Abstract.
Histidine-rich protein 2 of Plasmodium falciparum (PfHRP2) forms the basis of many current malaria rapid diagnostic tests (RDTs). It is concerning that there are parasites that lack part or all of the pfhrp2 gene, and thus do not express the PfHRP2 protein; such parasites are not identifiable by PfHRP2-detecting RDTs. Very limited information is available regarding pfhrp2 genetic variation in Papua New Guinea (PNG). In the present study, this gene variation was evaluated using 169 samples previously collected from the Wosera area in East Sepik Province of PNG. Molecular diagnosis of these samples showed that 81% were infected, and P. falciparum was present in 91% of those infected samples. One hundred and twenty samples were amplified for pfhrp2 exon-2, from which 12 randomly selected amplicons were sequenced, yielding 18 sequences, all of which were unique. Baker repeat type 2 × type 7 numbers ranged from 0 to 108. Epitope mapping analysis revealed that three major epitopes, DAHHAHHA, AHHAADAHHA, and AHHAADAHH, were present in high prevalence and frequencies. These major epitopes have been shown to be recognized by the monoclonal antibodies 3A4 and PTL-3 (DAHHAHHA), C1-13 (AHHAADAHHA), and S2-5 and C2-3 (AHHAADAHH). This study provides further information on the high genetic variation of pfhrp2 and its unclear relationship with prediction of RDT detection sensitivity, and identifies major epitopes in this gene from PNG. These results could be relevant and useful to understand the genetic diversity of this gene and the performance of current and future RDTs in this malarious region of the world.
INTRODUCTION
Histidine-rich protein 2 (HRP2) is a unique protein produced exclusively by Plasmodium falciparum, and thus has been used as a biomarker for falciparum malaria infection.1 In addition, it forms the basis of many current rapid diagnostic tests (RDTs).2 Over the past 10 years, HRP2-detecting RDTs have become a widely used diagnostic tool for P. falciparum, especially in endemic areas that may not have appropriate microscopy capacity available. Several such HRP2-based combination RDTs have been tested in Papua New Guinea (PNG) in various populations: ParaSight-F dipstick in adults3 and children (0.5–5 years)4 from the Wosera area (Wosera-Gawi district) in East Sepik Province; ICT (HRP2 and pLDH) in infants (3–27 months) from Mugil (Madang Province) and Maprik (East Sepik Province)5; ICT (HRP2 and aldolase) in children (0.5–10 years) from Madang Province6; and CareStart™ in pregnant women from Madang Province.7 In these studies, the sensitivities of the RDTs for P. falciparum detection varied from 45.6%7 to 96%.6
A number of host and parasite factors may influence the accuracy and sensitivity of RDTs detecting P. falciparum–specific HRP2 (PfHRP2).8 Most importantly, parasites lacking part or all of the pfhrp2 gene do not express the PfHRP2 protein and are, therefore, not identifiable by PfHRP2-detecting RDTs. Recent studies have reported pfhrp2 gene deletions in field isolates of P. falciparum from South and Central America, Asia, and Africa, resulting in false-negative test results.8 Given the emphasis on the World Health Organization test and treat guidelines (http://www.who.int/malaria/publications/atoz/9789241549127/en/), P. falciparum parasites lacking pfhrp2 raise concerns that antimalarial treatment could be withheld from infected patients, and this scenario could potentially undermine malaria elimination efforts.
Studies of the pfhrp2 gene worldwide show that polymorphism within this gene is extensive, and the correlation between this genetic variation and RDT detection sensitivity remains unclear.9,10 Initially, testing ParaCheck Pf and ICT Malaria Pf RDTs against 16 cultured lines/isolates from Africa and Asia-Pacific, Baker et al.10 developed a binary logistic regression model that was able to predict detection sensitivity of these two RDTs based on pfhrp2 sequence structure. The model predicted an isolate to be detected at ≤ 250 parasites/μL if the number of amino acid repeats type 2 × type 7 was > 43, with an accuracy of 87.5%. Subsequently, using this analysis, 34 PfHRP2-detecting RDTs were tested against 79 global isolates at 200 parasites/μL.9 In this testing, the regression model did not show a correlation between pfhrp2 sequence structure and the overall RDT detection rates. Thus, the genetic variation in pfhrp2 does not appear to affect RDT detection sensitivity at ≥ 200 parasites/μL.9 Consistent with Baker et al.’s subsequent conclusion, similar results have been reported by Kumar Bharti et al.11 using P. falciparum–positive blood samples collected from 15 sites in eight malaria-endemic states in India and by Willie et al.12 using samples collected from three health centers in the Ampasimpotsy area in Madagascar.
Previous studies included a very limited number of samples from PNG.9,10 In one study, nine samples from PNG (three, Bougainville; six, collection area not defined) were included.10 Plasmodium falciparum histidine-rich protein 2 exon-2 sequencing revealed high genetic diversity among these samples (89%, eight unique sequences). In the second study, 17 samples from PNG (collection area not defined) were included, which also showed extensive genetic variation (71%, 12 unique sequences).9 Although one parasite line (D10) from PNG has been reported to have pfhrp2 gene deletion,13 neither of these two studies found any evidence of pfhrp2 deletion in PNG.9,10 Finally, the studies that have tested PfHRP2-detecting RDTs did not analyze pfhrp2 variation.3–7
The primary aim of the present study was to provide further insights into pfhrp2 variation in PNG. For this aim, 169 samples were randomly selected from a previous malaria epidemiological study conducted in the Wosera area.14 Using these samples, molecular assays to detect Plasmodium spp. infection and analyze pfhrp2 gene sequences were performed.
