Abstract.
In the Americas, 8 million people are infected with Chagas disease, and an additional 90 million people are at risk for infection. Little is known about the role bats play in the sylvatic transmission cycle of Trypanosoma cruzi, the parasite causing Chagas disease. Here, we captured bats in the villages of Palmiche, Pachacutec, Nuevo San Martin, and Mayuriaga located in the Datem del Marañon Province in Loreto, Peru. Venous blood samples were collected by cardiac puncture or from the upper extremities, and trypanosomatids were identified by microscopy and molecularly. We collected blood samples from 121 bats on filter paper for molecular studies and 111 slides for microscopic examination of thin and thick blood smears from 16 different bat species. The prevalence of trypanosomatids in all bats species was 34.7% (42/121) and the prevalence of T. cruzi was 4.1% (5/121). In hematophagous bat species, the prevalence of trypanosomatids and T. cruzi was 36.9% (27/73) and 2.7% (2/73), respectively. In non-hematophagous bats, the prevalences of trypanosomatids and T. cruzi were 31.2% (15/48) and 6.2% (3/48), respectively. Also, we confirm the presence of T. cruzi in salivary glands of hematophagous bats Diaemus youngi. These results suggest a sylvatic cycle of trypanosomatid transmission in which bats may harbor infectious T. cruzi parasites that could be transmitted to humans via hematophagous bat bites or salivary contamination by non-hematophagous bats of vegetables consumed by humans.
INTRODUCTION
American trypanosomiasis, also known as Chagas disease, is an important vector-borne zoonosis caused by the kinetoplastid parasite Trypanosoma cruzi.1,2 There is evidence that supports the passage of a clade from a common ancestor of T. cruzi and Trypanosoma rangeli from bats to terrestrial mammals with subsequent diversification and extension. The independent passage of this group gave rise to T. cruzi facilitated by the exchange of niches between bats, terrestrial mammals, and invertebrate vectors.3 In South America, T. cruzi is prevalent in countries between 40° North and 45° South latitudes and is transmitted by insects of the family Reduviidae to mammals of seven different orders, including humans, in both urban and rural settings.2,4,5 Chagas disease is endemic in 21 countries in the Americas, infecting 8 million people and posing an additional risk of infection to 90 million people. Because of its widespread distribution in the Americas, the World Health Organization has considered this disease for control and elimination.6,7
Studies demonstrate that many mammalian species can serve as hosts in the T. cruzi life cycle.8 Molecular characterization of the parasite reveals high genetic diversity with two recognized subspecies (T. c. cruzi and T. c. marinkellei)9 and up to seven T. cruzi genotypes or discrete typing units (DTUs: TcI to Tc VI and Tcbat).10,11
Understanding Chagas disease epidemiology is important for designing effective control strategies. In recent years, T. cruzi diagnosis has been greatly improved with the use of molecular biology, which permits clarification of the diversity of T. cruzi genotypes and their affinity to specific host mammals.11 Through molecular studies, bats have been found to be likely natural hosts of T. cruzi11 with some species harboring T. marinkellei and Tcbat.11–13 Although T. marinkellei is restricted to bats, evidence that the Tcbat DTU can infect both bats and humans suggests that bats could be part of a sylvatic T. cruzi transmission cycle by serving as natural hosts of the parasite.11–13 Studies in the coastal and Andean regions of Peru have described the domestic life cycle of T. cruzi in guinea pig, dog, goat, and cat hosts, and in Triatoma infestans and Panstrongylus herreri vectors.14–18 In the Peruvian Amazon Basin, the sylvatic life cycle of T. cruzi has been described in monkeys (Saimiri boliviensis), Andean white-eared opossum (Didelphis pernigra), and lesser spear-nosed bat (Phyllostomus elongatus) hosts, and in Panstrongylus geniculatus, P. herreri, Panstrongylus chinai, Rhodnius ecuadoriensis, and R. pictipes vectors.14–20
In this study, we aimed to assess natural T. cruzi infection in bats from different native communities located in the Datem del Marañon Province within the Peruvian Amazon Basin to evaluate the potential role of these species in the transmission of Chagas disease.
METHODS
Ethics.
This study was approved by the Institutional Animal Care and Use Committee of the U.S. Naval Medical Research Unit No. 6 (NAMRU-6). Experiments were conducted in compliance with the Animal Welfare Act and in accordance with the principles set forth in the “Guide for the Care and Use of Laboratory Animals,” Institute of Laboratory Animals Resources, National Research Council, National Academy Press, 1996. The Peruvian government agency “Dirección de Gestión Forestal y Fauna Silvestre del Peru (N° 0156)” approved the collection of bat specimens as described. As required by Peruvian regulations, a special entry permit was obtained to enter and perform scientific research in native indigenous territories in the Peruvian Amazon Basin.
