Abstract
Background
When developing CRISPR/Cas9 systems for crops, it is crucial to invest time characterizing the genome editing efficiency of the CRISPR/Cas9 cassettes, especially if the transformation system is difficult or time-consuming. Cotton is an important crop for the production of fiber, oil, and biofuel. However, the cotton stable transformation is usually performed using Agrobacterium tumefaciens taking between 8 and 12 months to generate T0 plants. Furthermore, cotton is a heterotetraploid and targeted mutagenesis is considered to be difficult as many genes are duplicated in this complex genome. The application of CRISPR/Cas9 in cotton is severely hampered by the long and technically challenging genetic transformation process, making it imperative to maximize its efficiency.
Results
In this study, we provide a new system to evaluate and validate the efficiency of CRISPR/Cas9 cassettes in cotton using a transient expression system. By using this system, we could select the most effective CRISPR/Cas9 cassettes before the stable transformation. We have also optimized the existing cotton CRISPR/Cas9 system to achieve vastly improved mutagenesis efficiency by incorporating an endogenous GhU6 promoter that increases sgRNA expression levels over the Arabidopsis AtU6-29 promoter. The 300 bp GhU6.3 promoter was cloned and validated using the transient expression system. When sgRNAs were expressed under the control of the GhU6.3 promoter in CRISPR/Cas9 cassettes, expression levels were 6–7 times higher than those provided by the AtU6-29 promoter and CRISPR/Cas9-mediated mutation efficiency was improved 4–6 times.
Conclusions
This study provides essential improvements to maximize CRISPR/Cas9-mediated mutation efficiency by reducing risk and workload for the application of CRISPR/Cas9 approaches in the targeted mutagenesis of cotton.
Electronic supplementary material
The online version of this article (10.1186/s13007-018-0353-0) contains supplementary material, which is available to authorized users.
Keywords: CRISPR/Cas9, U6 promoter, Genome editing, Target mutagenesis, Transient expression
Background
Targeted genome editing is an extremely useful tool for basic and applied plant research and a number of techniques such as meganucleases, zinc finger nucleases (ZFNs) and transcription activator-like effectors nucleases (TALENs) had been available for some time [1–3]. Nevertheless, the technical complexity of the above mentioned technologies precluded its adoption by the wider scientific community until the advent of CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9). The intrinsic versatility, simplicity and high efficiency of CRISPR/Cas9 has resulted in an explosion of research using genome-editing as the preferred method to generate precise alterations in the genome of numerous plant species [4–10]. CRISPR/Cas9 derives from a microbial adaptive immune system and its major components are the Cas9 nuclease capable of producing double strand breaks and a small guide RNA (sgRNA) which directs the Cas9 protein to the target site. A number of factors influence the efficiency of the CRISPR/Cas9 system with strong expression of Cas9 and sgRNA being essential to obtain high mutation rates [9, 10].
U6 small nuclear RNAs (snRNAs) are non-coding RNAs involved in intron splicing during the production of mature mRNA molecules in eukaryotic cells. The U6 promoter is a class III RNA polymerase III promoter that has been frequently used to drive high expression levels of small RNAs in plants and animals [11, 12] and has been the preferred choice to drive sgRNA expression in CRISPR/Cas9 vectors [13, 14]. In addition, the U6 promoter has a highly conserved transcription start site starting with a guanine nucleotide, which helps to improve the homogeneity of the transcribed sgRNA molecule and reduce off-target effects [12]. CRISPR/Cas9 vector systems using the U6 promoter to drive sgRNA expression have been successfully used in various plant species, with the OsU6a, OsU6b and OsU6c promoters from rice being the most commonly used for monocotyledons, and the Arabidopsis AtU6-1 and AtU6-29 promoters being the preferred ones for dicotyledons [10, 15, 16]. It has nevertheless become clear that the use of species-specific U6 promoters can result in increased sgRNA expression and thus enhanced editing efficiency [9, 17]. In soybean, for example, sgRNA levels driven by the endogenous GmU6 promoter were twice higher than those obtained using the Arabidopsis AtU6-26 promoter, resulting in massive improvements in gene editing efficiency (14.7–20.2% for GmU6 vs. 3.2–9.7% for AtU6-26) [17]. It is also important to keep in mind that plant genomes contain multiple U6 genes with different expression levels with the corollary that not all U6 promoters are equally efficient in driving gene expression [18, 19]. When developing CRISPR/Cas9 systems for new species it is important to invest time characterizing exogenous and endogenous U6 promoters to choose the optimal one, especially if the transformation system is difficult or time consuming.
Cotton is an important crop for the production of fiber, oil and biofuel. The genome of both sea-island and upland cotton were sequenced in 2015, paving the way for the use of tools such as CRISPR/Cas9 in genetic improvement programs [20–22]. Recent studies using protoplast transient transformation or transgenic plants through stable transformation have demonstrated that CRISPR/Cas9 can be used for gene editing in cotton [23–28]. However, despite the great potential shown by these reports, cotton is a specially difficult crop with many obstacles that must be overcome before large scale genetic improvement programs can be implemented through gene editing. Upland cotton (Gossypium hirsutum) is a heterotetraploid and targeted mutagenesis is considered to be difficult as many genes are duplicated in this complex genome [20, 21]. In addition, CRISPR/Cas9 applications are hindered by the labor-intensive and lengthy transformation protocols. Cotton stable transformation is usually performed using Agrobacterium tumefaciens taking between 8 to 12 months to generate T0 plants [23, 29]. Maximizing CRISPR/Cas9-mediated mutation efficiency is critical in order to reduce workload and facilitate genome editing approaches in cotton.
