in this study (Pearson’s correlation coefficient r= 0.93, p<0.00001). D) CD3+ T-cell density (cells/mm2) on tissue microarray spot correlates significantly with CD3+ T-cell density (cells/mm2) on deeper level of the same tissue microarray spot, when assessed by automated digital quantification. (Pearson’s correlation coefficient r= 0.83, p<0.00001). E) CD8+ T-cell density (cells/mm2) on tissue microarray spot correlates significantly with CD8+ T-cell density (cells/mm2) on deeper level of the same tissue microarray spot, when assessed by automated digital quantification (Pearson’s correlation coefficient r= 0.79, p<0.00001)
Next, we assessed whether automated T-cell density measurements (Figure 1A) correlated with visual quantification of T-cell density in the same tissue microarray spots. Here, the correlation was very high across 59 cases from the high grade African-American tissue microarray set (r=0.93, p<0.00001), validating the automated quantification methodology (Figure 1C). Finally, we also evaluated whether CD3+ or CD8+ T-cell density assessed on one level from the tissue microarray block was correlated with the same marker assessed on a deeper level (at least 100 µm deeper) of the tissue microarray block. For CD3+ T-cell density assessed across 85 cases, there was strong correlation between the two tissue microarray levels (r=0.83, p<0.00001) with a similar correlation for CD8+ T-cell density assessed across 72 cases from the same cohort (r=0.79, p<0.00001) (Figure 1D and 1E).
Association of T-cell density with clinical-pathologic variables
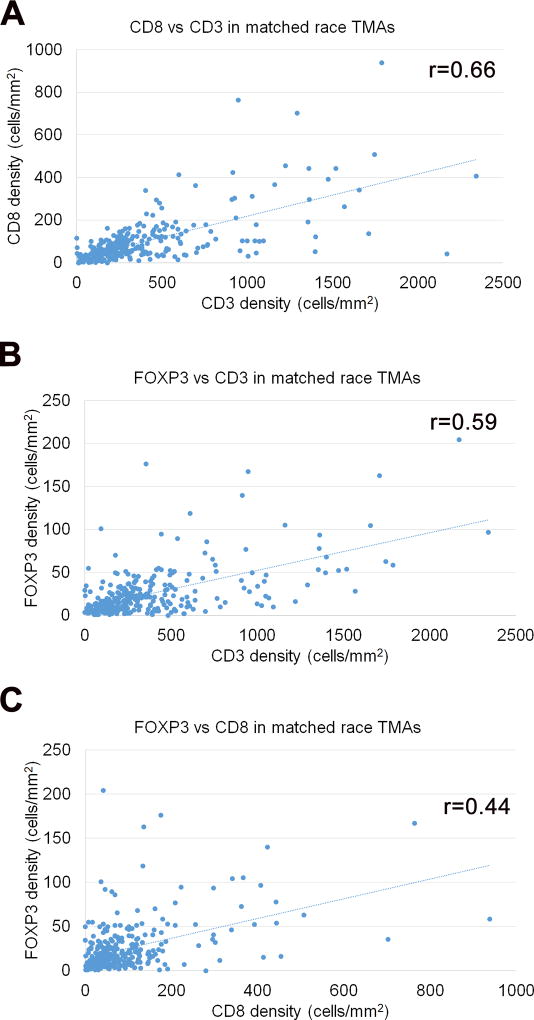
We first assessed the correlation of clinical-pathologic parameters with T-cell densities using the matched race set of tissue microarrays. These tissue microarrays contained patients of self-identified European-American or African-American ancestry matched by most clinical-pathologic parameters as reported previously (25), with the exception of prostate specific antigen levels (slightly higher in African-American patients) and age (slightly higher in European-American patients) (Supplementary Table S1). In addition, to enrich for adverse oncologic outcomes in subsequent analyses, we also examined a separate group of 85 high-grade African-American cases with Gleason score 4+3=7 or higher, considering these cases on their own or when combined with the African-American cases from the matched race tissue microarray sets (Supplementary Table S1). In the overall matched race tissue microarray set, quantified CD3+ and CD8+ T-cell densities were correlated (r=0.66, p<0.00001) as were CD3+ and FOXP3+ T-cell densities (r=0.59, p<0.00001). CD8+ and FOXP3+ T-cell density were also correlated, though more weakly (r=0.45, p<0.00001) (Figure 2). CD3+, CD8+ and FOXP3+ median T-cell densities in the matched race tissue microarray set did not significantly differ by patient racial ancestry (Table 1). In addition, there were no statistically significant differences in median T-cell densities by tumor stage or Gleason grade in either racial group considered separately from the matched race tissue microarrays (Table 1), nor in the high grade African-American set (Supplementary Table S2) or in the combined African-American set (Table 1). All median T-cell densities were comparable when compared overall, by race, and within strata of race by pathologic stage and grade.
Figure 2. Correlation of T-cell density measurements across various subsets in matched race tissue microarray (TMA) cohort.
A) Density of CD8+ T-cells (cells/mm2) correlates with density of CD3+ T-cells (cells/mm2) in prostate tumor on tissue microarray spots (Pearson’s correlation coefficient r= 0.66, p<0.00001) B) Density of FOXP3+ T-cells (cells/mm2) correlates with density of CD3+ T-cells (cells/mm2) in prostate tumor on tissue microarray spots (Pearson’s correlation coefficient r= 0.59, p<0.00001) C) Density of FOXP3+ T-cells (cells/mm2) correlates with density of CD8+ infiltrating lymphocytes (cells/mm2) in prostate tumor on tissue microarray spots (Pearson’s correlation coefficient r= 0.45, p<0.00001)
Table 1.
