Here, Priesnitz and Becker review the pathways that adjust mitochondrial protein synthesis and import of nuclear-encoded subunits to the assembly of respiratory chain complexes and outline how defects in protein import into mitochondria affect nuclear gene expression to maintain protein homeostasis under physiological and stress conditions.
Keywords: mitochondria, respiratory chain, protein sorting, mitochondrial gene expression
Abstract
Mitochondria contain their own genome that encodes for a small number of proteins, while the vast majority of mitochondrial proteins is produced on cytosolic ribosomes. The formation of respiratory chain complexes depends on the coordinated biogenesis of mitochondrially encoded and nuclear-encoded subunits. In this review, we describe pathways that adjust mitochondrial protein synthesis and import of nuclear-encoded subunits to the assembly of respiratory chain complexes. Furthermore, we outline how defects in protein import into mitochondria affect nuclear gene expression to maintain protein homeostasis under physiological and stress conditions.
Mitochondria originated from the incorporation of a prokaryote similar to existing α-proteobacteria by a eukaryotic ancestor cell >1.5 billion years ago (Zimorski et al. 2014; Archibald 2015). During the course of evolution, the vast majority of genetic information of the endosymbiont was transferred to the host nuclear genome. Mitochondria contain ∼1000 proteins in baker's yeast Saccharomyces cerevisiae and 1500 proteins in humans (Sickmann et al. 2003; Pagliarini et al. 2008; Morgenstern et al. 2017). About 99% of the mitochondrial proteins are synthesized as precursors on cytosolic ribosomes and imported into the target organelle by dedicated protein translocases. Mitochondria retain their own genome that encodes eight proteins in yeast (Table 1) and 13 proteins in humans (Hällberg and Larsson 2014; Ott et al. 2016). Remarkably, about one quarter of the mitochondrial proteins in yeast (∼250 proteins) are involved in expression and maintenance of the mitochondrial genome, reflecting the central role of the gene products for mitochondrial function (Sickmann et al. 2003; Morgenstern et al. 2017).
Table 1.
Translational regulators in yeast mitochondria
Mitochondria fulfill many functions for cellular metabolism. Biosynthesis of lipids and amino acids, degradation of fatty acids, formation of iron-sulfur clusters, and reaction steps of heme biosynthesis and the urea cycle occur within mitochondria. The most prominent function of mitochondria is the production of the bulk of cellular ATP by oxidative phosphorylation. In this process, respiratory chain complexes of the inner membrane transport electrons from the reducing equivalents NADH and FADH2 to oxygen to produce water. The respiratory chain complexes use the released energy from the electron transport process to establish a proton gradient across the inner membrane. The proton gradient drives the activity of the F1Fo-ATP synthase (ATP synthase) to generate ATP from ADP and phosphate by a complex molecular mechanism. In humans, the respiratory chain is composed of four multisubunit protein complexes: the NADH dehydrogenase (complex I), succinate dehydrogenase (complex II), cytochrome bc1 complex (complex III), and cytochrome c oxidase (complex IV). Mitochondria of baker's yeast S. cerevisiae lack a canonical complex I but contain three alternative NADH dehydrogenases in the inner membrane that do not pump protons across the inner membrane: The internal NADH dehydrogenase (Ndi1) oxidizes NADH produced inside mitochondria, whereas two external NADH dehydrogenases (Nde1 and Nde2) oxidize cytosolic NADH (Marres et al. 1991; Luttik et al. 1998; Small and McAlister-Henn 1998). A typical feature of the mitochondrial respiratory chain is the organization of the cytochrome bc1 complex, cytochrome c oxidase, and mammalian NADH dehydrogenase in respiratory chain supercomplexes (Cruciat et al. 2000; Schägger and Pfeiffer 2000; Böttinger et al. 2012; Gu et al. 2016; Letts et al. 2016).
Respiratory chain complexes I, III, and IV as well as the ATP synthase contain proteins of dual genetic origin. Mitochondrially encoded proteins constitute reactive centers and associate with many nuclear-encoded proteins in functional protein machineries. The assembly of these protein complexes is a complicated process involving a large number of assembly factors as well as the coordinated expression of nuclear and mitochondrial genes (Fox 2012; Smith et al. 2012; Dennerlein et al. 2017; Song et al. 2018; Timón-Gómez et al. 2018). Malfunctions in this process cause an accumulation of unassembled subunits or vestigial complexes, which in turn leads to increased levels of reactive oxygen species that are harmful to the cell (Khalimonchuk et al. 2007; Zara et al. 2007; He et al. 2018). Defects in the formation of respiratory chain complexes have been linked to several diseases such as neurodegenerative disorders and cardiomyopathies (Shoubridge 2001; Smeitink et al. 2006). A recent study revealed that cytosolic protein synthesis and mitochondrial protein synthesis are synchronized in yeast cells (Couvillion et al. 2016). How the expression of the two different genomes is coordinated is poorly understood. In this review, we highlight recent findings that shed light into this fundamental biological question. We describe molecular circuits that adjust mitochondrial protein production to the assembly stage of respiratory chain complexes and summarize the novel concept that the rate of mitochondrial translation can be adjusted to the import of nuclear-encoded proteins. Finally, we outline the critical role of protein import pathways in the regulation of mitochondrial protein homeostasis during cellular signaling processes.
