Abstract
Accurate and comprehensive characterization of genetic variation is essential for deciphering the genetic basis of diseases and other phenotypes. A vast amount of genetic variation stems from large-scale sequence changes arising from the duplication, deletion, inversion, and translocation of sequences. In the past 10 years, high-throughput short reads have greatly expanded our ability to assay sequence variation due to single nucleotide polymorphisms. However, a recent de novo assembly of a second Drosophila melanogaster reference genome has revealed that short read genotyping methods miss hundreds of structural variants, including those affecting phenotypes. While genomes assembled using high-coverage long reads can achieve high levels of contiguity and completeness, concerns about cost, errors, and low yield have limited widespread adoption of such sequencing approaches. Here we resequenced the reference strain of D. melanogaster (ISO1) on a single Oxford Nanopore MinION flow cell run for 24 hr. Using only reads longer than 1 kb or with at least 30x coverage, we assembled a highly contiguous de novo genome. The addition of inexpensive paired reads and subsequent scaffolding using an optical map technology achieved an assembly with completeness and contiguity comparable to the D. melanogaster reference assembly. Comparison of our assembly to the reference assembly of ISO1 uncovered a number of structural variants (SVs), including novel LTR transposable element insertions and duplications affecting genes with developmental, behavioral, and metabolic functions. Collectively, these SVs provide a snapshot of the dynamics of genome evolution. Furthermore, our assembly and comparison to the D. melanogaster reference genome demonstrates that high-quality de novo assembly of reference genomes and comprehensive variant discovery using such assemblies are now possible by a single lab for under $1,000 (USD).
Keywords: Drosophila, genome assembly, nanopore sequencing
The characterization of comprehensive genetic variation is crucial for the discovery of mutations affecting phenotypes. In the last 10 years, the exponential decline in cost of high-throughput short-read sequencing has revolutionized our ability to assay genome-wide sequence variation. Short reads excel at identifying single nucleotide polymorphisms (SNPs) and short insertion-deletion (indel) polymorphisms in unique genomic regions. However, the majority of the sequence difference between individuals is caused by duplications, deletions, inversions, or translocation of sequences—collectively known as structural variants (SVs) (Feuk et al. 2006). SVs are often longer than short sequencing reads (generally 50–150 bp), meaning genotyping of SVs is indirect and relies on features of alignments to a reference genome such as divergent read mappings, split reads, or elevated read coverage (Medvedev et al. 2009; Alkan et al. 2011b). However, comparisons of extremely contiguous de novo genome assemblies from humans (Huddleston and Eichler 2016; Chaisson et al. 2017) and Drosophila melanogaster (Chakraborty et al. 2018) revealed that short reads miss 40–80% of SVs. Consequently, methods that are not susceptible to the shortcomings of short-read sequencing are essential to obtain a more complete view of genome variation (Alkan et al. 2011b). We propose one approach—comparison of contiguous and accurate de novo genome assemblies—that would overcome these limitations and drastically improve our understanding of genetic variation (Alkan et al. 2011b).
Although short reads have been used extensively for de novo genome assembly, they fail to resolve repetitive regions in genomes, leaving errors and gaps while assembling such regions (Paszkiewicz and Studholme 2010; Treangen and Salzberg 2011; Bradnam et al. 2013). Such fragmented draft-quality assemblies are therefore poorly suited for identification of SVs (Alkan et al. 2011b) and lead to incomplete and/or missing gene models (Gordon et al. 2016). Theoretical considerations of the genome assembly problem predict that, with sufficient read depth and length, genome assemblies can resolve even difficult regions (Motahari et al. 2013; Lam et al. 2014; Bresler et al. 2013; Shomorony et al. 2016). Consistent with this, long reads produced by Single Molecule Real-Time sequencing from Pacific Biosciences (PacBio) and Nanopore sequencing from Oxford Nanopore Technologies (ONT) provide data capable of achieving remarkably contiguous de novo genome assemblies (Kim et al. 2014; Berlin et al. 2015; Michael et al. 2018; Jain et al. 2018; Koren et al. 2017). However, due to high error rates (∼10–15%), generation of reliable assemblies with these reads requires non-trivial coverage (generally 30x or greater) (Koren et al. 2017; Chakraborty et al. 2016). Nevertheless, until recently, long-read methods have required prohibitively expensive reagents, and technologies like PacBio also require substantial capital investment related to the housing and maintenance of equipment necessary to perform the sequencing. Combined with concerns about high error rates, widespread adoption of long-molecule sequencing for de novo assembly and variant detection has been tentative.
Sequencing using ONT may produce reads that are hundreds of kilobases in length (Jain et al. 2018), though their application to de novo assembly of reference-grade multicellular eukaryotic genomes is not yet routine. To understand how effective assembly using ONT is when applied to de novo genome assembly of a metazoan like Drosophila, we measured the contiguity, completeness, and accuracy of a de novo assembly constructed with ONT reads. To accomplish this, we resequenced the D. melanogaster reference genome strain (ISO1) using the ONT MinION and compared the resulting assembly with the latest release of the D. melanogaster reference assembly (Hoskins et al. 2015), which is arguably the best metazoan reference genome available. We followed an assembly merging approach (Chakraborty et al. 2016) to combine modest long-read coverage from a single ONT MinION flow cell (30x depth of coverage with an average read length of 7,122 bp) and Illumina short-read data. This assembly resulted in a highly contiguous and accurate genome assembly. Notably, with this approach, the majority of the euchromatin of each chromosome arm is represented by a single contiguous sequence (contig). Collectively, the assembly recovered 97.7% of Benchmarking Universal Single Copy Orthologs (BUSCOs). This is similar to the 98.3% BUSCOs recovered in the most recent release of the D. melanogaster genome (version 6.16). Scaffolding of the assembly with Bionano optical maps led to further improvements in contiguity. Finally, we examined the structural differences between our assembly and the published assembly of the same strain and observed several candidate SVs, the majority of which are transposable element (TE) insertions and copy-number variants (CNVs). These are mutations that must be either: 1) recent mutations that occurred in the genome strain since it was sequenced; 2) segregating in the genome strain due to incomplete isogeny; or 3) errors in one of the assemblies.
Overall, we show that high-quality de novo genome assembly of D. melanogaster genomes is feasible using low-cost ONT technology, enabling an assembly strategy that can be applied broadly to metazoan genomes. This strategy will make high-quality reference assemblies obtainable for species lacking reference genomes. Moreover, de novo assemblies for population samples of metazoan species is now feasible, opening the door for studying evolutionary and functional consequences of structural genetic variation in large populations.
Methods
Stocks
The ISO1 D. melanogaster reference stock used for both Nanopore and Illumina sequencing was obtained from the BDGP in 2014 (Hoskins et al. 2015). All flies were kept on standard cornmeal-molasses medium and were maintained at 25°.
DNA isolation and quantification
DNA for Nanopore sequencing was isolated from males and females using the Qiagen Blood & Cell Culture DNA Mini Kit. Briefly, 60–80 flies were placed in two 1.5-mL Eppendorf Lo-Bind tubes and frozen in liquid nitrogen before being homogenized using a pestle in 250 µL of Buffer G2 with RNAse. 750 µL of Buffer G2 and 20 µL of 20 mg/mL proteinase K was then added to each tube and incubated at 50° for 2 hr. After 2 hr, each tube was spun at 5,000 RPM for 5 min, and the supernatant was removed and placed in a new 1.5-mL Lo-Bind tube and vortexed for 10 sec. The supernatant from both tubes was then transferred onto the column and allowed to flow through via gravity. The column was washed 3x with wash buffer and eluted twice with 1 mL of elution buffer into two 1.5-mL Lo-Bind tubes. 700 µl of isopropanol was added and mixed via inversion before being spun at 14,000 RPM for 15 min and 4°. The supernatant was removed and the pellet was washed with 70% ethanol, then spun at 14,000 RPM for 10 min at 4°. The supernatant was removed and 25 µL of ddH2O was added to each tube and allowed to sit at room temperature for 1 hr. Both tubes were then combined into one. DNA was quantified on a Nanodrop and Qubit. DNA for Illumina sequencing was isolated from males and females using the Qiagen DNeasy Blood and Tissue Kit according to the manufacturer’s instructions and quantified on a Qubit.