MATERIALS AND METHODS
Study samples.
Samples previously collected for a malaria epidemiological study conducted in collaboration with the PNG Institute of Medical Research at Case Western Reserve University, Cleveland, Ohio, were used.14 In this study, there were four serial cross-sectional surveys conducted between August 2001 and June 2003 in 29 villages in the Wosera area. From these surveys, a total of 16,209 blood samples were collected from 8,793 villagers aged < 2 (2.1%) to ≥ 40 (16.7%) years. In all surveys, most participants were asymptomatic. All these blood samples were analyzed for Plasmodium blood-stage infections using standard blood smear microscopy and molecular Plasmodium species–specific, post–polymerase chain reaction (PCR)/ligase detection reaction–fluorescent microsphere assay (LDR-FMA).15 It is to be noted that the samples collected in this epidemiological study were before the RDTs were made widely available at the point-of-care in the country, and, therefore, RDT-based diagnosis was not performed. From this study,14 169 already extracted genomic DNA samples, which represented 10% of all samples, were randomly selected, which belonged to five of the 29 villages. These were Kumunugum 1 (also called Kuminkum 1), Kumunugum 2 (also called Kuminkum 2), Kumunugum 3 (also called Kuminkum 3), Mikau 1 (also called Miko 1), and Mikau 2 (also called Miko 2).
Molecular diagnosis of Plasmodium spp. infections.
Molecular diagnosis of Plasmodium spp. was performed using PCR followed by LDR-FMA. All methods for PCR amplification of small subunit rRNA target sequences and Plasmodium species–specific detection by LDR-FMA have been described in detail by McNamara et al.15 Genomic DNA extracted from P. falciparum, Plasmodium vivax, Plasmodium malariae, and Plasmodium ovale–infected blood samples, provided by the Malaria Research and Reference Reagent Resource Center and Dr. W. E. Collins (Centers for Disease Control and Prevention), served as positive controls. DNA was extracted from 50 to 100 μL of each sample using a QIAamp® DNA Micro Kit (QIAGEN, Valencia, CA).
Amplification and sequence analysis of pfhrp2 gene.
The pfhrp2 exon-2 (expected size 836 base pairs [bp]) was amplified using the primers and amplification conditions described elsewhere.12,16 Genomic DNA from the P. falciparum strains 3D7 and HB3 were used as positive controls, whereas that from the Dd2 strain, which lacks the pfhrp2 gene,17 was used as a negative control in these amplification reactions. Amplicons were purified using a QIAquick® PCR Purification Kit (QIAGEN). The nucleotide sequences in all purified amplicons were determined by Sanger sequencing, in both forward and reverse directions, which was performed using the BigDye® Terminator v3.1 Cycle Sequencing Kit protocol from Applied Biosystems (Foster City, CA).
CodonCode Aligner program (v.6.0.2; CodonCore Corporation, Centerville, MA) was used for the alignment and base-calling of the raw sequences. Geneious software (v.10.0.2; Biomatters Ltd., Auckland, New Zealand) was used for the alignment of sequences, virtual construction of the pfhrp2 gene, translation into protein sequences, grouping of specific amino acid repeats, identification of insertions/deletions, and comparison with the pfhrp2 sequences from previous studies. The sequences generated in this study (N = 18, see Results) were compared with a total of 15 sequences from the two previous studies (GenBank accession numbers AY816241.1–AY816243.1, AY816253.1–AY816258.110, FJ871161.1–FJ871163.1, FJ871241.1, FJ871294.1, and FJ871358.19).
The pfhrp2 gene sequences were translated into protein sequences and 24 amino acid repeat types (1–24) were classified as described by Baker et al.9,10 Of these 24 amino acid repeat types (called Baker repeats), 20 (repeats 1–14, 19–24) are present in pfhrp2, whereas four (repeats 15–18) are present in the pfhrp2 paralog gene pfhrp3.9 Because RDT-based diagnosis was not performed on these samples,14 both scenarios of the predictive model, based on the number of type 2 × type 7 repeats, were used to predict the sensitivity of an RDT detecting PfHRP2: (a) an isolate would be detectable at ≤ 250 parasites/μL if the number of repeat types is > 4310 and (b) an isolate would be detectable at ≥ 200 parasites/μL regardless of the number of repeat types.9 In addition, the sequences from this study (N = 18) and previous studies (N = 15) were analyzed for the identification and distribution of 13 major epitopes, ranging 8–15 amino acids, that are recognized by 11 PfHRP2-specific commercially available monoclonal antibodies (MAbs).18
The prevalence and frequency of each amino acid repeat type or epitope were calculated as described.12
Ethics statement.
This study was conducted under the protocols approved by the PNG Medical Research Advisory Committee and University Hospitals of Cleveland Institutional Review Board.
RESULTS
Diagnosis of Plasmodium spp. infections.
