Abstract
The presequence translocase of the mitochondrial inner membrane (TIM23 complex) facilitates anterograde precursor transport into the matrix and lateral release of precursors with stop-transfer signal into the membrane (sorting). Sorting requires precursor exit from the translocation channel into the lipid phase through the lateral gate of the TIM23 complex. How the two transport modes are regulated and balanced against each other is unknown. Here we show that the import motor J-protein Pam18, which is essential for matrix import, controls lateral protein release into the lipid bilayer. Constitutively translocase-associated Pam18 obstructs lateral precursor transport. Concomitantly, Mgr2, implicated in precursor quality control, is displaced from the translocase. We conclude that during motor-dependent matrix protein transport, the transmembrane segment of Pam18 closes the lateral gate to promote anterograde polypeptide movement. This finding explains why a motor-free form of the translocase facilitates the lateral movement of precursors with a stop-transfer signal.
The mitochondrial TIM23-complex facilitates anterograde precursor transport across the inner membrane into the matrix and lateral release of precursors into the membrane. Here authors show that the import motor J-protein Pam18 controls lateral protein release into the lipid bilayer.
Introduction
Protein transport machineries of the endoplasmic reticulum, chloroplasts, and mitochondria facilitate anterograde protein translocation across membranes as well as lateral protein transport of precursors with hydrophobic transmembrane regions into the lipid phase. Proteinaceous channels enable passage of the precursors across membranes and provide lateral gates for lipid phase access. In case of mitochondria, the majority of precursors are targeted by N-terminal signals, termed presequences1,2. The precursors enter mitochondria through the translocase of the outer membrane (TOM) from which they are handed over to the presequence translocase (TIM23 complex) in the inner membrane3–6. The membrane potential (∆ψ) across the inner membrane acts on the positively charged presequence to drive initial translocation of the N terminus through the TIM23 complex7. The TIM23 complex facilitates the transport of two basic types of precursors (i) matrix proteins that are fully translocated across the inner membrane and depend on the activity of the ATP-driven import motor for transport; (ii) inner membrane proteins that are stalled during the process of inner membrane translocation and laterally released into the lipid phase5,6,8–10. The latter process requires the existence of a lateral gate in the translocation channel that provides access to the lipid phase for the translocating polypeptide chain.
Inner membrane proteins contain a hydrophobic stop-transfer signal, also referred to as sorting signal, that stalls translocation allowing preproteins to be released into the lipid bilayer11. The current concept on this transport process by the TIM23 complex postulates that the import channel can open toward the lipid phase by an unknown mechanism to allow precursor exit12. The TIM23 complex contains six integral membrane proteins, which could be involved in this process. Pam17 is only transiently associated with the complex and supports import of ∆ψ-sensitive proteins but is especially important for translocation of soluble proteins13. Tim50 exposes a receptor domain to the intermembrane space (IMS) and regulates channel opening14–16. Tim21 interacts with Tim50 and couples the TIM23 complex to the respiratory chain16,17. Tim23 and probably Tim17 form the channel unit of the translocase. While Tim23 also contains a receptor domain and is the main channel constituent, Tim17 plays a regulatory role by recruiting the import motor and regulating gating activity of the channel18–20. Mgr2 couples Tim21 to the translocase and has been suggested to act as a quality-control factor for the sorting process12,21.
Protein transport into the matrix requires the import motor. The central mtHsp70 chaperone drives proteins into the aqueous matrix at the expense of ATP. The ATPase activity of mtHsp70 is stimulated by the J-protein Pam18, which is recruited to the translocase and regulated by the membrane-associated J-like protein Pam1622–25. Tim44 represents the scaffold protein of the motor that recruits mtHsp70 to the import channel26. Similar to Pam16, Tim44 is peripherally associated with the membrane27. Both interactions with the inner membrane and direct protein–protein interactions with channel components couple the import motor to the translocase28. However, Pam18 is the sole motor component that has a membrane-spanning segment and interacts on both sites of the membrane with translocase components. Interestingly, during translocation of a polypeptide, the import motor is not a stable entity but undergoes subunit exchange with Pam18 displaying the strongest dynamics29.
The question on whether the observed dynamics of the import motor is critical for the transport process is controversially discussed. Two distinct states of the TIM23 complex have been observed. A Tim21-containing (TIM23SORT) form that is motor free and able to insert proteins laterally into the membrane upon reconstitution and a TIM23MOTOR form lacking Tim21, but instead associated with the motor complex to enable matrix translocation30,31. However, it remains unknown, if the motor-containing form of the translocase is solely capable of anterograde transport toward the matrix or also lateral transport into the lipid phase.
Here we find that by constitutively associating Pam18 to the TIM23 complex, lateral transport of translocating polypeptides into the lipid phase is compromised. Our analyses show that Pam18 seals the lateral gate when the motor complex drives anterograde protein transport. These findings now explain why the presequence translocase switches between a motor-bound and motor-free states and as to why the motor-free form of the translocase promotes inner membrane sorting. Our findings suggest that translocase complexes, which utilize lateral gates for membrane insertion of precursors, could regulate lateral movement by recruitment of membrane domains of dynamically associating proteins.
