SUMMARY
Tissue-specific mRNA stability is important for cell fate and physiology, but the mechanisms involved are not fully understood. We found that zygotic mRNA stability in Drosophila correlates with codon content: optimal codons are enriched in stable transcripts associated with metabolic functions like translation, while non-optimal codons are enriched in unstable transcripts, including those associated with neural development. Bioinformatic analyses and reporter assays revealed that similar codons stabilize or destabilize mRNAs in the nervous system and other tissues, but the link between codon content and stability is attenuated in the nervous system. We confirmed that optimal codons are decoded by abundant tRNAs while non-optimal codons are decoded by less abundant tRNAs in embryos and in the nervous system. We conclude that codon optimality is a general determinant of zygotic mRNA stability, and attenuation of codon optimality allows trans-acting factors to exert greater influence over mRNA decay in the nervous system.
In Brief
Burow et al. report that codon optimality is a general determinant of zygotic mRNA stability in Drosophila embryos, but the link between codons and stability is weak in the nervous system. Bioinformatics, reporter transcript assays, and tRNA quantitation show that the attenuation of codon optimality establishes neuralspecific mRNA decay.
Graphical Abstract
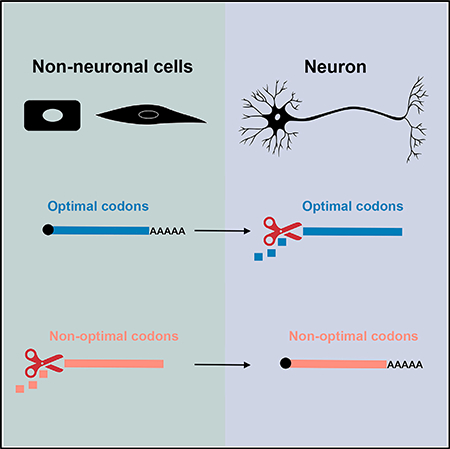
INTRODUCTION
Messenger RNA decay is important for precise temporal and spatial regulation of gene expression during development (Alonso, 2012). Studies in multiple systems have revealed a broad range of mRNA half-lives and coordinate decay of transcripts encoding functionally related proteins (Neff et al., 2012; Munchel et al., 2011; Thomsen et al., 2010). While RNA binding proteins and microRNAs regulate the stability of many mRNAs, these mechanisms are unlikely to account for the full range of half-lives in a cell (Radhakrishnan and Green, 2016). In contrast, codon usage has recently been shown to be a robust determinant of global mRNA half-lives. Pioneering work in budding yeast found that distinct codons are enriched in stable versus unstable mRNAs (Presnyak et al., 2015). This effect can be explained by the concept of codon optimality: codons with abundant cognate tRNAs (optimal codons) support rapid ribosome translocation and mRNA stability, while codons with less abundant cognate tRNAs (non-optimal codons) slow ribosome translocation and favor mRNA decay. The mechanism linking codon optimality and mRNA decay in yeast depends on the RNA helicase Dhh1p (Radhakrishnan et al., 2016). Dhh1p associates with slow ribosomes at nonoptimal codons and triggers mRNA decapping followed by co-translational 5’−3’ decay.
Codon optimality also affects mRNA stability in metazoans. Rapidly decayed maternal mRNAs in zebrafish contain a high frequency of non-optimal codons (Mishima and Tomari, 2016) and the role of codon optimality during the maternal to zygotic transition (MZT) is conserved across multiple vertebrate species and Drosophila (Bazzini et al., 2016). The extent to which codon optimality influences mRNA decay at developmental stages beyond the MZT is not known, although codon usage correlates with steady-state mRNA levels in vertebrate tissues (Bazzini et al., 2016). A potential role for codon optimality in tissue-specific programs of mRNA decay is also suggested by studies of differential tRNA expression. For example, analysis of tRNA abundance across multiple human tissues revealed that tRNA levels vary widely and correlate with codon usage in highly expressed tissue-specific mRNAs (Dittmar et al., 2006). While correlations between tRNA levels and codon usage suggest a role for codon optimality in tissue-specific mRNA decay, direct evidence is lacking.
Given the importance of codon usage in post-transcriptional regulation of gene expression, we hypothesized that codon optimality may influence zygotic mRNA stability in Drosophila embryos. We previously obtained transcriptome-wide mRNA decay measurements for late stage Drosophila embryos, including neural-specific measurements (Burow et al., 2015). This work showed that ~25% of broadly expressed mRNAs have altered stability in the nervous system. Here, we report that codon optimality explains much of the neural-specific changes in mRNA stability. While codon optimality is a strong determinant of mRNA stability when measured across all embryonic tissues, the link between codon usage and mRNA stability is attenuated in the nervous system. Our work provides a framework to understand how mRNA metabolism may be altered to establish tissue-specific programs of mRNA decay.
