Abstract
MET and epidermal growth factor receptor (EGFR) tyrosine kinases are crucial for liver regeneration and normal hepatocyte function. Recently, we demonstrated that in mice, combined inhibition of these two signaling pathways abolished liver regeneration after hepatectomy, with subsequent hepatic failure and death at 15 to 18 days after resection. Morbidity was associated with distinct and specific alterations in important downstream signaling pathways that led to decreased hepatocyte volume, reduced proliferation, and shutdown of many essential hepatocyte functions, such as fatty acid synthesis, urea cycle, and mitochondrial functions. Herein, we explore the role of MET and EGFR signaling in resting mouse livers that are not subjected to hepatectomy. Mice with combined disruption of MET and EGFR signaling were noticeably sick by 10 days and died at 12 to 14 days. Mice with combined disruption of MET and EGFR signaling mice showed decreased liver/body weight ratios, increased apoptosis in nonparenchymal cells, impaired liver metabolic functions, and activation of distinct downstream signaling pathways related to inflammation, cell death, and survival. The present study demonstrates that, in addition to controlling the regenerative response, MET and EGFR synergistically control baseline liver homeostasis in normal mice in such a way that their combined disruption leads to liver failure and death.
Signaling pathways associated with epidermal growth factor receptor (EGFR) and hepatocyte growth factor (HGF) with its receptor MET are crucial for both normal hepatocyte function and liver regeneration. It has long been known that HGF and the various EGFR-related ligands are potent mitogens for cultured hepatocytes.1 Several studies have also explored the crucial effects of these two distinct pathways on liver regeneration after partial hepatectomy (PHx)2, 3 because HGF/MET and EGF/EGFR signaling pathways are among the earliest to be activated after removing two thirds of the liver.4 EGFR, which dimerizes with other erythroblastic leukemia viral oncogene B (ErbB) family members, has also been suggested to play an essential role for hepatic development and function, whereas disruption of HGF production is known to be embryonic lethal.3, 5, 6 Injection of HGF and/or various EGF family members can cause enhanced proliferation and increased liver weight in normal mice.7 These two receptor tyrosine kinases trigger downstream signaling affecting many targets, including RAS–extracellular signal-regulated kinase (ERK)–mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K)–AKT pathways that are important for MET-mediated cell proliferation and survival, respectively.8 In addition, shortly after PHx, the activation of transcription factors, such as STAT3 and NF-κB, as well as an increased release of cytokines, such as IL-6,9, 10 have been noted.
We have recently shown that in animals subjected to PHx after combined disruption of signaling from both mitogenic receptors, there is impaired hepatocyte function, blocked regeneration, and liver decompensation/failure.11 Ours was the first study to report that combined disruption of these two key pathways abolishes liver regeneration and leads to severe decompensation of crucial hepatocyte functions, causing ascites and high plasma ammonia levels in mice. To date, there is no other extracellular signaling to hepatocytes, whose elimination completely inhibits liver regeneration. Hepatectomy initiates a strong mitogenic stimulus for hepatocytes, altering many fundamental gene expression patterns shortly after removal of two thirds of liver.12 After PHx, a cascade of genes, encoding transcription factors and cell cycle regulators, is induced,10, 13, 14 whereas hepatocytes simultaneously exit G0, enter the cell cycle, and replicate.15, 16, 17 In contrast, most cells in the resting liver are quiescent, with a long half-life.
In the current study, we explored the changes that occur in the resting livers of normal mice after genomic disruption of the c-Met gene (ΔMET), chemical elimination of EGFR signaling (EGFRi), or both (ΔMET + EGFRi). Surprisingly, the doubly disrupted mice were clearly sick at day 10 after initiating EGFR inhibition via canertinib diet and were dead by days 12 to 14, whereas their matched cohorts (control, EGFRi only, or ΔMET only) appeared healthy. Herein, we describe the structural, biochemical, and functional changes that we observed in the 10 days before the death of these animals. Relative to the other cohorts, ΔMET + EGFRi mice showed decreased liver/body weight ratios, decreased hepatocellular Ki-67 staining, increased parenchymal and nonparenchymal cell death, impaired liver metabolic functions, and distinct alterations in a variety of downstream signaling pathways, including activation of several caspases. Although not examined in great detail compared with liver, no histologically demonstrable abnormalities were observed in any other tissues (pancreas, kidney, lung, spleen, or intestine). These results indicate that an intricate synchronization between the EGFR and MET signaling pathways is essential for basic hepatic function and animal survival, independent of their coordinate roles during the regenerative response.
Materials and Methods
Generation of the METff/ff:TamCre+/+ Mice
METff/ff mice with a targeted deletion of c-Met exon 16 were a kind gift from Dr. Snorri Thorgeirsson (National Cancer Institute, NIH, Rockville, MD),18 and are now available at The Jackson Laboratory (Bar Harbor, ME) as 016974, Mettm1Sst. Deletion of exon 16 leads to a MET protein missing the ATP-binding region. To systemically remove the exon, the METff/ff animals were crossbred with mice purchased from The Jackson Laboratory carrying a tamoxifen-inducible Cre recombinase gene [Gt (ROSA) 26Sortm1 (cre/ERT2) Tyj], to ultimately produce METff/ff:TamCre+/+ mice that we now carry as a stable line. To generate ΔMET animals, male METff/ff:TamCre+/+ mice (average age, 24 weeks) were injected intraperitoneally for 5 consecutive days with tamoxifen (100 μL of a 10 mg/mL solution in corn oil), followed by a 4-day resting period to eliminate the effects of the drug. There are no adverse effects of tamoxifen on hepatocyte proliferation in wild-type animals under these conditions.11 In experiments using ΔMET mice alone, animals were sacrificed at 2, 7, and 10 days after the 4-day tamoxifen clearance (ie, days 6, 11, and 14 after the last dose of tamoxifen) as a means of coordinating timing with administration of the canertinib diet (see below). Controls consisted of METff/ff:TamCre+/+ males, where only corn oil (control vehicle, no tamoxifen) was administered. To remain consistent, the animals were allowed a 4-day resting period, and then their livers were harvested 2 to 10 days later.
Canertinib Diet
Canertinib, a pan-ErbB EGFR inhibitor with higher affinity for EGFR compared with the other ErbB family members, binds to the ATP-binding site, inhibits the tyrosine kinase activity, and is then thought to direct the receptor into intracellular trafficking routes for degradation,11 although there is also evidence that EGFR is not necessarily degraded.19 Canertinib was administered in the diet as previously described.11 Briefly, canertinib dihydrochloride salt (LC Laboratories, Woburn, MA; alias CI-1033) was added to a normal mouse diet at 480 mg/kg, with the assumption that mice weighing 30 g would consume 5 g of food/day (80 mg/kg body weight daily). METff/ff:TamCre+/+ animals (EGFRi) or ΔMET animals (ΔMET + EGFRi) were placed on the diet and then harvested at 2, 7, and 10 days after the diet began. Controls consisted of mice from each of these two groups fed the diet without canertinib.
Animal Protocols and Handling
All procedures performed on mice were approved under the University of Pittsburgh (Pittsburgh, PA) Institutional Animal Care and Use Committee protocols 14053713 and 17040392, conducted according to NIH's Guide for the Care and Use of Laboratory Animals,20 and performed as described. At defined time points, the animals were sacrificed according to Association for Assessment and Accreditation of Laboratory Animal Care International guidelines.
Protein Analysis
Tissue homogenization and Western blot analyses were performed as previously described.21 Band density for each time point (three or four mice per time point) was standardized to Ponceau Red, as previously described.21
Antibodies
The following primary antibodies (Abs) were used at a 1:1000 dilution. Antibodies obtained from Cell Signaling Technology (Danvers, MA) are the following: EGFR retinoblastoma (Rb) Ab (number 2640); phosphorylated (phospho)-EGFR (Y1068) Rb Ab (number 3777); phospho-MET (Y1349) Rb Ab (number 3121); MET mouse Ab (number 4560) (although it had not been tested on mice, it worked well in our samples); PI3K p110α Rb Ab (number 4249); PI3K p85 Rb Ab (number 4292); phospho-AKT (S473) Rb Ab (number 9271); phospho-AKT (T308) Rb Ab (number 9275); mammalian target of rapamycin (mTOR) Rb Ab (number 2983); phospho-mTOR (S2448) Rb Ab (number 5536); phospho-mTOR (S2481) Rb Ab (number 2974); non–phospho-phosphatase and tensin homolog (PTEN) Rb Ab (number 7960); PTEN Rb Ab (number 9188); cyclin D1 Rb Ab (number 2922); STAT3 Rb Ab (number 4904); phospho-STAT3 (Tyr705) Rb Ab (number 9131); phospho–glycogen synthetase kinase (GSK)-3β (Ser9) Rb Ab (number 9323); phospho-p44/42 mitogen-activated protein kinase (extracellular signal-regulated kinase 1/2) (T202/Y204) Rb Ab (number 9101); p44/42 mitogen-activated protein kinase (extracellular signal-regulated kinase 1/2) mouse Ab (number 4696); phospho-AMPKα (Thr172) Rb Ab (number 2535); AMPKα Rb Ab (number 5831); T-cell protein tyrosine phosphatase (TC45) Rb Ab (number 58935); and Apoptosis Antibody Sampler Kit (Mouse Preferred; number 9930). The following antibodies were obtained from Abcam Biotechnology (Cambridge, MA): IL-6 Rb Ab (number 7737); NF-κB p65 Rb Ab (number 31481); and DEP1 Rb Ab (number 202958). Phospho-MET (Y1234/1235) Rb Ab (number 101736) was purchased from Santa Cruz Biotechnology (Dallas, TX). Antiglutamine synthase (number G2781) was purchased from Sigma (St. Louis, MO). Secondary antibodies used for this project are anti-rabbit IgG, horseradish peroxidase–linked antibody (Cell Signaling Technology; number 7074) and anti-mouse IgG, horseradish peroxidase–linked antibody (Cell Signaling Technology; number 7076) at 1:50,000 dilution.
