Arongaus et al. show that genetic mutation of REPRESSOR OF UV-B PHOTOMORPHOGENESIS 2 (RUP2) renders the facultative long day plant Arabidopsis thaliana a day-neutral plant (specifically under light conditions that include UV-B radiation) and dependent on the UV RESISTANCE LOCUS 8 (UVR8) UV-B photoreceptor.
Keywords: sun simulator, plant–environment interaction, photoperiodism, flowering, UV-B photoreceptor, UVR8, Arabidopsis
Abstract
Plants have evolved complex photoreceptor-controlled mechanisms to sense and respond to seasonal changes in day length. This ability allows plants to optimally time the transition from vegetative growth to flowering. UV-B is an important part intrinsic to sunlight; however, whether and how it affects photoperiodic flowering has remained elusive. Here, we report that, in the presence of UV-B, genetic mutation of REPRESSOR OF UV-B PHOTOMORPHOGENESIS 2 (RUP2) renders the facultative long day plant Arabidopsis thaliana a day-neutral plant and that this phenotype is dependent on the UV RESISTANCE LOCUS 8 (UVR8) UV-B photoreceptor. We provide evidence that the floral repression activity of RUP2 involves direct interaction with CONSTANS, repression of this key activator of flowering, and suppression of FLOWERING LOCUS T transcription. RUP2 therefore functions as an essential repressor of UVR8-mediated induction of flowering under noninductive short day conditions and thus provides a crucial mechanism of photoperiodic flowering control.
Timely and synchronous flowering is important to optimize pollination and allow seed maturation during favorable environmental conditions. In addition to being adaptive traits for plants in natural environments, synchronous flowering and maximal seed yields are also crucial in horticulture and agricultural production systems. In recent decades, the genetic pathways and regulatory proteins that promote flowering in response to changes in day length (photoperiod) were largely defined in the model species Arabidopsis thaliana, a facultative long day (LD) plant (i.e., flowers early in LDs but will eventually also flower under short days [SDs]) (Song et al. 2015). Photoperiodic flowering in Arabidopsis is due to the suppression of flowering in SDs, which is released under LD conditions. Flowering under inductive LD photoperiods is activated by the CONSTANS (CO) transcription factor, a master regulator of FLOWERING LOCUS T (FT) expression (Putterill et al. 1995; Samach et al. 2000; Turck et al. 2008; Andres and Coupland 2012; Song et al. 2015). FT is a major component of the florigen, a systemic signal that moves through the vasculature from the leaves into the apical meristem, where it induces flowering in response to the inductive photoperiod (Wigge et al. 2005; Corbesier et al. 2007; Jaeger and Wigge 2007; Mathieu et al. 2007; Turck et al. 2008; Song et al. 2015). Regulation of CO activity is complex and takes place at many different levels (Romera-Branchat et al. 2014; Song et al. 2015; Shim et al. 2017). A prominent component of this regulation under noninductive SD conditions is CO ubiquitination during the night period by the CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1)–SUPPRESSOR OF PHYTOCHROME A-105 (SPA) E3 ubiquitin ligase complex followed by degradation in the 26S proteasome (Laubinger et al. 2006; Jang et al. 2008; Liu et al. 2008). Consistently, cop1 and spa1 plants flower early under SD conditions compared with wild type (McNellis et al. 1994; Laubinger et al. 2006). In LDs, the COP1–SPA complex is inhibited during the day period by cryptochrome 2 (cry2), which is required for early flowering under these conditions (Guo et al. 1998; Zuo et al. 2011). COP1 is also a well-known molecular player directly interacting with the UV-B photoreceptor UV RESISTANCE LOCUS 8 (UVR8) (Favory et al. 2009; Rizzini et al. 2011; Cloix et al. 2012; Yin et al. 2015; Jenkins 2017; Podolec and Ulm 2018). However, despite this and the fact that UV-B is an intrinsic part of sunlight, our molecular understanding of photoperiodic flowering regulation in Arabidopsis is basically based on growth chamber experiments in the absence of UV-B. Thus, the role of UVR8 signaling in photoperiodic control of flowering time has not been investigated previously.
The seven-bladed β-propeller protein UVR8 forms homodimers in the absence of UV-B (Favory et al. 2009; Rizzini et al. 2011). UVR8 monomerizes upon UV-B absorption by specific intrinsic tryptophan residues, which is followed by interaction with COP1 (Favory et al. 2009; Rizzini et al. 2011). As a result of this UV-B-dependent interaction, the COP1 target protein ELONGATED HYPOCOTYL 5 (HY5) is stabilized (Favory et al. 2009; Huang et al. 2013; Binkert et al. 2014). HY5 is a bZIP transcription factor that plays a central role in light signaling (Lau and Deng 2012), including UVR8-mediated UV-B signaling (Ulm et al. 2004; Brown et al. 2005; Stracke et al. 2010; Binkert et al. 2014). The UVR8 photocycle involves negative feedback regulation by REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 (RUP1) and RUP2, which are UVR8-interacting proteins that facilitate the ground state reversion of UVR8 via redimerization (Gruber et al. 2010; Heijde and Ulm 2013). RUP1 and RUP2 act largely redundantly for all UV-B responses characterized to date, and their role is to establish UVR8 homodimer/monomer equilibrium under diurnal conditions (Gruber et al. 2010; Heijde and Ulm 2013; Findlay and Jenkins 2016). A recent report has suggested that an apparently UV-B-independent role of RUP1 and RUP2 in flowering time regulation exists (note that EARLY FLOWERING BY OVEREXPRESSION 1 [EFO1] = RUP1 and EFO2 = RUP2) (Wang et al. 2011). However, the underlying molecular mechanism and the role of RUP1 and RUP2 in photoperiodic flowering regulation have remained enigmatic. Here we report how RUP2 functions as a key repressor of UVR8-mediated induction of flowering through regulation of CO activity and that this function is crucial to distinguish noninductive SDs from inductive LDs, thus enabling photoperiodic flowering.
Results
RUP2 is a repressor of flowering under SD conditions containing UV-B
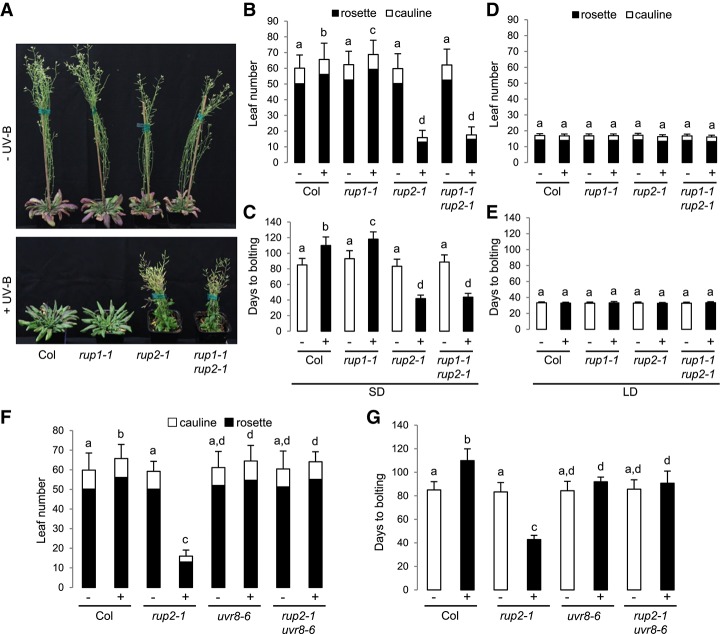
Flowering time regulation in natural ecological settings is complex and often distinct from that under laboratory conditions (Weinig et al. 2002; Wilczek et al. 2009; Brachi et al. 2010). UV-B is an important part of the sunlight spectrum that is usually lacking in controlled growth chamber environments. To better understand the potential roles of UV-B and RUP1/RUP2 in the regulation of flowering, we grew wild-type, rup1, rup2, and rup1 rup2 plants under LD (16-h/8-h light/dark) and SD (8-h/16-h light/dark) conditions. In contrast to a previous report (Wang et al. 2011), the flowering time and leaf number at flowering for rup2 as well as rup1 rup2 were comparable with those in wild type under standard laboratory growth conditions; i.e., in the absence of UV-B (LD − UV and SD − UV) (Fig. 1A–E). Strikingly, however, rup2 as well as rup1 rup2 flowered much earlier than wild type in SDs in the presence of UV-B (SD + UV) (Fig. 1A–C). This early flowering phenotype was specific to rup2, as rup1 flowered similarly to wild type (Fig. 1A–C). Moreover, the early flowering phenotype of rup2 and rup1 rup2 in SD + UV was indistinguishable and, importantly, dependent on the UV-B photoreceptor UVR8, as rup2 uvr8 and rup1 rup2 uvr8 plants flowered as late as wild type and uvr8 (Fig. 1F,G; Supplemental Fig. S1). Of note, the striking early flowering phenotype of rup2 under SD + UV was rescued by transgenic expression of the genomic RUP2 locus with an ∼1.5-kb promoter region (rup2-1/ProRUP2:RUP2) and was also observed in rup2-2 plants carrying a different T-DNA insertion in RUP2 than rup2-1 (Supplemental Fig. S2). Under LD conditions, the flowering phenotype of rup1, rup2, and rup1 rup2 was not different from that of wild type in both the absence and presence of UV-B (Fig. 1D,E). In fact, rup2 plants under SD + UV flowered with as few leaves as wild type and rup2 under LD conditions (Fig. 1B,D), indicating that RUP2 mutation rendered Arabidopsis from a facultative LD to a day-neutral plant. We conclude that RUP2 is essential to inhibit flowering under noninductive SD conditions, specifically in the presence of UV-B perceived by the UVR8 photoreceptor.