Expression of the mitochondrial genome
Depending on the organism and cell type, eukaryotic cells may contain up to many thousands of copies of mitochondrial DNA (mtDNA). Due to the presence of noncoding sequences, the size of the mitochondrial genome is extremely variable between species. Human and yeast mitochondrial genomes contain 16.5 and 75 kb, respectively (Hällberg and Larsson 2014; Ott et al. 2016). The two strands of mtDNA can be separated by density centrifugation and are therefore termed heavy and light strands (Battey and Clayton 1978). The circular mtDNA is densely packed into nucleoids with proteins such as human transcription factor A (TFAM). Replication, repair, and transcription of mtDNA take place within the nucleoid structure (Hällberg and Larsson 2014; Pearce et al. 2017). Transcription of the light and heavy strands of mtDNA by the mitochondrial RNA polymerase results in the formation of two polycistronic transcripts. In human mitochondria, maturation of the primary transcripts occurs in RNA granules. Here, the RNA transcripts are processed by RNase P at the 5′ end and by ELAC protein 2 at the 3′ end to form monocistronic and bicistronic transcripts that are further modified by polyadenylation and nucleotide modification (Hällberg and Larsson 2014; Pearce et al. 2017).
Distinct mechanisms control transcription of the mitochondrial genes. Several specific transcriptional factors localize to mitochondria to regulate expression of the mitochondrial genome (Quirós et al. 2016; Pearce et al. 2017). Recently, subunits of the nonspecific lethal (NSL) complex, including the acetyl transferase MOF (males absent on the first), were found in mitochondria of human cell lines (Chatterjee et al. 2016). The NSL complex binds to the promoter region of thousands of genes to regulate their expression in the nucleus (Raja et al. 2010). Human knockdown cell lines of central NSL components such as MOF contained reduced levels of mitochondrial transcripts and displayed a decreased translation rate of mitochondrial genes in vitro. Expression of a mitochondrion targeted MOF variant restores mitochondrial transcript levels and respiratory activity in the MOF knockdown cell lines (Chatterjee et al. 2016). The dual localization of the NSL complex points to a novel mechanism to fine-tune nuclear and mitochondrial gene expression. Finally, the stability of mRNA molecules can be modulated. Transcripts that are not used for translation are rapidly degraded. Several factors increase the stability of mRNA to promote protein synthesis. For instance, the Leu-rich pentatricopeptide motif-containing protein LRPPRC together with its partner protein, stem–loop-interacting RNA-binding protein (SLIRP), binds to mRNA molecules to prevent their degradation and stimulate their polyadenylation. In the absence of LRPPRC, orphan mRNAs are destabilized, leading to a reduction in gene expression and the neurodegenerative disease French Canadian type of Leigh syndrome (Sasarman et al. 2010; Chujo et al. 2012; Siira et al. 2017).
Mitochondrial protein biosynthesis
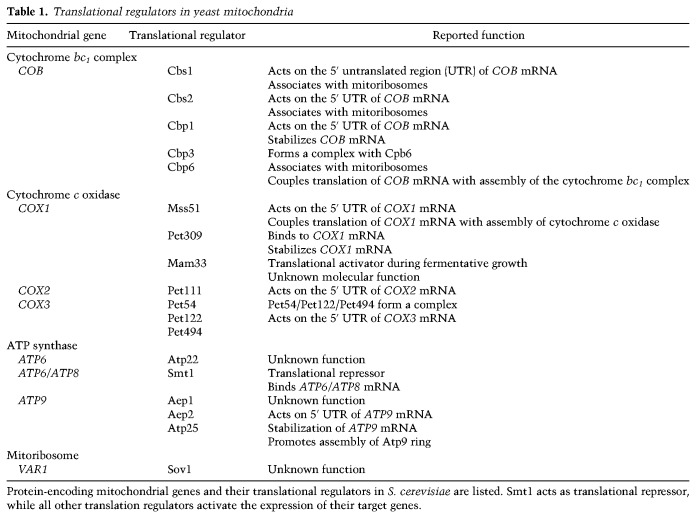
Mitochondria derived ribosomes from their bacterial progenitor. Recent structural analysis revealed several specific features of these mitoribosomes that are distinct from their bacterial counterparts (Amunts et al. 2015; Greber et al. 2015; Desai et al. 2017). Human mitoribosomes are composed of a 39S large subunit and a 28S small subunit and contain an unusually low ratio of ribonucleotides to proteins (30:70). They are composed of ∼80 proteins and two ribosomal RNA (rRNA) molecules. Remarkably, about half of the mitoribosomal proteins are not present in bacterial ribosomes (Ott et al. 2016; Dennerlein et al. 2017). Proteomic and superresolution microscopy studies revealed that yeast mitoribosomes and proteins involved in post-transcriptional maturation of RNA molecules associate in large molecular structures, termed MIOREX (mitochondrial organization of gene expression) (Kehrein et al. 2015). Mitoribosomes are specialized for the synthesis of the membrane-bound respiratory chain subunits and therefore predominantly associate with the inner membrane. Specific receptor proteins mediate their attachment to the membrane. In yeast, Mba1, Oxa1 (oxidase assembly), and Mdm38 contribute to docking of mitoribosomes to the inner membrane (Jia et al. 2003; Szyrach et al. 2003; Frazier et al. 2006; Ott et al. 2006; Bauerschmitt et al. 2010; Pfeffer et al. 2015), while MRPL45 forms the inner membrane tether of human mitoribosomes (Greber et al. 2014). The membrane anchor brings the polypeptide exit tunnel of the mitoribosome in close proximity to the protein insertase Oxa1 that cotranslationally integrates mitochondrially encoded proteins into the inner membrane (Fig. 1; Hell et al. 2001).
Figure 1.
Biogenesis of mitochondrial respiratory chain subunits. The majority of nuclear-encoded subunits of the respiratory chain is synthesized by cytosolic ribosomes as precursors and imported into mitochondria via the translocase of the mitochondrial outer membrane (TOM complex) and the presequence translocase (TIM23 complex). Transport into the mitochondrial matrix additionally requires the ATP-dependent activity of the presequence translocase-associated motor (PAM). The membrane potential (Δψ) across the inner membrane drives protein translocation via the presequence pathway. The OXA1 complex inserts mitochondrially encoded subunits into the inner membrane in a cotranslational manner. Finally, mitochondrially encoded and nuclear-encoded subunits assemble into functional respiratory chain complexes. (IM) Inner membrane; (IMS) intermembrane space; (OM) outer membrane.
