Abstract
Termination of mRNA translation occurs when a stop codon enters the A site of the ribosome, and in eukaryotes is mediated by release factors eRF1 and eRF3, which form a ternary eRF1/eRF3–guanosine triphosphate (GTP) complex. eRF1 recognizes the stop codon, and after hydrolysis of GTP by eRF3, mediates release of the nascent peptide. The post-termination complex is then disassembled, enabling its constituents to participate in further rounds of translation. Ribosome recycling involves splitting of the 80S ribosome by the ATP-binding cassette protein ABCE1 to release the 60S subunit. Subsequent dissociation of deacylated transfer RNA (tRNA) and messenger RNA (mRNA) from the 40S subunit may be mediated by initiation factors (priming the 40S subunit for initiation), by ligatin (eIF2D) or by density-regulated protein (DENR) and multiple copies in T-cell lymphoma-1 (MCT1). These events may be subverted by suppression of termination (yielding carboxy-terminally extended read-through polypeptides) or by interruption of recycling, leading to reinitiation of translation near the stop codon.
OVERVIEW OF TRANSLATION TERMINATION AND RECYCLING
Translation is a cyclical process that comprises initiation, elongation, termination, and ribosome recycling stages (Jackson et al. 2010). Termination is triggered when a stop codon enters the A site of the ribosome, and is mediated by the release factors eRF1 and eRF3 (Dever and Green 2012; Jackson et al. 2012). eRF1 is omnipotent, that is, it is responsible for recognition of all three stop codons, and induces release of the nascent polypeptide from the P-site peptidyl-transfer RNA (tRNA), whereas eRF3 is a GTPase that enhances polypeptide release. The resulting post-termination complex (post-TC) is recycled by splitting of the ribosome, which is mediated by ABCE1. This step is followed by release of deacylated tRNA and messenger RNA (mRNA) from the 40S subunit via redundant pathways involving initiation factors, ligatin (eIF2D) or density-regulated protein (DENR), and multiple copies in T-cell lymphoma-1 (MCT1). Recycling enables ribosomes and mRNAs to participate in multiple rounds of translation.
Here, I focus on recent advances in understanding of mechanisms of termination and ABCE1-mediated ribosomal splitting, discuss unresolved aspects of recycling, and consider how termination or recycling is subverted to allow translation of carboxy-terminally extended “read-through” polypeptides or of downstream open reading frames (ORFs).
TERMINATION
eRF1–eRF3 Interactions and Ternary Complex Formation
Termination is mediated by eRF1 and eRF3 (Alkalaeva et al. 2006). eRF1 has an amino-terminal domain (N) that is responsible for recognition of the stop codon in the A site (Bertram et al. 2000); a middle domain (M) containing a universally conserved apical GGQ motif that induces release of the nascent polypeptide from peptidyl-tRNA in the ribosomal P site (Frolova et al. 1999); and a carboxy-terminal domain (C) that binds to eRF3 and ABCE1, and contains a mini-domain that affects stop codon specificity (Fig. 1A) (Song et al. 2000; Mantsyzov et al. 2010). eRF3 consists of (1) a nonconserved amino-terminal domain that is not required for eRF3’s function in termination, but binds the poly(A)-binding protein (PABP) (Hoshino et al. 1999; Kozlov and Gehring 2010) and the nonsense-mediated decay (NMD) factor UPF3b (Neu-Yilik et al. 2017), and (2) a canonical guanosine triphosphate (GTP)-binding domain (G) and two β-barrel domains (2 and 3), which are homologous to GTP-binding translation factors such as EF-Tu, eEF1A, and the carboxy-terminal region of the ribosome rescue factor Hbs1 (Fig. 1B) (Kong et al. 2004; Atkinson et al. 2008; van den Elzen et al. 2010). As in all GTPases, the G domain of eRF3 contains “Switch I” and “Switch II” elements that are essential for binding and hydrolysis of GTP and that regulate the nucleotide-dependent conformational status of the factor. Switch I and Switch II are disordered in free eRF3 (Kong et al. 2004) but become ordered on binding to the γ-phosphate of GTP in the presence of eRF1, with which they also interact (Cheng et al. 2009; des Georges et al. 2014; Preis et al. 2014; Shao et al. 2016). There are two isoforms of eRF3 that are encoded by different genes and have different amino-terminal domains; both bind eRF1 and are functional termination factors (Hoshino et al. 1998; Chauvin et al. 2005). eRF3b is predominantly expressed in brain tissue, whereas eRF3a is ubiquitously expressed (Hoshino et al. 1998; Chauvin et al. 2005).
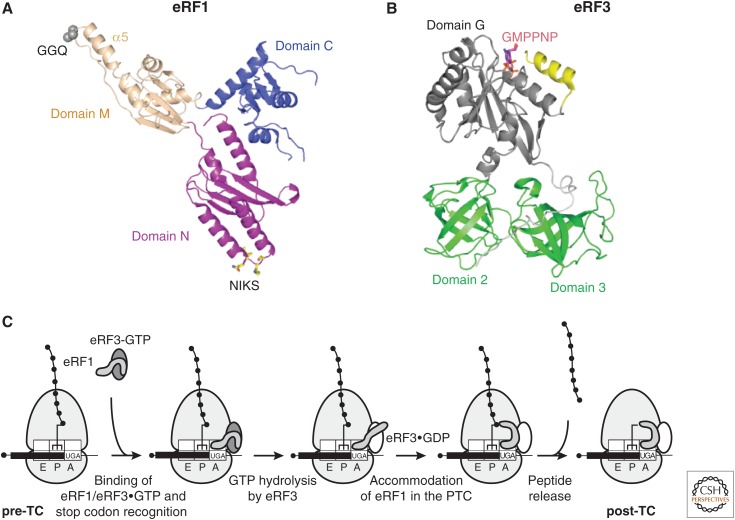
Figure 1.
Translation termination in eukaryotes. Ribbon representations of (A) human eRF1 (protein data bank [PDB]: 1DT9), with the Cα atoms of the GGQ motif in domain M and the NIKS motif in domain N shown in CPK and stick models, respectively, and (B) Schizosaccharomyces pombe eRF3 (amino acids 215–662, and thus lacking the nonconserved amino-terminal domain) (PDB: 1R5B), with bound GMPPNP shown in a stick model. (C) Outline of the termination process. The pretermination complex (pre-TC) contains peptidyl-transfer RNA (tRNA) in the P site. The eRF1/eRF3–guanosine triphosphate (GTP) complex binds to the A site of the pre-TC, and eRF1 recognizes the stop codon, which, with the +4 nucleotide, binds in a pocket formed by eRF1 and the 40S subunit. eRF1’s M domain dissociates from eRF3’s Switch I/Switch II elements, and after GTP hydrolysis by eRF3, accommodates in the peptidyl-transferase center (PTC), inducing peptide release. eRF1 and possibly eRF3-guanosine diphosphate (GDP) remain associated with post-termination complexes (post-TCs).
eRF1 enhances binding of GTP to eRF3 by acting as a GTP dissociation inhibitor, promoting formation of a stable eRF1/eRF3•GTP complex (Fig. 1C) (Hauryliuk et al. 2006; Mitkevich et al. 2006; Pisareva et al. 2006). eRF1 and eRF3 interact extensively with each other on and off the ribosome (Cheng et al. 2009; Taylor et al. 2012; des Georges et al. 2014; Preis et al. 2014; Shao et al. 2016). In pretermination TCs (pre-TCs) containing eRF1/eRF3 before GTP hydrolysis, eRF1’s M domain packs against eRF3, inserting into a cleft between domain 2 and the G domain such that the GGQ motif is fixed close to Switch I. The M domain may engage with Switch II (Cheng et al. 2009; des Georges et al. 2014; Preis et al. 2014; Shao et al. 2016). The “preaccommodation” conformation of the M domain before GTP hydrolysis is incompatible with peptide-release activity because the catalytic GGQ motif is sequestered >80 Å from the P-site peptidyl-tRNA’s ester bond in the peptidyl-transferase center (PTC) of the 60S subunit. eRF1’s N domain extends into the 40S subunit’s decoding center (see below), while domain C interacts with the stalk base of the 60S subunit and, via its mini-domain, with the 40S subunit beak. Like other translational GTPases, eRF3 binds to the GTPase-associated center (GAC), between the sarcin–ricin loop (SRL) of the 60S subunit and helices h5 and h14 of 18S ribosomal RNA (rRNA) on the 40S subunit (des Georges et al. 2014; Preis et al. 2014; Shao et al. 2016).
Stop Codon Recognition
The canonical genetic code has three stop codons (UAA, UAG, and UGA), and the efficiency of termination is enhanced by a purine residue in the +4 position and by a +5 purine if the +4 residue is a pyrimidine (McCaughan et al. 1995). In a few organisms, including some ciliate protists, green algae, and diplomonads, UGA is reassigned as a sense codon and UAA and UAG are retained as stop codons, UAA and UAG are reassigned as sense codons with UGA as the sole termination codon, or UAG is reassigned as a sense codon and UAA and UGA function as stop codons (Keeling 2016; Pánek et al. 2017). In extreme examples, such as the ciliate protist Condylostoma magnum and a member of the proposed Blastocrithidia clade of trypanosomatids (Heaphy et al. 2016; Swart et al. 2016; Záhonová et al. 2016), all three stop codons serve as sense codons at internal positions but specify termination when located close to the 3′-end of mRNA. A hypothesis concerning position-dependent termination is discussed below. Stop codon reassignment depends in part on alterations in eRF1’s sequence, and mutational studies coupled with analysis of eRF1 from “variant code” organisms identified highly conserved motifs in the N domain that influence stop codon recognition (Jackson et al. 2010; Blanchet et al. 2015). These motifs include GTS31-33, E55, TASNIKS58-64, and YxCxxxF125-130 (numbering for human eRF1). Recent cryoelectron microscopy (cryo-EM) studies (Brown et al. 2015; Matheisl et al. 2015; Shao et al. 2016) have established a molecular framework for this process.