Immunohistochemistry and Analysis
Paraffin-embedded liver tissue sections (4 μm thick) were used for immunohistochemical staining. Antigen retrieval was achieved by heating the slides in the microwave at high power in 1× citrate buffer for 10 minutes. The tissue sections were blocked in Ultra Vision Protein Block from Thermo Fisher (Waltham, MA; TA-125-PBQ), followed by incubation with Ki-67 primary antibody from Thermo Fisher (RM 9106S1) at room temperature for 20 minutes. The primary antibody was then linked to biotinylated secondary goat anti-rabbit antibody from Vector Laboratories (Burlingame, CA; BA-1000) at 1:250 dilution, followed by routine avidin-biotin complex method. Diaminobenzidine was used as the chromogen, which resulted in a brown reaction product. Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) staining was performed using ApopTag-Peroxidase In Situ Apoptosis Detection Kit (EMD Millipore, Burlington, MA; S7100). To count the Ki-67– and/or TUNEL-positive cells, three different microscopic images were taken from each mouse (n = 3 to 4) in the various categories using the 20× objective, for a total of 9 to 12 analyzed fields/category. All animals harvested in the three control groups were combined in the analyses. For hepatocellular volume, the number of hepatocytes in the images were counted within each group. The number of cells in each of the experimental groups was then divided by the control number, and the square root of the resultant ratios was determined. This number was then raised to the power of 3.
Statistical Analysis
Ordinary one-way analysis of variance was first performed using Prism 7.0a (GraphPad Software, La Jolla, CA) to determine experimental significance. Given the short term (10 days) of the experiments, post-test analyses indicated no variation between the three control time points in any experiments. Subsequently, when significant results were identified in an experimental group by analysis of variance, data from the three control groups were combined (total n = 9) and unpaired t-tests were performed for each time point versus the total control animals. Although significant differences were also observed within and between different date/time points of each treatment group, these differences have not been highlighted in this article.
Immunoprecipitation Studies
Total pooled cell lysates from each sample (500 μg) were mixed with either 10 μL of primary antibody (anti-p1349; Cell Signaling Technology; number 3121 for MET) or 5 μL of control IgG (Santa Cruz Biotechnology; number SC-2027). Samples were incubated overnight, with mixing at 4°C. Protein A/G plus agarose beads (20 μL; Santa Cruz Biotechnology; number SC-2003) were then added, and the samples were incubated an additional 24 hours, with mixing at 4°C. Samples were centrifuged at 1000 × g for 5 minutes, 4°C, and the initial supernatants were saved for use as a control. The beads were then washed three times with 200 μL ice-cold radioimmunoprecipitation assay buffer (0.1% SDS and 1% NP-40 in phosphate-buffered saline) containing a cocktail of protease inhibitors. The final pellets were then resuspended in 30 μL radioimmunoprecipitation assay with an additional 7.5 μL NuPAGE LDS sample buffer (Life Technologies, Carlsbad, CA; number NP0007) and 3 μL NuPAGE sample reducing agent (Life Technologies, number NP0009). To run the gels, samples were heated at 100°C for 10 minutes, and then 15 μL was loaded onto a 4% to 12% Bis-Tris gel (Life Technologies; number NP0322) with MES SDS running buffer (Life Technologies; number NP0002). Gels were run at 180 V for 1 hour on the X Cell Surelock mini (Life Technologies) and then transferred onto Immobilon-P (EMD Millipore) using the X Cell II Bolt Module (Life Technologies) using NuPAGE transfer buffer (NP0006) with 0.02% SDS added to effect transfer of high–molecular-weight proteins. To assess transfer efficiency, blots were stained with Ponceau and then subjected to Western blot analyses using a total MET (Cell Signaling Technology; number 4560), as previously described.21
Microarray and Data Analysis
Microarray analysis was performed on RNAs isolated from pooled liver samples from the various groups (control, day 10; ΔMET + EGFRi, days 7 and 10; n = 3 per group) using the GeneChip Mouse Genome 430 2.0 Array Set 430 (Affymetrix, Santa Clara, CA). cRNA synthesis, cDNA synthesis, and Affymetrix chip hybridization were done as previously described.11 Signal intensities were logarithmically (base 2) transformed and quantile normalized. Probes were mapped to corresponding genes, and if more than one probe was matched to the same gene, only the probe with the largest interquartile range will be the representative one. Genes with low expression and low variance were filtered out. Tight clustering algorithm was applied to group the genes, with consistent expression pattern across the samples.22 The tight clustering method was used to group genes into five clusters with distinct expression patterns, and these clustered genes were presented as a heat map. Enriched biological processes (gene ontology terms) were determined in each cluster compared with the Mus musculus reference gene list using the PANTHER Overrepresentation Test (PANTHER version 13.0; http://www.pantherdb.org). Statistical analysis was performed using Fisher's exact test with false discovery rate multiple test correction. Significantly expressed genes were also analyzed using Ingenuity Pathways Analysis version 01-10 (IPA; Ingenuity Systems, Redwood City, CA). Sets of genes up-regulated or down-regulated (twofold cutoff) in ΔMET + EGFRi versus the control group, along with the respective fold change values, were uploaded to the IPA software tool. Significantly altered canonical signaling pathways and upstream transcription factors were predicted on the basis of changes in downstream gene expression patterns. The microarray data discussed in this publication were deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus and are accessible through GEO Series (https://www.ncbi.nlm.nih.gov/geo; accession number GSE116307).
Results
Growth-Related Parameters in ΔMET, EGFRi, and ΔMET + EGFRi Mice
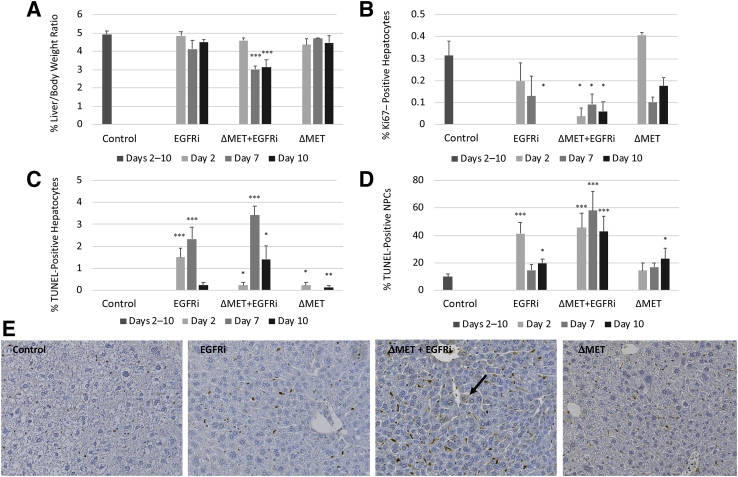
After the specified treatments for each group, it was observed that ΔMET + EGFRi mice appeared sick by day 10 and were dead by day 14. Subsequently, experiments were designed so that the animals on the various protocols were harvested on the days that coincide with days 2, 7, and 10 after initiation of the canertinib diet (Materials and Methods). ΔMET or EGFRi only mice had no statistically significant differences in liver/body weight ratio compared with control animals (Figure 1A). In contrast, in the ΔMET + EGFRi mice, there was a dramatic decrease in the liver/body weight ratio by day 7 (Figure 1A). To understand why, we began by measuring known parameters related to cell proliferation and death. When Ki-67 staining was performed to determine the number of dividing hepatocytes, it was noted that hepatocyte proliferation in control mice was approximately 0.3% (Figure 1B). This is consistent with the fact that the liver is a slowly self-renewing organ, and most hepatocytes are in G0 with an estimated lifespan of 200 to 400 days. Despite this low rate of self-renewal, the EGFRi mice were found to have a decreasing hepatocyte proliferation rate over time, which reached a statistically significant zero proliferation rate at day 10. In contrast, hepatocyte proliferation in the ΔMET mice was statistically within range of normal at all time points, whereas in ΔMET + EGFRi mice, there was a significant decrease in baseline hepatocyte proliferation beginning at day 2 that persisted through day 10 (Figure 1B). The hepatocyte death rate was generally low in the various experimental conditions, although a statistically significant increase in hepatocellular death was still observed in most groups (Figure 1C). Most treatments also resulted in an increase in TUNEL staining for the nonparenchymal cells (NPCs) compared with control (Figure 1D). This finding was particularly pronounced at day 2 in the EGFRi animals and at all time points in the ΔMET + EGFRi mice, with the latter cohort displaying the highest elevation in the number of TUNEL-positive NPCs (Figure 1, D and E).
