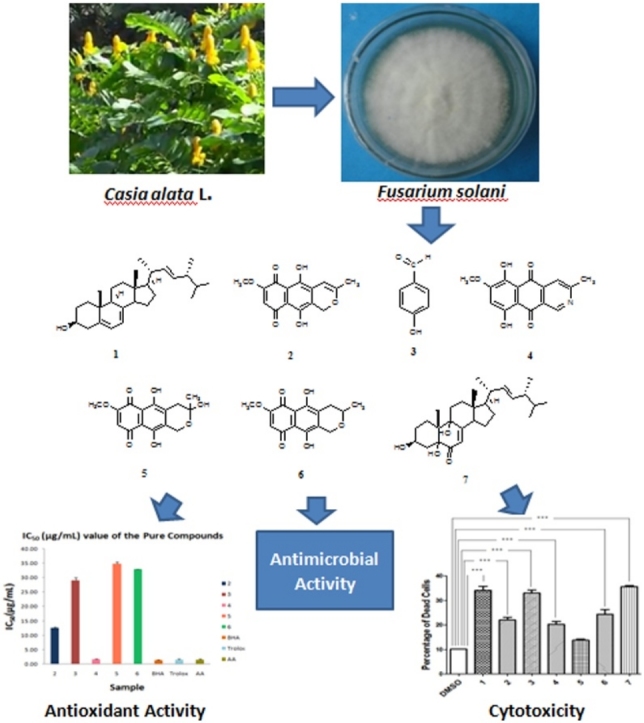
Graphical abstract
Keywords: Endophytic fungi, Fusarium solani, Cytotoxicity, Antimicrobial activity
Highlights
-
•
Endophytric fungi are natural reservoir for bioactive secondary metabolites.
-
•
Fusarium solani, an endophytic fungus, is a rich source for secondary metabolites specially for quinone groups.
-
•
Napthaquinone and aza-anthraquinone derivatives have significant antimicrobial, antioxidant and cytotoxic effect.
-
•
Napthaquinone and aza-anthraquinone derivatives have the potentiality to be lead for anticancer and antimicrobial agent.
-
•
Aza-anthraquinone derivatives are more bioactive than napthaquinone group due to the presence of nitrogen group.
Abstract
This study reports the chemical investigation and bioactivity of the secondary metabolites produced by the endophytic fungus Fusarium solani isolated from Cassia alata Linn. growing in Bangladesh. This plant was collected from conservation forest in Bangladesh and belongs to the Caesalpiniaceae family. The endophytic fungus Fusarium solani was isolated from the tissue of root of this plant. The fungal strain was identified by morphological characters and DNA sequencing. The crude organic extract of the fungal strain was proven to be active when tested for cytotoxicity against Brine Shrimp Lethality Bioassay, antimicrobial and antioxidant activity. The bioactivity guided fractionation of the ethyl acetate extract leads to the isolation of seven secondary metabolites in pure form. The structures of the isolated compounds were determined by the analysis of NMR and mass spectroscopic data. Bioassay investigation of the isolated secondary metabolites suggested aza-anthraquinones are more potent bioactive compounds as anticancer and antimicrobial agent.
1. Introduction
Endophytes are microorganisms that inhabit the interior of fresh plant tissues and show no apparent harm to their host [1]. Recently, endophytes are growing attention to the scientist as they are a good reservoir of outstanding secondary metabolites that are thought to be useful to the plant for their survival [2]. These secondary metabolites are potential sources of new leads to introduce new drugs, pesticides and antibiotics [3]. The prospects of searching new drugs that may be effective candidates for treating newly invading diseases in humans, animals and plants are highly demanding in the world [4].
The intention of the present investigation was to extract, explore and characterize the secondary metabolites produced by the endophytic fungus Fusarium solani isolated from roots of Cassia alata Linn. growing wild in Bangladesh and to evaluate their bioactivity. Cassia alata (Bengali name: Dadmurdan, Family: Caesalpiniaceae) is perennial, large spreading shrub [5], used extensively in traditional herbal medicine for various ailments. Historically, it is familiar for its fungicidal, insecticidal and antimicrobial properties [[6], [7], [8]]. An expectorant in bronchitis, astringent and laxative properties was also reported from this plant [[9], [10], [11]]. Previous phytochemical investigations resulted in the isolation of rhein, emodin, aloeemodin, crysophanol, isocrysophanol, glycosides of aloeemodin, sitosterol, sennosides A, B and C, physcion, kaempferol3gentiobioside, chrysophanic acid, adenine and flavonoids [[9], [10], [11]].
2. Materials and methods
2.1. General experimental procedures
The NMR spectra were recorded on a Bruker 400 MHz NMR spectrometer (Bruker, Switzerland) using CDCl3. The MS spectra were recorded on an Exactive Orbitrap by a Thermo Scientific Mass Spectrometer (Bruker, Switzerland) at King's College London,(London, UK) and the data was processed by Thermo XCalibur 2.2. Infrared spectra (IR) were measured on a SHIMADZU FTIRspectrometer at Jahangirnagar University, Savar, Dhaka. Ultraviolet (UV) spectra were obtained on a SPECORD 250 PLUS UV–vis spectrophotometer using methanol. Column chromatography (CC) was carried out on silica gel (70–230 and 230–400 mesh, Merck, Germany).
2.2. Isolation of fungal material
The fungus Fusarium solani was isolated from fresh healthy roots of Cassia alata Linn. The plant samples were collected from conservation forest of Gazipur, Bangladesh in August, 2013. The plant material was identified and authenticated by a taxonomist of Bangladesh National Herbarium (BNH), Dhaka, Bangladesh. A voucher specimen of this collection was maintained at BNH under the accession number DACB – 38,723. Endophytic fungi were isolated from the fresh plant parts following the procedure, established at Pharmaceutical Sciences Research Division, BCSIR Laboratories, Dhaka, Bangladesh [12,13].
2.3. Taxonomical identification of fungal cultures
2.3.1. Morphological identification
For the identification of endophytic fungal strains, slides prepared from cultures were stained with lactophenol cotton blue examined with a bright-field and phase contrast microscope [14]. Morphological identification was done according to the standard taxonomic key such as growth pattern, hyphae, the color of the colony and medium, margin character, aerial mycelium, surface, texture, sporulation and production of acervuli, the size and coloration of the conidia [15,16].
