Abstract
Selenium (Se) is a redox-active environmental mineral that is converted to only a small number of metabolites and required for a relatively small number of mammalian enzymes. Despite this, dietary and environmental Se has extensive impact on every layer of omics space. This highlights a need for global network response structures to provide reference for targeted, hypothesis-driven Se research. In this review, we survey the Se research literature from the perspective of the responsive physical and chemical barrier between an organism (functional genome) and its environment (exposome), which we have previously termed the redox interface. Recent advances in metabolomics allow molecular phenotyping of the integrated genome-metabolome-exposome structure. Use of metabolomics with transcriptomics to map functional network responses to supplemental Se in mice revealed complex network responses linked to dyslipidemia and weight gain. Central metabolic hubs in the network structure in liver were not directly linked to transcripts for selenoproteins but were, instead, linked to transcripts for glucose transport and fatty acid β-oxidation. The experimental results confirm the survey of research literature in showing that Se interacts with the functional genome through a complex network response structure. The results imply that systematic application of data-driven integrated omics methods to models with controlled Se exposure could disentangle health benefits and risks from Se exposures and also serve more broadly as an experimental paradigm for exposome research.
Keywords: dietary supplement, environmental toxicity, integrative omics, nutritional requirements, oxidative stress, selenium metabolism
Graphical abstract
I. Introduction
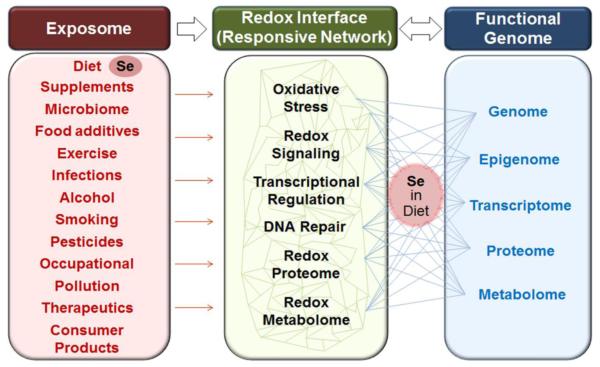
Selenium (Se) is a redox-active metalloid with complex roles in redox biology and medicine. Properties of Se allow stable structures with multiple oxidation states (−2, +2, +4, and +6), covalent bonding with non-metals (e.g., carbon) and also strong coordination with metals [e.g., cadmium Cd)]. These properties provide biological value for Se in enzymes with relatively specific functions in detoxification and also in systems with very broad integrative functions. These uses of Se share a common characteristic of being components of the redox interface through which an individual utilizes environmental resources and responds to challenges to maintain cellular and molecular organization. The notion of a redox interface was introduced as a way to map out the multiple, and often overlapping, functional systems that serve to protect relatively reduced intracellular redox elements from relatively oxidizing (O2-rich) environmental conditions [1]. This can be defined as the components of the physical, chemical, enzymatic and sequestration/chelation barrier system that protect redox elements of a living organism from environmental challenges. This is shown conceptually for Se in Fig 1 based upon an earlier depiction of the redox interface between the exposome and functional genome [1]. In this schematic, Se is included as part of the exposome (left) because this is the primary exposure for most individuals. In addition, exposure from dietary supplements, occupational, therapeutic, consumer product and pollution sources can occur. On the right-hand side of this figure, the functional genome is represented by the omics layers, each of which requires or responds to Se exposure in the diet. Response at each level impacts multiple nodes within the network structure (Fig 1, center) which serves as the responsive physical and chemical barrier between the individual and his/her environment. Additional description of the roles of Se in this network structure are provided below. Delineation of this organizational structure will provide a systems level understanding of Se biology, including Se nutrition as well as Se toxicology, and ultimately lead to improved Se-related therapeutics in cancer and age-related disease.
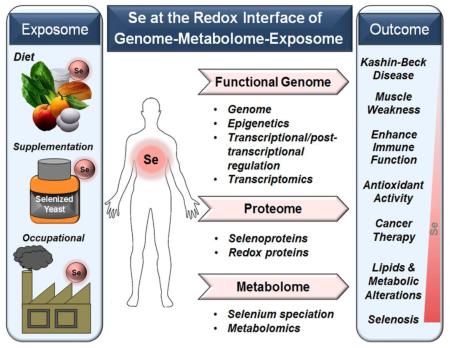
Fig. 1. Selenium at the Redox Interface.
The exposome (left) includes environmental exposures that complement the genome as determinants of health and disease. Se is an essential redox-active mineral present in mammalian food and environment. Se-dependent enzymes play a role in maintenance of macromolecular structure and function and also participate in the responsive physical and chemical barrier between an organism and its environment termed the redox interface. The redox interface (center) is a network of systems that function together to enable beneficial effects and protect against harmful environmental exposures. Se has a relatively limited number of metabolites and is required for only a small number of proteins, yet variation in Se impacts every level of omics space (right) and broadly impacts the redox interface network structure. Development of an understanding of this Se-related network structure will improve evaluation of health benefits and risks from Se and also serve as an example to more broadly develop systematic knowledge of the broad range of environmental exposures and also their interactions with variable Se intake.
The present review on Se at the redox interface is founded upon recent advances in redox theory, omics technologies and data-driven omics analyses, which have converged to create new opportunities to understand the organization and function of Se in complex systems. Redox theory was developed to complement genetics with four simple principles for molecular organization and function of complex living systems [2]. These principles provide a central logic to how cells maintain bioenergetics and metabolism and link these to macromolecular structure and function through reversible protein switches. Activation and deactivation of these protein switches in turn enables spatial and temporal organization within cells and cell populations. The collective function of these connections within an organism creates an adaptive network through which a genome utilizes environmental resources and protects against environmental threats (Fig 1).
Maturation of omics technologies has resulted in recognition that omics layers do not exist in isolation and that individual omics layers did not evolve in isolation. Indeed, data-driven omics integration now shows that transcripts can serve as central hubs for metabolites and metabolites can serve as central hubs for transcripts [3-5]. Thus, science has moved rapidly from reductionist models with a single cause and single effect to network models in which complex inter-omic interactions can be visualized and studied [6-9]. The reductionist and global approaches are complementary, with improved integrative tools being catalysts for transition in approaches and interpretations.
Consideration of Se at the redox interface offers an important prototype for broader consideration of genome-exposome interactions. Se is a component of the exposome, with human exposures ranging from nutritional deficiencies to excess environmental exposures which result in toxicities. Adaptation to variation in Se requires a broad range of compensatory responses. At the same time, only a very limited number of genes encode Se-containing proteins (25 selenoproteins in humans and 24 in mice), so only a small number of proteins are directly affected. The recent development of high-resolution metabolomics (HRM), with detection of tens of thousands chemical signals in biological extracts [10, 11], allows deep phenotyping of responses to variation in Se exposure [12]. HRM uses liquid chromatography (LC) or gas chromatography (GC) with ultra-high-resolution mass spectrometry and advanced data extraction algorithms to extend metabolic coverage beyond that obtained with commonly used targeted metabolomics platforms [11-13]. This detailed metabolic phenotyping enables connection of genetic responses to environmental Se exposures, i.e., a genome × metabolome × exposome (G × M × E) analysis. Together with the extensive knowledgebase of Se nutrition, molecular biology, biochemistry and toxicology, an integrated systems view of Se biology is beginning to emerge, allowing greater understanding and precision for the nutritional requirements and toxicity of Se.
In this review, we start with an overview of Se as an integral component of the functional genome, follow with a summary of the Se metabolome, continue with a review of Se exposures through the human exposome, and finish with recent data-driven integration of the G × M × E of Se. The results show that an integrated omics approach can help define optimal Se intake. The results also indicate that integrated omics tools can help address complex questions of metal-metal interactions in nutrition and environmental health.
II. Se as an Integral Component of the Functional Genome
The functional genome is a global term encompassing the complex relationship between the genotype and phenotype in an organism. Epigenetic regulation, gene transcription, translation, and protein function are studied as components of functional genomics research in an attempt to understand biological outcome. Functional genomics thereby includes the integration of vast information obtained by high throughput methodologies, approaching a comprehensive view of the function of a cell. Se, an essential dietary metal, forms an integral component of this functional genome. As illustrated in Fig 2, the effect of Se is pronounced not only at the gene level but at multiple layers of functional genomics including pre-transcriptional, post-transcriptional and post-translational as well. This section highlights the role of Se at the different layers of the functional genome.
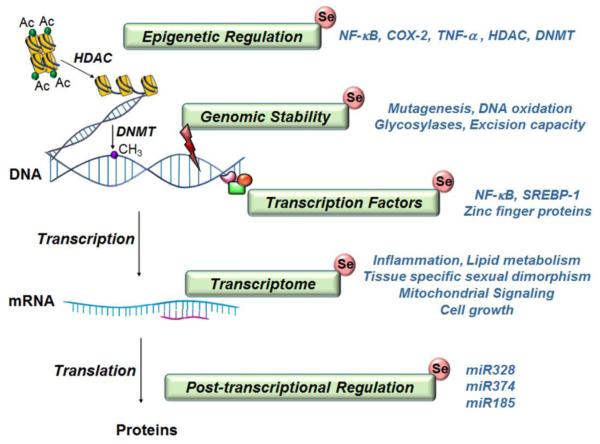
Fig. 2. Se has impacts on functional genomics.
Se mediates epigenetic regulation through changes in histone acetylation and DNA methylation potentially by alterations in histone deacetylases (HDAC) and DNA methyltransferases (DNMT). Low levels of Se lead to DNA damage and thereby impact genomic stability. Seleno-compounds decrease mutagenesis and DNA oxidation and increase DNA repair through glycosylases and excision repair. Se modulates transcription factors and thereby influences their function; examples include NF-κB for oxidative stress related pathways, zinc finger proteins for genomic stability and cell cycle, and SREBP-1 for lipid regulation. Extensive alterations in the global transcriptome occur in multiple tissue types and across multiple species in response to Se deficiency or excess selenium and affects immunity, cell growth and lipid metabolism. Post-transcriptional regulation involves influence of Se on multiple miRNAs responsible for protein synthesis and abundace in cell apoptosis or regulation of selenoproteins.
A. Selenoprotein genes
Se is co-translationally inserted in protein as the 21st amino acid, selenocysteine (Sec) and accounts for a vast majority of the biological activities of Se. The UGA codon in RNA serves as both a termination codon and, in selected contexts, as a Sec codon. Twenty-five selenoprotein genes have been identified in sequenced mammalian genomes [14]. These genes encode selenoproteins participating in multiple aspects of the cells such as redox signaling (GPX1, GPX3, GPX4, TRXRD1, TRXRD2) [15-18], protein folding and degradation (SEP15, SELS) and metabolism (SEPP1, SPS1, SPCS), which in turn alter regulation and gene expression [19, 20].
While Se incorporation into these proteins is essential for function, it is also well known that Se deficiency or excess regulates the transcription of these selenoproteins [21, 22]. Depending on Se dose, diverse effects of Se have been observed on different cellular functions. Hence Se affects several cellular and genetic functions associated with immunity [21], energy metabolism [23, 24] and cancer [25, 26]. Some of the diseases associated with selenoprotein gene polymorphisms include Kashin-Beck disease [27, 28], thyroid disease [29] and cancers, e.g., colorectal [30, 31], prostate [32, 33] and breast [34]. These effects may be related to Se incorporation into protein when Se replaces sulfur in the biosynthesis of cysteine or methionine, thereby replacing these natural amino acids [35, 36].
The use of Se for glutathione peroxidase and thioredoxin reductase activities in redox homeostasis has been extensively studied [37-39], especially concerning the benefit in reactivity to incorporation of selenolate compared to the typical thiolate in biological systems [40-42]. Recent studies describe how factors such as the increased electrophilicity and reduced pKa of Sec (~5.2) versus Cys (~8.3) contribute to the effectiveness of Sec in enzyme catalysis [43-47]. In redox-regulated proteins, the use of Se instead of sulfur is thought to be favored to tune redox processes due to its orders of magnitude faster kinetics [48]. Ultimately this benefits the oxidoreductase catalysis activities that depend on the presence of Sec at the enzymatic active site (reviewed in [44]). Se is also incorporated into proteins as selenomethionine (SeMet), but this is generally believed to be a non-specific incorporation into protein. Alternatively, such Se-containing proteins could provide a way to sense excess Se and/or contribute to cellular dysfunction and toxicity.
