Abstract
The tumor necrosis factor (TNF) cytokine family and the TNF/nerve growth factor (NGF) family of their cognate receptors together control numerous immune functions, as well as tissue-homeostatic and embryonic-development processes. These diverse functions are dictated by both shared and distinct features of family members, and by interactions of some members with nonfamily ligands and coreceptors. The spectra of their activities are further expanded by the occurrence of the ligands and receptors in both membrane-anchored and soluble forms, by “re-anchoring” of soluble forms to extracellular matrix components, and by signaling initiation via intracellular domains (IDs) of both receptors and ligands. Much has been learned about shared features of the receptors as well as of the ligands; however, we still have only limited knowledge of the mechanistic basis for their functional heterogeneity and for the differences between their functions and those of similarly acting cytokines of other families.
The study of protein families and their individual members contribute cooperatively to the assembly of knowledge, providing insights into the features shared by family members as well as their distinctive features. The benefit of such a cooperative endeavor is lavishly demonstrated by the huge advances in understanding the mechanisms of action of the tumor necrosis factor (TNF) ligand and TNF/nerve growth factor receptor (NGFR) families. The founding members of these families—the cytokine TNF and the low-affinity NGFR—were isolated and cloned three decades ago (Old 1985; Johnson et al. 1986; Radeke et al. 1987). In this review, I will present a brief overview of the knowledge of common structural, mechanistic, and functional features of the TNF ligand and TNF/NGFR families. I will also refer to interactions that are known for only a few family members, but whose occurrence raises the possibility that other family members participate in similar associations.
THE WIDE RANGE OF FUNCTIONS OF THE TNF FAMILY
There are 18 known human genes for the TNF ligand family and 29 for the TNF/NGFR family (see genenames.org/genefamilies/TNFSF and genenames.org/genefamilies/TNFRSF; see Table 1 for their major known functions). Practically, all cells in the body express receptors, and many also express some of the ligands of these families. Each receptor–ligand pair controls a wide range of cellular activities. Assessing the impact of functional arrest of specific members of the TNF and TNF/NGF families has revealed some key physiological roles served exclusively by them. Listed below and briefly discussed are the functions of family members that have attracted the greatest attention, either because they appear to be unique to the family or because of known pathological consequences of their deficiency or hyperactivity.
Table 1.
Principal known functions of receptors of the tumor necrosis factor (TNF)/nerve growth factor receptor (NGFR) family and ligands of the TNF family
| Receptor | Ligand | |||||
|---|---|---|---|---|---|---|
| Widely used name | Formal name | Other names | Widely used name | Formal name | Other names | Main known functions of the receptor |
| TNFR1 | TNFRSF1A | p55, CD120a | TNF | Tumor necrosis factor | Orchestration of inflammation; cell killing | |
| LT | Lymphotoxin α | |||||
| TNFR2 | TNFRSF1A | p75, CD120b | TNF | Tumor necrosis factor | Costimulation of T lymphocytes (information on various other functions is sporadic) | |
| LT | Lymphotoxin | |||||
| Fn14 | TNFRSF12A | TweakR | TWEAK | TNFSF12 | APO3L | Control of tissue regeneration in response to injury; inflammation |
| DR3 | TNFRSF12 | TRAMP, WSL-1, APO-3 | TL1 | TNFSF15 | TL1A, VEGI | Costimulation of T lymphocytes; inflammation |
| FAS | FAS | APO-1, CD95 | Fas ligand | FASL, Fas ligand | CD178 | Cell killing; cell growth; inflammation |
| DcR3 | TNFRSF6B | TR6 | Fas ligand, LIGHT, TL1 | Secreted decoy receptor that blocks the function of the three indicated ligands; seems also capable of triggering signaling through its binding to HSPG | ||
| TRAIL-R1 | TNFRSF10A | Apo2, DR4 | TRAIL | TNFSF10 | Apo-2L, CD253 | Cell killing |
| TRAIL-R2 | TNFRSF10B | DR5 | TRAIL | TNFSF10 | Cell killing | |
| DcR1 | TNFRSF10C | TRAILR3 | TRAIL | TNFSF10 | GPI-linked decoy receptor restraining the function of TRAIL | |
| DcR2 | TNFRSF10D | TRAILR4 | TRAIL | TNFSF10 | Transmembrane truncated decoy receptor restraining the function of TRAIL | |
| LTβR | LTβR (lymphotoxin β receptor) | LTα1β2 (lymphotoxin β [LTβ] in complex with lymphotoxin α), LIGHT | LTβ TNFSF14 | CD258 | Development and function of lymphoid organs; amplification of antiviral response and various other innate and adaptive immune functions | |
| HVEM | TNFRSF14 | ATAR, CD270 | LIGHT | TNFSF14 | Costimulation in T and B lymphocytes and in NK cells; triggering of immunosuppression through BTLA, and immunosuppression, inflammation, and NK activation through CD160 | |
| LT (weak binding) | ||||||
| OX40 | TNFRSF4 | CD134 | OX40 ligand (OX40L) | TNFSF4 | CD252 | Costimulation of T lymphocytes |
| CD27 | TNFRSF7 | CD70 | CD70 | CD27 ligand | Costimulation of T lymphocytes | |
| CD30 | TNFRSF8 | CD30 ligand (CD30L) | TNFSF8 | Costimulation of T lymphocytes | ||
| 4-1BB | TNFRSF9 | CD137 | 4-1BB ligand (4-1BBL) | TNFSF9 | Costimulation of T lymphocytes | |
| GITR | TNFRSF18 | AITR, CD357 | GITR ligand (GITRL) | TNFSF18 | AITR | Costimulation of T lymphocytes |
| RELT | RELT | ? | Unknown | |||
| CD40 | TNFRSF5 | CD40 ligand (CD40L) | CD40LG | CD154 | Wide range of effects contributing to initiation and progression of adaptive immunity | |
| TACI | TNFRSF13B | CD267 | BAFF APRIL | TNFSF20 TNFSF13 | BLYS, CD257 CD256 | B-lymphocyte growth, survival, and differentiation; costimulation of T lymphocytes |
| BAFFR | TNFRSF13C | CD268 | BAFF | B-lymphocyte survival, growth, and differentiation | ||
| BCMA | TNFRSF17 | CD269 | BAFF APRIL | B-lymphocyte survival, growth, and differentiation | ||
| RANK | TNFRSF11A | RANK ligand (RANKL) | TNFSF11 | RANKL, TRANCE, OPGL, CD254 | Osteoclast differentiation and activation; mammary gland development; development of lymph nodes; various immune functions | |
| OPG | TNFRSF11B | OCIF, TR1 | RANK ligand TRAIL | Secretory decoy receptor that blocks the functions of RANKL and TRAIL; seems also capable of triggering signaling through its binding to HSPG | ||
| EDAR | EDAR (ectodysplasin A receptor) | EDA1R | EDA | EDA (ectodysplasin A) | Embryonic development of ectodermal appendages | |
| XEDAR | EDA2R (ectodysplasin A2 receptor) | EDA-A2 (a shorter splice variant of the EDA gene) | Unknown | |||
| NGFR | Nerve growth factor receptor | p75NTR, CD271 | Neurotrophins; myelin-associated inhibitory factors; fragments of β-amyloid precursor protein; prion peptide | Neuronal growth and death; arrest of axonal degeneration; pain sensation | ||
| TROY | TNFRSF19 | TAJ | Myelin-associated inhibitory factors | Repression of axonal regeneration | ||
| DR6 | TNFRSF21 | CD358 | ? | Probably contributes to amyloid-β-induced axonal pruning | ||
TWEAK, TNF-related weak inducer of apoptosis; HSPG, heparan sulfate proteoglycan; TRAIL, TNF-related apoptosis-inducing ligand; GPI, glycosylphosphatidylinositol; HVEM, herpesvirus entry mediator; NK, natural killer; BTLA, B- and T-lymphocyte attenuator; GITR, glucocorticoid-induced TNFR-related protein; AITR, activation-inducible TNFR family receptor; RELT, receptor expressed in lymphoid tissue; TACI, transmembrane activator and calcium-modulating cyclophilin ligand interactor; BAFF, B-cell-activating factor of the tumor necrosis factor family; APRIL, a proliferation-inducing ligand; BCMA, B-cell maturation antigen; RANK, receptor activator of nuclear factor κB; OPG, osteoprotegerin; EDA, ectodermal dysplasia.
Control of Cell Survival
Stimulation of Cell Death
The ability of certain ligands of the TNF family to trigger death of cells independently of protein synthesis was the family’s first cellular effect to be described (Granger and Kolb 1968; Ruddle and Waksman 1968), and to this day it remains the only function not known to be shared with other cytokines. Several TNF family receptors (TNFR1, DR3, FAS, TNF-related apoptosis-inducing ligand [TRAIL]-R1, TRAIL-R2) trigger rapid cell death. Others signal for cell death less effectively, either through induction of TNF (Grell et al. 1999; Burkly 2014) or by poorly defined cell-autonomous mechanisms (Force et al. 2000; Georgopoulos et al. 2006; Elmetwali et al. 2010).
The cytotoxic functions of the TNF family contribute to immune-mediated cell killing. They also seem to contribute to the control of expansion and to the duration of activities of immune-cell populations and to the shaping of leukocyte repertoires (Falschlehner et al. 2009; Strasser et al. 2009).
Providing Survival Signaling
One way in which the TNF family members facilitate maintenance and amplification of immune responses is by providing the relevant cells with survival signals. Best documented are the crucial roles of several members of the family in maintaining the survival of B and T lymphocytes (Croft 2014; Figgett et al. 2014). TNF family members are also capable of inducing resistance of cells to the cytotoxic activities that they themselves activate (e.g., Wallach 1984; Hahn et al. 1985; Blomberg et al. 2008; Chen et al. 2010; Jeon et al. 2015).
Orchestration of Inflammation
The best-documented pivotal role of a ligand of the TNF family in pathological disorders is the contribution of TNF to chronic inflammatory diseases. This has been demonstrated by the therapeutic effects of TNF-blocking agents observed in millions of patients, as well as in experimental animal models for both chronic and acute inflammatory diseases (Tracey et al. 1987; Apostolaki et al. 2010; Sfikakis 2010). TNF contributes to the initiation, progression, and termination of inflammation, while displaying antagonistic effects: for example, induction of cell death but also cell growth and resistance to cell death, and obstruction and destruction of capillaries but also stimulation of angiogenesis. TNF is also an important player in systemic manifestations of inflammation, for example, in activating the acute-phase response in the liver (Wallach and Kovalenko 2016).
Also contributing to inflammation, although in more restricted ways, are several TNF family ligands, including TKA1, TNF-related weak inducer of apoptosis (TWEAK), TRAIL, CD40L, LIGHT, and receptor activator of nuclear factor (NF)-κB ligand (RANKL), which are known primarily for other functions.
Tissue Modeling
Tissue Remodeling in Response to Injury
Like TNF, TWEAK exerts a wide range of antagonistic effects on cell functions; it contributes to both inflammation and its arrest, as well as to both destruction and regeneration of tissues. It thus serves important roles in coordinating tissue remodeling in response to injury (Burkly 2014).
Bone Homeostasis
Calcified bone matter undergoes constant turnover as a result of the antagonistic effects of osteoblasts, which construct bones, and osteoclasts, which resorb them. Local inflammation in the bone facilitates bone destruction. RANKL serves a crucial role in maintaining the constitutive activity of the osteoclasts. Its soluble receptor, osteoprotegerin (OPG), blocks RANKL function by competing for the binding of RANK (Boyce and Xing 2007; Walsh and Choi 2014). CD40L potentiates this inhibition via several effects, including induction of OPG in B lymphocytes (Li et al. 2007). In contrast, TNF enhances bone dissolution. It does this by triggering the egress of osteoclast precursors from the bone marrow as well as by enhancing RANKL generation and triggering osteoclast differentiation and activation synergistically with this ligand as well as with the non-TNF family cytokine interleukin (IL)-1 (Li et al. 2004).
Control of the Development of Ectodermal Tissues
Ectodysplasin (EDA) and its receptor EDAR are the only known ligand–receptor pair within the TNF families that seems to make no contribution to immune regulation. They signal for embryonic development of ectodermal appendages such as the hair, teeth, and sweat glands. This role is evolutionarily conserved in other metazoan phyla in which EDA and its receptor control the development of ectodermal appendages such as feathers, scales, and fins (Lefebvre and Mikkola 2014).
