Abstract
Facioscapulohumeral muscular dystrophy, known in genetic forms FSHD1 and FSHD2, is associated with D4Z4 repeat array chromatin relaxation and somatic derepression of DUX4 located in D4Z4. A complete copy of DUX4 is present on 4qA chromosomes, but not on the D4Z4-like repeats of chromosomes 4qB or 10. Normally, the D4Z4 repeat varies between 8 and 100 units, while in FSHD1 it is only 1–10 units. In the rare genetic form FSHD2, a combination of a 4qA allele with a D4Z4 repeat size of 8–20 units and heterozygous pathogenic variants in the chromatin modifier SMCHD1 causes DUX4 derepression and disease. In this study, we identified 11/79 (14%) FSHD2 patients with unusually large 4qA alleles of 21–70 D4Z4 units. By a combination of Southern blotting and molecular combing, we show that 8/11 (73%) of these unusually large 4qA alleles represent duplication alleles in which the long D4Z4 repeat arrays are followed by a small FSHD-sized D4Z4 repeat array duplication. We also show that these duplication alleles are associated with DUX4 expression. This duplication allele frequency is significantly higher than in controls (2.9%), FSHD1 patients (1.4%) and in FSHD2 patients with typical 4qA alleles of 8–20 D4Z4 units (1.5%). Segregation analysis shows that, similar to typical 8–20 units FSHD2 alleles, duplication alleles only cause FSHD in combination with a pathogenic variant in SMCHD1. We conclude that cis duplications of D4Z4 repeats explain DUX4 expression and disease presentation in FSHD2 families with unusual long D4Z4 repeats on 4qA chromosomes.
Introduction
Facioscapulohumeral dystrophy (FSHD; OMIM 158900 and 158901) is a common inherited myopathy recently estimated to affect 1: 15 000–1: 8 500 individuals (1,2). Two clinically almost identical forms of the disease have been described, FSHD1 and FSHD2, which are both associated with somatic partial chromatin relaxation of the D4Z4 repeat array on chromosome 4 and derepression of the DUX4 retrogene (3–8). A copy of DUX4 is embedded in each D4Z4 unit. DUX4 encodes a transcription factor that activates cleavage stage embryonic transcriptional programs in mice and humans (9–11) DUX4 is also expressed in the luminal cells of the testis (12). It is believed to be normally silenced in somatic tissues and its inappropriate presence in skeletal muscle eventually causes muscle cell death (12,13). D4Z4 chromatin relaxation leads to stable DUX4 protein expression in a small number of myonuclei from the most distal copy of the D4Z4 repeat array and the region immediately distal to this copy where a 3′UTR region with a DUX4 polyadenylation signal (DUX4-PAS) can be found (6,14). Due to ancient duplication events, highly homologous and equally polymorphic D4Z4 arrays can be found in the subtelomeres of chromosomes 4qA, 4qB and 10q (15–17). The homologous region between these 3 chromosomes starts immediately distal to a single inverted D4Z4 unit 40 kb proximal to the D4Z4 array on chromosomes 4qA and 4qB (18) (Fig. 2A). The DUX4-PAS is located immediately distal to the D4Z4 repeat array on 4qA chromosomes, but not on 4qB and 10q chromosomes (6). The nearly unique linkage of the DUX4-PAS to 4qA explains the almost exclusive linkage of FSHD to 4qA chromosomes (19).
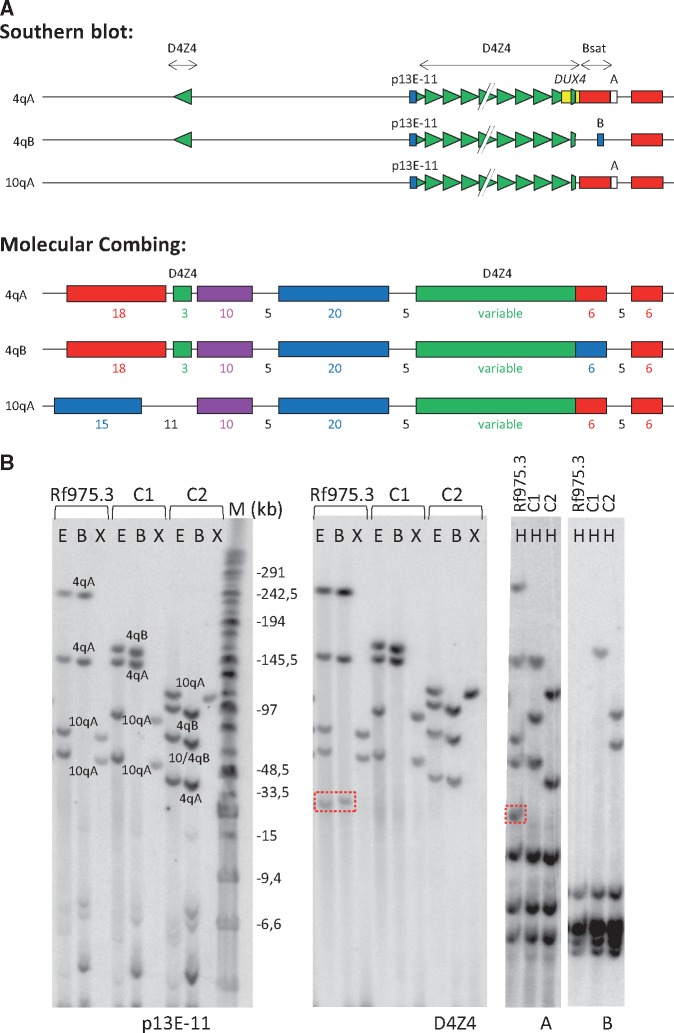
Figure 2.
(A) Schematic representation of D4Z4 locus on chromosomes 4qA, 4qB and 10qA for both Southern blot (top) and molecular combing (bottom) analysis. For both methods, D4Z4 units and the highly homologous inverted D4Z4 repeat unit on chromosomes 4qA and 4qB are indicated in green. For Southern blot analysis, the position of probes p13E-11 (blue), D4Z4 (green), A (white) and B (blue) are indicated, as well as position of the DUX4 gene (yellow). For molecular combing, the position and colour of the fluorescence probe which generate the unique genetic Morse code for each chromosome are indicated with sizes in kb. (B) Representative images of the Southern blot analysis in FSHD2 patient Rf975.3 and two unrelated controls (C1 and C2). Rf975.3 carries a D4Z4 duplication fragment, not visible by probe p13E-11, resistant to BlnI and hybridizing to the D4Z4 and A probes (hence 4qA-derived) (boxed repeat fragment). For the left two blots, DNA was digested by EcoRI and HindIII (E); EcoRI and BlnI (B) and XapI (X) and hybridized with probes p13E-11 and D4Z4, respectively. On the right two blots, DNA was digested with HindIII (H) and hybridized with probes A and B, respectively. Marker lane (M) in kb is indicated.
The D4Z4 repeat array is polymorphic in size ranging between 8 and 100 units on chromosome 4 in the population (20,21). The most common form of this disease, FSHD type 1 (FSHD1), is caused by contractions of the D4Z4 repeat array on chromosome 4qA to a size of 1–10 units (19,22). In the rare FSHD type 2 (FSHD2), D4Z4 repeat arrays on 4qA chromosomes are typically of intermediate size (8–20 units) and derepression of the D4Z4 repeat array is most often caused by mutations in chromatin modifiers that are necessary to establish and/or maintain a repressive D4Z4 chromatin structure in somatic cells (7,23). FSHD2 is heterogenic with the majority of patients having a heterozygous pathogenic variant in the structural maintenance of chromosomes flexible hinge domain containing 1 (SMCHD1) gene in combination with the presence of an intermediate sized D4Z4 repeat array on 4qA chromosome (7).