2.3.2. Molecular identification
Fungal strains were identified using a molecular biological protocol by DNA amplification and sequencing of the internal transcribed spacer (ITS) region [17]. A section of fungal hyphae (0.5-1.0 cm2) was collected from the petridish and lyophilized in a eppendorf tube (2 mL) (Eppendorf, Germany). The lyophilized fungal mycelia were pulverized and disrupted with the help of glass beads. Fungal DNA isolation was achieved by using DNeasy Plant Mini Kit (QIAgen, USA) according to the manufacturer’s protocol. The procedures were included cell lysis, digestion of RNA by RNase A, removal of precipitates and cell debris, DNA shearing, precipitation and purification. The isolated DNA was then amplified by polymerase chain reaction (PCR). The PCR was carried out using HotStarTaq Master Mix Kit (QIAgen, USA). ITS 1 (with base sequences TCCGTAGGTGAACCTGCGG) and ITS 4 (with base sequences TCCTCCGCTTATTGATATGC) (Invitrogen, USA), as primers, were mixed with HotStarTaq Master Mix Kit and DNA template in a total volume of 50 μL. The mixture was then applied to the thermal cycler (BioRad, USA) using the programmed PCR cycle as outlined below:
Initial activation step in 95 °C for 15 min to activate HotStarTaq DNA polymerase, cycling steps which were repeated 35 times, denaturing: 1 min at 95 °C, annealing: 1 min at 56 °C, extension: 1 min at 72 °C, final extension for 10 min in 72 °C.
The PCR product was purified using 2% Agarose-Gel-Electrophoresis at 75 V for 60 min in 1 X TBE buffer. The agarose gel was then stained using 1% ethidium bromide. An approximate size 550 bp stained DNA fragment was then excised from the agarose gel. The next step of PCR product purification was performed using Perfect Prep Gel Cleanup Kit (Eppendorf, USA) by following manufacturer’s protocol. The amplified fungal DNA (PCR product) was then submitted for sequencing by a commercial service and the base sequence was compared with publicly available databases such as GenBank with the help of Blast-Algoririthmus.
2.4. Isolation and structure elucidation of secondary metabolites
Fusarium solani species (internal strain no. CARE-1), which had been isolated following surface sterilization from the root of the plant C. alata were cultivated at 28 ± 2 °C for 28 days on potato dextrose agar medium. After the completion of the incubation, the cultured media were extracted three times with ethyl acetate to obtain the crude extracts [18]. The crude extract (4.2 g) was subjected to column chromatography for fractionation on silica gel by using a gradient of petroleum ether, followed by mixtures of petroleum ether-ethyl acetate, ethyl acetate and finally with the mixtures of ethyl acetate-methanol with increasing polarity. A total of 44 fractions of 100 mL each were collected and combined into 8 fractions according to their TLC patterns for further purification. Different solvent treatments were used to get the pure compounds from the 8 fractions. The fraction 3 was treated with few mL of n-hexane to remove coloring materials to get compound 1 (39.7 mg) as white needle shaped crystals in pure form. Compound 2 (8.5 mg) and 3 (5.2 mg) were isolated in pure form as violet and light brown crystals respectively by eluting with n-hexane, n-hexane-dichloromethane, dichloromethane and dichloromethane-methanol in gradient manner and treating with few mL of methanol. The fraction 5 was subjected to PTLC (stationary phase: silica gel F254, mobile phase: toluene -10% ethyl acetate, thickness of plates: 0.5 mm). From the developed plates two sets of bands were separately scrapped and extracted with ethyl acetate. After evaporation of solvent one band (Rf = 0.44 with 10% ethyl acetate in toluene) gave purple colored crystals which was designated as compound 4 (6.7 mg) and the other band (Rf = 0.34 with 10% ethyl acetate in toluene) gave orange red crystal which was designated as compound 5 (25.8 mg). The fraction 6 was dried by evaporation of solvent gave impure red amorphous solid which on further purification by treatment with n-hexane to get compound 6(6.8 mg) in pure form. Compound 7 (8.5 mg) was isolated from column fraction 7 and purified by using PTLC method (stationary phase: silica gel F254, mobile phase: chloroform-5% methanol, thickness of plates: 0.5 mm).The compound was found as brown colored solid (Rf = 0.35with 5% methanol in chloroform). The structures of the pure compounds were determined by NMR data analysis (1H, 13C, COSY, HSQC and HMBC).
Compound 1 (Ergosterol)
White crystalline solid; mp 149–150 °C; IR (neat): ν3422 (br.O-H), 3043, 2954, 2870, 1654 (C = C), 1458, 1369, 1055 cm−1; 1H NMR (CDCl3):δ 5.55–5.57 (1H, m, 6Hor 7 H), 5.37–5.38 (1H, m, 6H or 7 H), 5.14–5.25 (2H, m, H-22 &H-23), 3.60–3.66 (1H, m, H-3), 2.44–2.45 (1H, m), 2.80–2.82 (1H, m), 1.94–2.07 (3H, m), 1.82–1.90 (3H, m), 1.53–1.77 (6H, m), 1.44–1.51 (2H, m), 1.22–1.40 (5H, m), 1.03 (3H, d, J = 6.4 Hz, H-21 or H-28), 0.94 (3H, s, H-18), 0.93 (3H, d,J = 6.8, H-21 or H-28), 0.84 (3H, d,J = 6.4 Hz, H-26 or H-27), 0.82 (3H, d,J = 6.4 Hz, H-26 or H-27), 0.62 (3H, s, H-19);13C NMR (CDCl3):δ 141.3 (C-5 or C-8), 139.7 (C-5 or C-8), 135.5, 131.9, 119.6, 116.3, 70.4 (C-3), 55.7, 54.5, 46.2, 42.8 (2C), 40.8, 40.4, 39.1, 38.3, 37.0, 33.1, 32.0, 28.2, 23.0, 21.1 (2C), 19.9, 19.6, 17.6, 16.2, 12.0; MS: m/z 396 (M+), 395 (base peak), 378, 354, 335, 295, 212.
Compound 2 (Anhydrofusarubin)
Violet Solid; mp 202–204 °C; UVλmax232, 273, 415 nm; IR (neat): ν3430 (O-H), 2930, 2857, 1712 (C = O), 1603 (ar. C = C), 1459, 1413, 1265, 1251, 1220 (C—O) cm−1; 1H NMR (CDCl3):δ13.06 (1H, s, —OH), 12.68 (1H, s, —OH), 6.19 (1H, s, H-8), 6.01 (1H, s, H-4), 5.29 (1H, unr. s, Ha-1), 5.23 (1H, unr. s, Hb-1), 3.92 (3H, s, —OCH3), 2.01 (3H, s, —CH3); 13C NMR (CDCl3):δ 182.8 (C = 0), 177.8 (C O), 161.5 (C-7), 160.0 (C-3), 158.0 (C-10) 157.2 (C-5), 133.1 (C-11), 122.7 (C-12), 122.5 (C-13), 122.2 (C-14), 110.0 (C-8), 94.7 (C-4), 62.9 (C-1), 56.6 (—OCH3), 20.9 (-CH3); MS: m/z 289 (M++H), 288 (M+).