EDA-A2, a slightly shorter splice variant encoded by the Eda gene, binds to a distinct receptor of the TNF/NGF family, XEDAR. Although expressed in the developing hair follicle, this receptor does not seem to be required for hair growth. Limited evidence suggests that it might serve to control skeletal muscle homeostasis (Lefebvre and Mikkola 2014). TROY, an orphan receptor of the TNF/NGF family, is also coexpressed with EDAR in hair follicles and in embryonic skin, but its function at these sites is not known (Kojima et al. 2000).
Control of Adaptive Immunity
Elicitation of an adaptive immune response necessitates fine-tuning of the development of lymphocytes and of their interaction with antigen-presenting cells as well as regulation of the development and function of specific organs in which these processes occur. In all of these functions, members of the TNF family play pivotal roles.
Control of the Generation and Maintenance of Lymphoid Organs
Embryonic development of the secondary lymphoid organs crucially depends on signaling by lymphotoxin β receptor (LTβR) and RANK. Both LTβR-triggered and TNF-triggered signaling are required for maintenance of the microarchitecture of the lymphoid organs and their appropriate function in the adult (Ware 2005; McCarthy et al. 2006). Both are also required for the neogenesis of lymphoid assemblages at sites of chronic inflammation (Drayton et al. 2006). A similar “morphogenic” role is served by TNF in dictating the generation of granuloma (Kindler et al. 1989).
Control of the Development and function of T Lymphocytes
Signaling by several TNF/NGF family receptors, including TNFR1, LTβR, RANK, herpesvirus entry mediator (HVEM), OX40, and CD40, controls the migration, maturation, and activation of dendritic cells (Ware 2005; Summers deLuca and Gommerman 2012; Walsh and Choi 2014). Signaling by receptors of the TNF/NGF family contributes to the selection of lymphocytes in the thymus. TNFR1, TNFR2, DR3, HVEM, OX40, CD27, CD30, 4-1BB, B-cell-activating factor of the tumor necrosis factor family (BAFF), and transmembrane activator and calcium-modulating cyclophilin ligand interactor (TACI) provide costimulatory signals in antigen-stimulated T lymphocytes. Some of those receptors also initiate stimulatory signals in T lymphocytes whose antigen receptors have not been activated. These stimulatory signals enhance lymphocyte survival, growth, and effector functions. The various costimulatory family members apparently serve distinct roles at different phases of T-lymphocyte response, and the relative contribution of their costimulatory effects to defense varies depending on the nature of the particular pathogenic challenge (Croft 2014; Mbanwi and Watts 2014).
Control of the Development and Function of B Lymphocytes
Activation of the TNF/NGF family receptors BAFFR, B-cell maturation antigen (BCMA), TACI, and CD40 provide B lymphocytes with survival and growth signals at distinct stages of their development. CD40 signaling is crucially required also for antibody isotope switching and for the generation of memory B lymphocytes. Various other TNF/NGF family members also affect B-cell biology (Bishop and Hostager 2003; Dillon et al. 2006; Elgueta et al. 2009; Figgett et al. 2014).
Functions of the TNF Family in the Brain
At times of injury or autoimmune response within the brain, the affected cells display the same modes of TNF family–induced regulation as those observed in such situations elsewhere in the body (Akassoglou et al. 1999; Shohami et al. 1999). Some of these cytokine effects result in tissue damage. However, TNF also provides protective and survival signals in nervous tissue, in part through TNFR2 (Bruce et al. 1996; Fontaine et al. 2002). Effects of TNF on brain functions contribute to several behavioral responses to disease, including enhanced slow-wave sleep (Shoham et al. 1987), fever (Dinarello et al. 1986), anorexia (Cerami et al. 1985; Plata-Salaman et al. 1988), increased pain perception (Hess et al. 2011), and others (Dantzer 2001). Fever induction by TNF, as well as by several other inflammatory cytokines, is triggered indirectly through induction of RANK-mediated signaling in astrocytes (Hanada et al. 2009).
Emerging knowledge indicates that TNF family members also contribute to brain functions unrelated to immune defense. TNF produced by glial cells stabilizes neuronal circuits by dictating homeostatic synaptic scaling (Stellwagen and Malenka 2006). Some evidence indicates that FAS signals for neurogenesis in the adult brain (Corsini et al. 2009). The receptor DR3 is expressed in motor neurons and its tonic signaling seems to be necessary for their survival (Richard et al. 2015).
The function of NGFR, a TNF/NGF family receptor for which no ligand of the TNF family is known, serves cooperatively with several coreceptors to control the growth and survival of neurons in response to simulation by several ligands and also to control pain sensation (Hempstead 2002; Chao 2003; Ibanez and Simi 2012). Limited evidence suggests that TROY and DR6, additional members of the TNF/NGFR family not known to bind any ligand of the TNF family, also serve to signal for neuronal death (Shao et al. 2005; Nikolaev et al. 2009; Olsen et al. 2014).
TRIGGERING OF SIGNALING: INDUCED JUXTAPOSITION
Members of the TNF ligand and TNF/NGFR families share conserved extracellular motifs by which they bind each other. With the exception of lymphotoxin (LT), a secretory protein, all ligands are produced as type II transmembrane (TM) proteins in which the receptor-binding motif, whose structure consists of two packed sheets of eight antiparallel β strands, is located at the carboxyl terminus. Most ligands also occur in soluble forms, generated by proteolytic processing of the TM forms to yield soluble ligand-binding molecules. Both in their membrane-bound and in their soluble forms, the ligand molecules associate constitutively in trimers.
The receptors are produced as type I TM proteins, whose amino-terminal ligand-binding motif consists of a variable number of two conserved modules that together form a 40-amino-acid structure containing several cysteines, the “cysteine-rich domain” (CRD).
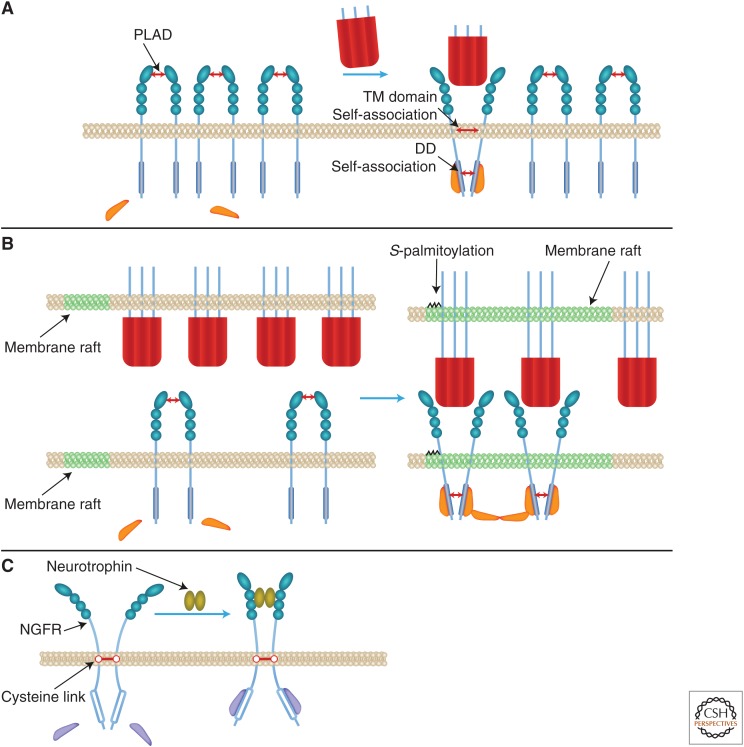
Distinct parts of receptors’ extracellular domains (EDs) serve opposing roles in controlling signaling. The amino-terminal CRD serves as a “pre-ligand assembly domain” (PLAD), which safeguards against ligand-independent signaling (Chan 2007). In the absence of ligands, the PLADs in the EDs of three or (more likely) two (Naismith et al. 1995) receptor molecules associate in a way that keeps their intracellular domains (IDs) apart. In contrast, ligand binding, which occurs downstream from the PLAD, imposes juxtaposition of the receptor IDs. In receptors whose IDs contain a death domain (DD) (see below), such juxtaposition is fostered by the propensity of the DD to self-associate (Boldin et al. 1995). Proline-containing motifs found in the transmembrane domain (TD) in FAS, and apparently also in the other receptors of the family, also tend to self-associate (forming homotrimers), further strengthening the ligand-binding effect (Fu et al. 2016).
Juxtaposition of the IDs of the receptors exposes their binding surfaces to signaling proteins. Their binding to the receptors and consequent juxtaposition triggers their enzymatic activity and/or their association with downstream signaling proteins (Fig. 1A).
Figure 1.
Signaling triggering: proposed mechanisms. (A) Triggering by intracellular domain (ID) juxtaposition imposed by tumor necrosis factor (TNF) family ligands, and its restriction by the pre-ligand assembly domain (PLAD). (B) Triggering potentiation by anchorage of ligands to membranes. (C) Triggering by ID distancing: A fraction of the nerve growth factor receptor (NGFR) molecules occurs as dimers of receptor molecules that are covalently linked through a conserved transmembrane (TM) cysteine residue. In these dimers, the intracellular death domains (DDs) seem to be constitutively associated. Neurotrophin-induced triggering of these dimers to signal for death was suggested to occur by distancing of the IDs, imposed rather like a snail tongue, with the TM cysteine link serving as a fulcrum (Vilar et al. 2009). (See discussion of these findings in the last paragraph of this review.)
The self-association of four of the receptors (CD40, NGFR, CD27, and the soluble receptor OPG) is facilitated by constitutive cysteine links (found in the ID of CD40, in the TD of NGFR, at the carboxyl terminus of OPG, and apparently in the ED of CD27) (Van Lier et al. 1987; Schneeweis et al. 2005; Vilar et al. 2009; Nadiri et al. 2015). Constitutive dimerization of the soluble receptor OPG is also dictated by two DD motifs found at its carboxyl terminus (Schneeweis et al. 2005).
Structural studies of glucocorticoid-induced TNFR-related protein (GITR) suggest that the trimeric ligands of the TNF family may tend to undergo further clustering through noncovalent homotypic self-association (Zhou et al. 2008). However, formation of a soluble supercluster (of 20 trimers) has been documented only in the case of one TNF family member, BAFF (Liu et al. 2002; Cachero et al. 2006). The trimers of EDA are exceptional in that they are constitutively dimerized by homotypic self-association of an amino-terminal collagen-like domain unique to this ligand (Swee et al. 2009).
SIGNALING FACILITATION BY MEMBRANE ANCHORAGE
The ability of membrane-anchored forms of the ligands to induce clustering of the receptors that they bind is greater than that of the soluble forms. This ability is enhanced by positioning of both the membrane-bound ligands and their receptors within membrane rafts (Legler et al. 2003; Muppidi et al. 2004; So and Croft 2013) and by S-palmitoylation of the ligands and their receptors (Chakrabandhu et al. 2007; Feig et al. 2007; Rossin et al. 2009; Poggi et al. 2013). Formation of larger aggregates stabilizes binding of the ligands to the receptors. It can also dictate higher-order organization of signaling proteins (Fig. 1B).
The various signaling functions activated by the TNF family differ in the extents of their dependence on such higher-order signaling-protein organization. Accordingly, membrane and soluble forms of the ligands have differential abilities to trigger different cellular responses. For example, although TNFR1 can be activated both by soluble and by membrane-anchored TNF, TNFR2 is activated only by the latter. FAS can be triggered to signal for death by a dimer of FASL trimers, but not by a single trimer. Fn14, when massively aggregated, activates both the canonical and the alternative NF-κB transcription factor pathways, but only the alternative one when mildly aggregated (Grell et al. 1995; Schneider et al. 1998; Muhlenbeck et al. 2000; Bishop and Hostager 2003; Holler et al. 2003; Stone et al. 2006; O’Reilly et al. 2009; Wyzgol et al. 2009; Burkly 2014).