Figure 1.
Liver weight, cell proliferation, and apoptosis in EGFRi, ΔMET + EGFRi, and ΔMET mice. A: The percentage liver/body weight ratio. B: The percentage of Ki-67–positive hepatocytes. Labeled and total hepatocytes from three different microscopic fields/mouse were counted to determine the percentage labeling; each treatment group contained a minimum of three mice; a minimum of 2000 total hepatocytes were counted for each condition tested. C: The number of terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL)–positive hepatocytes per high-power optic field (20× objective) was counted. Labeled hepatocytes from three different microscopic fields/mouse were counted, with a minimum of three mice/condition. D: The number of TUNEL-positive nonparenchymal cells (NPCs) per high-power optic field (20× objective) was counted. Labeled NPCs from three different microscopic images/mouse were counted, with a minimum of three mice/condition. E: Representative TUNEL staining photomicrographs from day 10 control, EGFRi, ΔMET + EGFRi, and ΔMET mice. Arrow depicts a TUNEL-positive hepatocyte. Images taken using a 20× objective. Data are expressed as means ± SEM (A–D). ∗P ≤ 0.05, ∗∗P ≤ 0.01, and ∗∗∗P ≤ 0.001 versus control animals.
Cell Volume in ΔMET, EGFRi, and ΔMET + EGFRi Mice
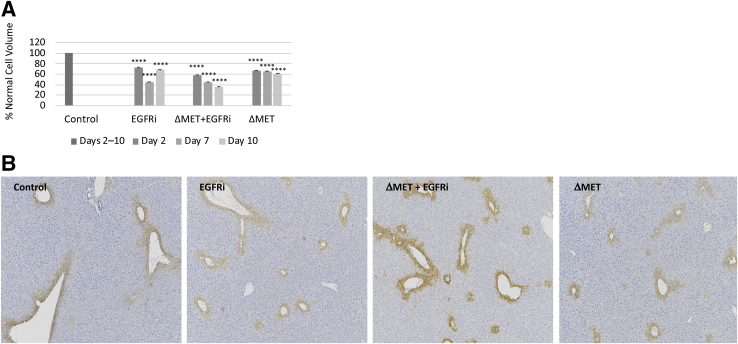
ΔMET + EGFRi animals subjected to hepatectomy display an overall reduction in hepatocyte volume.11 Therefore, it was determined whether the observed reduction in the ΔMET + EGFRi liver/body weight might similarly correlate with cell size. Surprisingly, the calculated hepatocyte volume was less in all tested conditions compared with control animals; however, the difference was particularly remarkable in the ΔMET + EGFRi mice (Figure 2A). An overall decrease was observed in the distance between hepatic lobules, particularly in the ΔMET + EGFRi group, consistent with previous findings, as evidenced by the sparse distances between the glutamine synthase staining that was used to depict the central veins (Figure 2B).11 No evident histologic or weight changes were noticed in other organs, including pancreas, kidneys, spleen, lung, and intestine.
Figure 2.
Lobular size and cell volume in EGFRi, ΔMET + EGFRi, and ΔMET mice. A: The mean average hepatocyte volume in EGFRi and/or ΔMET mouse compared with control. B: Representative glutamine synthase immunohistochemistry left to right from day 10 control, EGFRi, ΔMET + EGFRi, and ΔMET mice. The one layer of cells surrounding the central vein of each lobule is marked as positive for glutamine synthase. The distance between the marked areas demonstrates the decreased lobular dimensions in the ΔMET + EGFRi mice at day 10. Images taken using a 5× objective. Data are expressed as means ± SEM (A). ∗∗∗∗P ≤ 0.0001 versus control.
Metabolic Parameters in ΔMET, EGFRi, and ΔMET + EGFRi Mice
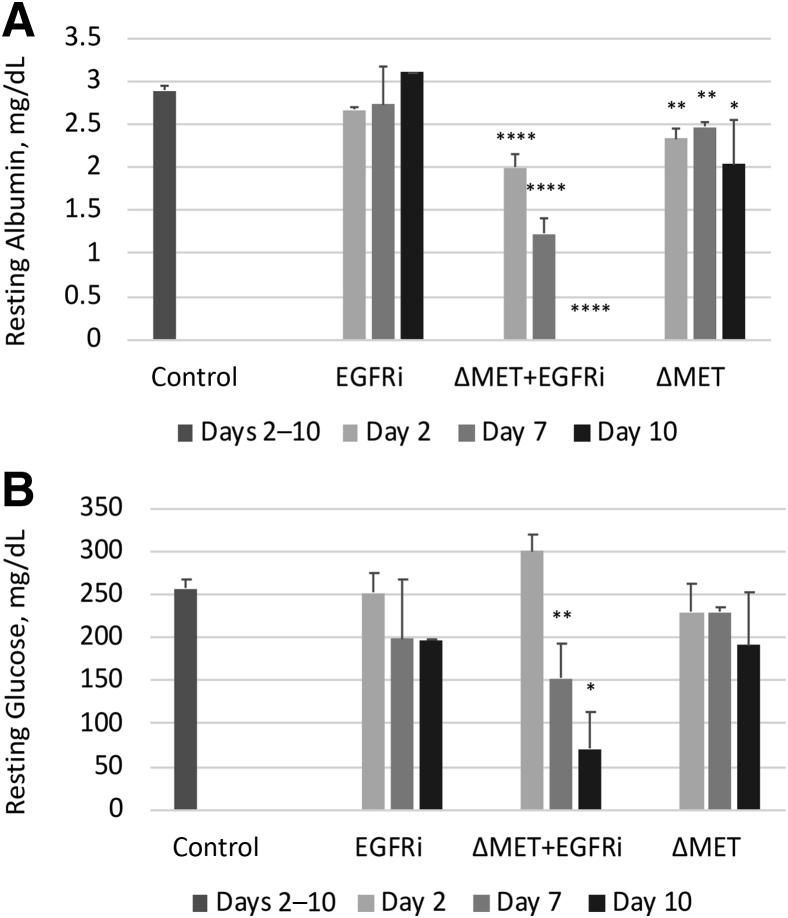
Next, it was determined if these changes in cell turnover and volume were associated with alterations in some of the essential metabolic parameters controlled by the liver. Serum albumin levels were deeply decreased in the ΔMET + EGFRi mice, reaching <0.001 mg/dL by day 10 compared with normal (average, 2.9 mg/dL) (Figure 3A). In the EGFRi mice, the albumin levels at all three time points were close to normal (averages ranged from 2.7 to 3.1 mg/dL). Interestingly, at all time points for the ΔMET only mice, there was a decrease in albumin, although small (average values ranged from 2.0 to 2.5 mg/dL), it was statistically significant. In contrast, serum glucose levels in both the EGFRi and ΔMET only cohorts were in the same range as the glucose levels observed in the control animals; however, strikingly, in the ΔMET + EGFRi mice, relative hypoglycemia was observed by day 7 that was further reduced by day 10 (Figure 3B). Subsequent testing for serum insulin levels in the day 7 and 10 ΔMET + EGFRi animals indicated that insulin levels were not significantly altered from normal. Serum parameters for alanine aminotransferase, aspartate aminotransferase, and bilirubin were also normal and similar across all groups, indicating these changes were independent of hepatic damage. Finally, staining for fat deposition by Oil Red O was unremarkable, with the exception of the day 10 ΔMET animals, in which mild microvesicular deposits were noted (data not shown).
Figure 3.
Levels of circulating albumin and blood glucose in EGFRi, ΔMET + EGFRi, and ΔMET mice. A: Serum albumin levels in all three treatment groups and control. B: Serum glucose levels in all three treatment groups and control. Data are expressed as means ± SEM (A and B). n = 3 (A, for each group, with two exceptions); n = 1 (A, EGFRi day 10); n = 2 (A, ΔMET + EGFRi day 10). ∗P ≤ 0.05, ∗∗P ≤ 0.01, and ∗∗∗∗P ≤ 0.0001 versus controls.
Receptor Activation in ΔMET, EGFRi, and ΔMET + EGFRi Mice
To determine the timing of the mechanisms leading to the observed biological changes, the activation of MET and EGFR was next examined in various cohorts. Normally, after HGF binding, MET receptor conformation is altered, the receptors dimerize, and a transient trans-phosphorylation of the two catalytic tyrosine residues (Y1234 and Y1235) occurs within the activation loop.23 Subsequently, this causes tyrosine residues Y1349 and Y1356 at the docking sites in the carboxy-terminal tail to become phosphorylated and recruit the signaling effectors.24 This latter phosphorylation drives the biological activity of the MET receptor.25 EGFR is more complex because it can be activated variably by many ligands, including EGF and transforming growth factor-α. On activation through these growth factor ligands, EGFR forms active homodimers26 or heterodimers through pairing with other ErbB receptor family members. As a result, autophosphorylation at various tyrosine residues occurs, activating downstream signaling pathways. In EGFR, similar to Y1049 in MET, Y1068 is the site where signaling effectors bind.