In a small proportion of FSHD2 families the presence of such repeat array is combined with heterozygous pathogenic variants in the DNA methyl transferase 3 Beta (DNMT3B) gene, with some patients still remaining genetically unresolved (23). Thus, both forms of the disease molecularly converge at D4Z4 chromatin relaxation in somatic tissue and DUX4 derepression in skeletal muscle. Hence, FSHD1 and FSHD2 are clinically largely identical being characterized by progressive and irreversible weakness of the facial-, shoulder girdle- and upper arm muscles (24). With disease progression also other muscles may become affected and studies suggest that FSHD2 has generally a milder disease progression than FSHD1. In both forms of the disease extramuscular tissues are rarely involved but the disease, especially in early-onset FSHD1 cases, may present with retinovasculopathy, hearing loss and intellectual disability in severe cases, although these extramuscular features have yet to be described in FSHD2 patients (2,24,25).
In the European control population the mean length of the D4Z4 repeat array on a permissive 4qA chromosomes is 38 units. The D4Z4 repeat array size of the shortest permissive 4qA allele (hereafter called SPA) in FSHD2 patients typically varies between 8 and 20 units, with a mean size of 12 units (26). The repeat size range from 8 to 10 units is particularly interesting as it overlaps between the FSHD1 population and the control population (20,21). Although originally the minimal size for FSHD2 was defined as 11 units and FSHD1 as 1–10 units, these cut-offs are likely not so rigorous with recent evidence suggesting that FSHD1 and FSHD2 form a continuum (27). Hence, D4Z4 repeat array sizes between 8 and 10 units are overlapping between control individuals and patients with FSHD1 or FSHD2. This suggests that in this size range the predisposition to DUX4 expression in skeletal muscle and disease presentation depends on genetic (the D4Z4 repeat array size), epigenetic (the ability to repress DUX4) and most likely also environmental factors. In this context, we proposed that heterozygous pathogenic variants in SMCHD1 or DNMT3B can only effectively derepress DUX4 in skeletal muscle when combined with a small, but normal-sized D4Z4 repeat array of 8–20 units on chromosome 4qA.
Recently, we performed a detailed genetic and clinical study on 79 different FSHD2 families (28). In this study, the majority of the patients have a pathogenic SMCHD1 variant combined with a SPA of 8–20 units. We also identified several mildly, or unaffected family members that carried the SMCHD1 variant in combination with a SPA >20 units or with only non-permissive (4qB) alleles. However, we identified a small number of FSHD2 patients with a moderate to severe phenotype while having a > 20 units SPA, which prompted us to investigate these patients in greater detail. With the recently reported duplications of D4Z4 in controls and FSHD patients (29) we studied the contribution of D4Z4 duplications in FSHD2 families.
Results
Genetic analysis of FSHD2 patients with an exceptional long SPA
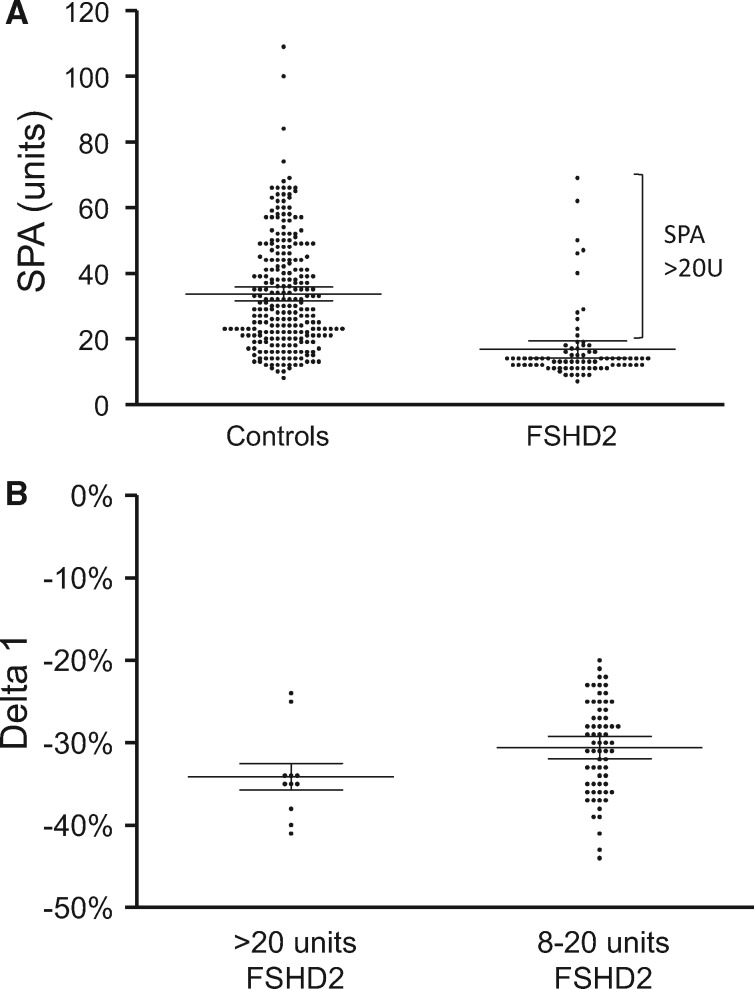
From our earlier study (28), we could perform more detailed genetic analysis in 74 FSHD2 families. From these families we included all FSHD2 patients with a unique SPA, i.e. not only single cases from each family but sometimes also affected relatives if having a unique SPA. If we identified multiple family members with an identical SPA, we selected the oldest patient, totalling 79 FSHD2 patients. We identified 68 FSHD2 patients with a SPA of 8–20 units and 11 FSHD2 patients from different families with >20 units SPA (Fig. 1A and Supplementary Material, Table S1). As FSHD2 disease presentation seems dependent on the D4Z4 repeat array size in the permissive allele and the level of D4Z4 methylation, we anticipated that D4Z4 CpG methylation would be extremely low in patients with a > 20 units SPA. The CpG methylation at D4Z4 is dependent on variants in epigenetic modifiers and on the size of the D4Z4 repeat. Therefore, we used the delta1 score, which is the methylation value corrected for the D4Z4 repeat array sizes on chromosomes 4 and 10 (26). However, the delta1 methylation score was not significantly different between FSHD2 samples with a larger SPA and the FSHD2 samples with SPAs between 8 and 20 units (Fig. 1B).
Figure 1.
(A) D4Z4 array size of the SPA in 79 FSHD2 patients (mean 16, 8 units) and in 252 controls having one permissive allele (mean 33, 7 units) shows a significant difference (P < 0.0001). 97/349 control individuals had two non-permissive alleles and were therefore excluded. All FSHD2 patients carrying >20 units SPA are marked. (B) Methylation levels plotted in delta1 values for FSHD2 patients with a SPA of 8–20 units (n = 66, for two samples no delta1 value was available) versus patients with a SPA >20 units (n = 11) (P = 0.0561).
