Abstract
Purpose
Bevacizumab is a VEGF-specific angiogenesis inhibitor indicated as an adjunct to chemotherapy for the treatment of multiple cancers. Hypertension is commonly observed during bevacizumab treatment, and high-grade toxicity can limit therapy or lead to cardiovascular complications. The factors that contribute to interindividual variability in blood pressure rise during bevacizumab treatment are not well understood.
Experimental Design
To identify genomic regions associated with bevacizumab-induced hypertension risk, sequencing of candidate genes and flanking regulatory regions was performed on 61 bevacizumab-treated patients (19 cases developed early-onset grade 3 hypertension and 42 controls had no reported hypertension in the first six cycles of treatment). SNP-based tests for common variant associations and gene-based tests for rare variant associations were performed in 174 candidate genes.
Results
Four common variants in independent linkage disequilibrium blocks between SLC29A1 and HSP90AB1 were among the top associations. Validation in larger bevacizumab-treated cohorts supported association between rs9381299 with early grade 3+ hypertension (P = 0.01, OR 2.4) and systolic blood pressure > 180 mmHg (P = 0.02, OR 2.1). rs834576 was associated with early grade 3+ hypertension in CALGB 40502 (P = 0.03, OR 2.9). These SNP regions are enriched for regulatory elements that may potentially increase gene expression. In vitro overexpression of SLC29A1 in human endothelial cells disrupted adenosine signaling and reduced nitric oxide levels that were further lowered upon bevacizumab exposure.
Conclusions
The genomic region between SLC29A1 and HSP90AB1 and its role in regulating adenosine signaling are key targets for further investigation into the pathogenesis of bevacizumab-induced hypertension.
Keywords: Bevacizumab, hypertension, sequencing, SLC29A1, HSP90AB1
Introduction
Bevacizumab is a recombinant humanized monoclonal antibody that targets human VEGF, preventing its binding to VEGF receptor 2 (VEGFR2); its modes of action are thought to include the inhibition of angiogenesis (1) as well as enhanced antigen presentation. Bevacizumab has been approved by the U.S. Food and Drug Administration for the treatment of metastatic colorectal cancer, advanced nonsquamous non-small cell lung cancer, metastatic renal cell carcinoma, recurrent glioblastoma, advanced cervical cancer, and platinum-resistant ovarian cancer (2).
The development of hypertension is frequently observed during treatment with bevacizumab. Hypertension of all grades has been observed in up to 36% of patients treated with bevacizumab (3), with grade 3–4 hypertension reported in 1.8 to 22% of treated patients (4). Generally, the blood pressure increase can be controlled with standard antihypertensive medications. However, unmanaged hypertension may lead to serious cardiovascular complications and can occasionally be life-threatening. In the setting of severe hypertension, bevacizumab is either held temporarily or discontinued, thereby limiting therapy that may otherwise be beneficial. There are currently no validated biomarkers to predict bevacizumab toxicity, and the factors that contribute to interindividual variability in blood pressure rise during bevacizumab treatment are not well understood.
Prior studies have evaluated and identified associations between hypertension incidence during bevacizumab treatment and common single nucleotide polymorphisms (SNPs) in candidate genes encoding VEGF (5–9) and VEGFR2 (10); however, the directions of effect for several of these findings are discordant. More recent studies utilizing expanded candidate gene and genome-wide strategies have identified risk variants in SV2C (11) and FIP200 (12) and modest associations in EGLN3, EGF, and WNK1 (13). Given the heritable but complex nature of primary hypertension, the genetic architecture underlying bevacizumab-induced hypertension is also likely to be polygenic. Additional examination of genetic variation in non-VEGF pathways and of rare variants with potentially large phenotypic effects may identify novel mechanisms contributing to bevacizumab-induced hypertension.
In the present study, candidate genes were sequenced in bevacizumab-treated subjects with the objective of identifying genes harboring multiple variants associated with severe, early-onset bevacizumab-induced hypertension. Functional studies in human endothelial cells provide evidence for the potential involvement of the most significant genomic region in this dose-limiting toxicity.
Materials and Methods
Patient population
The patient cohort for this study was selected from the bevacizumab arm of Cancer and Leukemia Group B (CALGB) 80405, a phase III trial conducted to determine if the addition of cetuximab to FOLFIRI or FOLFOX chemotherapy prolongs survival compared to FOLFIRI or FOLFOX with bevacizumab in patients with untreated advanced or metastatic colorectal cancer who have KRAS wild type tumors. This trial has been previously described in detail (14). CALGB is now part of the Alliance for Clinical Trials in Oncology. A total of 899 patients accrued into the bevacizumab arm and received 5 mg/kg bevacizumab every two weeks followed by FOLFOX or FOLFIRI every two weeks; treatment was continued until disease progression, unacceptable toxicity, or surgery with curative intent. DNA was available from 581 bevacizumab-treated patients who were also enrolled in a pharmacogenetic companion study (CALGB 60501) embedded within CALGB 80405. The protocol was approved by the National Cancer Institute Adult Central IRB and by local institutional review boards. All participants provided written informed consent for pharmacogenetic sample procurement and analysis. All studies were conducted in accordance with guidelines outlined in the Declaration of Helsinki.
Phenotype
Eligibility criteria required that patients with preexisting hypertension must be well controlled (blood pressure < 160/90 mmHg) on a regimen of antihypertensive therapy. Blood pressure was measured prior to randomization, at day 1 of each eight-week treatment cycle, and every two weeks during treatment. Serious adverse events, including hypertension, were reported during each cycle. The severity of hypertension was assessed on a scale of 1–5 according to the National Cancer Institute Common Terminology Criteria for Adverse Events version 3.
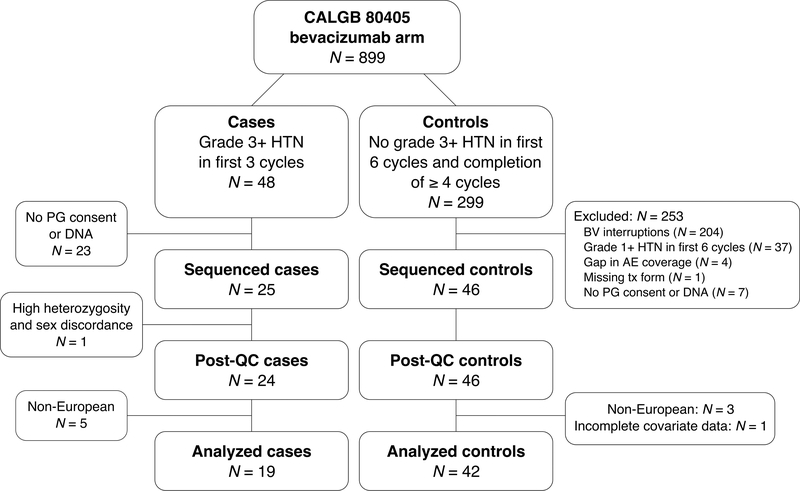
Cases and controls were selected using an extreme phenotype design to compare severe, early-onset hypertension patients to those with no reported hypertension (Fig. 1). Cases were defined as having at least one grade 3 or higher hypertension event during the first three treatment cycles. Forty-eight cases were confirmed by extensive chart reviews, with 25 having consent and available DNA. Controls were defined as having no reported hypertension during the first six treatment cycles while completing a minimum of four uninterrupted cycles with no gaps in adverse event form coverage; a cut-off of six cycles was chosen to limit the number of subjects who develop bevacizumab-induced hypertension only after longer treatment. After identifying 299 potential controls, 53 remained after exclusion for one or more criteria (Fig. 1). Forty-six controls who had both pharmacogenetic consent and available DNA were sequenced.