Results
TIM23 reorganization by constitutive association of Pam18
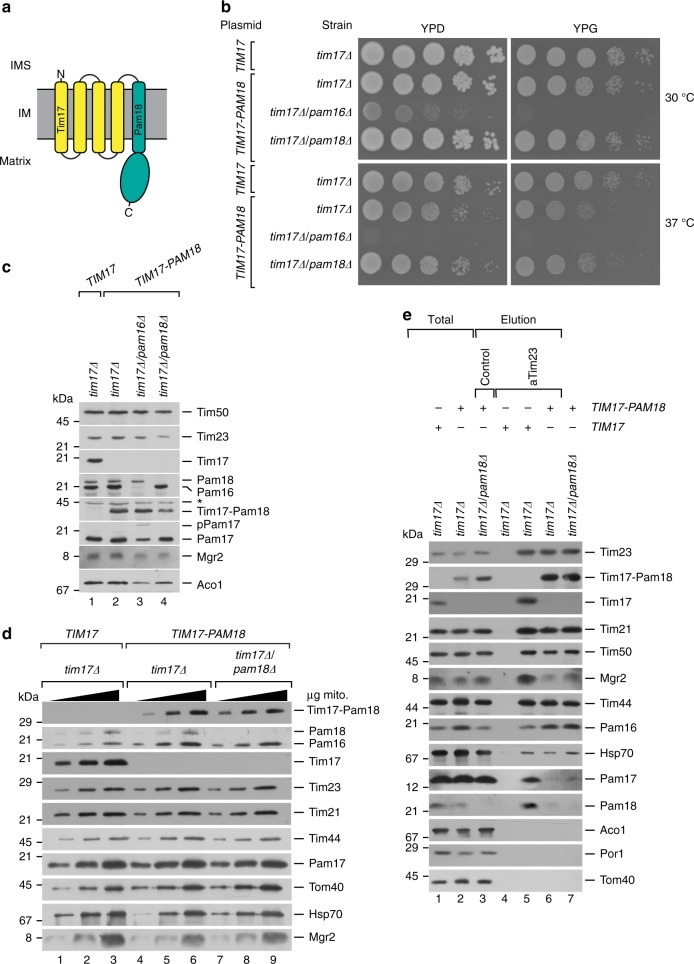
Pam18 is the presequence translocase-associated J-protein with a single transmembrane span. Its intermembrane space localized N-terminal domain associates to the translocation channel constituent Tim17. However, Pam18 is not permanently translocase associated but undergoes a replenishment cycle during import. Here we aimed to study the functional relevance of the dynamic association of Pam18 for protein translocation. To this end, we generated yeast strains in which the N terminus of Pam18 was covalently fused to the C terminus of Tim17 in a hybrid protein. Since the intermembrane space exposed C-terminal domain of Tim17 transiently interacts with the N-terminal domain of Pam18, we reasoned that this fusion protein would closely resemble the physiological translocase-associated state (Fig. 1a). In fact, expression of the Tim17–Pam18 fusion protein complemented the lethal growth defect of yeast cells deleted for TIM17 as well as a TIM17/PAM18 double deletion. The complemented cells displayed only a very mild growth defect on both fermentable and non-fermentable medium (Fig. 1b). Strikingly, expression of the Tim17–Pam18 fusion protein also allowed for deletion of the essential PAM16 gene. Pam16 is implicated in recruiting and regulating Pam18 at the translocase. These two functions can obviously, at least partially, be overcome by constitutive Pam18 association at the translocase. Nevertheless, these cells displayed a strong growth defect, indicating that a tight control of Pam18 function by Pam16 is crucial for proper import motor function (Fig. 1b). We assessed the expression of the Tim17–Pam18 fusion construct in all tested cells. Importantly, no free Tim17 or Pam18 were detected in the corresponding deletion backgrounds, demonstrating that the fusion construct remained stable in the tested strains (Fig. 1c). Due to the strong growth defect and the apparent import defect in vivo, indicated by the presence of the Pam17 precursor (Fig. 1c, lane 3), the tim17∆/pam16∆ strain was excluded from further analysis. Most importantly, expression of the Tim17–Pam18 fusion protein did not affect the amounts of other subunits of the translocase or the associated motor in mitochondria (Fig. 1d).
Fig. 1.
A functional Tim17–Pam18 fusion causes reorganization of the TIM23 complex. a Schematic representation of the Tim17–Pam18 fusion. N, N terminus; C, C terminus; IMS, intermembrane space; IM inner membrane. b Tenfold serial dilutions of wild-type and indicated Tim17–Pam18 strains were spotted on YPD and YPG plates and grown at indicated temperatures. c Western blot analysis of cell lysates from wild-type and Tim17–Pam18 strains. Asterisk, unspecific signal of antibody. Tim17–Pam18 protein was detected using Pam18 antibody. d Western blot analysis of steady-state protein using isolated mitochondria (5, 10, and 20 µg). e Digitonin-solubilized mitochondria were subjected to anti-Tim23 immunoisolation and samples were analyzed by western blotting. Total, 10%; elution, 100%
The TIM23 complex can be purified in two forms. The Tim21-containing TIM23SORT form, which lacks the import motor, and the TIM23MOTOR form, lacking Tim21 but that is bound to the motor complex. These two forms are in equilibrium and isolation of the TIM23 complex via the channel-forming Tim23 subunits leads to copurification of both states. As expected, immunoisolation with anti-Tim23 antibodies co-purified the Tim17–Pam18 fusion protein (Fig. 1e). In the context of the Tim17–Pam18 fusion protein, Tim50, Tim44, and Hsp70 co-isolated with Tim23 in wild-type-like amounts. While the amount of Tim21 was slightly reduced, Pam16, which forms a stable complex with Pam18, displayed increased copurification. In contrast, Pam17 and Mgr2 were not efficiently recovered with Tim23, even though their mitochondrial protein levels were not altered (Fig. 1e). Accordingly, covalent association of Pam18 to Tim17 affected the organization of the presequence translocase leading to the loss of the auxiliary subunits Pam17 and Mgr2. Interestingly, free Pam18 was almost completely lost from the TIM23 complex when the C terminus of Tim17 was occupied by the fused Pam18, indicating that this is the primary binding site for Pam18 at the complex. However, the constitutively Pam18-associated translocase must be capable to transport proteins since cells did not display a severe growth phenotype.
Import motor function of the Pam18-associated translocase
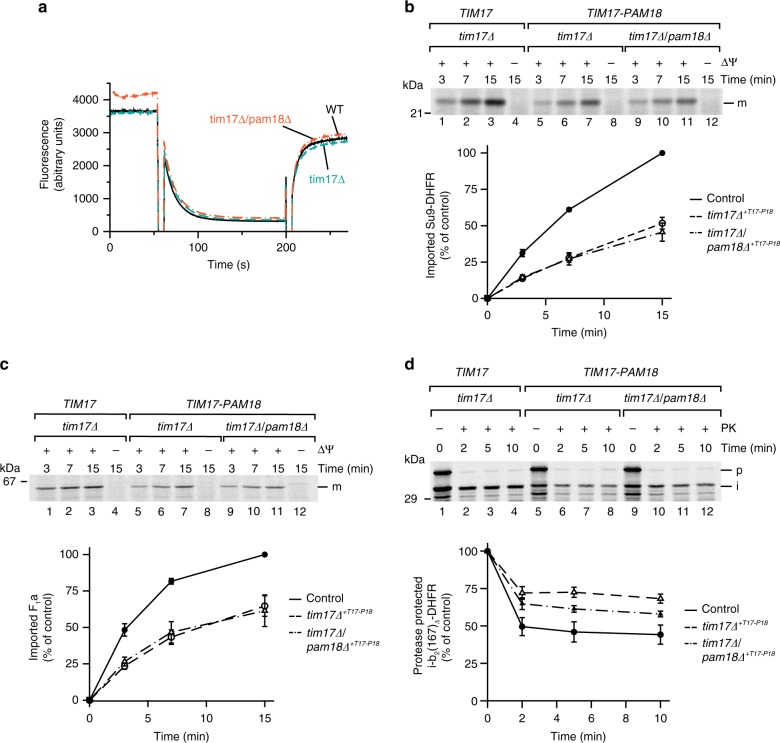
Since Pam18 is essential for motor-driven protein import into the matrix23,25,32, we aimed to analyze protein transport into mitochondria directly. To this end, we assessed the ∆ψ across the inner membrane that drives the initial presequence translocation. Wild-type and mutant mitochondria containing the Tim17–Pam18 fusion displayed similar magnitudes of ∆ψ (Fig. 2a). Based on the integrity of the ∆ψ, we assessed the matrix import capacity of wild-type and mutant mitochondria by in vitro import analyses. Import of the matrix protein F1α (a soluble subunit of the F1Fo-ATP synthase) and the model matrix protein Su9-DHFR were moderately affected in both mutants (Fig. 2b, c). An obvious explanation for this phenotype would be that motor function was partially affected.