Library preparation, sequencing, and basecalling
For Nanopore sequencing, 1.5 µg of DNA (5.49 µL of 273 ng/uL DNA) was used to prepare a 1D sequencing library (SQK-LSK108) according to the manufacturer’s instructions, including the FFPE repair step. The 75-µl library was then immediately loaded onto a R9.5 flow cell prepared according to the manufacturer’s instructions and run for approximately 24 hr. Basecalling was completed using ONT Albacore Sequencing Pipeline Software version 2.0.2. Reads collected during the mux phase of sequencing were not included.
For Illumina sequencing, ∼600-bp fragments were generated from 500 ng of DNA using a Covaris S220 sonicator. Fragments of 500–700 bp were selected using a Pippin and libraries were prepared using a KAPA High Throughput Library Preparation kit and Bioo Scientific NEXTflex DNA Barcodes. The library was pooled with others and run as a 150-bp paired-end run on a single flow cell of an Illumina NextSeq 500 in medium-output mode using RTA version 2.4.11. bcl2fastq2 v2.14 was then run in order to demultiplex reads and generate FASTQ files.
Genome assemblies
Canu (Koren et al. 2017) release v1.5 was used to assemble the ONT reads. Canu was run with default parameters in grid mode (Sun Grid Engine) using ONT reads >1 kb and a genome size of 130 Mb. To generate the De Bruijn graph contigs for the hybrid assembly, we used Platanus (Kajitani et al. 2014) v1.2.4 with default settings to assemble 67x of Illumina paired-end reads obtained from the DPGP (http://www.dpgp.org/dpgp2/DPGP2.html) (Pool et al. 2012). The hybrid assembly was generated with DBG2OLC (Ye et al. 2016) using contigs from the Platanus assembly and the longest 30x ONT reads. DBG2OLC settings (options: k 25 AdaptiveTh 0.01 KmerCovTh 2 MinOverlap 35 RemoveChimera 1) were similar to those used for PacBio hybrid assembly of ISO1 (Chakraborty et al. 2016), except that the k-mer size was increased to 25 and the MinOverlap to 35 to minimize the number of misassemblies. The consensus stage of DBG2OLC was run with PBDAG-Con (Chin et al. 2013) and BLASR (M. J. Chaisson and Tesler 2012). Separately, minimap v0.2-r123 (using a minimizer size window of 5, FLOAT fraction of minimizers of 0, and min matching length of 100) and miniasm v0.2-r123 (using default settings) were also used to assemble only the ONT reads (Li 2016).
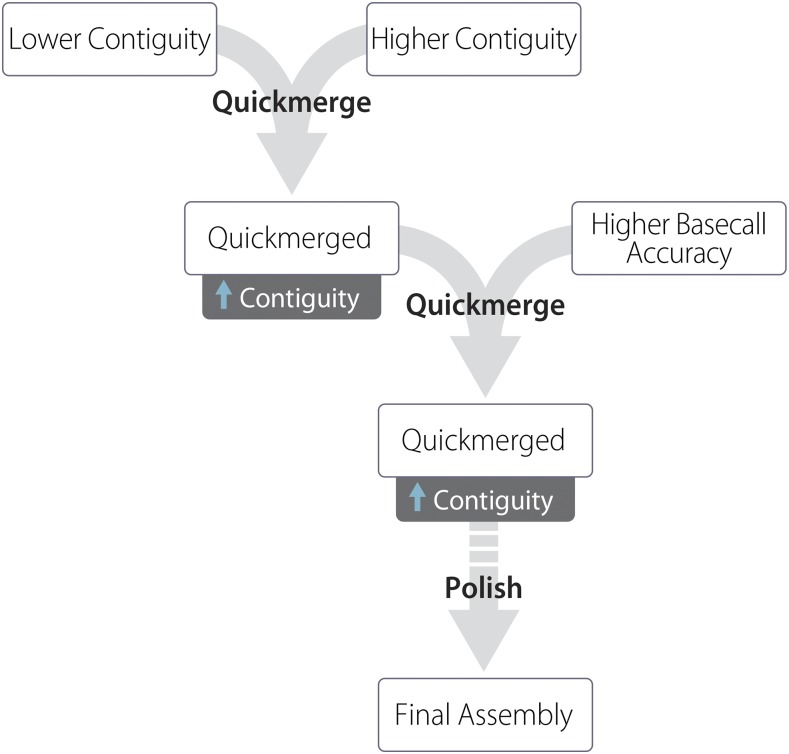
The Canu and DBG2OLC assemblies were merged using quickmerge (Chakraborty et al. 2016). First, the two assemblies were merged using the DBG2OLC assembly as the query and the Canu assembly as the reference. Thus, the first quickmerge run (options: hco 5.0 c 1.5 l 2900000 ml 20000) filled gaps in the DBG2OLC assembly using sequences from the Canu assembly, giving preference to the Canu assembly sequences at the homologous sequence junctions. The contigs that are unique to the Canu assembly were incorporated in the final assembly by a second round of quickmerge. In the second quickmerge run (options: hco 5.0 c 1.5 l 2900000 ml 20000), the merged assembly from the previous step was used as the reference assembly, and the Canu assembly was used as the query assembly (Figure 1).
Figure 1.
Assembly strategy used in this manuscript. A lower-contiguity assembly (Canu) is merged with a higher-contiguity assembly (DBG2OLC). The resulting assembly is again merged with the Canu assembly. The genome is then polished one or more times, here with nanopolish followed by Pilon.
Assembly polishing
Assembly polishing was performed two ways. First, nanopolish version 0.7.1 (Loman, Quick, and Simpson 2015) was run using the recommended settings with reads longer than 1 kb. Prior to running nanopolish, the merged genome assembly was indexed using bwa, and ONT reads were aligned to the genome using bwa mem. The resulting bam file was then sorted, indexed, and filtered for alignments of size larger than 1 kb using samtools 1.3 (Li et al. 2017). Nanopolish was then run with a minimum candidate frequency of 0.1. Following nanopolish, we polished the assembly twice with Pilon (Walker et al. 2014) v1.16. For Pilon, we aligned 44x 150-bp and 67x 100-bp Illumina paired-end reads to the assembly using bowtie2 (Langmead and Salzberg 2012) and then ran Pilon on the sorted bam files using default settings.
Bionano scaffolding
Bionano optical map data were collected following Chakraborty et al. (Chakraborty et al. 2018). ISO1 embryos less than 12 h old were collected on apple juice/agar Petri dishes, dechorionated using 50% bleach, rinsed with water, then stored at -80°. DNA was extracted from frozen embryos using the Animal Tissue DNA Isolation kit (Bionano Genomics, San Diego, CA). Bionano Irys optical data were generated and assembled with IrysSolve 2.1 at Bionano Genomics. We then merged the Bionano assembly with the final merged assembly contigs using IrysSolve, retaining Bionano assembly features when the two assemblies disagreed.
BUSCO analysis
We used BUSCO v1.22 to evaluate completeness and accuracy of all ISO1 assemblies (Simão et al. 2015). We used the Diptera database, which contains 2,799 highly conserved genes, to estimate assembly completeness.
Assembly comparison and quality metrics
Assemblies were compared using alignment dot plots. For dot plots, assembled genomes were aligned to the reference D. melanogaster genome (r6.16) using nucmer (using options: minmatch 100, mincluster 1000, diagfactor 10, banded, diagdiff 5) (Kurtz et al. 2004). The resulting delta alignment files were used to create the dot plots either with mummerplot (using options:–fat–filter–ps) or ggplot. QUAST v4.5.0 was used to compare each generated assembly to the contig reference genome assembly (D. melanogaster r6.16) for completeness and errors. QUAST was run in GAGE mode on contigs larger than 1 kb and with the reference assembly as fragmented, as it was originally the scaffolded assembly (Salzberg et al. 2012; Gurevich et al. 2013). GAGE was run on two types of reference assembly: the full reference assembly, and only the part of the reference assembly comprising ordered and oriented contigs on chromosome arms (i.e., Muller elements) and the mitochondrial sequence. Quality score analysis was performed by aligning our Illumina short reads against each genome assembly using Bowtie. The number of variants were summed and divided by the total number of bases (following Berlin et al. 2015) and also by the total number of bases aligned: Perror = Variants/(Total Bases), where “Total bases” represents the total number of bases in the assembly or the total aligned bases (following Koren et al. 2017). These values are proxies for the probability of error and are used to calculate the QV score for each assembly according to the relation: QV=-10*log10Perror.
