SUMMARY
How organisms perceive and respond to their surroundings is one of the great questions in biology. It is clear that RNA plays key roles in sensing. Cellular and environmental cues that RNA responds to include temperature, ions, metabolites, and biopolymers. Recent advances in next-generation sequencing and in vivo chemical probing have provided unprecedented insights into RNA folding in vivo and genome-wide. Patterns of chemical reactivity have implicated control of gene expression by RNA and aided prediction of RNA structure. Central to these advances has been development of molecular biological and chemical techniques. Key advances are improvements in the quality, cost, and throughput of library preparation; availability of a wider array of chemicals for probing RNA structure in vivo; and robustness and user friendliness of data analysis. Insights from probing transcriptomes and future directions are provided.
1. INTRODUCTION
This review focuses on the development of biological and chemical techniques for studying RNA folding in vivo. However, technique development in and of itself can ring hollow. Regarding the mere development of technique, a leader of the American studio glass movement, Harvey Littleton famously said, “Technique is cheap” (Byrd 2011). To give value to technique development in probing RNA folding genome-wide and in vivo, we root our motivation in the desire to understand RNA biology.
In many organisms, the majority of the genome appears to be transcribed, a concept known as “pervasive transcription” (Lybecker et al. 2014), leading to an astonishing but daunting array of RNA species, with hundreds of thousands of different RNAs in a living organism. For instance, in humans, 75%–85% of the genome is transcribed with just 1.5%–2% being protein coding (Pennisi 2012; Hangauer et al. 2013), and in Escherichia coli the majority of the genome is transcribed (Wade 2015) with pervasive transcription mostly occurring antisense to protein-coding genes (Lybecker et al. 2014). Adding to the challenge, the many varied functions of RNA typically manifest themselves in a meaningful way only in vivo. The RNA biologist is thus faced with the difficulty of probing vast numbers of RNA molecules in vivo and genome-wide to make sense of the natural world. This has been the primary driver of technique development in the RNA structural genomics field. To motivate probing of RNA structure in vivo and genome-wide, we describe some of the myriad roles RNA plays in nature, many of which remain to be explored, and then briefly discuss its range of structures and the intermolecular interactions.
Two broad functions of RNA are coding genetic information and regulating gene expression. Messenger RNAs (mRNAs) are unique in that they contain both coding and noncoding regions and so can partake in both roles. Standing as the intermediary between DNA and proteins, mRNAs comprise an ideal point in the flow of genetic information to control gene expression. That control often manifests itself in the 5′ and 3′ untranslated regions (UTRs) of mRNAs, which regulate translation initiation and mRNA turnover (Babitzke et al. 2009; Sonenberg and Hinnebusch 2009; Schoenberg and Maquat 2012). In addition, the coding sequence (CDS) itself can be a point of regulation; indeed, protein levels are often not directly proportional to mRNA levels (Ghaemmaghami et al. 2003; Taniguchi et al. 2010; Walley et al. 2016), suggesting additional functions for mRNAs, and splicing isoforms offer another type of regulatory control. Recent studies suggest that mRNAs can chaperone protein folding through RNA structure–based control of translation rate (Watts et al. 2009; Tang et al. 2016) or by interacting with proteins themselves to help them fold or retain their functional conformations (Docter et al. 2016; Horowitz and Bardwell 2016).
RNAs that do not encode proteins, referred to as noncoding RNAs (ncRNAs), make up all the RNAs in an organism other than mRNA. This broad class of RNAs is the majority both in terms of mass and diversity. The ncRNAs include familiar highly structured and abundant RNAs such as ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs), as well as introns (Lambowitz and Belfort 2015), long noncoding RNAs (lncRNAs) (Kung et al. 2013), and small RNAs such as the ∼20–30-nt microRNAs (miRNAs), small interfering RNAs (siRNAs), and Piwi-interacting RNAs (piRNAs) in eukaryotes (Grosshans and Filipowicz 2008; Axtell 2013), and the ∼50–500-nt small RNAs in bacteria (Storz et al. 2011). Multiple lines of support, including sequence conservation and crystal structures, have shown that introns have complex structures that include catalytic domains and engender complex RNA–protein interactions (Marcia and Pyle 2012). Assessing structure–function relationships of lncRNAs is an active area of research. Some studies have concluded that lncRNAs have conserved and biologically important structures (Maenner et al. 2010; Novikova et al. 2012a; Fang et al. 2015; Somarowthu et al. 2015; Hawkes et al. 2016), although others have not found such support using tests for statistical significance (Rivas et al. 2017). This is an important topic in which more data are needed before universal paradigms can be considered (Novikova et al. 2012b). Other RNAs such as miRNAs and siRNAs form extensive base-pairing with mRNAs, leading to double-stranded regions that control RNA stability and translation.
Because it is single stranded, RNA can form an astounding array of structures that are more protein-like than DNA-like in terms of their complexity and hierarchical nature. These structures are often modular and can comprise motifs as simple as base-paired double-stranded regions that form in trans or in cis (Bloomfield et al. 2000; Leontis and Westhof 2001), and as complex as intricate tertiary structures such as pseudoknots, ribose zippers, and kink turns (Westhof and Auffinger 2006). These motifs can also lead to independent folding domains in sufficiently large RNAs like the group I intron and the ribosome (Noller and Woese 1981; Murphy and Cech 1993). Each of these structural classes has important biological functions. For example, in humans and other higher organisms, double-stranded RNA (dsRNA) is recognized as a pathogen-associated molecular pattern (PAMP) in innate immunity by proteins such as protein kinase R (PKR) and retinoic acid-inducible gene I (RIG-I) (Heinicke et al. 2011; Jensen and Thomsen 2012). Indeed, dsRNA structures and certain RNA modifications allow discrimination between self and nonself (Nallagatla et al. 2011). Other biological roles of dsRNA in eukaryotes include base-pairing by miRNA, siRNA, and piRNA, in which structures form in trans primarily by base-pairing with mRNA, leading to RNA silencing, regulation of gene expression, and epigenetic events (Grosshans and Filipowicz 2008; Axtell 2013). In prokaryotes, helical regions, typically in the 5′ UTR or early in the CDS, are critical to controlling transcription and translation (Merino and Yanofsky 2005; Babitzke et al. 2009). These regions comprise the expression platform of riboswitches (Fig. 1) and can switch their base-pairing between mutually exclusive folds to sequester or expose critical elements that interact with ligands to dictate transcription, termination, or initiation of translation (Nudler and Mironov 2004; Tucker and Breaker 2005; Smith et al. 2010). Additionally, simple helices often form upstream of heat shock proteins in microorganisms and serve as RNA thermometers, melting out at higher temperatures to expose the Shine–Dalgarno sequence and drive translation (Narberhaus et al. 2006).
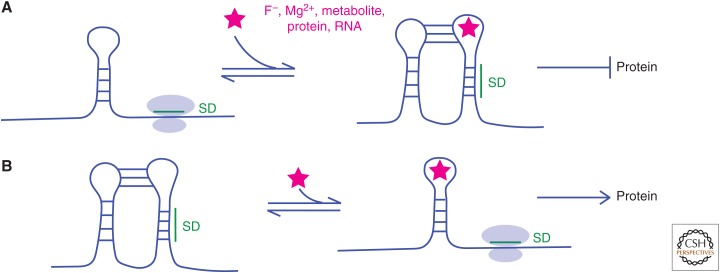
Figure 1.
