In this study, Wan et al. report that budding yeast mitochondrial outer membrane (MOM) proteins Tom70, Tom22, and Sam37 are required for efficient tRNA splicing. They show that defective tRNA splicing in MOM mutants is due not to loss of respiratory metabolism but instead inefficient targeting/tethering of tRNA splicing endonuclease (SEN) subunits to mitochondria.
Keywords: tRNA splicing endonuclease, RNA processing on the mitochondrial surface, Tom70, Sam37
Abstract
The mitochondrial cytoplasmic surface serves as a processing site for numerous RNAs from budding yeast to metazoans. We report that budding yeast mitochondrial outer membrane (MOM) proteins that are subunits of the translocase of the outer mitochondrial membrane (Tom70 and Tom 22) and sorting and assembly machinery (Sam37) are required for efficient pretransfer RNA (pre-tRNA) splicing. Defective pre-tRNA splicing in MOM mutants is due not to loss of respiratory metabolism but instead inefficient targeting/tethering of tRNA splicing endonuclease (SEN) subunits to mitochondria. Schizosaccharomyces pombe SEN subunits also localize to mitochondria, and Tom70 is required for this localization and pre-tRNA splicing. Thus, the role of MOM protein in targeting/tethering SEN subunits to mitochondria has been conserved for >500 million years.
Transfer RNAs (tRNAs) deliver amino acids to ribosomes during translation and are essential for protein synthesis in all kingdoms of life. All eukaryotic primary tRNA transcripts—synthesized in the nucleus by RNA polymerase III—contain 5′ leader and 3′ trailer sequences, and some contain introns (Hopper and Phizicky 2003). Removal of introns from pre-tRNA is essential to generate a complete set of tRNAs for protein synthesis in eukaryotes because tRNA genes encoding at least one tRNA family are exclusively intron-containing (Chan and Lowe 2009).
In yeast and vertebrates, intron removal from pre-tRNA is catalyzed by the heterotetrameric tRNA splicing endonuclease (SEN) complex consisting of subunits Sen2, Sen15, Sen34, and Sen54 (TSEN in humans) (Trotta et al. 1997; Paushkin et al. 2004). Conserved Sen2 and Sen34 function as the catalytic subunits and cleave intron-containing tRNAs at 5′ and 3′ splice sites, respectively (Trotta et al. 2006). Sen15 and Sen54 are poorly conserved but are thought to help establish the pre-tRNA cleavage sites (Trotta et al. 1997). Interactions between budding yeast Sen2 and Sen54 and between Sen15 and Sen34 have been shown by two hybrid studies, but interactions between the other subunits have not been reported (Trotta et al. 1997). Thus, how the SEN subunits form a heterotetrameric SEN complex remains unknown. Each of the genes encoding Saccharomyces cerevisiae SEN subunits is essential. Similarly, TSEN2 and TSEN54 are essential in multiple human haploid cancer cell lines, and TSEN15 and TSEN34 are essential in some, but not all, cell lines (Boone and Andrews 2015).
Although the SEN complex configuration and mechanism of catalysis are conserved in eukaryotes, the subcellular localizations differ among organisms. The SEN complex is located in the nucleus in human cells and Xenopus oocytes (Melton et al. 1980; Lund and Dahlberg 1998; Paushkin et al. 2004) and on the mitochondrial surface in S. cerevisiae (Yoshihisa et al. 2003). The mitochondrial surface appears to function as a “processing plant” for numerous RNA processing steps (Chatterjee et al. 2018), including pre-tRNA splicing (Yoshihisa et al. 2003) and cleavage of an mRNA in yeast (Tsuboi et al. 2015), specific tRNA modifications in yeast (Huh et al. 2003; Miyauchi et al. 2012) and mice (Boland et al. 2009), and piRNA (PIWI-interacting RNA) 5′ and/or 3′ maturation in mice (Choi et al. 2006; Watanabe et al. 2011), flies (Huang et al. 2011), silk worm (Izumi et al. 2016), and Caenorhabditis elegans (Tang et al. 2016). Studies of pre-tRNA splicing in yeast may provide understanding of how and why other RNA processing steps occur on the mitochondrial surface.
A comprehensive genome-wide screen for proteins with novel roles in tRNA biology identified two mitochondrial outer membrane (MOM) proteins that are subunits of the translocase of the outer mitochondrial membrane (Tom70) and sorting and assembly machinery (Sam37). Deletion of TOM70 or SAM37 causes accumulation of intron-containing tRNAIleUAU (Wu et al. 2015). Tom70 is a subunit of the TOM complex, and Sam37 is a subunit of the SAM complex. The TOM and SAM complexes are responsible for initial import steps of many mitochondrial proteins and aid insertion of α-helical and β-barrel proteins into the MOM (Stojanovski et al. 2012). tom70Δ and sam37Δ cells are defective in mitochondrial protein import and respiratory metabolism (Stojanovski et al. 2012).