Study sites.
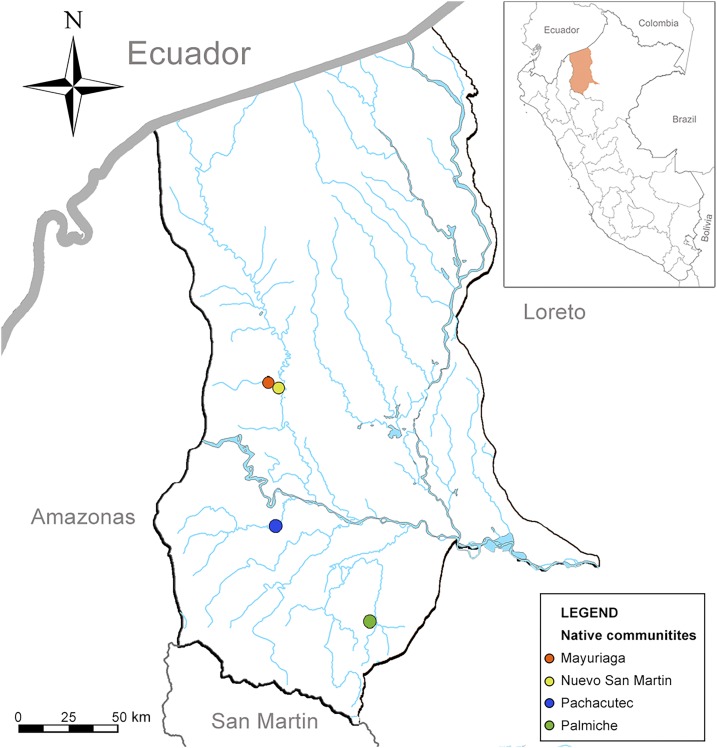
The collection of bats was performed in the Loreto Region, Datem del Marañon Province located in the Peruvian Amazon between August 11 and September 3, 2014. Study sites with high and moderate bat bite rates in humans, as reported by the Peruvian General Directorate of Epidemiology, were selected after considering logistical issues, such as accessibility. The selected study sites were Palmiche village (Cahuapanas district), Mayuriaga and Nuevo San Martín villages (Morona district), and Pachacutec village (Barranca district) (Figure 1).
Figure 1.
Map of study sites in Loreto-Peru. This figure appears in color at www.ajtmh.org.
Specimen collection and identification.
Chiropters were captured in each site during three to four consecutive nights using mist nets around houses and henhouses near villages. Mist nets were numbered consecutively at each site, placed at dusk, and checked at half-hour intervals until midnight. All captured bats were removed from the net and placed inside a canvas bag (one per animal) until processing the following morning in the field laboratory. Once in the field laboratory setting, bats were gently transferred to a Ziploc bag, sedated with isoflurane, euthanized by cervical dislocation, and finally we collected blood samples from the cardiac puncture.
Bats were weighed, measured, sexed, and morphologically identified by genus or species by a mammalogist in the field. Confirmatory identification was subsequently performed based on cranial and dental morphology at the San Marcos Natural History Museum’s Mammalogy Laboratory in Lima, Peru.
Sample size was not calculated because of the surveillance nature of the study. However, we aimed to target vampire bats such as Desmodus rotundus in a proportion of 2:1 hematophagous versus non-hematophagous bats at each collection site. Animals captured in excess of this ratio were released.
Sample collection.
Blood samples were collected from anesthetized animals by cardiac puncture for vampire bats and from the upper extremity veins (small peripheral and median veins) using capillary tubes for insectivorous or frugivorous species. A small amount of blood was also collected on filter paper (Whatman 3MM) for genotype analysis by polymerase chain reaction (PCR) and for microscopic examination of thick and thin blood smears for T. cruzi. These samples were transported at room temperature to the NAMRU-6 facilities in Lima and stored at −20°C and 4°C, respectively. Cryovials with blood and tissue samples (kidneys, liver, spleen, lungs, brain, salivary glands, and urine from the bladder) were collected and sent to the NAMRU-6 laboratory in Lima for analysis. All tissue samples were maintained in liquid nitrogen in the field and at −80°C in NAMRU-6, Lima, Peru.
Microscopic diagnosis of trypanosomatids.
Thick and thin blood smears were stained with Giemsa and examined under an Olympus BX53 microscopy at 100× magnification for the identification of blood-borne parasites. For morphological diagnosis, measurements were performed and pictures were taken using an 18 MP Canon EOS 60D digital camera.
Molecular diagnosis of trypanosomatids.