Figure 1. Subject selection from bevacizumab-induced hypertension extreme phenotypes in CALGB 80405.
Abbreviations used: HTN, hypertension; PG, pharmacogenetic; BV, bevacizumab; AE, adverse event; tx, treatment; QC, quality control.
Sequencing
Genomic DNA extracted from blood samples was provided by the Alliance Pathology Coordinating Office, and sequencing was performed at the UCSF Genomics Core Facility. Probes were designed to target the whole exome and intronic, UTR, and 50 kb regions upstream and downstream of selected candidate genes (Supplementary Table S1) for a total target size of 85 Mb (64 Mb standard exome + 21 Mb custom regions). Candidate genes were selected based on their documented role in VEGF signaling, endothelial cell biology, nitric oxide signaling or hypertension, as defined by assignment to a curated gene set or pathway in Gene Ontology Consortium (GO, http://www.geneontology.org/), Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/), or BioCarta (https://cgap.nci.nih.gov/Pathways/BioCarta_Pathways). GO annotations were required to be supported by experimental evidence, computational evidence, author statements, or curator statements. The gene sets and pathways that were examined for candidate gene selection are listed in Supplementary Table S2. Additional sequencing and quality control methods are described in the Supplementary Methods.
SNP-based association testing
Variants were filtered for MAF ≥ 0.10 in the study cohort (threshold chosen based on limited sample size) and LD-pruned at r2 > 0.8 before testing each variant independently for case-control association using logistic regression under an additive genetic model. Tests were adjusted for sex, age, body mass index (BMI) ≥ 25, preexisting hypertension, and preexisting diabetes, based on published data describing clinical predictors of the toxicity (15). Associations with Bonferroni-adjusted P-values < 0.05 were considered statistically significant; unadjusted P-values are reported. Top SNPs were analyzed by in silico methods described in the Supplementary Methods.
Gene-based association testing
Association testing was conducted at the gene level to evaluate combined effects from multiple rare and low frequency variants (MAF < 0.03) in each gene. Only genes containing more than one observed rare variant allele were tested. SKAT-O (16), which combines the sequence kernel association test (SKAT) and a standard burden test into a single framework and adaptively selects the best linear combination of test statistics, was used to test each gene for case-control association. All tests were adjusted for sex, age, BMI ≥ 25, preexisting hypertension, and preexisting diabetes. Associations with Bonferroni-adjusted P-values < 0.05 were considered statistically significant; unadjusted P-values are reported.
Replication analysis of top SNP associations
Top SNP associations were tested for replication in two larger, independent cohorts of bevacizumab-treated patients from clinical trials CALGB 40502 (17) and CALGB 90401 (18). Associations with systolic blood pressure (SBP) > 180 mmHg for available SNPs were also looked up in the GWAS results of a third independent cohort in the ECOG-5103 trial (11) (Supplementary Table S5).
rs3734704 and rs6902226 (proxy for rs6929249 at a physical distance of 346 bp and r2 = 0.98 in 1000G EUR) were individually genotyped in both CALGB populations. rs9381299 genotypes were extracted from existing GWAS data. rs6929249 and rs834576 genotypes in CALGB 40502 were imputed using genome-wide genotyping data and sequencing data from the 1000 Genomes Project (Michigan Imputation Server). SNPs were tested for association with early grade 3+ hypertension using logistic regression under an additive genetic model. Tests were adjusted for the same covariates as in the discovery study, where available. Early hypertension was defined as hypertension occurring within the number of treatment cycles equaling the same total exposure of bevacizumab (60 mg/kg) in the first three cycles of CALGB 80405. Associations with Bonferroni-adjusted P-values < 0.05 were considered statistically significant; unadjusted P-values are reported.
Cell culture
Human umbilical vein endothelial cells (HUVEC; C0035C; Thermo Fisher Scientific) were mycoplasma free upon purchase and maintained in endothelial cell growth medium-2 (EGM-2; Lonza) at 37 °C with 5% CO2. Cells within passages 2–5 were seeded on 24-well collagen-coated plates for experiments.
Pharmacological treatment
HUVEC were serum-starved overnight in endothelial basal medium-2 (EBM-2; Lonza) with 0.5% FBS. Cells were then treated for one hour at 37 °C with adenosine (Sigma-Aldrich) and human VEGF165 (50 ng/mL; Sigma-Aldrich). Where indicated, bevacizumab (provided by Genentech) was added at a 10X molar ratio of VEGF during the 1-hour treatment period.
For inhibitor experiments, cells were pre-incubated for 30 minutes at 37 °C with non-selective adenosine receptor antagonist CGS-15943 (1 μM; Tocris Bioscience) or ENT1 inhibitor S-(4-Nitrobenzyl)-6-thioinosine (NBTI, 1 μM; Sigma-Aldrich). Inhibitors were added at the same concentrations during the 1-hour treatment period.
Overexpression of SLC29A1 in HUVEC
At approximately 90% confluency, HUVEC were transiently transfected according to the manufacturer’s protocol with 0.75 μL/well Lipofectamine 3000 (Thermo Fisher), 1 μL/well P3000 reagent (Thermo Fisher), and 500 ng/well plasmid DNA in EGM-2. Cells were transfected with pcDNA3.1(+) vector containing human SLC29A1 cDNA (ORF clone ID OHu16500D; GenScript). HUVEC transfected with empty pcDNA3.1(+) vector (Thermo Fisher) were used as negative controls. Cells were incubated at 37 °C for 20 hours following transfection, then serum-starved for four hours (instead of overnight as in untransfected cells) prior to pharmacological treatment.
To confirm SLC29A1 overexpression, RNA was isolated using the RNeasy Mini Kit (Qiagen) and reverse transcribed using the iScript cDNA Synthesis Kit (Bio-Rad), according to the manufacturers’ protocols. Quantitative real-time PCR reactions prepared with Maxima SYBR Green/ROX qPCR Master Mix (Thermo Fisher) were run on the 7900HT Fast Real-time PCR System (Applied Biosystems) at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. qRT-PCR data were analyzed by SDS software v2.4 (Applied Biosystems).
Measurement of cyclic AMP and nitric oxide
Intracellular cyclic AMP levels were measured with a Human cAMP ELISA kit (ab133051; Abcam) following sample acetylation. Total nitrate and nitrite levels, as a measure of nitric oxide, were measured with a colorimetric Nitric Oxide Assay kit (ab65328; Abcam).
Statistical analysis
SNP-based association tests in the discovery sequencing analysis were performed using PLINK/SEQ (http://atgu.mgh.harvard.edu/plinkseq/). Gene-based SKAT-O analyses, implemented through the SKAT package (https://CRAN.R-project.org/package=SKAT), and logistic regression analyses in replication cohorts were performed using the R statistical environment (19).
To analyze functional data, linear trends across adenosine concentration levels were assessed by the Cochran-Armitage test. Differences between control and treatment groups (CGS-15943-treated vs. vehicle-treated, NBTI-treated vs. vehicle-treated, SLC29A1-transfected vs. empty vector-transfected, bevacizumab-treated vs. vehicle-treated) were analyzed using unpaired t-test or three-way ANOVA, as appropriate. Differences with two-sided P-values < 0.05 were considered statistically significant. Statistical analyses and visualization were conducted by the Kroetz laboratory and Alliance statisticians using R (19), on a data set frozen on February 26, 2013.
Results
Patient characteristics and selection
Seventy-one samples (25 cases and 46 controls) were selected for sequencing. Twelve cases developed grade 3 hypertension in the first treatment cycle, ten cases in cycle 2, and three cases in cycle 3. Following quality control procedures, one sample was excluded due to sample contamination (detected by high heterozygosity, low concordance with GWAS data, and discordant gender). A total of 61 European samples (19 cases and 42 controls) with complete sequence and phenotype data were retained for analysis. Demographic and clinical characteristics of the analyzed cohort are listed in Table 1 and, with the exception of preexisting hypertension, are similar between cases and controls and reflect the larger study population.
