Abstract
Background
Accurate interindividual comparability of deep brain stimulation (DBS) lead locations in relation to the surrounding anatomical structures is of eminent importance to define and understand effective stimulation areas. The objective of the current work is to compare the accuracy of the DBS lead localisation relative to the STN in native space with four recently developed three-dimensional subcortical brain atlases in the MNI template space. Accuracy is reviewed by anatomical and volumetric analysis as well as intraoperative electrophysiological data.
Methods
Postoperative lead localisations of 10 patients (19 hemispheres) were analysed in each individual patient based on Brainlab software (native space) and after normalization into the MNI space and application of 4 different human brain atlases using Lead-DBS toolbox within Matlab (template space). Each patient's STN was manually segmented and the relation between the reconstructed lead and the STN was compared to the 4 atlas-based STN models by applying the Dice coefficient. The length of intraoperative electrophysiological STN activity along different microelectrode recording tracks was measured and compared to reconstructions in native and template space. Descriptive non-parametric statistical tests were used to calculate differences between the 4 different atlases.
Results
The mean STN volume of the study cohort was 153.3 ± 40.3 mm3 (n = 19). This is similar to the STN volume of the DISTAL atlas (166 mm3; p = .22), but significantly larger compared to the other atlases tested in this study. The anatomical overlap of the lead-STN-reconstruction was highest for the DISTAL atlas (0.56 ± 0.18) and lowest for the PD25 atlas (0.34 ± 0.17). A total number of 47 MER trajectories through the STN were analysed. There was a statistically significant discrepancy of the electrophysiogical STN activity compared to the reconstructed STN of all four atlases (p < .0001).
Conclusion: Lead reconstruction after normalization into the MNI template space and application of four different atlases led to different results in terms of the DBS lead position relative to the STN. Based on electrophysiological and imaging data, the DISTAL atlas led to the most accurate display of the reconstructed DBS lead relative to the DISTAL-based STN.
Keywords: Deep brain stimulation, Lead localisation, Human brain atlas, MNI space
Abbreviations: Deep Brain Stimulation, (DBS); microelectrode recording, (MER); Parkinson's Disease, (PD); Subthalamic Nucleus, (STN); MNI152, (Montreal Neurological Institute 152)
Highlights
-
•
DBS leads are projected differently onto different subcortical brain atlases after normalization
-
•
DBS electrodes seem to be laterally shifted after normalization and atlas application
-
•
Anatomical and electrophysiologcal accuracy was highest for the DISTAL atlas
1. Introduction
Deep Brain Stimulation (DBS) is a surgical treatment option to alleviate symptoms of movement disorders such as Parkinson's Disease (PD) (Deuschl et al., 2006; Schuepbach et al., 2013). Different subcortical targets within the basal ganglia and thalamus are typically chosen for DBS lead placement. For instance, the Subthalamic Nucleus (STN) is a common target for DBS of PD. Evidence suggests accurate lead placement to be of paramount importance for postoperative outcome (Welter et al., 2014; Wodarg et al., 2012). Therefore, exact localisation of the DBS lead within the target and in relationship to surrounding anatomical structures may be helpful to analyse the underlying reason for poor stimulation efficacy. Moreover, with advanced lead design offering directional stimulation and current steering, programming can be adjusted according to the DBS lead location and the intended target structure to minimize stimulation induced side-effects (Pollo et al., 2014; Schupbach et al., 2017; Tinkhauser et al., 2018). Apart from DBS lead localisation in an individual patient (native space), comparison of DBS lead positions across subjects is an important and powerful tool for scientific approaches, e.g., to define efficacious stimulation areas (“sweet spots”) within well-known target structures (Accolla et al., 2016; Akram et al., 2017).
The undertaking of comparing anatomical data across subjects is challenging because of anatomical heterogeneity. Both position and sizes of anatomical structures in one brain must correspond to positions and sizes in another to make meaningful comparisons (Brett et al., 2002). With advances in imaging technologies and deformable registration algorithms, projection of DBS leads onto three-dimensional subcortical atlases has been established more recently to compare DBS lead locations of different subjects (Cheung et al., 2014; Welter et al., 2014). The basic underlying workflow usually consists of registering brains of individuals to a common template by applying non-rigid, deformable registration algorithms (normalization into template space) (Klein et al., 2009). Currently, the MNI152 (Montreal Neurological Institute) space, which is based on anatomical high-resolution images of 152 subjects, is broadly used as a template space. Implementation of brain-atlases into the template space then enables further anatomical analyses and comparison of interindividual data. Today, many different atlases that have been defined on the basis of magnetic resonance imaging (MRI) and/or histology are available (Chakravarty et al., 2006; Welter et al., 2014; Xiao et al., 2015; Yelnik et al., 2007). A major drawback of many atlases is that their spatial definition of anatomical structures does not correspond inherently to the MNI template (summarised in Ewert et al. (Ewert et al., 2017)). However, this is a crucial step for the analysis of lead locations in the context of surrounding structures. Inaccurate registrations will ultimately lead to wrong conclusions about DBS lead placements and the stimulated structures and can therefore render group comparisons erroneous. Another drawback is that most atlases are based on young and healthy individuals, whereas the study groups of interest in the field of DBS are usually old and their brains atrophic (Dalaker et al., 2011). To overcome these limitations, different groups have recently presented atlases that match the MNI space or which are based on PD patients instead of healthy subjects (Ewert et al., 2017; Pauli et al., 2017; Wang et al., 2016; Xiao et al., 2015).
The objective of the current work is to compare the accuracy of the DBS lead location relative to the STN in native space with four recently developed three-dimensional subcortical brain atlases in the MNI space. Accuracy is reviewed by anatomical and volumetric analysis as well as intraoperative electrophysiological data.