Figure 1.
rup2 flowers early in SDs with UV-B, which is dependent on the UVR8 photoreceptor. (A) Representative images of 100-d-old wild-type (Col), rup1-1, rup2-1, and rup1-1 rup2-1 Arabidopsis plants grown with (+) or without (−) UV-B. (B–E) Quantification of flowering time of wild-type (Col), rup1-1, rup2-1, and rup1-1 rup2-1 plants grown in SDs (B,C) and LDs (D,E) with (+) or without (−) UV-B. (F,G) Quantification of flowering time of wild-type (Col), rup2-1, uvr8-6, and rup2-1 uvr8-6 plants grown in SDs with (+) or without (−) UV-B. The flowering time is represented by total leaf number (rosette and cauline leaves; B,D,F) and days to bolting (C,E,G). Error bars represent standard deviation. n = 30. Shared letters indicate no statistically significant difference in the means. P > 0.05.
We further tested whether RUP2 overexpression represses flowering under LD conditions. However, RUP2 overexpression plants flowered as early as wild-type plants in both LD − UV and LD + UV (Supplemental Fig. S3A,B) despite strongly elevated RUP2 levels (Supplemental Fig. S3C). It should be noted that RUP2 overexpression is associated with a strong UV-B hyposensitive phenotype, resembling the “UV-B blindness” of uvr8-null mutants (Gruber et al. 2010). We thus conclude that RUP2 overexpression cannot repress flowering under LD conditions. However, blocking UVR8 activation precludes analysis of a distinct effect of RUP2 overexpression on the UVR8-induced flowering pathway. Moreover, in contrast to the results in a previous study (Wang et al. 2011), we did not observe an early flowering phenotype for the RUP2 overexpression line in SDs (Supplemental Fig. S3D,E).
It has been shown previously that UVR8 overexpression lines at the seedling stage display a UV-B phenotype enhanced similarly to rup2 and rup1 rup2 (Favory et al. 2009; Gruber et al. 2010). To test whether overactivation of the UV-B signaling pathway leads to early flowering under SD + UV, we used an established UVR8 overexpression line (Favory et al. 2009). As expected, the UVR8 overexpression line displayed a similar morphology in response to UV-B exposure compared with that of rup2, such as smaller rosettes (Supplemental Fig. S4A). However, UVR8 overexpression did not affect the flowering time in comparison with that in wild type (Supplemental Fig. S4B,C). It is of note that UVR8 overexpression was associated with strongly enhanced RUP2 levels (Supplemental Fig. S4D). Our data suggest that overactivation of the UVR8 signaling pathway is not sufficient to induce early flowering, likely due to the balancing effect of elevated RUP2 activity as a repressor of flowering.
We further tested the importance of RUP2 repression of early flowering in SD + UV in sun simulators that allow growth under a natural spectral balance from ultraviolet to infrared (Thiel et al. 1996). Under these more realistic irradiation conditions, rup2 plants maintained an early flowering phenotype, which is in contrast to that of wild-type, rup1, uvr8, and rup2 uvr8 plants (Fig. 2), thus confirming and further strengthening the results generated using plants grown in growth chambers containing UV-B. Therefore, we conclude that a major role of RUP2 concerns the repression of UVR8-induced flowering in SD + UV, which is an activity crucial for photoperiodic flowering under natural irradiation conditions, including UV-B.
Figure 2.
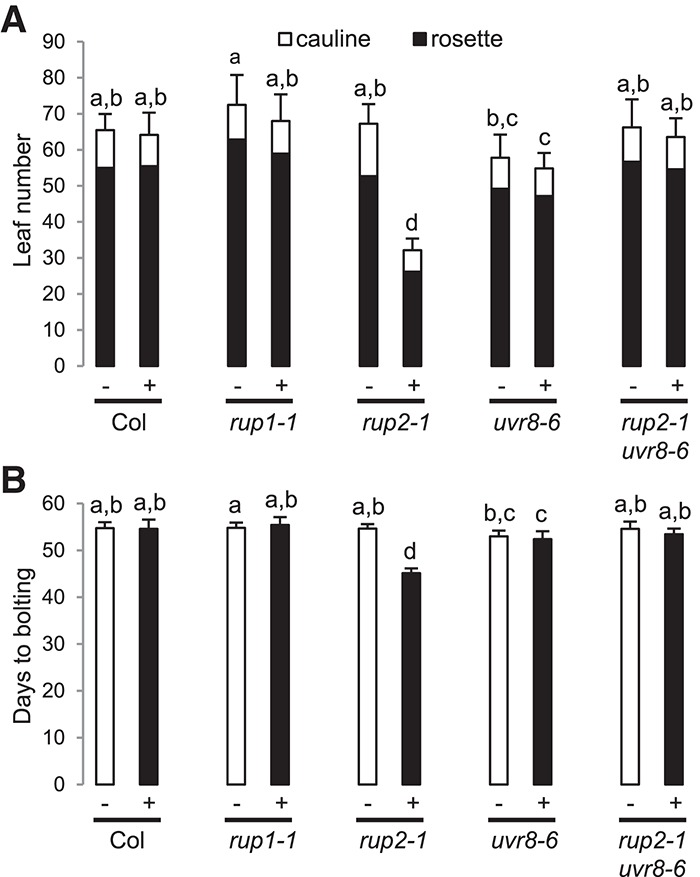
rup2-1 flowers early under realistic irradiation conditions in a sun simulator. Quantification of flowering time of wild-type (Col), rup1-1, rup2-1, uvr8-6, and rup2-1 uvr8-6 plants grown in SDs with (+) or without (−) UV. The flowering time is represented by total leaf number (rosette and cauline leaves; A) and days to bolting (B). Error bars represent standard deviation. n = 20. Shared letters indicate no statistically significant difference in the means. P > 0.05.
RUP2 interacts with CO
To better understand the role of RUP2 as a repressor of flowering, we performed a yeast two-hybrid screen, which identified the B-box proteins CO-LIKE 1 (COL1)/BBX2, COL2/BBX3, and COL5/BBX6 as RUP2-interacting partners (Supplemental Fig. S5). As rup2 shows an early flowering phenotype (Fig. 1) and the COL family members are highly related to the eponymous key flowering time regulator CO/BBX1 (Putterill et al. 1995; Khanna et al. 2009), we assessed the direct interaction between RUP2 and CO in yeast. Indeed, yeast two-hybrid growth assays indicated that RUP2 interacts with full-length CO (Fig. 3A). In contrast to the CO–COP1 interaction (Fig. 3A; Liu et al. 2008), the N-terminal 183 amino acids of CO are sufficient for the interaction with RUP2, whereas the C-terminal CCT domain of CO is not required for interaction with RUP2 (Fig. 3A).
Figure 3.
