Abstract
Background
Rotavirus is a leading cause of severe diarrheal disease, and one of the common causes of death in children aged under five years old. The dominant epidemic strains may change in different years in the same area. In order to provide evidence for rotavirus epidemic control and inform vaccine development, we analyzed epidemiological patterns and genetic characteristics of rotavirus in Beijing during 2011–2016.
Methods
Stool specimens of outpatient children under five years old were collected from three children’s hospitals on a weekly basis. Group A rotavirus antigens were detected using enzyme-linked immunosorbent assay (ELISA) kit. The partial VP4 genes and VP7 genes of rotavirus were both amplified and sequenced. Genotyping and phylogenetic analyses were performed. Logistic regression and Chi-square tests were performed to determine differences across age groups, districts and years in rotavirus prevalence and genotype distribution.
Results
A total of 3668 stool specimens from children with acute diarrhea identified through hospital-based surveillance were collected from 2011 to 2016 in Beijing. A total of 762 (20.8%) specimens tested positive for rotavirus. The rotavirus-positive rate was highest among the 1–2 years old age group (29.0%, 310/1070). November, December and January were the highest rotavirus-positive rate months each year. G9 was the most common G genotype (64.4%, 461/716), and P [8] was the most common P genotype (87.0%, 623/716) among the 716 rotavirus-positive specimens. G9P [8], G3P [8] and G2P [4] were the most common strains. The rotavirus-positive rates of samples in 2012 and 2013 were higher than that in 2011, and the dominant genotype changed from G3P [8] to G9P [8] in 2012 and 2013. VP7 gene sequences of G9 strains in this study clustered into two main lineages. Most of the G9 strains exhibited the highest nucleotide similarity (99.1%~ 100.0%) to the strain found in Japan (MI1128). VP4 gene sequences of P [8] strains were almost P[8]b.
Conclusions
Rotavirus accounted for more than one fifth of childhood diarrhea in Beijing during the study period. Targeted measures such as immunization with effective rotavirus vaccines should be carried out to reduce the morbidity and mortality due to rotavirus.
Keywords: Diarrhea, Children under five years old, Group A rotavirus, Genotype
Background
Infection with group A rotavirus is the main cause of severe diarrheal disease in children worldwide [1], and one of the common causes of death in children aged under 5 years old [2, 3]. According to World Health Organization (WHO), approximately 215,000 children aged under 5 years old died from rotavirus infection in 2013 [4]. In China, 40% of diarrhea-related hospitalizations and 30% of diarrhea-related outpatient visits among children younger than 5 years old from 1994 through 2014 were caused by group A rotavirus [5]. From 2003 to 2012, rotavirus infection contributed to 42% of diarrhea-associated death among Chinese children under 5 years old [6].
According to the sequence diversity of VP7 and VP4 genes, group A rotavirus can be divided into several G and P genotypes [7, 8]. So far, there have been 27 G genotypes and 37 P genotypes identified. Other new G and P genotypes are expected to be identified in the future [9–12]. G3P [8], G1P [8], G2P [4] and G4P [8] have been the four dominant strains from 1989 through 2004 globally [13–15]. Since 2004, G9P [8] was detected as one of the major epidemic strains, gradually increasing worldwide [16]. However, the major epidemic strains of group A rotavirus varies by countries and regions. Moreover, the major epidemic strains may change in different years in the same area [7, 17, 18]. In China, G1P [8] was the dominant strain from1983–2000 [19, 20]. The proportion of G3 increased dramatically from 2000 to 2007, and G3P [8] became the dominant strain [21–23]. In 2008, G9 was reported to be a highly prevalent genotype in Xinjiang in the northwest part of China like other countries [24–26].
There is no specific drug for the rotavirus diarrhea, but effective vaccines are available and can protect children from illness and death caused by rotavirus. In China, only the LLR (Lanzhou Institute of Biological Products, Lanzhou, China) vaccine is licensed for use, which is sourced from G10P [12] of sheep. However, this vaccine has not yet been included in the national immunization program, and there are no available clinical trial data so far. Further understanding the distribution of rotavirus strains may provide evidence for the use of vaccines in China. Previous data in China, usually based on a short-term observational studies, are limited. However, in this study we presented the results of 6-years of surveillance data to further elucidate the changing epidemiology, and thus inform targeted control measures and vaccine development.
Methods
Data source
Data were collected on children with acute diarrhea identified through hospital-based surveillance from 2011 to 2016 in Beijing. Acute diarrhea cases were defined as outpatient children aged under 5 years old who had diarrhea (3 or more loose stools within 24 h). Children who were not residents in Beijing were excluded. Sentinel hospital surveillance for rotavirus diarrhea among children under 5 years old was established in three children’s hospitals located in Xicheng, Chaoyang and Tongzhou districts respectively. Stool specimens and socio-demographic data were collected from children under 5 years old who were newly diagnosed with acute diarrhea at the first medical consultation. Fifteen specimens were collected every month from each sentinel hospital. Socio-demographic information includes district, surveillance month, age and sex. District centers for disease control (CDC) are responsible for sending specimens and information of patients to Beijing CDC each month.
Laboratory testing
Detection of group A rotaviruses
Rotavirus antigen detection was performed by enzyme linked immunosorbent assay (ELISA) using ProSpecT Rotavirus Microplate Assay (Oxoid Ltd., Basingstoke Hants, UK), according to the manufacturer’s instructions.
Viral RNA extraction
According to the manufacturer’s protocol, QIAamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany) was used to extract Viral RNA from 140 μL of a 10% fecal suspension in phosphate-buffered saline. Extracted RNA was stored at − 20 °C until further use.
Genotyping of group A rotaviruses
Rotavirus G and P genotyping were determined by multiplex semi-nested reverse transcription polymerase chain reaction (RT-PCR) [27, 28]. A total of 5 μL rotavirus RNA was mixed with 5 μL universal primers, primers VP7F/VP7R for G typing and primers VP4F/VP4R for P typing. The mixture was denatured at 98 °C for 5 min and cooled on ice for 5 min, then the one-step RT-PCR kit (QIAGEN, Hilden, Germany) was used to amplify VP7 and VP4 genes in a 50 μL reaction volume. RT-PCR was performed at 50 °C for 30 min and 95 °C for 15 min followed by 30 cycles of 94 °C for 30 s, 42 °C for 30 s, and 72 °C for 60 s. A final extension was run at 72 °C for 7 min.
The second-round PCR was performed using AmpliTaq Gold® 360 PCR Master Mix (Life Tech, Foster City, CA, USA). For G genotyping, primers aBT1, aCT2, G3, aDT4, aAT8, G9, G12 and VP7R were used, generating amplicons of 618 bp, 521 bp, 682 bp, 452 bp, 754 bp, 179 bp and 387 bp for G1, G2, G3, G4, G8, G9 and G12, respectively. For P genotyping, primers VP4F, 1 T-1D, 2 T-1, 3 T-1, 4 T-1, 5 T-1, and P [11] were used, generating amplicons of 362 bp, 146 bp, 224 bp, 270 bp, 462 bp and 191 bp for P [4], P [6], P [8], P [9], P [10] and P [11], respectively. PCR was performed at 95 °C for 10 min followed by 35 cycles of 95 °C for 30 s, 42 °C for 30 s, and 72 °C for 60 s; a final extension was run at 72 °C for 7 min. The PCR products were analyzed using a QIAxcel Advanced Instrument with a QIAxcel DNA Screening Kit (QIAGEN, Hilden, Germany).