2. Material and Methods
2.1. Patients
We retrospectively analysed 24 patients undergoing DBS lead implantation into the STN for advanced PD between October 2015 and April 2017 at our department. Inclusion criteria were age older than 18 years, availability of intraoperative electrophysiological testing results by specialized movement disorder neurologists, microelectrode recording results available along at least two parallel trajectories of each patient's hemisphere. All patients provided written prior consent for the procedure. The study was approved by the institutional review board.
2.2. Operative procedure
For detailed description of our targeting approach and operative procedure we refer to previously published reports of our group (Nowacki et al., 2017). In summary, each patient underwent preoperative MRI to target the STN. Imaging was performed with a 12-channel head coil for signal reception on a 3 T MRI system (MAGNETOM Trio™ Tim, Siemens, Germany). Imaging included T2-weighted sequences with an echo time of 380 ms, a repetition time of 3000 ms and a slice thickness of 1 mm. Targets and trajectories were identified using Brainlab Elements software (Brainlab AG, Germany). On the day of surgery a Leksell G frame (Elekta instruments, Sweden) was placed and a high-resolution, stereotactic CT scan was performed and co-registered with the preoperative MRI (Brainlab AG, Germany).
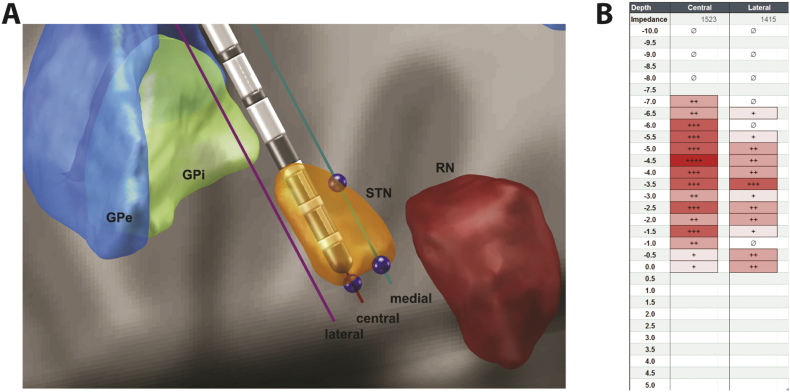
Intraoperative microelectrode recording (MER) was performed as described previously. Here, recording was performed on two to three parallel trajectories. From 10 mm to 5 mm above the target, electrophysiological activity was recorded in 1 mm-steps and from 5 mm above the target in 0.5 mm-steps. Electrophysiological activity was evaluated by a simple grading system, ‘+’ for little activity to ‘+++’ for strong activity, to determine entry and exit of the STN (Fig. 1). This grading was done live intraoperatively by an experienced rater and not ad-hoc nor in a blinded fashion.
Fig. 1.
Determination of STN lengths in an example patient set. (A) Image-based measurements with Lead DBS and microelectrode tool that places microelectrodes in specific trajectories and allows for measurement of STN length (blue points representing entry and exit, entry point for central trajectory covered by lead). The panel shows the DISTAL atlas. GPe – external globus pallidus, GPi – internal globus pallidus, STN – subthalamic nucleus, RN – red nucleus. (B) Intraoperative assessment of electrophysiological recording at our centre. Number of ‘+’ signs indicate STN activity rated by one experienced rater live intraoperatively at different positions of the microelectrode.
Postoperative high-resolution CT was performed for assessment of correct electrode placement on the day after surgery.
2.3. Postoperative DBS lead location
2.3.1. Native space
Postoperative DBS lead localisation was performed based on Brainlab Elements software in patient's native space. AC-PC coordinates of the intended target, the DBS lead tip as well as the targeting error were calculated as described previously (Nowacki et al., 2015). The integrated workflow was used to fuse preoperative MR images and post-operative CT images. The STN was manually segmented by the junior (AN) and senior neurosurgeon (CP) according to the hypointense signal based on T2-weighted images. The DBS lead was reconstructed according to the CT artefact in an automated way by the Brainlab software and its position manually adjusted, if necessary.
2.3.2. Normalized Template space
Lead-DBS toolbox version 2.0.0.6 was used within Matlab 2016b (The MathWorks, USA) for DBS lead visualisation (Horn and Kuhn, 2015). Preoperative MRI and postoperative CT were co-registered using SPM12 and Advanced Normalization Tools (Avants et al., 2008). Normalization to the MNI 152 2009b space (Montreal Neurological Institute) was also performed by applying Advanced Normalization Tools. The algorithm's accuracy and effectiveness were evaluated elsewhere (Klein et al., 2009). The lead was then automatically pre-localised on CT images with PaCER and manually adjusted, if the automatic localisation did not match the CT artefact (typical adjustments of less than half of the lead's diameter or 0.65 mm) (Husch et al., 2018). Lead-DBS provided a three-dimensional display of nuclei according to a selected atlas. Four atlases were evaluated: the DISTAL atlas combining MRI from the 2009b MNI152 template (normative young adult population), histology and structural connectivity data (Ewert et al., 2017); the CIT168 atlas derived from data of the Human Connectome Project based on 168 young adult subjects between the age of 22 and 35 (Pauli et al., 2017); the Wang atlas based on high-resolution 7 T MR scans of twelve healthy subjects; and the PD25 atlas derived from 3 T MRI scans of 25 Parkinson's disease patients (Xiao et al., 2015). Our rationale behind the selection of these four atlases was to cover a broad spectrum of modalities, e.g., different MRI data, histology and subject groups.
2.4. Anatomical comparison of STN in native and template space
To assess the accuracy of the four different brain atlases, we compared each patient's individual STN anatomy based on the hypointense T2-artefact on their MRI and the postoperative DBS lead location (native space) with the reconstructed DBS lead and STN according to the four brain atlases after normalization to the MNI space (template space).
Furthermore, we compared the volumes of each patient's individually segmented STN (left and right) according to the MRI-based T2 artefact and the atlas-based STN volume (supplementary fig. 1). In Lead-DBS, the segmented STNs according to the four atlases were exported and then analysed in Matlab 2016b to compute the volume.
