Abstract
The role of myeloid cells as regulators of tumor progression that significantly impact the efficacy of cancer immunotherapies makes them an attractive target for inhibition. Here we explore the effect of a novel, potent, and selective inhibitor of serine/threonine protein kinase CK2 on modulating myeloid cells in the tumor microenvironment. Although inhibition of CK2 caused only a modest effect on dendritic cells in tumor-bearing mice, it substantially reduced the amount of polymorphonuclear myeloid-derived suppressor cells (PMN-MDSC) and tumor-associated macrophages (TAM). This effect was not caused by the induction of apoptosis, but rather by a block of differentiation. Our results implicated downregulation of CCAAT-enhancer binding protein-α (C/EBPα) in this effect. Although CK2 inhibition did not directly affect tumor cells, it dramatically enhanced the antitumor activity of immune checkpoint receptor blockade using anti-CTLA-4 antibody. These results suggest a potential role of CK2 inhibitors in combination therapies against cancer.
Introduction
It is now evident that the ultimate success of cancer immunotherapy depends on its ability to overcome limitations imposed by tumors. Moreover, the same is probably true for other types of oncologic therapies. Myeloid cells are critical components of the tumor microenvironment widely implicated in suppressing antitumor immunity (1–3). These cells are comprised of tumor-associated macrophages (TAM), dendritic cells (DC), neutrophils, and pathologically activated immature myeloid cells with the potent ability to suppress immune responses, termed myeloid-derived suppressor cells (MDSC). In recent years, it has become clear that MDSC are not only an important negative regulator of immune responses, but also contribute to other aspects of tumor growth, namely tumor angiogenesis, tumor cell invasion, and formation of pre-metastatic niches (4). In tumor-bearing (TB) mice, the total population of MDSC consists of two large groups of cells; the most abundant (>75%) population are immature, pathologically activated neutrophils, termed polymorphonuclear MDSC (PMN-MDSC) and the less abundant (<20%) population are pathologically activated monocytes, termed monocytic MDSC (M-MDSC).
We have previously demonstrated that defective DC differentiation in cancer and the accumulation of PMN-MDSC was mediated by down-regulation of Notch signaling (5). These results were later confirmed by others (6). Inhibition of Notch signaling was partially caused by phosphorylation of Notch by casein kinase 2 (CK2) (5). This suggested that targeting CK2 could be potentially beneficial for improving immune responses in cancer.
CK2 is a serine/threonine protein kinase with numerous intracellular protein substrates. CK2 phosphorylates its substrates by utilizing either ATP or GTP as a phosphate donor. CK2 expression and activity are elevated in many types of cancer. Functionally, it is has been observed that activation of CK2 is associated with suppression of apoptosis of cancer cells (7–9). Conversely, downregulation of CK2 expression or inhibition of its activity facilitates cell death triggered upon drug exposure, ligation of death receptors, and ionizing radiation (8, 10). Inhibition of CK2 sensitized T-lymphoblastic cells to drug-induced apoptosis by increasing cellular uptake of the drug (11). CK2 inhibition also increased the sensitivity of rhabdomyosarcoma and colon carcinoma cells to TRAIL-induced apoptosis (12), and induced ROS-mediated apoptosis of leukemia cells (8). CK2 regulates a number of signaling pathways involved in tumor progression and also is critical for cell differentiation. Specifically, the phosphatase, PTEN, regulates cell survival by blocking the PI3K-AKT axis and has been shown to be phosphorylated by CK2 (13), which facilitates its proteasomal degradation (14). As a result of the inhibition of PTEN by CK2, the AKT survival signal is prolonged (15). In addition, CK2 can directly phosphorylate AKT at Ser129 (16). CK2 has also been implicated in direct targeting of the NF-κB and Wnt signaling pathways (16–18).
In this study we tested the effects of novel potent and selective CK2 inhibitors and found that in tumor-bearing, but not in tumor-free mice, they caused a substantial decrease in PMN-MDSC and macrophage differentiation, and dramatically enhanced the effect of anti-CTLA-4 immune-based therapy.
MATERIALS AND METHODS
Human samples
Cord blood samples were collected at Helen F. Graham Cancer Center. All patients signed written informed consent form. The studies were conducted in accordance with recognized ethical guidelines and were approved by Institutional Review Board of the Christiana Care Health System at the Helen F. Graham Cancer Center and The Wistar Institute.
Mice
All procedures were performed accordance with the NIH Guide for the Care and Use of Laboratory Animal guidelines and approved by the Institutional Animal Care and Use Committee (IACUC) at The Wistar Institute. Female five- to six- week old C57BL/6 mice and Balb/c mice (Charles River Labs and Envigo) were maintained in a temperature controlled room with a 12/12 hour light/dark schedule and food provided ad libitum.
CK2 inhibitors
BMS-211 (FW: 825.24) is a prodrug of the parent pan-CK2 inhibitor, BMS-699 (Details to be published). Suspension formulations for BMS-211 were prepared in 0.5% methocel-A4m, 0.1% TWEEN-80. Particle size of the suspension was typically ~10um (D50). Lower concentration formulations were prepared by appropriate dilution (v/v) of the highest concentration formulation. BMS-595 (FW: 532.44) is also pan-CK2 inhibitor and was solved with vehicle (Kolliphor TPGS:EtOH:PEG300 = 1:1:8) for in vivo experiments.
Combination therapy
LLC lung carcinoma, CT26 colon carcinoma, 4T1 breast carcinoma and EL4 lymphoma were obtained from ATCC and cultured in DMEM (Corning Incorporated) supplemented with 10% FBS (Atlanta Biologicals, Inc.) and 1% antibiotics (Thermo Fisher Scientific Inc.). Cells were incubated in a 37°C and 5% CO2. 70–80% confluent cells were harvested using 0.25% Trypsin (Thermo Fisher Scientific Inc.) and passaged or used for experiments. Cells were tested on mycoplasma contamination every three months.
Balb/c mice were subcutaneously implanted with 1×106 4T1 or CT26 cells in 0.2 mL PBS on Day 0 and C57BL/6 mice with 1×106 MC38 or 5×105 LLC cells in 0.2 mL PBS on Day 0. Six or seven days post implantation, TB mice were randomly divided into 4 groups (n = 7 – 10). Mice were treated with BMS-211 or BMS-595 solution orally daily for 21 days at 20 mg/kg or 60 mg/kg, respectively. In the 4T1 model, anti-CTLA-4 (clone 9D9) mIgG2a antibody or the isotype control was i.p. injected into the mice at 20 ug/mouse on Days 6, 13 and 20. In MC38 model, anti-CTLA-4 mIgG2a antibody or the isotype control was injected i.p. into the mice at 200 μg/mouse on Days 6, 13 and 20. In CT26 model, anti-CTLA-4 mIgG2a antibody was i.p. injected at 5 μg/mouse on Days 7, 14 and 21, and anti-CTLA-4 mIgG2b antibody was i.p. injected at 200 μg/mouse on Days 7, 14 and 21. In LLC model, anti-CTLA-4 mIgG2a antibody was i.p. injected at 200 μg/mouse on Day 7 and 14. For the CD8 T cell depletion study, LLC tumor bearing mice were i.p. injected with 100 μg of anti-CD8 alpha antibody (BioXcell, clone 53–6.7) on Day -3, 0, 3, 7, 10 and 14.