DNA was extracted from filter paper imprints, liver, and salivary gland tissues using the QIAamp DNA Mini kit (Qiagen, Hilden, Germany) according to manufacturer’s procedures. A total of ∼30 µL DNA was obtained from each sample and stored at −20°C. A nested PCR was used for amplifying the region specific for 24S α-ribosomal DNA gene of trypanosomatids (∼280 bp) and T. cruzi (∼110–130 bp) using the primers D75 (5′-GCA GAT CTT GGT TGG CGT AG-3′) and D76 (5′-GGT TCT CTG TTG CCC CTT TT-3′), and D71 (5′-AAG GTG CGT CGA CAG TGT GG-3′) and D72 (5′-TCA GAA TGG CCG AAC AGT-3′), respectively.21 The PCR products were subsequently analyzed by 2% agarose gel electrophoresis and visualized with GelRed stain. The secondary PCR products with the expected size were purified using QIAquick PCR Purification Kit and sequenced using an ABI 3130xL automated sequencer and compared with other sequences of The National Center for Biotechnology Information (NCBI) Genome database (accession numbers: AF288665, M28885, AY367119, AY367115, L14468, GQ303145, AY367116, and RU73612).22
RESULTS
A total of 121 bats belonging to 16 species (families: Vespertilionidae, Phyllostomidae, and Molossidae) were collected during the course of the study (Tables 1 and 2). The family Phyllostomidae predominated with 118 bats identified in the four communities: 36 (30.5%) in Pachacutec, 30 (25.4%) in Mayuriaga, 26 (22.0%) in Palmiche, and 26 (22.0%) in Nuevo San Martin. Only one specimen from the Vespertilionidae family was captured (Pachacutec village) and two from the Molossidae family (one in Mayuriaga and one in Palmiche). Of the 121 bats studied, 54 (44.6%) were males and 67 (55.4%) were females, with a similar proportion in each of the four surveillance sites (Table 2). The distribution of male bats was higher in Carollia perspicillata 8/10 (80%), Diphylla ecaudata 9/16 (56.2%), and Diaemus youngi 4/5 (80%) than other species; the distribution of female bats was greater in D. rotundus 37/52 (71.1%), which was the most prevalent species collected, with 52 animals (42.9%) (Table 1).
Table 1.
Taxonomic identification of hosts and trypanosomatids
| Microscopy | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristics | Sex | PCR | Density of parasites | ||||||||
| Family | Subfamily | Genus | Specie | n | Male | Female | Trypanosomatids | Trypanosoma cruzi | Mean | Median | Minimum to maximum |
| Molossidae | Molossinae | Molossus | molossus | 2 | 1 | 1 | 1 | – | 2 | 2 | 2 |
| Vespertilionidae | Vespertilioninae | Eptesicus | brasiliensis | 1 | 1 | – | – | – | – | – | – |
| Phyllostomidae | Stenodermatinae | Platyrrhinus | brachycephalus | 2 | – | 2 | 1 | – | 1 | 1 | 1 |
| – | Artibeus | planirostris | 15 | 6 | 9 | 3 | – | 2 | 1 | 1–4 | |
| – | – | lituratus | 4 | 3 | 1 | 1 | – | 2 | 2 | 2 | |
| – | – | obscurus | 4 | 3 | 1 | 2 | – | 8 | 8 | 8 | |
| – | Chiroderma | villosum | 1 | – | 1 | – | – | – | – | – | |
| – | Sturnira | lilium | 3 | 1 | 2 | – | – | – | – | – | |
| Carolliinae | Carollia | perspicillata | 10 | 8 | 2 | 4 | – | 4.5 | 3.5 | 1–10 | |
| – | – | castanea | 1 | – | 1 | – | – | – | – | – | |
| Phyllostominae | Phyllostomus | hastatus | 2 | – | 2 | 1 | 1 | 1 | 1 | 1 | |
| – | Lophostoma | silvicolum | 1 | 1 | – | – | – | – | – | – | |
| – | Trachops | cirrhosus | 2 | 2 | – | 2 | 2 | 10.5 | 1.5 | 1–20 | |
| Non-hematophagous | 48 | 26 | 22 | 15 | 3 | 4.2 | 2 | 1–20 | |||
| Desmodontinae | Diphylla | ecaudata | 16 | 9 | 7 | 7 | – | 4.3 | 3 | 1–14 | |
| – | Diaemus | youngi | 5 | 4 | 1 | 2 | 1 | 2 | 2 | 1–3 | |
| – | Desmodus | rotundus | 52 | 15 | 37 | 18 | 1 | 5.7 | 3.5 | 1–25 | |
| Hematophagous | 73 | 28 | 45 | 27 | 2 | 5.1 | 3 | 1–25 | |||
| Total | 121 | 54 | 67 | 42 | 5 | 4.8 | 3 | 1–25 | |||
(–) Represents a value of zero.
