Abstract
Exposure to intrauterine heat stress during late gestation affects offspring performance into adulthood. However, underlying mechanistic links between thermal insult in fetal life and postnatal outcomes are not completely understood. We examined morphology, DNA methylation, and gene expression of liver and mammary gland for bull calves and heifers that were gestated under maternal conditions of heat stress or cooling (i.e. in utero heat stressed vs. in utero cooled calves). Mammary tissue was harvested from dairy heifers during their first lactation and liver from bull calves at birth. The liver of in utero heat stressed bull calves contained more cells and the mammary glands of in utero heat stressed heifers were comprised of smaller alveoli. We identified more than 1,500 CpG sites differently methylated between maternal treatment groups. These CpGs were associated with approximately 400 genes, which play a role in processes, such as development, innate immune defense, cell signaling, and transcription and translation. We also identified over 100 differentially expressed genes in the mammary gland with similar functions. Interestingly, fifty differentially methylated genes were shared by both bull calf liver and heifer mammary gland. Intrauterine heat stress alters the methylation profile of liver and mammary DNA and programs their morphology in postnatal life, which may contribute to the poorer performance of in utero heat stressed calves.
Introduction
Early life experiences can program physiological function and health and disease outcomes later in life1,2. Perturbations in the intrauterine environment, from maternal malnutrition and stress to elevated body temperature during pregnancy, can induce structural and functional changes in the fetus that may persist through adulthood3–5. In several species, maternal heat stress (i.e. high ambient temperature) during gestation affects the growth and physiology of the developing young6–8. In dairy cattle, calves born to cows exposed to high ambient temperature during late gestation are lighter and shorter at birth and through puberty, have higher morbidity associated with depressed immune competence, and exhibit alterations in energy metabolism and reproductive physiology compared to those born to cows that were actively cooled6,9–13. Moreover, the in utero heat stressed female calves (i.e. heifers) produce significantly less milk during their first lactation, approximately two years after the developmental insult10.
Late gestation is characterized by enhanced muscle growth and fat deposition that contribute to rapid fetal weight gain14. In fact, 60% of calf birth weight is attained during the last 2 months of gestation15 and, during this time, organs undergo functional maturation in preparation for sustaining life outside the womb16. The liver and mammary gland are key organs mediating in utero heat stress effects on physiology and performance in dairy cattle17,18. Development of the mammary gland begins in the conceptus and although there is little formation of mammary parenchymal tissue in utero, the rudimentary ductal system upon which the functional alveoli will later form, develops prior to birth19,20. The liver is a key metabolic organ, orchestrating carbohydrate, lipid, and amino acid homeostasis to support the energetic demands of growth, maintenance, and production. Moreover, the liver is recognized as an important immune organ21–23.
The mechanisms through which in utero heat stress in late gestation affects postnatal liver and mammary function have yet to be elucidated, but could be attributed, in part, to impairment of tissue growth during the organ maturation stage of fetal development. Insults during late gestation can depress cell proliferation in tissues thereby reducing cell number and stunting tissue growth24,25. Ahmed et al.26 reported lower liver weight among in utero heat stressed bull calves at birth. Furthermore, acute exposure to high temperatures in mice induced hepatocyte apoptosis and reduced cell proliferation27. In rats, the density of hepatocytes increased concurrent with the number of consecutive days of heat stress exposure, despite general hepatic degeneration28. In terms of mammary development, exposure of multiparous dry cows to heat stress depressed mammary epithelial cell proliferation, potentially reducing the total number of cells in the mammary gland capable of producing milk18.
Influences of the early life environment on phenotypic variation can be mediated by epigenetic modifications that regulate gene expression. The most stable and well-studied epigenetic mark is DNA methylation. In mammals, methylation occurs predominately at CpG dinucleotides29 that can be methylated or demethylated by developmental signals or environmental cues30,31. Aberrant methylation patterns occurring during early development may be maintained throughout the life of the animal and permanently alter tissue-specific gene expression and function32. The liver and mammary gland are regulated by epigenetic mechanisms that may be altered by heat stress. Milk synthesis and mammary development are regulated by epigenetic processes in the dairy cow33,34; global DNA methylation within mammary epithelial cells appears to play a role in mammary cell differentiation and maintenance33. In the lactating cow, remethylation of the typically hypomethylated casein gene promoter drastically reduces casein mRNA expression and milk protein synthesis during acute udder infection35. DNA methylation also plays an integral role in hepatic differentiation and hepatocyte proliferation36. Subjecting male guinea pigs to chronic heat stress alters DNA methylation patterns in the liver of both the F0 and F1 generations37. However, the mechanistic links between developmental insult and mammary and liver function have yet to be fully elucidated. Our hypothesis is that in utero heat stress drives changes in phenotype by altered DNA methylation of key regulatory gene pathways in the liver and mammary gland. Specific objectives of this study were to, 1) examine effects of fetal exposure to intrauterine heat stress during late gestation on the postnatal mammary gland and liver tissue microstructure and, 2) to characterize changes in DNA methylation and gene expression that may explain the observed phenotypic outcomes.
Results
Liver and mammary gland histology
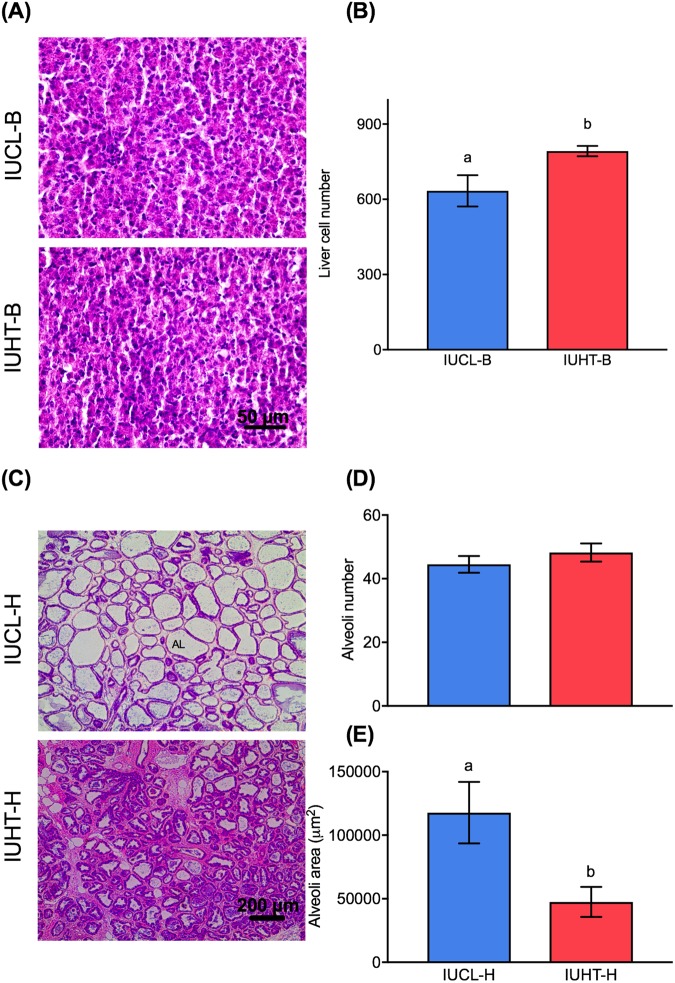
The liver of in utero heat stressed bull calves (IUHT-B) contained significantly more cells than the liver of in utero cooled bull calves (IUCL-B; 792.2 ± 20.5 vs. 634 ± 62.4, respectively; t = 2.41, df = 8, P = 0.04; Fig. 1A,B). The mammary glands of in utero heat stressed heifers (IUHT-H) and in utero cooled heifers (IUCL-H) had a similar number of alveoli (t = 0.95, df = 5, P = 0.38), but the area of IUCL-H alveoli was significantly larger than IUHT-H (t = 2.61, df = 5, P = 0.05; Fig. 1C–E).
Figure 1.
Histological evaluation of liver and mammary gland tissue. Liver was collected from in utero heat stressed (IUHT-B) and in utero cooled (IUCL-B) bull calves at birth. Mammary tissue was collected from in utero heat stressed (IUHT-H) and in utero cooled (IUCL-H) heifers at 21 days into their first lactation. (A) Photomicrograph of hematoxylin and eosion (H&E) stained liver tissue at 40x. (B) Difference in the number of liver cells between IUHT-B and IUCL-B. The liver of IUHT-B contained more cells than the liver of IUCL-B (P = 0.04). (C) Photomicrograph of H&E stained mammary tissue at 20x. AL = alveolar lumen. (D) Difference in the number of alveoli in the mammary glands of IUHT-H and IUCL-H. Alveoli number was similar between treatment groups (P = 0.38). (E) Difference in area of alveoli in the mammary glands of IUHT-H and IUCL-H. Alveoli area was larger for IUCL-H relative to IUHT-H (P = 0.05). Data are presented as mean ± SEM. Different letters indicate a statistically significant difference.