Fig. 2.
Mitochondria containing a Tim17–Pam18 fusion display mild-matrix import defects. a Membrane potential (Δψ) of mitochondria was assessed using the Δψ-sensitive dye DiSC3(5). Fluorescence quenching by mitochondria was recorded before and after addition of valinomycin. b, c [35S]-labeled precursor proteins were imported into the mitochondria and import stopped after indicated time points using antimycin A, valinomycin, and oligomycin (AVO). Samples were Proteinase K (PK)-treated and analyzed by SDS-PAGE and digital autoradiography. Results are presented as mean ± SEM, n = 3. The longest import time of the WT sample was set to 100%. m, mature protein; tim17∆+T17-P18 tim17∆ + TIM17–PAM18. d The inward-driving force of the import motor was assessed using [35S]-labeled b2(167)∆-DHFR in the presence of methotrexate (MTX). After 15 min import, Δψ was dissipated using valinomycin. In a second step, the precursor was chased for indicated time before PK addition. The amount of processed intermediate was quantified (100%: amount of processed intermediate without protease treatment). Results are shown as mean ± SEM, n = 4. p, precursor; i, intermediate
Motor activity can be monitored by assessing its inward-driving force with a model matrix protein b2(167)∆-DHFR. Import in the presence of the substrate analog methotrexate (MTX) prevents translocation of the C-terminal, stable-folded DHFR moiety leading to a translocation intermediate that spans both TOM and TIM23 complexes and engages with the import motor on the matrix side of the inner membrane. Depletion of ∆ψ after precursor accumulation and subsequent proteinase K digestion allows for degradation of the portion of b2(167)∆-DHFR that is not strongly pulled against the TOM complex by the import motor. Remarkably, mutant strains expressing the Tim17–Pam18 fusion displayed reduced protease-sensitivity indicative of increased motor activity (Fig. 2d). This effect appears to increase with the absence of free Pam18, as increased motor activity was slightly less apparent in the tim17Δ background compared to the tim17Δ/pam18Δ double mutant. The fact that constitutive association of Pam18 to Tim17 led to increased motor activity, but at the same time a moderately reduced matrix import efficiency suggested that affecting dynamics of Pam18 is not crucial for regulation of mtHsp70 but rather affects transport in a different manner.
Translocase association of Pam18 affects lateral sorting
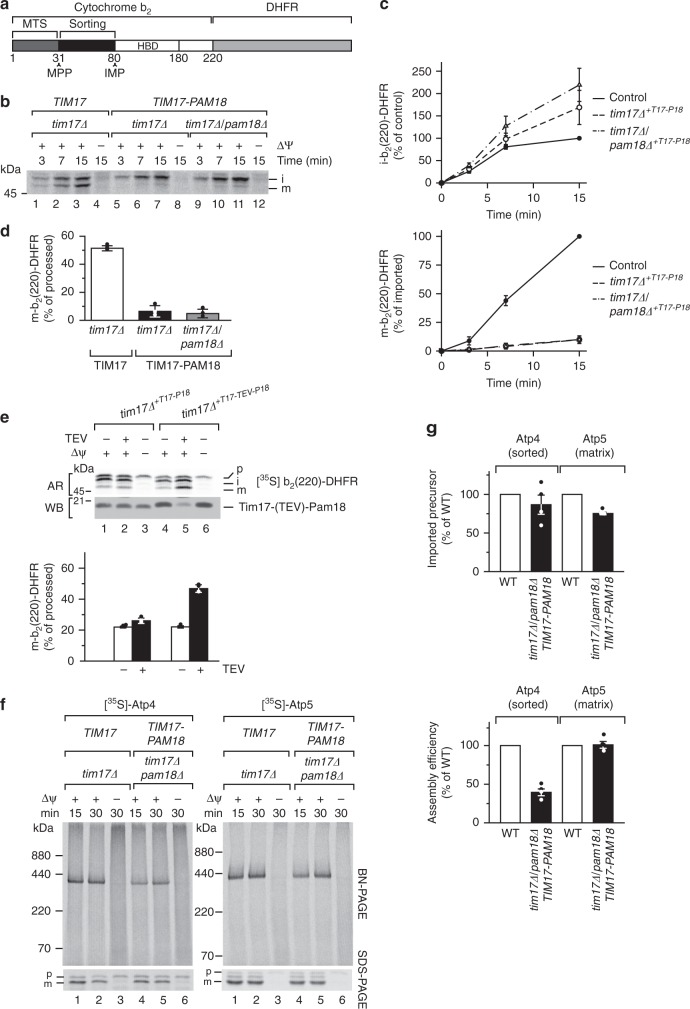
To investigate the import phenotype, we utilized the sorted inner membrane model protein b2(220)-DHFR (Fig. 3a). This protein is processed twice during import. In the matrix, the presequence is cleaved by the matrix processing peptidase (MPP). Subsequently, after the precursor has been inserted into the inner membrane by the TIM23 complex, the intermembrane space protease (IMP) cleaves the precursor a second time33. This processing pattern allows us to discriminate and kinetically resolve anterograde precursor movement by N-terminal presequence processing and succeeding lateral membrane insertion indicated by the second processing event. Upon import of the radiolabeled precursor into wild -type mitochondria, both processed forms, referred to as intermediate and mature form, were readily detectable revealing both transport events (Fig. 3b). In tim17∆ and tim17∆/pam18∆-mutant mitochondria containing the Tim17–Pam18 fusion, the intermediate form was efficiently generated, demonstrating precursor transport across the inner membrane by the TIM23 complex. Astonishingly, the second processing step was almost completely blocked (Fig. 3b). Quantification of both forms revealed that while the intermediate form accumulated in both mutants, compared to the wild-type, less than 10% of the mature form was generated (Fig. 3c). In line with this, while in the wild type situation roughly 50% of the precursor were processed twice, this was only true for around 5% in the mutant strains (Fig. 3d). To exclude that processing of cytochrome b2 by IMP was affected due to an indirect effect on the activity of this enzyme, we generated a yeast strain in which a Tobacco Etch Virus (TEV) cleavage site was introduced between Tim17 and Pam18 in the fusion construct. When the outer membrane of mitochondria was ruptured by osmotic swelling, processing to the mature form was improved in the TEV-treated sample, but not in the strain lacking the TEV site (Fig. 3e). Accordingly, the observed strong reduction of the second processing step in the presence of the Tim17–Pam18 fusion was not due to impaired IMP activity. Based on these observations, we concluded that the constitutive association of Pam18 to Tim17 did not affect anterograde protein transport of the polypeptide but rather lateral transport of the precursor into the membrane.
Fig. 3.