Compound 3 (4-hydroxybenzaldehyde)
Brown solid; mp 112–116 °C;IR (neat): ν3168 (br., O-H), 1668 (C = O), 1600 (ar. C = C), 1517, 1454, 1161 (C—O) cm−1; 1H NMR (CDCl3+2 drops of CD3OD):δ 9.71 (1H, s, -CHO), 7.68 (2H, d, J = 8.4 Hz, H-2 &H-6), 6.85 (2H, d, J = 8.4 Hz, H-3 &H-5); 13C NMR (CDCl3+2 drops of CD3OD):δ 191.4 (C O), 163.3 (C-4), 132.3 (C-3 &C-5), 128.8 (C-1), 115.8 (C-2 &C-6).
Compound 4 (Bostrycoidin)
Purple crystal; mp 200–202 °C; IR (neat): ν3447 (O-H str.), 2922, 2850, 1623 (C = O), 1586 (ar. C = C), 1465, 1383, 1292, 1198, 1136 cm−1; 1H NMR (CDCl3):δ 13.48 (1H, s, -OH), 13.20 (1H, s, —OH), 9.49 (1H, s, H-1), 7.95 (1H, s, H-4), 6.75 (1H, s,H-7), 4.02 (3H, s, —OCH3), 2.79 (3H, s, —CH3); 13C NMR (CDCl3):δ 186.4 (C=O), 183.6 (C=O), 165.3 (C-3), 161.3 (C-8), 157.7 (C-6), 151.2 (C-5), 149.2 (C-1), 139.2 (C-14), 138.7 (C-13), 124.5 (C-11), 117.9 (C-4), 114.0 (C-12), 107.9 (C-7), 56.7 (-OCH3), 25.2 (-CH3); MS: m/z 285 (M+).
Compound 5 (Fusarubin)
Orange red crystal; mp 218–220 °C; UV λmax238,292,547 nm;IR (neat): ν3451 (O-H), 3065, 2921, 2850 (C–H), 1599 (C = 0), 1575 (ar. C = C), 1447, 1384, 1216, 1149, 1080 cm−1; 1H NMR (CDCl3):δ 12.93 (1H, s, —OH), 12.66 (1H, s, —OH), 6.18 (1H, s, H-8), 4.89 (2H, s, H-1), 3.93 (3H, s, —OCH3), 3.03 (1H, d, J = 18 Hz, Ha-4), 2.71 (1H, d, J = 18 Hz, Hb-4), 1.65 (3H, s, —CH3); 13C NMR (CDCl3):δ 184.6 (C = 0), 178.1 (C = 0), 160.8 (C-7), 160.7 (C-5), 157.3 (C-10), 137.1 (C-12), 132.9 (C-11), 140.0 (C-13), 109.6 (C-8), 107.6 (C-14), 94.2 (C-3), 58.5 (C-1), 56.7 (-OCH3), 32.1 (C-4), 29.4 (-CH3); MS: m/z 329 (M++Na), 306 (M+).
Compound 6 (3-deoxyfusarubin)
red solid;IR (neat): ν3315 (O-H), 2955, 2918, 1701 (C = O), 1600 (ar. C = C), 1458, 1394, 1241 (C–O), 1189, 1060 cm−1;1H NMR (CDCl3):δ 12.93 (1H, s, —OH), 12.67 (1H, s, —OH), 6.18 (1H, s, H-8), 4.89 (2H, s, H-1), 3.93 (3H, s, -OCH3), 3.60 (1H, m, H-3), 2.34 (2H, t, J = 7.6 Hz, H-4), 1.64 (3H, d, J = 9.6 Hz, —CH3); 13C NMR (CDCl3):δ 174.4 (2C, C = 0), 160.9(C-7), 158.3 (C-5), 153.8 (C-10), 130.3 (C-12), 127.9 (C-11), 116.9 (C-13), 115.3 (C-14), 109.6 (C-8), 70.0 (C-3), 65.0 (C-1), 63.2 (—OCH3), 31.8 (C-4), 22.6 (—CH3).
Compound 7 (3,5,9-trihydroxyergosta-7,22-diene-6-one)
Brown gummy solid; mp 226–230 °C; IR (neat): ν3446 (br.O-H), 2926, 2856, 1680 (C = O), 1600 (C = C) 1458, 1255, 1161 (C—O) cm−1;1H NMR (CDCl3):δ 5.67 (1H, d, J = 1.6 Hz, H-7), 5.17–5.28 (2H, m, H-22 &H-23), 4.07 (1H, m, H-3),2.75–2.80 (m), 2.04–2.15 (m), 1.7–2.0 (m), 1.55–1.70 (m), 1.53–1.69 (m), 1.10–1.52 (m), 0.83–1.08 (15H, m), 0.63 (3H, s); 13C NMR (CDCl3):δ 197.9 (C O), 135.0, 132.6, 119.8, 114.0, 79.6, 72.3, 67.2, 56.0, 51.7, 45.3, 42.8, 41.7, 40.2, 34.9, 33.0, 31.9, 31.7, 29.3, 22.6, 21.0, 20.4, 19.9, 19.6, 17.6, 14.1, 14.0, 12.2; MS: (m/z) 444 (M+), 396, 395 (base peak), 335, 295, 288, 265, 208.
2.5. Bioactivity screening
2.5.1. Antimicrobial screening
The antimicrobial and antifungal activities were investigated by the method described by Bauer et al. [19]. The test microorganisms used in the antimicrobial study included four pathogenic bacterial strains Staphylococcus aureus (ATCC 25,923), Escherichia coli (ATCC 28,739), Bacillus megaterium (ATCC 18), Pseudomonas aeruginosa (ATCC 27,833), two fungal strains Aspergillus niger and A. flavus. The zones of growth inhibition around the discs were measured after 18–24 hrs of incubation at 37 °C for bacteria and 48–96 hrs of incubation at 28 °C for fungi. The sensitivities of the microorganism species to the fungal extract were determined by measuring the diameter of inhibitory zones in millimeter compared to kanamycin (30 μg/disc) and ketoconazole (30 μg/disc) as standard antibiotics for antibacterial and antifungal screening, respectively.
2.5.2. Antioxidant activity
The antioxidant activity of the fungal endophyte’s extracts was measured by the discoloration of methanol solution of DPPH (1,1-diphenyl-2-picryl hydrazyl) radical according to Brand-Willium et al. [20]. The scavenging ability of fungal extract was measured by spectroscopically (UV–vis Spectrophotometer, Analytikjena, Germany) at 517 nm. Butylated hydroxyanisole (BHA) and ascorbic acid (Vit C) were used as positive controls. The measurement was carried out in triplicate and averaged. The scavenging ability was calculated by the following method:
| Scavenging ability (%) = (A517 of control – A517 of sample/A517 of control) × 100 |
Results were presented as an IC50 (Concentration of the sample that showed 50% scavenging of the DPPH radical).