Yeast translational activators regulate mitochondrial translation
In yeast, specific translational activators have been identified for all mitochondrial mRNA molecules (Table 1). The molecular mechanisms underlying how the translational activators stimulate protein synthesis are not well understood. Different functions have been assigned to the individual translational activators. Yeast mitochondrial transcripts contain a 5′ untranslated region (UTR) that is a target for translational regulation by translational activators. It has been shown that several translational activators act on the 5′ UTR of the transcript to promote translation of mitochondrial mRNAs, while other translational activators stabilize transcripts or interact with ribosomes (Herrmann et al. 2013; Ott et al. 2016; Dennerlein et al. 2017; Timón-Gómez et al. 2018). Strikingly, superresolution microscopy and cryo-immunogold electron microscopy revealed that the translational activators of the cytochrome bc1 complex, the cytochrome c oxidase, and the ATP synthase colocalize with early assembly intermediates at the mitochondrial inner membrane (Stoldt et al. 2018). This observation indicates that the production of mitochondrial proteins is spatially linked to their assembly into mature protein machineries. Indeed, some translational activators are involved in the coordination of mitochondrial translation with the assembly of respiratory chain complexes as outlined below.
The cytochrome bc1 complex
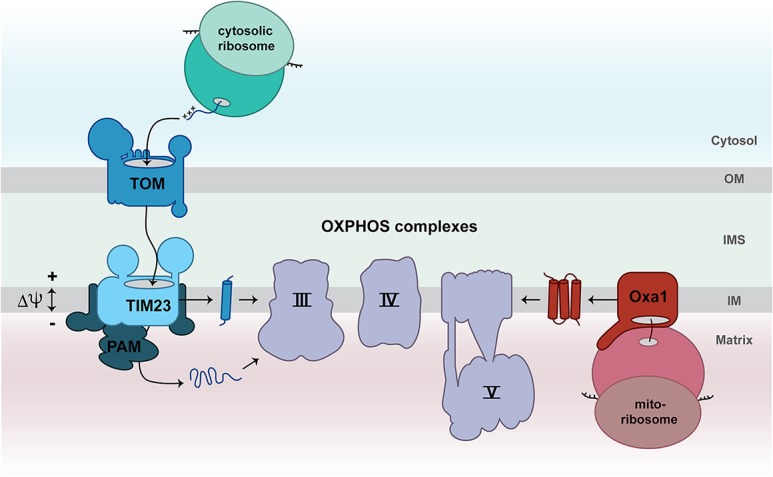
The cytochrome bc1 complex consists of 10 subunits in yeast and 11 subunits in human mitochondria. Crystal structures of the mammalian and yeast complex III have been reported (Xia et al. 1997; Iwata et al. 1998; Lange and Hunte 2002). The only mitochondrially encoded subunit, cytochrome b, is embedded via eight transmembrane spans into the inner membrane and contains two heme b molecules that form reactive centers of the cytochrome bc1 complex. The biogenesis of cytochrome b is critical for the formation of mature complex III. The translational activators Cbs1, Cbs2, Cbp1, and the Cpb3/Cbp6 complex control the translation of COB mRNA encoding cytochrome b in yeast mitochondria (Table 1; Ott et al. 2016; Dennerlein et al. 2017). Interestingly, the Cbp3/Cbp6 complex is present in two pools. One population of Cbp3/Cbp6 interacts with mitoribosomes in close proximity to the polypeptide exit tunnel to stimulate synthesis of cytochrome b by an unknown mechanism (Gruschke et al. 2011; García-Guerrero et al. 2018). A second pool of Cbp3/Cbp6 remains associated with cytochrome b after its synthesis is completed to keep the protein in a conformation that allows insertion of the first heme b molecule and binding of the assembly factor Cbp4 (Fig. 2; Gruschke et al. 2012; Hildenbeutel et al. 2014). The integration of the second heme b molecule into cytochrome b and the association of the first two nuclear-encoded complex III subunits (Qcr7 and Qcr8) contribute to the dissociation of Cbp3/Cbp6 from the assembly intermediate (Gruschke et al. 2012; Hildenbeutel et al. 2014). Subsequently, further subunits assemble in a stepwise manner to form the mature cytochrome bc1 complex (Fox 2012; Smith et al. 2012). The released Cbp3/Cbp6 complex in turn can stimulate a new round of translation of COB mRNA (Fig. 2; Gruschke et al. 2012). Upon failure of hemylation of cytochrome b or impaired assembly with nuclear-encoded partner proteins, Cbp3/Cbp6 remains associated with the intermediate and is not available to stimulate a further round of COB mRNA translation (Hildenbeutel et al. 2014). The Cbp3/Cbp6-mediated regulatory feedback loop is an elegant process to adapt cytochrome b biosynthesis to the availability of heme and nuclear-encoded subunits of the cytochrome bc1 complex.
Figure 2.
Feedback loop regulation of cytochrome b synthesis. The translational activators Cbp1, Cbs1, Cbs2, and Cbp3/Cbp6 promote the translation of COB mRNA to produce cytochrome b (Cyt b) in yeast mitochondria. The Cbp3/Cbp6 complex remains bound to membrane-inserted cytochrome b and facilitates insertion of heme b and association of the assembly factor Cbp4. Binding of the nuclear-encoded subunits Qcr8 and Qcr7 as well as the insertion of the second heme b molecule into cytochrome b promote dissociation of the Cbp3/Cbp6 complex. The released Cbp3/Cbp6 complex can stimulate further rounds of translation of COB mRNA. (b) Heme b.