RNA can adopt diverse and multiple folds. Shown is an RNA transcript with a Shine–Dalgarno sequence (SD). The RNA can bind a ligand, which could be a cation, anion, metabolite, protein, or other RNA, depicted as a star. (A) RNA binds a ligand, shown here in a kissing loop/pseudoknot tertiary structure, that sequesters the SD and prevents ribosome binding resulting in the absence of protein production. This is the more common type of riboswitch found for negative feedback. (B) A different RNA binds a ligand in a loop structure that releases the SD and allows ribosome binding resulting in protein production. This less common type of riboswitch might be for a toxic ligand and the protein produced could pump the ligand out of the cell. Not shown is the effect of temperature, which might melt the right hairpin in B (left) and expose the SD to translate a heat shock protein. The examples here are for prokaryotes but RNAs adopt multiple base-paired and tertiary structures in eukaryotes as well. The diversity of RNA fold and function motivates development of in vivo genome-wide probing technologies.
The first unveiling of the complexity of RNA tertiary structures came with the structure of tRNA (Kim et al. 1973). This structure revealed that RNA can form globular shapes with diverse interactions including non-Watson–Crick base pairs and interhelical interactions. Since that time, hundreds of RNA structures have appeared, including aptamers, riboswitches, ribozymes, the ribosome, and the spliceosome. The details of these structures are beyond the scope of this review, but collectively they show that RNA can achieve positioning of atoms that allows it to be an enzyme and to recognize small molecules specifically and tightly to regulate gene expression.
The versatility of RNA in forming both simple and complex structures allows it to engage in diverse interactions with other molecules, ranging from base-pairing with small RNAs to forming binding pockets for diminutive cations and anions (e.g., Mg2+, Mn2+, and F−) and complex metabolites (e.g., vitamins, coenzymes, nucleotide derivatives, and amino acids), as well as binding structural, processing and signaling proteins (e.g., ribosomal, capping/decapping, spliceosomal, and P-body proteins) (Grosshans and Filipowicz 2008; Re et al. 2014; McCown et al. 2017). Of course, mRNA and rRNA also form intermolecular interactions with each other and with tRNA to decode the genome. It is this dizzying array of structures and intermolecular interactions whose biology only makes sense in vivo that makes RNA an enticing yet challenging molecule for in-depth study. Uncovering the folds of RNAs in vivo and genome-wide has inspired our laboratories and those of others to develop new techniques that span biology and chemistry.
2. TECHNIQUE: THE CHEMISTRY OF PROBING RNA STRUCTURE
The structure of RNA can be determined by a variety of experimental methods. X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy offer atomic resolution views, whereas electron cryomicroscopy (cryoEM) is playing an increasingly important role in determining the structures of large and important RNAs at moderate resolution (Nguyen et al. 2016). Although deep insights have been generated from these approaches, they are low-throughput and cannot keep pace with the generation of genomic data, which can now be obtained in a matter of days via next-generation sequencing (Shendure et al. 2017). Moreover, crystallography cannot generally resolve dynamic molecules or all the states of structure-switching RNAs. Finally, most crystallography, NMR, and cryoEM structures obtained are not in the cell and so do not reflect cellular crowding, metabolites, and other RNA-interacting molecules (Leamy et al. 2016) or the potential kinetic effects of cotranscriptional folding (Watters et al. 2016a).
Methods for probing RNA structure with chemicals and enzymes have been known for nearly 40 years (Peattie and Gilbert 1980; Ehresmann et al. 1987). We begin by tracing the development of RNA structure probing, starting with the chemistry used; we then describe how studies moved from in vitro to in vivo; and finally we present more recent advances that have moved the field from gene-specific to genome-wide. Because enzymes have not generally been used to query RNA structure in vivo, our focus here is on chemical probes.
Before we describe the various chemical reactions and their specificities, we consider how the chemical modifications can be detected. Modifications can be read out several different ways. For chemicals that cleave the backbone—often accomplished via workup with a second reagent—RNA can be fractionated by polyacrylamide gel electrophoresis (PAGE) and analyzed alongside sequencing lanes. For chemicals that modify the RNA on the Watson–Crick face, as well as those that cleave the backbone, primer extension by reverse transcription (RT) can be used to query the RNA according to resultant RT termination or runoff (Inoue and Cech 1985; Ehresmann et al. 1987; Bloomfield et al. 2000). When combined with next-generation sequencing, primer extension is typically achieved genome-wide with the aid of random hexamers and can be used not only with RT stops but also with mutational profiling wherein polymerase readthrough of the modification generally leaves behind a mutation (Siegfried et al. 2014; Zubradt et al. 2017) (see Section 3). Reverse transcription has proven to be the most important of these readout methods for in vivo and genome-wide probing of RNA structures because the resulting complementary DNA (cDNA) libraries can be interrogated by next-generation sequencing methods.
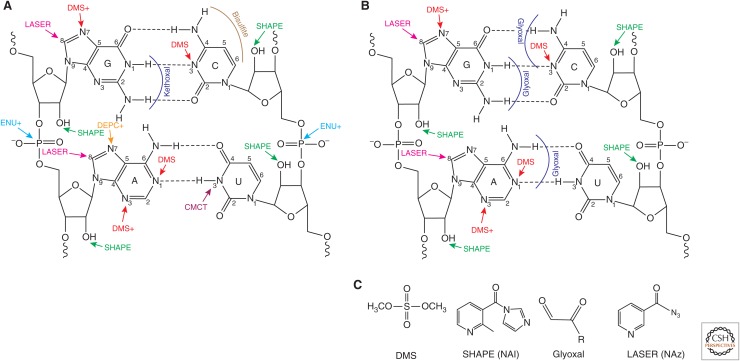
Chemistry has been established that queries nearly every aspect of RNA structure. Figure 2 shows this chemistry and also denotes whether the reactions directly result in a stop during reverse transcription or whether additional reactions are needed to read out the base-pairing status of the nucleotide. Probes have been identified that react with the Watson–Crick, Hoogsteen, and sugar faces of the base, the ribose sugar, and the phosphate, as reviewed in Ehresmann et al. (1987) with recent advances presented in Merino et al. (2005), Spitale et al. (2013), Umeyama and Ito (2017), Feng et al. (2018), and Mitchell et al. (2018). Dimethyl sulfate (DMS) reacts with A(N1) and C(N3) on the Watson–Crick face, as well as G(N7) and A(N3) in the major and minor grooves, respectively, where it has also been used to probe dsDNA (Lawley and Brookes 1963; Umeyama and Ito 2017). Kethoxal is used to probe the Watson–Crick face of G, whereas selective 2′-hydroxyl (2′-OH) acylation analyzed by primer extension (SHAPE) reacts with the 2′-OH (Merino et al. 2005) and light-activated structural examination of RNA (LASER) modifies the C8 position of purines (G>A) (Feng et al. 2018). Importantly, the above reactions can all be used in vivo as well (Fig. 2B), except that glyoxal has been used more than kethoxal in vivo (Mitchell et al. 2018). In addition, the following reactions occur exclusively in vitro: diethylpyrocarbonate (DEPC) with A(N7), 1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide metho-p-toluene sulfonate (CMCT) with U(N3), bisulfite with C(C4–C6), and ethylnitrosourea (ENU) with phosphates.