Differentially methylated genes and gene set enrichment analysis
Bull calf liver
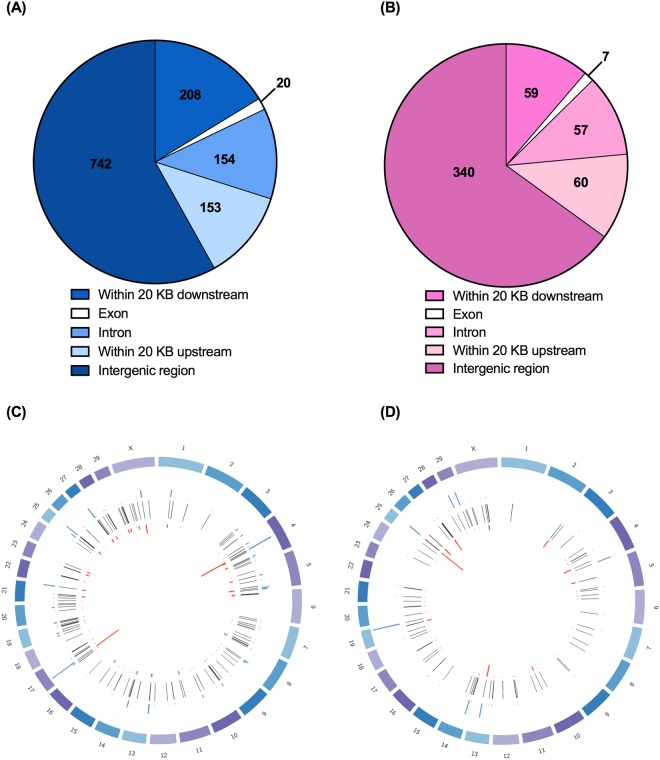
In liver tissue of IUCL-B, 421 million cytosines were identified in the CpG context and 78.1% of those cytosines were methylated. For IUHT-B, there were 243 million cytosines in the CpG context, 79.5% of which were methylated. Based on our cut-off criteria of ≥15% methylation difference and q-value ≤ 20%, there were 1,277 differentially methylated cytosines (DMCs) between IUCL-B and IUHT-B. Principal component analysis based on DMCs showed clear separation into in utero heat stressed and in utero cooled groups (Supplementary Fig. S1). Location of DMCs were identified as within a gene (intron or exon), within 20 Kb upstream of the transcription start site of a gene, or within 20 Kb downstream of the gene end. DMCs beyond 20 KB upstream or downstream of the gene were considered to be in the intergenic region. The majority of DMCs resided in the intergenic regions (n = 744), but a substantial number were associated with other genomic features (Fig. 2A). Of the 153 DMCs within 20 Kb upstream of the transcription start site, 68 are within 10 Kb upstream and 35 were within 5 Kb upstream. Of the 208 DMCs downstream of the gene end, 152 were within 10 Kb downstream and 91 were within 5 Kb downstream. The 1,277 DMCs comprised 239 differentially methylated genes (DMGs) that were distributed across most of the bovine chromosomes (Fig. 2C, Supplementary Table S1). DMGs included 16 non-coding RNAs, 38 encoding rRNAs (30 code for 5S rRNA and 8 code for 5.8S rRNA), and 43 novel bovine genes. Seven of the novel bovine genes are pseudogenes and the rest are protein-coding genes. A few interesting DMGs between IUHT-B and IUCL-B liver are advanced glycosylation end-product specific receptor (AGER), mediator complex subunit 1 (MED1), and BCL2 associated athanogene 1 (BAG1).
Figure 2.
Genomic locations of differentially methylated cytosines (DMCs) and genes (DMGs). Liver tissue was collected from in utero heat stressed (IUHT-B) and in utero cooled (IUCL-B) bull calves at birth. Mammary tissue was collected from in utero heat stressed (IUHT-H) and in utero cooled (IUCL-H) heifers at 21 days into their first lactation. (A) Genomic locations of DMCs for bull calf liver; (B) Genomic locations of DMCs for heifer mammary gland. Cut-off criteria for defining DMCs was 15% methylation difference between treatment groups and q-value < 0.2. DMCs were located within 20 KB upstream of the transcription start site of a gene, within introns, within exons, or within 20 KB downstream of the gene end. DMCs more than 20 KB upstream or more than 20 KB downstream of a gene were considered to reside in the intergenic region. (C) Chromosomal location of DMGs and DMCs for bull calf liver; (D) Chromosomal location of DMGs and DMCs for heifer mammary gland. Outer circle represents the chromosomes where DMCs and DMGs are located. The middle circle of black bars represents DMGs. DMGs contain at least 1 DMC. The red and blue bars indicate cytosines that are hypo-and hypermethylated in the heat stressed group relative to the cooled group, respectively. Height of the bars corresponds to the number of DMCs.
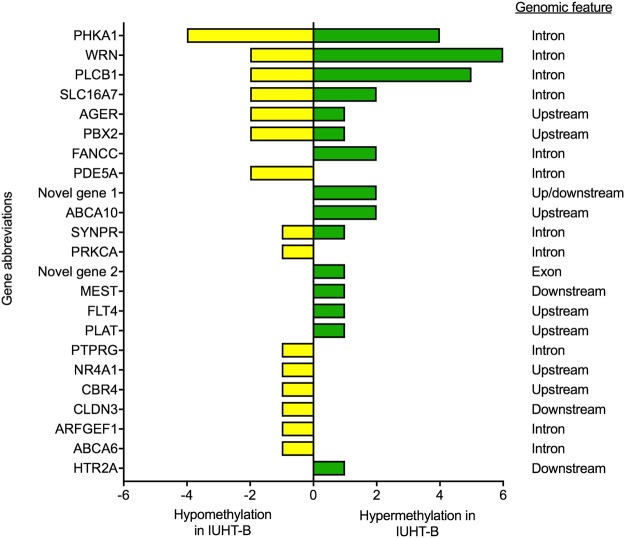
Based on enrichment analyses, the significant pathways and functions for DMGs included transcription, immune function, cell signaling, enzyme activity, cell cycle, and development (Table 1). Among DMGs in these pathways, some genes contained cytosines that were hypomethylated in IUHT-B compared to IUCL-B, some only hypermethylated cytosines, and some contained both hypo- and hypermethylated cytosines (Fig. 3, Supplementary Table S2). For example, there were 3 DMCs upstream of the AGER gene; 1 DMC was hypermethylated whereas 2 cytosines were hypomethylated in the liver of IUHT-B relative to IUCL-B.
Table 1.
Significant pathways, biological functions, and molecular processes of differentially methylated genes in the liver of in utero heat stressed (IUHT-B) compared to in utero cooled bull calves (IUCL-B).
| Pathways and functions | Genes in pathway | Differentially methylated genes | P-value |
|---|---|---|---|
| Transcription factors | 188 | PTPRG, NOVEL GENE 1, NOVEL GENE 2, SYNPR, PDE5A | 0.03 |
| Immunologic receptors | 48 | AGER, PRKCA, SYNPR, PTPRG | <0.001 |
| Positive regulation of MAPK cascade | 105 | PDE5A, HTR2A, PLCB1, PRKCA | 0.02 |
| Calcium signaling pathway | 163 | HTR2A, PHKA1, PLCB1, PRKCA | 0.07 |
| Gap junction | 83 | PRKCA, PLCB1, HTR2A | 0.04 |
| Regulation of nucleotide metabolic process | 62 | PRKCA, HTR2A, PDE5A | 0.02 |
| Fat cell differentiation | 57 | NR4A1, PLCB1, HTR2A | 0.02 |
| Cell cycle proteins | 67 | FANCC, WRN, NOVEL GENE 2 | 0.02 |
| ABC transporters | 41 | ABCA6, ABCA10 | 0.06 |
| Positive regulation of glycoprotein biosynthetic pathway | 6 | PLCB1, ARFGEF1 | 0.001 |
| Protein heterooligomerization | 13 | CBR4, CLDN3 | 0.007 |
| Mesoderm development | 20 | MEST, NOVEL GENE 2 | 0.02 |
| Phosphoric diester hydrolase activity | 32 | PLCB1, PDE5A | 0.04 |
| Major histocompatibility complex | 20 | AGER, PBX2 | 0.01 |
| Zinc fingers | 33 | SYNPR, PTPRG | 0.03 |
| RNA splicing | 37 | PRKCA, NOVEL GENE 2 | 0.04 |
| Sodium-hydrogen antiporter | 8 | PLCB1, SLC16A7 | 0.002 |
| Phosphatidylinositols | 12 | PLCB1, HTR2A | 0.005 |
| Urokinase-type plasminogen activator | 15 | PLAT, FLT4 | 0.008 |
| Type C phospholipase | 23 | PLCB1, AGER | 0.02 |
| Interleukin-8 | 29 | SYNPR, PTPRG | 0.03 |
Note: Enrichment analysis of differentially methylated genes was achieved through the Kyoto Encyclopedia for Genes and Genomes (KEGG) database, Medical Subject Headings (MeSH) database, and Gene Ontology Consortium database. P-values are derived from Fisher’s exact tests and indicate significance of enrichment. Italics indicate tendency for enrichment.
Figure 3.
Number and direction of differentially methylated cytosines (DMCs) corresponding to differentially methylated genes (DMGs) in the liver of in utero heat stressed (IUHT-B) compared to in utero cooled (IUCL-B) bull calves. DMGs are of the significant pathways and biological functions (from Table 1), identified through enrichment analysis. Hypomethylated cytosines are indicated by the yellow bars. Green bars denote hypermethylated cytosines. Hypo- or hypermethylation of cytosines refers to the in utero heat stressed group relative to the in utero cooled group.
