Abstract
Morbidity and mortality associated with retinoblastoma have decreased drastically in recent decades, in large part owing to better prediction of high-risk disease and appropriate treatment stratification. High-risk histopathologic features and severe anaplasia both predict the need for more aggressive treatment; however, not all centers are able to assess tumor samples easily for the degree of anaplasia. Instead, identification of genetic signatures that are able to distinguish among anaplastic grades and thus predict high- versus low-risk retinoblastoma would facilitate appropriate risk stratification in a wider patient population. A better understanding of genes dysregulated in anaplasia also would yield valuable insights into pathways underlying the development of more severe retinoblastoma. Here, we present the histopathologic and gene expression analysis of 28 retinoblastoma cases using microarray analysis. Tumors of differing anaplastic grade show clear differential gene expression, with significant dysregulation of unique genes and pathways in severe anaplasia. Photoreceptor and nucleoporin expression in particular are identified as highly dysregulated in severe anaplasia and suggest particular cellular processes contributing to the development of increased retinoblastoma severity. A limited set of highly differentially expressed genes also are able to predict severe anaplasia accurately in our data set. Together, these data contribute to the understanding of the development of anaplasia and facilitate the identification of genetic markers of high-risk retinoblastoma.
Retinoblastoma is the most common intraocular cancer of childhood, accounting for 6.1% of all cancers in children younger than 5 years of age.1 Advances in surveillance and vision-sparing techniques over the past several decades have greatly improved outcomes,2 with 5-year survival rates now greater than 95% in developed countries.3 Depending on the level of risk and stage of retinoblastoma at presentation, eyes with retinoblastoma may be salvaged using chemoreduction, in which chemotherapy is systemically infused for five to six cycles and the primary retinoblastoma is consolidated by laser photocoagulation in the operating room.4 Alternatively, super-selective ophthalmic artery endovascular infusion of chemotherapy via intra-arterial chemotherapy is used in some centers.5, 6, 7, 8 Both chemoreduction and intra-arterial chemotherapy have reduced enucleation rates to approximately 10% of retinoblastoma cases in developed countries. Such improvement in treatments and clinical outcomes stems from a better understanding of the risk factors associated with metastasis and mortality, which have in turn enabled better patient risk stratification and tailoring of care.
To date, estimates of clinical risk in retinoblastoma have been determined by a combination of clinical and histopathologic features. Clinical features such as older age at presentation, symptom duration longer than 6 months, poor visual acuity at presentation, buphthalmos, secondary glaucoma, iris neovascularization, ectropion uveae, orbital cellulitis, and group E tumors (the most severe according to the International Classification of Intraocular Retinoblastoma9) have been shown to be associated strongly with high-risk retinoblastoma.10 High-risk histopathologic features (HRPFs) known to be associated with worse prognosis include tumor invasion of the optic nerve, choroid, or anterior chamber and indicate the need for adjuvant chemotherapy after enucleation.11, 12, 13 However, a small percentage of retinoblastoma cases that progress to metastasis or death are not captured by these well-validated clinical and histopathologic indicators, prompting a search for additional histologic and genetic markers capable of more accurately predicting high-risk retinoblastoma and the need for chemotherapy.14
Cellular anaplasia recently was identified as an additional histopathologic indicator complementary to HRPFs in detecting high-risk retinoblastoma.15 Anaplasia, an entity similar to but distinct from cellular differentiation, is defined by specific and measurable cytologic features such as pleomorphism, hyperchromatic nuclei, and a high nucleus-to-cytoplasm ratio.15 Anaplasia is commonly used in the classification of pediatric medulloblastoma, another embryonal central nervous system tumor, which is known to be associated with aggressive clinical behavior.16 We recently determined that cellular anaplasia similarly can indicate high-risk retinoblastoma and serve as a predictor of metastases even in the absence of HRPFs.15 However, there are a limited number of ophthalmic pathology laboratories worldwide that are capable of properly examining enucleated eyes for anaplasia and HRPFs, preventing many clinical centers from taking advantage of this predictive tool.
The goal of the present study was to determine the gene expression profiles of different grades of anaplasia. Identifying differentially expressed genes may lend insight into the pathways contributing to increased anaplastic severity and also serve as the first step in defining genetic markers of anaplastic grade. Determining the gene expression profiles that distinguish anaplastic grades may lead to a gene-based clinical test to facilitate risk stratification and treatment management of retinoblastoma patients. Here, we present the gene expression profiles of 28 retinoblastoma samples of mild, moderate, and severe grade anaplasia in addition to normal retina and retinocytoma, a benign precursor of retinoblastoma. We furthermore identified genes that are differentially expressed in severe versus mild and moderate anaplasia and found that although mild and moderate anaplasia are highly similar in terms of gene expression, severe anaplasia showed significant changes in the expression of nonoverlapping genes and pathways. We describe these genetic pathways that appear to distinguish severe versus mild and moderate anaplasia and discuss how they may underlie increased severity in retinoblastoma. Finally, we determined particular genes that are able to predict severe anaplasia among our retinoblastoma samples. These data comprise the first genetic characterization of anaplasia in retinoblastoma and advance our understanding of this important predictor of severity in this childhood cancer.
Materials and Methods
Samples and Patient Data
Enucleation and exenteration specimens with a diagnosis of retinoblastoma were identified from the LF Montgomery Laboratory at the Emory Eye Center from January 1, 1940, to August 31, 2013. Exclusion criteria included any treatment before enucleation, fewer than two low-power (20×) fields of tumor, or retinoblastoma with diffuse growth patterns.17, 18 Patient medical records were reviewed for RB1 mutational status (heritable or sporadic) and demographic parameters, including age at presentation, age at enucleation, sex, and race. Clinical findings including laterality, symptoms, ophthalmologic findings, Reese-Ellsworth Classification,19 International Classification of Retinoblastoma,20 length of follow-up evaluation, and treatments also were reviewed. Clinical outcomes of interest as determined from chart review were local recurrence and the presence of distant metastasis and secondary tumors. Study approval was obtained from the Emory University Institutional Review Board.
Histopathologic Grading
Histopathologic evaluation of enucleation specimens was conducted as previously described to determine tumor size, growth pattern, level of differentiation, degree of apoptosis, grade of anaplasia, tumor seeding, extent of tissue invasion, and presence of retinocytoma.15 Two ophthalmic pathologists (C.S. and H.E.G.) blinded to patient data reviewed standard pupil–optic nerve sections and transverse optic nerve sections at the surgical margin of each specimen according to American Joint Committee on Cancer pathologic classifications.21, 22 Calottes were obtained and examined for all cases collected subsequent to protocol standardization [retinoblastoma (RB) 7, RB21-24, RB30, RB32, RB33].21 Calotte examination provides more thorough and conclusive determination of HRPFs; however, all cases lacking calottes were examined according to standard contemporary practices as described earlier in this paragraph.
High-risk features were determined as defined by the International Retinoblastoma Staging Working Group and included any invasion of the postlaminar optic nerve, massive choroidal invasion, anterior segment invasion, or prelaminar or laminar optic nerve invasion in conjunction with nonmassive choroidal invasion.21, 23 Anaplastic grade was determined by two independent ophthalmic pathologists (C.S. and H.E.G.) blinded to patient data and included assessment of cell shape, cell wrapping, mitosis, and nuclear size, contour, and chromaticity.15 Specifically, retinocytoma was defined as cells showing unenlarged nuclei, abundant eosinophilic cytoplasm and evenly dispersed chromatin, proper differentiation as reflected by fleurettes, and the absence of mitotic figures and pleomorphism. Mild anaplasia was defined by unenlarged nuclei, differentiation (Flexner-Wintersteiner and Homer Wright rosettes), and the presence of rare mitotic figures and mild pleomorphism. Moderate anaplasia was defined by enlarged nuclei, moderate to poor differentiation, frequent mitotic figures, and moderate pleomorphism. Severe anaplasia was defined by very large hyperchromatic nuclei, poor differentiation, numerous mitotic figures, extreme pleomorphism (angular, rhomboid, or fusiform), and cell wrapping. Grade was assigned to a sample according to the highest grade identified in any focal area.