BIDIRECTIONAL SIGNALING: MULTIPLE FORMS AND FUNCTIONS OF SOLUBLE AND MEMBRANE-ANCHORED LIGANDS AND RECEPTORS
Similar to the IDs of TNF/NGF family receptors, the IDs of membrane-anchored TNF family ligands are found to recruit and activate signaling proteins on receptor–ligand interaction. Thus, they trigger “reverse signaling” within the ligand-producing cells (Stuber et al. 1995; Arens et al. 2004; Eissner et al. 2004; Grohmann et al. 2007; Kang et al. 2007; Sun et al. 2007; Juhasz et al. 2013). Various other similarities in action of the TNF and TNF/NGF families further blur the distinction between their identities as ligands and as receptors. As with the ligands, many of the receptors can be proteolytically cleaved, yielding ligand-binding “soluble receptors.” Two of these receptors, OPG and DcR3, occur only in soluble forms, while two others, DcR1 and DcR2, despite their anchorage to the membrane, are devoid of the molecular structures within the IDs that are required for signaling.
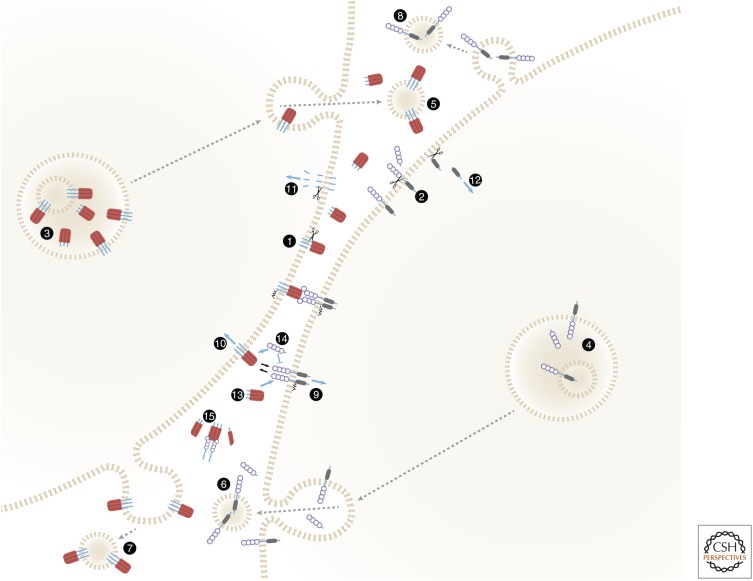
The various forms of soluble and membrane-anchored ligands and receptors, and the functions served by the transitions in these forms, are shown in Figure 2.
Figure 2.
Membrane-anchored and soluble forms of ligands and receptors of the tumor necrosis factor (TNF) families, and their functional roles. The figure shows an intriguing symmetry of the spectra of soluble and membrane-anchored forms attained by TNF family ligands and TNF/nerve growth factor (NGF) family receptors, and of the functions of these forms. Both ligands (1) and receptors (2) are proteolytically cleaved to yield soluble forms (Kriegler et al. 1988; Engelmann et al. 1990; Nophar et al. 1990; Black et al. 1997; Moss et al. 1997). The cleavage occurs constitutively or inducibly, either on the cell surface as illustrated (Black et al. 1997; Moss et al. 1997; Becker-Pauly and Rose-John 2013) or within the cell (Lopez-Fraga et al. 2001). (3,4) Some of the ligands (Gordon and Galli 1990; Bossi and Griffiths 1999; Koguchi et al. 2007) and receptors (Wang et al. 2003) accumulate within intracellular vesicles from which they are secreted in response to specific stimuli, thus supplementing either the cell-surface-expressed or the soluble pools. Some are released while anchored to membranes that might correspond either to exosomes that have accumulated in intracellular multivesicular bodies (5,6) or to microvesicles exfoliating from the cell surface (7,8) (Albanese et al. 1998; Martinez-Lorenzo et al. 1999; Islam et al. 2007). Binding of membrane-anchored ligands of the TNF family to their receptors, besides triggering receptor signaling (9), also triggers “reverse signaling” by the ligand molecules (10) (Stuber et al. 1995; Arens et al. 2004; Eissner et al. 2004; Grohmann et al. 2007; Kang et al. 2007; Sun et al. 2007; Juhasz et al. 2013). One mechanism contributing to this reverse signaling is intramembrane proteolytic cleavage, yielding ligand intracellular domain (ID) fragments that apparently mediate signaling following their translocation to the nucleus (Domonkos et al. 2001; Fluhrer et al. 2006; Friedmann et al. 2006; Kirkin et al. 2007) (11). Intramembrane cleavage that apparently contributes to signaling has also been reported for two receptors of the TNF/NGF family, nerve growth factor receptor (NGFR) and TNFR1 (Kanning et al. 2003; Kenchappa et al. 2006; Chhibber-Goel et al. 2016) (12). Accumulation of the soluble forms of both receptors and ligands interferes with the binding of membrane-bound forms to the ligands and the receptors, respectively, and can thus block signaling (13,14). To the extent that the soluble forms are incapable of triggering signaling, they may also interfere with signaling activation by their membrane-bound forms. However, the association of soluble receptors with soluble ligands also stabilizes the trimeric structures of the soluble ligands, so that they function not as mere inhibitors but rather as buffering agents. While decreasing the intensity of signaling activation, they also extend its duration (15) (Aderka et al. 1992; Eliaz et al. 1996).
“RE-ANCHORING” OF SOLUBLE FORMS OF LIGANDS AND RECEPTORS VIA THEIR BINDING TO EXTRACELLULAR MATRIX COMPONENTS
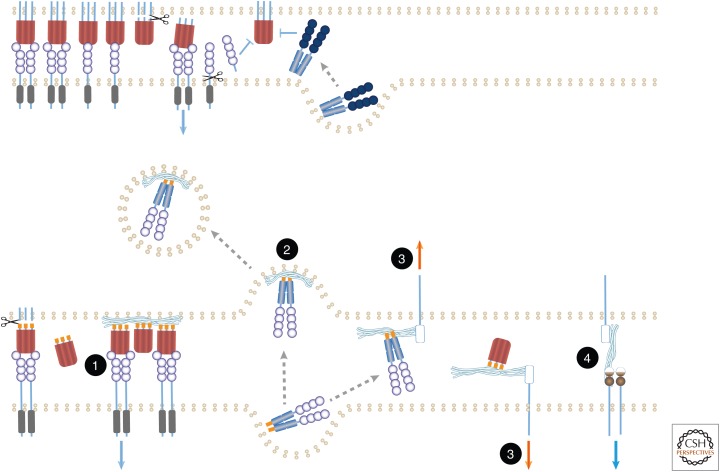
Several ligands of the TNF family and the two TNF-family receptors that occur only in soluble form (OPG and DCR3) contain sites that bind extracellular matrix components such as heparan sulfate proteoglycans (HSPGs) and fibronectin. Their associations with these extracellular compounds have several functional consequences. (The numbering that follows corresponds to that shown in Fig. 3.) (1) Binding of TNF (Alon et al. 1994) and of FASL (Aoki et al. 2001; Zanin-Zhorov et al. 2003) to extracellular matrix proteins such as fibronectin, and of APRIL to HSPGs (Ingold et al. 2005; Kimberley et al. 2009), facilitate signaling activation by these ligands, apparently by imposing an oligomeric state on them. Such binding can also impose local restrictions. The anchorage of APRIL to HSPGs in mucosal lymphoid tissues not only augments its function but also restricts this ligand to target cells at this site (Huard et al. 2008), while the binding of EDA to proteoglycan interferes with EDA function by preventing its access to EDAR. (2) Binding of OPG to HSPGs blocks the function of OPG by also causing its uptake into the cell and its subsequent degradation (Standal et al. 2002). (3) Binding of OPG, APRIL, or DcR3 to HSPG appears to also trigger signaling by cell-surface molecules such as syndecans, which are linked to HSPG moieties, or by cell-surface molecules with which these moieties associate noncovalently (Couchman 2003; Mosheimer et al. 2005; Chang et al. 2006; Dillon et al. 2006; You et al. 2008). (4) Conversely, binding of cell-associated HSPG to TACI triggers TACI signaling, apparently independently of any ligand of the TNF family (Bischof et al. 2006). Because both APRIL and its receptor TACI bind HSPG, they probably form tripartite complexes in which the three proteins modulate one another’s function (Moreaux et al. 2009).
Figure 3.
“Re-anchoring” of soluble forms of ligands and receptors by their binding to extracellular matrix components. Various reported effects of extracellular matrix components on the functions of soluble ligands and receptors, shown in the lower part of the figure, are compared with the functions of soluble ligands and receptors in the absence of such components, as shown in the upper part. See text for details and for numbering of the illustrated effects. Undulating lines correspond to extracellular matrix components. Orange forms correspond to structural motifs that dictate binding to such components.
MECHANISMS OF SIGNALING ACTIVATION BY THE TNF FAMILY
The cellular responses initiated by the TNF family reflect a gamut of molecular changes, including altered expression of many proteins, and extensive changes in their patterns of phosphorylation, ubiquitination, association, localization, and rates of turnover, as well as alterations in the expression of various lipid metabolites. Because neither the IDs of TNF/NGF family receptors nor those of TNF family ligands possess enzymatic activity, they need to bind to signaling proteins that do express such activities, which are stimulated as a consequence of ligand–receptor association.
Protein-Binding Motifs in the Receptors and Ligands
The receptors of the TNF family can be subclassified into two groups with distinct binding motifs in their IDs. The IDs of TNFR1, DR3, FAS, TRAIL-R1, TRAIL-R2, NGFR, and EDAR each contain a DD motif of about 80 amino acids that bind adapter proteins containing the same motif. All other receptors contain short (five to eight amino acid residues) motifs of binding to adapter proteins of the TNF receptor–associated factors (TRAFs) family. NGFR is the only receptor that contains both an (atypical) DD and a TRAF-binding motif. The TRAF-binding receptors are incapable of triggering signaling pathways activated by the DD adapter proteins. In contrast, because two of the DD-containing adapter proteins—TNF receptor-associated death domain (TRADD) and EDAR-associated death domain (EDARADD)—also contain a TRAF-binding motif, the DD-containing receptors are capable of triggering at least part of the signaling pathways activated by the TRAF-binding receptors.
Binding of both DD-containing adapter proteins and the TRAFs to their receptors dictates the formation of large complexes containing several receptor molecules and multiple adapter molecules. As to the associations of the receptors with TRAFs (associations whose structural basis has so far been explored only for TRAF2 and TRAF6), assembly of the signaling complexes is fostered by constitutive trimeric oligomerization of the carboxyl-terminal leucine zipper TRAF motifs and by induced dimeric association of the TRAFs’ amino-terminal regions (McWhirter et al. 1999; Park et al. 1999; Yin et al. 2009a). In the initiation of signaling through DD association, an event so far explored only for FAS association with the adapter protein FADD/MORT1, oligomerization is fostered by homotypic associations of the DDs of both FAS and FADD/MORT1 as well as by their heterotypic associations (Scott et al. 2009; Wang et al. 2010; Kersse et al. 2011; Li et al. 2013).
These associations between receptors and their adapter proteins initiate signaling by exposing, within the adapter proteins, binding surfaces for signaling proteins. With the exception of TRAF1, and—as was recently indicated, probably also with the exception of TRAF2 (Yin et al. 2009b)—members of the TRAF adapter protein family possess ubiquitin ligase activity, which is triggered by their induced associations with receptors.
Some adapter proteins initiate the above associations and activities while still bound to receptors expressed on the cell surface. Others do so only after uptake of the receptors into the cells (Schutze et al. 2008; Ganeff et al. 2011). Some of the signaling complexes that they generate remain associated with the receptors. Others dissociate from the receptors and generate signaling complexes in the cytoplasm (Sessler et al. 2013).
Sporadic evidence indicates that various other evolutionarily conserved amino acid residues within the IDs of receptors and of ligands serve—either without modification or after undergoing phosphorylation or ubiquitination—as binding sites for additional cytoplasmic proteins. In so doing, they contribute to the initiation of additional signaling pathways and to the control of trafficking of the receptors and ligands (e.g., Pocsik et al. 1995; Adam Klages et al. 1996; Watts et al. 1999; Eissner et al. 2004; Kimura et al. 2004; Sun et al. 2007; Juhasz et al. 2013; Ma et al. 2013; Fritsch et al. 2014; Chakrabandhu et al. 2016).