DNA sequencing and phylogenetic analysis
The first-round PCR products were purified and sequenced directly on an ABI 3730xl DNA Analyzer using a BigDye Terminator v3.1 Cycle Sequencing Kit (ABI, Austin, TX, USA). All sequences were prepared and aligned by BioEdit (version 7.0.9.0) with the Clustal W program. The phylogenetic tree was constructed using the neighbor-joining method with MEGA software (version 6.06) and bootstrap analysis was performed with 1000 replications. The nucleotide sequences used in this study were submitted in GenBank under accession numbers MH631063-MH631154, MH625726-MH625776 and MH625777-MH625915. The reference sequences used in plotting the phylogeny tree from the GenBank database are shown in Table 1.
Table 1.
Reference sequences used in plotting the phylogeny tree of this study
| Strains | Accession numbers | genotypes | Starains | Accession numbers | genotypes |
|---|---|---|---|---|---|
| MI1128 | LC228408 | G9 | CNMC3 | KT920808 | P [8] |
| BJ-Q322 | KF673479 | G9 | CK00095 | JX027933 | P [8] |
| VU12-13-101 | KT919508 | G9 | UR14-26 | LC105523 | P [8] |
| MRC-DPRU1102 | KJ753473 | G9 | X1302 | KJ794102 | P [8] |
| Km15064 | KX033644 | G9 | H1301 | KJ794103 | P [8] |
| Hon-JK17–6-14 | KU312101 | G9 | Z1557 | KF372006 | P [8] |
| BJ-Q532 | KF673480 | G9 | WZ7 | KU243615 | P [8] |
| 99-SP1542VP7 | AB091753 | G9 | AC314 | KT025876 | P [8] |
| 99-TK2082VP7 | AB091755 | G9 | To14–18 | LC105271 | P [8] |
| JP13-3 | AB176679 | G9 | SI-R13 | KJ432798 | P [8] |
| LL51695 | KC242226 | G9 | D | EF672570 | P [8] |
| WZ660 | KU243678 | G9 | ISO42 | DQ355958 | P [8] |
| km15118 | KX778607 | G9 | Nov10-N709 | HQ537506 | P [8] |
| MRC-DPRU5123 | KP752521 | G9 | LY238-9 | KU174038 | P [4] |
| F45 | AB180970 | G9 | N14-29 | LC342710 | P [4] |
| AU32 | AB045372 | G9 | RMRC-11-03-0614 | MF373697 | P [4] |
| N12–55 | LC380583 | G1 | O1157 | JX156413 | P [4] |
| N10–17 | LC348885 | G1 | 07–96 s-98 | KY489875 | P [4] |
| CK20027 | KC443768 | G2 | WZ193 | KU243633 | P [4] |
| km15028 | KX033592 | G2 | |||
| VU12–13-27 | MF167970 | G2 | |||
| E3239 | KF371889 | G3 | |||
| E2432 | KF371856 | G3 | |||
| 1RIZE2016 | MF494811 | G3 |
Statistical analysis
Socio-demographical data and laboratory data were double entered and checked. Data analyses were performed using SPSS (version 20.0). Univariate logistic regression analyses were conducted to identify factors predictive of rotavirus infection. Multivariate logistic regression was conducted to adjust for confounders. Chi-square test was used to compare the distribution of GP genotypes between different genders, age groups, living areas and years. All hypothesis tests were 2-tailed, and a probability of P < 0.05 was considered statistically significant.
Results
Patients with diarrhea
A total of 3668 stool specimens of children with acute diarrhea were collected from 3668 cases, including 2296 specimens from boys, 1363 specimens from girls, 9 cases of unknown gender. A total of 762 stool specimens tested positive for rotavirus with the positive rate of 20.8%.
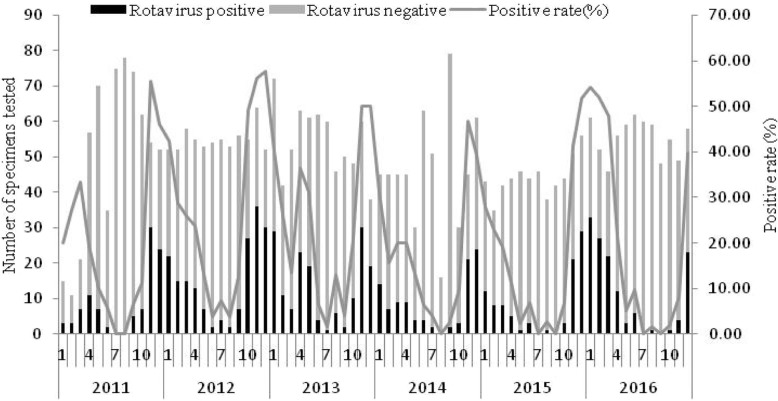
In 22 children, age was unknown. The rotavirus-positive rate in children aged less than 6 months was the lowest (9.0%). Children aged 1–2 years old had the highest rotavirus-positive rate (29.0%), followed by those aged 6–11 months old (24.0%), and children aged 2–3 years old (17.9%) (Table 2). Compared with children 0–5 months, age groups 6–11 months, 1–2 and 2–3 years old were at higher risk for rotavirus-positive. The rotavirus-positive rate in urban children was 23.9% (585/2480). Children who lived in rural areas were less likely to be rotavirus-positive (OR 0.57, 95% CI 0.47–0.68). From 2011 to 2016, the rotavirus-positive rates were 16.4% (99/604), 27.3% (180/659), 24.6% (161/654), 17.8% (99/555), 17.1% (91/531), and 19.8% (132/665), respectively each year. The rotavirus-positive rates in 2012 and 2013 were higher, being > 20%. Controlled for age and urban residence, rotavirus-positive rates were significantly different between different years (P < 0.001). The rotavirus-positive rates in 2012 and 2013 were higher, compared with 2011. Adjusted OR was 1.90 (95% CI 1.43–2.54) in 2012 and 1.60 (95% CI 1.19–2.15) in 2013. The largest number and highest rotavirus-positive rate of specimens were in November, December and the following January each epidemic season (Fig. 1).
Table 2.