B. Se affects epigenetic regulation
Se has long-term effects on gene expression through epigenetic mechanisms, such as acetylation/deacetylation of histone proteins (Fig 2, Epigenetic Regulation). Such epigenetic mechanisms modulate gene expression while the genetic sequence remains intact. Dysregulation of histone acetyltransferases and histone deacetylases have been implicated in a wide variety of diseases like cancer and inflammation [10–13]. Se supplementation in the form of selenite, which is a strong oxidant, in primary and immortalized macrophages, decreased histone acetylation in inflammation related genes such as NF-κB, COX-2 and TNF-α promoters. A similar decrease in histone acetylation by dietary levels of Se was also seen in colonic extracts from mice with inflammation induced by dextran sulfate [49]. The results suggest that the decrease in histone acetylation was due to inhibition in HAT activity (histone acetyltransferase). In addition to selenite, a number of other selenocompounds have been demonstrated as inhibitors of HDAC activity, as observed in human prostate cancer and melanoma, and in HeLa cell culture studies [50-52]. Long-term adaptation to dietary or environmental Se is also indicated by strong correlation of maternal Se levels with methylation patterns in offspring [53].
DNA methylation is another common epigenetic mechanism by which gene expression is regulated. Selenium in the form of SeMet increases global hepatic DNA methylation potentially by effects on one carbon metabolism (methyl donor for DNA methylation), including higher SAM/SAH ratio and serine hydroxymethyltransferase mRNA level [54]. In contrast, in a majority of cancer cells, Se compounds such as selenite and methylselininic acid (MSA) cause inhibition of DNMT activity or protein levels and decreased DNA methylation [49, 55-57]. The multiple types of Se compounds and differences in activities create a range of possible approaches so that epigenetic regulation by Se is an attractive approach for new cancer therapeutics. At the same time, these effects on DNA methylation suggest possible roles in cancer prevention which could be independent of previously studied antioxidant effects.
C. Se alters genomic stability
Low Se levels have been linked to cancer with several lines of evidence indicating an effect on genomic stability (Fig 2, Genomic Stability), and human interventional studies have suggested Se supplementation to be useful as a preventive strategy [58-60]. Lower Se levels have been associated with higher accumulated gene damage in high risk group for prostate cancer [61]. DNA damage measured by comet assay show a nonlinear dose response with increased damage induced by both low and high Se levels [62]. In further support of possible anti-cancer activity, Se protects against DNA damage from ionizing radiation [63] and UV light [64]. Selenite and SeMet are the most widely used compounds with extensive evidence for inhibition of mutagenesis, increase in excision capacity, increase in activity of DNA repair enzymes like glycosylases and reduction in DNA oxidation [65-67]. Collectively, the results show that Se influences the genome through maintenance of genomic stability. The complexity of the processes and forms of Se often preclude precise understanding of the mechanistic details. Application of integrated network approaches has the potential to improve this understanding.
D. Se modulates transcription factors
Se contributes to regulation of the functional genome through activation and repression of transcription factors controlling gene expression (Fig 2, Transcription Factors). Se at physiological levels blocks activation of the transcription factor NF-κB that regulates inflammatory gene expression [68, 69]. NF-κB, a cellular responder to oxidative stress, also regulates selenoprotein gene expression such as SELENOS, GPX4 and DIO2 [70]. This property of Se is widely being explored in cancer prevention [71]. Redox-active Se compounds modulate zinc finger motifs through oxidation. This influences gene expression and genomic stability because zinc finger motifs are an important component of multiple transcription factors, cell cycle and DNA repair proteins [72]. In the mouse liver, Se upregulates sterol regulatory element-binding transcription factor 1 (Srebf1) and thereby regulates lipid-associated genes [54]. While redox mechanisms are indicated, whether these occur by non-specific oxidative reactions of inorganic Se or specific biomolecular interactions of Se is difficult to resolve. It is noteworthy, however, that cellular studies suggest selenoproteins such as thioredoxins and GPX1 also regulate transcription factors, potentially through their antioxidant effect [73, 74].
E. Se alters the transcriptome
Transcriptomic analysis of selenoprotein genes has revealed extensive effects on differential gene expression pattern based on Se concentration and tissue of study (Fig 2, Transcriptome). Studies performed in 8 founder strains of mice, showed significant down-regulation of GPX1, SELENOW, and SELENOH transcripts in Se-deficient mice [75-78]. The regulation of selenoprotein transcripts by Se is not known, but for GPX1 and GPX4 mRNA, regulation is suggested to occur post-transcriptionally through nonsense-mediated decay, potentially due to the insufficiency of Sec-tRNA, but may occur through other factors as well [79, 80]. Global analysis of the seleno-transcriptome expression in human breast cancer MCF-7 and MDA-MB231 cell lines compared with healthy breast MCF-10A cells revealed downregulation of GPX1, GPX4, GPX5 and GPX7 and upregulation of DIO2, GPX2 and GPX3 genes [81].
While targeted transcriptomic pathways provide key information on Se-dependent responses, untargeted transcriptomics pathways reveal a comprehensive view of alterations in cellular patterns and insights into regulation of non-selenoprotein transcripts and related function. Transcriptomics analysis in human rectal mucosa in response to suboptimal Se levels showed cytoskeleton remodeling and reduction in inflammatory and immune responses. The differentially expressed genes belonged to cancer (80%), gastrointestinal diseases (58%), and inflammatory disease (39%) functional pathways [82]. Transcriptome analysis of goose T cells exposed to sodium selenite for 24 h showed that multiple selenoproteins involved in promotion of immune function and lysosome pathway related genes were promoted by Se stimulation, while the heat-shock proteins (HSP), interleukins (IL) and interferons were mainly down-regulated, suggesting weakened cytokine responses [22]. In relation to dietary Se and metabolism in juvenile rainbow trout, hepatic transcriptomic data further demonstrate that dietary Se increases the expression of networks for fatty acid synthesis and growth-related signaling cascades [23]. As discussed in more detail below, Se supplementation in mice to increase from an adequate intake (AI) to a four-fold higher intake approaching the tolerable upper intake level (UL) showed similar effects on liver transcription.
Selenium is also known to mediate responses through post transcriptional regulation as noted by its influence on microRNAs (Fig 2, Post-transcriptional regulation). Selenium deficiency in human colon adenocarcinoma cells (Caco-2) demonstrated altered expression of miR185, which in turn is responsible for expression of selenoprotein GPX2 and SEPSH2 [83]. Sodium selenite supplementation decreased miR328 levels (involved in cell apoptosis) in H9c2 cardiomyocyte cells [84]. In rat cardiomyocytes, miRNA microarray analysis showed highest expression change in miR374 in response to selenium deficiency [85]. Together with the transcriptome data above, these results show that variation in Se exposure has widespread effects on the functional genome. Such a range of molecular responses implies that effects occur through a global network structure response rather than through simple cause-effect relationships.
III. Se in the Metabolome
The quantitatively most important connection between the proteome and metabolome of animals occurs through two key Se-containing aminoacids, Sec and SeMet (Fig 3) [86]. Sec has the highest biologically activity and is different from Cys in several aspects. Sec contains Se in the selenol form; at physiologic pH, the selenol group is largely ionized to the selenolate, making it a more reactive nucleophile than the thiol of Cys. Selenols also have more negative redox potentials than thiols, and selanyl radicals are more easily produced than thiyl radicals. Free Sec exists at very low levels and at excess can cause toxicity to animal cells (IC50 in the micromolar range) via mechanisms involving oxidative stress, disruption in protein degradation, unfolded protein response and endoplasmic reticulum (ER) stress [87].
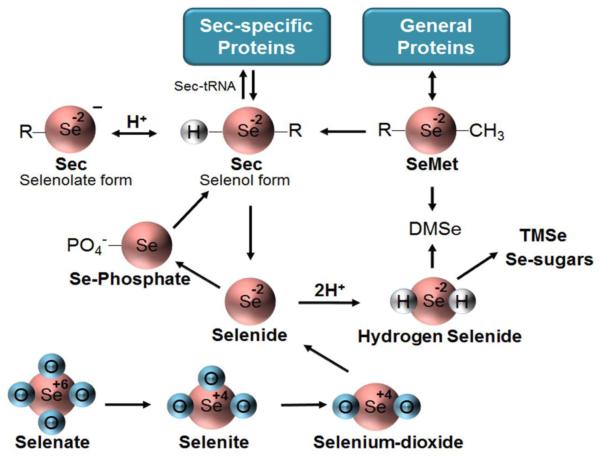
Fig. 3. Bioactive Selenochemicals.
Se is present in biological systems in many forms, both inorganic and organic, with Se at different oxidation states (shown inside circles) and the chemicals present at different ionization states (charge given outside circle). Inorganic selenooxyanions (bottom), such as selenate, selenite, and selenium dioxide, are reduced to hydrogen selenide through the glutathione and thioredoxin systems. Selenide is present as HSe- at physiologic pH, converted to Se-phosphate and then to selenocysteine (Sec), existing mostly as the selenolate at physiologic pH. Sec is then incorporated into proteins (Sec-specific Proteins) via Sec-tRNA. Selenocysteine lysase rapidly converts Sec back to inorganic selenium quickly yielding selenide restarting the cycle. Selenomethionone (SeMet), a seleno-based nutrient absorbed from mammalian diets, can be incorporated into proteins in a non-specific manner (General Proteins), and can be converted to Sec or eliminated via conversion to dimethylselenide (DMSe). Hydrogen selenide also undergoes methylation to form either DMSe or trimethyl-selenide (TMSe) or can undergo conjugation with a sugar to be excreted from the biological system.
Unlike Sec, SeMet is relatively inactive and without apparent biomedical function distinct from Met because ribosomal machinery for protein synthesis does not distinguish between SeMet and Met. Consequently, the content of SeMet in proteins is proportional to the relative abundance of SeMet and Met in the organism. The dose of SeMet necessary to elicit toxicity in cultured cells varies vastly from micromolar to millimolar range and is highly dependent on the Met concentrations [59, 88]. Toxicity from SeMet is thought to arise from metabolism to other species of Se, most importantly Sec [87]. To gain a better understanding of the range of naturally occurring Se compounds in animals, a broader consideration of Se in microorganisms, plants and fungi is necessary.
A. Se in microorganisms, fungi and plants
The requirement for Se and distribution of selenoproteins are widespread in all forms of life. Sec-containing proteins have been identified in bacteria, archaea and eukaryotes, and the number of annotated selenoproteins in sequenced genome of diverse species has increased dramatically in recent years (provided in http://www.selenodb.org) [89]. The number and function of selenoproteins varies extensively across individual species [90-92]. Microorganisms require Se for Sec in selenoenzymes, mainly in glycine reductase, formate dehydrogenase and NiFeSe hydrogenase [93, 94]. The contribution of Se to support functions of selenoproteins has received growing interest in the study of gut microbiome homeostasis in mammals [95]. Se intervention studies with germ-free mice show that dietary Se increases the diversity of the gastrointestinal microbiota and affects composition and colonization of the gastrointestinal microflora which in turn changes the host Se status and selenoproteome [96]. Oxidized forms of Se, i.e., Se oxyanions, can also serve as electron acceptors in anaerobic respiration, forming nanoparticles of elemental Se [93] (Fig 3). An additional common metabolic transformation of Se in prokaryotes involves methylation, catalyzed by several methyltransferases [97].
Since the 1950s, the metabolism of Se in yeast has been extensively studied to elucidate molecular mechanisms of Se metabolism and toxicity. This research has yielded Se enriched yeast as both a model for Se research and an important form for dietary Se supplementation. In consideration of yeast for dietary supplementation, it is important to keep in mind that selenoproteins are completely absent in yeast and higher plants [91]. This means that yeast and plants do not have specific mechanisms to directly incorporate Sec into proteins. As a result, in yeasts and higher plants, inorganic selenium is used in the sulfur assimilation pathway to form selenoamino acids which are incorporated non-specifically into proteins in the place of methionine and cysteine. The differences of yeast and animal metabolism have been reviewed and also compared to plant metabolism [87]. In addition to the lack of specific mechanisms to incorporate Sec into proteins, differences exist in the trans-sulfuration (and trans-selenylation) pathway, with animals having unidirectional conversion of Met to Cys, plants having unidirectional conversion of Cys to Met and yeast having bidirectional interconversions. Other differences include de-selenylation mechanisms and in some plants, capacity to generate S-methyl-Sec.
Rao et al. [98] used high-resolution mass spectrometry to map out various oxidized and conjugated forms of Se metabolites in Se-enriched yeast. Results showed that the spectrum of selenocompounds includes many small molecules with Se replacing sulfur, such as analogues of glutathione, γ-glutamylcysteine, homocysteine and respective diseleno-compounds. Further high-resolution mass spectrometry analysis based upon selenium isotopic pattern demonstrated 64 Se metabolites, including selenoethers, Sec-containing di- and tripeptides, and selenoadenosine metabolites [99]. The results show that widespread substitution of Se for S occurs in yeast metabolism and raises the possibility that these Se metabolites may also be found in humans consuming Se-enriched yeast.