Heifer mammary gland
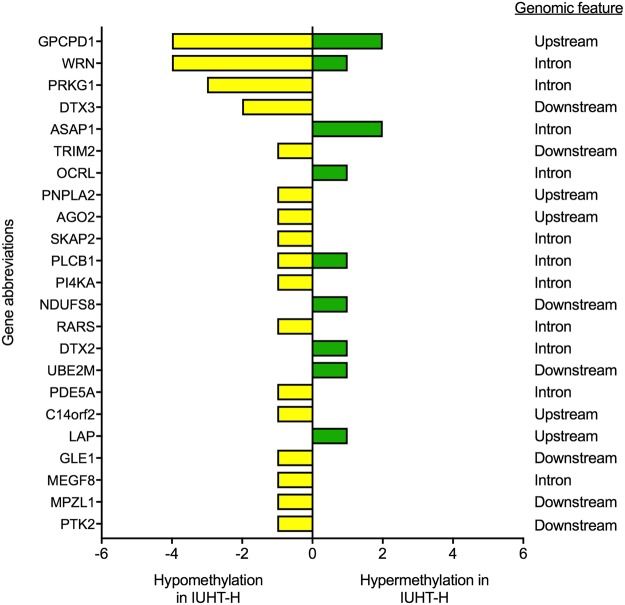
We observed 245 million and 237 million cytosines in CpG enriched areas of mammary DNA for IUCL-H and IUHT-H groups, respectively. The percent of cytosine methylation at CpGs was similar between IUCL-H and IUHT-H (IUCL-H, 74.1%; IUHT-H, 74.8%). There were 523 DMCs between treatment groups (methylation difference ≥15%, q-value ≤ 20%). Principle components analysis showed a slight separation between IUCL-H and IUHT-H groups (Supplementary Fig. S1). DMCs were found within the gene body, and upstream or downstream of the gene, but mostly occurred in the intergenic regions (n = 340; Fig. 2B). Of the 60 DMCs within 20 Kb upstream of the transcription start site, 34 were within 10 Kb upstream and 17 were within 5 Kb upstream. Of the 59 DMCs downstream of the gene end, 40 were within 10 Kb downstream and 18 were within 5 Kb downstream. The 523 DMCs corresponded to 135 DMGs spread across the genome (Fig. 2D, Supplementary Table S1), including 7 non-coding RNAs, 19 genes for 5S rRNA, 8 genes for 5.8S rRNA, and 23 novel bovine genes. Three of the novel genes are non-coding RNAs, 4 are pseudogenes, and 16 are protein coding. A few interesting DMGs between IUHT-H and IUCL-H mammary gland were cGMP dependent protein kinase 1 (PRKG1), phospholipase C beta 1 (PLCB1), and tripartite motif genes (TRIM 28 and TRIM 37). Significant pathways and biological functions identified through enrichment analysis included protein binding, phosphorylation, enzyme and cell activation, and cell signaling (Table 2). DMGs within these pathways varied in the number of DMCs they contained and the direction of methylation difference (Fig. 4, Supplementary Table S2).
Table 2.
Significant pathways, biological functions, and molecular processes of differentially methylated genes in the mammary gland of in utero heat stressed (IUHT-H) versus in utero cooled heifers (IUCL-H).
| Pathways and functions | Genes in pathway | Differentially methylated genes | P-value |
|---|---|---|---|
| Protein binding | 625 | ASAP1, C14orf2, PDE5A, PI4KA, PRKG1, WRN, MEGF8, NOVEL GENE 3 | 0.006 |
| Phosphorylation | 351 | PDE5A, PRKG1, ASAP1, MPZL1, PTK2 | 0.02 |
| Enzyme activation | 273 | PRKG1, PTK2, ASAP1, PLCB1 | 0.03 |
| Cell activation | 177 | PLCB1, PRKG1, SKAP2, PDE5A | 0.02 |
| Hydrolase activity, acting on ester bonds | 243 | AGO2, PNPLA2, PLCB1, PDE5A | 0.04 |
| Inositol phosphate metabolism | 54 | PLCB1, PIK4A, OCRL | 0.003 |
| Tyrosine | 88 | MPZL1, PTK2, NDUFS8 | 0.007 |
| Adenosine triphosphate | 120 | C14orf2, PRKG1, WRN | 0.01 |
| Ligase activity | 111 | TRIM2, UBE2M, RARS | 0.02 |
| Regulation of GTPase activity | 112 | ASAP1, PLCB1, PRKG1 | 0.02 |
| Notch signaling pathway | 39 | DTX2, DTX3 | 0.02 |
| Gap junction | 83 | PLCB1, PRKG1 | 0.08 |
| Negative regulation of cell-cell adhesion | 19 | PDE5A, PRKG1 | 0.005 |
| Regulation of translational initiation | 31 | AGO2, GLE1 | 0.01 |
| cGMP binding | 10 | PDE5A, PRKG1 | 0.001 |
| Lipase activity | 29 | PLCB1, PNPLA2 | 0.01 |
| Allosteric site | 16 | PDE5A, PRKG1 | 0.002 |
| Src homology domains | 17 | MPZL1, ASAP1 | 0.002 |
| Cation transport proteins | 21 | PLCB1, MEGF8 | 0.004 |
| Phosphotyrosine | 25 | PTK2, ASAP1 | 0.005 |
| Phosphoric diester hydrolases | 26 | GPCPD1, PDE5A | 0.006 |
| Src-family kinases | 34 | PTK2, ASAP1 | 0.01 |
| Cyclic GMP | 39 | PDE5A, PRKG1 | 0.01 |
| Calcium channels | 43 | PLCB1, PRKG1 | 0.02 |
Note: Enrichment analysis of differentially methylated genes was achieved through the Kyoto Encyclopedia for Genes and Genomes (KEGG) database, Medical Subject Headings (MeSH) database, and Gene Ontology Consortium database. P-values are derived from Fisher’s exact tests and indicate significance of enrichment. Italics indicate tendency for enrichment.
Figure 4.
Number and direction of differentially methylated cytosines (DMCs) corresponding to differentially methylated genes (DMGs) in the mammary gland of in utero heat stressed (IUHT-H) compared to in utero cooled (IUCL-H) heifers. DMGs are of the significant pathways and biological functions (from Table 2), identified through enrichment analysis. Hypomethylated cytosines are indicated by the yellow bars. Green bars denote hypermethylated cytosines. Hypo- or hypermethylation of cytosines refers to the in utero heat stressed group relative to the in utero cooled group.
Shared DMGs between bull calf liver and heifer mammary tissue
There were 50 common genes that were differentially methylated both between IUHT-B and IUCL-B and between IUHT-H and IUHT-CL (Fig. 5). Functions of the DMGs included mitochondrial function and oxidative defense (IMMP2L, C14orf2), DNA repair (WRN), cell signaling (PLCB1, PDE5A), transcription (CUX1, ZNF395, MED1), intracellular transport (KIF19, HOOK1), and cell adhesion and migration (UNC5D, KIRREL3). Additionally, 12 of the shared DMGs code for 5S rRNA, 8 code for 5.8S rRNA, and 10 are novel bovine genes (Supplementary Table S3). Of the novel bovine genes, 8 are protein-coding genes and 2 are pseudogenes.
Figure 5.
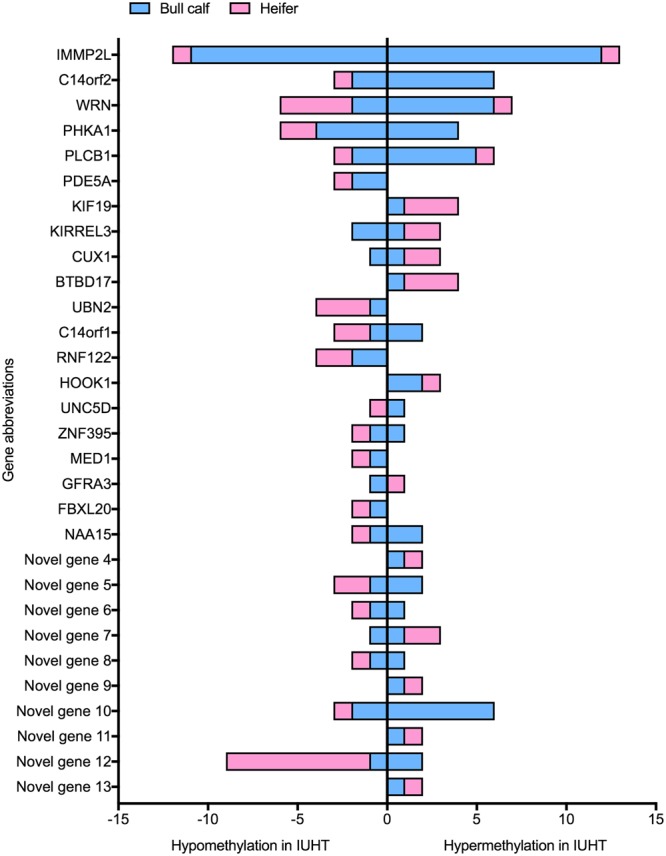
Number and direction of differentially methylated cytosines (DMCs) corresponding to 30 shared differentially methylated genes (DMGs) between liver and mammary gland tissue. Liver was collected from in utero heat stressed (IUHT-B) and in utero cooled (IUCL-B) bull calves at birth. Mammary tissue was collected from in utero heat stressed (IUHT-H) and in utero cooled (IUCL-H) heifers at 21 days into their first lactation. Methylation comparison was made between IUHT and IUCL groups for bull calves and heifers separately. Hypo- or hypermethylation of cytosines refers to IUHT relative to IUCL. Pink bars denote heifer mammary gland, blue bars indicate bull calf liver. Novel genes are genes that have not yet been characterized in the bovine genome. In addition to the 30 shared DMGs shown here, there were an additional 20 shared DMGs that code for rRNAs.
Differentially expressed genes and gene set enrichment analysis
We identified 15,366 genes expressed in the mammary gland of heifers, 117 of which were differentially expressed between IUHT-H and IUCL-H (P ≤ 0.01; Supplementary Table S4). Major functions of the differentially methylated genes (DEGs) included immunity, metabolism, cell signaling, and development (Supplementary Table S5). One of the DEGs is a non-coding RNA and 16 are novel bovine genes. Of the novel bovine genes, 15 are protein-coding genes and 1 is a pseudogene.
Correlation between methylation and gene expression
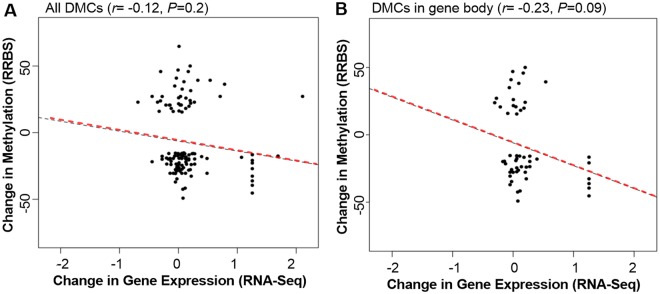
There was a non-significant negative correlation between change in DNA methylation and change in gene expression between IUHT-H and IUCL-H (r = −0.12, P = 0.2; Fig. 6A) and a tendency for a significant negative correlation between change in DNA methylation and change in gene expression when only including DMCs located in the gene body (r = −0.23, P = 0.09; Fig. 6B). There was no significant relationship between differential methylation of DMCs upstream or downstream of the gene and gene expression.
