In a whole-exome sequencing study of multiplex Alzheimer's disease (AD) families, we investigated three neuronal ceroid lipofuscinosis genes that have been linked to retromer, an intracellular trafficking pathway associated with AD: ceroid lipofuscinosis 3 (CLN3), ceroid lipofuscinosis 5 (CLN5), and cathepsin D (CTSD). We identified a missense variant in CLN5 c.A959G (p.Asn320Ser) that segregated with AD.
KEYWORDS: Alzheimer's disease, CLN5, endosomes, NCL, retromer
ABSTRACT
In a whole-exome sequencing study of multiplex Alzheimer's disease (AD) families, we investigated three neuronal ceroid lipofuscinosis genes that have been linked to retromer, an intracellular trafficking pathway associated with AD: ceroid lipofuscinosis 3 (CLN3), ceroid lipofuscinosis 5 (CLN5), and cathepsin D (CTSD). We identified a missense variant in CLN5 c.A959G (p.Asn320Ser) that segregated with AD. We find that this variant causes glycosylation defects in the expressed protein, which causes it to be retained in the endoplasmic reticulum with reduced delivery to the endolysosomal compartment, CLN5's normal cellular location. The AD-associated CLN5 variant is shown here to reduce the normal processing of cathepsin D and to decrease levels of full-length amyloid precursor protein (APP), suggestive of a defect in retromer-dependent trafficking.
INTRODUCTION
Lysosomal storage diseases (LSDs) are a group of inherited disorders that typically cause neurodegeneration early in life (1). Recent observations have established that genetic variants that cause one type of LSD, Gaucher's disease, can act as a risk factor for developing a late-onset neurodegenerative disease, Parkinson's disease (2). This observation suggests that genes that cause other LSDs might act as risk factors for other late-onset neurodegenerative disorders, notably, Alzheimer's disease (AD). While over the past decade over 25 genes that increase the risk of AD have been identified, a large part of the genetic contribution to AD remains to be clarified (3).
With this question in mind, we focused on a select group of genes that cause another LSD, neuronal ceroid lipofuscinosis (NCL), because they have been directly or indirectly associated with retromer trafficking (4, 5). Retromer is a multimodular protein assembly that has been linked to the pathogenesis of late-onset AD (6, 7) and is now considered the master conductor of endosomal sorting and trafficking (8). Among the group of NCL genes, we focused on these three: ceroid lipofuscinosis 3 (CLN3), whose expressed protein functions in trafficking the mannose-6-phosphate receptor (M6PR), a key cargo of retromer (9); CLN5, whose expressed protein is located at endosomal membranes and has been shown to function in the recruitment of retromer to endosomal membranes (4); and CTSD, whose expressed protein, cathepsin D, requires the normal retromer-dependent trafficking of M6PR to deliver procathepsin D to the endosome, during which it is processed to the mature form of cathepsin D.
To explore whether genetic variation in any of these three genes increases the risk of AD, we capitalized on data from a whole-exome sequencing study of multiplex Alzheimer's disease (AD) families. To validate any variant(s) identified in these analyses, we then turned to cell culture to determine whether the variant(s) has deleterious effects on normal function. Finally, because all three genes converge on cathepsin D, we tested whether the abnormal function caused by an identified variant(s) would affect the normal processing of this protein.
RESULTS
Whole-exome sequencing of multiplex AD families.
Whole-exome sequencing was performed in 31 Caribbean Hispanic families (98 affected and 12 unaffected relatives) from the Estudio Familiar de Influencia Genetica en Alzheimer (EFIGA). Families selected for sequencing had at least four affected individuals meeting standard criteria of the National Institute of Neurological and Communicative Disorders and Stroke and Alzheimer's Disease and Related Disorders Association (NINCDS-ADRDA) (10) for AD and were free of known mutations in APP, PSEN1, PSEN2, GRN, and MAPT. Analysis of the sequence data of the CLN5 gene identified a rare missense variant (rs199609750, c.A959G; population frequency, 7.418e−05) that segregated with AD status in one multiplex family and was present in two affected but no unaffected individuals (frequency in affected individuals, 2.0%; frequency in unaffected individuals, 0%). This variant was also significantly associated with AD when genotyped in all family members and compared to 438 internally genotyped controls of similar ancestry in which the mutant allele was entirely absent (P < 0.0001) or to population Latino controls in the Exome Aggregation Consortium (ExAC) database (allele frequency, 0.0006; P < 0.0001). No variants in CLN3 or CTSD segregated with AD status in these families.
Altered posttranslational processing in the CLN5 c.A959G (N320S) variant.