We therefore further analyzed the composition of the D4Z4 alleles in all FSHD2 patients and their family members. Southern blots that were previously hybridized with the diagnostic FSHD probe p13E-11 (recognizing D4F104S1 proximal to the D4Z4 array), were re-hybridized with a probe that is complementary to the D4Z4 repeat array (35), enabling the identification of all D4Z4 repeat arrays, including those that are not linked to p13E-11 (Fig. 2A).
By the comparison of the number of fragments containing D4Z4 repeat arrays upon hybridization with probes p13E-11 and D4Z4 we identified in 70/79 (88.6%) FSHD2 patients the same number of fragments. In 9 cases we identified one or more additional D4Z4 repeat array containing fragments by D4Z4 hybridization that were not visible with p13E-11 hybridizations(Fig. 2B). This finding indicates the presence of a D4Z4 repeat array duplication somewhere in the genome. For 8/9 of these duplication fragments the extra D4Z4 repeats are chromosome 4-derived as they are resistant to digestion with BlnI (Table 1; Supplementary Material, Fig. S1). The sizes of these non-p13E-11-linked duplicated D4Z4 repeat arrays ranged from 5 to 28 D4Z4 units. Interestingly, 8/9 duplications were identified in FSHD2 patients with an unusual long SPA (>20 units).
Table 1.
Presence and origin of D4Z4 duplication fragments in the FSHD2 population, the control and the FSHD1 population (only FSHD2 individuals with a unique SPA are included)
| Non-p13E-11-linked D4Z4 fragments |
||||||
|---|---|---|---|---|---|---|
| Individuals | Total number of duplication alleles | Frequency | 4qA | 4q0 | 10qA | |
| FSHD2 (8–20 U) | 68 | 1 | 1.5% | 1 | – | – |
| FSHD2 (>20U) | 11 | 8 | 72.7% | 6 | 1 | 1 |
| FSHD2 (total) | 79 | 9 | 11.4% | 7 | 1 | 1 |
| Controls | 349 | 10 | 2.9% | 4 | 2 | 4 |
| FSHD1 | 282 | 4 | 1.4% | – | 3 | 1 |
| Group 1 | Group 2 | P-value* | |
|---|---|---|---|
| FSHD2 (>20 U) | vs | Controls | <2.2E-16 |
| FSHD2 (>20U) | vs | FSHD1 | <2.2E-16 |
| FSHD2 (>20U) | vs | FSHD2 (8-20U) | <1.7E-10 |
| Controls | vs | FSHD1 | 0.34 |
| Controls | vs | FSHD2 (8-20U) | 0.81 |
| FSHD1 | vs | FSHD2 (8-20U) | 1.00 |
| FSHD2 (total) | vs | Controls | 2.5E-03 |
| FSHD2 (total) | vs | FSHD1 | 1.1E-04 |
Individual numbers are indicated in the first column. The proximal D4Z4 array columns show the haplotype and size of the repeat array in D4Z4 units that are vizualized upon p13E-11 hybridization. The extra D4Z4 array(s) columns show the size and allelotype (A/B) upon D4Z4 hybridization or by MC. The type of the analysis (SB: Southern blot, or MC: molecular combing) used to determine the composition of the duplication allele is indicated in the last column.
Significant after Bonferroni correction if P < 6.3E-03 (0.05/8).
Detailed genetic analysis of duplication alleles
Detailed 4qA/4qB genotyping of the eight 4q-derived (BlnI-resistant) duplication fragments in FSHD2 patients with unusually long SPA showed that in 7/8 cases the extra D4Z4 repeat array was of the 4qA-type. Only in DNA from patient Rf1666.1 the extra D4Z4 repeat array did not hybridize to probes A or B (Table 2 and Supplementary Material, Table S1). In 4/8 cases we were able to analyze multiple family members, and the Southern blot data suggested that the extra D4Z4 repeat array co-segregated with a 4qA allele. We further studied the two major 4qA haplotypes, 4A161S and 4A161L, which differ in size in their distal partial D4Z4 repeat unit and DUX4 gene structure without affecting the DUX4 open reading frame (28). To ensure specificity, we only selected individuals in whom the other chromosome 4 is of a non-4A161 haplotype. The FSHD2 patients Rf874.3, Rf1666.1 and Rf1727.2, and family members of the probands of FSHD2 families Rf696, Rf392 and Rf975 fulfilled these criteria. Surprisingly, 4A161L/4A161S analysis and segregation analysis showed that 6/7 analyzed 4qA duplication alleles were of the 4A161L haplotype, which can only be found in approximately 20% of the European control population (28). Interestingly, the duplication fragment for which the extra D4Z4 repeat array did not hybridize to probes A and B, was of the 4A161S haplotype (Rf1666.1). In all duplication cases in whom the homologous allele was 4qB, we identified by PCR (28) only 4A161S or 4A161L variants from the duplication alleles, and never a mixture of both variants, suggesting that the distal 4qA endings of the standard D4Z4 repeat array and the duplicated D4Z4 repeat array were always of the same haplotype. However, we cannot exclude that for one of the distal 4qA sequences due to the duplication event essential sequences in the 3′ untranslated region (3’UTR) of the DUX4 gene got lost, preventing PCR amplification.
Table 2.
Composition of all duplication alleles identified in controls, FSHD1 and FSHD2 patients
| Proximal D4Z4 array |
Extra D4Z4 array(s) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Individual | Haplotype | D4Z4 array 1 (U) | A,B,0 | D4Z4 array 2 (U) | A,B,0 | D4Z4 array 3 (U)> | A,B,0 | Analysis | |||||
| FSHD2 | Rf1021.2 | 10A166 | 19 | D4Z4(10) | A | 21 | D4Z4(10) | A | 2 | D4Z4(10) | A | SB+MC | |
| Rf1666.1 | 4A161S | 100 | D4Z4S | A | 5 | DUX4S | 0 | SB+MC | |||||
| Rf696.1 | 4A161L | 26 | D4Z4L | A | 10 | DUX4L | A | 5 | DUX4L | A | SB+MC | ||
| Rf392.2 | 4A161L | 29 | D4Z4L | A | 28 | DUX4L | A | 5 | DUX4L | A | SB+MC | ||
| Rf1727.2 | 4A161L | 46 | D4Z4L | A | 6 | DUX4L | A | SB+MC | |||||
| Rf975.3 | 4A161L | 74 | D4Z4L | A | 6 | DUX4L | A | SB+MC | |||||
| Rf878.2 | 4A161L | 72 | D4Z4L | A | 6 | DUX4L | A | SB+MC | |||||
| Rf874.3 | 4A161L | 69 | D4Z4L | A | 6 | DUX4L | A | SB+MC | |||||
| Rf844.1 | 4A161L | 68 | D4Z4L | A | 6 | DUX4L | A | SB+MC | |||||
| Controls | Rf1600.1 | 10A166 | 22 (or 40) | D4Z4(10) | A | 2 | D4Z4(10) | A | SB | ||||
| Rf1816.2 | 10A166 | 22 (or 26) | D4Z4(10) | A | 4 | D4Z4(10) | A | SB | |||||
| Rf1883.2 | 10A166 | 12 (or 39) | D4Z4(10) | A | 4 | D4Z4(10) | A | SB | |||||
| Rf1598.2 | 10A166 | 24 (or 29) | D4Z4(10) | A | 6 | D4Z4(10) | A | SB | |||||
| Rf1577.1 | 4A161S | 15 | D4Z4S | A | 6 | DUX4S | 0 | SB | |||||
| Rf1563.1 | 4A161S | 22(or 43) | D4Z4S | A | 28 | DUX4S | 0 | SB | |||||
| Rf1216.1 | 4A161L | 47 | D4Z4L | A | 4 | DUX4L | A | SB | |||||
| Rf1590.1 | 4A161L | 60 | D4Z4L | A | 4 | DUX4L | A | SB | |||||
| Rf741.1 | 4A161L | 46 (or 22) | D4Z4L | A | 18 | DUX4L | A | 6 | DUX4L | A | SB | ||
| Rf1900.1 | 4A161L | 48 | D4Z4L | A | 9 | DUX4L | A | SB | |||||
| FSHD1 | Rf650.1 | 10A166 | 15 | D4Z4(10) | A | 10 | D4Z4(10) | A | SB | ||||
| Rf887.1 | 4B168 | 23 | D4Z4(B) | B | 3 | D4Z4(B) | 0 | SB+MC | |||||
| Rf897.1 | 4B163 | 32 | D4Z4(B) | B | 3 | D4Z4(B) | 0 | SB+MC | |||||
| Rf903.1 | 4B163 | 46 | D4Z4(B) | B | 3 | D4Z4(B) | 0 | SB+MC | |||||
We find significantly more duplication alleles in FSHD2 patients with a usually long SPA compared to controls, FSHD1 patients and 8–20 units FSHD2 patients.