In vitro cytotoxic activity
The tumor cell lines were plated at 2×103 cells/well into 96 well plates and cultured 16h. BMS-595 was dissolved with DMSO and diluted with media, and added into the wells and incubated for 3 days. The cell viability was measured using MTS assay (Promega). IC50 values were calculated using linear regression analysis.
Flow cytometry
Single-cell suspensions were prepared from spleens and bone marrow. Tumor tissues were cut into small pieces, and digested with the mouse tumor dissociation kit (Miltenyi Biotec). Red blood cells in the cell suspensions were lysed using ammonium chloride lysis buffer. The list of antibodies is provided in Supplemental Table 1. Flow cytometry data were acquired using a BD LSR II flow cytometer and analyzed using FlowJo software (Tree Star).
Mouse hematopoietic progenitor cells culture
Lineage negative cells were purified from C57BL/6 naïve bone marrow using the lineage cell depletion kit (Miltenyi). Lineage negative cells were cultured in RPMI (Corning Incorporated) supplemented with 10% FBS, 1% antibiotics, and 50 μM 2-mercaptoethanol (Thermo Fisher Scientific Inc.) with 20 ng/mL of recombinant GM-CSF (Invitrogen), at 50,000 cells/well in 24-well plates. Tumor explant supernatants (TES) were obtained by culturing small pieces of EL4 tumors with complete RPMI media for 24 hours. On Day 1, TES or media with DMSO or BMS-595 solution was added into the wells at 10% to get the final concentration of 10 or 100 nM BMS-595. On Day 3, half of the culture supernatant was exchanged with fresh media supplemented with 20 ng/mL of GM-CSF, with or without 10% TES. On Day 6, the cells were collected and analyzed by flow cytometry. To assess proliferation, HPCs were cultured with 10 μM of BrdU for 24h, and then analyzed by flow cytometry using the BrdU detection kit (BD Pharmingen).
Human hematopoietic progenitor cells culture
CD34+ hematopoietic progenitor cells were purified from human cord blood by Ficoll-based density gradient centrifugation followed by CD34 magnetic beads selection (Miltenyi Biotec). OP9 mouse bone marrow stromal cells were cultured in the 6-well plate at 104 cells/well the day before CD34+ purification. On Day 0, CD34+ cells were co-cultured with OP9 cells at 105 cells/well in Iscove’s modified Dulbecco’s medium supplemented with 20% FBS, 1% antibiotics, 20 ng/mL human GM-CSF and 100 ng/mL human G-CSF (PeproTech). After overnight culture, tumor-conditioned media (TCM) from RPMI8226 human myeloma cell lines was added at 10% with DMSO or 100 nM BMS-595. On Day 3 and 6, fresh media containing 20 ng/mL GM-CSF, 100 ng/mL G-CSF, 10% TCM, and DMSO or 100 nM BMS-595 was added in the wells. On Day 9, the cells were collected and analyzed by flow cytometry.
Suppression assay
Ly6G+ cells were purified from spleen cells or tumor cells by positive selection using biotinylated Ly6G antibody and streptavidin microbeads (Miltenyi). The purity of the cell populations was >95%. CD11b+Ly6ChighLy6G− cells were isolated from spleen cells by cell sorting on a FACSAria cell sorter (BD Biosciences). CD8+ T cells from PMEL mice that recognize the gp100-derived peptide, were used as responders. Splenocytes from PMEL mice were mixed with splenocytes from naïve mice at 1:4 ratio in complete RPMI media and plated into 96-well U-bottom plates at 105 cells/well. Ly6G+ or CD11b+Ly6ChighLy6G− cells were added to the wells at 1:16–1:1 ratios. Murine gp100 peptide (25–33) EGSRNQDWL (AnaSpec, Inc.) was added into the wells at the final concentration of 0.1 μg/mL. After 48 hours, cells were pulsed with 3H-thymidine (1 μCi/well; GE healthcare) for 16 hours. 3H-thymidine uptake was counted using a liquid scintillation counter as counts per minute (cpm) and calculated the percentage of proliferation to the positive control (the wells with responder cells and peptide but without suppressive cells).
Apoptosis
Ly6G+ cells and CD11b+Ly6G−F4/80+Ly6Clow macrophages were treated with BMS-595 for 24 to 72h. The cells were stained with FITC-AnnexinV (BD Pharmingen) in AnnexinV binding buffer (BD Pharmingen) for 30 min at room temperature, and then suspended with DAPI containing MACS buffer and analyzed by flow cytometry. HPCs were cultured under the same condition as previously described with BMS-595 and 12.5 and 25μM of Z-VAD-FMK (Calbiochem), and then analyzed using flow cytometry.
Quantitative RT-PCR
RNA was extracted with a total RNA extraction kit (Omega Bio-tek). cDNA was synthesized (cDNA reverse transcriptase kit; Applied Biosystems), and PCR was performed in triplicate for each sample. To detect expression of Notch-, IRF8-, C/EBPα- or C/EBPβ-regulated genes, qRT-PCR was performed with 10 μl SYBR Master Mixture (Applied Biosystems) and the list of the primers is provided in Supplemental Table 2. Expressions of the different genes were normalized to β-actin. Relative expression was calculated using the 2–ΔΔCt method.
Western blot
Nuclear and cytoplasm extract were obtained from HPC treated with DMSO or 100 nM of BMS-595 for 24 h. Nuclear protein was extracted using CelLytic NuCLEAR Extraction Kit (Sigma) with phosphatase inhibitor cocktail (Sigma). To retrieve the cytoplasm lysate, the cells were lysed with RIPA buffer (Sigma) supplemented with a protease inhibitor cocktail (Sigma) and a phosphatase inhibitor cocktail. Denatured protein samples were separated by electrophoresis with 25 μg/lane and transferred onto PVDF membrane. The list of antibodies provided in Supplemental Table 1. The detected proteins were visualized by the ECL system (GE Healthcare Life Sciences).
RNA-Seq
LLC-TB mice were treated with vehicle or 60 mg/kg of BMS-595 for 2 weeks. HPC was isolated from the bone marrow of the vehicle- or BMS-595-treated mice. Total RNA was extracted from the HPC using the Qiagen’s Rneasy Mini Kit and RNA quality was assessed using the Agilent RNA ScreenTape Assay on Agilent’s 2200 TapeStation. RNA-seq library was prepared using TruSeq Stranded Total RNA kit (Illumina) and ran in a 75bp paired end run, 40 million reads on Illumina NextSeq instrument.
RNA-seq data was aligned using bowtie2 (19) against mm10 version of the human genome and RSEM v1.2.31 software (20) was used to estimate raw read counts and FPKM values using transcriptome information from Ensemble v84 set. DESeq2 (21) was used to estimate significance of differential expression between 5 replicates of untreated and BMS-595-treated samples. Gene expression changes were considered significant if passed FDR<5% threshold and had expression for all replicates from one group higher or lower than any replicates of another group. Gene set enrichment analysis of gene sets was done using QIAGEN’s Ingenuity® Pathway Analysis software (IPA®, QIAGEN Redwood City,www.qiagen.com/ingenuity) using “Canonical Pathways”, “Diseases & Functions” and “Upstream Analysis” options. Only significantly enriched at p<0.05 functions and regulators with significantly predicted activation state (Z-score |Z|>2) were considered. Pathways significantly affected at FDR<5% with predicted activation stated z-score of at least 1 were reported. The data was submitted to the Gene Expression Omnibus (GEO) database and can be accessed using accession number: GSE117712
Statistical analysis
Statistical analyses were performed using 2-tailed Student’s t test or Mann-Whitney U test and GraphPad Prism 5 software (GraphPad Software Inc.). Paired t test was used since data were normally distributed. All the data are presented as mean ± SD and p value less than 0.05 was considered significant.