Polymerase chain reaction diagnosis showed that 81% of all samples (137/169) were infected by Plasmodium spp. The LDR-FMA analysis showed that P. falciparum was present in 91% (125/137), P. vivax in 32% (44/137), P. malariae in 23% (31/137), and P. ovale in 2% (3/137) of the infected samples. Among those infected samples, 37% (51/137) showed mixed infections, predominantly including P. falciparum (36%, 50/137). On the other hand, only 26% of all samples (44/169) were positive by microscopy, all of them for P. falciparum, the majority (95%) as single infections. Plasmodium falciparum parasitemia ranged from 40 to 39,960 parasites/μL, and 32% (14/44) samples had parasitemia < 200 parasites/μL.
Amplification of the pfhrp2 gene.
Amplification of pfhrp2 exon-2 showed band sizes of 450–900 bp on 1% agarose gels for 120 samples. A comparison between pfhrp2 PCR and P. falciparum detection by LDR-FMA, used as the gold standard, is presented in Table 1. This analysis showed that among the 120 samples that were pfhrp2 PCR positive, 109 samples were P. falciparum positive and 11 samples were P. falciparum negative. Thus, these 11 samples were considered false positives. Of these, only one sample was P. falciparum positive by microscopy (parasitemia 1,120 parasites/μL), whereas 10 samples were P. falciparum negative by microscopy. Thus, these 10 samples, positive for pfhrp2 but P. falciparum negative by both LDR-FMA and microscopy, were considered to be “true” false positives. In addition, 16 P. falciparum–positive samples did not amplify for the pfhrp2 gene. Thus, these 16 samples were considered false negatives. Of these, 14 samples were P. falciparum negative by microscopy, whereas two samples were P. falciparum positive by microscopy (parasitemia 40 and 840 parasites/μL). Thus, these two samples, negative for pfhrp2 but P. falciparum positive by both LDR-FMA and microscopy, were considered to be “true” false negatives.
Table 1.
Comparison of pfhrp2 polymerase chain reaction and Plasmodium falciparum ligase detection reaction–fluorescent microsphere assay
pfhrp2 = P. falciparum histidine-rich protein 2 gene.
1, microscopy+; 10, microscopy−.
2, microscopy+; 14, microscopy−.
The overall concordance between pfhrp2 PCR and P. falciparum LDR-FMA molecular assays for this set of samples was 84%. Combining all three diagnostic assays, namely, pfhrp2 PCR, P. falciparum LDR-FMA, and microscopy, the sensitivity and specificity of pfhrp2 gene detection were 98% and 82%, respectively. Rapid diagnostic test–based diagnosis of malaria was not available for these samples.
Sequence analysis of the pfhrp2 gene.
Of the 109 pfhrp2 amplicons from the samples that were P. falciparum positive (Table 1), 12 were randomly selected for sequencing. From these 12 amplicons, a total of 18 exon-2 sequences were generated, ranging 468–858 bp (155–285 amino acids). A comparison among these 18 sequences revealed multiple-strain infections, inferred by the presence of two different sequences, in six of the 12 samples. It also showed that all 18 sequences were unique. Finally, a comparison between these 18 sequences and the 15 previous sequences from PNG showed that no sequence was shared between these two groups. Exon-2 sequences from this analysis were submitted to GenBank (accession numbers MF673786–MF673803).
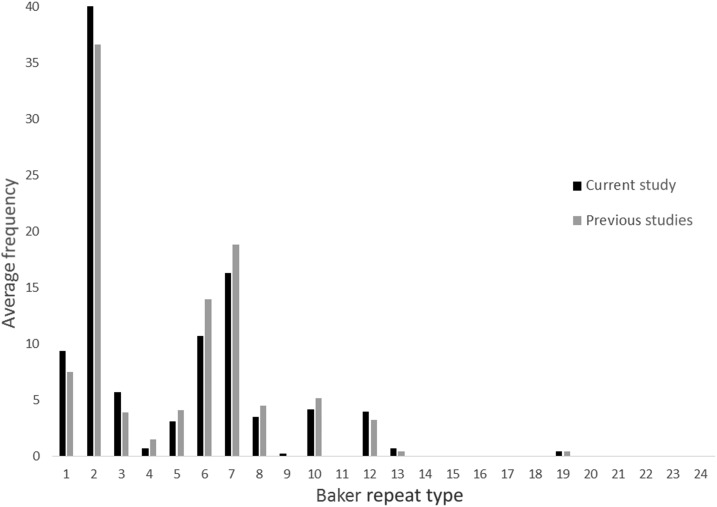
Of the 20 amino acid repeats present in pfhrp2 (repeats 1–14, 19–24), 13 repeats in our sequences and 12 repeats in previous PNG sequences were identified (Table 2). Repeat types 1, 2, 6, 7, and 12 were present in almost all sequences (89–100%). Repeat types 3, 5, 8, and 10 were also highly prevalent in all sequences (56–100%). Prevalence of repeat types 4, 13, and 19 was low to moderate in all samples (11–33%). The average frequency of each repeat within our and previous sequences is presented in Figure 1. Repeat types 2 and 7 were the most frequent within our (40%, range = 4–16, mean = 10; 16%, range = 0–9, mean = 4, respectively) and previous (37%, range = 7–15, mean = 11; 19%, range = 3–9, mean = 6, respectively) sequences.