Producing highly efficient and abundant sgRNA transcripts in planta is crucial for genome editing. Previously, we described a transient transformation system to rapidly validate the efficiency of sgRNAs [23]. In this study, we provide an additional alternative method to evaluate the efficiency of target sequences in CRISPR/Cas9-mediated target mutagenesis. Furthermore, we optimized the cotton CRISPR/Cas9 system by enhancing sgRNA expression using an endogenous U6 promoter. The GhU6.3 promoter produced higher sgRNA transcript levels than the Arabidopsis AtU6-29, leading to a 4–6 times increase in genome editing efficiency when used in CRISPR/Cas9 cassettes. Our work provides a faster and more efficient pipeline for the use of genome editing in cotton basic and applied research.
Methods
Plant materials and growth conditions
Gossypium hirsutum L. variety ‘TM-1’ seeds were imbibed in deionized water for 3 h then incubated at 28 °C for germination. The germinated seeds were transferred into soil and grown at 25/28 °C (night/day) and 16 h/8 h light/dark cycle. Seedlings were grown for 10 days before being used for Agrobacterium transformation [30]. Tobacco plants were grown in a greenhouse (25 °C at 16 h/8 h light/dark cycle) for 3 weeks before leaves were used for transformation [31].
Promoter sequences analysis and vector construction
The U6 snRNA (small nuclear RNA) sequences from Arabidopsis were used to identify the cotton U6 gene using the genome of ‘TM-1’ as reference (http://mascotton.njau.edu.cn/info/1054/1118.htm). 1 kb fragments upstream of the predicted cotton U6 genes were isolated and cloned for sequencing. Predicted promoter sequences were sub-cloned into pGWB433 by Gateway cloning (Invitrogen, USA). The AtU6-29 promoter was cloned in to pGWB433 as positive control.
To construct the non-functional GUS vector, a 23 bp fragment that targets sgRNA-PDS [23] was inserted behind the GUS start codon. The modified GUS gene was called fsGUS, and cloned into pK2GW7.0 [31].
CRISPR/Cas9 vector construction was performed as previously described [23, 32]. Briefly, the U6 promoter and sgRNA scaffold were integrated by PCR and then ligated into the CRISPR/Cas9 expression cassette by Golden Gate cloning (NEB, USA).
Transient transformation in tobacco and cotton
All vectors were introduced into A. tumefaciens GV3101 for transient transformation, and GV3101 strains carrying the vector were grown in selection media at 28 °C. Agrobacterium cells were collected by centrifugation and suspended in infiltration medium [10 mM magnesium chloride, 10 mM 2-(N-morpholine) ethyl sulfonic acid, and 200 μm acetosyringone]. After incubating at room temperature for 3 h, the Agrobacterium suspension was infiltrated into tobacco leaves or cotton cotyledons. The experiments were repeated at least three times with more than 8 leaves per experiment [23, 33, 34].
GUS staining and enzyme activity determination
GUS staining was performed by incubating infiltrated plant leaves in GUS staining solution for 10 h at 37 °C. Leaves were then incubated in 75% ethanol to remove chlorophyll. Stained samples were analyzed using a Leica microscope (USA) and GUS activity was determined as described in previous study [35].
RT-PCR and qPCR analysis
Total RNA was isolated from cotton cotyledons 2 days after infiltration using the Aidlab RNA extraction kit (Aidlab Biotechnologies, China). First strand cDNA was synthesized from 1 μg of total RNA using the M-MLV reverse transcript system (Promega, USA). RT-PCR was performed at 95 °C for 3 min followed by 28–35 cycles of amplification (95 °C for 20 s, 55–60 °C for 20 s and 72 °C for 20 s). qRT-PCR was performed on an ABI 7500 Real Time PCR system (Applied Biosystems, USA) with SYBR green (Bio-Rad, USA). Relative gene expression levels were calculated using the 2−∆∆Ct method with the cotton Ubiquitin 7 gene (UB7) as the reference gene [33, 34]. Primers used for PCR amplification are listed in Additional file 1.