Pam18 affects lateral insertion of membrane-targeted precursors. a Schematic presentation of b2(220)-DHFR precursor. MTS, mitochondrial targeting signal; MPP, mitochondrial processing peptidase cleavage site; IMP, inner membrane protease cleavage site; HBD, heme-binding domain. b [35S]-labeled b2(220)-DHFR was imported as described in Fig. 2. c Quantification of intermediate (upper) and mature (lower) form of b2(220)-DHFR from b. d Quantification of the b2(220)-DHFR import after 15 min from b (processed = i + m); i, intermediate, m, mature; tim17∆+T17-P18 tim17∆ + TIM17–PAM18. Results are shown as mean ± SEM, n = 3. e Import of [35S]-labeled b2(220)-DHFR into Tim17–Pam18 and Tim17-TEV–Pam18 (TEV cleavage site was introduced between Tim17 and Pam18). Mitochondria were swollen and TEV protease treated. Subsequently, precursors were imported for 15 min. Samples were analyzed by SDS-PAGE and digital autoradiography and western blotting. Results are shown as mean ± SEM, n = 3. tim17∆+T17-P18 tim17∆/pam18∆ + TIM17–PAM18, AR, autoradiography; WB, western blot. f After import of [35S]-labeled Atp4 (membrane protein) and Atp5 (soluble matrix protein), samples were analyzed by blue-native PAGE/SDS-PAGE and subjected to digital autoradiography. g Quantification from e, upper panel, imported precursor after 30 min (SDS-PAGE), lower panel assembly efficiency normalized to import. Results are shown as mean ± SEM, n = 4
To support this conclusion, we utilized native substrates of the presequence translocase. Atp4 and Atp5 are subunits of the peripheral stalk of the F1Fo-ATP synthase. While Atp4 is an integral membrane protein, Atp5 is a soluble matrix exposed component. In tim17∆/pam18∆-mutant mitochondria, containing the Tim17–Pam18 fusion, both proteins were imported and processed by MPP with only a slight reduction (Fig. 3f, g). To assess completion of the membrane insertion process, we analyzed protein assembly into the F1Fo-ATP synthase by blue-native PAGE (BN). After 30 min of assembly, the soluble Atp5 integrated into the F1Fo-ATPsynthase with the same efficiency in control and mutant mitochondria (corrected for the mild import defect in the quantification). In contrast, assembly of Atp4 was reduced by 60% in mutant mitochondria. Hence, while the initial translocation across the TIM23 complex was not affected, Atp4 did not assemble efficiently into the ATPase, likely due to the inner membrane sorting defect.
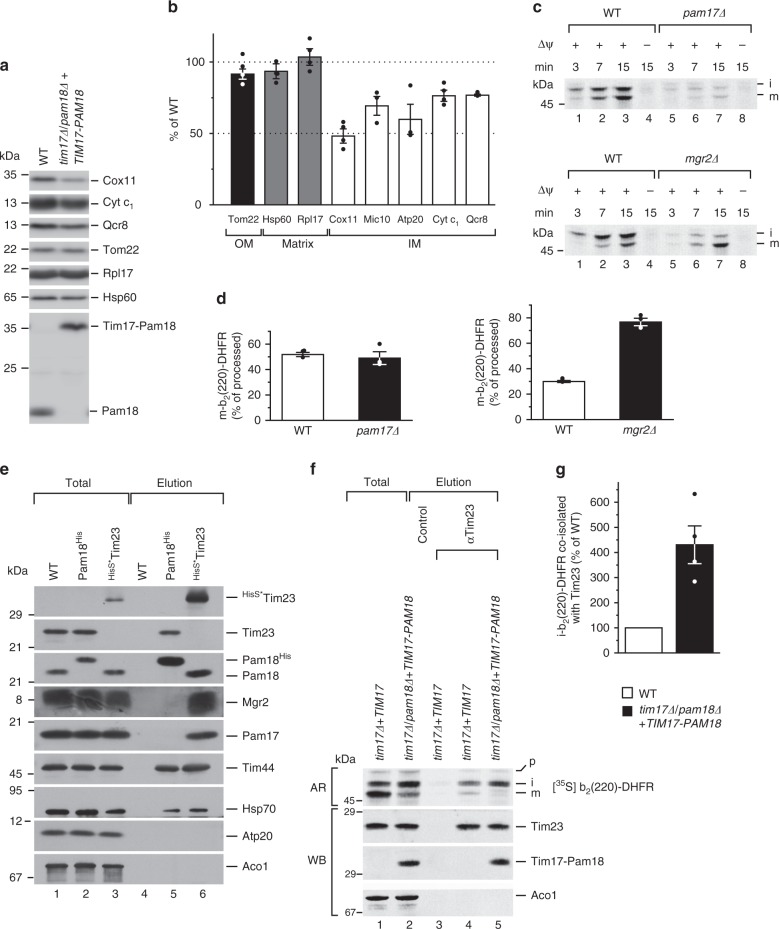
Next, we addressed whether the observed phenotype in membrane protein insertion in organello was recapitulated in the presence of the Tim17–Pam18 hybrid protein in intact cells. To this end, yeast cells were grown on fermentable medium and shifted to non-fermentable carbon source to stimulate mitochondrial biogenesis. Indeed, we found that membrane proteins of different inner membrane protein complexes were specifically reduced, while both outer membrane and matrix control proteins were unaffected. This finding confirms that Pam18 affects membrane protein insertion in vivo (Fig. 4a, b).
Fig. 4.
Pam18 regulates the lateral gate. a Yeasts were shifted to glycerol medium and grown for 4 h (Mic10 and Atp20) or 6 h (all other tested proteins). Cell lysates were analyzed for levels of indicated outer membrane (OM), matrix or inner membrane (IM) proteins by western blot. b Quantification of blots from a. Amounts of proteins in percentage of WT control. Results are shown as mean ± SEM, n = 3 (Mic10 and Atp20) n = 4 for all others. c, d Import of [35S]-labeled b2(220)-DHFR in mgr2∆ and pam17∆ and corresponding wild-type as described in Fig. 2. e WT mitochondria or mitochondria containing Pam18His or HisS*Tim23 were solubilized in digitonin buffer and subjected to Ni-NTA affinity chromatography. Samples were analyzed by SDS-PAGE and western blotting. HisS*, His-SUMOstar. f After import of [35S]-labeled b2(220)-DHFR without proteinase K treatment, mitochondria were solubilized in digitonin buffer and subjected to anti-Tim23 immunoisolation. Samples were analyzed by SDS-PAGE, western blotting, and digital autoradiography. AR, autoradiography; WB, western blot. g Quantification of co-isolated i-b2(220)-DHFR normalized to isolated Tim23. Results are shown as mean ± SEM, n = 4; p, precursor; I, intermediate;m, mature
Translocase association of Pam18 controls lateral transport
Our analyses showed that constitutive association of Pam18 to the translocase did not impact motor function per se. However, lateral transport of the precursor into the lipid bilayer was strongly affected. In addition, presence of Pam18 at the TIM23 complex lead to a loss of Mgr2 and Pam17 from the translocase. While Pam17 supports anterograde transport of ∆ψ-hypersensitive precursors into the matrix but is dispensable for lateral protein transport, Mgr2 has been suggested to provide a quality control on laterally released membrane proteins. To exclude that loss of these components indirectly caused the observed lateral sorting defect, b2(220)-DHFR was imported into mgr2∆ and pam17∆ mitochondria. In agreement with previous observations, import was strongly reduced for both the intermediate and mature forms in pam17∆ (Fig. 4c upper, d left). Compared to the phenotype of the Tim17–Pam18 fusion, loss of Mgr2 had the opposite transport effect displaying accelerated generation of mature b2(220)-DHFR (Fig. 4c lower, d right and ref. 12). Accordingly, neither loss of Pam17 nor Mgr2 explained the reduction of sorting efficiency in the presence of the Tim17–Pam18 fusion.