Table 2.
Prevalence of Baker repeat types in pfhrp2 sequences
| Repeat type | Sequence | Prevalence | |
|---|---|---|---|
| This study (N = 18) | Previous studies (N = 15) | ||
| % (n) | % (n) | ||
| 1 | AHHAHHVAD | 94 (17) | 100 (15) |
| 2 | AHHAHHAAD | 100 (18) | 100 (15) |
| 3 | AHHAHHAAY | 89 (16) | 100 (15) |
| 4 | AHH | 17 (3) | 33 (5) |
| 5 | AHHAHHASD | 78 (14) | 93 (14) |
| 6 | AHHATD | 89 (16) | 100 (15) |
| 7 | AHHAAD | 94 (17) | 100 (15) |
| 8 | AHHAAY | 83 (15) | 93 (14) |
| 9 | AAY | 6 (1) | 0 (0) |
| 10 | AHHAAAHHATD | 56 (10) | 93 (14) |
| 11 | AHN | 0 (0) | 0 (0) |
| 12 | AHHAAAHHEAATH | 100 (18) | 100 (15) |
| 13 | AHHASD | 17 (3) | 13 (2) |
| 14 | AHHAHHATD | 0 (0) | 0 (0) |
| 15* | AHHAHHAAN | 0 (0) | 0 (0) |
| 16* | AHHAAN | 0 (0) | 0 (0) |
| 17* | AHHDG | 0 (0) | 0 (0) |
| 18* | AHHDD | 0 (0) | 0 (0) |
| 19 | AHHAA | 11 (2) | 13 (2) |
| 20 | SHHDD | 0 (0) | 0 (0) |
| 21 | AHHAHHATY | 0 (0) | 0 (0) |
| 22 | AHHAHHAGD | 0 (0) | 0 (0) |
| 23 | ARHAAD | 0 (0) | 0 (0) |
| 24 | AHHTHHAAD | 0 (0) | 0 (0) |
pfhrp2 = Plasmodium falciparum histidine-rich protein 2 gene.
Not present in pfhrp2, present in pfhrp3 (Baker et al.9).
Figure 1.
Average frequency of amino acid repeat types (Baker repeat types) in the sequences from the current study (N = 18) and previous studies (N = 15) from Papua New Guinea (PNG).
Prediction of RDT detection sensitivity.
To predict the detection of an isolate by a PfHRP2-detecting RDT, an analysis of Baker repeat type 2 × type 7 numbers and microscopy diagnosis for the 18 sequences are presented in Table 3. The model, which predicts that the number of repeat types > 43 would be needed to detect ≤ 250 parasites/μL (scenario [a]),10 showed one such sequence (77 repeats, 200 parasites/μL). There were seven sequences (39%) where the number of repeat types was < 43 and the parasite densities were ≤ 250 parasites/μL; five of these sequences were from the samples that had submicroscopic infections (SMI, pfhrp2+ P. falciparum+ microscopy−). In addition, three sequences with the number of repeat types > 43 were also from the samples that had SMI. The model, which predicts that ≥ 200 parasites/μL would be detected regardless of the number of repeat types (scenario [b]),9 showed four such sequences (0–32 repeats, 280–600 parasites/μL). Finally, there were three sequences where the number of repeat types was > 43 and the parasite densities were well over 250 parasites/μL. Among these results, those where the number of repeat types was < 43 and the parasite densities were ≤ 250 parasites/μL, and those with SMI, regardless of the number of repeat types, make any prediction difficult.
Table 3.
Baker repeat types and Plasmodium falciparum infection diagnosis
| Sample* | Repeat type number (2 × 7) | LDR-FMA | Microscopy | Parasites/μL | RDT detection prediction |
|---|---|---|---|---|---|
| DFA4927 | 32 (16 × 2) | + | + | 280 | Yes† |
| DFA4929 | 80 (10 × 8) | + | − | − | SMI |
| DFA4934 | 77 (11 × 7) | + | + | 200 | Yes‡ |
| DFA4938 | 22 (11 × 2) | + | − | − | SMI |
| DFA4940A | 66 (11 × 6) | + | − | − | SMI |
| DFA4940B | 42 (6 × 7) | + | − | − | SMI |
| DFA4942 | 55 (11 × 5) | + | − | − | SMI |
| DFA5008A | 108 (12 × 9) | + | + | 8,440 | Yes |
| DFA5008B | 60 (12 × 5) | + | + | 8,440 | Yes |
| DFB0007A | 24 (8 × 3) | + | − | − | SMI |
| DFB0007B | 21 (7 × 3) | + | − | − | SMI |
| DFB0013A | 39 (13 × 3) | + | + | 40 | ? |
| DFB0013B | 11 (11 × 1) | + | + | 40 | ? |
| DFB0014A | 22 (11 × 2) | + | + | 360 | Yes† |
| DFB0014B | 16 (4 × 4) | + | + | 360 | Yes† |
| DFB0031A | 0 (7 × 0) | + | + | 600 | Yes† |
| DFB0013B | 48 (8 × 6) | + | + | 600 | Yes |
| DFB0051 | 30 (15 × 2) | + | − | − | SMI |
LDR-FMA = ligase detection reaction–fluorescent microsphere assay; RDT = rapid diagnostic test; SMI = submicroscopic infections.