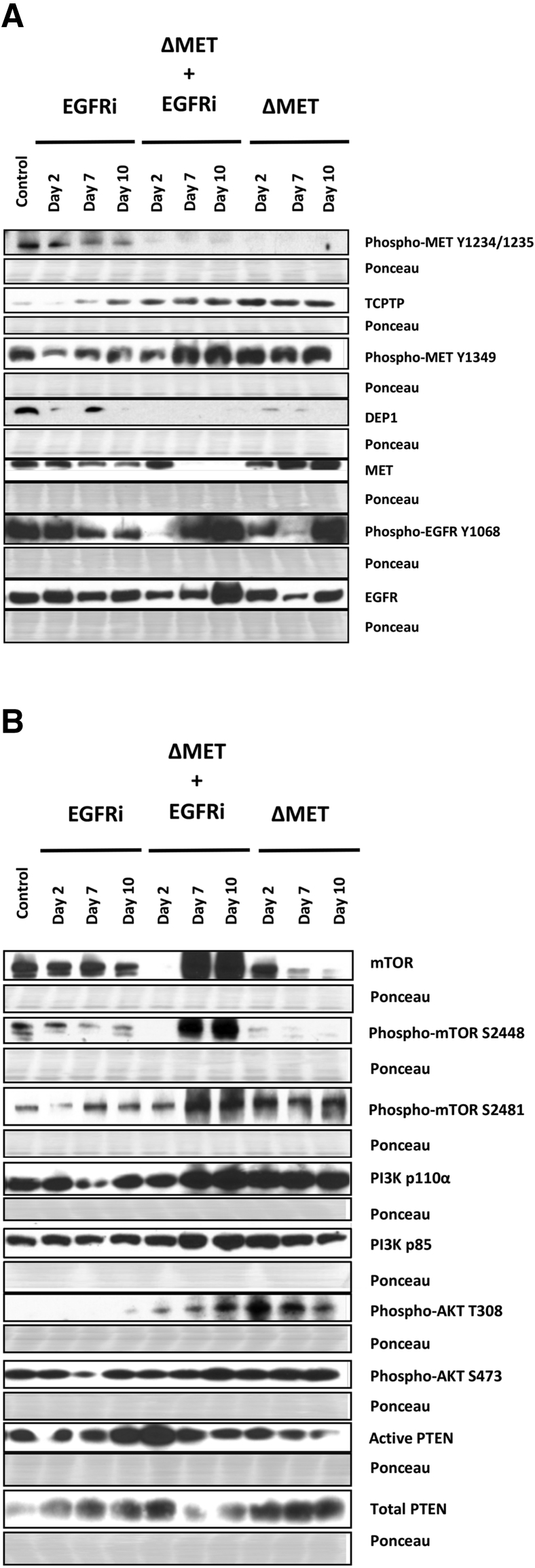
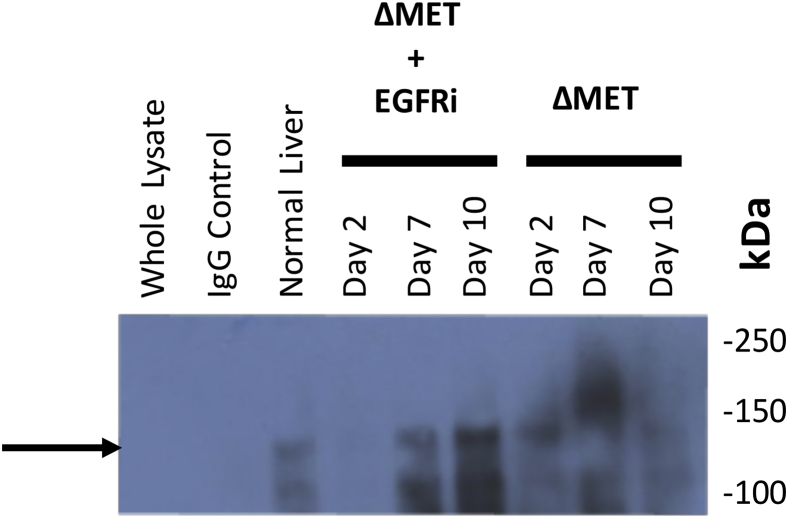
As anticipated, phospho-MET Y1234/1235 was greatly decreased in ΔMET mice at all time points compared with control and EGFRi mice, indicating that active transient signaling through MET was diminishing over time; however, surprisingly, phospho-MET Y1349 remained active and even increased in ΔMET mice (Figure 4). It is possible that as the gene was being deleted with tamoxifen, the effector portion of the gene, phospho-MET Y1349, that was previously activated by phospho-MET Y1234/1235 (see resting normal liver) remained active longer than the transiently activated Tyr1234/1235. Phosphatases are known to tightly regulate MET phosphorylation; the receptor-like protein tyrosine phosphatase DEP1 (alias PTPRJ) and the T-cell protein tyrosine phosphatase dephosphorylate Y134927 and Y1234/Y1235,28 respectively. Western blot analyses showed that DEP1 was almost completely absent when Y1349 remained phosphorylated, whereas T-cell protein tyrosine phosphatase was consistently elevated in the ΔMET mice when phosphorylation at Y1234/1235 was deficient. Interestingly, in the EGFRi mice, an inverse relationship was also observed over time for both phosphatases, relative to the phosphorylation status of their corresponding MET receptor target sites. When the total amount of MET protein was examined, an overall increase in total MET protein was observed over time in the ΔMET only mice. In contrast, in EGFRi only and ΔMET + EGFRi mice, the total amount of MET protein decreased over time, with little discernible MET protein as early as day 7 in the ΔMET + EGFRi mice. Despite the decrease of MET detectable by Western blot in the ΔMET + EGFRi samples, indicating consumption of the MET protein, some MET was, in fact, still detected using immunoprecipitation experiments to enrich for MET-Y1349, before probing for total MET (Supplemental Figure S1).
Figure 4.
EGFR, MET, and AKT signaling pathways in EGFRi, ΔMET + EGFRi, and ΔMET mice. A: Western immunoblot analysis of key signaling proteins and their phosphorylated (activated) derivatives downstream of MET and EGFR signaling pathways. B: The phosphatidylinositol 3-kinase (PI3K)/AKT effector pathway. Ponceau staining was used as a loading control; regions shown are at the molecular weight region of the protein shown in the Western blot immediately above. mTOR, mammalian target of rapamycin; PTEN, phosphatase and tensin homolog; TCPTP, T-cell protein tyrosine phosphatase.
The EGFR receptor was examined using an antibody against the EGFR Grb2 binding site (Y1068). As anticipated, EGFRi mice exhibited a decrease in phosphorylation of phospho-EGFR at Y1068 over time compared with control; however, the levels of EGFR protein remained constant. In the ΔMET + EGFRi mice, although phospho-EGFR Y1068 was almost absent at day 2, there was a subsequent increase in phosphorylation over time, with the strongest signal observed at day 10. This is likely because EGFR dephosphorylation at this site is also under the control of DEP1.29 This increase in phosphorylation was reflected by an overall increase in total EGFR protein, with the highest levels also observed at day 10. In the ΔMET only mice, although the levels of Y1068 fluctuated, they fluctuated in tandem with the levels of total EGFR. Combined, these data further support our previous findings that the functions of the MET and EGFR receptors are intimately related.30, 31
Downstream Signaling in ΔMET, EGFRi, and ΔMET + EGFRi Mice
To further understand why there was signaling interference despite the maintenance of markers indicating receptor activation, changes were determined in potential downstream targets of EGFR and MET. A classic pathway common to both receptors that is involved in cell growth and survival is PI3K/AKT/mTOR. In the canonical pathway, when PI3K is active, it generates phosphatidylinositol (3,4,5)-trisphosphate (PIP3), which binds to AKT, making it amenable to phosphorylation by PDK1 at T308, and eventually facilitating phosphorylation of mTOR at S2448 via ribosomal protein S6 kinase (S6K). mTOR activity is performed in two complexes: mTOR complex (mTORC) 1 and mTORC2. S2448 is predominantly found in the mTORC1 complex and is associated with cell growth, whereas phosphorylation of mTOR at S2481 is indicative of mTORC2 and associated with cell survival. The mTORC2 complex can further facilitate activation of AKT via phosphorylation at S473. PTEN negatively regulates induction of the AKT pathway by converting PIP3 to PIP2, so that AKT is unable to signal.
Phospho-mTOR S2448 (representing activated mTORC1 complex and cell growth) decreased over time in the ΔMET only mice; however, in mice treated with canertinib, the pattern mimicked Y1068 EGFR, with a great elevation in the ΔMET + EGFRi mice at days 7 and 10. Interestingly, phospho-mTOR S2481 (representing activated mTORC2 complex and cell survival) followed a pattern similar to Y1349 MET that was independent of canertinib treatment. The latter mTOR protein also paralleled the same expression patterns as PI3K (both the regulatory p85 and catalytic p110α subunits), phospho-AKT T308, and phospho-AKT S473; all were also increased in both the ΔMET and ΔMET + EGFRi mice, relative to control. Levels of active (dephosphorylated) PTEN were fairly constant over the 10-day time period in the EGFRi and ΔMET only mice; however, in the ΔMET + EGFRi cohort, there was an initial increase at day 2 that steadily declined over time.