Results
CK2 inhibitor has potent antitumor effects in combination with anti-CTLA4 antibody
Potent and selective, ATP-competitive CK2 inhibitors (BMS-595, BMS-699 and BMS-211) were developed by Bristol-Myers Squibb (Fig. S1A). Inhibition of proliferation of various mouse cell lines (LLC lung carcinoma, CT26 colon carcinoma, EL4 lymphoma) were at relatively high IC50 values between 250 nM and 1 μM, and the 4T1 breast adenocarcinoma cells were even less sensitive (Fig. S1B). The treatment of immunocompetent C57BL/6 mice bearing LLC tumors (Fig. 1A) or immunodeficient NSG mice bearing 4T1 or MC38 tumors with CK2 inhibitors demonstrated modest direct antitumor effects of the compounds (Fig. S1C,D). BMS-595 inhibited the in vitro proliferation of human colorectal and lung cancer cell lines with an IC50 ranging from 10 nM to greater than 1 μM (Fig. S2). In sensitive cell lines, anti-proliferative effects of BMS-595 and its structurally related analog strongly correlated with CK2 kinase inhibition.
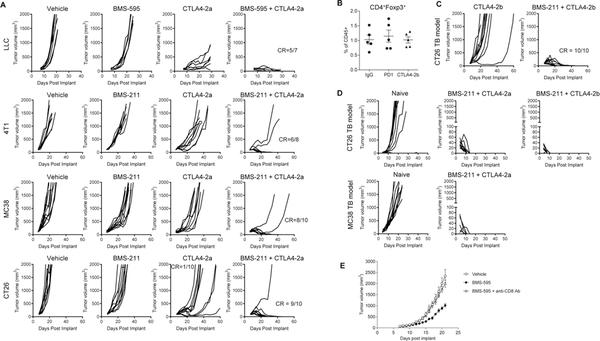
Figure 1: BMS-211 and BMS-595 enhanced the effect of immunotherapy in combination with anti-CTLA-4 antibody.
A. Individual tumor growth curves in 4 subcutaneous murine syngeneic tumor models treated with CK2 inhibitor, anti-CTLA-4 mIgG2a antibody or the combination as indicated. CR, complete response. B. 4T1 tumor bearing mice were treated with anti-CTLA-4 mIgG2b, anti-PD1 antibody or control mIgG at 20 μg/mouse on Day 6 and 10. On Day 12, the tumor cells were analyzed by flow cytometry. C. CT26 tumor bearing mice were treated with anti-CTLA-4 mIgG2b alone or in combination with CK2 inhibitor. Doses and treatment schedule are described in Materials and Methods. D. Individual tumor growth curve followed by CT26 or MC38 re-challenge. Mice from indicated groups, who rejected CT26 or MC38 tumors, were re-challenged s.c. 90 days later in the opposite site with 1×106 CT26 or MC38 tumor cells, respectively. As control, naive mice were also inoculated with the same cells (n = 10). E. LLC tumor bearing mice were treated with BMS-595 and anti-CD8 antibody. (n = 7)
However, although as a single-agent neither the CK2 inhibitor nor the immune check-point inhibitor, anti-CTLA-4-mIgG2a antibody at low dose, potently impacted antitumor activity, in combination, they showed remarkable antitumor effects in all tested tumor models (LLC, 4T1, MC38, and CT26) (Fig. 1A). Complete rejection (CR) was observed in 60–90% of mice treated with the combination therapy (Fig. 1A). Since anti-CTLA-4-mIgG2a has the potential to deplete Treg cells in tumors, in a separate set of experiments, we tested anti-CTLA-4-mIgG2b antibody, which was reported not to deplete intratumoral Treg cells in the CT26 model (22). In 4T1 model, anti-CTLA-4-mIgG2b did not deplete Treg cells in the tumors (Fig. 1B). Combination of the CK2 inhibitor with anti-CTLA-4-mIgG2b resulted in complete tumor rejection in all 10 mice used in the experiment (Fig. 1C), suggesting that Treg depletion is not involved in the effects mediated by CK2 inhibition. Mice that completely rejected tumor in the combination treatment groups were re-challenged with the same tumor cells after 90 days. All of these mice rejected a secondary tumor challenge indicating that immune memory was generated in the initial treatment (Fig. 1D). Anti-CD8 antibody injection of LLC tumor-bearing (TB) mice completely cancelled the anti-tumor efficacy of CK2 inhibitor, which suggested that CD8+ T cells were major effector cells in CK2 inhibitor-induced anti-tumor effect (Fig. 1E). Thus, although the CK2 inhibitor had limited antitumor effects, it has potent antitumor activity in combination with CTLA-4 immune check-point blockade and this effect was mediated by an immunological mechanism.
Effect of CK2 inhibition on host cells in the tumor microenvironment
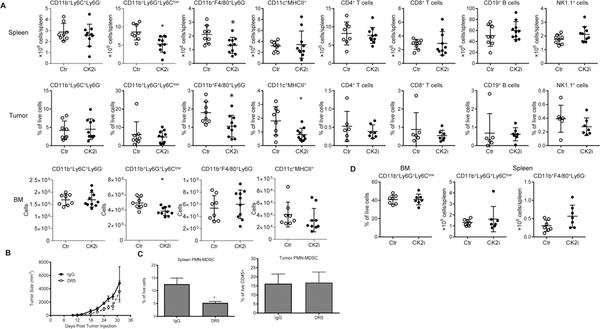
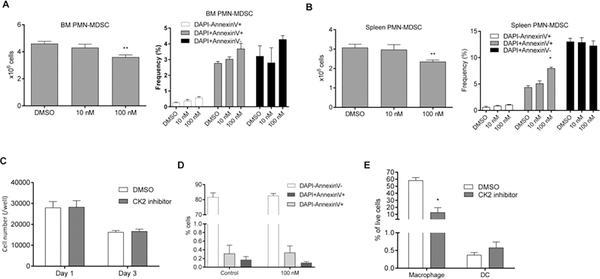
To better understand the mechanism of action of the CK2 inhibitor, we investigated the presence of myeloid cells and lymphocytes in LLC TB mice treated for 2 weeks with BMS-595. The treatment did not affect the presence of T, B, or NK cells in spleens or tumors of TB mice. It also did not affect populations of CD11b+Ly6G−Ly6Chi M-MDSC and CD11c+I-Abhi DC. However, substantial decreases in the presence of CD11b+Ly6G+Ly6Clo PMN-MDSC and CD11b+Ly6G−F4/80hi macrophages (MΦ) were observed in the spleen (Fig. 2A). In tumors, the frequency of PMN-MDSC was not significantly changed, whereas populations of tumor-associated macrophages (TAM) and DC were decreased (Fig. 2A). CK2 inhibition caused significant, albeit a modest decrease in granulocytic cells in the BM (Fig. 2A). The discrepancy between a decreased presence of granulocytic cells in the bone marrow and spleen after CK2 inhibitor treatment and the lack of changes in the tumor site could be explained by the fact that the decrease of granulocytic cells in the periphery may not be sufficient to prevent the migration of substantial numbers of PMN-MDSC to the tumor site. To verify this possibility, we depleted MDSC from spleens and blood using agonistic DR5 (TRAIL-R) antibody previously shown to selectively deplete a substantial portion of PMN-MDSC in TB mice (23). Consistent with previous observations, treatment with DR5 antibody alone had minor antitumor activity (Fig. 2B), while causing a significant reduction in PMN-MDSC in the spleens of mice (Fig. 2C). However, treatment with DR5 antibody did not change the presence of PMN-MDSC in the tumor site (Fig. 2C).