T-cell density stratified by race and clinical-pathologic parameters in matched race tissue microarrays and combined African-American set
| Matched race tissue microarrays |
CD3 Density | CD8 Density | FOXP3 Density | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| N | Median | p* | N | Median | p* | N | Median | p* | |
| Overall | 224 | 232.1 | 227 | 55.7 | 227 | 15.9 | |||
| European-American | 127 | 241.4 | 0.5 | 127 | 61.9 | 0.2 | 127 | 15.0 | 0.8 |
| African-American | 97 | 220.2 | 100 | 52.2 | 100 | 15.9 | |||
| European-American | |||||||||
| T2N0 | 59 | 236.1 | 0.6 | 59 | 63.7 | 0.9 | 59 | 16.5 | 0.7 |
| T3N0 | 55 | 246.7 | 55 | 59.2 | 55 | 13.3 | |||
| N1 | 12 | 215.3 | 12 | 56.1 | 12 | 19.0 | |||
| Gleason= 6 | 26 | 235.6 | 0.3 | 26 | 57.0 | 0.6 | 26 | 15.8 | 0.7 |
| Gleason=3+4 | 35 | 311.2 | 35 | 69.0 | 35 | 17.7 | |||
| Gleason=4+3 | 32 | 227.2 | 32 | 55.7 | 32 | 10.2 | |||
| Gleason=8 | 21 | 157.4 | 21 | 56.6 | 21 | 9.7 | |||
| Gleason=9 | 13 | 191.9 | 13 | 67.2 | 13 | 15.0 | |||
| African-American | |||||||||
| T2N0 | 40 | 233.9 | 0.1 | 41 | 55.7 | 0.1 | 41 | 13.3 | 0.1 |
| T3N0 | 41 | 176.8 | 42 | 33.2 | 42 | 14.1 | |||
| N1 | 9 | 352.8 | 10 | 64.5 | 10 | 31.4 | |||
| Gleason= 6 | 25 | 233.4 | 0.4 | 25 | 66.3 | 0.3 | 25 | 16.5 | 0.9 |
| Gleason=3+4 | 28 | 190.1 | 30 | 53.1 | 30 | 16.8 | |||
| Gleason=4+3 | 24 | 179.5 | 24 | 35.8 | 24 | 15.9 | |||
| Gleason=8 | 16 | 202.0 | 16 | 32.3 | 16 | 8.4 | |||
| Gleason=9 | 4 | 477.9 | 5 | 39.8 | 5 | 27.4 | |||
|
| |||||||||
| Combined African American** | |||||||||
| Overall | 181 | 246.8 | 182 | 60.1 | 185 | 15.9 | |||
| T2N0 | 78 | 241.9 | 0.3 | 79 | 60.1 | 0.4 | 79 | 14.1 | 0.1 |
| T3N0 | 81 | 242.4 | 81 | 54.8 | 83 | 15.9 | |||
| N1 | 13 | 352.8 | 14 | 81.1 | 14 | 31.4 | |||
| Gleason= 6 | 25 | 233.4 | 0.5 | 25 | 66.3 | 0.5 | 25 | 16.5 | 0.7 |
| Gleason=3+4 | 28 | 190.1 | 30 | 53.1 | 30 | 16.8 | |||
| Gleason=4+3 | 84 | 248.6 | 82 | 68.1 | 84 | 15.9 | |||
| Gleason=8 | 27 | 242.4 | 27 | 46.0 | 27 | 15.9 | |||
| Gleason=9 | 17 | 382.0 | 18 | 84.5 | 19 | 15.9 | |||
Kruskal-Wallis test
African-Americans from matched race tissue microarrays (above) and high grade African-American tissue microarray (see Supplementary Table S2)
T-cell densities were not significantly correlated with age overall or among European-American men in the matched race tissue microarrays (Table 2). CD3+ T-cell density was negatively correlated with age among African-American men in the matched race tissue microarray set (r=−0.22, p=0.03), but this correlation was not present in the high grade African-American set (Supplementary Table S3) and was attenuated in the combined African-American set (Table 2). CD3+ and FOXP3+ T-cell densities were not significantly correlated with prostate specific antigen levels overall or by race (Table 2, Supplementary Table S3). CD8+ T-cell density was negatively correlated with prostate specific antigen levels overall in the matched race tissue microarray set (r=−0.13, p=0.05; Table 2). Among European-American men, the correlation coefficient between CD8+ T cell density and prostate specific antigen levels was comparable, but not statistically significant, and among African-American men CD8+ T-cell density and prostate specific antigen levels were not correlated (Table 2, Supplementary Table S3).
Table 2.
Correlation of T-cell density with age and prostate specific antigen (PSA) in matched race tissue microarrays and combined African-American set
| Matched race tissue microarrays |
CD3 Density | CD8 Density | FOXP3 Density | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| N | r* | p | N | r* | p | N | r* | p | |
| Overall | |||||||||
| Age | 224 | −0.07 | 0.3 | 227 | −0.10 | 0.1 | 227 | −0.07 | 0.3 |
| PSA | 224 | −0.02 | 0.8 | 227 | −0.13 | 0.05 | 227 | −0.01 | 0.9 |
| European-American | |||||||||
| Age | 127 | 0.02 | 0.8 | 127 | −0.08 | 0.3 | 127 | 0.004 | 0.9 |
| PSA | 127 | −0.08 | 0.4 | 127 | −0.13 | 0.2 | 127 | 0.0001 | 0.9 |
| African-American | |||||||||
| Age | 97 | −0.22 | 0.03 | 100 | −0.15 | 0.1 | 100 | −0.17 | 0.1 |
| PSA | 97 | 0.09 | 0.4 | 100 | −0.09 | 0.4 | 100 | 0.01 | 0.9 |
|
| |||||||||
| Combined African American ** | |||||||||
| Overall | |||||||||
| Age | 181 | −0.08 | 0.3 | 182 | −0.08 | 0.3 | 185 | −0.05 | 0.5 |
| PSA | 181 | 0.01 | 0.9 | 182 | −0.03 | 0.7 | 185 | −0.02 | −.8 |
Spearman correlation coefficient
African-Americans from matched race tissue microarrays (above) and high grade African-American tissue microarray (see Supplementary Table S3)
Association of T-cell density with molecular prostate cancer subtypes
Next, we tested the association of ERG and PTEN status with tumor-infiltrating T-cell densities using the combined matched race tissue microarray set. All T-cell markers were significantly higher among men with PTEN loss as compared to men with PTEN intact tumors (CD3: 65% higher median density, CD8: 38% higher median density, and FOXP3: 127% higher median density) and significantly higher among men with ERG expression positive tumors as compared to tumors without ERG expression (CD3: 64% higher median density, CD8: 61% higher median density, and FOXP3: 71% higher median density; Table 3). Similar patterns were observed for European-American and African-American men analyzed separately (Table 3, Supplementary Table S4), though not all comparisons remained statistically significant. Although CD8+ T-cell density appeared higher for tumors with PTEN loss as compared to tumors with PTEN intact, in race-stratified analyses, the difference was no longer statistically significant. Similarly, among African-American men (combined set) CD8+ T-cell density appeared higher for men with ERG expression as compared to men without ERG expression, but the difference was not statistically significant. Because PTEN loss is more common among cases with ERG rearrangement (28–30), we jointly classified cases by both alterations and observed that there was a significant difference in median lymphocyte densities for all markers across categories overall; and men with both PTEN loss and ERG rearrangement had the highest median lymphocyte densities for all markers. When stratified by race, all patterns were consistent for European-American men and for African-American men (combined set), though the difference in median CD8+ T-cell densities was no longer statistically significant for either race considered individually (Table 3 and Supplementary Table S4).