A modified pipeline originally published in McCoy et al. (2014) and modified following Berlin et al. (2015) and Koren et al. (2017) was used (this pipeline relies on release 5.57 naming conventions) for calculating total genome reconstruction. Nucmer was used to align each assembly to the FlyBase 5.57 reference, and then separated and merged by euchromatic and heterochromatic regions for each chromosome arm using BEDtools (Quinlan and Hall 2010). Gene and TE reconstruction was completed similar to Berlin et al. (2015). For measuring the accuracy of gene models and TE reconstruction, both releases 5.57 and 6.16 of the reference were used. Fasta files containing all genes and separately, TEs for both releases were downloaded from FlyBase. In total, 17,730 gene models and 5,392 transposons from release 6.16, as well as 17,294 gene models and 5,409 transposons from release 5.57 were aligned to each genome assembly independently using nucmer. Nucmer was used to align each gene and transposon fasta file to each assembly independently. Alignments greater than 0%, 99% and equal to 100% similarity were reported.
Structural variant detection
Large (>100 bp) SVs were detected by aligning the Bionano scaffolds to the FlyBase (dos Santos et al. 2015) reference assembly using MUMmer v3.23 (nucmer -maxmatch) (Kurtz et al. 2004) and then annotating the disagreements between the assemblies as indels and duplications using SVMU v0.2beta (commit 4e65e95) (Chakraborty et al. 2018). Insertions overlapping with Repeatmasker 4.0.7 (Smit et al. 2013) annotated TEs were annotated as TE insertions. SVs were validated with at least two ONT reads spanning the entire genomic feature containing the SVs plus 200 bp on both sides of the SV. TEs were inferred to be segregating when corrected long reads supporting the TE insertions were contradicted by other reads showing absence of the TE. For validation with spanning long reads, we aligned Canu-corrected ONT reads to the FlyBase and Bionano assemblies using BLASR (v 1.3.1) (Chaisson and Tesler 2012) and the sorted alignment bam files were visually examined with IGV (Thorvaldsdóttir et al. 2013).
Mitochondrial genome identification
The mitochondrial genome was identified by using BLAST (Altschul et al. 1990) to compare our final assembly (the Bionano assembly) against the mitochondrial genome from r6.16 of the D. melanogaster genome. A single contig was identified (tig00000438_pilon_pilon_obj) with 99% identity to the reference mitochondrial genome. This contig contained two copies of the mitochondrial genome in tandem (assembly duplications of circular genomes are not uncommon when doing assembly with long-read data), therefore the first 16,806 and last 2,104 nucleotides were removed from the contig. We used MITOS (Bernt et al. 2013) with default settings, metazoan reference, and invertebrate genetic code to annotate both the reference mitochondrial genome and our assembled mitochondrial genome.
Data availability
The Illumina and basecalled ONT data generated in this study has been uploaded to the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/) under bioproject PRJNA433573. Genomes assembled in this study are available at https://github.com/danrdanny/Nanopore_ISO1. Releases 6.16 and 5.57 of the D. melanogaster genome used in this study are available on FlyBase (http://www.flybase.org). Bioinformatic scripts used in this pipeline are available at https://github.com/esolares/DMI1. Original data underlying this manuscript can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb-1268. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6813398.
Results
Sequencing results
A 1D sequencing library was loaded onto a release 9.5 flow cell and run for approximately 24 hr (see Methods), generating a total of 663,784 reads. Basecalling was performed with Albacore 2.0.2, with 593,354 (89%) of all reads marked as “pass” (reads having a quality score ≥7) and an average fragment length of 7,122 bp (Table 1, Figure 2). The read N50 for those that passed filter was 11,840, with 41 reads longer than 50 kb and a maximum read length of 379,978 bp (Figure S1).
Table 1. Statistics of reads used for genome assembly*.
| Total reads | 593,354 |
| Average read length | 7,122 bp |
| Total bases sequenced | 4,184,159,334 |
| Genome Coverage | 30.2x |
| Reads > 1 kb | 530,466 |
| Genome coverage in reads > 1 kb | 29.9x |
| Reads > 10 kb | 145,634 |
| Genome coverage in reads > 10 kb | 17.5x |
Only reads with quality scores ≥ 7 were used. A genome size of 140 Mb was used for all calculations.
Figure 2.
Read length distribution for reads with quality scores greater than or equal to 7. Length is the sequence length after basecalling by Albacore, not the length that aligned to the genome. (A) Distribution of read lengths less than 50 kb. (B) Distribution of reads 50 kb or greater. The longest read that passed quality filtering was 380 kb.
Genome assembly
ONT-only assembly using minimap/miniasm:
To evaluate ONT data for de novo genome assembly, we performed an ONT-only assembly using minimap and miniasm (Li 2016). Together, these programs allow for rapid assembly and analysis of long, error-prone reads. We generated an assembly with a total size of 132 Mb, with 208 contigs and a contig N50 of 3.8 Mb (N50 is the length of the contig such that 50% of the genome is contained within contigs that are equal to or longer than this contig) (Table 2). Evaluation of an alignment dot plot between this assembly and the D. melanogaster reference genome revealed high correspondence between our assembly and the reference genome (Figure S2). However, BUSCO analysis of the minimap assembly found only 0.5% of expected single-copy genes present—much lower than the BUSCO score of 98.3% obtained from the current release of the D. melanogaster genome (Table 3). BUSCO analysis evaluates the presence of universal single-copy orthologs as a proxy of completeness. Such a low BUSCO score as found in the minimap assembly is unlikely to be measuring low completeness, but rather suggests a high rate of errors that disrupt the genes, making them difficult to properly assay.
Table 2. Genome assembly statistics*.
| Name | Genome size (bp) | Contigs | Largest contig (bp) | N50 (bp) | L50 |
|---|---|---|---|---|---|
| FlyBase r6.16* | 142,573,024 | 2,442 | 27,905,053 | 21,485,538 | 3 |
| MiniMap | 131,856,353 | 208 | 16,991,501 | 3,866,686 | 9 |
| DBG2OLC | 131,359,678 | 339 | 13,129,070 | 9,907,730 | 6 |
| Canu | 139,205,737 | 295 | 14,326,064 | 2,971,262 | 11 |
| QuickMerge 2x | 138,130,519 | 250 | 25,434,901 | 18,616,266 | 4 |
| QM2x Nanopolish | 139,303,903 | 250 | 25,367,201 | 18,818,677 | 4 |
| QM2x NP + Pilon x2 | 140,153,080 | 250 | 25,783,280 | 18,923,871 | 4 |
| QuickMerge 2x Bionano | 142,817,829 | 231 | 28,580,427 | 21,305,147 | 3 |
Values are for scaffolds, not contigs.
Table 3. Busco scores demonstrating genome quality before or after polishing.
| Name | Single copy | Duplicate | Fragmented | Missing | Complete |
|---|---|---|---|---|---|
| FlyBase r6.16 | 2,749 (98.2%) | 14 (0.5%) | 22 (0.8%) | 14 (0.5%) | 2,763 |
| MiniMap | 14 (0.5%) | 0 (0.0%) | 31 (1.1%) | 2,754 (98.4%) | 14 |
| DBG2OLC | 1,332 (47.6%) | 3 (0.1%) | 557 (19.9%) | 907 (32.4%) | 1,335 |
| Canu | 1,884 (67.3%) | 11 (0.2%) | 557 (19.9%) | 347 (12.4%) | 1,895 |
| QuickMerge (QM) 2x | 1,623 (58.0%) | 6 (0.3%) | 560 (20.0%) | 610 (21.8%) | 1,629 |
| QM 2x Nanopolish (NP) | 2,189 (78.2%) | 8 (0.3%) | 400 (14.3%) | 202 (7.2%) | 2,197 |
| QM 2x NP + Pilon x2 | 2,726 (97.4%) | 14 (0.5%) | 39 (1.6%) | 20 (0.7%) | 2,740 |
| QM 2x Pilon x2 | 2,718 (97.1%) | 14 (0.5%) | 45 (1.4%) | 22 (0.8%) | 2,732 |
| QM 2x Bionano | 2,715 (97.0%) | 15 (0.5%) | 40 (1.4%) | 29 (1.0%) | 2,730 |
| QM 2x Bionano All | 2,720 (97.2%) | 16 (0.6%) | 40 (1.4%) | 23 (0.8%) | 2,736 |
ONT-only assemblies using Canu:
We also generated an ONT-only assembly with Canu using only reads longer than 1 kb. Alignment dot plots for the assembled genome and the D. melanogaster reference genome also revealed large colinear blocks between this assembly and the reference, indicating only one large misassembly on chromosome 2L (this misassembly was broken prior to merging and polishing) (Figure 3A). The Canu assembly was marginally less contiguous (contig N50 = 3.0 Mb) than the minimap assembly, but resulted in a higher BUSCO score of 67.7% (Table S1). The errors in the Canu assembly and the low BUSCO score are consequences of inherently high error rate of ONT reads. However, due to the higher accuracy and completeness of the Canu assembly compared to the minimap assembly, we used the ONT-only Canu assembly for the remainder of our analysis.