We report that the pre-tRNA splicing defects of tom70Δ and sam37Δ cells are due not to the loss of mitochondrial respiratory metabolism but instead defective association of SEN subunits with the mitochondrial surface. Using budding yeast strains, each with individual SEN subunits tagged with GFP at their endogenous loci in wild-type, tom70Δ, and sam37Δ backgrounds, we show that SEN subunit levels on mitochondria are reduced in the mutant cells. Thus, Tom70 and Sam37 are required for proper localization, assembly, and function of the SEN subunits on mitochondria in S. cerevisiae. We also report that Tom70 is required for pre-tRNA splicing and SEN subunit protein levels and localization to the mitochondrial surface in Schizosaccharomyces pombe. Thus, the mitochondrial localization of the SEN subunits and the role of Tom70 in this localization have been conserved for at least 500 millions years (Rhind et al. 2011).
Results and Discussion
Mutations of MOM protein-encoding genes affect tRNA splicing after tRNA nuclear export
Deletion of the budding yeast tRNA exportin LOS1 causes accumulation of end-processed unspliced pre-tRNAs (Hopper et al. 1980). Splicing defects occur because tRNA end processing is catalyzed in the nucleus, but splicing occurs in the cytoplasm on the surface of mitochondria (Yoshihisa et al. 2003), and, in los1Δ cells, intron-containing tRNAs are retained in the nucleus away from the splicing machinery (Sarkar and Hopper 1998). Similar to los1Δ cells, cells harboring deletions of TOM70 or SAM37 genes accumulate more than fourfold and threefold, respectively, end-processed intron-containing tRNAIleUAU than do wild-type cells (Wu et al. 2015; Fig. 1). A yeast strain harboring a temperature-sensitive (ts) allele of TOM22 (tom22-ts) encoding an essential TOM complex subunit also accumulates end-processed unspliced tRNAIleUAU upon incubation for 2 h at the nonpermissive temperature (37°C) (Supplemental Fig. S1A,B). The tom70Δ, sam37Δ, and tom22-ts mutants also accumulate end-processed intron-containing tRNATyrGUA (Supplemental Fig. S1C,D). Thus, Tom70, Sam37, and Tom22 are required for efficient pre-tRNA splicing.
Figure 1.
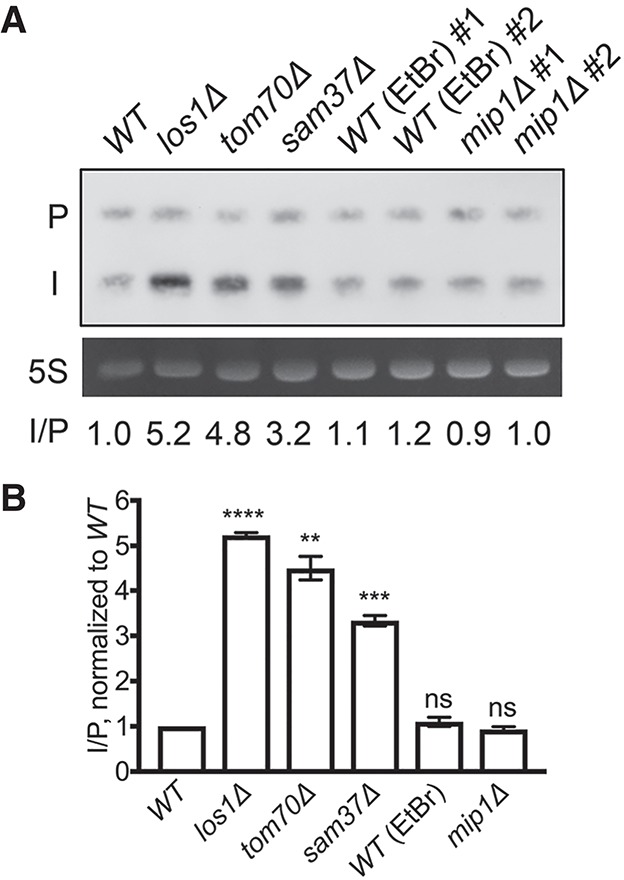
Respiratory metabolism is not required for efficient pre-tRNA splicing in S. cerevisiae. (A) Northern analysis of tRNAIleUAU (probe JW0041) in cells lacking mitochondrial DNA. Wild-type cells lacking mitochondrial DNA via treatment with ethidium bromide [wild-type (EtBr)] and mip1Δ cells. (P) Primary tRNA transcript; (I) end-processed intron-containing tRNA; (5S) 5S rRNA stained with EtBr; (I/P) signal intensity of end-processed intron-containing tRNA/primary tRNA transcript, normalized to wild type. (B) Statistical analysis of end-processed intron-containing tRNA/primary tRNA transcript ratios compared with wild type from three biological repeats. (ns) P > 0.05; (**) P ≤ 0.01; (***) P ≤ 0.001; (****) P ≤ 0.0001.