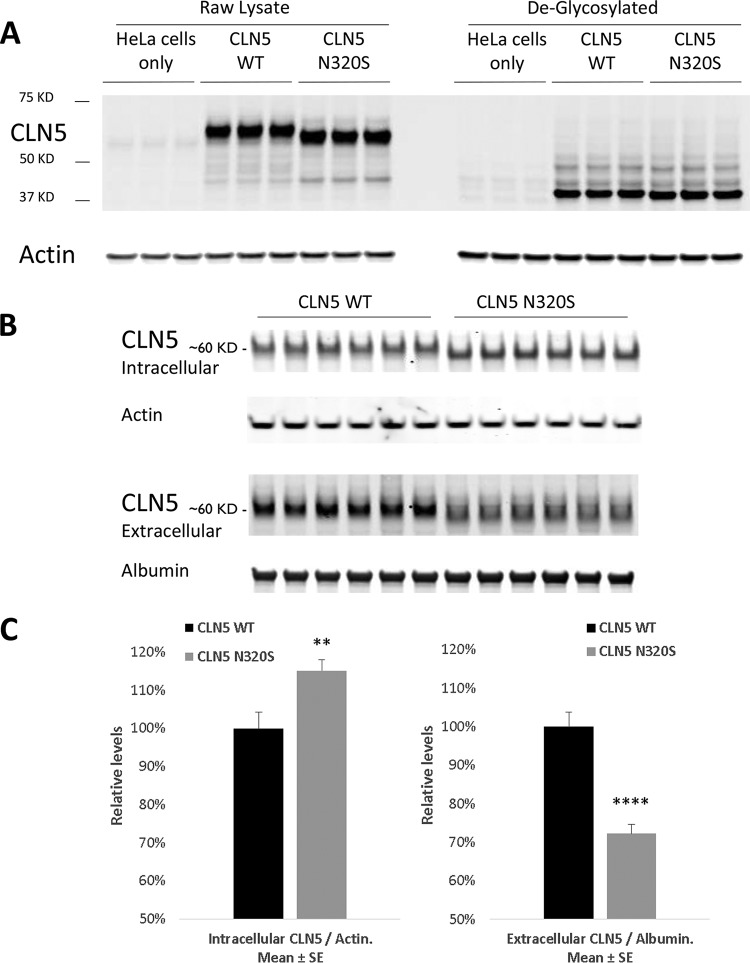
To validate possible biological effects of the CLN5 c.A959G (N320S, rs199609750) AD variant, we began by expressing wild-type (WT) and c.A959G (N320S) CLN5 in HeLa cells. The WT CLN5 displayed an expected molecular weight of 60 kDa; however, we observed a slight shift of ∼2.5 kDa in the c.A959G (N320S) variant (Fig. 1A). This shift indicates the possibility of an aberrant posttranslational modification, as c.A959G is a missense mutation and should not result in any form of premature truncation of the protein.
FIG 1.
Glycosylation deficits in AD variant CLN5 N320S. CLN5 WT and N320S AD variant plasmids were transfected into HeLa cells. After 24 h the cells were lysed and probed for CLN5. (A) CLN5 migrates to ∼60 kDa with a difference of approximately 2.5 kDa between the WT and AD variants; however, when sugar moieties were removed using the enzyme endoglycosidase H, CLN5 resolved to a lower location on the gel at ∼37 kDa, and the molecular weight difference disappeared. (B) The relative level of CLN5 inside the cell was compared to its level in the medium after 24 h of transfection. The WT CLN5 band was observed at ∼60 kDa in both intra- and extracellular compartments. (C) Quantification showed a significant increase and decrease in intracellular and extracellular levels of the CLN5 (N320S) variant, respectively. SE, standard error. **, P < 0.01; ****, P < 0.0001.
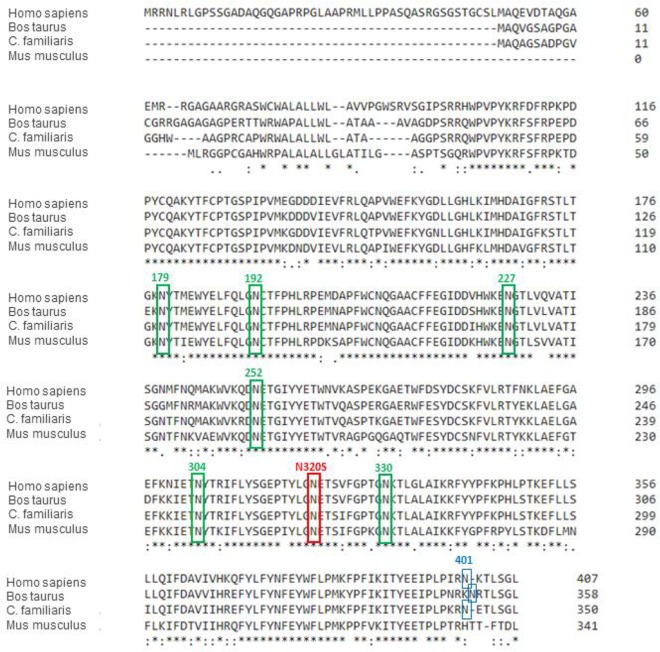
CLN5 has eight asparagine (N) glycosylation sites, and all of these sites are glycosylated in WT human CLN5 (11). The c.A959G missense mutation results in the replacement of the amino acid at position 320, an asparagine, with serine (N320S); asparagine 320 is one of the glycosylation sites (Fig. 2). Moharir et al. also indicated that lack of N-glycosylation on certain sites in CLN5, including N320, impairs CLN5 trafficking and function (11). Site-directed mutagenesis of individual asparagine residues to glutamines on each of the N-glycosylation consensus sites, followed by colocalization studies, suggested that there are functional differences in various N-glycosylation sites of CLN5 which differentially affect folding, trafficking, and lysosomal function of CLN5 and identified three groups of mutants based on function: (i) folding of the protein without which CLN5 is retained in the endoplasmic reticulum (ER) (N179Q, N252Q, N304Q, and N320Q), (ii) glycosylation involved in endosome/lysosome trafficking without which CLN5 is accumulated in the Golgi compartment (N401Q), and (iii) glycosylation involved in lysosomal function (N192Q and N227Q). We hypothesized therefore that this novel amino acid substitution (N320S) should also interfere with N-glycosylation at this particular site. To confirm this glycosylation deficit, sugar moieties were stripped away from these proteins before they were subjected to gel electrophoresis. After the deglycosylation reaction, both proteins, the WT and the N320S mutant, resolved at a location close to 37 kDa, and the difference in molecular weights between them disappeared. We also noticed a 15% increase in the levels of the intracellular CLN5 N320S variant (Fig. 1B and C). This increase in levels can be explained by the glycosylation defect as it can lead to abnormal folding of CLN5, resulting in endoplasmic reticulum entrapment (11, 12). If so, and since CLN5 can be secreted into the extracellular space (11, 13), ER retention of the CLN5 N320S variant should be associated with its diminished secretion. Accordingly, we analyzed CLN5 levels in cell culture medium, and, confirming the hypothesis, we found a significant decrease in the CLN5 N320S variant released into the medium compared to the level of WT CLN5 (Fig. 1C). These experiments establish that the novel AD-linked CLN5 variant is aberrantly glycosylated and provide biochemical evidence that suggests that the variant might be trapped in the ER.