Higher plants and fungi do not require Se yet metabolize Se as part of mechanisms to protect against Se toxicity [86]. Plants can readily absorb the inorganic Se forms (e.g. selenate and selenite) (Fig 3) from soil and incorporate Se into Sec and SeMet. A detailed description of Se metabolism in plants was provided by Pilon-Smits [100]. Excess SeMet is converted to volatile dimethylselenide (DMSe), while Sec can be converted to elemental Se and Se-methylselenocysteine (SeMSC). The later may be converted to Se-methylselenomethionine and then to volatile dimethyldiselenide (DMDSe) for elimination. Se can also be accumulated to high levels by members of the genus Astragalus (Fabaceae). Animals that ingest these hyperaccumulator plants may convert methyl-Sec to Sec leading to Se poisoning in animals consuming these plants [101]. Other forms identified in plants include selenocystathionine (converted from Sec), γ-glutamyl-Se-methyl-Sec (from SeMSC), Se-adenosyl-SeMet and Se-adenosyl-Sec (both from SeMet), γ-glutamyl-selenocystathionine, selenopeptides and selenohomocysteine [100]. A few of the forms are especially of interest, such as SeMSC and γ-glutamyl-Se-methyl-Sec, due to their therapeutic application in cancer treatment. However, despite the complex pathways of Se metabolism in plants, network analyses are not available and most of the understanding in Se metabolism is limited to the few major dietary forms (e.g. SeMet).
B. Metabolism of dietary Se in animals
Central reactions in the metabolism of Se in animal cells are well understood and have been extensively reviewed [87, 102-105]). The KEGG database provides a list of enzymes and selenocompounds that have been identified in these metabolic pathways (http://www.genome.jp/kegg-bin/show_pathway?map00450). Briefly, the predominant dietary form of Se, SeMet, is converted to Se-adenosylmethionine and undergoes methyl transfer to yield Se-adenosylhomocysteine (SeAH). SeAH is then hydrolyzed to selenohomocysteine (SeHCys) which is converted to Sec by the trans-sulfuration pathway. Excess SeMet is nonspecifically incorporated into body proteins in place of Met. Sec is obtained from the diet or from the catabolism of selenoproteins. Sec can release Se as hydrogen selenide (Fig 3). Inorganic Se forms, selenite and selenate, are reduced to hydrogen selenide through the glutathione and thioredoxin systems. For selenoprotein synthesis, hydrogen selenide is converted to selenophosphate, an active precursor of Sec. Hydrogen selenide can also be metabolized and excreted in the form of selenosugars and trimethylselenonium ions (TMSe). At extremely high levels of Se intake, volatile endproducts of Se are formed and exhaled as dimethyl selenide (known as the garlic odor) [105].
Compared to the selenoprotein biosynthesis precursors, less is known about the levels and other possible fates of hydrogen selenide and selenophosphate; a large gap has been consistently reported between the sum of identified urinary Se species and total urinary Se [102, 106], indicating a need for additional research in this area. Application of the high-resolution mass spectrometry methods used for yeast [98] could potentially improve understanding of the spectrum of selenometabolites and their nutritional and toxicological importance in mammalian systems.
C. Improved analytical methods for Se speciation
Despite the high-resolution mass spectrometry methods described above, the availability of analytical procedures for the determination of total Se and speciation in different environmental media [107], the complete speciation of Se remains a major challenge due to selenoamino acid incorporation and release during protein turnover. The mostly widely implemented analytical technique consists of a separation chromatography coupled with ICP-MS (inductively coupled plasma mass spectrometer), and often supplemented with other molecular mass spectrometry [107-110]. The compartmentalization of Se species can influence the serum Se levels for SeMet compared to inorganic Se supplement [86, 111]. Se in plasma is mainly present as selenoprotein P (SEPP1; 68 ± 7%), glutathione peroxidase (GPX3; 25 ± 4%) and associated to albumin (7 ± 4%). Sec and other selenols also undergo oxidation, so an additional challenge is avoidance of oxidation artifacts. Encinar et al. used a procedure where the protein-bound Sec was reduced and derivatized with iodoacetamide and separated from SeMet. The species were then detected by ICP-MS equipped with a collision cell [112].
In serum, other low molecular weight Se compounds are minor and can be difficult to detect. Selenosugars are major urinary metabolites, and trimethylselenonium ion (TMSe) and selenite are present as minor products [102]. The responses of human urinary and serum Se metabolites are summarized in Table 1. Notably, selenosugar 1 responds most strongly to Se supplementation while TMSe only increases by a small amount suggesting that TMSe is only a byproduct of the methylation enzyme instead of a significant source of detoxification [106].
TABLE 1.
Se metabolites idenfied from human urinary and serum.
| Se metabolite | Structure | Monoisoto pic mass |
Baseline abundance |
Response | Ref |
|---|---|---|---|---|---|
| TMSe (trimethylselenoniu m ion) |
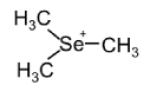
|
124.987 | -Trace in urine and serum. -Varied between TMSe producers (major) and non-TMSe producers (minor or non- detectable) in urine |
-Increase as a minor urinary product after SeMet supplement -Increase moderately in urine after selenite supplement -Minor product in urine |
[106, 180, 181] |
| Selenosugar 1 (methyl-2- acetamido-2-deoxy- 1-seleno-b-D- galactopyranoside) |
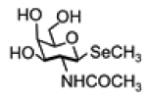
|
299.027 | -Small amount in urine and serum -Major metabolite in urine non-TMSe producers -Major urinary metabolite |
-Increase to the major product after SeMet treatment -Respond strongly to SeMet treatment as the major metabolite |
[102, 180, 181] |
| Selenosugar 3 (methyl-2-amino-2- deoxy-1-seleno-b-D- galactopyranoside) |
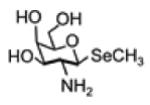
|
257.0164 | -Small amount in urine and serum -2nd major metabolite in non- TMSe producers -Major metabolite |
-Increase to the 2nd major product after SeMet treatment -Increase moderately after selenite supplement |
[106, 180, 181] |
| SeMet (selenomethionine) |
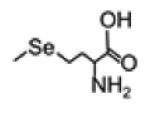
|
196.9955 | -Below detection limit in serum and urine |
-Increase after L- SeMet supplement; - No response to other supplement |
[180, 181] |
| SeMSC (Se-methyl- selenocysteine) |
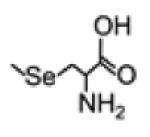
|
182.9799 | -Expected but not detected |
-Trace in serum. Increase in Se excreted in urine after supplementat ion with SeMSC |
[182] |
| Selenite |
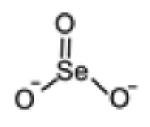
|
127.9013 | -Low amount in serum -Below detection limit in urine |
-Unchanged in serum -Below detection limit in urine |
[106, 180] |
| Selenate |
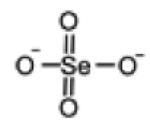
|
143.8962 | -Below detection limit in urine |
-Increase moderately after selenite supplement in urine |
[106] |
| Selenocystathionine |
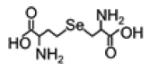
|
270.0119 | -detected not quantified in the urine |
-Increased in epithelial ovarian cancer |
[183] |
| Selenocystine |
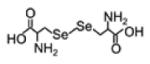
|
335.9128 | -detected and quantified in the serum |
-decrease in cystic fibrosis patients compared to healthy controls |
[184] |
D. Metabolome-wide association study (MWAS) of Se
The development of orbitrap mass spectrometry provides capabilities to overcome analytical challenges for Se metabolism by providing improved sensitivity and specificity [87]. The application of orbitrap MS in yeast, described above, enabled mapping of minor species of Se metabolic pathways otherwise impossible to detect using traditional analytical procedures.
This approach has been expanded to metabolome-wide association study (MWAS) of Se in mice to provide information on metabolic pathways and functional networks which respond to variation in Se. Se supplementation (Na2SeO4, 20 μmol/L in drinking water) for 16 weeks in mice showed that many liver metabolites vary with Se supplementation [24]. These included acylcarnitines, triglycerides and glycerophospholipid, long-chain acyl-CoAs, phosphatidylcholines and sterols (Supplemental Fig 1 and Fig 3 in [24]). Changes in lipid metabolism pathways were also observed in lung and plasma. Harup et al. [113] used urinary metabolomics to investigate the effects of Se nanoparticles and selenite in rats and also found that excess Se intake altered fatty acid metabolism.
E. The impact of Se intake on the whole metabolome: integrated omics
Mounting evidence suggests that Se intake over the range of adequate to excess levels significantly impacts the transcriptome and metabolome beyond effects on Se metabolism and selenoprotein abundance or activity. High doses of Se cause increased body weight and hyperinsulinemia, hyperglycemia, insulin resistance, glucose intolerance, and altered lipid metabolism in mice, rats and pigs [114-116]. Possible mechanisms involve a more reduced state and elevated glutathione peroxidase activity, with over-scavenging of H2O2, a second messenger for insulin signaling [75, 114, 117]. Research also shows, however, that plasma and tissue selenoprotein concentrations and activities reach a plateau after slightly exceeding adequate intake [75, 118, 119], and Se up to 20 times the required intake level does not change the transcript levels of 22 detected selenoproteins [119]. Thus, supplemental Se disrupts energy metabolism by mechanisms other than those controlling the abundance or activities of selenoproteins.
We recently developed integrated omics methods [120-122], which allow use of HRM data with other omics data to define central hubs in complex systems [123, 124]. In particular, transcriptome-metabolome wide association study (TMWAS) [120] provides an approach to address complexity in nutrition and health, as this can show how specific nutrients interact with genes, proteins and metabolites [125]. By comparing Se intakes near the upper limit (UL) to those at adequate intake (AI) or recommended dietary allowance (RDA) levels, the central organizational structure of biologic responses to Se over an otherwise adequate range of intake was obtained [126] [120]. The research showed that Se supplementation induced changes in hepatic gene expression and associated metabolites in fatty acid metabolism, sterol and glucose homeostasis (Supplemental Fig 1) with no detectable differences in liver Se content as measured by ICP-MS, and no detectable differences in transcripts or activities for representative liver selenoproteins. Accumulation of acylcarnitines and other lipid metabolites and decreased bile acid metabolites suggested that Se supplementation altered fatty acid oxidation and created an imbalance with downstream energy metabolism dependent upon acetyl-CoA. Such a metabolic disruption could contribute to an increased body mass observed with supplemental Se [24].
IV. Se as a Ubiquitous Component of the Exposome
Such a systems level response is expected when one considers the natural history of the interactions of living systems with Se in the environment. Se is commonly found within the earth so living organisms have always had some level of exposure. In modern times, Se is used in commercial activities such as ore manufacturing and coal burning, with production of gaseous products such as hydrogen selenide and Se dioxide [127]. In non-occupational settings, individuals are exposed to Se through the diet, especially including the more organo-seleno compounds like Sec, SeMet, and seleno-phosphate [128] (Fig 3). The many biological roles, such as maintenance of antioxidant response, thiol/disulfide-like exchange, gut microbiome expression, and normal development, have occurred as a consequence of evolution of complex organisms with continual Se exposures [38, 44, 96].
As noted above, inorganic forms of Se from the soil are accumulated in plants and converted to more bioactive forms such as Sec or SeMet. In plants, Se also stimulates the synthesis of phytochelatins, oligomers of glutathione produced by phytochelatin synthase, which chelate Se and a range of metals for detoxification [129-131]. Se concentration in the soil varies widely upon geographic location, ranging anywhere from 0.01 mg/kg up to 1200 mg/kg [132, 133]. SeMet is the primary form of Se absorbed from plants. Highest intake of Sec occurs through consumption of animal tissues, especially organ meats [134, 135].
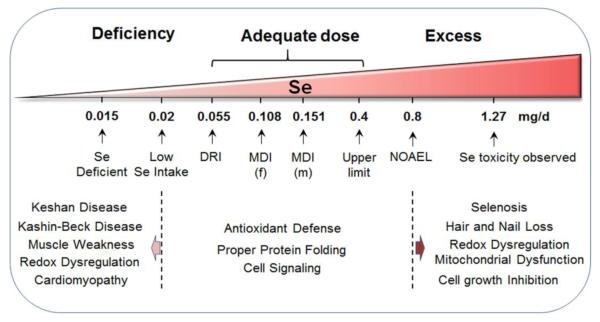
Importantly, the window between sufficient and toxic levels of Se intake in mammals is quite narrow (Fig 4). The minimum amount of dietary Se required for normal human function is 0.055 mg/d, while the majority of Americans tend to consume higher amounts, typically within the mean range of 0.108-0.151 mg/d depending on gender and Se supplement status [136, 137]. This range emphasizes the critical need to understand the adaptive systems which support normal function despite wide variation. This nutrient intake range also provides an excellent window to study Se in balancing the pro-oxidant and pro-reductant states within individuals, especially because either extreme state could cause deleterious effects. In principle, a large number of genetic and epigenetic variations could impact how an individual responds to this variation, either in terms of health benefit or disease.
Fig. 4. Se dose range for individuals.