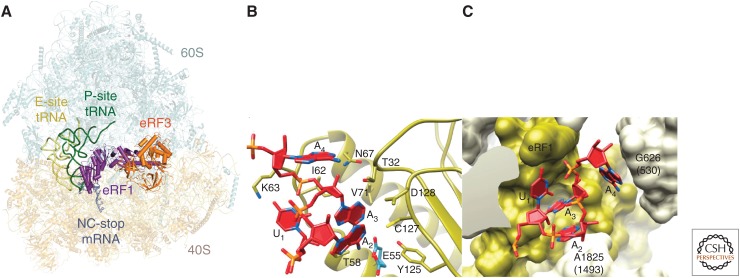
In eRF1-bound ribosomal complexes, the N domain reaches into the A site, forming a pocket that accommodates the stop codon and the +4 nucleotide in a compacted conformation (Fig. 2). This compacted state and eRF1’s interactions with the stop codon are likely maintained throughout the termination process until eRF1 dissociates (Shao et al. 2016). Stabilization of the compacted state requires a +1U, which is thus a determinant of stop codon recognition, as are the stacking interactions of the +2, +4, and +5 nucleotides with A1825, G626, and C1698 of 18S rRNA, respectively, as well as the stop codon’s multiple interactions with eRF1. The stabilizing interaction of the +1 uridine with N61 and K63 of the TASNIKS motif would not be possible for cytidine at this position, and steric hindrance discriminates against +1 purines. Interactions of the YxCxxxF motif and E55 with +2 and +3 nucleotides are possible only with purines, and T32 of the GTS motif can hydrogen bond with the +3 nucleotide of UAG but not UGA or UGG codons. The specificity of the flexible GTS motif, and the mutual repulsion of G residues at +2 and +3 positions, account for eRF1’s discrimination against UGG codons. These observations are consistent with site-directed cross-linking analysis of stop codon/eRF1 interactions (Chavatte et al. 2002; Bulygin et al. 2010) and mutational analyses of stop codon recognition and specificity (Cheng et al. 2009; Jackson et al. 2010; Conard et al. 2012; Blanchet et al. 2015). Recognition of stop codons, particularly UGA, likely occurs via multiple steps involving RNA compaction in the A site, conformational changes in TASNIKS, and GTS motifs in eRF1 and other localized eRF3-induced changes (Fan-Minogue et al. 2008; Wong et al. 2012; Kryuchkova et al. 2013).
Figure 2.
Structure of the mammalian termination complex. (A) The ribosomal termination complex assembled with eRF1 (purple) and eRF3 (orange). (Panel A from Shao et al. 2016; reprinted, with permission, under the terms of Creative Commons Attribution License CC-BY). (B) Close-up view of the stop codon binding pocket of the eRF1 N domain bound to a UAA(A) stop codon (dark red), showing residues important for stop codon recognition, including GTS(31–33), E55, TASNIKS(58–64), N67, V71, and YxCxxxF(125–131). (C) Surface representation of the UAA(A)-binding cavity formed by the eRF1 N domain and 18S ribosomal RNA (rRNA) in the decoding center of the 40S ribosomal subunit, showing A1825 and G626, which engage in stacking interactions with the +2 and +4 nucleotides of the stop codon. (Panel C from Matheisl et al. 2015; reprinted, with permission, from Oxford University Press © 2015.)
The Mechanism of GTP Hydrolysis by eRF3
eRF3 belongs to a family of translational GTPases, including eEF1A, EF-Tu, and Hbs1, which deliver aminoacyl-tRNA, eRF1 or the related protein Pelota to the A site (Atkinson et al. 2008; Pisareva et al. 2011). In the case of ribosome-bound eEF1A/EF-Tu, establishment of complementarity between cognate aminoacyl-tRNA and the A-site codon induces GTP hydrolysis and is required for release of the tRNA “cargo” so that it can accommodate in the PTC. The decoding center on the 40S subunit is >70 Å from the active site of these GTPases, so that activation of these factors requires long-range signaling. Binding of cognate aminoacyl-tRNA to the A-site codon leads to domain closure in the small ribosomal subunit, which moves EF-Tu’s GTPase domain so that it binds to the SRL (Loveland et al. 2017). This is an obligatory step in the activation of EF-Tu and its hydrolysis of GTP (Maracci and Rodnina 2016). Binding of cognate aminoacyl-tRNA to the A site of eukaryotic ribosomal complexes leads to a similar domain closure and activation of eEF1A (Shao et al. 2016).
The mechanism of activation of eRF3’s GTPase activity is somewhat different. eRF3’s GTPase activity requires eRF1’s M and C domains and is ribosome-dependent, and although eRF1 can stimulate this activity in the absence of its N domain and an A-site termination codon (Frolova et al. 1996; Kononenko et al. 2008), stop codon recognition by eRF1 is thought to accelerate GTP hydrolysis by eRF3 (e.g., Wada and Ito 2014). eRF3, eEF1A, and EF-Tu all engage in similar interactions with the shoulder of the small subunit (Voorhees et al. 2010; des Georges et al. 2014; Shao et al. 2016). Termination complexes containing eRF1 and eRF3 at a stage just before hydrolysis of eRF3-bound GTP have not been visualized. However, several observations suggest that the SRL is required for activation of eRF3, as for other translational GTPases. Thus, eRF3’s Switch I interacts with G4600 of the SRL (Shao et al. 2016), and termination defects are caused by a substitution in the Saccharomyces cerevisiae SRL (Liu and Liebman 1996) and by substitutions in eRF3 that are close to the SRL in pre-TCs (Wada and Ito 2014). Domain closure in the 40S subunit is not induced by binding of eRF1 or Pelota to the A site (Hilal et al. 2016; Shao et al. 2016), however, indicating that a different mechanism must be responsible for the relative repositioning of eRF3 so that Switch I and Switch II can interact with the SRL. This necessary dissociation of eRF1’s M domain from Switch I/Switch II to remove a steric block to GTP hydrolysis might be induced by the establishment of interactions between h14 of the 40S subunit and the conserved R192 in eRF1’s M domain, and of h5 with eRF3 domain 2 following binding of eRF1/eRF3•GTP to the pre-TC (des Georges et al. 2014). These interactions are important for eRF1-mediated stimulation of eRF3’s GTP hydrolysis activity (Cheng et al. 2009) and mimic a step in EF-Tu activation that leads to displacement of the 3′-end of tRNA from the Switch I loop (Voorhees et al. 2010), respectively.
Conformational Rearrangements following Hydrolysis of eRF3-Bound GTP
The timing of events after hydrolysis of eRF3-bound GTP has not been established, but by analogy with EF-Tu (Pape et al. 1998), rapid GTP hydrolysis would be followed by slower accommodation of eRF1 in the PTC and dissociation of eRF3. eRF3-guanosine diphosphate (GDP) appears to remain on the ribosome for an extended period (Bulygin et al. 2016).
After hydrolysis of eRF3-bound GTP, eRF1 adopts an extended conformation that allows the catalytic GGQ motif at the tip of domain M to accommodate in the PTC (Matheisl et al. 2015; Muhs et al. 2015; Shao et al. 2016). This transition results from domain M undergoing a 140° rotation relative to domain N, which remains bound to the stop codon in the A site, as well as rotation of domain C. The motive force for the reorientation of M and C domains may be relaxation of an eRF3-enforced kink between α-helix 8 (in domain M) and α-helix 9 (in domain C), enabling them to form a single continuous α-helix (Shao et al. 2016). The inhibition of eRF1-mediated peptide release by eRF3•GMPPNP (Alkalaeva et al. 2006; Fan-Minogue et al. 2008) is likely because it “locks” eRF1 so that it cannot switch from the compact to the extended conformation. Conformational changes in eRF1 domain C and ribosomal protein uL11 disrupt the interaction of the eRF1 mini-domain with the head of the 40S subunit and lead to establishment of new interactions with uL11 and the L7/L12 stalk base that may stabilize binding of domain C. Consistently, termination is impaired in an S. cerevisiae uL11 deletion strain (Salas-Marco and Bedwell 2005).
Although eRF1 alone can induce peptide release, this activity is strongly increased by eRF3 (Alkalaeva et al. 2006), confirming suggestions that eRF3’s GTPase activity couples stop codon recognition and peptidyl-tRNA hydrolysis by eRF1 (Salas-Marco and Bedwell 2004). This effect is apparent even when recycling of eRF1 is not required, and it can therefore not be solely attributed to enhanced release of eRF1 from post-TCs or from ribosomal subunits after dissociation of post-TCs (e.g., Eyler et al. 2013). Enhancement by eRF3 of peptide release could be the result of increased eRF1 recruitment to pre-TCs (by eRF3 escorting it to the A site in an EF-Tu-like manner) or by augmentation of the catalytic rate of peptidyl-tRNA hydrolysis. The characteristic +2 nt toeprint shift induced by binding of eRF1 to pre-TCs (Alkalaeva et al. 2006) correlates with the compaction of the extended stop codon sequence (Brown et al. 2015; Matheisl et al. 2015). eRF3 enhances the shift induced by wild-type eRF1 and leads to its appearance in the presence of some eRF1 mutants (Kryuchkova et al. 2013), suggesting that eRF3 promotes stop codon recognition or stabilizes the resulting pre-TC. As well as enhancing the rate of termination, eRF3 could potentially increase its fidelity by kinetic proofreading, by introducing an irreversible GTP hydrolysis step between stop codon recognition and peptidyl-tRNA hydrolysis.