Defects in pre-tRNA splicing in tom70Δ and sam37Δ cells could be due to reduced pre-tRNA nuclear export or defects in splicing after the tRNAs have been exported to the cytoplasm. To distinguish between these possibilities, we conducted fluorescence in situ hybridization (FISH) to visualize the subcellular localization of intron-containing tRNAs. FISH probe SRIM03 hybridizes to the intron of pre-tRNAIleUAU and detects the primary tRNA transcripts and end-processed intron-containing tRNAs. Because tRNAs are transcribed in the nucleus, there are basal nuclear signals in wild-type cells overlapping with DAPI staining of nuclear DNA (Fig. 2). Only low cytoplasmic FISH signals are detected in wild-type cells because intron-containing tRNAs are efficiently spliced upon nuclear export. Due to the defect in tRNA nuclear export, there are elevated nuclear signals in los1Δ cells (Fig. 2). In contrast, unspliced tRNAs are elevated in the cytoplasm of tom70Δ and sam37Δ cells (Fig. 2). Quantification and normalization of the average FISH signals from 10 randomly chosen cells in each of three biological repeat experiments (Supplemental Fig. S2) document that, compared with wild type, tom70Δ and sam37Δ cells have 1.5-fold and 1.4-fold increases in the cytoplasmic versus nuclear signals, respectively. In contrast, los1Δ cells have decreased cytoplasmic versus nuclear signals compared with wild-type cells (Fig. 2). Similar results were obtained by using a different quantification method that assesses every cell in a field (Supplemental Fig. S3). Thus, tom70Δ and sam37Δ cells accumulate intron-containing tRNAs in the cytoplasm; the data are consistent with Tom70 and Sam37 serving a role in tRNA splicing at a step after tRNA nuclear export.
Figure 2.
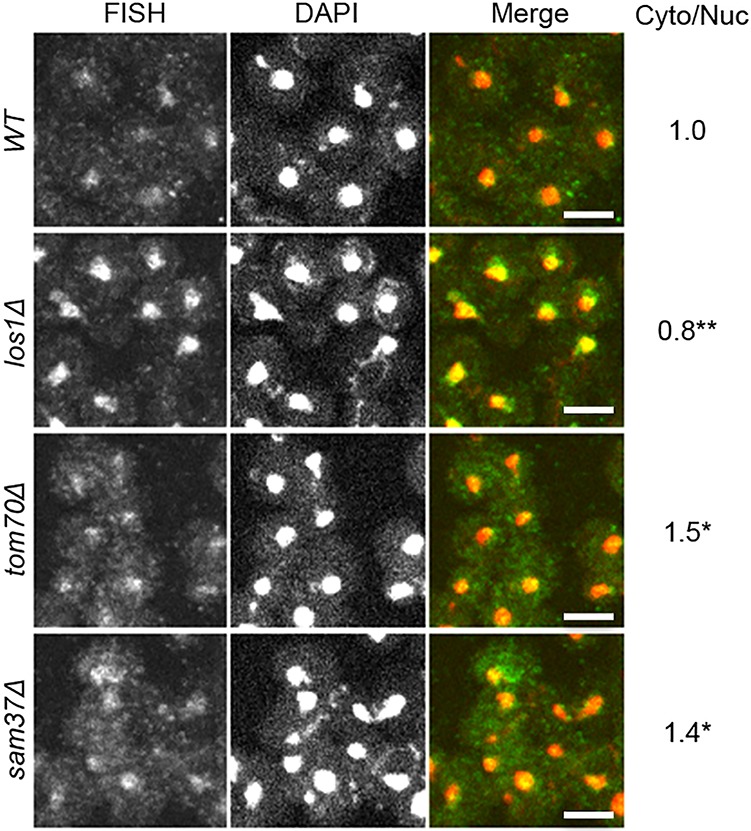
S. cerevisiae tom70Δ and sam37Δ cells accumulate end-processed intron-containing tRNAs in the cytoplasm. FISH analysis of the subcellular localization of tRNAIleUAU using intron probe SRIM03 (green). Nuclei were visualized by DAPI staining (red). Bar, 4 µm. The average signal intensities from the cytoplasm and nucleus were obtained from 10 cells, and statistical analysis was performed from three biological repeats (Supplemental Fig. S2; Supplemental Material). The ratio between signal intensity of cytoplasm versus nucleus was calculated and normalized to wild type (Cyto/Nuc). (*) P ≤ 0.05; (**) P ≤ 0.01.
The absence of respiratory metabolism does not affect tRNA splicing
Because tom70Δ and sam37Δ cells are defective in respiratory metabolism, we explored the possibility that efficient pre-tRNA splicing is coupled with efficient respiratory metabolism. If inefficient pre-tRNA splicing in the cytoplasm results from defective respiratory metabolism of tom70Δ and sam37Δ cells, then one would predict that cells unable to conduct respiratory metabolism via a different mechanism would also evidence inefficient pre-tRNA splicing. To test this, we used two methods to create strains lacking mitochondrial DNA (mtDNA). Cells lacking mitochondrial genomes are unable to grow on nonfermentable carbon sources but possess the mitochondrial organelle. MIP1 encodes the mtDNA polymerase, and thus mip1Δ cells are unable to replicate mtDNA (Genga et al. 1986). Likewise, treatment of cells with ethidium bromide (EtBr) causes loss of mtDNA (Ferguson and von Borstel 1992).
Using DAPI staining that detects nuclear DNA and mtDNA, independent isolates of mip1Δ cells and cells treated with EtBr evidenced no detectable mtDNA, and the cells are unable to grow on medium with nonfermentable glycerol as the carbon source (Supplemental Fig. S4). Thus, these strains have defective respiratory metabolism. As assessed by Northern analysis, mip1Δ and EtBr-treated strains splice intron-containing tRNA as efficiently as do wild-type cells (Fig. 1). Therefore, efficient tRNA splicing does not require mitochondrial respiratory metabolism, and Tom70 and Sam37 must affect tRNA splicing by a mechanism independent of respiratory metabolism.