Se absorption is critical for normal cellular function and processes. Se-deficient individuals, clinically defined as people who have levels of Se lower than 0.015 mg/d, often undergo increased oxidative stress and are diagnosed with pathologies such as Keshan disease and Kashin-Beck disease as well as frequently experience moderate to severe muscle weakness and cardiomyopathy. The daily recommended intake (DRI) level for Se in the US is 0.055 mg/d, while other countries have differing ranges for basal and normal levels of Se status. In the US, the mean daily intake (MDI) has been found to be 0.108 mg/d for females and 0.151 mg/d for males after accounting for Se supplementation and Se in the diet. Within this range, normal cellular processes such as cell-signaling, protein folding, and anti-oxidant defense are performed. However, hyper-abundance of Se often is defined as being above the tolerable upper intake level (UL), which is set at 0.4 mg/d, while individuals with known cases of Se toxicity have Se serum levels of 1.27 mg/d. Se toxicity is found to dysregulate mitochondrial function and produce increased oxidative stress. Pathologies of Se toxicity, commonly defined as selenosis, involve symptoms such as the loss of hair and nails, halitosis, nausea, lesions and peripheral neuropathy.
With appropriate dietary intake, Se is sufficient to maintain cellular homeostasis through proper protein folding, cellular signaling, and maintenance of antioxidant defenses (Fig 4). Se deficiency, though rare in many countries, leads to changes in GSH metabolism, inactivation of several CYP-450’s, and loss of many selenoproteins which ultimately leads to cellular dysregulation [138-144]. These cellular changes are often exhibited in symptoms which range from muscle weakness, cardiomyopathies, to more serious conditions such as Keshan disease and Kashin-Beck disease, which left untreated ultimately lead to death [145-148] (Fig 4).
Conversely, when the upper limit of Se levels of 0.4 mg/d is exceeded, Se toxicity can also occur. Pathologies such as selenosis, which involve factors such as increased oxidative stress and redox dysregulation can lead to cell growth inhibition [135] (Fig 4). Physical symptoms often include hair and nail loss, as well as nausea, skin lesions, and neurophathy [134]. To determine which species of Se is most toxic, yeast models have been used to examin toxicological profiles of seleno-compounds. These results showed that Sec and SeMet were both significantly less toxic than inorganic form of selenite [149].
Due to the interaction of Se with genome, transcriptome, proteome, metabolome, and microbiome, Se is an archetype for the study of genome-exposome interactions. The exposome represents the sum of all exposures for individuals and includes the cumulative biologic responses, both beneficial and noxious, as they relate to health [150, 151]. The inherent complexity of cumulative environmental exposures, in combination with the role of Se as an essential cog in eukaryotic cellular function, make Se attractive for a conceptual, “top-down”, look at mechanisms of Sec in the function systems protecting living organisms from their environments.
V. Se at the Redox Interface of the Functional Genome, Metabolome and Exposome
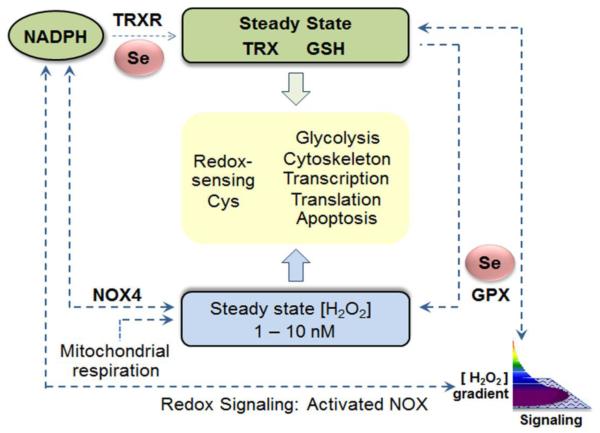
The redox interface between a individual genome and respective exposome was presented as a collection of proteomic and metabolic pathways by which an organism responds to and protects against oxidative stress [1]. The original concept was founded upon the observation that the steady-state oxidation of cysteine residues in proteins mapped according to functional pathways rather than to subcellular compartments [126]. This led to consideration of possible mechanisms by which Cys residues in multiple proteins could be coordinately controlled [152]. Experimental studies with differential inhibition of thioredoxin reductase- and glutathione-dependent redox systems [153], as well toxicologic studies with cadmium [154], showed that either stimulated oxidation or impaired function of reductase systems resulted in pathway-specific changes in steady-state oxidation of Cys residues. A bilateral model to maintain stable steady-state oxidation of Cys residues contains Se-dependent systems at central control points for both reduction and oxidation (Fig 5). For reduction, the selenoenzymes, thioredoxin reductases, have a central role in reducing oxidized proteins. For oxidation, the selenoenzymes GSH peroxidases, as well as the thioredoxin-dependent peroxiredoxins, function to regulate the H2O2 supply for protein oxidation. The implication of this structure is that the entire redox proteome functions as a network with an inherent stability to changes in oxidant burden [155]. This network is well suited to maintain cell functions at a relatively reducing redox poise despite the organism existing within a relatively oxidizing environment.
Fig. 5. Selenium-dependent enzymes support redox systems controlling the redox proteome.
The redox proteome includes reversible sulfur switches which serve to coordinate and regulate functional systems. The system operates with kinetic control through steady-state oxidation of protein Cys residues by H2O2 and other oxidants counterbalanced by steady-state reduction by thioredoxins (TRX) and GSH. The H2O2 availability is kinetically controlled by Se-dependent GSH peroxidases (GPX) and other control systems. The TRX system is maintained by Se-dependent thioredoxin reductases (TRXR). Figure based upon [2].
A. Energy extraction, bioenergetics; NAD, ATP and other energy currencies
The redox proteomics network structure is maintained by relatively low-flux redox pathways linked to NADP within an overall high-flux structure supporting energy metabolism. The high-flux pathways are near-equilibrium reactions linked to NAD [2]. In these high-flux reactions, metabolic fuels are oxidized to provide energy currencies, including transmembranal electrochemical gradients, ATP, acetyl-CoA and other chemicals used to support physical, chemical, electrical and osmotic work. NADPH is also used to support work, especially for reduction reactions in biosynthesis and detoxification; NADPH is coupled to the NAD system through the mitochondrial transhydrogenase. This organizational structure allows bioenergetics to be stabilized by fuel acquisition, storage and use for work while metabolic and macromolecular organization are maintained by kinetically controlled switches within the proteome [2, 156]. The central bioenergetics systems do not directly depend upon Se, but instead, depends upon an orthogonal system of redox regulation [157] which is critically dependent upon selenoenzymes. While all of the bioenergetic machinery is susceptible to oxidative stress, and selenoenzymes protect against oxidative stress, a more fundamental role for selenoenzymes may lie in maintenance of the metabolic and structural organization through reversible redox switches in proteins as described in the second principle of the redox code [2, 156]. In this operational structure (Fig 5), NADPH is used to generate H2O2 and other oxidants to maintain low-flux oxidation of sulfur switches in proteins and also to maintain thioredoxins and GSH to reduce the sulfur switches. With this stable Se-dependent redox structure in place, the organism has maximal capacity to tolerate variations in exogenous oxidative threats.
B. Se role in redox regulation; pro-oxidative and anti-oxidative redox properties
The four common oxidation states of Se (−2, +2, +4, +6) and the concentrations of chemicals with these oxidation states are important factors affecting cellular redox systems. Numerous studies show that Se has both anti-oxidative and pro-oxidative activities, depending on forms of the Se species [91, 158, 159]. Selenoproteins display anti-oxidative properties of Se, specifically via catalytically active Sec in their chemical structure [91]. Of the 25 human selenoproteins, reducing redox functions are shown by representative examples of redox reactions: 1) Sec in GPX1 scavenges H2O2 [160] by reducing it to H2O, and the redox state of Sec is restored in the active site by involving glutathione reductase which consumes NADPH as a source of reducing equivalents. 2) Sec in TRXRD1 is present within the Gly-Cys-Sec-Gly redox active center and shows higher redox reactivity than Cys in thiol-disulfide exchange reactions [161]. 3) Sec in methionine-R-sulfoxide reductase-1 attacks the sulfur atom of methionine-R-sulfoxide and releases reduced methionine by forming a seleninic acid [162]. These anti-oxidative properties of Se have provided strong justification for use of Se compounds for the prevention of cancer and protection against cardiovascular, infectious and neurological diseases.
In contrast to these antioxidant activities, selenite, selenocysteine, selenolates, hydrogen selenide and monomethylselenol (CH3SeH), are highly reactive and have pro-oxidative properties (Fig 3). These compounds generate reactive oxidants, superoxide anion and H2O2 upon reaction with thiols [163, 164], and therefore are cytotoxic. Selenite (Se+4) (Fig 3), a major inorganic Se compound, functions as an oxidant and is reduced to selenium dioxide and hydrogen selenide (Se−2) by substrates such as NADPH [165]; hydrogen selenide functions as a potent reductant yielding elemental Se or seleno-phosphate in the presence of a proper electron acceptor [166, 167]. GSH and selenite spontaneously react to generate oxidants as well as several species of selenium intermediates including glutathioselenol, hydrogen selenide, selenodiglutathione, and elemental Se [166]. Selenite can similarly react with protein thiols and/or GSH to cause protein malfunction or misfolding as associated with cardiovascular disease, cancer, viral and bacterial infections, and other multiple diseases. The selenolates (RSe-) and hydrogen selenide (HSe-) (Fig 3) react with oxygen and thiols efficiently resulting in non-stoichiometric consumption of thiols and NADPH, increased levels of oxidants, and eventually stimulation of cytotoxic signaling and cell death mechanism [168]. In addition, selenium compounds, including SeMet and 1,4-anhydro-5-seleno-D-talitol (SeTal), react rapidly with oxidants to form selenoxides (SeMetO and SeTalO) [169].
Proteomics analyses have been used to study both beneficial and toxic effects of Se. In rice, lower doses of Se (2 and 6 mg/L sodium selenite) activated an anti-oxidative system and enhanced photosynthesis while higher Se treatment (10 mg/L sodium selenite) damaged photosynthesis apparatus and inhibited photosynthesis [170]. In embryonic lung fibroblasts, a strong correlation between Se and replicative senescence was observed, with increased oxidants and decreased antioxidant defense and cell metabolism in senescent cells [171]. In PC-3 prostate cancer cells treated with methaneseleninic acid (MSA) [172], MSA was rapidly reduced to methylselenol [173], a cytotoxic Se species suppressing tumorigenesis in animal models. A total of 194 proteins responding to MSA were identified as redox-sensitive. These proteins include p53, nestin and heat shock protein 70 (HSP70). HSP70 was one of the most redox sensitive in response to MSA from an early state (0.5 h) as well as late state (24 h). The results suggested that MSA and selenite were converted to the reactive monomethylated metabolite more efficiently than SeMet and resulted in rapid redox changes in proteins without affecting the expression levels [172].
C. Integration of signaling, control and responses to environmental challenges
The numbers of Se-containing metabolites and proteins with specific incorporation of Se are relatively small, yet Se broadly impacts all aspects of cell function. Effects on endogenous systems can be readily studied by targeted manipulations. In contrast, studies of interactions within the spectrum of environmental exposures are much more difficult because diets are complex and exposures are variable in frequency (continuous, repetitive, intermittent), variable intensity, and presence of co-exposures. Additionally, responses of individuals to exposures can be cumulative due to the irreversible nature of some biologic changes (e.g., mutations, cell selection in immunity, physical scarring). Thus, elaboration of a systems level understanding of Se will require development of a strategy for systematic study and development of a redox selenobiology atlas.
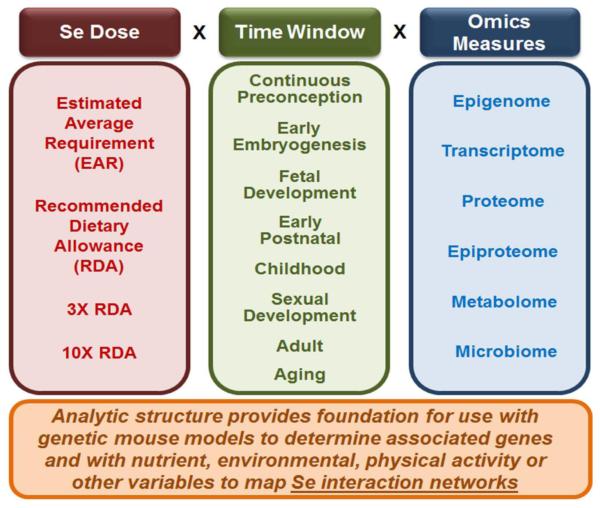
Practical aspects could include studies with varied Se exposure within a standardized grid of other exposures (Fig 6). For this, Se dosing could be standardized relative to Dietary Recommended Intakes (DRI) by using the Estimated Average Requirement (EAR), the Recommended Dietary Allowance (RDA), 3 × RDA and 10 × RDA (Fig 6). This would cover a range from marginally suboptimal to somewhat excessive levels. Temporal exposures could be continuous over lifespan and also sequentially varied for different exposure windows, e.g., preconception, early embryogenesis, fetal development, early postnatal, childhood and sexual maturation, adult and aging (Fig 6). With these doses and time windows for exposure, application of available omics technologies to 36 dose-time windows would provide an initial look at the time-resolved integrated genome-metabolome-exposome network structure. This would identify central interactive hubs and communities of functional networks. The data would also provide a reference data set for more detailed time resolution and dose response studies.