Table 3.
T-cell density stratified by race and molecular classification in matched race tissue microarrays and combined African-American set
| CD3 Density
|
CD8 Density
|
FOXP3 Density
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Matched race tissue microarrays |
N | Median | p* | N | Median | p* | N | Median | p* |
| Overall | 224 | 232.1 | 227 | 55.7 | 227 | 15.9 | |||
| PTEN Intact | 168 | 192.3 | 0.001 | 171 | 49.5 | 0.04 | 171 | 11.5 | 0.0004 |
| PTEN Loss | 56 | 317.4 | 56 | 68.5 | 56 | 26.1 | |||
| ERG Negative | 137 | 188.3 | 0.0004 | 140 | 43.3 | 0.001 | 140 | 11.9 | 0.001 |
| ERG Positive | 87 | 308.6 | 87 | 69.9 | 87 | 20.3 | |||
| PTEN Intact & ERG Negative | 118 | 184.8 | 0.0001 | 121 | 42.4 | 0.01 | 121 | 10.6 | <0.0001 |
| PTEN Intact & ERG Positive | 50 | 229.5 | 50 | 74.7 | 50 | 12.8 | |||
| PTEN Loss & ERG Negative | 19 | 244.0 | 19 | 59.2 | 19 | 20.3 | |||
| PTEN Loss & ERG Positive | 37 | 365.2 | 37 | 69.0 | 37 | 30.9 | |||
| European-American | |||||||||
| PTEN Intact | 91 | 192.8 | 0.005 | 91 | 50.4 | 0.2 | 91 | 12.4 | 0.01 |
| PTEN Loss | 36 | 305.8 | 36 | 69.4 | 36 | 25.2 | |||
| ERG Negative | 64 | 190.1 | 0.005 | 64 | 42.4 | 0.01 | 64 | 10.2 | 0.01 |
| ERG Positive | 63 | 299.5 | 63 | 69.9 | 63 | 19.5 | |||
| PTEN Intact & ERG Negative | 54 | 181.7 | 0.004 | 54 | 42.0 | 0.1 | 54 | 9.3 | 0.004 |
| PTEN Intact & ERG Positive | 37 | 224.6 | 37 | 77.8 | 37 | 13.3 | |||
| PTEN Loss & ERG Negative | 10 | 237.0 | 10 | 68.1 | 10 | 20.3 | |||
| PTEN Loss & ERG Positive | 26 | 379.8 | 26 | 69.4 | 26 | 31.8 | |||
| African-American | |||||||||
| PTEN Intact | 77 | 191.9 | 0.1 | 80 | 46.0 | 0.1 | 80 | 11.5 | 0.03 |
| PTEN Loss | 20 | 337.8 | 20 | 59.2 | 20 | 29.2 | |||
| ERG Negative | 73 | 188.3 | 0.04 | 76 | 44.2 | 0.1 | 76 | 12.8 | 0.03 |
| ERG Positive | 24 | 311.2 | 24 | 70.3 | 24 | 28.7 | |||
| PTEN Intact & ERG Negative | 64 | 187.0 | 0.1 | 67 | 43.3 | 0.3 | 67 | 11.5 | 0.02 |
| PTEN Intact & ERG Positive | 13 | 234.3 | 13 | 71.6 | 13 | 9.7 | |||
| PTEN Loss & ERG Negative | 9 | 306.8 | 9 | 53.9 | 9 | 16.5 | |||
| PTEN Loss & ERG Positive | 11 | 352.8 | 11 | 69.0 | 11 | 30.9 | |||
|
| |||||||||
| Combined African American** | |||||||||
| PTEN Intact | 149 | 234.3 | 0.03 | 150 | 54.8 | 0.1 | 153 | 14.2 | 0.05 |
| PTEN Loss | 32 | 355.9 | 32 | 71.6 | 32 | 28.3 | |||
| ERG Negative | 144 | 235.6 | 0.01 | 147 | 53.9 | 0.1 | 148 | 15.0 | 0.04 |
| ERG Positive | 37 | 313.9 | 35 | 78.7 | 37 | 28.3 | |||
| PTEN Intact & ERG Negative | 127 | 230.0 | 0.03 | 130 | 50.9 | 0.2 | 131 | 14.2 | 0.1 |
| PTEN Intact & ERG Positive | 22 | 287.1 | 20 | 87.1 | 22 | 14.2 | |||
| PTEN Loss & ERG Negative | 17 | 306.8 | 17 | 93.8 | 17 | 16.8 | |||
| PTEN Loss & ERG Positive | 15 | 359.0 | 15 | 69.0 | 15 | 30.1 | |||
Kruskal-Wallis test
African-Americans from matched race tissue microarrays (above) and high grade African-American tissue microarray (see Supplementary Table S4)
Association of T-cell density with prostate cancer outcomes among African-American men (combined set)
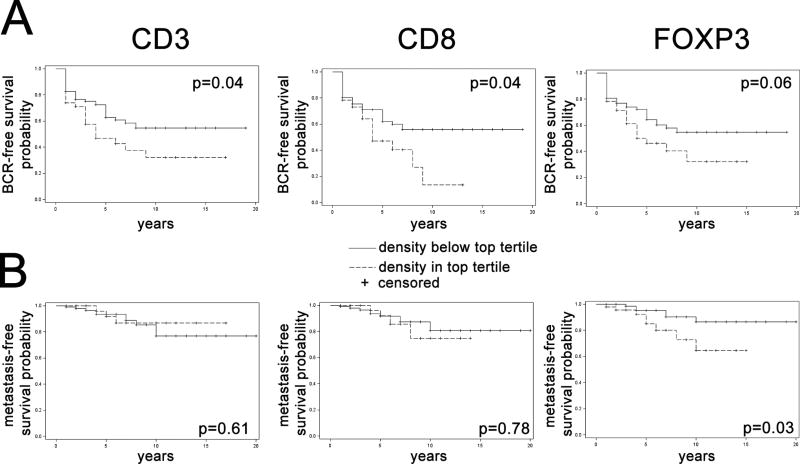
We had adequate numbers of adverse oncologic events to examine the association of tumor infiltrating T-cell density with prostate cancer outcomes only in the combined African-American set, since it was highly enriched for high Gleason score tumors by design (25). We utilized biochemical recurrence and metastasis-free survival as the primary clinical endpoints because the number of events for cancer specific survival was low in this cohort (Supplementary Table S5, Supplementary Figure S4) and because metastasis-free survival is now recognized as a reasonable surrogate endpoint for overall survival in localized prostate cancer (31). We dichotomized T-cell densities into the top tertile compared to the bottom two tertiles for each marker (Figure 3). On Kaplan-Meier and univariable Cox regression models, there was a trend towards higher risk of biochemical recurrence or metastasis for patients in the top tertile of lymphocyte densities compared to lower two tertiles, though statistical significance was not reached for all markers and all endpoints and p-values differed slightly depending on whether log-rank test (for Kaplan-Meier) or Cox regression models (for univariable analysis) were used (Table 4, Figure 3). On multivariable analysis, we generally did not observe significant associations between biochemical recurrence and being in the highest tertile of lymphocyte density as compared to being in the lower two tertiles of lymphocyte density for any T-cell markers, adjusting for patient age, prostate specific antigen