Abstract
Current variant discovery approaches often rely on an initial read mapping to the reference sequence. Their effectiveness is limited by the presence of gaps, potential misassemblies, regions of duplicates with a high-sequence similarity and regions of high-sequence divergence in the reference. Also, mapping-based approaches are less sensitive to large INDELs and complex variations and provide little phase information in personal genomes. A few de novo assemblers have been developed to identify variants through direct variant calling from the assembly graph, micro-assembly and whole-genome assembly, but mainly for whole-genome sequencing (WGS) data. We developed SGVar, a de novo assembly workflow for haplotype-based variant discovery from whole-exome sequencing (WES) data. Using simulated human exome data, we compared SGVar with five variation-aware de novo assemblers and with BWA-MEM together with three haplotype- or local de novo assembly-based callers. SGVar outperforms the other assemblers in sensitivity and tolerance of sequencing errors. We recapitulated the findings on whole-genome and exome data from a Utah residents with Northern and Western European ancestry (CEU) trio, showing that SGVar had high sensitivity both in the highly divergent human leukocyte antigen (HLA) region and in non-HLA regions of chromosome 6. In particular, SGVar is robust to sequencing error, k-mer selection, divergence level and coverage depth. Unlike mapping-based approaches, SGVar is capable of resolving long-range phase and identifying large INDELs from WES, more prominently from WGS. We conclude that SGVar represents an ideal platform for WES-based variant discovery in highly divergent regions and across the whole genome.
Keywords: de Bruijn graph, exome sequencing, human leukocyte antigen, de novo assembly, string graph, variant discovery
Introduction
Complete and accurate detection of sequence variations is a key prerequisite for deciphering the genetic etiology of disease [1, 2]. Mapping-based approaches currently dominate the field of variant discovery through whole-genome sequencing (WGS) or whole-exome sequencing (WES), but with limitations in several key aspects. First, the human reference sequence is not perfect [3], containing misassemblies [4] and gaps [5]. In addition, some regions show high-sequence divergence or complex structural variations between haplotypes, or represent recent duplications with high-sequence similarity, causing ambiguity in short-read mapping [6, 7]. The 1000 Genomes Project Consortium estimated 171 Mb (5.5%) of the human genome reference GRCh37 as being inaccessible to the short-read sequencing technologies and mapping algorithms [8], including 130 Mb of recent segmental duplications between which a reliable differentiation is often difficult [9]. Importantly, some of the missing, highly divergent and inaccessible regions are associated with human disease [2]. Current standard practice is less effective in variation discovery from these regions [2]. For example, a recent study revealed that mapping bias led to 19% error rate in assigning single nucleotide polymorphism (SNP) genotypes for five critical genes in human leukocyte antigen (HLA), a highly divergent region associated with over 100 diseases [10].
Also, mapping efficiency often biases toward the reference allele in the presence of INDELs [6], reducing the detection of INDELs, especially large ones and those located within microsatellites [11, 12]. Even though numerous tools have been developed specifically for INDEL detection, an initial read mapping is still needed [13]. Finally, phasing facilitates the inference of causal mutations, as haplotype structure provides pertinent information as to whether two or more deleterious variants are located on the same allele [14]. In organ transplantation, phase information is useful for predicting the donor–recipient match [15]. Nevertheless, current mapping-based approaches report only unphased genotypes or limited phasing information from local de novo assembly-based callers. Without genotype information from trio or reference population, it will be difficult to infer haplotype and uncover transmitted variants in personal genomes.
The variation-aware de novo assemblers represent an attractive alternative. In principle, they are similar to the consensus sequence assemblers through the implementation of either de Bruijn graph [3] or string graph [4, 16]. In de Bruijn graph-based assembly paradigm, reads are split into substrings of length k called k-mers that are often error-corrected before used to build contigs. Thus, the performance of an assembler is correlated with the k-mer coverage rather than the base coverage, which highlights the importance of selecting appropriate k-mers, especially in cases when long reads are used. In string graph-based assembly, contigs represent paths of string graph built from overlapped reads. In this way, read coherence is fully retained in string graph but lost in de Bruijn graph. Most importantly, there is a key distinction between variation-aware and consensus sequence assemblers when applied to non-haploid organisms. While the former tries to preserve heterozygotes at polymorphic sites, the latter collapses them into consensus bases [17].
The variation-aware assemblers are capable of assembling reads into haplotype contigs [18], facilitating the identification of long INDELs and structural variations [2, 4, 12, 19]. In fact, a few packages have been developed for direct variant calling from the graph [3], local assembly (micro-assembly) using mapped reads [1, 12], or whole-genome de novo assembly [4, 20, 21].
Cortex is the first de novo assembly-based algorithm for direct variant calling from short reads. Cortex implements colored de Bruijn graph for single- or multi-sample variant calling, using the reference sequence if available [3]. In the graph, the nodes and edges are colored by the samples having them, thus allowing population-based variant discovery. Variants are called directly from the graph through the function ‘bubble-calling’ (for simple variants) or ‘path-divergence calling’ (for complex variants). However, the current version has a low sensitivity, with nearly 40% false-negative rate [1].
The String Graph Assembler (SGA) was originally developed for consensus sequence assembly of large genomes [16]. It creates overlapping graphs by first performing Burrows–Wheeler Transform and Ferragina–Manzini (FM) indexing of reads [16]. With the implementation of such a data structure, SGA can efficiently compute the path of string graph in large genomes, alleviating the requirement of high computing, a limitation inherent to string graph [22]. The ‘graph-diff’ module, a supplement to the SGA package, provides two modes to call variants directly from string graph and de Bruijn graph, respectively. However, the efficiency of this development version has yet to be assessed.
Scalpel [12] and DISCOVAR [1] perform local assembly based on de Bruijn graph. Reads are first mapped to the genome reference, and pairs with at least one mapped read are selected. Scalpel was designed solely for INDEL detection from WGS or WES data [12]. DISCOVAR was developed for assembling longer (250 bp) reads from WGS of polymerase chain reaction (PCR)-free libraries; its error correction and variant detection algorithms might not work well on the typical shorter (76–150 bp) reads that are often generated from libraries involving PCR amplification [1]. Importantly, their performance in highly divergent regions remains unknown.
Finally, Fermi and FermiKit (fermi2) are string graph-based whole-genome assemblers. Fermi implements Ferragina–Manzini DNA (FMD) index, a variety of FM index used in SGA, for forward–backward extension of DNA sequences [4]. It outputs unitigs that preserve SNPs, short INDELs and structural variations, without subsequent unitig mapping and variant calling. FermiKit is an updated version of Fermi [20], which uses the BFC algorithm [23] for less greedy error correction compared with the k-mer frequency used by Fermi, BWA-MEM [24] for mapping unitigs to the reference and HTSBox (https://github.com/lh3/htsbox) for variant detection. Nevertheless, both versions lack the flexibility needed for tweaking parameters when applied to WES or targeted gene panel sequencing data.
Except for Scalpel that was designed exclusively for INDEL detection in WES and WGS [12], the other packages described above were developed for WGS. Unlike in WGS, de novo assembly of WES data is complicated by the variability in coverage because of capture, sequencing and mapping bias [7, 25, 26]. De novo assembly of WES in complex regions is particularly challenging. Short-read assembly often implements de Bruijn graph, a data structure tending to be less effective compared with string graph when complex regions are assembled [27].
Understanding the advantages and limitations of individual methods is critical for optimizing variant discovery through de novo assembly. However, a detailed comparison of de novo assemblers is lacking in both WGS and WES. Using simulated exome data as well as real WES and WGS, we assessed SGVar, a chromosome-level de novo assembly pipeline we developed for haplotype-based variant discovery, along with five other de novo assembly-based and three mapping-based variant discovery methods. SGVar demonstrates excellence in both sensitivity and precision, which is largely independent of variability in coverage and divergence. SGVar can achieve long-range phasing over some of the most divergent HLA genes. It is powerful in detecting large INDELs, SNP clusters and complex structural variations.
Methods
Test data sets
We used simulated exome data and publicly available 76–150 bp whole-exome data from NA12878 (Supplementary Table S1). A 250 bp PCR-free WGS data set (∼60 × coverage) from a CEU trio including NA12878, NA12891 and NA12892 was also used, which was generated by the 1000 Genomes Project (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase3/data).