Figure 2: BMS-595 induced the changes in myeloid cell populations.
A. LLC TB mice were orally treated with vehicle or 60 mg/kg of BMS-595 for 2 weeks (n = 8–10). Single cell suspensions were prepared from spleen, tumor and bone marrow and analyzed by flow cytometry. *p < 0.05 in two-tailed Student’s t-test. B, C. LLC TB mice were treated for 10 days with DR5 antibody. Tumor growth (B) and the proportion of spleen and tumor PMN-MDSC (C) was measured. Each group included 4 mice. *p < 0.05 in two-tailed Student’s t-test. D. Tumor-free naïve C57BL/6 mice were orally treated with vehicle or 60 mg/kg of BMS-595 for 2 weeks (n = 7). Then spleen and bone marrow were processed to single cells and analyzed by flow cytometry.
Since the CK2 inhibitor affected populations of granulocytic cells and MΦ in TB mice, we tested its effect on these cells in naïve, tumor-free mice. Two-week treatment with BMS-595 did not change the proportion of neutrophils in the BM or the absolute number of these cells in the spleen. Similarly, no changes were seen in the presence of splenic MΦ (Fig. 2D).
We verified that the populations of granulocytic and monocytes cells in the spleen and tumor of TB mice were immune suppressive and can be indeed identified as PMN-MDSC and M-MDSC, respectively (Fig. S3). Treatment with the CK2 inhibitor did not abrogate suppressive activity of these cells (Fig. S3). Thus, the CK2 inhibitor caused a significant reduction in the presence of PMN-MDSC in the BM and spleens of TB mice and decreased the population of splenic MΦ and TAMs. It did not, however, affect these cells in tumor-free mice.
Mechanisms regulating the effect of CK2 inhibition on PMN-MDSC and macrophages
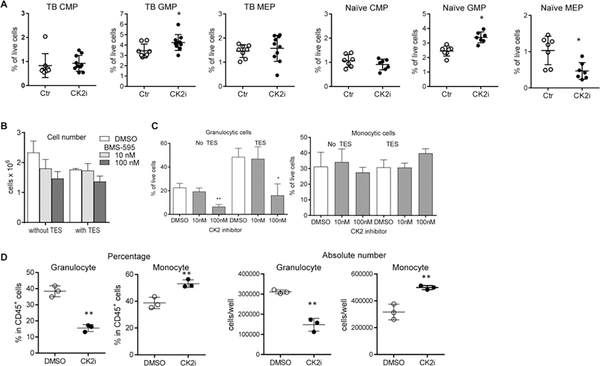
We then evaluated the effect of CK2 inhibition on the generation of myeloid cells. Treatment of LLC TB mice with BMS-595 did not affect common myeloid (CMP) and megakaryocyte–erythroid (MEP) progenitor cells in the BM, but slightly increased the proportion of granulocyte-macrophage progenitors (GMP) (Fig. 3A). This increase in GMP likely reflects compensatory changes resulting from the decreased presence of granulocytes and macrophages in tissues. In the naïve mice, BMS-595 treatment increased the proportion of GMP but decreased that of MEP (Fig. 3A). We assessed the differentiation of myeloid cells in vitro from enriched BM-derived hematopoietic progenitor cells (HPC) with and without tumor explant supernatant (TES). BMS-595 had a modest effect on the total number of cells generated from HPC during 6-day culture with GM-CSF (Fig. 3B). However, it caused a dramatic (almost 3-fold) decrease in the proportion of granulocytic cells (Fig. 3C). We evaluated the effect of BMS-595 on the myeloid cell differentiation from CD34+ progenitor cells from human cord blood. After 9 days of culture, BMS-595 decreased the proportion and the absolute number of granulocytic cells (Fig. 3D). In contrast, the proportion and the number of monocytic cells were increased by BMS-595.
Figure 3: BMS-595 decreased differentiation of granulocytic cells from mouse and human progenitor cells.
A.LLC TB mice or naïve mice were treated with vehicle or 60 mg/kg of BMS-595 for 2 weeks (n = 8–10). Bone marrow cells were processed and analyzed by flow cytometry. *- p < 0.05 in two-tailed Student’s t-test. B,C. HPCs were isolated from naïve bone marrow and cultured with or without TES and 10 or 100 nM BMS-595 for 6 days. B. Total number of cells; C. Proportion of granulocytic and monocytes cells. D. Human CD34+ progenitor cells were isolated from cord blood and cultured with TCM and 100 nM BMS-595 for 9 days. Three independent experiments were performed in triplicates. Representative data are shown. *-p < 0.05; **-p<0.01 from DMSO control in two-tailed Student’s t-test. n=3.
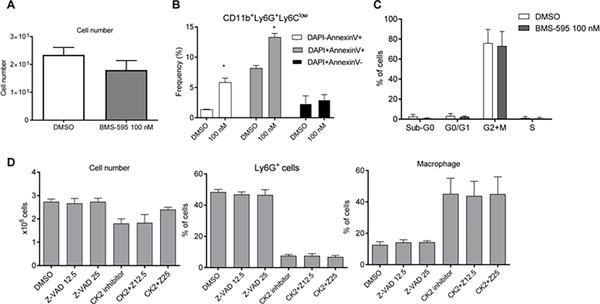
Such a dramatic decrease in granulocytic cells could be the result of BMS-595 induced apoptosis, a block of proliferation of the precursors, or inhibition of differentiation. Ly6G+ cells were isolated after 3 day-culture of HPC with GM-CSF and were treated, in the presence of GM-CSF, with BMS-595 for 24 hr. This time frame was selected because a 24 hr incubation of granulocytes with GM-CSF sustained viability of granulocytes above 70%. After 48 hrs, the viability dropped below 40% and any effect of CK2 inhibition on apoptosis is difficult to establish. The CK2 inhibitor caused a very small decrease in the total number of cells recovered after incubation (Fig. 4A), which was associated with a modest (5%) up-regulation of apoptosis (Fig. 4B). No difference in cell proliferation was observed (Fig. 4C).
Figure 4: BMS-595 did not induce apoptosis in granulocytes.
A,B. HPCs were cultured with 20 ng/mL of GM-CSF for 1 day and treated with 10% of TES. On Day 3, Ly6G+ cells were purified and treated with 100 nM of BMS-595 for 24h. A. Total number of cells collected from the culture. B. Apoptosis in the cells was analyzed by Annexin V staining using flow cytometry. C. Cell cycle of cells was analyzed after 24 hr culture of HPCs with 20 ng/mL of GM-CSF in the presence of 10% of TES, 100 nM of BMS-595, and 10 mM of BrdU. Anti-BrdU antibody and 7AAD were used in flow cytometry to evaluate cell cycle (n=3). D. HPCs were cultured with 20 ng/mL of GM-CSF, 10% of TES, 100 nM of BMS-595 and 12.5 or 25 μM of Z-VAD-FMK. On Day 6, the cells were collected and analyzed by flow cytometry.