Tom70 and Sam37 are important for SEN subunit mitochondrial localization in S. cerevisiae
Tom70 and Sam37 are integral MOM proteins that function in mitochondrial protein import as well as distribution of other mitochondrial proteins to the MOM (Stojanovski et al. 2012). The SEN subunits are localized on the surface of mitochondria as nonintegral proteins (Yoshihisa et al. 2003). One way that Tom70 and Sam37 might be required for efficient pre-tRNA splicing would be for these proteins to function in the localization and/or assembly of the SEN complex on mitochondria.
To study a putative role for Tom70 and Sam37 in the localization of the SEN subunits on mitochondria, we tagged each SEN subunit with GFP at its endogenous chromosomal locus in wild-type, tom70Δ, and sam37Δ strains. As each SEN subunit must interact with the other subunits of the heterotetramer, GFP tagging may interfere with the SEN complex assembly and/or catalysis. In fact, a C-terminal GFP-tagged SEN34 strain was not obtained in the genome-wide GFP tagging study (Huh et al. 2003). We verified that C-terminal GFP-tagged Sen34 results in lethality when expressed from the SEN34 promoter in a single-copy plasmid (Supplemental Fig. S5A). Therefore, to assure that GFP tagging does not affect SEN complex function, we generated yeast in which each gene encoding a SEN subunit was endogenously tagged with GFP to either the N terminus or the C terminus and then assessed the consequences on viability, growth rate, and pre-tRNA splicing. Surprisingly, only Sen15 could be C-terminally GFP-tagged and maintain full function (Supplemental Fig. S5B; Supplemental Table S1). The levels of tRNA splicing for the resulting N-terminally or C-terminally tagged SEN subunits, as assessed by Northern analyses, are similar to the untagged strains (Supplemental Fig. S6).
The subcellular localization of each endogenously GFP-tagged fully functional SEN subunit was assessed by live-cell confocal microscopy using DAPI to detect nuclear DNA and mtDNA. The GFP-Sen2 signals overlap with the mtDNA DAPI staining (Fig. 3) in wild-type cells, but the signal on mitochondria is highly reduced in tom70Δ cells and moderately reduced in sam37Δ cells. To quantify GFP-Sen2 levels associated with mitochondria, we fractionated yeast into whole-cell lysate, mitochondrial-enriched, and cytoplasm-excluding mitochondria-enriched fractions and assessed protein levels by Western blot analysis. As shown by antibodies to mitochondrial MTCO1 (detects Cox1) and cytoplasmic Pgk1 markers, there is no detectable cross-contamination between the subcellular fractions (Fig. 4A). We chose Cox1 as a mitochondrial marker because it is encoded by the mitochondrial genome and does not require Tom70 or Sam37 for mitochondrial protein import, as do other imported mitochondrial proteins such as Porin, which is decreased in sam37Δ cells (Wenz et al. 2015). However, even MTCO1 levels are reduced ∼30% in tom70Δ and sam37Δ cells (Supplemental Fig. S7E) compared with wild-type cells, possibly due to defective mitochondrial import of other cytochrome c oxidase subunits. Compared with MTCO1, GFP-Sen2 is significantly reduced in tom70Δ cells and trends less in sam37Δ cells (Supplemental Fig. S7A). To obtain a quantitative analysis of the roles of Tom70 and Sam37 in the mitochondrial location of SEN subunits that is not biased by the import/stability of single proteins, the ratios of GFP-Sen2 versus total protein cofractioning with mitochondria were calculated from three biological repeat experiments. Consistent with the live-cell imaging, GFP-Sen2 is decreased approximately fivefold and approximately twofold in the mitochondrial fractions of tom70Δ and sam37Δ cells, respectively (Fig. 4A). Thus, Tom70 and Sam37 are required for efficient localization of Sen2 to the mitochondria.
Figure 3.
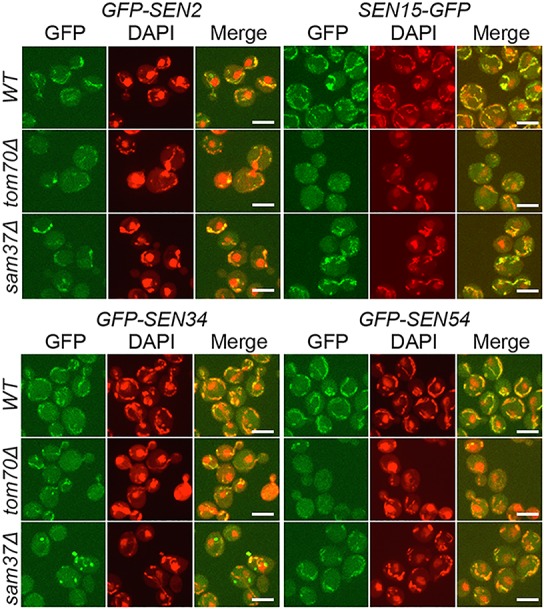
Deletion of TOM70 or SAM37 causes mislocalization of SEN subunits from the mitochondrial surface. Cells with endogenously GFP-tagged SEN2, SEN15, SEN34, and SEN54 in wild-type, tom70Δ, and sam37Δ strains were subjected to live-cell fluorescence microscopy; DAPI staining was used to locate nuclear DNA and mtDNAs (red). Bar, 4 µm.
Figure 4.