Figure 3.
Dot plots showing colinearity of our assembled genomes with the current version of the D. melanogaster reference genome. Red dots represent regions where the assembly and the reference aligned in the same orientation; blue dots represent regions where the genomes are inverted with respect to one another. The vertical grid lines represent boundaries between chromosome scaffolds in the reference assembly. Horizontal grid lines represent boundaries between contig (A-C) or scaffolds (D) in the assemblies reported here. (A) Plot of the Canu-only assembly against the reference genome. (B) Plot of the hybrid DBG2OLC Nanopore and Illumina assembly against the reference. (C) Plot of merged DBG2OLC and Canu assemblies showing a more contiguous assembly than either of the component assemblies. (D) Bionano scaffolding of the merged assembly resolves additional gaps in the merged assembly.
Hybrid assembly using ONT and Illumina reads:
Modest coverage assemblies of PacBio long reads can exhibit high contiguity when a hybrid assembly method involving Illumina reads is used (Chakraborty et al. 2016). Therefore, we examined whether such assembly contiguity improvements also occur when ONT long reads are supplemented with Illumina paired-end reads. We performed a hybrid assembly using DBG2OLC with the longest 30x ONT reads and contigs from a De Bruijn graph assembly constructed with 67x of Illumina paired-end data. To optimize the parameters, we performed a gridsearch on four parameters (AdaptiveTh, KmerSize, KCovTh and MinOverlap), yielding 36 genome assemblies. We used a range of values recommended by the authors for low-coverage assemblies and verified our KmerSize by looking at meryl’s kmer histogram and found it coincided with a value that represented a 99% fraction of all k-mers (http://kmer.sourceforge.net/wiki/index.php/Getting_Started_with_Meryl). We selected the best genome based on colinearity and largest N50. The best hybrid assembly was substantially more contiguous (contig N50 = 9.9 Mb) than the ONT-only Canu assembly (contig N50 = 3.0 Mb), had large blocks of contiguity with the reference (Figure 3B), yet had lower BUSCO scores (47.7% compared to the 67.7% observed in the Canu assembly) (Table 3).
Merging of Canu and DBG2OLC assemblies:
We have previously shown that merging assemblies constructed using only PacBio long reads with hybrid assemblies constructed with PacBio long reads and Illumina paired-end short reads results in a considerably more contiguous assembly than either of the two component assemblies alone (Chakraborty et al. 2016). To examine the effect of assembly merging on assemblies created with ONT long reads, we merged the ONT-only Canu assembly with the DBG2OLC assembly with two rounds of quickmerge (see Methods). The merged assembly (contig N50 = 18.6 Mb) was more contiguous than both the Canu and the DBG2OLC assemblies alone (Table 2), exhibiting large-scale colinearity with the reference genome as seen in the dot plot (Figure 3C). As expected, the BUSCO score of the merged assembly (58.2%) fell between the two component assemblies (Table 3).
Assembly quality
Polishing:
A high number of SNP and indel polymorphisms in all of our assemblies is consistent with other de novo assemblies created with noisy long reads with high error rates (Loman et al. 2015; Chakraborty et al. 2016). Such errors typically lead to error-ridden genic sequence that can give the appearance that many important genes are missing (Table 3; Table S1). Many of these errors can be fixed via assembly polishing, and a number of assembly polishing tools exist. We chose to employ two: nanopolish, which performs consensus-based error correction using ONT reads (Loman et al. 2015), and Pilon, which performs error correction using Illumina reads (Walker et al. 2014). This approach of applying long-read consensus correction followed by short-read polishing has resulted in BUSCO scores comparable to that of the FlyBase reference genome (e.g., Chakraborty et al. 2016 and Chakraborty et al. 2018).
To evaluate this approach for ONT data, we evaluated three different polishing approaches: one using nanopolish alone, one using only two rounds of Pilon, and one using one round of nanopolish followed by two rounds of Pilon (see Methods). Applying nanopolish alone recovered only 79%, 79.2%, and 78.5% complete BUSCOs for the hybrid, ONT-only, and the merged assemblies, respectively (Table S2), suggesting that polishing with ONT reads alone only partially improved assembly quality. On the other hand, polishing all three assemblies twice with Pilon alone fixed a large number of errors as evidenced by improved BUSCO scores of the resulting assemblies (Simão et al. 2015) (Table 3). The merged assembly was polished once with nanopolish, then twice with Pilon, resulting in nearly all complete BUSCOs (97.9%) being present in our polished assembly, comparable to that of the reference assembly (98.3%) (Table S2). Variation in BUSCO scores generally tracked the method of polishing more than the type of assembly. However, the hybrid assembly did recover a slightly different subset of BUSCOs than the ONT-only assembly, leading to a slightly higher number of BUSCOs being recovered in the final merged assembly.