RESULTS
Codon Usage Correlates with Zygotic mRNA Stability in Drosophila Embryos
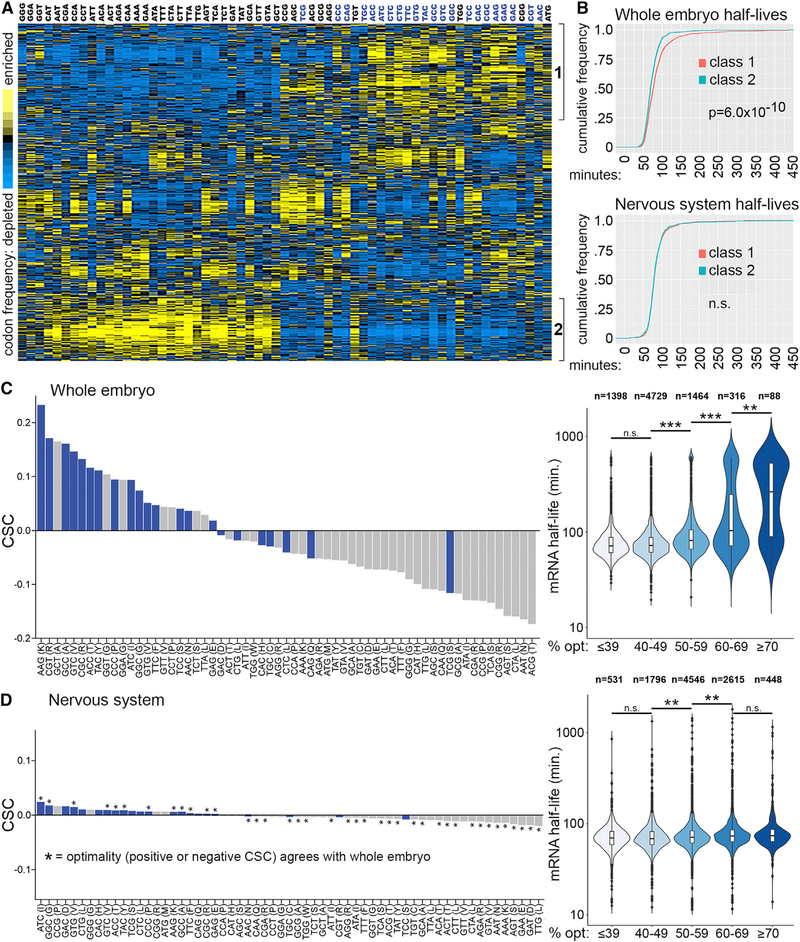
Our previous work focused on the identification of cis-regulatory elements and trans-acting factors that determine mRNA stability in the nervous system and did not address the mechanisms underlying differential stability in the nervous system versus other tissues. For this study, we searched for enrichment or depletion of potential cis-regulatory elements (RNA-binding protein [RBP] motifs, microRNA seed sequences, and any hexameric sequences) that might explain differential transcript stability in the nervous system compared to whole embryos. These analyses failed to identify any sequence features that were significantly enriched or depleted among 1,038 transcripts with ≥ 1.5-fold altered stability in the nervous system. For example, out of 49 previously defined RBP motifs in Drosophila (Paz et al., 2014), none were significantly enriched or depleted in the 3’ UTR of transcripts with decreased stability in the nervous system (Figure S1A). Based on the absence of strong candidate cis-regulatory elements, we next asked if codon usage may explain half-life differences in the whole embryo versus neural-specific data. To test for correlations between codon usage and mRNA stability, we clustered 3,312 mRNAs (those with whole embryo and neural-specific half-life measurements), according to codon usage. We also identified potential preferred codons based on codon frequency in abundant mRNAs (Akashi, 1994; Duret and Mouchiroud, 1999), codon bias across all coding sequences (Powell and Moriyama, 1997), and tRNA gene copy number (Vicario et al., 2007). This clustering revealed two classes of mRNAs with nearly opposite codon usage (Figure 1A; Table S1). Class 1 mRNAs have an enrichment of preferred codons (blue codons in Figure 1A), while class 2 mRNAs are depleted of preferred codons. We next asked if there is any difference in the half-life distribution of class 1 versus class 2 transcripts. Analysis of whole embryo mRNA decay data revealed that class 1 mRNAs are significantly more stable than class 2 mRNAs (Figure 1B). In contrast, analysis of neural-specific mRNA decay data revealed no significant difference in the half-life distributions of class 1 and class 2 transcripts (Figure 1B). These observations provided an initial clue that relationships between codon usage and mRNA stability are distinct in our neural-specific data.
Figure 1. Codon Usage Correlates with Embryo mRNA Stability but Not Neural mRNA Stability.
(A) 3,312 mRNAs clustered according to codon usage. Each row is a different mRNA and each column represents the frequency of the codon listed on top.Codons in blue type are predicted preferred codons.
(B) Distribution of mRNA half-lives for class 1 versus class 2 mRNAs based on whole embryo decay measurements (top panel) and neural-specific decay measurements (bottom panel). p value determined by Kolmogorov-Smirnov test, n.s., no significant difference between classes.
(C) Codon stabilization coefficients (left) and mRNA half-lives categorized by percent optimal codon content (right) based on whole embryo measurements(7,995 mRNAs).
(D) Codon stabilization coefficients for all codons (left) and mRNA half-lives categorized by optimal codon content (right) based on neural-specific measurements (9,699 mRNAs). Codons with the same CSC direction (positive or negative) in the whole embryo and neural-specific datasets are indicted with an asterisk. Significant differences among categories in the violin plots of (C) and (D) were detected by Kruskal-Wallis test and p values (***p < 1 × 1010, **p < 0.001, or no significant difference [n.s.]) for the indicated pairwise comparisons are based on Dunn’s test. See also Table S1.