Gene Expression Analysis
Tissue cores were obtained from formalin-fixed, paraffin-embedded tissue blocks for RNA preparation. Samples with less than 70% tumor content determined by histology were excluded from analysis. Total RNA was extracted using the AllPrep DNA/RNA formalin-fixed, paraffin-embedded tissue kit (Qiagen, Valencia, CA) and Mag-Bind XP formalin-fixed, paraffin-embedded tissue RNA kit (Omega Bio-tek, Norcross, GA), and RNA integrity was assessed using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA).
First- and second-strand cDNA synthesis, labeling, and hybridization to Affymetrix (Santa Clara, CA) Human Gene 2.0ST arrays were performed according to standard Affymetrix protocols by the Emory University Integrated Genomics Core. Data were deposited to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo; accession number GSE110811). Samples that failed amplification (three retinoblastoma and two normal retina samples) were eliminated from the study. Microarray expression data were processed, normalized by robust multiarray average, and log2 transformed using the Oligo Bioconductor R package version 1.36.1.24, 25
Microarray Data Analysis
Differentially expressed genes were identified using t-tests for each gene between groups of interest (retinoblastoma versus normal retina, severe anaplasia versus mild and moderate anaplasia, and mild and moderate anaplasia versus normal retina), and P values were corrected for multiple comparisons using the Benjamini–Hochberg false discovery rate procedure. Principal component analysis (PCA) was performed on normalized expression data using the FactoMineR version 1.3926 package in R. For hierarchal clustering, affinity propagation clustering was performed with the APCluster package version 1.4.4 in R for all differentially expressed genes (P < 0.05, |log2 fold change| > 0.6).27, 28 Gene set enrichment analysis (GSEA) was performed with the GSEA preranked algorithm with rank determined by −log10(padj) * sign (fold change).29 Gene set collections searched included the hallmark, curated (C2), computational (C4), GO (C5), and oncogenic (C6) gene sets from the molecular signatures database,30 as well as gene sets generated from differentially expressed genes described by Kapatai et al.31 Linear discriminant analysis and leave-one-out cross-validation were used to identify and test genes for the prediction of anaplastic grade.28 Data were visualized using the Plotly32 and ggplot2 packages in R.33
Results
Patient Demographics and Retinoblastoma Histopathologic Analysis
There were 28 retinoblastoma samples (3 with matched normal retina and 3 with retinocytoma components) that met criteria for the present study, for a total of 34 samples. The majority of patients were male (64.2%), with age at diagnosis ranging from 1 to 54 months (Tables 1 and 2). Nineteen retinoblastoma cases (67.9%) presented with unilateral disease, and 6 cases developed secondary tumors (7.1%) or distant metastases (14.2%) (Tables 1 and 2). RB1 mutational status was determined in 17 (60.7%) retinoblastoma cases. Of these, six cases were found to have germline mutations. One sample had neither RB1 mutations nor MYCN amplification, which drives a unique subset of unilateral retinoblastoma cases,34 but instead showed 100% hypermethylation of the RB1 promoter (sample RB31).35, 36 All patients with high-risk histology received adjuvant chemotherapy.
Table 1.
Patient Demographics
| Patient characteristics (N = 28) | N (%) |
|---|---|
| Sex | |
| Male | 18 (64.2) |
| Female | 10 (35.8) |
| Laterality | |
| Unilateral | 19 (67.9) |
| Bilateral | 9 (32.1) |
| Age at diagnosis (months) | |
| Mean | 17.7 |
| Range | 1–54 |
| RB1 mutational status | |
| Not known or test not performed | 11 (39.2) |
| Known | 17 (60.7) |
| Germline | 6/17 (35.3) |
| Nongermline | 10/17 (58.8) |
| No RB1 mutation identified | 1/17 (5.9) |
| Secondary tumors | 2 (7.1) |
| Distant metastasis | 4 (14.2) |
Table 2.
Retinoblastoma Characteristics
| Anaplastic grade | Sample | HRPF | Distant metastasis | Secondary tumors | RB1 mutation | Laterality | Age at diagnosis (months) | Follow-up period (months) |
|---|---|---|---|---|---|---|---|---|
| Mild | RB 3 | No | No | No | ND | Bilateral | 3 | 120 |
| RB 5 | Postlaminar ON | No | Pinealoma | G | Bilateral | 1 | 56 | |
| RB 6 | No | No | No | ND | Bilateral | 2 | 69 | |
| RB 7 | Postlaminar ON, massive choroidal invasion | No | No | G | Bilateral | 1 | 10 | |
| RB 19 | No | No | No | ND | Unilateral | 11 | 81 | |
| RB 23 | No | No | No | NG | Unilateral | 17 | 42 | |
| Moderate | RB 1 | No | No | RCC | ND | Bilateral | 3 | 212 |
| RB 2 | No | No | No | ND | Bilateral | 10 | 179 | |
| RB 4 | No | No | No | ND | Bilateral | 4 | 186 | |
| RB 8 | No | No | No | G | Bilateral | 4 | 141 | |
| RB 9 | No | No | No | G | Unilateral | 11 | 96 | |
| RB 11 | Postlaminar ON | No | No | ND | Unilateral | 4 | 71 | |
| RB 14 | Postlaminar ON, massive choroidal invasion | Brain | No | ND | Unilateral | 28 | 22 | |
| RB 16 | Massive choroidal invasion | Bone | No | ND | Unilateral | 24 | 140 | |
| RB 17 | No | No | No | G | Unilateral | 21 | 76 | |
| RB 18 | No | No | No | NG | Unilateral | 2 | 91 | |
| RB 22 | No | No | No | NG | Unilateral | 11 | 16 | |
| RB 26 | No | No | No | ND | Bilateral | 26 | 138 | |
| RB 31 | No | No | No | None | Unilateral | 23 | 64 | |
| Severe | RB 12 | Postlaminar ON | No | No | ND | Unilateral | 18 | 45 |
| RB 13 | Postlaminar ON, massive choroidal invasion | No | No | G | Unilateral | 27 | 78 | |
| RB 21 | Postlaminar ON, massive choroidal invasion | No | No | NG | Unilateral | 34 | 35 | |
| RB 24 | No | Bone, Liver | No | NG | Unilateral | 19 | 19 | |
| RB 25 | No | No | No | NG | Unilateral | 28 | 40 | |
| RB 28 | Laminar ON, nonmassive choroidal invasion | No | No | NG | Unilateral | 46 | 58 | |
| RB 30 | No | Liver, Bone marrow | No | NG | Unilateral | 27 | 9 | |
| RB 32 | No | No | No | NG | Unilateral | 54 | 43 | |
| RB 33 | Massive choroidal invasion | No | No | NG | Unilateral | 37 | 12 |
G, germline; ND, not determined; NG, nongermline; ON, optic nerve invasion; RB, retinoblastoma; RCC, renal cell carcinoma.
Anaplastic grade and the presence of HRPFs were determined and compared with demographic and genetic data (Table 2). Ten samples were found to have HRPFs. Nine samples (32.1%) had severe anaplasia, 13 samples had moderate anaplasia (46.4%), and 6 samples had mild anaplasia (21.4%). Interestingly, HRPFs were identified in samples of each anaplastic grade from mild to severe, as described by Mendoza et al15 (Figure 1).
Figure 1.
Anaplastic grades in retinoblastoma. A–D: Hematoxylin-eosin staining showing retinocytoma (A) and mild (B), moderate (C), and severe (D) anaplasia. Original magnification, ×100.