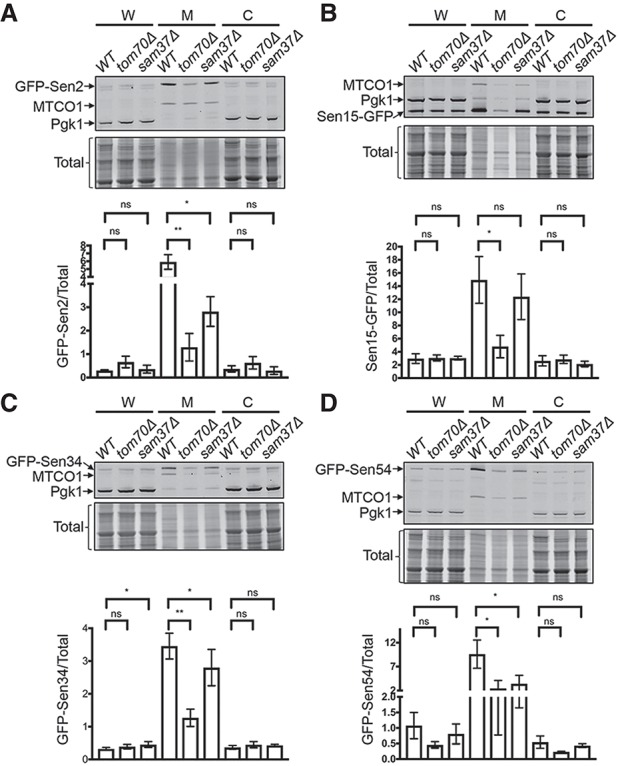
Subcellular fractionation of endogenously tagged S. cerevisiae SEN subunits. (A–D) Subcellular fractionations for GFP-Sen2 (A), Sen15-GFP (B), GFP-Sen34 (C), and GFP-Sen54 (D). Whole-cell lysate (W), mitochondria-enriched (M), and cytoplasm-excluding mitochondria-enriched (C) fractions were isolated, and extracted proteins were analyzed by Western blots. Anti-MTCO1 and anti-Pgk1 were used to assess mitochondrial and cytoplasmic proteins, respectively. Total protein stain was used as the loading control. Signal intensities were quantified from three biological repeats. The mean and standard deviation (SD) of GFP-tagged SEN subunits versus total protein are plotted. (ns) Not significant (P > 0.05); (*) P ≤ 0.05; (**) P ≤ 0.01. Note that for the two closely migrating bands for GFP-Sen2, only the top band was associated with the mitochondria-enriched fraction and used for quantification.
We conducted similar studies for the other three SEN subunits. Sen15, Sen34, and Sen54 evidence reduced mitochondrial localization in tom70Δ cells as assessed by microscopy, although to different extents (Fig. 3). For sam37Δ cells, the mitochondrial localization of Sen2 and Sen54 is moderately reduced, whereas Sen15 does not appear to be affected. Interestingly, Sen34 localizes to distinct unknown foci in sam37Δ cells that do not colocalize with mitochondria (Fig. 3). The results from the subcellular fractionation experiments are consistent with the microscopy data (Fig. 4B–D; Supplemental Fig. S7B–D).
Although deletion of TOM70 or SAM37 causes reduced association of the SEN subunits with mitochondria and inefficient pre-tRNA splicing, total SEN subunit levels are not reduced in tom70Δ or sam37Δ cells (Fig. 4; Supplemental Fig. S7). The data support the notion that the mitochondrial surface may serve as a platform for assembly of the heterotetrameric SEN complexes. SEN subunits are low in abundance—estimated to range from 25 to 530 molecules per cell for Sen2, Sen15, and Sen54 and ∼1300 molecules per cell for Sen34 (Ghaemmaghami et al. 2003; Kulak et al. 2014). Therefore, it may be difficult for SEN subunits to efficiently “find each other” in the dilute cytoplasm; targeting SEN subunits to the mitochondrial surface may aid their efficient interactions. Even though pre-tRNA splicing is essential, Tom70 and Sam37 are unessential. This could be due to the existence of TOM71 (a TOM70 paralog), residual SEN complexes on the mitochondrial surface, or low levels of SEN complex assembly in the cytoplasm in tom70Δ and sam37Δ cells.
The SEN subunits may interact either directly with Tom70 and Sam37 or indirectly by interacting with unidentified MOM proteins inserted by Tom70 or Sam37. The latter seems more likely, since we identified three separate proteins of the TOM and SAM complexes (Tom70, Tom22, and Sam37) that are important for pre-tRNA splicing.
Mitochondrial localization of SEN subunits in S. pombe and a conserved role for Tom70 in localization and the protein level of SEN subunits
The subcellular localization of pre-tRNA splicing surprisingly differs among organisms: Splicing occurs on mitochondria in budding yeast but is nucleoplasmic in Xenopus oocytes and human cells (Melton et al. 1980; Lund and Dahlberg 1998; Paushkin et al. 2004). However, data from genetic studies indicate that pre-tRNA splicing outside of the nucleus may not be unique to S. cerevisiae. In S. pombe and Arabidopsis thaliana, deletions of the S. cerevisiae LOS1 homologs XpoT and PAUSED, respectively, cause intron-containing tRNA accumulation (Park et al. 2005; Cherkasova et al. 2012). The data are consistent with the hypothesis that pre-tRNA splicing may occur in the cytoplasm—and perhaps on the mitochondrial surface—in evolutionally diverged yeasts and plants. However, imaging studies of the subcellular localizations of SEN subunits have led to different conclusions. Data from a genome-wide protein localization study using overexpression by the inducible nmt1 promoter in S. pombe reported that Sen2 localizes to undefined cytoplasmic foci, Sen15 localizes to the nucleus, and Sen54 localizes to mitochondria (Matsuyama et al. 2006). For A. thaliana, AtSen1 and AtSen2 (putative yeast Sen2 orthologs) locate to the nucleus and cytoplasm when transiently overexpressed via the strong 35S Ω promoter in Allium epidermal and Vicia guard cells (Englert et al. 2007).