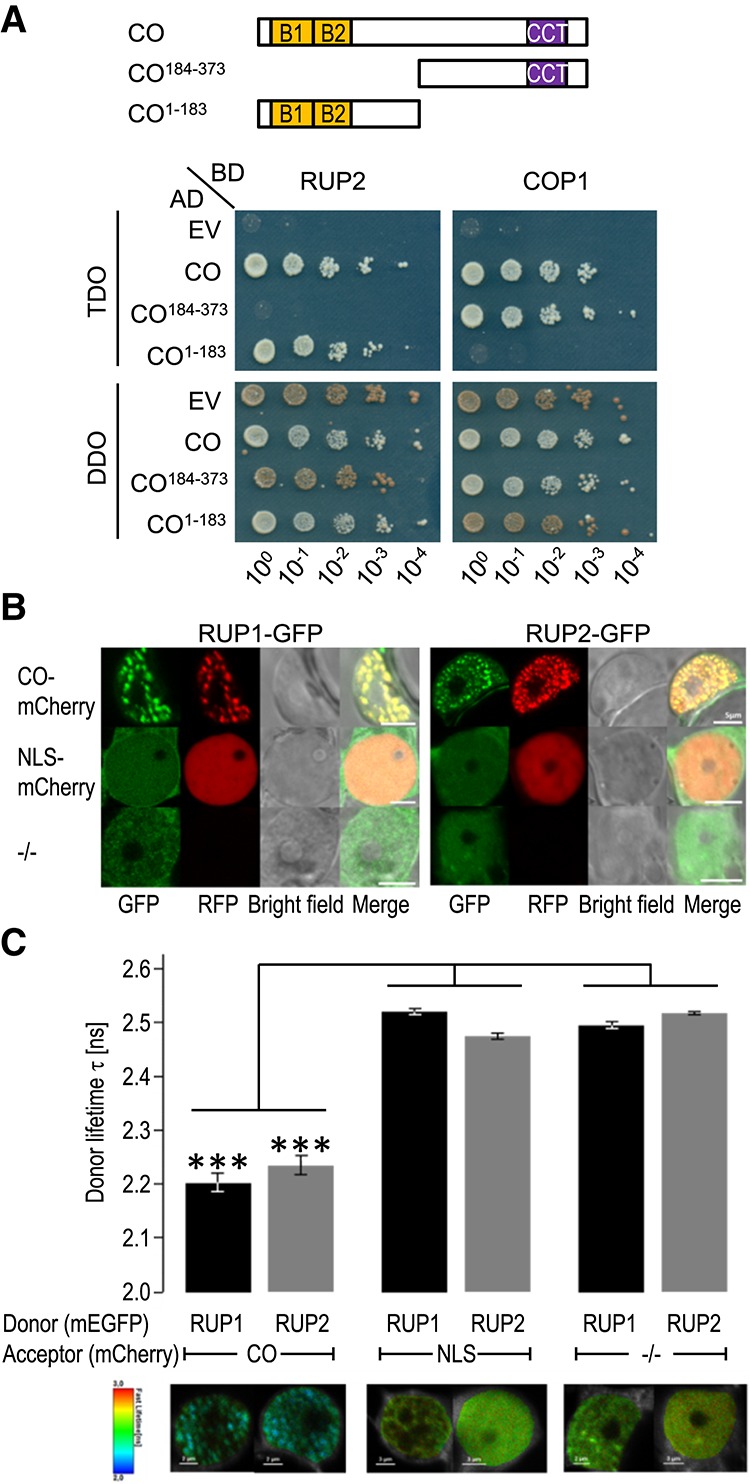
RUP1 and RUP2 interact with CO. (A) Interaction of RUP1 and RUP2 with CO in a yeast two-hybrid growth assay. (Top) Schematic representation of full-length and truncated CO used in interaction analysis. (Bottom) Tenfold serial dilutions of transformed yeast spotted on SD/−Trp/−Leu (DDO; nonselective for interaction) and SD/−Trp/−Leu/−His (TDO; selective) plates. (AD) Activation domain; (BD) binding domain; (EV) empty vector. (B) Colocalization analysis of RUP1-mEGFP and RUP2-mEGFP with either CO-mCherry or NLS-mCherry or without a mCherry fusion protein (−/−) in transiently transformed Nicotiana benthamiana epidermal leaf cells. Shown are confocal images in the GFP and RFP channel as well as the corresponding bright-field and merged images. Bars, 5 µm. (C) Fluorescence lifetime imaging microscopy (FLIM) analyses comparing the different Förster resonance energy transfer (FRET) pairs. (Top) FLIM measurements of transiently transformed N. benthamiana epidermal leaf cells expressing RUP1-mEGFP or RUP2-mEGFP donors in the presence of CO-mCherry or NLS-mCherry acceptor fusion or without a mCherry acceptor (−/−). Error bars indicate standard deviation. n ≥ 20. (***) P ≤ 0.001, a significant difference. (Bottom) Heat maps of representative nuclei used for FLIM measurements. Donor lifetimes of RUP1-mEGFP and RUP2-mEGFP are color-coded according to the scale at the left.
CO was found to be highly unstable in protein extracts, which precluded coimmunoprecipitation experiments. We thus resorted to Förster resonance energy transfer (FRET)-fluorescence lifetime imaging microscopy (FLIM) as a cell biological assay for protein–protein association in transiently transformed Nicotiana benthamiana epidermal leaf cells. First, we observed that RUP1-GFP and RUP2-GFP localized to the nucleus in a diffuse manner when expressed alone or together with an NLS-mCherry but aggregated in nuclear speckles when coexpressed with CO-mCherry (Fig. 3B). Further supporting CO–RUP interaction in yeast, our in planta FRET-FLIM analysis detected highly significant changes in the lifetime of the donor RUP1-GFP and RUP2-GFP fusions in the nucleus when coexpressed with CO-mCherry (Fig. 3C). In contrast, we did not observe significant GFP fluorophore lifetime changes when RUP1-GFP and RUP2-GFP were expressed alone or with NLS-mCherry (Fig. 3C). We thus conclude that RUP1 and RUP2 are closely associated with the key flowering regulator CO in plant cells.
Early flowering of rup2 in SD + UV depends on the flowering time regulator CO and its target, FT
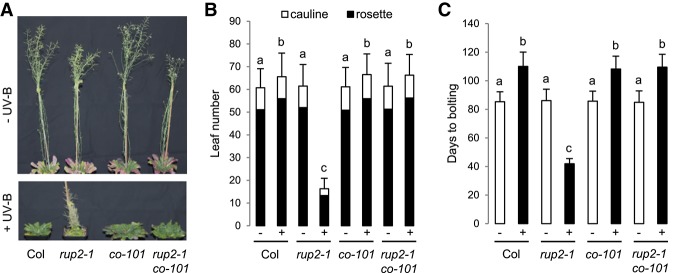
Our finding that RUP2 interacts with CO suggests that rup2 early flowering may depend on CO activity. Indeed, the early flowering phenotype of rup2 in SD + UV was completely suppressed in rup2 co double mutants (Fig. 4). CO is an activator of FT expression that encodes the florigen FT, a major positive regulator of flowering time (Turck et al. 2008). In agreement with the rup2 early flowering phenotype under SD + UV, FT expression was indeed up-regulated in rup2 and rup1 rup2 compared with that in wild-type, rup1, and rup1 rup2 uvr8 plants (Fig. 5A). Furthermore, FT promoter-driven GUS expression (ProFT:GUS) in the leaf vasculature under SD + UV was enhanced in the rup2 background in comparison with that in wild-type, uvr8, and rup2 uvr8 backgrounds (Fig. 5B). Our findings suggest that rup2 early flowering depends on enhanced CO-regulated FT expression and thus FT activity. Indeed, the early flowering phenotype of rup2 under SD + UV was completely suppressed in rup2 ft double mutants (Fig. 5C–E). We thus conclude that FT expression is deregulated in rup2 due to enhanced CO activity and that active FT is required for early flowering of rup2 under SD + UV.
Figure 4.
Early flowering of rup2 in SDs supplemented with UV-B depends on the key flowering regulator CO. (A) Representative images of 100-d-old wild-type (Col), rup2-1, co-101, and rup2-1 co-101 Arabidopsis plants grown with (+) or without (−) UV-B. (B,C) Quantification of flowering time of wild-type (Col), rup2-1, co-101, and rup2-1 co-101 plants grown in SD with (+) or without (−) UV-B. The flowering time is represented by total leaf number (rosette and cauline leaves; B) and days to bolting (C). Error bars represent standard deviation. n = 21. Shared letters indicate no statistically significant difference in the means. P > 0.05.
Figure 5.