Table 2.
Distribution of chiropters by collection site
| Collection site | ||||||||
|---|---|---|---|---|---|---|---|---|
| Family | Subfamily | Genus | Specie | Pachacutec | Mayuriaga | Palmiche | Nuevo San Martin | Total |
| Molossidae | Molossinae | Molossus | molossus | – | 1 | 1 | – | 2 |
| Vespertilionidae | Vespertilioninae | Eptesicus | brasiliensis | 1 | – | – | – | 1 |
| Phyllostomidae | Stenodermatinae | Platyrrhinus | brachycephalus | – | 2 | – | – | 2 |
| – | Artibeus | planirostris | 2 | – | 2 | 11 | 15 | |
| – | – | lituratus | 1 | 1 | 1 | 1 | 4 | |
| – | – | obscurus | – | 4 | – | – | 4 | |
| – | Chiroderma | villosum | – | 1 | – | – | 1 | |
| – | Sturnira | lilium | – | – | 3 | 3 | ||
| Carolliinae | Carollia | perspicillata | 5 | 2 | 3 | – | 10 | |
| – | – | castanea | 1 | – | – | – | 1 | |
| Phyllostominae | Phyllostomus | hastatus | – | 2 | – | – | 2 | |
| – | Lophostoma | silvicolum | – | 1 | – | – | 1 | |
| – | trachops | cirrhosus | 2 | – | – | – | 2 | |
| Desmodontinae | Diphylla | ecaudata | 11 | 3 | 2 | 16 | ||
| – | Diaemus | youngi | 2 | 1 | – | 2 | 5 | |
| – | Desmodus | rotundus | 12 | 13 | 18 | 9 | 52 | |
| Sex | – | Males | – | 17 | 16 | 12 | 9 | 54 |
| – | Females | – | 20 | 15 | 15 | 17 | 67 | |
| Age | – | Subadult | – | 1 | 1 | – | – | 2 |
| – | Adult | – | 36 | 30 | 27 | 26 | 119 | |
| Total | 37 | 31 | 27 | 26 | 121 | |||
(–) Represents a value of zero.
The prevalence of Trypanosoma sp. and T. cruzi by microscopy was 37.8% (42/111) and 1.8% (2/111), respectively. The prevalence of Trypanosoma sp. was similar between hematophagous (38.8%, 26/67) and non-hematophagous (36.3%, 16/44) bats. For T. cruzi, the prevalence in hematophagous bats was 3.4% (1/26) versus 7.6% (1/16) for non-hematophagous bats.
The mean microscopically measured trypanosome mean body length was 21.2 μm (range: 11.9–37.0 μm; SD = 6.74 μm) and the mean body width was 3.5 μm (range: 2.5–5.1 μm; SD = 0.6 μm) (Table 3).
Table 3.
Intrinsic characteristics of Trypanosoma sp.
| Measurements | n | Mean | Median | DS | Minimum to maximum | IC 95% |
|---|---|---|---|---|---|---|
| TL | 39 | 21.2 | 20.5 | 6.74 | 11.9–37.0 | 19.0–23.3 |
| W | 38 | 3.5 | 3.5 | 0.63 | 2.5–5.1 | 3.3–3.7 |
| NP | 37 | 9.3 | 8.02 | 4.16 | 2.9–18.1 | 7.9–10.7 |
| NT | 37 | 8.3 | 7.9 | 2.96 | 3.5–15.0 | 7.3–9.3 |
| LN | 39 | 3.3 | 3.1 | 0.93 | 1.7–6.1 | 3.0–3.6 |
| LK | 39 | 1 | 1 | 0.05 | 1.0–1.3 | 0.9–1.0 |
| KN | 33 | 2.4 | 2 | 1.94 | 0.7–9.6 | 1.7–3.1 |
| KP | 35 | 11 | 10.3 | 4.87 | 0–19.6 | 9.2–12.6 |
| KT | 35 | 5.6 | 5.4 | 3 | 1.3–13.7 | 4.5–6.6 |
TL = total length; W = width; NP = nucleus to the previous part; NT = nucleus to the terminal part; LN = length of the nucleus; LK = length of the kinetoplast; KN = length between the kinetoplast and the nucleus; KP = kinetoplast to the previous part; KT = nucleus to the terminal part.