Incidence of D4Z4 duplications in control and FSHD1 individuals
We next analyzed the incidence of D4Z4 duplications in the general population. Previously, Nguyen and colleagues (29) identified by Molecular Combing (MC) one 4q-like duplication allele in DNA from 50 control individuals (2.0%). We studied p13E-11 and D4Z4-hybridized Southern blots of 349 Caucasian control individuals (28) and found D4Z4 duplications in 10/349 (2.9%) controls (Table 1). Assuming that these duplications are also in cis with the D4Z4 repeat array, six of the duplication alleles identified in controls were associated with a 4qA haplotype and three of these alleles carried an FSHD1-sized D4Z4 array ≤ 6 units (identified by the D4Z4 probe) distal to the longer p13E-11-linked longer D4Z4 array (Table 2). In addition, we analyzed 282 unrelated FSHD1 patients, and identified 4 duplications. Thus, with a frequency of 8 out of 11 (73%), we identified a significantly higher proportion of D4Z4 duplications among FSHD2 patients with a SPA >20 units compared to controls, FSHD1 patients and FSHD2 patients with a 8–10 units SPA (Tables 1 and 2). Therefore, duplications likely explain the increased susceptibility to DUX4 expression and disease presentation in FSHD2 patients with unusually long SPA. When eliminating all duplication alleles in the FSHD2 patients from Figure 1A, only two exceptional long SPA (28 and 50 units) remain and the mean SPA repeat size drops from 16.8 to 14.1 units (Supplementary Material, Fig. S2). While most (7/8) of the duplications identified in FSHD2 patients with a SPA of >20 units were of the 4qA-type, D4Z4 repeat array duplications found in controls and FSHD1 patients were more often chromosome 4qB- or chromosome 10-derived (Table 2).
Molecular combing of duplication alleles
In order to study the exact composition of the D4Z4 duplication fragments, we applied MC on all FSHD2 samples with a SPA >20 units since MC allows for the visualization of D4Z4 repeat arrays together with the region proximal and distal to D4Z4 (Fig. 2A). A representative MC result for all chromosomes 4 and 10 D4Z4 loci is shown in Figure 3A for patient Rf975.3 from Figure 2B. MC confirmed the Southern blot finding and showed that the duplication is in cis with a D4Z4 repeat array on chromosome 4 (Fig. 3B). For all 4q-like duplication alleles, MC showed that they are composed of a rather large (>20 units) D4Z4 repeat array and a 5 to 6 units non-p13E-11 linked D4Z4 repeat array both ending with a beta-satellite repeat sequence (red signal), with the FSHD-sized D4Z4 repeat array always situated at the distal end (Supplementary Material, Fig. S1). In some cases, like Rf392.2, Rf696.1 and Rf1021.2, the composition of the locus was more complex with evidence for two duplicated D4Z4 repeat arrays (Fig. 3B). The non-p13E-11 linked D4Z4 arrays of the duplication allele lack the blue MC signal proximal to the array which labels D4F104S1 (p13E-11) consistent with the Southern blot data. MC analysis was also performed on DNA from three FSHD1 cases, who have an extra 4q0-like (i.e. not hybridizing with probes A or B) allele based on Southern blotting. For all these cases, detailed analysis did not reveal a D4Z4 duplication, but instead showed an expansion of the inverted D4Z4 repeat unit 40 kb proximal to the D4Z4 array (Fig. 2A). This expansion did not occur on the 4A161S-type FSHD1 allele, but instead on the homologous 4qB chromosome (Fig. 3B andSupplementary Material, Fig. S1). The Southern blot and MC results for all duplication alleles identified in FSHD2 patients and controls is summarized in Table 2.
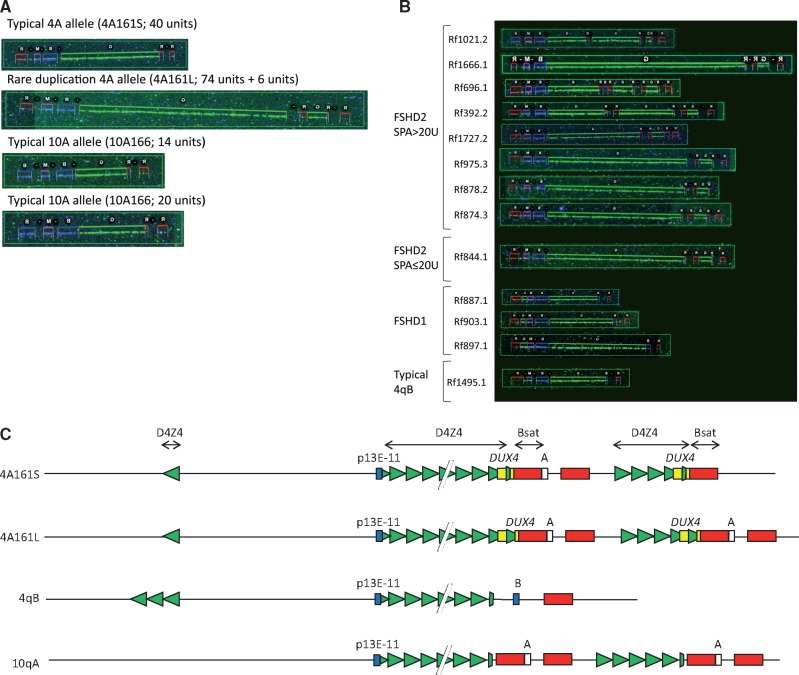
Figure 3.
(A) Representative MC images of the chromosome 4q and 10q alleles identified in FSHD patient Rf975.3. The MC alleles are matching to the Southern blot results (B), with the largest chromosome 4 allele carrying a 6 D4Z4 units duplication. The other alleles have a standard composition according to Figure 2A. (B) Representative MC images of the duplication alleles identified in the FSHD1 and FSHD2 cohort. The upper 8 alleles have been identified in FSHD2 patients with a SPA > 20 units. The next duplication allele was found in SPA ≤20 units) FSHD2 patient (Rf844.1). Below three unusual 4qB alleles having an expansion of the inverted D4Z4 repeat unit are shown. These alleles were identified in FSHD1 patients. As a reference a MC image of a typical 4qB allele with a single (standard) inverted D4Z4 repeat unit is shown at the bottom. (C) Composition of the three main groups of duplication alleles (4A161S, 4A161L and 10A166) and one group of unusual 4qB alleles that have been identified.