Fig. 6. Development of a data-driven foundation for redox systems biology of selenium.
A minimal analytic framework to develop a reference omics dataset for Se exposures includes systematic variation in dose of exposure (left), time window of exposures (middle) and omics measures of biologic responses (right). The Recommended Dietary Allowance (RDA), set at the amount of Se expected to be adequate in about 97% of the population, provides a useful starting point. The estimated average requirement (EAR) is a useful value for comparison because this level is expected to be inadequate in 50% of the population. A value 3X the RDA is useful to determine effects of excess Se within the adequate intake range, while a value of 10X the RDA is somewhat above the UL and expected to contribute to adverse outcomes. Omics data for continuous exposure to these levels would show the extent of epigenetic, transcriptomic, proteomic, metabolomic and microbiome responses to this spectrum of exposures from moderate deficiency to moderate excess exposures. This reference would provide a basis for comparison to effects from varied exposures in specific time windows (middle), with 1X RDA intake at other times. The continuous exposure data would also provide a reference to test for genetic differences in responses in genetic mouse models and to test for interactions of Se exposures with other dietary and environmental exposures (bottom).
This structure to map integrated Se network responses could be replicated with each of the nearly 40 essential nutrients, thereby elaborating the genome-metabolome-exposome network structure for all essential nutrients. For instance, Se level is known to impact functions of vitamin E, and levels of vitamin E impact function of Se [174, 175]. Vitamin E can at least partially compensate for loss of Gpx4 by protecting cells against deleterious lipid peroxidation [175]. Overlapping roles of Se and vitamin E show synergistic improvement of oxidative status of liver [176]. Some studies in animal models or cells report that most selenoprotein transcripts levels are altered by Se deficiency [177, 178]; whereas, in studies that supplement with vitamin E, the number of selenoprotein transcripts altered by Se status is considerably reduced [175, 179]. Thus, elaboration of the integrated network response structure for Se with other nutrients such as vitamin E will substantially improve understanding of system responses to oxidative stress.
This approach can be further used as a foundation to map the time-resolved interactions of Se with other essential nutrients during development and aging. The maps could also be extended to time-resolved interaction of Se with other exposures, such as exercise, environmental chemicals or infections. Such a structure naturally creates opportunity to evaluate any disease biomarkers, drug response or disease model in the context of Se exposures, thereby creating a universal structure to test responses to nutritional, pharmacologic and toxicologic challenges.
V. Summary and Perspective
Se is a widely distributed environmental mineral that is redox active and present in many forms in the human diet. Se is present in few metabolites and required for only a small number of selenoproteins, but every level of functional organization within mammals responds to Se exposure. Available research using metabolomics and transcriptomics shows that functional network responses to Se in mouse liver are linked to dyslipidemia and weight gain without detectable changes in Se content or selenoprotein transcript abundances or selenoenzyme activities. Application of such data-driven integrated omics methods to systematically study Se exposure will allow the biologic responses of Se to be disentangled from other exposures and enable precise understanding of health benefits and risks from dietary and supplemental Se at an individual level. Such a research strategy would provide a prototype for molecular phenotyping of the integrated genome-metabolome-exposome structure for any dietary or environmental exposure.
Supplementary Material
Highlights.
-
✓
Selenium (Se) is required for a small number of enzymes but dietary and environmental Se impacts every layer of omics space
-
✓
Metabolome and transcriptome interactions in mice reveal complex network responses linked to dyslipidemia and weight gain
-
✓
Central metabolic hubs in the network structure were not directly linked to selenoproteins but were linked to transcripts for glucose transport and fatty acid β-oxidation
-
✓
Global network response structures are needed to provide focus for targeted, hypothesis-driven Se research
Acknowledgements
This study was supported by NIEHS Grants R01 ES023485 (DPJ and YMG), R21 ES025632 (DPJ and YMG), and NIH S10 OD018006 (DPJ).
Abbreviations
- AI
adequate intake
- Cys
cysteine
- DNMT, DIO
iodothyronine deiodinase
- DNA
methyltransferase
- EAR
estimated average requirement
- HDAC
histone deacetylase
- HSP
heat shock protein
- HRM
high-resolution metabolomics
- ICP-MS
inductively coupled plasma mass spectrometer
- G × M × E
genome × metabolome × exposome
- GPX
glutathione peroxidase
- KEGG
Kyto Encyclopedia of Genes and Genomes
- Met
methionine
- MSA
methaneseleninic acid
- RDA
recommended dietary allowance
- Se
selenium
- Sec
selenocysteine
- SeMet
selenomethionine
- SEP
selenoprotein
- SREBF
sterol regulatory element-binding transcription factor
- TMSe
trimethylselenonium
- TMWAS
transcriptome-metabolome-wide association study
- TRXR
thioredoxin reductase
- UL
tolerable upper intake level.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Go YM; Jones DP Redox biology: interface of the exposome with the proteome, epigenome and genome. Redox Biol 2:358–360; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Jones DP; Sies H The Redox Code. Antioxid Redox Signal 23:734–746; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Go YM; Fernandes J; Hu X; Uppal K; Jones DP Mitochondrial network responses in oxidative physiology and disease. Free Radic Biol Med 116:31–40; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Roede JR; Uppal K; Park Y; Tran V; Jones DP Transcriptome–metabolome wide association study (TMWAS) of maneb and paraquat neurotoxicity reveals network level interactions in toxicologic mechanism. Toxicology Reports 1:435–444; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wang Y; Xu L; Shen H; Wang J; Liu W; Zhu X; Wang R; Sun X; Liu L Metabolomic analysis with GC-MS to reveal potential metabolites and biological pathways involved in Pb &Cd stress response of radish roots. Sci Rep 5:18296; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kim M; Rai N; Zorraquino V; Tagkopoulos I Multi-omics integration accurately predicts cellular state in unexplored conditions for Escherichia coli. Nat Commun 7:13090; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Uppal K; Ma C; Go YM; Jones DP; Wren J xMWAS: a data-driven integration and differential network analysis tool. Bioinformatics 34:701–702; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Loscalzo J; Barabasi AL Systems biology and the future of medicine. Wiley Interdiscip Rev Syst Biol Med 3:619–627; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Menche J; Sharma A; Kitsak M; Ghiassian SD; Vidal M; Loscalzo J; Barabasi AL Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science 347:1257601; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Go YM; Walker DI; Liang Y; Uppal K; Soltow QA; Tran V; Strobel F; Quyyumi AA; Ziegler TR; Pennell KD; Miller GW; Jones DP Reference Standardization for Mass Spectrometry and High-resolution Metabolomics Applications to Exposome Research. Toxicol Sci 148:531–543; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Uppal K; Walker DI; Liu K; Li S; Go YM; Jones DP Computational Metabolomics: A Framework for the Million Metabolome. Chem Res Toxicol 29:1956–1975; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Walker DI; Go Y-M; Liu K; Pennell K; D. Jones DP Population Screening for Biological and Environmental Properties of the Human Metabolic Phenotype: Implications for Personalized Medicine. Elsevier; 2016. [Google Scholar]
- [13].Go YM; Uppal K; Walker DI; Tran V; Dury L; Strobel FH; Baubichon-Cortay H; Pennell KD; Roede JR; Jones DP Mitochondrial metabolomics using high-resolution Fourier-transform mass spectrometry. Methods Mol Biol 1198:43–73; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kryukov GV; Castellano S; Novoselov SV; Lobanov AV; Zehtab O; Guigo R; Gladyshev VN Characterization of mammalian selenoproteomes. Science 300:1439–1443; 2003. [DOI] [PubMed] [Google Scholar]
- [15].Locy ML; Rogers LK; Prigge JR; Schmidt EE; Arner ES; Tipple TE Thioredoxin reductase inhibition elicits Nrf2-mediated responses in Clara cells: implications for oxidant-induced lung injury. Antioxid Redox Signal 17:1407–1416; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hatfield DL; Tsuji PA; Carlson BA; Gladyshev VN Selenium and selenocysteine: roles in cancer, health, and development. Trends in biochemical sciences 39:112–120; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ingold I; Berndt C; Schmitt S; Doll S; Poschmann G; Buday K; Roveri A; Peng X; Porto Freitas F; Seibt T; Mehr L; Aichler M; Walch A; Lamp D; Jastroch M; Miyamoto S; Wurst W; Ursini F; Arner ESJ; Fradejas-Villar N; Schweizer U; Zischka H; Friedmann Angeli JP; Conrad M Selenium Utilization by GPX4 Is Required to Prevent Hydroperoxide-Induced Ferroptosis. Cell 172:409–422 e421; 2018. [DOI] [PubMed] [Google Scholar]
- [18].Lei XG; Zhu JH; Cheng WH; Bao Y; Ho YS; Reddi AR; Holmgren A; Arner ES Paradoxical Roles of Antioxidant Enzymes: Basic Mechanisms and Health Implications. Physiol Rev 96:307–364; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hatfield DL; Tsuji PA; Carlson BA; Gladyshev VN Selenium and selenocysteine: roles in cancer, health, and development. Trends Biochem Sci 39:112–120; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Meplan C Selenium and chronic diseases: a nutritional genomics perspective. Nutrients 7:3621–3651; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Goldson AJ; Fairweather-Tait SJ; Armah CN; Bao Y; Broadley MR; Dainty JR; Furniss C; Hart DJ; Teucher B; Hurst R Effects of selenium supplementation on selenoprotein gene expression and response to influenza vaccine challenge: a randomised controlled trial. PLoS One 6:e14771; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Cao N; Li W; Li B; Tian Y; Xu D Transcriptome profiling reveals the immune response of goose T cells under selenium stimuli. Anim Sci J 88:2001–2009; 2017. [DOI] [PubMed] [Google Scholar]
- [23].Knight R; Marlatt VL; Baker JA; Lo BP; deBruyn AMH; Elphick JR; Martyniuk CJ Dietary selenium disrupts hepatic triglyceride stores and transcriptional networks associated with growth and Notch signaling in juvenile rainbow trout. Aquat Toxicol 180:103–114; 2016. [DOI] [PubMed] [Google Scholar]
- [24].Hu X; Chandler JD; Orr ML; Hao L; Liu K; Uppal K; Go Y-M; Jones DP Selenium Supplementation Alters Hepatic Energy and Fatty Acid Metabolism in Mice. Journal of Nutrition 148:675–684; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Meplan C; Hesketh J The influence of selenium and selenoprotein gene variants on colorectal cancer risk. Mutagenesis 27:177–186; 2012. [DOI] [PubMed] [Google Scholar]
- [26].Penney KL; Li H; Mucci LA; Loda M; Sesso HD; Stampfer MJ; Ma J Selenoprotein P genetic variants and mrna expression, circulating selenium, and prostate cancer risk and survival. Prostate 73:700–705; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Huang L; Shi Y; Lu F; Zheng H; Liu X; Gong B; Yang J; Lin Y; Cheng J; Ma S; Lin H; Yang Z Association study of polymorphisms in selenoprotein genes and Kashin-Beck disease and serum selenium/iodine concentration in a Tibetan population. PLoS One 8:e71411; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Xiong YM; Mo XY; Zou XZ; Song RX; Sun WY; Lu W; Chen Q; Yu YX; Zang WJ Association study between polymorphisms in selenoprotein genes and susceptibility to Kashin-Beck disease. Osteoarthritis Cartilage 18:817–824; 2010. [DOI] [PubMed] [Google Scholar]