Results
Identification of U6 promoters in upland cotton
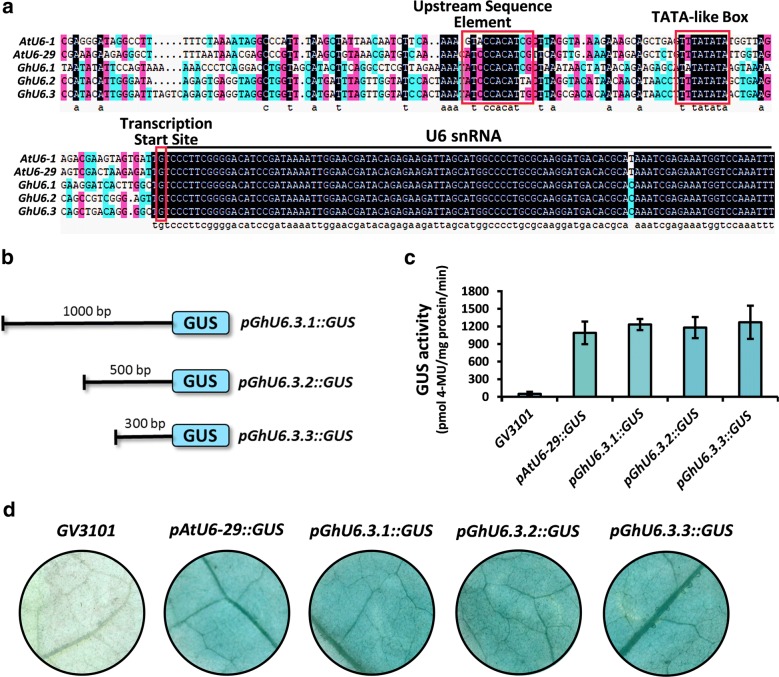
Arabidopsis thaliana U6 promoters are commonly used in dicot CRISPR/Cas9 cassettes to drive the expression of sgRNAs as they are thought to achieve high expression levels. We have successfully used AtU6-29 to mutagenize the Cloroplastos alterados 1 (CLA1) gene in transgenic G. hirsutum plants [23]. In an effort to optimize the CRISPR /Cas9 system in G. hirsutum, we performed BLAST searches of the cotton genome using AtU6-29 as query and identified three most homologous sequences showing high homology which were named GhU6.1, GhU6.2, and GhU6.3 (Fig. 1a). Although the U6 snRNA transcript sequences in cotton and Arabidopsis were almost identical, the promoter regions were very divergent except for the two motifs required for transcription, the upstream sequence element (USE) and the TATA-like Box (Fig. 1a). Among the G. hirsutum promoter sequences, GhU6.2, and GhU6.3 showed strong conservation in the first 467 bp upstream from the start of transcription while GhU6.1 was quite divergent (Additional file 2). The 1 kb sequences upstream of the transcription initiation site of three promoters were cloned into the pGWB433 vector upstream of the β-glucuronidase (GUS) reporter gene. Transient transformation experiments were performed in tobacco leaves by Agrobacterium infiltration using all promoter constructs. All the three promoters can promote GUS expression, and the GhU6.3 promoter shows the most stable and strong promoting ability than GhU6.1 and GhU6.2 promoter (Additional file 3). Therefore, the GhU6.3 promoter was chosen for further study (noted as proGhU6.3.1) and two genomic fragments of 500 bp and 300 bp (proGhU6.3.2, proGhU6.3.3) upstream of the transcription initiation site were cloned to build the vectors to drive GUS expression (Fig. 1b). An additional construct containing the Arabidopsis AtU6-29 promoter was also prepared for comparison purposes. GUS staining and quantitative GUS activity assays showed similar staining intensities and GUS activity values for pGhU6.3.1::GUS, pGhU6.3.2::GUS, pGhU6.3.3::GUS and pAtU6-29::GUS (Fig. 1c, d). We therefore used the shortest fragment, ProGhU6.3.3 (300 bp), to test its efficiency for CRISPR/Cas9-mediated gene editing in cotton.
Fig. 1.
Identification and validation of cotton U6 promoters. a Multiple alignments of cotton and Arabidopsis U6 gene and promoter sequences. Black line denotes the U6 snRNA transcript. USE (upstream sequence element), TATA-like Box and the transcription start site are labeled with red boxes. b Schematic representation of GUS expression constructs using different GhU6.3 promoter fragments. c GUS activity levels and d GUS staining in tobacco leaves infiltrated with Agrobacterium carrying different promoter constructs. The error bars indicate the standard deviation estimated from the eight replicates
The endogenous GhU6.3.3 promoter drives higher sgRNA expression levels than the Arabidopsis AtU6-29 promoter resulting in higher CRISPR/Cas9-mediated mutation efficiency in cotton
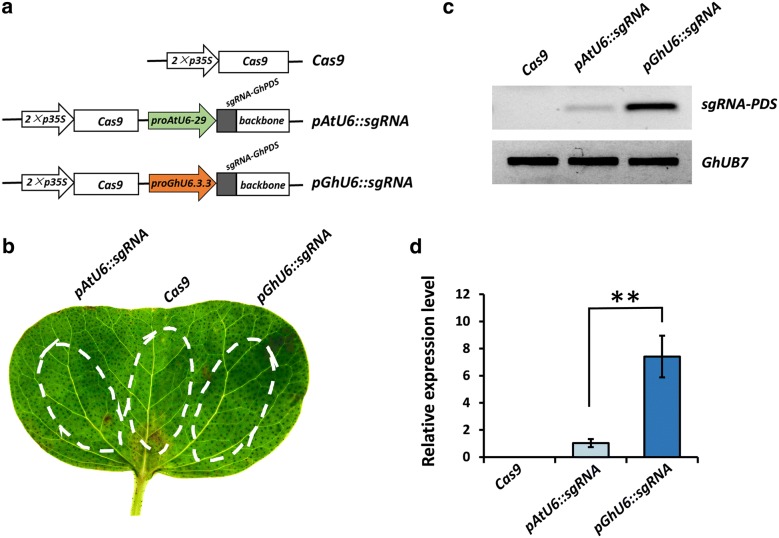
To compare the levels of sgRNA expression achieved by the proGhU6.3.3 and proAtU6-29 in cotton, we constructed two CRISPR vectors using each promoter to drive a previously designed sgRNA targeting the Phytoene desaturase (PDS) gene [23] while an empty vector (Cas9) was used as negative control (Fig. 2a). All constructs were introduced into A. tumefaciens GV3101 and transient expression experiments performed in cotton cotyledons by infiltrating each of the constructs in a different region of the same cotyledon (Fig. 2b). Cotyledons were harvested 48 h after infiltration and the levels of sgRNA-PDS transcripts quantified by RT-PCR and qRT-PCR. Our results showed that proGhU6.3.3 generated 6–7 times higher levels of sgRNA-PDS expression than proAtU6-29, suggesting that it could be a better choice for CRISPR/Cas9 applications in upland cotton (Fig. 2c, d).