The Mechanism of Peptide Release
In the extended conformation of eRF1, domain M positions the GGQ motif in the PTC close to the CCA end of the P-site peptidyl-tRNA, with Q135 adjacent to the ester bond that links the tRNA to the nascent polypeptide (Preis et al. 2014; Brown et al. 2015; Muhs et al. 2015). The similar conformations of the GGQ motif of eRF1 and the GGQ motifs of the otherwise unrelated bacterial RF1/RF2 release factors, and the interactions of the eRF1 M domain and domain 3 of bacterial RF1/RF2 with conserved elements of the ribosome (e.g., Laurberg et al. 2008; Jin et al. 2010) suggest that they function analogously to promote peptidyl-tRNA hydrolysis. Placement of the bacterial GGQ motif in the PTC induces conformational changes that expose the peptidyl-tRNA ester bond to nucleophilic attack by water and stabilize the tetrahedral transition state, promoting cleavage and releasing the nascent polypeptide (Jin et al. 2010; see the discussion of models for this process in Rodnina 2018).
RECYCLING
Recycling of post-termination ribosomes (Fig. 3) is initiated by the highly conserved, essential protein ABCE1, which also recycles vacant 80S ribosomes and stalled ribosomal elongation complexes after their recognition by Hbs1/Pelota (Pisarev et al. 2010, 2011; Franckenberg et al. 2012; Jackson et al. 2012). ABCE1-mediated recycling of post-TCs depends on the presence of eRF1 in the A site (Pisarev et al. 2010). eRF1 therefore participates in two successive stages of protein synthesis: termination and recycling.
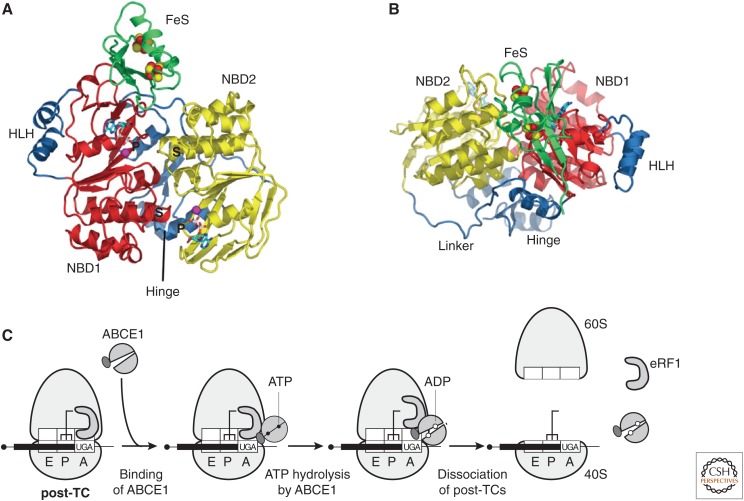
Figure 3.
Ribosome recycling in eukaryotes. (A,B) Ribbon representations of ABCE1 from Pyrococcus abyssi bound to ADP (protein data bank [PDB]: 3BK7). Top (A) and front (B) views, showing nucleotide-binding domain NBD1 (red), with its helix–loop–helix insertion (HLH), linked to NBD2 (yellow) by the hinge domain. The FeS domain includes two [4Fe-S] clusters (red and yellow spheres), and binds to the lateral opening of the nucleotide-binding cleft. Nucleotides bind to the two composite binding sites formed by P-loop/Walker A (“P”) and signature (S) motifs. (From Karcher et al. 2008; reprinted, with permission, from The American Society for Biochemistry and Molecular Biology © 2008.) (C) Model for ribosome recycling (Pisarev et al. 2010), in which ABCE1 binds to eRF1 on the post-termination complex (post-TC), and ATP hydrolysis leads to a power stroke that splits the post-termination ribosome, yielding a 60S subunit, eRF1, ABCE1, and a 40S subunit still bound to messenger RNA (mRNA) and deacylated transfer RNA (tRNA).
The Structure of ABCE1 and Its Interactions with eRF1
ABCE1 has twin nucleotide-binding domains (NBDs) with two composite nucleotide-binding sites formed by motifs from both domains, a helix–loop–helix (HLH) motif in NBD1 and a unique amino-terminal FeS domain containing two [4Fe-4S]2+ clusters, which is connected to the NBD core by a hinged cantilever arm (Fig. 3A,B) (Barthelme et al. 2007, 2011; Karcher et al. 2008). The NBDs adopt an “open” state when bound to ADP or nucleotide-free (Karcher et al. 2008; Barthelme et al. 2011), but transition to a closed form with an extensive interface between NBDs on binding ATP (Heuer et al. 2017). ABCE1 has been observed on the 80S ribosome in an intermediate semi-closed state (Becker et al. 2012; Preis et al. 2014; Brown et al. 2015), but must undergo domain closure to manifest its ATPase activity and functionality in recycling.
ABCE1 binds in the ribosomal intersubunit space, interacting extensively via the HLH and hinge elements with sites on the 40S subunit (h5–h15 and h8–h14), which constitute contact points for ribosome-bound translational GTPases (such as eRF3) and via NBD2 at a single site (rpL9) on the 60S subunit. The amino-terminal FeS domain binds NBD2 and eRF1’s C domain. Given that ABCE1 binds sites on eRF1 (Preis et al. 2014; Shao et al. 2016) and the 40S subunit that interact with eRF3, ABCE1’s association with post-TCs requires prior dissociation of eRF3.
The Mechanism of ABCE1-Mediated Ribosomal Splitting
Cycles of binding of ATP to the twin nucleotide-binding sites in ABC proteins (yielding a “closed” state), hydrolysis of ATP and release of ADP (leading to an “open” state) induce conformational changes in these proteins that are thought to generate power strokes that drive structural changes in associated domains or macromolecules (Rees et al. 2009). In ribosome-bound complexes, ABCE1 is in an intermediate semi-closed state, in which NBD2 has rotated, leading to repositioning of the FeS domain (Becker et al. 2012; Franckenberg et al. 2012). These changes are accompanied by movement of the eRF1 C domain, because of its close association with the FeS domain, and possibly result in further conformational shifts in eRF1. Subsequent conformational changes in ABCE1 could destabilize intersubunit bridges, leading to ribosomal splitting (Fig. 3C). Movement of the FeS domain has been observed, albeit in complexes that had been assembled in vitro from archaeal 30S subunits or yeast 40S subunits and ABCE1•AMP-PNP without eRF1 (Kiosze-Becker et al. 2016; Heuer et al. 2017). However, a complete structural outline of the mechanism of recycling, linking specific conformational changes in ABCE1, eRF1, and the ribosome to binding and hydrolysis of ATP in the twin nucleotide-binding sites of ABCE1, remains to be established.
ATP hydrolysis by ABCE1 is required for it to split eRF1-bound post-TCs and Pelota-associated ribosomal complexes (Pisarev et al. 2010, 2011; Shoemaker and Green 2011; Becker et al. 2012). In one report, splitting of 80S ribosomes lacking bound mRNA and P-site peptidyl-tRNA was dependent on ATP binding but not its hydrolysis, which was, however, required for release of ABCE1 from the small ribosomal subunit (Barthelme et al. 2011). The basis for this discrepancy remains unclear. This latter alternative model proposes that retention of ABCE1 on the small subunit primes the next round of initiation by enhancing recruitment of initiation factors (Heuer et al. 2017), but neither the stage at which ABCE1 is released in this hypothetical process, nor the trigger for ATP hydrolysis by 40S-bound ABCE1 that would induce release have been determined. Whereas ABCE1’s intrinsic ATPase activity is strongly activated by eRF1-bound ribosomes, stimulation by 40S subunits is weak, even though they bind avidly to ABCE1 (Pisarev et al. 2010).
Release of mRNA and Deacylated tRNA from Post-Termination Ribosomal Complexes
Deacylated tRNA and mRNA remain bound to the 40S subunit after release of the 60S subunit, and can be released by eIF1, eIF1A, eIF3 and its weakly associated eIF3j subunit (Pisarev et al. 2007, 2010). The multi-subunit eIF3 binds to the 40S subunit’s solvent side, extending from the mRNA entrance to the exit (des Georges et al. 2015), interacting with eIF3j at the former site and with eIF1 at the latter. eIF1 and eIF1A bind to the subunit’s interface, on the platform adjacent to P-site initiator tRNA and in the A site, respectively. eIF1 discriminates against noninitiator tRNAs in the P site (Lomakin et al. 2006) and destabilizes their binding in a manner that is augmented by eIF1A and particularly eIF3 (Pisarev et al. 2010). Release of P-site tRNA leads to dissociation of mRNA in the absence of eIF3 and its partial retention in its presence. eIF3j enhances release of mRNA. At concentrations below 1 mm free Mg2+, these factors can recycle post-TCs, eIF3 being primarily responsible for ribosome splitting (Pisarev et al. 2007). Interestingly, eIF3-mediated splitting is inhibited by eIF4F (Skabkin et al. 2013).