To address the potential role of apoptosis in the substantial loss of granulocytic cells during HPC differentiation, experiments were repeated in the presence of the apoptosis inhibitor z-VAD. z-VAD at optimal doses (12–25μM) did not prevent inhibition of granulocytic cell differentiation by BMS-595 (Fig. 4D).
To clarify the effect of the CK2 inhibitor on differentiated PMN-MDSC, these cells were isolated from BM and spleens of LLC TB mice. BMS-595 caused a small decrease in the number of cells recovered after a 24 hr culture of BM PMN-MDSC, and no increase in apoptosis was observed (Fig. 5A). Similar results were observed with treatment of splenic PMN-MDSC with the CK2 inhibitor (Fig. 5B). Altogether, these results indicate that cell death plays a relatively minor role in the decrease of PMN-MDSC by CK2 inhibition in TB mice. It raised the possibility that the CK2 inhibitor may interfere with granulocytic differentiation.
Figure 5: BMS-595 did not induce apoptosis of macrophage but inhibited their differentiation from monocytes.
A,B. Ly6G+ cells were purified from bone marrow (A) and spleen (B) of LLC TB mice and cultured with 10 or 100 nM of BMS-595 for 24hr. The cells were collected, and total number of cells was counted (left panel). In parallel, apoptosis was measured by AnnexinV staining using flow cytometry. C,D. Macrophages (CD11b+Ly6G−F4/80+Ly6Clow) were sorted from tumors of LLC TB mice and cultured for 1 or 3 days with 100 nM BMS-595. C. Total number of cells.; D. Proportion of apoptotic cells measured by Annexin V on Day 3 (n=3). E. Monocytic cells (CD11b+Ly6ChighLy6G−) were sorted from naïve bone marrow cells and cultured with 20 ng/mL GM-CSF, TES and 100 nM BMS-595 for 3 days. The cells were collected, and proportion of live cells were evaluated by flow cytometry with AnnexinV staining (n=3).
In parallel, we evaluated the effect of the CK2 inhibitor on MΦ. First, we tested the hypothesis that the CK2 inhibitor causes TAM cell death. CD11b+Ly6CloLy6G−F4/80hi MΦ were sorted from the spleens and tumors of LLC TB mice and then were treated with the CK2 inhibitor. BMS-595 did not affect the total number of MΦ (Fig. 5C) or cause apoptosis in these cells (Fig. 5D). Next, we evaluated the effect of the CK2 inhibitor on MΦ differentiation. We tested the possibility that the CK2 inhibitor can block differentiation of M-MDSC to MΦ. Monocytic cells were isolated from bone marrow of naive mice and were cultured for 3 days with BMS-595 in the presence of TES. The CK2 inhibitor significantly reduced the proportion of macrophages generated from monocytic cells. The proportion of DC generated from monocytic cells was not changed (Fig. 5E). These results indicate that BMS-595 inhibited differentiation of PMN-MDSC and macrophages from their precursors without causing apoptosis of these cells.
Molecular mechanisms regulating the effect of the CK2 inhibitor on myeloid cells
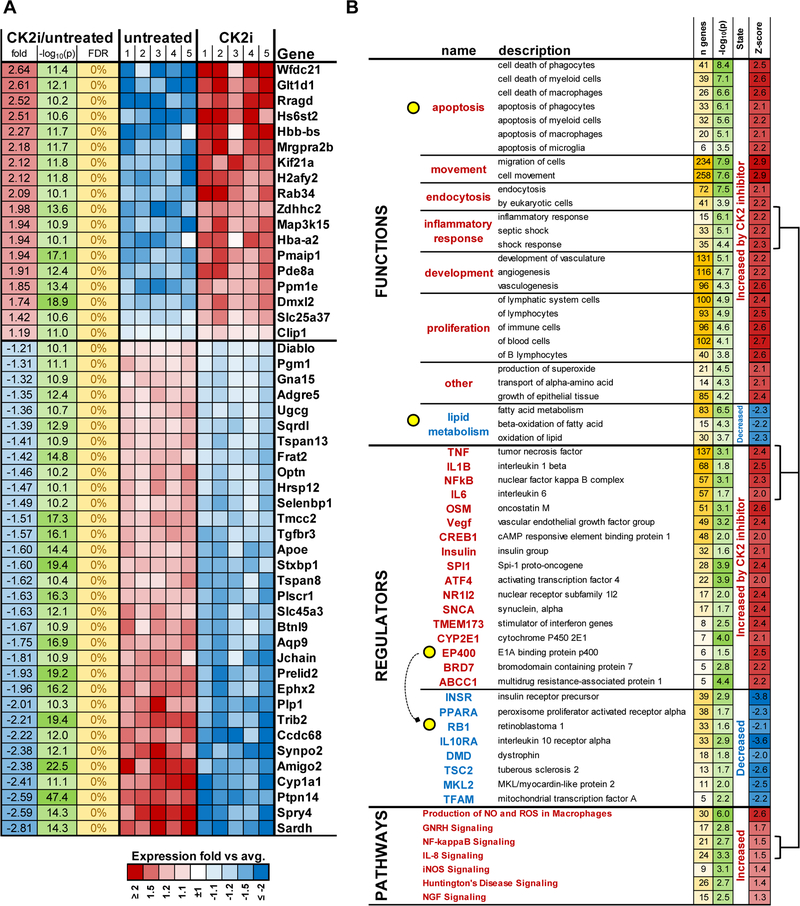
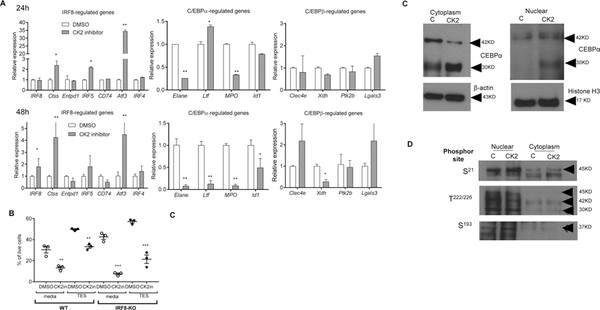
While the pro-apoptotic effects of CK2 inhibition on tumor cells is well established (7, 8, 24), the role of CK2 in the inhibition of granulocyte and macrophage differentiation is a novel finding. To elucidate this mechanism, LLC TB mice were treated for 2 weeks with BMS-595. HPC were isolated from the BM and transcriptome was evaluated by RNAseq. CK2 inhibitors caused changes in 2633 genes with FDR<5%. Most significantly changed genes are presented in Fig. 6A. Although no changes in apoptosis were determined in these cells, genes associated with apoptosis were up-regulated in HPC in mice treated with the CK2 inhibitor while down-regulated pathways included oxidation of lipids (Fig. 6B). The CK2 inhibitor caused increased expression of genes regulated by pro-inflammatory mediators: LPS, IL-1β, TNF, NF-kB, and IL-6. It also included E1A-binding protein P400, which is involved in transcriptional activation of E2F1, which is regulated by the retinoblastoma (Rb) protein. Not surprisingly, Rb1 was one of the regulators that were prominently inhibited by BMS-595 in HPC (Fig. 6). Consistent with previous results, the CK2 inhibitor caused increased expression of genes regulated by Notch signaling (Fig. S4). However, it did not prevent the substantial decrease in PMN-MDSC. Therefore, we further studied genes regulated by transcription factors that were previously directly implicated in PMN-MDSC differentiation: IRF8, C/EBPα, and C/EBPβ. BM-derived HPC from TB mice were treated with GM-CSF and BMS-595 for 24 and 48 hrs. CK2 inhibition caused a significant increase in the expression of only few genes regulated by IRF8 (Fig. 7A). To test whether this up-regulation was sufficient to block granulocytic differentiation, we used HPC isolated from the BM of IRF8 KO mice. CK2 inhibitor caused similar reduced differentiation of granulocytes from WT and IRF8 KO HPC (Fig. 7B), which exclude possible involvement of IRF8 in CK2 inhibitor-mediated effects. Treatment of HPC with BMS-595 did not affect genes regulated by C/EBPβ, however, it caused down-regulation of many genes controlled by C/EBPα (Fig. 7A). C/EBPα is a critical regulator of myeloid differentiation. Active form of C/EBPα is C/EBPα-p42 protein. C/EBPα-p30 protein is produced by alternative transcription initiation. It retains the DNA-binding domain but lacks the N terminal transactivation domain of the longer form of C/EBPα-p42. C/EBPα-p30 exhibits a dominant negative function over C/EBPα-p42 (25, 26). Treatment of HPC with the CK2 inhibitor reduced the presence of the functionally active p42 subunit of C/EBPα in the cytoplasm and increased the dominant negative p30 subunit of C/EBPα in both the cytoplasm and nucleus (Fig. 7C). No differences were found in phosphorylation of three major sites of C/EBPα (S21, T222/226, S193) (Fig. 7D). These results suggest possible mechanism regulating function of C/EBPα by CK2 inhibition.