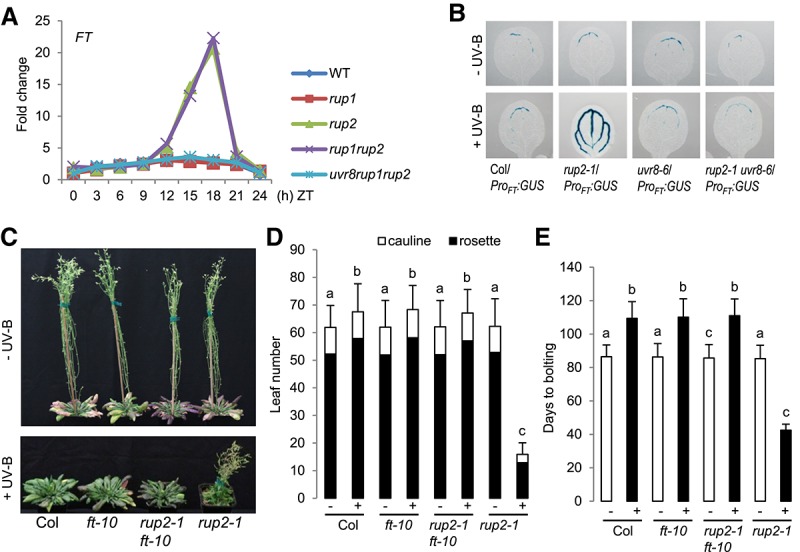
Early flowering of rup2 in SDs with UV-B depends on the florigen FT. (A) Quantitative RT–PCR (qRT–PCR) analysis of FT expression in 30-d-old wild-type, rup1-1, rup2-1, rup1-1 rup2-1, and uvr8-6 rup1-1 rup2-1 plants grown under SD + UV on soil. Samples were collected every 3 h; a representative experiment is shown. (ZT) Zeitgeber time; (ZT0) lights on; (ZT8) lights off. (B) GUS assays representing FT promoter activity in 5-d-old wild-type (Col/ProFT:GUS), rup2-1/ProFT:GUS, uvr8-6/ProFT:GUS, and rup2-1 uvr8-6/ProFT:GUS seedlings grown in SDs with (+) or without (−) UV-B. (C) Representative images of 100-d-old wild-type (Col), ft-10, rup2-1 ft-10, and rup2-1 Arabidopsis plants grown with (+) or without (−) UV-B. (D,E) Quantification of flowering time of wild-type (Col), ft-10, rup2-1 ft-10, and rup2-1 plants grown in SDs with (+) or without (−) UV-B. The flowering time is represented by total leaf number (rosette and cauline leaves; D) and days to bolting (E). Error bars represent standard deviation. n = 21. Shared letters indicate no statistically significant difference in the means. P > 0.05.
RUP2 represses CO binding to the FT promoter
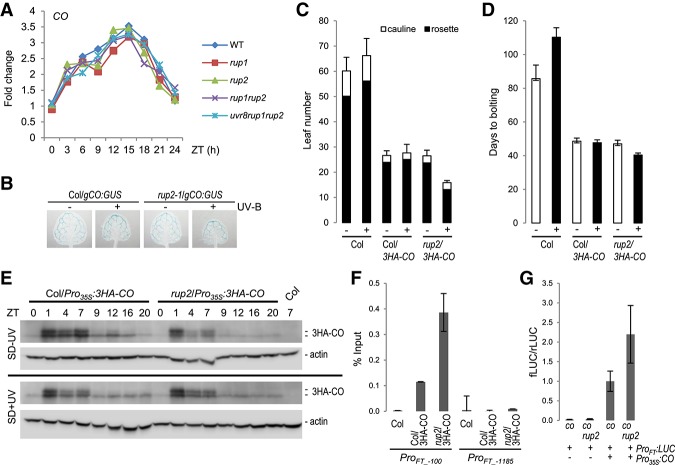
Our findings that mutation of RUP2 affects flowering in a CO-dependent manner and that RUP2 interacts with CO suggest that RUP2 may regulate CO post-transcriptionally. In agreement, the expression pattern of CO was not altered in rup2 compared with that in wild type during a 24-h time course under SD + UV conditions, excluding any effect on the diurnal regulation of CO mRNA levels (Fig. 6A,B). As endogenous CO levels have never been detected in wild type, we expressed a Pro35S:3HA-CO transgene in rup2 plants. As described before (Song et al. 2012), HA-tagged CO was detectable on protein immunoblots, and its expression in a wild-type background resulted in accelerated flowering in SDs (Fig. 6C–E). This effect was also detectable in the rup2 mutant background, thus strongly diminishing the effect of RUP2 mutation on flowering time under SD + UV (Fig. 6C,D). Although this caveat has to be taken into consideration, regulation of diurnal protein dynamics of overexpressed HA-CO was not affected by RUP2 loss of function when compared with wild-type (Fig. 6E). We further tested whether RUP2 has an effect on CO activity. CO associates with CO-responsive elements (COREs; with CCACA core motif) located at −220 and −161 base pairs relative to the start codon that are essential for CO-mediated FT activation (Tiwari et al. 2010; Song et al. 2012; Bu et al. 2014; Gnesutta et al. 2017). Indeed, chromatin immunoprecipitation (ChIP) assays of HA-CO showed specific and strongly enhanced binding to the FT promoter region in close vicinity to the CORE sequences (ProFT-100 fragment) in rup2/3HA-CO compared with that in the wild-type background (Col/3HA-CO) in plants grown under UV-B (Fig. 6F). The specificity of the ChIP data was demonstrated by the negative controls provided by the nontransgenic Col wild type as well as by a distal FT promoter region (ProFT-1185 fragment) not bound by CO (Fig. 6F; Song et al. 2012; Bu et al. 2014). In agreement with enhanced CO activity and thus FT expression, transient transcription activity assays revealed enhanced FT promoter activation by CO in protoplasts deficient of RUP2 compared with those with wild-type RUP2 (Fig. 6G). We thus conclude that RUP2 represses CO activity on FT expression by interfering with its FT promoter-binding capacity.
Figure 6.
RUP2 represses CO binding to the FT promoter and inhibits CO-mediated FT expression. (A) qRT–PCR analysis of CO expression in 30-d-old wild-type, rup1-1, rup2-1, rup1-1 rup2-1, and uvr8-6 rup1-1 rup2-1 plants grown under SD + UV on soil. Samples were collected every 3 h; a representative experiment is shown. (ZT) Zeitgeber time; (ZT0) lights on; (ZT8) lights off. (B) GUS assays representing CO promoter activity in 5-d-old wild-type (Col/gCO:GUS) and rup2-1/gCO:GUS seedlings grown in SDs with (+) or without (−) UV-B. (C,D) Quantification of flowering time of wild-type (Col), Col/Pro35S:3HA-CO, and rup2-1/Pro35S:3HA-CO plants grown in SDs with (+) or without (−) UV-B. The flowering time is represented by total leaf number (rosette and cauline leaves; C) and days to bolting (D). Error bars represent standard deviation. n = 16. (E) RUP2 does not affect the diurnal regulation of CO stability in Pro35S:3HA-CO overexpression lines. Immunoblot analysis of 3HA-CO protein level at the indicated Zeitgeber time in 10-d-old Col/Pro35S:3HA-CO and rup2/Pro35S: 3HA-CO plants grown in the absence (SD − UV; top panel) or presence (SD + UV; bottom panel) of UV-B. Actin levels are shown as a loading control; wild type (Col) at ZT7 was added as a control sample for anti-HA specificity. (F) HA-CO ChIP-qPCR using 12-d-old wild type (Col), Col/Pro35S:3HA-CO, and rup2/Pro35S:3HA-CO seedlings grown in SD + UV (ZT8). The numbers of the analyzed DNA fragments indicate the positions of the 5′ base pair of the amplicon relative to the translation start site. ChIP efficiency of DNA associated with HA-CO is presented as the percentage recovered from the total input DNA (% Input). A representative experiment is shown; error bars represent standard deviation of three technical replicates. (G) Relative LUC activity of protoplasts isolated from co-101 and co-101 rup2-1 plants growing under SD + UV. After protoplast transfection with ProFT:fLUC and Pro35S:CO, chemiluminescence was measured at ZT3–ZT4. Error bars represent standard deviation of four independent experiments, each consisting of at least two independent protoplast transfections.
Discussion
Seasonal patterns of flowering are of great importance for the reproductive success of many plants in natural ecosystems as well as in horticulture and agricultural production systems. The impact of day length on flowering has been studied since the discovery of photoperiodism in 1920 (Garner and Allard 1920). In recent decades, the genetic pathways and regulatory proteins that promote flowering in response to photoperiod were largely defined in the model species A. thaliana (Turck et al. 2008; Andres and Coupland 2012; Song et al. 2015). However, most of the work was and still is performed in growth chambers whose light spectrum does not include UV-B, an intrinsic portion of sunlight. Here, using controlled growth environments containing UV-B, we identified and characterized the unanticipated role of RUP2 in photoperiodic flowering control as a crucial repressor of CO activity associated with UVR8-inducible flowering in SDs. RUP2-mediated prevention of flowering thus contributes to the perception of day length by allowing discrimination of SDs from LDs in the presence of UV-B.