Simulated reads provide a simple system to evaluate the performance of variant discovery methods over key factors, such as sequencing error, coverage and divergence level, as both ‘true’ (pre-placed by simulation) and ‘false’ variants are known [28]. We generated two simulated data sets, with and without sequencing error, from exonic regions of chromosome 6. These regions were compiled from hg19 refGene exon annotation and the capture regions interrogated by four Agilent SureSelectXT All Exon kits [29]. Dwgsim (v0.1.11) was used to simulate 100 bp paired-end reads at each of the seven mutation rates between 0.05 and 15%, with eight coverage depths (20 – 200×) for error-free reads and four coverage depths (40 – 200×) for reads simulated with error (Supplementary Note).
Overall analytical strategy
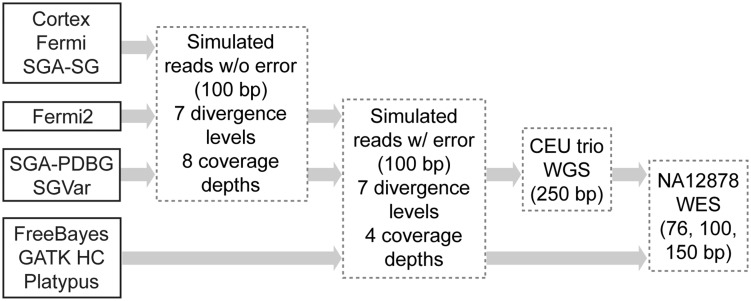
We developed SGVar, a string graph-based de novo assembly pipeline for variant discovery. It adopts the k-mer-based error-correction and string graph assembly modules from the SGA consensus assembly framework. We set up rules toward haplotype assembly by performing stringent read preprocessing, k-mer-based rather than reads overlap-based error correction, and by requiring perfect matches over overlapped bases in merging reads or sequences. To select high-quality reads for de novo assembly, we kept those if they were at least 65 bp long, contained only called bases (A, T, G and C) and no low-complexity sequences, and had Phred-scale quality scores of ≥20 (for 150 bp reads) or 30 (for 76 and 100 bp reads) for over half of the bases; the low-quality bases were then trimmed from the 3′ end until the base with a Phred-scale quality score of ≥30 was reached. More details are provided in the Supplementary Note. Here, we compared SGVar with five other de novo assemblers (Figure 1, Supplementary Table S2) and three mapping-based methods.
Figure 1.
Flowchart illustrating variant discovery methods and test data sets used in the study. All six de novo assemblers were first assessed on simulated reads without error across seven divergence levels (0.05, 0.1, 0.5, 1, 5, 10 and 15%) and eight coverage depths (20, 40, 60, 80, 100, 120, 160 and 200×). Three of them, SGA-PDBG, SGVar and fermi2, were further assessed using reads simulated with error (0.01% error rate at the 5′ and 1% at the 3′ end of reads) at the above seven divergence levels and four coverage depths (40, 60, 100 and 200×). SGA-PDBG and SGVar as representatives of de Bruijn graph-based and string graph-based assemblers were finally assessed on the CEU trio (NA12878, NA12891 and NA12892) WGS and eight NA12878 WES data (see Supplementary Table S1). Arrows point to the data sets on which a method was assessed. As controls, three callers together with BWA or BWA-MEM as the mapper were also tested on simulated reads with error and NA12878 WES data.
Of the six variation-aware assembly-based methods, the two methods in the SGA graph-diff module [16], which are SGA paired de Bruijn graph (SGA-PDBG) and SGA string graph (SGA-SG), and Cortex [3] perform direct variant calling from the assembly graph; Fermi [4], FermiKit [20] and SGVar assemble reads into contigs (unitigs), followed by contig mapping to the reference and variant calling. The initial assessment used simulated error-free reads. SGA-PDBG, SGVar and fermi2 that showed relatively better performance were then assessed using simulated reads with error. The first two were finally assessed on whole-genome data from a CEU (Utah residents with Northern and Western European ancestry) trio including NA12878, NA12891 and NA12892 and on whole-exome data from NA12878.
De novo assembly- and mapping-based variant detection
The detailed procedure and parameter settings for each of the six assemblers are provided in Supplementary Note. To use SGVar in the detection of variants from Chr6, NA12878 WES reads were mapped to the reference using BWA (v0.6.2). We kept pairs including at least one uniquely mapped read (to Chr6) with a mapping quality score of ≥20. As we focused on the highly divergent HLA region (see below), unmapped pairs were also included. These reads were preprocessed by trimming off low-quality bases from 3′ end and filtering out low-quality reads. The retained reads were assembled into haplotype contigs using the ‘sga assemble’ command. Contigs were filtered based on A-statistic score [30, 31] and contig size. Retained contigs were mapped to the hg19 reference, and variants were identified using SAMtools (v0.1.8, with the pileup function). We also tested SAMtools v0.1.12a with the mpileup function. The mpileup function missed some positions with SNPs or INDELs, which are real variants supported by the manual inspection of the contig–reference alignments. Key SGVar parameters were provided in Supplementary Table S3.
For SGA-SG and SGA-PDBG, variants were identified using the commands ‘sga graph-diff’ and ‘sga graph-diff - -paired-debruijn’, respectively. In using Cortex to call variants from simulated reads without error, we followed the instructions in the user manual (http://cortexassembler.sourceforge.net/cortex_var_user_manual.pdf). To assemble error-free reads with Fermi, we ran the commands ‘run-fermi.pl -e fermi’ and ‘make’, which perform k-mer frequency-based error correction and construct the FMD index. Unitigs were built and cleaned by the ‘fermi unitig’ and ‘fermi clean’ commands, respectively. Unitig mapping and variant detection followed the procedure used by SGVar. Finally, we used FermiKit (fermi2) to call variants from simulated reads with or without error, following the online user manual (https://github.com/lh3/fermikit).
De novo assembly-based approaches are expected to be effective in regions with high levels of divergence and heterozygosity, as a high density of SNPs should help resolve haplotypes. We thus focused on the highly divergent HLA region, which encodes antigen-presenting molecules that play essential roles in the immune system [32]. We analyzed the impact of coverage and divergence on assembly-based variant detection. Using SGA-PDBG and SGVar as representatives, we also sought to investigate how sequencing error and k-mer selection might impact de Bruijn graph-based versus string graph-based assemblers. Reads preprocessing is a key step in de novo assembly. For the data simulated with sequencing error, the dumpy base quality score does not fully reflect the characteristic error profile in real Illumina sequencing data. Considering this, we performed a simple preprocessing by arbitrarily trimming 10 bases from the 3′ end. For whole-genome data from the CEU trio and exome data from NA12878, systematic, quality-based read filtering and end trimming were applied (Supplementary Note).
As controls, BWA-backtrack [33] (referred to as BWA) or BWA-MEM (v0.7.12) [24] together with three callers were also tested on simulated reads with error and NA12878 exome data (Figure 1). We previously found that local realignment and base quality score recalibration, which are recommended by the Genome Analysis Toolkit (GATK) Best Practices [34, 35], had little benefit for the selected methods (Figure 1) [36]. Therefore, after duplicate marking, we only performed local realignment for NA12878 WES using the Mills and 1000G gold standard INDELs (Mills_and_1000G_gold_standard.indels.hg19.vcf.gz). Variants were identified with FreeBayes (v9.9.2-27) [37], GATK HaplotypeCaller (GATK HC) (v2.7-2) [34, 38] and Platypus (v0.5.2) [7], following our previous study [36].
Quality metrics
For simulated data, pre-placed variants were treated as the ‘true positive’, and the performance was assessed based on sensitivity, precision and overall genotype concordance, as previously described [29]. For NA12878 WGS and WES data, we estimated the sensitivity using the union of two public call lists below as the proxy for reference call set (‘true positive’). The high-confidence call set was generated from 11 WGS and 3 WES data using seven mappers and three callers [39], which is available at ftp://ftp-trace.ncbi.nih.gov/giab/ftp/data/NA12878/analysis/GIAB_integration/NIST_RTG_PlatGen_merged_highconfidence_v0.2.primitives.vcf.gz. The second call set was merged from three separate variant lists, which were generated by Cortex, DISCOVAR and BWA-MEM + GATK HC from 250 bp paired-end reads of a PCR-free genomic library [1]. The list is available at ftp://ftp.broadinstitute.org/pub/crd/DiscovarManuscript/vcf/.