Figure 6.
Relationship between differential cytosine methylation and differential gene expression in the mammary gland of in utero heat stressed (IUHT-H) and in utero cooled (IUCL-H) heifers. (A) Overall DMCs regardless of their genomic location. There was a non-significant negative correlation between differential methylation and differential gene expression when including DMCs regardless of genomic location (r = −0.12, P = 0.2). (B) Includes only DMCs located in the gene body. There was a tendency for a significant negative correlation between differential methylation and differential gene expression when including only DMCs located in the gene body (r = −0.23, P = 0.09).
Discussion
In dairy cattle, exposure of the pregnant cow to environmental heat stress impacts her calf up to at least two years after the insult. In particular, calves born to heat stressed dams have reduced immune function, altered systemic metabolism, and reduced milk production during the first lactation6,9–13. The underlying mechanisms responsible for phenotypic effects of in utero heat stress, however, are still unknown. Herein we examined DNA methylation and gene expression changes in the mammary gland as potential mechanisms modulating in utero heat stress effects on offspring. We identified approximately 400 genes that were differentially methylated and approximately 100 genes that were differentially expressed between in utero heat stressed and in utero cooled cattle, corroborating previous documentation of heat induced changes in DNA methylation. Two paternal imprinted genes in mouse blastocysts were demethylated upon acute exposure to high temperature38. Wild guinea pig males exposed to chronic high temperature showed changes in hundreds of methylated regions in the liver after heat exposure37. Furthermore, differences in methylation levels in liver and testis were detected in sons conceived before and after paternal exposure to heat, indicative of transgenerational epigenetic inheritance37.
Based on the differentially methylated genes in bull calf liver, enrichment analysis revealed several pathways and biological functions potentially epigenetically altered by in utero exposure to heat stress, such as immune function, gene and protein expression, including posttranscriptional and posttranslational modifications, and cell processes. Immune related pathways, functions, and terms included immunologic receptors, major histocompatibility complex, and interleukin-8. The AGER gene appears in multiple immune related pathways and encodes a receptor for advanced glycation end products (RAGE), a pattern recognition receptor belonging to the immunoglobulin superfamily39–41. RAGE is involved in a number of intracellular signaling pathways that regulate cell functions, such as proliferation, apoptosis, autophagy, cell migration, and inflammation42. Binding of certain ligands to RAGE can also generate reactive oxygen species by activation of NADPH oxidase43 and results in the cellular depletion of glutathione and ascorbic acid, two important antioxidants44, which may contribute to oxidative stress. A recent proteomics analysis of the liver of periparturient cows heat stressed during late gestation found evidence of oxidative stress17. In the present study, genes contributing to superoxide metabolism, such as NOXO1, GSTT1, and SOD2, were upregulated in the mammary gland of IUHT-H relative to IUCL-H. Higher levels of pro-inflammatory cytokines have been detected in peripheral blood mononuclear cells of heat stressed pregnant cows and in utero heat stressed calves have higher circulating acute phase proteins, whose production is induced by pro-inflammatory cytokines45,46. Together, these results suggest that altered DNA methylation patterns may contribute to heightened inflammatory responses and oxidative stress among in utero heat stressed cattle.
Several genes involved in chromatin structure and the transcriptional machinery were differentially methylated in the liver of IUHT-B versus IUCL-B, including MED1 encoding a subunit of a transcription activator47, H2AY encoding a core histone48, and EPOP, which regulates chromatin structure at actively transcribed genes49. Three genes in the zinc finger gene family, ZMAT5, ZNF608, and ZNF395 were differentially methylated in the liver. Many zinc fingers function as transcription factors, binding specific long sequences of DNA to control gene transcription50–52. ZNF395, specifically, appears to play a role in innate immunity, through a hypoxia-induced inflammatory response and through an antiviral pathway mediated by NF-κB activation. Interestingly, both NF-κB activation and ZNF395 expression are induced by hypoxia and ZNF395 is a target gene of HIFα, an integral mediator of the cellular response to hypoxia53–56. Heat stress during gestation reduces placental mass and vascularization, leading to lower oxygen and nutrient delivery to the conceptus and intrauterine growth restriction8,57–59. Thus, it is conceivable that fetal methylation of ZNF395 in the liver is altered by heat stress-induced intrauterine hypoxia, which may have implications for innate immune function.
Calves born to dams heat stressed during late gestation had a greater number of cells in the liver relative to those born to cooled dams. Similarly, rats exposed to heat stress over several consecutive days had greater hepatocyte density28. Differential methylation of genes involved in apoptosis and proliferation, such as BAG1 and PRKCA60–63, may be associated with heat stress-induced effects on tissue and organ growth as organ size is determined by regulation of these cell processes. The weight of several organs, including the liver, was lighter at birth in utero heat stressed bull calves26. Given that organ mass is a product of cell number as well as cell mass and the mass of the extracellular compartment64, the in utero heat stressed calves likely have smaller cells and/or reduced extracellular matrix. Although hepatocytes account for approximately 80% of liver mass65,66, it is unclear if the greater hepatic cell number in heat stressed bull calves in our study is due to the presence of more hepatocytes or other cell populations, such as stellate cells or immune cells.
The majority of pathways and functions of differentially methylated genes in heifer mammary gland were related to protein activity and cell signaling. Terms included phosphorylation, hydrolase activity, ligase activity, GTPase activity, lipase activity, enzyme activation, and protein binding. DMGs in these pathways, such as PRKG1, PI4KA, PLCB1, ASAP1, and PTK2, regulate a variety of physiological processes involved in mammary morphogenesis and milk synthesis67–73. PRKG1, PI4KA, and PLCB1 regulate intracellular calcium concentration. Calcium is an essential ion for functions such as milk synthesis, immune activation, cell proliferation, and signal transduction. PI4KA and PLCB1 encode enzymes in phosphoinositol biosynthetic pathways, indirectly affecting intracellular calcium release through formation of inositol 1,4,5-trisphosphate (IP3), an integral second messenger67,68. PLCB1 also catalyzes the formation of diacylglycerol (DAG), another important second messenger67. The phosphoinositide pathway is one pathway mediating the mitogenic activity of PRL in mammary epithelial cells69. On the other hand, PRKG1 encodes cGMP dependent protein kinase 1, an intracellular receptor for cGMP, which negatively regulates the release of intracellular calcium by inhibiting IP3-mediated Ca release70. Results of RNA sequencing and histological evaluation of the mammary gland lend further support for in utero heat stress effects on these pathways and on mammary development. Cell signaling and protein activity were main functions enriched in our dataset of DEGs and included hydrolase activity, G-protein couple receptor signaling pathway, signaling receptor binding, and phospholipase C activity. Several genes involved in development and organogenesis (HOXA9, ELLE3, SFRP1, RIPPLY3, and FR2B) and in the phosphoinositide pathway (SDCBP2 and INPP5E), were upregulated in the mammary gland of IUHT-H compared to IUCL-H. We also found evidence of blunted mammary development associated with in utero heat stress; IUHT-H had significantly smaller mammary alveoli relative to IUCL-H at 21 days into the first lactation. Our results suggest that changes in DNA methylation of genes involved in mammary development and milk synthesis may be associated with ultrastructural differences in the mammary gland and with differential milk production observed between IUHT-H and IUCL-H10.
For mammary tissue, genes involved in transcription, translation, and gene regulation were differentially methylated between IUHT-H compared to IUCL-H. DMGs with these functions included the zinc finger genes ZNF395, ZC3H6, MZF1, and ZNF75D AGO2, and tripartite motif (TRIM) genes (TRIM28 and TRIM37). The AGO2 gene plays a role in transcriptional and post-transcriptional gene silencing74,75 and TRIM genes regulate gene expression through a variety of mechanisms, such as recruiting transcription coregulators, histone deacetylases, and other chromatin modifiers76,77. Several zinc finger genes were also differentially expressed between IUHT-H and IUCL-H; one was downregulated (ZBTB47) and three were upregulated (ZNF555, ZNF134, ZNF177) in IUHT-H relative to IUCL-H.
Although immune function was not a term significantly enriched in our DMG dataset for mammary tissue, many of the identified DEGs are involved in immune related functions (immune system process, immunoglobulin production, antigen processing and presentation, etc). Most DEGs in these pathways were upregulated (BLA-DQB, JCHAIN, DEFB, GRO1, HP) in the mammary glands of IUHT-H compared to IUCL-H, although a couple were downregulated (BOLA-DQB, BOLA). These results are in concordance with immune deficits and elevated acute phase protein production observed among in utero heat stressed cattle26,45,78, but suggest that DNA methylation is not contributing substantially to regulation of immune-related genes in the mammary gland.
Fifty genes were differentially methylated between IUHT-B and IUCL-B liver and also between IUHT-H and IUCL-H mammary gland. Among these were the DNA repair gene, WRN, and two genes encoding mitochondrial proteins, IMMP2L and C14orf2, important for oxidative defense and ATP synthesis, respectively79,80. Furthermore, ZNF395, MED1, and the developmentally important homeobox gene, CUX1, were differentially methylated in both liver and mammary tissue. In both the heifer and bull calf, genes in the notch signaling pathway were differentially methylated (heifer: DTX2, DTX3; bull calf: Notch3, Notch4), which is involved in cell differentiation and development of both liver and mammary gland81,82. Twenty differentially methylated genes shared between bull calf liver and heifer mammary gland were rRNA genes (5S rDNA and 5.8S rDNA). Bull calf liver had more DMCs associated with rDNA and the majority were hypermethylated in IUHT-B relative to IUCL-B. Expression of rDNA is regulated in part by CpG methylation; hypomethlyation of CpGs is associated with gene activation whereas transcriptionally silent genes are hypermethylated83,84. However, there can be hundreds of copies of rRNA genes across the genome, only a fraction of which are actively transcribed85,86. How methylation changes in a few copies of rRNA genes, as seen in the present study, impact rRNA transcription, ribosome biogenesis, and ultimately polypeptide formation is unclear. Thus, there are common patterns of DNA methylation changes induced by heat stress regardless of animal sex, age, or tissue type that may affect fundamental processes including cellular repair, oxidative defense, energy metabolism, and development.