Figure 2.
Current state of chemical probing of RNA structure in vitro and in vivo. Shown are the standard GC and AU Watson–Crick base pairs. Reaction of the Watson–Crick face will only occur if the base is not protected by base-pairing or interacting with a protein or small molecule. Sites of reaction of RNA structures (A) in vitro and (B) in vivo are provided. The following chemical reagents are common to in vitro and in vivo approaches: Dimethyl sulfate (DMS) reacts with A(N1) and C(N3) on the Watson–Crick face, as well as the G(N7) and A(N3); the latter two require additional workup (denoted with a plus sign) of the reaction in the presence of an amine to induce depurination (Ehresmann et al. 1987; Bloomfield et al. 2000; Gates 2009). Glyoxal (Kethoxal in vitro, see below) reacts with the amidine functionality on the Watson–Crick face of G, C, and A, with a preference for G, and can be directly detected by reverse transcription. The SHAPE reagents acylate the 2′-hydroxyl group (2′-OH) of the sugar of all four nucleosides, whereas the LASER reagent reacts with purines at the C8 position, with a preference for G>A. Both SHAPE and LASER reactions can be directly detected by reverse transcription. The following reagents are exclusive to in vitro reactions at this point: diethylpyrocarbonate (DEPC) reacts with A(N7); 1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide metho-p-toluene sulfonate (CMCT) reacts with U(N3); bisulfite reacts with C(C4–C6), and ethylnitrosourea (ENU) reacts with the phosphates. Generally, kethoxal has been used in vitro and is reported to be G-specific. Detection of DEPC and ENU require additional chemical treatment (denoted with a plus sign), whereas CMCT and bisulfite can be directly detected by reverse transcription. (These panels are adapted from Ehresmann et al. 1987.) (C) Four chemicals used to date for in vivo chemical probing of RNA. Note that these chemicals are small, neutral, and relatively hydrophobic (R = H, CH3, phenyl).
Early work probed the structures of individual RNAs in vitro and allowed secondary structure models to be developed (Ehresmann et al. 1987; Bloomfield et al. 2000). The importance of measuring RNA folding in vivo was realized relatively early on and applied to single, abundant transcripts. Seminal studies probed DNA–protein and RNA–protein structures in vivo, taking advantage of the ability of DMS to modify the Hoogsteen face of G in fully dsDNA (Nick and Gilbert 1985), as well as the Watson–Crick faces of A and C in single-stranded RNA (ssRNA) (Climie and Friesen 1988; Ares and Igel 1990; Senecoff and Meagher 1992). Subsequent studies by Zaug and Cech on three different RNA species in living Tetrahymena thermophila revealed that RNA structure is often altered by proteins (Zaug and Cech 1995): In the case of the U2 small nuclear RNA (snRNA) and telomerase RNA, very different folds were found in vivo and in vitro that could be explained in terms of protein binding in vivo, whereas the self-splicing rRNA intron had the same fold in vivo and in vitro supporting it acting alone as an enzyme.
The above in vitro chemistry has been applied in vivo, albeit in a more limited way, with four chemicals reported to date (Fig. 2B). Small, neutral, and somewhat hydrophobic chemicals seem to work best, as they can permeate the cell membrane/cell wall (Fig. 2C). At present, DMS, SHAPE (2-methylnicotinic acid imidazolide, NAI), glyoxal, and LASER (nicotinoyl azide, NAz) have been used in vivo in organisms that include plants, human cells, and bacteria. Sometimes, small alterations to the reagents improve their performance. For instance, probing with SHAPE has been reported in living cells (Spitale et al. 2013; Smola et al. 2015a), with optimal probing and permeability reported with the SHAPE reagent NAI (Lee et al. 2016), whereas methyl- and phenyl-glyoxal provide stronger signals than glyoxal in vivo (Mitchell et al. 2018). For LASER, the small molecule NAz leads to a highly reactive nitrene species on exposure to ultraviolet (UV) light, which is detectable by reverse transcription (Feng et al. 2018), perhaps because it drives the syn conformation of the base.
Some of the above four in vivo chemical reagents are sensitive to base-pairing, although others are not. For instance, reaction of A(N1) and C(N3) with DMS, as well as reaction of G, C, and A with glyoxal are blocked by Watson–Crick base-pairing. Additionally, reaction of SHAPE is greatly reduced on base-pairing (McGinnis et al. 2012). On the other hand, reaction of G(N7) and A(N3) with DMS is not sensitive to Watson–Crick base-pairing but only to unusual base pairs and triples. Additionally, because LASER modification occurs at a non-hydrogen bonding position (C8 of G or A), it is not generally sensitive to any base-pairing (Feng et al. 2018). The latter two sets of reactions are potentially powerful because their reactivity should be impaired at nucleotides engaged in protein binding or tertiary structure, and thus absence of chemical modification should provide a readout of the presence of those phenomena.
Deeper insights into biology require a more holistic view than that provided by just one or a few transcripts. It was the confluence of the above seminal experiments, which worked out nucleic acid–probing chemistry and reverse transcription in the test tube and then living cells, and next-generation sequencing that set the stage for the development of techniques to query RNA folding in vivo and genome-wide. The next section provides the rationale behind the approaches taken at the level of library preparation. Each of these approaches relies on at least one of the above chemical modifications of the RNA, with readout by reverse transcription stops or mutational profiling.
3. TECHNIQUE: THE MOLECULAR BIOLOGY OF PROBING RNA STRUCTURE IN VIVO AND GENOME-WIDE
The previous section considered the various chemistries that allow RNA structure to be probed in vivo. We presented a growing list of RNA-reactive membrane-permeant chemicals. Probing RNA structure in vivo first began with the application of DMS 30 years ago (Climie and Friesen 1988), and more recently SHAPE, glyoxal, and LASER approaches were added (Spitale et al. 2013; Smola et al. 2015a; Feng et al. 2018; Mitchell et al. 2018). These four reagents have opened the door to current efforts at probing RNA structure genome-wide and in vivo. During a given treatment of a living organism, all of the RNAs in vivo to which the reactive base is exposed are amenable to modification by these chemicals. The challenge then becomes to decode the sites of reactivity and to score them quantitatively.
In this section we shift our focus from the chemistry of RNA to the reading out of the reactivities, which involves application of molecular biology and nucleic acid sequencing approaches. These techniques are multistep and generally quite complex, perhaps not unlike a many-step natural product synthesis for an organic chemist. The goal of this section of the review is not to provide a step-by-step accounting of these methods or to present protocols on how to perform these experiments; indeed, our laboratory and others have published such reviews and protocols previously and we direct the interested reader to these (Low and Weeks 2010; Weeks 2010; Wan et al. 2011; Spitale et al. 2014; Kwok et al. 2015; Bevilacqua et al. 2016; Ritchey et al. 2018; Tack et al. 2018). Rather, we wish to trace the rationale behind the development of the molecular biology aspects of the techniques, giving consideration to how the various approaches differ. We hope that this point of view will facilitate the future growth of this field, which is still ripe for innovation. However, before we describe in vivo probing of RNA genome-wide, it is important to consider early foundational studies that allowed RNA structure to be probed genome-wide in vitro.