The overall prevalence by PCR was 34.7% (42/121) for trypanosomatids and 4.1% (5/121) for T. cruzi. In hematophagous bats, the PCR prevalence of trypanosomatids and T. cruzi were 36.9% (27/73) and 2.7% (2/73), respectively. In non-hematophagous bats, the prevalence of trypanosomatids was 31.2% (15/48) and that of T. cruzi was 6.2% (3/48) (Table 1). Compared with females, male bats demonstrated a higher prevalence of Trypanosoma sp. and T. cruzi with 48.1% (26/54, P < 0.005) and 7.4% (4/54, P = 0.104) versus 23.8% (16/67) and 1.4% (1/67). Among the bat species identified, the largest crude number of trypanosomatid-positive bats belonged to D. rotundus 18/52 (34.6%), followed by D. ecaudata 7/16 (43.7%), whereas the highest proportion of trypanosomatid-positive bats belonged to Trachops cirrhosus 2/2 (100%), and these two samples were also positive for T. cruzi. Also, only the salivary glands of a D. youngi specimen were positive by PCR and confirmed by sequencing.
Because the biological material collected from bats did not have sufficient quality for sequencing, we only obtained two sequences in blood samples belonging to two Trachops cirrhosus. Phylogenetic analysis of the sequence of the 24 alpha ribosomal gene revealed that these samples had a 98% similarity with the T. cruzi lineage TcV (AY367121). However, the sequence obtained from the salivary gland of D. youngi had a greater similarity with T. cruzi strain Sylvio X10/1 (ADWP02003259).
DISCUSSION
In Peru, research oriented toward the epidemiology and prevention of Chagas disease in humans and wild mammals is still scarce. Although there are reports of T. cruzi in bats from other Latin American countries, to our knowledge, this is the first report of T. cruzi in bats from Peru, which hosts a unique ecologic and epidemiologic environment. The presence of T. cruzi in bats suggests a bat-associated risk for Chagas disease transmission. Because of their abundance and distribution, bats infected with T. cruzi could represent a focus of dissemination of Chagas disease in the Amazonian region of Peru, posing risk to indigenous human populations and mobile populations working and traveling in the Peruvian Amazon Basin.
The detection of trypanosomatids in D. rotundus, a hematophagous bat that feeds on blood from pigs, hens, and humans, suggests a pathway for Chagas disease transmission via close contact between the bat body and its fomites with the human victim during blood feeding. This risk is further supported by data showing that the Datem del Marañon Province, where our study was conducted, demonstrates the highest incidence of D. rotundus bites in humans in south America reported in the scientific literature.23,24
The genus Trypanosoma can infect a variety of mammals, including bats, which represent important pathogen reservoirs because of their longevity, variety of shelters, and aggregate population.11,25 Moreover, it has been suggested that some trypanosomatids, such as T. cruzi, may have adapted first to bat hosts before infecting other terrestrial mammals,3 making them likely reservoirs with a potentially active role in T. cruzi dissemination.
From all studied communities, 34.7% of collected bats from six different subfamilies (Stenodermatinae, Carolliinae, Phyllostominae, Molossinae, Vespertilioninae, and Desmodontinae) were infected with trypanosomatids with a T. cruzi prevalence of 4.1%. This T. cruzi prevalence was lower than that of an Argentinian study, which reported an 8% prevalence of T. cruzi in bats.26 Also, a study from Bolivia reported 15.6% of bats infected with Trypanosoma genus but without detection of T. cruzi.27 In Ecuador, an estimated prevalence of 36.5% for T. cruzi and 1.3% for T. rangeli was reported.11
These differences in Trypanosoma prevalence underscore the need for further surveillance to assess changes in parasite distribution across different settings. These epidemiological studies will contribute to integration of opportunities for control and surveillance of Chagas disease and help to characterize the transmission of this parasite in animals and humans within South America.
The relatively uniform prevalence of T. cruzi in the four villages in this study (trypanosomatids: Mayuriaga 35.4%, Palmiche 37%, Pachacutec 32.4%, and Nuevo san Martin 34.9%; T. cruzi: Mayuriaga 3.23%, Palmiche 3.7%, Pachacutec 5.4%, and Nuevo san Martin 3.8%) suggests that the villages may have common features that contribute to similar parasite distributions. These characteristics that may include weather, geography, diversity of host mammals, and vectors should be further explored.