Figure 6: BMS-595 caused changes in gene expressions.
HPCs were isolated from the LLC TB mice treated with vehicle or 60 mg/kg of BMS-595 for 2 weeks, and used for RNAseq analysis. A. most changed genes between the groups. B. changes in pathways, functions, and regulators between treated and not treated groups.
Figure 7: BMS-595 increased dominant negative p30 subunit of C/EBPα.
A. HPCs were cultured with 20 ng/mL of GM-CSF overnight and then treated with 10% TES, and 100 nM BMS-595 for 24hr and 48h. Total RNA was extracted and gene expression was analyzed by qRT-PCR. n=3, * - p<0.05; ** - p<0.01 in two-tailed Student’s t-test. B. HPCs from WT or IRF-8-KO mice (n =3) were cultured with TES and 100 nM BMS-595 for 6 days, and analyzed by flow cytometry. ** - p<0.01, ***-p<0.001 in Student’s t-test B. C,D. HPCs were cultured overnight with 20 ng/mL of GM-CSF and then treated with 10% TES and 100 nM BMS-595 for 24hr. Cells were collected and lysed to obtain cytoplasm and nuclear extracts. C. C/EBPα, β-actin and Histone H3 were detected by western blot. D. Phosphorylations on C/EBPα (S21, T222/226 and S193) were detected by western blot.
Discussion
This study was based on previous observations that CK2 may be involved in the regulation of MDSC and DC differentiation in cancer via inhibition of Notch signaling (5). We suggested that inhibition of CK2 activity with potent and selective inhibitors could improve differentiation of DC and potentially antitumor immune responses. In TB mice, the effect of the CK2 inhibitors on DC was modest. Unexpectedly, we observed a dramatic decrease in the presence PMN-MDSC and TAM. This effect was not observed in tumor-free mice. It is possible CK2 activity is more critical for abnormal myelopoiesis observed in TB mice. CK2 inhibitors dramatically enhanced the antitumor activity of anti-CTLA-4 antibody in several mouse tumor models. Since direct antitumor effects of CK2 inhibitor are marginal, it suggested that regulation of myeloid cells could be responsible for the observed synergistic antitumor effect. CK2 inhibition has a known pro-apoptotic effect in various solid and hematologic malignancies (7, 8, 24). However, its effect on myeloid cells was described in only several studies. CK2 was implicated in the regulation of phagocytosis by macrophages (27). It was also involved in the regulation of p47phox and p40phox components of NADPH oxidase in neutrophils and macrophages (28–30). No information existed on the effect of CK2 inhibition on myeloid cell differentiation.
Based on known mechanisms of action, we expected that the CK2 inhibitors would cause apoptosis of myeloid cells. However, this was not the case. Instead, its effect was associated with a block of differentiation from the precursors of granulocytes and macrophages. Since M-MDSC differentiate into TAM in tumors and MΦ in the spleens, this may explain the observed substantial decrease of MΦ after the treatment. PMN-MDSC don’t differentiate from precursors in tumors and accumulate there after migration from the circulation. The decrease of PMN-MDSC caused by CK2 inhibition was observed in the BM and spleen but not in tumors. This can be explained by the fact that despite depletion, there was still enough PMN-MDSC in circulation to migrate to the tumor site. The biological effect of CK2 inhibition apparently involves a systemic decrease of PMN-MDSC that attenuates the suppression of T cells expanded in peripheral lymphoid organs and depletion of TAM in tumors that decrease local suppression.
In the search for a mechanism of how CK2 inhibition could affect myeloid cell differentiation, we found that CK2 inhibition significantly reduced the expression of genes regulated by the C/EBPα transcription factor, a leucine zipper transcription factor mainly involved in monopoiesis and granulopoiesis. It also can affect gene expression independent of DNA-binding via interaction with E2F1 (31, 32), HDAC1 or HDAC3 (33, 34). A low level of Cebpa RNA expression is detected in Lin−Sca-1+c-Kit+ (LSK) cells; Cebpa expression increases twofold in CMP and tenfold in GMP (35). Reduced levels of C/EBPα may contribute to monopoiesis by hetero-dimerizing with AP-1 proteins such as c-Jun and c-Fos (36, 37). This may explain the fact that we did not observe changes in monocytic cells generated from HPC in the presence of the CK2 inhibitor. C/EBPα induces transcription of several regulatory proteins required for subsequent lineage maturation. This includes the transcription factors C/EBPε, Gfi-1, KLF5 (38). C/EBPα cooperates with PU.1, c-Myb, and RUNX1 to activate genes such as myeloperoxidase, neutrophil elastase, lysozyme, lactoferrin, CSF1R, CSF2R, CSF3R in immature granulocytic or monocytic cells (39). While the role of C/EBPα in granulocytic differentiation is well established, it is only in recent years that evidence has emerged suggesting it has a possible regulatory role in the differentiation of MΦ. Transient expression of C/EBPα and PU.1 in THP-1 cells synergistically promoted differentiation of monocytes to MΦ (40). C/EBPα enabled the induction of a monocytic cell differentiation program (41). Our results suggest that activation of CK2 in cancer can contribute to accumulation of PMN-MDSC and MΦ. The specific molecular mechanisms by which CK2 influences their accumulation warrants further investigation.