The Dice coefficient was applied as a measure of the degree of similarity between the native space STN and the atlas-based STN. It was calculated according to Eq. (1), where A and B are the STN volumes in native and template space, respectively.
| (1) |
2.5. STN MER-MRI trajectory length comparison
One further pertinent way to assess the accuracy of the reconstructed DBS lead relative to STN anatomy was to compare the position of the DBS lead with intraoperative MER results. Therefore, we determined the length of electrophysiological STN activity along the two to three intraoperatively used parallel trajectories and compared it to image-based reconstructed trajectories across the STN according to the four atlases (template space) as well as to the T2-hypointense signal of each patient's STN visualised on MRI (native space).
Native-space based analysis was performed with Brainlab software. Reconstructed trajectories of MER adjacent to the DBS lead were manually placed in inline view with a fixed 2 mm distance to the lead according to known features of the five-channel Ben-Gun. The entry and exit points of the STN were then measured manually in the software to calculate the resulting STN length. Lead-DBS provided a microelectrode tool to determine STN lengths along the different microelectrode trajectories such as central, lateral or medial (Fig. 1).
We then calculated the difference of the image- or atlas-based STN trajectory length (SMRI/atlas) and the MER-based trajectory length (SMER) in each patient (ΔS = SMRI/atlas - SMER). If no electrophysiological activity was recorded for a given trajectory, SMER was set to zero. Similarly, if the image- or atlas-based trajectory was projected outside the STN, SMRI/atlas was set to zero for that trajectory.
2.6. Clinical outcome assessment
Preoperative motor subscores of the Unified Parkinson's Disease Rating Scale (UPDRS part III) in “medication-off” state and postoperative UPDRS III scores in the “stimulation-on, medication-off” state (overnight withdrawal of PD medication) were systematically determined 12 months after DBS implantation. DBS outcome was assessed by analysing the differences of respective preoperative and postoperative UPDRS III scores.
2.7. Statistical analysis
For both anatomical and trajectory length comparisons, measurements were done separately for the left and right hemispheres to account for asymmetries. Finally, data were then analysed together with descriptive/nonparametric statistics using Prism software (GraphPad Prism 6, USA). One-way ANOVA (Friedman test) was applied to test for statistical differences between the image-based and electrophysiological-based STN lengths. Post-hoc analysis was performed by Dunn's multiple comparisons test. A P-value <.05 was considered statistically significant.
3. Results
According to our inclusion criteria we included a total number of ten patients (19 hemispheres) in the final analysis. Altogether, a total number of 47 MER trajectories through the STN were analysed (cf. Table I, 10 along a medial trajectory, 19 along a central trajectory, 18 along a lateral trajectory). The mean targeting error was 1.01 ± 0.57 mm (range 0.40–1.95 mm) indicating precise targeting and no cases of clinically relevant lead displacement.
3.1. Clinical outcome data
Preoperative mean UPDRS III score was 36.9 ± 9.75. Postoperative mean UPDRS III score was significantly reduced to 10.55 ± 6.40 (p < .0001). The mean percentage improvement was 71.72 ± 14.96%. One patient was lost to clinical follow-up as he died (unrelated to DBS surgery).
3.2. DBS lead relative to STN anatomy in native and template space
The mean AC-PC coordinates of the right lead tip were LAT 11.40 ± 1.10 mm, ANT -3.31 ± 0.39 mm and VERT -4.43 ± 0.55 mm. After transformation of the DBS leads into the MNI space the mean MNI coordinates were LAT 9.38 ± 1.33 mm, AP -4.44 ± 0.83 mm and VERT -9.35 ± 1.2 mm. Table 1 summarizes patients´ individual DBS lead tip position relative to the MCP in native space and after normalization in the MNI space.
Table 1.
Coordinates of right lead tip in native and MNI space with reference to the mid-commissural point (MCP).
| ID |
MCP Coordinates in native space |
MCP coordinates in MNI space |
||||
|---|---|---|---|---|---|---|
| LAT | AP | VERT | LAT | AP | VERT | |
| 1 | 9.49 | −3.73 | −5.73 | 7.49 | −3.68 | −10.57 |
| 2 | 11.47 | −3.63 | −4.59 | 8.44 | −5.82 | −10.73 |
| 3 | 12.93 | −3.65 | −3.98 | 11.17 | −5.2 | −7.33 |
| 4 | 11.74 | −3.51 | −4 | 9.95 | −4.4 | −8.44 |
| 5 | 12.02 | −3.03 | −4.12 | 10.72 | −4.28 | −8.14 |
| 6 | 12.38 | −3.41 | −4.4 | 10.02 | −5.05 | −10.72 |
| 7 | 10.43 | −2.6 | −4.78 | 9.67 | −3.35 | −9.83 |
| 8 | 12.04 | −3.6 | −3.92 | 9.18 | −4.26 | −9.17 |
| 9 | 10.03 | −2.91 | −4.67 | 7.11 | −3.36 | −10.02 |
| 10 | 11.51 | −3.01 | −4.15 | 10.09 | −5.06 | −8.55 |
| mean | 11.40 | −3.31 | −4.43 | 9.38 | −4.44 | −9.35 |
| SD | 1.10 | 0.39 | 0.55 | 1.33 | 0.83 | 1.20 |
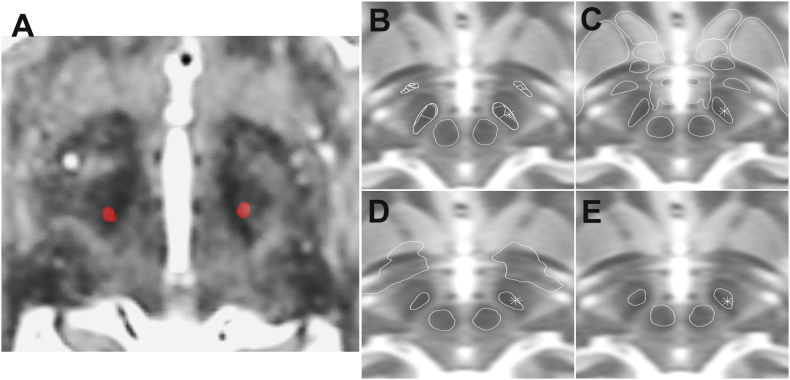
Electrode reconstruction after normalization and application of the four different atlases led to different results with respect to the lead position within the STN and compared to the native space. Fig. 2 visualises the DBS lead and STN delineation of the manually segmented STN compared to the different atlases in one representative patient of the study cohort. In general, we could observe that the DBS lead was projected more laterally within the STN after applying the DISTAL and CIT atlas in comparison to the native space. No such shift could be observed in case of the PD25 and Wang atlases, in which cases, however, the DBS lead was frequently located outside the atlas-based STN borders.