DMGs in the present study consisted of a few to only 1 DMC, and those DMCs appeared to be associated with phenotype and function. Likewise, several studies have linked methylation changes at just one or two CpG sites with transcription activity and phenotype87–90. For instance, degenerate bovine embryos that failed to develop from the morula to the blastocyst stage had a higher level of methylation at a single CpG site upstream of the imprinted PHLDA2 gene relative to embryos that reached the blastocyst stage, which was associated with upregulation of PHLDA288. In another study, methylation of one specific CpG site within a 400 bp region of the oxytocin receptor gene (Oxtr) promoter inhibited transcription in an immortalized hypothalamic cell line91. A study on the role of epigenetics in developmental programming reported differences in methylation levels at specific CpG sites within the hippocampal glucocorticoid receptor (GR) gene between offspring reared by mothers with disparate maternal care behaviors87. Changes in methylation were associated with differences in histone H3-K9 acetylation and transcription factor binding to the GR promoter, with consequences for gene expression and offspring stress response87. These studies indicate that methylation patterns of one or a few critical CpG sites can drive changes in gene expression that can impact phenotype.
Although the general biological functions of differentially methylated genes and differentially expressed genes were similar in the present study, we found weak correlations between differential methylation and gene expression. Only two genes were both differentially methylated and differentially expressed, one of which is a novel bovine gene and another, TCIRG1, which is involved in adaptive immunity as a T cell regulator92. The precise regulatory role of DNA methylation in transcription is a current subject of debate. Methylation was originally considered to be transcriptionally repressive, particularly when occurring in promoter and enhancer regions, however the role of methylation in modulating transcription has proven to be substantially more complex29,93. For example, methylation of CpG rich promoters, which includes the majority of gene promoters, inhibits expression due to the binding of methylation-sensitive binding proteins and transcriptional repressors94,95. However, CpG poor promoters are often methylated irrespective of transcription activity96–98. It has also been noted that transcribed genes generally appear to be hypermethylated within the gene body96,99,100. The slight negative correlation between differential methylation of cytosines in the gene bodies and differential gene expression in our study seemingly contradicts these reports, but may be explained by the variability of gene expression relative to DNA methylation. According to common dogma, DNA methylation may be relatively stable throughout life, whereas patterns of gene expression vary drastically31. Thus, performing RNA sequencing at a particular time point provides a single snapshot of gene expression that may not necessarily correlate with DNA methylation patterns at that time. It is also possible that heat-induced changes in DNA methylation may not play a significant regulatory role in gene expression. Rather, heat stress may alter gene expression through other epigenetic modifications, such as histone modifications, chromatin remodeling, or microRNAs.
Conclusions
Despite a few limitations of our study, including the 2-year interval between fetal exposure to heat stress and mammary tissue collection and the high culling rate of in utero heat stressed heifers which limited our pool of eligible biological replicates, we identified a substantial number of DMGs and DEGs between in utero heat stressed and cooled cattle. Those were involved in functions ranging from development, transcription regulation, and immunity to cell signaling, ATP synthesis, and oxidative defense. Dysregulation of these pathways may be associated with the observed morphological differences in the liver and mammary gland between in utero heat stressed and in utero cooled cattle and might contribute to the higher postnatal morbidity and reduced lactation performance of in utero heat stressed animals. A common pattern of DMGs between bull calf liver and heifer mammary gland was discovered despite obvious disparities in sex, tissue type, and age, suggesting that in utero heat stress may program different organs in a similar manner. Future studies evaluating the protein expression of DMGs and DEGs will provide further insight into the functional relevance of our findings.
Materials and Methods
Animals and experimental design
Trials were conducted at the University of Florida Dairy Unit (Hague, FL) in the summer months of 2014–2016 on a herd of Holstein cows. In 2014 and 2015, multiparous pregnant cows were dried off 46 days prior to expected calving date and randomly assigned to one of two groups; cooled (CL) and heat stressed (HT), based on mature equivalent milk production in the previous lactation. The CL and HT dams were housed on similar sized adjacent pens of the same shaded, sand-bedded, free-stall barn, but the cooled pen was equipped with fans and water soakers whereas the heat stressed pen lacked active cooling. Thus, the heat stressed group was exposed to the ambient environment with no heat abatement whereas the cooled group was exposed to the ambient environment but also provided heat abatement through fans and water soakers to facilitate evaporative cooling. In the cooled pen, fans ran continuously and water soakers turned on for 1.5 min in 6 min intervals when ambient temperature rose above 21.1 °C. The temperature humidity index (THI, Dikmen et al.101) of both sides of the barn was above 70 for the duration of the experiments in both 2014 and 20156. Elevated respiration rate and rectal temperature are physiological indicators of heat stress in dairy cattle and other animals and were therefore used to assess treatment effectiveness6,102,103. Respiration rate (at 1400 h) and rectal temperature (at 1430 h) were recorded 3 times per week. Respiration rate was measured by counting the number of flank movements in one minute. Respiration rate and rectal temperature were significantly lower for CL relative to HT dams, confirming heat stress abatement in the cooled group and heat stress in the heat stressed group6,102.
Ten bull calves (n = 7 born in 2014, n = 3 born in 2015) and 7 heifers (all born in 2014) gestated by CL or HT dams were used for this study. These calves were exposed to the maternal treatment through the intrauterine environment during the last 6–7 weeks (~46 days) of fetal development. Thus calves belonged to one of four groups; in utero heat stressed heifers (IUHT-H, n = 3), in utero cooled heifers (IUCL-H, n = 4), in utero heat stressed bulls (IUHT-B, n = 5), and in utero cooled bulls (IUCL-B, n = 5). Bull calves were sacrificed by captive bolt stunning and exsanguination within 4 hours of birth, prior to first colostrum feeding. Heifers born in 2014 remained in the herd and were housed together as a group through their first lactation in summer 2016. The heifers all experienced the same environmental conditions and the same management regime, including separation from the dam at birth, routine vaccinations, and diet. Animal studies were approved by the Institutional Animal Care and Use Committee at the University of Florida (#201408505 and #201609371), and all experiments were conducted in accordance with their rules and regulations.
Liver and mammary gland collection
Liver tissue was harvested from bull calves immediately after sacrifice, snap frozen in liquid nitrogen, and stored at −80 °C until methylation sequencing. Mammary gland biopsies were collected from heifers on day 21 of lactation, coinciding with the rising milk yield phase of the lactation cycle. Biopsies were performed using a stainless steel biopsy tool attached to a drill following the method described by104, with modifications by our group18. Mammary tissue was snap frozen in liquid nitrogen and stored at −80 °C until methylation sequencing. An additional 5 mm piece of liver and mammary tissue was placed in tissue cassettes overnight at 4 °C in 4% paraformaldehyde, subsequently rinsed in a graded series of ethanol, and paraffin embedded. Tissues were sectioned at 5 μm onto slides coated with poly-L-lysine and stained with hematoxylin and eosin to visualize morphology.
Quantification of histological sections
Three images per tissue section were photographed with an EVOS XL Core imaging system (Advanced Microscopy Group, Bothell, WA) using a 20X or 40X objective (total tissue area was 573,573.619 μm2 and 95,908.32 μm2, respectively). The number of alveoli in mammary tissue (at 20X) and the number of cells in liver tissue (at 40X) were quantified for each tissue image using the Point Picker plugin (Biomedical Imaging Group, Swiss Federal Institute of Technology Lausanne, Lausanne, Switzerland) for Image J software (National Institutes of Health, Bethesda, MD). Alveoli were discriminated from other structures by their rounded hollow lumen enclosed by a single layer of mammary epithelial cells. Alveoli area (at 20X) was quantified using the free-hand drawing tool in Image J and expressed as μm2. Mammary ducts were not included in alveoli counts and area measurements. Differences between IUHT and IUCL groups were analyzed by two-tailed unpaired T-tests using SAS v. 9.4 (SAS Institute, Cary, NC). Heifers and bulls were analyzed separately. P-values < 0.05 were considered statistically significant. Data are presented as the mean ± standard error of the mean (SEM).
DNA isolation and methylation sequencing
DNA was isolated from 50 mg of liver and mammary tissue with the FitAmpTM General Tissue Section DNA Isolation Kit (Epigentek, P-1003). DNA methylation was analyzed by double restriction enzyme reduced representation bisulfite sequencing (RRBS). RRBS provides methylation resolution at the single nucleotide level from a subset of the genome105. Despite low genome coverage, on average 2%106,107, RRBS enriches for CpG rich areas, including the majority of CpG islands and gene promoters and to a lesser extent introns, exons, CpG shores, and CpG shelves106. Thus, RRBS is a powerful method that is broadly used for the assessment of DNA methylation patterns.
First, DNA was digested with MspI restriction enzyme for 2 hrs at 37 °C followed by digestion with TaqαI restriction endonuclease at 65 °C. DNA fragments < 300 bp were selected and used for bisulfite treatment with the Methylamp DNA Conversion Kit (Epigentek, P-1001). Conversion efficiency was determined by rtPCR using two primer pairs; the first for β-actin against bisulfite converted DNA and the second for GAPDH against unconverted DNA. The same bisulfite converted DNA sample was run with both primer pairs. DNA was >98% converted. Standard lllumina sequencing adaptors were ligated onto the bisulfite treated DNA fragments using a random probing method. Library amplification was achieved using indexed primers and analyzed on a Bioanalyzer and with a KAPA Library Quantification Kit according to the manufacturer’s protocol (KapaBiosystems, MA, USA). Purified DNA (15 nM of sample) libraries were sequenced using the Illumina HiSeq. 4000 system, generating 50 bp single-end reads. Reads from one bull calf (IUHT-B) could not be aligned to the reference genome and was therefore excluded from further analysis. RRBS data can be accessed by NCBI GEO with accession number GSE119445.