IRF8 is an essential transcription factor for the development of myeloid cells. It plays an important role in the switch between monocytic and granulocytic differentiation (42, 43) and its down-regulation was implicated in the accumulation of PMN-MDSC (44–46). It was previously shown that IRF8 can physically interact with C/EBPα and prevents its binding to chromatin blocking the ability of C/EBPα to stimulate transcription and neutrophil differentiation. A partial inhibition of C/EBP activity in irf8−/− haematopoietic progenitors alleviates the neutrophil overproduction in vivo (47). In our experiments, CK2 inhibition did not reverse granulocyte hyperproduction from irf8−/− progenitors in vitro suggesting that its effect was independent of Irf8. C/EBPα is phosphorylated at several sites, indicating a potential role for post-translational modifications in mediating C/EBPα activity. Several kinases were implicated in phosphorylation of C/EBPα. S21 phosphorylation is mediated by Erk1/2 and interferes with granulocytic differentiation (48). Phosphorylation of C/EBPα at S248 is mediated by activated Ras leading to the enhanced ability of C/EBPα to upregulate the expression of the granulocyte-colony stimulating factor receptor (G-CSFR) (49). Phosphorylation at S193 blocks proliferation of hepatocytes (50). Dephosphorylation of S193 promotes proliferation by preferentially binding the retinoblastoma protein (Rb) in the repressive Rb–E2F complex. A role for S193 in modulating granulopoiesis has not been reported. Finally, phosphorylation at T222 was associated with glycogen synthase kinase 3 (GSK3 kinase) activity, which regulates preadipocyte differentiation (51). We did not observe changes in phosphorylation of C/EBPα caused by the CK2 inhibitor. It is possible, however, that CK2 utilizes different sites for phosphorylation. In our study, we observed up-regulation of the 30 kDa dominant-negative isoform of C/EBPα-p30 by the CK2 inhibitor. It is known that p30 can compete for C/EBPα specific promoters with the active p42 isoform (52). Our data suggest that CK2 may affect the balance of these isoforms and thus inhibit C/EBPα activity. It is possible that the sensitivity of monocytic cells to CK2 inhibitors is different at different stages of maturation or CK2 inhibitors may affect other cell types (for instance, fibroblasts) that support myeloid cell differentiation. These possibilities require further elucidation.
Thus, our study demonstrates a novel effect of CK2 inhibition, resulting in a decrease of immune suppressive and tumor-promoting population of PMN-MDSC and TAM in TB mice. This resulted in a very substantial augmentation of antitumor activity of immune therapy. Our results suggest a novel mechanism by which CK2 may interfere with myeloid cell differentiation, leading to the inhibition of C/EBPα activity. Taken together, small molecule inhibitors targeting CK2 should be considered as a potentially valuable addition to immune-based combination therapies.
Supplementary Material
Acknowledgements
This work was supported in part by grant from Bristol-Myers Squibb and by NIH grant CA 084488 to D. Gabrilovich
Footnotes
Conflict of interest disclosure CG, AVP, HD, SW, JH, and MJK are employee of Bristol-Myers Squibb.
References
- 1.Coussens LM, Pollard JW. Leukocytes in mammary development and cancer. Cold Spring Harbor perspectives in biology. 2011;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galdiero MR, Garlanda C, Jaillon S, Marone G, Mantovani A. Tumor associated macrophages and neutrophils in tumor progression. J Cell Physiol. 2013;228:1404–12. [DOI] [PubMed] [Google Scholar]
- 3.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12:253–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Condamine T, Ramachandran I, Youn JI, Gabrilovich DI. Regulation of tumor metastasis by myeloid-derived suppressor cells. Annu Rev Med. 2015;66:97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng P, Kumar V, Liu H, Youn JI, Fishman M, Sherman S, et al. Effects of notch signaling on regulation of myeloid cell differentiation in cancer. Cancer Res. 2014;74:141–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang SH, Lu QY, Guo YH, Song YY, Liu PJ, Wang YC. The blockage of Notch signalling promoted the generation of polymorphonuclear myeloid-derived suppressor cells with lower immunosuppression. Eur J Cancer. 2016;68:90–105. [DOI] [PubMed] [Google Scholar]
- 7.Ahmad KA, Harris NH, Johnson AD, Lindvall HC, Wang G, Ahmed K. Protein kinase CK2 modulates apoptosis induced by resveratrol and epigallocatechin-3-gallate in prostate cancer cells. Mol Cancer Ther. 2007;6:1006–12. [DOI] [PubMed] [Google Scholar]
- 8.Hanif IM, Ahmad KA, Ahmed K, Pervaiz S. Involvement of reactive oxygen species in apoptosis induced by pharmacological inhibition of protein kinase CK2. Ann N Y Acad Sci. 2009;1171:591–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morgillo F, Kim WY, Kim ES, Ciardiello F, Hong WK, Lee HY. Implication of the insulin-like growth factor-IR pathway in the resistance of non-small cell lung cancer cells to treatment with gefitinib. Clin Cancer Res. 2007;13:2795–803. [DOI] [PubMed] [Google Scholar]
- 10.Yamane K, Kinsella TJ. CK2 inhibits apoptosis and changes its cellular localization following ionizing radiation. Cancer Res. 2005;65:4362–7. [DOI] [PubMed] [Google Scholar]
- 11.Di Maira G, Brustolon F, Bertacchini J, Tosoni K, Marmiroli S, Pinna LA, et al. Pharmacological inhibition of protein kinase CK2 reverts the multidrug resistance phenotype of a CEM cell line characterized by high CK2 level. Oncogene. 2007;26:6915–26. [DOI] [PubMed] [Google Scholar]
- 12.Izeradjene K, Douglas L, Delaney A, Houghton JA. Casein kinase II (CK2) enhances death-inducing signaling complex (DISC) activity in TRAIL-induced apoptosis in human colon carcinoma cell lines. Oncogene. 2005;24:2050–8. [DOI] [PubMed] [Google Scholar]
- 13.Miller SJ, Lou DY, Seldin DC, Lane WS, Neel BG. Direct identification of PTEN phosphorylation sites. FEBS Lett. 2002;528:145–53. [DOI] [PubMed] [Google Scholar]
- 14.Torres J, Pulido R. The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. J Biol Chem. 2001;276:993–8. [DOI] [PubMed] [Google Scholar]
- 15.Scaglioni PP, Yung TM, Cai LF, Erdjument-Bromage H, Kaufman AJ, Singh B, et al. A CK2-dependent mechanism for degradation of the PML tumor suppressor. Cell. 2006;126:269–83. [DOI] [PubMed] [Google Scholar]
- 16.Di Maira G, Salvi M, Arrigoni G, Marin O, Sarno S, Brustolon F, et al. Protein kinase CK2 phosphorylates and upregulates Akt/PKB. Cell death and differentiation. 2005;12:668–77. [DOI] [PubMed] [Google Scholar]
- 17.Seldin DC, Landesman-Bollag E, Farago M, Currier N, Lou D, Dominguez I. CK2 as a positive regulator of Wnt signalling and tumourigenesis. Mol Cell Biochem. 2005;274:63–7. [DOI] [PubMed] [Google Scholar]
- 18.Homma MK, Li D, Krebs EG, Yuasa Y, Homma Y. Association and regulation of casein kinase 2 activity by adenomatous polyposis coli protein. Proc Natl Acad Sci U S A. 2002;99:5959–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selby MJ, Engelhardt JJ, Quigley M, Henning KA, Chen T, Srinivasan M, et al. Anti-CTLA-4 antibodies of IgG2a isotype enhance antitumor activity through reduction of intratumoral regulatory T cells. Cancer Immunol Res. 2013;1:32–42. [DOI] [PubMed] [Google Scholar]