Cytochrome c oxidase
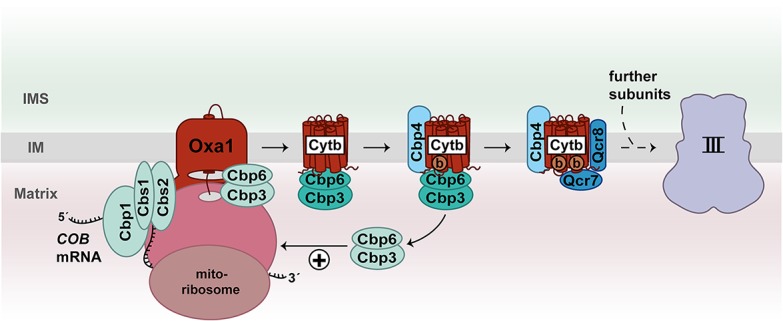
Three mitochondrially encoded subunits (Cox1, Cox2, and Cox3) form the reactive core of complex IV and associate with nuclear-encoded proteins to form a functional cytochrome c oxidase (Tsukihara et al. 1996). In yeast, specific translational activators control the synthesis of Cox1 (Mss51, Pet309, and Mam33), Cox2 (Pet111), and Cox3 (Pet54, Pet122, and Pet494) (Table 1; Herrmann et al. 2013; Ott et al. 2016; Dennerlein et al. 2017). The association of Cox1, Cox2, and Cox3 with nuclear-encoded subunits during formation of the cytochrome c oxidase is a complicated process and involves >30 assembly factors (Fox 2012; Dennerlein et al. 2017; Timón-Gómez et al. 2018). The biogenesis of Cox1 is a key starting point for the de novo formation of complex IV. The protein is embedded into the inner membrane via 12 transmembrane segments and contains two heme a molecules and one copper as prosthetic groups. Mss51 is a central regulator of Cox1 biogenesis. One pool of Mss51 binds to the 5′ UTR of the COX1 mRNA to stimulate its translation, while a second Mss51 fraction associates with newly synthesized Cox1 that is already integrated into the inner membrane (Fig. 3; Decoster et al. 1990; Pérez-Martínez et al. 2003; Barrientos et al. 2004; Mick et al. 2010). Binding of Mss51 and the assembly factors Coa3 and Cox14 protects the nascent Cox1 from degradation. Upon inhibition of downstream assembly steps of complex IV, this intermediate form accumulates and sequesters Mss51 to down-regulate further translation of COX1 mRNA (Barrientos et al. 2004; Pérez-Martínez et al. 2009; Mick et al. 2010; Fontanesi et al. 2011). How Mss51 is released from the Cox1-containing intermediate is not entirely understood. According to current models, the Shy1-mediated insertion of heme a and the assembly of nuclear-encoded complex IV subunits Cox5a and Cox6 contribute to dissociation of Mss51 (Pérez-Martínez et al. 2009; Khalimonchuk et al. 2010; Mick et al. 2010; Fontanesi et al. 2011). The dynamic distribution of Mss51 couples mitochondrial translation of COX1 mRNA to the assembly status of the cytochrome c oxidase (Fig. 3). Strikingly, the function of Mss51 as a translational activator of Cox1 depends on its bound heme b cofactor (Soto et al. 2012). Thus, Mss51 could sense the mitochondrial heme content to fine-tune translation of COX1 mRNA (Soto et al. 2012). Altogether, synthesis of Cox1 is tightly controlled by the availability of heme and its assembly with partner proteins.
Figure 3.
Feedback loop regulation of Cox1 synthesis. The translational activators Pet309, Mam33, and Mss51 promote translation of COX1 mRNA in yeast mitochondria. Mss51 as well as the assembly factors Cox14 and Coa3 bind to membrane-inserted Cox1 in an early assembly intermediate of complex IV. Both populations of Mss51 bind to mtHsp70. The biogenesis factors Coa1 and Shy1 are added to the intermediate complex followed by association of the first nuclear-encoded subunits. At this stage, Mss51/mtHsp70 is released and can stimulate a new round of Cox1 translation. (b) Heme b.
ATP synthase
The ATP synthase consists of a matrix-located soluble F1 domain and membrane-integrated Fo domain. The F1 domain contains the catalytic centers and is linked via a central and peripheral stalk to the Fo rotor domain (Rubinstein et al. 2003; Lau et al. 2008; Hahn et al. 2016; Guo et al. 2017). The overall composition of the ATP synthase in human and yeast mitochondria is largely similar (He et al. 2018; Song et al. 2018). In yeast, the mitochondrial genome encodes ATP synthase subunits Atp6, Atp8, and Atp9. Ten Atp9 subunits form the Fo rotor domain. The association of the Fo rotor domain with Atp6 leads to the formation of the proton-conducting channel (Stock et al. 1999) and represents a critical step in the assembly of the ATP synthase (Rak et al. 2011; Naumenko et al. 2017). According to current models, formation of the proton-conducting channel of ATP synthase occurs in intermediate forms that contain the F1 domain and peripheral stalk to prevent unimpeded proton leakage across the inner membrane. Consequently, the assembly process of the ATP synthase has to be strictly coordinated with the synthesis of mitochondrially encoded subunits (Rak et al. 2009; Fox 2012; Rühle and Leister 2015; Dennerlein et al. 2017). So far, little is known about the underlying molecular pathway. Interestingly, unassembled F1 domains stimulate the production of Atp6 and Atp8 by an unknown mechanism (Rak and Tzagoloff 2009). The protein Smt1 binds to the bicistronic ATP6/ATP8 mRNA to repress its translation (Rak et al. 2016). It was speculated that the unassembled F1 domain could remove Smt1 from the mRNA, enabling the association of Atp22 that in turn stimulates synthesis of Atp6 and Atp8 (Fig. 4; Helfenbein et al. 2003; Rak et al. 2016). Future experimental work has to provide insights into the molecular mechanism by which Smt1 regulates translation of the ATP6/ATP8 mRNA. The INA complex promotes the final formation of the proton-conducting channel by assembling the Fo rotor domain with an Atp6/Atp8-containing module of the ATP synthase (Naumenko et al. 2017). Interestingly, in the absence of the INA complex, the in organello synthesis of Atp9 is reduced (Naumenko et al. 2017). Whether a molecular switch exists that controls the translation of ATP9 mRNA in response to the assembly stage of the ATP synthase remains to be investigated.