Fig. 2.
The GhU6.3.3 promoter drives strong sgRNA expression in cotton. a Diagram of CRISPR/Cas9 expression vectors: Cas9, vector lacking sgRNA cassette used as negative control; pAtU6::sgRNA, CRISPR/Cas9 construct with sgRNA-PDS driven by proAtU6-29; pGhU6::sgRNA, CRISPR/Cas9 construct with sgRNA-PDS driven by proGhU6.3.3. b Schematic diagram of Agrobacterium-mediated transient expression in cotton cotyledon with different constructs. c sgRNA expression levels determined by RT-PCR and d qPCR (n > 8, **P < 0.01, t-test)
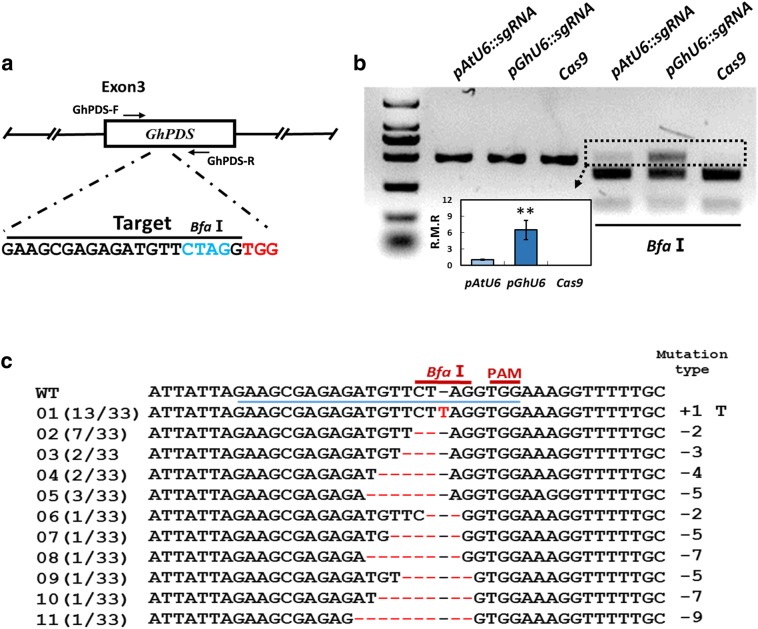
To determine whether the increased sgRNA expression levels driven by the GhU6.3.3 promoter resulted in improved genome editing efficiency in cotton, we performed transient expression experiments with the binary vectors described above (Fig. 2a). The target sequence for the sgRNA contains a BfaI restriction site adjacent to the protospacer adjacent motif (PAM), thus CRISPR/Cas9-mediated genome-editing events were expected to alter the BfaI recognition sequence (Fig. 3a). Cotyledons were infiltrated with Agrobacterium containing each of the three CRISPR/Cas9 cassettes, pGhU6::sgRNA, pAtU6::sgRNA and Cas9 (Fig. 2a), and tissue collected 3 days after infiltration. Genomic DNA was purified and a 486 bp genomic fragment containing the target sequence in the PDS gene amplified by PCR (Fig. 3b, lanes 1–3). Digestion of the amplicon with BfaI should yield two fragments of 340 bp and 146 bp respectively in WT sequences while genome editing events modifying the BfaI restriction site should result in the appearance of an uncut product. Electrophoretic analysis of the BfaI digested amplicons showed the existence of uncut products in the proGhU6.3.3 or proAtU6-29 samples (pAtU6::sgRNA and pGhU6::sgRNA, Fig. 3b, lane 4, 5), but not in the control containing a vector lacking sgRNA (Fig. 3b, lane 6). Densitometry analysis showed that the intensity of the uncut band in amplicons from pGhU6::sgRNA samples was approximately 4–6 times stronger than those obtained in pAtU6::sgRNA samples (Fig. 3b).
Fig. 3.