To investigate the seeming contradictions between the genetic and imaging data for the localization of the SEN subunits in S. pombe and A. thaliana cells, we hypothesized that imaging studies using overexpressed constructs may not accurately report protein localization. Specifically, overexpression of a single subunit would have aberrant stoichiometry for a heterotetrameric complex and may not achieve the same localization as an endogenously expressed subunit. To test this, we determined the subcellular localizations of overexpressed GAL1-inducible GFP-tagged SEN subunits in S. cerevisiae. Upon 1 h of galactose induction, overexpressed Sen2, Sen15, and Sen34 accumulate in the cytoplasm, whereas Sen54 maintains its mitochondrial localization (Supplemental Fig. S8). Since overexpression of SEN subunits causes their mislocalization in budding yeast, we re-examined S. pombe SEN subunit localizations by tagging S. pombe SEN subunits with C-terminal mECitrine at their endogenous loci. Sen2, Sen34, and Sen54 endogenously tagged at their C termini locate to mitochondria, as indicated by the overlapping signal with the mitochondrion stain MitoView Blue (Fig. 5A). However, we were unable to create a strain with Sen15 C-terminally tagged with mECitrine, perhaps because the C-terminal mECitrine tag affects its function for S. pombe. Because the SEN complex is a heterotetramer and we showed that three SEN subunits expressed from endogenous loci locate to mitochondria, it is very likely that the SEN complex in S. pombe is also located on mitochondria.
Figure 5.
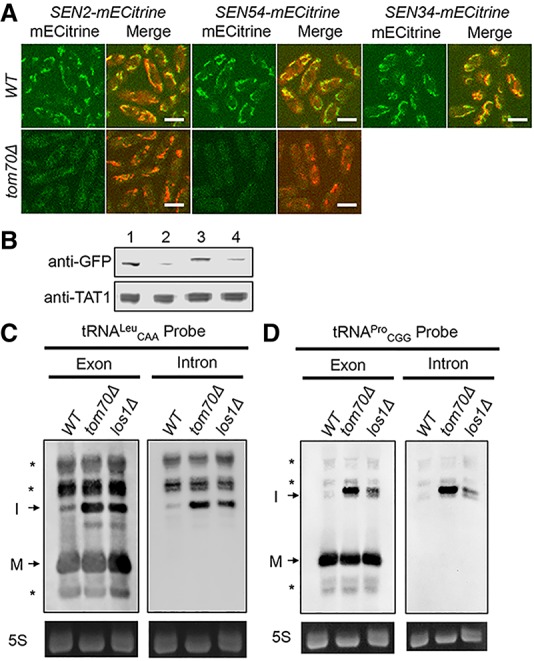
S. pombe SEN subunits localize to mitochondria, and TOM70 deletion causes mislocalization, reduced SEN subunit levels, and inefficient tRNA splicing. (A) Cells with endogenously mECitrine-tagged SEN2, SEN54, and SEN34 in wild-type and tom70Δ strains were subjected to live-cell fluorescence microscopy (green). S. pombe mitochondria were stained with MitoView Blue (red). Bar, 6 µm. (B) Proteins were extracted from cells with endogenously mECitrine-tagged SEN2 or SEN54 in wild-type and tom70Δ backgrounds and subjected to Western analysis. Anti-GFP was used to detect mECitrine-tagged SEN subunits. Anti-TAT1 was used to detect α-tubulin as the loading control. (Lane 1) Wild-type SEN2-mECitrine. (Lane 2) tom70Δ SEN2-mECitrine. (Lane 3) Wild-type SEN54-mECitrine. (Lane 4) tom70Δ SEN54-mECitrine. (C) Northern analysis of tRNALeuCAA from wild-type, tom70Δ, or los1Δ cells using exon or intron probes. Unknown RNAs are indicated by an asterisk. (5S) 5S rRNA stained with EtBr as the loading control. (D) Northern analysis of tRNAProCGG from wild-type, tom70Δ, and los1Δ cells as in C.
Tom70 is conserved in eukaryotes; BLASTp alignments of S. pombe and S. cerevisiae Tom70 showed 32% identity and 49% similarity. Sam37 appears not to be conserved. We assessed whether S. pombe Tom70 serves a function for fission yeast in locating the SEN complex on mitochondria similar to what it does in budding yeast. To accomplish this, we deleted the TOM70 gene in the S. pombe strains that contain endogenously mECitrine-tagged SEN subunits. Although we obtained TOM70 deletions in SEN2-mECitrine and SEN54-mECitrine strains, the SEN34-mECitrine/tom70Δ strain was not obtained. By confocal microscopy, Sen2-mECitrine and Sen54-mECitrine signals were very weak in tom70Δ cells, with the residual signals located in the cytoplasm and some located on mitochondria (Fig. 5A).