levels and body mass index (BMI) as well as pathologic stage and grade. However, men in the highest tertile of CD3+ T-cell density had a suggestive, though not statistically significant, higher risk of biochemical recurrence as compared to men in the lowest two tertiles on multivariable analysis, (Hazard Ratio:1.63; 95% Confidence Interval: 0.84–3.03, Table 4). We did not observe significant associations between risk of metastases and being in the highest tertile of CD8 or CD3+ T-cell density as compared to being in the lower two tertiles of CD8+ or CD3+ T-cell density, respectively. However, men in the highest tertile of FOXP3 density had a significantly higher risk of metastases as compared to men in the lower two tertiles of FOXP3 (Hazard Ratio: 12.89; 95% Confidence Interval:1.59–104.4). Given the imprecision of this estimate based on the width of the confidence interval, we interpret this finding with caution, and as possibly hypothesis-generating for future studies.
Figure 3. Kaplan-Meier analysis stratified by T-cell density for combined African-American set.
Kaplan-Meier analysis for (A) biochemical recurrence (BCR)-free survival and (B) metastasis-free survival stratified by T-cell density for CD3, CD8, FOXP3. Patients in the top tertile of T-cell densities are shown by the hashed line, while those in the bottom two tertiles are shown by the unbroken line. P-values by log-rank test.
Table 4.
Association of T-cell density with oncologic outcomes in combined African-American set* by Cox Analysis
| Biochemical Recurrence
|
Metastasis
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N (case/control) |
Univariable Analysis |
Multivariable Analysis** |
N (case/control) |
Univariable Analysis |
Multivariable Analysis** |
|||||
|
|
|
|||||||||
| HR (95% CI) |
p value | HR (95% CI) |
p value | HR (95% CI) | p value | HR (95% CI) |
p value | |||
| CD3 Density | ||||||||||
| group1*** | 36/74 | Ref | Ref | 11/99 | Ref | Ref | ||||
| group2*** | 23/23 | 1.68 | 0.05 | 1.63 | 0.15 | 4/42 | 0.72 | 0.61 | 1.15 | 0.88 |
| (0.84–3.03) | (0.18–7.29) | (0.20–2.61) | (0.996–2.84) | |||||||
|
| ||||||||||
| CD8 Density | ||||||||||
| group1*** | 38/74 | Ref | Ref | 12/100 | Ref | Ref | ||||
| group2*** | 23/23 | 1.65 | 0.06 | 1.21 | 0.56 | 4/42 | 1.18 | 0.78 | 0.89 | 0.88 |
| (0.64–2.26) | (0.98–2.79) | (0.18–4.33) | (0.37–3.81) | |||||||
|
| ||||||||||
| FOXP3 Density | ||||||||||
| group1*** | 36/73 | Ref | Ref | 7/102 | Ref | Ref | ||||
| group2*** | 25/26 | 1.57 | 0.08 | 1.12 | 0.71 | 9/42 | 3.10 | 0.04 | 12.89 | 0.02 |
| (0.94–2.63) | (0.60–2.11) | (1.07–8.97) | (1.59–104.40) | |||||||
African-Americans from matched race tissue microarrays and high grade African-American tissue microarray
adjusted for age, stage, grade, PSA and body mass index
group1, below the top tertile; group2, in the top tertile
HR: Hazard Ratio
CI: Confidence Interval
Discussion
Our study is the first to correlate different subsets of tumor infiltrating T-cells with clinical outcome and molecular features across patients of varying racial ancestry. One strength of this work is that we carefully validated our automated T-cell density measurements, ensuring that our results on tissue microarray spots--which sample only a small area of the tumor (~1 mm2)--were correlated with the measurements on standard tissue sections and manually counted T-cell densities. Importantly, we did not find any evidence of differences in intratumoral T-cell densities when comparing tumors from patients of African-American or European-American ancestry, matched by most pathologic and clinical parameters. There were also only a few differences in T-cell densities when stratified by most clinical-pathologic parameters in both racial groups, with T-cell densities negatively correlated with patient age and prostate specific antigen levels in some comparisons for some races. Intriguingly, we did observe a statistically significant association between ERG expression or PTEN loss with higher intratumoral lymphocyte infiltration, irrespective of racial ancestry. Among African-Americans, where we had adequate power to examine associations between T-cell densities and outcomes, we observed that higher FOXP3+ cell density was associated with an increased propensity for metastasis (HR= 12.89, p=0.02) in multivariable analysis, however a similar association was not observed for CD3+ and CD8+ T-cell density. In addition, the confidence intervals on this hazard ratio estimate were quite wide indicating that it is imprecise and requires independent validation in additional cohorts of African-American tumors.