Retinoblastoma, Retinocytoma, and Normal Retina Show Distinct Gene Expression Profiles
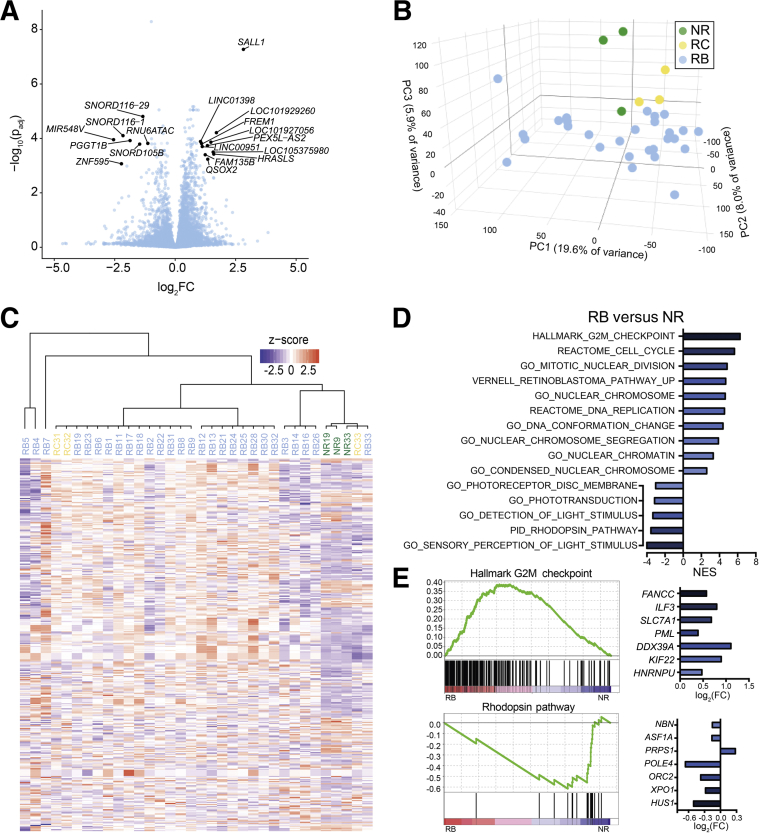
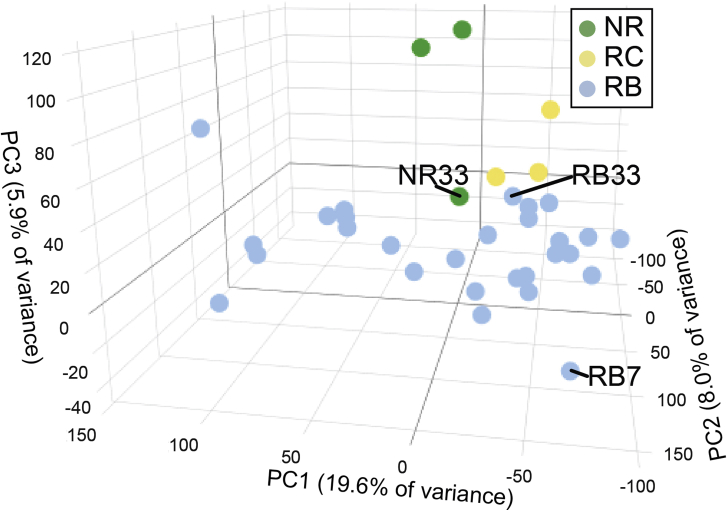
To determine whether increasing degree of anaplasia in retinoblastoma is associated with different patterns of gene expression, microarray analysis was performed on the 28 retinoblastoma specimens in addition to the 3 retinocytoma and 3 normal retina specimens (Figure 2A, Supplemental Table S1). A total of 783 genes were differentially expressed in retinoblastoma compared with normal retina (583 down-regulated, 200 up-regulated) (|log2 fold change| > 0.6; P < 0.05). In retinocytoma versus normal retina, 90 genes were differentially expressed (76 down-regulated, 14 up-regulated) (P < 0.05). Further analysis showed clear separation of samples into retinoblastoma, retinocytoma, and normal retina groups using both PCA (Figure 2B) and hierarchical clustering (Figure 2C). One normal retina sample (Supplemental Figure S1) did not cluster with the other normal retina samples, possibly because of inclusion of some tumor cells.
Figure 2.
Gene expression profiles separate retinoblastoma, retinocytoma, and normal retina. A: Volcano plot showing differential expression of genes in retinoblastoma (RB) relative to normal retina (NR) [plot of −log10 of the P value, adjusted for multiple comparisons (padj), and log2 of the fold change (FC)]. Genes with FC >2 and P < 0.001 are labeled and shown in gray. B: Principal component (PC) analysis showing sample separation into three groups: RB (blue), retinocytoma (RC; yellow), and NR (green). C: Affinity propagation clustering of all samples using genes with |log2FC| > 0.6 and P < 0.05 in RB versus NR. D: Normalized enrichment scores (NES) of selected gene sets significantly enriched in genes up-regulated and down-regulated in RB versus NR. E: Selected enrichment plots showing gene sets enriched in genes up-regulated (hallmark G2M checkpoint) and down-regulated (rhodopsin pathway) in RB versus NR. Associated bar graphs show log2FC (RB versus NR) of the genes with the greatest ranked scores in their respective gene set.
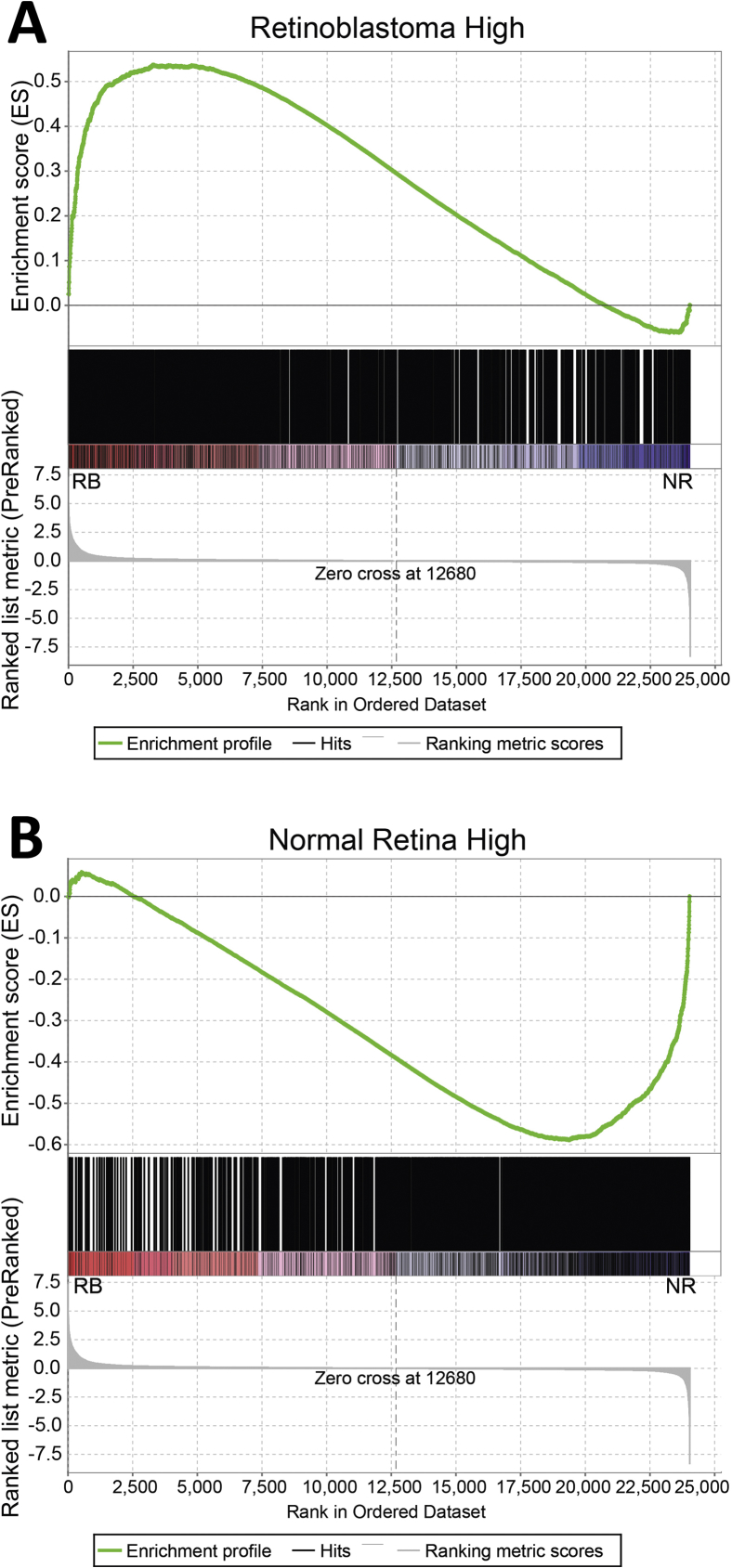
GSEA showed that genes up-regulated in retinoblastoma versus normal retina were strongly enriched for those identified previously by Kapatai et al31 as retinoblastoma-associated genes, whereas down-regulated genes were enriched for normal retina-associated genes (Supplemental Figure S2). GSEA of retinoblastoma versus normal retina using the molecular signatures database showed enrichment of gene sets related to cell-cycle checkpoint, DNA replication, and nuclear chromatin among genes up-regulated in retinoblastoma versus normal retina, whereas gene sets such as the rhodopsin pathway were enriched among down-regulated genes (Figure 2, D and E, Supplemental Tables S2 and S3). This suggests that the samples not only separated into distinct groups, but also showed expected patterns of gene expression.