Fig. 2.
Lead position in relation to STN. (A) Postoperative CT lead artefact (red) superimposed on preoperative MR T2 images. (B)-(E) Right lead marked as asterisk in atlas segmented STN for DISTAL (B), CIT (C), Wang (D) and PD25 (E), respectively.
The mean STN volume of the study cohort was 153.3 ± 40.3 mm3 (n = 19). This is similar to the STN volume of the DISTAL atlas (166 mm3; p = .22), but significantly larger compared to the CIT atlas (136 mm3; p = .04), the atlas of Wang et al. (85 mm3; p < .001) and the PD25 atlas (102 mm3; p < .001).
The Dice coefficient of the native space STN volume compared to each of the different atlas-based STN was determined in each patient's hemisphere. The overlap was highest for the DISTAL atlas (0.56 ± 0.18). Less similar results were found for the CIT atlas (0.50 ± 0.15). Furthermore, we found significant differences for the atlas of Wang et al. (0.42 ± 0.12) as well as the PD25 (0.34 ± 0.17).
3.3. STN MER-MRI trajectory comparison in native space and template space
Based on all three trajectories, there was no significant difference between ΔS in native space, however there was a statistically significant difference of ΔS for all four atlases (p < .0001).
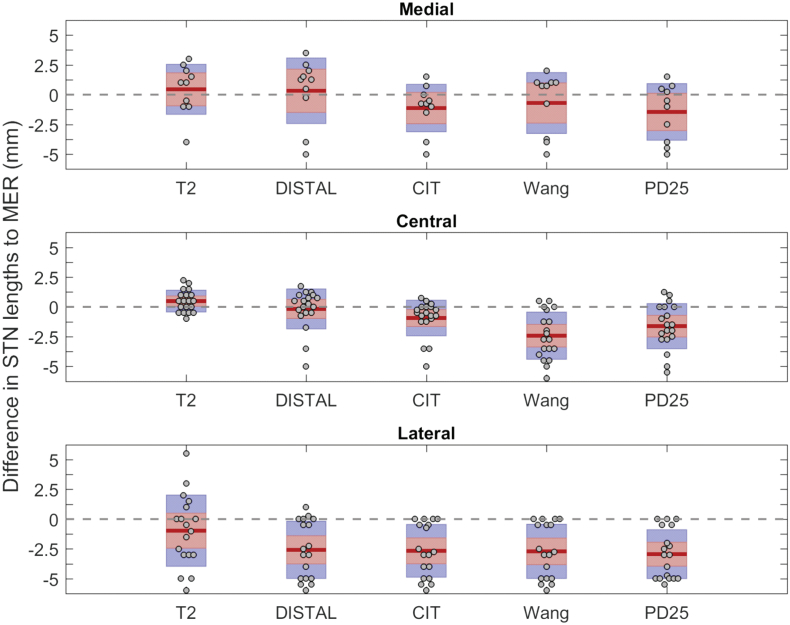
Fig. 3 shows the differences of the electrophysiological and image-based STN trajectory lengths separately for the central, medial and lateral trajectories.
Fig. 3.
Differences in measured STN lengths between manually segmented, atlas based STN and intraoperative electrophysiology data. Thick red lines indicate mean; red boxes represent the 95% confidence interval of the mean; blue box represents the standard deviation; gray dots represent individual data.
The mean differences (ΔS) between the reconstructed MRI-based trajectory length in native space and intraoperatively measured MER-based STN length was 0.49 ± 0.91 mm for the central trajectory, 0.45 ± 2.10 mm for the medial and − 0.97 ± 2.99 mm for the lateral trajectory.
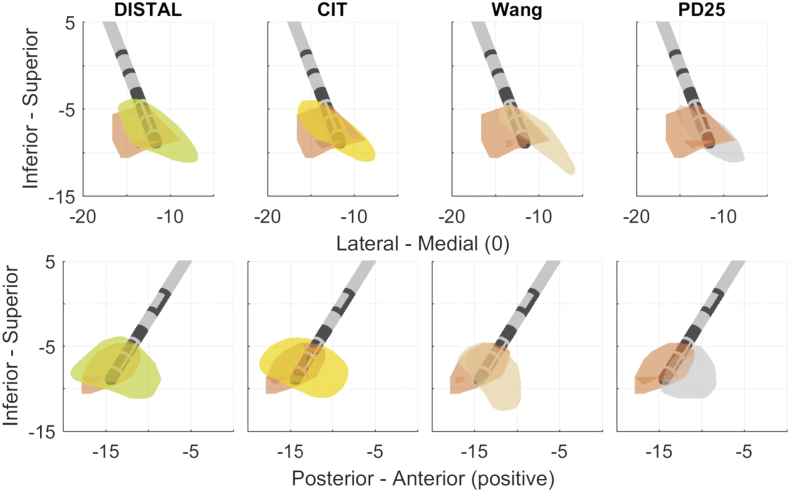
The mean differences (ΔS) between the reconstructed DISTAL atlas-based STN length and MER-based STN length was −0.17 ± 1.68 mm for the central trajectory, 0.33 ± 2.75 mm for the medial and − 2.58 ± 2.40 mm for the lateral trajectory. Frequently, the lateral microelectrode trajectory was displayed outside the DISTAL atlas-based STN, while there was marked electrophysiological activity recorded intraoperatively. This led to negative ΔS-values on the lateral trajectory in those cases. On the other hand medial trajectories were displayed within the DISTAL atlas-based STN, even though no electrophysiological activity was measured (Fig. 4).