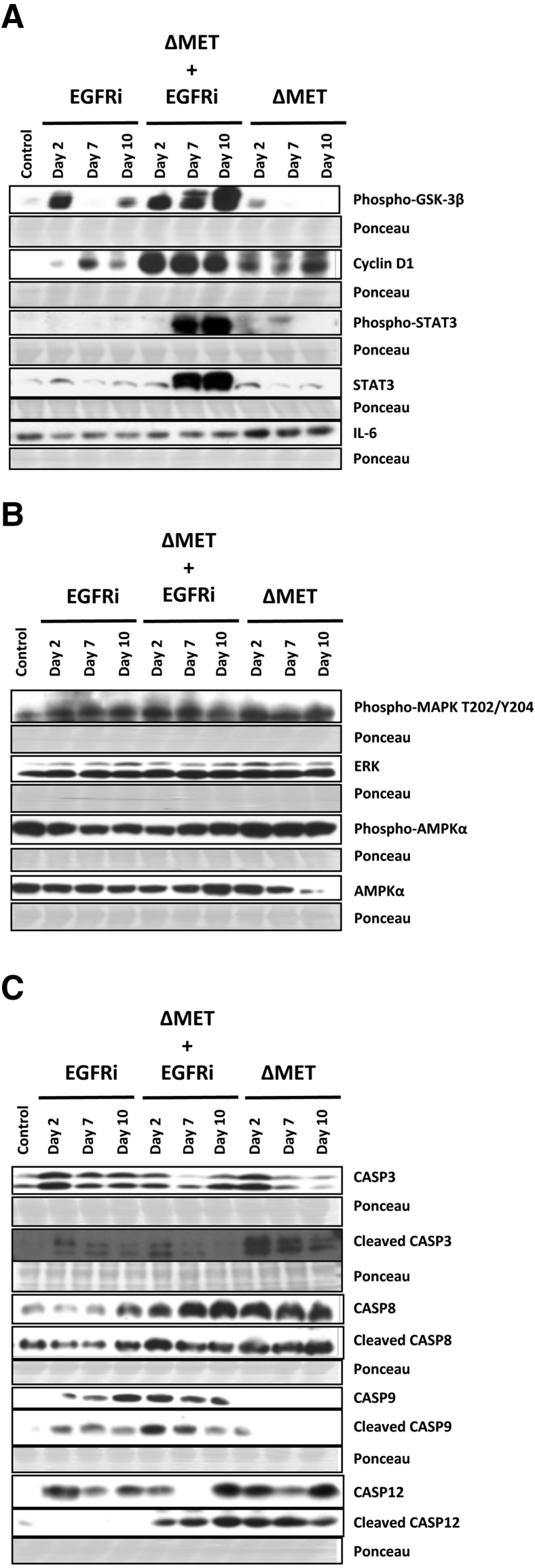
As noted above, various members of the PI3K-AKT pathway are associated with either cell growth and/or cell survival. With regard to the former, PI3K-AKT is known to positively regulate G1/S cell cycle progression via phosphorylation and inactivation of GSK-3β.32 In the ΔMET + EGFRi mice, GSK-3β was inactive (phosphorylated), whereas cyclin D1, STAT3, and phosphorylated STAT3 were all increased (Figure 5A). Interestingly, in the ΔMET mice, although GSK-3β was unaffected, cyclin D1 was also increased, along with IL-6, a protein generally associated with cell survival that, in stimulated livers, can be made by hepatocytes as well as macrophages.21 p44/42 Mitogen-activated protein kinase (extracellular signal-regulated kinase 1/2) levels, often elevated during active growth and/or during cell migration, were not greatly affected in any of the mouse categories (Figure 5B). Phospho-AMPKα, an indicator of cell stress and ATP depletion,33 was decreased in the EGFRi and ΔMET + EGFRi mice, relative to total AMPKα levels, whereas the ratio was increased in the ΔMET only mice.
Figure 5.
Signaling proteins related to key cell growth, survival, migration, stress, and death pathways in EGFRi, ΔMET + EGFRi, and ΔMET mice. A: Western immunoblot analysis of glycogen synthetase kinase (GSK)-3β, cyclin D1, STAT3, and IL-6 signaling proteins. B: Western immunoblot analysis of mitogen-activated protein kinase (MAPK) and AMPKα. C: Western immunoblot analysis of various caspases. Ponceau staining was used as a loading control; regions shown are at the molecular weight region of the protein shown in the Western blot immediately above. ERK, extracellular signal-regulated kinase.
Finally, the role of caspase-induced apoptosis was explored in EGFRi and/or ΔMET mice. The classic (Fas-induced) apoptosis pathway was seemingly activated in all ΔMET mice, as demonstrated by the increased expression of caspase 8 and cleaved caspase 8 (Figure 5C). On the other hand, in all EGFRi mice, the mitochondrial apoptosis pathway prevailed, as indicated by an increased activation of caspase 9 and cleaved caspase 9. Caspase 12 has been implicated in endoplasmic reticulum stress–induced apoptotic pathways34; cleaved caspase 12 was also increased in all mice with ΔMET. Hence, in the ΔMET + EGFRi mice, all three pathways of apoptosis were simultaneously activated, potentially providing an explanation for the increase in TUNEL staining shown in Figure 1. Interestingly, cleaved caspase 3 was initially elevated at day 2 in the EGFRi, ΔMET + EGFRi, and ΔMET mice, before receding.
Gene Array Analysis
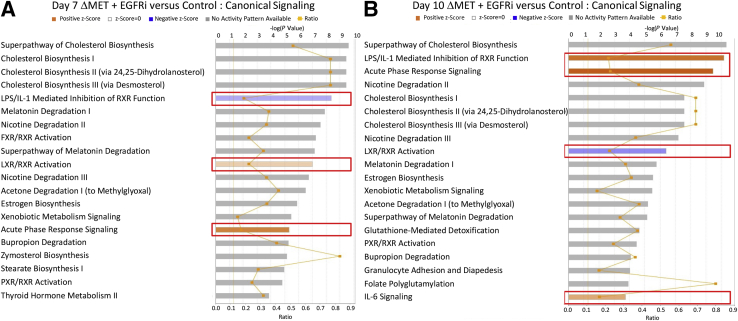
Because the animals did not show severe distress until day 10, RNA changes occurring between days 7 and 10 in the ΔMET + EGFRi mice may provide further insight as to increased morbidity. Gene array analyses were performed, and two approaches were taken to analyze the data of the array (Materials and Methods). First, IPA was used to help identify the top 20 canonical signaling pathways that differed between ΔMET + EGFRi mice and controls at days 7 and 10. Many of the same pathways were commonly affected at days 7 and 10, such as the pathway of cholesterol biosynthesis, which was the most significantly changed pathway at both time points; however, in most cases, there was no clear direction regarding an activation or inhibition of the signaling pathway (Figure 6). There were, however, some notable exceptions. At day 7, the lipopolysaccharide/IL-1 pathway inhibiting retinoid X receptor (RXR) function was clearly inhibited, whereas the liver X receptor (LXR)/RXR and acute phase response pathways were both activated. On the other hand, by day 10, both the lipopolysaccharide/IL-1 inhibition of RXR function and acute phase response pathways were activated, suggesting a further increase in inflammatory-mediated signaling, whereas the LXR/RXR pathway was instead inhibited. Interestingly, the farnesoid X receptor (FXR)/RXR pathway was also only in the top 20 for the day 7 samples, indicating suppression of RXR signaling by induction of the lipopolysaccharide/IL-6 inhibition of RXR function pathway is likely to be important as the animals begin to succumb. Upstream regulator analysis was performed on array data using IPA to predict the most significant transcription factors that were either activated or inhibited at day 10. Many of the pathways that are predicted to be altered provide a good confirmation of the other data (Table 1). For example, inhibition of the hepatic nuclear factor 4 alpha (HNF4A), peroxisome proliferator-activated receptor, and LXR transcription factor pathways would not be an unexpected finding with the severe metabolic changes observed in these animals35 (Figure 3), whereas the increase in activated STAT3 observed by Western blot analyses (Figure 5A) was verified at the RNA level as well.
Figure 6.
Analysis of global gene expression profile in ΔMET + EGFRi mice using Ingenuity Pathway Analysis (IPA). IPA was used to identify the top altered canonical signaling pathways in ΔMET + EGFRi mice at days 7 (A) and 10 (B) compared with normal controls. The top 20 significantly altered pathways are depicted for each. The orange (positive z-score) and blue (negative z-score) colored bars indicate predicted pathway activation or predicted inhibition, respectively, on the basis of z-scores. Boxed areas identify pathways that are consistently changed in either a positive or a negative direction. Gray bars indicate pathways where no prediction on directionality of activity can be made. Primary y axis displays negative logarithm of P value calculated by Fisher's exact test (threshold for –log P value was 1.3). Ratio in the secondary y axis represents number of genes in a given pathway in the data set, divided by the total number of genes that make up that pathway and that are in the reference gene set. FXR, farnesoid X receptor; LPS, lipopolysaccharide; LXR: liver X receptor; PXR: pregnane X receptor; RXR: retinoid X receptor.