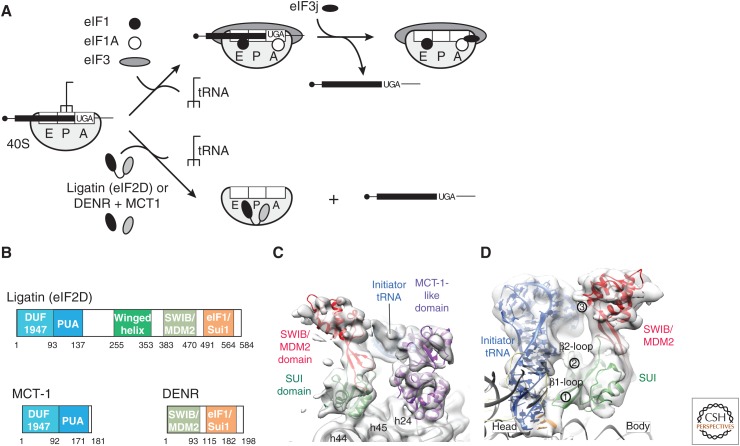
There is significant redundancy in the process of tRNA/mRNA release from 40S subunits after ABCE1-mediated dissociation of post-TCs (Fig. 4A). This process can also be mediated by ligatin (eIF2D) and, less effectively, by MCT-1 and DENR, which are interacting proteins that are homologs of the amino- and carboxy-terminal regions of ligatin (eIF2D) (Fig. 4B) (Skabkin et al. 2010). MCT-1 and ligatin’s (eIF2D) amino-terminal region contains DUF1947 and PUA domains (Tempel et al. 2013), whereas DENR and ligatin’s (eIF2D) carboxy-terminal region contains SWIB/MDM2 and SUI1/eIF1 domains (Fig. 4C) (Vaidya et al. 2017; Weisser et al. 2017). Ligatin (eIF2D) also contains a central winged-helix domain (WHD). The location of the MCT1-like and WHD domains on the interface surface of the 40S subunit may be incompatible with binding of peripheral elements of eIF3a and eIF3c or eIF3b (Lomakin et al. 2017; Weisser et al. 2017), and the SUI1/eIF1 domains of DENR and ligatin (eIF2D) could clash with the anticodon stem-loop of deacylated P-site tRNA and eject it.
Figure 4.
Release of messenger RNA (mRNA) and deacylated transfer RNA (tRNA) from recycled 40S subunits. (A) Outline of the release process mediated by initiation factors 1, 1A, 3, and 3j, by ligatin (eIF2D), or by multiple copies in T-cell lymphoma-1 (MCT1) and density-regulated protein (DENR). (B) Domain organization of human ligatin (eIF2D), MCT-1, and DENR. (C) The SUI domain (green) and the MCT-1-like domain (purple) of ligatin (eIF2D) bound to the 40S subunit (gray) and interacting with P-site tRNA, seen from the foot of the 40S subunit. (D) A model derived by docking the crystal structure of the SWIB/MDM2-SUI domain of ligatin (eIF2D) into the cryo-EM map of a ligatin (eIF2D)-bound ribosomal complex containing P-site tRNA and seen from the A site, showing interactions of the SUI loop b1 with the codon–anticodon duplex (1), loop b2 with C11 and G12 in the D-loop (2), and the SWIB/MDM2 domain with C66, G67, and G68 of the acceptor stem (3). (Panels B–D from Weisser et al. 2017; reprinted, with permission, from Elsevier © 2017.)
REGULATION OF TERMINATION
Various trans-acting factors regulate termination and integrate it with processes such as NMD (see Karousis and Mühlemann 2018). Termination may be regulated to arrest ribosomes at the stop codon of an upstream ORF, thereby reducing translation of downstream ORFs, or to modulate read-through, in which a near-cognate or natural suppressor tRNA decodes the stop codon, allowing translation to proceed in the same reading frame (e.g., Beier and Grimm 2001). Stop codon suppression allows the expression of carboxy-terminally extended forms of a protein, potentially in a tissue-specific or developmentally regulated manner, and is used by some viruses to modulate the relative levels of expression of structural and nonstructural polyproteins (Firth and Brierley 2012). In yeast, insertion of near-cognate tRNA at premature stop codons is determined by the ability of the ribosomal A site to accommodate mispairing, leading to insertion of Trp, Arg, and Cys at UGA codons and Gln, Tyr, and Lys at UAA and UAG codons (Blanchet et al. 2014; Roy et al. 2015). Flanking residues, particularly the +4 nucleotide, influence selection of near-cognate aminoacyl-tRNAs in yeast and humans (Beznosková et al. 2016; Xue et al. 2017).
Regulation of Termination by trans-Acting Factors
mRNAs harboring premature termination codons are targeted for destruction by NMD, which is thought to be activated because termination at such codons is slower and less efficient than at “normal” stop codons (reviewed in He and Jacobson 2015). Termination at premature termination codons is thought to be impaired both by attenuation of the activity of termination enhancers and by negative regulator(s). PABP binds to eRF3 (Hoshino et al. 1999) and promotes termination in humans (Ivanov et al. 2008, 2016). Impaired termination might thus reflect attenuation of PABP’s stimulatory influence caused by the greater spacing between the PABP-bound 3′-poly(A) tail and premature vs. “normal” stop codons. A similar PABP-mediated position-dependent effect may underlie termination on mRNAs lacking dedicated termination codons in which UAA, UAG, and UGA codons at internal positions are decoded as sense codons, whereas those near the 3′-end of mRNAs are recognized by eRF1 as stop codons (Heaphy et al. 2016; Swart et al. 2016).
Various observations suggest that NMD factors also influence termination, possibly in an organism-specific manner. UPF1, UPF2, and UPF3 interact with eRF1 and eRF3 at the A-site of terminating ribosomes (Kashima et al. 2006), but whereas deletion of UPF genes in yeast increased stop codon read-through (e.g., Wang et al. 2001), small-interfering RNA (siRNA)-mediated depletion of UPF1, UPF2, and UPF3B in human cells had the opposite effect (Ivanov et al. 2008; Jia et al. 2017). ATP hydrolysis by UPF1 is required for efficient termination at PTCs in yeast (Serdar et al. 2016), whereas human UPF1 had no influence on termination in a fully reconstituted translation termination system (Neu-Yilik et al. 2017). However, UPF3B, which binds directly to eRF3 and eRF1, impaired stop codon recognition and peptide release in this system, and dissociated post-termination complexes, by as-yet-undetermined mechanisms.
eIF5A promotes elongation, particularly on polyproline-encoding sequences (Saini et al. 2009). Ribosomal profiling determined that termination is also significantly impaired in eIF5A-depleted S. cerevisiae (Pelechano and Alepuz 2017; Schuller et al. 2017). Estimates of eIF5A’s stimulation of the termination rate range from 2- to 17-fold (Saini et al. 2009; Schuller et al. 2017). eIF5A binds between the ribosomal P and E sites, and interacts with the CCA-end of P-site tRNA, potentially stabilizing it in an orientation that favors peptidyl-tRNA hydrolysis (Melnikov et al. 2016; Schmidt et al. 2016). Depletion of eIF5A in HeLa cells (Hoque et al. 2017) and a temperature-sensitive mutation in yeast eIF5A (Schrader et al. 2006) both attenuated NMD, hinting that some termination complexes may interact with eIF5A in a manner that targets them for NMD.
ABCE1 has a critical role in post-termination ribosome recycling, but in S. cerevisiae also promotes eRF1-mediated peptide release in a manner that is independent of ATP hydrolysis by ABCE1 and that is enhanced by but not dependent on eRF3 (Shoemaker and Green 2011). ABCE1’s activity in recycling depends on prior dissociation of eRF3 from eRF1, but its enhancement of termination is not caused by stabilization of binding of eRF1 to ribosomes (Shoemaker and Green 2011). The basis for this activity remains unknown.
Trans-acting factors may also down-regulate termination by sequestering release factors. The Moloney murine leukemia virus (MuLV) reverse transcriptase (RT) binds to the carboxy-terminal domain of eRF1, outcompeting eRF3 and thereby promoting stop codon read-through (Tang et al. 2016). Suppression of the gag UAG stop codon permits read-through to the in-frame pol gene and synthesis of replicase proteins (including RT) as a Gag-Pol fusion protein.
Regulation of Termination by cis-Acting RNA Elements
In MuLV, read-through is also promoted by a pseudoknot downstream from the gag stop codon (Houck-Loomis et al. 2011) by an unknown mechanism that might involve impairment of ribosome function or release factor binding. Numerous viruses use read-through, in many instances promoted by secondary structures immediately downstream from the stop codon (Firth et al. 2011; Napthine et al. 2012). Until a decade ago, read-through was thought to be exceedingly rare in nonviral mRNAs, but is now known to be pervasive in many eukaryotes (Jungreis et al. 2011, 2016; Dunn et al. 2013; Eswarappa et al. 2014; Loughran et al. 2014). Several suppressible stop codons are associated with downstream structural elements, which in the case of the Drosophila hdc stop codon, is a hairpin that can function in heterologous mRNAs (Steneberg and Samakovlis 2001; Jungreis et al. 2011). However, other mRNAs lack such elements, and read-through is instead promoted by a downstream element that binds hnRNP A2/B1 (Eswarappa et al. 2014), by a downstream GUAC motif (Loughran et al. 2014) or by as-yet-unidentified effectors, in all instances by unknown mechanisms.
Regulation of Termination by Post-Translational Modification
Components of the translation apparatus, including eRF1, eRF3, and the ribosome, are post-translationally modified, but the functional consequences of these modifications remain largely uncharacterized. Hydroxylation of mammalian eRF1 at K63 in the TASNIKS motif enhances termination efficiency (Feng et al. 2014), possibly by establishing an additional hydrogen-bonding interaction with mRNA (Brown et al. 2015). OGFOD1 catalyzes hydroxylation of a conserved prolyl residue in rps23 at a site that projects into the decoding center. This modification has varying effects on termination in yeast and human cells, and at stop codons with different contexts (e.g., Keeling et al. 2006; Loenarz et al. 2014; Singleton et al. 2014). It could potentially regulate read-through on specific mRNAs in specific circumstances. The Gln of the GGQ motif of yeast eRF1 is methylated by a methyltransferase that consists of the catalytic Mtq2 subunit and the zinc finger protein Trm112 (Graille et al. 2012). It is not known whether this modification enhances eRF1’s function in promoting peptide release, as has been reported in bacteria for RF1 and RF2 (Pierson et al. 2016).