Genome-wide in vitro studies took advantage of the favorable properties of ribonucleases (RNases) as reagents because the membrane impermeability inherent to enzymes is not an issue in vitro. Fragmentation sequencing, or FragSeq, probed RNA structure with the ss-specific RNase P1 and analyzed sites of reaction with next-generation sequencing (Underwood et al. 2010). In the same year, parallel analysis of RNA structure (PARS) was developed, which uses a combination of ss- and ds-specific RNases (Kertesz et al. 2010). This technique enabled development of parallel analysis of RNA structure with temperature elevation (PARTE), a temperature-dependent version of PARS (Wan et al. 2012). The unique feature of PARS and PARTE was the use of RNase V1, which allows direct detection of double-stranded regions of the RNA (Lockard and Kumar 1981; Lowman and Draper 1986). Protein footprinting was assessed genome-wide in vivo with the invention of protein interaction profile sequencing (PIP-seq) in which RNAs and proteins are first cross-linked in vivo followed by extraction and treatment with ss- and ds-specific RNases with and without treatment by proteinase K, which degrades proteins (Silverman et al. 2014). The issue of protein binding in vivo is an important one and this approach is notable for taking it on. The in vitro RNA structure-determining aspects of PIP-seq have several concerns, however, including buffer mismatch between RNase treatments and the potential for RNA refolding on protein degradation. Finally, at least one genome-wide in vitro study has taken advantage of membrane impermeant chemicals to probe RNA structure. In chemical interference of RNA structure sequencing (CIRS-Seq), CMCT, which does not react in vivo (D Mitchell, SM Assmann, and PC Bevilacqua, unpubl.), likely because its size and charge prevent cell membrane permeation, was used along with DMS to offer chemical information on all four bases in vitro and genome-wide because CMCT can react with G/U and DMS reacts on the Watson–Crick face with A/C (Incarnato et al. 2014). This provided an information-rich view of RNA folding in vitro. In sum, the above methods have taken advantage of a warehouse of known RNA-reactive enzymes and chemicals (Bloomfield et al. 2000), which allows extensive analysis of RNA structure genome-wide but in vitro. These studies have inspired, and continue to inspire, development of in vivo reagents that have the desirable properties of reacting with diverse functionalities on the RNA as well as with unique RNA structural motifs.
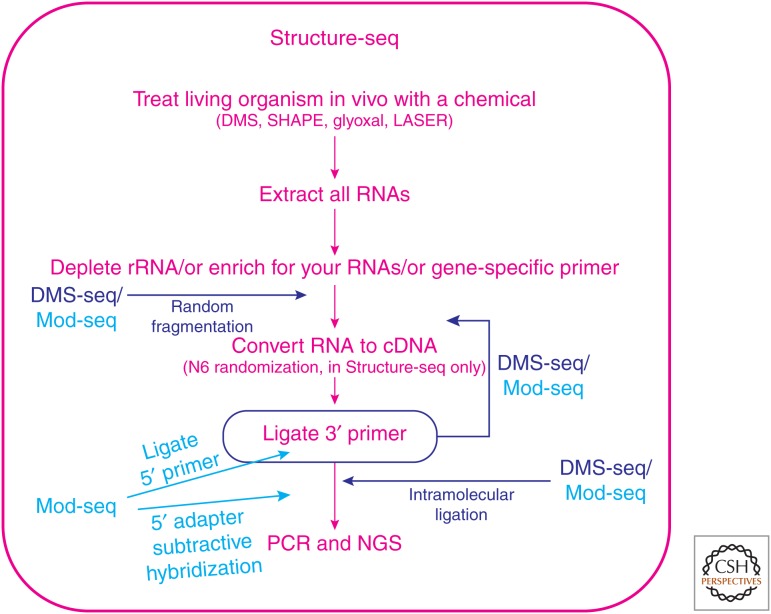
The first two reports of probing RNA structure genome-wide and in vivo appeared in a single journal issue in 2014. These studies included the Structure-seq method from our laboratories and the DMS-seq approach from the Weissman laboratory (Ding et al. 2014; Rouskin et al. 2014). Shortly thereafter, modification sequencing (Mod-seq) was reported from the McManus laboratory (Talkish et al. 2014). A diagram comparing these methods is provided in Figure 3. After in vivo treatment with the structure-probing chemical, the process of analyzing RNA structure genome-wide and in vivo begins with extraction of the treated RNA, typically using a commercial kit. Unless it is the focus of the study, rRNA needs to be removed from the extracted RNA because rRNA accounts for ∼80% of all of the RNA in a cell and will otherwise waste an enormous number of next-generation sequencing reads. Removal of rRNA can be accomplished by a subtractive method such as Ribo-Zero, in which the rRNA binds to complementary oligonucleotides and is pulled down. The Ribo-Zero approach is critical in cases in which there is no common sequence, like a poly(A) tail, to capture the RNAs of interest. Alternatively, the RNA of interest can be pulled down leaving behind rRNA. The most common method uses an oligonucleotide deoxythymine (oligo-dT) column to collect polyadenylated mRNA (Ding et al. 2014; Rouskin et al. 2014), although one could alternatively extract only a single RNA or a specific collection of RNAs using appropriate capture oligonucleotides. It is also possible to use gene-specific primers to focus in on a single RNA without first removing rRNA or enriching for the RNA of interest. For instance, our laboratory used gene-specific primers to target single RNAs (U12 snRNA, 25S rRNA, 5.8S rRNA, and PSBA) in plants (Arabidopsis) and to probe their in vivo structures with both DMS and the SHAPE reagent NAI. Using a ligation-mediated polymerase chain reaction (LMPCR) approach, we increased the sensitivity of the method to attomole level detection (Kwok et al. 2013). Simons and colleagues probed the structure of lncRNA Xist in mouse cells by tiling 87 different gene-specific primers spaced ∼200 nt apart along the ∼17,000 nt RNA (Fang et al. 2015). They dubbed this method “targeted Structure-seq” because it is based on Structure-seq (Ding et al. 2014) and targets a single RNA. In addition, Lucks and colleagues have reported a pipeline for applying SHAPE in vitro or in vivo to a collection of RNAs and generating restraints for RNA folding via SHAPE-Seq (Watters et al. 2016b).
Figure 3.
Comparison of the Structure-seq, DMS-seq, and modification sequencing (Mod-seq) in vivo genome-wide RNA structure probing techniques. Structure-seq is shown in magenta font. Additional steps present in DMS-seq and Mod-seq are depicted in dark blue and light blue font, respectively. Note the extra steps before conversion of RNA to cDNA in DMS-seq and Mod-seq as well as the different points of enhancing coverage in the various techniques. Mod-seq has the most steps of all techniques. PCR, Polymerase chain reaction; NGS, next-generation sequencing.
