Abstract
Two male siblings ages 15 and 10 yr old had similar features of intellectual disability, developmental delay, severe speech impairment, microcephaly, prematurity, and transient elevation of liver enzymes in infancy. Exome sequencing revealed one novel (c.65C>A; p.Ala22Asp) and one ultra-rare (c.3214T>C; p.Phe1072Leu) predicted damaging missense variant in trans in the gene encoding cytoplasmic valyl-tRNA synthetase (VARS). Biallelic variants in VARS have previously been associated with a neurodevelopmental disorder characterized by microcephaly, seizures, and cortical atrophy (NDMSCA; MIM #617802). Although our patients have no history of seizures or cortical atrophy, we suggest that the biallelic variants in VARS p.Ala22Asp and p.Phe1072Leu in this family are likely pathogenic and associated with NDMSCA, expanding the clinical phenotype of the condition.
Keywords: absent speech; appendicular hypotonia; intellectual disability, moderate; microcephaly; premature birth following premature rupture of fetal membranes
CASE PRESENTATIONS
Index case (II-1, Supplemental Fig. 1) is a 15-yr-old male with intellectual disability (ID), severe speech impairment, acquired microcephaly, and hypotonia (Table 1). The pregnancy was unremarkable except for elevated α-fetoprotein (AFP) levels on maternal serum screening, which were later found normal in amniotic fluid analysis. He was born at 31 wk of gestation with normal weight (1.47 kg [25%–50%]), length (40.6 cm [50%]), and head circumference (28 cm [25%–50%]). He was admitted to the neonatal intensive care unit (NICU) because of prematurity and spent 45 d in the hospital with no major complications. He was noted to have neonatal tooth eruption. At 3 mo of age he was severely hypotonic. He had a transient mild elevation of his liver enzymes without an identifiable etiology during infancy, which resolved spontaneously. He sat without support at 4 yr of age, started crawling at ∼4.5 yr of age, and now can walk with a gait trainer but usually scoots around on his bottom. He is nonverbal. He has had no regression of his milestones nor has he had problems with weight gain or short stature, but his head circumference fell >2 SDs below the mean at ∼18 mo of age, and he remained microcephalic afterward. He has no problems with feeding and sleeping. He has repetitive movements and rocks his legs back and forth when seated and kicks his legs repeatedly when sitting on the floor. There is no history of seizures and an overnight ambulatory EEG at age 4–5 yr showed no epileptiform discharges. A brain MRI was also found normal. Previous genetic studies including karyotype, chromosomal microarray, fragile X, MECP2 sequencing, and metabolic screening (urine organic acids, very long chain fatty acids, and lysosomal storage disorders) were all normal.
Table 1.
Clinical findings of patients with biallelic variants in VARS
| This study | Karaca et al. 2015 | Stephen et al. 2018 | ||||
|---|---|---|---|---|---|---|
| Patient ID | II-1 | II-2 | BAB3643 | BAB3186 | BAB3187 | II-2 |
| Gender | Male | Male | Female | Female | Female | Male |
| Age | 15-yr-old | 10-yr-old | ? | ? | ? | 32-mo-old |
| Genotype | p.Ala22Asp p.Phe1072Leu |
p.Ala22Asp p.Phe1072Leu |
p.Arg1058Gln p.Arg1058Gln |
p.Leu885Phe p.Leu885Phe |
p.Leu885Phe p.Leu885Phe |
c.1577-2A>G p.Met1064Ile |
| Intellectual disability | Yes | Yes | Yes | Yes | Yes | Yes |
| Speech | Nonverbal | Nonverbal | NA | NA | NA | Nonverbal |
| Microcephaly (congenital vs. acquired) | Yes (acquired) | Yes (NA) | Yes (NA) | Yes (NA) | Yes (NA) | Yes (congenital) |
| Seizures | No | No | Yes | Yes | Yes | Yes |
| Hypotonia | Yes | Yes | NA | NA | NA | Yes |
| Cortical atrophy | No | NA | Yes | Yes | Yes | Yes |
| Other | ||||||
| Prematurity | Yes | Yes | NA | NA | NA | No |
| Elevated liver enzymes | Yes | Yes | NA | NA | NA | No |
| Stereotypic behavior | Yes | No | NA | NA | NA | No |
| Neonatal tooth | Yes | No | NA | NA | NA | No |
| Volvulus | No | Yes | NA | NA | NA | No |
| Dysmorphic features | Yes | No | NA | NA | NA | No |
NA, not available.