Figure 4.
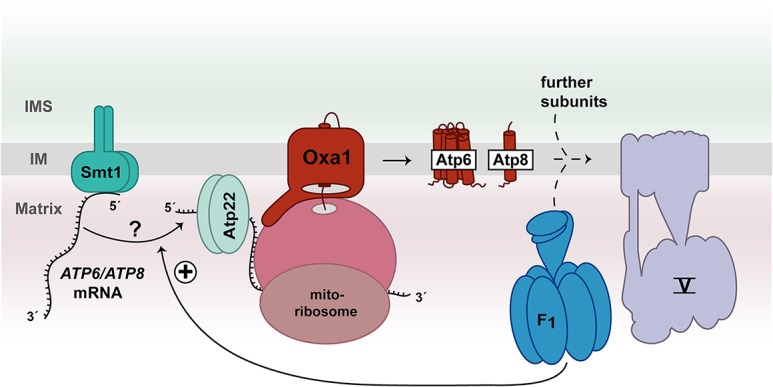
Model of feedback loop regulation of Atp6/Atp8 synthesis. Yeast Smt1 represses the translation of the bicistronic ATP6/ATP8 mRNA. The unassembled F1 part of complex V may stimulate the release of Smt1 from ATP6/ATP8 mRNA by an unknown mechanism. Subsequently, Atp22 can bind to the mRNA to promote the translation of Atp6/Atp8 subunits.
Regulation of protein production in human mitochondria
Mitochondrial translational plasticity
The control of mitochondrial protein synthesis in human cells is poorly understood. The transcripts in human mitochondria either lack or contain only a short 5′ UTR (Hällberg and Larsson 2014). The only known mitochondrial translational activator, TACO1, stimulates the synthesis of COX1 by an unknown mechanism. In contrast to yeast Mss51, TACO1 does not interact with COX1, which is present in assembly intermediates (Weraarpachai et al. 2009). Consequently, different modes exist to adjust protein production to the assembly of complex IV in human mitochondria. Recently, a novel mode of translational regulation of human COX1 mRNA was reported (Richter-Dennerlein et al. 2016). The assembly factor C12ORF62 (human COX14) binds to COX1, while it is produced on mitoribosomes. The binding of COX14 stalls the translation of COX1 mRNA (Richter-Dennerlein et al. 2016). Subsequently, mitochondrial translation regulation assembly intermediate of the cytochrome c oxidase 12 (MITRAC12; human COA3) associates with COX14 to stabilize the nascent chain of COX1 within the MITRAC complex (Fig. 5; Richter-Dennerlein et al. 2016; Bourens and Barrientos 2017). The translation of COX1 mRNA is paused within this complex until the first nuclear-encoded subunit, COX4-1 (homolog of yeast Cox6), is added to the assembly line (Richter-Dennerlein et al. 2016). The ability of mitoribosomes to adjust protein production to the assembly process of complex IV is termed translational plasticity (Richter-Dennerlein et al. 2016).
Figure 5.
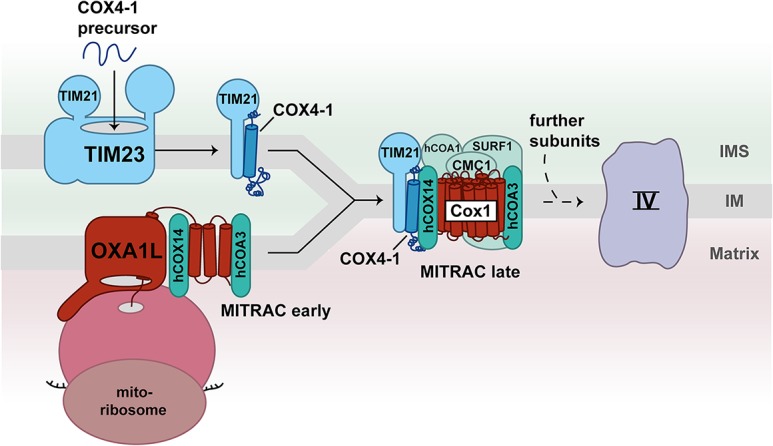
Coupling of protein import to the assembly of human complex IV. C12ORF62 (human COX14) and MITRAC12 (human COA3) bind to the nascent chain of COX1, forming the early MITRAC assembly intermediate in human mitochondria. The translation of COX1 mRNA on mitoribosomes is paused within this complex. The assembly of the first nuclear-encoded subunit, COX4-1, leads to completion of COX1 translation. The presequence translocase subunit TIM21 delivers the COX4-1 precursor from the TIM23 complex to the MITRAC assembly intermediate. Additional assembly factors such as MITRAC15 (human COA1), CMC1, and SURF1 (homolog to yeast Shy1) are added to form the late MITRAC assembly intermediate.