Considering the difficulty in defining the exact INDEL boundary, for simulated data, INDELs identified within 10 bp of the pre-placed ones were counted as matches. For real WES and WGS, exact matching in genomic position is required. As the reference-quality sequence is not available for NA12878, we inferred the precision by (i) checking alignments around method-specific variants from WES; and (ii) checking long haplotype contigs assembled with the 2 × 250 bp WGS data in the CEU trio.
Results
The six de novo assemblers showed marked differences on simulated reads
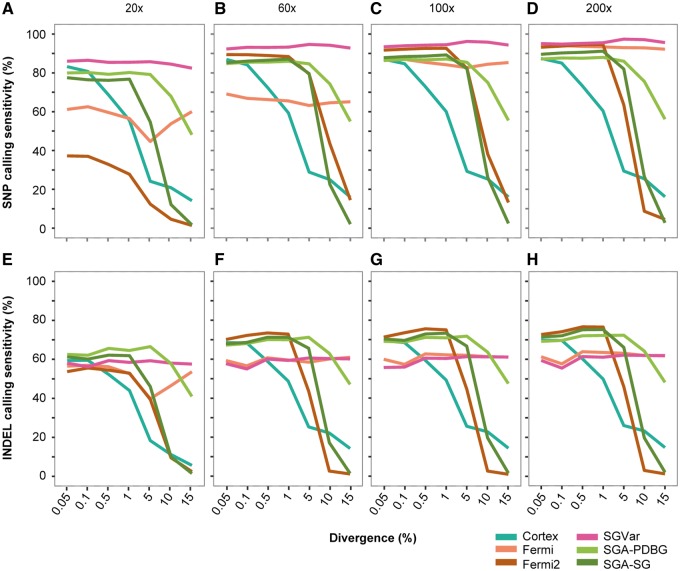
Of the six methods, SGVar, Fermi and fermi2 first assemble reads into contigs or unitigs, while Cortex and SGA graph-diff call variant directly from the assembly graph. We first assessed them (Supplementary Table S2) on simulated error-free data sets. We plotted sensitivity (Figure 2A–H, Supplementary Figure S1A–E), precision (Supplementary Figure S1F–J) and genotype concordance (Supplementary Figure S1K–O) over eight coverage depths and seven divergence levels.
Figure 2.
Sensitivity in SNP and INDEL detection by six de novo assemblers. (A–D) SNP sensitivity at four coverage depths. (E–H) INDEL sensitivity at four coverage depths. Reads were simulated without introducing error. In estimating the number of matches in INDELs, we extended the coordinates of the simulated INDELs by ± 10 bp before compared with the called INDELs. SGA-PDBG=SGA paired de Bruijn graph; SGA-SG=SGA string graph.
Overall, Cortex had the lowest SNP (Figure 2A–D) and INDEL (Figure 2E–H) calling sensitivity in a majority of the cases, supporting the previous finding [1]. It also had low precision (Supplementary Figure S1F–J) and genotype concordance (Supplementary Figure S1K–O) at high divergence. Remarkably, SGVar performed similarly well across different divergence levels, with the highest SNP calling sensitivity (median 94.3%, Supplementary Figure S1A–E) and genotype concordance (median 98.6%, Supplementary Figure S1K–O) across all coverage depths and divergence levels, although it had slightly lower precision (median 96.8%, Supplementary Figure S1F–J). Like SGVar, Fermi also had sensitivity largely independent of divergence level; in part this may be because of the fact that we used the same tools implemented in SGVar to map unitigs from Fermi and to identify variants. Fermi and fermi2 required high coverage to achieve a high sensitivity (Figure 2A–H), especially in SNP calling. Unlike Fermi, fermi2 showed a marked reduction in sensitivity at high divergence, resembling the two SGA graph-diff modes SGA-PDBG and SGA-SG (Figure 2A–H).
INDELs are more difficult to detect than SNPs. Also, it is challenging to define the exact boundary of INDELs, especially for those located within microsatellites [12]. Considering this limitation, we counted the called INDELs as true positives if they are within ±10 bp of simulated INDELs. At ≤5% divergence, SGVar and Fermi are generally less sensitive than fermi2, SGA-PDBG and SGA-SG (Figure 2E–H). However, the first two methods determined the exact boundary for a larger proportion (54.7–66.6%) of the identified INDELs, compared with 31.7–51.9% by the other three methods (Supplementary Table S4).
Error correction is a critical component in assembly-based variant calling. Based on the outcome from error-free reads, we further assessed SGVar, SGA-PDBG and fermi2 on reads simulated with error. We arbitrarily trimmed 10 bases from the 3′ portion of the reads that were simulated at a higher error rate than the 5′ bases. For each of the three methods, the sensitivity was comparable between error-free reads (Figure 2A–H) and reads simulated with error after the 10 bp trimming (Supplementary Figure S2A–H). In SNP calling, overall SGVar had the highest sensitivity across all the divergence levels (Supplementary Figure S2A–D). In INDEL calling, SGVar had the lowest sensitivity at low divergence but the highest sensitivity at high divergence (Supplementary Figure S2E–H).
For both SGA-PDBG and SGVar in SNP calling, the sensitivity was highly comparable among error-free reads and error-containing reads with and without 3′ end trimming (data not shown). However, sequencing errors reduced the precision, more obviously for SGA-PDBG at high coverage (Supplementary Figure S3). For 200× simulated reads with error, compared with error-free reads, SGA-PDBG and SGVar lost 23.9 and 16.2% in precision on average without 3′ end trimming versus 15.3 and 4.1% when 3′ end trimming was applied (Supplementary Figure S3). This is consistent with the finding that de Bruijn graph is more sensitive to sequencing errors than string graph [27]. Also, for both SGVar and SGA-PDBG, there was a bigger loss of precision at 200×, compared with the lower coverage data (Supplementary Figure S3). It has been found that the assembly quality would decrease once the sequencing depth goes too high [40–42], likely because of the accumulation of uncorrected sequencing errors.
In parallel, we also tested three haplotype (FreeBayes) or local de novo assembly-based callers (GATK HC and Platypus) on simulated reads with error. As we previously found [29], BWA is not suitable for mapping reads at high divergence (Figure 1). We thus focused on the methods with BWA-MEM as the mapper. In SNP calling, Platypus performed poorly at ≥1% divergence; the other two callers had a higher sensitivity (96.4% on average) than SGVar (92.9%) at ≤1% divergence but performed less well (44.8–84.3 versus 94.8%) at ≥5% divergence (Supplementary Figure S2A–D). Overall SGVar was less sensitive in INDEL detection (Supplementary Figure S2E–H), in particular when compared with GATK HC and Platypus that are known to be ideal for INDEL calling [29]. Considering the limitation of simulated reads, we further assessed SGVar and SGA-PDBG on real WGS and WES data.
Variant calling in real WGS data
We have investigated six de novo assembly-based variant calling methods on simulated reads. However, real sequencing reads are far more complex. In particular, they contain platform- and sequence context-specific bias (like GC bias) and error. Such bias may lead to low or no coverage in some regions [43]. For WES, the actual coverage also depends on the exome enrichment platforms [44]. In addition, de novo assembly is extremely sensitive to sequencing error. Without proper correction, sequencing error could markedly reduce the assembly quality [45, 46]. Sequencing error, read length, divergence level and coverage depth are key factors in de novo assembly-based variant calling. It will be critical to fully assess the performance of a method over these key factors using real sequencing data.
The most comprehensive call set in NA12878 was generated from 250 bp paired-end reads of a PCR-free genomic library [1], using two assemblers (Cortex and DISCOVAR) together with BWA-MEM + GATK HC, a widely used mapping-based pipeline. This list accounts for a majority of the reference call set compiled in this study. The same type of WGS data is available in a CEU trio (NA12878, NA12891 and NA12892), allowing us to first assess these methods and the quality of the reference call set on WGS before applied to WES data.