Gene Expression Analysis Separates Retinoblastoma Samples by Anaplastic Grade But Not Presence of HRPFs
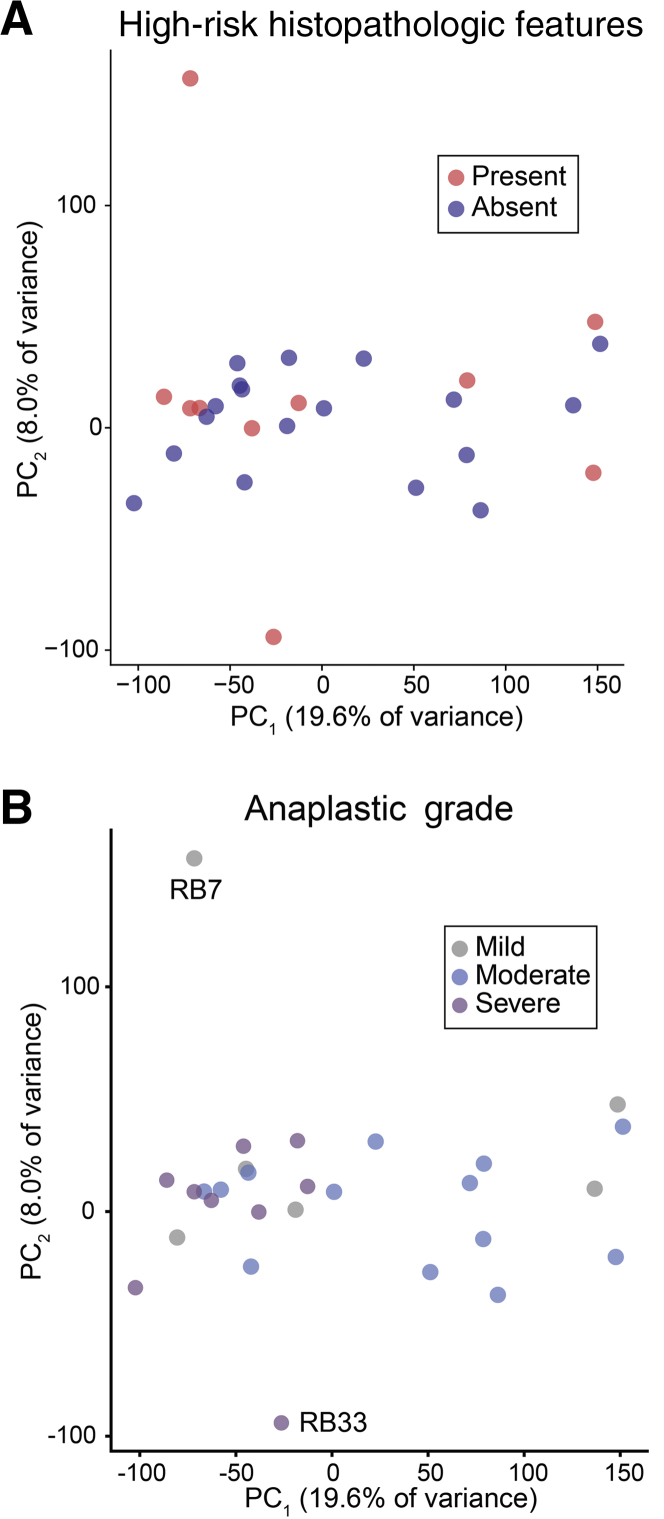
It was next determined whether retinoblastoma with different histopathologic patterns could be distinguished by gene expression. Of interest, PCA of the 28 retinoblastoma samples did not show a clear separation of tumors with versus without HRPFs (Figure 3A). In contrast, PCA of retinoblastoma samples showed grouping of tumors with severe anaplasia, suggesting a gene expression profile distinct from mild and moderate anaplastic grade tumors (Figure 3B). Interestingly, although RB33 did not group with other severe anaplasia samples (Figures 3B and Supplemental Figure S1), it showed tumor invasion of the choroid, an HRPF. RB7 similarly did not group with any samples, possibly because it possessed HRPFs but only mild anaplasia (although affinity propagation clustering shows similarity of RB7 with other mild anaplasia samples in Figure 4A). As described by Mendoza et al,15 some mild anaplasia cases can show invasion of the optic nerve and choroid whereas other tumors with severe anaplasia lack HRPFs, highlighting that HRPFs and anaplasia appear to be distinct features of retinoblastoma pathology.
Figure 3.
Gene expression can distinguish samples by anaplasia but not high-risk histopathologic features. A and B: Principal component (PC) analysis showing retinoblastoma samples by the presence of high-risk histopathologic features (A) and anaplastic grade (B).
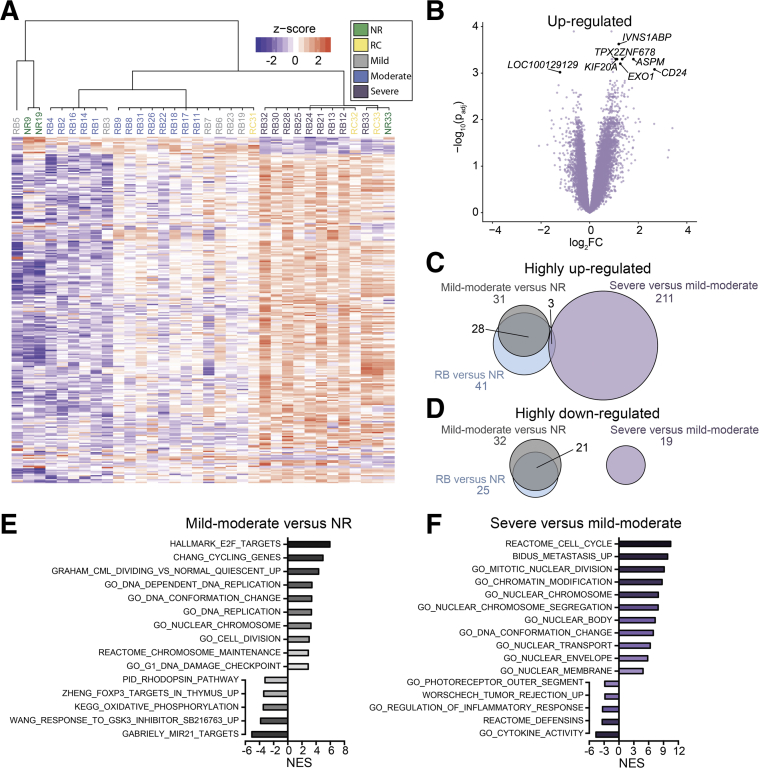
Figure 4.