In addition to their ligand–receptor interaction motifs, the EDs of both the ligands and the receptors contain, in closer proximity to their TDs, specific sequences that determine their mode of shedding. Several ligands, including EDA, BAFF, and APRIL, contain consensus sequences for furin cleavage and are constitutively shed by intracellularly located furin. Other ligands as well as several receptors, including TNF and its receptors, are shed inducibly after they reach the cell membrane, mainly via the effects of metalloproteinases such as TACE/ADAM17. Little is known of the sequence determinants that dictate this selective shedding (Brakebusch et al. 1994; Thomas 2002; Hayashida et al. 2010).
Signaling Pathways
Studies of the mechanisms of signaling for apoptotic and for necrotic cell death as well as for activation of NF-κB transcription factors by TNF family members provided the foundation for exploring the regulation of these functions by external inducers. These activities of the TNF family are the most extensively studied to date. However, several others are known, including signaling for activation of the extracellular signal-regulated kinase (ERK), c-Jun amino terminal kinase (JNK), and p38 mitogen-activated protein (MAP) kinase cascades, other serine/threonine kinases, the phosphoinositide 3-kinase (PI3K)/Akt pathway, superoxide generation, soluble Src-family tyrosine kinases, the neutral and acidic sphingomyelinases, and phospholipase C. Space constraints preclude further attention in this review to these signaling mechanisms, which have been extensively reviewed elsewhere (Eissner et al. 2004; Hayden and Ghosh 2012; Juhasz et al. 2013; Li et al. 2013; Sessler et al. 2013; So and Croft 2013; Sabio and Davis 2014; Wallach 2016b).
FOREIGN ENCOUNTERS: NONCANONICAL CORECEPTORS AND LIGANDS
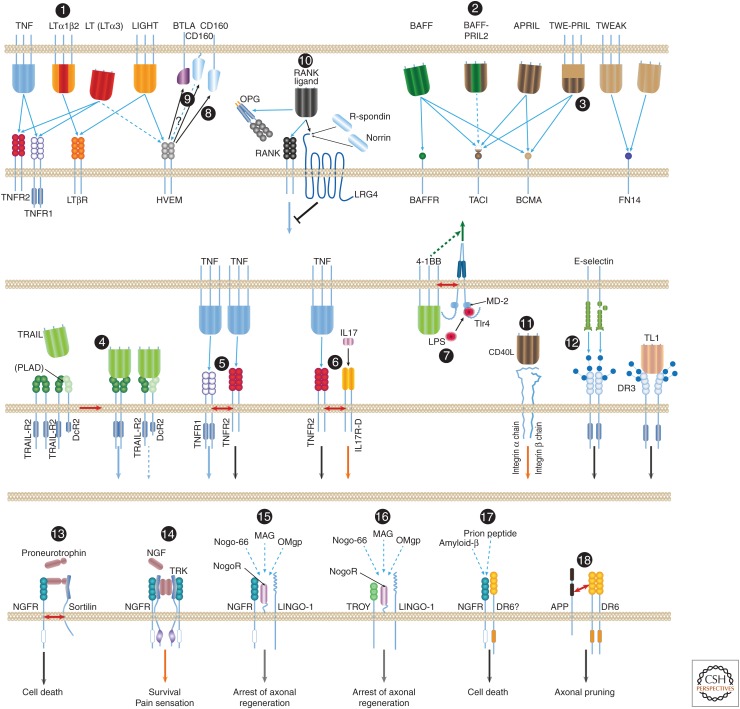
The information presented above delineates a shared set of mechanistic principles by which the different members of the TNF ligand and receptor families associate. However, several members of the families can also participate in unique interactions with coreceptors and ligands, including some that do not belong to the TNF ligand and TNF/NGFR families. Examples of these noncanonical associations, some extensively documented and others for which the evidence is limited and requires confirmation, are presented in Figure 4. Although such interactions have been noticed for only a few members of the TNF ligand and receptor families up to now, they might be found to occur with other members as well.
Figure 4.
Noncanonical associations of the tumor necrosis factor (TNF) ligand and the TNF/ nerve growth factor receptor (NGFR) families. Most of the interactions of ligands and receptors of the TNF ligand and TNF/NGFR families are known to occur between homotrimeric molecules of a particular ligand and molecules of a particular receptor. A few exceptions, however, are known: (1) Lymphotoxin β (LTβ) functions only within heterotrimers that it forms with lymphotoxin α (LTα). Whereas homotrimers of LT bind to the TNF receptors, the LTα1β2 heterotrimer binds to the LTβ receptor (LTβR) (Ware 2005). (2) B-cell-activating factor of the tumor necrosis factor family (BAFF) and a proliferation-inducing ligand (APRIL) also form heterotrimers (so far discerned only in patients with autoimmune diseases), and these heterotrimers apparently activate only transmembrane activator and calcium-modulating cyclophilin ligand interactor (TACI) (Roschke et al. 2002; Dillon et al. 2006; Schuepbach-Mallepell et al. 2015). (3) Although APRIL is constitutively shed and its trimers therefore occur only in soluble form, its gene also yields a fused joint splice variant with the neighboring TWEAK gene. The protein encoded by this transcript—TWE-PRIL—is anchored to the cell membrane (Pradet-Balade et al. 2002). (4) Association of molecules of TRAIL-R2 and the membrane-anchored truncated receptor DcR2 through their preligand assembly domain (PLAD) dictate association of TRAIL with the two receptors in mixed complexes wherein TRAIL-R2 signaling is suppressed by DcR2 (Clancy et al. 2005). (5) Molecules of the two receptors for TNF (TNFR1 and TNFR2) reportedly also associate on their binding to ligand. However, this association does not occur through binding of the two receptors to the same ligand molecule. In fact, the two receptor species are incapable of binding simultaneously to the same TNF molecule; molecules of the two receptors rather associate only after binding independently to distinct TNF molecules (Pinckard et al. 1997). (6) Molecules of TNFR2 reportedly also associate with molecules of IL-17 receptor D, a member of an unrelated cytokine-receptor family. This association, which occurs on triggering by the ligands of the two receptors, imposes assembly of aggregates of the two receptors and functional cooperation between them (Yang et al. 2015). (7) Another example of functional interaction with a structurally unrelated receptor is the association of the ligand for 4-1BB with a coexpressed TLR4–MD2 complex. This apparently occurs through the TMs of 4-1BBL and Tlr4 and the consequent potentiation of lipopolysaccharide (LPS)-induced Tlr4 signaling in a way that depends on the function of the intracellular domain (ID) of 4-1BBL but independently of the association of 4-1BBL with 4-1BB (Kang et al. 2007; Ma et al. 2013). (8) A different kind of noncanonical association is observed in the function of herpesvirus entry mediator (HVEM), a receptor of the TNF family. Besides its association with two TNF family ligands (with LIGHT, and [weakly] with LT homotrimers) it also binds to B- and T-lymphocyte attenuator (BTLA) and CD160, two cell-surface proteins of the immunoglobulin superfamily, thereby triggering inhibitory signaling by those two proteins (Shui et al. 2011). (9) Whether this association also triggers signaling by the ID of HVEM is not known. (10) RANKL, besides binding to RANK and to the soluble receptor osteoprotegerin (OPG), also binds to the seven-transmembrane (TM), leucine-rich repeat containing G protein-coupled receptor 4 (LGR4), which also serves as receptor for R-spondins and for Norrin. It thus triggers signaling antagonistic to that initiated by RANK (Luo et al. 2016). (11) Binding of soluble CD40L to integrin αIIbβ3 and to integrin α5β1 triggers signaling by those two membrane–protein complexes (Andre et al. 2002; Leveille et al. 2007). (12) Binding of the cell-adhesion lectin, E-selectin, to DR3-linked sialic-acid-linked sugar chains triggers signaling by DR3 (Porquet et al. 2011). Juxtaposition and activation of the two TRAIL receptors, DR4 and DR4, by TRAIL also depends, for a reason not yet clear, on glycosylation of these receptors (Wagner et al. 2007). The most elaborate known set of noncanonical associations is observed in the function of the NGFR, a receptor of the TNF/NGF family for which no ligand of the TNF family is known. NGFR contributes to signaling for different effects in response to different inducers through association with a series of different coreceptors. (13) It contributes to signaling for death in response to proneurotrophins, to which it binds in association with a sortilin family receptor. (14) When associating with a Trk tyrosine kinase receptor, apparently through the TMs and IDs of both receptors (Esposito et al. 2001), NGFR contributes to high-affinity binding of NGF, and signals for cell survival and for pain sensation. (15) NGFR is also found to form, through extracellular domain (ED) associations, a ternary complex with the glycosylphosphatidylinositol (GPI)-linked Nogo-66 receptor (NogoR) and the TM receptor LINGO-1. In response to myelin-associated inhibitory factors (a 66-amino-acid fragment of the oligodendrocyte-derived growth inhibitory protein Nogo, the oligodendrocyte myelin glycoprotein [OMgp] or the myelin-associated glycoprotein [MAG]), this complex signals for arrest of axonal regeneration following injury (Hempstead 2002; Chao 2003; Ibanez and Simi 2012). (16) In that complex, NGFR can be replaced by the orphan TNF family receptor TROY (Shao et al. 2005). (17) Finally, direct binding of amyloid-β to NGFR (Hempstead 2002; Chao 2003; Ibanez and Simi 2012), and probably also to a complex of NGFR and another orphan receptor of the TNF/NGF family, DR6 (Hu et al. 2013), reportedly triggers signaling for neuronal death. (18) DR6 was found to bind to a carboxyl-terminal region in the ED of the amyloid precursor protein (APP). APP and DR6 cooperate in the induction of axonal pruning. The mechanism underlying this cooperation is not clear, nor is it known whether the cooperation occurs between proteins expressed in the same cell, as illustrated in the figure, or in distinct cells (Olsen et al. 2014). BCMA, B-cell maturation antigen.
HOW IS REGULATION BY THE TNF FAMILY REGULATED?
As in the case of other cytokines, signaling by members of the TNF family is triggered when ligand and receptor molecules are brought close enough to allow their binding to each other. This approximation was found to be dictated at multiple mechanistic levels. In most cases, it occurs by induced up-regulation of the ligand molecules, allowing them to bind to receptor molecules that are constitutively expressed. Such is the case with TNF, whose induction mostly occurs in a transient manner, although TNFR1 to which it binds is widely expressed constitutively. However, the inverse type of modulation is also observed. TWEAK, for example, is a ligand that is constitutively expressed in macrophages, whereas the expression of its receptor Fn14 is induced in injured tissues. Up-regulation of the expression of both ligands and receptors is induced by specific signaling pathways that are activated in response to insults or developmental cues. The best-documented mode of up-regulation is by enhanced transcription. However, it also occurs on other mechanistic levels, including splicing, RNA transport, and altered messenger RNA (mRNA) stability and mRNA translation rates. Also contributing to this regulation are associations of proteins, microRNAs and long noncoding RNAs with transcripts (Wallach 2016a; Wallach and Kovalenko 2016). Once synthesized, the ligands and receptors can be subjected to further regulation by mechanisms controlling their translocation to the cell surface (Bossi and Griffiths 1999) and, in the case of their soluble forms, by the proteolytic activity through which these forms are generated. For some family members, this proteolytic activity is exerted constitutively (Lopez-Fraga et al. 2001), whereas for others it is activated by specific signals (Black et al. 1997; Moss et al. 1997; Becker-Pauly and Rose-John 2013). Approximation of receptors to membrane-bound ligands can also be dictated merely by induced juxtapositioning of the cells that express the receptors and ligands.
Some of the ligands accumulate within intracellular reservoirs. Signaling initiation by these ligands is thus dictated by their exocytotic secretion (Gordon and Galli 1990; Bossi and Griffiths 1999; Koguchi et al. 2007).
Increased expression of the receptors or ligands, either artificially (Boldin et al. 1995) or in response to natural stimuli (Lu et al. 2014), is in some situations sufficient to allow the IDs of these molecules to encounter one another and hence to trigger signaling independently of ligand–receptor association.