We first assessed SGVar and SGA-PDBG on the 250 bp PCR-free WGS from NA12878. SGVar had a much higher SNP and INDEL sensitivity than SGA-PDBG in the HLA region, particularly in the six highly divergent genes (HLA-A, HLA-B, HLA-C, HLA-DQA1, HLA-DQB1 and HLA-DRB1) (Table 1). For example, in identifying known INDELs, SGA-PDBG had a sensitivity of only ∼10%, compared with nearly 70% by SGVar. While SGVar performed similarly between simulated reads (Figure 2E–H, Supplementary Figure S2E–H) and the 250 bp WGS (Table 1) in INDEL calling, SGA-PDBG was much worse in WGS (Table 1) than in simulated reads (Figure 2E–H, Supplementary Figure S2E–H). Obviously, SGA-PDBG is not suitable for INDEL detection.
Table 1.
Percentage of variant calling sensitivity from NA12878 WGS
| Type | SGVar |
SGA-PDBG |
||
|---|---|---|---|---|
| HLA | 6 genes | HLA | 6 genes | |
| Known SNP | 94.4 | 97.2 | 88.5 | 85.8 |
| Novel SNP | 61.8 | 79.3 | 47.9 | 54.1 |
| Total | 92.2 | 96 | 85.7 | 83.7 |
| Known INDEL | 69.7 | 67.6 | 8.9 | 14.5 |
| Novel INDEL | 39 | 25.7 | 8.6 | 10.8 |
| Total | 55 | 49.4 | 9.3 | 14 |
Note: Contigs generated by SGVar were mapped to the hg19 reference using BWA-MEM. Known variants are those in dbSNP v138. The 6 genes are HLA-A, HLA-B, HLA-C, HLA-DQA1, HLA-DQB1 and HLA-DRB1. The 250 bp PCR-free WGS is from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase3/data.
DISCOVAR is a de Bruijn graph-based assembler developed specifically for variant detection from long (250 bp) paired-end reads of PCR-free library [1]. To understand how SGVar performs relative to DISCOVAR, we intersected the list of known variants (SNPs and INDELs) identified by SGVar with the two public lists by DISCOVAR and BWA-MEM + GATK HC [1]. In the HLA region, 39.3% (10 266 of 26 149) of the known variants identified by SGVar were not in the DISCOVAR list, compared with only 7.7% (2005 of 26 141) not in the GATK HC list (Table 2). The percentages of SGVar-specific calls were nearly doubled in the six highly divergent HLA genes (Table 2). The results suggested that DISCOVAR is much less sensitive than SGVar in the HLA region.
Table 2.
Shared and unique known variants from NA12878 WGS and WES
| Data | Type | Region | SGVar versus DISCOVAR |
SGVar versus GATK HC |
||||
|---|---|---|---|---|---|---|---|---|
| Shared | SGVar only | DISCOVAR only | Shared | SGVar only | GATK HC only | |||
| WGS | SNP | HLA | 14 985 | 9701 | 423 | 22 830 | 1857 | 1178 |
| SNP | 6 genes | 660 | 2169 | 7 | 2458 | 369 | 64 | |
| INDEL | HLA | 898 | 565 | 285 | 1306 | 148 | 545 | |
| INDEL | 6 genes | 23 | 112 | 8 | 111 | 20 | 56 | |
| WES | SNP | HLA | 614 | 346 | 28 | 860 | 98 | 53 |
| SNP | 6 genes | 159 | 248 | 6 | 361 | 45 | 26 | |
| SNP | Non-HLA | 1320 | 105 | 73 | 1358 | 67 | 66 | |
| INDEL | HLA | 15 | 11 | 5 | 21 | 5 | 11 | |
| INDEL | 6 genes | 2 | 9 | 2 | 8 | 3 | 8 | |
| INDEL | Non-HLA | 79 | 27 | 32 | 80 | 26 | 28 | |
Note: Known variants (in dbSNP v138) detected by SGVar were compared with two public call sets identified by DISCOVAR and BWA-MEM with GATK HC [1]. The 6 HLA genes (6 genes) are HLA-A, HLA-B, HLA-C, HLA-DQA1, HLA-DQB1 and HLA-DRB1. The WGS data are from 250 bp paired-end sequencing of a PCR-free library (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase3/data). For WES, the average number of variants identified from the two 150 bp data sets (Supplementary Table S1) is shown.
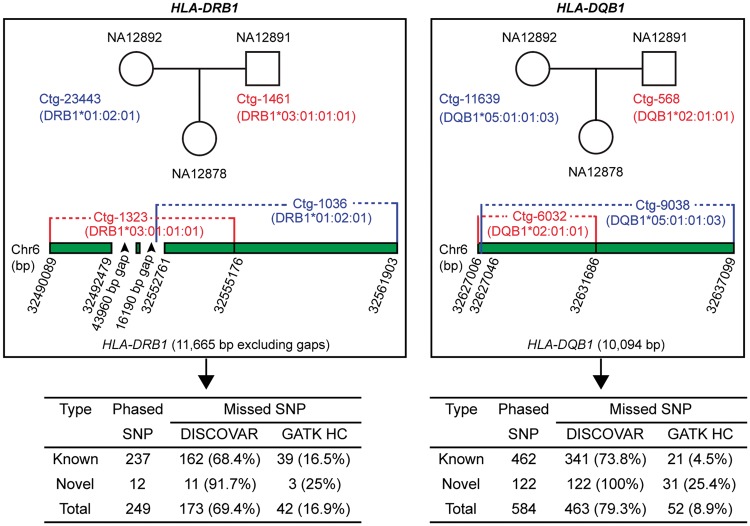
We next assessed whether SGVar can achieve long-range phasing, using PCR-free 250 bp WGS data from the CEU trio. We focused on HLA-DQB1 and HLA-DRB1, two of the most highly divergent HLA genes. We first used BLAT to identify two longest contigs from NA12878 (daughter) that best matched the alleles from each of the two genes in the international ImMunoGeneTics (IMGT)/HLA database (ftp://ftp.ebi.ac.uk/pub/databases/ipd/imgt/hla/fasta/). The identified four contigs (4794–10 094 bp) were used to pull out the best hits from contigs assembled in NA12891 (father) and NA12892 (mother) via BLAT. For both genes, SGVar successfully separated the two alleles in NA12878 (Table 3, Figure 3). For HLA-DRB1, allele DRB1*03:01:01:01 (represented by contig Ctg-1323) in NA12878 is from NA12891 (Ctg-1461); the two contigs perfectly matched HLA haplotype 6_qbl_hap6 but showed only 93% similarity with the hg19 reference. The second allele DRB1*01:02:01 (Ctg-1036) is from NA12892 (Ctg-23443), with the first 7439 bp from Ctg-1036 fully matching Ctg-23443. For HLA-DQB1, allele DQB1*02:01:01 (matches 6_qbl_hap6) is from NA12891. The second allele DQB1*05:01:01:03 (Ctg-9038, 10 042 bp) fully matched Ctg-11639 (14 010 bp) from NA12892. These results strongly suggest that SGVar can achieve long-range phasing in highly divergent regions.
Table 3.