GenBank accession numbers MF673786–MF673803.
Scenario (b) (Baker et al.9).
Scenario (a) (Baker et al.10).
Major epitopes in pfhrp2 exon-2 targeted by MAbs in RDTs.
Epitope identification analysis on all sequences revealed that eight of the 13 major epitopes were present in majority of the sequences (78–100%) (Table 4). Among these, three major epitopes, DAHHAHHA, AHHAADAHHA, and AHHAADAHH, were present within those sequences at much higher average frequencies (18–26%) than the other five major epitopes (2–8%). These three major epitopes have been shown to be recognized by PfHRP2-specific MAbs 3A4 and PTL-3 (DAHHAHHA), C1-13 (AHHAADAHHA), and S2-5 and C2-3 (AHHAADAHH).18
Table 4.
Distribution of major epitopes in pfhrp2 sequences
| Major epitope | This study (N = 18) | Previous studies (N = 15) | MAb* | ||
|---|---|---|---|---|---|
| Prevalence | Average frequency | Prevalence | Average frequency | ||
| % (n) | % | % (n) | % | ||
| DAHHAHHA | 100 (18) | 18 | 100 (15) | 18 | 3A4, PTL-3 |
| DAHHAADAHH | 94 (17) | 8 | 100 (15) | 7 | 2G12-1C12 |
| DAHHVADAHH | 0 (0) | 0 | 0 (0) | 0 | 2G12-1C12 |
| YAHHAHHA | 94 (17) | 4 | 100 (15) | 4 | 1E1-A9, PTL-3 |
| DAHHAHHV | 89 (16) | 5 | 100 (15) | 3 | 1E1-A9 |
| HATDAHHAAD | 61 (11) | 2 | 67 (10) | 3 | A6-4 |
| HATDAHHAAA | 67 (12) | 2 | 100 (15) | 2 | A6-4 |
| AHHAADAHHA | 100 (18) | 26 | 100 (15) | 26 | C1-13 |
| DAHHAADAHHA | 94 (17) | 8 | 100 (15) | 7 | N7 |
| AHHAADAHH | 100 (18) | 26 | 100 (15) | 26 | S2-5, C2-3 |
| AHHASDAHH | 78 (14) | 2 | 100 (15) | 2 | S2-5 |
| TDAHHAADAHHAADA | 50 (9) | 1 | 53 (8) | 1 | TC-10 |
| AAYAHHAHHAAY | 0 (0) | 0 | 0 (0) | 0 | Genway |
MAb = monoclonal antibody; pfhrp2 = Plasmodium falciparum histidine-rich protein 2 gene.
Lee et al.18
DISCUSSION
Similar to previous malaria epidemiological studies in PNG, which used both standard blood smear microscopy and PCR-based diagnosis,19,20 we noticed a large difference in the overall prevalence of Plasmodium infections determined by using these methods (26% vs. 81%, respectively). These results reflect the well-known low concordance between microscopy and LDR-FMA diagnostic methods and are also consistent with the malaria infection characteristics of the Wosera area reported previously.14,19,21
The overall concordance between pfhrp2 PCR and P. falciparum post-PCR/LDR-FMA was 84%, suggesting variation in amplification of two different target sequences in this set of samples. In our recent study conducted on samples from Madagascar, the overall concordance between these two molecular assays was 97%.12 The Wosera samples were collected and processed in 2001–2003, 15 or more years ago, and have been used for a number of molecular investigations since then. Thus, their storage and usage over the years, likely leading to some level of degradation, may be a factor to consider for the lower concordance. Regarding the 10 “true” false-positive samples (pfhrp2+ P. falciparum− microscopy−), there could be other possible reasons, such as very low-level infections or, less likely, amplification of the pfhrp2 paralog gene pfhrp3, which has a sequence homology of more than 75% in the tandem repeat region to pfhrp2.18,22 None of these 10 samples were sequenced. There were two “true” false-negative samples (pfhrp2− P. falciparum+ microscopy+). As RDTs were not performed on these samples,14 pfhrp2 deletion status of these two samples could not be determined (see Limitations).