The Structure-seq, DMS-seq, and Mod-seq methodologies all proceed to form libraries but use quite different approaches. In an effort to minimize the universally negative impacts of RNA degradation, we designed Structure-seq to have its RT process of converting RNA into cDNA relatively early on in the pipeline (Ding et al. 2014). We convert RNA to cDNA immediately after poly(A) selection, whereas DMS-seq and Mod-seq have two or four additional RNA-handling steps, respectively, before RT (Fig. 3) (Kwok et al. 2015). To increase coverage in preparing the library, we conduct RT using a primer whose 3′ end is composed of a library of random hexanucleotide sequences (N6) to facilitate random priming, but whose 5′ end has an adapter sequence to bind to TruSeq reverse primers that barcode for a given condition and allow Illumina sequencing (Ding et al. 2014). It is then necessary to append a fixed sequence to the other end of the cDNA to enable PCR, which we do via intermolecular ligation of a ssDNA adapter. This sample then undergoes PCR and next-generation sequencing. Recently, we introduced “Structure-seq2”—a significantly improved version of the original Structure-seq (Ritchey et al. 2017).
DMS-seq and Mod-seq enhance coverage not via a randomized section of a primer, but with fragmentation of the RNA, conducted via Zn2+-mediated hydrolysis (Rouskin et al. 2014; Talkish et al. 2014). Both approaches then ligate on a 3′-RNA adapter, followed by RT and a subsequent intramolecular ligation to form cDNA circles that are subject to PCR amplification and next-generation sequencing. The Mod-seq technique has an additional step of 5′-RNA adapter ligation that allows a later 5′-adapter subtractive hybridization to remove samples that have no chemical modification wherein the RT reads through to the 5′-adapter.
There are several key differences in the libraries produced by these three methods. In Structure-seq2 and Mod-seq, reads from a minus chemical lane are used to correct for background stops, which can arise from RNA secondary structure, covalent modification, or strand scission attributable to degradation or processing. In addition, both Structure-seq and Mod-seq use single-hit kinetics in which only about one in 200 RNA nucleotides is modified; this is generally desired because modification can refold the RNA and reduce reads near the 5′ end of the RNA. The DMS-seq method does not use a minus DMS correction nor single-hit kinetics. In addition, although Mod-seq libraries have fewer unmodified reads owing to its adapter subtractive hybridization, as a result it also does not have an accurate tally on the total number of reads in a given RNA and so may not wholly account for transcript abundance.
Several other improvements in the technique of library preparation have recently appeared. Chang, Spitale, and coworkers introduced an improvement to their NAI SHAPE reagent in the method of in vivo click SHAPE, or icSHAPE (Spitale et al. 2015). In this technique, their NAI reagent is fused to an azido group, which can be attached to a biotin functionality via copper-free click chemistry to enrich for SHAPE-modified RNAs via a streptavidin pulldown. This technique achieves a similar result as Mod-seq—removal of unmodified RNAs from the library—although it is achieved for Mod-seq by capturing the undesired RNAs, whereas for icSHAPE it is accomplished by capturing the desired RNAs.
Two other improvements in preparation of genome-wide in vivo RNA libraries involve the reading out of multiple modifications from a single sequencing read. Weeks and coworkers introduced mutational profiling in SHAPE-MaP, in which SHAPE-modified nucleotides induce a mutation during RT rather than a stop (Siegfried et al. 2014; Smola et al. 2015b; Busan and Weeks 2017). The mutation is achieved by altering the RT conditions, specifically using Mn2+ with the reverse transcriptase Superscript II (SSII). The primary advantage of SHAPE-MaP is that correlated modifications in a single RNA can be identified, essentially turning the method into a single-molecule experiment. For instance, two alleles can be monitored simultaneously, potentially overcoming sequence and structural polymorphism. Weeks and coworkers also used SHAPE-MaP correlations to predict RNA pseudoknots (Siegfried et al. 2014). Rouskin and coworkers adapted mutational profiling to DMS modifications in DMS-MaPseq, wherein a thermostable group II intron reverse transcriptase (TGIRT) is used to obtain mutational data (Zubradt et al. 2017). As compared with the SSII-Mn2+ method, these investigators achieved significantly higher detection of adenine methylation (48% vs. 1.4% in one instance) and significantly lower rate of insertions/deletions (indels) (6% vs. ∼33%), which is helpful for accurate mapping. Another technical advantage of mutational approaches is that library preparation is simplified and more closely follows standard RNA-seq protocols.
It is worth briefly considering some of the biological insights that have been gained via these techniques. These methods have identified RNA reactivity features in plants associated with the CDS and start and stop codons, alternative polyadenylation and alternative splice sites, and stress-related genes (Ding et al. 2014), as well as relationships of protein and mRNA structure (Watts et al. 2009; Tang et al. 2016). Studies in mammalian and yeast cells revealed that RNAs are more unfolded in vivo than in vitro, and that adenosine triphosphate (ATP)-dependent processes, such as helicase unwinding, are at least partially responsible for this (Rouskin et al. 2014). Additional studies in mouse embryonic stem cells revealed that certain RNA features are the same in vitro and in vivo, such as structural features prevailing at translational start and pause sites, whereas RNA-binding proteins and RNA modifications rearrange RNA structure in vivo and can provide reactivity patterns whose molecular features agree with crystal structures (Spitale et al. 2015). In other cases, identification of known and new RNA motifs in human immunodeficiency virus 1 (HIV-1) has been achieved, including several pseudoknots, and polymorphic RNAs have been studied in a single experiment (Siegfried et al. 2014). Translation efficiency in E. coli has been related to open reading frame structures (Burkhardt et al. 2017). Finally, functional structures involved in noncanonical translation initiation have been characterized, along with comparison of spliced and unspliced isoforms (Zubradt et al. 2017). This brief summary illustrates that these structure-probing methods are organism-agnostic and suggests that there is nothing inherent to any of these methods that would prevent their application, singly or multiplexed, to any organism.
All of the above in vivo and genome-wide techniques generate big datasets and so require careful computational analysis. A full review of computational techniques is beyond the scope of this review, but we point out some important advances. Each of the experimental methods described above had a computational approach published as part of the initial study. Some studies later published additional stand-alone companion computational studies and advances (Talkish et al. 2014; Tang et al. 2015; Busan and Weeks 2017; Tack et al. 2018). Key features of computational studies are subtraction of background reaction and normalization for RNA abundance. In addition, all of the methods require benchmarking, which is most commonly performed with structurally characterized RNAs such as rRNA (Spitale et al. 2013; Ding et al. 2014; Rouskin et al. 2014; Talkish et al. 2014). One important feature of computational analysis is consideration of the tendency of an RNA sequence to adopt more than one fold. Recently, several advances have been made in this arena, both experimentally with mutational profiling (Siegfried et al. 2014; Zubradt et al. 2017) and computationally by modeling folding ensembles (Li and Aviran 2018; Spasic et al. 2018). In addition, efforts are afoot to make the software accessible to scientists without extensive expertise in bioinformatics and to incorporate more downstream analysis (Tack et al. 2018). As the field moves forward, it will become increasingly important for the computational pipelines to have the capacity to handle datasets involving more than one chemical and multiple conditions.