A number of previous studies have examined the correlation between tumor infiltrating lymphocytes and clinical-pathologic parameters and outcomes in prostate cancer (Supplementary Table S6). Importantly, most of these studies have been conducted using visual scoring systems and all in predominantly European or European-American cohorts, which contrasts with our digital scoring and the enrichment for African-American ancestry in our cohort. The results of these previous studies have suggested that both extremes of intratumoral lymphocyte densities-- high and low--may be associated with worse outcomes, with more studies finding that high densities are associated with poor outcomes than low densities. One of the earliest studies, by Vesalainen et al, used H&E standard histologic sections visually scored lymphocyte infiltrates in primary prostate cancers, binning into three groups: rare, moderate and dense lymphocyte infiltrate (17). They found that absent or rare lymphocyte infiltrates were associated with a high risk of tumor progression and fatal disease (p=0.016). In contrast, Irani et al reported that patients with high-grade inflammation surrounding malignant glands in radical prostatectomy specimens had significantly more postoperative biochemical recurrence than patients with low-grade inflammation (32).
Since these early investigations, more detailed studies have been performed using immunohistochemistry to classify lymphocyte infiltration and to categorize lymphocyte location as epithelial and stromal. One of the largest studies, by Flammiger et al, found that high and low levels of intratumoral T- but not B-lymphocytes were associated with poor clinical outcomes in primary prostate cancer (21). In this study, patients with very low and very high numbers of CD3+ T-cells per tissue microarray spot had a significantly shorter prostate specific antigen biochemical recurrence-free survival compared to patients with intermediate numbers of T-cells (p=0.0188). Another study observed that patients with metastatic primary cancers (either prostatectomies, needle biopsies or transurethral resection of prostate specimens) showed higher levels of CD3+ lymphocytes compared with those that did not metastasize (20). Similar to our study, this is one of the few prior studies to use digital image analysis for CD3 quantification. Finally, a recent study by Ness et al showed that higher levels of manually scored CD3+ T-cells in tumor epithelial areas on tissue microarray were associated with a higher risk of biochemical recurrence (24).
One confounder in these prior studies is that all CD3+ T-cells or even all lymphocytes were lumped together, without considering the effects of different T-cell subsets. However, we found that most T-cell markers were strongly correlated in primary prostate tissue, and CD8+ and FOXP3+ cell densities tracked together, similar to other recent studies examining CD8, CD4 and even CD20 (19). Consistent, with this, at least one prior study showed that both CD3+ and CD8+ T-cell levels were associated with a higher risk of biochemical recurrence when measured on tissue microarray (24). Only a few studies have examined regulatory T-cell immune markers and their correlation with clinical outcome and clinical-pathological variables. In 2013, Flammiger found that a higher number of intratumoral FOXP3+ regulatory T-cells was associated with a more advanced tumor stage and higher Ki67 labelling index, but in multivariable analysis, FOXP3 density was not an independent predictor of prostate specific antigen biochemical recurrence (22). In the same year, Davidsson et al found that patients with higher number of regulatory T-cells (CD4+ FOXP3+) had an increased risk of dying of prostate cancer, with every additional CD4+ FOXP3+ cell associated with a 12% increase in the odds of dying of prostate cancer (23).
Our finding that higher FOXP3+ cell density may be associated with a higher risk of metastasis among African-American patients appears generally consistent with these prior studies but requires validation in a larger independent cohort. Mechanistically, it remains unclear which immunomodulatory/immunosuppressive pathways may play the most significant role in prostate cancer. The recent finding from our group that PD-L1 is only expressed in a small minority of primary prostate tumors (<8%) is intriguing (33), though this study was performed in a predominantly white cohort using tissue microarrays. Recent work from other groups using standard histologic sections to detect generally patchy PD-L1 positivity has intriguingly suggested that African-American ancestry may be associated with higher PD-L1 expression (34). Finally, a number of groups are actively investigating alternative immune checkpoints that may be more important than PD-L1 in primary prostate cancer, such as B7-H3, which is much more highly expressed in primary prostate cancer (35). Finally, since there is a significant correlation between CD3+, CD8+ and FOXP3+ T-cell densities in primary prostate tissues, it seems plausible that there is a general association of higher T-lymphocyte levels with adverse outcomes in prostate cancer and the association only reaches statistical significance for some T-cell subsets in a subset of studies for stochastic reasons. By this reasoning, there may not be any particular significance for FOXP3 immunosuppressive T-cell density in prostate cancer.
We did not find any associations of T-cell densities with racial ancestry in this study. This is interesting in light of the fact that there is mounting evidence for differing immune microenvironments by race in primary prostate tumors. The incidence of prostatic inflammation may be higher in the benign prostates and prostate tumors of African-American men compared to white men, though most studies have not distinguished between types of inflammatory cells and have not used digital quantification (36). There is also some evidence that inflammation-related gene expression, in particular, that of cytokines, may differ in primary prostate tumors by race (37–39). Because our study is restricted to analysis of T-cells, it certainly may have missed other important differences in the inflammatory microenvironment of African-American versus European-American tumors. Future studies will examine other immune cell subsets in this cohort.
Another important line of inquiry in our study was the comparison of tumor molecular subtypes with T-cell densities in primary prostate cancers. There is mounting evidence that somatic genomic alterations may be associated with changes in the immune microenvironment, in prostate as well as other tumor types. As an extreme example, genomic defects in the mismatch repair pathway are associated with increased immune response in both colorectal cancers (11–13) and prostate cancer (14), likely due to the increased mutational load in these tumors facilitating generation of neoantigens. Similarly, a recent study in mouse models of prostate cancer demonstrated remarkably diverse immune responses to tumors with differing somatic alterations, including PTEN deletion (40). Intriguingly, we found that tumors with ERG gene rearrangements or PTEN deletion, each detected by genetically validated protein expression assays, are each associated with an increased density of tumor infiltrating T-cells compared to tumors without these alterations. That this association is fairly strong was evidenced by the fact that it could be observed in tumors from both African-American and European-American patients, despite the fact that ERG gene rearrangements are only about half as common in the former compared to the latter group (25, 41–45).