Table 1.
Characteristics of extreme phenotype patient subgroups
| Cases | Controls | CALGB 80405 bevacizumab arm | CALGB 80405 total population | |
|---|---|---|---|---|
| Subjects (N) | 19 | 42 | 899 | 2334 |
| Female (N, %) | 9 (47%) | 14 (33%) | 364 (40%) | 976 (42%) |
| Age, years (median, range) | 59 (31–80) | 61 (37–82) | 59 (22–85) | 59 (21–90) |
| BMI, kg/m2 (median, range) | 25.5 (17.7–40.8) | 26.9 (18.6–58.4) | 26.8 (14.7–70.7) | 27.1 (14.5–70.7) |
| Preexisting diabetes (N, %) | 3 (16%) | 4 (10%) | 139 (15%) | 347 (15%) |
| Preexisting hypertension (N, %)a | 14 (74%) | 12 (29%) | 361 (40%) | 932 (40%) |
Difference between cases vs. controls: P = 0.002.
Candidate gene analysis
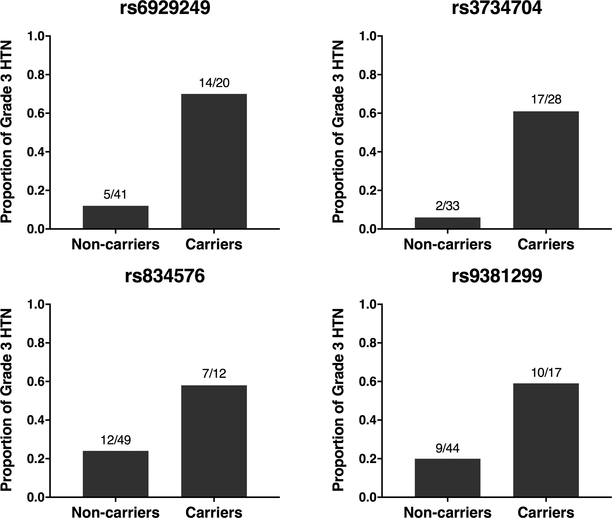
In a targeted analysis of 174 prespecified candidate genes (Supplementary Table S1) for which additional intronic, UTR, and flanking regions were sequenced, association testing was performed on 92,886 coding and noncoding variants (Supplementary Fig. S1). Common variants (MAF ≥ 0.10, N = 9,356) were analyzed in SNP-based association tests (Table 2); no variants reached Bonferroni-corrected statistical significance (P < 5.3 x 10−6). Within the ten strongest associations, four SNPs were located in the same intergenic region downstream of SLC29A1 and upstream of HSP90AB1: rs6929249, rs3734704, rs834576, and rs9381299 (Supplementary Fig. S2). For all four SNPs, the proportion of early grade 3 hypertension in minor allele carriers was consistently higher (58–70%) than in non-carriers (6–24%) (Fig. 2).
Table 2.
SNP-based analysis of common variants in candidate genes
| SNP (ref/var) | Case genotype countsa | Control genotype countsa | Case MAF | Control MAF | OR (95% CI) | Pb | Candidate genec | Function |
|---|---|---|---|---|---|---|---|---|
| rs6929249 (A/G) | 5/13/1 | 36/6/0 | 0.39 | 0.07 | 37.8 (5.8–247) | 1 x 10−4 | HSP90AB1 | upstream |
| rs3734704 (A/C) | 2/14/3 | 31/10/1 | 0.53 | 0.14 | 28.9 (4.2–199) | 6 x 10−4 | HSP90AB1 | upstream |
| rs834576 (C/A) | 12/6/1 | 37/5/0 | 0.21 | 0.06 | 19.3 (2.9–127) | 0.002 | HSP90AB1 | upstream |
| rs59189065 (G/A) | 7/11/1 | 35/7/0 | 0.34 | 0.08 | 9.9 (2.2–44.3) | 0.003 | PRKCA | intronic |
| rs2470417 (C/T) | 6/12/1 | 33/9/0 | 0.37 | 0.10 | 12.7 (2.4–67.5) | 0.003 | CACNA1C | upstream |
| rs9381299 (T/C) | 9/8/2 | 35/7/0 | 0.32 | 0.08 | 8.8 (2.0–37.8) | 0.004 | HSP90AB1 | upstream |
| rs11651806 (C/G) | 13/6/0 | 11/27/4 | 0.16 | 0.41 | 0.1 (0.0–0.5) | 0.004 | CCL2 | upstream |
| rs72869749 (C/T) | 9/9/1 | 35/7/0 | 0.29 | 0.08 | 26.1 (2.8–239) | 0.004 | PDE3B | intronic |
| rs142385484 (C/T) | 12/7/0 | 38/3/1 | 0.18 | 0.06 | 13.9 (2.3–84.2) | 0.004 | NOSIP | downstream |
| rs11739214 (G/C) | 10/6/3 | 33/9/0 | 0.32 | 0.10 | 10.1 (2.1–48.9) | 0.004 | FLT4 | downstream |
Genotype counts: Homozygous reference / heterozygous / homozygous variant.
Unadjusted P-value from logistic regression under an additive genetic model and adjusted for sex, age, BMI ≥ 25, preexisting hypertension, and preexisting diabetes.
Represents candidate gene ± 50 kb flanking region containing the sequenced variant. In some cases, this may not be the nearest gene to the variant.
CI, confidence interval
Figure 2. Proportion of early grade 3 hypertension in bevacizumab-treated patients stratified by top SLC29A1-HSP90AB1 SNP carrier status.
Variant alleles of each SNP are associated with higher incidence of early grade 3 hypertension (HTN) in analyzed subjects. Fractions represent the number of HTN cases over the total number of subjects for each carrier status.
To test the possibility that individual genes harboring multiple rare and low frequency variants contribute to toxicity risk, 35,472 variants with MAF < 0.03 in the 174 candidate gene regions were tested using SKAT-O. No gene associations met statistical significance after Bonferroni correction (P < 2.9 x 10−4; Supplementary Table S6).
In silico functional analysis
The four SNPs between SLC29A1 and HSP90AB1 identified in the candidate gene analysis are noncoding, and all proxy SNPs (r2 > 0.8) are also in noncoding regions. These variants were examined for overlap with regulatory elements inferred from functional data generated in HUVEC from the ENCODE Project (Supplementary Fig. S3). rs6929249 and rs3734704 are located within 1 kb downstream of the 3’ end of SLC29A1 in a moderately transcribed region enriched for histone modifications linked to gene activation (H3K4me1, H3K27ac, H3K36me3, H3K79me2) and binding of CTCF, which can act as an insulator. rs834576 is in a region approximately 3.8 kb downstream of SLC29A1 that is also enriched for H3K4me1, H3K27ac, and CTCF as well as additional activating histone modifications (H3K4me2, H3K4me3, H3K9ac, H2A.Z) and binding of RNA polymerase II; the variant site also maps to a CpG island, a feature often associated with promoters. rs3734704 and rs834576 also overlap with DNase I hypersensitivity peaks representing regions of open chromatin and are in genomic segments predicted to be strong enhancers. rs6929249 is in a predicted region of transcriptional transition, and rs9381299 in a predicted region of weak transcription. Similar data from the Roadmap Epigenomics Project for a diversity of cell lines corroborated the ENCODE predictions for HUVEC and support a more global role for the regions containing the rs6929249, rs3734704 and rs834576 variants as enhancers or promoters. In a search of previously published eQTL analyses, the risk allele of rs9381299 was found to be associated with increased SLC29A1 expression in monocytes (P = 3.8 x 10−5, β = 0.18) (20).