Retinoblastoma samples show differential patterns of gene expression by anaplastic grade. A: Hierarchical clustering using genes with |log2FC| > 0.6 and P < 0.05 in severe anaplasia versus mild and moderate anaplasia. B: Volcano plot showing differential expression of genes in severe anaplasia relative to mild and moderate anaplasia (plot of −log10 of the P value [adjusted for multiple comparisons (padj)] and log2FC). C and D: Venn diagrams showing the number of highly up-regulated (C) and highly down-regulated (D) genes (P < 0.05, |log2FC| > 1) shared in all retinoblastoma (RB) versus normal retina (NR) (blue), mild-moderate anaplasia versus NR (gray), and severe versus mild-moderate anaplasia (purple). E and F: Plots of the normalized enrichment score (NES) of gene sets enriched in comparisons of mild-moderate anaplasia versus NR (E) and severe versus mild-moderate anaplasia (F).
The fact that mild and moderate anaplasia samples did not show clear separation from each other suggests a similar gene expression profile in these two groups. Indeed, Mendoza et al15 previously showed clinical outcomes associated with mild and moderate anaplasia to be highly similar, with >99% and 97% 10-year survival in cases with mild and moderate anaplasia, respectively, in contrast to 75% survival in cases with severely anaplastic tumors. Both mild and moderate anaplasia were thus considered as a single group in subsequent statistical analyses.
Hierarchical clustering using genes with |log2 fold change| > 0.6 and P < 0.05 in severe anaplasia versus mild and moderate anaplasia furthermore showed clustering according to severe versus mild-moderate anaplasia (Figure 4A). Analysis of gene expression in severe versus mild-moderate anaplasia identified several highly differentially expressed genes associated with proliferation and apoptosis of tumor cells such as CD24 (Figure 4B).37
Further analysis explored the relative number of genes dysregulated in mild-moderate anaplasia versus normal retina and severe versus mild-moderate anaplasia (Figure 4, C and D). Although the ability to detect gene expression changes may have been limited by the small number of normal retina samples and the potential for some genes to be down-regulated in mild anaplasia and up-regulated in severe anaplasia (or vice versa), two clear patterns emerged. First, the majority of the highly dysregulated genes in retinoblastoma are characteristic of mild and moderate anaplasia (Figure 4, C and D). Second, few of these genes were shared among genes highly dysregulated in severe anaplasia. Together, these data suggest that unique cellular processes drive the development of severe anaplasia rather than further dysregulation of processes occurring in mild and moderate grade anaplasia.
Genes Associated with Nuclear Morphology Are Differentially Expressed in Severe Anaplasia
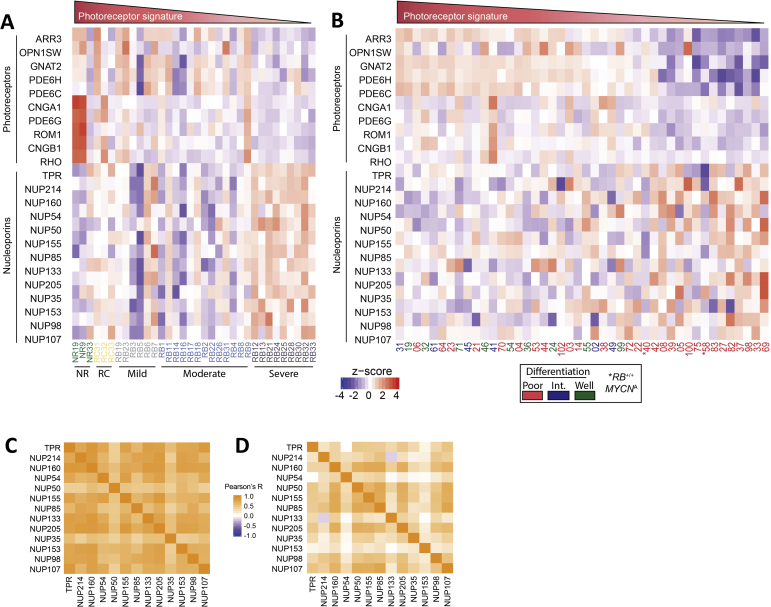
Given the separation of severely anaplastic from mild-moderate anaplastic grade retinoblastoma, the gene sets enriched in mild-moderate anaplasia versus normal retina (Figure 4E) and in severe versus mild-moderate anaplasia (Figure 4F) were analyzed. As expected, processes related to cell cycle and proliferation were enriched among up-regulated genes in both mild-moderate anaplasia versus normal retina and severe versus mild-moderate anaplasia. Of interest, only one gene set associated with the nucleus and chromatin was significantly enriched in mild-moderate anaplasia versus normal retina, whereas several such gene sets were enriched among up-regulated genes in severe versus mild-moderate anaplasia (nuclear body, chromatin modification, DNA conformation change, and so forth). Leading edge analysis of selected gene sets related to nuclear morphology showed altered expression of many nucleoporins and other nuclear morphology-related genes (Supplemental Table S4). In fact, expression of nucleoporins identified by leading edge analysis clearly distinguished those samples with severe anaplasia (Supplemental Figure S3A), with only one additional sample (RB6, mild anaplasia) also showing marked up-regulation of nucleoporin genes. Such enrichment of nuclear morphology–related genes among severe anaplasia samples is not unexpected given the pattern of nuclear condensation observed on histologic analysis of severe versus mild and moderate anaplasia. Indeed, severe anaplasia is defined by very large hyperchromatic nuclei and extreme pleomorphism on nuclear morphometric analysis.15 Enrichment of genes associated with nuclear shape and trafficking in severe anaplasia, then, may account for such morphologic changes observed on histology.
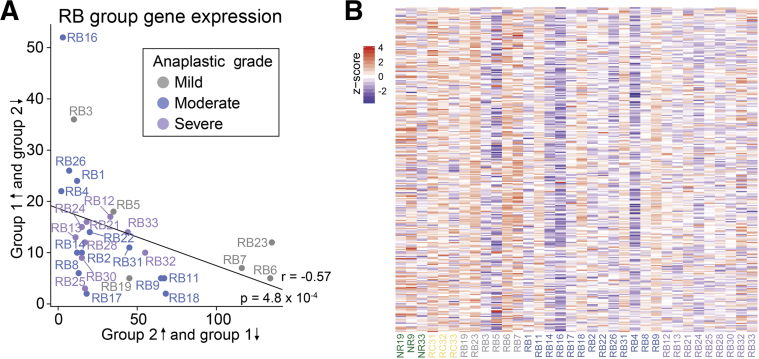
Severe Anaplasia Shows Some Association with Previously Identified Group 2 Retinoblastoma
Previous work by Kapatai et al31 identified two gene sets associated with retinoblastoma of differing severities and postulated cells of origin (group 1 was associated with invasive growth and possible derivation from retinal progenitor cells, and group 2 was derived from cone photoreceptors). To determine whether anaplastic grades correlated with these previously reported gene sets,31 the expression of group 1 and group 2 genes was analyzed in each retinoblastoma sample (Figure 5). Overall, a negative correlation existed between group 1–like gene expression (sample overexpression of group 1 genes and underexpression of group 2 genes relative to the mean of all samples) and group 2–like gene expression (overexpression of group 2 genes and underexpression of group 1 genes) (r = −0.57, P = 4.8 × 10−4).
Figure 5.
Anaplastic grade and group 1 and group 2 retinoblastoma. A: Plot of the number of previously reported group 1 genes31 overexpressed (z-score > 1) and group 2 genes underexpressed (z-score < −1) (and vice versa) relative to the mean of all samples for each retinoblastoma (RB) sample. B: Heat map of group 2 gene expression in each sample.
GSEA analysis using the group 1 and group 2 gene lists showed significant enrichment of group 2 genes in severe versus mild-moderate anaplasia (false discovery rate-q value = 0.01), but no significant associations with either group in mild-moderate anaplasia versus normal retina. Thus, although there was an overall negative correlation of group 1 and group 2 genes within our sample set, anaplastic grades did not appear to clearly segregate into either group.