Our findings show that T. cruzi has a wild life cycle in the Peruvian Amazon, involving bats as its host. It is important to note that the occurrence of T. cruzi in insectivorous and fruit bats can have public health implications, as the bats could contaminate fruits and vegetables, leading to oral transmission of the Chagas disease as suggested by previous studies.28,29
Infection in hematophagous bats (trypanosomatids: 36.9% and T. cruzi: 2.7%) could be the result of infection during the bat’s feeding on another infected mammal. This pathway is possible in Trypanosoma evansi on D. rotundus, which acts as both reservoirs and vectors and that could explain the presence of T. cruzi in the salivary glands of D. youngi. Another mechanism of infection might occur when regurgitated blood is consumed by juvenile bats from infected adults.24,26 In the case of non-hematophagous bats (trypanosomatids: 31.2% and 6.2% T. cruzi), infection may be caused by multiple factors such as the ingestion of infected triatomines or other vertebrates, including other bats; vertical transmission from mother to offspring during gestation or breastfeeding; or oral infection. Most of these suggested mechanisms have been confirmed by transmission experiments.26,27,30–33 T. cruzi and trypanosomatid infection in bats was higher for males (48.1% trypanosomatids, P < 0.005 and 7.4% T. cruzi, P = 0.104) than for females (23.8% trypanosomatids and 1.4% in T. cruzi). This could be due to aggressive behavior during mating season that could lead to an increased risk of injury-associated infections. Like many mammals, male bats migrate to other territories once they reach sexual maturity. This could contribute to disease spread among different colonies, in addition to generating outbreaks in different locations.34
For a long time, T. cruzi was the only Trypanosoma genus involved in human infections. However, recent reports describe human infections caused by other trypanosomatid species that circulate in Latin America such as T. rangeli and Trypanosoma lewisi.35,36 For this reason, epidemiological studies are needed to detect potential vectors and hosts involved in the wild life cycle of the Trypanosoma genus.
Between 2006 and 2009, six cases of American trypanosomiasis were identified in the province of Datem del Marañón, which were passively diagnosed through laboratory malaria surveillance. The Mayuriaga village that was evaluated in our study also reported a case of human Chagas disease.37 Future studies will assist to develop strategies for prevention, control, and diagnosis of American trypanosomiasis in the Peruvian Amazon Basin which presents a new and a different scenario for Chagas disease in Peru, where populations often live in remote and isolated locations with serious health-care access limitations.
This article is the first exploratory study in Peru that aims to show the presence of potential pathogenic agents for humans in bats. Therefore, additional studies are needed to incriminate bats in the transmission of T. cruzi to humans.
LIMITATIONS
As it was not an exclusive study for T. cruzi, we could not collect the data needed to confirm direct transmission of T. cruzi from bats to humans. Also, the blood samples in filter paper collected from bats did not have sufficient quality for sequencing of multiple genes because of their storage and transport at room temperature.
Acknowledgments:
We would like to express our gratitude to Dr. Hugo Rodriguez and Dr. Cristiam Carey from “Dirección Regional de Salud Loreto,” Neyser Satalaya Reategui, Víctor Manuel Iñipe, Juan Ramón Meza Velásquez, and Ismael Perez Petza from “Red de Salud del Datem del Marañon” for their logistic support, and Nelson Kuji Chimpa and Ismael Perez Petza from CORPI (Coordinadora Regional de los Pueblos Indígenas de San Lorenzo) for their support in providing consultation for the study and in executing the field work. We thank the Apus and the Pachacutec, Palmiche, Mayuriaga, and Nuevo San Martin communities for allowing us to perform bat captures in their jurisdiction. We also thank Arturo Chavéz, Víctor Manuel Iñipe, Christhian Álava Rios, Wilber López Cariajano, Taner Chanchari Púa, Rusvell Ushiñahua del Aguila, Otto Torres Chumbe, Weninger Mozombite Gonzales, and Diomedes Yalta Guela for their work in the bats capture. We would also like to thank Andres (Willy) Lescano for his methodological support, and Hugo Valdivia and Cesar Náquira for their careful review of the manuscript. This material is based upon work supported by the Naval Medical research Unit-Six (NAMRU-6) under Contract No. N62654-16-F-0066.
Disclaimer: The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, and the U.S. Government.
REFERENCES
- 1.Hotez PJ, Bottazzi ME, Franco-Paredes C, Ault SK, Periago MR, 2008. The neglected tropical diseases of Latin America and the Caribbean: a review of disease burden and distribution and a roadmap for control and elimination. PLoS Negl Trop Dis 2: e300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Lana M, Menezes Machado E, 2010. Biology of Trypanosoma cruzi and Biological Diversity. American Trypanosomiasis, Chagas Disease: One Hundred Years of Research. Amsterdam, The Netherlands: Elsevier, 339–363. [Google Scholar]
- 3.Hamilton PB, Teixeira MM, Stevens JR, 2012. The evolution of Trypanosoma cruzi: the ‘bat seeding’hypothesis. Trends Parasitol 28: 136–141. [DOI] [PubMed] [Google Scholar]
- 4.Abad-Franch F, et al. 2015. On palms, bugs, and Chagas disease in the Americas. Acta Trop 151: 126–141. [DOI] [PubMed] [Google Scholar]
- 5.Burkholder J, Allison T, Kelly V, 1980. Trypanosoma cruzi (Chagas) (Protozoa: Kinetoplastida) in invertebrate, reservoir, and human hosts of the lower Rio Grande valley of Texas. J Parasitol 66: 305–311. [PubMed] [Google Scholar]
- 6.Crompton DWT, Daumerie D, Peters P, Savioli L, Marinelli A, Marinelli GJA, March GJ, Heymann D, March GJ, Marinelli AD, 2010. Working to Overcome the Global Impact of Neglected Tropical Diseases: First WHO Report on Neglected Tropical Diseases. First WHO report on neglected tropical diseases: Organización Mundial de la Salud. Geneva, Switzerland: World Health Organization, 75 pp.