Replication analysis of top SNP associations
rs6929249 (or a proxy SNP, rs6902226), rs3734704, rs834576, and rs9381299 were tested for association with early grade 3+ hypertension in independent bevacizumab-treated cohorts within CALGB 40502 and CALGB 90401; imputed rs834576 genotypes were available only in CALGB 40502. Association of rs9381299 with SBP > 180 mmHg was also examined in ECOG-5103 GWAS data, with SBP > 180 being an even more extreme phenotype than grade 3+ hypertension, which typically occurs at SBP > 160. Other SNPs were not available for lookup in ECOG-5103. Descriptions of these studies and clinical characteristics of the cohorts used in this study are summarized in Supplementary Tables S5 and S7.
rs9381299 significantly associated with early grade 3+ hypertension in CALGB 40502 (P = 0.01, odds ratio (OR) 2.4) and with SBP > 180 in ECOG-5103 (P = 0.02, OR 2.1) with a concordant direction of effect, but did not associate with grade 3+ hypertension in CALGB 90401 (Table 3). The proportion of hypertension was consistently higher in risk variant carriers for rs9381299: 5% vs. 14% in CALGB 40502 and 7% vs. 16% in ECOG-5103 (Supplementary Fig. S4). rs834576 also associated with early grade 3+ hypertension in CALGB 40502 (P = 0.03, OR 2.9); the proportion of rs834576 risk variant carriers with hypertension was 16%, compared to 6% of those with the homozygous reference genotype (Supplementary Fig. S4). The associations of rs6929249 and rs3734704 with grade 3+ hypertension were not validated in either CALGB cohort
Table 3.
Replication analysis of top SLC29A1-HSP90AB1 SNP associations in independent bevacizumab-treated cohorts
| Study | Phenotype | SNP | Case genotype countsa | Control genotype countsa | Case MAF | Control MAF | OR (95% CI) | Pb |
|---|---|---|---|---|---|---|---|---|
| CALGB 40502 | Early grade 3+ hypertensionc |
rs6929249d | 19/10/0 | 278/98/10 | 0.17 | 0.15 | 1.1 (0.5–2.2) | 0.81 |
| rs3734704 | 11/17/1 | 222/134/30 | 0.33 | 0.25 | 1.5 (0.8–2.6) | 0.20 | ||
| rs834576d | 23/6/0 | 355/30/1 | 0.10 | 0.04 | 2.9 (1.0–7.6) | 0.03 | ||
| rs9381299 | 16/13/0 | 303/76/7 | 0.22 | 0.12 | 2.4 (1.2–4.9) | 0.01 | ||
| CALGB 90401 | Early grade 3+ hypertensionc | rs6902226e | 8/5/0 | 214/82/5 | 0.19 | 0.15 | 1.4 (0.4–3.6) | 0.57 |
| rs3734704 | 6/5/2 | 164/120/17 | 0.35 | 0.26 | 1.6 (0.6–3.8) | 0.29 | ||
| rs9381299 | 8/5/0 | 225/73/3 | 0.19 | 0.13 | 1.7 (0.5–4.7) | 0.36 | ||
| ECOG-5103 | SBP > 180 mmHg | rs9381299 | 23/16/0 | 308/79/4 | 0.21 | 0.11 | 2.1 (1.1–3.7) | 0.02 |
Genotype counts: Homozygous reference / heterozygous / homozygous variant.
Unadjusted P-value from logistic regression under an additive genetic model and adjusted for covariates listed in Supplementary Table S5.
Early hypertension is defined as hypertension occurring within the number of treatment cycles equaling the same total exposure of bevacizumab (60 mg/kg) in the first three treatment cycles of CALGB 80405.
rs6929249 and rs834576 genotypes were imputed. The imputation accuracy for these two SNPs was excellent: rs6929249: R2 = 0.95; rs834576: R2 = 0.94.
rs6902226 is a proxy for rs6929249 (r2 = 0.98).
CI, confidence interval
Functional characterization of ENT1 inhibition and SLC29A1 overexpression
SLC29A1 encodes ENT1, a member of the equilibrative nucleoside transporter family. ENT1 is responsible for regulating circulating levels of adenosine, which increases endothelial nitric oxide synthase (eNOS)-dependent activity via adenosine receptor signaling (21). eNOS also lies downstream of VEGF signaling, and activation of eNOS produces NO, a potent vasodilator. To follow up on our genetic findings and the eQTL association with SLC29A1 expression, the functional effects of SLC29A1 expression on VEGF signaling were evaluated in HUVEC.
Cyclic AMP (cAMP) levels, as a measure of adenosine receptor signaling, increased as a function of adenosine concentration (P = 0.0002; Supplementary Fig. S5). Treatment with the non-selective adenosine receptor antagonist CGS-15943 almost completely blocked adenosine-stimulated cAMP formation (P = 0.005; Supplementary Fig. S6), demonstrating the selectivity of cAMP as a measure of adenosine receptor signaling. Levels of NO, which acts as a vasodilatory signaling molecule, also increased as a function of adenosine concentration (P = 3 x 10−5; Supplementary Fig. S5) and decreased by ~50% when adenosine receptor signaling was blocked (P = 0.002; Supplementary Fig. S6), indicating that NO is also generated from pathway(s) distinct from adenosine signaling.
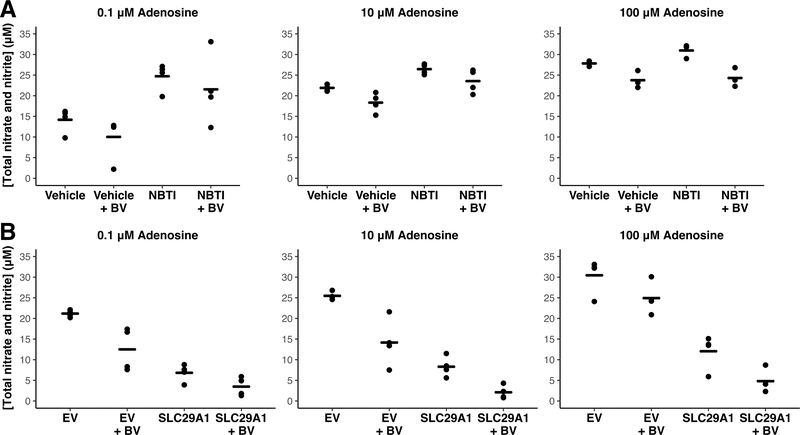
HUVEC treated with the ENT1 inhibitor NBTI showed increased cAMP levels compared to vehicle-treated cells (P = 1 x 10−10; Supplementary Fig. S7), indicating an increase in adenosine receptor signaling. NBTI-treated HUVEC also showed increased NO levels compared to controls (P = 6 x 10−13; Fig. 3A). Addition of bevacizumab resulted in significant decreases in NO levels both in the presence and absence of NBTI (P = 5 x 10−6; Fig. 3A), without an effect on cAMP levels (Supplementary Fig. S7). These results are consistent with a specific effect of bevacizumab on VEGF-mediated NO production without affecting adenosine signaling.
Figure 3. Effect of ENT1 inhibition and SLC29A1-overexpression on nitric oxide levels in HUVEC.