Photoreceptor Gene Expression Decreases in Severe Anaplasia
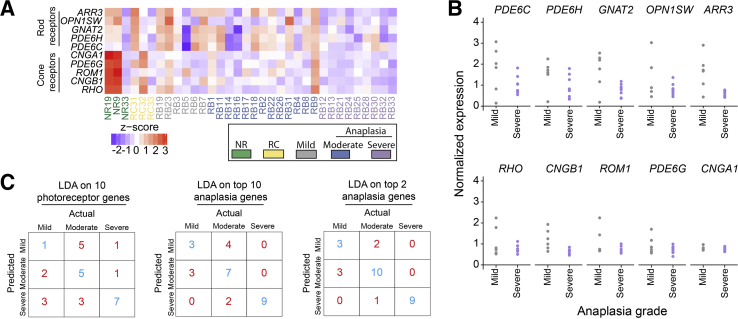
Kooi et al38 recently identified a decrease in photoreceptorness that was associated with the increasing severity of retinoblastoma. In the present study, expression of cone photoreceptors indeed appeared to be somewhat decreased in mild and moderate anaplasia (Figure 6, A and B). Severely anaplastic samples showed further decreased expression of both rod and cone photoreceptors relative to normal retina and mild-moderate anaplasia. Moreover, linear discriminant analysis using the 10 photoreceptors identified by Kooi et al38 accurately predicted severe anaplasia in seven of nine cases (Figure 6C). Although the original article did not intend to analyze anaplasia in retinoblastoma, it is interesting that this small gene set is capable of distinguishing severe versus mild-moderate anaplasia and that the present study again finds photoreceptor expression to be negatively correlated with retinoblastoma severity.
Figure 6.
Photoreceptor genes are expressed differentially by anaplastic grade. A: Heat map of the expression of the previously reported 10 rod and cone photoreceptor genes within each sample.38B: Plot of normalized expression of photoreceptor genes in samples with mild and severe anaplasia. C: Linear discriminant analysis (LDA) was used to predict anaplastic grade from the genes indicated, with results of leave-one-out cross-validation shown (top anaplasia genes selected by P value ranked lowest to highest in severe versus mild-moderate anaplasia).
A Limited Gene Set Can Accurately Predict Severe Anaplasia in the Present Study
Finally, it was determined whether genes significantly differentially expressed between severe and mild-moderate anaplasia could accurately predict anaplastic grade within our data set. In fact, four genes perfectly separated severe from mild and moderate anaplasia samples (Table 3).39, 40, 41, 42 Linear discriminant analysis followed by leave-one-out cross-validation using the 10 most highly differentially expressed genes in severe versus mild-moderate anaplasia accurately predicted all nine cases of severe anaplasia (Figure 6C). Even using only the top two most differentially expressed genes maintained this accuracy. In contrast, prediction of mild versus moderate anaplasia was much less reliable using either 10 or 2 genes, as expected given their overall similarity in gene expression (Figures 2 and 3).
Table 3.
Four Genes Perfectly Separate Severe from Mild and Moderate Anaplasia
| Symbol | Name | log2FC | padj | Function |
|---|---|---|---|---|
| EXOC8 | Exocyst complex component 8 | 0.896602312 | 0.000127061 | Regulates exocytosis by tethering post-Golgi vesicles to the plasma membrane Associated with Ras-mediated oncogenesis39 |
| CHTOP | Chromatin target of PRMT1 | 0.937928804 | 0.000500989 | Small nuclear protein involved in glioblastomagenesis40 |
| NUCKS1 | Nuclear casein kinase and cyclin-dependent kinase substrate 1 | 1.126599037 | 0.051952553 | Chromatin-associated protein involved in DNA repair and homologous recombination41 |
| ADSS | Adenylosuccinate synthase | 0.931391858 | 0.000574213 | Catalyzes the conversion of inosine monophosphate to adenosine monophosphate42 |
FC, fold change; padj, P value adjusted for multiple comparisons.
Discussion
Although novel therapies have greatly decreased both morbidity and mortality in retinoblastoma over recent decades, their full potential can be realized only through highly accurate identification of patients at risk for poor clinical outcome.2 Current means of risk stratification and determination of the need for additional therapy in retinoblastoma include assessment of clinical features, including patient age, iris neovascularization, and orbital cellulitis,10 as well as histopathologic features such as invasion of the optic nerve or choroid11, 12, 13 and degree of cellular anaplasia.15
Numerous oncologic studies have indicated that gene-based analyses may provide more powerful and easily applicable tools to predict the need for adjuvant therapy. Gene-based tests such as that already in use for uveal melanoma facilitate rapid diagnosis and risk stratification for these diseases.43 However, such an application currently is lacking for retinoblastoma, the most common ocular tumor of childhood.1 The present study evaluated the gene expression patterns associated with a known histopathologic indicator of risk, cellular anaplasia, to determine whether gene expression profiles can be used as a surrogate for histopathologic indicators of disease severity.
Results of our study indeed showed differential gene expression among retinoblastoma anaplastic grades, although interestingly there was no continuum of changes from mild to moderate to severe anaplasia. Although mild and moderate anaplasia can be distinguished from each other and severe anaplasia histopathologically,15 gene expression profiling failed to clearly separate these three grades into individual groups. Instead, mild and moderate grade anaplasia showed significant overlap, with no clear separation by either PCA or hierarchical clustering (Figures 2B and 3A). Such results are not surprising given the known similarity of clinical outcomes for mild and moderate anaplasia: 10-year retinoblastoma survival rates associated with mild and moderate anaplasia are >99% and 97%, respectively, but this number significantly decreases with severe anaplasia (75%).15
The disparity in gene expression between severe and mild-moderate anaplasia enabled the identification of a limited number of highly differentially expressed genes able to accurately predict all severe anaplasia samples in our data set (Figure 6C). This establishes proof of principle that particular genes could serve as surrogate markers of severe anaplasia and thus indicate high-risk retinoblastoma in a gene-based assay. However, further studies are needed to cross-validate the ability of these genes to predict severe anaplasia in additional cases of retinoblastoma.
The differences in clinical outcomes, morphology, and gene expression between severe and mild-moderate anaplasia suggest a significant shift in cellular processes between these two overarching groups. Indeed, the number of genes differentially expressed in severe compared with mild-moderate anaplasia was greater than the number of genes differentially expressed between all retinoblastoma samples versus normal retina (Figure 4, C and D). Furthermore, genes dysregulated in severe anaplasia appeared to be a separate subset from those differentially expressed in mild and moderate anaplasia. Thus, it appears that a unique set of cellular processes underlies the distinction in anaplastic grades rather than up-regulation or down-regulation of processes on a continuum from mild and moderate to severe anaplasia.
Specific gene pathways underlying this gap between severe and mild-moderate anaplasia appear to be related to changes in photoreceptor expression and nuclear morphology. Indeed, all samples with severe anaplasia in our study showed down-regulation of both rod and cone photoreceptors, consistent with findings by Kooi et al38 that severe retinoblastoma is characterized by decreased photoreceptorness. Given that severe anaplasia is defined by hyperchromatic nuclei and extreme pleomorphism on nuclear morphometric analysis,15 it is of particular interest that severe anaplasia showed significant up-regulation of genes related to the regulation of nuclear morphology and chromatin condensation (Figure 4). Up-regulation of nucleoporins in particular distinguished samples with severe anaplasia (Supplemental Figure S3A). Interestingly, nucleoporin expression also was increased in a subset of poorly differentiated retinoblastoma samples from Kooi et al38 that showed the lowest photoreceptor expression (Supplemental Figure S3B). An exception to this pattern, however, appears to be the two Kooi et al38 RB1+/+MYCNA tumors, which showed low photoreceptor and nucleoporin expression. Nucleoporins with increased expression in severe anaplasia furthermore appeared to be co-regulated, with high correlation among nucleoporins in both the present data set (Supplemental Figure S3C) and in the study by Kooi et al38 (Supplemental Figure S3D). Together, these data show that increased nucleoporin expression, in combination with decreased photoreceptor expression, is characteristic of both severe anaplasia and the related, but distinct, phenotype of poor differentiation.