Bioinformatics analysis
Quality control of the Illumina raw reads was performed using FastQC software (version 0.11.2, Babraham Bioinformatics, UK). Low quality base removal and adapter trimming was carried out using Trim Galore (version 0.2.7, Babraham Bioinformatics, UK) in the RRBS-specific mode. A Sanger Phred score of 20 was used as the cut-off criterion for removal of low quality reads. The 3′ Illumina adapter was trimmed and any reads shorter than 20 bp were removed. Trimmed reads were mapped to the bovine genome assembly UMD3.1.1 using Bismark software (version 0.16.1, Babraham Bioinformatics, UK), which utilizes the Bowtie short read aligner108. The options –n 1 and pbat were selected, which permits up to one mismatch in the seed region and indicates the sequencing library was constructed following a post-bisulfite (PBAT) protocol, respectively. Methylation information was extracted using the Bismark methylation extractor (version 0.16.1, Babraham Bioinformatics, UK) at the base resolution. The ignore 6 option was selected to ignore the first 6 bp of each read to reduce methylation bias typically observed in PBAT libraries. A minimum read coverage of 5 and minimum quality score of 20 at each base position was applied. Sequencing read counts and levels of methylation were calculated with the methylKit package in R. Only cytosines with more than 10 reads were analyzed further. Differentially methylated cytosines (DMCs) between IUHT-H and IUCL-H and between IUHT-B and IUCL-B were identified via logistic regression with a cut-off value of 15% methylation difference and a q-value < 0.2. DMCs that had a lower level of methylation in the IUHT groups relative to the IUCL groups were considered to be hypomethylated. Conversely, DMCs that had a higher level methylation in the IUHT groups relative to the IUCL groups were considered to be hypermethylated. Principal component analysis was used to distinguish groups based on intra- and inter-group variability in methylation levels. Differentially methylated genes (DMGs) have at least 1 differentially methylated cytosine. Pathways, biological processes, and molecular functions of DMGs were identified using Kyoto Encyclopedia for Genes and Genomes (KEGG) database, Medical Subject Headings (MeSH) database, and Gene Ontology database (Gene Ontology Consortium).
RNA isolation and RNA-sequencing
In order to explore regulation of gene expression by DNA methylation we conducted correlations between differential methylation at the cytosine level and differential gene expression using the mammary gland tissue. A portion of the mammary biopsies from all IUHT-H and IUCL-H were stored at −80 °C in RNALater until RNA extraction for RNA sequencing. Briefly, RNA was extracted from mammary tissue using the RNeasy Plus Universal Mini Kit (Qiagen, Valencia, CA) and RNA concentration was determined using a NanoDrop instrument (ND-2000, ThermoFisher Scientific, Waltham, MA). RNA quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA). RNA with 28S/18S > 1 and an RNA integrity number ≥7 were used for library construction. RNA-Seq library was constructed using NEBNext® Ultra™ RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA) following manufacturer’s recommendations. Libraries were sequenced on the Illumina HiSeq. 3000 platform generating 100 paired-end reads.
Quality control of sequencing reads was conducted using FastQC software. Tophat (v2.0.13) software was used to map reads to the bovine reference genome (bosTau7). Identification of differentially expressed genes (DEGs) was accomplished using the R package edgeR (v.3.4.2), using a P-value cut-off of 0.01. Gene set enrichment analysis for differentially expressed genes was performed in R using the Gene Ontology database (Gene Ontology Consortium). Relationships between differentially expressed genes and differentially methylated cytosines associated with those genes were analyzed through Pearson correlation in R software. Separate correlations were run for the overall DMCs and a subset of the DMCs based on genomic location (upstream, downstream, and gene body) to determine if DMCs associated with particular genomic regions have different influences on gene expression. RNA-seq data can be accessed by NCBI GEO with the accession number GSE119216.
Electronic supplementary material
Acknowledgements
The authors would like to thank the staff at the University of Florida (UF) Dairy Unit for animal care and Joyce Hayen for her contributions to this project. Funding was provided by a UF-Institute of Food and Agricultural Sciences Experiment Station Seed Grant (J. Laporta), and National Institute of Food and Agriculture-Agriculture and Food Research Initiative Grant (NIFA-AFRI; #2015-67015-23409; G.E. Dahl).
Author Contributions
J.L., A.S. and G.D. conceived and designed the project. A.S. and B.A. completed the on-farm trials and harvested mammary and liver tissue samples. R.A. and F.P. conducted bioinformatics analysis for methylation and gene expression data. A.S. ran statistical analyses of histological data. A.S., J.L., G.D. and F.P. contributed to interpretation of results. A.S. wrote the manuscript. All authors read, edited and approved the final version of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
G. E. Dahl, Email: gdahl@ufl.edu
J. Laporta, Email: jlaporta@ufl.edu
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-32975-1.
References
- 1.Barker BJP, Eriksson JG, Forsén T, Osmond C. Fetal origins of adult disease: strength of effects and biological basis. International Journal of Epidemiology. 2002;31:1235–1239. doi: 10.1093/ije/31.6.1235. [DOI] [PubMed] [Google Scholar]
- 2.Lucas A. Programming by early nutrition in man. Ciba Foundation Symposium. 1991;156:38–55. [PubMed] [Google Scholar]
- 3.Lindström J. Early development and fitness in birds and mammals. Trends in Ecology and Evolution. 1999;14:343–348. doi: 10.1016/S0169-5347(99)01639-0. [DOI] [PubMed] [Google Scholar]
- 4.McMillen IC, Robinson JS. Developmental origins of the metabolic syndrome: prediction, plasticity, and programming. Physiological Reviews. 2005;85:571–633. doi: 10.1152/physrev.00053.2003. [DOI] [PubMed] [Google Scholar]
- 5.Fowden AL, Guissani DA, Forhead AJ. Intrauterine programming of physiological systems: causes and consequences. Physiology. 2006;21:29–37. doi: 10.1152/physiol.00050.2005. [DOI] [PubMed] [Google Scholar]
- 6.Monteiro APA, et al. Effect of maternal heat stress during the dry period on growth and metabolism of calves. Journal of Dairy Science. 2016;99:3896–3907. doi: 10.3168/jds.2015-10699. [DOI] [PubMed] [Google Scholar]
- 7.Flouris AD, Spiropoulos Y, Sakellariou GJ, Koutedakis Y. Effect of seasonal programming on fetal development and longevity: links with environmental temperature. American Journal of Human Biology. 2009;21:214–216. doi: 10.1002/ajhb.20818. [DOI] [PubMed] [Google Scholar]
- 8.Bell AW, McBride BW, Slepetis R, Early RJ, Currie WB. Chronic heat stress and prenatal development in sheep: I. Conceptus growth and maternal plasma hormones and metabolites. Journal of Animal Science. 1989;67:3289–3299. doi: 10.2527/jas1989.67123289x. [DOI] [PubMed] [Google Scholar]
- 9.Tao S, Monteiro APA, Thompson IM, Hayen MJ, Dahl GE. Effect of late-gestation maternal heat stress on growth and immune function of dairy calves. Journal of Dairy Science. 2012;95:7128–7136. doi: 10.3168/jds.2012-5697. [DOI] [PubMed] [Google Scholar]
- 10.Monteiro APA, Tao S, Thompson IMT, Dahl GE. In utero heat stress decreases calf survival and performance through the first lactation. Journal of Dairy Science. 2016;99:8443–8450. doi: 10.3168/jds.2016-11072. [DOI] [PubMed] [Google Scholar]
- 11.do Amaral BC, et al. Heat-stress abatement during the dry period: does cooling improve transition into lactation? Journal of Dairy Science. 2009;92:5988–5999. doi: 10.3168/jds.2009-2343. [DOI] [PubMed] [Google Scholar]
- 12.Tao S, Monteiro APA, Hayen MJ, Dahl GE. Maternal heat stress during the dry period alters postnatal whole-body insulin response of calves. Journal of Dairy Science. 2014;97:897–901. doi: 10.3168/jds.2013-7323. [DOI] [PubMed] [Google Scholar]
- 13.Tao S, et al. Effect of cooling heat-stressed dairy cows during the dry period on insulin response. Journal of Dairy Science. 2012;95:5035–5046. doi: 10.3168/jds.2012-5405. [DOI] [PubMed] [Google Scholar]
- 14.Mao WH, et al. Growth- and breed-related changes of fetal development in cattle. Asian-Astralasian Journal of Animal Science. 2008;21:640–647. doi: 10.5713/ajas.2008.70293. [DOI] [Google Scholar]
- 15.Bauman DE, Currie WB. Partitioning of nutrients during pregnancy and lactation: a review of mechanisms involving homeostasis and homeorhesis. Journal of Dairy Science. 1980;63:1514–1529. doi: 10.3168/jds.S0022-0302(80)83111-0. [DOI] [PubMed] [Google Scholar]
- 16.Sangild PT, et al. Blood chemistry, nutrient metabolism, and organ weights in fetal and newborn calves derived from in vitro-produced bovine embryos. Biology of Reproduction. 2000;62:1495–1504. doi: 10.1095/biolreprod62.6.1495. [DOI] [PubMed] [Google Scholar]
- 17.Skibiel AL, Zachut M, do Amaral BC, Levin Y, Dahl GE. Liver proteomic analysis of postpartum Holstein cows exposed to heat stress or cooling conditions during the dry period. Journal of Dairy Science. 2018;101:705–716. doi: 10.3168/jds.2017-13258. [DOI] [PubMed] [Google Scholar]
- 18.Tao S, et al. Effect of heat stress during the dry period on mammary gland development. Journal of Dairy Science. 2011;94:5976–5986. doi: 10.3168/jds.2011-4329. [DOI] [PubMed] [Google Scholar]
- 19.Akers, R. M. Lactation and the mammary gland. (Blackwell Publishing, 2002).