SGVar assembly of HLA-DQB1 and HLA-DRB1 from WGS in a CEU trio
| Sample | Gene | Contig | Length (bp) | Best hit in IMGT/HLA |
Best hit in eight haplotypes |
||||
|---|---|---|---|---|---|---|---|---|---|
| Allele | Match (bp) | Mismatch (bp) | Best hit | Span (bp) | Sim (%) | ||||
| NA12892 | HLA-DQB1 | Ctg-11639 | 14 010 | DQB1*05:01:01:03 | 7090 | 0 | 6 | 14 020 | 97.4 |
| NA12891 | HLA-DQB1 | Ctg-4703 | 5373 | DQB1*06:02:01:01 | 4556 | 0 | 6 | 5373 | 100.0 |
| NA12878 | HLA-DQB1 | Ctg-9038 | 10 042 | DQB1*05:01:01:03 | 7090 | 0 | 6 | 10 054 | 96.5 |
| NA12891 | HLA-DQB1 | Ctg-568 | 4724 | DQB1*02:01:01 | 3963 | 0 | 6_qbl_hap6 | 4724 | 100.0 |
| NA12878 | HLA-DQB1 | Ctg-6032 | 4794 | DQB1*02:01:01 | 3985 | 0 | 6_qbl_hap6 | 4794 | 100.0 |
| NA12892 | HLA-DRB1 | Ctg-23443 | 11 257 | DRB1*01:02:01 | 3654 | 1 | 6 | 13 910 | 99.2 |
| NA12891 | HLA-DRB1 | Ctg-14574 | 10 605 | DRB1*15:01:01:02 | 6292 | 0 | 6 | 10 605 | 100.0 |
| NA12878 | HLA-DRB1 | Ctg-1036 | 10 094 | DRB1*01:02:01 | 6305 | 5 | 6 | 10 087 | 97.9 |
| NA12892 | HLA-DRB1 | Ctg-23796 | 9266 | RB1*01:02:01 | 7764 | 6 | 6 | 9615 | 96.6 |
| NA12891 | HLA-DRB1 | Ctg-1461 | 5305 | DRB1*03:01:01:01 | 5305 | 0 | 6_qbl_hap6 | 5305 | 100.0 |
| NA12878 | HLA-DRB1 | Ctg-1323 | 5004 | DRB1*03:01:01:01 | 5004 | 0 | 6_qbl_hap6 | 5004 | 100.0 |
Note: Contigs were generated at 31-mer. BLAT was used to search contig sequences against the IMGT/HLA database (ftp://ftp.ebi.ac.uk/pub/databases/ipd/imgt/hla/fasta/) and the eight different haplotypes. The 250 bp PCR-free WGS is from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase3/data. Note that the first 7439 bp from Ctg-1036 perfectly match Ctg-23443 in HLA-DRB1 and Ctg-9038 perfectly matches a 10 042 bp region from Ctg-11639 in HLA-DQB1. NA12878, daughter; NA12892, mother; NA12891, father; Sim, similarity.
Figure 3.
SGVar phasing of SNPs in HLA-DRB1 and HLA-DQB1. Haplotype contigs were assembled from 250 bp PCR-free WGS in a CEU trio. Contig sequences were compared with the IMGT/HLA database (ftp://ftp.ebi.ac.uk/pub/databases/ipd/imgt/hla/fasta/) and with hg19 reference sequence via BLAT (Table 3), by which the matching between alleles from NA12878 and those in NA12892 or NA12891 was inferred. To identify shared and method-specific SNP calls, phased SNPs in the indicated contigs from NA12878 were intersected with two public variant lists identified by DISCOVAR and GATK HC, also from 250 bp PCR-free WGS [1].
A closer examination of a 11.7 kb phased region within HLA-DRB1 of NA12878 revealed that DISCOVAR missed nearly 70% (173 of 249) of the SNPs carried by alleles DRB1*03:01:01:01 and DRB1*01:02:01 and BWA-MEM + GATK HC missed 17% (42 of 249) of them (Figure 3). In another 10.1 kb phased region from HLA-DQB1, DISCOVAR and BWA-MEM + GATK HC missed 79.3% (463 of 584) and 9% (52 of 584) of the SNPs identified by SGVar (Figure 3). Therefore, while the public list is likely of high quality in ordinary regions [1], it is less complete in the HLA region. As reliable assessment of variant discovery methods strongly depends on the quality of the reference call set, there is a need to generate a more complete and accurate reference call set for the HLA region.
Using WGS data from the CEU trio, we found that some contigs were partially aligned to the reference by BWA-MEM, showing stretches of hard- or soft-clipped bases at the contig ends (represented by ‘H’ or ‘S’ in the CIGAR string of the BAM file). We argued that some of the clipped bases likely represent extremely divergent bases, large INDELs or structural variation. Based on extensive manual inspection of alignments between contigs from the six HLA genes (Table 1) in NA12878 and the human genome reference as well as the eight HLA haplotype sequences available in the University of California, Santa Cruz genome browser, we tested a two-tier contig mapping strategy aiming to improve alignment accuracy. All contigs were first mapped by BWA-MEM, and those showing hard- or soft-clipping were remapped by BLAT. As 99.4% of the known INDELs [in Single Nucleotide Polymorphism Database (dbSNP) v138] from chromosome 6 have a length of 20 bp or shorter, the known INDELs identified by BWA-MEM and BWA-MEM + BLAT were mostly <20 bp in size (Supplementary Figure S4A and B). However, remapping with BLAT identified large (>90 bp) novel INDELs missed by BWA-MEM alone (Supplementary Figure S4C and D).
Variant calling in real WES data
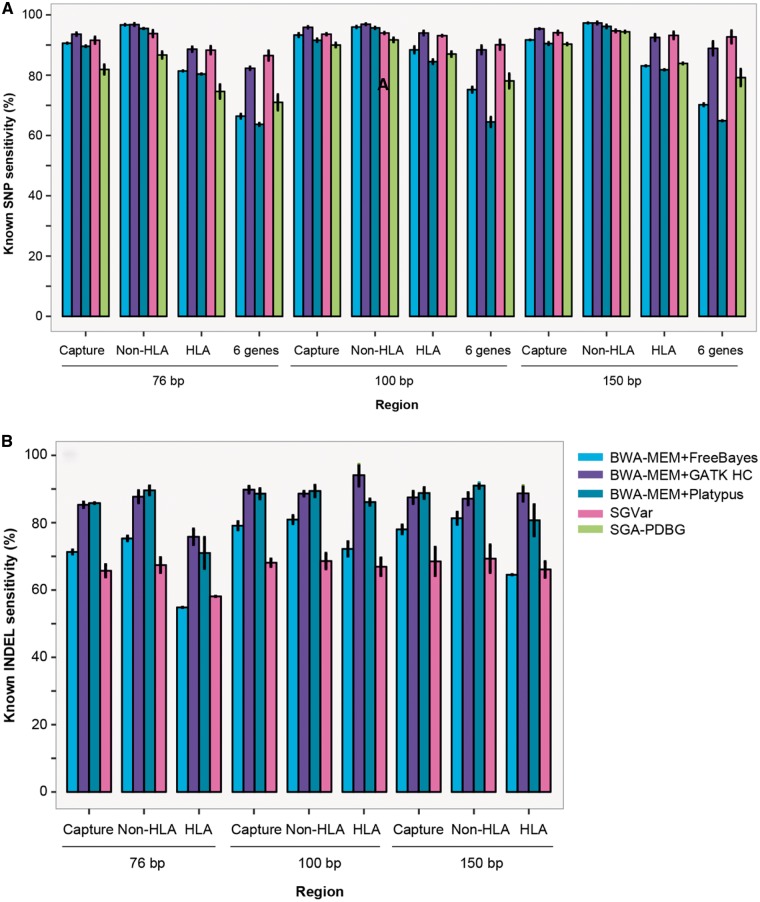
To develop a robust workflow across different capture platforms, read lengths and genomic regions, we finally assessed SGVar and SGA-PDBG, together with the three mapping-based pipelines, on eight NA12878 WES data sets. These data sets were generated using Roche (100 bp) and Illumina (76 and 150 bp) exome capture kits (Supplementary Table S1). Analysis of the 150 bp WES data indicated that DISCOVAR missed a significant portion of variants in the HLA region, as demonstrated in the WGS data (Table 2). In contrast, in the non-HLA region, over 90% of the known variants were shared between DISCOVAR and SGVar. Therefore, unlike SGVar that performs well across a wide range of divergence (Figure 2A–H, Figure 4A and B, Supplementary Figure S2A–H), DISCOVAR is much less sensitive in the HLA region than in the non-HLA regions (Table 2). On the other hand, SGVar is more sensitive than SGA-PDBG (Figure 4A and B), especially in INDEL detection (data not shown), consistent with the finding from WGS (Table 1).
Figure 4.
Sensitivity of known variant detection in NA12878 WES. (A) Known SNP sensitivity. (B) Known INDEL sensitivity. The mean and SD were plotted for WES data listed in Supplementary Table S1. Chr6 variants were identified by SGVar, SGA-PDBG and three mapping-based approaches and separated into known (in dbSNP v138) and novel ones. In mapping-based approaches, reads were mapped to hg19 reference using BWA-MEM. We required exact match in genomic coordinates when calculating INDEL sensitivity. SGA-PDBG had a low sensitivity in INDEL detection and was not shown here. The six HLA genes (‘6 gene’) are HLA-A, HLA-B, HLA-C, HLA-DQA1, HLA-DQB1 and HLA-DRB1.