The loss of Mgr2 from the TIM23 complex when Pam18 was constitutively present suggested that these two proteins bind to the transport channel in a mutually exclusive manner during lateral transport of membrane proteins. To test this, we isolated the TIM23 complex via His-tagged Tim23 (HisSUMOstar-Tim23) and His-tagged Pam18. In both samples, channel and motor components (Tim44, Hsp70, and Tim23) were efficiently isolated. However, while Tim23 co-purified Mgr2 and Pam17, neither of these could be detected when the translocase was isolated via Pam18. Thus, similar to Pam17, which recently was shown to only bind to Tim23 in the absence of the import motor, Mgr2 is only found translocase associated in the motor-free form (Fig. 4e). Accordingly, Pam18 and Mgr2 define two distinct states of the translocase and potentially associate through overlapping binding sites.
Our data are in agreement with a model in which Pam18 controls the release of membrane proteins through the lateral gate into the lipid bilayer. What happens to sorted precursors that are not released into the inner membrane? Our results can be interpreted in two ways. The precursors could either be mistargeted into the mitochondrial matrix or be trapped in the TIM23 complex. To test this, we imported the inner membrane protein Ina22 into wild-type and Tim17–Pam18 mitochondria, ruptured the outer membrane, and assessed mistargeting to the matrix by protease sensitivity of the substrate. Similar minute amounts of Ina22 were found protected in both strains (Supplementary Fig. 1a). Accordingly, in the presence of Tim17–Pam18, Ina22 was not mistargeted to the mitochondrial matrix, which would lead to an increase in protease protection.
To assess an arrest in the translocase channel, we imported b2(220)-DHFR in the presence of MTX into mitochondria containing the constitutively translocase-associated Pam18. Subsequently, we isolated the TIM23 complex. As expected, in the mutant strain four times more of the intermediate form of the precursor was isolated with the translocase compared to the wild-type control (Fig. 4f, g). We conclude that Pam18 prevents the lateral precursor transport from the translocase and thus traps the polypeptide in a translocase-spanning manner.
Discussion
In order to maintain organelle homeostasis, proteins have to be targeted to and imported into organelles, which includes anterograde translocation across and lateral transport into membranes. In most cases, membrane-embedded protein complexes mediate transport of proteins to their target destinations and, with a few exceptions, these complexes can transport both soluble proteins across or hydrophobic proteins into membranes. Different modes of protein insertion into membranes exist, depending on the nature of the precursor. One of the most common principle is transport through a lateral gate, which involves lateral opening of the import channel toward and concomitant release of the precursor into the lipid bilayer. This has been described extensively for the SecYEG and Sec61 translocon in the bacterial and mammalian ER, respectively, but was also proposed for other systems including the mitochondrial TIM23 complex34–38.
The TIM23 complex exists in at least two forms, the TIM23SORT form, which is dedicated to insertion of membrane proteins and the TIM23MOTOR form, which drives full protein translocation at the expense of ATP. It is established that matrix transport of soluble proteins cannot occur in the absence of the import motor, however, if the import motor also affects transport of membrane proteins remained elusive. Here we show that in addition to its function in motor-dependent precursor translocation, the J-protein Pam18 also regulates lateral membrane proteins insertion into the lipid phase. A covalent linkage between Pam18 and the channel component Tim17 delays lateral release of hydrophobic proteins into the mitochondrial inner membrane. Pam18 is the only motor component that contains a membrane-spanning α-helix and is connected to the import channel on both sides of the translocase. This raises the possibility that the transmembrane domain of Pam18 is directly involved in closing the lateral gate when associated with the TIM23 complex. In line with this, Mgr2, which is involved in precursor quality control and affects membrane protein insertion, is displaced from the translocase when Pam18 is covalently fused to the channel component Tim17. This finding suggests overlapping binding sites of these proteins at the lateral gate. Interestingly, Mgr2 was not co-purified with Pam18His, indicating that both proteins define different states of the translocase.
We propose that during transport of matrix-targeted proteins, recruitment of Pam18 blocks the lateral gate thereby promoting anterograde transport of the precursors into the mitochondrial matrix. Once a hydrophobic transmembrane span gets arrested in the TIM23 channel, Pam18 dissociates allowing Mgr2 recruitment, which mediates quality control during membrane protein insertion. This would also explain why in mgr2∆ mitochondria, the dynamic recruitment of Pam18 is reduced during precursor translocation. This is especially interesting for membrane proteins that have N-terminal matrix domains which have to be imported by the motor complex but are lateral released into the inner membrane such as Cox5a in yeast. For such precursors, the TIM23 complex has to identify the transmembrane segment, which leads to a transfer arrest, probably associated with motor dissociation and Mgr2 association. Strikingly, the transmembrane segment of Pam18 has a cluster of only mildly hydrophobic residues, which could be exposed into the water-filled transport channel when Pam18 is TIM23 associated. Our findings explain a surprising previous observation. The deletion of the IMS domain of Pam18 was found to increase import of membrane proteins39. Since the IMS domain of Pam18 couples the protein to Tim17, abrogating this interactions site affects association of Pam18 with the import channel and concomitantly allows for a more efficient lateral transport.
Interestingly, despite the clear import defect for membrane proteins in vivo after switching yeast to non-fermentable carbon sources, we did not observe a decrease in membrane potential in isolated mitochondria (Fig. 2a). On the one hand, this could be due to the fact that the constitutive presence of Pam18 at the lateral gate does not completely block lateral sorting (Fig. 4a, b). On the other hand, a recent study showed that in response to accumulation of membrane-targeted TIM23 substrates, yeast cells mount a transcriptional response, which ultimately helps to remove stalled precursors and thereby improve import efficiency40. It would be interesting to address if this process also supports the transport process in the Tim17–Pam18 mutant.
To what extent the dynamics of Pam18 (and Pam16) is important for protein translocation is not known. In the Tim17–Pam18 fusion construct, Pam18 cannot be exchanged at the translocase, even if endogenous Pam18 is still present. Nevertheless, yeast cells are viable and only display a mild growth defect while transport of soluble proteins is reduced by roughly 50%, which might be attributed to reduced motor dynamics. Interestingly, when motor-depended backsliding of a stalled precursor was assessed in this strain, mitochondria displayed an increase in protease-protected precursor, which points toward increased motor function. Since this in contrast to the import phenotype for matrix proteins, we suggest that in the Tim17–Pam18 strain reduced motor dynamics lead to a prolonged stimulation of Hsp70, which is bound to both precursor and translocase. As a result, we observe a decrease in anterograde movement of the precursor while backsliding is prevented.