At least for ERG, these data appear to be reproducible in that they are similar to results from two previously published studies in European or European-American cohorts, both of which demonstrated a similar association between ERG expression and CD3+ T-cell density (21, 46), as well as a separate study reporting a similar association with FOXP3+ cell density (47). However, the mechanism of this association remains unclear. In vitro evidence suggests that ERG gene fusions—a very early and likely initiating event in prostatic tumorigenesis--may be caused, at least in part, by inflammation-induced oxidative stress (48). Thus, it is conceivable that this association of ERG-positive tumors with increased inflammation persists during later stages of tumor progression. This notion is supported, at least in principle, by recent epidemiologic evidence. In a population-based case-control study of patients who did or did not use aspirin, use of aspirin (an anti-inflammatory drug) was associated with a significant reduction in the relative risk of ERG fusion-positive prostate cancers, but a similar association was not detected for ERG-negative prostate tumors (49).
Alternatively, it is possible that ERG expression itself drives transcriptional programs in tumor cells that increase the recruitment of inflammatory cells to the tumor microenvironment. ERG fusions increase NF-κB transcriptional activity in prostate epithelial cells and human prostate tumors via toll-like receptor pathways (50), which may in turn promote T-cell immunosurveillance in the tumor microenvironment (51). ERG expression is also associated with increased expression of HLA-DMB in prostate cancer, a MHC class II molecule involved in antigen presentation and T-cell activation (46, 52, 53). Finally, gene fusions in prostate cancer can result in the formation of unique chimeric amino acid sequences that span the breakpoint of the two fused gene products and potentially generating fusion proteins with neoepitopes. In a recent computational study, type VI TMPRSS2-ERG gene fusions contained high-affinity HLA-restricted epitopes and were recognized by CD8+ T-cells in vitro (54). Though these mechanistic possibilities are intriguing, it is also possible that the association of higher lymphocyte densities with ERG rearrangement is due to a confounding additional molecular alteration, which is itself associated with ERG fusion.
Some limitations of the current study that warrant discussion. First, all tumor-infiltrating lymphocyte densities were measured on tissue microarray spots, comprising only about 1 mm2 of total tissue for analysis per patient. While we demonstrated a significant correlation between the T-cell densities measured on tissue microarrays and those measured on standard histologic sections of the dominant tumor nodule, lymphocyte infiltrates are quite heterogeneous in prostate tumors and sampling error could certainly mask significant differences between groups in this setting. Second, we examined only T-cells in the current manuscript, using a single color immunohistochemistry assay rather than a multiplex assay. Studies of tumor immune microenvironments are increasingly using multispectral immunofluorescence, which enable more accurate immunophenotyping and require less tissue input. However, multispectral immunofluorescence assays have proven challenging to quantify and many of the digital analysis platforms for this technology are not easily adaptable to high throughput analysis of tissue microarrays. Third, we used a relatively crude definition of tumor-infiltrating lymphocytes, as any lymphocyte in a tissue microarray spot containing tumor epithelium. We did not distinguish between lymphocytes within the tumor epithelium (quite rare in prostate, see Figure 1) and those in the stroma, nor did we distinguish lymphocytes within the invasive front of the tumor from those in the middle of tumor nodule. Future studies may use a more refined definition of tumor-infiltrating lymphocytes since lymphocyte location, rather than T-cell density, is increasingly recognized as a potential biomarker of prognosis (2) and response to immunotherapies, supporting the biological relevance of this measure. Finally, the relatively small size of our cohorts matched on racial ancestry is a definite limitation, and all findings in the current study must be replicated in larger independent cohorts in the future. In these studies, it may also be helpful to include genetic definitions of racial ancestry rather than simple self-identification as we have done here.
In conclusion, this is among the first studies in primary prostate cancer to examine associations between digitally quantified tumor infiltrating T-cell densities, tumor molecular subtype and racial ancestry. While we did not find any variations in T-cell density by racial ancestry, we did find that T-cell density varies significantly with tumor ERG and PTEN status, in both patients of European-American and African-American decent. In addition, we are the first to find some evidence of potential associations between higher FOXP3+ T-cell densities and adverse oncologic outcomes among African-American patients, though this must be validated in future work. Ultimately, integrating clinical-pathologic, somatic molecular subtyping and immune microenvironment data is becoming increasingly important in the era of precision medicine and many future studies of this subject are warranted.
Supplementary Material
Acknowledgments
Financial Support: Funding for this research was provided in part by two Health Disparity Research Awards from the CDMRP-PCRP (W81XWH-15-1-0661 to SAT, EMS and TLL and W81XWH-17-1-0286 to KSS, TLL, LBG, and HBK). Additional funding and resources were provided by the NIH/NCI Prostate SPORE P50CA58236 and the NCI Cancer Center Support Grant 5P30CA006973-52. LR and LM were supported by fellowships from the CUPID (Cancer in the Under-Privileged Indigent or Disadvantaged) Program.
Footnotes
Disclosure/Conflict of Interest: TLL has received research support from Ventana Medical Systems for other studies.