HUVEC were incubated in 50 ng/mL VEGF and adenosine (0.1, 10, 100 μM) for one hour. A) Treatment with the ENT1 inhibitor NBTI (1 μM) increased levels of nitric oxide (NO; P = 6 x 10−13) compared to vehicle-treated cells at all adenosine concentrations. Exposure to bevacizumab (BV; 10X molar ratio of VEGF) decreased NO levels (P = 5 x 10−6). There was no difference in the effect of BV on NBTI-treated versus vehicle-treated cells. B) Prior to pharmacological treatment, an expression vector encoding SLC29A1 cDNA was transfected into HUVEC. Empty vector (EV) was transfected into HUVEC as a negative control. SLC29A1 overexpression decreased the release of nitric oxide when compared to EV-transfected HUVEC (P < 2 x 10−16). Exposure to bevacizumab reduced NO levels in both groups and had a greater inhibitory effect in the SLC29A1-transfected cells (P = 0.02 compared to BV effect in EV). Points represent means from three or four independent experiments; lines represent group means. For each study, the effects of either NBTI treatment or SLC29A1 overexpression and the comparison of BV exposure at each adenosine concentration were analyzed by three-way ANOVA.
HUVEC transfected with SLC29A1 cDNA were confirmed to have substantially increased SLC29A1 mRNA expression compared to empty vector-transfected control cells (difference of relative expression: P = 0.0002). SLC29A1-overexpressing HUVEC, in the presence of adenosine, decreased the generation of cAMP when compared to empty vector-transfected controls at all tested adenosine concentrations (P < 2 x 10−16; Supplementary Fig. S8), indicating a decrease in adenosine receptor signaling when SLC29A1 expression is increased. SLC29A1-overexpressing HUVEC showed decreased NO levels compared to empty vector-transfected cells (P < 2 x 10−16; Fig. 3B). Addition of bevacizumab had no effect on cAMP levels (Supplementary Fig. S8) but reduced NO levels. The inhibitory effect of bevacizumab on NO levels ranged from an 18–44% decrease in empty vector-transfected cells compared to a 50–75% decrease in SLC29A1-transfected cells, demonstrating a greater reduction in NO following bevacizumab treatment under conditions of high SLC29A1 expression (P = 0.02; Fig. 3B).
Discussion
The present study used a sequencing analysis to discover genes that potentially contribute to bevacizumab-induced hypertension. Although no SNP- or gene-based associations achieved genome-wide significance, we identified SNPs with modest associations that may have regulatory effects on biological pathways related to the phenotype.
A targeted analysis examined variants within and flanking preselected candidate genes. The top ten variant associations with bevacizumab-induced hypertension included four SNPs located in the same genomic region between SLC29A1 and HSP90AB1. These SNPs are not in strong LD (r2 < 0.6, Supplementary Fig. S2 and S9), suggesting that they could have independent effects on the toxicity. Genetic variation in SLC29A1 or HSP90AB1 has not been previously associated with hypertension or other cardiovascular phenotypes. While the discovery of common variants was not the original strategy of this sequencing analysis, other than rs9381299, these SNPs are not tagged on any standard commercial GWAS or exome arrays, demonstrating the added potential of examining variants of all allele frequencies identified from sequencing data in the setting of prospectively identified clinical outcomes.
SLC29A1 encodes ENT1, which regulates adenosine levels. Increased circulating adenosine levels have been observed in ENT1 knockout mice (22), and increased extracellular adenosine concentrations are associated with reduced ENT1 expression in HUVEC (23). ENT1 also regulates eNOS-dependent NO production via adenosine receptor signaling (21). eNOS lies downstream of VEGF and has been implicated in hypertension resulting from VEGF inhibition (24).
To investigate whether SLC29A1 may influence an endothelial cell’s response to bevacizumab, functional studies were performed to determine a potential role for ENT1 and adenosine in the regulation of VEGF-dependent NO production. Under conditions of increased extracellular adenosine from pharmacological inhibition of ENT1, bevacizumab selectively inhibited a portion of NO formation without altering cAMP production, suggesting that the effects of VEGF inhibition are not simply an off-target effect on adenosine signaling. Consistent with these findings, overexpression of SLC29A1 in HUVEC caused a decrease in adenosine receptor signaling that is insensitive to VEGF inhibition while NO synthesis from the parallel VEGF pathway has enhanced sensitivity to bevacizumab under these conditions. While these observations must be further validated, we hypothesize that variants causing increased ENT1 expression and activity may reduce extracellular adenosine levels and cause vasoconstriction by impairing the endothelium’s ability to maintain a necessary amount of adenosine-stimulated NO production while under conditions of reduced VEGF signaling. Patients with increased ENT1 expression would thereby be more sensitive to changes in eNOS signaling and NO production from VEGF inhibition by bevacizumab. Similar changes in sensitivity to bevacizumab treatment could occur following changes in ENT1 expression or function as a result of epigenetic regulation of SLC29A1 or drug interactions.
Variation in other nucleoside transport and adenosine signaling pathway genes may have similar effects on the toxicity. Changes in ENT1 expression have been reported to directly affect adenosine deaminase and adenosine A2A receptor expression (25), and decreased SLC29A1 expression and ENT1 transport activity have been described during preeclampsia (26,27), a complication of pregnancy that is characterized by high blood pressure. Such changes are accompanied by observations of increased adenosine concentrations, increased adenosine-generating enzyme activity (26), reduced A2A receptor expression and enhanced A2B receptor signaling (27,28), and upregulation of soluble VEGFR1 (sFlt-1) (29,30), which binds to VEGF as bevacizumab does. Although the association of decreased SLC29A1 expression with hypertension contradicts our hypothesized mechanism, our findings still support the idea of dysregulated adenosine transport having an effect on blood pressure. Expression and activity of these proteins and of other nucleoside transporters should be considered in future studies of bevacizumab-induced hypertension, as multiple aberrations of the adenosine signaling pathway could result in a more sensitive response. Changes in adenosine transport rates, receptor binding affinities, or enzyme kinetics modulated by genetic effects or drug interactions should also be considered during functional studies of bevacizumab treatment.
Although there was more bioinformatic evidence supporting the association of the identified SLC29A1-HPS90AB1 SNPs with SLC29A1 expression and function, HSP90AB1 is also of biological interest. This gene encodes a constitutively expressed, cytosolic isoform of Hsp90, a molecular chaperone that facilitates normal folding, stability, activation, function, and proteolytic turnover of many key regulators of cell growth and survival, including eNOS (31). Reduced Hsp90 expression in primary HUVEC has been observed during preeclampsia (32), and hypertension has presented as a common adverse event during early clinical trials of Hsp90 inhibitor treatment (33,34). Hsp90 binds to eNOS and enhances its NO-synthesizing activity (35); at least in part, VEGF itself signals the binding of Hsp90 with eNOS (36,37). Disruption of Hsp90 signaling, either by chemical inhibition or mutagenesis, has been shown to attenuate VEGF-stimulated binding of Hsp90 to eNOS, NO production, and endothelium-dependent relaxation of isolated blood vessels (35,37–39). Thus, variants reducing the expression or function of Hsp90 may have similar effects as increased ENT1 function. Specifically, reduced baseline eNOS activity and NO production could amplify the effects of reduced VEGF signaling during bevacizumab treatment and contribute to the observed dysregulation of vascular tone and increased blood pressure. Future studies should explore this possibility and the complex interaction between VEGF, Hsp90 and adenosine signaling.
Based on data from our in silico analyses, the identified SNPs are located in putative regulatory regions in HUVEC. Three of the four SNPs are in moderately transcribed regions just downstream of SLC29A1 that are enriched for epigenetic marks, RNA polymerase II binding, and other genomic features associated with transcriptional activity. Together, these data suggest that this region may be part of an unannotated SLC29A1 splice variant or contain an enhancer that regulates expression of a nearby gene. Variants that alter CTCF binding sites may disrupt insulator activity and permit promoter-enhancer interactions in this region. Detailed analysis of the ENCODE data used for our in silico analyses found an abundance of disease-associated SNPs in predicted strong enhancers (40), supporting a role for the regulatory SNPs identified in these studies in the bevacizumab-induced hypertension phenotype. The fourth SNP, rs9381299, was found to be associated with increased SLC29A1 expression in monocytes but not in other more relevant tissues like the artery. Functional studies in vascular endothelial cell models are needed to better assess the SNP effects on both SLC29A1 and HSP90AB1 expression and function.