Regulation of Termination by Nascent Peptides
Translation of a subset of mRNAs is regulated by the peptide encoded by an upstream ORF (uORF) in the 5′-leader region, causing ribosomes to stall at the uORF stop codon. The human cytomegalovirus uORF2-encoded peptide impairs translation of the gp48 gene (Janzen et al. 2002), and, in fungi, the uORF-encoded arginine attenuator peptide stalls ribosomes at the uORF termination codon of the arg-2 gene when arginine levels are high, resulting in feedback inhibition of synthesis of the first enzyme specific for arginine biosynthesis (Wang and Sachs 1997). These regulatory nascent peptides bind to the ribosomal tunnel and perturb the PTC so that although the eRF1 GGQ-loop is appropriately positioned, it cannot promote peptide release (Bhushan et al. 2010; Matheisl et al. 2015). Systematic analysis of uORFs in Arabidopsis thaliana suggests that this mechanism is prevalent in all eukaryotes (Ebina et al. 2015). Stalling in A. thaliana is responsive to small molecules such as boric acid (Tanaka et al. 2016) and sucrose (Yamashita et al. 2017), allowing for feedback metabolic control of gene expression.
Therapeutic Enhancement of Stop Codon Read-Through
Nonsense mutations change sense codons to premature termination codons, leading to a loss of function because of the synthesis of defective truncated proteins and/or decreased mRNA stability (Keeling et al. 2014). They account for ∼11% of the mutations that cause inherited human diseases (Mort et al. 2008), and PTC read-through therefore has the potential to ameliorate numerous genetic disorders. Termination suppression occurs at ∼10-fold higher levels at PTCs than at naturally occurring stop codons and is augmented by aminoglycoside antibiotics, which bind to the ribosomal decoding center, promoting misincorporation of near-cognate aminoacyl-tRNAs at premature termination codons without significantly affecting the fidelity of elongation (Keeling et al. 2014). The small molecule drug ataluren (PTC124), which is being developed to ameliorate diseases caused by nonsense mutations, has similarly been reported to enhance insertion of near-cognate aminoacyl-tRNAs (Roy et al. 2016), although doubts about its efficacy have been raised (McElroy et al. 2013).
REINITIATION OF TRANSLATION FOLLOWING INTERRUPTED RIBOSOME RECYCLING
Reinitiation after translation of a uORF, first reported more than 30 years ago (Kozak 1984, 1987), is now recognized as a key regulatory process in post-transcriptional control of eukaryotic gene expression. Genome-wide ribosomal profiling, bioinformatics, and proteomic analyses indicate that ∼50% of mammalian mRNAs contain a uORF, many of which are translated (Calvo et al. 2009; Ingolia et al. 2011; Plaza et al. 2017). Only 13% of yeast mRNAs contain uORFs (Lawless et al. 2009). Reinitiation of translation can occur by different mechanisms depending on how far the recycling process has progressed.
Reinitiation of Translation Mediated by Canonical Initiation Factors
Reinitiation after ABCE1-mediated splitting of post-TCs occurs in circumstances that prevent dissociation of mRNA from 40S subunits. Efficient reinitiation usually occurs only after translation of short ORFs, and the level of reinitiation drops with uORF length (Luukkonen et al. 1995; Kozak 2001). The realization that reinitiation efficiency is determined by the time taken to translate a uORF rather than by its length led to the hypothesis that reinitiation depends on ribosomal retention of a critical factor during elongation and termination (Kozak 2001). This factor would dissociate stochastically, and only those ribosomes that retained it would be reinitiation-competent. Notably, initiation factors that bind to the 40S subunit’s interface surface are displaced during subunit joining (Unbehaun et al. 2004). Analysis of reinitiation on mRNAs that use different sets of factors for initiation indicated that reinitiation depends on the initial involvement of eIF4F, presumably retained on ribosomes via its interaction with eIF3 (Pöyry et al. 2004). Consistently, eIF3 binds primarily on the solvent surface of the 40S subunit (des Georges et al. 2015); cross-linking showed that eIF3 is retained on ribosomes in yeast during elongation on uORFs (Mohammad et al. 2017) and, in the absence of eIF3j, eIF3 impairs release of mRNA from recycled post-TCs (Pisarev et al. 2007), reflecting its activity in stabilizing 40S/mRNA association (Kolupaeva et al. 2005). In vitro reconstitution experiments (Skabkin et al. 2013) yielded a more complete overview of the process, showing that eIF1, eIF1A, eIF2, eIF3, and Met-tRNAiMet must be present for reinitiation to occur; eIF2•GTP/Met-tRNAiMet can be reacquired during scanning (Hinnebusch 2005). eIF4F imposes 5′-3′ directionality on scanning, but in its absence 40S subunits move bidirectionally on unstructured regions of mRNA and initiate upstream or downstream from the stop codon.
Reinitiation of Translation Mediated by Reinitiation Factors
In addition to their involvement in recycling, ligatin (eIF2D) and MCT1/DENR promote reinitiation downstream from uORFs in in vitro reconstituted reactions (Skabkin et al. 2013) in Drosophila and in human cells (Schleich et al. 2014, 2017). These in vivo reports indicated an extreme dependence on uORF length, with reinitiation mediated by MCT1/DENR occurring in human cells only after translation of uORFs containing one or two sense codons. However, in vitro, ligatin (eIF2D) promoted reinitiation after translation of longer ORFs (Skabkin et al. 2013; Zinoviev et al. 2015) and in NIH3T3 cells, several mRNAs containing significantly longer uORFs are translated in a DENR-dependent manner (Janich et al. 2015). They encode proteins involved in establishing circadian rhythms and, consistently, silencing of DENR led to shortened circadian periods (Janich et al. 2015).
Structural studies indicated that rather than delivering Met-tRNAiMet to 40S subunits, ligatin (eIF2D) and MCT1/DENR engage in multiple interactions with P-site tRNA that could anchor it on the 40S subunit (Fig. 4D), showing that the functionally important phosphorylation of MCT1 (Schleich et al. 2014) may modulate positioning of the Met-tRNAiMet acceptor stem, and suggesting that these factors could sterically prevent binding of multiple canonical eIFs to the 40S subunit interface surface (Lomakin et al. 2017; Vaidya et al. 2017; Weisser et al. 2017). Many aspects of the mechanism of reinitiation promoted by these noncanonical factors remain to be established, including what determines whether they act to promote recycling or reinitiation, to what degree their functions are compatible with scanning, whether they can promote reinitiation with or discriminate against specific elongator tRNAs, whether and how they may function in conjunction with other factors such as eIF3, and how ligatin (eIF2D)/MCT-1 dissociate from the aminoacylated -CCA end of Met-tRNAiMet before its accommodation into the P site of the 60S subunit.
Mobility of Post-Termination 80S Ribosomes and Reinitiation of Translation
Post-termination ribosomes are usually weakly anchored to mRNA, and if they are not split by ABCE1 in in vitro reconstituted mammalian translation reactions (Skabkin et al. 2013; Zinoviev et al. 2015), in yeast cells in which it has been depleted (Young et al. 2015) or in primary human platelets and reticulocytes in which ABCE1 has been degraded during terminal differentiation (Mills et al. 2016), can migrate by sliding upstream and downstream from the stop codon. They then stop at triplets that are cognate to the P-site deacylated tRNA and reinitiate translation. In a model (Skabkin et al. 2013) that accounts for these phenomena, dissociation of eRF1 from post-TCs allows P-site tRNA to adopt the P/E hybrid state, disrupting P-site codon–anticodon base pairing, which enables the ribosomes to migrate bidirectionally by sliding in a manner that may be biased by local mRNA structure. Binding of cognate eIF1A•GTP/aa-tRNA to the A-site sense codon is thought to be followed by pseudo-translocation, leading to resumption of translation and accounting for reinitiation occurring in vivo without codon preference. This mechanism is not known to be used for the synthesis of functional gene products, likely because of the semi-random nature of start codon selection, but it may account for the translation of rare peptides from 3′UTR ORFs lacking AUG codons that are presented to major histocompatibility complex (MHC) class I molecules for immune surveillance (Schwab et al. 2003).
There are circumstances in which post-TCs become tethered to mRNA in a manner that promotes reinitiation near the stop codon. For example, the bicistronic subgenomic mRNAs of caliciviruses encode major and minor capsid proteins, and translation of ORF2 occurs by reinitiation. Although reinitiation generally does not occur after long ORFs, likely caused by dissociation of factors (eIF3, eIF4F) that bind to the ribosome’s solvent surface, calicivirus mRNAs can support this process because they contain an essential 40- to 80-nt-long structured “termination upstream ribosome-binding site” (TURBS) upstream of the restart AUG. The TURBS binds to eIF3 and base pairs with the apical loop of h26 of 18S rRNA, likely to trap 80S post-termination ribosomes, and this leads to reinitiation being favored over ribosomal dissociation (Pöyry et al. 2007; Luttermann and Meyers 2009; Zinoviev et al. 2015). The TURBS also supports reinitiation by eIFs or ligatin (eIF2D) with 40S subunits generated by ABCE1-mediated splitting of post-TCs; by arresting the ribosome, it also promotes initiation at near-cognate and even at noncognate codons, in the latter case, either by post-termination 80S ribosomes or by 40S subunits and ligatin (eIF2D) (Zinoviev et al. 2015).
In conclusion, recycling can be interrupted at distinct steps to allow post-termination ribosomes to reinitiate translation by diverse mechanisms. This allows access of ribosomes to the principal ORF in an mRNA to be regulated, but also enables the coding capacity of viral mRNAs to be maximized and, as suggested previously (Skabkin et al. 2013), could lead to the translation of sequences that could ultimately evolve into novel genes by permitting ribosomal access to 3′UTR sequences or to alternative ORFs overlapping the stop codon (Carvunis et al. 2012).
ACKNOWLEDGMENTS
We thank M. Weisser and N. Ban for the figures. Research in the author’s laboratory is supported by National Institutes of Health (NIH) Grant AI123406.