Founder alleles
Seven of the nine duplication alleles identified in FSHD2 patients are of the less common 4A161L haplotype. Four of these duplication alleles have a very similar composition: in Rf975.3, Rf878.2, Rf874.3 and Rf844.1 it starts with D4Z4 repeat arrays of 74, 72, 69 and 68 units, respectively, followed by a six D4Z4 units duplication at the distal end. Most of the duplication alleles identified in controls were also of the 4A161L haplotype and Southern blot analysis showed each D4Z4 array in all identified 4A161L duplication alleles ended with a 4qA sequence. In contrast, three 4A161S duplication alleles identified in controls and FSHD2 patients terminate with a sequence that is neither 4qA nor 4qB. Finally, the duplication-like 4qB alleles identified FSHD1 individuals were shown to have an expansion of the inverted D4Z4 repeat unit rather than a D4Z4 repeat array duplication (Fig. 3B). These uniformity within each of the classes of D4Z4 duplication alleles suggest that duplication alleles are likely derivatives of a limited number of founder alleles (Fig. 3C).
Role of SMCHD1 variants in duplication alleles
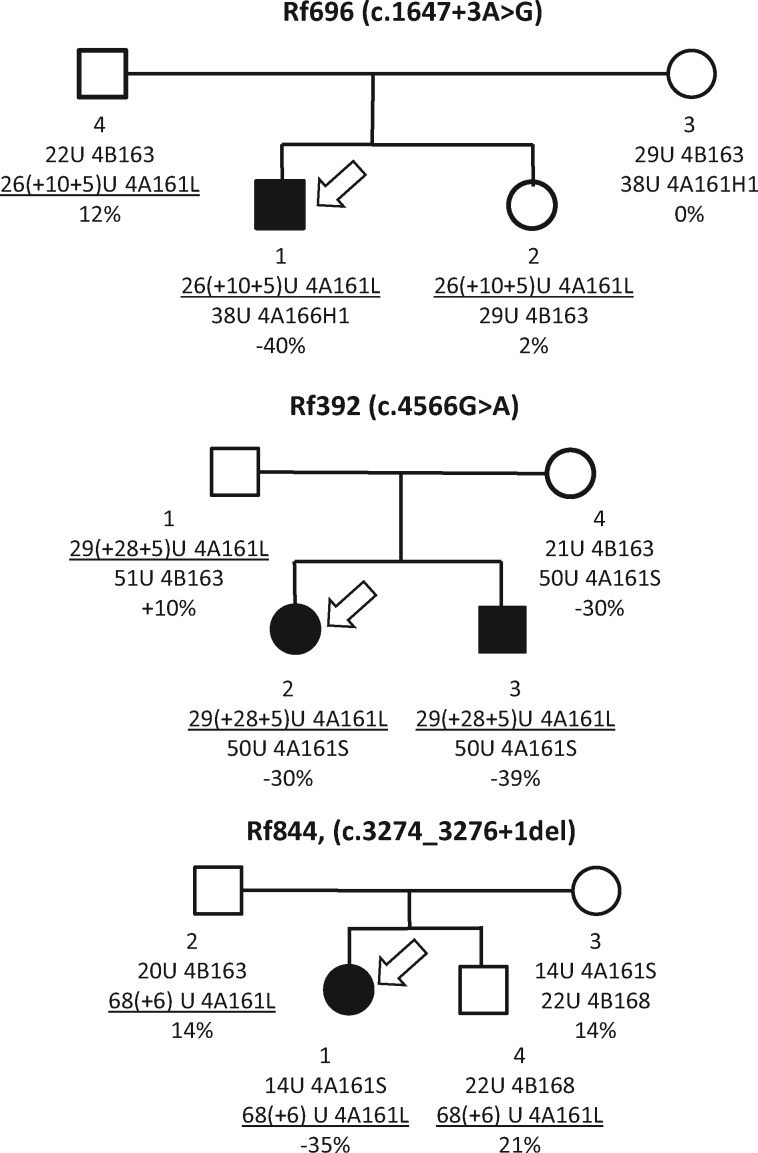
To study the role of SMCHD1 in disease penetrance of duplication alleles we analyzed the family members of FSHD2 patients with duplication alleles. In 3 of the 11 families we identified family members that carried the duplication allele in the absence of the pathogenic SMCHD1 variant. In all three families we observed that the duplication allele in the absence of the SMCHD1 variant, and hence D4Z4 hypomethylation, did not result in FSHD (Fig. 4). This suggests that, for these duplication alleles, a pathogenic SMCHD1 variant is required for disease presentation.
Figure 4.
Pedigrees of three FSHD2 families in which the index patient (indicated with arrow) has a duplication allele (underlined). For all families, the duplication allele only cause FSHD in combination with a pathogenic SMCHD1 variant and not without the variant. Below each individual the identifier is shown, the genetic information about both chromosomes 4, and the delta1 score.
Transcription studies
In order to study transcription from duplication alleles we re-analyzed the data previously described from differentiated myoblasts obtained from FSHD2 patients Rf696.1 and Rf844.1 (28). Patient Rf696.1 has a 38 D4Z4 units 4A161S allele and a 4A161L duplication allele that consist of 26, 10 and 5 D4Z4 units long repeat arrays. Patient Rf844.1 has a 14 D4Z4 unit 4A161S allele combined with a 4A161L duplication allele that consist of 68 and 6 D4Z4 units long repeat arrays. For myotubes from Rf696.1 we observed rather high DUX4 levels (Supplementary Material, Fig. S3), which originates exclusively from the 4A161L duplication allele. In Rf844.1 DUX4 expression could be observed from both alleles consistent with a relatively severe phenotype (Ricci 7 at age 31, ACSS 226). This suggests that duplication alleles can produce DUX4 protein, possibly from the distal short duplicated repeat array as this is in the size range that is susceptible to pathogenic SMCHD1 variants.
Discussion
Over the years a mechanistic model for FSHD has emerged by which the disease in most patients can be molecularly explained by inappropriate expression of the DUX4 retrogene in skeletal muscle (27,36). Normally DUX4 is not expressed, or expressed at very low levels in skeletal muscle. In FSHD, as a consequence of partial chromatin relaxation of the D4Z4 repeat array in somatic cells, DUX4 repression is incomplete, leading to the presence DUX4 protein and downstream DUX4 targets and transcriptional programs in skeletal muscle. Many studies have addressed the consequences of DUX4 in muscle. Recently, consensus has been reached that DUX4 is toxic to muscle in many ways, eventually leading to muscle cell death. However, the exact pathways and their relative contribution to muscle pathology are still under intense study (12,13,37).