Demographic characteristics of the outpatient children under five years old with diarrhea in Beijing, China, 2011–2016
| Demographicharacteristics | Total number | Number of rotavirus positive(%) | OR | 95%CI | P-value |
|---|---|---|---|---|---|
| Gendera | |||||
| Male | 2296 | 482 (21.0%) | 1.00 | Reference | |
| Female | 1363 | 271 (19.9%) | 0.93 | 0.79–1.10 | 0.422 |
| Age, yearsa | |||||
| 0–5 months | 837 | 75 (9.0%) | 1.00 | Reference | |
| 6–11 months | 1179 | 283 (24.0%) | 3.21 | 2.44–4.21 | < 0.01 |
| 1- | 1070 | 310 (29.0%) | 4.14 | 3.16–5.44 | < 0.01 |
| 2- | 285 | 51 (17.9%) | 2.21 | 1.51–3.25 | < 0.01 |
| 3- | 150 | 18 (12.0%) | 1.39 | 0.80–2.39 | 0.242 |
| 4- | 125 | 12 (9.6%) | 1.08 | 0.57–2.05 | 0.816 |
| Living area | |||||
| Urban | 2480 | 585 (23.9%) | 1.00 | Reference | |
| Rural | 1188 | 177 (14.9%) | 0.57 | 0.47–0.68 | < 0.01 |
| Years | |||||
| 2011 | 604 | 99 (16.4%) | 1.00 | Reference | |
| 2012 | 659 | 180 (27.3%) | 0.79 | 0.59–1.06 | 0.111 |
| 2013 | 654 | 161 (24.6%) | 1.52 | 1.17–1.96 | 0.001 |
| 2014 | 555 | 99 (17.8%) | 1,32 | 1.02–1.71 | 0.038 |
| 2015 | 531 | 91 (17.1%) | 0.88 | 0.66–1.17 | 0.372 |
| 2016 | 665 | 132 (19.8%) | 0.84 | 0.62–1.12 | 0.232 |
aThere are missing data
Fig. 1.
Monthly distribution of the number and positive rate of rotavirus among outpatient children under five years old with diarrhea in Beijing, China, 2011–2016
Among 762 rotavirus positive specimens, 716 (94.0%) were genotyped successfully for G and P (Table 3). G9 was the most common G genotype, accounting for 64.4% (461/716). P [8] was the most common P genotype, accounting for 87.0% (623/716).
Table 3.
The distribution of G and P genotypes of rotavirus positive specimens isolated from outpatient children under five years old with diarrhea in Beijing, China, 2011–2016
| Total number | P [4] | P [8] | P [9] | P Mix | UT | |
|---|---|---|---|---|---|---|
| G1 | 35 | 0 | 35 | 0 | 0 | 0 |
| G2 | 57 | 47 | 8 | 0 | 1 | 1 |
| G3 | 68 | 1 | 64 | 0 | 3 | 0 |
| G8 | 1 | 0 | 1 | 0 | 0 | 0 |
| G9 | 461 | 5 | 446 | 0 | 4 | 6 |
| G Mix | 67 | 5 | 55 | 0 | 6 | 1 |
| UT | 27 | 0 | 14 | 1 | 1 | 11 |
| Total | 716 | 58 | 623 | 1 | 15 | 19 |
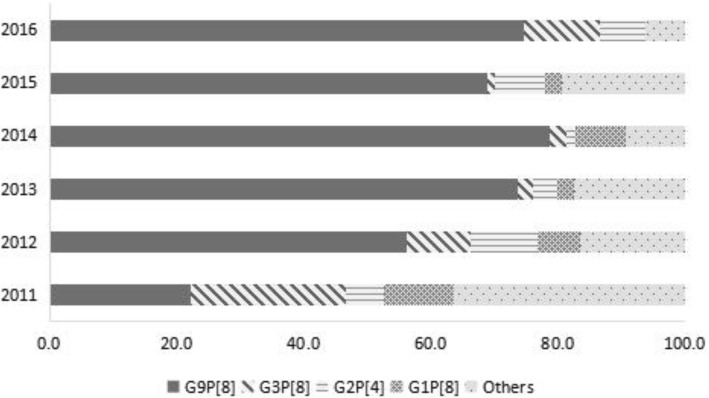
The most common GP combined genotype was G9P [8] (62.3%, 446/716), followed by G3P [8] and G2P [4]. G3P [8] and G9P [8] mixed accounted for 48.4% (46/95) of the others. There was no statistically significant difference in distribution of genotype by gender (χ2 = 4.492, P = 0.344), age group (χ2 = 15.036, P = 0.774) or living area (χ2 = 2.906, P = 0.574). In 2011, G3P [8] and G9P [8] were both the dominant genotypes, accounting for 24.2 (24/99) and 22.2% (22/99) of samples respectively. In 2012, G9P [8] became the dominant genotype, accounting for 56.1% (101/180) of samples. From 2013 to 2016, G9P [8] was the dominant genotype, accounting for 70%~ 80% of samples (Fig. 2). There was a statistically significant difference in distribution of genotype by year (χ2 = 141.254, P < 0.001).
Fig. 2.
The change of dominant genotypes of rotavirus-positive specimens isolated from outpatient children under five years old with diarrhea in Beijing, China, 2011–2016
Phylogenetic analysis of VP7 genes
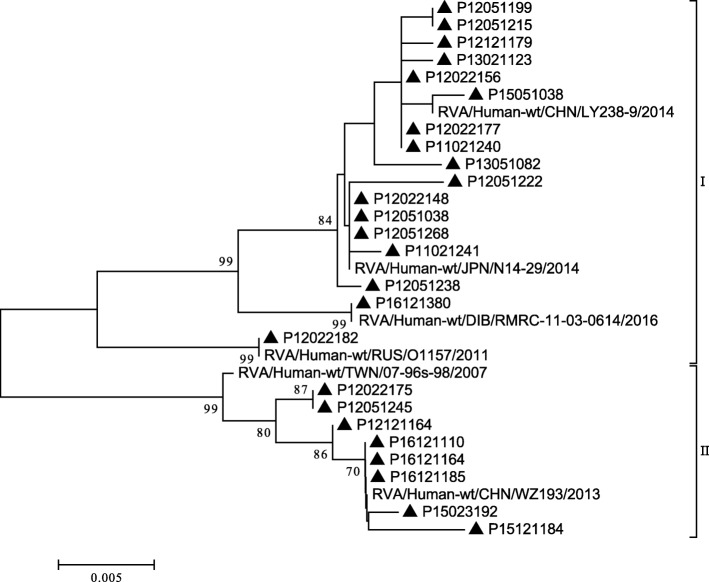
G genotypes were identified by phylogenetic analysis of VP7 genes. In this study, the G9 strains isolated were clustered into two minor lineages (I and II) (Fig. 3). Most of the G9 strains were in lineage I, which exhibited the highest nucleotide similarity (99.1%~ 100.0%) to the strain found in Japan (MI1128). In lineage II, only three strains in 2011 and one strain in 2013 were clustered separately. Lineage II exhibited the highest nucleotide similarity (98.8%~ 100.0%) to the strain found in China (WZ660). Moreover, the phylogenetic analysis of VP7 genes indicated that strains were all clustered correctly for genotypes G1, G2 and G3 (Fig. 4).
Fig. 3.

Phylogenetic analysis based on partial VP7 genes (790 bp) of G9 that were identified in Beijing. The trees were generated using the neighbor-joining method with the Kimura 2-parameter model. Bootstrap values estimated with 1000 replicate data sets were indicated at each node. The scale bar indicated the number of nucleotide substitutions per site. Bootstrap values lower than 70% are not shown
Fig. 4.