Codon Content Is Not a Strong Predictor of mRNA Stability in the Nervous System
To define codon optimality for each dataset (whole embryo dataset [7,995 mRNAs] and neural-specific dataset [9,699 mRNAs], as opposed to only the 3,312 overlapping mRNAs clustered in Figure 1A), we calculated whole embryo and neural-specific codon stabilization coefficients (CSCs). CSC is a measure of the Pearson correlation between codon usage and transcript stability (Presnyak et al., 2015). The distribution of CSCs based on whole embryo measurements revealed strong correlations between certain codons and mRNA stability (Figure 1C). In addition, 15 of the 22 predicted preferred codons have positive CSCs. As expected, mRNAs with similar optimal codon content have similar decay rates and codon content is a significant predictor of half-life (Figure 1C; Table S2). We also found that CSCs correlate well with a standard measure of codon preference, the tRNA adaptive index (tAI), and the correlation with tAI is lost when CSCs are calculated using +1 frameshifted coding sequences (Figure S2A). The range of CSCs and proportion of optimal versus non-optimal codons we observed is similar to that seen in Saccharomyces cerevisiae (Presnyak et al., 2015) (22 optimal and 39 non-optimal in Drosophila versus 23 optimal and 38 non-optimal in yeast), although the optimal and nonoptimal sets of codons are distinct for each organism. Next, we calculated CSCs based on neural-specific mRNA decay measurements. As predicted by the similar half-life distributions of class 1 and class 2 transcripts in the nervous system (Figure 1B), correlations between codons and transcript stability were much weaker than those obtained for whole embryos (Figure 1D; Table S3). However, 18 of the 22 predicted preferred codons have positive CSC values, similar codons are optimal (positive CSC) and non-optimal (negative CSC) in the whole embryo and neural-specific datasets (Figure 1D), and there is a positive correlation between CSC values in whole embryos and the nervous system (Figure S2B). As expected, given the low magnitude of neural-specific CSCs, neural-specific optimal codon content is a less significant predictor of mRNA half-life compared to whole embryo optimal codon content (compare violin plots and statistics in Figures 1C and 1D). The relationship between percent optimal codons and neural mRNA half-life was similar when whole embryo CSCs were used to calculate optimal codon content (Figure S2D). There is a weak correlation between neural CSCs and tRNA adaptive index, and this correlation is lost using +1 frameshifted coding sequences (Figure S2C). Taken together, these CSC calculations suggest that codon optimality is a general determinant of zygotic mRNA stability throughout embryonic tissues, but codon content alone is less likely to explain neural-specific mRNA decay rates.
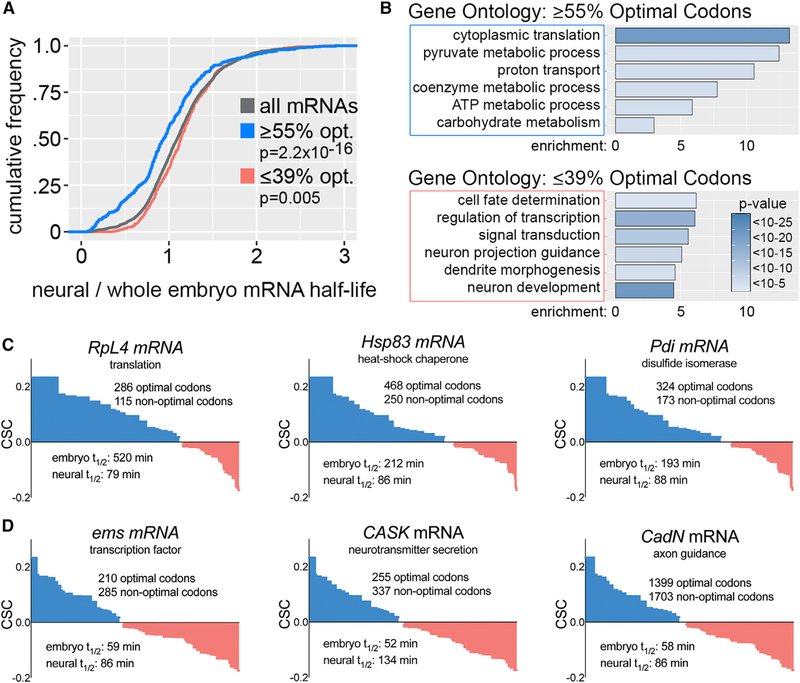
Given that similar codons are optimal and non-optimal based on whole embryo and neural-specific measurements but correlations with stability are much stronger for whole embryo CSCs, we used whole embryo CSCs to define optimal codons (positive CSC) and non-optimal codons (negative CSC) for subsequent analyses. These codon optimality assignments largely explain the properties of class 1 and class 2 transcripts in the overlapping mRNA dataset (the 3,312 transcripts with whole embryo half-life measurements and neural-specific measurements). Transcripts within the 90th percentile of optimal codon content (≥55% of codons have a positive CSC) are primarily class 1 mRNAs (82% are class 1 mRNAs, 2% are class 2 mRNAs) (Table S1). Transcripts in this high optimality group have a significant trend toward decreased stability in the nervous system (Figure 2A). Similarly, ≥ 55% optimal codon content as a sequence feature is significantly enriched in transcripts with decreased stability in the nervous system (Figure S1B). Gene ontology analysis revealed that transcripts in this high optimality group encode proteins associated with metabolic functions, particularly cytoplasmic translation (Figure 2B). For example, transcripts encoding the ribosomal protein RpL4, the chaperone Hsp83, and the disulfide isomerase Pdi all have ≥65% optimal codon content and are very stable in embryos but have shorter half-lives based on neural-specific measurements (Figure 2C). Transcripts within the 10th percentile of optimal codon content (≤39% of codons have a positive CSC) are primarily class 2 mRNAs (54% are class 2 mRNAs, 6% are class 1 mRNAs) (Table S1). Transcripts in this low optimality group have a weak but significant trend toward increased stability in the nervous system (Figure 2A). Gene ontology analysis revealed that low optimality transcripts encode proteins associated with cell fate determination and neural-specific functions (Figure 2B). For example, transcripts encoding the cell adhesion protein CadN, the transcription factor Ems, and the scaffold protein CASK have low optimal codon content and low stability when measured across all tissues but above average half-lives in the nervous system (Figure 2D). We also tested if transcript length differed between the ≥55% and ≤39% optimal codon content mRNAs. The ≤39% optimal codon content mRNAs have slightly longer 5’and 3’UTRs, but the most significant difference is increased coding sequence length in the ≤39% optimal codon content mRNAs (Figure S2E). This agrees with the previous finding that non-optimal codon frequency increases with coding sequence length in Drosophila mRNAs (Duret and Mouchiroud, 1999).