Although initially considered two clinically largely identical, but genetically distinct subtypes, recent studies suggest that with regard to the genetic mechanism FSHD1 and FSHD2 represent a continuum: the combined contribution of D4Z4 repeat array size, and the ability to epigenetically repress DUX4 in skeletal muscle determines FSHD disease presentation (27). For FSHD1 patients having a repeat of 1–7 D4Z4 units, the size of the repeat array is likely the major contribution to DUX4 expression. For 8–10 D4Z4 units 4qA alleles, other epigenetic factors contribute to the pathogenicity, as these repeat sizes are associated with marked clinical variability, and with a relatively high prevalence of asymptomatic or non-penetrant mutation carriers (26,38,39). These 8–10 D4Z4 units 4qA alleles are also encountered in the control population (20,21). On the other hand, incomplete repressive activity of SMCHD1, DNMT3B or other epigenetic modifiers of the D4Z4 repeat array, contribute to DUX4 expression in affected muscle of FSHD2 patients (7,23). In addition, similarly as in FSHD1, the D4Z4 repeat size is likely to contribute to disease presentation since repeat array sizes in these patients are generally shorter than in the general population (8–20 versus 8–100 units). The overlap in the 8–10 unit range between controls, FSHD1 and FSHD2 further supports a disease model in which both repeat size and chromatin regulators of the D4Z4 repeat array collectively determine the DUX4 repressive capacity in skeletal muscle (27).
It was therefore surprising that we previously encountered FSHD2 in families that have a damaging SMCHD1 variant combined with an unusual long SPA sizes (with sizes which we predicted to be insensitive to incomplete repressive activity of SMCHD1) (33). This finding suggested that other mechanisms, such as mutations in other D4Z4 chromatin modifiers, or unrecognized small D4Z4 repeat arrays, are at play in these families.
In 8/11 FSHD2 cases with an unusually long SPA, we indeed identified the presence of an additional D4Z4 hybridizing fragment on Southern blots suggesting that these families have a duplication of the D4Z4 repeat array that might explain the disease. We confirmed the D4Z4 duplication and in 6 of them this allele is of the 4A161L haplotype. In these cases the telomeric duplicated D4Z4 array of 5 or 6 units terminates as 4qA suggesting that they are permissive to DUX4 expression. In patient Rf1666.1, the duplication allele is associated to the 4A161S. The shorter distal array in this duplication allele contained 5 D4Z4 units but was not recognized by probes A or B on the Southern blot. 4qA alleles have a beta-satellite repeat sequence distal to the 3’UTR of DUX4 (Fig. 2A) (19), and as this beta-satellite repeat is still visible in the duplication allele of Rf1666.1 by MC, it probably contains a complete DUX4 gene in the distal array. In all FSHD2 cases the distal D4Z4 duplication was FSHD-sized, and much shorter than the proximal D4Z4 repeat array that was seen by Southern blot analysis using probe p13E-11. For two 4A161L duplication cases, a myotube sample was available allowing us to confirm transcription of 4A161L-specific DUX4, i.e. originating from the duplication allele.
We identified one FSHD2 patient with a duplication fragment on chromosome 10. This might be a coincidence, as we identified these 10qA-like duplication alleles also in 4/349 control individuals where it does not contribute to disease. Probably, the standard rather long (and therefore less pathogenic) 4qA allele is here pathogenic. This is supported by a Ricci score of 4 at 67 years, which is within the selection criteria but is the lowest of all patients. Indeed, mild right scapular winging was only observed upon clinical examination, the patient herself did not report symptoms of FSHD (40).
Duplication alleles are infrequently found in the control population and in FSHD1 patients. In particular 4qA-type duplication alleles, where the distal D4Z4 duplication is FSHD-sized, are very uncommon (4/349 in controls and 0/282 FSHD1 patients). We did identify one such duplication allele in 68 FSHD2 patients with a standard SPA of 8–20 D4Z4 units (1/68; 1.5%), a frequency similar as in the control population (4/349). With a frequency of 3/282, we less often observed a duplication in FSHD1 patients than in controls and never on the pathogenic chromosome. This difference might be due to a selection bias as the FSHD1 allele is a standard allele in each case. When only taking into account non-FSHD1 alleles the frequencies are similar (10/698 in controls and 3/282 in FSHD1 patients). The three duplications identified in FSHD1 patients are on 4qB chromosomes and were actually expansions of the inverted D4Z4. Thus, the high frequency of in cis duplications found in FSHD2 patients with unusually long SPA suggest that these alleles are pathogenic due to the FSHD-sized distal repeat duplication. Segregation analysis in three families indeed confirm that these duplication alleles are pathogenic, but only in combination with a pathogenic SMCHD1 variant.
We wondered why most of the duplication alleles were 4A161L-type and not 4A161S-type, while in Europe 4A161S is five times more common than 4A161L (28). Previously, we have shown that chromosome 4qA, 4qB, 10qA and 10qB haplotypes arose by four discrete interchromosomal sequence transfers during recent human evolution (17). Based on similarities in the composition of the duplication alleles found in this study, we propose that most duplication alleles are part of four founder groups and it is tempting to speculate that these duplication alleles arose by major intrachromosomal sequence transfers on chromosomes 4qA, 4qA-L and 10qA, and one expansion on chromosome 4qB.
Recently, a MC study was performed on 586 samples, for which in 230 cases a D4Z4 repeat array of 1–10 units was identified confirming FSHD1. In 14 cases from 10 different families, a duplication allele was identified similar to the alleles that we found (29). In 3/230 independent FSHD1 patients a duplication allele was identified on the non-affected allele, which is comparable to the frequency that we found in FSHD1. For 4 of the other 7 families the duplication was found in combination with D4Z4 hypomethylation and a pathogenic SMCHD1 variant, consistent with our suggestion that these duplication alleles are more likely to be disease causing when combined with reduced repressive activity at D4Z4.
Previously, we identified in approximately 2% of FSHD patients an FSHD1 allele in which the partial deletion of the D4Z4 repeat extends proximally and includes D4F104S1 (41). For Southern blot-based genetic diagnosis the identification of these deletions is challenging as the diagnostic probe p13E-11 recognizes D4F104S1. Successive hybridization with probe D4Z4 can reveal the missing allele enabling genetic diagnosis in these situations. This study shows that the D4Z4 probe can also be used in the detection of duplication fragments. However, this study also suggest that these D4Z4 duplication alleles are only pathogenic in combination with pathogenic SMCHD1 variants. Thus, genetic counselors should be able to discriminate D4F104S1 deletion alleles from duplication alleles, especially when applying Southern blotting without PFGE.
In conclusion, we show that D4Z4 duplications can be identified with relative high frequency in the FSHD2 population, and that these duplications likely explain the increased susceptibility to DUX4 expression and disease presentation in those FSHD2 patients with unusually long SPA. This finding strengthens the concept that pathogenic variants in SMCHD1 only cause FSHD when combined with SPAs <20 D4Z4 units.
Materials and Methods
Patients and controls
This study was approved by the Medical Ethical Committees from participating institutions. From a previous study, we included 74 FSHD2 families for which we were able to obtain detailed genotyping results based on hybridizations with probes p13E-11 and D4Z4 (28). All FSHD2 families have one or more FSHD2 patients with an age corrected severity score (ACSS) (30) ≥ 50 or a clear FSHD phenotype without documented ACSS. They have significant D4Z4 hypomethylation in combination with a damaging variant in SMCHD1. We included 282 FSHD1 patients from different families, and for non-FSHD-related samples (n = 349) we studied 208 unaffected controls (unaffected individuals and siblings/spouses of individuals with FSHD1) and 141 individuals with a different established genetic condition. All patients and controls have a European background to avoid confounding effects from population specific distributions of D4Z4 haplotypes (17).