Table 1.
Top Transcription Factors Predicted to be Inhibited/Activated in ΔMET + EGFRi (Day 10) Compared with Control Group Analyzed Using the Upstream Regulator Analysis Feature of Ingenuity Pathway Analysis
| Upstream regulator | Expression ratio | Predicted activation state | Activation z-score | P value of overlap |
|---|---|---|---|---|
| HNF4A | Inhibited | −3.276 | 1.04 × 10−5 | |
| LHX1 | Inhibited | −3.023 | 2.13 × 10−3 | |
| ZBTB20 | −2.16 | Inhibited | −3 | 2.66 × 10−8 |
| PPARG | Inhibited | −2.95 | 6.31 × 10−8 | |
| PPARA | −2.16 | Inhibited | −2.5 | 2.32 × 10−28 |
| PPARGC1A | 3.69 | Inhibited | −2.428 | 3.09 × 10−4 |
| ZFP36 | Inhibited | −2.392 | 2.17 × 10−3 | |
| MLXIPL | Inhibited | −2.254 | 2.35 × 10−5 | |
| BCL3 | 3.44 | Inhibited | −2.2 | 3.62 × 10−2 |
| TP73 | Inhibited | −2.041 | 4.97 × 10−2 | |
| NR1H2 (LXRβ) | Inhibited | −2.022 | 2.62 × 10−5 | |
| STAT 3 | 3.62 | Activated | 4.342 | 2.72 × 10−7 |
| XBP1 | 3.36 | Activated | 3.494 | 7.32 × 10−8 |
| EGR1 | Activated | 2.95 | 1.6 × 10−3 | |
| NFKB1 | Activated | 2.815 | 3.11 × 10−4 | |
| SPI1 | Activated | 2.738 | 5.19 × 10−4 | |
| STAT 1 | Activated | 2.621 | 1.24 × 10−3 | |
| CREB1 | Activated | 2.565 | 6.38 × 10−3 | |
| PDX1 | Activated | 2.492 | 6.79 × 10−3 | |
| STAT 4 | Activated | 2.445 | 6.9 × 10−5 | |
| GATA1 | Activated | 2.373 | 5.68 × 10−3 | |
Top transcription factors whose signaling is predicted to be activated at day 10.
Data on gene expression from Affymetrix arrays were used to assess the signature (expression of transcription factor–dependent signaling proteins) downstream of specific transcription factors. The overall effect on the expression of dependent signaling proteins for each transcription factor is expressed by z-score, with positive z-score representing predicted activation and negative z-score representing predicted inhibition of transcription factor activity (absolute z-score > 2 considered as significant). P values signify the extent of overlap between set of downstream target genes of a given transcription factor in data set compared with all known downstream target genes of a given transcription factor in reference genome. Expression ratio represents actual gene expression (fold change of ΔMET + EGFRi versus control) of the transcription factor (specified if greater than twofold change in the absolute value).
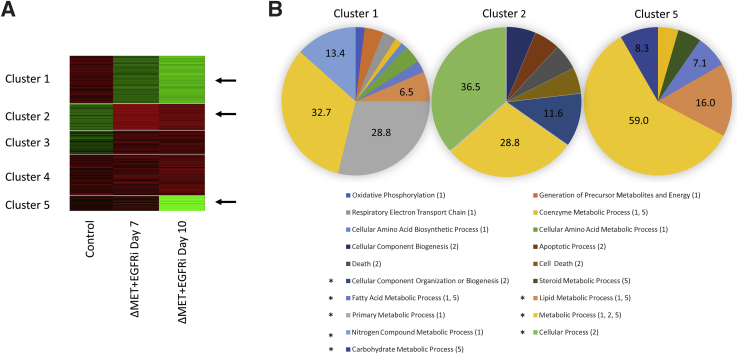
Gene clustering analysis was also performed (Materials and Methods). Gene expression analyses of microarray data in ΔMET + EGFRi mice at days 7 and 10 versus control mice revealed five tight gene clusters (Figure 7A). Each gene cluster is represented in a row block, with column 1 representing control and columns 2 and 3 representing days 7 and 10, respectively. Cluster 1 showed a gradual decrease in gene expression over time versus control, whereas clusters 2 and 3 went from low to high expression. Notably, cluster 5 greatly decreased from day 7 to day 10 in the ΔMET + EGFRi mice, suggesting the loss of genes within this cluster might be an indicator of impending death. To determine the importance of the genes in these various clusters, PANTHER gene ontology analyses were performed to predict biological processes significantly represented as participants in these clusters (Table 2). Only clusters 1, 2, and 5 showed obvious differences in several categories (Table 2 and Figure 7B). Genes related to metabolic processes were represented to a great degree in all three clusters, whereas cluster 2 was uniquely enriched for genes related to cellular process and cell death. Interestingly, cluster 1 was uniquely enriched for genes related to oxidative phosphorylation and respiratory electron transport chain (Supplemental Table S1).
Figure 7.
Gene tight clustering analysis of gene expression in ΔMET + EGFRi mice. A: Heat map identifying five clusters where there was either increased RNA expression (red) or decreased expression (green) of a subset of genes, relative to control. B: PANTHER gene ontology analysis identifying uppermost biological processes identified in clusters 1, 2, and 5 (arrows in A) compared with Mus musculus reference genome. An asterisk on the left of the process indicates there was a minimum of at least 6.5% of the genes in one or more of the three clusters. Numbers in parenthesis indicate which of the clusters had genes in the identified pathways.
Table 2.
Biological Process (GO Terms) Significantly Enriched in Clusters 1, 2, and 5 (Top to Bottom) Analyzed Using PANTHER Overrepresentation Test and Compared with the Mus musculus Reference List
| PANTHER GO biological process | Reference gene no. | Cluster gene no. | Fold enrichment | P value |
|---|---|---|---|---|
| Cluster 1 | ||||
| Oxidative phosphorylation | 48 | 10 | 11.62 | 5.97 × 10−6 |
| Generation of precursor metabolites and energy | 161 | 21 | 7.28 | 9.11 × 10−10 |
| Respiratory electron transport chain | 124 | 15 | 6.75 | 2.95 × 10−6 |
| Coenzyme metabolic process | 67 | 8 | 6.66 | 8.26 × 10−3 |
| Cellular amino acid biosynthetic process | 61 | 7 | 6.4 | 3.23 × 10−2 |
| Cellular amino acid metabolic process | 222 | 20 | 5.03 | 1.60 × 10−6 |
| Fatty acid metabolic process | 210 | 13 | 3.45 | 3.25 × 10−2 |
| Lipid metabolic process | 620 | 33 | 2.97 | 1.04 × 10−5 |
| Primary metabolic process | 4367 | 146 | 1.87 | 5.67 × 10−13 |
| Metabolic process | 5460 | 166 | 1.7 | 1.25 × 10−11 |
| Nitrogen compound metabolic process | 2357 | 68 | 1.61 | 1.45 × 10−2 |
| Cluster 2 | ||||
| Cellular component biogenesis | 496 | 18 | 3.87 | 3.18 × 10−4 |
| Apoptotic process | 445 | 16 | 3.83 | 1.40 × 10−3 |
| Death | 466 | 16 | 3.66 | 2.47 × 10−3 |
| Cell death | 466 | 16 | 3.66 | 2.47 × 10−3 |
| Cellular component organization or biogenesis | 1655 | 33 | 2.12 | 8.11 × 10−3 |
| Metabolic process | 5460 | 82 | 1.6 | 4.33 × 10−4 |
| Cellular process | 8341 | 104 | 1.33 | 4.54 × 10−2 |
| Cluster 5 | ||||
| Coenzyme metabolic process | 67 | 7 | 9.12 | 3.50 × 10−3 |
| Steroid metabolic process | 120 | 8 | 5.82 | 2.04 × 10−2 |
| Fatty acid metabolic process | 210 | 11 | 4.57 | 8.98 × 10−3 |
| Lipid metabolic process | 620 | 25 | 3.52 | 1.73 × 10−5 |
| Carbohydrate metabolic process | 340 | 13 | 3.34 | 4.27 × 10−2 |
| Metabolic process | 5460 | 92 | 1.47 | 6.01 × 10−3 |
Statistical analysis was performed using Fisher's exact test with false discovery rate multiple test correction.
GO, gene ontology.