CONCLUDING THEOLOGICAL REMARKS
Members of the TNF cytokine family and of the TNF/NGFR family are used by almost all metazoan phyla. Some of their activities have been well preserved over more than 500 million years of evolution (Igaki and Miura 2014; Quistad et al. 2014). Their major proximal signaling enzymes originated even earlier (Uren et al. 2000; Zapata et al. 2007; Yuan et al. 2009; Zmasek and Godzik 2013; Sakamaki et al. 2014). Nature, it would seem, had good reason to preserve this cytokine family so well and for so long. At face value, however, the known activities of this family do not seem to be sufficiently unique to warrant such preservation. Other than in the case of the cytotoxic activity of a few family members, the various individual cellular effects of the TNF family and the signaling mechanisms that account for them do not seem to differ radically from those of various other cytokines. The pattern of cellular effects of IL-1 in inflammation, for example, as well as the signaling mechanisms that it activates, greatly resemble those of TNF. Why would nature preserve two cytokines that seem to serve the same set of functions, and why would it choose to use them differentially in different situations?
Also puzzling is the remarkable similarity among the ranges of signaling activities triggered by the various family members. How can the heterogeneous and distinct biological activity patterns of the different family members be explained in terms of a set of mechanisms so limited and so invariant? These enigmas suggest that the mechanisms of action of the TNF families possess a greater degree of sophistication than has so far met the eye.
An example of the insight gained in attempting to deepen our perception of this area was the discovery that, although all TNF family members signal for activation of NF-κB, some are able to do so via a distinct route, the so-called “alternative pathway,” which yields molecular targets and functional consequences that differ from those of the more widely used “canonical pathways” (Hayden and Ghosh 2012). The alternative pathway, and the protein kinase NF-κB-inducing kinase (NIK) that initiates it, appear to have evolved relatively late in the history of the TNF family, contemporaneously with emergence of the vertebrates and of their adaptive immunity that this pathway regulates. Other signaling pathways shared by different TNF family members may likewise be found to have evolved into several different forms that are affected differentially by these different members and serve distinct functions.
The various “noncanonical” interactions of the TNF and TNF/NGF families (Fig. 4) doubtless also endow individual members of the family with unique mechanisms of action. An example of a unique mechanism that might depend on such noncanonical interactions is presented in Figure 1C. As opposed to the usual mode of signaling initiation by the TNF/NGF family (by induced juxtaposition), NGFR—which participates in several such noncanonical associations—was suggested rather to trigger signaling by imposing separation of the receptor IDs. This mechanism is dictated by covalent linkage of a conserved cysteine in the TM of NGFR. Conserved cysteines also occur in the TDs of several other TNF/NGF family members. It was therefore suggested that portions of these other receptors also occur as covalently linked dimers and serve similarly to mediate unique functions, which likewise may depend on some noncanonical associations (Vilar et al. 2009).
We have come a long way in clarifying some general mechanisms of action of the various families of cytokines, including the TNF family. It is now time to focus on clarifying the mechanisms that endow each family, and each family member, with uniqueness. Progress in this regard will undoubtedly increase our ability to design selective therapeutic modulation of specific functions of these fascinating molecules.
Footnotes
Editors: Warren J. Leonard and Robert D. Schreiber
Additional Perspectives on Cytokines available at www.cshperspectives.org
REFERENCES
- Adam Klages S, Adam D, Wiegmann K, Struve S, Kolanus W, Schneider Mergener J, Kronke M. 1996. FAN, a novel WD-repeat protein, couples the p55 TNF-receptor to neutral sphingomyelinase. Cell 86: 937–947. [DOI] [PubMed] [Google Scholar]
- Aderka D, Engelmann H, Maor Y, Brakebusch C, Wallach D. 1992. Stabilization of the bioactivity of tumor necrosis factor by its soluble receptors. J Exp Med 175: 323–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akassoglou K, Bauer J, Kassiotis G, Lassmann H, Kollias G, Probert L. 1999. Transgenic models of TNF induced demyelination. Adv Exp Med Biol 468: 245–259. [DOI] [PubMed] [Google Scholar]
- Albanese J, Meterissian S, Kontogiannea M, Dubreuil C, Hand A, Sorba S, Dainiak N. 1998. Biologically active Fas antigen and its cognate ligand are expressed on plasma membrane-derived extracellular vesicles. Blood 91: 3862–3874. [PubMed] [Google Scholar]
- Alon R, Cahalon L, Hershkoviz R, Elbaz D, Reizis B, Wallach D, Akiyama SK, Yamada KM, Lider O. 1994. TNF-α binds to the N-terminal domain of fibronectin and augments the β1-integrin-mediated adhesion of CD4+ T lymphocytes to the glycoprotein. J Immunol 152: 1304–1313. [PubMed] [Google Scholar]
- Andre P, Prasad KS, Denis CV, He M, Papalia JM, Hynes RO, Phillips DR, Wagner DD. 2002. CD40L stabilizes arterial thrombi by a β3 integrin-dependent mechanism. Nat Med 8: 247–252. [DOI] [PubMed] [Google Scholar]
- Aoki K, Kurooka M, Chen JJ, Petryniak J, Nabel EG, Nabel GJ. 2001. Extracellular matrix interacts with soluble CD95L: Retention and enhancement of cytotoxicity. Nat Immunol 2: 333–337. [DOI] [PubMed] [Google Scholar]
- Apostolaki M, Armaka M, Victoratos P, Kollias G. 2010. Cellular mechanisms of TNF function in models of inflammation and autoimmunity. Curr Dir Autoimmun 11: 1–26. [DOI] [PubMed] [Google Scholar]
- Arens R, Nolte MA, Tesselaar K, Heemskerk B, Reedquist KA, van Lier RA, van Oers MH. 2004. Signaling through CD70 regulates B cell activation and IgG production. J Immunol 173: 3901–3908. [DOI] [PubMed] [Google Scholar]
- Becker-Pauly C, Rose-John S. 2013. TNFα cleavage beyond TACE/ADAM17: Matrix metalloproteinase 13 is a potential therapeutic target in sepsis and colitis. EMBO Mol Med 5: 902–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof D, Elsawa SF, Mantchev G, Yoon J, Michels GE, Nilson A, Sutor SL, Platt JL, Ansell SM, von Bulow G, et al. 2006. Selective activation of TACI by syndecan-2. Blood 107: 3235–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GA, Hostager BS. 2003. The CD40–CD154 interaction in B cell–T cell liaisons. Cytokine Growth Factor Rev 14: 297–309. [DOI] [PubMed] [Google Scholar]
- Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S, et al. 1997. A metalloproteinase disintegrin that releases tumour-necrosis factor-α from cells. Nature 385: 729–733. [DOI] [PubMed] [Google Scholar]
- Blomberg J, Ruuth K, Santos D, Lundgren E. 2008. Acquired resistance to Fas/CD95 ligation in U937 cells is associated with multiple molecular mechanisms. Anticancer Res 28: 593–599. [PubMed] [Google Scholar]
- Boldin MP, Mett IL, Varfolomeev EE, Chumakov I, Shemer AY, Camonis JH, Wallach D. 1995. Self-association of the “death domains” of the p55 tumor necrosis factor (TNF) receptor and Fas/APO1 prompts signaling for TNF and Fas/APO1 effects. J Biol Chem 270: 387–391. [DOI] [PubMed] [Google Scholar]
- Bossi G, Griffiths GM. 1999. Degranulation plays an essential part in regulating cell surface expression of Fas ligand in T cells and natural killer cells. Nat Med 5: 90–96. [DOI] [PubMed] [Google Scholar]
- Boyce BF, Xing L. 2007. Biology of RANK, RANKL, and osteoprotegerin. Arthritis Res Ther 9: S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakebusch C, Varfolomeev EE, Batkin M, Wallach D. 1994. Structural requirements for inducible shedding of the p55 tumor necrosis factor receptor. J Biol Chem 269: 32488–32496. [PubMed] [Google Scholar]
- Bruce AJ, Boling W, Kindy MS, Peschon J, Kraemer PJ, Carpenter MK, Holtsberg FW, Mattson MP. 1996. Altered neuronal and microglial responses to excitotoxic and ischemic brain injury in mice lacking TNF receptors. Nat Med 2: 788–794. [DOI] [PubMed] [Google Scholar]
- Burkly LC. 2014. TWEAK/Fn14 axis: The current paradigm of tissue injury-inducible function in the midst of complexities. Semin Immunol 26: 229–236. [DOI] [PubMed] [Google Scholar]
- Cachero TG, Schwartz IM, Qian F, Day ES, Bossen C, Ingold K, Tardivel A, Krushinskie D, Eldredge J, Silvian L, et al. 2006. Formation of virus-like clusters is an intrinsic property of the tumor necrosis factor family member BAFF (B-cell-activating factor). Biochemistry 45: 2006–2013. [DOI] [PubMed] [Google Scholar]
- Cerami A, Ikeda Y, Le Trang N, Hotez PJ, Beutler B. 1985. Weight loss associated with an endotoxin-induced mediator from peritoneal macrophages: The role of cachectin (tumor necrosis factor). Immunol Lett 11: 173–177. [DOI] [PubMed] [Google Scholar]
- Chakrabandhu K, Herincs Z, Huault S, Dost B, Peng L, Conchonaud F, Marguet D, He HT, Hueber AO. 2007. Palmitoylation is required for efficient Fas cell death signaling. EMBO J 26: 209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabandhu K, Huault S, Durivault J, Lang K, Ta Ngoc L, Bole A, Doma E, Derijard B, Gerard JP, Pierres M, et al. 2016. An evolution-guided analysis reveals a multi-signaling regulation of Fas by tyrosine phosphorylation and its implication in human cancers. PLoS Biol 14: e1002401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan FK. 2007. Three is better than one: Pre-ligand receptor assembly in the regulation of TNF receptor signaling. Cytokine 37: 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Chan YH, Jackson DG, Hsieh SL. 2006. The glycosaminoglycan-binding domain of decoy receptor 3 is essential for induction of monocyte adhesion. J Immunol 176: 173–180. [DOI] [PubMed] [Google Scholar]
- Chao MV. 2003. Neurotrophins and their receptors: A convergence point for many signalling pathways. Nat Rev Neurosci 4: 299–309. [DOI] [PubMed] [Google Scholar]
- Chen L, Park SM, Tumanov AV, Hau A, Sawada K, Feig C, Turner JR, Fu YX, Romero IL, Lengyel E, et al. 2010. CD95 promotes tumour growth. Nature 465: 492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhibber-Goel J, Coleman-Vaughan C, Agrawal V, Sawhney N, Hickey E, Powell JC, McCarthy JV. 2016. γ-Secretase activity is required for regulated intramembrane proteolysis of tumor necrosis factor (TNF) receptor 1 and TNF-mediated pro-apoptotic signaling. J Biol Chem 291: 5971–5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy L, Mruk K, Archer K, Woelfel M, Mongkolsapaya J, Screaton G, Lenardo MJ, Chan FK. 2005. Preligand assembly domain-mediated ligand-independent association between TRAIL receptor 4 (TR4) and TR2 regulates TRAIL-induced apoptosis. Proc Natl Acad Sci 102: 18099–18104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsini NS, Sancho-Martinez I, Laudenklos S, Glagow D, Kumar S, Letellier E, Koch P, Teodorczyk M, Kleber S, Klussmann S, et al. 2009. The death receptor CD95 activates adult neural stem cells for working memory formation and brain repair. Cell Stem Cell 5: 178–190. [DOI] [PubMed] [Google Scholar]