Figure 2. Codon Optimality Establishes Decay of Functionally Related Transcripts.
(A) Relative stability of mRNAs based on neural-specific versus whole embryo measurements. x axis is the neural-specific half-life divided by whole embryohalf-life. p values are based on Kolmogorov-Smirnov test comparing all mRNAs to high optimality or low optimality mRNAs.
(B) Gene ontology categories enriched in high optimality versus low optimality mRNAs.
(C) Codon content and differential stability of representative high optimality mRNAs. The codon content of each gene is plotted in order of optimality (high to low) as defined by whole embryo CSCs.
(D) Codon content and differential stability of representative low optimality mRNAs. Codon content is plotted as in (C). See also Table S1.
Tests of Potential Influence from trans-Acting Factors Suggest that Codon Optimality Is Intrinsically Weakened in Neural mRNAs
One potential explanation for the weak correlations between codon content and mRNA stability in our neural-specific data is that neural-specific factors that promote mRNA decay may override the effects of codon optimality. In contrast, any influence of tissue-specific trans-acting factors is less likely to be observed in the whole embryo dataset because decay measurements were obtained from a mixture of all tissues. We tested if trans-acting factors may “mask” codon optimality in the neural-specific dataset by calculating neural-specific CSCs for transcripts that are not predicted to be targets of the RBPs Pumilio, Fmrp, and Orb2B or the microRNAs miR-124 and miR-315. These factors were selected because good target datasets are available (Gerber et al., 2006; McMahon et al., 2016; Stepien et al., 2016; Schnall-Levin et al., 2010), they are known to induce decay of their targets, and they are largely neural-specific at the stages we investigated. These calculations resulted in modest changes in CSCs, both for individual trans-acting factors (data not shown) and for the set of mRNAs that are not predicted to be targets of any of these factors (Figure S3). CSC directionality was largely maintained (optimal codon CSCs became more positive and non-optimal codon CSCs became more negative) and agreed with optimal and non-optimal assignments based on the complete neural-specific dataset. However, the magnitude of the non-target CSCs was small compared to whole embryo CSCs and similar changes in CSC magnitude were obtained using a randomly selected set of genes (Figure S3), suggesting that non-target CSC changes are not significant. We conclude that, at least for this set of RBPs and microRNAs, the activity of neural-specific decay-promoting factors does not mask otherwise strong correlations between codon content and mRNA stability in the nervous system.
We next predicted that if codon optimality is attenuated in the nervous system, correlations between targeting by decay factors and mRNA stability should be largely insensitive to codon content. Conversely, in embryos we expect antagonism between high optimal codon content and destabilization by trans-acting factors. To test this relationship using whole embryo data, we focused on mRNAs that contain an AU-richelement(ARE) intheir 3′ UTR(Cairrao et al., 2009). Tis11 is a RNA-binding protein that promotes mRNA decay via ARE binding (Choi et al., 2014) and is broadly expressed in embryos (Ma et al., 1994). We therefore considered ARE-containingmRNAsaslikelytargetsofTis11-mediateddecay. ARE-containing mRNAs are significantly less stable than mRNAs that lack this element (Figure S4A). As predicted, when ARE-containing mRNAs are grouped according to percent optimal codons, high optimal codon content mRNAs are significantly more stable than intermediate and low optimal codon content mRNAs. This suggests there is competition between the destabilizing effect of Tis11 and the stabilizing effect of optimal codons.
The stability of ARE-containing mRNAs is not significantly different from non-ARE mRNAs in the neural-specific dataset. Therefore, to test the relationship between codon content and trans-acting factors in the nervous system we focused on targets of Pumilio and miR-315. These are the only factors from the list above whose targets are significantly less stable than non-targets (Figures S4B and S4C). In both cases, when target mRNAs were grouped according to codon content (defined by either neural-specific data or whole embryo data) there were no significant differences in the half-lives of high, intermediate and low optimal codon content mRNAs. This suggests that Pumilio and miR-315 are the primary determinants of target mRNA half-life and that codon content has little or no influence. These analyses provided an additional clue that codon optimality is intrinsically weakened in the nervous system.
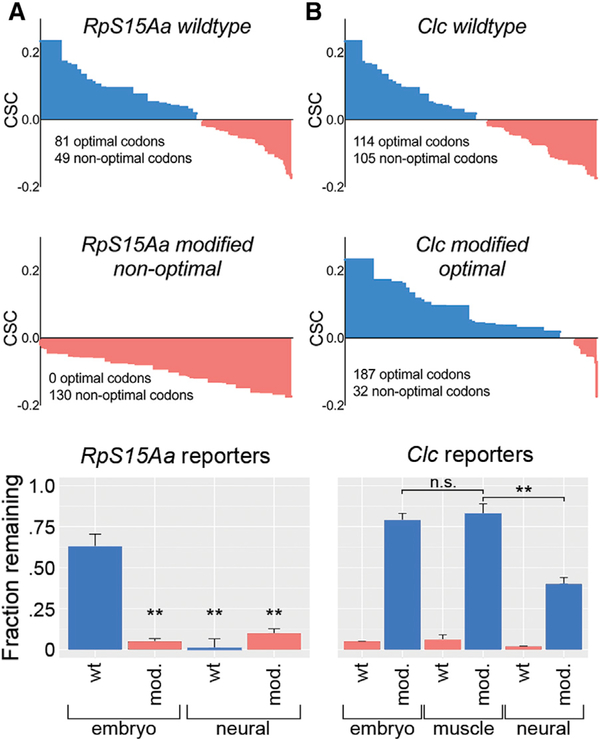
The Stabilizing Effects of Optimal Codons Are Attenuated in the Nervous System
To test our hypothesis that optimal codons are intrinsically less stabilizing in the nervous system (independent of any effects of trans-acting factors), we used synonymous codon substitutions to make reporter constructs with varying optimal codon content. All reporters contained hsp70Bb 5’ and 3’ UTRs to normalize effects of UTR length or sequence and to rule out targeting by trans-acting factors because these UTRs lack any predicted microRNA or RBP binding sites. The reporters were expressed ubiquitously or in specific tissues using the Gal4/UAS system (Brand and Perrimon, 1993) and reporter mRNA decay was measured by TU-tagging (Burow et al., 2015). First, we made a wild-type RpS15Aa reporter (63% optimal codons) and a nonoptimal RpS15Aa reporter (0% optimal codons) (Figure 3A). As predicted, wild-type RpS15Aa was stable while non-optimal RpS15Aa was strongly destabilized in whole embryos (Figure 3A). In contrast, codon content had little effect on RpS15Aa stability in the nervous system: both the optimal and non-optimal versions of RpS15Aa were unstable.