References
- 1.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–70. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 2.Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 3.Eerola AK, Soini Y, Paakko P. Tumour infiltrating lymphocytes in relation to tumour angiogenesis, apoptosis and prognosis in patients with large cell lung carcinoma. Lung Cancer. 1999;26:73–83. doi: 10.1016/s0169-5002(99)00072-0. [DOI] [PubMed] [Google Scholar]
- 4.Al-Shibli K, Al-Saad S, Andersen S, et al. The prognostic value of intraepithelial and stromal CD3-, CD117- and CD138-positive cells in non-small cell lung carcinoma. APMIS. 2010;118:371–82. doi: 10.1111/j.1600-0463.2010.02609.x. [DOI] [PubMed] [Google Scholar]
- 5.Bogunovic D, O’Neill DW, Belitskaya-Levy I, et al. Immune profile and mitotic index of metastatic melanoma lesions enhance clinical staging in predicting patient survival. Proc Natl Acad Sci U S A. 2009;106:20429–34. doi: 10.1073/pnas.0905139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bromwich EJ, McArdle PA, Canna K, et al. The relationship between T-lymphocyte infiltration, stage, tumour grade and survival in patients undergoing curative surgery for renal cell cancer. Br J Cancer. 2003;89:1906–8. doi: 10.1038/sj.bjc.6601400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakano O, Sato M, Naito Y, et al. Proliferative activity of intratumoral CD8(+) T-lymphocytes as a prognostic factor in human renal cell carcinoma: clinicopathologic demonstration of antitumor immunity. Cancer Res. 2001;61:5132–6. [PubMed] [Google Scholar]
- 8.Jass JR, Love SB, Northover JM. A new prognostic classification of rectal cancer. Lancet. 1987;1:1303–6. doi: 10.1016/s0140-6736(87)90552-6. [DOI] [PubMed] [Google Scholar]
- 9.Ropponen KM, Eskelinen MJ, Lipponen PK, Alhava E, Kosma VM. Prognostic value of tumour-infiltrating lymphocytes (TILs) in colorectal cancer. J Pathol. 1997;182:318–24. doi: 10.1002/(SICI)1096-9896(199707)182:3<318::AID-PATH862>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 10.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 11.Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–20. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee V, Murphy A, Le DT, Diaz LA., Jr Mismatch Repair Deficiency and Response to Immune Checkpoint Blockade. Oncologist. 2016;21:1200–11. doi: 10.1634/theoncologist.2016-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dudley JC, Lin MT, Le DT, Eshleman JR. Microsatellite Instability as a Biomarker for PD-1 Blockade. Clin Cancer Res. 2016;22:813–20. doi: 10.1158/1078-0432.CCR-15-1678. [DOI] [PubMed] [Google Scholar]
- 14.Guedes LB, Antonarakis ES, Schweizer MT, et al. MSH2 Loss in Primary Prostate Cancer. Clin Cancer Res. 2017;23:6863–74. doi: 10.1158/1078-0432.CCR-17-0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sfanos KS, Yegnasubramanian S, Nelson WG, De Marzo AM. The inflammatory microenvironment and microbiome in prostate cancer development. Nat Rev Urol. 2018;15:11–24. doi: 10.1038/nrurol.2017.167. [DOI] [PubMed] [Google Scholar]
- 16.Miyahira AK, Roychowdhury S, Goswami S, et al. Beyond Seed and Soil: Understanding and Targeting Metastatic Prostate Cancer; Report From the 2016 Coffey-Holden Prostate Cancer Academy Meeting. Prostate. 2017;77:123–44. doi: 10.1002/pros.23260. [DOI] [PubMed] [Google Scholar]
- 17.Vesalainen S, Lipponen P, Talja M, Syrjanen K. Histological grade, perineural infiltration, tumour-infiltrating lymphocytes and apoptosis as determinants of long-term prognosis in prostatic adenocarcinoma. Eur J Cancer. 1994;30A:1797–803. doi: 10.1016/0959-8049(94)e0159-2. [DOI] [PubMed] [Google Scholar]
- 18.McArdle PA, Canna K, McMillan DC, et al. The relationship between T-lymphocyte subset infiltration and survival in patients with prostate cancer. Br J Cancer. 2004;91:541–3. doi: 10.1038/sj.bjc.6601943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karja V, Aaltomaa S, Lipponen P, et al. Tumour-infiltrating lymphocytes: A prognostic factor of PSA-free survival in patients with local prostate carcinoma treated by radical prostatectomy. Anticancer Res. 2005;25:4435–8. [PubMed] [Google Scholar]
- 20.Richardsen E, Uglehus RD, Due J, Busch C, Busund LT. The prognostic impact of M-CSF, CSF-1 receptor, CD68 and CD3 in prostatic carcinoma. Histopathology. 2008;53:30–8. doi: 10.1111/j.1365-2559.2008.03058.x. [DOI] [PubMed] [Google Scholar]
- 21.Flammiger A, Bayer F, Cirugeda-Kuhnert A, et al. Intratumoral T but not B lymphocytes are related to clinical outcome in prostate cancer. APMIS. 2012;120:901–8. doi: 10.1111/j.1600-0463.2012.02924.x. [DOI] [PubMed] [Google Scholar]
- 22.Flammiger A, Weisbach L, Huland H, et al. High tissue density of FOXP3+ T cells is associated with clinical outcome in prostate cancer. Eur J Cancer. 2013;49:1273–9. doi: 10.1016/j.ejca.2012.11.035. [DOI] [PubMed] [Google Scholar]
- 23.Davidsson S, Ohlson AL, Andersson SO, et al. CD4 helper T cells, CD8 cytotoxic T cells, and FOXP3(+) regulatory T cells with respect to lethal prostate cancer. Mod Pathol. 2013;26:448–55. doi: 10.1038/modpathol.2012.164. [DOI] [PubMed] [Google Scholar]
- 24.Ness N, Andersen S, Valkov A, et al. Infiltration of CD8+ lymphocytes is an independent prognostic factor of biochemical failure-free survival in prostate cancer. Prostate. 2014;74:1452–61. doi: 10.1002/pros.22862. [DOI] [PubMed] [Google Scholar]
- 25.Tosoian JJ, Almutairi F, Morais CL, et al. Prevalence and Prognostic Significance of PTEN Loss in African-American and European-American Men Undergoing Radical Prostatectomy. Eur Urol. 2017;71:697–700. doi: 10.1016/j.eururo.2016.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lotan TL, Wei W, Morais CL, et al. PTEN Loss as Determined by Clinical-grade Immunohistochemistry Assay Is Associated with Worse Recurrence-free Survival in Prostate Cancer. Eur Urol Focus. 2016;2:180–8. doi: 10.1016/j.euf.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahearn TU, Pettersson A, Ebot EM, et al. A Prospective Investigation of PTEN Loss and ERG Expression in Lethal Prostate Cancer. J Natl Cancer Inst. 2016:108. doi: 10.1093/jnci/djv346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshimoto M, Joshua AM, Cunha IW, et al. Absence of TMPRSS2:ERG fusions and PTEN losses in prostate cancer is associated with a favorable outcome. Mod Pathol. 2008;21:1451–60. doi: 10.1038/modpathol.2008.96. [DOI] [PubMed] [Google Scholar]