Nucleoporins, components of the nuclear pore complex, are known to be up-regulated in poorly differentiated cancers such as breast, ovarian, colorectal, and hematopoietic malignancies.44, 45, 46 These proteins play a central role in the dysregulation of gene expression and nuclear morphology in cancer through multiple avenues, including their ability to restrict trafficking of cell-cycle regulators and transcription factors, their roles in chromosomal tethering during the G2/M cell-cycle transition, and their effects on cellular motility that are hypothesized to affect metastatic potential.46 Given the up-regulation of nucleoporins in severe anaplasia in retinoblastoma in this study, it is interesting to speculate whether dysregulation of these genes contributes to both changes in nuclear morphology and the rapid and dysregulated cell cycling in more severe retinoblastoma.
In sum, this study defined the gene expression patterns of anaplastic grade in retinoblastoma. Although mild and moderate grade anaplasia are similar in terms of both gene expression and clinical outcome, severe anaplasia shows significant differences in both the number and type of dysregulated genes. Such disparity in anaplastic grades enabled the identification of specific genes able to predict severe anaplasia within the present data set, serving as a first step in determining whether particular genes could be used for clinical risk stratification. Cellular processes related to increased anaplastic severity include decreased photoreceptor expression, as previously identified,38 and markedly increased nucleoporin expression relative to mild and moderate grade anaplasia. Thus, we present here the first description of gene pathways that may underlie the increased nuclear pleomorphism and poor clinical outcomes associated with severe anaplasia in retinoblastoma. Corroboration of these findings in future retinoblastoma studies and further exploration of gene pathways underlying increased anaplastic severity will yield valuable insights into our understanding and management of this disease.
Acknowledgments
We thank Dr. J. William Harbour (Bascom Palmer Eye Institute) for helpful discussions and critical review of the manuscript, the Emory University Integrated Genomics Core for assistance in microarray data collection, and the Winship Biostatistics and Bioinformatics Shared Resource of Emory University for assistance in data analysis.
Footnotes
Supported by National Eye Institute grant NEI P30EY06360 (H.E.G.) and a grant from Research to Prevent Blindness.
Disclosures: None declared.
Supplemental material for this article can be found at https://doi.org/10.1016/j.ajpath.2018.06.013.
Supplemental Data
Supplemental Figure S1.
Principal component analysis (PCA) of all samples. PCA showing sample separation into retinoblastoma (RB, blue), retinocytoma (RC, yellow), and normal retina (NR, green), with numbers corresponding to the samples listed in Table 2.
Supplemental Figure S2.
Gene set enrichment analysis based on genes reported by Kapatai et al.31A and B: Enrichment plots showing enrichment of the previously reported retinoblastoma (A) and normal retina (B) gene sets versus genes up-regulated and down-regulated in retinoblastoma (RB) versus normal retina (NR) in the present study, respectively.
Supplemental Figure S3.
Photoreceptor and nucleoporin genes are expressed differentially in severe retinoblastoma. A: Heat maps showing expression of photoreceptor genes and the nucleoporins identified by leading edge analysis for each sample according to anaplastic grade. B: Heat map showing expression of the same photoreceptors and nucleoporins in previously reported retinoblastoma samples38 for which differentiation data were available. C and D: Correlation of nucleoporin expression in the present data set (C) and that previously reported38 (D). Int., intermediate differentiation; NR, normal retina, RC, retinocytoma.
References
- 1.Broaddus E., Topham A., Singh A.D. Incidence of retinoblastoma in the USA: 1975-2004. Br J Ophthalmol. 2008;93:21–23. doi: 10.1136/bjo.2008.138750. [DOI] [PubMed] [Google Scholar]
- 2.Grossniklaus H.E. Retinoblastoma. Fifty years of progress. The LXXI Edward Jackson Memorial Lecture. Am J Ophthalmol. 2014;158:875–881.e1. doi: 10.1016/j.ajo.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Canturk S., Qaddoumi I., Khetan V., Ma Z., Furmanchuk A., Antoneli C.B., Sultan I., Kebudi R., Sharma T., Rodriguez-Galindo C., Abramson D.H., Chantada G.L. Survival of retinoblastoma in less-developed countries impact of socioeconomic and health-related indicators. Br J Ophthalmol. 2010;94:1432–1436. doi: 10.1136/bjo.2009.168062. [DOI] [PubMed] [Google Scholar]
- 4.Shields C.L., Honavar S.G., Meadows A.T., Shields J.A., Demirci H., Singh A., Friedman D.L., Naduvilath T.J. Chemoreduction plus focal therapy for retinoblastoma: factors predictive of need for treatment with external beam radiotherapy or enucleation. Am J Ophthalmol. 2002;133:657–664. doi: 10.1016/s0002-9394(02)01348-x. [DOI] [PubMed] [Google Scholar]
- 5.Abramson D.H., Dunkel I.J., Brodie S.E., Kim J.W., Gobin Y.P. A phase I/II study of direct intraarterial (ophthalmic artery) chemotherapy with melphalan for intraocular retinoblastoma initial results. Ophthalmology. 2008;115:1398–1404. doi: 10.1016/j.ophtha.2007.12.014. 1404.e1. [DOI] [PubMed] [Google Scholar]
- 6.Shields C.L., Bianciotto C.G., Jabbour P., Ramasubramanian A., Lally S.E., Griffin G.C., Rosenwasser R., Shields J.A. Intra-arterial chemotherapy for retinoblastoma: report no. 1, control of retinal tumors, subretinal seeds, and vitreous seeds. Arch Ophthalmol. 2011;129:1399–1406. doi: 10.1001/archophthalmol.2011.150. [DOI] [PubMed] [Google Scholar]
- 7.Abramson D.H. Super selective ophthalmic artery delivery of chemotherapy for intraocular retinoblastoma: ‘chemosurgery’ the first Stallard lecture. Br J Ophthalmol. 2010;94:396–399. doi: 10.1136/bjo.2009.174268. [DOI] [PubMed] [Google Scholar]
- 8.Gobin Y.P., Dunkel I.J., Marr B.P., Brodie S.E., Abramson D.H. Intra-arterial chemotherapy for the management of retinoblastoma: four-year experience. Arch Ophthalmol. 2011;129:732–737. doi: 10.1001/archophthalmol.2011.5. [DOI] [PubMed] [Google Scholar]
- 9.Shields C.L., Mashayekhi A., Au A.K., Czyz C., Leahey A., Meadows A.T., Shields J.A. The international classification of retinoblastoma predicts chemoreduction success. Ophthalmology. 2006;113:2276–2280. doi: 10.1016/j.ophtha.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 10.Kaliki S., Shields C.L., Rojanaporn D., Al-Dahmash S., McLaughlin J.P., Shields J.A., Eagle R.C. High-risk retinoblastoma based on international classification of retinoblastoma: analysis of 519 enucleated eyes. Ophthalmology. 2013;120:997–1003. doi: 10.1016/j.ophtha.2012.10.044. [DOI] [PubMed] [Google Scholar]
- 11.Shields C.L., Shields J.A., Baez K., Cater J.R., De Potter P. Optic nerve invasion of retinoblastoma. Metastatic potential and clinical risk factors. Cancer. 1994;73:692–698. doi: 10.1002/1097-0142(19940201)73:3<692::aid-cncr2820730331>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 12.Shields C.L., Shields J.A., Baez K.A., Cater J., De Potter P.V. Choroidal invasion of retinoblastoma: metastatic potential and clinical risk factors. Br J Ophthalmol. 1993;77:544–548. doi: 10.1136/bjo.77.9.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haik B.G., Dunleavy S.A., Cooke C., Ellsworth R.M., Abramson D.H., Smith M.E., Karcioglu Z.A. Retinoblastoma with anterior chamber extension. Ophthalmology. 1987;94:367–370. doi: 10.1016/s0161-6420(87)33437-2. [DOI] [PubMed] [Google Scholar]