Figure 3. The Stabilizing Effects of Optimal Codons Are Attenuated in the Nervous System.
(A) RpS15Aa wild-type or RpS15Aa non-optimal coding sequences are illustrated in order of their codon optimality. Reporter transcript abundance after a 2-hr chase is plotted for RpS15Aa wild-type (blue) and RpS15Aa non-optimal (salmon) as measured in whole embryos or in the nervous system.
(B) Clc wild-type (salmon) or Clc optimized (blue) reporter transcript decay is plotted as described for (A), with the addition of mesoderm/muscle-specific measurements. Data in (A) and (B) are mean ± SEM. Significant differences in reporter construct decay were identified by t tests, **p value < 0.001.
We next asked if re-coding a low stability transcript to contain an exceptionally high frequency of optimal codons could increase mRNA stability in whole embryos, the nervous system, and an additional tissue: the developing muscular system (mesoderm and muscle). We focused on the clathrin light chain (Clc) transcript, which normally contains 53% optimal codons and has intermediate stability according to whole embryo (t1/2 = 75 min) and neural-specific (t1/2 = 62 min) measurements. We made a highly optimized Clc reporter containing 85% optimal codons (Figure 3B). Optimized Clc was strongly stabilized in whole embryos and the muscular system but this stabilizing effect was significantly weaker in the nervous system (Figure 3B). These results suggest that a core set of optimal codons have a general stabilizing effect in embryos but the strength of this stabilization is attenuated in the nervous system.
The Relative Abundance of tRNAs that Decode Codons Enriched in Ribosomal Protein mRNAs Is Conserved in the Nervous System
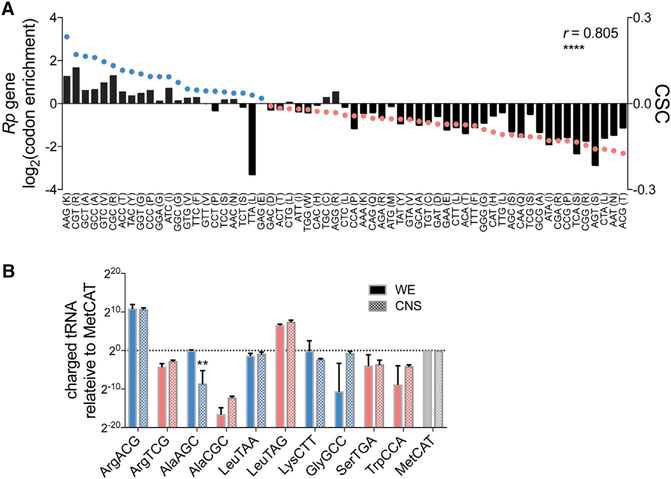
A majority (63%) of functionally annotated mRNAs with high optimal codon content and strong destabilization in the nervous system (neural/whole embryo half-life ≤0.33) encode ribosomal proteins (Table S1). This suggests that codon optimality outside the nervous system may largely be shaped by selection for stabilization of transcripts supporting ribosome production. To test this hypothesis, we compared codon usage frequency and CSC for ribosomal protein transcripts (Rp mRNAs). This analysis revealed a strong correlation between codon usage in Rp mRNAs and CSCs (Figure 4A). No such correlation exists for Rp mRNA codon usage and neural-specific CSCs (data not shown). We interpret these results as evidence that ribosomal protein mRNA stability is a strong selector for codon optimality, and attenuation of codon optimality contributes to the decreased stability of Rp mRNAs in the nervous system.
Figure 4. The Relative Abundance of Charged tRNAs that Decode Codons Enriched in Ribosomal Protein mRNAs Is Conserved in Embryos and the Nervous System.
(A) Codon enrichment or depletion in Rp genes is plotted as bars, embryo CSC is plotted as dots. Codons are arranged from left to right in order of their optimality in embryos (optimal CSC in blue and non-optimal CSC in salmon). Codons corresponding to tRNAs analyzed in (B) are shown in blue (optimal CSC) or salmon (non-optimal CSC).
(B) Whole embryo and CNS charged tRNA levels are plotted relative to tRNA-MetCAT, with optimal codons blue and non-optimal codons salmon. Data are represented as mean ± SD. **p < 0.001 for AlaAGC embryo versus AlaAGC neural based on t test with Bonferroni multiple testing correction.