- [29].Santos LR; Duraes C; Mendes A; Prazeres H; Alvelos MI; Moreira CS; Canedo P; Esteves C; Neves C; Carvalho D; Sobrinho-Simoes M; Soares P A polymorphism in the promoter region of the selenoprotein S gene (SEPS1) contributes to Hashimoto’s thyroiditis susceptibility. J Clin Endocrinol Metab 99:E719–723; 2014. [DOI] [PubMed] [Google Scholar]
- [30].Meplan C; Hughes DJ; Pardini B; Naccarati A; Soucek P; Vodickova L; Hlavata I; Vrana D; Vodicka P; Hesketh JE Genetic variants in selenoprotein genes increase risk of colorectal cancer. Carcinogenesis 31:1074–1079; 2010. [DOI] [PubMed] [Google Scholar]
- [31].Sutherland A; Kim DH; Relton C; Ahn YO; Hesketh J Polymorphisms in the selenoprotein S and 15-kDa selenoprotein genes are associated with altered susceptibility to colorectal cancer. Genes Nutr 5:215–223; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Meplan C; Rohrmann S; Steinbrecher A; Schomburg L; Jansen E; Linseisen J; Hesketh J Polymorphisms in thioredoxin reductase and selenoprotein K genes and selenium status modulate risk of prostate cancer. PLoS One 7:e48709; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Steinbrecher A; Meplan C; Hesketh J; Schomburg L; Endermann T; Jansen E; Akesson B; Rohrmann S; Linseisen J Effects of selenium status and polymorphisms in selenoprotein genes on prostate cancer risk in a prospective study of European men. Cancer Epidemiol Biomarkers Prev 19:2958–2968; 2010. [DOI] [PubMed] [Google Scholar]
- [34].Meplan C; Dragsted LO; Ravn-Haren G; Tjonneland A; Vogel U; Hesketh J Association between polymorphisms in glutathione peroxidase and selenoprotein P genes, glutathione peroxidase activity, HRT use and breast cancer risk. PLoS One 8:e73316; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Burk RF; Hill KE; Motley AK Plasma selenium in specific and non-specific forms. Biofactors 14:107–114; 2001. [DOI] [PubMed] [Google Scholar]
- [36].Turanov AA; Xu XM; Carlson BA; Yoo MH; Gladyshev VN; Hatfield DL Biosynthesis of selenocysteine, the 21st amino acid in the genetic code, and a novel pathway for cysteine biosynthesis. Adv Nutr 2:122–128; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Baker RD; Baker SS; LaRosa K; Whitney C; Newburger PE Selenium regulation of glutathione peroxidase in human hepatoma cell line Hep3B. Arch Biochem Biophys 304:53–57; 1993. [DOI] [PubMed] [Google Scholar]
- [38].Brigelius-Flohe R; Kipp AP Selenium in the redox regulation of the Nrf2 and the Wnt pathway. Methods Enzymol 527:65–86; 2013. [DOI] [PubMed] [Google Scholar]
- [39].Gromer S; Urig S; Becker K The thioredoxin system--from science to clinic. Med Res Rev 24:40–89; 2004. [DOI] [PubMed] [Google Scholar]
- [40].Nauser T; Steinmann D; Grassi G; Koppenol WH Why selenocysteine replaces cysteine in thioredoxin reductase: a radical hypothesis. Biochemistry 53:5017–5022; 2014. [DOI] [PubMed] [Google Scholar]
- [41].Ossowski J; Wachter T; Silies L; Kind M; Noworolska A; Blobner F; Gnatek D; Rysz J; Bolte M; Feulner P; Terfort A; Cyganik P; Zharnikov M Thiolate versus Selenolate: Structure, Stability, and Charge Transfer Properties. ACS Nano 9:4508–4526; 2015. [DOI] [PubMed] [Google Scholar]
- [42].Cardey B; Enescu M A computational study of thiolate and selenolate oxidation by hydrogen peroxide. Chemphyschem 6:1175–1180; 2005. [DOI] [PubMed] [Google Scholar]
- [43].Conant JB; Kirner WR; Hussey RE The relation between the structure of organic halides and the speeds of their reaction with inorganic iodides. III. The influence of unsaturated groups. J Am Chem Soc 47:488–501; 1925. [Google Scholar]
- [44].Hondal RJ; Marino SM; Gladyshev VN Selenocysteine in thiol/disulfide-like exchange reactions. Antioxid Redox Signal 18:1675–1689; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Huang JK; Li CB; Nolan SP; Petersen JL Solution calorimetric investigation of oxidative addition of HEAr (E = O, S, Se; Ar = C6H4X, X = CH3, H, Cl, NO2) to (PMe3)(4)Ru(C2H4): Relationship between HEAr acidity and enthalpy of reaction. Organometallics 17:3516–3521; 1998. [Google Scholar]
- [46].Snider GW; Ruggles E; Khan N; Hondal RJ Selenocysteine confers resistance to inactivation by oxidation in thioredoxin reductase: comparison of selenium and sulfur enzymes. Biochemistry 52:5472–5481; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lothrop AP; Snider GW; Flemer S Jr.; Ruggles EL; Davidson RS; Lamb AL; Hondal RJ Compensating for the absence of selenocysteine in high-molecular weight thioredoxin reductases: the electrophilic activation hypothesis. Biochemistry 53:664–674; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Nauser T; Steinmann D; Koppenol WH Why do proteins use selenocysteine instead of cysteine? Amino acids 42:39–44; 2012. [DOI] [PubMed] [Google Scholar]
- [49].Narayan V; Ravindra KC; Liao C; Kaushal N; Carlson BA; Prabhu KS Epigenetic regulation of inflammatory gene expression in macrophages by selenium. J Nutr Biochem 26:138–145; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Desai D; Salli U; Vrana KE; Amin S SelSA, selenium analogs of SAHA as potent histone deacetylase inhibitors. Bioorg Med Chem Lett 20:2044–2047; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Gowda R; Madhunapantula SV; Desai D; Amin S; Robertson GP Selenium-containing histone deacetylase inhibitors for melanoma management. Cancer Biol Ther 13:756–765; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lee JI; Nian H; Cooper AJ; Sinha R; Dai J; Bisson WH; Dashwood RH; Pinto JT Alpha-keto acid metabolites of naturally occurring organoselenium compounds as inhibitors of histone deacetylase in human prostate cancer cells. Cancer Prev Res (Phila) 2:683–693; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Simpkin AJ; Hemani G; Suderman M; Gaunt TR; Lyttleton O; McArdle WL; Ring SM; Sharp GC; Tilling K; Horvath S; Kunze S; Peters A; Waldenberger M; Ward-Caviness C; Nohr EA; Sorensen TI; Relton CL; Smith GD Prenatal and early life influences on epigenetic age in children: a study of mother-offspring pairs from two cohort studies. Hum Mol Genet 25:191–201; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Speckmann B; Schulz S; Hiller F; Hesse D; Schumacher F; Kleuser B; Geisel J; Obeid R; Grune T; Kipp AP Selenium increases hepatic DNA methylation and modulates one-carbon metabolism in the liver of mice. J Nutr Biochem 48:112–119; 2017. [DOI] [PubMed] [Google Scholar]
- [55].de Miranda JX; Andrade Fde O; Conti A; Dagli ML; Moreno FS; Ong TP Effects of selenium compounds on proliferation and epigenetic marks of breast cancer cells. J Trace Elem Med Biol 28:486–491; 2014. [DOI] [PubMed] [Google Scholar]
- [56].Fiala ES; Staretz ME; Pandya GA; El-Bayoumy K; Hamilton SR Inhibition of DNA cytosine methyltransferase by chemopreventive selenium compounds, determined by an improved assay for DNA cytosine methyltransferase and DNA cytosine methylation. Carcinogenesis 19:597–604; 1998. [DOI] [PubMed] [Google Scholar]
- [57].Xiang N; Zhao R; Song G; Zhong W Selenite reactivates silenced genes by modifying DNA methylation and histones in prostate cancer cells. Carcinogenesis 29:2175–2181; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Clark LC; Combs GF Jr.; Turnbull BW; Slate EH; Chalker DK; Chow J; Davis LS; Glover RA; Graham GF; Gross EG; Krongrad A; Lesher JL Jr.; Park HK; Sanders BB Jr.; Smith CL; Taylor JR Effects of selenium supplementation for cancer prevention in patients with carcinoma of the skin. A randomized controlled trial. Nutritional Prevention of Cancer Study Group. JAMA 276:1957–1963; 1996. [PubMed] [Google Scholar]
- [59].Redman C; Scott JA; Baines AT; Basye JL; Clark LC; Calley C; Roe D; Payne CM; Nelson MA Inhibitory effect of selenomethionine on the growth of three selected human tumor cell lines. Cancer Lett 125:103–110; 1998. [DOI] [PubMed] [Google Scholar]
- [60].Schrauzer GN; White DA; Schneider CJ Cancer mortality correlation studies--III: statistical associations with dietary selenium intakes. Bioinorg Chem 7:23–31; 1977. [DOI] [PubMed] [Google Scholar]
- [61].Karunasinghe N; Ryan J; Tuckey J; Masters J; Jamieson M; Clarke LC; Marshall JR; Ferguson LR DNA stability and serum selenium levels in a high-risk group for prostate cancer. Cancer Epidemiol Biomarkers Prev 13:391–397; 2004. [PubMed] [Google Scholar]
- [62].Waters DJ; Shen S; Glickman LT; Cooley DM; Bostwick DG; Qian J; Combs GF Jr.; Morris JS Prostate cancer risk and DNA damage: translational significance of selenium supplementation in a canine model. Carcinogenesis 26:1256–1262; 2005. [DOI] [PubMed] [Google Scholar]
- [63].Cekan E; Tribukait B; Vokal-Borek H Protective effect of selenium against ionizing radiation-induced malformations in mice. Acta Radiol Oncol 24:267–271; 1985. [DOI] [PubMed] [Google Scholar]
- [64].Overvad K; Thorling EB; Bjerring P; Ebbesen P Selenium inhibits UV-light-induced skin carcinogenesis in hairless mice. Cancer Lett 27:163–170; 1985. [DOI] [PubMed] [Google Scholar]
- [65].Kumar MS; Pollok KE; Smith ML Selenomethionine or methylseleninic acid inhibits mutagenesis of a reporter gene in mouse bone marrow. Anticancer Res 30:291–293; 2010. [PubMed] [Google Scholar]
- [66].Laffon B; Valdiglesias V; Pasaro E; Mendez J The organic selenium compound selenomethionine modulates bleomycin-induced DNA damage and repair in human leukocytes. Biol Trace Elem Res 133:12–19; 2010. [DOI] [PubMed] [Google Scholar]
- [67].Seo YR; Sweeney C; Smith ML Selenomethionine induction of DNA repair response in human fibroblasts. Oncogene 21:3663–3669; 2002. [DOI] [PubMed] [Google Scholar]
- [68].Maehira F; Miyagi I; Eguchi Y Selenium regulates transcription factor NF-kappaB activation during the acute phase reaction. Clin Chim Acta 334:163–171; 2003. [DOI] [PubMed] [Google Scholar]
- [69].Youn HS; Lim HJ; Choi YJ; Lee JY; Lee MY; Ryu JH Selenium suppresses the activation of transcription factor NF-kappa B and IRF3 induced by TLR3 or TLR4 agonists. Int Immunopharmacol 8:495–501; 2008. [DOI] [PubMed] [Google Scholar]
- [70].Hatfield DL; Schweizer U; Tsuji PA; Gladyshev VN Selenium: Its Molecular Biology and Role in Human Health. Springer International Publishing; 2016. [Google Scholar]
- [71].Galli F; Tew KD Selenium and Selenoproteins in Cancer. Elsevier Science; 2017. [Google Scholar]
- [72].Blessing H; Kraus S; Heindl P; Bal W; Hartwig A Interaction of selenium compounds with zinc finger proteins involved in DNA repair. Eur J Biochem 271:3190–3199; 2004. [DOI] [PubMed] [Google Scholar]
- [73].Schenk H; Klein M; Erdbrugger W; Droge W; Schulze-Osthoff K Distinct effects of thioredoxin and antioxidants on the activation of transcription factors NF-kappa B and AP-1. Proc Natl Acad Sci U S A 91:1672–1676; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Li Q; Sanlioglu S; Li S; Ritchie T; Oberley L; Engelhardt JF GPx-1 gene delivery modulates NFkappaB activation following diverse environmental injuries through a specific subunit of the IKK complex. Antioxid Redox Signal 3:415–432; 2001. [DOI] [PubMed] [Google Scholar]
- [75].Barnes KM; Evenson JK; Raines AM; Sunde RA Transcript Analysis of the Selenoproteome Indicates That Dietary Selenium Requirements of Rats Based on Selenium-Regulated Selenoprotein mRNA Levels Are Uniformly Less Than Those Based on Glutathione Peroxidase Activity. The Journal of Nutrition 139:199–206; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Barnes KM; Evenson JK; Raines AM; Sunde RA Transcript analysis of the selenoproteome indicates that dietary selenium requirements of rats based on selenium-regulated selenoprotein mRNA levels are uniformly less than those based on glutathione peroxidase activity. J Nutr 139:199–206; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Sunde RA Selenium regulation of selenoprotein enzyme activity and transcripts in a pilot study with Founder strains from the Collaborative Cross. PLoS One 13:e0191449; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Sunde RA; Raines AM; Barnes KM; Evenson JK Selenium status highly regulates selenoprotein mRNA levels for only a subset of the selenoproteins in the selenoproteome. Biosci Rep 29:329–338; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Moriarty PM; Reddy CC; Maquat LE Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol Cell Biol 18:2932–2939; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Weiss SL; Sunde RA Cis-acting elements are required for selenium regulation of glutathione peroxidase-1 mRNA levels. RNA 4:816–827; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Rusolo F; Capone F; Pasquale R; Angiolillo A; Colonna G; Castello G; Costantini M; Costantini S Comparison of the seleno-transcriptome expression between human non-cancerous mammary epithelial cells and two human breast cancer cell lines. Oncol Lett 13:2411–2417; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Meplan C; Johnson IT; Polley AC; Cockell S; Bradburn DM; Commane DM; Arasaradnam RP; Mulholland F; Zupanic A; Mathers JC; Hesketh J Transcriptomics and proteomics show that selenium affects inflammation, cytoskeleton, and cancer pathways in human rectal biopsies. FASEB J 30:2812–2825; 2016. [DOI] [PubMed] [Google Scholar]