Fig. 4.
Overlay of manually segmented STN (orange) and atlas based STN according to Distal (green), CIT (yellow), Wang (rose) and PD25 (gray) of an example patient. The lead tip was used as anchor to align manually segmented and atlas-based STNs.
The mean differences (ΔS) between the reconstructed CIT atlas-based STN length and MER-based STN length was −0.93 ± 1.50 mm for the central trajectory, −1.13 ± 1.99 mm for the medial and − 2.67 ± 2.21 mm for the lateral trajectory.
The mean differences (ΔS) between the reconstructed Wang atlas-based STN length and MER-based STN length was −2.42 ± 1.98 mm for the central trajectory, −0.7 ± 2.56 mm for the medial and − 2.71 ± 2.28 mm for the lateral trajectory.
The mean differences (ΔS) between the reconstructed PD25 atlas-based STN length and MER-based STN length was −1.62 ± 1.90 mm for the central trajectory, −1.45 ± 2.38 mm for the medial and − 2.94 ± 2.05 mm for the lateral trajectory.
These difference calculations were repeated in native space by transforming the four atlases from MNI space to native patient space. There we also observed the smallest differences for the DISTAL atlas (supplementary fig. 2).
4. Discussion
Our study results show that lead reconstruction after normalization into the MNI template space and application of four different atlases led to different results in terms of the DBS lead position relative to the STN. The STN volume and delineation of the four different atlases as well as the relation to the reconstructed DBS lead were compared to the T2-hypointense MRI signal in each individual patient and to the STN electrophysiological profile recorded during intraoperative MER. We demonstrated that the process of normalization into the MNI template space and application of any of the four tested atlases resulted in significant differences between the electrophysiological profile and the atlas-based profile of the STN. Furthermore, significant differences were found between atlases regarding volume and similarity index compared to native space. Based on electrophysiological and imaging data, the DISTAL atlas led to the most accurate display of the reconstructed DBS lead relative to the DISTAL-based STN. Notably, after applying the DISTAL atlas the reconstructed DBS leads were displayed too lateral within the STN compared to native space and to the electrophysiological profile.
Accurate display of the DBS lead in relation to the surrounding anatomical structures is of paramount importance in clinical routine as well as for scientific purposes such as cohort comparisons. Understanding the relation of the lead to the surrounding anatomical structures may help clinicians to adapt stimulation parameters to avoid side effects and optimize stimulation. Importantly, given the fact that the target structures for DBS are usually small in size with only a few millimetres of diameter, accurate DBS lead reconstruction is crucial when it comes to comparison of DBS lead placements across subjects to infer optimal stimulation sites (Horn et al., 2017). Three-dimensional subcortical human brain atlases (Cheung et al., 2014; Welter et al., 2014) in conjunction with computational models of volume of tissue activated or fibre tractography are helpful tools to address these questions (Gunalan et al., 2018). Different subcortical atlases have been developed to enable microstructural analysis of brain anatomy. Some atlases are based on histologic data of single or multiple brains, while others are defined solely through MRI data. Some more recent atlases, such as the DISTAL atlas studied herein, combine several modalities to improve accuracy (Ewert et al., 2018).
From each patient's data set, we concluded that the DBS lead artefact on postoperative CT reflected the actual lead position within the STN reliably as we found no difference between the reconstructed image-based trajectories through the STN and their corresponding electrophysiological activity. This is in line with previous work by our and other groups (Hamani et al., 2005; Kocabicak et al., 2013; Nowacki et al., 2017; Schlaier et al., 2011).
On the other hand, normalization of individual MRI patient data into the MNI space and application of the four atlases tested inevitably led to certain shifts of the reconstructed DBS lead in relation to the STN anatomy. One factor is the normalization to the MNI space. A comparison of patients lead tip position with reference to MCP in native space and after normalization into the MNI template space shows a difference in all coordinates especially in the vertical direction. A thorough discussion and probabilistic conversion of AC/PC coordinates to MNI coordinates is provided by Horn et al. (Horn et al., 2017). In addition, MNI space defines the origin close to the AC point, not quite on the AC-PC line, but a few millimetres more inferior.
Comparison between atlas-based reconstructed MER trajectories through the STN showed significant differences compared to the electrophysiological activity. However, the degree of discrepancy between imaging and electrophysiological data was considerably different between the four atlases tested. Whereas there was a good correspondence of electrophysiological activity and the reconstructed MER trajectory through the STN of the DISTAL and CIT atlases along the central trajectory, there was a low correspondence in case of the Wang and PD25 atlas along each of the tested trajectories. It is important to note that, frequently, lateral trajectories were displayed outside the DISTAL and CIT atlas STN, while there was marked electrophysiological activity leading to negative ΔS-values. On the other hand, medial trajectories were displayed within the STN even though no electrophysiological activity was measured. It highlights the tendency of a lateral shift of the reconstructed DBS lead relative to all four atlas-based STN. This shift might be due to the normalization algorithm, specifically as the algorithm transforms brain images of Parkinson's disease patients – which typically show ventricular enlargement but smaller basal ganglia structures than normal controls – to the MNI template brain (Geng et al., 2006). This warrants further investigation in order to adapt the normalization algorithm for DBS research.