Based on the standard model of codon optimality, tRNAs that decode codons enriched in Rp mRNAs should be more abundant in Rp mRNAs. Wetested these predictions by measuring the charged tRNA abundance of three isoacceptor pairs: tRNA-ArgACG (positive CSC, enriched in Rp mRNA) and tRNA-ArgTCG (negative CSC, depleted in Rp mRNA), tRNA-AlaAGC (positive CSC, enriched in Rp mRNA) and tRNA-AlaCGC (negative CSC, depleted in Rp mRNA), and tRNA-LeuTAA (positive CSC, highly depleted in Rp mRNA) and tRNA-LeuTAG (negative CSC, depleted but more frequent than tRNA-LeuTAA in Rp mRNA) (Figure 4B). For the Arg and Ala tRNAs, abundance closely corresponded to codon frequency and optimality: tRNA-ArgACG and tRNA-AlaAGC were significantly more abundant than tRNA-ArgTCG and tRNA-AlaCGC. For the Leu tRNAs, codon frequency in Rp mRNAs was a stronger predictor of relative tRNA abundance than CSC: tRNA-LeuTAG was more abundant than tRNA-LeuTAA. These results suggest that codon frequency in Rp mRNAs is shaped by tRNA abundance and that codon stabilization coefficients, as indicated by the Leu isoacceptor results, may be influenced by factors other than tRNA abundance.
One explanation for the decreased stability of Rp mRNAs in the nervous system could be neural-specific changes in the abundance of tRNAs that decode codons enriched in Rp mRNAs. We tested this possibility by quantifying tRNA levels in the CNS dissected from newly hatched larvae (the early larval CNS closely resembles the late embryonic CNS used for mRNA decay measurements). We found that tRNA levels in the CNS are very similar to tRNA levels in whole embryos (Figure 4B). The only significant difference was a decrease in the total abundance of tRNA-AlaAGC, but the relative abundance of the Ala isoacceptors is the same (tRNA-AlaAGC is more abundant than tRNA-AlaCGC). While the decreased abundance of tRNAAlaAGC in the nervous system may weaken the stabilizing effect of the corresponding GCT codon (GCT has a weak negative CSC value based on neural-specific measurements), these tRNA abundance data suggest that decreased stability of Rp mRNAs in the nervous system is not solely due to changes in the abundance of tRNAs that decode optimal codons.
DISCUSSION
Implications of the Relationship between Codon Content and mRNA Decay in Drosophila
We discovered a role for codon optimality in determining Drosophila zygotic mRNA stability, plus a surprising additional level of regulation: the attenuation of codon optimality in the nervous system. This work was motivated by our interest in identifying stability determinants that explain half-life differences in whole embryo and neural-specific mRNA decay measurements. Our bioinformatic analyses and reporter assays all point to tissue-specific differences in codon optimality as a major determinant of differential stability and explain why we were unable to find any strong candidate cis-regulatory features that, on their own, explain differential stability.
One of the most dramatic examples of differential stability in the nervous system is the destabilization of Rp mRNAs. Previous work has shown that Rp mRNA stability decreases in differentiated human fibroblasts compared to mitotic progenitors (Johnson et al., 2017), and maturing neurons undergo a sharp decrease in ribosome biogenesis (Slomnicki et al., 2016). Destabilization of Rp mRNAs might be particularly important in neurons, where protein metabolism needs are distinct from those of mitotic cells and localized translation occurs in axon growth cones and dendrites (Holt and Schuman, 2013). Our reporter transcript assays support a model in which the stability of Rp mRNAs and other high optimal codon content transcripts is decreased in the nervous system due to the attenuation, not the absence, of stabilizing effects of optimal codons. Transcripts like RpS15Aa that have ~60% optimal codons are generally stabilized, but this level of codon optimality does not have a strong stabilizing effect in the nervous system. The Clc results show that increasing optimal codon content to exceptionally high levels dramatically increases stability throughout embryos, but this stabilizing effect is weaker in the nervous system. We consider 85% optimal codon content to be exceptionally high because only 2 genes in our datasets have optimal codon content >80%, and only 74 genes have optimal codon content >70%. Importantly, the optimized Clc reporter was highly stabilized in another tissue type, the mesoderm and muscle, arguing that the attenuation of the stabilizing effects of optimal codons may be unique to the nervous system. We do not propose that attenuation of codon optimality is the sole determinant of differential mRNA stability in the nervous system but rather that attenuated codon optimality synergizes with the translation dampening and decay promoting activity of trans-acting factors.
Relationships between codon content and trans-acting stability determinants were recently described in Drosophila S2 cells using reporter transcripts containing microRNA binding sites and varying optimal codon content (Cottrell et al., 2017). This work showed that transcripts with high or low optimal codon content (defined by tAI) were weakly repressed by microRNAs, while transcripts with intermediate optimal codon content were strongly repressed by microRNAs. We observed a similar relationship for ARE-containing mRNAs based on whole embryo data: ARE-containing mRNAs with high optimal codon content are more stable than those with intermediate or low optimal codon content. The absence of such relationships for Pumilio and miR-315 targets in our neural-specific data suggests that the intersection of trans-acting factors and codon optimality is distinct in the nervous system. Multiple lines of evidence suggest that post-transcriptional regulation of gene expression is particularly important in the nervous system (Loya et al., 2010; Pilaz and Silver, 2015). We propose that attenuation of the link between translation rates and mRNA decay evolved to enable RBPs and microRNAs to exert a stronger influence on mRNA half-life in the nervous system than they would in other tissues.
Potential Mechanisms of Attenuated Codon Optimality in the Nervous System
Our measurements of charged tRNA abundance support two conclusions. First, these data confirm the predicted link between codon usage and tRNA availability in embryos: codons decoded by abundant charged tRNAs are enriched in highly stable Rp mRNAs. Second, these data rule out differential charged tRNA abundance as the primary cause of attenuated codon optimality in the nervous system. While it is possible that the abundance of other charged tRNAs might differ in the nervous system or that neural-specific post-transcriptional modifications of tRNAs could affect the link between translation and decay, such effects would likely be revealed by our neural-specific CSC analyses. Instead, there are no codons that strongly correlate with increased or decreased stability in the nervous system and those that have modest correlations largely agree with the optimal/ non-optimal assignments in whole embryos.