Genetic analysis D4Z4 repeats
For the genetic analysis high molecular weight DNA was obtained by pronase and Sarkosyl treatment of agarose embedded white blood cells of approximately 1 million cells per agarose block. The agarose plugs were digested with informative restriction enzymes and DNA was separated by pulsed field gel electrophoresis (PFGE) followed by Southern blotting and sequential hybridization with radioactive labeled probes p13E-11, D4Z4, 4qA and 4qB, and completed with haplotype analysis using the SSLP PCR as described previously (31). For the discrimination between 4A161S and 4A161L haplotypes we used the previously described PCR (28). Hybridization with probe p13E-11 was performed in Church buffer supplemented with 1% BSA, D4Z4 hybridization in phosphate buffer with 50% formamide and 4qA and 4qB hybridization in phosphate buffer with polyethylene glycol 6000 (31,32). For all members of the FSHD2 families and most of the other samples, we determined the methylation at the FseI site in D4Z4 and calculated the delta1 methylation score (26). All FSHD2 patients had D4Z4 CpG methylation values below the previously defined threshold (FseI value ≤ 27%, or delta1 value ≤ −21%) (33).
Molecular combing
Sarkosyl and pronase treated agarose blocks generated for PFGE analysis were equilibrated with TE−4 after which they were treated for 16 h with 500 ug proteinase K and Sarkosyl. Subsequent steps to prepare the combed glass slides were according to the manufacturers protocols (Genomic Vision) (34). Slides were hybridized with antibody-labeled FSHD-specific probes and after several wash steps slides were scanned overnight by the Fibervision HeliXScan. D4Z4 alleles on chromosomes 4 and 10 were selected and counted using the general procedure by Fiberstudio 0.9.12 software, after which alleles with an unusual composition could be acquired from ‘unusual’ and ‘extra’ category. The detailed composition of the unusual D4Z4 duplication alleles was confirmed by visual inspection and editing of 6 to 17 images per allele.
SMCHD1 variant testing
The SMCHD1 variants have previously been identified with Sanger sequencing or with whole exome or whole genome sequencing followed by confirmation by Sanger sequencing (7,26).
DUX4 expression analysis in FSHD2 myoblast
DUX4 expression was studied in two FSHD2 myoblast lines (Rf696.1 and Rf844.1), which originated from the University of Rochester Medical Center bio repository. Culturing and myogenic differentiation of these primary myoblasts, isolation of total RNA and the generation of cDNA was similar as described before. DUX4 expression analysis between 4A161S and 4A161L haplotypes in Rf696.1 and Rf844.1 was done by previously described PCR conditions and primer pairs (28).
Statistical analysis
The D4Z4 array size of the SPA between controls and FSHD2 patients (Fig. 1A) and the delta1 methylation score in FSHD2 patients with SPA between 8 and 20 units and >20 units (Fig. 1B) were compared using the unpaired t-test in Graphpad Prism 7. For the comparisons shown in Table 1, we used a Pearson chi-square test with Yates’ continuity correction in R 3.3.2 (https://www.R-project.org/; date last accessed July 2, 2018).
Supplementary Material
Acknowledgements
This study was supported by funds from the National Institute of Neurological Disorders and Stroke award number P01 NS069539, the Prinses Beatrix Spierfonds (award numbers W.OP14-01 and W.OB17-01) and Spieren voor Spieren.
Conflict of Interest statement. None declared.
References
- 1. Deenen J.C., Arnts H., van der Maarel S.M., Padberg G.W., Verschuuren J.J., Bakker E., Weinreich S.S., Verbeek A.L., van Engelen B.G. (2014) Population-based incidence and prevalence of facioscapulohumeral dystrophy. Neurology, 83, 1056–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Padberg G.W. (1982) Facioscapulohumeral disease, PhD thesis, The Netherlands: Leiden University. https://openaccess.leidenuniv.nl/handle/1887/25818
- 3. Dixit M., Ansseau E., Tassin A., Winokur S., Shi R., Qian H., Sauvage S., Matteotti C., van Acker A.M., Leo O. (2007) DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc. Natl. Acad. Sci. U.S.A., 104, 18157–18162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gabriels J., Beckers M.C., Ding H., De Vriese A., Plaisance S., van der Maarel S.M., Padberg G.W., Frants R.R., Hewitt J.E., Collen D.. et al. (1999) Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene, 236, 25–32. [DOI] [PubMed] [Google Scholar]
- 5. Hewitt J.E., Lyle R., Clark L.N., Valleley E.M., Wright T.J., Wijmenga C., van Deutekom J.C., Francis F., Sharpe P.T., Hofker M.. et al. (1994) Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Hum. Mol. Genet., 3, 1287–1295. [DOI] [PubMed] [Google Scholar]
- 6. Lemmers R.J., van der Vliet P.J., Klooster R., Sacconi S., Camano P., Dauwerse J.G., Snider L., Straasheijm K.R., Jan vO., Padberg G.W.. et al. (2010) A unifying genetic model for facioscapulohumeral muscular dystrophy. Science, 329, 1650–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lemmers R.J., Tawil R., Petek L.M., Balog J., Block G.J., Santen G.W., Amell A.M., van der Vliet P.J., Almomani R., Straasheijm K.R.. et al. (2012) Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat. Genet., 44, 1370–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Snider L., Geng L.N., Lemmers R.J.L.F., Kyba M., Ware C.B., Nelson A.M., Tawil R., Filippova G.N., van der Maarel S.M., Tapscott S.J.. et al. (2010) Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet., 6, e1001181.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. De Iaco A., Planet E., Coluccio A., Verp S., Duc J., Trono D. (2017) DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet., 49, 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hendrickson P.G., Dorais J.A., Grow E.J., Whiddon J.L., Lim J.W., Wike C.L., Weaver B.D., Pflueger C., Emery B.R., Wilcox A.L.. et al. (2017) Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet., 49, 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Whiddon J.L., Langford A.T., Wong C.J., Zhong J.W., Tapscott S.J. (2017) Conservation and innovation in the DUX4-family gene network. Nat. Genet., 49, 935–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Geng L.N., Yao Z., Snider L., Fong A.P., Cech J.N., Young J.M., van der Maarel S.M., Ruzzo W.L., Gentleman R.C., Tawil R.. et al. (2012) DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell, 22, 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bosnakovski D., Xu Z., Gang E.J., Galindo C.L., Liu M., Simsek T., Garner H.R., Agha-Mohammadi S., Tassin A., Coppee F.. et al. (2008) An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J., 27, 2766–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Snider L., Asawachaicharn A., Tyler A.E., Geng L.N., Petek L.M., Maves L., Miller D.G., Lemmers R.J., Winokur S.T., Tawil R.. et al. (2009) RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum. Mol. Genet., 18, 2414–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bakker E., Wijmenga C., Vossen R.H., Padberg G.W., Hewitt J., van der Wielen M., Rasmussen K., Frants R.R. (1995) The FSHD-linked locus D4F104S1 (p13E-11) on 4q35 has a homologue on 10qter. Muscle Nerve, 2, 39–44. [PubMed] [Google Scholar]