2.5.3. Cytotoxicity test
Antiproliferative activity was examined against African Green Monkey Kidney cell (Vero cell) line (CLS 605372, Germany) using the Trypan Blue Exclusion Method [21]. From the test samples, stock solutions in DMSO (1.0 mg/mL) were prepared. Vero cells were cultured in 75cm2 sterile flasks in Dulbecco’s Modified Eagles Medium (DMEM), supplemented with 10% (v/v) fetal bovine serum (FBS), 1% penicillin/streptomycin/neomycin (100 U/mL), (0.1 mg/mL) and 25 mM HEPES in 5% (v/v) CO2, pH 7.4 at 37 °C [22]. For every treatment, cells were split into 3 groups according to experimental design and all the groups had three replica. Treatment groups were compared with vehicle group. The day before the experiments, vero cells were split and a number of 2.5 × 106 cells were cultured into T-25 flask. The prepared doses were administered into 1 day old T-25 cell culture flasks and doses were prepared just before using. Flasks were incubated for 24 h. Cells were then harvested by trypsin and 1.0 mL cell suspension was collected for cell counting. The number of viable cells was determined by using trypan blue and automated cell counter (LUNA-II™, analytikjena, South Korea) [23]. Two drops of cell suspension with 0.4% w/v trypan blue (1:1) were added on the surface of haemocytometer and was placed on automated cell counter and the number of unstained (viable) cells and stained (non-viable) cells were counted. Cells were calculated as percentage of dead cells by the following way:
2.5.4. Statistical analysis
IC50 values between gro ups were compared using independent student’s t- tests. For the analysis of the cytotoxicity results for each secondary metabolite, continuous variables between groups were compared with one-way analysis of variance (ANOVA) with post hoc tukey’s test. Mean values between groups were compared using independent student t-test for equality of variances. Statistical significance was accepted when P < 0.001.
3. Results and discussions
3.1. Identification of fungal strain
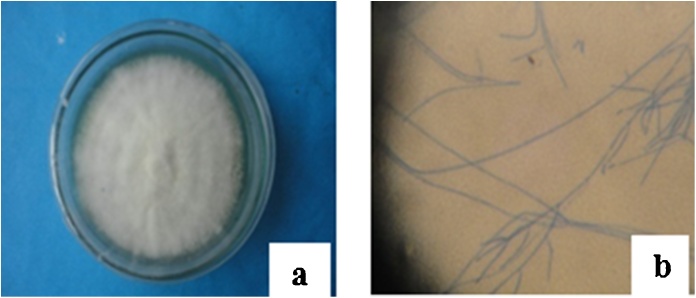
Endophytic fungal strains were identified based on morphological characteristics such as growth pattern, hyphae, the color of the colony and medium, surface texture, margin character, aerial mycelium, sporulation and production of acervuli and the size and coloration of the conidia using standard identification manuals [[15], [16], [17]] (Fig. 1). Molecular analysis of the fungus (CARE-1) based on 5.8 s rRNA gene revealed 99% similarity to another fungal isolate of accession number KF918581.1 that itself was identified as Fusarium solani, similarly to other related texa, for example, Fusarium solani (99%, accession numbers KF897913.1, KF897909.1, KF897904.1, DQ535183.1, KC254048.1, KX349467.1, KC254052.1, KC808240.1 etc.) deposited in the U.S. National Center for Biotechnology Information (NCBI).
Fig. 1.
Macroscopic (a) and Microscopic (b) view of Fusarium solani sp. The pictures represent macroscopic view of Fusarium solani from roots of Casia alata after 10 days of cultivation on PDA media and also represent microscopic view (40X).
>Seq1 [organism = Fusarium solani] 5.8 s rRNA
CCCTTTGGTAAGCGGAAGGACATTACCGAGTTATACAACTCATCAACCCTGTGAACATACCTAAAACGTTGCTTCGGCGGGAACAGACGGCCCCGTAACACGGGCCGCCCCCGCCAGAGGACCCCATAACTCTGTTTCTATTATGTTTCTTCTGAGTAAAACAAGCAAATAAATTAAAACTTTCAACAACGGATCTCTTGGCTCTGGCATCGATGAAGAACGCAGCGAAATGCGATAAGTAATGTGAATTGCAGAATTCAGTGAATCATCGAATCTTTGAACGCACATTGCGCCCGCCAGTATTCTGGCGGGCATGCCTGTTCGAGCGTCATTACAACCCTCAGGCCCCCGGGCCTGGCGTTGGGGATCGGCGAGGCGCCCCATGCGGGCACACGCCGTCCCCCAAATACAGTGGCGGTCCCGCCGCAGCTTCCATTGCGTAGTAGCTAACACCTCGCAACTGGAGAGCGGCGCGGCCACGCCGTAAAACACCCAACTTCTGAATGTTGACCTCGAATCAGGTAGGAATACCCGCTGAACTTAAGCATATCAATAAGCGGAGGAACTT
3.2. Structure elucidation of isolated compounds
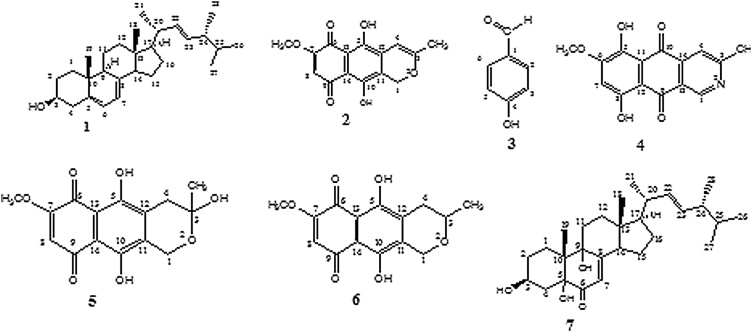
Large scale cultivation of endophytic fungus Fusarium solani on PDA medium followed by extraction with ethyl acetate and various chromatographic separation process yielded three naphthoquinones anhydrofusarubin (2) (8.5 mg), fusarubin (5) and 3-deoxyfusarubin (6) (6.8 mg), one aza-anthraquinone bostrycoidin (4) (6.7 mg), two sterols ergosterol (1) (39.7 mg) and 3,5,9-trihydroxyergosta-7,22-diene-6-one (7) (8.5 mg) and 4-hydroxybenzaldehyde (3) (5.2 mg) (Fig. 2). This is the first report of their isolation from endophytic fungus Fusarium solani obtained from the root of C. alata. The structures were confirmed by IR, 1H NMR, 13C NMR and mass spectroscopic data. As the metabolites are known compounds they were identified by comparison of their spectroscopic data with those reported in the literature.
Fig. 2.
Secondary metabolites produced from Fusarium solani isolated from C. alata - Ergosterol (1), Anhydrofusarubin (2), 4-hydroxybenzaldehyde (3), Bostrycoidin (4), Fusarubin (5), 3-deoxyfusarubin (6) and 3,5,9-trihydroxyergosta-7,22-6-one (7).