CO is a B-box family transcriptional regulator that is a key activator of flowering by inducing FT expression. Thus, the activity of CO is regulated at many levels, including transcription, phosphorylation status, protein stability, and activity (Romera-Branchat et al. 2014; Song et al. 2015; Shim et al. 2017). Under inductive LD conditions, CO accumulates toward the end of the day, forming a complex with the histone-fold domain containing dimeric B and C subunits of nuclear factor Y (NF-Y) (Ben-Naim et al. 2006; Wenkel et al. 2006; Jang et al. 2008; Gnesutta et al. 2017). The CCT domain of CO within the heterotrimeric NF–CO complex conveys binding specificity to the CORE in the FT promoter, thereby promoting FT expression near dusk (Gnesutta et al. 2017). Here, we provide evidence that RUP2 is a major repressor of CO activity under noninductive SD + UV conditions, since rup2 plants flower very early under SD + UV conditions. Moreover, as this early flowering phenotype is suppressed in rup2 uvr8 and rup2 co double mutants, it is thus UVR8- and CO-dependent. RUP2 apparently does not affect CO transcription or CO protein levels, but its repressive activity involves direct interaction with CO. Indeed, CO transcriptional activity is repressed by RUP2, and this effect is detectable at the level of reduced FT expression, FT promoter activity in transient reporter assays, and CO association with the FT promoter in ChIP assays. Interestingly, several CO-interacting proteins were described recently as negative regulators of CO transcriptional activity, acting through recruitment of TOPLESS repressor proteins or through inhibition of CO binding to target genes (Wang et al. 2014, 2016; Nguyen et al. 2015; Zhang et al. 2015; Graeff et al. 2016; Xu et al. 2016; Ordonez-Herrera et al. 2018), the latter of which is similar to our findings for RUP2 activity. It is interesting to note that RUP2 binds to the N-terminal part of CO, which is comprised of two tandem B-box domains. This interaction could directly affect binding of CO to target promoters. Alternately, this interaction may facilitate the binding of a presently unknown repressor of CO and/or may prevent interaction with a positive regulatory interaction partner by blocking the interaction site.
If RUP2 is a general repressor of CO activity in the absence of UV-B, we would expect delayed flowering in RUP2 overexpression lines particularly under LD − UV conditions and early flowering in rup2 plants in SD − UV. Previous work has suggested that overexpression of RUP2/EFO2 results in early flowering in both SDs and LDs (Wang et al. 2011), a phenotype that we, however, did not observe in our experimental conditions using lines for which RUP2 overexpression was clearly confirmed by immunoblot analysis. Furthermore, we did not observe delayed flowering of RUP2 overexpression lines in LD − UV or early flowering of rup2 in SD − UV. This suggests that RUP2 affects photoperiodic flowering very specifically for a distinct UVR8-induced CO activation mechanism. As CO–FT regulation is largely localized to phloem companion cells in the leaf vasculature (Takada and Goto 2003; Turck et al. 2008; Song et al. 2015), the tissue specificity of UVR8 and RUP2 activity in the regulation of flowering remains to be determined as well as the exact mechanism by which UVR8 activates CO.
Interpretation of the lack of a RUP2 overexpression effect in LD + UV is complicated due to the fact that UVR8 activity is fully repressed by RUP2 overexpression (Gruber et al. 2010; Heijde and Ulm 2013). Indeed, RUP2 overexpression lines mimic the phenotype of uvr8-null mutants, and, indeed, no UVR8 monomers and no physiological response were detected in these lines upon UV-B treatment (Gruber et al. 2010; Heijde and Ulm 2013). It is thus clear that UVR8-mediated activation of flowering is impaired at the level of photoreceptor regulation in RUP2 overexpression lines, and an independent effect on CO activity cannot be investigated, as no UVR8-mediated signaling occurs with RUP2 overexpression. Notwithstanding this, it is of note that the role of RUP2 in flowering time regulation seems independent of its role in the regulation of UVR8 activity. This is particularly highlighted by the fact that UVR8 overexpression plants do not show early flowering, although they display a UV-B hypersensitivity similar to that in rup2, as determined by the rosette phenotype. This is further supported by the interaction of RUP2 with CO and its effect on CO transcriptional activity and FT promoter binding.
It is noteworthy that wild type developed slower and flowered later under SD + UV than under SD − UV conditions (e.g., Figs. 1, 4A–C, 5C–E), which is in agreement with a recent report (Dotto et al. 2018). Interestingly, this delayed flowering was partially UVR8-dependent (Fig. 1F,G; Supplemental Fig. S1) and has been linked previously to the age pathway of flowering (Dotto et al. 2018). The potential interplay between the effects of UVR8 signaling on the age and photoperiod pathways remains to be determined; however, it is clear that the effect of RUP2 mutation on the photoperiodic pathway overrides the potential effect of UVR8 hyperactivity in rup2 on the age pathway. Moreover, it is of note that the delay in flowering under UV-B was not detectable in the sun simulator experiment, but the repressor function of RUP2 clearly was (Fig. 2).
Seasonal responses of flowering time assessed in field trials are not always as anticipated based on experiments performed under laboratory conditions (Weinig et al. 2002; Wilczek et al. 2009; Brachi et al. 2010; Andres and Coupland 2012). In part, the absence of UV-B in most laboratory experiments may contribute to this phenomenon; however, such a notion needs to be experimentally further verified. Independent of this, we show that RUP2 loss of function renders the facultative LD species A. thaliana into a day-neutral plant in the presence of UV-B, suggesting that RUP2 is required for flowering time regulation by day length under natural conditions. Thus, although UV-B seems to play a rather minor role in Arabidopsis wild-type flowering, loss of RUP2 exposes an existing UVR8-activated pathway that can efficiently promote flowering in noninductive SDs. It is intriguing to speculate why wild-type Arabidopsis has a pathway to flower in response to UV-B but apparently does not make use of it. A possibility is that the Arabidopsis rup2 mutant revealed a UVR8 flowering pathway that is indeed active in other plant species but repressed in Arabidopsis as a (facultative) LD plant. Alternatively, it remains to be investigated whether RUP2 may integrate other environmental factors to regulate flowering in the field under sunlight, with its intrinsic UV-B. For example, it can be envisaged that RUP2 degradation may be a potent inducer of flowering in noninductive photoperiods, a possibility that deserves further investigation.
Materials and methods
Plant material and growth conditions
The mutants and overexpression lines used in this study were in the A. thaliana Columbia (Col) accession and were described previously as follows: uvr8-6 (Favory et al. 2009), rup1-1, rup2-1, rup2-1/Pro35S:RUP2 (Gruber et al. 2010), cop1-4 (Deng et al. 1992), ft-10 (Yoo et al. 2005), co-101 (Takada and Goto 2003), and Pro35S:3HA-CO line #7 (Song et al. 2012). rup2-2 (SALK_139836) (Alonso et al. 2003) was characterized in this study (Supplemental Fig. S6). The GUS reporter lines used were ProFT:GUS (Takada and Goto 2003), which was introgressed into rup2-1, uvr8-6, and rup2-1 uvr8-6 mutants by genetic crossing, and gCO:GUS (Takada and Goto 2003), which was introgressed into rup2-1. The RUP2 (At5g23730) genomic locus, including an ∼1.5-kb promoter region, was amplified with primers RUP2pFW (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCCACGTATGACTCGTCCTTACTTTGC-3′; the attB1 site is in italic, and the gene-specific sequence is underlined) and RUP2pREV (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCATGAAAACAGAGTAATGACTGTTG C-3′; the attB2 site is in italic, and the gene-specific sequence is underlined), cloned into pDONR207 using Gateway technology (Invitrogen), and sequenced to confirm the integrity of the cloned fragment. The genomic clone was inserted into the binary destination vector pMDC163 (Curtis and Grossniklaus 2003). rup2-1 plants were transformed by Agrobacterium using the floral dip method (Clough and Bent 1998).
For flowering time experiments, quantitative RT–PCR (qRT–PCR), GUS reporter assays, and transient expression assays, seeds were stratified for 2 d at 4°C in the dark, and plants were grown with a day/night temperature cycle of 22°C/18°C in GroBanks (CLF Plant Climatics) with Philips Master TL-D 58W/840 white light fluorescent tubes (120 µmol m−2 s−1, measured with a LI-250 light meter; LI-COR Biosciences) supplemented or not with UV-B from Philips TL40W/01RS narrowband UV-B tubes (0.07 mW cm−2; measured with a VLX-3W ultraviolet light meter equipped with a CX-312 sensor; Vilber Lourmat). Plants were grown under 8-h/16-h light/dark SD or 16-h/8-h light/dark LD conditions, as indicated.
For immunoblot analysis, ChIP, hypocotyl length measurement, and anthocyanin quantification, seeds were surface-sterilized with 70% ethanol and 0.005% Tween 20 and plated on half-strength MS medium (Duchefa) containing 1% sucrose and 0.8% agar. For hypocotyl length measurement and anthocyanin quantification, seedlings were grown as described previously (Oravecz et al. 2006; Favory et al. 2009). For immunoblot analysis, qRT–PCR, and ChIP, seedlings were grown in GroBanks under SD − UV or SD + UV conditions, as indicated.