If altered availability of charged tRNAs is unlikely to explain attenuated codon optimality in the nervous system, what is the likely mechanism? One possibility is that the molecular machinery linking codon usage, ribosome translocation, and mRNA decay is altered in the nervous system. The Drosophila ortholog of Dhh1p, Me31b, is broadly expressed in embryos (data not shown) and co-purifies with ribosome complexes (Antic et al., 2015). Me31b-mediated mRNA decay may be altered in the nervous system due to differential expression of co-factors that affect Me31b activity, similar to the changes in Me31b-mediated translation repression and mRNA decay that occur during the maternal to zygotic transition in Drosophila embryos (Wang et al., 2017). Weak correlations between codon usage and mRNA stability could also be caused by the uncoupling of translation repression and decay for transcripts that are localized to axon growth cones or dendrites (Holt and Schuman, 2013). Regardless of the underlying mechanisms, altered codon optimality in the nervous system likely serves two functions. First, it allows neural-specific mRNA decay mechanisms, such as targeting by RNA binding proteins, to function in the absence of strong competition with the codon optimality pathway. Second, it helps shape neural-specific mRNA decay rates to meet the unique biological needs of neurons.
EXPERIMENTAL PROCEDURES
Computational Analysis
RNA-binding protein motif searches were performed using RBPMap (Paz et al., 2014). MicroRNA seed sequences and enriched or depleted hexameric sequences were sought using FIRE (Elemento et al., 2007). Codon clustering and CSC calculations were performed as previously described (Presnyak et al., 2015). For codon clustering, the relative frequency of occurrence of the 61 codons was computed within each mRNA. These values were then ranked (using RANK.AVG function in Excel). The obtained matrix was then clustered using Cluster3 (Spearman distance metric and k-means clustering) and visualized by Java Treeview. The CSC values were determined for each codon as the Pearson correlation coefficient between the frequency of occurrence of the codon in the transcripts and the half-lives of these transcripts. Drosophila melanogaster tRNA gene copy number is based on the Genomic tRNA database (GtRNAdb 2.0) (Chan and Lowe, 2016) and the Berkeley Drosophila Genome Project release 6 (dm6). tRNA adaptive index (tAI) values were calculated using CUA, an open-source program available on CPAN (Zhang, 2015). Gene ontology analysis was performed using GO-Term Finder (Boyle et al., 2004).
Fly Lines and Reporter Assays
Chemically synthesized RpS15Aa-RD wild-type, RpS15Aa-RD non-optimal, Clc-RA wild-type, or Clc-RA optimal DNA (Integrated DNA Technologies) was ligated into the pUAST-emGFP-hsp70BbUTR-attB backbone and injected into P{nos-phi-C31int.NLS}X, P{CaryP}attP40 embryos as previously described (Burow et al., 2015). Reporter lines were crossed to either the neural-specific prospero-GAL4 (Burow et al., 2015) that labels neural progenitors, post-mitotic neurons and glia, the ubiquitous R45H06-Gal4 (Bloomington Drosophila Stock Center), or the mesoderm and muscle-specific how24B-Gal4 (Bloomington Drosophila Stock Center). TU-tagging-based decay measurements and realtime qPCR were performed as previously described (Burow et al., 2015) with normalization to 5S rRNA. All experiments were performed in stage 12–15 embryos in order to match the developmental timing used in Burow et al. (2015).
Charged tRNA Quantitation
RNA from Canton-S stage 12–15 embryos and dissected newly hatched larval central nervous system tissues (0–4 hr after larval hatching) was size selected (<200 bp) using RNeasy Mini spin columns. Charged tRNAs were selected via oxidation with 10 mM NaIO4 on ice for 40 min, precipitation and re-suspension in 20 mM TrisHCl (pH 9.0) for 40 min at 37C to remove amino acids, then final precipitation with ethanol and linear polyacrylamide. Charged tRNAs were used in “four leaf clover” real-time qPCR reactions (Honda et al., 2015) with Ct values normalized to 5S rRNA.
Statistical Analysis
Statistical tests and random gene sampling were performed using the computing environment R (R Core Team, 2017). Relevant tests are named in the figure legends or text. For reporter assays and tRNA quantitation, duplicate qPCR measurements were taken from cDNA prepared from two or three independent biological replicates: input RNA from independent TU-taggings for reporter decay measurements, input RNA from independent embryo and larval CNS preparations for tRNA quantitation.
Supplementary Material
Highlights.
Codon content is a general determinant of mRNA stability in Drosophila embryos
Codon content is a weak determinant of mRNA stability in the nervous system
The relative abundance of tRNAs is conserved in embryos and neural tissue
Attenuation may augment regulation of neural mRNA stability by trans-acting factors
ACKNOWLEDGMENTS
This work was supported by the NIH (R01 HD076927 to M.C. and GM118018 and GM125086 to J.C.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures and three tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.07.039.