- Couchman JR. 2003. Syndecans: Proteoglycan regulators of cell-surface microdomains? Nat Rev Mol Cell Biol 4: 926–937. [DOI] [PubMed] [Google Scholar]
- Croft M. 2014. The TNF family in T cell differentiation and function—Unanswered questions and future directions. Semin Immunol 26: 183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzer R. 2001. Cytokine-induced sickness behavior: Mechanisms and implications. Ann NY Acad Sci 933: 222–234. [DOI] [PubMed] [Google Scholar]
- Dillon SR, Gross JA, Ansell SM, Novak AJ. 2006. An APRIL to remember: Novel TNF ligands as therapeutic targets. Nat Rev Drug Discov 5: 235–246. [DOI] [PubMed] [Google Scholar]
- Dinarello CA, Cannon JG, Wolff SM, Bernheim HA, Beutler B, Cerami A, Figari IS, Palladino MA Jr, O’Connor JV. 1986. Tumor necrosis factor (cachectin) is an endogenous pyrogen and induces production of interleukin 1. J Exp Med 163: 1433–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domonkos A, Udvardy A, Laszlo L, Nagy T, Duda E. 2001. Receptor-like properties of the 26 kDa transmembrane form of TNF. Eur Cytokine Netw 12: 411–419. [PubMed] [Google Scholar]
- Drayton DL, Liao S, Mounzer RH, Ruddle NH. 2006. Lymphoid organ development: From ontogeny to neogenesis. Nat Immunol 7: 344–353. [DOI] [PubMed] [Google Scholar]
- Eissner G, Kolch W, Scheurich P. 2004. Ligands working as receptors: Reverse signaling by members of the TNF superfamily enhance the plasticity of the immune system. Cytokine Growth Factor Rev 15: 353–366. [DOI] [PubMed] [Google Scholar]
- Elgueta R, Benson MJ, de Vries VC, Wasiuk A, Guo Y, Noelle RJ. 2009. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev 229: 152–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliaz R, Wallach D, Kost J. 1996. Long-term protection against tumor necrosis factor (TNF) effects by controlled delivery of the soluble p55 tumor necrosis factor receptor. Cytokine 8: 482–487. [DOI] [PubMed] [Google Scholar]
- Elmetwali T, Young LS, Palmer DH. 2010. CD40 ligand-induced carcinoma cell death: A balance between activation of TNFR-associated factor (TRAF) 3-dependent death signals and suppression of TRAF6-dependent survival signals. J Immunol 184: 1111–1120. [DOI] [PubMed] [Google Scholar]
- Engelmann H, Novick D, Wallach D. 1990. Two tumor necrosis factor-binding proteins purified from human urine. Evidence for immunological cross-reactivity with cell surface tumor necrosis factor receptors. J Biol Chem 265: 1531–1536. [PubMed] [Google Scholar]
- Esposito D, Patel P, Stephens RM, Perez P, Chao MV, Kaplan DR, Hempstead BL. 2001. The cytoplasmic and transmembrane domains of the p75 and Trk A receptors regulate high affinity binding to nerve growth factor. J Biol Chem 276: 32687–32695. [DOI] [PubMed] [Google Scholar]
- Falschlehner C, Schaefer U, Walczak H. 2009. Following TRAIL’s path in the immune system. Immunology 127: 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig C, Tchikov V, Schutze S, Peter ME. 2007. Palmitoylation of CD95 facilitates formation of SDS-stable receptor aggregates that initiate apoptosis signaling. EMBO J 26: 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figgett WA, Vincent FB, Saulep-Easton D, Mackay F. 2014. Roles of ligands from the TNF superfamily in B cell development, function, and regulation. Semin Immunol 26: 191–202. [DOI] [PubMed] [Google Scholar]
- Fluhrer R, Grammer G, Israel L, Condron MM, Haffner C, Friedmann E, Bohland C, Imhof A, Martoglio B, Teplow DB, et al. 2006. A γ-secretase-like intramembrane cleavage of TNFα by the GxGD aspartyl protease SPPL2b. Nat Cell Biol 8: 894–896. [DOI] [PubMed] [Google Scholar]
- Fontaine V, Mohand-Said S, Hanoteau N, Fuchs C, Pfizenmaier K, Eisel U. 2002. Neurodegenerative and neuroprotective effects of tumor necrosis factor (TNF) in retinal ischemia: Opposite roles of TNF receptor 1 and TNF receptor 2. J Neurosci 22: RC216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force WR, Glass AA, Benedict CA, Cheung TC, Lama J, Ware CF. 2000. Discrete signaling regions in the lymphotoxin-β receptor for tumor necrosis factor receptor-associated factor binding, subcellular localization, and activation of cell death and NF-κB pathways. J Biol Chem 275: 11121–11129. [DOI] [PubMed] [Google Scholar]
- Friedmann E, Hauben E, Maylandt K, Schleeger S, Vreugde S, Lichtenthaler SF, Kuhn PH, Stauffer D, Rovelli G, Martoglio B. 2006. SPPL2a and SPPL2b promote intramembrane proteolysis of TNFα in activated dendritic cells to trigger IL-12 production. Nat Cell Biol 8: 843–848. [DOI] [PubMed] [Google Scholar]
- Fritsch J, Stephan M, Tchikov V, Winoto-Morbach S, Gubkina S, Kabelitz D, Schutze S. 2014. Cell fate decisions regulated by K63 ubiquitination of tumor necrosis factor receptor 1. Mol Cell Biol 34: 3214–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q, Fu TM, Cruz AC, Sengupta P, Thomas SK, Wang S, Siegel RM, Wu H, Chou JJ. 2016. Structural basis and functional role of intramembrane trimerization of the Fas/CD95 death receptor. Mol Cell 61: 602–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganeff C, Remouchamps C, Boutaffala L, Benezech C, Galopin G, Vandepaer S, Bouillenne F, Ormenese S, Chariot A, Schneider P, et al. 2011. Induction of the alternative NF-κB pathway by lymphotoxin αβ (LTαβ) relies on internalization of LTβ receptor. Mol Cell Biol 31: 4319–4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos NT, Steele LP, Thomson MJ, Selby PJ, Southgate J, Trejdosiewicz LK. 2006. A novel mechanism of CD40-induced apoptosis of carcinoma cells involving TRAF3 and JNK/AP-1 activation. Cell Death Differ 13: 1789–1801. [DOI] [PubMed] [Google Scholar]
- Gordon JR, Galli SJ. 1990. Mast cells as a source of both preformed and immunologically inducible TNF-α/cachectin. Nature 346: 274–276. [DOI] [PubMed] [Google Scholar]
- Granger GA, Kolb WP. 1968. Lymphocyte in vitro cytotoxicity: Mechanisms of immune and non-immune small lymphocyte mediated target L cell destruction. J Immunol 101: 111–120. [PubMed] [Google Scholar]
- Grell M, Douni E, Wajant H, Löhden M, Clauss M, Baxeiner B, Georgopoulos S, Lesslauer W, Kollias G, Pfizenmaier K, et al. 1995. The transmembrane form of tumor necrosis factor is the prime activating ligand of the 80 kDa tumor necrosis factor receptor. Cell 83: 793–802. [DOI] [PubMed] [Google Scholar]
- Grell M, Zimmermann G, Gottfried E, Chen CM, Grunwald U, Huang DC, Wu Lee YH, Durkop H, Engelmann H, Scheurich P, et al. 1999. Induction of cell death by tumour necrosis factor (TNF) receptor 2, CD40 and CD30: A role for TNF-R1 activation by endogenous membrane-anchored TNF. EMBO J 18: 3034–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann U, Volpi C, Fallarino F, Bozza S, Bianchi R, Vacca C, Orabona C, Belladonna ML, Ayroldi E, Nocentini G, et al. 2007. Reverse signaling through GITR ligand enables dexamethasone to activate IDO in allergy. Nat Med 13: 579–586. [DOI] [PubMed] [Google Scholar]
- Hahn T, Toker L, Budilovsky S, Aderka D, Eshhar Z, Wallach D. 1985. Use of monoclonal antibodies to a human cytotoxin for its isolation and for examining the self-induction of resistance to this protein. Proc Natl Acad Sci 82: 3814–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada R, Leibbrandt A, Hanada T, Kitaoka S, Furuyashiki T, Fujihara H, Trichereau J, Paolino M, Qadri F, Plehm R, et al. 2009. Central control of fever and female body temperature by RANKL/RANK. Nature 462: 505–509. [DOI] [PubMed] [Google Scholar]
- Hayashida K, Bartlett AH, Chen Y, Park PW. 2010. Molecular and cellular mechanisms of ectodomain shedding. Anat Rec (Hoboken) 293: 925–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. 2012. NF-κB, the first quarter-century: Remarkable progress and outstanding questions. Genes Dev 26: 203–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempstead BL. 2002. The many faces of p75NTR. Curr Opin Neurobiol 12: 260–267. [DOI] [PubMed] [Google Scholar]
- Hess A, Axmann R, Rech J, Finzel S, Heindl C, Kreitz S, Sergeeva M, Saake M, Garcia M, Kollias G, et al. 2011. Blockade of TNF-α rapidly inhibits pain responses in the central nervous system. Proc Natl Acad Sci 108: 3731–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler N, Tardivel A, Kovacsovics-Bankowski M, Hertig S, Gaide O, Martinon F, Tinel A, Deperthes D, Calderara S, Schulthess T, et al. 2003. Two adjacent trimeric Fas ligands are required for Fas signaling and formation of a death-inducing signaling complex. Mol Cell Biol 23: 1428–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Lee X, Shao Z, Apicco D, Huang G, Gong BJ, Pepinsky RB, Mi S. 2013. A DR6/p75(NTR) complex is responsible for β-amyloid-induced cortical neuron death. Cell Death Dis 4: e579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huard B, McKee T, Bosshard C, Durual S, Matthes T, Myit S, Donze O, Frossard C, Chizzolini C, Favre C, et al. 2008. APRIL secreted by neutrophils binds to heparan sulfate proteoglycans to create plasma cell niches in human mucosa. J Clin Invest 118: 2887–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez CF, Simi A. 2012. p75 neurotrophin receptor signaling in nervous system injury and degeneration: Paradox and opportunity. Trends Neurosci 35: 431–440. [DOI] [PubMed] [Google Scholar]
- Igaki T, Miura M. 2014. The Drosophila TNF ortholog Eiger: Emerging physiological roles and evolution of the TNF system. Semin Immunol 26: 267–274. [DOI] [PubMed] [Google Scholar]
- Ingold K, Zumsteg A, Tardivel A, Huard B, Steiner QG, Cachero TG, Qiang F, Gorelik L, Kalled SL, Acha-Orbea H, et al. 2005. Identification of proteoglycans as the APRIL-specific binding partners. J Exp Med 201: 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam A, Shen X, Hiroi T, Moss J, Vaughan M, Levine SJ. 2007. The brefeldin A-inhibited guanine nucleotide-exchange protein, BIG2, regulates the constitutive release of TNFR1 exosome-like vesicles. J Biol Chem 282: 9591–9599. [DOI] [PubMed] [Google Scholar]
- Jeon YJ, Middleton J, Kim T, Lagana A, Piovan C, Secchiero P, Nuovo GJ, Cui R, Joshi P, Romano G, et al. 2015. A set of NF-κB-regulated microRNAs induces acquired TRAIL resistance in lung cancer. Proc Natl Acad Sci 112: E3355–E3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D, Lanahan A, Buck C R, Sehgal A, Morgan C, Mercer E, Bothwell M, Chao M. 1986. Expression and structure of the human NGF receptor. Cell 47: 545–554. [DOI] [PubMed] [Google Scholar]
- Juhasz K, Buzas K, Duda E. 2013. Importance of reverse signaling of the TNF superfamily in immune regulation. Exp Rev Clin Immunol 9: 335–348. [DOI] [PubMed] [Google Scholar]
- Kang YJ, Kim SO, Shimada S, Otsuka M, Seit-Nebi A, Kwon BS, Watts TH, Han J. 2007. Cell surface 4-1BBL mediates sequential signaling pathways “downstream” of TLR and is required for sustained TNF production in macrophages. Nat Immunol 8: 601–609. [DOI] [PubMed] [Google Scholar]