Discussion
The combined disruption of the MET and EGFR signaling pathways abolishes liver regeneration after PHx.11 This finding was associated with suppression of a variety of specific pathways that are normally associated with effective liver regeneration. In this study, it has been demonstrated that the unanticipated finding that combined interference with both MET and EGFR signaling is itself sufficient to result in the death of these animals, even when the animals are not subjected to loss of hepatic mass through PHx. Although the cause of death may be multifactorial, a severe depression of circulating albumin and glucose levels was observed (Figure 3), suggesting there was a complete shutdown of normal liver function despite the absence of overt hepatocellular damage. Many downstream signaling changes in the resting livers were different from those elicited in the ΔMET + EGFRi mouse livers after PHx. For example, previously, the AKT/PI3K pathway was suppressed after PHx, whereas in the present study, it was instead elevated. Hence, induction of the PI3K survival pathway was insufficient for sustained hepatic function.11
Although the persistence of phosphorylated MET at Y1349 in this study is intriguing, it correlates with a decrease in at least one of the phosphatases (DEP1) known to dephosphorylate this site. Regulation of MET phosphorylation at Y1349 is also known to be controlled by other parameters besides activation by Y1234/1235. A recent study demonstrated that phosphorylation of MET Y1349 is directly dependent on SRC kinase activity,36 and there is a well-studied relationship between the EGFR, MET, and SRC phosphorylation pathways in cancer.37
In the individual ΔMET and EGFRi animals, there was an induction of differing apoptotic pathways (Figure 5C). At day 2, in ΔMET animals, there was an increase in cleaved caspases 8 and 12, whereas the EGFRi animals had an increase in cleaved caspase 9. In both cases, there was also an initial elevation in cleaved caspase 3 at day 2 that gradually decreased over time. Interestingly, although the caspase 8, 9, and 12 pathways were persistently elevated in the ΔMET + EGFRi animals (Figure 5C), caspase 3 remained near baseline levels for all three time points. Staining for TUNEL reflected these findings; although there was a small increase in hepatocellular apoptosis with the individual and combined treatments (Figure 1C), most cell death was observed in the NPCs (Figure 1D), indicating that hepatocellular death is not the reason for hepatic failure. On the other hand, the severe loss and altered functions in NPCs may be a crucial contributor to altered hepatocyte and overall hepatic loss of function. Multiple studies have determined that NPC interactions with hepatocytes are key to normal hepatocellular function,38, 39 although signaling exchanges between hepatocytes and nonparenchymal cells are complex and it is beyond the scope of this study to specifically determine which pathways in NPCs were especially disturbed. Notably, the gene array analyses also identified increased expression of mRNAs of genes related to cell death at day 10 (Figure 7B).
The severe decrease in both serum albumin and glucose also likely contributed to animal death (Figure 3). For albumin, in the ΔMET only animals, there was already a decrease in albumin levels, suggesting activation of the acute phase response, before administration of the canertinib; the simultaneous increase of IL-6 production in these ΔMET only animals (Figure 5A) is another indicator of an already activated acute phase response.21 Although IL-6 returned to baseline levels after the administration of canertinib, the albumin levels continued to steadily decline until albumin was virtually undetectable in the blood, suggesting there was a continued activation of the acute phase response. Consistent with this interpretation is the finding that both Western blot (Figure 5A) and IPA analyses (Table 1) display significant activation of STAT3. The decrease in serum albumin also correlates with the ascites that progressively increased in the affected mice.
The loss of glucose observed is consistent with the aberrant changes in metabolism that the gene array analyses uncovered. With regard to glucose specifically, the decrease in glucose primarily coincided with the loss of GSK-3β activity (ie, phosphorylation of GSK-3β) over time (Figure 5A). Normally, active (nonphosphorylated) GSK-3β phosphorylates glycogen synthase, rendering the enzyme inactive, so that glucose is not converted to glycogen. Hence, loss of GSK-3β activity leads to constitutively active glycogen synthase, consumption of glucose, and concomitant production of glycogen.40 However, when the presence of increased glycogen was tested, the levels were not altered in these animals (data not shown), indicating that either glycogen is not being produced or, alternatively, that any glycogen initially synthesized is subsequently used because of an overactive metabolism. In this regard, it is notable that the AKT/PI3K pathway, which gradually becomes active in the ΔMET + EGFRi mice (Figure 5B), is able to phosphorylate and inactivate GSK-3β41 and that the PI3K pathway is, itself, downstream of MET Y1349 and EGFR Y1068.
These results present new insights into pathways that lead to liver failure. The role and function of these two receptors in conjunction with liver failure in a variety of toxic or infectious conditions have not been assessed. Although there are multiple pathways that may be affected, normally, hepatocytes are constantly exposed to high levels of both HGF and EGF. HGF is present in high concentrations in the hepatic extracellular matrix,42 whereas EGF is constantly supplied to the liver through the portal circulation, produced by the Brunner glands of the duodenum.43 Both growth factors deliver a constant signaling to their receptors, as indicated by the activated state of both MET (phospho-MET Y1234/1235, phospho-MET Y1349) and EGFR (phospho-EGFR Y1068) in all control animals. A fine balance in the combined signaling of the two receptors is fundamental for normal hepatic function, and signaling from other pathways is not sufficient to maintain the viability of mice when there is interference in these two crucial signaling pathways.
Acknowledgments
We thank Gina Coudriet and Jon Piganelli for performing the mouse insulin assays and Dr. Snorri Thorgeirsson for providing METff/ff mice with a targeted deletion of c-Met exon 16.
Footnotes
Supported by the NIH/National Institute of Diabetes and Digestive and Kidney Diseases Program Project grant P01 DK096690 (G.K.M.), the Cleveland Foundation (G.K.M.), and the Menten Endowment Foundation of the University of Pittsburgh (G.K.M.).
A.T. and W.M.M. contributed equally to this work.
Disclosures: None declared.
Supplemental material for this article can be found at https://doi.org/10.1016/j.ajpath.2018.06.009.
Supplemental Data
Supplemental Figure S1.
Samples were immunoprecipitated using a polyclonal antibody against MET p-Y1349 and then probed with a monoclonal antibody against MET. The first three lanes from left to right were from the same pool of normal liver lysates, with 10 μg of whole lysate (not immunoprecipitated) used in lane 1 as a control. Lane 2 is an immunoprecipitation control using normal IgG. The black arrow points to the precipitated MET β subunit (approximately 145 kDa).
References
- 1.Block G.D., Locker J., Bowen W.C., Petersen B.E., Katyal S., Strom S.C., Riley T., Howard T.A., Michalopoulos G.K. Population expansion, clonal growth, and specific differentiation patterns in primary cultures of hepatocytes induced by HGF/SF, EGF and TGF alpha in a chemically defined (HGM) medium. J Cell Biol. 1996;132:1133–1149. doi: 10.1083/jcb.132.6.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paranjpe S., Bowen W.C., Tseng G.C., Luo J.H., Orr A., Michalopoulos G.K. RNA interference against hepatic epidermal growth factor receptor has suppressive effects on liver regeneration in rats. Am J Pathol. 2010;176:2669–2681. doi: 10.2353/ajpath.2010.090605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Natarajan A., Wagner B., Sibilia M. The EGF receptor is required for efficient liver regeneration. Proc Natl Acad Sci U S A. 2007;104:17081–17086. doi: 10.1073/pnas.0704126104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stolz D.B., Mars W.M., Petersen B.E., Kim T.H., Michalopoulos G.K. Growth factor signal transduction immediately after two-thirds partial hepatectomy in the rat. Cancer Res. 1999;59:3954–3960. [PubMed] [Google Scholar]
- 5.Uehara Y., Minowa O., Mori C., Shiota K., Kuno J., Noda T., Kitamura N. Placental defect and embryonic lethality in mice lacking hepatocyte growth factor/scatter factor. Nature. 1995;373:702–705. doi: 10.1038/373702a0. [DOI] [PubMed] [Google Scholar]
- 6.Carver R.S., Stevenson M.C., Scheving L.A., Russell W.E. Diverse expression of ErbB receptor proteins during rat liver development and regeneration. Gastroenterology. 2002;123:2017–2027. doi: 10.1053/gast.2002.37060. [DOI] [PubMed] [Google Scholar]
- 7.Michalopoulos G.K. Liver regeneration. J Cell Physiol. 2007;213:286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shiota G., Wang T.C., Nakamura T., Schmidt E.V. Hepatocyte growth factor in transgenic mice: effects on hepatocyte growth, liver regeneration and gene expression. Hepatology. 1994;19:962–972. [PubMed] [Google Scholar]