REFERENCES
- Akashi H (1994). Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics 136, 927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso CR (2012). A complex ‘mRNA degradation code’ controls gene expression during animal development. Trends Genet. 28, 78–88. [DOI] [PubMed] [Google Scholar]
- Antic S, Wolfinger MT, Skucha A, Hosiner S, and Dorner S (2015). General and microRNA-mediated mRNA degradation occurs on ribosome complexes in Drosophila cells. Mol. Cell. Biol 35, 2309–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Del Viso F, Moreno-Mateos MA, Johnstone TG, Vejnar CE, Qin Y, Yao J, Khokha MK, and Giraldez AJ (2016). Codon identity regulates mRNA stability and translation efficiency during the maternal-tozygotic transition. EMBO J. 35, 2087–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, and Sherlock G (2004). GO:TermFinder–open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20, 3710–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, and Perrimon N (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Burow DA, Umeh-Garcia MC, True MB, Bakhaj CD, Ardell DH, and Cleary MD (2015). Dynamic regulation of mRNA decay during neural development. Neural Dev. 10, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairrao F,Halees AS, Khabar KS,Morello D,andVanzo N(2009).AU-rich elements regulate Drosophila gene expression. Mol. Cell. Biol 29, 2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, and Lowe TM (2016). GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44 (D1), D184–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YJ, Lai WS, Fedic R, Stumpo DJ, Huang W, Li L, Perera L, Brewer BY, Wilson GM, Mason JM, and Blackshear PJ (2014). The Drosophila Tis11 protein and its effects on mRNA expression in flies. J. Biol. Chem 289, 35042–35060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell KA, Szczesny P, and Djuranovic S (2017). Translation efficiency is a determinant of the magnitude of miRNA-mediated repression. Sci. Rep 7, 14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, Goodenbour JM, and Pan T (2006). Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2, e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, and Mouchiroud D (1999). Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc. Natl. Acad. Sci. USA 96, 4482–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elemento O, Slonim N, and Tavazoie S (2007). A universal framework for regulatory element discovery across all genomes and data types. Mol. Cell 28, 337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Luschnig S, Krasnow MA, Brown PO, and Herschlag D (2006). Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 103, 4487–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, and Schuman EM (2013). The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron 80, 648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S, Shigematsu M, Morichika K, Telonis AG, and Kirino Y (2015). Four-leaf clover qRT-PCR: A convenient method for selective quantification of mature tRNA. RNA Biol. 12, 501–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EL, Robinson DG, and Coller HA (2017). Widespread changes in mRNA stability contribute to quiescence-specific gene expression patterns in a fibroblast model of quiescence. BMC Genomics 18, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loya CM, Van Vactor D, and Fulga TA (2010). Understanding neuronal connectivity through the post-transcriptional toolkit. Genes Dev. 24, 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Wadleigh D, Chi T, and Herschman H (1994). The Drosophila TIS11 homologue encodes a developmentally controlled gene. Oncogene 9, 3329–3334. [PubMed] [Google Scholar]
- McMahon AC, Rahman R, Jin H, Shen JL, Fieldsend A, Luo W, and Rosbash M (2016). TRIBE: hijacking an RNA-editing enzyme to identify cell-specific targets of RNA-binding proteins. Cell 165, 742–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y, and Tomari Y (2016). Codon usage and 30 UTR length determine maternal mRNA stability in zebrafish. Mol. Cell 61, 874–885. [DOI] [PubMed] [Google Scholar]
- Munchel SE, Shultzaberger RK, Takizawa N, and Weis K (2011). Dynamic profiling of mRNA turnover reveals gene-specific and system-wide regulation of mRNA decay. Mol. Biol. Cell 22, 2787–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff AT, Lee JY, Wilusz J, Tian B, and Wilusz CJ (2012). Global analysis reveals multiple pathways for unique regulation of mRNA decay in induced pluripotent stem cells. Genome Res. 22, 1457–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz I, Kosti I, Ares M Jr., Cline M, and Mandel-Gutfreund Y (2014). RBPmap: a web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 42, W361–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilaz LJ, and Silver DL (2015). Post-transcriptional regulation in corticogenesis: how RNA-binding proteins help build the brain. Wiley Interdiscip. Rev. RNA 6, 501–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR, and Moriyama EN (1997). Evolution of codon usage bias in Drosophila. Proc. Natl. Acad. Sci. USA 94, 7784–7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnyak V, Alhusaini N, Chen YH, Martin S, Morris N, Kline N, Olson S, Weinberg D, Baker KE, Graveley BR, and Coller J (2015). Codon optimality is a major determinant of mRNA stability. Cell 160, 1111–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2017). R: A Language and Environment for Statistical Computing. https://www.R-project.org.
- Radhakrishnan A, and Green R (2016). Connections underlying translation and mRNA stability. J. Mol. Biol 428, 3558–3564. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan A, Chen YH, Martin S, Alhusaini N, Green R, and Coller J (2016). The DEAD-box protein Dhh1p couples mRNA decay and translation by monitoring codon optimality. Cell 167, 122–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnall-Levin M, Zhao Y, Perrimon N, and Berger B (2010). Conserved microRNA targeting in Drosophila is as widespread in coding regions as in 3’UTRs. Proc. Natl. Acad. Sci. USA 107, 15751–15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomnicki LP, Pietrzak M, Vashishta A, Jones J, Lynch N, Elliot S, Poulos E, Malicote D, Morris BE, Hallgren J, and Hetman M (2016). Requirement of neuronal ribosome synthesis for growth and maintenance of the dendritic tree. J. Biol. Chem 291, 5721–5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepien BK, Oppitz C, Gerlach D, Dag U, Novatchkova M, Krüttner S, Stark A, and Keleman K (2016). RNA-binding profiles of Drosophila CPEB proteins Orb and Orb2. Proc. Natl. Acad. Sci. USA, Published online October 24, 2016 10.1073/pnas.1603715113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen S, Anders S, Janga SC, Huber W, and Alonso CR (2010). Genome-wide analysis of mRNA decay patterns during early Drosophila development. Genome Biol. 11, R93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicario S, Moriyama EN, and Powell JR (2007). Codon usage in twelve species of Drosophila. BMC Evol. Biol 7, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Ly M, Lugowski A, Laver JD, Lipshitz HD, Smibert CA, and Rissland OS (2017). ME31B globally represses maternal mRNAs by two distinct mechanisms during the Drosophila maternal-to-zygotic transition. eLife 6, e27891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z (2015). Cua: a flexible and comprehensive codon usage analyzer. bioRxiv. 10.1101/022814. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.