- 16. Deidda G., Cacurri S., Grisanti P., Vigneti E., Piazzo N., Felicetti L. (1995) Physical mapping evidence for a duplicated region on chromosome 10qter showing high homology with the Facioscapulohumeral muscular dystrophy locus on chromosome 4qter. Eur. J. Hum. Genet., 3, 155–167. [DOI] [PubMed] [Google Scholar]
- 17. Lemmers R.J.L.F., van der Vliet P.J., van der Gaag K.J., Zuniga S., Frants R.R., de Knijff P., van der Maarel S.M. (2010) Worldwide population analysis of the 4q and 10q subtelomeres identifies only four discrete duplication events in human evolution. Am. J. Hum. Genet., 86, 364–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van Geel M., Dickson M.C., Beck A.F., Bolland D.J., Frants R.R., van der Maarel S.M., de Jong P.J., Hewitt J.E. (2002) Genomic analysis of human chromosome 10q and 4q telomeres suggests a common origin. Genomics, 79, 210–217. [DOI] [PubMed] [Google Scholar]
- 19. Lemmers R.J., de Kievit P., Sandkuijl L., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M. (2002) Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat. Genet., 32, 235–236. [DOI] [PubMed] [Google Scholar]
- 20. Lemmers R.J., Wohlgemuth M., van der Gaag K.J., van der Vliet P.J., van Teijlingen C.M., de Knijff P., Padberg G.W., Frants R.R., van der Maarel S.M. (2007) Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet., 81, 884–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Scionti I., Fabbri G., Fiorillo C., Ricci G., Greco F., D'Amico R., Termanini A., Vercelli L., Tomelleri G., Cao M.. et al. (2012) Facioscapulohumeral muscular dystrophy: new insights from compound heterozygotes and implication for prenatal genetic counselling. J. Med. Genet., 49, 171–178. [DOI] [PubMed] [Google Scholar]
- 22. Wijmenga C., Hewitt J.E., Sandkuijl L.A., Clark L.N., Wright T.J., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R.. et al. (1992) Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet., 2, 26–30. [DOI] [PubMed] [Google Scholar]
- 23. van den Boogaard M.L., Lemmers R.J., Balog J., Wohlgemuth M., Auranen M., Mitsuhashi S., van der Vliet P.J., Straasheijm K.R., van den Akker R.F., Kriek M.. et al. (2016) Mutations in DNMT3B modify epigenetic repression of the D4Z4 repeat and the penetrance of facioscapulohumeral dystrophy. Am. J. Hum. Genet., 98, 1020–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de Greef J.C., Lemmers R.J., Camano P., Day J.W., Sacconi S., Dunand M., van Engelen B.G., Kiuru-Enari S., Padberg G.W., Rosa A.L.. et al. (2010) Clinical features of facioscapulohumeral muscular dystrophy 2. Neurology, 75, 1548–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Statland J., Tawil R. (2014) Facioscapulohumeral muscular dystrophy. Neurol. Clin., 32, 721–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lemmers R.J., Goeman J.J., van der Vliet P.J., van Nieuwenhuizen M.P., Balog J., Vos-Versteeg M., Camano P., Ramos Arroyo M.A., Jerico I., Rogers M.T.. et al. (2015) Inter-individual differences in CpG methylation at D4Z4 correlate with clinical variability in FSHD1 and FSHD2. Hum. Mol. Genet., 24, 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Daxinger L., Tapscott S.J., van der Maarel S.M. (2015) Genetic and epigenetic contributors to FSHD. Curr. Opin. Genet. Dev., 33, 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lemmers R.J., van der Vliet P.J., Balog J., Goeman J.J., Arindrarto W., Krom Y.D., Straasheijm K.R., Debipersad R.D., Ozel G., Sowden J.. et al. (2018) Deep characterization of a common D4Z4 variant identifies biallelic DUX4 expression as a modifier for disease penetrance in FSHD2. Eur. J. Hum. Genet., 26, 94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nguyen K., Puppo F., Roche S., Gaillard M.C., Chaix C., Lagarde A., Pierret M., Vovan C., Olschwang S., Salort-Campana E.. et al. (2017) Molecular combing reveals complex 4q35 rearrangements in Facioscapulohumeral dystrophy. Hum. Mutat., 38, 1432–1441. [DOI] [PubMed] [Google Scholar]
- 30. van Overveld P.G., Enthoven L., Ricci E., Rossi M., Felicetti L., Jeanpierre M., Winokur S.T., Frants R.R., Padberg G.W., van der Maarel S.M. (2005) Variable hypomethylation of D4Z4 in facioscapulohumeral muscular dystrophy. Ann. Neurol., 58, 569–576. [DOI] [PubMed] [Google Scholar]
- 31. Lemmers R.J. (2017) Analyzing copy number variation using pulsed-field gel electrophoresis: providing a genetic diagnosis for FSHD1. Methods Mol. Biol., 1492, 107–125. [DOI] [PubMed] [Google Scholar]
- 32. Church G.M., Gilbert W. (1984) Genomic sequencing. Proc. Natl. Acad. Sci. U.S.A., 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lemmers R.J., van den Boogaard M.L., van der Vliet P.J., Donlin-Smith C.M., Nations S.P., Ruivenkamp C.A., Heard P., Bakker B., Tapscott S., Cody J.D.. et al. (2015) Hemizygosity for SMCHD1 in facioscapulohumeral muscular dystrophy type 2: consequences for 18p deletion syndrome. Hum. Mutat., 36, 679–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vasale J., Boyar F., Jocson M., Sulcova V., Chan P., Liaquat K., Hoffman C., Meservey M., Chang I., Tsao D.. et al. (2015) Molecular combing compared to Southern blot for measuring D4Z4 contractions in FSHD. Neuromuscul. Disord., 25, 945–951. [DOI] [PubMed] [Google Scholar]
- 35. Ehrlich M., Jackson K., Tsumagari K., Camano P., Lemmers R.J. (2007) Hybridization analysis of D4Z4 repeat arrays linked to FSHD. Chromosoma, 116, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Statland J.M., Tawil R. (2016) Facioscapulohumeral Muscular Dystrophy. Continuum, 22, 1916–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Block G.J., Narayanan D., Amell A.M., Petek L.M., Davidson K.C., Bird T.D., Tawil R., Moon R.T., Miller D.G. (2013) Wnt/beta-catenin signaling suppresses DUX4 expression and prevents apoptosis of FSHD muscle cells. Hum. Mol. Genet., 22, 4661–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gaillard M.C., Roche S., Dion C., Tasmadjian A., Bouget G., Salort-Campana E., Vovan C., Chaix C., Broucqsault N., Morere J.. et al. (2014) Differential DNA methylation of the D4Z4 repeat in patients with FSHD and asymptomatic carriers. Neurology, 83, 733–742. [DOI] [PubMed] [Google Scholar]
- 39. Wohlgemuth M., Lemmers R.J.L.F., Jonker M., van der Kooi E., Horlings C., van Engelen B.G., van der Maarel S.M., Padberg G., Voermans N. (2018) A family-based study into penetrance in facioscapulohumeral muscular dystrophy type 1. Neurology, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sacconi S., Lemmers R.J., Balog J., van der Vliet P.J., Lahaut P., van Nieuwenhuizen M.P., Straasheijm K.R., Debipersad R.D., Vos-Versteeg M., Salviati L.. et al. (2013) The FSHD2 gene SMCHD1 is a modifier of disease severity in families affected by FSHD1. Am. J. Hum. Genet., 93, 744–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lemmers R.J., Osborn M., Haaf T., Rogers M., Frants R.R., Padberg G.W., Cooper D.N., van der Maarel S.M., Upadhyaya M. (2003) D4F104S1 deletion in facioscapulohumeral muscular dystrophy: phenotype, size, and detection. Neurology, 61, 178–183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.