CRISPR/Cas9-induced targeted mutagenesis of GhPDS in cotton. a Position of the sgRNA-GhPDS target site in the GhPDS gene. GhPDS-F and GhPDS-R indicate primers used to amplify the target fragment. b Detection of CRISPR/Cas9 induced sgRNA-GhPDS mutations. Gel electrophoresis analysis of PCR amplicons of target fragments from Agrobacterium transient expression experiments using different CRISPR/Cas9 cassettes. 1–3: undigested PCR products; 4–6: PCR products digested with BfaI. The insert (column-value histogram) shows the relative mutation rates (n > 8, **P < 0.01, t-test). c Sequencing of mutated PCR products. The sgRNA target sequence is underlined in blue. Deletions are shown as red dashes, and insertions are denoted with red letters. The frequency of each mutation is shown on the left and the mutation types on the right
The uncut bands from samples transfected with pGhU6::sgRNA and pAtU6::sgRNA were purified from the gel and cloned into a plasmid vector to investigate the nature of the editing events in the GhPDS target site. Sequencing analysis of multiple colonies showed that the types and proportion of mutations were similar in samples transfected with pGhU6::sgRNA and pAtU6::sgRNA with 61% of clones harboring small deletions (Fig. 3c and Additional file 4).
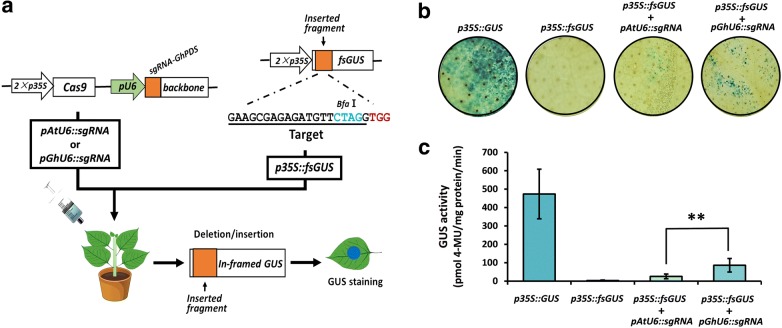
To confirm our results, we used the frame-shift GUS (fsGUS) reporter system [31] to further compare the efficiency of CRISPR/Cas9 cassettes containing either proGhU6.3.3 or proAtU6-29 to drive sgRNA expression. In this system, a 23 bp sequence including the sgRNA-PDS target was inserted into the GUS coding sequence, causing a frame shift and avoiding the production of functional GUS protein. CRISPR/Cas9-mediated gene editing typically generates a variety of mutations at the target site, some of which will correct the frame shift in fsGUS, resulting in a functional GUS protein that can be detected by staining and enzymatic activity assays (Fig. 4a). As shown in Fig. 4b, no GUS expression was observed in cotton cotyledons after infiltration with Agrobacterium harboring the p35S::fsGUS construct only. Cotyledons co-infiltrated with p35S::fsGUS and CRISPR/Cas9 constructs containing either pAtU6::sgRNA or pGhU6::sgRNA showed spots with blue coloration, suggesting the successful generation of mutations in fsGUS. In agreement with our previous results, co-infiltration with p35S::fsGUS and pGhU6::sgRNA produced a larger number of colored spots and a stronger color intensity than co-infiltration with p35S::fsGUS and pAtU6::sgRNA (Fig. 4b). Visual observations were complemented with GUS activity measures showing 4 times higher GUS activity values in the pGhU6::sgRNA samples (Fig. 4c).
Fig. 4.
Detection of CRISPR/Cas9-mediated mutation efficiency using the frame-shift GUS (fsGUS) system. a Diagram of the fsGUS reporting system. The 23 bp target sequence of sgRNA-PDS was inserted after the GUS start codon to generate fsGUS (p35S::fsGUS). CRISPR/Cas9 constructs (pAtU6::sgRNA and pGhU6::sgRNA) were co-expressed with p35S::fsGUS in cotton cotyledons, the p35S::GUS was used as positive control. b GUS staining of cotton cotyledons co-transfected with fsGUS and CRISPR/Cas9 constructs. c GUS activity assays of cotton cotyledons co-transfected with fsGUS and CRISPR/Cas9 constructs (n > 8, **P < 0.01, t-test)
Discussion
Cotton accounts for 40% of the international fiber market making it irreplaceable in the world economy. Traditional breeding has failed to keep up with the demand for yield and quality improvement gradually making molecular breeding the method of choice for cotton as it can shorten the breeding cycle while maintaining high-quality traits. The sequencing of the diploid and allotetraploid cotton genomes, combined with recent transcriptomics and proteomics work [20–22, 36–40], have provided an invaluable resource for genetic studies and the development of innovative biotechnological approaches for cotton improvement. The CRISPR/Cas9 system allows the production of precise targeted mutations in the genome and can generate transgene-free mutants, potentially avoiding costly regulatory requirements associated with genetically manipulated crops [4, 6–8, 10, 16]. Although CRISPR/Cas9 has now been used in many crop species, its application in cotton is severely hampered by the long and technically challenging genetic transformation process, making it imperative to maximize its efficiency.