4. THE FUTURE OF PROBING RNA STRUCTURE IN VIVO AND GENOME-WIDE
There are tremendous opportunities to advance RNA structure probing in vivo and genome-wide. First, there is the need to develop a more expansive chemical toolbox for probing RNA structure in vivo. As shown in Figure 2, there are several regions of the RNA that are presently not probed in vivo. These include the Watson–Crick face of uracil, as well as the phosphate backbone. Another area for advancement is increasing the sensitivity of probing. Although advances have been made (Kwok et al. 2013), simpler and more sensitive probing techniques might facilitate probing of smaller populations of RNA, such as might be found in biologically interesting areas of specific tissues and even single cells. The latter is an especially exciting area of modern biology and may simplify RNA structurome results, as the folding ensemble may be less diverse.
Another area ripe for development is prediction of RNA structure from sequence. Efforts have been made in this area using in vitro SHAPE chemical probing restraints, which generally manifest themselves as pseudo-free energy parameters (Deigan et al. 2009); however, prediction using in vivo chemical probing restraints is overall still inaccurate for many RNAs (Leamy et al. 2016). The increasing diversity of chemicals available for probing RNA structure in vivo will provide richer experimental data to help constrain RNA structure prediction. The recently developed psoralen cross-linking method (PARIS) from Chang and colleagues provides information on RNA duplexes and RNA–RNA interactions in vivo and genome-wide (Lu et al. 2016). Advances in predicting RNA 3D structure are being made (Miao et al. 2017), and with a plethora of experimental and proximity constraints it should become possible to model 3D structure of RNA genome-wide as it occurs in vivo. Finally, the ability of RNA to switch between conformations requires measuring and modeling folding ensembles (Siegfried et al. 2014; Zubradt et al. 2017; Li and Aviran 2018; Spasic et al. 2018), and further genome-wide advances in this area can be anticipated.
Yet another area for further advancement is probing protein–RNA interactions. Although high reactivity of a given site fully supports its single-stranded conformation, low reactivity observed in vivo does not necessarily support base-pairing, as protection can also be conferred by binding of proteins, small molecules, and ions; moreover, low reactivity does not distinguish intramolecular versus intermolecular base-pairing. Advances in protein protection have been reported in correlating SHAPE reactivities with RNA–protein 3D structures (Spitale et al. 2015) and in using uncertainties in SHAPE reactivity between states (Smola et al. 2015a). In addition, LASER probes the non-hydrogen bonding C8 position of purines and so is inherently sensitive to protein protection at this position (Feng et al. 2018), as is reactivity at other non-Watson–Crick positions (Umeyama and Ito 2017). Ultimately, a combination of these approaches, along with description of RNA-binding motifs (Ray et al. 2013) and selective deployment of cross-linking (Lu et al. 2016) may help with the assignment of protections.
Finally, there are a large number of organisms and conditions to probe. To date, in vivo genome-wide experiments have been conducted in living human and mouse cells, as well as in living whole organisms such as Arabidopsis, rice, yeast, and E. coli (Ding et al. 2014; Incarnato et al. 2014; Rouskin et al. 2014; Spitale et al. 2015; Ritchey et al. 2017; Deng et al. 2018). There is the potential to now perform comparative analysis of RNA structure between different states, for instance, between organisms that differ in genotype and phenotype, between closely or distantly related species, and indeed among all three domains of life. Moreover, there is the opportunity to perform comparative analysis of RNA structure in a single organism between two states, such as different environmental or stress conditions. This offers the chance to discover new functional RNAs and pathways. Finally, the issue of time comes into play, allowing one to ask how the structure of the transcriptome changes over long or even very short times. For example, the LASER reagent can react in less than a second and is self-quenching, allowing querying of rapid processes genome-wide (Feng et al. 2018). There have been tremendous advances in our understanding of RNA biology in vivo over just the last 5 years. All of these can be traced to advances in chemical and biological techniques. These advances have led to surprising new insights into the relationship of RNA structure and gene regulation. New biology awaits discovery. With additional technical advances, these discoveries will be accelerated, allowing greater insight into the natural world.
ACKNOWLEDGMENTS
We gratefully acknowledge the National Science Foundation (NSF)-Plant Genome Research Program (NSF-IOS-1339282) for support, and Dr. Elizabeth Jolley and Laura Ritchey for careful reading of the manuscript.
Footnotes
Editors: Thomas R. Cech, Joan A. Steitz, and John F. Atkins
Additional Perspectives on RNA Worlds available at www.cshperspectives.org
REFERENCES
- Ares M Jr, Igel AH. 1990. Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes Dev 4: 2132–2145. [DOI] [PubMed] [Google Scholar]
- Axtell MJ. 2013. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64: 137–159. [DOI] [PubMed] [Google Scholar]
- Babitzke P, Baker CS, Romeo T. 2009. Regulation of translation initiation by RNA binding proteins. Annu Rev Microbiol 63: 27–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua PC, Ritchey LE, Su Z, Assmann SM. 2016. Genome–wide analysis of RNA secondary structure. Annu Rev Genet 50: 235–266. [DOI] [PubMed] [Google Scholar]
- Bloomfield VA, Crothers DM, Tinoco I Jr. 2000. Nucleic acids: Structures, properties, and functions. University Science Books, Sausalito, CA. [Google Scholar]
- Burkhardt DH, Rouskin S, Zhang Y, Li GW, Weissman JS, Gross CA. 2017. Operon mRNAs are organized into ORF-centric structures that predict translation efficiency. eLife 6: e22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busan S, Weeks KM. 2017. Accurate detection of chemical modifications in RNA by mutational profiling (MaP) with ShapeMapper 2. RNA 24: 143–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd JF. 2011. Harvey K. Littleton: A life in glass. Rizzoli, New York. [Google Scholar]
- Climie SC, Friesen JD. 1988. In vivo and in vitro structural analysis of the rplJ mRNA leader of Escherichia coli Protection by bound L10-L7/L12. J Biol Chem 263: 15166–15175. [PubMed] [Google Scholar]
- Deigan KE, Li TW, Mathews DH, Weeks KM. 2009. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci 106: 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Cheema J, Zhang H, Woolfenden H, Norris M, Liu Z, Liu Q, Yang X, Yang M, Deng X, et al. 2018. Rice in vivo RNA structurome reveals RNA secondary structure conservation and divergence in plants. Mol Plant 11: 607–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. 2014. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505: 696–700. [DOI] [PubMed] [Google Scholar]
- Docter BE, Horowitz S, Gray MJ, Jakob U, Bardwell JC. 2016. Do nucleic acids moonlight as molecular chaperones? Nucleic Acids Res 44: 4835–4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehresmann C, Baudin F, Mougel M, Romby P, Ebel J-P, Ehresmann B. 1987. Probing the structure of RNAs in solution. Nucleic Acids Res 15: 9109–9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R, Moss WN, Rutenberg-Schoenberg M, Simon MD. 2015. Probing Xist RNA structure in cells using Targeted Structure-Seq. PLoS Genet 11: e1005668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C, Chan D, Joseph J, Muuronen M, Coldren WH, Dai N, Correa IR Jr., Furche F, Hadad CM, Spitale RC. 2018. Light-activated chemical probing of nucleobase solvent accessibility inside cells. Nat Chem Biol 14: 276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates KS. 2009. An overview of chemical processes that damage cellular DNA: Spontaneous hydrolysis, alkylation, and reactions with radicals. Chem Res Toxicol 22: 1747–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. 2003. Global analysis of protein expression in yeast. Nature 425: 737–741. [DOI] [PubMed] [Google Scholar]