FIG 2.
Sequence alignment of CLN5 (performed using Clustal Omega [http://www.ebi.ac.uk/Tools/msa/clustalo/]). The red box indicates the N-glycosylation site corresponding to human N320, which is conserved among different species and at which the identified variant rs199609750 (c.A959G) exerts its effect. The green boxes indicate the six additional N-glycosylation sites conserved among different species. The blue boxes indicate the N-glycosylation site corresponding to human N401, which is not conserved in rodents. Sequences and NCBI accession numbers used in this alignment are as follows: Homo sapiens, NP_006484.1; Bos taurus, ABD83352.1; Canis lupus familiaris (C. familiaris) NP_001011556.1; Mus musculus, NP_001028414.1. Asterisks indicate positions which have a single, fully conserved residue. A colon indicates conservation between groups of strongly similar properties. A period indicates conservation between groups of weakly similar properties.
Impaired cellular localization and retromer-mediated trafficking in the CLN5 N320S variant.
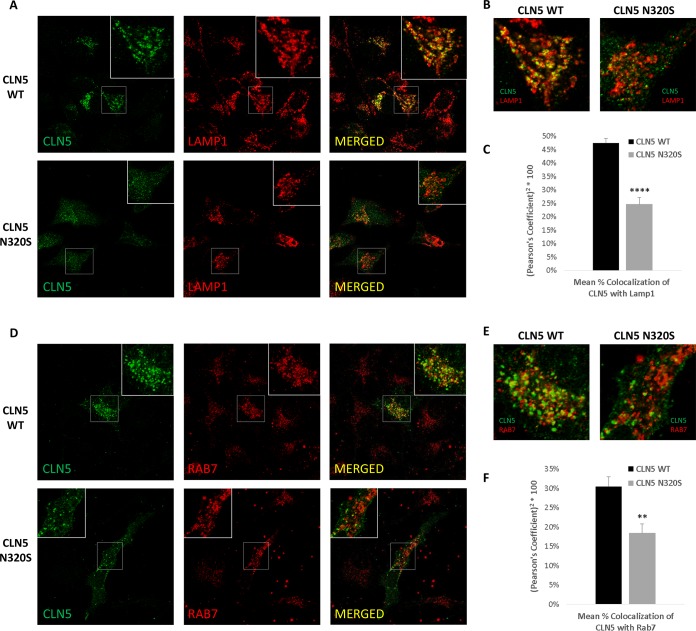
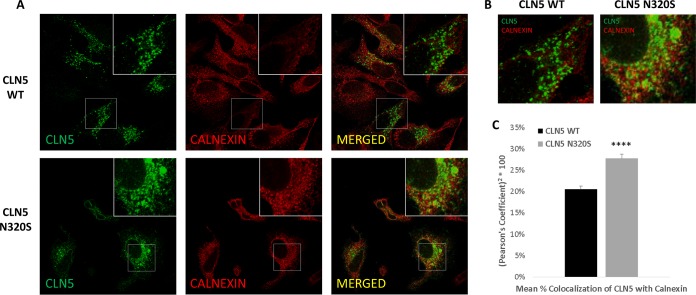
We then turned to confocal microscopy to further confirm this interpretation by comparing the intracellular localization of WT CLN5 to the AD-associated N320S variant. WT CLN5 is proteolytically cleaved and glycosylated prior to its transport to the endosomal-lysosomal compartments (13–15). As confirmation of the hypothesis, the AD-associated N320S variant showed increased colocalization with ER markers compared to the level with WT CLN5 but decreased colocalization with markers of the endosomes and lysosomes (Fig. 3 and 4; see also File S1 in the supplemental material).
FIG 3.