Footnotes
Editors: Michael B. Mathews, Nahum Sonenberg, and John W.B. Hershey
Additional Perspectives on Translation Mechanisms and Control available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. 2006. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 125: 1125–1136. [DOI] [PubMed] [Google Scholar]
- Atkinson GC, Baldauf SL, Hauryliuk V. 2008. Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components. BMC Evol Biol 8: 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelme D, Scheele U, Dinkelaker S, Janoschka A, Macmillan F, Albers SV, Driessen AJ, Stagni MS, Bill E, Meyer-Klaucke W, et al. 2007. Structural organization of essential iron-sulfur clusters in the evolutionarily highly conserved ATP-binding cassette protein ABCE1. J Biol Chem 282: 14598–14607. [DOI] [PubMed] [Google Scholar]
- Barthelme D, Dinkelaker S, Albers SV, Londei P, Ermler U, Tampé R. 2011. Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. Proc Natl Acad Sci 108: 3228–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, Armache JP, Sieber H, Ungewickell C, Berninghausen O, Daberkow I, et al. 2012. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature 482: 501–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier H, Grimm M. 2001. Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res 29: 4767–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram G, Bell HA, Ritchie DW, Fullerton G, Stansfield I. 2000. Terminating eukaryote translation: Domain 1 of release factor eRF1 functions in stop codon recognition. RNA 6: 1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beznosková P, Gunišová S, Valášek LS. 2016. Rules of UGA-N decoding by near-cognate tRNAs and analysis of readthrough on short uORFs in yeast. RNA 22: 456–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan S, Meyer H, Starosta AL, Becker T, Mielke T, Berninghausen O, Sattler M, Wilson DN, Beckmann R. 2010. Structural basis for translational stalling by human cytomegalovirus and fungal arginine attenuator peptide. Mol Cell 40: 138–146. [DOI] [PubMed] [Google Scholar]
- Blanchet S, Cornu D, Argentini M, Namy O. 2014. New insights into the incorporation of natural suppressor tRNAs at stop codons in Saccharomyces cerevisiae. Nucleic Acids Res 42: 10061–10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchet S, Rowe M, Von der Haar T, Fabret C, Demais S, Howard MJ, Namy O. 2015. New insights into stop codon recognition by eRF1. Nucleic Acids Res 43: 3298–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A, Shao S, Murray J, Hegde RS, Ramakrishnan V. 2015. Structural basis for stop codon recognition in eukaryotes. Nature 524: 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulygin KN, Khairulina YS, Kolosov PM, Ven’yaminova AG, Graifer DM, Vorobjev YN, Frolova LY, Kisselev LL, Karpova GG. 2010. Three distinct peptides from the N domain of translation termination factor eRF1 surround stop codon in the ribosome. RNA 16: 1902–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulygin KN, Bartuli YS, Malygin AA, Graifer DM, Frolova LY, Karpova GG. 2016. Chemical footprinting reveals conformational changes of 18S and 28S rRNAs at different steps of translation termination on the human ribosome. RNA 22: 278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Pagliarini DJ, Mootha VK. 2009. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci 106: 7507–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvunis AR, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, Charloteaux B, Hidalgo CA, Barbette J, Santhanam B, et al. 2012. Proto-genes and de novo gene birth. Nature 487: 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvin C, Salhi S, Le Goff C, Viranaicken W, Diop D, Jean-Jean O. 2005. Involvement of human release factors eRF3a and eRF3b in translation termination and regulation of the termination complex formation. Mol Cell Biol 25: 5801–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavatte L, Seit-Nebi A, Dubovaya V, Favre A. 2002. The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO J 21: 5302–5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Saito K, Pisarev AV, Wada M, Pisareva VP, Pestova TV, Gajda M, Round A, Kong C, Lim M, et al. 2009. Structural insights into eRF3 and stop codon recognition by eRF1. Genes Dev 23: 1106–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conard SE, Buckley J, Dang M, Bedwell GJ, Carter RL, Khass M, Bedwell DM. 2012. Identification of eRF1 residues that play critical and complementary roles in stop codon recognition. RNA 18: 1210–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- des Georges A, Hashem Y, Unbehaun A, Grassucci RA, Taylor D, Hellen CU, Pestova TV, Frank J. 2014. Structure of the mammalian ribosomal pre-termination complex associated with eRF1•eRF3•GDPNP. Nucleic Acids Res 42: 3409–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- des Georges A, Dhote V, Kuhn L, Hellen CU, Pestova TV, Frank J, Hashem Y. 2015. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature 525: 491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Green R. 2012. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol 4: a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JG, Foo CK, Belletier NG, Gavis ER, Weissman JS. 2013. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife 2: e01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina I, Takemoto-Tsutsumi M, Watanabe S, Koyama H, Endo Y, Kimata K, Igarashi T, Murakami K, Kudo R, Ohsumi A, et al. 2015. Identification of novel Arabidopsis thaliana upstream open reading frames that control expression of the main coding sequences in a peptide sequence-dependent manner. Nucleic Acids Res 43: 1562–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswarappa SM, Potdar AA, Koch WJ, Fan Y, Vasu K, Lindner D, Willard B, Graham LM, DiCorleto PE, Fox PL. 2014. Programmed translational readthrough generates antiangiogenic VEGF-Ax. Cell 157: 1605–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyler DE, Wehner KA, Green R. 2013. Eukaryotic release factor 3 is required for multiple turnovers of peptide release catalysis by eukaryotic release factor 1. J Biol Chem 288: 29530–29538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan-Minogue H, Du M, Pisarev AV, Kallmeyer AK, Salas-Marco J, Keeling KM, Thompson SR, Pestova TV, Bedwell DM. 2008. Distinct eRF3 requirements suggest alternate eRF1 conformations mediate peptide release during eukaryotic translation termination. Mol Cell 30: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng T, Yamamoto A, Wilkins SE, Sokolova E, Yates LA, Münzel M, Singh P, Hopkinson RJ, Fischer R, Cockman ME, et al. 2014. Optimal translational termination requires C4 lysyl hydroxylation of eRF1. Mol Cell 53: 645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth AE, Brierley I. 2012. Non-canonical translation in RNA viruses. J Gen Virol 93: 1385–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth AE, Wills NM, Gesteland RF, Atkins JF. 2011. Stimulation of stop codon readthrough: Frequent presence of an extended 3′ RNA structural element. Nucleic Acids Res 39: 6679–6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franckenberg S, Becker T, Beckmann R. 2012. Structural view on recycling of archaeal and eukaryotic ribosomes after canonical termination and ribosome rescue. Curr Opin Struct Biol 22: 786–796. [DOI] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Zhouravleva G, Davydova E, Philippe M, Kisselev L. 1996. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA 2: 334–341. [PMC free article] [PubMed] [Google Scholar]
- Frolova LY, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, Kisselev LL. 1999. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA 5: 1014–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graille M, Figaro S, Kervestin S, Buckingham RH, Liger D, Heurgué-Hamard V. 2012. Methylation of class I translation termination factors: Structural and functional aspects. Biochimie 94: 1533–1543. [DOI] [PubMed] [Google Scholar]
- Hauryliuk V, Zavialov A, Kisselev L, Ehrenberg M. 2006. Class-1 release factor eRF1 promotes GTP binding by class-2 release factor eRF3. Biochimie 88: 747–757. [DOI] [PubMed] [Google Scholar]
- He F, Jacobson A. 2015. Nonsense-mediated mRNA decay: Degradation of defective transcripts is only part of the story. Annu Rev Genet 49: 339–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaphy SM, Mariotti M, Gladyshev VN, Atkins JF, Baranov PV. 2016. Novel ciliate genetic code variants including the reassignment of all three stop codons to sense codons in Condylostoma magnum. Mol Biol Evol 33: 2885–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer A, Gerovac M, Schmidt C, Trowitzsch S, Preis A, Kötter P, Berninghausen O, Becker T, Beckmann R, Tampé R. 2017. Structure of the 40S-ABCE1 post-splitting complex in ribosome recycling and translation initiation. Nat Struct Mol Biol 24: 453–460. [DOI] [PubMed] [Google Scholar]
- Hilal T, Yamamoto H, Loerke J, Bürger J, Mielke T, Spahn CM. 2016. Structural insights into ribosomal rescue by Dom34 and Hbs1 at near-atomic resolution. Nat Commun 7: 13521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59: 407–450. [DOI] [PubMed] [Google Scholar]
- Hoque M, Park JY, Chang YJ, Luchessi AD, Cambiaghi TD, Shamanna R, Hanauske-Abel HM, Holland B, Pe’ery T, Tian B, et al. 2017. Regulation of gene expression by translation factor eIF5A: Hypusine-modified eIF5A enhances nonsense-mediated mRNA decay in human cells. Translation 5: e1366294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino S, Imai M, Mizutani M, Kikuchi Y, Hanaoka F, Ui M, Katada T. 1998. Molecular cloning of a novel member of the eukaryotic polypeptide chain-releasing factors (eRF). Its identification as eRF3 interacting with eRF1. J Biol Chem 273: 22254–22259. [DOI] [PubMed] [Google Scholar]
- Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T. 1999. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-Poly(A) tail of mRNA. Direct association of eRF3/GSPT with polyadenylate-binding protein. J Biol Chem 274: 16677–16680. [DOI] [PubMed] [Google Scholar]