In conclusion, for translocation of precursors across or into the inner mitochondrial membrane, opening of the lateral gate is tightly regulated by the import motor constituent Pam18. Hence, motor recruitment switches the direction of precursor movement not only by providing energy for anterograde transport, but also by restricting the access to the lateral gate (Fig. 5). It will be important to define if a similar regulation of lateral precursor movement by associated transmembrane segments exists in chloroplast and the endoplasmic reticulum. Interestingly, in both cases, for the mammalian Sec complex and the TIC complex, membrane-bound J-proteins have been reported.
Fig. 5.
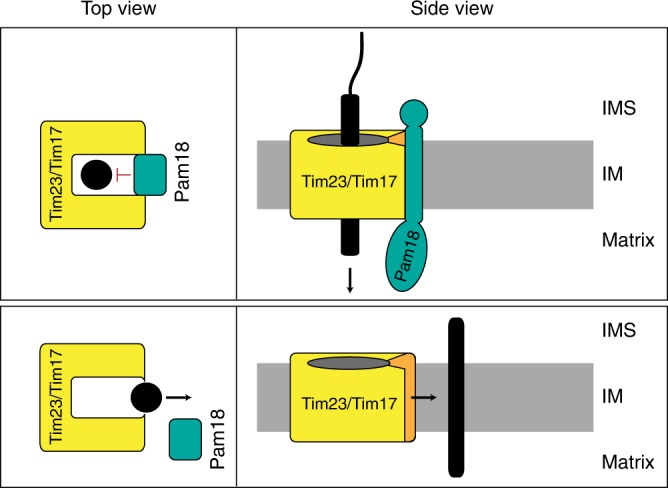
Pam18 controls the lateral gate promoting anterograde protein transport. Model of how Pam18 might affect release of membrane-targeted proteins into the lipid bilayer. When Pam18 is bound together with other motor components, the lateral gate is closed, which promotes matrix translocation of substrates. IMS, intermembrane space; IM, inner membrane
Methods
Yeast strains and handling
The wild-type yeast strain YPH499 (MATa, ade2-101, his3-∆200, leu2-∆1, ura3-52, trp1-∆63, and lys2-801) (ATCC® 204679™) and mutants derived from it were used in this study. mgr2∆, pam17∆, and Pam18His have been described previously21,31,41. Yeast strains were grown in YP medium (1% yeast extract, 2% peptone) containing 2% glucose (YPD) or 3% glycerol (YPG) at 30 °C. mgr2∆ and corresponding WT were grown at 25 °C. For generation of His-SUMOstar-Tim23 (HisS*Tim23), a His-tag (MTSKHHHHSGHHHTGHHHHSGSHHHGS) together with SUMOstar42 was fused to the TIM23 gene. This construct together with 511 bp upstream and 308 bp downstream of the open reading frame were cloned into pRS413. For generation of the Tim17–Pam18 strain and modifications thereof, TIM17 + 996 bp upstream were cloned with EcoRI and BamHI into pFL39. In a second round, PAM18 + 1000 bp downstream were cloned with BamHI and SalI into the vector. The resulting plasmid (pCS164) and wild-type TIM17 containing plasmids were transformed into YPH499 in which tim17 deletion was rescued with wild-type TIM17 expressed from URA3-containing plasmid. The URA3-containing plasmid was shuffled out by selecting yeast on 5-FOA-containing medium. In a second round, PAM18 was deleted using pFA6a HIS and PAM16 with pFA6a hphNT1. For generation of TEV cleavable Tim17–Pam18, the TEV site (ENLYFYG) was introduced by overlap PCR downstream of the BamHI site. Primers used in this study are listed in the Supplementary Table 1.
For preparation of whole cell lysates, yeast cells were grown in YPD medium at 30 °C over night, 5 OD600 harvested the next day, cells sedimented, washed with water, and suspended in 300 µl H2O. After addition of 100 µl 50% TCA, cells were incubated for 15 min on ice, precipitated proteins pelleted, washed with 80% ice-cold acetone, pelleted again and resuspended in buffer containing 1% SDS, 100 mM NaOH. For analysis of in vivo import defects, cells were grown over night in YPD medium at 30 °C. Next morning cells were diluted to OD600 = 0.3 in YPG medium, grown for 4–6 h and analyzed as described before.
Import of precursor proteins
Precursor proteins were radiolabeled by translation in the presence of [35S]-methionine using rabbit reticulocyte lysate (Promega). Isolated mitochondria were mixed in import buffer (250 mM sucrose, 10 mM MOPS/KOH pH 7.2, 80 mM KCl, 2 mM KH2PO4, 5 mM MgCl2, 5 mM methionine, and 3% fatty acid-free BSA) with 2 mM ATP and 2 mM NADH. Aliquots of 1 µM valinomycin, 8 µM antimycin A, and 20 µM oligomycin were used to dissipate the membrane potential to stop import reactions, after which Proteinase K (PK, 20 µg/ml) treatment was performed to remove, not imported proteins. After 10 min incubation on ice, 2 mM phenylmethylsuophonyl fluoride (PMSF) was added for 10 min on ice to inactivate PK. After pelleting mitochondria and washing with SEM (250 mM sucrose, 20 mM MOPS pH 7.2, and 1 mM EDTA), import was analyzed by SDS-PAGE or BN-PAGE (for BN samples, PK treatment was omitted) and autoradiography. Quantifications were performed using ImageQuant TL (GE Healthcare) using a rolling ball background subtraction.
For import after TEV cleavage, mitochondria were resuspended in import buffer lacking sucrose and incubated with 60 µg/ml TEV protease and kept on ice for 30 min. Subsequently, 2 mM NADH and ATP were added following the import reaction.
For swelling experiments, precursor was imported for 15 min, import stopped with AVO and samples split into two equal samples. After SEM wash, samples were treated with EM buffer (20 mm MPOS pH 7.2, and 1 mM EDTA) for 20 min on ice and 5 µg PK was added to 100 µl EM buffer. After 15 min incubation, 2 mM PMSF was added for 15 min, mitochondria were washed with SEM and analyzed by SDS-PAGE and autoradiography.
Membrane potential measurements
Mitochondrial membrane potential was assessed using the potential sensitive dye 3,3′-dipropylthiadicarbocyanine iodide (DiSC3(5)). For this, mitochondria were diluted in buffer containing 600 mM sorbitol, 1% (wt/vol) BSA, 10 mM MgCl2, and 20 mM KPi, pH 7.4 to a concentration of 166 µg/ml. Changes in fluorescence were recorded using a F-7000 fluorescence spectrophotometer (Hitachi) at 25 °C with excitation at 622 nm, emission at 670 nm, and slits of 5 nm. To 500 µl of buffer in a cuvette with DiSC3(5), 83 µg of mitochondria were added first and, after fluorescence signal reached a stable signal, 1 µM valinomycin was added. The difference in fluorescence before and after the addition of valinomycin was used to compare relative membrane potential between strains.