QUAST/GAGE metrics of quality:
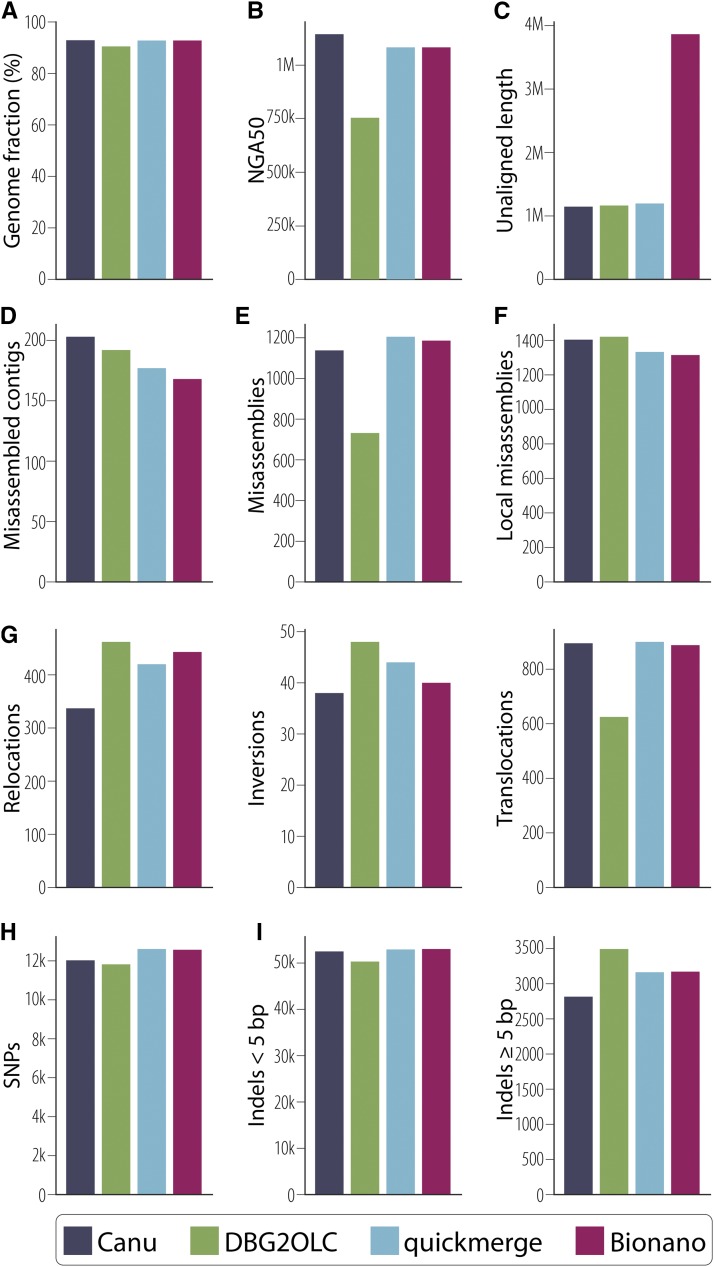
The QUAST output comparing the Canu, DBG2OLC, merged, and Bionano genome assemblies against the D. melanogaster reference shows that the quickmerge assembly resulted in intermediate error rates and discordance as compared to the component assemblies (Figure 4). All four assemblies exhibited approximately the same number of SNP and indel errors less than 5 bp, whereas the DBG2OLC assembly resulted in approximately 20% more indels greater than 5 bp in size than the Canu assembly (Figure 4H-I, Table S3, Table S4). Among ONT assemblies, the Canu assembly exhibited superior accuracy and fewer misassemblies, although our quickmerge assembly was nearly as good. When mismatches are measured on a phred scale, the ONT assemblies range in quality from 32 to 33, which is lower than the score of 37 from a PacBio assembly with threefold more data (Koren et al. 2017) (Table S3, Table S4). Canu also exhibited the lowest raw contiguity for an ONT assembly as measured by N50/NG50 and L50/LG50. The N50 for our merged assembly (19 Mb) exceeded that of Canu (3 Mb) and was nearly double that of DBG2OLC (10 Mb), while maintaining large-scale colinearity with the reference (Figure 3C). However, Canu outperforms the merging approach when considering the contiguity of error-free alignment blocks as demonstrated by the NA50 and NGA50 for Canu, which are greater than that of quickmerge (Table S3). We also observe that half of the assemblies accrue in fewer error-free alignments for Canu than for quickmerge, resulting in slightly lower values in LA50 and LGA50 for Canu than quickmerge (Table S3). As a consequence, when evaluating which approach to employ, we advise users to carefully weigh the tradeoffs between large-scale contiguity and local misassembly and sequence errors and to tailor their decision to the biological questions being addressed. For example, in creating a reference for QTL mapping, there might be a strong preference for high long-range contiguity even at the cost of local misassemblies. On the other hand, when avoiding misassemblies is paramount (as might be the case when characterizing the structure of individual loci), one could make the argument in favor of less long-range contiguity in exchange for fewer misassemblies.
Figure 4.
QUAST was used to compare each assembly to the D. melanogaster reference genome with selected statistics presented here. (A) Greater than 90% of bases in the reference genome were aligned to each of our four assemblies. (B) The contiguity of assembly blocks aligned to the reference. (C) Total unaligned length includes contigs that did not align to the reference as well as unaligned sequence of partially aligned contigs. (D) The number of contigs that contain misassemblies in which flanking sequences are 1 kb apart, flanking sequences overlap by 1 kb or more, or flanking sequences align to different reference scaffolds. (E) Total count of misassemblies as described in (D). (F) Local misassemblies include those positions in which a gap or overlap between flanking sequence is less than 1 kb [(D) and (E) show those greater than 1 kb] and larger than the maximum indel length of 85 bp on the same reference genome scaffold. (G) Misassemblies can be subdivided into relocations (a single assembled contig aligns to the same reference scaffold but in pieces at least 1 kb apart), inversions (at least one part of a single assembled contig aligns to the reference in an inverted orientation), or translocations (at least one part of a single assembled contig aligns to two different reference scaffolds). Not all misassemblies are captured in these three categories. (H) Total SNPs per assembly are shown and were not significantly different among assemblies. (I) Indels per 100 kb can be divided into small indels (those <5 bp) and large indels (≥ 5 bp). Indels >85 bp are considered misassemblies and are shown in panels D, E, or F.
Finally, the low-coverage ONT dataset presented here performed surprisingly well compared to a PacBio dataset three times larger (Kim et al. 2014; Koren et al. 2017) in terms of contiguity (N50, NA50, L50, LA50), completeness (genome fraction), and accuracy (identity, SNPs, InDels, translocations, etc.) (Table S3). In all measures, the merged assembly performed almost as well as the much higher coverage PacBio assembly from Koren et al. (2017). Importantly, however, the GAGE corrected N50 of Koren et al. (2017) was substantially larger than the ONT assemblies, likely due to its superior coverage (Table S3, Table S4).
Completeness: reconstructing genes, TEs, and chromosome arms:
We next evaluated the completeness of the assemblies by assessing the reconstruction of elements in the genome, including genes, TEs, and chromosome arms. For all following metrics, Canu and the final Quickmerge assembly performed similar to one another, with DBG2OLC performing somewhat worse. As a result, remaining comparisons will be made between the final Quickmerge ONT assembly and the PacBio assembly produced by Koren et al. 2017, which we use as a standard of comparison.
Our final assembly contained 96.37% of release 6.16 genes greater than 99% complete as compared to 98.4% for the Koren et al. (2017) assembly (Table S5). However, our final assembly was able to reconstruct only 72.71% of gene models compared to 88.14% for the Koren et al. (2017) assembly when only 100% complete gene models are considered (Table S5). Transposon reconstruction followed the same pattern with a 6.66% and a 15.51% difference in counts between the two assemblies for those with greater than 99% identity and complete reconstruction, respectively, with both favoring the higher-coverage PacBio assembly.
Muller element (chromosome arm) reconstruction was more similar between the ONT and PacBio assemblies for the euchromatic regions, with an average difference of 0.47% and a maximum difference of 0.94% in the X chromosome (Table S5). The largest difference was in the number of alignments. Bionano and quickmerge assemblies covered the Muller elements in fewer segments than the Koren et al. (2017) assembly, though they exhibited less total chromosome coverage. The differences were more apparent in the heterochromatic scaffolds, as our final assembly contained less coverage than the Koren et al. (2017) assembly, with an average deficit of 4.44% coverage and a maximum deficit of 7.89% across all heterochromatic scaffolds linked to Muller elements (Table S6). The difference in the Y chromosome was the largest, as expected, for our relatively low coverage of mixed sex flies compared to the high coverage of all males for Koren et al. (2017).
Base quality:
We aligned Illumina short reads to measure the sequencing error rate following Berlin et al. (2015). The proportion of reads successfully mapped to assemblies varied from 93.5 to 94.6%. The aligned reads resulted in QV scores ranging from 40.2 to 41.1. A slightly different approach used by Koren et al. (2017) yielded very similar QV scores in the range of 40.1–40.2 (Table S7), suggesting that our assembly was of relatively high quality in areas where reads align well.
Bionano scaffolding
Bionano fragments, which are substantially longer than ONT reads, can be used for scaffolding contigs across repetitive regions. We generated 81,046 raw Bionano fragments (20.5 Gb) with an average fragment length of 253 kb. To use these fragments for scaffolding, we first created a Bionano assembly (509 maps, haploid assembly size = 145 Mb) using 78,397 noise-rescaled fragments (19.9 Gb, mean fragment length 253 kb). At 145 Mb, the Bionano assembly is comparable to the reference assembly size (144 Mb). The Bionano maps were used to scaffold the merged assembly. The resulting scaffolded assembly was more contiguous (N50 = 21.3 Mb) than the unscaffolded contigs without substantially changing the number of SNP and indel errors (Figure 3D; Tables S1, S3-S4). Bionano scaffolding of our assembled genome did not change BUSCO scores (Table 3).