- 20.Knight CH, Sorenson A. Windows in early mammary development: critical or not? Reproduction. 2001;122:337–345. doi: 10.1530/rep.0.1220337. [DOI] [PubMed] [Google Scholar]
- 21.Zhou Z, Xu MJ, Gao B. Hepatocytes: a key cell type for innate immunity. Cell and Molecular Immunology. 2016;13:300–314. doi: 10.1038/cmi.2015.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ju C, Tacke F. Hepatic macrophages in homeostasis and liver diseases. Cell and Molecular Immunology. 2016;13:315–326. doi: 10.1038/cmi.2015.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng H, et al. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. The Journal of Clinical Investigation. 2013;123:1444–1456. doi: 10.1172/JCI66381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fowden AL, Li J, Forhead AJ. Glucocorticoids and the preparation for life after birth: are there long-term consequences of the life insurance. Proceedings of the Nutrition Society. 1998;57:113–122. doi: 10.1079/PNS19980017. [DOI] [PubMed] [Google Scholar]
- 25.Harding JE, Johnson B. Nutrition and fetal growth. Reproduction Fertility and Development. 1995;7:538–547. doi: 10.1071/rd9950539. [DOI] [PubMed] [Google Scholar]
- 26.Ahmed, B. M. S. et al. Maternal heat stress reduces body and organ growth in calves: relationship to immune tissue development. Journal of Dairy Science99 (Suppl. 1), 606 (Abstr.) (2016).
- 27.Li A-Q, et al. Systematical analysis of impacts of heat stress on the proliferation, apoptosis, and metabolism of mouse hepatocyte. Journal of Physiological Sciences. 2012;62:29–43. doi: 10.1007/s12576-011-0183-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma RK. Morphological and morphometric studies on liver in rats subjected to repetitive heat stress. Indian Journal of Medical Research. 1997;106:20–26. [PubMed] [Google Scholar]
- 29.Bird A. DNA methylation patterns and epigenetic memory. Genes & Development. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 30.Angers B, Castonguay E, Massicotte R. Environmentally induced phenotypes and DNA methylation: how to deal with unpredictable conditions until the next generation and after. Molecular Ecology. 2010;19:1283–1295. doi: 10.1111/j.1365-294X.2010.04580.x. [DOI] [PubMed] [Google Scholar]
- 31.Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nature Reviews Genetics. 2012;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- 32.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nature Reviews Genetics. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rijnkels M, et al. The epigenetic landscape of mammary gland development and functional differentiation. Journal of Mammary Gland Biology and Neoplasia. 2010;15:85–100. doi: 10.1007/s10911-010-9170-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh K, et al. Epigenetics: a possible role in acute and transgenerational regulation of dairy cow milk production. Animal. 2012;6:375–381. doi: 10.1017/S1751731111002564. [DOI] [PubMed] [Google Scholar]
- 35.Vanselow J, et al. DNA-remethylation around a STAT5-binding enhancer in the far distal alphaS1-casein promoter is associated with abrupt shut-down of alphaS1-casein synthesis during acute mastitis. Journal of Molecular Endocrinology. 2006;37:463–477. doi: 10.1677/jme.1.02131. [DOI] [PubMed] [Google Scholar]
- 36.Snykers S, et al. Role of epigenetics in liver-specific gene transcription, hepatocyte differentiation and stem cell reprogrammation. Journal of Hepatology. 2009;51:187–211. doi: 10.1016/j.jhep.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 37.Weyrich A, et al. Paternal intergenerational epigenetic response to heat exposure in male Wild guinea pigs. Molecular Ecology. 2016;25:1729–1740. doi: 10.1111/mec.13494. [DOI] [PubMed] [Google Scholar]
- 38.Zhu J-Q, et al. Heat stress causes aberrant DNA methylation of H19 and Igf-2r in mouse blastocysts. Molecules and Cells. 2008;25:211–215. [PubMed] [Google Scholar]
- 39.Yamagishi Sho-ichi, Matsui Takanori. Role of receptor for advanced glycation end products (RAGE) in liver disease. European Journal of Medical Research. 2015;20(1):15. doi: 10.1186/s40001-015-0090-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bierhaus A, et al. Understanding RAGE, the receptor for advanced glycation end products. Journal of Molecular Medicine. 2005;83:876–886. doi: 10.1007/s00109-005-0688-7. [DOI] [PubMed] [Google Scholar]
- 41.Neeper M, et al. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. Journal of Biological Chemistry. 1992;267:14998–15004. [PubMed] [Google Scholar]
- 42.Xie J, Méndez JD, Méndez-Valenzuela V, Aguilar-Hernández MM. Cellular signalling of the receptor for advanced glycation end products (RAGE) Cellular Signalling. 2013;25:2185–2197. doi: 10.1016/j.cellsig.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 43.Wautier M-P, et al. Activation of NADPH oxidase by AGE links oxidant stress to altered gene expression via RAGE. American Journal of Physiology-Endocrinology and Metabolism. 2001;280:E685–E694. doi: 10.1152/ajpendo.2001.280.5.E685. [DOI] [PubMed] [Google Scholar]
- 44.Bierhaus A., Chevion S., Chevion M., Hofmann M., Quehenberger P., Illmer T., Luther T., Berentshtein E., Tritschler H., Muller M., Wahl P., Ziegler R., Nawroth P. P. Advanced Glycation End Product-Induced Activation of NF- B is Suppressed by -Lipoic Acid in Cultured Endothelial Cells. Diabetes. 1997;46(9):1481–1490. doi: 10.2337/diab.46.9.1481. [DOI] [PubMed] [Google Scholar]
- 45.Skibiel AL, et al. Effects of feeding an immunomodulatory supplement to heat-stressed or actively cooled cows during late gestation on postnatal immunity, health, and growth of calves. Journal of Dairy Science. 2017;100:7659–7668. doi: 10.3168/jds.2017-12619. [DOI] [PubMed] [Google Scholar]
- 46.Tao S., Connor E.E., Bubolz J.W., Thompson I.M., do Amaral B.C., Hayen M.J., Dahl G.E. Short communication: Effect of heat stress during the dry period on gene expression in mammary tissue and peripheral blood mononuclear cells. Journal of Dairy Science. 2013;96(1):378–383. doi: 10.3168/jds.2012-5811. [DOI] [PubMed] [Google Scholar]
- 47.Pandey P, et al. Activation of TRAP/Mediator subunit TRAP220/Med1 is regulated by mitogen-activated protein kinase-dependent phosphorylation. Molecular and Cellular Biology. 2005;25:10695–10710. doi: 10.1128/MCB.25.24.10695-10710.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Angelov D, et al. The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Molecular Cell. 2003;11:1033–1041. doi: 10.1016/S1097-2765(03)00100-X. [DOI] [PubMed] [Google Scholar]
- 49.Liefke R, Karwacki-Neisius V, Shi Y. EPOP interacts with Elongin BC and USP7 to modulate the chromatin landscape. Molecular Cell. 2016;64:659–672. doi: 10.1016/j.molcel.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fedotova AA, Bonchuk AN, Mogila VA, Georgiev PG. C2H2 zinc finger proteins: the largest but poorly explored family of higher eukaryotic transcription factors. Acta Naturae. 2017;9:47–58. [PMC free article] [PubMed] [Google Scholar]
- 51.Najafabadi HS, et al. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nature Biotechnology. 2015;33:555–562. doi: 10.1038/nbt.3128. [DOI] [PubMed] [Google Scholar]
- 52.Iuchi S. Three classes of C2H2 zinc finger proteins. Cellular and Molecular Life Sciences CMLS. 2001;58:625–635. doi: 10.1007/PL00000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murat A, et al. Modulation of angiogenic and inflammatory response in glioblastoma by hypoxia. Plos One. 2009;4:e5947. doi: 10.1371/journal.pone.0005947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jordanovski D, et al. The hypoxia-inducible transcription factor ZNF395 is controlled by IκB kinase-signaling and activates genes involved in the innate immune response and cancer. Plos One. 2013;8:e74911. doi: 10.1371/journal.pone.0074911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herwartz C, Castillo-Juárez P, Schröder L, Barron BL, Steger G. The transcription factor ZNF395 is required for the maximal hypoxic induction of proinflammatory cytokines in U87-MG cells. Mediators of Inflammation. 2015;2015:1–9. doi: 10.1155/2015/804264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koong AC, Chen EY, Giaccia AJ. Hypoxia Causes the Activation of Nuclear Factor κB through the Phosphorylation of IκBα on Tyrosine Residues. Cancer Research. 1994;54:1425. [PubMed] [Google Scholar]
- 57.Fowden AL, Ward JW, Wooding FPB, Forhead AJ, Constancia M. Programming placental nutrient transport capacity. The Journal of Physiology. 2006;572:5–15. doi: 10.1113/jphysiol.2005.104141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Collier RJ, Doelger SG, Head HH, Thatcher WW, Wilcox CJ. Effects of heat stress during pregnancy on maternal hormone concentrations, calf birth weight and postpartum milk yield of Holstein cows. Journal of Animal Science. 1982;54:309–319. doi: 10.2527/jas1982.542309x. [DOI] [PubMed] [Google Scholar]
- 59.Reynolds LP, et al. Evidence for altered placental blood flow and vascularity in compromised pregnancies. Journal of Physiology. 2006;572:51–58. doi: 10.1113/jphysiol.2005.104430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bardelli A, et al. HGF receptor associates with the anti‐apoptotic protein BAG‐1 and prevents cell death. The EMBO Journal. 1996;15:6205–6212. doi: 10.1002/j.1460-2075.1996.tb01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takayama S, et al. Cloning and functional analysis of BAG-1: A novel Bcl-2-binding protein with anti-cell death activity. Cell. 1995;80:279–284. doi: 10.1016/0092-8674(95)90410-7. [DOI] [PubMed] [Google Scholar]