Import-driving activity
Import-driving activity was assessed as previously described43. In brief, mitochondria were mixed in import buffer supplemented with 5 mM creatine phosphate (CP) and 0.01 mg/ml creatine kinase (CK). Radiolabeled b2(167)∆-DHFR was imported at 25 °C for 15 min in the presence of 5 µM methotrexate (MTX) and membrane potential was dissipated using 1 µM valinomycin. A sample (∆t = 0) was taken prior to chase for 1, 5, and 10 min and PK was added for 15 min on ice. After protease inhibition with PMSF, samples were analyzed by SDS-PAGE and autoradiography. The amount of PK-resistant intermediate was quantified and normalized to the amount of generated intermediate (∆t = 0) for each strain.
Protein complex isolation
Isolation of TIM23 complex using Tim23-specific serum was carried out essentially as described44. For this, mitochondria were resuspended to 1 mg/ml in solubilization buffer (20 mM Tris/HCl pH 7.4, 150 mM NaCl, 10% glycerol (w/v), 1 mM PMSF, and 1% digitonin) and solubilized for 30 min on ice. The soluble fraction was loaded on Protein A sepharose beads cross-linked to Tim23 antibodies, incubated for 30 min to 1 h on a spinning wheel at 4 °C, washed five times (solubilization buffer with 0.3% digitonin) and eluted with double bed volume of 0.1 M glycine pH 2.8. For isolation by His-tags, mitochondria were solubilized as described earlier with 10 mM imidazole, supernatant was loaded on Ni-NTA beads, washed with washing buffer containing 10 mM imidazole and eluted with washing buffer containing 200 mM imidazole.
For TIM23 isolation after import, b2(220)-DHFR was imported in the presence of 5 mM methotrexate (MTX) and mitochondria solubilized after an SEM wash without addition of antimycin A, Valinomycin, oligomycin, or Proteinase K.
Isolation of mitochondria
Mitochondria were isolated as previously described45. Yeast were grown in YPG medium at 30 °C and harvested at OD600 = 2–3 if not stated otherwise. Pelleted cells were resuspended in buffer A (10 mM DTT, 100 mM Tris/H2SO4 pH 9.4) and incubated for 30 min at 30 °C. After a washing step, cell wall removed in zymolyase buffer (20 mM KPO4 pH 7.4, 1.2 M sorbitol, and 0.57 mg/l zymolyase) for 1–2 h at 30 °C and opened in ice-cold homogenization buffer (600 mM sorbitol, 10 mM Tris/HCl pH 7.4, 1 g/l BSA, 1 mM PMSF, and 1 mM EDTA) using a cell homogenizer. Mitochondrial fraction was obtained by differential centrifugation, resuspended in SEM buffer and frozen in liquid nitrogen.
Electronic supplementary material
Acknowledgements
We thank D. Mokranjac and D. Pacheu-Grau for the support, critical reading, and discussion. We thank D. Görlich for the His-SUMOstar construct. Supported by the Deutsche Forschungsgemeinschaft SFB860 project B01 (P.R.), the Max Planck Society (P.R.), the Copernicus Award (P.R. and A.C.), Boehringer Ingelheim Fonds (C.S.), Foundation for Polish Science First TEAM Program co-financed by the European Union under the European Regional Development Fund (P.B.), and the PhD program Molecular Biology – International Max Planck Research School and the Göttingen Graduate School for Neurosciences and Molecular Biosciences (GGNB; DFG grant GSC 226/1) (A.S., N.N., R.G., and C.S.).
Author contributions
A.S., A.C., and P.R. conceived the research and designed the experiments. A.S., R.G., C.S., P.B., N.N., and P.B. performed the experiments and prepared figures and tables. A.S. and P.R. wrote the manuscript with the input of B.G. and A.C. All authors discussed the results. All authors approved the final version of the manuscript.
Data availability
Data supporting the findings of this manuscript are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information accompanies this paper at 10.1038/s41467-018-06492-8.
References
- 1.Roise D, Schatz G. Mitochondrial presequences. J. Biol. Chem. 1988;263:4509–4511. [PubMed] [Google Scholar]
- 2.Vögtle FN, et al. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 2009;139:428–439. doi: 10.1016/j.cell.2009.07.045. [DOI] [PubMed] [Google Scholar]
- 3.Hill K, et al. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature. 1998;395:516–521. doi: 10.1038/26780. [DOI] [PubMed] [Google Scholar]
- 4.Chacinska A, et al. Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell. 2005;120:817–829. doi: 10.1016/j.cell.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 5.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schulz C, Schendzielorz A, Rehling P. Unlocking the presequence import pathway. Trends Cell Biol. 2015;25:265–275. doi: 10.1016/j.tcb.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Martin J, Mahlke K, Pfanner N. Role of an energized inner membrane in mitochondrial protein import. Delta psi drives the movement of presequences. J. Biol. Chem. 1991;266:18051–18057. [PubMed] [Google Scholar]
- 8.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 9.Mokranjac D, Neupert W. Energetics of protein translocation into mitochondria. Biochim. Biophys. Acta. 2008;1777:758–762. doi: 10.1016/j.bbabio.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 10.Wiedemann N, Pfanner N. Mitochondrial machineries for protein import and assembly. Annu. Rev. Biochem. 2017;86:12.1–12.30. doi: 10.1146/annurev-biochem-060815-014352. [DOI] [PubMed] [Google Scholar]
- 11.Glick BS, et al. Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell. 1992;69:809–822. doi: 10.1016/0092-8674(92)90292-K. [DOI] [PubMed] [Google Scholar]
- 12.Ieva R, et al. Mgr2 functions as lateral gatekeeper for preprotein sorting in the mitochondrial inner membrane. Mol. Cell. 2014;56:641–652. doi: 10.1016/j.molcel.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 13.Schendzielorz AB, et al. Two distinct membrane potential-dependent steps drive mitochondrial matrix protein translocation. J. Cell Biol. 2017;216:83–92. doi: 10.1083/jcb.201607066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian X, et al. Structural basis for the function of Tim50 in the mitochondrial presequence translocase. J. Mol. Biol. 2011;411:513–519. doi: 10.1016/j.jmb.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schulz C, et al. Tim50’s presequence receptor domain is essential for signal driven transport across the TIM23 complex. J. Cell Biol. 2011;195:643–656. doi: 10.1083/jcb.201105098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lytovchenko O, et al. Signal recognition initiates reorganization of the presequence translocase during protein import. EMBO J. 2013;32:886–898. doi: 10.1038/emboj.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Laan M, et al. A role for Tim21 in membrane-potential-dependent preprotein sorting in mitochondria. Curr. Biol. 2006;16:2271–2276. doi: 10.1016/j.cub.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 18.Bauer Matthias F, Sirrenberg Christian, Neupert Walter, Brunner Michael. Role of Tim23 as Voltage Sensor and Presequence Receptor in Protein Import into Mitochondria. Cell. 1996;87(1):33–41. doi: 10.1016/S0092-8674(00)81320-3. [DOI] [PubMed] [Google Scholar]