- 29.Carver BS, Tran J, Gopalan A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–24. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King JC, Xu J, Wongvipat J, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–6. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie W, Regan MM, Buyse M, et al. Metastasis-Free Survival Is a Strong Surrogate of Overall Survival in Localized Prostate Cancer. J Clin Oncol. 2017;35:3097–104. doi: 10.1200/JCO.2017.73.9987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Irani J, Goujon JM, Ragni E, et al. High-grade inflammation in prostate cancer as a prognostic factor for biochemical recurrence after radical prostatectomy. Pathologist Multi Center Study Group. Urology. 1999;54:467–72. doi: 10.1016/s0090-4295(99)00152-1. [DOI] [PubMed] [Google Scholar]
- 33.Haffner MC, Guner G, Taheri D, et al. Comprehensive Evaluation of Programmed Death-Ligand 1 Expression in Primary and Metastatic Prostate Cancer. Am J Pathol. 2018 doi: 10.1016/j.ajpath.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calagua C, Russo J, Sun Y, et al. Expression of PD-L1 in Hormone-naive and Treated Prostate Cancer Patients Receiving Neoadjuvant Abiraterone Acetate plus Prednisone and Leuprolide. Clin Cancer Res. 2017;23:6812–22. doi: 10.1158/1078-0432.CCR-17-0807. [DOI] [PubMed] [Google Scholar]
- 35.Benzon B, Zhao SG, Haffner MC, et al. Correlation of B7-H3 with androgen receptor, immune pathways and poor outcome in prostate cancer: an expression-based analysis. Prostate Cancer Prostatic Dis. 2017;20:28–35. doi: 10.1038/pcan.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eastham JA, May RA, Whatley T, et al. Clinical characteristics and biopsy specimen features in African-American and white men without prostate cancer. J Natl Cancer Inst. 1998;90:756–60. doi: 10.1093/jnci/90.10.756. [DOI] [PubMed] [Google Scholar]
- 37.Reams RR, Agrawal D, Davis MB, et al. Microarray comparison of prostate tumor gene expression in African-American and Caucasian American males: a pilot project study. Infect Agent Cancer. 2009;(4 Suppl 1):S3. doi: 10.1186/1750-9378-4-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wallace TA, Prueitt RL, Yi M, et al. Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer Res. 2008;68:927–36. doi: 10.1158/0008-5472.CAN-07-2608. [DOI] [PubMed] [Google Scholar]
- 39.Powell IJ, Dyson G, Land S, et al. Genes associated with prostate cancer are differentially expressed in African American and European American men. Cancer Epidemiol Biomarkers Prev. 2013;22:891–7. doi: 10.1158/1055-9965.EPI-12-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bezzi M, Seitzer N, Ishikawa T, et al. Diverse genetic-driven immune landscapes dictate tumor progression through distinct mechanisms. Nat Med. 2018;24:165–75. doi: 10.1038/nm.4463. [DOI] [PubMed] [Google Scholar]
- 41.Khani F, Mosquera JM, Park K, et al. Evidence for molecular differences in prostate cancer between African American and Caucasian men. Clin Cancer Res. 2014;20:4925–34. doi: 10.1158/1078-0432.CCR-13-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magi-Galluzzi C, Tsusuki T, Elson P, et al. TMPRSS2-ERG gene fusion prevalence and class are significantly different in prostate cancer of Caucasian, African-American and Japanese patients. Prostate. 2011;71:489–97. doi: 10.1002/pros.21265. [DOI] [PubMed] [Google Scholar]
- 43.Faisal FA, Sundi D, Tosoian JJ, et al. Racial Variations in Prostate Cancer Molecular Subtypes and Androgen Receptor Signaling Reflect Anatomic Tumor Location. Eur Urol. 2016;70:14–7. doi: 10.1016/j.eururo.2015.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang FW, Mosquera JM, Garofalo A, et al. Exome Sequencing of African-American Prostate Cancer Reveals Loss-of-Function ERF Mutations. Cancer Discov. 2017;7:973–83. doi: 10.1158/2159-8290.CD-16-0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindquist KJ, Paris PL, Hoffmann TJ, et al. Mutational Landscape of Aggressive Prostate Tumors in African American Men. Cancer Res. 2016;76:1860–8. doi: 10.1158/0008-5472.CAN-15-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roudier MP, Winters BR, Coleman I, et al. Characterizing the molecular features of ERG-positive tumors in primary and castration resistant prostate cancer. Prostate. 2016;76:810–22. doi: 10.1002/pros.23171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaczmarczyk-Sekula K, Galazka K, Glajcar A, et al. Prostate cancer with different ERG status may show different FOXP3-positive cell numbers. Pol J Pathol. 2016;67:313–7. doi: 10.5114/pjp.2016.65861. [DOI] [PubMed] [Google Scholar]
- 48.Mani RS, Amin MA, Li X, et al. Inflammation-Induced Oxidative Stress Mediates Gene Fusion Formation in Prostate Cancer. Cell Rep. 2016;17:2620–31. doi: 10.1016/j.celrep.2016.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wright JL, Chery L, Holt S, et al. Aspirin and NSAID use in association with molecular subtypes of prostate cancer defined by TMPRSS2:ERG fusion status. Prostate Cancer Prostatic Dis. 2016;19:53–6. doi: 10.1038/pcan.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Cai Y, Shao LJ, et al. Activation of NF-{kappa}B by TMPRSS2/ERG Fusion Isoforms through Toll-Like Receptor-4. Cancer Res. 2011;71:1325–33. doi: 10.1158/0008-5472.CAN-10-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hopewell EL, Zhao W, Fulp WJ, et al. Lung tumor NF-kappaB signaling promotes T cell-mediated immune surveillance. J Clin Invest. 2013;123:2509–22. doi: 10.1172/JCI67250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Massoner P, Kugler KG, Unterberger K, et al. Characterization of transcriptional changes in ERG rearrangement-positive prostate cancer identifies the regulation of metabolic sensors such as neuropeptide Y. PLoS One. 2013;8:e55207. doi: 10.1371/journal.pone.0055207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paulo P, Ribeiro FR, Santos J, et al. Molecular subtyping of primary prostate cancer reveals specific and shared target genes of different ETS rearrangements. Neoplasia. 2012;14:600–11. [Google Scholar]
- 54.Kalina JL, Neilson DS, Lin YY, et al. Mutational Analysis of Gene Fusions Predicts Novel MHC Class I-Restricted T-Cell Epitopes and Immune Signatures in a Subset of Prostate Cancer. Clin Cancer Res. 2017;23:7596–607. doi: 10.1158/1078-0432.CCR-17-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.