Compound 1: In the 1H NMR spectrum the two 1H multiplets at δ 5.55–5.57 and 5.37–5.38 ppm and two 2Hmultiplets at δ 5.14–5.25 and δ 3.60–3.66 ppm,four doublets and two singlets in the region at δ 0.62–1.03 ppm were found. In the 13C NMR spectrum six signals in the region at δ 116.3–141.3 ppm,a signal at δ 70.4 ppm and the presence of 28 carbons indicated by the 26 signals were found. The analysis of 13C NMR and DEPT-135 spectral data confirmed that the molecule contains six methyl, seven methylene, eleven methine and four quaternary carbons.The mass spectrum of the compound showed a molecular ion peak at m/z 396 which is corresponding to the molecular formula C28H44O. Finally, the structure of compound 1 was confirmed as ergosterol by comparison with the published data [24]. The structure of the compound 1 is given in Fig. 2.
Compound 2:1H NMR spectrum showed two 1H singlets at δ 13.06 and 12.68 ppm, two singlets at δ 6.19 (1 H) and 6.01 ppm (1 H) in the aromatic region, two 1H unresolved singlets at δ 5.29 and 5.23 ppm, two singlets at δ 3.92 and 2.01 ppm. In the 13C NMR spectrum methylene carbon at δ 62.9 ppm,15 signals for fifteen carbons, two carbonyl carbons at δ 182.8 and 177.8 ppm, three carbons attached to hydroxyl groups & methoxyl group ascertained by the three signals at δ 161.5, 158.0 and 157.2 ppm, oxygen attached carbon at δ 160.0 ppm were observed. The above analysis of all available spectral data strongly suggested that the compound 2 is an oxy-anthraquinone derivative containing two hydroxyls, one methoxyl and one methyl groups. The mass spectrum of the compound 2 showed a molecular ion peak at m/z 288 which is corresponding to the molecular formula C15H12O6. Finally, the structure of compound 2 was confirmed as anhydrofusarubin by comparison with the published data [25,26]. The structure of the compound 2 is given in Fig. 2.
Compound 3:1H NMR spectrum contained one1H singlet at δ 9.71 ppm, two 2H doublets at δ 7.68 and 6.85 ppm.In 13C NMR spectrum carbonyl carbon at δ 191.4 ppm, two pairs of equivalent carbons confirmed by the signals at δ 115.8 and 132.3 ppm, C-4signal at δ 163.3 ppm, C-1signal at δ 128.8 ppm were observed. Finally, the structure of compound 3 was confirmed as 4-hydroxybenzaldehyde by comparison with the published data [27]. The structure of the compound 3 is given in Fig. 2.
Compound 4: In the 1H NMR spectrum, the two 3H singlets at δ 2.79 and 4.02 ppm, three 1H singlets at δ 9.49, 7.95 and 6.75 ppm, two 1H singlets at δ 13.48 and 13.20 ppm were found. 13C NMR spectrum contained two signals at δ 25.2 and 56.7 ppm, 4 displayed 15 signals for 15 carbons, two signals at δ 186.4 and 183.6 ppm, five signals at δ 165.3 (C-3), 161.3 (C-8), 157.7 (C-6), 151.2 (C-5) and 149.2 ppm (C-1).The protons H-1 and H-4 were more deshielded because they are attached to the nitrogen containing aromatic ring. The peak at δ 9.49 ppm was due to the proton at C-1 which is directly attached to the nitrogen atom. The mass spectrum of the compound 4 showed a molecular ion peak at m/z 285 which is corresponding to the molecular formula C15H11NO5. All the above data suggested that the compound 4 is an aza-anthraquinone derivative containing two hydroxyl, one methoxyl and one methyl groups. Based on all spectroscopic data, literature values [28] of the compound 4, it was confirmed that the compound is bostrycoidin. The structure of the compound 4 is given in Fig. 2.
Compound 5: In the 1H NMR spectrum, two 1H singlets at δ 12.93 and 12.66 ppm,1H singlets at δ 6.18 and δ 4.89 ppm, two doublets at δ 3.03 and 2.71 ppm,two 3H singlets at δ 3.93 and 1.65 ppm were found. In the 13C NMR spectrum, signal at δ 58.5 ppm assigned for methylene carbon (C-1), signal at δ 32.1 ppm indicated methylene carbon (C-4), 15 signals for 15 carbons, two signals at δ 184.6 and 178.1 ppm indicated two carbonyl groups, three signals at δ 160.8, 160.7 and 157.3 ppm indicated three aromatic carbons, C-7, C-5 and C-10 which are directly attached to the oxygen atom, signal at δ 94.2 ppm indicated C-3with attachment of two oxygen atoms, signals at δ 56.7 and 29.4 ppm indicated methoxyl and methyl carbons. The mass spectrum of the compound 5 showed a molecular ion peak at m/z 329 [M + Na]+, which is corresponding to the molecular formula C15H14O7. Based on all spectroscopic data and literature values [26] of the compound 5, it was confirmed that the compound is fusarubin. The structure of the compound 5 is given in Fig. 2.
Compound 6:1H NMR spectrum showed two 1H singlets at δ 12.93 and 12.67 ppm, signal at δ 6.18 ppm assigned for the proton attached to C-8, 2H singlet at δ 4.89 ppm, 1H multiplet at δ 3.60 ppm, 2H triplet at δ 2.34 ppm, 3H singlet at δ 3.93 ppm, 3H doublet at δ 1.64 ppm. In the 13C NMR spectrum methylene carbons (C-1) could be assigned by the signal at δ 65.0 ppm,methylene carbon (C-4)signal at δ 31.8 ppm, carbonyl carbon signal atδ 174.4 ppm, three oxygen attached aromatic carbon could be assigned by the signals at δ 160.9 (C-7), 158.3 (C-5) and 153.8 ppm (C-10), methoxyl and methyl carbon signal atδ 63.2 and 22.6 ppm were found.The structure of 6 was confirmed as 3-deoxyfusarubin by comparison with the published NMR data [25,26]. The structure of the compound is given in Fig. 2.
Compound 7:1H NMR spectrum contained 1H singlet in the olefinic region at δ 5.67 ppm, multiplet at δ 5.17–5.28 ppm (2 H),1H multiplet at δ 4.07 ppm, a multiplet at δ 0.83–1.08 ppm (15 H) and a singlet at δ 0.63 ppm (3 H) indicated six methyl groups in the molecule, signals in the region at δ 1.10–2.15 ppm indicated methylene and methine protons. In the 13C NMR spectrum, four signals at δ 135.0, 132.6, 119.8 and 114.0 ppm, three signals at δ 79.6, 72.3 and 67.2 ppm indicated three carbon C-3, C-5 and C-9 those are directly attached to the hydroxyl group, the signal at δ 197.9 ppm indicated carbonyl carbon (C-6) were found. The mass spectrum of the compound showed a molecular ion peak at m/z 444 which is corresponding to a molecular formula C28H44O4. Based on all spectroscopic data and literature values [29] it was confirmed that the compound is 3,5,9-trihydroxyergosta-7,22-diene-6-one.The structure of the compound 7 is given in Fig. 2.