- 19.Ramesh A, et al. A disulfide bond in the TIM23 complex is crucial for voltage gating and mitochondrial protein import. J. Cell Biol. 2016;6:417–431. doi: 10.1083/jcb.201602074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Caballero S, Grigoriev SM, Herrmann JM, Campo ML, Kinnally KW. Tim17p regulates the twin pore structure and voltage gating of the mitochondrial protein import complex TIM23. J. Biol. Chem. 2007;282:3584–3593. doi: 10.1074/jbc.M607551200. [DOI] [PubMed] [Google Scholar]
- 21.Gebert M, et al. Mgr2 promotes coupling of the mitochondrial presequence translocase to partner complexes. J. Cell Biol. 2012;197:595–604. doi: 10.1083/jcb.201110047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frazier AE, et al. Pam16 has an essential role in the mitochondrial protein import motor. Nat. Struct. Mol. Biol. 2004;11:226–233. doi: 10.1038/nsmb735. [DOI] [PubMed] [Google Scholar]
- 23.Truscott KN, et al. A J-protein is an essential subunit of the presequence translocase-associated protein import motor of mitochondria. J. Cell Biol. 2003;163:707–713. doi: 10.1083/jcb.200308004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mokranjac D, Bourenkov G, Hell K, Neupert W, Groll M. Structure and function of Tim14 and Tim16, the J and J-like components of the mitochondrial protein import motor. EMBO J. 2006;25:4675–4685. doi: 10.1038/sj.emboj.7601334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D’Silva PD, Schilke B, Walter W, Andrew A, Craig EA. J protein cochaperone of the mitochondrial inner membrane required for protein import into the mitochondrial matrix. Proc. Natl Acad. Sci. USA. 2003;100:13839–13844. doi: 10.1073/pnas.1936150100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Q, D’Silva P, Walter W, Marszalek J, Craig EA. Regulated cycling of mitochondrial Hsp70 at the protein import channel. Science. 2003;300:139–141. doi: 10.1126/science.1083379. [DOI] [PubMed] [Google Scholar]
- 27.Marom M, et al. Interaction of the Tim44 C-terminal domain with negatively charged phospholipids. Biochemistry. 2009;48:11185–11195. doi: 10.1021/bi900998v. [DOI] [PubMed] [Google Scholar]
- 28.Schilke BA, Hayashi M, Craig EA. Genetic analysis of complex interactions among components of the mitochondrial import motor and translocon in Saccharomyces cerevisiae. Genetics. 2012;190:1341–1353. doi: 10.1534/genetics.112.138743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schulz C, Rehling P. Remodelling of the active presequence translocase drives motor-dependent mitochondrial protein translocation. Nat. Commun. 2014;5:1–9. doi: 10.1038/ncomms5349. [DOI] [PubMed] [Google Scholar]
- 30.van der Laan M, et al. Motor-free mitochondrial presequence translocase drives membrane integration of preproteins. Nat. Cell Biol. 2007;9:1152–1159. doi: 10.1038/ncb1635. [DOI] [PubMed] [Google Scholar]
- 31.Chacinska A, et al. Distinct forms of mitochondrial TOM-TIM supercomplexes define signal-dependent states of preprotein sorting. Mol. Cell. Biol. 2010;30:307–318. doi: 10.1128/MCB.00749-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mokranjac D, Sichting M, Neupert W, Hell K. Tim14, a novel key component of the import motor of the TIM23 protein translocase of mitochondria. EMBO J. 2003;22:4945–4956. doi: 10.1093/emboj/cdg485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voos W, Gambill BD, Guiard B, Pfanner N, Craig EA. Presequence and mature part of preproteins strongly influence the dependence of mitochondrial protein import on heat shock protein 70 in the matrix. J. Cell Biol. 1993;123:119–126. doi: 10.1083/jcb.123.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park E, Rapoport TA. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu. Rev. Biophys. 2012;41:21–40. doi: 10.1146/annurev-biophys-050511-102312. [DOI] [PubMed] [Google Scholar]
- 35.Shao S, Hegde RS. Membrane protein insertion at the endoplasmic reticulum. Annu. Rev. Cell. Dev. Biol. 2011;27:25–56. doi: 10.1146/annurev-cellbio-092910-154125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lycklama A Nijeholt JA, Driessen AJM. The bacterial Sec-translocase: structure and mechanism. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012;367:1016–1028. doi: 10.1098/rstb.2011.0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schleiff E, Becker T. Common ground for protein translocation: access control for mitochondria and chloroplasts. Nat. Rev. Mol. Cell Biol. 2011;12:48–59. doi: 10.1038/nrm3027. [DOI] [PubMed] [Google Scholar]
- 38.Oreb M, Tews I, Schleiff E. Policing Tic ‘n’ Toc, the doorway to chloroplasts. Trends Cell Biol. 2008;18:19–27. doi: 10.1016/j.tcb.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Popov-Celeketić D, Waegemann K, Mapa K, Neupert W, Mokranjac D. Role of the import motor in insertion of transmembrane segments by the mitochondrial TIM23 complex. EMBO Rep. 2011;12:542–548. doi: 10.1038/embor.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weidberg H, Amon A. MitoCPR-A surveillance pathway that protects mitochondria in response to protein import stress. Science. 2018;360:eaan4146. doi: 10.1126/science.aan4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Laan M, et al. Pam17 is required for architecture and translocation activity of the mitochondrial protein import motor. Mol. Cell. Biol. 2005;25:7449–7458. doi: 10.1128/MCB.25.17.7449-7458.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frey S, Görlich D. The Xenopus laevis Atg4B protease: insights into substrate recognition and application for tag removal from proteins expressed in pro- and eukaryotic hosts. PLoS ONE. 2015;10:e0125099. doi: 10.1371/journal.pone.0125099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voisine C, et al. The protein import motor of mitochondria: unfolding and trapping of preproteins are distinct and separable functions of matrix Hsp70. Cell. 1999;97:565–574. doi: 10.1016/S0092-8674(00)80768-0. [DOI] [PubMed] [Google Scholar]
- 44.Herrmann JM, Westermann B, Neupert W. Analysis of protein-protein interactions in mitochondria by coimmunoprecipitation and chemical cross-linking. Methods Cell Biol. 2001;65:217–230. doi: 10.1016/S0091-679X(01)65013-1. [DOI] [PubMed] [Google Scholar]
- 45.Meisinger C, Pfanner N, Truscott KN. Isolation of yeast mitochondria. Methods Mol. Biol. 2006;313:33–39. doi: 10.1385/1-59259-958-3:033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this manuscript are available from the corresponding author on reasonable request.