- 14.Mabtum E.D., Bonanomi M.T., Lima P.P., Almeida M.T. Orbital retinoblastoma: case report. Arq Bras Oftalmol. 2013;76:247–249. doi: 10.1590/s0004-27492013000400013. [DOI] [PubMed] [Google Scholar]
- 15.Mendoza P.R., Specht C.S., Hubbard G.B., Wells J.R., Lynn M.J., Zhang Q., Kong J., Grossniklaus H.E. Histopathologic grading of anaplasia in retinoblastoma. Am J Ophthalmol. 2015;159:764–776. doi: 10.1016/j.ajo.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eberhart C.G., Kepner J.L., Goldthwaite P.T., Kun L.E., Duffner P.K., Friedman H.S., Strother D.R., Burger P.C. Histopathologic grading of medulloblastomas. Cancer. 2002;94:552–560. doi: 10.1002/cncr.10189. [DOI] [PubMed] [Google Scholar]
- 17.Jijelava K.P., Grossniklaus H.E. Diffuse anterior retinoblastoma: a review. Saudi J Ophthalmol. 2013;27:135–139. doi: 10.1016/j.sjopt.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shields C.L., Ghassemi F., Tuncer S., Thangappan A., Shields J.A. Clinical spectrum of diffuse infiltrating retinoblastoma in 34 consecutive eyes. Ophthalmology. 2008;115:2253–2258. doi: 10.1016/j.ophtha.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 19.Reese A. ed 3. Harper and Row; Hagerstown, MD: 1976. Tumors of the Eye. [Google Scholar]
- 20.Murphree A.L. Intraocular retinoblastoma: the case for a new group classification. Ophthalmol Clin North Am. 2005;18:41–53. doi: 10.1016/j.ohc.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 21.Sastre X., Chantada G.L., Doz F., Wilson M.W., De Davila M.T.G., Rodríguez-Galindo C., Chintagumpala M., Chévez-Barrios P. Proceedings of the Consensus Meetings from the International Retinoblastoma Staging Working Group on the pathology guidelines for the examination of enucleated eyes and evaluation of prognostic risk factors in retinoblastoma. Arch Pathol Lab Med. 2009;133:1199–1202. doi: 10.5858/133.8.1199. [DOI] [PubMed] [Google Scholar]
- 22.Edge S.B., Compton C.C. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 23.Eagle R.C., Jr. High-risk features and tumor differentiation in retinoblastoma: a retrospective histopathologic study. Arch Pathol Lab Med. 2009;133:1203–1209. doi: 10.5858/133.8.1203. [DOI] [PubMed] [Google Scholar]
- 24.Carvalho B.S., Irizarry R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A.J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J.Y.H., Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80 doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lê S., Josse J., Husson F. FactoMineR: an R package for multivariate analysis. J Stat Softw. 2008;25:1–18. [Google Scholar]
- 27.Bodenhofer U., Kothmeier A., Hochreiter S. APCluster: an R package for affinity propagation clustering. Bioinformatics. 2011;27:2463–2464. doi: 10.1093/bioinformatics/btr406. [DOI] [PubMed] [Google Scholar]
- 28.Frey B.J., Dueck D. Clustering by passing messages between data points. Science. 2007;315:972–976. doi: 10.1126/science.1136800. [DOI] [PubMed] [Google Scholar]
- 29.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liberzon A., Subramanian A., Pinchback R., Thorvaldsdottir H., Tamayo P., Mesirov J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kapatai G., Brundler M.A., Jenkinson H., Kearns P., Parulekar M., Peet A.C., McConville C.M. Gene expression profiling identifies different sub-types of retinoblastoma. Br J Cancer. 2013;109:512–525. doi: 10.1038/bjc.2013.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plotly Technologies Inc. Plotly Technologies Inc.; Montreal, QC: 2015. Collaborative Data Science. [Google Scholar]
- 33.Wickham H. ed 2. Springer-Verlag; New York, NY: 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 34.Rushlow D.E., Mol B.M., Kennett J.Y., Yee S., Pajovic S., Thériault B.L., Prigoda-Lee N.L., Spencer C., Dimaras H., Corson T.W., Pang R., Massey C., Godbout R., Jiang Z., Zacksenhaus E., Paton K., Moll A.C., Houdayer C., Raizis A., Halliday W., Lam W.L., Boutros P.C., Lohmann D., Dorsman J.C., Gallie B.L. Characterisation of retinoblastomas without RB1 mutations: genomic, gene expression, and clinical studies. Lancet Oncol. 2013;14:327–334. doi: 10.1016/S1470-2045(13)70045-7. [DOI] [PubMed] [Google Scholar]
- 35.Ohtani-Fujita N., Fujita T., Aoike A., Osifchin N.E., Robbins P.D., Sakai T. CpG methylation inactivates the promoter activity of the human retinoblastoma tumor-suppressor gene. Oncogene. 1993;8:1063–1067. [PubMed] [Google Scholar]
- 36.Stirzaker C., Millar D.S., Paul C.L., Warnecke P.M., Harrison J., Vincent P.C., Frommer M., Clark S.J. Extensive DNA methylation spanning the Rb promoter in retinoblastoma tumors. Cancer Res. 1997;57:2229–2237. [PubMed] [Google Scholar]
- 37.Li J., Li C., Yuan H., Gong F. Clinical value of CD24 expression in retinoblastoma. J Biomed Biotechnol. 2012;2012:1–6. doi: 10.1155/2012/158084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kooi I.E., Mol B.M., Moll A.C., van der Valk P., de Jong M.C., de Graaf P., van Mil S.E., Schouten-van Meeteren A.Y.N., Meijers-Heijboer H., Kaspers G.L., te Riele H., Cloos J., Dorsman J.C. Loss of photoreceptorness and gain of genomic alterations in retinoblastoma reveal tumor progression. EBioMedicine. 2015;2:660–670. doi: 10.1016/j.ebiom.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Issaq S.H., Lim K.H., Counter C.M. Sec5 and Exo84 foster oncogenic Ras-mediated tumorigenesis. Mol Cancer Res. 2010;8:223–231. doi: 10.1158/1541-7786.MCR-09-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takai H., Masuda K., Sato T., Sakaguchi Y., Suzuki T., Suzuki T., Koyama-Nasu R., Nasu-Nishimura Y., Katou Y., Ogawa H., Morishita Y., Kozuka-Hata H., Oyama M., Todo T., Ino Y., Mukasa A., Saito N., Toyoshima C., Shirahige K., Akiyama T. 5-hydroxymethylcytosine plays a critical role in glioblastomagenesis by recruiting the CHTOP-methylosome complex. Cell Rep. 2014;9:48–60. doi: 10.1016/j.celrep.2014.08.071. [DOI] [PubMed] [Google Scholar]
- 41.Parplys A.C., Zhao W., Sharma N., Groesser T., Liang F., Maranon D.G., Leung S.G., Grundt K., Dray E., Idate R., Østvold A.C., Schild D., Sung P., Wiese C. NUCKS1 is a novel RAD51AP1 paralog important for homologous recombination and genome stability. Nucleic Acids Res. 2015;43:9817–9834. doi: 10.1093/nar/gkv859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stayton M.M., Rudolph F.B., Fromm H.J. Regulation, genetics, and properties of adenylosuccinate synthetase: a review. Current Topics in Cellular Regulation, vol. 22. In: Horecker B.L., Stadman E.R., editors. Academic Press; Cambridge, MA: 1983. pp. 103–141. [DOI] [PubMed] [Google Scholar]
- 43.Harbour J.W. A prognostic test to predict the risk of metastasis in uveal melanoma based on a 15-gene expression profile. Methods Mol Biol. 2014;1102:427–440. doi: 10.1007/978-1-62703-727-3_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kabachinski G., Schwartz T.U. The nuclear pore complex—structure and function at a glance. J Cell Sci. 2015;128:423–429. doi: 10.1242/jcs.083246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strambio-De-Castillia C., Niepel M., Rout M.P. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- 46.Chow K.-H., Factor R.E., Ullman K.S. The nuclear envelope environment and its cancer connections. Nat Rev Cancer. 2012;12:196–209. doi: 10.1038/nrc3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.