- Kanning KC, Hudson M, Amieux PS, Wiley JC, Bothwell M, Schecterson LC. 2003. Proteolytic processing of the p75 neurotrophin receptor and two homologs generates C-terminal fragments with signaling capability. J Neurosci 23: 5425–5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenchappa RS, Zampieri N, Chao MV, Barker PA, Teng HK, Hempstead BL, Carter BD. 2006. Ligand-dependent cleavage of the P75 neurotrophin receptor is necessary for NRIF nuclear translocation and apoptosis in sympathetic neurons. Neuron 50: 219–232. [DOI] [PubMed] [Google Scholar]
- Kersse K, Verspurten J, Vanden Berghe T, Vandenabeele P. 2011. The death-fold superfamily of homotypic interaction motifs. Trends Biochem Sci 36: 541–552. [DOI] [PubMed] [Google Scholar]
- Kimberley FC, van Bostelen L, Cameron K, Hardenberg G, Marquart JA, Hahne M, Medema JP. 2009. The proteoglycan (heparan sulfate proteoglycan) binding domain of APRIL serves as a platform for ligand multimerization and cross-linking. FASEB J 23: 1584–1595. [DOI] [PubMed] [Google Scholar]
- Kimura A, Naka T, Nagata S, Kawase I, Kishimoto T. 2004. SOCS-1 suppresses TNF-α-induced apoptosis through the regulation of Jak activation. Int Immunol 16: 991–999. [DOI] [PubMed] [Google Scholar]
- Kindler V, Sappino AP, Grau GE, Piguet PF, Vassalli P. 1989. The inducing role of tumor necrosis factor in the development of bactericidal granulomas during BCG infection. Cell 56: 731–740. [DOI] [PubMed] [Google Scholar]
- Kirkin V, Cahuzac N, Guardiola-Serrano F, Huault S, Luckerath K, Friedmann E, Novac N, Wels WS, Martoglio B, Hueber AO, et al. 2007. The Fas ligand intracellular domain is released by ADAM10 and SPPL2a cleavage in T-cells. Cell Death Differ 14: 1678–1687. [DOI] [PubMed] [Google Scholar]
- Koguchi Y, Thauland TJ, Slifka MK, Parker DC. 2007. Preformed CD40 ligand exists in secretory lysosomes in effector and memory CD4+ T cells and is quickly expressed on the cell surface in an antigen-specific manner. Blood 110: 2520–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima T, Morikawa Y, Copeland NG, Gilbert DJ, Jenkins NA, Senba E, Kitamura T. 2000. TROY, a newly identified member of the tumor necrosis factor receptor superfamily, exhibits a homology with Edar and is expressed in embryonic skin and hair follicles. J Biol Chem 275: 20742–20747. [DOI] [PubMed] [Google Scholar]
- Kriegler M, Perez C, DeFay K, Albert I, Lu SD. 1988. A novel form of TNF/cachectin is a cell surface cytotoxic transmembrane protein: Ramifications for the complex physiology of TNF. Cell 53: 45–53. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Mikkola ML. 2014. Ectodysplasin research—Where to next? Semin Immunol 26: 220–228. [DOI] [PubMed] [Google Scholar]
- Legler DF, Micheau O, Doucey MA, Tschopp J, Bron C. 2003. Recruitment of TNF receptor 1 to lipid rafts is essential for TNFα-mediated NF-κB activation. Immunity 18: 655–664. [DOI] [PubMed] [Google Scholar]
- Leveille C, Bouillon M, Guo W, Bolduc J, Sharif-Askari E, El-Fakhry Y, Reyes-Moreno C, Lapointe R, Merhi Y, Wilkins JA, et al. 2007. CD40 ligand binds to α5β1 integrin and triggers cell signaling. J Biol Chem 282: 5143–5151. [DOI] [PubMed] [Google Scholar]
- Li P, Schwarz EM, O’Keefe RJ, Ma L, Looney RJ, Ritchlin CT, Boyce BF, Xing L. 2004. Systemic tumor necrosis factor α mediates an increase in peripheral CD11bhigh osteoclast precursors in tumor necrosis factor α-transgenic mice. Arthritis Rheum 50: 265–276. [DOI] [PubMed] [Google Scholar]
- Li Y, Toraldo G, Li A, Yang X, Zhang H, Qian WP, Weitzmann MN. 2007. B cells and T cells are critical for the preservation of bone homeostasis and attainment of peak bone mass in vivo. Blood 109: 3839–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yin Q, Wu H. 2013. Structural basis of signal transduction in the TNF receptor superfamily. Adv Immunol 119: 135–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Xu L, Opalka N, Kappler J, Shu HB, Zhang G. 2002. Crystal structure of sTALL-1 reveals a virus-like assembly of TNF family ligands. Cell 108: 383–394. [DOI] [PubMed] [Google Scholar]
- Lopez-Fraga M, Fernandez R, Albar JP, Hahne M. 2001. Biologically active APRIL is secreted following intracellular processing in the Golgi apparatus by furin convertase. EMBO Rep 2: 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Lawrence DA, Marsters S, Acosta-Alvear D, Kimmig P, Mendez AS, Paton AW, Paton JC, Walter P, Ashkenazi A. 2014. Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science 345: 98–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Yang Z, Ma Y, Yue Z, Lin H, Qu G, Huang J, Dai W, Li C, Zheng C, et al. 2016. LGR4 is a receptor for RANKL and negatively regulates osteoclast differentiation and bone resorption. Nat Med 22: 539–546. [DOI] [PubMed] [Google Scholar]
- Ma J, Bang BR, Lu J, Eun SY, Otsuka M, Croft M, Tobias P, Han J, Takeuchi O, Akira S, et al. 2013. The TNF family member 4-1BBL sustains inflammation by interacting with TLR signaling components during late-phase activation. Sci Signal 6: ra87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lorenzo MJ, Anel A, Gamen S, Monle NI, Lasierra P, Larrad L, Pineiro A, Alava MA, Naval J. 1999. Activated human T cells release bioactive Fas ligand and APO2 ligand in microvesicles. J Immunol 163: 1274–1281. [PubMed] [Google Scholar]
- Mbanwi AN, Watts TH. 2014. Costimulatory TNFR family members in control of viral infection: Outstanding questions. Semin Immunol 26: 210–219. [DOI] [PubMed] [Google Scholar]
- McCarthy DD, Summers-Deluca L, Vu F, Chiu S, Gao Y, Gommerman JL. 2006. The lymphotoxin pathway: Beyond lymph node development. Immunol Res 35: 41–54. [DOI] [PubMed] [Google Scholar]
- McWhirter SM, Pullen SS, Holton JM, Crute JJ, Kehry MR, Alber T. 1999. Crystallographic analysis of CD40 recognition and signaling by human TRAF2. Proc Natl Acad Sci 96: 8408–8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreaux J, Sprynski AC, Dillon SR, Mahtouk K, Jourdan M, Ythier A, Moine P, Robert N, Jourdan E, Rossi JF, et al. 2009. APRIL and TACI interact with syndecan-1 on the surface of multiple myeloma cells to form an essential survival loop. Eur J Haematol 83: 119–129. [DOI] [PubMed] [Google Scholar]
- Mosheimer BA, Kaneider NC, Feistritzer C, Djanani AM, Sturn DH, Patsch JR, Wiedermann CJ. 2005. Syndecan-1 is involved in osteoprotegerin-induced chemotaxis in human peripheral blood monocytes. J Clin Endocrinol Metab 90: 2964–2971. [DOI] [PubMed] [Google Scholar]
- Moss ML, Jin SL, Milla ME, Bickett DM, Burkhart W, Carter HL, Chen WJ, Clay WC, Didsbury JR, Hassler D, et al. 1997. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-α. Nature 385: 733–736. [DOI] [PubMed] [Google Scholar]
- Muhlenbeck F, Schneider P, Bodmer JL, Schwenzer R, Hauser A, Schubert G, Scheurich P, Moosmayer D, Tschopp J, Wajant H. 2000. The tumor necrosis factor-related apoptosis-inducing ligand receptors TRAIL-R1 and TRAIL-R2 have distinct cross-linking requirements for initiation of apoptosis and are non-redundant in JNK activation. J Biol Chem 275: 32208–32213. [DOI] [PubMed] [Google Scholar]
- Muppidi JR, Tschopp J, Siegel RM. 2004. Life and death decisions: Secondary complexes and lipid rafts in TNF receptor family signal transduction. Immunity 21: 461–465. [DOI] [PubMed] [Google Scholar]
- Nadiri A, Jundi M, El Akoum S, Hassan GS, Yacoub D, Mourad W. 2015. Involvement of the cytoplasmic cysteine-238 of CD40 in its up-regulation of CD23 expression and its enhancement of TLR4-triggered responses. Int Immunol 27: 555–565. [DOI] [PubMed] [Google Scholar]
- Naismith JH, Devine TQ, Brandhuber BJ, Sprang SR. 1995. Crystallographic evidence for dimerization of unliganded tumor necrosis factor receptor. J Biol Chem 270: 13303–13307. [DOI] [PubMed] [Google Scholar]
- Nikolaev A, McLaughlin T, O’Leary DD, Tessier-Lavigne M. 2009. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature 457: 981–989. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nophar Y, Kemper O, Brakebusch C, Engelmann H, Zwang R, Aderka D, Holtmann H, Wallach D. 1990. Soluble forms of tumor necrosis factor receptors (TNF-Rs). The cDNA for the type I TNF-R cloned using amino acid data of its soluble form, encodes for both the cell surface and a soluble form of the receptor. EMBO J 9: 3269–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Old LJ. 1985. Tumor necrosis factor (TNF). Science 230: 630–632. [DOI] [PubMed] [Google Scholar]
- Olsen O, Kallop DY, McLaughlin T, Huntwork-Rodriguez S, Wu Z, Duggan CD, Simon DJ, Lu Y, Easley-Neal C, Takeda K, et al. 2014. Genetic analysis reveals that amyloid precursor protein and death receptor 6 function in the same pathway to control axonal pruning independent of β-secretase. J Neurosci 34: 6438–6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly LA, Tai L, Lee L, Kruse EA, Grabow S, Fairlie WD, Haynes NM, Tarlinton DM, Zhang JG, Belz GT, et al. 2009. Membrane-bound Fas ligand only is essential for Fas-induced apoptosis. Nature 461: 659–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YC, Burkitt V, Villa AR, Tong L, Wu H. 1999. Structural basis for self-association and receptor recognition of human TRAF2. Nature 398: 533–538. [DOI] [PubMed] [Google Scholar]
- Pinckard JK, Sheehan KC, Schreiber RD. 1997. Ligand-induced formation of p55 and p75 tumor necrosis factor receptor heterocomplexes on intact cells. J Biol Chem 272: 10784–10789. [DOI] [PubMed] [Google Scholar]
- Plata-Salaman CR, Oomura Y, Kai Y. 1988. Tumor necrosis factor and interleukin-1β: Suppression of food intake by direct action in the central nervous system. Brain Res 448: 106–114. [DOI] [PubMed] [Google Scholar]
- Pocsik E, Duda E, Wallach D. 1995. Phosphorylation of the 26 kDa TNF precursor in monocytic cells and in transfected HeLa cells. J Inflamm 45: 152–160. [PubMed] [Google Scholar]
- Poggi M, Kara I, Brunel JM, Landrier JF, Govers R, Bonardo B, Fluhrer R, Haass C, Alessi MC, Peiretti F. 2013. Palmitoylation of TNFα is involved in the regulation of TNF receptor 1 signalling. Biochim Biophys Acta 1833: 602–612. [DOI] [PubMed] [Google Scholar]
- Porquet N, Poirier A, Houle F, Pin AL, Gout S, Tremblay PL, Paquet ER, Klinck R, Auger FA, Huot J. 2011. Survival advantages conferred to colon cancer cells by E-selectin-induced activation of the PI3K-NF-κB survival axis downstream of death receptor-3. BMC Cancer 11: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradet-Balade B, Medema JP, Lopez-Fraga M, Lozano JC, Kolfschoten GM, Picard A, Martinez AC, Garcia-Sanz JA, Hahne M. 2002. An endogenous hybrid mRNA encodes TWE-PRIL, a functional cell surface TWEAK-APRIL fusion protein. EMBO J 21: 5711–5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quistad SD, Stotland A, Barott KL, Smurthwaite CA, Hilton BJ, Grasis JA, Wolkowicz R, Rohwer FL. 2014. Evolution of TNF-induced apoptosis reveals 550 My of functional conservation. Proc Natl Acad Sci 111: 9567–9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radeke MJ, Misko TP, Hsu C, Herzenberg LA, Shooter EM. 1987. Gene transfer and molecular cloning of the rat nerve growth factor receptor. Nature 325: 593–597. [DOI] [PubMed] [Google Scholar]
- Richard AC, Ferdinand JR, Meylan F, Hayes ET, Gabay O, Siegel RM. 2015. The TNF-family cytokine TL1A: From lymphocyte costimulator to disease co-conspirator. J Leukoc Biol 98: 333–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschke V, Sosnovtseva S, Ward CD, Hong JS, Smith R, Albert V, Stohl W, Baker KP, Ullrich S, Nardelli B, et al. 2002. BLyS and APRIL form biologically active heterotrimers that are expressed in patients with systemic immune-based rheumatic diseases. J Immunol 169: 4314–4321. [DOI] [PubMed] [Google Scholar]