The AD variant CLN5 N320S shows reduced localization in the endolysosomal system. HeLa cells grown on coverslips were transfected with CLN5 WT and N320S variant plasmids. Cells were fixed with paraformaldehyde and stained for CLN5 and LAMP1. (A and B) Colocalization of WT and N320S CLN5 with LAMP1. WT CLN5 staining is more punctate than that of the N320S variant and has a higher level of colocalization with the lysosomal marker LAMP1. (C) Analysis from three independent immunofluorescence experiments showing reduced colocalization of N320S CLN5 with LAMP1 (total no of cells analyzed, 75). (D to F) CLN5 colocalization with the late endosomal marker RAB7 also followed a pattern similar to that of LAMP1 colocalization (total no of cells analyzed, 27). For each experiment, the image/cell showing the Pearson correlation value closest to the mean was selected as a representative image. **, P < 0.01; ****, P < 0.0001.
FIG 4.
Impaired intracellular trafficking of CLN5 N320S in HeLa cells. (A and B) Colocalization of WT and N320S CLN5 with the ER marker calnexin. (C) Analysis from three independent immunofluorescence experiments showing increased colocalization of N320S CLN5 with the ER (total no of cells analyzed, 189). The image/cell showing the Pearson correlation value closest to the mean was selected as a representative image. ****, P < 0.0001.
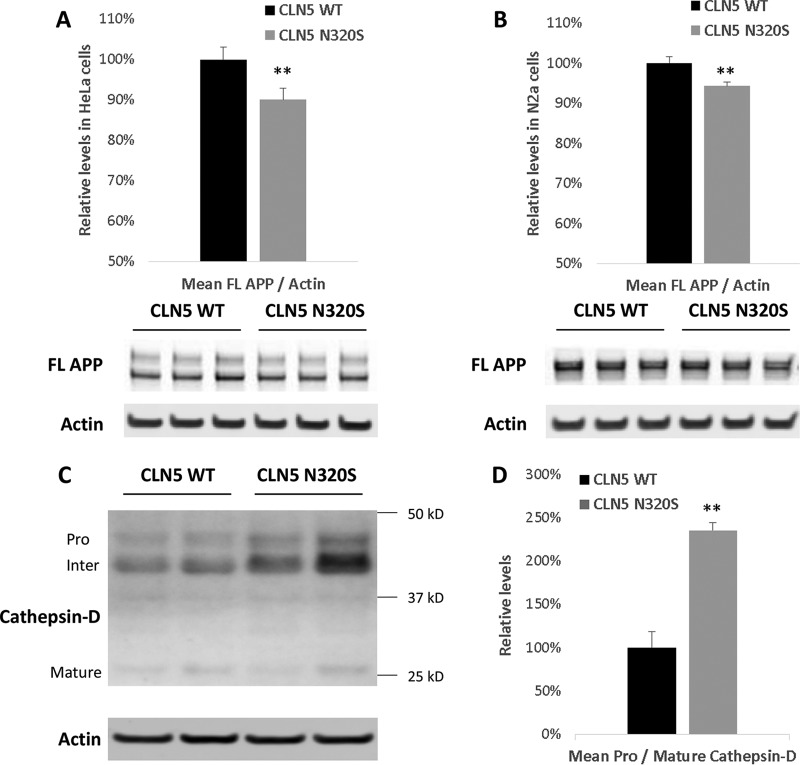
Upon further biochemical analysis of HeLa cells transiently expressing the WT and the N320S CLN5, we observed a small but consistent decrease in intracellular full-length amyloid precursor protein (APP) (∼10% decrease; P = 0.01) when N320S CLN5 was expressed. To confirm this finding, we repeated these experiments in mouse N2a cells and found a similar decrease in full-length APP in these neuroblastoma cells (Fig. 5A and B).
FIG 5.
Reduced levels of full-length APP and effects on cathepsin D processing. HeLa and N2a cells were transfected with CLN5 WT and N320S variant plasmids. After 24 h, the cells were lysed and probed for full-length APP. (A and B) Full-length APP levels were significantly reduced in CLN5 N320S-expressing HeLa and N2a cells. (C) Some wells of the HeLa cells expressing the CLN5 WT and N320S variant plasmids were selected with hygromycin to generate stable cell lines. Stable cell lines were plated in six-well plates, and after 48 h the cells were lysed and probed for cathepsin D. (D) Quantification revealed a significant increase in the ratio of procathepsin D to mature cathepsin D. **, P < 0.01.
The glycosylation deficiency and ER retention of the CLN5 N320S variant suggest that it is a loss-of-function mutation. One function assigned to CLN5 is its role in retromer trafficking—a pathway firmly linked to AD etiology by animal, cell biology, and genetic studies—based on a study that found that CLN5-depleted cells show evidence of retromer dysfunction (4). The study relied on previous observations establishing that a reliable cellular readout of retromer dysfunction is a defect in transport of cathepsin D to the endosomal-lysosomal compartments, a secondary consequence of the dependence on retromer for mannose-6-phosphate receptor recycling. During the transport to the endosomal-lysosomal compartments, procathepsin D is processed into its mature form (16), and retromer dysfunction is therefore associated with a relative increase in procathepsin D compared to the level of mature cathepsin D (17). We relied on this cellular readout to determine whether the AD-associated CLN5 N320S mutant causes a partial loss of CLN5 function. We measured procathepsin D and mature cathepsin D in HeLa cell lines stably expressing WT and N320S CLN5 constructs. Compared to the levels in cell lines expressing WT CLN5, a significant increase in procathepsin D levels in the cell lines expressing CLN5 N320S mutant was observed (Fig. 5C and D).