- 9.Yamada Y., Kirillova I., Peschon J.J., Fausto N. Initiation of liver growth by tumor necrosis factor: deficient liver regeneration in mice lacking type I tumor necrosis factor receptor. Proc Natl Acad Sci U S A. 1997;94:1441–1446. doi: 10.1073/pnas.94.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cressman D.E., Greenbaum L.E., DeAngelis R.A., Ciliberto G., Furth E.E., Poli V., Taub R. Liver failure and defective hepatocyte regeneration in interleukin-6-deficient mice. Science. 1996;274:1379–1383. doi: 10.1126/science.274.5291.1379. [DOI] [PubMed] [Google Scholar]
- 11.Paranjpe S., Bowen W.C., Mars W.M., Orr A., Haynes M.M., DeFrances M.C., Liu S., Tseng G.C., Tsagianni A., Michalopoulos G.K. Combined systemic elimination of MET and epidermal growth factor receptor signaling completely abolishes liver regeneration and leads to liver decompensation. Hepatology. 2016;64:1711–1724. doi: 10.1002/hep.28721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taub R., Greenbaum L.E., Peng Y. Transcriptional regulatory signals define cytokine-dependent and -independent pathways in liver regeneration. Semin Liver Dis. 1999;19:117–127. doi: 10.1055/s-2007-1007104. [DOI] [PubMed] [Google Scholar]
- 13.Servillo G., Penna L., Foulkes N.S., Magni M.V., Della Fazia M.A., Sassone-Corsi P. Cyclic AMP signalling pathway and cellular proliferation: induction of CREM during liver regeneration. Oncogene. 1997;14:1601–1606. doi: 10.1038/sj.onc.1200996. [DOI] [PubMed] [Google Scholar]
- 14.Behrens A., Sibilia M., David J.P., Mohle-Steinlein U., Tronche F., Schutz G., Wagner E.F. Impaired postnatal hepatocyte proliferation and liver regeneration in mice lacking c-jun in the liver. EMBO J. 2002;21:1782–1790. doi: 10.1093/emboj/21.7.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taub R. Liver regeneration 4: transcriptional control of liver regeneration. FASEB J. 1996;10:413–427. [PubMed] [Google Scholar]
- 16.Michalopoulos G.K., DeFrances M.C. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 17.Fausto N. Liver regeneration. J Hepatol. 2000;32:19–31. doi: 10.1016/s0168-8278(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 18.Huh C.G., Factor V.M., Sanchez A., Uchida K., Conner E.A., Thorgeirsson S.S. Hepatocyte growth factor/c-met signaling pathway is required for efficient liver regeneration and repair. Proc Natl Acad Sci U S A. 2004;101:4477–4482. doi: 10.1073/pnas.0306068101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu J., Chen W., Xia G., Zhang J., Shao J., Tan B., Zhang C., Yu W., Weng Q., Liu H., Hu M., Deng H., Hao Y., Shen J., Yu Y. Design, synthesis, and biological evaluation of novel conformationally constrained inhibitors targeting EGFR. ACS Med Chem Lett. 2013;4:974–978. doi: 10.1021/ml4002437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Academies Press; Washington, DC: 2011. Committee for the Update of the Guide for the Care and Use of Laboratory Animals; National Research Council: Guide for the Care and Use of Laboratory Animals: Eighth Edition. [Google Scholar]
- 21.Norris C.A., He M., Kang L.I., Ding M.Q., Radder J.E., Haynes M.M., Yang Y., Paranjpe S., Bowen W.C., Orr A., Michalopoulos G.K., Stolz D.B., Mars W.M. Synthesis of IL-6 by hepatocytes is a normal response to common hepatic stimuli. PLoS One. 2014;9:e96053. doi: 10.1371/journal.pone.0096053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tseng G.C., Wong H.W. Tight clustering: a resampling-based approach for identifying stable and tight patterns in data. Biometrics. 2005;61:10–16. doi: 10.1111/j.0006-341X.2005.031032.x. [DOI] [PubMed] [Google Scholar]
- 23.Trusolino L., Bertotti A., Comoglio P.M. MET signalling: principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol. 2010;11:834–848. doi: 10.1038/nrm3012. [DOI] [PubMed] [Google Scholar]
- 24.Organ S.L., Tsao M.S. An overview of the c-MET signaling pathway. Ther Adv Med Oncol. 2011;3:S7–S19. doi: 10.1177/1758834011422556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ponzetto C., Bardelli A., Zhen Z., Maina F., dalla Zonca P., Giordano S., Graziani A., Panayotou G., Comoglio P.M. A multifunctional docking site mediates signaling and transformation by the hepatocyte growth factor/scatter factor receptor family. Cell. 1994;77:261–271. doi: 10.1016/0092-8674(94)90318-2. [DOI] [PubMed] [Google Scholar]
- 26.Yarden Y., Schlessinger J. Epidermal growth factor induces rapid, reversible aggregation of the purified epidermal growth factor receptor. Biochemistry. 1987;26:1443–1451. doi: 10.1021/bi00379a035. [DOI] [PubMed] [Google Scholar]
- 27.Palka H.L., Park M., Tonks N.K. Hepatocyte growth factor receptor tyrosine kinase met is a substrate of the receptor protein-tyrosine phosphatase DEP-1. J Biol Chem. 2003;278:5728–5735. doi: 10.1074/jbc.M210656200. [DOI] [PubMed] [Google Scholar]
- 28.Sangwan V., Paliouras G.N., Abella J.V., Dube N., Monast A., Tremblay M.L., Park M. Regulation of the Met receptor-tyrosine kinase by the protein-tyrosine phosphatase 1B and T-cell phosphatase. J Biol Chem. 2008;283:34374–34383. doi: 10.1074/jbc.M805916200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tarcic G., Boguslavsky S.K., Wakim J., Kiuchi T., Liu A., Reinitz F., Nathanson D., Takahashi T., Mischel P.S., Ng T., Yarden Y. An unbiased screen identifies DEP-1 tumor suppressor as a phosphatase controlling EGFR endocytosis. Curr Biol. 2009;19:1788–1798. doi: 10.1016/j.cub.2009.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Runge D.M., Runge D., Dorko K., Pisarov L.A., Leckel K., Kostrubsky V.E., Thomas D., Strom S.C., Michalopoulos G.K. Epidermal growth factor- and hepatocyte growth factor-receptor activity in serum-free cultures of human hepatocytes. J Hepatol. 1999;30:265–274. doi: 10.1016/s0168-8278(99)80073-7. [DOI] [PubMed] [Google Scholar]
- 31.Presnell S.C., Stolz D.B., Mars W.M., Jo M., Michalopoulos G.K., Strom S.C. Modifications of the hepatocyte growth factor/c-met pathway by constitutive expression of transforming growth factor-alpha in rat liver epithelial cells. Mol Carcinog. 1997;18:244–255. [PubMed] [Google Scholar]
- 32.Liang J., Slingerland J.M. Multiple roles of the PI3K/PKB (Akt) pathway in cell cycle progression. Cell Cycle. 2003;2:339–345. [PubMed] [Google Scholar]
- 33.Yang M., Hao Y., Gao J., Zhang Y., Xu W., Tao L. Spinosad induces autophagy of Spodoptera frugiperda Sf9 cells and the activation of AMPK/mTOR signaling pathway. Comp Biochem Physiol C Toxicol Pharmacol. 2017;195:52–59. doi: 10.1016/j.cbpc.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 34.Ferri K.F., Kroemer G. Organelle-specific initiation of cell death pathways. Nat Cell Biol. 2001;3:E255–E263. doi: 10.1038/ncb1101-e255. [DOI] [PubMed] [Google Scholar]
- 35.Desvergne B., Michalik L., Wahli W. Transcriptional regulation of metabolism. Physiol Rev. 2006;86:465–514. doi: 10.1152/physrev.00025.2005. [DOI] [PubMed] [Google Scholar]
- 36.Baumann C., Ullrich A., Torka R. GAS6-expressing and self-sustaining cancer cells in 3D spheroids activate the PDK-RSK-mTOR pathway for survival and drug resistance. Mol Oncol. 2017;11:1430–1447. doi: 10.1002/1878-0261.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stabile L.P., He G., Lui V.W., Thomas S., Henry C., Gubish C.T., Joyce S., Quesnelle K.M., Siegfried J.M., Grandis J.R. c-Src activation mediates erlotinib resistance in head and neck cancer by stimulating c-Met. Clin Cancer Res. 2013;19:380–392. doi: 10.1158/1078-0432.CCR-12-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michalopoulos G.K., Bowen W.C., Mule K., Stolz D.B. Histological organization in hepatocyte organoid cultures. Am J Pathol. 2001;159:1877–1887. doi: 10.1016/S0002-9440(10)63034-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bale S.S., Geerts S., Jindal R., Yarmush M.L. Isolation and co-culture of rat parenchymal and non-parenchymal liver cells to evaluate cellular interactions and response. Sci Rep. 2016;6:25329. doi: 10.1038/srep25329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaidanovich-Beilin O., Eldar-Finkelman H. Long-term treatment with novel glycogen synthase kinase-3 inhibitor improves glucose homeostasis in ob/ob mice: molecular characterization in liver and muscle. J Pharmacol Exp Ther. 2006;316:17–24. doi: 10.1124/jpet.105.090266. [DOI] [PubMed] [Google Scholar]
- 41.Rayasam G.V., Tulasi V.K., Sodhi R., Davis J.A., Ray A. Glycogen synthase kinase 3: more than a namesake. Br J Pharmacol. 2009;156:885–898. doi: 10.1111/j.1476-5381.2008.00085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nejak-Bowen K., Orr A., Bowen W.C., Jr., Michalopoulos G.K. Conditional genetic elimination of hepatocyte growth factor in mice compromises liver regeneration after partial hepatectomy. PLoS One. 2013;8:e59836. doi: 10.1371/journal.pone.0059836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skov Olsen P., Boesby S., Kirkegaard P., Therkelsen K., Almdal T., Poulsen S.S., Nexo E. Influence of epidermal growth factor on liver regeneration after partial hepatectomy in rats. Hepatology. 1988;8:992–996. doi: 10.1002/hep.1840080503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.