A sun simulator of the Research Unit Environmental Simulation at the Helmhotz Zentrum München (Thiel et al. 1996) was used to study flowering time regulation under conditions simulating natural light and UV radiation conditions. The condition of the treatment in the sun simulator was similar to that described previously (Favory et al. 2009; Gruber et al. 2010; González Besteiro et al. 2011) with an 8-h day period with mean photosynthetically active radiation (PAR; 400–700 nm) of 600 μmol m−2 s−1 and 6 h of UV-B irradiance with a biologically effective radiation of 308 mW m−2 (weighted by the generalized plant action spectrum [Caldwell 1971] and normalized at 300 nm) (Supplemental Fig. S7). Controls were grown excluding the entire UV radiation spectrum. The temperature was maintained at 23°C during the day and 18°C at night. The relative humidity was kept constant at 60%.
PCR genotyping of mutants and isolation of double mutants
Single mutants were crossed, and the double mutants were identified by PCR genotyping in the F2 generation. rup1-1, rup2-1, and uvr8-6 were genotyped as described previously (Gruber et al. 2010). co-101, ft-10, and rup2-2 were genotyped as follows: co-101: CO101_LP (5′-AGCTCCCACACCATCAAACTTACTACATC-3′) + CO101_RP (5′-AGTCCATACTCGAGTTGTAATCCA-3′) = 0.6 kb for wild type, and CO101_LP + T-DNA primer LB3 (5′-TAGCATCTGAATTTCATAACCAATCTCGATACAC-3′) = 0.45 kb for co-101; ft-10 (GABI_290E08): FT10_LP (5′-ATATTG ATGAATCTCTGTTGTGG-3′) + FT10_RP (5′-AGGGTTGCTAGGACTTGGAACA-3′) = 0.3 kb for wild type, and T-DNA primer 8474 (5′-ATAATAACGCTGCGGACATCTACATTTT-3′) + FT_RP = 0.5 kb for ft-10; and rup2-2 (SALK_139836): RUP2_SALK_139836_LP (5′-TGTTTCGGTGTTACCATTACG-3′) + RUP2_SALK_139836_RP (5′-TCGGATCCCATACTTGCATAG-3′) = 1.0 kb for wild type, and T-DNA primer LBb1.3 (5′-ATTTTGCCGATTTCGGAAC-3′) + RUP2_SALK_139836_ RP = 0.5 kb for rup2-2.
Immunoblot analysis
Proteins were extracted in 50 mM Na-phosphate (pH 7.4), 150 mM NaCl, 10% glycerol, 5 mM EDTA, 1 mM DTT, 0.1% Triton X-100, 50 µM MG132, 2 mM Na3VO4, 2 mM NaF, and 1% (v/v) protease inhibitor mixture for plant extracts (Sigma-Aldrich, P9599). For immunoblot analysis, total cellular proteins were separated by electrophoresis in 10% (w/v) SDS–polyacrylamide gels and transferred to PVDF membranes according to the manufacturer's instructions (iBlot dry blotting system, Thermo Fisher Scientific).
Rabbit polyclonal antibodies were generated against synthetic peptides derived from the RUP2 protein sequence [amino acids 1–15 + C: MNTLHPHKQQQEQAQC; anti-RUP2(1–15)] and were affinity-purified against the peptide (Eurogentec). Anti-RUP2(1–15), anti-UVR8(426–440) (Favory et al. 2009), anti-HA.11 (BioLegend, 901513), and anti-actin (Sigma-Aldrich, A0480) were used as primary antibodies. Horseradish peroxidase (HRP)-conjugated anti-rabbit and anti-mouse immunoglobulins (Dako A/S) were used as the secondary antibodies. Chemiluminescent signals were generated with the ECL Plus Western detection kit and revealed with an ImageQuant LAS 4000 mini-CCD camera system (GE Healthcare).
Yeast two-hybrid interaction assays
A yeast two-hybrid screen was performed using RUP2 as bait fused to the GAL4-binding domain (Matchmaker Gold yeast two-hybrid system, Clontech). The screen was carried out following the standard protocol suggested by the manufacturer.
Arabidopsis RUP1-coding (At5g52250) and RUP2-coding sequences were cloned into yeast two-hybrid plasmid containing a DNA-binding domain (pGBKT7-GW) (Yin et al. 2015), and CO was cloned into plasmid containing an activation domain (pGADT7-GW). Bait and prey constructs were transformed into Saccharomyces cerevisiae strain Y2H Gold and Y187, respectively. To quantify protein–protein interaction, using CPRG as a substrate, yeast growth was carried out directly on the plates as described before (Rizzini et al. 2011), and the assay was performed according to the protocol described in the yeast protocol handbook from Clontech (version PR973283). The lacZ β-galactosidase activity is expressed as Miller units.
Anthocyanin extraction and measurement
Arabidopsis seedlings were grown for 4 d under low narrowband UV-B fields with the appropriated cutoff filters, as described previously (Oravecz et al. 2006; Favory et al. 2009). Fifty milligrams of seedlings was harvested from agar plates and immediately frozen in liquid nitrogen. Sample tissues were processed for 10 sec using a Silamat S5 mixer (Ivoclar Vivadent). Two-hundred-fifty microliters of acidic methanol (1% [w/v] HCl) was added to each sample that was homogenized and placed in an overhead shaker for 1 h at 4°C as described before (Yin et al. 2012). Samples were centrifuged at 14,000 rpm for 1 min, and the supernatant was used to quantify anthocyanin content in a spectrophotometer at 535 and 650 nm. Values are reported as A530 −0.25 (A657) per gram of fresh weight.
Hypocotyl length
Four-day-old Arabidopsis seedlings were grown in the appropriate light conditions, and their hypocotyl lengths were measured (n > 30) using ImageJ software as described previously (Oravecz et al. 2006).
Statistical analysis of flowering time experiments
ANOVA with post-hoc Tukey HSD statistical analyses were performed using the R software package. The means and standard deviations were derived from replicated independent biological samples unless stated otherwise. Shared letters indicate no statistically significant difference in the means (P > 0.05).
Confocal laser scanning microscopy (CLSM) and FLIM analyses
For CLSM and FLIM analyses, the binary 2in1 vectors were used (Hecker et al. 2015). The coding sequences of RUP1 or RUP2 were cloned into the donor plasmid (mEGFP), while CO was cloned into acceptor plasmid (mCherry) using the MultiSite Gateway Technology (Invitrogen). mCherry fused to an NLS was used as a negative control. These constructs were transformed into Agrobacterium tumefaciens strain GV3101 and infiltrated into N. benthamiana leaves as described previously (Hecker et al. 2015). Leaves were subjected to CLSM and FLIM analyses 1–2 d after infiltration.
The measurements were performed as described previously (Hecker et al. 2015). Briefly, all CLSM and FLIM measurements were performed using a Leica TCS SP8 confocal microscope (Leica Microsystems) equipped with a FLIM unit (PicoQuant). Images were acquired using a 63×/1.20 water immersion objective. For the excitation and emission of fluorescent proteins, the following settings were used: mEGFP at excitation 488 nm and emission 495–530 nm; and mCherry at excitation 561 nm and emission 580–630 nm.
FLIM data were derived from measurements of at least 20 nuclei for each fusion protein combination. To excite RUP1-mEGFP and RUP2-mEGFP for FLIM experiments, a 470-nm pulsed laser (LDH-P-C-470) was used, and the corresponding emission was detected with a SMD emission SPFLIM PMT 495–545 nm by time-correlated single-photon counting using a Picoharp 300 module (PicoQuant). Each time-correlated single-photon counting histogram was reconvoluted with the corresponding instrument response function and fitted against a monoexponential decay function for donor-only samples and a biexponential decay function for the other samples to unravel the mEGFP fluorescence lifetime of each nucleus.
The average mEGFP fluorescence lifetimes as well as the standard error values were calculated using Microsoft Excel 2013. Statistical analysis was performed with JMP (version 12.2.0). To test for homogeneity of variance, Levene's test (df = 5/140, F = 26.298, P < 0.0001) was used, and statistical significance was calculated by a two-tailed all-pair Kruskal-Wallis test followed by a Steel-Dwass post hoc correction.