These identified SNPs were tested for association in larger, independent bevacizumab-treated cohorts. The association of rs9381299 and rs834576 with early grade 3+ hypertension in CALGB 40502 and rs9381299 with SBP > 180 mmHg in ECOG-5103 supports the discovery results. Other SNPs were not validated. Limited statistical power, differences in eligibility, clinical trial design, bevacizumab dosing, and adverse event collection, or demographic and clinical differences between study populations may explain why some associations failed to replicate. In particular, CALGB 90401 did not have preexisting hypertension data available for adjustment of the models, which might contribute to the lack of replication in this cohort. Examination of all of these SNPs in additional bevacizumab-treated cohorts is warranted.
A major limitation of this study is sample size. Due to lack of consent or available DNA and a rigorous phenotypic review, many potential subjects could not be sequenced. An extreme phenotype design was utilized to enrich sampling of causal variants with large effect sizes and to increase statistical power (41). Such an approach has been used in cohorts of similar sizes to identify novel variants associated with complex disease and pharmacogenetic traits, such as P. aeruginosa infection (42), warfarin dosing (43), drug-induced long QT interval syndrome (44), gemcitabine/carboplatin-induced myelosuppression (45), and clopidogrel response (46). Considering sample size, we recognize the limitations of the study design and approach these analyses largely as a discovery study. Replication and sequencing are needed in additional cohorts to extend these current findings.
Another challenge of this study lies in the difficulty of identifying true drug-induced hypertension. We limited our phenotype to hypertension that occurred in the first three treatment cycles, as bevacizumab-induced hypertension has been shown to develop early (15), thus removing later cases of hypertension that are more likely to be attributed to other factors. Preexisting hypertension was significantly correlated with on-treatment hypertension (P = 0.002, OR 6.7), with 14 of 19 (74%) cases having preexisting hypertension compared to 12 of 42 (29%) controls. This association is possibly confounding, and genetic models were adjusted for preexisting hypertension to minimize this effect. However, this association has been previously shown (47,48) and may possibly be informative. Hypertensive patients were required to be normotensive upon study initiation, and even while on antihypertensive medications many still developed the toxicity. This suggests that some patients may be genetically predisposed to an especially sensitive response to bevacizumab, such that even pharmacologically controlled hypertension is further exacerbated upon bevacizumab treatment. Although the present study had too few subjects to test this hypothesis, a future study stratifying by the class of antihypertensive medication could elucidate a more specific mechanism of the toxicity. The inclusion of subjects from ECOG-5103 as a replication cohort provides some insight regarding the contribution of the identified SNPs in baseline hypertension versus bevacizumab-induced hypertension. The association of rs9381299 with bevacizumab-induced hypertension was replicated in ECOG-5103, a cohort that excluded all patients with preexisting hypertension, supporting our original discovery results with unbalanced preexisting hypertension status. Post hoc analyses of the 19 cases of bevacizumab-induced hypertension used in the discovery analysis matched for preexisting hypertension status with a new control set of 49 subjects receiving at least four cycles of bevacizumab gave a similar effect size and direction as the original results for the three SNPs that could be evaluated (data not shown), further supporting our discovery findings. Future genomic and functional studies are needed to more carefully define the involvement of SLC29A1 and HSP90AB1 in these phenotypes.
Hypertension also occurs during treatment with other VEGF pathway inhibitors (15), including aflibercept, a soluble decoy receptor that binds VEGF, and small molecule VEGF receptor tyrosine kinase inhibitors (e.g., sunitinib, sorafenib). The frequency of all-grade hypertension during treatment with these small molecule inhibitors ranges from 15 to 67%, with a greater incidence observed during treatment with more potent inhibitors such as axitinib, regorafenib, and cediranib, where incidence of grade 3–4 hypertension has been reported as high as 43% (49). Genetic associations with hypertension during sunitinib treatment have also been reported and include SNPs in VEGFA, KDR, and NOS3 (eNOS) (50). These pharmacogenetic findings and the correlation of potency and hypertension incidence suggest that the hypertension is primarily an on-target effect of VEGF and eNOS inhibition, as is hypothesized for bevacizumab-induced hypertension. Therefore, our results may also be extended to the study and treatment of other VEGF inhibitor toxicities.
A better understanding of the genetic architecture of bevacizumab-induced hypertension will advance the understanding of how genetic variability influences the action of angiogenesis inhibitors and the pathogenesis of drug-induced hypertension. Furthermore, identifying genetic predictors of this toxicity could support the development of improved and novel strategies to predict and treat pharmacologically induced hypertension and aid in the selection of appropriate treatment for cancer patients. The identification of sub-populations of patients at risk for bevacizumab toxicity will support more aggressive monitoring of blood pressure and prophylactic antihypertensive treatment in these patients. Since bevacizumab treatment that may otherwise be beneficial is held or discontinued upon development of hypertension, it is clinically important to manage complications to prolong effective therapy. Blood pressure elevation itself has been proposed as a marker for VEGF inhibitor efficacy (51); if true, this group may be at a therapeutic benefit if their hypertension can be well controlled.
In this study, potential regulatory bevacizumab-induced hypertension risk variants were identified near genes not previously associated with this toxicity. This is the first study to date using sequencing to examine bevacizumab-induced hypertension. While our association results were modest, the region between SLC29A1 and HSP90AB1 as well as other genes in the adenosine signaling pathway should be prioritized for follow-up in higher-powered replication studies and in functional studies to define the role of these genes in regulation of vasodilation upon bevacizumab exposure.
Supplementary Material
Translational Relevance.
Bevacizumab use in the treatment of cancer can lead to the development of hypertension in some patients. High-grade hypertension can lead to cardiovascular complications and require delay or discontinuation of therapy. Biomarkers for the prediction of bevacizumab-induced hypertension could inform personalized treatment decisions and minimize the number of patients experiencing this dose-limiting toxicity. A sequencing analysis of candidate genes identified a putative regulatory genomic region between SLC29A1 and HSP90AB1 containing several variants associated with hypertension in bevacizumab-treated colorectal cancer patients with extreme toxicity phenotypes. Functional experiments in human endothelial cells further suggest that the expression of SLC29A1 determines in part the cellular response to inhibition of VEGF signaling. The proposed association of novel genes with bevacizumab-induced hypertension may lead to improved prediction and management of this toxicity in patients at elevated hypertension risk.
Acknowledgments
Support: Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Numbers U10CA180821 and U10CA180882 (to the Alliance for Clinical Trials in Oncology), U10CA041287, U10CA045808, U10CA047559, U10CA047577, U24CA114725, U10CA138561, U10CA180836, U10CA180838, UG1CA189858, CA180830 (H.-J. Lenz), CA180826 (H.S. Hochster), CA180797, CA189828, Alliance Clinical Trial Foundation (D.L. Kroetz and A.P. Venook), and the Pharmacogenomics Development Fund to NIH award number P30CA082103 (D.L. Kroetz). M. Li was supported in part by NIH grants T32GM007175 and F31GM113350. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of interest disclosure: The authors declare no potential conflicts of interest.
ClinicalTrials.gov Identifiers: NCT00265850 (CALGB 80405); NCT00110214 (CALGB 90401); NCT00785291 (CALGB 40502); NCT00433511 (ECOG-5103)
Sequence data for the discovery cohort is available via the NCBI dbGaP repository under accession number phs001597.v1.p1.