- Grosshans H, Filipowicz W. 2008. Molecular biology: The expanding world of small RNAs. Nature 451: 414–416. [DOI] [PubMed] [Google Scholar]
- Hangauer MJ, Vaughn IW, McManus MT. 2013. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet 9: e1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes EJ, Hennelly SP, Novikova IV, Irwin JA, Dean C, Sanbonmatsu KY. 2016. COOLAIR antisense RNAs form evolutionarily conserved elaborate secondary structures. Cell Rep 16: 3087–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinicke LA, Nallagatla SR, Hull CM, Bevilacqua PC. 2011. RNA helical imperfections regulate activation of the protein kinase PKR: Effects of bulge position, size, and geometry. RNA 17: 957–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz S, Bardwell JC. 2016. RNAs as chaperones. RNA Biol 13: 1228–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incarnato D, Neri F, Anselmi F, Oliviero S. 2014. Genome-wide profiling of mouse RNA secondary structures reveals key features of the mammalian transcriptome. Genome Biol 15: 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Cech TR. 1985. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: A technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc Natl Acad Sci 82: 648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S, Thomsen AR. 2012. Sensing of RNA viruses: A review of innate immune receptors involved in recognizing RNA virus invasion. J Virol 86: 2900–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. 2010. Genome-wide measurement of RNA secondary structure in yeast. Nature 467: 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Quigley GJ, Suddath FL, McPherson A, Sneden D, Kim JJ, Weinzierl J, Rich A. 1973. Three-dimensional structure of yeast phenylalanine transfer RNA: Folding of the polynucleotide chain. Science 179: 285–288. [DOI] [PubMed] [Google Scholar]
- Kung JT, Colognori D, Lee JT. 2013. Long noncoding RNAs: Past, present, and future. Genetics 193: 651–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok CK, Ding Y, Tang Y, Assmann SM, Bevilacqua PC. 2013. Determination of in vivo RNA structure in low-abundance transcripts. Nat Commun 4: 2971. [DOI] [PubMed] [Google Scholar]
- Kwok CK, Tang Y, Assmann SM, Bevilacqua PC. 2015. The RNA structurome: Transcriptome-wide structure probing with next-generation sequencing. Trends Biochem Sci 40: 221–232. [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Belfort M. 2015. Mobile bacterial group II introns at the crux of eukaryotic evolution. Microbiol Spectr 40: MDNA3-0050-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley PD, Brookes P. 1963. Further studies on the alkylation of nucleic acids and their constituent nucleotides. Biochem J 89: 127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leamy KA, Assmann SM, Mathews DH, Bevilacqua PC. 2016. Bridging the gap between in vitro and in vivo RNA folding. Q Rev Biophys 49: e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Flynn RA, Kadina A, Guo JK, Kool ET, Chang HY. 2016. Comparison of SHAPE reagents for mapping RNA structures inside living cells. RNA 23: 169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Westhof E. 2001. Geometric nomenclature and classification of RNA base pairs. RNA 7: 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Aviran S. 2018. Statistical modeling of RNA structure profiling experiments enables parsimonious reconstruction of structure landscapes. Nat Commun 9: 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockard RE, Kumar A. 1981. Mapping tRNA structure in solution using double-strand-specific ribonuclease V1 from cobra venom. Nucleic Acids Res 9: 5125–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low JT, Weeks KM. 2010. SHAPE-directed RNA secondary structure prediction. Methods 52: 150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowman HB, Draper DE. 1986. On the recognition of helical RNA by cobra venom V1 nuclease. J Biol Chem 261: 5396–5403. [PubMed] [Google Scholar]
- Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, Davidovich C, Gooding AR, Goodrich KJ, Mattick JS, et al. 2016. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165: 1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybecker M, Bilusic I, Raghavan R. 2014. Pervasive transcription: Detecting functional RNAs in bacteria. Transcription 5: e944039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenner S, Blaud M, Fouillen L, Savoye A, Marchand V, Dubois A, Sanglier-Cianferani S, Van Dorsselaer A, Clerc P, Avner P, et al. 2010. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS Biol 8: e1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcia M, Pyle AM. 2012. Visualizing group II intron catalysis through the stages of splicing. Cell 151: 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCown PJ, Corbino KA, Stav S, Sherlock ME, Breaker RR. 2017. Riboswitch diversity and distribution. RNA 23: 995–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis JL, Dunkle JA, Cate JH, Weeks KM. 2012. The mechanisms of RNA SHAPE chemistry. J Am Chem Soc 134: 6617–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino E, Yanofsky C. 2005. Transcription attenuation: A highly conserved regulatory strategy used by bacteria. Trends Genet 21: 260–264. [DOI] [PubMed] [Google Scholar]
- Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. 2005. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc 127: 4223–4231. [DOI] [PubMed] [Google Scholar]
- Miao Z, Adamiak RW, Antczak M, Batey RT, Becka AJ, Biesiada M, Boniecki MJ, Bujnicki J, Chen SJ, Cheng CY, et al. 2017. RNA-Puzzles Round III: 3D RNA structure prediction of five riboswitches and one ribozyme. RNA 23: 655–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D, 3rd, Ritchey LE, Park H, Babitzke P, Assmann SM, Bevilacqua PC. 2018. Glyoxals as in vivo RNA structural probes of guanine base-pairing. RNA 24: 114–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy FL, Cech TR. 1993. An independently folding domain of RNA tertiary structure within the Tetrahymena ribozyme. Biochemistry 32: 5291–5300. [DOI] [PubMed] [Google Scholar]
- Nallagatla SR, Toroney R, Bevilacqua PC. 2011. Regulation of innate immunity through RNA structure and the protein kinase PKR. Curr Opin Struct Biol 21: 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narberhaus F, Waldminghaus T, Chowdhury S. 2006. RNA thermometers. FEMS Microbiol Rev 30: 3–16. [DOI] [PubMed] [Google Scholar]
- Nguyen TH, Galej WP, Fica SM, Lin PC, Newman AJ, Nagai K. 2016. CryoEM structures of two spliceosomal complexes: Starter and dessert at the spliceosome feast. Curr Opin Struct Biol 36: 48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick H, Gilbert W. 1985. Detection in vivo of protein-DNA interactions within the lac operon of Escherichia coli. Nature 313: 795–798. [DOI] [PubMed] [Google Scholar]
- Noller HF, Woese CR. 1981. Secondary structure of 16S ribosomal RNA. Science 212: 403–411. [DOI] [PubMed] [Google Scholar]
- Novikova IV, Hennelly SP, Sanbonmatsu KY. 2012a. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res 40: 5034–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova IV, Hennelly SP, Sanbonmatsu KY. 2012b. Sizing up long non-coding RNAs: Do lncRNAs have secondary and tertiary structure? Bioarchitecture 2: 189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E, Mironov AS. 2004. The riboswitch control of bacterial metabolism. Trends Biochem Sci 29: 11–17. [DOI] [PubMed] [Google Scholar]