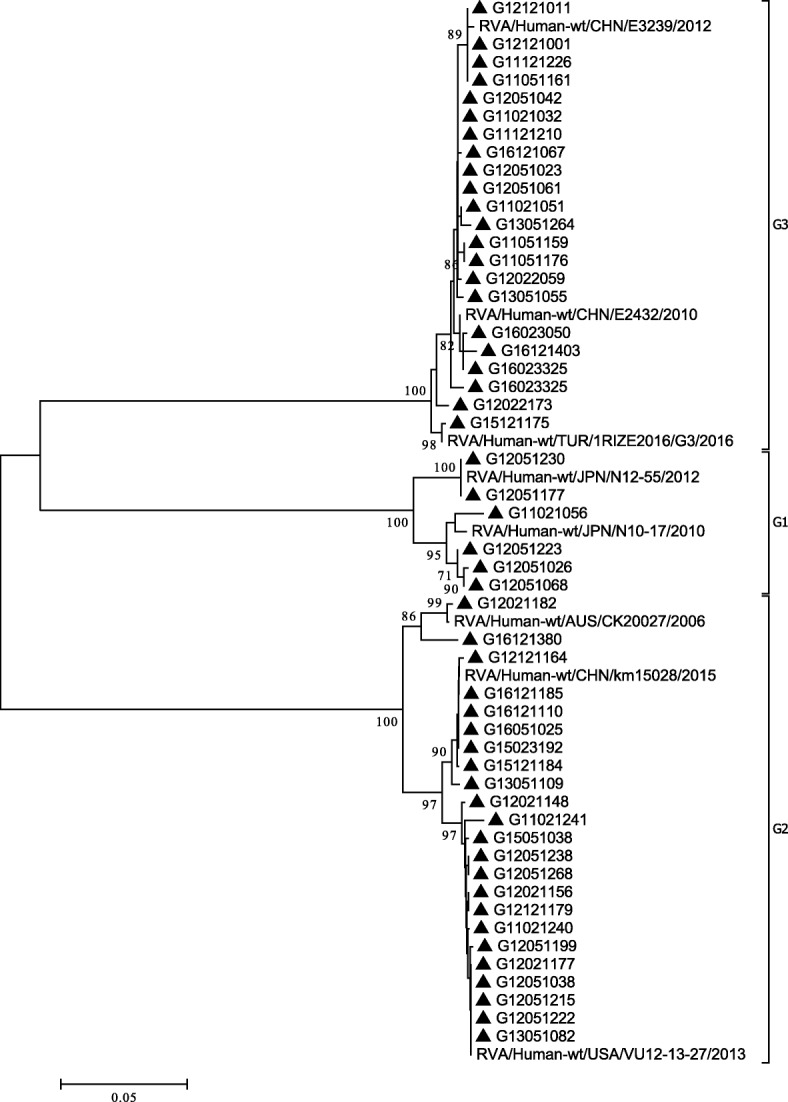
Phylogenetic analysis based on partial VP7 genes (790 bp) of G1, G2, and G3 that were identified in Beijing. The trees were generated using the neighbor-joining method with the Kimura 2-parameter model. Bootstrap values estimated with 1000 replicate data sets were indicated at each node. The scale bar indicated the number of nucleotide substitutions per site. Bootstrap values lower than 70% are not shown
Phylogenetic analysis of VP4 genes
P genotypes were identified by phylogenetic analysis of VP4 genes. In this study, the P [8] strains isolated were clustered into two minor lineages (P[8]b and P[8]c) (Fig. 5). Most of the P [8] strains were P[8]b, which exhibited the highest nucleotide similarity (97.5%~ 100.0%) to the strain found in Australia (CK00095). Fifteen strains in 2011 and 2012 were clustered separately, which exhibited 97.8%~ 99.5% nucleotide similarity to the strain found in Russia (Nov10-N709).
Fig. 5.
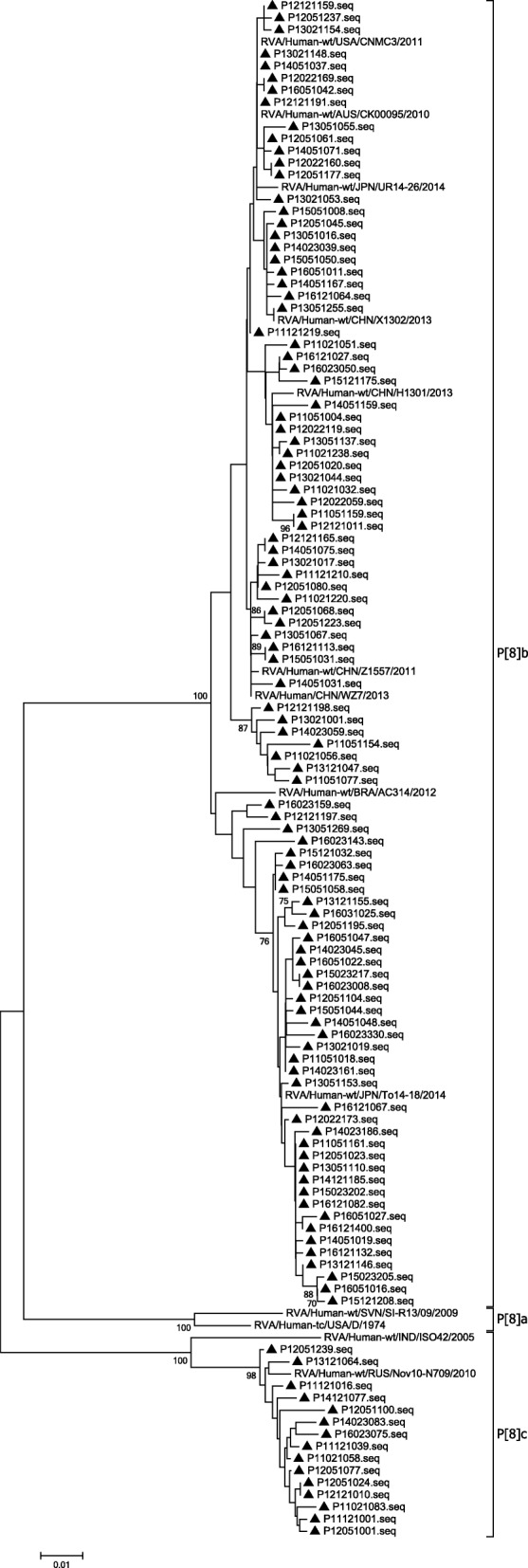
Phylogenetic analysis based on partial VP4 genes (615 bp) of P [8] that were identified in Beijing. The trees were generated using the neighbor-joining method with the Kimura 2-parameter model. Bootstrap values estimated with 1000 replicate data sets were indicated at each node. The scale bar indicated the number of nucleotide substitutions per site. Bootstrap values lower than 70% are not shown
The P [4] strains isolated were clustered into two minor lineages (I and II), similar to the results of Than VT et al. [29]. Phylogenetic analysis of P [4] genotypes in South Korea from 1989 to 2009 revealed that those strains clustered into two lineages, respectively. Lineage I of P [4] genotypes in our study exhibited 97.8%~ 100.0% nucleotide sequence similarity to the strain found in Dibrugarh (RMRC-11-03-0614). Only eight strains of P [4] strains were in lineage II, exhibited 99.3%~ 100.0% nucleotide sequence similarity to the strain found in China (WZ193) (Fig. 6).