We next compared SGVar with the three mapping-based methods. In SNP calling, SGVar was slightly less sensitive (1.5–3.0% lower) in the non-HLA regions but generally more sensitive in the HLA region (Figure 4A), consistent with the trend over different divergence levels observed in simulated data (Supplementary Figure S2A–D). For example, in the six HLA genes, SGVar was ∼3% more sensitive than GATK HC and 15–28% more sensitive than FreeBayes and Platypus (Figure 4A). In INDEL calling, SGVar was less sensitive in both HLA and non-HLA regions (Figure 4B). We found that some of the INDELs detected by SGVar extended beyond of the capture regions. As we limited the analysis to the capture regions, the chance of identifying long INDELs would have been reduced. Indeed, by targeting the entire chromosome 6 rather than just the capture regions, SGVar identified large INDELs up to 800 bp that were missed by the mapping-based methods. The result supports the notion that de novo assembly-based approach should be able to identify large (≥30 bp) INDELs [47]. Here, SGVar sensitivity is likely underestimated. In the HLA region, the reference call set is less complete (Figure 3). Also, the vast majority of the variants in the reference were generated by BWA-MEM together with GATK HC, which biases against SGVar in the comparison, as the three mapping-based approaches also used BWA-MEM as the mapper.
On the other hand, in SNP calling from the HLA region, SGVar was about 5% more sensitive on the 100 and 150 bp data sets, compared with the 76 bp data set (Figure 4A). The difference was much less obvious in the non-HLA regions, indicating that longer reads favor SGVar more in highly divergent regions. Next, we focused on one of the 150 bp WES data FC1_NA12878_S1_S4 (Supplementary Table S1). Compared with the public call set, SGVar had 80 unique known SNPs but missed 56 in the HLA region versus 63 and 75 in the non-HLA region. To get a rough estimation of the precision of SGVar, we assessed authenticity of the above 80 known SNPs by intersecting with those identified from 250 bp WGS in the CEU trio and further checking the BWA-MEM alignments between WES reads and the hg19 reference in the Integrative Genomics Viewer [48].
Of the 80 known SNPs unique to SGVar, about half (41) were also identified by SGVar in the WGS data from both NA12878 and one of the parents, supporting the fidelity of these calls (Supplementary Table S5). The 41 SNPs were from long (2.3–10 kb, WGS) haplotype contigs assembled in four of the highly divergent genes, including HLA-DQA1 (9), HLA-DQB1 (4), HLA-DRB1 (15) and HLA-DRB5 (13). The other 39 SNPs were missed in NA12878 WGS, including 7 that were fully supported by the alignments in WES. We thus believed that 60% (48 of 80) of the SGVar-specific known SNPs represent true variants. The remaining 32 SNPs are likely low-confidence calls or false positives. Nine of them were supported by only two reads (1), flanked by INDELs in low-complexity regions (2) or called from a contig erroneously mapped to the reference (6). The other 23 (32−9) were mostly supported by low-quality bases, with 16 in gene MUC21. These low-quality bases had >10× coverage and were located in the middle of the reads, remaining the same after quality-based 3′ end trimming and error correction. Apparently, the mapping-based approaches would not identify these SNPs, as they only consider bases with Q ≥ 17. Therefore, these calls may represent context-specific sequencing errors in Illumina reads [49].
We further checked the 56 known SNPs in the HLA region that are present in the public call set but missed by SGVar (Supplementary Table S5). Twenty-nine likely represent false positives in the public call set, as SGVar also missed them in NA12878 WGS. The other 27 that were identified by SGVar from the WGS but not from WES are probably false negatives. Of the 27, 12 were identified from WES with 31-mer, which we did not include in the final SGVar call list that was generated using 51-mer to 91-mer. Nine SNPs were in regions with no (3) or only 2–6× total coverage (6), likely reflecting low capture affinity. The other six were in regions with reasonable (≥ 4 ×) coverage for the alternative allele.
For the known INDELs detected from FC1_NA12878_S1_S4 in the capture regions, SGVar had 32 unique INDELs and missed 40 in the public call set. We manually checked the alignments around some of these INDELs (Supplementary Table S5). Of the 32, 19 were also identified by the mapping-based approaches and three were supported by the alignments (≥7 supporting reads). For another six, the two haplotype contigs differed at the polymorphic sites, with one haplotype having an INDEL and the second haplotype having an SNP (3), or one haplotype having an insertion and the second haplotype having a deletion (3). The remaining four INDELs showed strong allele bias, with only 2–5 supporting reads (of >50). Therefore, we believed that nearly 90% (28 of 32) of the SGVar-specific known INDELs are true variants.
On the other hand, SGVar missed 40 known INDELs in the public call set (Supplementary Table S5). Nineteen of them likely represent WGS-specific calls or false positives in the public call set, including 12 that overlapped the contigs without an INDEL and 7 from regions where no contig was assembled. For another five, SGVar reported the INDELs at a slightly different location or with a different genotype compared with the public call set. The remaining 16 were identified in contigs assembled at individual k-mers; however, they were not reported in the final list after merging the calls from 51-mer to 91-mer by the GATK tool ‘CombineVariants’, suggesting a need for improving the consolidation of INDEL calls. Collectively, our analysis supports the power of de novo assembly in the detection of INDELs [47], especially those in complex genomic regions.
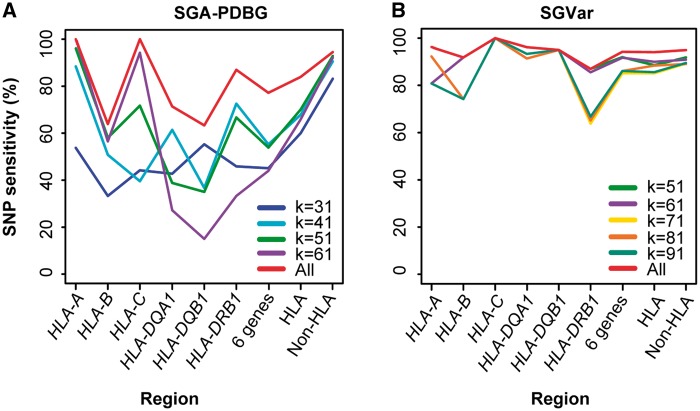
SGVar is less impacted by the k-mer selection than SGA-PDBG
In de Bruijn graph-based assembly, k-mer represents the error correction and overlap parameter. In string graph-based assembly, k-mer is only used in error correction. Therefore, k-mer as a key factor may impact the two approaches differently in variant discovery. To gain a better understanding of the possible differences, we tested different k-mers for SGA-PDBG and SGVar, using 250 bp WGS and 150 bp WES data (FC1_NA12878_S1_S4) from NA12878 (Figure 5A and B, Supplementary Figure S5A and B).
Figure 5.
Known SNP sensitivity in WES at different k-mers. The 150 bp WES data (FC1_NA12878_S1_S4, Supplementary Table S1) were used. (A) SGA-PDBG. (B) SGVar. All: the union of known SNPs identified from all k-mers; 6 genes: HLA-A, HLA-B, HLA-C, HLA-DQA1, HLA-DQB1 and HLA-DRB1.
In SGA-PDBG assembly of the HLA region from WGS, we observed a maximum difference of 13% in known SNP sensitivity among the four k-mers (Supplementary Figure S5A). In contrast, the variability was over 4-fold smaller for SGVar (Supplementary Figure S5B). A similar pattern was observed in WES (Figure 5A and B). We further checked overlap of known SNPs identified from WES at different k-mers (Supplementary Figure S6A–D). For SGVar, 81% (784 of 968) of the known SNPs were shared in the HLA region (Supplementary Figure S6A) versus only 38.6% (404 of 1046) for SGA-PDBG (Supplementary Figure S6C). Their difference dropped to only 6% in the non-HLA regions (Supplementary Figure S6B and D). These results indicate that, for SGA-PDBG, multiple k-mers are needed to maximize sensitivity in highly divergent regions like HLA, which will require more computing and extra effort to consolidate different variant lists. In this aspect, the less dependence on k-mer selection makes SGVar particularly appealing.