- 23.Condamine T, Kumar V, Ramachandran IR, Youn JI, Celis E, Finnberg N, et al. ER stress regulates myeloid-derived suppressor cell fate through TRAIL-R-mediated apoptosis. J Clin Invest. 2014;124:2626–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quotti Tubi L, Gurrieri C, Brancalion A, Bonaldi L, Bertorelle R, Manni S, et al. Inhibition of protein kinase CK2 with the clinical-grade small ATP-competitive compound CX-4945 or by RNA interference unveils its role in acute myeloid leukemia cell survival, p53-dependent apoptosis and daunorubicin-induced cytotoxicity. Journal of hematology & oncology. 2013;6:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pulikkan JA, Dengler V, Peer Zada AA, Kawasaki A, Geletu M, Pasalic Z, et al. Elevated PIN1 expression by C/EBPalpha-p30 blocks C/EBPalpha-induced granulocytic differentiation through c-Jun in AML. Leukemia. 2010;24:914–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pabst T, Mueller BU, Zhang P, Radomska HS, Narravula S, Schnittger S, et al. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat Genet. 2001;27:263–70. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki T Signal transduction mechanisms through Fc gamma receptors on the mouse macrophage surface. FASEB J. 1991;5:187–93. [DOI] [PubMed] [Google Scholar]
- 28.Kil IS, Lee JH, Yoon SH, Bae YS, Kim S, Shin SW, et al. S-Nitrosylation of p47(phox) enhances phosphorylation by casein kinase 2. Redox report : communications in free radical research. 2015;20:228–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bourdonnay E, Serezani CH, Aronoff DM, Peters-Golden M. Regulation of alveolar macrophage p40phox: hierarchy of activating kinases and their inhibition by PGE2. J Leukoc Biol. 2012;92:219–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park HS, Lee SM, Lee JH, Kim YS, Bae YS, Park JW. Phosphorylation of the leucocyte NADPH oxidase subunit p47(phox) by casein kinase 2: conformation-dependent phosphorylation and modulation of oxidase activity. Biochem J. 2001;358:783–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porse BT, Pedersen TA, Xu X, Lindberg B, Wewer UM, Friis-Hansen L, et al. E2F repression by C/EBPalpha is required for adipogenesis and granulopoiesis in vivo. Cell. 2001;107:247–58. [DOI] [PubMed] [Google Scholar]
- 32.Johansen LM, Iwama A, Lodie TA, Sasaki K, Felsher DW, Golub TR, et al. c-Myc is a critical target for c/EBPalpha in granulopoiesis. Mol Cell Biol. 2001;21:3789–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paz-Priel I, Cai DH, Wang D, Kowalski J, Blackford A, Liu H, et al. CCAAT/enhancer binding protein alpha (C/EBPalpha) and C/EBPalpha myeloid oncoproteins induce bcl-2 via interaction of their basic regions with nuclear factor-kappaB p50. Mol Cancer Res. 2005;3:585–96. [DOI] [PubMed] [Google Scholar]
- 34.Paz-Priel I, Ghosal AK, Kowalski J, Friedman AD. C/EBPalpha or C/EBPalpha oncoproteins regulate the intrinsic and extrinsic apoptotic pathways by direct interaction with NF-kappaB p50 bound to the bcl-2 and FLIP gene promoters. Leukemia. 2009;23:365–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang P, Iwasaki-Arai J, Iwasaki H, Fenyus ML, Dayaram T, Owens BM, et al. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity. 2004;21:853–63. [DOI] [PubMed] [Google Scholar]
- 36.Hong S, Skaist AM, Wheelan SJ, Friedman AD. AP-1 protein induction during monopoiesis favors C/EBP: AP-1 heterodimers over C/EBP homodimerization and stimulates FosB transcription. J Leukoc Biol. 2011;90:643–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friedman AD. C/EBPalpha in normal and malignant myelopoiesis. Int J Hematol. 2015;101:330–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007;26:6816–28. [DOI] [PubMed] [Google Scholar]
- 40.Kim MS, Kang JW, Park YS, Lee DH, Bak Y, Kwon T, et al. IL-32theta inhibits monocytic differentiation of leukemia cells by attenuating expression of transcription factor PU.1. Oncotarget. 2015;6:4394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koffel R, Meshcheryakova A, Warszawska J, Hennig A, Wagner K, Jorgl A, et al. Monocytic cell differentiation from band-stage neutrophils under inflammatory conditions via MKK6 activation. Blood. 2014;124:2713–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H, Yan M, Sun J, Jain S, Yoshimi R, Abolfath SM, et al. A reporter mouse reveals lineage-specific and heterogeneous expression of IRF8 during lymphoid and myeloid cell differentiation. J Immunol. 2014;193:1766–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holtschke T, Lohler J, Kanno Y, Fehr T, Giese N, Rosenbauer F, et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87:307–17. [DOI] [PubMed] [Google Scholar]
- 44.Stewart TJ, Greeneltch KM, Reid JE, Liewehr DJ, Steinberg SM, Liu K, et al. Interferon regulatory factor-8 modulates the development of tumour-induced CD11b+Gr-1+ myeloid cells. Journal of cellular and molecular medicine. 2009;13:3939–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waight JD, Netherby C, Hensen ML, Miller A, Hu Q, Liu S, et al. Myeloid-derived suppressor cell development is regulated by a STAT/IRF-8 axis. J Clin Invest. 2013;123:4464–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paschall AV, Zhang R, Qi CF, Bardhan K, Peng L, Lu G, et al. IFN Regulatory Factor 8 Represses GM-CSF Expression in T Cells To Affect Myeloid Cell Lineage Differentiation. J Immunol. 2015;194:2369–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kurotaki D, Yamamoto M, Nishiyama A, Uno K, Ban T, Ichino M, et al. IRF8 inhibits C/EBPalpha activity to restrain mononuclear phagocyte progenitors from differentiating into neutrophils. Nature communications. 2014;5:4978. [DOI] [PubMed] [Google Scholar]
- 48.Ross SE, Radomska HS, Wu B, Zhang P, Winnay JN, Bajnok L, et al. Phosphorylation of C/EBPalpha inhibits granulopoiesis. Mol Cell Biol. 2004;24:675–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Behre G, Singh SM, Liu H, Bortolin LT, Christopeit M, Radomska HS, et al. Ras signaling enhances the activity of C/EBP alpha to induce granulocytic differentiation by phosphorylation of serine 248. J Biol Chem. 2002;277:26293–9. [DOI] [PubMed] [Google Scholar]
- 50.Wang GL, Timchenko NA. Dephosphorylated C/EBPalpha accelerates cell proliferation through sequestering retinoblastoma protein. Mol Cell Biol. 2005;25:1325–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu H-K, Perrier S, Lipina C, Finlay D, McLauchlan H, Hastie CJ, et al. Functional characterisation of the regulation of CAAT enhancer binding protein alpha by GSK-3 phosphorylation of Threonines 222/226. BMC Molecular Biology. 2006;7:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Avellino R, Delwel R. Expression and regulation of C/EBPalpha in normal myelopoiesis and in malignant transformation. Blood. 2017;129:2083–91. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.