The pfhrp2 exon-2 sequence sizes (468–858 bp, 155–285 amino acids) were similar to those reported previously from PNG and globally.9,10 All 18 sequences (100%) were found to be unique. This observation is similar to previous observations from PNG, where 89%10 and 71%9 pfhrp2 sequence diversity was reported. In a global analysis, the pfhrp2 sequence diversity was found to be higher in countries with high transmission intensity.9 A strong correlation between malaria transmission intensity and pfhrp2 diversity may be expected, as malaria infections in high-transmission settings, including those in PNG, often involve multiple-strain infections.23 In the present study, using Sanger sequencing, two different sequences were identified in six of the 12 samples, implying multiple-strain infections. Sanger sequencing has limited ability to detect multiple parasite types in a mixed infection and can miss a significant proportion of minority variants. It is possible that the use of next-generation sequencing technology, which allows high-resolution analyses of a heterogeneous mixture of the parasites within the host,24,25 may have enabled more comprehensive detection of pfhrp2 sequences present in those samples. Although comparative analysis of the sequences showed that no two sequences were the same between our study and previous studies, the prevalence and frequency of each of the 12 Baker repeat types, common between these two groups, were comparable. This was also true for the repeat types 2 and 7, which were used in a model to predict RDT detection sensitivity.9,10
Detection sensitivity for different RDTs tested in different populations in PNG was as low as 45.6%7 to as high as 96%.6 Although the correlation between RDT performance and pfhrp2 variation remains unclear,9–12 the studies that have tested RDTs in PNG did not analyze pfhrp2 variation.3–7 All 18 sequences in the present study were generated from 12 samples that were P. falciparum positive by LDR-FMA. Thirty-nine percent (7/18) of these sequences had the number of type 2 × type 7 repeats < 43, ranging from 0 (7 × 0, respectively) to 42 (6 × 7, respectively). The parasitemia in these samples was from 40 parasites/μL to submicroscopic. This suggests that under scenario (a), these samples would not be detected, that is, they would be considered nonsensitive to RDT detection.10,16 In addition, there were samples where the sequences had > 43 repeats, but those samples had SMI. It is important to recognize that commercial malaria RDTs were approved for detecting symptomatic malaria cases and not for detecting submicroscopic cases. Therefore, it is difficult to predict whether all these samples would be detected or not by an RDT. Given the field sample–based observations made by Kumar Bharti et al.11 and Willie et al.,12 verification of these RDT detection predictions made on the basis of the repeat numbers is required, particularly for the samples that had very low (< 50 parasites/μL) or submicroscopic parasitemia levels.
Depending on the RDT used and study setting and design, RDT may26,27 or may not28,29 perform better than microscopy in detecting SMI. Such infections, if undetectable by the current generation of RDTs, may compromise the current focus on malaria elimination in PNG.30 Recently, funding agencies, manufacturers, and researchers have been working toward developing ultra-sensitive RDTs (uRDT) with limits of detection similar to those nucleic acid amplification–based methods. As a step in this direction, uRDT (Alere™ Malaria Ag P.f (05FK140), Standard Diagnostics/Alere, San Diego, CA) has been commercialized with claims of an analytical sensitivity approximately 10 times the detection limit of the best conventional RDTs. Under laboratory conditions, using different P. falciparum culture strains, uRDT showed a greater than 10-fold improvement over the SD Bioline Malaria Ag P.f RDT (05FK50); pfhrp2-positive strains ITG and HB3 were detected down to 49 parasites/μL by the SD Bioline Malaria Ag P.f RDT, whereas the same strains were detected down to 3.13 parasites/μL by uRDT.31 It may be valuable to test whether the use of such an RDT can improve detection of very low level infections/SMI in PNG.
Investigations of the effect of pfhrp2 sequence variation on the binding of MAbs to PfHRP21 have led to the characterization of 13 major epitopes recognized by 11 such antibodies (Table 4).18,22 The information regarding PfHRP2-specific MAbs being used in RDTs is generally not disclosed by the manufacturers. Epitope identification in all PNG sequences showed that epitopes DAHHAHHA, AHHAADAHHA, and AHHAADAHH were present in high prevalence and frequencies. Epitopes AHHAADAHHA and DAHHAHHA are recognized by MAbs C1-13 and PTL-3, respectively, both of which have shown the best potential (based on the prevalence and frequency of epitopes together with the heat durability profile) for use in an RDT.18 These epitope identification results should be helpful for studies comparing performances of different RDTs in PNG and for those evaluating new antibodies for the development of improved RDTs.32
LIMITATIONS
It is acknowledged that no RDT data were available pertaining to the samples analyzed in the present study. Based on recent field sample-based studies, where no direct relationship between pfhrp2 repeat types and the ability to detect low-density infections was observed,11,12 the detection predictions made in the present study should be verified with RDTs. This verification is also important given the finding that there was no correlation between pfhrp2 sequence length or repeat type and PfHRP2 plasma concentration in African children.33 In the present study, two samples, positive by P. falciparum LDR-FMA and microscopy, were pfhrp2 PCR negative (“true” false negatives in Table 1). In the absence of RDT data, pfhrp2 deletion status of these two samples could not be predicted. In addition, attempts to amplify the pfhrp2 paralog gene pfhrp3 and the pfhrp2 flanking genes PF3D7_0831900 and PF3D7_0831700, which provide confirmatory evidence of pfhrp2 deletion,8 could not be made because of exhaustion of the samples. HRP3 has a sequence homology of more than 75% in the tandem repeat region to HRP2 and is recognized by most anti-HRP2 MAbs including C1-13.18,22 Although HRP3 is less abundant than HRP2,34 in situations of low-level parasitemia or when parasites have pfhrp2 deletions, it could enhance the sensitivity of the RDTs.31,35,36 Finally, the samples in the present study were collected from a single area in PNG, before the RDTs were made widely available at the point-of-care and the selection pressure on the parasite population may have been exerted. Using more recent samples from multiple areas in PNG, further studies should evaluate the current status of pfhrp2 variation together with performance of the PfHRP2-detecting RDTs.