Our data further showed that the DISTAL atlas-based STN volume corresponded well to the mean STN volume in our cohort of patients. Application of the other atlases led to an underestimation of the STN volume, despite the fact that the volume of the MNI template brain tends to be larger than the average human brain (Horn et al., 2017). Similarity analysis revealed the best correspondence between the STN shape of the DISTAL atlas compared to each patient's individual STN. Poor anatomical overlap was found between the Wang and PD25 atlas STN and our cohort of patients.
Our study has some limitations. First, our relatively small sample size of ten patients that we have included in the final analysis prevents generalizing our findings to a broader population. Second, we only analysed four different atlases. Literature now abounds with different subcortical brain atlases. Analysing all of them was beyond the scope of this study. We focused on the four presented atlases here to compare and contrast the multimodality of the DISTAL atlas, the high resolution and high subject count (168) of the CIT atlas, the 7 T based Wang atlas and the disease-specific PD25 atlas based on Parkinson's disease patients. While the DISTAL atlas was specifically defined in the MNI space used herein, the other three atlases were each defined on custom templates. The CIT atlas was based on data from the Human Connectome Project, the 7 T based atlas from a groupwise template built from the data of the twelve healthy subjects scanned and the PD25 atlas was defined as an averaged atlas across the 25 Parkinson's disease patients. These three atlases were transformed to the MNI template space and the transformed registrations were provided within LEAD-DBS. This additional transformation may have affected the comparison. However, for better comparison we specifically intended to analyse all four atlases in the same template space. Third, analysis of the STN size and shape in native space was based on manual segmentation of the T2-hypointense STN signal based on MRI ignoring the fact that different MRI sequences have been proposed to display the STN, such as susceptibility weighted sequences or fluid attenuated inversion recovery sequences (Chandran et al., 2016; Heo et al., 2015; Senova et al., 2016). Furthermore, delineation of the STN borders by imaging is not always clear, which has been described previously by our and other groups (Nowacki et al., 2017; Starr et al., 2002). Especially the anterior-posterior axis of the STN was found to be prone to MRI distortions (Sumanaweera et al., 1994). Similarly, delineation of the STN with electrophysiological data was rated live intraoperatively by a single experienced rater and not in a blinded retrospective fashion with multiple raters or with an automised algorithm (Zaidel et al., 2010). Fourth, our analysis focused on STN length and did not study entry and exit points in detail that could be prone to error by different lead localization algorithms and manual corrections. Finally, we relied on previous evaluation of registration and normalization algorithms15, but we think that further improvements in this domain may be warranted. Furthermore, we performed an early post-operative CT scan as part of the clinical routine to rule out complications. Due to resorption of possible postoperative pneumencephalus, the lead position might slightly change over the postoperative period and introduce a potential bias into DBS lead position analysis. Furthermore, we did not measure the electrophysiological activity along an anterior or posterior trajectory, which would have added great value to evaluate the anatomical accuracy of the atlases with respect to the border between the anterior STN and internal capsule.
5. Conclusions
Normalization of individual MRI data into the MNI template space and application of different three-dimensional subcortical atlases inevitably led to distortions of the reconstructed lead in relation to the STN. Of the four different atlases tested in this work, the DISTAL atlas leads to the most accurate results. However, results must be cautiously interpreted as there seems to be a lateral shift of the reconstructed DBS lead relative to the STN.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2018.09.030.
Appendix A. Supplementary data
Supplementary material
References
- Accolla E.A., Herrojo Ruiz M., Horn A., Schneider G.H., Schmitz-Hubsch T., Draganski B., Kuhn A.A. Brain networks modulated by subthalamic nucleus deep brain stimulation. Brain. 2016;139:2503–2515. doi: 10.1093/brain/aww182. [DOI] [PubMed] [Google Scholar]
- Akram H., Sotiropoulos S.N., Jbabdi S., Georgiev D., Mahlknecht P., Hyam J., Foltynie T., Limousin P., De Vita E., Jahanshahi M., Hariz M., Ashburner J., Behrens T., Zrinzo L. Subthalamic deep brain stimulation sweet spots and hyperdirect cortical connectivity in Parkinson's disease. Neuroimage. 2017;158:332–345. doi: 10.1016/j.neuroimage.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants B.B., Epstein C.L., Grossman M., Gee J.C. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008;12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett M., Johnsrude I.S., Owen A.M. The problem of functional localization in the human brain. Nat Rev. Neurosci. 2002;3:243–249. doi: 10.1038/nrn756. [DOI] [PubMed] [Google Scholar]