- 7.Luna EJ, et al. 2016. Prevalence of Trypanosoma cruzi infection among Bolivian immigrants in the city of São Paulo, Brazil. Mem Inst Oswaldo Cruz 112: 70–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schofield CJ, Jannin J, Salvatella R, 2006. The future of Chagas disease control. Trends Parasitol 22: 583–588. [DOI] [PubMed] [Google Scholar]
- 9.Barnabe C, Brisse S, Tibayrenc M, 2003. Phylogenetic diversity of bat trypanosomes of subgenus Schizotrypanum based on multilocus enzyme electrophoresis, random amplified polymorphic DNA, and cytochrome b nucleotide sequence analyses. Infect Genet Evol 2: 201–208. [DOI] [PubMed] [Google Scholar]
- 10.Zingales B, et al. 2012. The revised Trypanosoma cruzi subspecific nomenclature: rationale, epidemiological relevance and research applications. Infect Genet Evol 12: 240–253. [DOI] [PubMed] [Google Scholar]
- 11.Lima L, Espinosa-Álvarez O, Ortiz PA, Trejo-Varón JA, Carranza JC, Pinto CM, Serrano MG, Buck GA, Camargo EP, Teixeira MM, 2015. Genetic diversity of Trypanosoma cruzi in bats, and multilocus phylogenetic and phylogeographical analyses supporting Tcbat as an independent DTU (discrete typing unit). Acta Trop 151: 166–177. [DOI] [PubMed] [Google Scholar]
- 12.Cottontail VM, Kalko EK, Cottontail I, Wellinghausen N, Tschapka M, Perkins SL, Pinto CM, 2014. High local diversity of Trypanosoma in a common bat species, and implications for the biogeography and taxonomy of the T. cruzi clade. PLoS One 9: e108603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramírez JD, Tapia-Calle G, Muñoz-Cruz G, Poveda C, Rendón LM, Hincapié E, Guhl F, 2014. Trypanosome species in neo-tropical bats: biological, evolutionary and epidemiological implications. Infect Genet Evol 22: 250–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vargas F, Córdova Paz Soldán O, Marín C, Jose Rosales M, Sánchez-Gutierrez R, Sánchez-Moreno M, 2007. Epidemiology of American trypanosomiasis in northern Peru. Ann Trop Med Parasitol 101: 643–648. [DOI] [PubMed] [Google Scholar]
- 15.Náquira C, Cabrera R, 2009. Breve reseña histórica de la enfermedad de Chagas, a cien años de su descubrimiento y situación actual en el Perú. Rev Peru Med Exp Salud Publica 26: 494–504. [Google Scholar]
- 16.Herrer A, 1960. Distribución geografica de la enfermedad de Chagas y de sus vectores en el Peril. Bol Oficina Sanit Panam 49: 572. [PubMed] [Google Scholar]
- 17.Guhl F, 2009. Enfermedad de Chagas: realidad y perspectivas. Rev Biomed 20: 228–234. [Google Scholar]
- 18.Ceballos LAG, 2010. Ciclo Silvestre de Transmision de Trypanosoma cruzi en el Noroeste de Argentina The Sylvatic Transmission Cycle of Trypanosoma cruzi in Northwest Argentina/Sylvatic Transmission Cycle of Trypanosoma cruzi in Northwest Argentina. e-libro, Corp., Biblioteca Digital FCEN-UBA (Tesis Doctoral). [Google Scholar]
- 19.Nancy C, Solís H, Pancorbo F, 2012. Reservorios silvestres de Trypanosoma cruzi: evaluación preliminar en la amazonia peruana. The Biologist 10: 59. [Google Scholar]
- 20.Solís HM, Carlos N, 2016. Reservorios silvestres de Trypanosoma cruzi en cuatro localidades de las regiones Amazonas y Loreto. Theorēma (Lima, Segunda época, En línea) 2: 63–73. [Google Scholar]
- 21.Souto RP, Zingales B, 1993. Sensitive detection and strain classification of Trypanosoma cruzi by amplification of a ribosomal RNA sequence. Mol Biochem Parasitol 62: 45–52. [DOI] [PubMed] [Google Scholar]