Previous results of our laboratory also showed that ethyl acetate extract of Fusarium solani strain isolated from the plant Aponogeton undulates produced seven secondary metabolites (four napthaquinones and two aza-anthraquinones namely, 9-desmethylherbarine, fusarubin, anhydrofusarubin, javanicin and 7-desmethylscorpinone, 7-desmethyl-6-methylbostrycoidin, respectively along with one sterol, cerevesterol [2]. Another evidence has been reported from a research group that a Fusarium solani strain isolated from the plant Glycyrrhiza glabra produced four quinone metabolites (3,6,9-trihydroxy-7-methoxy-4,4-dimethyl-3,4-dihydro-1H-benzo[g]isochromene-5,10-dione, fusarubin, 3-O-methylfusarubin and javanicin [30]. In 1979, a research group also reported that a Fusarium solani strain isolated from the soil produced four pigmented metabolites including fusarubin [31].
3.3. Bio-screening of isolated pure compounds
3.3.1. Antimicrobial test
In the antimicrobial screening, the zone of inhibition produced by compounds fusarubin (5), bostrycoidin (4), anhydrofusarubin (2) were found to be 21–32 mm, 12–16 mm and 10–17 mm, respectively (Table 1). The investigated compound fusarubin (5) exhibited highly significant activity against four tested pathogenic bacteria B. megaterium, S. aureus, P. aeruginosa and E. coli. Bostrycoidin (4) and anhydrofusarubin (2) exhibited prominent inhibition against the above tested pathogenic bacteria. Similar to our findings, another group of researchers also reported that anti-tuberculosis potential of bioactive molecules from endophytic F. solani isolated from Glycyrrhiza glabra evaluated against the virulent strain of Mycobacterium tuberculosis [30].
Table 1.
Antimicrobial Screening of Isolated Compounds.
| Sample (100 μg/disc) |
Diameter of Zone of Inhibition (mm) |
||||||
|---|---|---|---|---|---|---|---|
| Bacterial Strain |
Fungal Strain |
||||||
| Gram positive |
Gram negative |
||||||
|
B. megaterium |
S. aureus | P. aeruginosa | E. coli | A. niger | A. flavus | ||
| 1 | – | – | – | – | – | – | |
| 2 | 17 | 11 | 10 | 12 | – | – | |
| 3 | – | – | – | 7 | – | – | |
| 4 | 12 | 12 | 15 | 16 | – | – | |
| 5 | 32 | 24 | 22 | 21 | – | – | |
| 6 | 34 | 26 | 24 | 25 | |||
| 7 | – | – | – | – | – | – | |
| Positive control (30 μg/disc) |
KM | 48 | 30 | 30 | 35 | nd | nd |
| KC | nd | nd | nd | nd | 25 | 26 | |
KM – Kanamycin; KC – Ketoconazole; “-” indicates no sensitivity; nd: indicates not done
Quinone groups of various Fusarium species have been described to possess antimicrobial activity against bacteria and fungi [26,30,31]. Many microbial napthaquinones and aza-anthraquinones compounds exhibit antimicrobial activity with a specific mode of action [32]. These metabolites have been reported to inhibit various macromolecular syntheses by a secondary effect of the action on respiratory electron transport [32,33]. Bostrycoidin and fusarubin can stimulate cellular respiration and membrane NADH oxidation of bacteria, thereby, exhibit antimicrobial activity. Cytotoxicity of the quinones exhibited resulting from their interaction with NADH complex I and intercalation of DNA [2,33]. Further investigation is required to understand the mechanism of actions of the isolated napthaquinones and aza-anthraquinones groups from Fusarium solani.
3.3.2. Antioxidant test
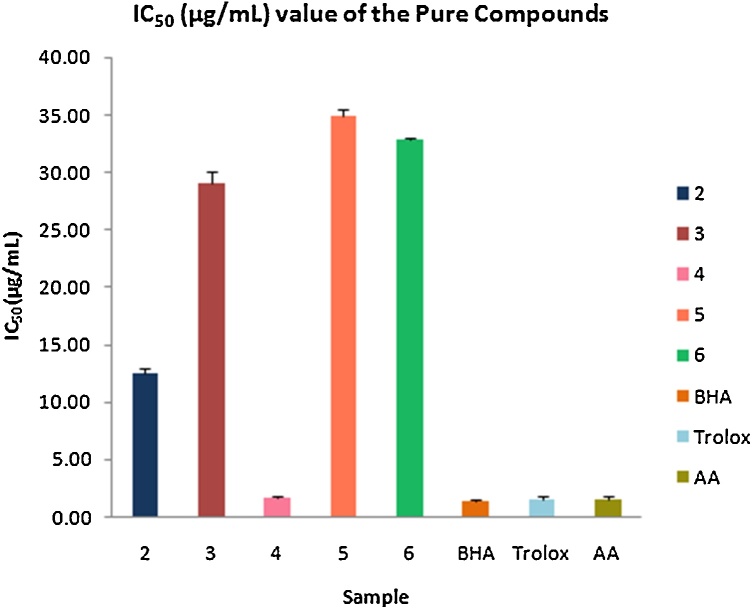
The free radical scavenging activity of the compounds was assayed by using DPPH scavenging method. The results showed that the compound bostrycoidin (4) exhibited significant antioxidant activity with IC50 value of 1.6 μg/mL which is very much comparable to the IC50 values of positive control 1.2, 1.3 and 1.5 for BHA, trolox and ascorbic acid, respectively (Fig. 3). Anhydrofusarubin (2), 4-hydroxybenzaldehyde (3) and fusarubin (5) exhibited good antioxidant activity with IC50 values of 12.4, 28.9 and 34.8 μg/mL, respectively. These results indicate that bostrycoidin (4) has high antioxidant capacity which may have potentiality to be a drug for preventing certain diseases, being either the cause or the consequence of reactive oxygen species. The other napthaquinone compounds anhydrofusarubin (2), fusarubin (5) and 4-hydroxybenzaldehyde (3) also have potentiality to be active for free radical scavenging.
Fig. 3.
Free radical scavenging activity of isolated compounds.
Antioxidants that can inhibit or delay the oxidation of an oxidizable substrate in a chain reaction, therefore, appear to be very important [2]. Thus, the isolated compounds from endophytic fungi Fusarium solaniderived from roots of C. alata can be a potential candidate to be explored for the treatment of damage caused by free radicals.