- Chakravarty M.M., Bertrand G., Hodge C.P., Sadikot A.F., Collins D.L. The creation of a brain atlas for image guided neurosurgery using serial histological data. Neuroimage. 2006;30:359–376. doi: 10.1016/j.neuroimage.2005.09.041. [DOI] [PubMed] [Google Scholar]
- Chandran A.S., Bynevelt M., Lind C.R. Magnetic resonance imaging of the subthalamic nucleus for deep brain stimulation. J Neurosurg. 2016;124:96–105. doi: 10.3171/2015.1.JNS142066. [DOI] [PubMed] [Google Scholar]
- Cheung T., Noecker A.M., Alterman R.L., McIntyre C.C., Tagliati M. Defining a therapeutic target for pallidal deep brain stimulation for dystonia. Ann Neurol. 2014;76:22–30. doi: 10.1002/ana.24187. [DOI] [PubMed] [Google Scholar]
- Dalaker T.O., Zivadinov R., Ramasamy D.P., Beyer M.K., Alves G., Bronnick K.S., Tysnes O.B., Aarsland D., Larsen J.P. Ventricular enlargement and mild cognitive impairment in early Parkinson's disease. Mov Disord. 2011;26:297–301. doi: 10.1002/mds.23443. [DOI] [PubMed] [Google Scholar]
- Deuschl G., Schade-Brittinger C., Krack P., Volkmann J., Schafer H., Botzel K., Daniels C., Deutschlander A., Dillmann U., Eisner W., Gruber D., Hamel W., Herzog J., Hilker R., Klebe S., Kloss M., Koy J., Krause M., Kupsch A., Lorenz D., Lorenzl S., Mehdorn H.M., Moringlane J.R., Oertel W., Pinsker M.O., Reichmann H., Reuss A., Schneider G.H., Schnitzler A., Steude U., Sturm V., Timmermann L., Tronnier V., Trottenberg T., Wojtecki L., Wolf E., Poewe W., Voges J., German Parkinson Study Group, N.S A randomized trial of deep-brain stimulation for Parkinson's disease. N Engl J Med. 2006;355:896–908. doi: 10.1056/NEJMoa060281. [DOI] [PubMed] [Google Scholar]
- Ewert S., Plettig P., Li N., Chakravarty M.M., Collins D.L., Herrington T.M., Kuhn A.A., Horn A. Toward defining deep brain stimulation targets in MNI space: A subcortical atlas based on multimodal MRI, histology and structural connectivity. Neuroimage. 2017;170:271–282. doi: 10.1016/j.neuroimage.2017.05.015. [DOI] [PubMed] [Google Scholar]
- Ewert S., Plettig P., Li N., Chakravarty M.M., Collins D.L., Herrington T.M., Kuhn A.A., Horn A. Toward defining deep brain stimulation targets in MNI space: A subcortical atlas based on multimodal MRI, histology and structural connectivity. Neuroimage. 2018;170:271–282. doi: 10.1016/j.neuroimage.2017.05.015. [DOI] [PubMed] [Google Scholar]
- Geng D.Y., Li Y.X., Zee C.S. Magnetic resonance imaging-based volumetric analysis of basal ganglia nuclei and substantia nigra in patients with Parkinson's disease. Neurosurgery. 2006;58:256–262. doi: 10.1227/01.NEU.0000194845.19462.7B. [DOI] [PubMed] [Google Scholar]
- Gunalan K., Howell B., McIntyre C.C. Quantifying axonal responses in patient-specific models of subthalamic deep brain stimulation. Neuroimage. 2018;172:263–277. doi: 10.1016/j.neuroimage.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamani C., Richter E.O., Andrade-Souza Y., Hutchison W., Saint-Cyr J.A., Lozano A.M. Correspondence of microelectrode mapping with magnetic resonance imaging for subthalamic nucleus procedures. Surg Neurol. 2005;63:249–253. doi: 10.1016/j.surneu.2004.05.036. discussion 253. [DOI] [PubMed] [Google Scholar]
- Heo Y.J., Kim S.J., Kim H.S., Choi C.G., Jung S.C., Lee J.K., Lee C.S., Chung S.J., Cho S.H., Lee G.R. Three-dimensional fluid-attenuated inversion recovery sequence for visualisation of subthalamic nucleus for deep brain stimulation in Parkinson's disease. Neuroradiology. 2015;57:929–935. doi: 10.1007/s00234-015-1555-z. [DOI] [PubMed] [Google Scholar]
- Horn A., Kuhn A.A. Lead-DBS: a toolbox for deep brain stimulation electrode localizations and visualizations. Neuroimage. 2015;107:127–135. doi: 10.1016/j.neuroimage.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Horn A., Kuhn A.A., Merkl A., Shih L., Alterman R., Fox M. Probabilistic conversion of neurosurgical DBS electrode coordinates into MNI space. Neuroimage. 2017;150:395–404. doi: 10.1016/j.neuroimage.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husch A., M V.P., Gemmar P., Goncalves J., Hertel F. PaCER - A fully automated method for electrode trajectory and contact reconstruction in deep brain stimulation. Neuroimage Clin. 2018;17:80–89. doi: 10.1016/j.nicl.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A., Andersson J., Ardekani B.A., Ashburner J., Avants B., Chiang M.C., Christensen G.E., Collins D.L., Gee J., Hellier P., Song J.H., Jenkinson M., Lepage C., Rueckert D., Thompson P., Vercauteren T., Woods R.P., Mann J.J., Parsey R.V. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46:786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocabicak E., Aygun D., Ozaydin I., Jahanshahi A., Tan S., Onar M., Boke O., Kurt M., Guz H., Terzi M., Alptekin O., Temel Y. Does probe's eye subthalamic nucleus length on T2W MRI correspond with microelectrode recording in patients with deep brain stimulation for advanced Parkinson's disease? Turk Neurosurg. 2013;23:658–665. doi: 10.5137/1019-5149.JTN.8328-13.0. [DOI] [PubMed] [Google Scholar]
- Nowacki A., Fiechter M., Fichtner J., Debove I., Lachenmayer L., Schupbach M., Oertel M.F., Wiest R., Pollo C. Using MDEFT MRI Sequences to Target the GPi in DBS Surgery. PLoS One. 2015;10 doi: 10.1371/journal.pone.0137868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki A., Debove I., Fiechter M., Rossi F., Oertel M.F., Wiest R., Schupbach M., Pollo C. Targeting Accuracy of the Subthalamic Nucleus in Deep Brain Stimulation Surgery: Comparison Between 3 T T2-Weighted Magnetic Resonance Imaging and Microelectrode Recording Results. Oper Neurosurg (Hagerstown) 2017;15(1):66–71. doi: 10.1093/ons/opx175. [DOI] [PubMed] [Google Scholar]