MicroRNA molecules coordinate mitochondrial translation
MicroRNAs such as miR-1 can regulate protein synthesis in human mitochondria (Zhang et al. 2014). In the cytoplasm, the microRNA miR-1 and the catalytic Argonaute protein AGO2 are part of the RNA-induced silencing complex (RISC) that binds target mRNAs to regulate their stability and translation. The partner protein GW182 recruits factors to the RISC that modify the bound mRNA to facilitate its degradation (Carthew and Sontheimer 2009; Czech and Hannon 2011). AGO2 and miR-1, but not GW182, have also been found in mitochondria. Here, the AGO2/miR-1 complex performs a different function. It binds to mitochondrial transcripts of several respiratory chain subunits to promote their translation. This microRNA-stimulated protein synthesis plays an important role in adjusting respiratory activity to the increased energy demand during muscle differentiation (Zhang et al. 2014). Interestingly, several additional microRNAs have been detected in mitochondria (Das et al. 2012; Sripada et al. 2012). Further studies are needed to investigate whether microRNAs play a more common role in regulating mitochondrial gene expression.
Import of nuclear-encoded proteins into mitochondria
Nuclear-encoded proteins destined for import into mitochondria are produced as precursors on cytosolic ribosomes. Molecular chaperones keep these preproteins in an unfolded import-competent state and guide them to receptors of the translocase of the outer mitochondrial membrane (TOM complex) (Young et al. 2003; Hoseini et al. 2016; Jores et al. 2018). The protein-conducting channel of the TOM complex mediates transport of the majority of precursor proteins across the outer membrane. Upon passage of the TOM channel, specific protein translocases sort the precursor proteins into the different mitochondrial subcompartments: the outer and inner membrane, the intermembrane space, and the matrix (Endo et al. 2011; Hewitt et al. 2011; Schleiff and Becker 2011; Neupert 2015; Wiedemann and Pfanner 2017). One exception is the integration of proteins with α-helical membrane anchors in the outer membrane that appear to occur independently of the TOM channel (Dukonovic and Rapaport 2011; Ellenrieder et al. 2015). In yeast, ∼70% of mitochondrial proteins, including many respiratory chain subunits, are produced with a cleavable presequence and imported via the presequence translocase (TIM23 complex) into the inner membrane or matrix (Vögtle et al. 2009). The membrane potential across the inner mitochondrial membrane drives protein transport via the presequence pathway. Transport of preproteins into the mitochondrial matrix additionally requires ATP-dependent action of the presequence translocase-associated motor (PAM). The core subunit of the PAM machinery is the mitochondrial Hsp70 (mtHsp70) that powers protein transport into the matrix by ATP hydrolysis (Endo et al. 2011; Hewitt et al. 2011; Schleiff and Becker 2011; Neupert 2015; Wiedemann and Pfanner 2017).
Coupling of protein import to respiratory chain biogenesis
The import of nuclear-encoded proteins is closely linked to the activity and assembly of respiratory chain complexes. In yeast, the supercomplex of the cytochrome bc1 complex and cytochrome c oxidase interacts with the TIM23 complex and the Pam16/Pam18 module (van der Laan et al. 2006; Wiedemann et al. 2007). According to the current model, the presequence translocase-associated respiratory chain complexes locally establish a high membrane potential to promote import of precursor proteins via the TIM23 complex (van der Laan et al. 2006). Interestingly, the import of some precursor proteins such as Atp2 of the ATP synthase appears to be hypersensitive toward perturbations of the membrane potential (Schendzielorz et al. 2017). Whether the coupling between the TIM23 complex and the respiratory chain in particular stimulates the import of these substrates remains to be shown.
Components of the PAM machinery promote assembly of respiratory chain subunits in yeast mitochondria as well. The mitochondrial chaperone mtHsp70 is involved in the formation of respiratory chain complexes via two independent mechanisms. First, mtHsp70 robustly interacts with Mss51 and thereby contributes to the regulation of COX1 mRNA translation (Fig. 2; Fontanesi et al. 2010). Second, the complex IV subunit Cox4 was identified as a major interaction partner of mtHsp70 in affinity purifications (Böttinger et al. 2013). The addition of Cox4 to assembly intermediates is a critical step in the formation of the mature cytochrome c oxidase (Frazier et al. 2006; Böttinger et al. 2013). Interestingly, Cox4 accumulates at mtHsp70 when the formation of the cytochrome c oxidase is blocked. Thus, mtHsp70-bound Cox4 represents a reservoir of free Cox4 that can be delivered into the assembly line when needed (Böttinger et al. 2013). Similarly, assembly intermediates of Cox1 and cytochrome b can be detected under physiological conditions (Mick et al. 2010; Hildenbeutel et al. 2014). The association of different assembly factors and chaperones keeps the core subunits in an assembly-competent state. Therefore, the presence of such key intermediates could ensure rapid formation of mitochondrial respiratory chain complexes. Such a mechanism could enable yeast cells to rapidly adapt to respiratory growth conditions.
In human mitochondria, TIM21 plays a direct role in the formation of respiratory chain complexes via its interaction with two protein machineries. On one hand, TIM21 is a subunit of the TIM23 translocase; on the other hand, it associates with MITRAC that forms a platform for the assembly of the cytochrome c oxidase (Mick et al. 2012). TIM21 shuttles nuclear-encoded subunits from the TIM23 complex to MITRAC to promote formation of complex IV (Fig. 5). Supporting this model, the assembly of COX4-1 was strongly affected in mitochondria from TIM21 knockdown cells (Mick et al. 2012). The addition of COX4-1 to the MITRAC intermediate is critical to complete the translation of COX1 mRNA as described above (Richter-Dennerlein et al. 2016). Therefore, the TIM21-mediated delivery of COX4-1 represents a crucial step in the formation of cytochrome c oxidase and establishes an intriguing connection between the protein import machinery and mitochondrial protein biosynthesis. Altogether, in both human and yeast mitochondria, components of the TIM23 complex are closely linked to the assembly pathway of the respiratory chain.