DISCUSSION
Numerous studies focusing on neuronal ceroid lipofuscinosis (NCL) have identified many NCL-related mutations (18). Our findings identify, we believe for the first time, a variant in one of these genes that is linked to late-onset AD. While CLN5's function is not fully understood, this transmembrane protein is normally located at endosomal membranes (11), and one study suggested that it plays a role in retromer trafficking (4). Retromer is a multimodular protein assembly that is now considered a master conductor (8) of endosomal sorting and trafficking. Each retromer module is made up of a group of proteins that serves a dedicated role, and the modules work together in support of retromer trafficking function (6). One key module is the membrane-recruiting module, which functions in recruiting retromer's cargo recognition module to the membrane of endosomes. The complete list of proteins that are part of the membrane-recruiting module remains unknown, but a previous study has provided strong evidence that CLN5, which normally resides in the endosome, functions in this module (4). This observation prompted us to include CLN5 in our genetic analysis.
In a whole-exome sequencing study we identified the missense N320S (p.Asn320Ser) variant in CLN5 to segregate with AD status in families multiply affected by the disease. This variant was also significantly associated with AD when genotyped in all family members and compared to 438 internally genotyped controls of similar ancestry in which the mutant allele was entirely absent or to population Latino controls in the ExAC database. Using a combination of molecular biology, biochemistry, and immunofluorescence experiments, we further validated this variant, functionally demonstrating that it is glycosylation deficient, which causes the expressed protein to be partially trapped in the ER and reduces its normal delivery to the endolysosomal system. Guided by a previous study (4) that showed that CLN5 deficiency affects retromer's function, we demonstrate that an effective deficiency in endosomal CLN5 caused by the missense variant results in a shift in the relative levels of procathepsin D, an established phenotype of retromer dysfunction (16, 17, 19, 20), and a reduction in full-length APP. Genomic and cell-biological findings have linked retromer dysfunction to AD pathogenesis (6). More than simply identifying a CLN5 variant genetically linked to AD and validating that it causes a loss of function, our results suggest that it converges with established pathophysiological mechanisms of disease.
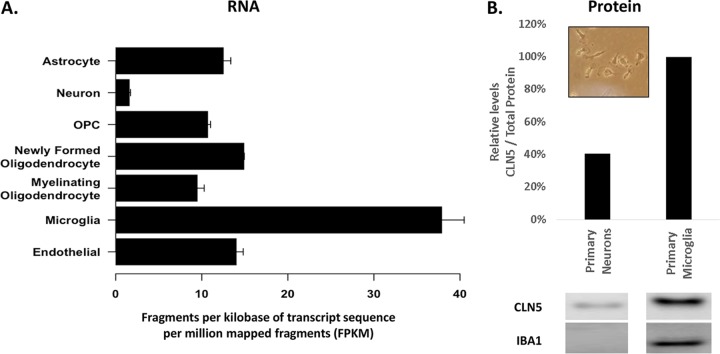
We postulate that the identified N320S missense mutant might mediate its AD-associated toxicity by affecting retromer function in microglia. CLN5 is highly expressed in the brain, and within the brain CLN5 is heavily enriched in microglia (Fig. 6) (21, 22), a cell type linked to AD. Microglia are activated upon tissue damage and are critical for brain homeostasis through clearance of cellular debris. The identification of CLN5 as a microglial gene associated with AD is in line with the implication of the microglial gene TREM2 (encoding a phagocytic receptor) as an AD susceptibility gene (23). Notably, retromer deficiency has been found in microglia of AD brains (24), but the mechanisms underlying this deficiency are still unclear. Thus, besides providing additional evidence for the role and molecular mechanisms of retromer dysfunction in AD, our findings provide critical support for the link between retromer, microglia, and AD, although we recognize that the missense N320S (p.Asn320Ser) variant should be further validated in additional independent AD data sets and that the CLN5 gene should be further scrutinized in various ethnic groups for additional potential disease-associated variants. Future studies relying on genetically engineered mice expressing the CLN5 mutation are required to better establish the functional consequence of this mutation on the brain and its contribution to various AD-related pathologies, including the potential effect on APP processing suggested by this study.
FIG 6.
CLN5 Expression in different brain cells. (A) CLN5 RNA expression measured in fragments per kilobase of transcript sequence per million mapped fragments (FPKM). OPC, oligodendrocyte precursor cell. (Reproduced with permission from http://Jiaqianwulab.org/resource [22].) (B) Lysates from primary mouse neurons and primary mouse microglia were probed with anti-CLN5 antibody and anti-IBA1 antibody. Protein levels were normalized to total protein by Ponceau stain. Primary microglia at day 7 of culture are shown in the inset.
MATERIALS AND METHODS
Study sample for whole-exome sequencing.