Discussion
De novo assembly is increasingly used in haplotype construction from WGS, more suitable for the detection of large INDELs and complex variants. In these instances, mapping-based approaches are less effective. Current assemblers often implement de Bruijn graph, a data structure operating on k-mers rather than on reads [50]. We developed SGVar, a string graph-based, chromosome-scale de novo assembly pipeline for variant discovery. To ensure haplotype assembly and minimize noise, we have implemented highly stringent reads filtering, reads trimming, k-mer-based error correction and contig filtering in SGVar. Using both reads mapped to Chr6 and unmapped reads, SGVar achieved high sensitivity and precision in WES. It outperformed all tested de Bruijn graph- and string graph-based assemblers, especially in highly divergent regions like HLA. There are over 100 highly divergent regions in the human genome [2, 51], from which variant discovery has been challenging. SGVar should be most effective in those regions.
The performance of SGVar is generally independent of divergence and coverage, making it superior to DISCOVAR, Cortex, SGA graph-diff and fermi2 that have a much lower sensitivity at high divergence, and to Fermi that requires high-sequencing depth. Critically, SGVar could achieve long-range phasing up to several kilobases and detect large INDELs in WES, more notably in WGS owing to the continuity in read coverage. In NA12878 WGS, a 10.1 kb region from HLA-DQB1 harbors 584 phased SNPs, and another 11.7 kb region within HLA-DRB1 contains 249 phased SNPs. This finding has significant implication in biomedical studies, as there is increasing evidence that some diseases are associated with multiple linked variants [14, 52, 53]. Although short reads carry limited phase information if they contain two or more variation sites [54], long-range phasing through de novo assembly increases the chance of identifying variants linked to complex diseases. In this aspect, the incorporation of mate-pair or chromosome conformation capture (3C)-based sequencing data into the assembly will help resolve haplotypes over large regions [14, 55]. We expect an increasing role for haplotype construction in genomic-based disease diagnosis [53].
Existing methods perform either whole-genome assembly or local assembly within a few kilobases. In local assembly, only pairs with at least one read mapped to particular regions are included [1, 12]. We argue that some of the unmapped pairs are likely derived from regions of high divergence, complex variation or large INDELs, or from regions currently missed in the reference. It is estimated that 5–40 Mb euchromatic sequences are missed from the human reference genome [2], including some that are associated with complex disease [56–58]. To circumvent this limitation, SGVar performs chromosome-level assembly, which takes pairs with one or both reads mapped to a given chromosome and unmapped pairs. The inclusion of unmapped pairs should be particularly helpful for de novo assembly in highly divergent regions. Also, the chromosome-level assembly approach provides the flexibility to target one or a few chromosomes, or to speed up whole-genome assembly through parallel computing.
The SGVar pipeline is designed for chromosome-level assembly. However, some studies may focus on specific regions or genes, like key genes within the HLA region. Also, targeted resequencing of clinical samples is widely used in biomedical studies. Thus, implementing a separate module for regional assembly will extend the application of SGVar. On the other hand, the detection of INDELs, especially large ones, has been challenging for most methods [12, 59]. We found that remapping of contigs with hard- or soft-clipping by BLAT improves the detection of large INDELs. Further, effort is needed to optimize SGVar in INDEL detection.
The power of de novo assembly depends on factors like read length and quality, sequencing depth, assembly algorithms and parameter settings [2, 60]. One of the key parameters is k-mer size. In de Bruijn graph, k-mer represents the overlap parameter. The choice of a longer k-mer, which will reduce the k-mer coverage, improves the precision at the cost of sensitivity. Conversely, a shorter k-mer would result in more false positives. It is recommended that de Bruijn graph-based assemblers should run multiple k-mers to increase the sensitivity, as implemented in Cortex [3]. However, extra effort is required to consolidate the multiple variant sets into a unified list. In string graph-based assemblers like SGVar, the k-mer is used in error correction but not in the assembly step. Despite the importance, the setting of key parameters is often less well justified. Therefore, a systematic assessment of k-mer size and other key parameters will be needed to provide recommendations for end user, which should enhance the future application of de novo assemblers in variant discovery.
Conclusion
Complete and accurate variant detection is critical, as we move into precision-based medical practice. We have developed the SGVar pipeline for variant discovery from WES data. SGVar took advantage of the SGA data structure and implemented key modules toward haplotype assembly. By splitting mapped reads over individual chromosomes and also including unmapped reads in the assembly, SGVar is memory efficient and outperforms other de novo assembly-based approaches, especially in regions of high genomic complexity like HLA. Specifically, SGVar is robust to sequencing errors, coverage and divergence variability and k-mer selection. Though limited by the design to target only exons and some other important genomic regions in WES, SGVar can detect large INDELs and achieve long-range phasing, which is more striking in WGS. However, the other de novo assemblers are less powerful at high divergence or low-sequencing depth. Future work will be needed to develop modules for local assembly of selected regions, such as those in the targeted gene sequencing panels, and to enhance SGVar in INDEL detection.
Key Points
Mapping-based approaches represent the major choice in variant discovery through WES or WGS, but they are less sensitive to large INDELs and variants in complex genomic regions.
We have developed SGVar, a string graph-based de novo assembly pipeline for variant discovery. In contrast with mapping-based approaches that provide little phasing information, SGVar achieves long-range phasing in the most divergent HLA region.
Comparison of SGVar with another five variation-aware de novo assemblers has revealed that SGVar is robust to sequencing error, k-mer size and variability in coverage and divergence.
Unlike SGVar, Cortex, fermi2 and SGA graph-diff are less sensitive at high divergence, and Fermi is less sensitive at low-sequencing depth.
NA12878 has been widely used in benchmark study of variant calling. While the public call set from NA12878 is of high quality in ordinary regions, it is less accurate and complete in the HLA region.
Supplementary Material
Funding
The National Institutes of Health (grant number CA118444 to S.L.S.) and Mayo CCaTS (grant number UL1TR000135 to S.L.S.).
Shulan Tian is a bioinformatics specialist in the Division of Biomedical Statistics and Informatics, Department of Health Sciences Research, Mayo Clinic, USA. She recently completed her PhD thesis ‘Identification of genetic variation in highly divergent regions using whole exome sequencing’ under the supervision of Susan Slager.
Huihuang Yan is an assistant professor in the Division of Biomedical Statistics and Informatics, Department of Health Sciences Research, Mayo Clinic, USA. His research focuses on cancer genomics and epigenetics, and the development of methods for analyzing next-generation sequencing data.
Eric W. Klee is an assistant professor in the Division of Biomedical Statistics and Informatics, Department of Health Sciences Research, and associated director of the Center for Individualized Medicine Bioinformatics Program, Mayo Clinic, USA. His research focused on the clinical applications of next-generation sequencing in diagnostic testing and the elucidation of genetic causes of rare Mendelian disease.
Michael Kalmbach is a senior analyst/programmer in the Division of Information Management and Analytics, Department of Information Technology, Mayo Clinic, USA. He collaborates with the Division of Biomedical Statistics and Informatics to develop and support bioinformatics pipelines and other tools used to analyze next-generation sequencing data.
Susan L. Slager is a professor and chair in the Division of Biomedical Statistics and Informatics, Department of Health Sciences Research, Mayo Clinic, USA. She studies the genetic basis of lymphoma and develops algorithms for variant discovery through next-generation sequencing.