- Rossin A, Derouet M, Abdel-Sater F, Hueber AO. 2009. Palmitoylation of the TRAIL receptor DR4 confers an efficient TRAIL-induced cell death signalling. Biochem J 419: 185–192. [DOI] [PubMed] [Google Scholar]
- Ruddle NH, Waksman BH. 1968. Cytotoxicity mediated by soluble antigen and lymphocytes in delayed hypersensitivity. III: Analysis of mechanisms. J Exp Med 128: 1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabio G, Davis RJ. 2014. TNF and MAP kinase signalling pathways. Semin Immunol 26: 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamaki K, Shimizu K, Iwata H, Imai K, Satou Y, Funayama N, Nozaki M, Yajima M, Nishimura O, Higuchi M, et al. 2014. The apoptotic initiator caspase-8: Its functional ubiquity and genetic diversity during animal evolution. Mol Biol Evol 31: 3282–3301. [DOI] [PubMed] [Google Scholar]
- Schneeweis LA, Willard D, Milla ME. 2005. Functional dissection of osteoprotegerin and its interaction with receptor activator of NF-κB ligand. J Biol Chem 280: 41155–41164. [DOI] [PubMed] [Google Scholar]
- Schneider P, Holler N, Bodmer JL, Hahne M, Frei K, Fontana A, Tschopp J. 1998. Conversion of membrane-bound Fas(CD95) ligand to its soluble form is associated with downregulation of its proapoptotic activity and loss of liver toxicity. J Exp Med 187: 1205–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuepbach-Mallepell S, Das D, Willen L, Vigolo M, Tardivel A, Lebon L, Kowalczyk-Quintas C, Nys J, Smulski C, Zheng TS, et al. 2015. Stoichiometry of heteromeric BAFF and APRIL cytokines dictates their receptor binding and signaling properties. J Biol Chem 290: 16330–16342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutze S, Tchikov V, Schneider-Brachert W. 2008. Regulation of TNFR1 and CD95 signalling by receptor compartmentalization. Nat Rev Mol Cell Biol 9: 655–662. [DOI] [PubMed] [Google Scholar]
- Scott FL, Stec B, Pop C, Dobaczewska MK, Lee JJ, Monosov E, Robinson H, Salvesen GS, Schwarzenbacher R, Riedl SJ. 2009. The Fas-FADD death domain complex structure unravels signalling by receptor clustering. Nature 457: 1019–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessler T, Healy S, Samali A, Szegezdi E. 2013. Structural determinants of DISC function: New insights into death receptor-mediated apoptosis signalling. Pharmacol Therapeut 140: 186–199. [DOI] [PubMed] [Google Scholar]
- Sfikakis PP. 2010. The first decade of biologic TNF antagonists in clinical practice: Lessons learned, unresolved issues and future directions. Curr Dir Autoimmun 11: 180–210. [DOI] [PubMed] [Google Scholar]
- Shao Z, Browning JL, Lee X, Scott ML, Shulga-Morskaya S, Allaire N, Thill G, Levesque M, Sah D, McCoy JM, et al. 2005. TAJ/TROY, an orphan TNF receptor family member, binds Nogo-66 receptor 1 and regulates axonal regeneration. Neuron 45: 353–359. [DOI] [PubMed] [Google Scholar]
- Shoham S, Davenne D, Cady AB, Dinarello CA, Krueger JM. 1987. Recombinant tumor necrosis factor and interleukin 1 enhance slow-wave sleep. Am J Physiol 253: R142–R149. [DOI] [PubMed] [Google Scholar]
- Shohami E, Ginis I, Hallenbeck JM. 1999. Dual role of tumor necrosis factor α in brain injury. Cytokine Growth Factor Rev 10: 119–130. [DOI] [PubMed] [Google Scholar]
- Shui JW, Steinberg MW, Kronenberg M. 2011. Regulation of inflammation, autoimmunity, and infection immunity by HVEM-BTLA signaling. J Leukoc Biol 89: 517–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So T, Croft M. 2013. Regulation of PI-3-kinase and Akt signaling in T lymphocytes and other cells by TNFR family molecules. Front Immunol 4: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standal T, Seidel C, Hjertner O, Plesner T, Sanderson RD, Waage A, Borset M, Sundan A. 2002. Osteoprotegerin is bound, internalized, and degraded by multiple myeloma cells. Blood 100: 3002–3007. [DOI] [PubMed] [Google Scholar]
- Stellwagen D, Malenka RC. 2006. Synaptic scaling mediated by glial TNF-α. Nature 440: 1054–1059. [DOI] [PubMed] [Google Scholar]
- Stone GW, Barzee S, Snarsky V, Kee K, Spina CA, Yu XF, Kornbluth RS. 2006. Multimeric soluble CD40 ligand and GITR ligand as adjuvants for human immunodeficiency virus DNA vaccines. J Virol 80: 1762–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser A, Jost PJ, Nagata S. 2009. The many roles of FAS receptor signaling in the immune system. Immunity 30: 180–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuber E, Neurath M, Calderhead D, Fell HP, Strober W. 1995. Cross-linking of OX40 ligand, a member of the TNF/NGF cytokine family, induces proliferation and differentiation in murine splenic B cells. Immunity 2: 507–521. [DOI] [PubMed] [Google Scholar]
- Summers deLuca L, Gommerman JL. 2012. Fine-tuning of dendritic cell biology by the TNF superfamily. Nat Rev Immunol 12: 339–351. [DOI] [PubMed] [Google Scholar]
- Sun M, Lee S, Karray S, Levi-Strauss M, Ames KT, Fink PJ. 2007. Cutting edge: Two distinct motifs within the Fas ligand tail regulate Fas ligand-mediated costimulation. J Immunol 179: 5639–5643. [DOI] [PubMed] [Google Scholar]
- Swee LK, Ingold-Salamin K, Tardivel A, Willen L, Gaide O, Favre M, Demotz S, Mikkola M, Schneider P. 2009. Biological activity of ectodysplasin A is conditioned by its collagen and heparan sulfate proteoglycan-binding domains. J Biol Chem 284: 27567–27576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G. 2002. Furin at the cutting edge: From protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol 3: 753–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracey KJ, Fong Y, Hesse DG, Manogue KR, Lee AT, Kuo GC, Lowry SF, Cerami A. 1987. Anti-cachectin/TNF monoclonal antibodies prevent septic shock during lethal bacteraemia. Nature 330: 662–664. [DOI] [PubMed] [Google Scholar]
- Uren AG, O’Rourke K, Aravind LA, Pisabarro MT, Seshagiri S, Koonin EV, Dixit VM. 2000. Identification of paracaspases and metacaspases: Two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol Cell 6: 961–967. [DOI] [PubMed] [Google Scholar]
- Van Lier RAW, Borst J, Vroom TM, Klein H, Van Mourik P, Zeijlemaker WP, Melief CJ. 1987. Tissue distribution and biochemical and functional properties of Tp55 (CD27), a novel T cell differentiation antigen. J Immunol 139: 1589–1596. [PubMed] [Google Scholar]
- Vilar M, Charalampopoulos I, Kenchappa RS, Simi A, Karaca E, Reversi A, Choi S, Bothwell M, Mingarro I, Friedman WJ, et al. 2009. Activation of the p75 neurotrophin receptor through conformational rearrangement of disulphide-linked receptor dimers. Neuron 62: 72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner KW, Punnoose EA, Januario T, Lawrence DA, Pitti RM, Lancaster K, Lee D, von Goetz M, Yee SF, Totpal K, et al. 2007. Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat Med 13: 1070–1077. [DOI] [PubMed] [Google Scholar]
- Wallach D. 1984. Preparations of lymphotoxin induce resistance to their own cytotoxic effect. J Immunol 132: 2464–2469. [PubMed] [Google Scholar]
- Wallach D. 2016a. The cybernetics of TNF: Old views and newer ones. Semin Cell Dev Biol 50: 105–114. [DOI] [PubMed] [Google Scholar]
- Wallach D. 2016b. TNFα. In Encyclopedia of inflammatory diseases (ed. Parnham M). Springer, Basel. [Google Scholar]
- Wallach D, Kovalenko A. 2016. Roles of TNF and other members of the TNF family in the regulation of innate immunity. In Encyclopedia of immunobiology (ed. Ratcliffe MJH), pp. 454–465. Academic, Oxford. [Google Scholar]
- Walsh MC, Choi Y. 2014. Biology of the RANKL-RANK-OPG system in immunity, bone, and beyond. Front Immunol 5: 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Al-Lamki RS, Zhang H, Kirkiles-Smith N, Gaeta ML, Thiru S, Pober JS, Bradley JR. 2003. Histamine antagonizes tumor necrosis factor (TNF) signaling by stimulating TNF receptor shedding from the cell surface and Golgi storage pool. J Biol Chem 278: 21751–21760. [DOI] [PubMed] [Google Scholar]
- Wang L, Yang JK, Kabaleeswaran V, Rice AJ, Cruz AC, Park AY, Yin Q, Damko E, Jang SB, Raunser S, et al. 2010. The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations. Nat Struct Mol Biol 17: 1324–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware CF. 2005. Network communications: Lymphotoxins, LIGHT, and TNF. Annu Rev Immunol 23: 787–819. [DOI] [PubMed] [Google Scholar]
- Watts AD, Hunt NH, Wanigasekara Y, Bloomfield G, Wallach D, Roufogalis BD, Chaudhri G. 1999. A casein kinase I motif present in the cytoplasmic domain of members of the tumour necrosis factor ligand family is implicated in “reverse signalling.” EMBO J 18: 2119–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyzgol A, Muller N, Fick A, Munkel S, Grigoleit GU, Pfizenmaier K, Wajant H. 2009. Trimer stabilization, oligomerization, and antibody-mediated cell surface immobilization improve the activity of soluble trimers of CD27L, CD40L, 41BBL, and glucocorticoid-induced TNF receptor ligand. J Immunol 183: 1851–1861. [DOI] [PubMed] [Google Scholar]
- Yang S, Wang Y, Mei K, Zhang S, Sun X, Ren F, Liu S, Yang Z, Wang X, Qin Z, et al. 2015. Tumor necrosis factor receptor 2 (TNFR2)·interleukin-17 receptor D (IL-17RD) heteromerization reveals a novel mechanism for NF-κB activation. J Biol Chem 290: 861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Lin SC, Lamothe B, Lu M, Lo YC, Hura G, Zheng L, Rich RL, Campos AD, Myszka DG, et al. 2009a. E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol 16: 658–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Lamothe B, Lamothe B, Wu H. 2009b. Structural basis for the lack of E2 interaction in the ring of TRAF2. Biocheminstry 48: 10558–10567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You RI, Chang YC, Chen PM, Wang WS, Hsu TL, Yang CY, Lee CT, Hsieh SL. 2008. Apoptosis of dendritic cells induced by decoy receptor 3 (DcR3). Blood 111: 1480–1488. [DOI] [PubMed] [Google Scholar]
- Yuan S, Liu T, Huang S, Wu T, Huang L, Liu H, Tao X, Yang M, Wu K, Yu Y, et al. 2009. Genomic and functional uniqueness of the TNF receptor-associated factor gene family in amphioxus, the basal chordate. J Immunol 183: 4560–4568. [DOI] [PubMed] [Google Scholar]
- Zanin-Zhorov A, Hershkoviz R, Hecht I, Cahalon L, Lider O. 2003. Fibronectin-associated Fas ligand rapidly induces opposing and time-dependent effects on the activation and apoptosis of T cells. J Immunol 171: 5882–5889. [DOI] [PubMed] [Google Scholar]
- Zapata JM, Martinez-Garcia V, Lefebvre S. 2007. Phylogeny of the TRAF/MATH domain. Adv Exp Med Biol 597: 1–24. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Song X, Berezov A, Zhang G, Li Y, Zhang H, Murali R, Li B, Greene MI. 2008. Human glucocorticoid-induced TNF receptor ligand regulates its signaling activity through multiple oligomerization states. Proc Natl Acad Sci 105: 5465–5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmasek CM, Godzik A. 2013. Evolution of the animal apoptosis network. Cold Spring Harb Perspect Biol 5: a008649. [DOI] [PMC free article] [PubMed] [Google Scholar]