Structural variants
One advantage of high quality de novo assemblies is that they permit comprehensive detection of large (>100 bp) SVs. Highly contiguous assemblies, such as the one generated here, allow comparisons between two or more assemblies, revealing novel SVs and facilitating the study of their functional and evolutionary significance. Although we sequenced the reference genome stock, structural differences between our stock and the published assembly are expected due to error, but also to new mutations—especially for TEs, which are the most dynamic structural components of the genome. Indeed, such mutations have been observed in substrains of ISO1 before (Zakharenko et al. 2007; Moschetti et al. 2010; Rahman et al. 2015). Similarly, our assembly revealed the presence of 34 new TE insertions and 12 TE losses compared to the reference genome assembly. Among the 34 TE insertions, 50% (17/34) are LTR TEs comprising chiefly of copia (5/17) and roo elements (6/17). Interestingly, 29% (10/34) of the TE insertions are defective hobo elements that lack an average of 1.7 kb of sequence (base pairs 905–2510 of full-length hobo are absent) from the middle segment of the element encoding the transposase. However, alignment of long reads to the assembly regions harboring the TE insertions revealed that 6/34 insertions are not fixed, but are segregating in the strain (Table S2). The high insertion rate of the LTR and hobo elements in this single strain mirrors the recent spread of LTR and hobo elements in D. melanogaster populations (Pascual and Periquet 1991; Periquet et al. 1994; Bowen and McDonald 2001). As expected, the majority (27/34) of the new TE insertions are located within introns, because insertions within exons generally result in gene disruption. Nonetheless, we found five genes (Ance, Pka-C1, CG31826, CG43446, and Ilp6) in which new TEs have inserted within exons (Table S2). We also found 12 TEs present in the reference assembly that are missing from our final scaffolded assembly, among which six are LTR TEs (five 297 elements and one roo element). The high rate of insertion and loss of LTR elements underscores their dynamic evolutionary history in the D. melanogaster genome. Because TEs can be locally unique, the presence or absence of such events does not pose a fundamental limitation to assembly when reads are long enough to span the TEs. Consequently, we predict most of these events to be new mutations rather than errors in assembly.
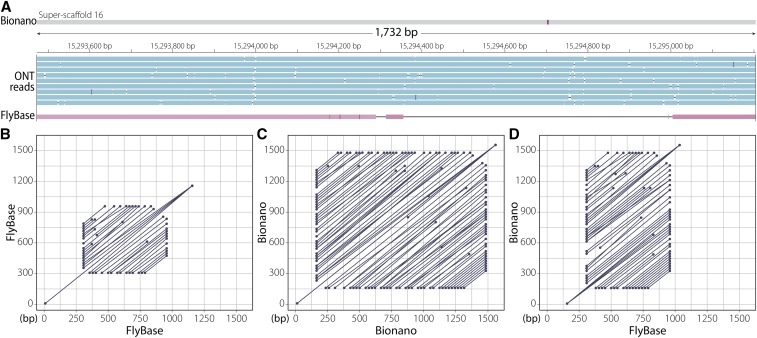
Additionally, we identified several duplications present in our assembly. For example, a tandem duplication of a ∼9-kb segment has created partial copies of the genes infertile crescent (ifc) and little imaginal discs (lid). Another ∼2-kb tandem duplication has created partial copies of the genes CG10137 and CG33116 (Table S8). Apart from CNVs affecting single copy sequences, our assembly also uncovered copy number increases in tandem arrays with potential functional consequences. For example, we observed copy number increase in a tandem array of a 207-bp segment within the third exon of the chitin-binding protein gene Muc26B. While CNVs such as this have been challenging to identify and validate in the past, at least two Nanopore reads spanning this entire tandem array support the presence of a tandem duplication at this position (Figure 5). Unlike TEs, classifying such tandem events as errors stemming from shorter Sanger reads or an actual mutation is difficult without the access to the original material from which the Sanger data were derived.
Figure 5.
Copy number increase in a 207-bp tandem array located inside the third exon of Muc26B. (A) Three tracks showing Bionano assembly (top) with Nanopore long reads (blue) and reference (Flybase) assembly (red) aligned to it. The alignment gap in the reference assembly is due to the extra sequence copies in the Bionano assembly. (B) Alignment dot plot between the reference sequence possessing the tandem array to itself. (C) Alignment dot plot between the genomic region possessing the tandem array in the Bionano assembly to itself. As evidenced by the dot plot, the Bionano assembly has more repeats in this region than the reference assembly in panel B. (D) Alignment dot plot between the reference genomic region (x axis) shown in (B) and the corresponding Bionano genomic region (y axis) shown in (C).
Mitochondrial genome identification and identity
After assembly, merging, polishing, and scaffolding, we used BLAST (Altschul et al. 1990) to identify the mitochondrial genome. Using the published D. melanogaster mitochondrial genome as the subject, we identified one 38,261-bp contig with nearly 100% identity to the reference mitochondrial genome. The first 16,806 and last 2,100 nucleotides of this contig were 99.6% identical to the reference mitochondrial genome, while the middle 19,228 nucleotides were 98% identical to the reference mitochondrial genome. This suggests that our assembled mitochondrial genome had been duplicated during assembly, which commonly occurs when assembling circular genomes using long sequencing reads. For the tandem assembled genomes, nearly all of the SNP and indel polymorphisms occurred in the last 4,000 nucleotides of the reference genome.
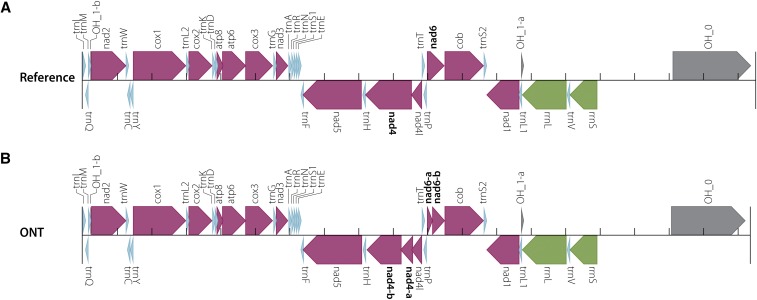
To determine if all genes and features that are present in the reference mitochondrial genome were present in our assembled genome, we annotated both genomes using MITOS (Bernt et al. 2013). Both assemblies were annotated nearly identically, with MITOS reporting that both assemblies were missing the origin of replication for the L region (OL) (Figure 6). The Nanopore assembly also resulted in two split genes not observed in the reference genome assembly: nad4 and nad6 were each annotated as two continuous genes rather than one single gene.
Figure 6.
Mitochondrial genome annotations generated by MITOS. (A) Annotation of the reference mitochondrial genome. (B) Annotation of the mitochondrial genome assembled in this project is identical to the reference except that nad4 and nad6 in the reference assembly were both annotated as two genes—nad4 as nad4-a and nad4-b, and nad6 as nad6-a and nad6-b.
Discussion
Drosophila melanogaster was the first genome assembled using a whole-genome shotgun (WGS) strategy (Myers et al. 2000; Adams et al. 2000). This successful proof-of-principle led to the prevalence of the WGS sequencing approach as a tool in virtually all subsequent metazoan genome assembly projects (Lander et al. 2001; Mouse Genome Sequencing Consortium et al. 2002; Goff et al. 2002; Yu et al. 2002; Aparicio et al. 2002; Gibbs et al. 2004; International Chicken Genome Sequencing Consortium 2004). While improvements in sequencing technology have led to a precipitous drop in the cost of sequencing (Anonymous 2018), stagnation and even regression of read lengths resulted in highly fragmented and incomplete assemblies (Alkan et al. 2011a). Furthermore, complementary approaches (like hierarchical shotgun sequencing and other clone-based approaches) were required to obtain nearly complete and highly contiguous reference genomes, which adds complexity, cost, and time to assembly projects.
Short reads provided by next generation sequencing technologies present limitations to what a pure WGS assembly approach can accomplish (Alkan et al. 2011a; Narzisi and Schatz 2015). The advent and development of long-read sequencing technologies has led to dramatic increases in read length, permitting assemblies that span previously recalcitrant repetitive regions. Early implementations of these technologies produced reads longer than previous short-read technologies yet still shorter than relatively common repeats, while the cost and error rate remained high compared to the short-read approaches. However, continued improvements in read length overcame many of these difficulties, permitting nearly complete, highly contiguous metazoan genome assemblies with only a WGS strategy (Kim et al. 2014; Berlin et al. 2015; Koren et al. 2017). Such approaches led to assemblies comparable in completeness and contiguity to release 6 of the D. melanogaster genome assembly (Hoskins et al. 2015) for approximately $10,000 USD (Chakraborty et al. 2016, 2018). However, even with the rapid development of these approaches, substantial capital investment in the form of expensive instrumentation and dedicated genome facility staff was required.