Case 2 (II-2, Supplemental Fig. 1) is the 10-yr-old brother of the proband, who is similarly affected with ID, severe speech impairment, microcephaly, hypotonia, transient elevation of liver enzymes in infancy, and prematurity. He was born at 27 wk of gestation with intestinal volvulus which was repaired surgically in the first week of life. He has always been underweight, and his normalized height has just recently started to decline. He is also nonverbal and nonambulatory.
There is no history of consanguinity between parents and the remaining family history is unremarkable. The parents are both of European ethnicity.
VARIANT INTERPRETATION
Our patients are compound heterozygous for one novel and one ultra-rare missense variant, NM_006295:c.65C>A:p.Ala22Asp and NM_006295:c.3214T>C:p.Phe1072Leu, in the gene encoding cytoplasmic valyl-tRNA synthetase protein (VARS). Although the p.Ala22Asp variant is not present in any population databases, p.Phe1072Leu is observed in one NonFinnish European allele in gnomAD. Computational prediction algorithms favor pathogenicity for both variants. Variants are evaluated as of uncertain significance (VUS) according to American College of Medical Genetics and Genomics (ACMG) variant interpretation guidelines fulfilling PM3_Supporting, PP1, and PP4 criteria (Richards et al. 2015), although because of the overlapping clinical features to the previously reported cases they are likely causative of the patients’ phenotypes (Table 2). No other likely pathogenic variants were identified that segregated with the phenotype in the family.
Table 2.
Genomic findings and variant interpretation
| Gene | Genomic location | HGVS cDNA | HGVS protein | Zygosity | Parent of origin | Variant interpretation |
|---|---|---|---|---|---|---|
| VARS | Chr 6:31762930 (hg19) Chr 6:31795153 (hg38) |
c.65C>A | p.Ala22Asp | Heterozygous | Maternal | Variant of uncertain significance |
| VARS | Chr 6:31747459 (hg19) Chr 6:31779682 (hg38) |
c.3214T>C | p.Phe1072Leu | Heterozygous | Paternal | Variant of uncertain significance |
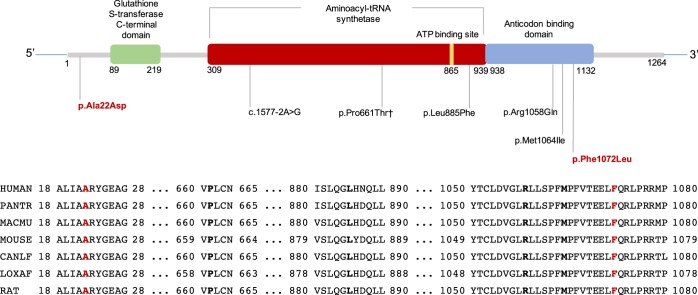
Aminoacyl-tRNA synthetases catalyze the aminoacylation of tRNA by their cognate amino acid. There are 37 aminoacyl-tRNA synthetases (18 cytoplasmic, 17 mitochondrial, 2 bifunctional) in human cells, and biallelic variants in many aminoacyl-tRNA synthetases have been reported in individuals with neurodevelopmental disorders (Meyer-Schuman and Antonellis 2017). Variants in VARS were first identified in patients with microcephaly, ID, and cortical atrophy (Karaca et al. 2015), and a recent study reported additional variants in patients with similar clinical findings associated with biallelic variants in VARS (Table 1; Stephen et al. 2018). VARS has three important conserved domains: glutathione S-transferase, carboxy-terminal domain (GST_C), tRNA synthetases class I (tRNA_synt_1), and anticodon-binding domain of tRNA (Anticodon_1). Four predicted likely pathogenic variants reported in the literature and in ClinVar are missense variants, and they all are located in either tRNA_synt_1 or Anticodon_1 domain. The only reported variant that is located outside of those domains is a canonical splice site variant (Fig. 1). The p.Phe1072Leu variant is also located in the Anticodon_1 domain (SIFT: Damaging; PolyPhen-2: Benign; CADD v1.3: 24.8). The p.Ala22Asp variant is not located in any of the domains, but the prediction scores for this variant are supportive of pathogenicity (SIFT: Damaging; Polyphen-2: Damaging; CADD v1.3: 32). All of the previously reported variants and the variants we report are located in highly conserved residues (Fig. 1).