References
- 1. Weisenfeld NI, Yin S, Sharpe T, et al.Comprehensive variation discovery in single human genomes. Nat Genet 2014;46:1350–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chaisson MJ, Wilson RK, Eichler EE.. Genetic variation and the de novo assembly of human genomes. Nat Rev Genet 2015;16:627–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Iqbal Z, Caccamo M, Turner I, et al.De novo assembly and genotyping of variants using colored de Bruijn graphs. Nat Genet 2012;44:226–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li H. Exploring single-sample SNP and INDEL calling with whole-genome de novo assembly. Bioinformatics 2012;28:1838–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chaisson MJ, Huddleston J, Dennis MY, et al.Resolving the complexity of the human genome using single-molecule sequencing. Nature 2015;517:608–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lunter G, Goodson M.. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res 2011;21:936–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rimmer A, Phan H, Mathieson I, et al.Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet 2014; 46:912–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abecasis GR, Auton A, Brooks LD, et al.An integrated map of genetic variation from 1,092 human genomes. Nature 2012;491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bishara A, Liu Y, Weng Z, et al.Read clouds uncover variation in complex regions of the human genome. Genome Res 2015;25:1570–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brandt DY, Aguiar VR, Bitarello BD, et al.Mapping bias overestimates reference allele frequencies at the HLA genes in the 1000 genomes project phase I data. G3 2015; 5:931–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu TD, Nacu S.. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 2010; 26:873–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Narzisi G, O'Rawe JA, Iossifov I, et al.Accurate de novo and transmitted INDEL detection in exome-capture data using microassembly. Nat Methods 2014;11:1033–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sudmant PH, Rausch T, Gardner EJ, et al.An integrated map of structural variation in 2,504 human genomes. Nature 2015;526:75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Selvaraj S, Dixon JR, Bansal V, et al.Whole-genome haplotype reconstruction using proximity-ligation and shotgun sequencing. Nat Biotechnol 2013;31:1111–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Petersdorf EW, Malkki M, Gooley TA, et al.MHC haplotype matching for unrelated hematopoietic cell transplantation. PLoS Med 2007;4:e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Simpson JT, Durbin R.. Efficient de novo assembly of large genomes using compressed data structures. Genome Res 2012;22:549–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bodily PM, Fujimoto M, Ortega C, et al.Heterozygous genome assembly via binary classification of homologous sequence. BMC Bioinformatics 2015;16 (Suppl 7):S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang WY, Hormozdiari F, Wang Z, et al.Leveraging reads that span multiple single nucleotide polymorphisms for haplotype inference from sequencing data. Bioinformatics 2013; 29:2245–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics 2014; 30:2843–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li H. FermiKit: assembly-based variant calling for Illumina resequencing data. Bioinformatics 2015;31:3694–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Simpson JT, Pop M.. The theory and practice of genome sequence assembly. Annu Rev Genomics Hum Genet 2015; 16:153–72. [DOI] [PubMed] [Google Scholar]
- 22. Myers EW. The fragment assembly string graph. Bioinformatics 2005;21 (Suppl 2):ii79–85. [DOI] [PubMed] [Google Scholar]
- 23. Li H. BFC: correcting Illumina sequencing errors. Bioinformatics 2015;31:2885–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv2013:1303.3997.
- 25. Lupski JR, Gonzaga-Jauregui C, Yang Y, et al.Exome sequencing resolves apparent incidental findings and reveals further complexity of SH3TC2 variant alleles causing Charcot-Marie-Tooth neuropathy. Genome Med 2013;5:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meynert AM, Ansari M, FitzPatrick DR, et al.Variant detection sensitivity and biases in whole genome and exome sequencing. BMC Bioinformatics 2014;15:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li Z, Chen Y, Mu D, et al.Comparison of the two major classes of assembly algorithms: overlap-layout-consensus and de-Bruijn-graph. Brief Funct Genomics 2012;11:25–37. [DOI] [PubMed] [Google Scholar]
- 28. Olson ND, Lund SP, Colman RE, et al.Best practices for evaluating single nucleotide variant calling methods for microbial genomics. Front Genet 2015;6:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tian S, Yan H, Neuhauser C, et al.An analytical workflow for accurate variant discovery in highly divergent regions. BMC Genomics 2016;17:703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Myers EW, Sutton GG, Delcher AL, et al.A whole-genome assembly of Drosophila. Science 2000;287:2196–204. [DOI] [PubMed] [Google Scholar]
- 31. Kelley DR, Salzberg SL.. Detection and correction of false segmental duplications caused by genome mis-assembly. Genome Biol 2010;11:R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gough SC, Simmonds MJ.. The HLA region and autoimmune disease: associations and mechanisms of action. Curr Genomics 2007;8:453–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li H, Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. DePristo MA, Banks E, Poplin R, et al.A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Van der Auwera GA, Carneiro MO, Hartl C, et al.From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics 2013;43:11.10.1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tian S, Yan H, Kalmbach M, et al.Impact of post-alignment processing in variant discovery from whole exome data. BMC Bioinformatics 2016;17:403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. 2014. http://arxiv.org/abs/1207.3907v2
- 38. McKenna A, Hanna M, Banks E, et al.The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zook JM, Chapman B, Wang J, et al.Integrating human sequence data sets provides a resource of benchmark SNP and INDEL genotype calls. Nat Biotechnol 2014;32:246–51. [DOI] [PubMed] [Google Scholar]
- 40. Mirebrahim H, Close TJ, Lonardi S.. De novo meta-assembly of ultra-deep sequencing data. Bioinformatics 2015;31:i9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lonardi S, Mirebrahim H, Wanamaker S, et al.When less is more: ′slicing′ sequencing data improves read decoding accuracy and de novo assembly quality. Bioinformatics 2015; 31:2972–80. [DOI] [PubMed] [Google Scholar]
- 42. Desai A, Marwah VS, Yadav A, et al.Identification of optimum sequencing depth especially for de novo genome assembly of small genomes using next generation sequencing data. PLoS One 2013;8:e60204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen YC, Liu T, Yu CH, et al.Effects of GC bias in next-generation-sequencing data on de novo genome assembly. PLoS One 2013;8:e62856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Meienberg J, Zerjavic K, Keller I, et al.New insights into the performance of human whole-exome capture platforms. Nucleic Acids Res 2015;43:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Miller JR, Koren S, Sutton G.. Assembly algorithms for next-generation sequencing data. Genomics 2010;95:315–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Paszkiewicz K, Studholme DJ.. De novo assembly of short sequence reads. Brief Bioinform 2010;11:457–72. [DOI] [PubMed] [Google Scholar]
- 47. Narzisi G, Schatz MC.. The challenge of small-scale repeats for INDEL discovery. Front Bioeng Biotechnol 2015;3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Robinson JT, Thorvaldsdottir H, Winckler W, et al.Integrative genomics viewer. Nat Biotechnol 2011;29:24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nakamura K, Oshima T, Morimoto T, et al.Sequence-specific error profile of Illumina sequencers. Nucleic Acids Res 2011;39:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pevzner PA, Tang H, Waterman MS.. An Eulerian path approach to DNA fragment assembly. Proc Natl Acad Sci USA 2001;98:9748–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McLure CA, Hinchliffe P, Lester S, et al.Genomic evolution and polymorphism: segmental duplications and haplotypes at 108 regions on 21 chromosomes. Genomics 2013;102:15–26. [DOI] [PubMed] [Google Scholar]
- 52. Fujimoto M, Bodily PM, Okuda N, et al.Effects of error-correction of heterozygous next-generation sequencing data. BMC Bioinformatics 2014;15 (Suppl 7):S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Glusman G, Cox HC, Roach JC.. Whole-genome haplotyping approaches and genomic medicine. Genome Med 2014;6:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Delaneau O, Howie B, Cox AJ, et al.Haplotype estimation using sequencing reads. Am J Hum Genet 2013;93:687–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vasilinetc I, Prjibelski AD, Gurevich A, et al.Assembling short reads from jumping libraries with large insert sizes. Bioinformatics 2015;31:3262–8. [DOI] [PubMed] [Google Scholar]
- 56. Falchi M, El-Sayed Moustafa JS, Takousis P, et al.Low copy number of the salivary amylase gene predisposes to obesity. Nat Genet 2014;46:492–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yang Y, Chung EK, Wu YL, et al.Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am J Hum Genet 2007;80:1037–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shen S, Pyo CW, Vu Q, et al.The essential detail: the genetics and genomics of the primate immune response. ILAR J 2013;54:181–95. [DOI] [PubMed] [Google Scholar]
- 59. Mose LE, Wilkerson MD, Hayes DN, et al.ABRA: improved coding INDEL detection via assembly-based realignment. Bioinformatics 2014;30:2813–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Compeau PE, Pevzner PA, Tesler G.. How to apply de Bruijn graphs to genome assembly. Nat Biotechnol 2011;29:987–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.