Here, we report an independent resequencing and assembly of the D. melanogaster reference strain, ISO1, for less than $1,000 USD in sequencing costs and without the need for extensive capital or personnel investment. The resulting assembly before scaffolding is comparable to release 6 of FlyBase in terms of contiguity and completeness (18.9 Mbp contig N50 and 97.1% complete single copy BUSCOs). We achieved this using 4.2 Gbp of sequence from a single Oxford Nanopore flow cell in conjunction with Illumina short-read data. Such reduction in complexity and cost permits a small team of scientists to shepherd a sequencing project from sample to near-reference-quality assembly in a relatively short amount of time. The addition of optical mapping data permitted ordering and orientation of the contigs, yielding an assembly nearly as contiguous as the published reference (scaffold N50 of 21.3 Mbp).
Comparing this assembly to the FlyBase reference genome shows that it is both accurate (21 mismatches/100 kbp, 36 indels/100 kbp) and colinear (Figure 3D, Figure 4, Table S3). Most of the small-scale differences are expected to be errors introduced by the noisy Oxford Nanopore reads that escaped correction via polishing. It is possible that some of these errors are SVs, which are expected to accumulate because the ISO1 stock has been maintained in the laboratory for approximately 350 generations (assuming 20 gen/year) since initial sequencing in 2000. This allows for the accumulation of new mutations by genetic drift, including ones reducing fitness (Assaf et al. 2017). Due to the high contiguity in the euchromatic region, our assembly facilitates detection of such euchromatic SVs. Several apparent “assembly errors” in our Bionano assembly are due to TE indels that are supported by spanning long reads. We found 28 homozygous euchromatic TE insertions, which are predominantly LTR and defective hobo elements, suggesting a high rate of euchromatic TE insertions (∼0.08 insertion/gen). That we observed a predominance of LTR and hobo elements among the new TE insertions mirrors their recent spread in D. melanogaster populations (Pascual and Periquet 1991; Periquet et al. 1994; Bowen and McDonald 2001; De Freitas Ortiz and Silva Loreto 2008). The abundance of defective hobo elements among the new insertions is particularly interesting given that these hobo elements lack the transposase enzyme necessary for mobilization.
Although most novel TE insertions were found in introns, five were found within exons: the 5′ UTR of Ance, CG31826, and Ilp6; the 3′ UTR of Pka-C1; and the coding region of CG43446. Similarly, TE loss primarily involved LTR TEs, including loss of TEs from the 5′ UTR of the genes Snoo and CG1358. We also observed copy number increases both in unique sequences as well as in tandem arrays (Table S8), with one duplication creating a new copy of the entire coding sequence of the gene lid. Collectively, our assembly provides a snapshot of ongoing genome structure evolution in a metazoan genome, which is often assumed to be approximately invariant for experimental genetics.
A crucial feature of this work is that it is performed in a strain used to generate one of the highest quality reference genomes available, ensuring that our inferences can be judged against a high-quality standard. This approach allowed us to demonstrate that assembly with modest amounts of long-molecule data paired with inexpensive short-read data can yield highly accurate and contiguous reference genomes with minimal expenditure of resources. This demonstration opens myriad opportunities for high-quality genomics in systems with limited resources for genome projects. Moreover, we can now conceive of studying entire populations with high-quality assemblies capable of resolving repetitive structural variants, something previously unattainable with short-read sequencing alone.
Acknowledgments
We would like to thank Christina Lin for assistance with the Bionano scaffolding, Melanie Oakes for help generating the Bionano data, and Angela Miller for assistance with editing and figure preparation. We thank Casey Bergman for providing examples of novel TEs in substrains of ISO1. DEM is part of the Oxford Nanopore Educational Initiative and has received free flow cells as part of that program. This material is based upon work supported by the National Science Foundation Bridge to the Doctorate Fellowship under Grant No. 1500284 and the National Science Foundation Graduate Research Fellowship under Grant No. DGE-1321846. MC is supported by US National Institutes of Health (NIH) grant R01 RR024862-01A6. JJE is supported by NIH grant R01GM123303-1 and University of California, Irvine setup funds. This work was made possible, in part, through access to the Genomics High-Throughput Facility Shared Resource of the Cancer Center Support Grant CA-62203 at the University of California, Irvine, and NIH shared-instrumentation grants 1S10RR025496-01, 1S10OD010794-01, and 1S10OD021718-01. RSH is an American Cancer Society Research Professor.
Note added in proof: See Miller et al. 2018 (pp. 3131–3141) in this issue, for a related work.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.6813398.
Communicating editor: S. Celniker
Literature Cited
- Adams M. D., Celniker S. E., Holt R. A., Evans C. A., Gocayne J. D., et al. , 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. 10.1126/science.287.5461.2185 [DOI] [PubMed] [Google Scholar]
- Alkan C., Sajjadian S., Eichler E. E., 2011a Limitations of next-generation genome sequence assembly. Nat. Methods 8: 61–65. 10.1038/nmeth.1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan C., Coe B. P., Eichler E. E., 2011b Genome structural variation discovery and genotyping. Nat. Rev. Genet. 12: 363–376. 10.1038/nrg2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Anonymous, 2018. DNA Sequencing Costs: Data. National Human Genome Research Institute, NHGRI; https://www.genome.gov/27565109/the-cost-of-sequencing-a-human-genome//the-cost-of-sequencing-a-human-genome [Google Scholar]
- Aparicio S., Chapman J., Stupka E., Putnam N., Chia J.-M., et al. , 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301–1310. 10.1126/science.1072104 [DOI] [PubMed] [Google Scholar]
- Assaf Z. J., Tilk S., Park J., Siegal M. L., Petrov D. A., 2017. Deep sequencing of natural and experimental populations of Drosophila melanogaster reveals biases in the spectrum of new mutations. Genome Res. 27: 1988–2000. 10.1101/gr.219956.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin K., Koren S., Chin C.-S., Drake J. P., Landolin J. M., et al. , 2015. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol. 33: 623–630 (corrigendum: Nat. Biotechnol. 33: 1109) 10.1038/nbt.3238 [DOI] [PubMed] [Google Scholar]
- Bernt M., Donath A., Jühling F., Externbrink F., Florentz C., et al. , 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69: 313–319. 10.1016/j.ympev.2012.08.023 [DOI] [PubMed] [Google Scholar]
- Bowen N. J., McDonald J. F., 2001. Drosophila euchromatic LTR retrotransposons are much younger than the host species in which they reside. Genome Res. 11: 1527–1540. 10.1101/gr.164201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradnam K. R., Fass J. N., Alexandrov A., Baranay P., Bechner M., et al. , 2013. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. Gigascience 2: 10 10.1186/2047-217X-2-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresler G., Bresler M., Tse D., 2013. Optimal assembly for high throughput shotgun sequencing. BMC Bioinformatics 14: S18 10.1186/1471-2105-14-S5-S18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson M. J., Tesler G., 2012. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13: 238 10.1186/1471-2105-13-238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson M. J. P., Sanders A. D., Zhao X., Malhotra A., Porubsky D., et al. , 2017. Multi-platform discovery of haplotype-resolved structural variation in human genomes. bioRxiv. 10.1101/193144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty M., Baldwin-Brown J. G., Long A. D., Emerson J. J., 2016. Contiguous and accurate de novo assembly of metazoan genomes with modest long read coverage. Nucleic Acids Res. 44: e147 10.1093/nar/gkw654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty M., VanKuren N. W., Zhao R., Zhang X., Kalsow S., et al. , 2018. Hidden genetic variation shapes the structure of functional elements in Drosophila. Nat. Genet. 50: 20–25. 10.1038/s41588-017-0010-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C.-S., Alexander D. H., Marks P., Klammer A. A., Drake J., et al. , 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10: 563–569. 10.1038/nmeth.2474 [DOI] [PubMed] [Google Scholar]
- De Freitas Ortiz M., Silva Loreto E. L., 2008. The hobo-related elements in the melanogaster species group. Genet. Res. 90: 243–252. 10.1017/S0016672308009312 [DOI] [PubMed] [Google Scholar]