- 62.Takayama S, et al. BAG‐1 modulates the chaperone activity of Hsp70/Hsc70. The EMBO Journal. 1997;16:4887–4896. doi: 10.1093/emboj/16.16.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakashima S. Protein kinase Cα (PKCα): regulation and biological function. The Journal of Biochemistry. 2002;132:669–675. doi: 10.1093/oxfordjournals.jbchem.a003272. [DOI] [PubMed] [Google Scholar]
- 64.Stanger BZ. Organ size determination and the limits of regulation. Cell Cycle. 2008;7:318–324. doi: 10.4161/cc.7.3.5348. [DOI] [PubMed] [Google Scholar]
- 65.Kmieć Z. Cooperation of liver cells in health and disease. Advances in Anatomy, Embryology and Cell Biology. 2001;161:1–151. doi: 10.1007/978-3-642-56553-3_1. [DOI] [PubMed] [Google Scholar]
- 66.Gao B. Basic liver immunology. Cellular and Molecular Immunology. 2016;13:265–266. doi: 10.1038/cmi.2016.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Toker A. The synthesis and cellular roles of phosphatidylinositol 4,5-bisphosphate. Current Opinion in Cell Biology. 1998;10:254–261. doi: 10.1016/S0955-0674(98)80148-8. [DOI] [PubMed] [Google Scholar]
- 68.Doughman RL, Firestone AJ, Anderson RA. Phosphatidylinositol Phosphate Kinases Put PI4,5P2 in Its Place. The Journal of Membrane Biology. 2003;194:77–89. doi: 10.1007/s00232-003-2027-7. [DOI] [PubMed] [Google Scholar]
- 69.Das R, Vonderhaar BK. Prolactin as a mitogen in mammary cells. Journal of Mammary Gland Biology and Neoplasia. 1997;2:29–39. doi: 10.1023/a:1026369412612. [DOI] [PubMed] [Google Scholar]
- 70.Ruth P, et al. Transfected cGMP-dependent protein kinase suppresses calcium transients by inhibition of inositol 1,4,5-trisphosphate production. Proceedings of the National Academy of Sciences. 1993;90:2623–2627. doi: 10.1073/pnas.90.7.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li C, et al. Genome wide association study identifies 20 novel promising genes associated with milk fatty acid traits in Chinese Holstein. Plos One. 2014;9:e96186. doi: 10.1371/journal.pone.0096186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sheridan JM, et al. A pooled shRNA screen for regulators of primary mammary stem and progenitor cells identifies roles for Asap1 and Prox1. BMC Cancer. 2015;15:221. doi: 10.1186/s12885-015-1187-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nagy T, et al. Mammary epithelial-specific deletion of the focal adhesion kinase gene leads to severe lobulo-alveolar hypoplasia and secretory immaturity of the murine mammary gland. Journal of Biological Chemistry. 2007;282:31766–31776. doi: 10.1074/jbc.M705403200. [DOI] [PubMed] [Google Scholar]
- 74.Janowski BA, et al. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nature Structural &Amp; Molecular Biology. 2006;13:787. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- 75.Adams BD, Claffey KP, White BA. Argonaute-2 expression Is regulated by epidermal growth factor receptor and mitogen-activated protein kinase signaling and correlates with a transformed phenotype in breast cancer cells. Endocrinology. 2009;150:14–23. doi: 10.1210/en.2008-0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cammas, F., Khetchoumian, K., Chambon, P. & Losson, R. In TRIM/RBCC Proteins (ed Germana Meroni) Ch. 5, 59–76 (Springer New York, 2012).
- 77.Bhatnagar S, et al. TRIM37 is a new histone H2A ubiquitin ligase and breast cancer oncoprotein. Nature. 2014;516:116–137. doi: 10.1038/nature13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Monteiro APA, Tao S, Thompson IM, Dahl GE. Effect of heat stress during late gestation on immune function and growth performance of calves: isolation of altered colostral and calf factors. Journal of Dairy Science. 2014;97:6426–6439. doi: 10.3168/jds.2013-7891. [DOI] [PubMed] [Google Scholar]
- 79.Lu B, et al. A mutation in the inner mitochondrial membrane peptidase 2-like gene (Immp2l) affects mitochondrial function and impairs fertility in mice. Biology of Reproduction. 2008;78:601–610. doi: 10.1095/biolreprod.107.065987. [DOI] [PubMed] [Google Scholar]
- 80.Fujikawa M, Ohsakaya S, Sugawara K, Yoshida M. Population of ATP synthase molecules in mitochondria is limited by available 6.8-kDa proteolipid protein (MLQ) Genes to Cells. 2014;19:153–160. doi: 10.1111/gtc.12121. [DOI] [PubMed] [Google Scholar]
- 81.Valenti L., Mendoza R. M., Rametta R., Maggioni M., Kitajewski C., Shawber C. J., Pajvani U. B. Hepatic Notch Signaling Correlates With Insulin Resistance and Nonalcoholic Fatty Liver Disease. Diabetes. 2013;62(12):4052–4062. doi: 10.2337/db13-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Macias H, Hinck L. Mammary gland development. Wiley Interdisciplinary Reviews: Developmental Biology. 2012;1:533–557. doi: 10.1002/wdev.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ghoshal K, et al. Role of Human Ribosomal RNA (rRNA) Promoter Methylation and of Methyl-CpG-binding Protein MBD2 in the Suppression of rRNA Gene Expression. Journal of Biological Chemistry. 2004;279:6783–6793. doi: 10.1074/jbc.M309393200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou H, et al. Overexpression of Ribosomal RNA in the Development of Human Cervical Cancer Is Associated with rDNA Promoter Hypomethylation. PLOS ONE. 2016;11:e0163340. doi: 10.1371/journal.pone.0163340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Conconi Antonio, Widmer Rosa M., Koller Theo, Sogo JoséM. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989;57(5):753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- 86.Torres-Machorro AL, Hernández R, Cevallos AM, López-Villaseñor I. Ribosomal RNA genes in eukarytoic microorganisms: witnesses of phylogeny? FEMS Microbiology Reviews. 2010;34:59–86. doi: 10.1111/j.1574-6976.2009.00196.x. [DOI] [PubMed] [Google Scholar]
- 87.Weaver ICG, et al. Epigenetic programming by maternal behavior. Nature Neuroscience. 2004;7:847. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 88.Driver AM, Huang W, Kropp J, Peñagaricano F, Khatib H. Knockdown of CDKN1C (p57kip2) and PHLDA2 results in developmental changes in bovine pre-implantation embryos. Plos One. 2013;8:e69490. doi: 10.1371/journal.pone.0069490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lan, X. et al. Maternal diet during pregnancy induces gene expression and DNA methylation changes in fetal tissues in sheep. Frontiers in Genetics4, 10.3389/fgene.2013.00049 (2013). [DOI] [PMC free article] [PubMed]
- 90.Namous H, et al. Integrative analysis of methylomic and transcriptomic data in fetal sheep muscle tissues in response to maternal diet during pregnancy. BMC Genomics. 2018;19:123. doi: 10.1186/s12864-018-4509-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mamrut S, et al. DNA methylation of specific CpG sites in the promoter region regulates the transcription of the mouse oxytocin receptor. PLOS ONE. 2013;8:e56869. doi: 10.1371/journal.pone.0056869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Makaryan V, et al. TCIRG1 associated congenital neutropenia. Human Mutation. 2014;35:824–827. doi: 10.1002/humu.22563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature Reviews Genetics. 2012;13:484. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 94.Jones PL, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nature Genetics. 1998;19:187. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 95.Nan X, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 96.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Edwards JR, et al. Chromatin and sequence features that define the fine and gross structure of genomic methylation patterns. Genome Research. 2010;20:972–980. doi: 10.1101/gr.101535.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bestor TH, Edwards JR, Boulard M. Notes on the role of dynamic DNA methylation in mammalian development. Proceedings of the National Academy of Sciences. 2015;112:6796–6799. doi: 10.1073/pnas.1415301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nature Reviews Genetics. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 101.Dikmen S, et al. Differences in thermoregulatory ability between slick-haired and wild-type lactating Holstein cows in response to acute heat stress. Journal of Dairy Science. 2008;91:3395–3402. doi: 10.3168/jds.2008-1072. [DOI] [PubMed] [Google Scholar]
- 102.Fabris TF, et al. Effect of nutritional immunomodulation and heat stress during the dry period on subsequent performance of cows. Journal of Dairy Science. 2017;100:6733–6742. doi: 10.3168/jds.2016-12313. [DOI] [PubMed] [Google Scholar]
- 103.Matsuzuka T, et al. Effects of heat stress on the redox status in the oviduct and early embryonic development in mice. Journal of Reproduction and Development. 2005;51:281–287. doi: 10.1262/jrd.16089. [DOI] [PubMed] [Google Scholar]
- 104.Farr VC, et al. An improved method for the routine biopsy of bovine mammary tissue. Journal of Dairy Science. 1996;79:543–549. doi: 10.3168/jds.S0022-0302(96)76398-1. [DOI] [PubMed] [Google Scholar]
- 105.Stevens M, et al. Estimating absolute methylation levels at single-CpG resolution from methylation enrichment and restriction enzyme sequencing methods. Genome Research. 2013;23:1541–1553. doi: 10.1101/gr.152231.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gu H, et al. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nature Protocols. 2011;6:468–481. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- 107.Choi M, et al. Genome-wide analysis of DNA methylation in pigs using reduced representation bisulfite sequencing. DNA Research. 2015;22:343–355. doi: 10.1093/dnares/dsv017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Langmead B, Trapnell C, Pop M, Salzberg SL. Ulfrafast and memory-efficient alignment of shortDNA sequences to the human genome. Genome Biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.