GUS staining
Arabidopsis leaves were fixed in 90% acetone for 30 min. After washing three times in ice-cold water, plant tissues were incubated with staining buffer (0.5 mg/mL 5-bromo-4-chromo-3-indolyl-β-d-glucuronide [X-Glc], 10 mM EDTA, 0.5 mM ferricyanide, 0.5 mM ferrocyanide, 0.1% Triton X-100 in phosphate buffer) for 5 min at 4°C followed by incubation at 37°C. After removal of staining solution, tissue was cleared by successive washes with 75% ethanol. Samples were mounted in glycerol and analyzed using a stereomicroscope (Leica MZ16, Leica Microsystems AG) or a differential interference contrast (DIC) microscope (Zeiss Axioscope II, Carl Zeiss AG, or Nikon Eclipse 80i, Nikon AG).
qRT–PCR
Arabidopsis total RNA was isolated with the plant RNeasy kit according to the manufacturer's instructions (Qiagen), followed by DNase I treatment. In order to inactivate DNase I, 20 mM EDTA was added, and samples were incubated for 10 min at 65°C. Synthesis of the first strand of cDNA was performed using the TaqMan reverse transcription reagent kit according to the manufacturer's standard protocol (Thermo Fisher Scientific). Each qRT–PCR reaction was composed of cDNA synthesized with a 1:1 mixture of oligo(dT) primers and random hexamers from 25 ng of total RNA. PCR reactions were performed using the ABsolute qPCR Rox mix kit (ABgene) and a QuantStudio 5 real-time PCR system (Thermo Fisher Scientific). The following primers were used: for CO (At5g15840), CO_qRT_fw (5′-CCTCAGGGACTCACTACAACG-3′) and CO_qRT_rv (5′-TCTTGGGTGTGAAGCTGTTG-3′), and for FT (At1g65480), FT_qRT_fw (5′-CCAAGAGTTGAGATTGGTGGA-3′) and FT_qRT_rv (5′-ATTGCCAAAGGTTGTTCCAG-3′). The levels of expression of 18S and UBQ10 (Czechowski et al. 2005) were used to normalize the concentrations of the various mRNA samples in which gene expression was analyzed using qbasePLUS real-time PCR data analysis software version 2.4 (Biogazelle). Each reaction was performed in technical triplicates; data shown are representative of at least two independent biological repetitions.
ChIP
Samples were cross-linked in 3% formaldehyde solution in PBS, and cross-linking was quenched with 0.2 M glycine. Nucleus enrichment was performed as described (Fiil et al. 2008). Samples were sonicated in lysis buffer (50 mM Tris-HCL at pH 8, 10 mM EDTA, 1% SDS) and further processed as described (Stracke et al. 2010; Binkert et al. 2014). The chromatin was immunoprecipitated with anti-HA antibody (ChIP-grade; Abcam, ab9110) overnight at 4°C, after which cross-linking was reversed for 2 h at 85°C. DNA was purified using QIAquick PCR purification kit (Qiagen) before analysis with a QuantStudio 5 real-time PCR system (Thermo Fisher Scientific) and the following primer sets: ProFT_-100-Fw (5′-AGAGGGTTCATGCCTATGATA C-3′), ProFT_-100-Rv (5′-CTTTGATCTTGAACAAACAGGTG-3′) (Bu et al. 2014), ProFT_-1185-Fw (5′-TTATCCTGGTCGTGCAAATG-3′), and ProFT_-1185-Rv (5′-CAAGCGGCCATATTATGGAA-3′) (Song et al. 2012). qPCR data were analyzed according to the percentage of input method (Haring et al. 2007). Each reaction was performed in technical triplicates; data shown are representative of three independent biological repetitions.
Transient expression assays in protoplasts
For the ProFT:fLUC reporter construct, the FT promoter region (−1 to −5722) was amplified with primers oVCG-475 (5′-CCCCCCTCGAGGTCGACATTTGCTGAACAAAAATCTATT-3′; the XhoI site is in italic, and the gene-specific sequence is underlined) and oVCG-476 (5′-GGTGGCGGCCGCTCTAGCTTTGATCTTGAACAAACAGGTG-3′; the NotI site is in italic, and the gene-specific sequence is underlined) from the BAC clone F5I14 and cloned into pGREENII 0800-LUC XhoI/NotI restriction sites (Hellens et al. 2005).
Protoplasts were isolated from 4- to 8-wk-old co-101 and rup2-1 co-101 plants growing under SD + UV. Expanded leaves were harvested, and protoplasts were prepared as described previously (Wu et al. 2009). Each protoplast transfection was performed with 5 µg of ProFT:fLUC and Pro35S:CO plasmids and incubated overnight in darkness at 21°C. Luciferase assay was performed with the dual-luciferase reporter assay system (Promega) at Zeitgeber time (ZT) 3–4 (ZT0 = lights on, ZT8 = lights off) following the manufacturer's instructions and a GloMax 96 Microplate Luminometer (Promega). Relative luciferase activity corresponds to normalized firefly/Renilla ratio.
Supplementary Material
Acknowledgments
We thank Takato Imaizumi and Koji Goto for providing plant material, Christopher Grefen for the binary 2in1 vectors, Hongtao Liu for the pGREENII 0800-LUC construct, Rodrigo S. Reis for technical assistance with protoplast transient expression assays, Isabelle Fleury for contributing some of the crosses, Stefanie Mühlhans for excellent technical assistance in sun simulator experiments, and Michael Hothorn for helpful comments on the manuscript. This work was supported by the University of Geneva, the Swiss National Science Foundation (grant nos. 31003A_175774 to R.U., and CRSII3_154438 to R.U. and C.F.), the European Research Council (ERC) under the European Union's Seventh Framework Programme (grant no. 310539 to R.U.), and the German Research Foundation (grant CRC 1101-D02 to K.H.). V.C.G. was supported by an EMBO long-term fellowship (ALTF 293-2013).
Author contributions: A.B.A. and R.U. conceived and designed the study. A.B.A. performed all of the experiments reported here except for the following: N.G. and K.H. contributed the FRET-FLIM data (Fig. 3B,C), A.A. and J.B.W. contributed the sun simulator data (Fig. 2; Supplemental Fig. S7), M.P. contributed the ChIP data (Fig. 6F), S.C. contributed the protein immunoblots (Fig. 6E; Supplemental Figs. S3C, S4D), and V.C.G. and C.F. contributed the transient expression assays in protoplasts (Fig. 6G). R.U. supervised the research, and A.B.A. and R.U. wrote the manuscript with input from all authors.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.318592.118.
Freely available online through the Genes & Development Open Access option.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Andres F, Coupland G. 2012. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639. [DOI] [PubMed] [Google Scholar]
- Ben-Naim O, Eshed R, Parnis A, Teper-Bamnolker P, Shalit A, Coupland G, Samach A, Lifschitz E. 2006. The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J 46: 462–476. [DOI] [PubMed] [Google Scholar]
- Binkert M, Kozma-Bognar L, Terecskei K, De Veylder L, Nagy F, Ulm R. 2014. UV-B-responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. Plant Cell 26: 4200–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachi B, Faure N, Horton M, Flahauw E, Vazquez A, Nordborg M, Bergelson J, Cuguen J, Roux F. 2010. Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genet 6: e1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BA, Cloix C, Jiang GH, Kaiserli E, Herzyk P, Kliebenstein DJ, Jenkins GI. 2005. A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci 102: 18225–18230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Z, Yu Y, Li Z, Liu Y, Jiang W, Huang Y, Dong AW. 2014. Regulation of Arabidopsis flowering by the histone mark readers MRG1/2 via interaction with CONSTANS to modulate FT expression. PLoS Genet 10: e1004617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell MM. 1971. Solar UV irradiation and the growth and development of higher plants. In Photophysiology (ed. Giese AC), pp. 131–177. Academic Press, New York. [Google Scholar]
- Cloix C, Kaiserli E, Heilmann M, Baxter KJ, Brown BA, O'Hara A, Smith BO, Christie JM, Jenkins GI. 2012. C-terminal region of the UV-B photoreceptor UVR8 initiates signaling through interaction with the COP1 protein. Proc Natl Acad Sci 109: 16366–16370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. 2007. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033. [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. 2003. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XW, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, Quail PH. 1992. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a Gβ homologous domain. Cell 71: 791–801. [DOI] [PubMed] [Google Scholar]
- Dotto M, Gomez MS, Soto MS, Casati P. 2018. UV-B radiation delays flowering time through changes in the PRC2 complex activity and miR156 levels in Arabidopsis thaliana. Plant Cell Environ 41: 1394–1406. [DOI] [PubMed] [Google Scholar]
- Favory JJ, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, Albert A, Cloix C, Jenkins GI, Oakeley EJ, et al. 2009. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J 28: 591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiil BK, Qiu JL, Petersen K, Petersen M, Mundy J. 2008. Coimmunoprecipitation (co-IP) of nuclear proteins and chromatin immunoprecipitation (ChIP) from Arabidopsis. CSH Protoc 2008: pdb prot5049. [DOI] [PubMed] [Google Scholar]
- Findlay KM, Jenkins GI. 2016. Regulation of UVR8 photoreceptor dimer/monomer photo-equilibrium in Arabidopsis plants grown under photoperiodic conditions. Plant Cell Environ 39: 1706–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner WW, Allard HA. 1920. Effect of the relative length of day and night and other factors of the environment on growth and reproduction in plants. J Agric Res 18: 553–606. [Google Scholar]