With a few notable exceptions, genetic transformation is a time consuming and labor intensive process emphasizing the need to optimize CRISPR/Cas9-mediated mutagenesis efficiency. The two core elements of the CRISPR/Cas9 system are the Cas9 nuclease and the associated sgRNA. Previous studies have shown that gene editing activity is strongly reliant on achieving strong expression of both elements and thus the choice of promoters is essential for the overall mutagenesis efficiency [7–10, 17]. Strong constitutive RNA polymerase II promoters such as the cauliflower mosaic virus 35S (CaMV 35S) and the ubiquitin promoters are typically used for the control of Cas9 expression in stably transformed plants obtained using tissue culture-based protocols [10, 32]. In contrast, the CaMV 35S promoter has proven to be sub-optimal for Arabidopsis transformed using the floral dipping method with germ line specific and cell division specific promoters proving vastly superior [41]. Improvements have also been achieved by optimizing Cas9 codon usage for the species of interest in order to enhance translation efficiency [42]. Transcription of the sgRNA molecule has been usually controlled by RNA polymerase III dependent promoters such as the U6 promoters. U6 promoters have a number of advantages such as their precise start of transcription and the tight control over the length of the transcript. In addition, U6 promoters are usually active in multiple species with the Arabidopsis U6 promoter able to direct strong transcription in tobacco, tomato, poplar and other dicotyledonous species [11, 12, 32]. Nevertheless, there are limitations to the ‘universal’ nature of U6 promoters as the Arabidopsis U6 promoter was inefficient in wheat and rice [43]. There is also evidence suggesting that the use of endogenous U6 promoters can improve the efficiency of CRISPR/Cas9 systems such as in soybean, where the GmU6-10 promoter produced 2–6 times higher mutation efficiency than the Arabidopsis U6-26 [17]. Finally, not all target sequences are equal, with different targets producing different mutation efficiencies perhaps due to secondary structure factors caused by the GC content [44].
In crops such as cotton, with lengthy and labor intensive transformation protocols it is essential to select the best possible CRISPR/Cas9 system before attempting stable transformation. We have previously developed a transient transformation protocol combined with restriction enzyme digestion of the targeted genomic loci to validate and assess the functionality of different sgRNAs in cotton [23]. Here, we provide a second independent method to further validate target sites using the fsGUS system. With this system, we validated different CRISPR/Cas9 constructs in 3 days using simple experimental techniques such as Agrobacterium infiltration combined with GUS staining and activity assays.
Achieving high sgRNA expression is essential for effective mutagenesis. We show here that a 300 bp GhU6.3 promoter fragment is enough to drive consistent gene expression in tobacco with similar expression levels than the Arabidopsis AtU6-29. In contrast, when both promoters were tested in cotton, we found that the sgRNA transcript levels driven by the endogenous GhU6.3 were 6–7 times higher than those driven by the Arabidopsis AtU6-29. Our results are consistent with previous reports suggesting that endogenous U6 promoters produce higher expression levels than non-endogenous promoters [17]. The increased expression levels were reflected on higher mutagenesis efficiencies with GhU6.3 resulting in 4–6 times higher mutagenesis rates than AtU6-26 measured with two independent methods, in agreement with pervious observations in other species [9, 17, 24]. As expected, the types mutation produced by both of promoters were similar. Even though we have previously used the Arabidopsis AtU6-29 promoter to successfully generate gene editing in cotton [23], the increased efficiency provided by GhU6.3 will be extremely useful to reduce the number of transgenic lines needed to ensure the generation of mutants, especially in the case of the heterotetraploid G. hirsutum or if multiple genes are simultaneously targeted.
Conclusion
In summary, we provide a fast and effective method to validate sgRNA mutagenesis efficiency in cotton using CRISPR/Cas9 and transient expression methods. We also provide an improved CRISPR/Cas9 cassette using an endogenous U6 promoter to drive sgRNA expression that generates improved mutagenesis efficiency over the existing one. Generating stable transformation of cotton is time-consuming and labor-intensive and thus the improvements should result in important savings for research groups using CRISPR/Cas9 in cotton.
Additional files
Additional file 1. List of primers used in this study.
Additional file 2. Multiple alignments of proGhU6.1, proGhU6.2 and proGhU6.3 sequences. The USE and TATA-box were boxed, the transcriptional start site was marked with star.
Additional file 3. The GUS staining in tobacco leaves infiltrated with Agrobacterium carrying different promoter constructs.
Additional file 4. Sequencing of mutated PCR products generated by pAtU6::sgRNA. The sgRNA target sequence is underlined in black. Deletions are shown as red dashes, and insertions are denoted with red letters. The frequency of each mutation is shown on the left and the mutation types on the right.
Authors’ contributions
CPS and JRB conceived and supervised the study; LL and WG designed experiments; LL, DDG, WWY and LPH performed experiments; XNM and YCM provided help in data analysis; LL, WG and JRB wrote and revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We are grateful to Prof. Y. G. Liu (South China Agricultural University, China) for generously providing the binary pYLCRISPR/Cas9 multiplex genome targeting vector system.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The authors are pleased to share analyzed/raw data and plant materials upon reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was financially supported by the National Natural Science Foundation of China (31601344, 31701473), the Ministry of Agriculture of China (2016ZX08009–003), the National Key R & D Project for Crop Breeding (2016YFD0101006) and the 111 project of China (D16014).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jose Ramon Botella, Phone: +61-7-336-51128, Email: j.botella@uq.edu.au.
Chun-Peng Song, Phone: + 86-371-23880002, Email: songcp@henu.edu.cn.