To assess levels of SEN subunits in wild-type and tom70Δ cells, we used Western analysis using a GFP antibody that detects the highly similar mECitrine (Supplemental Fig. S9). Using α-tubulin as the loading control, the levels of Sen2-mECitrine and Sen54-mECitrine are reduced in tom70Δ cells compared with wild type (Fig. 5B; Supplemental Fig. S9). We interpret these results to support the hypothesis that S. pombe Tom70 functions in the mitochondrial localization of SEN subunits; when mitochondrial localization is not achieved, the SEN subunits become unstable, although it is possible that SEN subunits are poorly expressed in tom70Δ cells. Thus, mitochondrial localization for RNA processing enzymes may serve to protect them from cytoplasmic degradation. There is precedence for this notion; for example, when individual S. cerevisiae ribosomal proteins are in excess and unable to assemble with ribosomes, the “free” ribosomal proteins are destabilized (Perry 2007). Likewise, S. cerevisiae Hrr25 becomes unstable when mislocalized from P bodies to the cytoplasm (Zhang et al. 2016).
If S. pombe Tom70 is important for function and/or stability of the SEN complex, then deletion of S. pombe TOM70 would be expected to cause defects in pre-tRNA splicing. We used Northern analysis with probes complementary to the 5′ exons or introns of S. pombe tRNALeuCAA and tRNAProCGG. tom70Δ cells accumulate more intron-containing tRNALeuCAA and tRNAProCGG than wild-type cells (Fig. 5C), documenting a role for S. pombe Tom70 in pre-tRNA splicing.
Summary
Several post-transcriptional RNA processing steps are catalyzed on the mitochondrial surface by mitochondria-localized enzymes (Chatterjee et al. 2018). Here, we report that the mitochondrial localizations of endogenous SEN subunits are conserved between S. cerevisiae and S. pombe, which diverged >500 million years ago (Rhind et al. 2011), and that there are novel and conserved roles for MOM proteins in the mitochondrial localization of SEN subunits (Supplemental Fig. S10). Mitochondrial localization appears to serve for assembly of the SEN complex in S. cerevisiae and for maintenance of SEN subunit levels in S. pombe. Mitochondrial localization of RNA processing enzymes may also serve additional functions, such as aiding substrate/enzyme interactions to mediate specificity for RNases such as piRNA 5′ processing Zucchini, which is nonspecific in vitro (Ipsaro et al. 2012). It will be interesting to learn whether the conserved role of Tom70 in tRNA processing is also required for the other RNA processing steps occurring on the mitochondrial surface or in other organisms.
Materials and methods
Strains
S. cerevisiae and S. pombe strains were created with SEN subunits tagged at their endogenous loci.
Reagents
Probes for Northern analysis and FISH, antibodies for Western analysis, and primers for plasmid constructs are detailed in the Supplemental Material.
Mitochondrion isolation
Mitochondria were isolated by yeast mitochondrion isolation kits (Sigma-Aldrich [MITOISO3] or BioVision [K259]).
Imaging
S. cerevisiae mitochondria were viewed by live-cell DAPI staining. S. pombe mitochondria were stained by MitoView Blue (Biotium, 70052).
Additional experimental details are in the Supplemental Material.
Supplementary Material
Acknowledgments
We are grateful to Dr. J.-Q. Wu and laboratory members for providing advice, strains, and plasmids for the S. pombe experiments. Special thanks to Dr. E. Grayhack, Dr. N.C. Martin, Dr. S. Osmani, and Dr. J.-Q. Wu for scientific discussions and insightful comments on the manuscript. We thank A.K.H. laboratory members for excellent scientific interactions and manuscript proofreading. This work was supported by National Institute of Health grants GM27930 (prior) and GM122884 (current) to A.K.H., and an Ohio State University Comprehensive Cancer Center Pelotonia Graduate Fellowship to Y.W.
Author contributions: Y.W. and A.K.H. designed the experiments. Y.W. conducted the experiments. Y.W. and A.K.H. prepared the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.316257.118.