- Feuk L., Carson A. R., Scherer S. W., 2006. Structural variation in the human genome. Nat. Rev. Genet. 7: 85–97. 10.1038/nrg1767 [DOI] [PubMed] [Google Scholar]
- Gibbs R. A., Weinstock G. M., Metzker M. L., Muzny D. M., Sodergren E. J., et al. , 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428: 493–521. 10.1038/nature02426 [DOI] [PubMed] [Google Scholar]
- Goff S. A., Ricke D., Lan T.-H., Presting G., Wang R., et al. , 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100. 10.1126/science.1068275 [DOI] [PubMed] [Google Scholar]
- Gordon D., Huddleston J., Chaisson M. J. P., Hill C. M., Kronenberg Z. N., et al. , 2016. Long-read sequence assembly of the gorilla genome. Science 352: aae0344 10.1126/science.aae0344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., Tesler G., 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29: 1072–1075. 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins R. A., Carlson J. W., Wan K. H., Park S., Mendez I., et al. , 2015. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 25: 445–458. 10.1101/gr.185579.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huddleston J., Eichler E. E., 2016. An Incomplete Understanding of Human Genetic Variation. Genetics 202: 1251–1254. 10.1534/genetics.115.180539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium , 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432: 695–716. 10.1038/nature03154 [DOI] [PubMed] [Google Scholar]
- Jain M., Koren S., Miga K. H., Quick J., Rand A. C., et al. , 2018. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 36: 338–345. 10.1038/nbt.4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajitani R., Toshimoto K., Noguchi H., Toyoda A., Ogura Y., et al. , 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24: 1384–1395. 10.1101/gr.170720.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. E., Peluso P., Babayan P., Yeadon P. J., Yu C., et al. , 2014. Long-read, whole-genome shotgun sequence data for five model organisms. Sci. Data 1: 140045 10.1038/sdata.2014.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S., Walenz B. P., Berlin K., Miller J. R., Bergman N. H., et al. , 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27: 722–736. 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Phillippy A., Delcher A. L., Smoot M., Shumway M., et al. , 2004. Versatile and open software for comparing large genomes. Genome Biol. 5: R12 10.1186/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam K.-K., Khalak A., Tse D., 2014. Near-optimal assembly for shotgun sequencing with noisy reads. BMC Bioinformatics 15: S4 10.1186/1471-2105-15-S9-S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., et al. , 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., 2016. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32: 2103–2110. 10.1093/bioinformatics/btw152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loman N. J., Quick J., Simpson J. T., 2015. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods 12: 733–735. 10.1038/nmeth.3444 [DOI] [PubMed] [Google Scholar]
- McCoy Rajiv C., Taylor R. W., Blauwkamp T. A., Kelley J. L., Kertesz M., et al. , 2014. Illumina TruSeq Synthetic Long-Reads Empower de Novo Assembly and Resolve Complex, Highly-Repetitive Transposable Elements. PLoS One 9: e106689 10.1371/journal.pone.0106689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedev P., Stanciu M., Brudno M., 2009. Computational methods for discovering structural variation with next-generation sequencing. Nat. Methods 6: S13–S20. 10.1038/nmeth.1374 [DOI] [PubMed] [Google Scholar]
- Michael T. P., Jupe F., Bemm F., Motley S. T., Sandoval J. P., et al. , 2018. High contiguity Arabidopsis thaliana genome assembly with a single nanopore flow cell. Nat. Commun. 9: 541 10.1038/s41467-018-03016-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschetti R., Dimitri P., Caizzi R., Junakovic N., 2010. Genomic instability of I elements of Drosophila melanogaster in absence of dysgenic crosses. PLoS One 5: e13142 10.1371/journal.pone.0013142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motahari A. S., Bresler G., Tse D. N. C., 2013. Information Theory of DNA Shotgun Sequencing. IEEE Trans. Inf. Theory 59: 6273–6289. 10.1109/TIT.2013.2270273 [DOI] [Google Scholar]
- Mouse Genome Sequencing Consortium; . Waterston R. H., Lindblad-Toh K., Birney E., Rogers J., Abril J. F., et al. , 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. 10.1038/nature01262 [DOI] [PubMed] [Google Scholar]
- Myers E. W., Sutton G. G., Delcher A. L., Dew I. M., Fasulo D. P., et al. , 2000. A whole-genome assembly of Drosophila. Science 287: 2196–2204. 10.1126/science.287.5461.2196 [DOI] [PubMed] [Google Scholar]
- Narzisi G., Schatz M. C., 2015. The challenge of small-scale repeats for indel discovery. Front. Bioeng. Biotechnol. 3: 8 10.3389/fbioe.2015.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual L., Periquet G., 1991. Distribution of hobo transposable elements in natural populations of Drosophila melanogaster. Mol. Biol. Evol. 8: 282–296. 10.1093/oxfordjournals.molbev.a040649 [DOI] [PubMed] [Google Scholar]
- Paszkiewicz K., Studholme D. J., 2010. De novo assembly of short sequence reads. Brief. Bioinform. 11: 457–472. 10.1093/bib/bbq020 [DOI] [PubMed] [Google Scholar]
- Periquet G., Lemeunier F., Bigot Y., Hamelin M. H., Bazin C., et al. , 1994. The evolutionary genetics of the hobo transposable element in the Drosophila melanogaster complex. Genetica 93: 79–90. 10.1007/BF01435241 [DOI] [PubMed] [Google Scholar]
- Pool J. E., Corbett-Detig R. B., Sugino R. P., Stevens K. A., Cardeno C. M., et al. , 2012. Population Genomics of sub-saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genet. 8: e1003080 10.1371/journal.pgen.1003080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman R., Chirn G.-W., Kanodia A., Sytnikova Y. A., Brembs B., et al. , 2015. Unique transposon landscapes are pervasive across Drosophila melanogaster genomes. Nucleic Acids Res. 43: 10655–10672. 10.1093/nar/gkv1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg S. L., Phillippy A. M., Zimin A., Puiu D., Magoc T., et al. , 2012. GAGE: a critical evaluation of genome assemblies and assembly algorithms. Genome Res. 22: 557–567. 10.1101/gr.131383.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos G., Schroeder A. J., Goodman J. L., Strelets V. B., Crosby M. A., et al. , 2015. FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 43: D690–D697. 10.1093/nar/gku1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomorony I., Courtade T., Tse D., 2016. Do Read Errors Matter for Genome Assembly? bioRxiv. 10.1101/014399 [DOI] [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Smit A. F. A., Hubley R., Green P.2013-2015 RepeatMasker Open-4.0. http://www.repeatmasker.org [Google Scholar]
- Thorvaldsdóttir H., Robinson J. T., Mesirov J. P., 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14: 178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen T. J., Salzberg S. L., 2011. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat. Rev. Genet. 13: 36–46 (corrigendum: Nat. Rev. Genet. 13: 146) 10.1038/nrg3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B. J., Abeel T., Shea T., Priest M., Abouelliel A., et al. , 2014. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS One 9: e112963 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C., Hill C. M., Wu S., Ruan J., Ma Z. S., 2016. DBG2OLC: Efficient Assembly of Large Genomes Using Long Erroneous Reads of the Third Generation Sequencing Technologies. Sci. Rep. 6: 31900 10.1038/srep31900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Hu S., Wang J., Wong G. K.-S., Li S., et al. , 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92. 10.1126/science.1068037 [DOI] [PubMed] [Google Scholar]
- Zakharenko L. P., Kovalenko L. V., Mai S., 2007. Fluorescence in situ hybridization analysis of hobo, mdg1 and Dm412 transposable elements reveals genomic instability following the Drosophila melanogaster genome sequencing. Heredity 99: 525–530. 10.1038/sj.hdy.6801029 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Illumina and basecalled ONT data generated in this study has been uploaded to the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/) under bioproject PRJNA433573. Genomes assembled in this study are available at https://github.com/danrdanny/Nanopore_ISO1. Releases 6.16 and 5.57 of the D. melanogaster genome used in this study are available on FlyBase (http://www.flybase.org). Bioinformatic scripts used in this pipeline are available at https://github.com/esolares/DMI1. Original data underlying this manuscript can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb-1268. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6813398.