References
- 1.Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malzahn A, Lowder L, Qi Y. Plant genome editing with TALEN and CRISPR. Cell Biosci. 2017;7:21. doi: 10.1186/s13578-017-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang K, Raboanatahiry N, Zhu B, Li M. Progress in genome editing technology and its application in plants. Front Plant Sci. 2017;8:177. doi: 10.3389/fpls.2017.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shan Q, Wang Y, Li J, Gao C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc. 2014;9(10):2395–2410. doi: 10.1038/nprot.2014.157. [DOI] [PubMed] [Google Scholar]
- 5.Bortesi L, Fischer R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv. 2015;33(1):41–52. doi: 10.1016/j.biotechadv.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Hilscher J, Burstmayr H, Stoger E. Targeted modification of plant genomes for precision crop breeding. Biotechnol J. 2017;12(1):201600173. doi: 10.1002/biot.201600173. [DOI] [PubMed] [Google Scholar]
- 7.Arora L, Narula A. Gene editing and crop improvement using CRISPR-Cas9 system. Front Plant Sci. 2017;8:1932. doi: 10.3389/fpls.2017.01932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V. Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods. 2013;9(1):39. doi: 10.1186/1746-4811-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng H, Dean N. Dramatic improvement of CRISPR/Cas9 editing in Candida albicans by increased single guide RNA expression. mSphere. 2017;2:e00385-16. doi: 10.1128/mSphere.00385-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41(20):e188. doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyagishi M, Taira K. U6 promoter-driven siRNAs with four uridine 3’ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat Biotechnol. 2002;20(5):497–500. doi: 10.1038/nbt0502-497. [DOI] [PubMed] [Google Scholar]
- 12.Li X, Jiang D, Yong K, Zhang D, Chen Y. Varied transcriptional efficiencies of multiple Arabidopsis U6 small nuclear RNA genes. J Integr Plant Biol. 2007;2:222–229. doi: 10.1111/j.1744-7909.2007.00393.x. [DOI] [Google Scholar]
- 13.Friedland AE, Tzur YB, Esvelt KM, Colaiácovo MP, Church GM, Calarco JA. Heritable genome editing in C elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10(8):741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol. 2013;31(8):688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Zhang D, Sheen J. Cas9-based genome editing in Arabidopsis and tobacco. Methods Enzymol. 2014;546:459–472. doi: 10.1016/B978-0-12-801185-0.00022-2. [DOI] [PubMed] [Google Scholar]
- 16.Mikami M, Toki S, Endo M. Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice. Plant Mol Biol. 2015;88(6):561–572. doi: 10.1007/s11103-015-0342-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun X, Hu Z, Chen R, Jiang Q, Song G, Zhang H, Xi Y. Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci Rep. 2015;5:10342. doi: 10.1038/srep10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang MB, Helliwell CA, Wu LM, Waterhouse PM, Peacock WJ, Dennis ES. Hairpin RNAs derived from RNA polymerase II and polymerase III promoter-directed transgenes are processed differently in plants. RNA. 2008;14(5):903–913. doi: 10.1261/rna.760908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domitrovich AM, Kunkel GR. Multiple, dispersed human U6 small nuclear RNA genes with varied transcriptional efficiencies. Nucleic Acids Res. 2003;31(9):2344–2352. doi: 10.1093/nar/gkg331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L acc TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015;33(5):531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- 21.Li F, Fan G, Lu C, Xiao G, Zou C, Kohel RJ, Ma Z, Shang H, Ma X, Wu J, et al. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol. 2015;33(5):524–530. doi: 10.1038/nbt.3208. [DOI] [PubMed] [Google Scholar]
- 22.Yuan D, Tang Z, Wang M, Gao W, Tu L, Jin X, Chen L, He Y, Zhang L, Zhu L, et al. The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci Rep. 2015;5:17662. doi: 10.1038/srep17662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao W, Long L, Tian X, Liu J, Singh PK, Botella JR, Song C. Genome editing in cotton with the CRISPR/Cas9 system. Front Plant Sci. 2017;8:1364. doi: 10.3389/fpls.2017.01364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang P, Zhang J, Sun L, Ma Y, Xu J, Liang S, Deng J, Tan J, Zhang Q, Tu L, et al. High efficient multisites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system. Plant Biotechnol J. 2018;16(1):137–150. doi: 10.1111/pbi.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Meng Z, Liang C, Meng Z, Wang Y, Sun G, Zhu T, Cai Y, Guo S, Zhang R, et al. Increased lateral root formation by CRISPR/Cas9-mediated editing of arginase genes in cotton. Sci China Life Sci. 2017;60(5):524–527. doi: 10.1007/s11427-017-9031-y. [DOI] [PubMed] [Google Scholar]
- 26.Chen X, Lu X, Shu N, Wang S, Wang J, Wang D, Guo L, Ye W. Targeted mutagenesis in cotton (Gossypium hirsutum L) using the CRISPR/Cas9 system. Sci Rep. 2017;7:44304. doi: 10.1038/srep44304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janga MR, Campbell LM, Rathore KS. CRISPR/Cas9-mediated targeted mutagenesis in upland cotton (Gossypium hirsutum L) Plant Mol Biol. 2017;94(4–5):349–360. doi: 10.1007/s11103-017-0599-3. [DOI] [PubMed] [Google Scholar]