References
- Boland C, Hayes P, Santa-Maria I, Nishimura S, Kelly VP. 2009. Queuosine formation in eukaryotic tRNA occurs via a mitochondria-localized heteromeric transglycosylase. J Biol Chem 284: 18218–18227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone C, Andrews BJ. 2015. The indispensable genome. Science 350: 1028–1029. [DOI] [PubMed] [Google Scholar]
- Chan PP, Lowe TM. 2009. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res 37: D93–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee K, Nostramo RT, Wan Y, Hopper AK. 2018. tRNA dynamics between the nucleus, cytoplasm and mitochondrial surface: location, location, location. Biochim Biophys Acta 1861: 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasova V, Maury LL, Bacikova D, Pridham K, Bähler J, Maraia RJ. 2012. Altered nuclear tRNA metabolism in La-deleted Schizosaccharomyces pombe is accompanied by a nutritional stress response involving Atf1p and Pcr1p that is suppressible by Xpo-t/Los1p. Mol Biol Cell 23: 480–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S-YY, Huang P, Jenkins GM, Chan DC, Schiller J, Frohman MA. 2006. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat Cell Biol 8: 1255–1262. [DOI] [PubMed] [Google Scholar]
- Englert M, Latz A, Becker D, Gimple O, Beier H, Akama K. 2007. Plant pre-tRNA splicing enzymes are targeted to multiple cellular compartments. Biochimie 89: 1351–1365. [DOI] [PubMed] [Google Scholar]
- Ferguson LR, von Borstel RC. 1992. Induction of the cytoplasmic ‘petite’ mutation by chemical and physical agents in Saccharomyces cerevisiae. Mutat Res 265: 103–148. [DOI] [PubMed] [Google Scholar]
- Genga A, Bianchi L, Foury F. 1986. A nuclear mutant of Saccharomyces cerevisiae deficient in mitochondrial DNA replication and polymerase activity. J Biol Chem 261: 9328–9332. [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. 2003. Global analysis of protein expression in yeast. Nature 425: 737–741. [DOI] [PubMed] [Google Scholar]
- Hopper AK, Phizicky EM. 2003. tRNA transfers to the limelight. Genes Dev 17: 162–180. [DOI] [PubMed] [Google Scholar]
- Hopper AK, Schultz LD, Shapiro RA. 1980. Processing of intervening sequences: a new yeast mutant which fails to excise intervening sequences from precursor tRNAs. Cell 19: 741–751. [DOI] [PubMed] [Google Scholar]
- Huang H, Gao Q, Peng X, Choi S-YY, Sarma K, Ren H, Morris AJ, Frohman MA. 2011. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev Cell 20: 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. [DOI] [PubMed] [Google Scholar]
- Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ. 2012. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature 491: 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi N, Shoji K, Sakaguchi Y, Honda S, Kirino Y, Suzuki T, Katsuma S, Tomari Y. 2016. Identification and functional analysis of the pre-piRNA 3′ trimmer in silkworms. Cell 164: 962–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. 2014. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods 11: 319–324. [DOI] [PubMed] [Google Scholar]
- Lund E, Dahlberg JE. 1998. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science 282: 2082–2085. [DOI] [PubMed] [Google Scholar]
- Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, et al. 2006. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 24: 841–847. [DOI] [PubMed] [Google Scholar]
- Melton DA, De Robertis EM, Cortese R. 1980. Order and intracellular location of the events involved in the maturation of a spliced tRNA. Nature 284: 143–148. [DOI] [PubMed] [Google Scholar]
- Miyauchi K, Kimura S, Suzuki T. 2012. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat Chem Biol 9: 105–111. [DOI] [PubMed] [Google Scholar]
- Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS. 2005. Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci 102: 3691–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin SV, Patel M, Furia BS, Peltz SW, Trotta CR. 2004. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3′ end formation. Cell 117: 311–321. [DOI] [PubMed] [Google Scholar]
- Perry RP. 2007. Balanced production of ribosomal proteins. Gene 401: 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, et al. 2011. Comparative functional genomics of the fission yeasts. Science 332: 930–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Hopper AK. 1998. tRNA nuclear export in Saccharomyces cerevisiae: in situ hybridization analysis. Mol Biol Cell 9: 3041–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D, Bohnert M, Pfanner N, van der Laan M. 2012. Mechanisms of protein sorting in mitochondria. Cold Spring Harb Perspect Biol 4: a011320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Tu S, Lee HC, Weng Z, Mello CC. 2016. The RNase PARN-1 trims piRNA 3′ ends to promote transcriptome surveillance in C. elegans. Cell 164: 974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta CR, Miao F, Arn EA, Stevens SW, Ho CK, Rauhut R, Abelson JN. 1997. The yeast tRNA splicing endonuclease: a tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell 89: 849–858. [DOI] [PubMed] [Google Scholar]
- Trotta CR, Paushkin SV, Patel M, Li H, Peltz SW. 2006. Cleavage of pre-tRNAs by the splicing endonuclease requires a composite active site. Nature 441: 375–377. [DOI] [PubMed] [Google Scholar]
- Tsuboi T, Yamazaki R, Nobuta R, Ikeuchi K, Makino S, Ohtaki A, Suzuki Y, Yoshihisa T, Trotta C, Inada T. 2015. The tRNA splicing endonuclease complex cleaves the mitochondria-localized Cbp1 mRNA. J Biol Chem 290: 16021–16030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, Toyoda A, Hoki Y, Fujiyama A, Shibata T, Sado T, et al. 2011. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell 20: 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz LS, Ellenrieder L, Qiu J, Bohnert M, Zufall N, van der Laan M, Pfanner N, Wiedemann N, Becker T. 2015. Sam37 is crucial for formation of the mitochondrial TOM–SAM supercomplex, thereby promoting β-barrel biogenesis. J Cell Biol 210: 1047–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Bao A, Chatterjee K, Wan Y, Hopper AK. 2015. Genome-wide screen uncovers novel pathways for tRNA processing and nuclear-cytoplasmic dynamics. Genes Dev 29: 2633–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihisa T, Yunoki-Esaki K, Ohshima C, Tanaka N, Endo T. 2003. Possibility of cytoplasmic pre-tRNA splicing: the yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol Biol Cell 14: 3266–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Shi Q, Varia SN, Xing S, Klett BM, Cook LA, Herman PK. 2016. The activity-dependent regulation of protein kinase stability by the localization to P-bodies. Genetics 203: 1191–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