References
- 1.Ferrara N, Hillan KJ, Gerber HP, Novotny W. Discovery and development of bevacizumab, an anti-VEGF antibody for treating cancer. Nat Rev Drug Discov. 2004;3:391–400. doi: 10.1038/nrd1381. [DOI] [PubMed] [Google Scholar]
- 2.Ferrara N, Adamis AP. Ten years of anti-vascular endothelial growth factor therapy. Nat Rev Drug Discov. 2016;15:385–403. doi: 10.1038/nrd.2015.17. [DOI] [PubMed] [Google Scholar]
- 3.Zhu X, Wu S, Dahut WL, Parikh CR. Risks of proteinuria and hypertension with bevacizumab, an antibody against vascular endothelial growth factor: systematic review and meta-analysis. Am J Kidney Dis. 2007;49:186–93. doi: 10.1053/j.ajkd.2006.11.039. [DOI] [PubMed] [Google Scholar]
- 4.Ranpura V, Pulipati B, Chu D, Zhu X, Wu S. Increased risk of high-grade hypertension with bevacizumab in cancer patients: a meta-analysis. Am J Hypertens. 2010;23:460–8. doi: 10.1038/ajh.2010.25. [DOI] [PubMed] [Google Scholar]
- 5.Schneider BP, Wang M, Radovich M, Sledge GW, Badve S, Thor A, et al. Association of vascular endothelial growth factor and vascular endothelial growth factor receptor-2 genetic polymorphisms with outcome in a trial of paclitaxel compared with paclitaxel plus bevacizumab in advanced breast cancer: ECOG 2100. J Clin Oncol. 2008;26:4672–8. doi: 10.1200/JCO.2008.16.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Etienne-Grimaldi MC, Formento P, Degeorges A, Pierga JY, Delva R, Pivot X, et al. Prospective analysis of the impact of VEGF-A gene polymorphisms on the pharmacodynamics of bevacizumab-based therapy in metastatic breast cancer patients. Br J Clin Pharmacol. 2011;71:921–8. doi: 10.1111/j.1365-2125.2010.03896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morita S, Uehara K, Nakayama G, Shibata T, Oguri T, Inada-Inoue M, et al. Association between bevacizumab-related hypertension and vascular endothelial growth factor (VEGF) gene polymorphisms in Japanese patients with metastatic colorectal cancer. Cancer Chemother Pharmacol. 2013;71:405–11. doi: 10.1007/s00280-012-2028-2. [DOI] [PubMed] [Google Scholar]
- 8.Sibertin-Blanc C, Mancini J, Fabre A, Lagarde A, Del Grande J, Levy N, et al. Vascular Endothelial Growth Factor A c.*237C>T polymorphism is associated with bevacizumab efficacy and related hypertension in metastatic colorectal cancer. Dig Liver Dis. 2015;47:331–7. doi: 10.1016/j.dld.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 9.Gampenrieder SP, Hufnagl C, Brechelmacher S, Huemer F, Hackl H, Rinnerthaler G, et al. Endothelin-1 genetic polymorphism as predictive marker for bevacizumab in metastatic breast cancer. Pharmacogenomics J. 2016 doi: 10.1038/tpj.2016.25. [DOI] [PubMed] [Google Scholar]
- 10.Jain L, Sissung TM, Danesi R, Kohn EC, Dahut WL, Kummar S, et al. Hypertension and hand-foot skin reactions related to VEGFR2 genotype and improved clinical outcome following bevacizumab and sorafenib. J Exp Clin Cancer Res. 2010;29:95. doi: 10.1186/1756-9966-29-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider BP, Li L, Shen F, Miller KD, Radovich M, O'Neill A, et al. Genetic variant predicts bevacizumab-induced hypertension in ECOG-5103 and ECOG-2100. Br J Cancer. 2014;111:1241–8. doi: 10.1038/bjc.2014.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger MD, Yamauchi S, Cao S, Hanna DL, Sunakawa Y, Schirripa M, et al. Autophagy-related polymorphisms predict hypertension in patients with metastatic colorectal cancer treated with FOLFIRI and bevacizumab: Results from TRIBE and FIRE-3 trials. Eur J Cancer. 2017;77:13–20. doi: 10.1016/j.ejca.2017.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambrechts D, Moisse M, Delmar P, Miles DW, Leighl N, Escudier B, et al. Genetic markers of bevacizumab-induced hypertension. Angiogenesis. 2014;17:685–94. doi: 10.1007/s10456-014-9424-7. [DOI] [PubMed] [Google Scholar]
- 14.Venook AP, Niedzwiecki D, Lenz H-J, Innocenti F, Fruth B, Meyerhardt JA, et al. Effect of first-line chemotherapy combined with cetuximab or bevacizumab on overall survival in patients with KRAS wild-type advanced or metastatic colorectal cancer: a randomized clinical trial. JAMA. 2017;317:2392–401. doi: 10.1001/jama.2017.7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maitland ML, Bakris GL, Black HR, Chen HX, Durand JB, Elliott WJ, et al. Initial assessment, surveillance, and management of blood pressure in patients receiving vascular endothelial growth factor signaling pathway inhibitors. J Natl Cancer Inst. 2010;102:596–604. doi: 10.1093/jnci/djq091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, et al. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012;91:224–37. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rugo HS, Barry WT, Moreno-Aspitia A, Lyss AP, Cirrincione C, Leung E, et al. Randomized phase III trial of paclitaxel once per week compared with nanoparticle albumin-bound nab-paclitaxel once per week or ixabepilone with bevacizumab as first-line chemotherapy for locally recurrent or metastatic breast cancer: CALGB 40502/NCCTG N063H (Alliance) J Clin Oncol. 2015;33:2361–9. doi: 10.1200/JCO.2014.59.5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly WK, Halabi S, Carducci M, George D, Mahoney JF, Stadler WM, et al. Randomized, double-blind, placebo-controlled phase III trial comparing docetaxel and prednisone with or without bevacizumab in men with metastatic castration-resistant prostate cancer: CALGB 90401. J Clin Oncol. 2012;30:1534–40. doi: 10.1200/JCO.2011.39.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.R Core Team. R . A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. https://www.R-project.org/ [Google Scholar]
- 20.Fairfax BP, Makino S, Radhakrishnan J, Plant K, Leslie S, Dilthey A, et al. Genetics of gene expression in primary immune cells identifies cell type–specific master regulators and roles of HLA alleles. Nat Genet. 2012;44:502–10. doi: 10.1038/ng.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.San Martín R, Sobrevia L. Gestational diabetes and the adenosine/L-arginine/nitric oxide (ALANO) pathway in human umbilical vein endothelium. Placenta. 2006;27:1–10. doi: 10.1016/j.placenta.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 22.Rose JB, Naydenova Z, Bang A, Ramadan A, Klawitter J, Schram K, et al. Absence of equilibrative nucleoside transporter 1 in ENT1 knockout mice leads to altered nucleoside levels following hypoxic challenge. Life Sci. 2011;89:621–30. doi: 10.1016/j.lfs.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Vásquez G, Sanhueza F, Vásquez R, González M, San Martín R, Casanello P, et al. Role of adenosine transport in gestational diabetes-induced L-arginine transport and nitric oxide synthesis in human umbilical vein endothelium. J Physiol. 2004;560:111–22. doi: 10.1113/jphysiol.2004.068288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Facemire CS, Nixon AB, Griffiths R, Hurwitz H, Coffman TM. Vascular endothelial growth factor receptor 2 controls blood pressure by regulating nitric oxide synthase expression. Hypertension. 2009;54:652–8. doi: 10.1161/HYPERTENSIONAHA.109.129973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bone DBJ, Choi DS, Coe IR, Hammond JR. Nucleoside/nucleobase transport and metabolism by microvascular endothelial cells isolated from ENT1−/− mice. Am J Physiol Heart Circ Physiol. 2010;299:H847–56. doi: 10.1152/ajpheart.00018.2010. [DOI] [PubMed] [Google Scholar]