Fig. 6.
Phylogenetic analysis based on partial VP4 genes (615 bp) of P [4] that were identified in Beijing. The trees were generated using the neighbor-joining method with the Kimura 2-parameter model. Bootstrap values estimated with 1000 replicate data sets were indicated at each node. The scale bar indicated the number of nucleotide substitutions per site. Bootstrap values lower than 70% are not shown
Discussion
Understanding the distribution and antigenic variation of rotavirus infection among children under 5 years old in China can inform vaccination policy in the future. Our study showed that rotavirus was responsible for 20.8% of diarrhea in outpatient children in Beijing, ranging from 16.4 to 27.3%. Children aged 1–2 years old had the highest rotavirus-positive rate (29.0%), followed by those aged 6–11 months old (24.0%), and children aged 2–3 years old (17.9%). The circulation of rotavirus was seasonal, peaking from November to January each year in Beijing. Urban children had higher rotavirus-positive rates compared with children who lived in rural areas, probably reflecting crowding and sanitation. The data also shows significantly different rotavirus-positive rates in different years. G9P [8] had become the dominant genotype in recent years. Some lineages of VP7 and VP4 gene sequences were discovered in previous years, but not in recent years.
Previous studies showed that, the rotavirus-positive rate varied among different regions and years in China. These studies showed that the rotavirus-positive rates of outpatient service ranged from 14.1 to 40.8%, with the rotavirus-positive of hospitalization was even higher in previous studies [30–33]. Our rate was lower than in Gansu Province (66/229, 28.8%) [26], but higher than Shanghai (138/1479, 9.3%) [25].
Most children infected with rotavirus at least once before the age of 5 years old [1, 34]. A previous study [28] also presented similar results which showed children aged 6 months to 3 years old were the most vulnerable to rotavirus infection. Rotavirus-positive rate of children under 6 months is lower (9.0%), possibly due to maternal antibodies remained and less contact with the outside world.
There was a strong association between rotavirus infection in childhood and climate factors, with highest rotavirus-positive rates during the cold season [35]. For every 1 °C increase in mean temperature, the rotavirus incidence reduced by 10% [36]. Data from 11 sentinel hospitals in China from August 2003 through July 2007 [23] showed rotavirus-positive peaks in winter in all regions. Our result was consistent with this finding.
Our research found that children who lived in rural areas had lower rotavirus-positive rates compared with urban children. This result was similar to a study in Northern Cameroon [37]. Less crowding and better sanitation in rural areas may explain this difference. However, medical consultations were the highest in Beijing hospitals, two of which were in the city, one in the suburbs. This might result in sampling bias. Further, children in rural areas may have less access to hospitals or to clinics.
Epidemic strains vary over time and place in both developed and developing countries. G12P [8] was the dominant genotype in children in the United States during 2011 to 2013 [38]. G1P [8] was the most predominant genotype in Japan during 2014 to 2015 [39]. However, G12P [8], G1P [8] and G12P [6] were the most common strains in Myanmar, Sir Lanka and Nepal respectively of south-east Asian from 2009 to 2015 [40]. In China, from 2000 through 2013, G3P [8] was the major stain in Wuhan, and there was genetic evolution [14]. In our study, G3P [8] and G9P [8] were both the dominant genotypes, accounting for 24.2 (24/99) and 22.2% (22/99) respectively in 2011. However, G9 became the main genotype through these years in China [41], accounting for 56.1% (101/180) in Beijing in 2012. Our results confirmed that G9P [8] was the dominant genotype from 2013 to 2016, accounting for more than 70% of samples. Many factors could influence the rotavirus-positive rate in surveillance studies, such as socioeconomic factors [42], access to health facilities, sampling, serotype [43], density of population, vaccination rate [3], crowding, education [44], malnutrition [45], sanitation, geographic and behavioral factors [35]. Our longstanding hospital-based surveillance provided insight into trends in rotavirus infection. Some uncommon strains such as G8P [8], G2P [8], G3P [4] and G9P [4] were detected in this study, but not G12 [46, 47].
Evolutionary trees in this study were used to verify that the G and P strains were divided into different genotypes correctly. There were at least ten branches of VP7 gene sequences of G9 type, originating from people or pigs. Our study showed that G9 strains clustered into two lineages. The main lineage of G9 strains were similar to that in Japan, the United States, China and other countries. In another lineage, there were only four strains. These were similar to strains found in USA (F45) in the past. But in recent 3 years, this strain has not been circulating. The VP4 gene sequences of P [8] clustered into most P[8]b and P[8]c lineages. Only fifteen strains were P[8]c mostly before 2014, similar to the strain found in Russia (Nov10-N709). However, P[8]b was widely prevalent in Beijing in recent years. The P[8]b in Beijing was similar to that in Japan (UR14–26, To14–18), the United States (CNMC3), China (X1302, Z1557) and other countries of the world in recent years. According to previous studies [48, 49], some P[8]b strains might have occurred through reassortment between P[8]a strains and unidentified strains with the P[8]b-VP4 gene. However, P[8]a were not detected in our study.
In conclusion, our study highlights that rotavirus causes a high disease burden in children under 5 years in Beijing, with a change in predominant strains over time. At present, G9P [8] is the dominant strain. Further research is needed to explore the related factors influencing genotypes selection, and correlation between genotypes with clinical illness severity in children. The economic burden of rotavirus-positive and the cost-effectiveness of a vaccination program are also should be considered. Currently there is no specific drug for the rotavirus diarrhea, but vaccines are available and can protect children from illness and death caused by rotavirus. In 2009, WHO recommended that all countries should promote the rotavirus vaccine [50]. RotaTeq® (RV5) and Rotarix® (RV1) were included in the national immunization program in 84 countries by the end of 2015 [4], and 33 African countries by 2016 [51], but not in China. Understanding the local and national distribution of rotavirus strains is vital for targeted research and development of vaccine for China. Available rotavirus vaccination could be considered in China to prevent rotavirus infection and reduce the burden of disease [52, 53].
Conclusions
Rotavirus was a major cause of childhood diarrhea in Beijing during 2011 to 2016. The rotavirus-positive rate of 2012 and 2013 were higher than 2011, the dominant genotype changed from G3P [8] to G9P [8] in that 2 years. At present, G9P [8] is the dominant strain. Targeted measures, such as available rotavirus vaccination could be considered in China to prevent rotavirus infection and reduce the morbidity and mortality.
Acknowledgements
We acknowledge the staffs in the Xicheng, Chaoyang and Tongzhou CDC and surveillance hospitals, who collected demographical data and stool specimens.