- Gnesutta N, Kumimoto RW, Swain S, Chiara M, Siriwardana C, Horner DS, Holt BF III, Mantovani R. 2017. CONSTANS imparts DNA sequence specificity to the histone fold NF-YB/NF-YC dimer. Plant Cell 29: 1516–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González Besteiro MA, Bartels S, Albert A, Ulm R. 2011. Arabidopsis MAP kinase phosphatase 1 and its target MAP kinases 3 and 6 antagonistically determine UV-B stress tolerance, independent of the UVR8 photoreceptor pathway. Plant J 68: 727–737. [DOI] [PubMed] [Google Scholar]
- Graeff M, Straub D, Eguen T, Dolde U, Rodrigues V, Brandt R, Wenkel S. 2016. MicroProtein-mediated recruitment of CONSTANS into a TOPLESS trimeric complex represses flowering in Arabidopsis. PLoS Genet 12: e1005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber H, Heijde M, Heller W, Albert A, Seidlitz HK, Ulm R. 2010. Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc Natl Acad Sci 107: 20132–20137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Yang H, Mockler TC, Lin C. 1998. Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363. [DOI] [PubMed] [Google Scholar]
- Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M. 2007. Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker A, Wallmeroth N, Peter S, Blatt MR, Harter K, Grefen C. 2015. Binary 2in1 vectors improve in planta (co)localization and dynamic protein interaction studies. Plant Physiol 168: 776–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijde M, Ulm R. 2013. Reversion of the Arabidopsis UV-B photoreceptor UVR8 to the homodimeric ground state. Proc Natl Acad Sci 110: 1113–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. 2005. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Ouyang X, Yang P, Lau OS, Chen L, Wei N, Deng XW. 2013. Conversion from CUL4-based COP1–SPA E3 apparatus to UVR8–COP1–SPA complexes underlies a distinct biochemical function of COP1 under UV-B. Proc Natl Acad Sci 110: 16669–16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA. 2007. FT protein acts as a long-range signal in Arabidopsis. Curr Biol 17: 1050–1054. [DOI] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G. 2008. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27: 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GI. 2017. Photomorphogenic responses to ultraviolet-B light. Plant Cell Environ 40: 2544–2557. [DOI] [PubMed] [Google Scholar]
- Khanna R, Kronmiller B, Maszle DR, Coupland G, Holm M, Mizuno T, Wu SH. 2009. The Arabidopsis B-box zinc finger family. Plant Cell 21: 3416–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Deng XW. 2012. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci 17: 584–593. [DOI] [PubMed] [Google Scholar]
- Laubinger S, Marchal V, Gentilhomme J, Wenkel S, Adrian J, Jang S, Kulajta C, Braun H, Coupland G, Hoecker U. 2006. Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133: 3213–3222. [DOI] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ. 2008. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Warthmann N, Kuttner F, Schmid M. 2007. Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr Biol 17: 1055–1060. [DOI] [PubMed] [Google Scholar]
- McNellis TW, von Arnim AG, Araki T, Komeda Y, Misera S, Deng XW. 1994. Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen KT, Park J, Park E, Lee I, Choi G. 2015. The Arabidopsis RING domain protein BOI inhibits flowering via CO-dependent and CO-independent mechanisms. Mol Plant 8: 1725–1736. [DOI] [PubMed] [Google Scholar]
- Oravecz A, Baumann A, Mate Z, Brzezinska A, Molinier J, Oakeley EJ, Adam E, Schafer E, Nagy F, Ulm R. 2006. CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18: 1975–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordonez-Herrera N, Trimborn L, Menje M, Henschel M, Robers L, Kaufholdt D, Hansch R, Adrian J, Ponnu J, Hoecker U. 2018. The transcription factor COL12 is a substrate of the COP1/SPA E3 ligase and regulates flowering time and plant architecture. Plant Physiol 176: 1327–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolec R, Ulm R. 2018. Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr Opin Plant Biol 45: 18–25. [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. 1995. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857. [DOI] [PubMed] [Google Scholar]
- Rizzini L, Favory JJ, Cloix C, Faggionato D, O'Hara A, Kaiserli E, Baumeister R, Schafer E, Nagy F, Jenkins GI, et al. 2011. Perception of UV-B by the Arabidopsis UVR8 protein. Science 332: 103–106. [DOI] [PubMed] [Google Scholar]
- Romera-Branchat M, Andres F, Coupland G. 2014. Flowering responses to seasonal cues: what's new? Curr Opin Plant Biol 21: 120–127. [DOI] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. 2000. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616. [DOI] [PubMed] [Google Scholar]
- Shim JS, Kubota A, Imaizumi T. 2017. Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol 173: 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T. 2012. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336: 1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Shim JS, Kinmonth-Schultz HA, Imaizumi T. 2015. Photoperiodic flowering: time measurement mechanisms in leaves. Annu Rev Plant Biol 66: 441–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Favory JJ, Gruber H, Bartelniewoehner L, Bartels S, Binkert M, Funk M, Weisshaar B, Ulm R. 2010. The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ 33: 88–103. [DOI] [PubMed] [Google Scholar]
- Takada S, Goto K. 2003. TERMINAL FLOWER2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell 15: 2856–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel S, Döhring T, Köfferlein M, Kosak A, Martin P, Seidlitz HK. 1996. A phytotron for plant stress research: how far can artificial lighting compare to natural sunlight? J Plant Physiol 148: 456–463. [Google Scholar]
- Tiwari SB, Shen Y, Chang HC, Hou Y, Harris A, Ma SF, McPartland M, Hymus GJ, Adam L, Marion C, et al. 2010. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol 187: 57–66. [DOI] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G. 2008. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59: 573–594. [DOI] [PubMed] [Google Scholar]
- Ulm R, Baumann A, Oravecz A, Mate Z, Adam E, Oakeley EJ, Schafer E, Nagy F. 2004. Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci 101: 1397–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yang D, Feldmann KA. 2011. EFO1 and EFO2, encoding putative WD-domain proteins, have overlapping and distinct roles in the regulation of vegetative development and flowering of Arabidopsis. J Exp Bot 62: 1077–1088. [DOI] [PubMed] [Google Scholar]
- Wang CQ, Guthrie C, Sarmast MK, Dehesh K. 2014. BBX19 interacts with CONSTANS to repress FLOWERING LOCUS T transcription, defining a flowering time checkpoint in Arabidopsis. Plant Cell 26: 3589–3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Pan J, Li Y, Lou D, Hu Y, Yu D. 2016. The DELLA–CONSTANS transcription factor cascade integrates gibberellic acid and photoperiod signaling to regulate flowering. Plant Physiol 172: 479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinig C, Ungerer MC, Dorn LA, Kane NC, Toyonaga Y, Halldorsdottir SS, Mackay TF, Purugganan MD, Schmitt J. 2002. Novel loci control variation in reproductive timing in Arabidopsis thaliana in natural environments. Genetics 162: 1875–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G. 2006. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18: 2971–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. 2005. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059. [DOI] [PubMed] [Google Scholar]
- Wilczek AM, Roe JL, Knapp MC, Cooper MD, Lopez-Gallego C, Martin LJ, Muir CD, Sim S, Walker A, Anderson J, et al. 2009. Effects of genetic perturbation on seasonal life history plasticity. Science 323: 930–934. [DOI] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS. 2009. Tape–Arabidopsis sandwich—a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Li T, Xu PB, Li L, Du SS, Lian HL, Yang HQ. 2016. DELLA proteins physically interact with CONSTANS to regulate flowering under long days in Arabidopsis. FEBS Lett 590: 541–549. [DOI] [PubMed] [Google Scholar]
- Yin R, Messner B, Faus-Kessler T, Hoffmann T, Schwab W, Hajirezaei MR, von Saint Paul V, Heller W, Schaffner AR. 2012. Feedback inhibition of the general phenylpropanoid and flavonol biosynthetic pathways upon a compromised flavonol-3-O-glycosylation. J Exp Bot 63: 2465–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R, Arongaus AB, Binkert M, Ulm R. 2015. Two distinct domains of the UVR8 photoreceptor interact with COP1 to initiate UV-B signaling in Arabidopsis. Plant Cell 27: 202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SK, Chung KS, Kim J, Lee JH, Hong SM, Yoo SJ, Yoo SY, Lee JS, Ahn JH. 2005. CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol 139: 770–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang L, Zeng L, Zhang C, Ma H. 2015. Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev 29: 975–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z, Liu H, Liu B, Liu X, Lin C. 2011. Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol 21: 841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.