- [83].Maciel-Dominguez A; Swan D; Ford D; Hesketh J Selenium alters miRNA profile in an intestinal cell line: evidence that miR-185 regulates expression of GPX2 and SEPSH2. Mol Nutr Food Res 57:2195–2205; 2013. [DOI] [PubMed] [Google Scholar]
- [84].Zheng X; Hu X; Ge T; Li M; Shi M; Luo J; Lai H; Nie T; Li F; Li H MicroRNA-328 is involved in the effect of selenium on hydrogen peroxide-induced injury in H9c2 cells. J Biochem Mol Toxicol 31; 2017. [DOI] [PubMed] [Google Scholar]
- [85].Xing Y; Liu Z; Yang G; Gao D; Niu X MicroRNA expression profiles in rats with selenium deficiency and the possible role of the Wnt/beta-catenin signaling pathway in cardiac dysfunction. Int J Mol Med 35:143–152; 2015. [DOI] [PubMed] [Google Scholar]
- [86].Ross AC Modern nutrition in health and disease. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2014. [Google Scholar]
- [87].Lazard M; Dauplais M; Blanquet S; Plateau P Recent advances in the mechanism of selenoamino acids toxicity in eukaryotic cells. Biomolecular concepts 8:93–104; 2017. [DOI] [PubMed] [Google Scholar]
- [88].Weiller M; Latta M; Kresse M; Lucas R; Wendel A Toxicity of nutritionally available selenium compounds in primary and transformed hepatocytes. Toxicology 201:21–30; 2004. [DOI] [PubMed] [Google Scholar]
- [89].Romagne F; Santesmasses D; White L; Sarangi GK; Mariotti M; Hubler R; Weihmann A; Parra G; Gladyshev VN; Guigo R; Castellano S SelenoDB 2.0: annotation of selenoprotein genes in animals and their genetic diversity in humans. Nucleic Acids Res 42:D437–443; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Gladyshev VN; Kryukov GV Evolution of selenocysteine-containing proteins: significance of identification and functional characterization of selenoproteins. BioFactors 14:87–92; 2001. [DOI] [PubMed] [Google Scholar]
- [91].Labunskyy VM; Hatfield DL; Gladyshev VN Selenoproteins: molecular pathways and physiological roles. Physiol Rev 94:739–777; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Mariotti M; Ridge PG; Zhang Y; Lobanov AV; Pringle TH; Guigo R; Hatfield DL; Gladyshev VN Composition and evolution of the vertebrate and mammalian selenoproteomes. PloS one 7:e33066; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Stolz JF; Basu P; Santini JM; Oremland RS Arsenic and selenium in microbial metabolism. Annu. Rev. Microbiol 60:107–130; 2006. [DOI] [PubMed] [Google Scholar]
- [94].Garcin E; Vernede X; Hatchikian EC; Volbeda A; Frey M; Fontecilla-Camps JC The crystal structure of a reduced [NiFeSe] hydrogenase provides an image of the activated catalytic center. Structure 7:557–566; 1999. [DOI] [PubMed] [Google Scholar]
- [95].Kudva AK; Shay AE; Prabhu KS Selenium and inflammatory bowel disease. American Journal of Physiology - Gastrointestinal and Liver Physiology 309:G71–G77; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Kasaikina MV; Kravtsova MA; Lee BC; Seravalli J; Peterson DA; Walter J; Legge R; Benson AK; Hatfield DL; Gladyshev VN Dietary selenium affects host selenoproteome expression by influencing the gut microbiota. The FASEB Journal 25:2492–2499; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Eswayah AS; Smith TJ; Gardiner PH Microbial Transformations of Selenium Species of Relevance to Bioremediation. Appl Environ Microbiol 82:4848–4859; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Rao Y; McCooeye M; Windust A; Bramanti E; D’Ulivo A; Mester Z Mapping of selenium metabolic pathway in yeast by liquid chromatography-Orbitrap mass spectrometry. Analytical chemistry 82:8121–8130; 2010. [DOI] [PubMed] [Google Scholar]
- [99].Arnaudguilhem C; Bierla K; Ouerdane L; Preud’homme H; Yiannikouris A; Lobinski R Selenium metabolomics in yeast using complementary reversed-phase/hydrophilic ion interaction (HILIC) liquid chromatography-electrospray hybrid quadrupole trap/Orbitrap mass spectrometry. Analytica chimica acta 757:26–38; 2012. [DOI] [PubMed] [Google Scholar]
- [100].Pilon-Smits EA; Quinn CF Selenium metabolism in plants Cell biology of metals and nutrients: Springer; 2010: 225–241. [Google Scholar]
- [101].Freeman JL; Zhang LH; Marcus MA; Fakra S; McGrath SP; Pilon-Smits EA Spatial imaging, speciation, and quantification of selenium in the hyperaccumulator plants Astragalus bisulcatus and Stanleya pinnata. Plant physiology 142:124–134; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Francesconi KA; Pannier F Selenium metabolites in urine: a critical overview of past work and current status. Clinical Chemistry 50:2240–2253; 2004. [DOI] [PubMed] [Google Scholar]
- [103].Dumont E; Vanhaecke F; Cornelis R Selenium speciation from food source to metabolites: a critical review. Analytical and bioanalytical chemistry 385:1304–1323; 2006. [DOI] [PubMed] [Google Scholar]
- [104].Suzuki KT Metabolomics of selenium: Se metabolites based on speciation studies. Journal of health science 51:107–114; 2005. [Google Scholar]
- [105].ATSDR Toxicological Profile for Selenium. In: Registry A. f. T. S. a. D., ed. Atlanta: US Department of Helath and Human Services, Public Health Service; 2003: 1–418. [Google Scholar]
- [106].Lajin B; Kuehnelt D; Francesconi KA Exploring the urinary selenometabolome following a multi-phase selenite administration regimen in humans. Metallomics 8:774–781; 2016. [DOI] [PubMed] [Google Scholar]
- [107].Kotrebai M; Birringer M; Tyson JF; Block E; Uden PC Selenium speciation in enriched and natural samples by HPLC-ICP-MS and HPLC-ESI-MS with perfluorinated carboxylic acid ion-pairing agents. The Analyst 125:71–78; 2000. [DOI] [PubMed] [Google Scholar]
- [108].Cavalli S; Cardellicchio N Direct determination of seleno-amino acids in biological tissues by anion-exchange separation and electrochemical detection. Journal of Chromatography A 706:429–436; 1995. [DOI] [PubMed] [Google Scholar]
- [109].Huang C; Hu B; He M; Duan J Organic and inorganic selenium speciation in environmental and biological samples by nanometer-sized materials packed dual-column separation/preconcentration on-line coupled with ICP-MS. Journal of mass spectrometry : JMS 43:336–345; 2008. [DOI] [PubMed] [Google Scholar]
- [110].Li H-F; Lombi E; Stroud JL; McGrath SP; Zhao F-J Selenium Speciation in Soil and Rice: Influence of Water Management and Se Fertilization. Journal of Agricultural and Food Chemistry 58:11837–11843; 2010. [DOI] [PubMed] [Google Scholar]
- [111].Burk RF; Norsworthy BK; Hill KE; Motley AK; Byrne DW Effects of Chemical Form of Selenium on Plasma Biomarkers in a High-Dose Human Supplementation Trial. Cancer Epidemiology Biomarkers & Prevention 15:804–810; 2006. [DOI] [PubMed] [Google Scholar]
- [112].Encinar JR; Schaumlöffel D; Ogra Y; Lobinski R Determination of Selenomethionine and Selenocysteine in Human Serum Using Speciated Isotope Dilution-Capillary HPLC–Inductively Coupled Plasma Collision Cell Mass Spectrometry. Analytical Chemistry 76:6635–6642; 2004. [DOI] [PubMed] [Google Scholar]
- [113].Hadrup N; Loeschner K; Skov K; Ravn-Haren G; Larsen EH; Mortensen A; Lam HR; Frandsen HL Effects of 14-day oral low dose selenium nanoparticles and selenite in rat—as determined by metabolite pattern determination. PeerJ 4:e2601; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Cheng J; Zhao W; Wang Q; Liu X; Wang W Accumulation of mercury, selenium and PCBs in domestic duck brain, liver and egg from a contaminated area with an investigation of their redox responses. Environ Toxicol Pharmacol 35:388–394; 2013. [DOI] [PubMed] [Google Scholar]
- [115].Zeng M-S; Li X; Liu Y; Zhao H; Zhou J-C; Li K; Huang J-Q; Sun L-H; Tang J-Y; Xia X-J; Wang K-N; Lei XG A high-selenium diet induces insulin resistance in gestating rats and their offspring. Free Radical Biology and Medicine 52:1335–1342; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Chandler JD; Wongtrakool C; Banton SA; Li S; Orr ML; Barr DB; Neujahr DC; Sutliff RL; Go YM; Jones DP Low-dose oral cadmium increases airway reactivity and lung neuronal gene expression in mice. Physiol Rep 4; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Labunskyy VM; Lee BC; Handy DE; Loscalzo J; Hatfield DL; Gladyshev VN Both maximal expression of selenoproteins and selenoprotein deficiency can promote development of type 2 diabetes-like phenotype in mice. Antioxid Redox Signal 14:2327–2336; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Bendich A Dietary reference intakes for vitamin C, vitamin E, selenium, and carotenoids institute of medicine washington, DC: National Academy Press, 2000 ISBN: 0-309-06935-1. Elsevier; 2001. [Google Scholar]
- [119].Sunde RA; Raines AM Selenium regulation of the selenoprotein and nonselenoprotein transcriptomes in rodents. Advances in Nutrition: An International Review Journal 2:138–150; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Go YM; Roede JR; Orr M; Liang Y; Jones DP Integrated redox proteomics and metabolomics of mitochondria to identify mechanisms of cd toxicity. Toxicol Sci 139:59–73; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Uppal K; Go Y-M; Jones DP xMWAS: an R package for data-driven integration and differential network analysis. bioRxiv; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Go YM; Kim CW; Walker DI; Kang DW; Kumar S; Orr M; Uppal K; Quyyumi AA; Jo H; Jones DP Disturbed flow induces systemic changes in metabolites in mouse plasma: a metabolomics study using ApoE(-)/(-) mice with partial carotid ligation. Am J Physiol Regul Integr Comp Physiol 308:R62–72; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Chandler JD; Hu X; Ko EJ; Park S; Lee YT; Orr M; Fernandes J; Uppal K; Kang SM; Jones DP; Go YM Metabolic pathways of lung inflammation revealed by high-resolution metabolomics (HRM) of H1N1 influenza virus infection in mice. Am J Physiol Regul Integr Comp Physiol 311:R906–R916; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Cribbs SK; Uppal K; Li S; Jones DP; Huang L; Tipton L; Fitch A; Greenblatt RM; Kingsley L; Guidot DM; Ghedin E; Morris A Correlation of the lung microbiota with metabolic profiles in bronchoalveolar lavage fluid in HIV infection. Microbiome 4:3; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Ohlhorst SD; Russell R; Bier D; Klurfeld DM; Li Z; Mein JR; Milner J; Ross AC; Stover P; Konopka E Nutrition research to affect food and a healthy life span. The Journal of Nutrition 143:1349–1354; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Go YM; Duong DM; Peng J; Jones DP Protein Cysteines Map to Functional Networks According to Steady-state Level of Oxidation. J Proteomics Bioinform 4:196–209; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].EPA Air Pollution Aspects of Selenium and its compounds. In: Agency EP, ed. Litton Systems, INC; 1969. [Google Scholar]
- [128].Lobanov AV; Hatfield DL; Gladyshev VN Selenoproteinless animals: selenophosphate synthetase SPS1 functions in a pathway unrelated to selenocysteine biosynthesis. Protein Sci 17:176–182; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Gupta M; Gupta S An Overview of Selenium Uptake, Metabolism, and Toxicity in Plants. Front Plant Sci 7:2074; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Kumar A; Singh RP; Singh PK; Awasthi S; Chakrabarty D; Trivedi PK; Tripathi RD Selenium ameliorates arsenic induced oxidative stress through modulation of antioxidant enzymes and thiols in rice (Oryza sativa L.). Ecotoxicology 23:1153–1163; 2014. [DOI] [PubMed] [Google Scholar]