Funding
Not applicable. There is no support or funding to report.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Authors’ contributions
YT wrote the first draft and conducted the bioinformatics analysis. AC and DH were in charge of modifying the manuscript. YC and LJ carried out the epidemiological investigation. ZG and HY conducted the detection of specimens. BL collected the data and set up the database, QW and RM reviewed and edited the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Institutional Review Board and Human Research Ethics Committee of Beijing Center for Disease Prevention and Control. The agreement and verbal informed consent was obtained. The children and their families about the study prospectively were informed and asked for their consent.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yi Tian, Email: tianyixingfu_1@163.com.
Abrar Ahmad Chughtai, Email: abrar.chughtai@unsw.edu.au.
Zhiyong Gao, Email: zhiyonggao1@163.com.
Hanqiu Yan, Email: hqyan907@sina.com.
Yanwei Chen, Email: chen1988yanwei@sina.com.
Baiwei Liu, Email: yy0728@sohu.com.
Da Huo, Email: howardhuo@yahoo.com.
Lei Jia, Email: lailajia@126.com.
Quanyi Wang, Phone: (86) 10 6440 7109, Email: bjcdcxm@126.com.
Chandini Raina MacIntyre, Email: r.macintyre@unsw.edu.au.
References
- 1.Najafi A, Kargar M, Jafarpour T. Burden and typing of rotavirus group a in children with acute gastroenteritis in shiraz, southern Iran. Iran Red Crescent Med J. 2012;14(9):531–540. [PMC free article] [PubMed] [Google Scholar]
- 2.Operario DJ, Platts-Mills JA, Nadan S, Page N, Seheri M, Mphahlele J, Praharaj I, Kang G, Araujo IT, Leite JPG, Cowley D, Thomas S, Kirkwood CD, Dennis F, Armah G, Mwenda JM, Wijesinghe PR, Rey G, Grabovac V, Berejena C, Simwaka CJ, Uwimana J, Sherchand JB, Thu HM, Galagoda G, Bonkoungou IJO, Jagne S, Tsolenyanu E, Diop A, Enweronu-Laryea C, Borbor SA, Liu J, McMurry T, Lopman B, Parashar U, Gentsch J, Steele AD, Cohen A, Serhan F, Houpt ER. Etiology of severe acute watery diarrhea in children in the global rotavirus surveillance network using quantitative polymerase chain reaction. J Infect Dis. 2017;216(2):220–227. doi: 10.1093/infdis/jix294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewnard JA, Lopman BA, Parashar UD, Bar-Zeev N, Samuel P, Guerrero ML, Ruiz-Palacios GM, Kang G, Pitzer VE. Naturally acquired immunity against rotavirus infection and gastroenteritis in children: paired Reanalyses of birth cohort studies. J Infect Dis. 2017;216(3):317–326. doi: 10.1093/infdis/jix310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization. (2016). Media centre: Immunization coverage. [online] Available: http://www.who.int/mediacentre/factsheets/fs378/en/ (July 2017)
- 5.Wu D, Yen C, Yin ZD, Li YX, Liu N, Liu YM, Wang HQ, Cui FQ, Gregory CJ, Tate JE, Parashar UD, Yin DP, Li L. The public health burden of rotavirus disease in children younger than five years and considerations for rotavirus vaccine introduction in China. Pediatr Infect Dis J. 2016;35(12):e392–e3e8. doi: 10.1097/INF.0000000000001327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J, Duan Z, Payne DC, Yen C, Pan X, Chang Z, Liu N, Ye J, Ren X, Tate JE, Jiang B, Parashar UD. Rotavirus-specific and overall diarrhea mortality in Chinese children younger than 5 years: 2003 to 2012. Pediatr Infect Dis J. 2015;34(10):e233–e237. doi: 10.1097/INF.0000000000000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babji S, Arumugam R, Priyahemavathy R, Sriraman A, Sarvanabhavan A, Manickavasagam P, Simon A, Aggarwal I, Moses PD, Arora R, Kang G. Vaccine. 2017. Genotype distribution of group a rotavirus from southern India, 2005-2016. [DOI] [PubMed] [Google Scholar]
- 8.Coluchi N, Munford V, Manzur J, Vazquez C, Escobar M, Weber E, Marmol P, Racz ML. Detection, subgroup specificity, and genotype diversity of rotavirus strains in children with acute diarrhea in Paraguay. J Clin Microbiol. 2002;40(5):1709–1714. doi: 10.1128/JCM.40.5.1709-1714.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desselberger U. Rotaviruses. Virus Res. 2014;190:75–96. doi: 10.1016/j.virusres.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 10.Pacilli M, Cortese MM, Smith S, Siston A, Samala U, Bowen MD, Parada JP, Tam KI, Rungsrisuriyachai K, Roy S, Esona MD, Black SR. Outbreak of gastroenteritis in adults due to rotavirus genotype G12P[8] Clin Infect Dis. 2015;61(4):e20–e25. doi: 10.1093/cid/civ294. [DOI] [PubMed] [Google Scholar]
- 11.Jain S, Thakur N, Vashistt J, Grover N, Krishnan T, Changotra H. Predominance of unusual rotavirus G1P[6] strain in North India: an evidence from hospitalized children and adult diarrheal patients. Infect Genet Evol. 2016;46:65–70. doi: 10.1016/j.meegid.2016.10.021. [DOI] [PubMed] [Google Scholar]
- 12.Do LP, Kaneko M, Nakagomi T, Gauchan P, Agbemabiese CA, Dang AD, Nakagomi O. Molecular epidemiology of rotavirus a, causing acute gastroenteritis hospitalizations among children in Nha Trang, Vietnam, 2007-2008: identification of rare G9P[19] and G10P[14] strains. J Med Virol. 2017;89(4):621–631. doi: 10.1002/jmv.24685. [DOI] [PubMed] [Google Scholar]
- 13.Santos N, Hoshino Y. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol. 2005;15(1):29–56. doi: 10.1002/rmv.448. [DOI] [PubMed] [Google Scholar]
- 14.Wang YH, Pang BB, Ghosh S, Zhou X, Shintani T, Urushibara N, Song YW, He MY, Liu MQ, Tang WF, Peng JS, Hu Q, Zhou DJ, Kobayashi N. Molecular epidemiology and genetic evolution of the whole genome of G3P[8] human rotavirus in Wuhan, China, from 2000 through 2013. PLoS One. 2014;9(3):e88850. doi: 10.1371/journal.pone.0088850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alam MM, Khurshid A, Shaukat S, Suleman RM, Sharif S, Angez M, Malik SA, Ahmed TM, Aamir UB, Naeem M, Zaidi SS. Epidemiology and genetic diversity of rotavirus strains in children with acute gastroenteritis in Lahore, Pakistan. PLoS One. 2013;8(6):e67998. doi: 10.1371/journal.pone.0067998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Page N, Esona M, Armah G, Nyangao J, Mwenda J, Sebunya T, Basu G, Pyndiah N, Potgieter N, Geyer A, Steele AD. Emergence and characterization of serotype G9 rotavirus strains from Africa. J Infect Dis. 2010;202(Suppl):S55–S63. doi: 10.1086/653551. [DOI] [PubMed] [Google Scholar]