- 26.Salsoso R, Farías M, Gutiérrez J, Pardo F, Chiarello DI, Toledo F, et al. Adenosine and preeclampsia . Mol Aspects Med. 2017 doi: 10.1016/j.mam.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Escudero C, Casanello P, Sobrevia L. Human equilibrative nucleoside transporters 1 and 2 may be differentially modulated by A2B adenosine receptors in placenta microvascular endothelial cells from pre-eclampsia. Placenta. 2008;29:816–25. doi: 10.1016/j.placenta.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 28.Acurio J, Troncoso F, Bertoglia P, Salomon C, Aguayo C, Sobrevia L, et al. Potential role of A2B adenosine receptors on proliferation/migration of fetal endothelium derived from preeclamptic pregnancies. Biomed Res Int. 2014;2014:274507. doi: 10.1155/2014/274507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Powe CE, Levine RJ, Karumanchi SA. Preeclampsia, a disease of the maternal endothelium: the role of antiangiogenic factors and implications for later cardiovascular disease. Circulation. 2011;123:2856–69. doi: 10.1161/CIRCULATIONAHA.109.853127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iriyama T, Sun K, Parchim NF, Li J, Zhao C, Song A, et al. Elevated placental adenosine signaling contributes to the pathogenesis of preeclampsia. Circulation. 2015;131:730–41. doi: 10.1161/CIRCULATIONAHA.114.013740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–28. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 32.Gu Y, Lewis DF, Zhang Y, Groome LJ, Wang Y. Increased superoxide generation and decreased stress protein Hsp90 expression in human umbilical cord vein endothelial cells (HUVECs) from pregnancies complicated by preeclampsia. Hypertens Pregnancy. 2006;25:169–82. doi: 10.1080/10641950600912950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yong K, Cavet J, Johnson P, Morgan G, Williams C, Nakashima D, et al. Phase I study of KW-2478, a novel Hsp90 inhibitor, in patients with B-cell malignancies. Br J Cancer. 2016;114:7–13. doi: 10.1038/bjc.2015.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maddocks K, Hertlein E, Chen TL, Wagner AJ, Ling Y, Flynn J, et al. A phase I trial of the intravenous Hsp90 inhibitor alvespimycin (17-DMAG) in patients with relapsed chronic lymphocytic leukemia/small lymphocytic lymphoma. Leuk Lymphoma. 2016;57:2212–5. doi: 10.3109/10428194.2015.1129536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.García-Cardeña G, Fan R, Shah V, Sorrentino R, Cirino G, Papapetropoulos A, et al. Dynamic activation of endothelial nitric oxide synthase by Hsp90. Nature. 1998;392:821–4. doi: 10.1038/33934. [DOI] [PubMed] [Google Scholar]
- 36.Brouet A, Sonveaux P, Dessy C, Balligand J-L, Feron O. Hsp90 ensures the transition from the early Ca2+-dependent to the late phosphorylation-dependent activation of the endothelial nitric-oxide synthase in vascular endothelial growth factor-exposed endothelial cells. J Biol Chem. 2001;276:32663–9. doi: 10.1074/jbc.M101371200. [DOI] [PubMed] [Google Scholar]
- 37.Duval M, Le Bœuf F, Huot J, Gratton J-P. Src-mediated phosphorylation of Hsp90 in response to vascular endothelial growth factor (VEGF) is required for VEGF receptor-2 signaling to endothelial NO synthase. Mol Biol Cell. 2007;18:4659–68. doi: 10.1091/mbc.E07-05-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papapetropoulos A, García-Cardeña G, Madri JA, Sessa WC. Nitric oxide production contributes to the angiogenic properties of vascular endothelial growth factor in human endothelial cells. J Clin Invest. 1997;100:3131–9. doi: 10.1172/JCI119868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miao RQ, Fontana J, Fulton D, Lin MI, Harrison KD, Sessa WC. Dominant-negative Hsp90 reduces VEGF-stimulated nitric oxide release and migration in endothelial cells. Arterioscler Thromb Vasc Biol. 2008;28:105–11. doi: 10.1161/ATVBAHA.107.155499. [DOI] [PubMed] [Google Scholar]
- 40.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–9. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barnett IJ, Lee S, Lin X. Detecting rare variant effects using extreme phenotype sampling in sequencing association studies. Genet Epidemiol. 2013;37:142–51. doi: 10.1002/gepi.21699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emond MJ, Louie T, Emerson J, Zhao W, Mathias RA, Knowles MR, et al. Exome sequencing of extreme phenotypes identifies DCTN4 as a modifier of chronic Pseudomonas aeruginosa infection in cystic fibrosis. Nat Genet. 2012;44:886–9. doi: 10.1038/ng.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daneshjou R, Gamazon ER, Burkley B, Cavallari LH, Johnson JA, Klein TE, et al. Genetic variant in folate homeostasis is associated with lower warfarin dose in African Americans. Blood. 2014;124:2298–305. doi: 10.1182/blood-2014-04-568436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weeke P, Mosley JD, Hanna D, Delaney JT, Shaffer C, Wells QS, et al. Exome sequencing implicates an increased burden of rare potassium channel variants in the risk of drug-induced long QT interval syndrome. J Am Coll Cardiol. 2014;63:1430–7. doi: 10.1016/j.jacc.2014.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gréen H, Hasmats J, Kupershmidt I, Edsgärd D, de Petris L, Lewensohn R, et al. Using whole-exome sequencing to identify genetic markers for carboplatin and gemcitabine-induced toxicities. Clin Cancer Res. 2016;22:366–73. doi: 10.1158/1078-0432.CCR-15-0964. [DOI] [PubMed] [Google Scholar]
- 46.Scott SA, Collet J-P, Baber U, Yang Y, Peter I, Linderman M, et al. Exome sequencing of extreme clopidogrel response phenotypes identifies B4GALT2 as a determinant of on-treatment platelet reactivity. Clin Pharmacol Ther. 2016;100:287–94. doi: 10.1002/cpt.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wicki A, Hermann F, Prêtre V, Winterhalder R, Kueng M, von Moos R, et al. Pre-existing antihypertensive treatment predicts early increase in blood pressure during bevacizumab therapy: the prospective AVALUE cohort study. Oncol Res Treat. 2014;37:230–6. doi: 10.1159/000362376. [DOI] [PubMed] [Google Scholar]
- 48.Hamnvik OP, Choueiri TK, Turchin A, McKay RR, Goyal L, Davis M, et al. Clinical risk factors for the development of hypertension in patients treated with inhibitors of the VEGF signaling pathway. Cancer. 2015;121:311–9. doi: 10.1002/cncr.28972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brinda BJ, Viganego F, Vo T, Dolan D, Fradley MG. Anti-VEGF-induced hypertension: a review of pathophysiology and treatment options. Curr Treat Options Cardiovasc Med. 2016;18:33. doi: 10.1007/s11936-016-0452-z. [DOI] [PubMed] [Google Scholar]
- 50.Semeniuk-Wojtaś A, Lubas A, Stec R, Szczylik C, Niemczyk S. Influence of tyrosine kinase inhibitors on hypertension and nephrotoxicity in metastatic renal cell cancer patients. Int J Mol Sci. 2016;17:2073. doi: 10.3390/ijms17122073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maitland ML, Moshier K, Imperial J, Kasza KE, Karrison T, Elliott W, et al. Blood pressure (BP) as a biomarker for sorafenib (S), an inhibitor of the vascular endothelial growth factor (VEGF) signaling pathway. J Clin Oncol (Meeting Abstracts) 2006;24:2035. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.