- [131].Simmons DBD; Emery RJN Phytochelatin Induction by Selenate in Chlorella Vulgaris, and Regulation of Effect by Sulfate Levels. Environ Toxicol Chem 30:469–476; 2011. [DOI] [PubMed] [Google Scholar]
- [132].Dhillon KS; Dhillon SK Distribution and management of seleniferous soils. Adv Agron 79:119–184; 2003. [Google Scholar]
- [133].Fernández-Martínez A; Charlet L Selenium environmental cycling and bioavailability: a structural chemist point of view. Reviews in Environmental Science and Bio/Technology 8:81–110; 2009. [Google Scholar]
- [134].Burk RF; Levander O. A. Selenium. In: Shils ME SM, and Ross CA ed. Modern Nutrition in Health and Disease. Philadelphia: Lippincott Williams & Wilkins; 2005: 312–325. [Google Scholar]
- [135].Sunde R Selenium. In: Ross CA CB, Cousins RJ, Tucker KL, and Ziegler TR., ed. Modern Nutrition in Health and Disease. Philadelphia: Wolters Kluwer and Lippincott Williams & Wilkins; 2012. [Google Scholar]
- [136].ATSDR Priority List of Hazardous Substances. In: Control C. f. D., ed. Atlanta GA: US Department of Helath and Human Services, Public Health Service; 2015. [Google Scholar]
- [137].Laclaustra M; Stranges S; Navas-Acien A; Ordovas JM; Guallar E Serum selenium and serum lipids in US adults: National Health and Nutrition Examination Survey (NHANES) 2003-2004. Atherosclerosis 210:643–648; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Burk RF; Hill KE; Nakayama A; Mostert V; Levander XA; Motley AK; Johnson DA; Johnson JA; Freeman ML; Austin LM Selenium deficiency activates mouse liver Nrf2-ARE but vitamin E deficiency does not. Free Radic Biol Med 44:1617–1623; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Hill KE; Burk RF; Lane JM Effect of selenium depletion and repletion on plasma glutathione and glutathione-dependent enzymes in the rat. J Nutr 117:99–104; 1987. [DOI] [PubMed] [Google Scholar]
- [140].Hill KE; Taylor MA; Burk RF Influence of selenium deficiency on glutathione disulfide metabolism in isolated perfused rat heart. Biochim Biophys Acta 923:431–435; 1987. [DOI] [PubMed] [Google Scholar]
- [141].Prohaska JR; Ganther HE Interactions between selenium and methylmercury in rat brain. Chem Biol Interact 16:155–167; 1977. [DOI] [PubMed] [Google Scholar]
- [142].Reiter R; Wendel A Selenium and drug metabolism--I. Multiple modulations of mouse liver enzymes. Biochem Pharmacol 32:3063–3067; 1983. [DOI] [PubMed] [Google Scholar]
- [143].Suzuki T; Kelly VP; Motohashi H; Nakajima O; Takahashi S; Nishimura S; Yamamoto M Deletion of the selenocysteine tRNA gene in macrophages and liver results in compensatory gene induction of cytoprotective enzymes by Nrf2. J Biol Chem 283:2021–2030; 2008. [DOI] [PubMed] [Google Scholar]
- [144].Thompson KM; Haibach H; Sunde RA Growth and plasma triiodothyronine concentrations are modified by selenium deficiency and repletion in second-generation selenium-deficient rats. J Nutr 125:864–873; 1995. [DOI] [PubMed] [Google Scholar]
- [145].Beck MA Selenium and vitamin E status: impact on viral pathogenicity. J Nutr 137:1338–1340; 2007. [DOI] [PubMed] [Google Scholar]
- [146].Sheridan PA; Zhong N; Carlson BA; Perella CM; Hatfield DL; Beck MA Decreased selenoprotein expression alters the immune response during influenza virus infection in mice. J Nutr 137:1466–1471; 2007. [DOI] [PubMed] [Google Scholar]
- [147].Smith AG; Sheridan PA; Harp JB; Beck MA Diet-induced obese mice have increased mortality and altered immune responses when infected with influenza virus. J Nutr 137:1236–1243; 2007. [DOI] [PubMed] [Google Scholar]
- [148].Stone R Diseases. A medical mystery in middle China. Science 324:1378–1381; 2009. [DOI] [PubMed] [Google Scholar]
- [149].Letavayova L; Vlasakova D; Spallholz JE; Brozmanova J; Chovanec M Toxicity and mutagenicity of selenium compounds in Saccharomyces cerevisiae. Mutat Res 638:1–10; 2008. [DOI] [PubMed] [Google Scholar]
- [150].Dennis KK; Auerbach SS; Balshaw DM; Cui Y; Fallin MD; Smith MT; Spira A; Sumner S; Miller GW The Importance of the Biological Impact of Exposure to the Concept of the Exposome. Environ Health Perspect 124:1504–1510; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Dennis KK; Jones DP The Exposome: A New Frontier for Education. Am Biol Teach 78:542–548; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Go YM; Jones DP Thiol/disulfide redox states in signaling and sensing. Crit Rev Biochem Mol Biol 48:173–181; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Go YM; Roede JR; Walker DI; Duong DM; Seyfried NT; Orr M; Liang Y; Pennell KD; Jones DP Selective targeting of the cysteine proteome by thioredoxin and glutathione redox systems. Mol Cell Proteomics 12:3285–3296; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Go YM; Orr M; Jones DP Actin cytoskeleton redox proteome oxidation by cadmium. Am J Physiol Lung Cell Mol Physiol 305:L831–843; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Go YM; Jones DP The redox proteome. J Biol Chem 288:26512–26520; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Go YM; Jones DP Redox theory of aging: implications for health and disease. Clin Sci (Lond) 131:1669–1688; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Jones DP Redox sensing: orthogonal control in cell cycle and apoptosis signalling. J Intern Med 268:432–448; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Lee KH; Jeong D Bimodal actions of selenium essential for antioxidant and toxic prooxidant activities: the selenium paradox (Review). Mol Med Rep 5:299–304; 2012. [DOI] [PubMed] [Google Scholar]
- [159].Misra S; Boylan M; Selvam A; Spallholz JE; Bjornstedt M Redox-active selenium compounds--from toxicity and cell death to cancer treatment. Nutrients 7:3536–3556; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Lubos E; Loscalzo J; Handy DE Glutathione peroxidase-1 in health and disease: from molecular mechanisms to therapeutic opportunities. Antioxid Redox Signal 15:1957–1997; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Arner ES Selenoproteins-What unique properties can arise with selenocysteine in place of cysteine? Exp Cell Res 316:1296–1303; 2010. [DOI] [PubMed] [Google Scholar]
- [162].Kim HY; Gladyshev VN Methionine sulfoxide reductases: selenoprotein forms and roles in antioxidant protein repair in mammals. Biochem J 407:321–329; 2007. [DOI] [PubMed] [Google Scholar]
- [163].Brigelius-Flohe R; Flohe L Selenium and redox signaling. Arch Biochem Biophys 617:48–59; 2017. [DOI] [PubMed] [Google Scholar]
- [164].Spallholz JE On the nature of selenium toxicity and carcinostatic activity. Free Radic Biol Med 17:45–64; 1994. [DOI] [PubMed] [Google Scholar]
- [165].Bjornstedt M; Kumar S; Holmgren A Selenite and selenodiglutathione: reactions with thioredoxin systems. Methods in enzymology 252:209–219; 1995. [DOI] [PubMed] [Google Scholar]
- [166].Tarze A; Dauplais M; Grigoras I; Lazard M; Ha-Duong NT; Barbier F; Blanquet S; Plateau P Extracellular production of hydrogen selenide accounts for thiol-assisted toxicity of selenite against Saccharomyces cerevisiae. J Biol Chem 282:8759–8767; 2007. [DOI] [PubMed] [Google Scholar]
- [167].Lobanov AV; Hatfield DL; Gladyshev VN Selenoproteinless animals: selenophosphate synthetase SPS1 functions in a pathway unrelated to selenocysteine biosynthesis. Protein science : a publication of the Protein Society 17:176–182; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Kumar S; Bjornstedt M; Holmgren A Selenite is a substrate for calf thymus thioredoxin reductase and thioredoxin and elicits a large non-stoichiometric oxidation of NADPH in the presence of oxygen. Eur J Biochem 207:435–439; 1992. [DOI] [PubMed] [Google Scholar]
- [169].Carroll L; Pattison DI; Fu S; Schiesser CH; Davies MJ; Hawkins CL Catalytic oxidant scavenging by selenium-containing compounds: Reduction of selenoxides and N-chloramines by thiols and redox enzymes. Redox Biol 12:872–882; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Wang YD; Wang X; Wong YS Proteomics analysis reveals multiple regulatory mechanisms in response to selenium in rice. J Proteomics 75:1849–1866; 2012. [DOI] [PubMed] [Google Scholar]
- [171].Hammad G; Legrain Y; Touat-Hamici Z; Duhieu S; Cornu D; Bulteau AL; Chavatte L Interplay between Selenium Levels and Replicative Senescence in WI-38 Human Fibroblasts: A Proteomic Approach. Antioxidants (Basel) 7; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Lee JS; Ma YB; Choi KS; Park SY; Baek SH; Park YM; Zu K; Zhang H; Ip C; Kim YH; Park EM Neural network-based analysis of thiol proteomics data in identifying potential selenium targets. Prep Biochem Biotechnol 36:37–64; 2006. [DOI] [PubMed] [Google Scholar]
- [173].Ip C; Thompson HJ; Zhu Z; Ganther HE In vitro and in vivo studies of methylseleninic acid: evidence that a monomethylated selenium metabolite is critical for cancer chemoprevention. Cancer Res 60:2882–2886; 2000. [PubMed] [Google Scholar]
- [174].Friedmann Angeli JP; Schneider M; Proneth B; Tyurina YY; Tyurin VA; Hammond VJ; Herbach N; Aichler M; Walch A; Eggenhofer E; Basavarajappa D; Radmark O; Kobayashi S; Seibt T; Beck H; Neff F; Esposito I; Wanke R; Forster H; Yefremova O; Heinrichmeyer M; Bornkamm GW; Geissler EK; Thomas SB; Stockwell BR; O’Donnell VB; Kagan VE; Schick JA; Conrad M Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol 16:1180–1191; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Carlson BA; Tobe R; Yefremova E; Tsuji PA; Hoffmann VJ; Schweizer U; Gladyshev VN; Hatfield DL; Conrad M Glutathione peroxidase 4 and vitamin E cooperatively prevent hepatocellular degeneration. Redox Biol 9:22–31; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Sobajic SS; Mihailovic MB; Miric MO The effects of selenium deficiency, dietary selenium, and vitamin E supplementation on the oxidative status of pig liver. J Environ Pathol Toxicol Oncol 17:265–270; 1998. [PubMed] [Google Scholar]
- [177].Zhang JL; Xu B; Huang XD; Gao YH; Chen Y; Shan AS Selenium Deficiency Affects the mRNA Expression of Inflammatory Factors and Selenoprotein Genes in the Kidneys of Broiler Chicks. Biological trace element research 171:201–207; 2016. [DOI] [PubMed] [Google Scholar]
- [178].Liu CP; Fu J; Lin SL; Wang XS; Li S Effects of dietary selenium deficiency on mRNA levels of twenty-one selenoprotein genes in the liver of layer chicken. Biological trace element research 159:192–198; 2014. [DOI] [PubMed] [Google Scholar]
- [179].Saito Y; Yoshida Y; Akazawa T; Takahashi K; Niki E Cell death caused by selenium deficiency and protective effect of antioxidants. J Biol Chem 278:39428–39434; 2003. [DOI] [PubMed] [Google Scholar]
- [180].Kokarnig S; Tsirigotaki A; Wiesenhofer T; Lackner V; Francesconi KA; Pergantis SA; Kuehnelt D Concurrent quantitative HPLC-mass spectrometry profiling of small selenium species in human serum and urine after ingestion of selenium supplements. Journal of trace elements in medicine and biology : organ of the Society for Minerals and Trace Elements 29:83–90; 2015. [DOI] [PubMed] [Google Scholar]
- [181].Kuehnelt D; Kienzl N; Traar P; Le NH; Francesconi KA; Ochi T Selenium metabolites in human urine after ingestion of selenite, L-selenomethionine, or DL-selenomethionine: a quantitative case study by HPLC/ICPMS. Analytical and bioanalytical chemistry 383:235–246; 2005. [DOI] [PubMed] [Google Scholar]
- [182].Marshall JR; Burk RF; Payne Ondracek R; Hill KE; Perloff M; Davis W; Pili R; George S; Bergan R Selenomethionine and methyl selenocysteine: multiple-dose pharmacokinetics in selenium-replete men. Oncotarget 8:26312–26322; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Zhang T; Wu X; Ke C; Yin M; Li Z; Fan L; Zhang W; Zhang H; Zhao F; Zhou X; Lou G; Li K Identification of potential biomarkers for ovarian cancer by urinary metabolomic profiling. J Proteome Res 12:505–512; 2013. [DOI] [PubMed] [Google Scholar]
- [184].Michalke B Selenium speciation in human serum of cystic fibrosis patients compared to serum from healthy persons. J Chromatogr A 1058:203–208; 2004. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.