- 17.Phan TG, Khamrin P, Quang TD, Dey SK, Takanashi S, Okitsu S, Maneekarn N, Ushijima H. Detection and genetic characterization of group a rotavirus strains circulating among children with acute gastroenteritis in Japan. J Virol. 2007;81(9):4645–4653. doi: 10.1128/JVI.02342-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nirwati H, Wibawa T, Aman AT, Wahab A, Soenarto Y. Detection of group a rotavirus strains circulating among children with acute diarrhea in Indonesia. Springerplus. 2016;5:97. doi: 10.1186/s40064-016-1724-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang ZY, Yang H, Qi J, Zhang J, Sun LW, Tang JY, Ma L, Du ZQ, He A, Xie JP, Lu YY, Ji ZZ, Zhu BQ, Wu HY, Lin SE, Xie HP, Griffin DD, Ivanoff B, Glass RI, Gentsch JR. Diversity of rotavirus strains among children with acute diarrhea in China: 1998-2000 surveillance study. J Clin Microbiol. 2002;40(5):1875–1878. doi: 10.1128/JCM.40.5.1875-1878.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orenstein EW, Fang ZY, Xu J, Liu C, Shen K, Qian Y, Jiang B, Kilgore PE, Glass RI. The epidemiology and burden of rotavirus in China: a review of the literature from 1983 to 2005. Vaccine. 2007;25(3):406–413. doi: 10.1016/j.vaccine.2006.07.054. [DOI] [PubMed] [Google Scholar]
- 21.Wang YH, Kobayashi N, Zhou DJ, Yang ZQ, Zhou X, Peng JS, Zhu ZR, Zhao DF, Liu MQ, Gong J. Molecular epidemiologic analysis of group a rotaviruses in adults and children with diarrhea in Wuhan city, China, 2000-2006. Arch Virol. 2007;152(4):669–685. doi: 10.1007/s00705-006-0904-y. [DOI] [PubMed] [Google Scholar]
- 22.Li DD, Liu N, Yu JM, Zhang Q, Cui SX, Zhang DL, Yang SH, Cao DJ, Xu ZQ, Duan ZJ. Molecular epidemiology of G9 rotavirus strains in children with diarrhoea hospitalized in mainland China from January 2006 to December 2007. Vaccine. 2009;27(Suppl 5):F40–F45. doi: 10.1016/j.vaccine.2009.08.073. [DOI] [PubMed] [Google Scholar]
- 23.Duan ZJ, Liu N, Yang SH, Zhang J, Sun LW, Tang JY, Jin Y, Du ZQ, Xu J, Wu QB, Tong ZL, Gong ST, Qian Y, Ma JM, Liao XC, Widdowson MA, Jiang B, Fang ZY. Hospital-based surveillance of rotavirus diarrhea in the People's Republic of China, august 2003-July 2007. J Infect Dis. 2009;200(Suppl 1):S167–S173. doi: 10.1086/605039. [DOI] [PubMed] [Google Scholar]
- 24.Rahman Mustafizur, Yang Xue-lei, Sun He, Mahzebin Khandaker, Verstappen Natasha Willemina, Novo Luís, Matthijnssens Jelle, Van Ranst Marc. Emerging G9 rotavirus strains in the northwest of China. Virus Research. 2008;137(1):157–162. doi: 10.1016/j.virusres.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Shen Z, Wang G, Zhang W, Qian F, Li Y, Zhang M, Gu S, Wang M, Lin F, Hu Y, Yuan Z, Zhang J. Rotavirus infection and its genetic characterization in non-hospitalized adults with acute gastroenteritis in Shanghai, China. Archives of virology. 2013;158(8):1671–1677. doi: 10.1007/s00705-013-1663-1. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Liu H, Jia L, Payne DC, Hall AJ, Xu Z, Gao Z, Chang Z, Jiang B, Parashar UD, Meng L, Yu H, Duan Z. Active, population-based surveillance for rotavirus gastroenteritis in Chinese children: Beijing municipality and Gansu Province, China. Pediatr Infect Dis J. 2015;34(1):40–46. doi: 10.1097/INF.0000000000000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kheyami AM, Nakagomi T, Nakagomi O, Dove W, Hart CA, Cunliffe NA. Molecular epidemiology of rotavirus diarrhea among children in Saudi Arabia: first detection of G9 and G12 strains. J Clin Microbiol. 2008;46(4):1185–1191. doi: 10.1128/JCM.02244-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuccotti G, Meneghin F, Dilillo D, Romano L, Bottone R, Mantegazza C, Giacchino R, Besana R, Ricciardi G, Sterpa A, Altamura N, Andreotti M, Montrasio G, Macchi L, Pavan A, Paladini S, Zanetti A, Radaelli G. Epidemiological and clinical features of rotavirus among children younger than 5 years of age hospitalized with acute gastroenteritis in northern Italy. BMC Infect Dis. 2010;10:218. doi: 10.1186/1471-2334-10-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Than VT, Kim W. Prevalence of rotavirus genotypes in South Korea in 1989-2009: implications for a nationwide rotavirus vaccine program. Korean J Pediatr. 2013;56(11):465–473. doi: 10.3345/kjp.2013.56.11.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou N, Lv D, Wang S, Lin X, Bi Z, Wang H, Wang P, Zhang H, Tao Z, Hou P, Song Y, Xu A. Continuous detection and genetic diversity of human rotavirus a in sewage in eastern China, 2013-2014. Virol J. 2016;13(1):153. doi: 10.1186/s12985-016-0609-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu J, Jing H, Lai S, Xu W, Li M, Wu J, Liu W, Yuan Z, Chen Y, Zhao S, Wang X, Zhao Z, Ran L, Wu S, Klena JD, Feng L, Li F, Ye X, Qiu Y, Yu H, Li Z, Yang W. Etiology of diarrhea among children under the age five in China: results from a five-year surveillance. J Inf Secur. 2015;71(1):19–27. doi: 10.1016/j.jinf.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu XH, Tian L, Cheng ZJ, Liu WY, Li S, Yu WT, Zhang WQ, Xiang X, Sun ZY. Viral and bacterial etiology of acute diarrhea among children under 5 years of age in Wuhan, China. Chin Med J. 2016;129(16):1939–1944. doi: 10.4103/0366-6999.187852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu L, Qian Y, Zhang Y, Zhao L, Jia L, Dong H. Epidemiological aspects of rotavirus and adenovirus in hospitalized children with diarrhea: a 5-year survey in Beijing. BMC Infect Dis. 2016;16(1):508. doi: 10.1186/s12879-016-1829-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jadali F, Karimi A, Fallah F, Zahraei M, Esteghamati A, Navidinia M, Adabian S. A survey on rotavirus associated diarrhea in 5 Main cities of Iran. Arch Pediatr Infect Dis. 2013;1(1):23–26. doi: 10.5812/pedinfect.6431. [DOI] [Google Scholar]