3.3.3. Cytotoxicity test
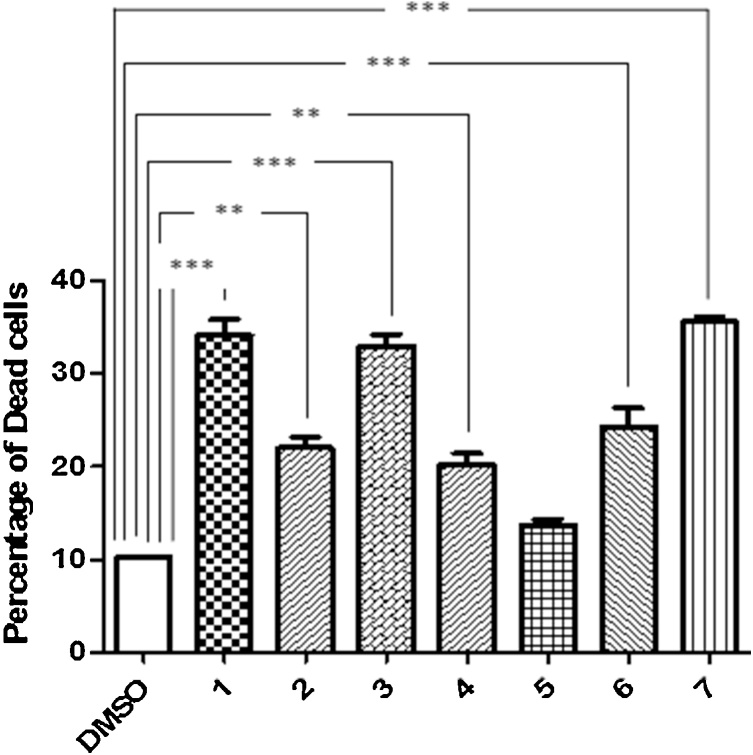
The cytotoxic potential of the pure compounds isolated from endophytic fungus Fusarium solani was determined by African Green Monkey Kidney Cell (vero cell) line. The percentage of dead cells was nearly 25 in 4-hydroxybenzaldehyde (3) and bostrycoidin (4) and about 35 in anhydrofusarubin(2) and 3,5,9-trihydroxy ergosta-7,22-diene-6-one (7), respectively. Results showed that 4-hydroxybenzaldehyde, bostrycoidin, anhydrofusarubin and 3,5,9-trihydroxyergosta-7,22-diene-6-one have significant cytotoxic effects on vero cells (Fig. 4).
Fig. 4.
Cytotoxic effects of isolated compounds on vero cell line after 24 hour incubation. Results are compared with vehicle (DMSO). The results are expressed as mean ± SEM (n = 3). Degrees of significance determined using ANOVA with Post Hoc Tukey’s test for comparison of isolated pure compounds with vehicle are *** P < 0.001 and 0.01.
These results indicate that 4-hydroxybenzaldehyde, bostrycoidin, anhydrofusarubin and 3,5,9-trihydroxyergosta-7,22-diene-6-one compounds may inhibit cell proliferation activity or activate apoptosis or necrosis in transformed kidney cells or kidney (renal) cancer cells. These inhibitory activities would be beneficial to introduce anticancer drugs for the kidney (renal) cancer patients to reduce tumor growth in Kidney. Similar to this observation, other investigation of this laboratory also found that other two aza-anthraquinones metabolites 7-desmethylscorpinone and 7-desmethyl-6-methylbostrycoidin showed better bioactivity against cytotoxicity and further evident by molecular docking [2].
4. Conclusion
This study supports the growing evidence that bioactive substances specially napthaquinones and aza-anthraquinones are produced by fungal endophytes Fusarium solani. To the best of our knowledge, this is the first study that reports napthaquinones and aza-anthraquinones are produced by a Fusarium solani strain isolated from Casia alata. In a continuation of searching bioactive secondary metabolites from natural sources at our laboratory, chemical investigation of ethyl acetate extract of Fusarium solani strain isolated from Casia alata led to the isolation of seven known secondary metabolites (three naphthaquinones anhydrofusarubin, fusarubin and 3-deoxyfusarubin, one aza-anthraquinone bostrycoidin, two sterols ergosterol and 3,5,9-trihydroxyergosta-7,22-diene-6-one and 4-hydroxybenzaldehyde). Results of this investigation showed that napthaquinones and aza-anthraquinones have potentiality as bioactive compounds against cytotoxicity, antimicrobial and antioxidant test. Compare to bioactivity of napthaquinones, aza-anthraquinone (bostrycoidin) showed better result. Thus, evidences indicated that Fusarium solani strain is a good source of napthaquinones and aza-anthraquinones metabolites.
Conflict of interest
The authors declared there is no conflict of interest in this published materials.
Transparency document
Acknowledgment
The authors are thankful to the authority of BCSIR for providing laboratory facilities and also thankful to the authority of University Grant Commission (UGC) for UGC Scholarship. The authors gratefully acknowledge Dr. Khondaker Miraz Rahman, Institute of Pharmaceutical Sciences, King’s College London for taking some NMR spectroscopy data.
References
- 1.Azevedo J.L., Maccheroni Jr W., Pereira J.O., de Araújo W.L. Endophytic microorganisms: a review on insect control and recent advances on tropical plants. Electron. J. Biotechnol. 2000;3(1):15–16. [Google Scholar]
- 2.Chowdhury N.S., Sohrab M.H., Rana M.S., Hasan C.M., Jamshidi S., Rahman K.M. Cytotoxic Naphthoquinone and Azaanthraquinone derivatives from an endophytic Fusarium solani. J. Nat. Prod. 2017;80(4):1173–1177. doi: 10.1021/acs.jnatprod.6b00610. [DOI] [PubMed] [Google Scholar]
- 3.Debbab A., Aly H.A., Edrada-Ebel R.A., Müller W.E., Mosaddak M., Hakiki A., Proksch P. Bioactive secondary metabolites from the endophytic Fungus Chaetomium sp. isolated from Salvia officinalis growing in Morocco. Biotechnol. Agron. Soc. Environ. 2009;13(2):229–234. [Google Scholar]
- 4.Strobel G., Daisy B. Bioprospecting for microbial endophytes and their natural products. Microbiol. Mol. Biol. Rev. 2003;67(4):491–502. doi: 10.1128/MMBR.67.4.491-502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faruq Z.U., Rahman U.A., Bello M., Obianke M., Atiku F.A. Antibacterial activity of the active Component of Cassia alata (Linn) leaves. Niger. J. Basic Appl. Sci. 2010;18(1):97–100. [Google Scholar]
- 6.Kumar M.P. Isolation and characterization of a compound from the leaves of Cassia alata linn. EC Chemistry. 2016;2:138–144. [Google Scholar]
- 7.El-Mahmood A.M., &Doughari J.H. Phytochemical screening and antibacterial evaluation of the leaf and root extracts of Cassia alata Linn. Afr. J. Pharm. Pharmacol. 2008;2(7):124–129. [Google Scholar]
- 8.Makinde A.A., Igoli J.O., Ta’Ama L., Shaibu S.J., Garba A. Antimicrobial activity of Cassia alata. Afr. J. Biotechnol. 2007;6(13) [Google Scholar]
- 9.Moriyama H., Iizuka T., Nagai M., Hoshi K. Adenine, an inhibitor of platelet aggregation, from the leaves of Cassia alata. Biol. Pharm. Bull. 2003;26(9):1361–1364. doi: 10.1248/bpb.26.1361. [DOI] [PubMed] [Google Scholar]