- 28.Li C, Unver T, Zhang B. A high-efficiency CRISPR/Cas9 system for targeted mutagenesis in cotton (Gossypium hirsutum L) Sci Rep. 2017;7:43902. doi: 10.1038/srep43902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin S, Zhang X. Factors affecting stable transformation and plant regeneration during transforming embryogenic callus of upland cotton (Gossypium hirsutum L) via Agrobacterium tumefaciens. Plant Cell Tissue Organ Cult. 2005;81(2):229–237. doi: 10.1007/s11240-004-5209-9. [DOI] [Google Scholar]
- 30.Li H, Li K, Guo Y, Guo J, Miao K, Botella JR, Song C, Miao Y. A transient transformation system for gene characterization in upland cotton (Gossypium hirsutum) Plant Methods. 2018;14:50. doi: 10.1186/s13007-018-0319-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin K, Han T, Liu G, Chen T, Wang Y, Yu A, Liu Y. A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci Rep. 2015;5:14926. doi: 10.1038/srep14926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant. 2015;8(8):1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 33.Gao W, Long L, Xu L, Lindsey K, Zhang X, Zhu L. Suppression of the homeobox gene HDTF1 enhances resistance to Verticillium dahliae and Botrytis cinerea in cotton. J Integr Plant Biol. 2016;58(5):503–513. doi: 10.1111/jipb.12432. [DOI] [PubMed] [Google Scholar]
- 34.Gao W, Xu F, Guo D, Zhao J, Liu J, Guo Y, Singh P, Ma X, Long L, Botella JR, et al. Calcium-dependent protein kinases in cotton: insights into early plant responses to salt stress. BMC Plant Biol. 2018;18(1):15. doi: 10.1186/s12870-018-1230-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng F, Tu L, Tan J, Li Y, Nie Y, Zhang X. GbPDF1 is involved in cotton fiber initiation via the core cis-element HDZIP2ATATHB2. Plant Physiol. 2012;158(2):890–904. doi: 10.1104/pp.111.186742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li F, Fan G, Wang K, Sun F, Yuan Y, Song G, Li Q, Ma Z, Lu C, Zou C, et al. Genome sequence of the cultivated cotton Gossypium arboretum. Nat Genet. 2014;46(6):567–572. doi: 10.1038/ng.2987. [DOI] [PubMed] [Google Scholar]
- 37.Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, Jin D, Llewellyn D, Showmaker KC, Shu S, Udall J, et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature. 2012;492(7429):423–427. doi: 10.1038/nature11798. [DOI] [PubMed] [Google Scholar]
- 38.Wang M, Wang P, Lin M, Ye Z, Li G, Tu L, Shen C, Li J, Yang Q, Zhang X. Evolutionary dynamics of 3D genome architecture following polyploidization in cotton. Nat Plants. 2018;4(2):90–97. doi: 10.1038/s41477-017-0096-3. [DOI] [PubMed] [Google Scholar]
- 39.Hu H, Wang M, Ding Y, Zhu S, Zhao G, Tu L, Zhang X. Transcriptomic repertoires depict the initiation of lint and fuzz fibres in cotton (Gossypium hirsutum L) Plant Biotechnol J. 2018;16(5):1002–1012. doi: 10.1111/pbi.12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao W, Long L, Zhu L, Xu L, Gao W, Sun L, Liu L, Zhang X. Proteomic and virus-induced gene silencing (VIGS) Analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahlia. Mol Cell Proteomics. 2013;12(12):3690–3703. doi: 10.1074/mcp.M113.031013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mao Y, Zhang Z, Feng Z, Wei P, Zhang H, Botella JR, Zhu J. Development of germ-line-specific CRISPR-Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol J. 2016;14(2):519–532. doi: 10.1111/pbi.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson RA, Gurevich V, Filler S, Samach A, Levy AA. Comparative assessments of CRISPR-Cas nucleases’ cleavage efficiency in planta. Plant Mol Biol. 2015;87(1–2):143–156. doi: 10.1007/s11103-014-0266-x. [DOI] [PubMed] [Google Scholar]
- 43.Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi J, Qiu J, et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol. 2013;31(8):686–688. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- 44.Liu X, Homma A, Sayadi J, Yang S, Ohashi J, Takumi T. Sequence features associated with the cleavage efficiency of CRISPR/Cas9 system. Sci Rep. 2016;6:19675. doi: 10.1038/srep19675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. List of primers used in this study.
Additional file 2. Multiple alignments of proGhU6.1, proGhU6.2 and proGhU6.3 sequences. The USE and TATA-box were boxed, the transcriptional start site was marked with star.
Additional file 3. The GUS staining in tobacco leaves infiltrated with Agrobacterium carrying different promoter constructs.
Additional file 4. Sequencing of mutated PCR products generated by pAtU6::sgRNA. The sgRNA target sequence is underlined in black. Deletions are shown as red dashes, and insertions are denoted with red letters. The frequency of each mutation is shown on the left and the mutation types on the right.
Data Availability Statement
The authors are pleased to share analyzed/raw data and plant materials upon reasonable request.