Families included in the whole-exome sequencing were part of the Estudio Familiar de Influencia Genetica en Alzheimer (EFIGA). All participants were recruited after providing written informed consent and with approval by the relevant institutional review boards at Columbia University Medical Center, Pontificia Universidad Catolica Madre y Maestra, and University of Puerto Rico (Medical Sciences Campus).
DNA isolation and sequencing.
High-molecular-weight DNA was isolated from either fresh or frozen blood stored at −80°C using Gentra Puregene and FlexiGene kits (Qiagen). When high-quality DNA from blood was unavailable (13 probands), DNA was isolated from lymphocyte cell lines. TruSeq DNA Preparation and Exome Enrichment kits (Illumina, San Diego, CA) were used to prepare indexed genomic DNA (gDNA) libraries and isolate exonic regions for high-throughput sequencing. Multiplexed DNA samples were sequenced in batches of up to 12 samples on Illumina's Genome Analyzer IIx, HiSeq 2000, and MiSeq platforms (Illumina, San Diego, CA). Paired-end reads were performed over 82 to 307 sequencing cycles, yielding high coverage at an average depth of >60× per sample and interval region captured.
Downstream bioinformatics analysis of sequence data.
Using the Burrows-Wheeler Aligner (25), the reads obtained from the pooled sequencing were aligned to the human reference genome build 37 (http://bio-bwa.sourceforge.net/). Quality control of the sequencing data was done using established pipelines, including base alignment quality calibration and refinement of local alignment around putative indels using the Genome Analysis Toolkit (GATK) (26). Variants were called and recalibrated using multisample calling with GATK's UnifiedGenotyper and VariantRecalibrator modules. Reliably called variants were annotated by ANNOVAR (27) including in silico functional prediction using POLYPHEN (28) and extent of cross-species conservation using PHYLOP (29).
Statistical analyses of sequence data.
We tested segregation and AD association of individual single nucleotide variants (SNVs) in the sequenced samples. To validate SNVs prioritized from these analyses, we genotyped them in all family members of the families in which they were discovered and a in set of 438 unrelated, unaffected population controls of similar ancestry (68.1% women; mean age at examination, 82.0 ± 7.3 years; APOE ε4 allele frequency, 9.8%) using the Sequenom MassARRAY platform. We compared their allele frequencies in affected individuals with those in samples from unaffected individuals from this follow-up genotyping using Fisher's exact test. The 438 unaffected, unrelated controls were determined to be of the same ethnic background as the familial cases using methods described previously (30). We also compared the allele frequencies in affected individuals with the publicly available ExAC data. Because of the lack of an optimal ethnically matched control data set for Caribbean Hispanics, we used the ExAC Latino cohort for an estimate of the population allele frequencies of identified variants.
Design and preparation of the CLN5 constructs.
Plasmids expressing the WT and c.A959G (N320S) CLN5 proteins (human) tagged with Flag at the C terminus were designed using ThermoFisher's GeneArt portal. A flexible polyglycine linker (6×G) was inserted between the protein and the tag. mRNA sequence for CLN5 was acquired from the National Center for Biotechnology Information (NCBI). The constructs were then subcloned into pcDNA3.1/Hygro(+) vectors with a cytomegalovirus (CMV) promoter.
Cell culture.
HeLa cells were cultured using Dulbecco's modified Eagle's medium (DMEM) plus 10% fetal bovine serum (FBS) and GlutaMAX, with penicillin, streptomycin, and amphotericin B to prevent microbial contamination. Mouse neuroblastoma (N2a) cells were cultured in 50% DMEM (high glucose) and 50% Opti-MEM plus 10% FBS and glutamine (2 mM) with penicillin and streptomycin to prevent microbial contamination. Primary mouse cortical neuron cultures were performed as described previously (31). Primary microglia were cultured as described previously with slight modifications (32). Briefly brain homogenate from day 1 pups was plated onto poly-d-ornithine-coated flasks in DMEM–F-12 medium plus 10% FBS and GlutaMAX with macrophage colony-stimulating factor (mCSF). At day 7 microglia were dislodged from the bottom of the flask by tapping the flask gently. Careful tapping results in floating of microglia prior to other cells/debris. Medium containing microglia was passed through a cell strainer to remove remaining debris. The suspension was spun down at 500 × g, and the pellet was resuspended in fresh medium, plated in six-well plates, and allowed to grow and stabilize for 7 days prior to harvesting.
Transfection of cells with CLN5 WT and N320S variant and biochemistry.
WT and N320S CLN5-expressing plasmids were transfected into HeLa and N2a cells using Lipofectamine. Cells were harvested 24 h after transfection; culture medium was also collected at the same time. Lysates from the samples were run on NuPAGE Bis-Tris 4 to 12% gels, transferred onto nitrocellulose membranes using iblot, and probed with antibodies against CLN5 (ab170899, 1:500; Abcam), actin (NB600-535, 1:2,000; Novus), albumin (ab3781, 1:2,000; Abcam), Iba1 (NB100-1028, 1:500; Novus), amyloid precursor protein (ab32136, 1:10,000; Abcam), and cathepsin D (ab75852, 1:500; Abcam). For the deglycosylation experiments an endoglycosidase H (Endo H) kit from New England BioLabs (P0702L) was used with the recommended protocol (https://www.neb.com/protocols/2012/10/18/endo-hf-protocol); briefly, lysates were denatured in glycoprotein denaturing buffer at 100°C for 10 min and then incubated in Glycobuffer 3 and Endo H at 37°C for 1 h. To generate stable cell lines, some wells containing HeLa cells expressing the CLN5 WT and N320S variant plasmids were selected with hygromycin B for ∼14 days. The surviving stably transfected cells were plated in six-well plates and harvested after 48 h for biochemistry.