- Peattie DA, Gilbert W. 1980. Chemical probes for higher-order structure in RNA. Proc Natl Acad Sci 77: 4679–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. 2012. ENCODE project writes eulogy for junk DNA. Science 337: 1159–1161. [DOI] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499: 172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Re A, Joshi T, Kulberkyte E, Morris Q, Workman CT. 2014. RNA–protein interactions: An overview. In: RNA sequence, structure, and function: Computational and bioinformatic methods (ed. Gorodkin J, Ruzzo WL), pp. 491–521. Humana, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- Ritchey LE, Su Z, Tang Y, Tack DC, Assmann SM, Bevilacqua PC. 2017. Structure-seq2: Sensitive and accurate genome-wide profiling of RNA structure in vivo. Nucleic Acids Res 45: e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchey LE, Su Z, Assmann SM, Bevilacqua PC. 2018. In vivo genome-wide RNA structure probing with Structure-seq2. Methods Mol Biol (in press). [DOI] [PubMed] [Google Scholar]
- Rivas E, Clements J, Eddy SR. 2017. A statistical test for conserved RNA structure shows lack of evidence for structure in lncRNAs. Nat Methods 14: 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. 2014. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505: 701–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg DR, Maquat LE. 2012. Regulation of cytoplasmic mRNA decay. Nat Rev Genet 13: 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senecoff JF, Meagher RB. 1992. In vivo analysis of plant RNA structure: Soybean 18S ribosomal and ribulose-1,5-bisphosphate carboxylase small subunit RNAs. Plant Mol Biol 18: 219–234. [DOI] [PubMed] [Google Scholar]
- Shendure J, Balasubramanian S, Church GM, Gilbert W, Rogers J, Schloss JA, Waterston RH. 2017. DNA sequencing at 40: Past, present and future. Nature 550: 345–353. [DOI] [PubMed] [Google Scholar]
- Siegfried NA, Busan S, Rice GM, Nelson JA, Weeks KM. 2014. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat Methods 11: 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman IM, Li F, Alexander A, Goff L, Trapnell C, Rinn JL, Gregory BD. 2014. RNase-mediated protein footprint sequencing reveals protein-binding sites throughout the human transcriptome. Genome Biol 15: R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Fuchs RT, Grundy FJ, Henkin TM. 2010. Riboswitch RNAs: Regulation of gene expression by direct monitoring of a physiological signal. RNA Biol 7: 104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola MJ, Calabrese JM, Weeks KM. 2015a. Detection of RNA-protein interactions in living cells with SHAPE. Biochemistry 54: 6867–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola MJ, Rice GM, Busan S, Siegfried NA, Weeks KM. 2015b. Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat Protoc 10: 1643–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somarowthu S, Legiewicz M, Chillon I, Marcia M, Liu F, Pyle AM. 2015. HOTAIR forms an intricate and modular secondary structure. Mol Cell 58: 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. 2009. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136: 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spasic A, Assmann SM, Bevilacqua PC, Mathews DH. 2018. Modeling RNA secondary structure folding ensembles using SHAPE mapping data. Nucleic Acids Res 46: 314–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale RC, Crisalli P, Flynn RA, Torre EA, Kool ET, Chang HY. 2013. RNA SHAPE analysis in living cells. Nat Chem Biol 9: 18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale RC, Flynn RA, Torre EA, Kool ET, Chang HY. 2014. RNA structural analysis by evolving SHAPE chemistry. Wiley Interdiscip Rev RNA 5: 867–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, et al. 2015. Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519: 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G, Vogel J, Wassarman KM. 2011. Regulation by small RNAs in bacteria: Expanding frontiers. Mol Cell 43: 880–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tack DC, Tang Y, Ritchey LE, Assmann SM, Bevilacqua PC. 2018. StructureFold2: Bringing chemical probing data into the computational fold of RNA structural analysis. Methods 10.1016/j.ymeth.2018.01.018. [DOI] [PubMed] [Google Scholar]
- Talkish J, May G, Lin Y, Woolford JL Jr., McManus CJ. 2014. Mod-seq: High-throughput sequencing for chemical probing of RNA structure. RNA 20: 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Bouvier E, Kwok CK, Ding Y, Nekrutenko A, Bevilacqua PC, Assmann SM. 2015. StructureFold: Genome-wide RNA secondary structure mapping and reconstruction in vivo. Bioinformatics 31: 2668–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Assmann SM, Bevilacqua PC. 2016. Protein structure is related to RNA structural reactivity in vivo. J Mol Biol 428: 758–766. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. 2010. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329: 533–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker BJ, Breaker RR. 2005. Riboswitches as versatile gene control elements. Curr Opin Struct Biol 15: 342–348. [DOI] [PubMed] [Google Scholar]
- Umeyama T, Ito T. 2017. DMS-seq for in vivo genome-wide mapping of protein–DNA interactions and nucleosome centers. Cell Rep 21: 289–300. [DOI] [PubMed] [Google Scholar]
- Underwood JG, Uzilov AV, Katzman S, Onodera CS, Mainzer JE, Mathews DH, Lowe TM, Salama SR, Haussler D. 2010. FragSeq: Transcriptome-wide RNA structure probing using high-throughput sequencing. Nat Methods 7: 995–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade JT. 2015. Where to begin? Mapping transcription start sites genome-wide in Escherichia coli. J Bacteriol 197: 4–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley JW, Sartor RC, Shen Z, Schmitz RJ, Wu KJ, Urich MA, Nery JR, Smith LG, Schnable JC, Ecker JR, et al. 2016. Integration of omic networks in a developmental atlas of maize. Science 353: 814–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Kertesz M, Spitale RC, Segal E, Chang HY. 2011. Understanding the transcriptome through RNA structure. Nat Rev Genet 12: 641–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Qu K, Ouyang Z, Kertesz M, Li J, Tibshirani R, Makino DL, Nutter RC, Segal E, Chang HY. 2012. Genome-wide measurement of RNA folding energies. Mol Cell 48: 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters KE, Strobel EJ, Yu AM, Lis JT, Lucks JB. 2016a. Cotranscriptional folding of a riboswitch at nucleotide resolution. Nat Struct Mol Biol 23: 1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters KE, Yu AM, Strobel EJ, Settle AH, Lucks JB. 2016b. Characterizing RNA structures in vitro and in vivo with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq). Methods 103: 34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW Jr, Swanstrom R, Burch CL, Weeks KM. 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460: 711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks KM. 2010. Advances in RNA structure analysis by chemical probing. Curr Opin Struct Biol 20: 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E, Auffinger P. 2006. RNA tertiary structure. In Encyclopedia of analytical chemistry (ed. Meyers RA), pp. 5222–5232. Wiley, New York. [Google Scholar]
- Zaug AJ, Cech TR. 1995. Analysis of the structure of Tetrahymena nuclear RNAs in vivo: Telomerase RNA, the self-splicing rRNA intron, and U2 snRNA. RNA 1: 363–374. [PMC free article] [PubMed] [Google Scholar]
- Zubradt M, Gupta P, Persad S, Lambowitz AM, Weissman JS, Rouskin S. 2017. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat Methods 14: 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]