Figure 1.
Distribution of reported VARS variants in the literature and ClinVar over 2D representation of VARS domains given on InterPro for P26640 (SYVC_HUMAN) and the mutated residues’ sequence alignment across species. Mutated residues are shown in bold black font for previously reported variants and in bold red font for variants reported in our study. HUMAN (P26640); Homo sapiens, PANTR (H2QSM5); Pan troglodytes (chimpanzee), MACMU (G7MRK4); Rhesus macaque; MOUSE (G3UY93); Mus musculus; CANLF (E2RTJ7), Canis lupus familiaris (dog); LOXAF (G5E7F5), African elephant; RAT (Q04462), Rattus norvegicus. †This variant is deposited into ClinVar but not reported in any publication.
SUMMARY
Here we report biallelic missense variants in VARS in two similarly affected siblings with ID, severe speech impairment, acquired microcephaly, hypotonia, prematurity, and transient elevation of liver enzymes in infancy, consistent with previously reported findings in patients with biallelic VARS variants. Our patients did not have seizures or cortical atrophy, and this is different from some other reported patients in the literature, expanding the phenotypic spectrum of NDMSCA. Our patients share overlapping clinical features reported in other autosomal recessively inherited aminoacyl-tRNA synthetase disorders (i.e., IARS; MIM #617093, LARS; MIM #615438) including elevated liver enzymes along with microcephaly and hypotonia. Future clinical and functional studies are needed to better define the genotype–phenotype correlations and assess molecular mechanism of disease.
METHODS
Clinical exome sequencing was performed with peripheral blood DNA samples from the two affected brothers and their mother and was nondiagnostic. Raw data was requested from the diagnostic laboratory and reanalysis was performed. Variants were filtered by their population allele frequencies in Exome Aggregation Consortium (ExAC) (Lek et al. 2016), Exome Sequencing Project (ESP; http://evs.gs.washington.edu/EVS/), 1000 Genomes Samples (Auton et al. 2015), and Genome Aggregation Database (gnomAD) (Lek et al. 2016) using 1% and 3% thresholds for autosomal dominant and autosomal recessive inheritance models, respectively. Biallelic predicted pathogenic variants that fit to an autosomal recessive inheritance pattern were identified. Variants were confirmed by Sanger sequencing, and they segregated with the affected individuals in the family (Supplemental Fig. 1).
ADDITIONAL INFORMATION
Data Deposition and Access
All sequence data and interpreted variants have been deposited in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) under accession numbers SCV000803173 (for c.65C>A) and SCV000803174 (for c.3214T>C).
Ethics Statement
Written informed consent was obtained for all individuals in this study. The study is approved by the Institutional Review Board of Columbia University under protocol #AAAJ8651.
Acknowledgments
We thank the patients and the family for participating in this study. We thank Patricia Lanzano, Jiangyuan Hu, and Charles LeDuc for study management and Sanger sequencing confirmations.
Author Contributions
V.O. and M.G. analyzed the data and drafted and critically reviewed the manuscript. A.W. provided the clinical data and critically reviewed the manuscript. W.K.C. conceived of the study, provided clinical data, analyzed the data, and drafted and critically reviewed the manuscript.
Funding
This work was supported in part by a grant from Simons Foundation and the JPB Foundation.
Competing Interest Statement
The authors have declared no competing interest.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. 2015. A global reference for human genetic variation. Nature 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaca E, Harel T, Pehlivan D, Jhangiani SN, Gambin T, Coban Akdemir Z, Gonzaga-Jauregui C, Erdin S, Bayram Y, Campbell IM, et al. 2015. Genes that affect brain structure and function identified by rare variant analyses of Mendelian neurologic disease. Neuron 88: 499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Schuman R, Antonellis A. 2017. Emerging mechanisms of aminoacyl-tRNA synthetase mutations in recessive and dominant human disease. Hum Mol Genet 26: R114–R127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. 2015. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17: 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen J, Nampoothiri S, Banerjee A, Tolman NJ, Penninger JM, Elling U, Agu CA, Burke JD, Devadathan K, Kannan R, et al. 2018. Loss of function mutations in VARS encoding cytoplasmic valyl-tRNA synthetase cause microcephaly, seizures, and progressive cerebral atrophy. Hum Genet 137: 293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.