- 22.Hwang WS, Zhang G, Maslov D, Weirauch C, 2010. Infection rates of Triatoma protracta (Uhler) with Trypanosoma cruzi in southern California and molecular identification of trypanosomes. Am J Trop Med Hyg 83: 1020–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert AT, Petersen BW, Recuenco S, Niezgoda M, Gómez J, Laguna-Torres VA, Rupprecht C, 2012. Evidence of rabies virus exposure among humans in the Peruvian Amazon. Am J Trop Med Hyg 87: 206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desquesnes M, Holzmuller P, Lai D-H, Dargantes A, Lun Z-R, Jittaplapong S, 2013. Trypanosoma evansi and surra: a review and perspectives on origin, history, distribution, taxonomy, morphology, hosts, and pathogenic effects. Biomed Res Int 2013: 194176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cramer MJ, Willig MR, Jones C, 2001. Trachops cirrhosus Mamm Species 656: 1–6. [Google Scholar]
- 26.Argibay H, Orozco M, Cardinal M, Rinas M, Arnaiz M, Mena SC, Gürtler R, 2016. First finding of Trypanosoma cruzi II in vampire bats from a district free of domestic vector-borne transmission in northeastern Argentina. Parasitology 143: 1–11. [DOI] [PubMed] [Google Scholar]
- 27.García L, Ortiz S, Osorio G, Torrico MC, Torrico F, Solari A, 2012. Phylogenetic analysis of Bolivian bat trypanosomes of the subgenus Schizotrypanum based on cytochrome B sequence and minicircle analyses. PLoS One 7: e36578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.da Silva Valente SA, da Costa Valente V, das Neves Pinto AY, de Jesus Barbosa César M, dos Santos MP, Miranda COS, Cuervo P, Fernandes O, 2009. Analysis of an acute Chagas disease outbreak in the Brazilian Amazon: human cases, triatomines, reservoir mammals and parasites. Trans R Soc Trop Med Hyg 103: 291–297. [DOI] [PubMed] [Google Scholar]
- 29.Steindel M, Pacheco LK, Scholl D, Soares M, de Moraes MH, Eger I, Kosmann C, Sincero TCM, Stoco PH, Murta SMF, 2008. Characterization of Trypanosoma cruzi isolated from humans, vectors, and animal reservoirs following an outbreak of acute human Chagas disease in Santa Catarina State, Brazil. Diagn Microbiol Infect Dis 60: 25–32. [DOI] [PubMed] [Google Scholar]
- 30.Rueda K, Trujillo JE, Carranza JC, Vallejo GA, 2014. Transmisión oral de Trypanosoma cruzi: una nueva situación epidemiológica de la enfermedad de Chagas en Colombia y otros países suramericanos. Biomedica 34: 631–641. [DOI] [PubMed] [Google Scholar]
- 31.Ferrer A, Lew D, Lasso C, Carlos A, Nota sobre depredación por Trachops cirrhosus Spix, 1823. (Chiroptera, Phyllostomidae) en Venezuela. Mem Soc Cienc Nat La Salle 58: 145–147. [Google Scholar]
- 32.Thomas ME, Rasweiler I, John J, D’Alessandro A, 2007. Experimental transmission of the parasitic flagellates Trypanosoma cruzi and Trypanosoma rangeli between triatomine bugs or mice and captive neotropical bats. Mem Inst Oswaldo Cruz 102: 559–565. [DOI] [PubMed] [Google Scholar]
- 33.Añez N, Crisante G, Soriano PJ, 2009. Trypanosoma cruzi congenital transmission in wild bats. Acta Trop 109: 78–80. [DOI] [PubMed] [Google Scholar]
- 34.Streicker DG, Winternitz JC, Satterfield DA, Condori-Condori RE, Broos A, Tello C, Recuenco S, Velasco-Villa A, Altizer S, Valderrama W, 2016. Host–pathogen evolutionary signatures reveal dynamics and future invasions of vampire bat rabies. Proc Natl Acad Sci U S A 113: 10926–10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Sousa MA, 2014. On opportunist infections by Trypanosoma lewisi in humans and its differential diagnosis from T. cruzi and T. rangeli. Parasitol Res 113: 4471–4475. [DOI] [PubMed] [Google Scholar]
- 36.Stevens J, Teixeira M, Bingle L, Gibson W, 1999. The taxonomic position and evolutionary relationships of Trypanosoma rangeli. Int J Parasitol 29: 749–757. [DOI] [PubMed] [Google Scholar]
- 37.Cabrera R, Vega S, Valderrama Y, Cabanillas K, Fernandez C, Rodriguez O, Del Aguila C, Hernandez J, Mendoza L, Ramon Meza J, 2013. New focus of active transmission of Chagas disease in indigenous populations in the Peruvian Amazon basin. Rev Soc Bras Med Trop 46: 367–372. [DOI] [PubMed] [Google Scholar]