- Houck-Loomis B, Durney MA, Salguero C, Shankar N, Nagle JM, Goff SP, D’Souza VM. 2011. An equilibrium-dependent retroviral mRNA switch regulates translational recoding. Nature 480: 561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. 2008. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J 27: 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A, Mikhailova T, Eliseev B, Yeramala L, Sokolova E, Susorov D, Shuvalov A, Schaffitzel C, Alkalaeva E. 2016. PABP enhances release factor recruitment and stop codon recognition during translation termination. Nucleic Acids Res 44: 7766–7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. 2012. Termination and post-termination events in eukaryotic translation. Adv Protein Chem Struct Biol 86: 45–93. [DOI] [PubMed] [Google Scholar]
- Janich P, Arpat AB, Castelo-Szekely V, Lopes M, Gatfield D. 2015. Ribosome profiling reveals the rhythmic liver translatome and circadian clock regulation by upstream open reading frames. Genome Res 25: 1848–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen DM, Frolova L, Geballe AP. 2002. Inhibition of translation termination mediated by an interaction of eukaryotic release factor 1 with a nascent peptidyl-tRNA. Mol Cell Biol 22: 8562–8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Werkmeister E, Gonzalez-Hilarion S, Leroy C, Gruenert DC, Lafont F, Tulasne D, Lejeune F. 2017. Premature termination codon readthrough in human cells occurs in novel cytoplasmic foci and requires UPF proteins. J Cell Sci 130: 3009–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Kelley AC, Loakes D, Ramakrishnan V. 2010. Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release. Proc Natl Acad Sci 107: 8593–8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungreis I, Lin MF, Spokony R, Chan CS, Negre N, Victorsen A, White KP, Kellis M. 2011. Evidence of abundant stop codon readthrough in Drosophila and other metazoa. Genome Res 21: 2096–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungreis I, Chan CS, Waterhouse RM, Fields G, Lin MF, Kellis M. 2016. Evolutionary dynamics of abundant stop codon readthrough. Mol Biol Evol 33: 3108–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcher A, Schele A, Hopfner KP. 2008. X-ray structure of the complete ABC enzyme ABCE1 from Pyrococcus abyssi. J Biol Chem 283: 7962–7971. [DOI] [PubMed] [Google Scholar]
- *.Karousis ED, Mühlemann O. 2018. Nonsense-mediated mRNA decay begins where translation ends. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032862. [DOI] [PMC free article] [PubMed]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. 2006. Binding of a novel SMG-1–Upf1–eRF1–eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 20: 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ. 2016. Genomics: Evolution of the genetic code. Curr Biol 26: R851–R853. [DOI] [PubMed] [Google Scholar]
- Keeling KM, Salas-Marco J, Osherovich LZ, Bedwell DM. 2006. Tpa1p is part of an mRNP complex that influences translation termination, mRNA deadenylation, and mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol 26: 5237–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling KM, Xue X, Gunn G, Bedwell DM. 2014. Therapeutics based on stop codon readthrough. Annu Rev Genomics Hum Genet 15: 371–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiosze-Becker K, Ori A, Gerovac M, Heuer A, Nürenberg-Goloub E, Rashid UJ, Becker T, Beckmann R, Beck M, Tampé R. 2016. Structure of the ribosome post-recycling complex probed by chemical cross-linking and mass spectrometry. Nat Commun 7: 13248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Unbehaun A, Lomakin IB, Hellen CU, Pestova TV. 2005. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA 11: 470–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong C, Ito K, Walsh MA, Wada M, Liu Y, Kumar S, Barford D, Nakamura Y, Song H. 2004. Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe. Mol Cell 14: 233–245. [DOI] [PubMed] [Google Scholar]
- Kononenko AV, Mitkevich VA, Dubovaya VI, Kolosov PM, Makarov AA, Kisselev LL. 2008. Role of the individual domains of translation termination factor eRF1 in GTP binding to eRF3. Proteins 70: 388–393. [DOI] [PubMed] [Google Scholar]
- Kozak M. 1984. Selection of initiation sites by eucaryotic ribosomes: Effect of inserting AUG triplets upstream from the coding sequence for preproinsulin. Nucleic Acids Res 12: 3873–3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1987. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol Cell Biol 7: 3438–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 2001. Constraints on reinitiation of translation in mammals. Nucleic Acids Res 29: 5226–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov G, Gehring K. 2010. Molecular basis of eRF3 recognition by the MLLE domain of poly(A)-binding protein. PLoS ONE 5: e10169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryuchkova P, Grishin A, Eliseev B, Karyagina A, Frolova L, Alkalaeva E. 2013. Two-step model of stop codon recognition by eukaryotic release factor eRF1. Nucleic Acids Res 41: 4573–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. 2008. Structural basis for translation termination on the 70S ribosome. Nature 454: 852–857. [DOI] [PubMed] [Google Scholar]
- Lawless C, Pearson RD, Selley JN, Smirnova JB, Grant CM, Ashe MP, Pavitt GD, Hubbard SJ. 2009. Upstream sequence elements direct post-transcriptional regulation of gene expression under stress conditions in yeast. BMC Genomics 10: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Liebman SW. 1996. A translational fidelity mutation in the universally conserved sarcin/ricin domain of 25S yeast ribosomal RNA. RNA 2: 254–263. [PMC free article] [PubMed] [Google Scholar]
- Loenarz C, Sekirnik R, Thalhammer A, Ge W, Spivakovsky E, Mackeen MM, McDonough MA, Cockman ME, Kessler BM, Ratcliffe PJ, et al. 2014. Hydroxylation of the eukaryotic ribosomal decoding center affects translational accuracy. Proc Natl Acad Sci 111: 4019–4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Shirokikh NE, Yusupov MM, Hellen CU, Pestova TV. 2006. The fidelity of translation initiation: Reciprocal activities of eIF1, IF3 and YciH. EMBO J 25: 196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Stolboushkina EA, Vaidya AT, Zhao C, Garber MB, Dmitriev SE, Steitz TA. 2017. Crystal structure of the human ribosome in complex with DENR-MCT-1. Cell Rep 20: 521–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughran G, Chou MY, Ivanov IP, Jungreis I, Kellis M, Kiran AM, Baranov PV, Atkins JF. 2014. Evidence of efficient stop codon readthrough in four mammalian genes. Nucleic Acids Res 42: 8928–8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveland AB, Demo G, Grigorieff N, Korostelev AA. 2017. Ensemble cryo-EM elucidates the mechanism of translation fidelity. Nature 546: 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttermann C, Meyers G. 2009. The importance of inter- and intramolecular base pairing for translation reinitiation on a eukaryotic bicistronic mRNA. Genes Dev 23: 331–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luukkonen BG, Tan W, Schwartz S. 1995. Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J Virol 69: 4086–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantsyzov AB, Ivanova EV, Birdsall B, Alkalaeva EZ, Kryuchkova PN, Kelly G, Frolova LY, Polshakov VI. 2010. NMR solution structure and function of the C-terminal domain of eukaryotic class 1 polypeptide chain release factor. FEBS J 277: 2611–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maracci C, Rodnina MV. 2016. Review: Translational GTPases. Biopolymers 105: 463–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheisl S, Berninghausen O, Becker T, Beckmann R. 2015. Structure of a human translation termination complex. Nucleic Acids Res 43: 8615–8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughan KK, Brown CM, Dalphin ME, Berry MJ, Tate WP. 1995. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc Natl Acad Sci 92: 5431–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy SP, Nomura T, Torrie LS, Warbrick E, Gartner U, Wood G, McLean WH. 2013. A lack of premature termination codon read-through efficacy of PTC124 (Ataluren) in a diverse array of reporter assays. PLoS Biol 11: e1001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov S, Mailliot J, Shin BS, Rigger L, Yusupova G, Micura R, Dever TE, Yusupov M. 2016. Crystal structure of hypusine-containing translation factor eIF5A bound to a rotated eukaryotic ribosome. J Mol Biol 428: 3570–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EW, Wangen J, Green R, Ingolia NT. 2016. Dynamic regulation of a ribosome rescue pathway in erythroid cells and platelets. Cell Rep 17: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitkevich VA, Kononenko AV, Petrushanko IY, Yanvarev DV, Makarov AA, Kisselev LL. 2006. Termination of translation in eukaryotes is mediated by the quaternary eRF1•eRF3•GTP•Mg2+ complex. The biological roles of eRF3 and prokaryotic RF3 are profoundly distinct. Nucleic Acids Res 34: 3947–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad MP, Munzarová Pondelícková V, Zeman J, Gunišová S, Valášek LS. 2017. In vivo evidence that eIF3 stays bound to ribosomes elongating and terminating on short upstream ORFs to promote reinitiation. Nucleic Acids Res 45: 2658–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mort M, Ivanov D, Cooper DN, Chuzhanova NA. 2008. A meta-analysis of nonsense mutations causing human genetic disease. Hum Mutat 29: 1037–1047. [DOI] [PubMed] [Google Scholar]
- Muhs M, Hilal T, Mielke T, Skabkin MA, Sanbonmatsu KY, Pestova TV, Spahn CM. 2015. Cryo-EM of ribosomal 80S complexes with termination factors reveals the translocated cricket paralysis virus IRES. Mol Cell 57: 422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napthine S, Yek C, Powell ML, Brown TD, Brierley I. 2012. Characterization of the stop codon readthrough signal of Colorado tick fever virus segment 9 RNA. RNA 18: 241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu-Yilik G, Raimondeau E, Eliseev B, Yeramala L, Amthor B, Deniaud A, Huard K, Kerschgens K, Hentze MW, Schaffitzel C, et al. 2017. Dual function of UPF3B in early and late translation termination. EMBO J 36: 2968–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pánek T, Žihala D, Sokol M, Derelle R, Klimeš V, Hradilová M, Zadrobílková E, Susko E, Roger AJ, Čepička I, et al. 2017. Nuclear genetic codes with a different meaning of the UAG and the UAA codon. BMC Biol 15: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina MV. 1998. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J 17: 7490–7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Alepuz P. 2017. eIF5A facilitates translation termination globally and promotes the elongation of many non polyproline-specific tripeptide sequences. Nucleic Acids Res 45: 7326–7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson WE, Hoffer ED, Keedy HE, Simms CL, Dunham CM, Zaher HS. 2016. Uniformity of peptide release is maintained by methylation of release factors. Cell Rep 17: 11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Hellen CU, Pestova TV. 2007. Recycling of eukaryotic posttermination ribosomal complexes. Cell 131: 286–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV. 2010. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell 37: 196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva VP, Pisarev AV, Hellen CU, Rodnina MV, Pestova TV. 2006. Kinetic analysis of interaction of eukaryotic release factor 3 with guanine nucleotides. J Biol Chem 281: 40224–40235. [DOI] [PubMed] [Google Scholar]