Regulation of mitochondrial protein import
Many signaling pathways mediate the communication between mitochondria and the nucleus under different physiological and stress conditions (Ryan and Hoogenraad 2007; Quirós et al. 2016). Protein translocases represent a major target for the control of mitochondrial function. More than 30 phosphorylation sites have been identified in the TOM complex of yeast mitochondria (Schmidt et al. 2011). Phosphorylation by proteinase A (PKA) modulates protein import in response to metabolic changes. Shifting yeast cells from respiratory to fermentable growth leads to activation of PKA. It has been reported that PKA phosphorylates precursors of TOM components and thereby impairs the formation of the TOM complex (Rao et al. 2012; Gerbeth et al. 2013). Furthermore, PKA phosphorylates the import receptor Tom70, which affects import of hydrophobic precursor proteins such as metabolite carriers (Schmidt et al. 2011; Gerbeth et al. 2013). Cyclin-dependent kinase Cdc28 phosphorylates the small TOM subunit Tom6 during the G2–M transition in the cell cycle, which in turn modulates the assembly of the TOM complex (Harbauer et al. 2014). Altogether, phosphorylation of TOM proteins regulates function and assembly of the TOM complex to adapt protein import to specific requirements during metabolic changes or the cell cycle (Schmidt et al. 2011; Gerbeth et al. 2013; Harbauer et al. 2014). Moreover, the efficiency of TIM23-mediated protein import can be modulated upon cellular stress signaling. In human cells, arsenite-induced stress causes a reduction of TIM17A levels. Since TIM17A is an essential component of the TIM23 complex, decreased content of this subunit impairs protein transport into mitochondria (Rainbolt et al. 2013). These observations established protein translocases as central targets for regulation of protein import into mitochondria in response to cellular signaling.
Role of protein import in mitochondrial stress responses
Different pathways have been described to address how impaired protein import modulates cytosolic protein homeostasis by retrograde signaling. Yeast mutants that are defective in protein import display an increased proteasomal activity to degrade nonimported precursor proteins, which is termed unfolded protein response (UPR) activated by mistargeting of proteins (UPRam) (Wrobel et al. 2015). In addition, cytosolic protein biosynthesis is modulated upon mitochondrial precursor overaccumulation stress (mPOS). The CAP-dependent translation on cytosolic ribosomes is decreased (Wang and Chen 2015; Wrobel et al. 2015), while the Gis2-promoted CAP-independent translation of specific mRNAs is stimulated to maintain cell survival (Wang and Chen 2015). Reactive oxygen could play an important role in this response pathway. Defects in protein import lead to increased levels of reactive oxygen species, which in turn affect protein synthesis by modification of cytosolic ribosomes (Topf et al. 2018). Finally, a control mechanism at the mitochondrial surface has been reported. Overexpression of mitochondrial precursor proteins leads to a partial block of protein import and activation of the mitochondrial compromised protein import response (mitoCPR). In the mitoCPR pathway, the transcription factor Pdr3 induces expression of CIS1 (Weidberg and Amon 2018). The cytosolic protein Cis1 binds to the Tom70 receptor and Msp1. The AAA ATPase Msp1 removes accumulated precursor proteins from the mitochondrial outer membrane for proteasomal degradation (Weidberg and Amon 2018). Msp1 also extracts mistargeted ER and peroxisomal tail-anchored proteins from the mitochondrial outer membrane (Chen et al. 2014; Okreglak and Walter 2014; Weir et al. 2017). Thus, Msp1 plays a central role to prevent an overload of mitochondria with mislocalized proteins. Altogether, distinct protective mechanisms take place in the cytosol and on the mitochondrial surface to alleviate damage due to mitochondrial protein import. The challenge of future research will be the identification of molecular players and mechanisms of the different protective stress response pathways.
In the nematode Caenorhabditis elegans, an elegant mechanism has been identified demonstrating how mitochondrial damage induces the mitochondrial UPR (UPRmt) (Nargund et al. 2012; Higuchi-Sanabria et al. 2018; Samluk et al. 2018; Shpilka and Haynes 2018). The cellular localization of the transcription factor ATFS-1 is pivotal for the induction of the UPRmt. ATFS-1 contains a mitochondrial and a nuclear targeting signal. In intact mitochondria, the protein is imported into the matrix and degraded by the protease LON. Upon mitochondrial damage, the import of ATFS-1 is impaired, and the transcription factor relocalizes to the nucleus (Nargund et al. 2012). In the nucleus, ATFS-1 and the transcriptional regulators DVE-1 and UBL-5 induce the expression of genes encoding mitochondrial chaperones and proteases to protect mitochondria against oxidative damage (Fig. 6; Benedetti et al. 2006; Haynes et al. 2007; Tian et al. 2016). In addition, ATFS-1 down-regulates the expression of oxidative phosphorylation (OXPHOS) genes in the nucleus and in mitochondria to facilitate recovery of the cell organelle (Fig. 6; Nargund et al. 2015). In mammals, the three transcription factors ATFS-4, ATFS-5, and CHOP are involved in the induction of the UPRmt by an unknown mechanism (Higuchi-Sanabria et al. 2018; Samluk et al. 2018; Shpilka and Haynes 2018). Expression of human ATFS-5 can compensate for the loss of ATFS-1 in worms, indicating that basic mechanisms to induce the UPRmt are conserved between worms and humans (Fiorese et al. 2016). All of these different examples illustrate that protein import exerts a dual role as a target of regulation and as a sensor for mitochondrial damage to induce cellular stress responses by retrograde signaling.
Figure 6.