- 35.Patel MM, Pitzer VE, Alonso WJ, Vera D, Lopman B, Tate J, Viboud C, Parashar UD. Global seasonality of rotavirus disease. Pediatr Infect Dis J. 2013;32(4):e134–e147. doi: 10.1097/INF.0b013e31827d3b68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levy K, Hubbard AE, Eisenberg JN. Seasonality of rotavirus disease in the tropics: a systematic review and meta-analysis. Int J Epidemiol. 2008;38(6):1487–1496. doi: 10.1093/ije/dyn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ndze VN, Akum AE, Kamga GH, Enjema LE, Esona MD, Banyai K, Therese OA. Epidemiology of rotavirus diarrhea in children under 5 years in northern Cameroon. Pan Afr Med J. 2012;11:73. [PMC free article] [PubMed] [Google Scholar]
- 38.Payne DC, Currier RL, Staat MA, Sahni LC, Selvarangan R, Halasa NB, Englund JA, Weinberg GA, Boom JA, Szilagyi PG, Klein EJ, Chappell J, Harrison CJ, Davidson BS, Mijatovic-Rustempasic S, Moffatt MD, McNeal M, Wikswo M, Bowen MD, Morrow AL, Parashar UD. Epidemiologic association BetweenFUT2Secretor status and severe rotavirus gastroenteritis in children in the United States. JAMA Pediatr. 2015;169(11):1040. doi: 10.1001/jamapediatrics.2015.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khandoker N, Thongprachum A, Takanashi S, Okitsu S, Nishimura S, Kikuta H, Yamamoto A, Sugita K, Baba T, Kobayashi M, Hayakawa S, Mizuguchi M, Ushijima H. Molecular epidemiology of rotavirus gastroenteritis in Japan during 2014-2015: characterization of re-emerging G2P[4] after rotavirus vaccine introduction. J Med Virol. 2018;90(6):1040–1046. doi: 10.1002/jmv.25067. [DOI] [PubMed] [Google Scholar]
- 40.Giri S, Priya Hemavathy R, Arumugam R, Sherchand JB, Thu HM, Galagoda G, Myat TW, Abeysinghe N, Gunasekara M, Janakan N, Pradhan R, Bura V, Wijesinghe P, Kang G. Vaccine. 2018. Molecular epidemiology of rotaviruses in the south-east Asian region from 2009 to 2015. [DOI] [PubMed] [Google Scholar]
- 41.Zhang S, Yin J, Yang J, Tian L, Li D, Zhang Q, Chen J, Xu W, Zhou X. Epidemiology and genetic diversity of group a rotavirus in acute diarrhea patients in pre-vaccination era in Southwest China. J Med Virol. 2017;89(1):71–78. doi: 10.1002/jmv.24606. [DOI] [PubMed] [Google Scholar]
- 42.Wang X, Wang J, Sun H, Xia S, Duan R, Liang J, Xiao Y, Qiu H, Shan G, Jing H. Etiology of childhood infectious diarrhea in a developed region of China: compared to childhood diarrhea in a developing region and adult diarrhea in a developed region. PLoS One. 2015;10(11):e0142136. doi: 10.1371/journal.pone.0142136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saluja T, Dhingra MS, Sharma SD, Gupta M, Kundu R, Kar S, Dutta AK, Silveira MD, Singh JV, Kamath VG, Chaudhary A, Rao V, Ravi MD, Murthy K, Arumugam R, Moureau A, Prasad R, Patnaik BN. Association of rotavirus strains and severity of gastroenteritis in Indian children. Hum Vaccin Immunother. 2017;13(3):711–716. doi: 10.1080/21645515.2016.1238994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al-Badani A, Al-Areqi L, Majily A, Al-Sallami S, Al-Madhagi A, Amood A-KM. Rotavirus diarrhea among children in Taiz, Yemen: Prevalence-Risk Factors and Detection of Genotypes. Int J Pediatr. 2014;2014:928529. doi: 10.1155/2014/928529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Das Sumon Kumar, Chisti Mohammod Jobayer, Sarker Mohammad Habibur Rahman, Das Jui, Ahmed Shawnawaz, Shahunja K. M., Nahar Shamsun, Gibbons Nora, Ahmed Tahmeed, Faruque Abu Syed Golam, Rahman Mustafizur, J Fuchs George, Al Mamun Abdullah, John Baker Peter. Long-term impact of changing childhood malnutrition on rotavirus diarrhoea: Two decades of adjusted association with climate and socio-demographic factors from urban Bangladesh. PLOS ONE. 2017;12(9):e0179418. doi: 10.1371/journal.pone.0179418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pun SB, Nakagomi T, Sherchand JB, Pandey BD, Cuevas LE, Cunliffe NA, Hart CA, Nakagomi O. Detection of G12 human rotaviruses in Nepal. Emerg Infect Dis. 2007;13(3):482–484. doi: 10.3201/eid1303.061367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ansari S, Sherchand JB, Rijal BP, Parajuli K, Mishra SK, Dahal RK, Shrestha S, Tandukar S, Chaudhary R, Kattel HP, Basnet A, Pokhrel BM. Characterization of rotavirus causing acute diarrhoea in children in Kathmandu, Nepal, showing the dominance of serotype G12. J Med Microbiol. 2013;62(Pt 1):114–120. doi: 10.1099/jmm.0.048124-0. [DOI] [PubMed] [Google Scholar]
- 48.Wang YH, Zhou X, Ghosh S, Zhou DJ, Pang BB, Peng JS, Hu Q, Kobayashi N. Prevalence of human rotavirus genotypes in Wuhan, China, during 2008-2011: changing trend of predominant genotypes and emergence of strains with the P[8]b subtype of the VP4 gene. Arch Virol. 2011;156(12):2221–2231. doi: 10.1007/s00705-011-1122-9. [DOI] [PubMed] [Google Scholar]
- 49.Cashman O, Collins PJ, Lennon G, Cryan B, Martella V, Fanning S, Staines A, O'Shea H. Molecular characterization of group a rotaviruses detected in children with gastroenteritis in Ireland in 2006-2009. Epidemiol Infect. 2012;140(2):247–259. doi: 10.1017/S0950268811000306. [DOI] [PubMed] [Google Scholar]
- 50.World Health Organization. WHO position paper-January 2013. Wkly Epidemiol Rec. 2013;88(5):49–64.
- 51.Shah MP, Tate JE, Mwenda JM, Steele AD, Parashar UD. Estimated reductions in hospitalizations and deaths from childhood diarrhea following implementation of rotavirus vaccination in Africa. Expert Rev Vaccines. 2017;16(10):987–995. doi: 10.1080/14760584.2017.1371595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burnett E, Jonesteller CL, Tate JE, Yen C, Parashar UD. Global impact of rotavirus vaccination on childhood hospitalizations and mortality from diarrhea. J Infect Dis. 2017;215(11):1666–1672. doi: 10.1093/infdis/jix186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zaman K, Sack DA, Neuzil KM, Yunus M, Moulton LH, Sugimoto JD, Fleming JA, Hossain I, Arifeen SE, Azim T, Rahman M, Lewis KDC, Feller AJ, Qadri F, Halloran ME, Cravioto A, Victor JC. Effectiveness of a live oral human rotavirus vaccine after programmatic introduction in Bangladesh: a cluster-randomized trial. PLoS Med. 2017;14(4):e1002282. doi: 10.1371/journal.pmed.1002282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.