In conclusion, the present study provides in-depth information regarding pfhrp2 genetic variation in a malaria-endemic area of PNG, which could be a valuable starting point for testing and interpreting the results of RDTs. As heavy reliance is placed on RDTs for point-of-care malaria diagnosis in PNG,5–7 it is essential from a public health perspective to perform routine monitoring, where the use of RDT is combined with the analysis of pfhrp2 variation. Such monitoring may help alert the authorities to potential diagnostic failure risks and the need to explore complimentary/alternative diagnostic approaches in this malarious region of the world.
Acknowledgments:
We thank all study participants and field technicians for their participation and support, and are grateful to Rosalind Howes for her valuable time and critical evaluation of this manuscript.
REFERENCES
- 1.Parra ME, Evans CB, Taylor DW, 1991. Identification of Plasmodium falciparum histidine-rich protein 2 in the plasma of humans with malaria. J Clin Microbiol 29: 1629–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moody A, 2002. Rapid diagnostic tests for malaria parasites. Clin Microbiol Rev 15: 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Genton B, Hii J, Paget S, Alpers MP, 1996. Rapid manual diagnosis of Plasmodium falciparum malaria using ParaSight-F dipsticks applied to human blood and urine. J Travel Med 3: 172–173. [DOI] [PubMed] [Google Scholar]
- 4.Genton B, Paget S, Beck HP, Gibson N, Alpers MP, Hii J, 1998. Diagnosis of Plasmodium falciparum infection using ParaSight®-F test in blood and urine of Papua New Guinean children. Southeast Asian J Trop Med Public Health 29: 35–40. [PubMed] [Google Scholar]
- 5.Senn N, Rarau P, Manong D, Salib M, Siba P, Robinson LJ, Reeder J, Rogerson S, Mueller I, Genton B, 2012. Rapid diagnostic test-based management of malaria: an effectiveness study in Papua New Guinean infants with Plasmodium falciparum and Plasmodium vivax malaria. Clin Infect Dis 54: 644–651. [DOI] [PubMed] [Google Scholar]
- 6.Manning L, Laman M, Rosanas-Urgell A, Turlach B, Aipit S, Bona C, Warrell J, Siba P, Mueller I, Davis TM, 2012. Rapid antigen detection tests for malaria diagnosis in severely ill Papua New Guinean children: a comparative study using Bayesian latent class models. PLoS One 7: e48701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Umbers AJ, et al. 2015. Accuracy of an HRP-2/panLDH rapid diagnostic test to detect peripheral and placental Plasmodium falciparum infection in Papua New Guinean women with anaemia or suspected malaria. Malar J 14: 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng Q, Gatton ML, Barnwell J, Chiodini P, McCarthy J, Bell D, Cunningham J, 2014. Plasmodium falciparum parasites lacking histidine-rich protein 2 and 3: a review and recommendations for accurate reporting. Malar J 13: 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker J, et al. 2010. Global sequence variation in the histidine-rich proteins 2 and 3 of Plasmodium falciparum: implications for the performance of malaria rapid diagnostic tests. Malar J 9: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker J, McCarthy J, Gatton M, Kyle DE, Belizario V, Luchavez J, Bell D, Cheng Q, 2005. Genetic diversity of Plasmodium falciparum histidine-rich protein 2 (PfHRP2) and its effect on the performance of PfHRP2-based rapid diagnostic tests. J Infect Dis 192: 870–877. [DOI] [PubMed] [Google Scholar]
- 11.Kumar Bharti P, Singh Chandel H, Krishna S, Nema S, Ahmad A, Udhayakumar V, Singh N, 2017. Sequence variation in Plasmodium falciparum histidine rich proteins 2 and 3 in Indian isolates: implications for malaria rapid diagnostic test performance. Sci Rep 7: 1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willie N, Mehlotra RK, Howes RE, Rakotomanga TA, Ramboarina S, Ratsimbasoa AC, Zimmerman PA, 2018. Insights into the performance of SD Bioline Malaria Ag P.f/Pan rapid diagnostic test and Plasmodium falciparum histidine-rich protein 2 gene variation in Madagascar. Am J Trop Med Hyg 98: 1683–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kemp DJ, Thompson JK, Walliker D, Corcoran LM, 1987. Molecular karyotype of Plasmodium falciparum: conserved linkage groups and expendable histidine-rich protein genes. Proc Natl Acad Sci USA 84: 7672–7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasehagen LJ, et al. 2006. Changing patterns of Plasmodium blood-stage infections in the Wosera region of Papua New Guinea monitored by light microscopy and high throughput PCR diagnosis. Am J Trop Med Hyg 75: 588–596. [PMC free article] [PubMed] [Google Scholar]
- 15.McNamara DT, Kasehagen LJ, Grimberg BT, Cole-Tobian J, Collins WE, Zimmerman PA, 2006. Diagnosing infection levels of four human malaria parasite species by a polymerase chain reaction/ligase detection reaction fluorescent microsphere-based assay. Am J Trop Med Hyg 74: 413–421. [PMC free article] [PubMed] [Google Scholar]
- 16.Mariette N, Barnadas C, Bouchier C, Tichit M, Menard D, 2008. Country-wide assessment of the genetic polymorphism in Plasmodium falciparum and Plasmodium vivax antigens detected with rapid diagnostic tests for malaria. Malar J 7: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walker-Jonah A, Dolan SA, Gwadz RW, Panton LJ, Wellems TE, 1992. An RFLP map of the Plasmodium falciparum genome, recombination rates and favored linkage groups in a genetic cross. Mol Biochem Parasitol 51: 313–320. [DOI] [PubMed] [Google Scholar]