- Pisareva VP, Skabkin MA, Hellen CU, Pestova TV, Pisarev AV. 2011. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J 30: 1804–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza S, Menschaert G, Payre F. 2017. In search of lost small peptides. Annu Rev Cell Dev Biol 33: 391–416. [DOI] [PubMed] [Google Scholar]
- Pöyry TA, Kaminski A, Jackson RJ. 2004. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev 18: 62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöyry TA, Kaminski A, Connell EJ, Fraser CS, Jackson RJ. 2007. The mechanism of an exceptional case of reinitiation after translation of a long ORF reveals why such events do not generally occur in mammalian mRNA translation. Genes Dev 21: 3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preis A, Heuer A, Barrio-Garcia C, Hauser A, Eyler DE, Berninghausen O, Green R, Becker T, Beckmann R. 2014. Cryoelectron microscopic structures of eukaryotic translation termination complexes containing eRF1-eRF3 or eRF1-ABCE1. Cell Rep 8: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees DC, Johnson E, Lewinson O. 2009. ABC transporters: The power to change. Nat Rev Mol Cell Biol 10: 218–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Rodnina MV. 2018. Translation in prokaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032664. [DOI] [PMC free article] [PubMed]
- Roy B, Leszyk JD, Mangus DA, Jacobson A. 2015. Nonsense suppression by near-cognate tRNAs employs alternative base pairing at codon positions 1 and 3. Proc Natl Acad Sci 112: 3038–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Friesen WJ, Tomizawa Y, Leszyk JD, Zhuo J, Johnson B, Dakka J, Trotta CR, Xue X, Mutyam V, et al. 2016. Ataluren stimulates ribosomal selection of near-cognate tRNAs to promote nonsense suppression. Proc Natl Acad Sci 113: 12508–12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini P, Eyler DE, Green R, Dever TE. 2009. Hypusine-containing protein eIF5A promotes translation elongation. Nature 459: 118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Marco J, Bedwell DM. 2004. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol Cell Biol 24: 7769–7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Marco J, Bedwell DM. 2005. Discrimination between defects in elongation fidelity and termination efficiency provides mechanistic insights into translational readthrough. J Mol Biol 348: 801–815. [DOI] [PubMed] [Google Scholar]
- Schleich S, Strassburger K, Janiesch PC, Koledachkina T, Miller KK, Haneke K, Cheng YS, Kuechler K, Stoecklin G, Duncan KE, et al. 2014. DENR–MCT-1 promotes translation re-initiation downstream of uORFs to control tissue growth. Nature 512: 208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleich S, Acevedo JM, Clemm von Hohenberg K, Teleman AA. 2017. Identification of transcripts with short stuORFs as targets for DENR•MCTS1-dependent translation in human cells. Sci Rep 7: 3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C, Becker T, Heuer A, Braunger K, Shanmuganathan V, Pech M, Berninghausen O, Wilson DN, Beckmann R. 2016. Structure of the hypusinylated eukaryotic translation factor eIF-5A bound to the ribosome. Nucleic Acids Res 44: 1944–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader R, Young C, Kozian D, Hoffmann R, Lottspeich F. 2006. Temperature-sensitive eIF5A mutant accumulates transcripts targeted to the nonsense-mediated decay pathway. J Biol Chem 281: 35336–35346. [DOI] [PubMed] [Google Scholar]
- Schuller AP, Wu CC, Dever TE, Buskirk AR, Green R. 2017. eIF5A functions globally in translation elongation and termination. Mol Cell 66: 194–205.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab SR, Li KC, Kang C, Shastri N. 2003. Constitutive display of cryptic translation products by MHC class I molecules. Science 301: 1367–1371. [DOI] [PubMed] [Google Scholar]
- Serdar LD, Whiteside DL, Baker KE. 2016. ATP hydrolysis by UPF1 is required for efficient translation termination at premature stop codons. Nat Commun 7: 14021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, Murray J, Brown A, Taunton J, Ramakrishnan V, Hegde RS. 2016. Decoding mammalian ribosome-mRNA states by translational GTPase complexes. Cell 167: 1229–1240.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. 2011. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci 108: E1392–E1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton RS, Liu-Yi P, Formenti F, Ge W, Sekirnik R, Fischer R, Adam J, Pollard PJ, Wolf A, Thalhammer A, et al. 2014. OGFOD1 catalyzes prolyl hydroxylation of RPS23 and is involved in translation control and stress granule formation. Proc Natl Acad Sci 111: 4031–4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV. 2010. Activities of ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev 24: 1787–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Hellen CUT, Pestova TV. 2013. Reinitiation and other unconventional post-termination events during eukaryotic translation. Mol Cell 51: 249–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D. 2000. The crystal structure of human eukaryotic release factor eRF1—Mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100: 311–321. [DOI] [PubMed] [Google Scholar]
- Steneberg P, Samakovlis C. 2001. A novel stop codon readthrough mechanism produces functional headcase protein in Drosophila trachea. EMBO Rep 2: 593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart EC, Serra V, Petroni G, Nowacki M. 2016. Genetic codes with no dedicated stop codon: Context-dependent translation termination. Cell 166: 691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Sotta N, Yamazumi Y, Yamashita Y, Miwa K, Murota K, Chiba Y, Hirai MY, Akiyama T, Onouchi H, et al. 2016. The minimum open reading frame, AUG-Stop, induces boron-dependent ribosome stalling and mRNA degradation. Plant Cell 28: 2830–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Zhu Y, Baker SL, Bowler MW, Chen BJ, Chen C, Hogg JR, Goff SP, Song H. 2016. Structural basis of suppression of host translation termination by Moloney murine leukemia virus. Nat Commun 7: 12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D, Unbehaun A, Li W, Das S, Lei J, Liao HY, Grassucci RA, Pestova TV, Frank J. 2012. Cryo-EM structure of the mammalian eukaryotic release factor eRF1–eRF3-associated termination complex. Proc Natl Acad Sci 109: 18413–18418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempel W, Dimov S, Tong Y, Park HW, Hong BS. 2013. Crystal structure of human multiple copies in T-cell lymphoma-1 oncoprotein. Proteins 81: 519–525. [DOI] [PubMed] [Google Scholar]
- Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. 2004. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev 18: 3078–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya AT, Lomakin IB, Joseph NN, Dmitriev SE, Steitz TA. 2017. Crystal structure of the C-terminal domain of human eIF2D and its implications on eukaryotic translation initiation. J Mol Biol 429: 521–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Elzen AM, Henri J, Lazar N, Gas ME, Durand D, Lacroute F, Nicaise M, van Tilbeurgh H, Séraphin B, Graille M. 2010. Dissection of Dom34–Hbs1 reveals independent functions in two RNA quality control pathways. Nat Struct Mol Biol 17: 1446–1452. [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. 2010. The mechanism for activation of GTP hydrolysis on the ribosome. Science 330: 835–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada M, Ito K. 2014. A genetic approach for analyzing the co-operative function of the tRNA mimicry complex, eRF1/eRF3, in translation termination on the ribosome. Nucleic Acids Res 42: 7851–7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Sachs MS. 1997. Ribosome stalling is responsible for arginine specific translational attenuation in Neurospora crassa. Mol Cell Biol 17: 4904–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Czaplinski K, Rao Y, Peltz SW. 2001. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J 20: 880–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisser M, Schäfer T, Leibundgut M, Böhringer D, Aylett CHS, Ban N. 2017. Structural and functional insights into human re-initiation complexes. Mol Cell 67: 447–456.e7. [DOI] [PubMed] [Google Scholar]
- Wong LE, Li Y, Pillay S, Frolova L, Pervushin K. 2012. Selectivity of stop codon recognition in translation termination is modulated by multiple conformations of GTS loop in eRF1. Nucleic Acids Res 40: 5751–5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X, Mutyam V, Thakerar A, Mobley J, Bridges RJ, Rowe SM, Keeling KM, Bedwell DM. 2017. Identification of the amino acids inserted during suppression of CFTR nonsense mutations and determination of their functional consequences. Hum Mol Genet 26: 3116–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y, Takamatsu S, Glasbrenner M, Becker T, Naito S, Beckmann R. 2017. Sucrose sensing through nascent peptide-meditated ribosome stalling at the stop codon of Arabidopsis bZIP11 uORF2. FEBS Lett 591: 1266–1277. [DOI] [PubMed] [Google Scholar]
- Young DJ, Guydosh NR, Zhang F, Hinnebusch AG, Green R. 2015. Rli1/ABCE1 recycles terminating ribosomes and controls translation reinitiation in 3′UTRs in vivo. Cell 162: 872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Záhonová K, Kostygov AY, Ševčíková T, Yurchenko V, Eliáš M. 2016. An unprecedented non-canonical nuclear genetic code with all three termination codons reassigned as sense codons. Curr Biol 26: 2364–2369. [DOI] [PubMed] [Google Scholar]
- Zinoviev A, Hellen CU, Pestova TV. 2015. Multiple mechanisms of reinitiation on bicistronic calicivirus mRNAs. Mol Cell 57: 1059–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]