Immunocytochemistry.
HeLa cells were transfected with CLN5 plasmids in a 24-well plate (with coverslips) using Lipofectamine 2000. Twenty-four hours after transfection, cells were fixed using 4% paraformaldehyde for 10 min and permeabilized using digitonin (0.01%) for 10 min. To block nonspecific staining, cells were incubated in 5% donkey serum overnight. Cells were probed for CLN5 (ab170899, 1:250 [Abcam], or SAB1412697, 1:200 [Sigma]), LAMP1 (AF4800, 1:500; R&D Systems), Rab7 (SC-376362, 1:50), and calnexin (A01240, 1:500; GeneScript) primary antibodies prepared in 1% donkey serum. Secondary antibodies (Life Technologies) conjugated with Alexa Fluor dyes were used. Images were taken using a Zeiss LSM 700 META confocal microscope equipped with a 63× plan-apochromat objective and HeNe1, HeNe2, and argon lasers. Colocalization analysis was performed using ImageJ's JACoB plug-in.
Supplementary Material
ACKNOWLEDGMENT
We thank all participants from the Estudio Familiar de Influencia Genetica en Alzheimer (EFIGA) for taking part in EFIGA.
This work was supported by grants from the National Institutes of Health (RF1AG054080, RF1AG015473, and P50AG008702).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00011-18.
REFERENCES
- 1.Plotegher N, Duchen MR. 2017. Mitochondrial dysfunction and neurodegeneration in lysosomal storage disorders. Trends Mol Med 23:116–134. doi: 10.1016/j.molmed.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Stirnemann J, Belmatoug N, Camou F, Serratrice C, Froissart R, Caillaud C, Levade T, Astudillo L, Serratrice J, Brassier A, Rose C, Billette de Villemeur T, Berger MG. 2017. A review of Gaucher disease pathophysiology, clinical presentation and treatments. Int J Mol Sci 18:E441. doi: 10.3390/ijms18020441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thorton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Moron FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fievet N, Huentelman MW, Gill M, Brown K, et al. . 2013. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet 45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mamo A, Jules F, Dumaresq-Doiron K, Costantino S, Lefrancois S. 2012. The role of ceroid lipofuscinosis neuronal protein 5 (CLN5) in endosomal sorting. Mol Cell Biol 32:1855–1866. doi: 10.1128/MCB.06726-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kama R, Kanneganti V, Ungermann C, Gerst JE. 2011. The yeast Batten disease orthologue Btn1 controls endosome-Golgi retrograde transport via SNARE assembly. J Cell Biol 195:203–215. doi: 10.1083/jcb.201102115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Small SA, Petsko GA. 2015. Retromer in Alzheimer disease, Parkinson disease and other neurological disorders. Nat Rev Neurosci 16:126–132. doi: 10.1038/nrn3896. [DOI] [PubMed] [Google Scholar]
- 7.Muhammad A, Flores I, Zhang H, Yu R, Staniszewski A, Planel E, Herman M, Ho L, Kreber R, Honig LS, Ganetzky B, Duff K, Arancio O, Small SA. 2008. Retromer deficiency observed in Alzheimer's disease causes hippocampal dysfunction, neurodegeneration, and Abeta accumulation. Proc Natl Acad Sci U S A 105:7327–7332. doi: 10.1073/pnas.0802545105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burd C, Cullen PJ. 2014. Retromer: a master conductor of endosome sorting. Cold Spring Harb Perspect Biol 6:a016774. doi: 10.1101/cshperspect.a016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Metcalf DJ, Calvi AA, Seaman M, Mitchison HM, Cutler DF. 2008. Loss of the Batten disease gene CLN3 prevents exit from the TGN of the mannose 6-phosphate receptor. Traffic 9:1905–1914. doi: 10.1111/j.1600-0854.2008.00807.x. [DOI] [PubMed] [Google Scholar]
- 10.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. 1984. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology 34:939–944. doi: 10.1212/WNL.34.7.939. [DOI] [PubMed] [Google Scholar]
- 11.Moharir A, Peck SH, Budden T, Lee SY. 2013. The role of N-glycosylation in folding, trafficking, and functionality of lysosomal protein CLN5. PLoS One 8:e74299. doi: 10.1371/journal.pone.0074299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lebrun AH, Storch S, Ruschendorf F, Schmiedt ML, Kyttala A, Mole SE, Kitzmuller C, Saar K, Mewasingh LD, Boda V, Kohlschutter A, Ullrich K, Braulke T, Schulz A. 2009. Retention of lysosomal protein CLN5 in the endoplasmic reticulum causes neuronal ceroid lipofuscinosis in Asian sibship. Hum Mutat 30:E651–E661. doi: 10.1002/humu.21010. [DOI] [PubMed] [Google Scholar]