- Pauli W.M., Nili A.N., Tyszka J.M. A High-Resolution Probabilistic In Vivo Atlas of Human Subcortical Brain Nuclei. bioRxiv. 2017:211,201. doi: 10.1038/sdata.2018.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollo C., Kaelin-Lang A., Oertel M.F., Stieglitz L., Taub E., Fuhr P., Lozano A.M., Raabe A., Schupbach M. Directional deep brain stimulation: an intraoperative double-blind pilot study. Brain. 2014;137:2015–2026. doi: 10.1093/brain/awu102. [DOI] [PubMed] [Google Scholar]
- Schlaier J.R., Habermeyer C., Warnat J., Lange M., Janzen A., Hochreiter A., Proescholdt M., Brawanski A., Fellner C. Discrepancies between the MRI- and the electrophysiologically defined subthalamic nucleus. Acta Neurochir (Wien) 2011;153:2307–2318. doi: 10.1007/s00701-011-1081-7. [DOI] [PubMed] [Google Scholar]
- Schuepbach W.M., Rau J., Knudsen K., Volkmann J., Krack P., Timmermann L., Halbig T.D., Hesekamp H., Navarro S.M., Meier N., Falk D., Mehdorn M., Paschen S., Maarouf M., Barbe M.T., Fink G.R., Kupsch A., Gruber D., Schneider G.H., Seigneuret E., Kistner A., Chaynes P., Ory-Magne F., Brefel Courbon C., Vesper J., Schnitzler A., Wojtecki L., Houeto J.L., Bataille B., Maltete D., Damier P., Raoul S., Sixel-Doering F., Hellwig D., Gharabaghi A., Kruger R., Pinsker M.O., Amtage F., Regis J.M., Witjas T., Thobois S., Mertens P., Kloss M., Hartmann A., Oertel W.H., Post B., Speelman H., Agid Y., Schade-Brittinger C., Deuschl G., Group, E.S Neurostimulation for Parkinson's disease with early motor complications. N Engl J Med. 2013;368:610–622. doi: 10.1056/NEJMoa1205158. [DOI] [PubMed] [Google Scholar]
- Schupbach W.M.M., Chabardes S., Matthies C., Pollo C., Steigerwald F., Timmermann L., Visser Vandewalle V., Volkmann J., Schuurman P.R. Directional leads for deep brain stimulation: Opportunities and challenges. Mov Disord. 2017;32:1371–1375. doi: 10.1002/mds.27096. [DOI] [PubMed] [Google Scholar]
- Senova S., Hosomi K., Gurruchaga J.M., Gouello G., Ouerchefani N., Beaugendre Y., Lepetit H., Lefaucheur J.P., Badin R.A., Dauguet J., Jan C., Hantraye P., Brugieres P., Palfi S. Three-dimensional SPACE fluid-attenuated inversion recovery at 3 T to improve subthalamic nucleus lead placement for deep brain stimulation in Parkinson's disease: from preclinical to clinical studies. J Neurosurg. 2016;125:472–480. doi: 10.3171/2015.7.JNS15379. [DOI] [PubMed] [Google Scholar]
- Starr P.A., Christine C.W., Theodosopoulos P.V., Lindsey N., Byrd D., Mosley A., Marks W.J., Jr. Implantation of deep brain stimulators into the subthalamic nucleus: technical approach and magnetic resonance imaging-verified lead locations. J Neurosurg. 2002;97:370–387. doi: 10.3171/jns.2002.97.2.0370. [DOI] [PubMed] [Google Scholar]
- Sumanaweera, T.S., Adler, J.R., Jr., Napel, S., Glover, G.H., 1994. Characterization of spatial distortion in magnetic resonance imaging and its implications for stereotactic surgery. Neurosurgery 35, 696–703; discussion 703–694. [DOI] [PubMed]
- Tinkhauser G., Pogosyan A., Debove I., Nowacki A., Shah S.A., Seidel K., Tan H., Brittain J.S., Petermann K., di Biase L., Oertel M., Pollo C., Brown P., Schuepbach M. Directional local field potentials: A tool to optimize deep brain stimulation. Mov Disord. 2018;33:159–164. doi: 10.1002/mds.27215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.T., Poirier S., Guo T., Parrent A.G., Peters T.M., Khan A.R. SPIE Medical Imaging. SPIE; 2016. Generation and evaluation of an ultra-high-field atlas with applications in DBS planning; p. 10. [Google Scholar]
- Welter M.L., Schupbach M., Czernecki V., Karachi C., Fernandez-Vidal S., Golmard J.L., Serra G., Navarro S., Welaratne A., Hartmann A., Mesnage V., Pineau F., Cornu P., Pidoux B., Worbe Y., Zikos P., Grabli D., Galanaud D., Bonnet A.M., Belaid H., Dormont D., Vidailhet M., Mallet L., Houeto J.L., Bardinet E., Yelnik J., Agid Y. Optimal target localization for subthalamic stimulation in patients with Parkinson disease. Neurology. 2014;82:1352–1361. doi: 10.1212/WNL.0000000000000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarg F., Herzog J., Reese R., Falk D., Pinsker M.O., Steigerwald F., Jansen O., Deuschl G., Mehdorn H.M., Volkmann J. Stimulation site within the MRI-defined STN predicts postoperative motor outcome. Mov Disord. 2012;27:874–879. doi: 10.1002/mds.25006. [DOI] [PubMed] [Google Scholar]
- Xiao Y., Fonov V., Beriault S., Al Subaie F., Chakravarty M.M., Sadikot A.F., Pike G.B., Collins D.L. Multi-contrast unbiased MRI atlas of a Parkinson's disease population. Int J Comput Assist Radiol Surg. 2015;10:329–341. doi: 10.1007/s11548-014-1068-y. [DOI] [PubMed] [Google Scholar]
- Yelnik J., Bardinet E., Dormont D., Malandain G., Ourselin S., Tande D., Karachi C., Ayache N., Cornu P., Agid Y. A three-dimensional, histological and deformable atlas of the human basal ganglia. I. Atlas construction based on immunohistochemical and MRI data. Neuroimage. 2007;34:618–638. doi: 10.1016/j.neuroimage.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Zaidel A., Spivak A., Grieb B., Bergman H., Israel Z. Subthalamic span of beta oscillations predicts deep brain stimulation efficacy for patients with Parkinson's disease. Brain. 2010;133:2007–2021. doi: 10.1093/brain/awq144. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material