- 18.Lee N, Gatton ML, Pelecanos A, Bubb M, Gonzalez I, Bell D, Cheng Q, McCarthy JS, 2012. Identification of optimal epitopes for Plasmodium falciparum rapid diagnostic tests that target histidine-rich proteins 2 and 3. J Clin Microbiol 50: 1397–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehlotra RK, Kasehagen LJ, Baisor M, Lorry K, Kazura JW, Bockarie MJ, Zimmerman PA, 2002. Malaria infections are randomly distributed in diverse holoendemic areas of Papua New Guinea. Am J Trop Med Hyg 67: 555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mehlotra RK, Lorry K, Kastens W, Miller SM, Alpers MP, Bockarie M, Kazura JW, Zimmerman PA, 2000. Random distribution of mixed species malaria infections in Papua New Guinea. Am J Trop Med Hyg 62: 225–231. [DOI] [PubMed] [Google Scholar]
- 21.Genton B, al-Yaman F, Beck HP, Hii J, Mellor S, Narara A, Gibson N, Smith T, Alpers MP, 1995. The epidemiology of malaria in the Wosera area, East Sepik Province, Papua New Guinea, in preparation for vaccine trials. I. Malariometric indices and immunity. Ann Trop Med Parasitol 89: 359–376. [DOI] [PubMed] [Google Scholar]
- 22.Lee N, Baker J, Andrews KT, Gatton ML, Bell D, Cheng Q, McCarthy J, 2006. Effect of sequence variation in Plasmodium falciparum histidine- rich protein 2 on binding of specific monoclonal antibodies: implications for rapid diagnostic tests for malaria. J Clin Microbiol 44: 2773–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fola AA, Harrison GLA, Hazairin MH, Barnadas C, Hetzel MW, Iga J, Siba PM, Mueller I, Barry AE, 2017. Higher complexity of infection and genetic diversity of Plasmodium vivax than Plasmodium falciparum across all malaria transmission zones of Papua New Guinea. Am J Trop Med Hyg 96: 630–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lalremruata A, et al. 2017. Species and genotype diversity of Plasmodium in malaria patients from Gabon analysed by next generation sequencing. Malar J 16: 398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levitt B, Obala A, Langdon S, Corcoran D, O’Meara WP, Taylor SM, 2017. Overlap extension barcoding for the next generation sequencing and genotyping of Plasmodium falciparum in individual patients in western Kenya. Sci Rep 7: 41108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Golassa L, Enweji N, Erko B, Aseffa A, Swedberg G, 2013. Detection of a substantial number of sub-microscopic Plasmodium falciparum infections by polymerase chain reaction: a potential threat to malaria control and diagnosis in Ethiopia. Malar J 12: 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris I, et al. 2010. A large proportion of asymptomatic Plasmodium infections with low and sub-microscopic parasite densities in the low transmission setting of Temotu Province, Solomon Islands: challenges for malaria diagnostics in an elimination setting. Malar J 9: 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tripura R, et al. 2017. Submicroscopic Plasmodium prevalence in relation to malaria incidence in 20 villages in western Cambodia. Malar J 16: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaw MT, et al. 2017. Asymptomatic and sub-microscopic malaria infection in Kayah State, eastern Myanmar. Malar J 16: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosewell A, Makita L, Muscatello D, John LN, Bieb S, Hutton R, Ramamurthy S, Shearman P, 2017. Health information system strengthening and malaria elimination in Papua New Guinea. Malar J 16: 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das S, Peck RB, Barney R, Jang IK, Kahn M, Zhu M, Domingo GJ, 2018. Performance of an ultra-sensitive Plasmodium falciparum HRP2-based rapid diagnostic test with recombinant HRP2, culture parasites, and archived whole blood samples. Malar J 17: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leow CH, Jones M, Cheng Q, Mahler S, McCarthy J, 2014. Production and characterization of specific monoclonal antibodies binding the Plasmodium falciparum diagnostic biomarker, histidine-rich protein 2. Malar J 13: 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramutton T, et al. 2012. Sequence variation does not confound the measurement of plasma PfHRP2 concentration in African children presenting with severe malaria. Malar J 11: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker J, Gatton ML, Peters J, Ho MF, McCarthy JS, Cheng Q, 2011. Transcription and expression of Plasmodium falciparum histidine-rich proteins in different stages and strains: implications for rapid diagnostic tests. PLoS One 6: e22593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beshir KB, Sepulveda N, Bharmal J, Robinson A, Mwanguzi J, Busula AO, de Boer JG, Sutherland C, Cunningham J, Hopkins H, 2017. Plasmodium falciparum parasites with histidine-rich protein 2 (pfhrp2) and pfhrp3 gene deletions in two endemic regions of Kenya. Sci Rep 7: 14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gamboa D, et al. 2010. A large proportion of P. falciparum isolates in the Amazon region of Peru lack pfhrp2 and pfhrp3: implications for malaria rapid diagnostic tests. PLoS One 5: e8091. [DOI] [PMC free article] [PubMed] [Google Scholar]