- 13.Isosomppi J, Vesa J, Jalanko A, Peltonen L. 2002. Lysosomal localization of the neuronal ceroid lipofuscinosis CLN5 protein. Hum Mol Genet 11:885–891. doi: 10.1093/hmg/11.8.885. [DOI] [PubMed] [Google Scholar]
- 14.Schmiedt ML, Bessa C, Heine C, Ribeiro MG, Jalanko A, Kyttala A. 2010. The neuronal ceroid lipofuscinosis protein CLN5: new insights into cellular maturation, transport, and consequences of mutations. Hum Mutat 31:356–365. doi: 10.1002/humu.21195. [DOI] [PubMed] [Google Scholar]
- 15.Holmberg V, Jalanko A, Isosomppi J, Fabritius AL, Peltonen L, Kopra O. 2004. The mouse ortholog of the neuronal ceroid lipofuscinosis CLN5 gene encodes a soluble lysosomal glycoprotein expressed in the developing brain. Neurobiol Dis 16:29–40. doi: 10.1016/j.nbd.2003.12.019. [DOI] [PubMed] [Google Scholar]
- 16.Benes P, Vetvicka V, Fusek M. 2008. Cathepsin D—many functions of one aspartic protease. Crit Rev Oncol Hematol 68:12–28. doi: 10.1016/j.critrevonc.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rojas R, van Vlijmen T, Mardones GA, Prabhu Y, Rojas AL, Mohammed S, Heck AJ, Raposo G, van der Sluijs P, Bonifacino JS. 2008. Regulation of retromer recruitment to endosomes by sequential action of Rab5 and Rab7. J Cell Biol 183:513–526. doi: 10.1083/jcb.200804048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams RE, Mole SE. 2012. New nomenclature and classification scheme for the neuronal ceroid lipofuscinoses. Neurology 79:183–191. doi: 10.1212/WNL.0b013e31825f0547. [DOI] [PubMed] [Google Scholar]
- 19.Miura E, Hasegawa T, Konno M, Suzuki M, Sugeno N, Fujikake N, Geisler S, Tabuchi M, Oshima R, Kikuchi A, Baba T, Wada K, Nagai Y, Takeda A, Aoki M. 2014. VPS35 dysfunction impairs lysosomal degradation of alpha-synuclein and exacerbates neurotoxicity in a Drosophila model of Parkinson's disease. Neurobiol Dis 71:1–13. doi: 10.1016/j.nbd.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 20.Follett J, Norwood SJ, Hamilton NA, Mohan M, Kovtun O, Tay S, Zhe Y, Wood SA, Mellick GD, Silburn PA, Collins BM, Bugarcic A, Teasdale RD. 2014. The Vps35 D620N mutation linked to Parkinson's disease disrupts the cargo sorting function of retromer. Traffic 15:230–244. doi: 10.1111/tra.12136. [DOI] [PubMed] [Google Scholar]
- 21.Schmiedt ML, Blom T, Blom T, Kopra O, Wong A, von Schantz-Fant C, Ikonen E, Kuronen M, Jauhiainen M, Cooper JD, Jalanko A. 2012. Cln5-deficiency in mice leads to microglial activation, defective myelination and changes in lipid metabolism. Neurobiol Dis 46:19–29. doi: 10.1016/j.nbd.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, Wu JQ. 2014. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ulland TK, Song WM, Huang SC, Ulrich JD, Sergushichev A, Beatty WL, Loboda AA, Zhou Y, Cairns NJ, Kambal A, Loginicheva E, Gilfillan S, Cella M, Virgin HW, Unanue ER, Wang Y, Artyomov MN, Holtzman DM, Colonna M. 2017. TREM2 maintains microglial metabolic fitness in Alzheimer's disease. Cell 170:649–663.e13. doi: 10.1016/j.cell.2017.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lucin KM, O'Brien CE, Bieri G, Czirr E, Mosher KI, Abbey RJ, Mastroeni DF, Rogers J, Spencer B, Masliah E, Wyss-Coray T. 2013. Microglial beclin 1 regulates retromer trafficking and phagocytosis and is impaired in Alzheimer's disease. Neuron 79:873–886. doi: 10.1016/j.neuron.2013.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K, Li M, Hakonarson H. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. 2010. A method and server for predicting damaging missense mutations. Nat Methods 7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. 2010. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JH, Cheng R, Barral S, Reitz C, Medrano M, Lantigua R, Jimenez-Velazquez IZ, Rogaeva E, St George-Hyslop PH, Mayeux R. 2011. Identification of novel loci for Alzheimer disease and replication of CLU, PICALM, and BIN1 in Caribbean Hispanic individuals. Arch Neurol 68:320–328. doi: 10.1001/archneurol.2010.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhalla A, Vetanovetz CP, Morel E, Chamoun Z, Di Paolo G, Small SA. 2012. The location and trafficking routes of the neuronal retromer and its role in amyloid precursor protein transport. Neurobiol Dis 47:126–134. doi: 10.1016/j.nbd.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Floden AM, Combs CK. 2007. Microglia repetitively isolated from in vitro mixed glial cultures retain their initial phenotype. J Neurosci Methods 164:218–224. doi: 10.1016/j.jneumeth.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.