- 10.Moriyama H., Iizuka T., Nagai M., Miyataka H., Satoh T. Anti-inflammatory activity of heat-treated Cassia alata leaf extract and its flavonoidglycoside. Yakugaku Zasshi. 2003;123(7):607–611. doi: 10.1248/yakushi.123.607. [DOI] [PubMed] [Google Scholar]
- 11.Moriyama H., Iizuka T., Nagai M., Murata Y. HPLC quantification of kaempferol-3-O-gentiobioside in Cassia alata. Fitoterapia. 2003;74:425–430. doi: 10.1016/s0367-326x(03)00058-3. [DOI] [PubMed] [Google Scholar]
- 12.Chowdhury N.S., Sohrab M.H., Rony S.R., Sharmin S., Begum M.N., Rana M.S., Hasan C.M. Identification and bioactive potential of endophytic fungi from Monochoriahastata (L.) Solms. Bangladesh J. Bot. 2016;45(1):187–193. [Google Scholar]
- 13.Khan M.I.H., Sohrab M.H., Rony S.R., Tareq F.S., Hasan C.M., &Mazid M.A. Cytotoxic and antibacterial naphthoquinones from an endophytic fungus, Cladosporium sp. Toxicol. Rep. 2016;3:861–865. doi: 10.1016/j.toxrep.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sadananda T.S., Govindappa M., Ramachandra Y.L. 2014. In Vitro Antioxidant Activity of Lectin from Different Endophytic Fungi of Viscum album L. [Google Scholar]
- 15.Devi N.N., &Prabakaran J.J. Bioactive metabolites from an endophytic fungus Penicillium sp. isolated from Centellaasiatica. Curr. Res. Environ. Appl. Mycol. J. Fungal Biol. 2014;4(1):34–43. [Google Scholar]
- 16.Barnett H.L., Hunter B.B. 3rd ed. 1972. Illustrated Genera of Imperfect Fungi. [Google Scholar]
- 17.Cappiccino J.G., Sherman N. 6th ed. The Benjamin/ Cummings Publishing Company, Redwood City; CA, USA: 1996. Microbiology a Laboratory Manual. 1996. [Google Scholar]
- 18.Alzoreky N.S., Nakahara K. Antibacterial activity of extracts from some edible plants commonly consumed in Asia. Int. J. Food Microbiol. 2003;80(3):223–230. doi: 10.1016/s0168-1605(02)00169-1. [DOI] [PubMed] [Google Scholar]
- 19.Bauer A.W., Kirby W.M., Sherris J.C., Truck M. Antibiotic susceptibility testing by a standard single disc method. Am. J. Clin. Pathol. 1966;45:493–496. [PubMed] [Google Scholar]
- 20.Brand-Williams W., Cuvelier M.E., &Berset C.L.W.T. Use of a free radical method to evaluate antioxidant activity. LWT-Food Sci. Technol. 1995;28(1):25–30. [Google Scholar]
- 21.Strober W. Trypan blue exclusion test of cell viability. Curr. Protoc. Immunol. 2001;A3-B doi: 10.1002/0471142735.ima03bs111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson C.H., Ali E.S., Scrimgeour N., Martin A.M., Hua J., Tallis G.A., Barritt G.J. Steatosis inhibits liver cell store-operated Ca2+ entry and reduces ER Ca2+ through a protein kinase C-dependent mechanism. Biochem. J. 2015;466(2):379–390. doi: 10.1042/BJ20140881. [DOI] [PubMed] [Google Scholar]
- 23.Berry M.N., Barritt G.J., Edwards A.M. Elsevier; 1991. Isolated Hepatocytes: Preparation, Properties and Applications. [Google Scholar]
- 24.Goulston G., Mercer E.I., Goad L.J. The identification of 24-methylene-24, 25-dihydrolanosterol and other possible ergosterol precursors in Phycomyces blakesleeanus and Agaricuscampestris. Phytochemistry. 1975;14(2):457–462. [Google Scholar]
- 25.Tatum J.H., Baker R.A. Naphthoquinones produced by Fusarium solani isolated from citrus. Phytochemistry. 1983;22(2):543–547. [Google Scholar]
- 26.Kurobane I., Zaita N., Fukuda A. New metabolites of Fusarium martii related to dihydrofusarubin. J. Antibiot. 1986;39(2):205–214. doi: 10.7164/antibiotics.39.205. [DOI] [PubMed] [Google Scholar]
- 27.Jang D.S., Han A.R., Park G., Jhon G.J., &Seo E.K. Flavonoids and aromatic compounds from the rhizomes of Zingiber zerumbet. Arch. Pharm. Res. 2004;27(4):386–389. doi: 10.1007/BF02980078. [DOI] [PubMed] [Google Scholar]
- 28.Dame Z.T., Silima B., Gryzenhout M., van Ree T. Bioactive compounds from the endophytic fungus Fusarium proliferatum. Nat. Prod. Res. 2016;30(11):1301–1304. doi: 10.1080/14786419.2015.1053089. [DOI] [PubMed] [Google Scholar]
- 29.Kawagishi H., Katsumi R., Sazawa T., Mizuno T., Hagiwara T., Nakamura T. Cytotoxic steroids from the mushroom Agaricus blazei. Phytochemistry. 1988;27(9):2777–2779. [Google Scholar]
- 30.Shah A., Rather M.A., Hassan Q.P., Aga M.A., Mushtaq S., Shah A.M., Ahmad Z. Discovery of anti‐microbial and anti‐tubercular molecules from Fusarium solani: an endophyte of Glycyrrhiza glabra. J. Appl. Microbiol. 2017;122(5):1168–1176. doi: 10.1111/jam.13410. [DOI] [PubMed] [Google Scholar]
- 31.Ammar M.S., Gerber N.N., McDaniel L.E. New antibiotic pigments related to fusarubin from Fusarium solani (MART.) SACC. J. Antibiot. 1979;32(7):679–684. doi: 10.7164/antibiotics.32.679. [DOI] [PubMed] [Google Scholar]
- 32.Marumo H., Kitaura K., Morimoto M., Tanaka H., &Omura S. The mode of action of nanaomycin A in Gram-positive bacteria. J. Antibiot. 1980;33(8):885–890. doi: 10.7164/antibiotics.33.885. [DOI] [PubMed] [Google Scholar]
- 33.Haraguchi H., Yokoyama K., Oike S., Ito M., Nozaki H. Respiratory stimulation and generation of superoxide radicals in Pseudomonas aeruginosa by fungal naphthoquinones. Arch. Microbiol. 1997;167(1):6–10. doi: 10.1007/s002030050409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.