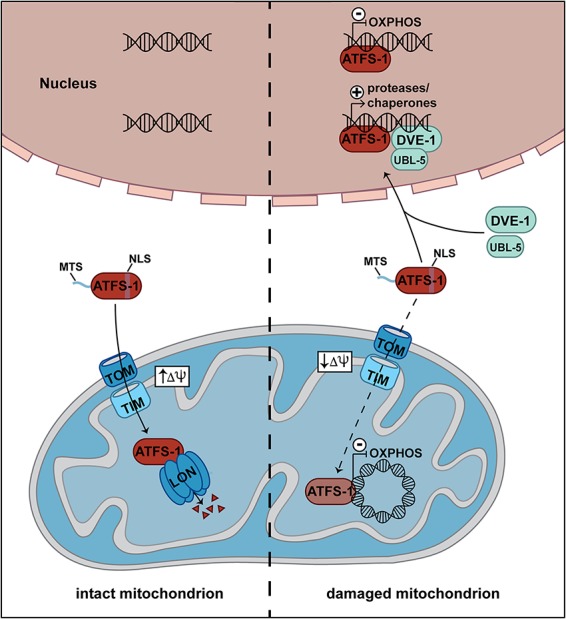
Defects in mitochondrial protein import induce the UPRmt. In C. elegans, the transcription factor ATFS-1 contains a mitochondrial (MTS) and a nuclear (NLS) targeting signal. The TOM and TIM23 complexes transport ATFS-1 into the matrix of intact mitochondria, where it is degraded by the LON protease. In damaged mitochondria, the membrane potential (Δψ) is diminished, and the import of ATFS-1 into mitochondria is impaired. Instead, a fraction of ATFS-1 relocalizes to the nucleus. Together with the transcription factors DVE-1 and UBL-5, it induces the transcription of genes involved in the UPRmt. In addition, uncharacterized fractions of ATFS-1 silence the expression of nuclear and mitochondrial OXPHOS genes.
Conclusion
Balancing mitochondrial and cytoplasmic protein production is crucial for building up respiratory chain complexes and is therefore of central importance for cellular metabolism. Mitochondrial protein biogenesis and import of nuclear-encoded proteins are central targets of regulation. A few mechanisms such as translational plasticity and regulatory feedback loops via translational activators have been discovered that fine-tune mitochondrial protein biogenesis to the assembly process of respiratory chain complexes. The presence of additional translation factors in yeast mitochondria leads to the question of whether similar negative feedback loops such as those for Cox1 and cytochrome b exist for all mitochondrially encoded proteins. Regulation is likely even more complex, since cross-talk between the different assembly lines appears to occur (Mick et al. 2012; Mayorga et al. 2016). This observation indicates that a dynamic network of biogenesis factors may balance the formation of different respiratory chain complexes. In addition, import of nuclear-encoded proteins is closely linked to the assembly lines of respiratory chain complexes. Supporting this view, the initial assembly steps of complex III and complex IV occur primarily in the inner boundary membrane, where TIM23 complexes are also enriched (Vogel et al. 2006; Stoldt et al. 2018). We propose that coupling of protein import and mitochondrial protein biogenesis is a key mechanism to coordinate the assembly of respiratory chain complexes. We further suggest that import of nuclear-encoded proteins plays a dual role in controlling mitochondrial protein homeostasis. On one hand, protein translocases are a central target for regulation via phosphorylation; on the other hand, import pathways are critical to induce stress response pathways upon mitochondrial damage. In conclusion, recent studies indicate that a number of different pathways coordinate the protein biosynthesis in mitochondria and the cytosol.
Acknowledgments
This work was funded by grants of the Deutsche Forschungsgemeinschaft (BE4679/2-1), Sonderforschungsbereich 746, Research Training Group 2202, and the Excellence Initiative of the German Federal and State Governments (EXC 294 BIOSS).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.316547.118.
References
- Amunts A, Brown A, Toots J, Scheres SHW, Ramakrishnan V. 2015. The structure of the human mitochondrial ribosome. Science 348: 95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald JM. 2015. Endosymbiosis and eukaryotic cell evolution. Curr Biol 25: 911–921. [DOI] [PubMed] [Google Scholar]
- Barrientos A, Zambrano A, Tzagoloff A. 2004. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J 23: 3472–3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battey J, Clayton DA. 1978. The transcription map of mouse mitochondrial DNA. Cell 14: 143–156. [DOI] [PubMed] [Google Scholar]
- Bauerschmitt H, Mick DU, Deckers M, Vollmer C, Funes S, Kehrein K, Ott M, Rehling P, Herrmann JM. 2010. Ribosome-binding proteins Mdm38 and Mba1 display overlapping functions for regulation of mitochondrial translation. Mol Biol Cell 21: 1937–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti C, Haynes CM, Yang Y, Harding HP, Ron D. 2006. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics 174: 229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttinger L, Horvath SE, Kleinschroth T, Hunte C, Daum G, Pfanner N, Becker T. 2012. Phosphatidylethanolamine and cardiolipin differently affect the stability of mitochondrial respiratory chain supercomplexes. J Mol Biol 423: 677–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttinger L, Guiard B, Oeljeklaus S, Kulawiak B, Zufall N, Wiedemann N, Warscheid B, van der Laan M, Becker T. 2013. A complex of Cox4 and mitochondrial Hsp70 plays an important role in the assembly of the cytochrome c oxidase. Mol Biol Cell 24: 2609–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourens M, Barrientos A. 2017. A CMC1-knockout reveals translation-independent control of human mitochondrial complex IV biogenesis. EMBO Rep 18: 477–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Seyfferth J, Lucci J, Gilsbach R, Preissl S, Böttinger L, Mårtensson CU, Panhale A, Stehle T, Kretz O, et al. 2016. MOF acetyl transferase regulates transcription and respiration in mitochondria. Cell 167: 722–738. [DOI] [PubMed] [Google Scholar]
- Chen YC, Umanah GK, Dephoure N, Andrabi SA, Gygi SP, Dawson TM, Rutter J. 2014. Msp1/ATAD1 maintains mitochondrial functions by facilitating the degradation of mislocalized tail-anchored proteins. EMBO J 33: 1548–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo T, Ohira T, Sakaguchi Y, Goshima N, Nomura N, Nagao A, Suzuki T. 2012. LRPPRC/SLIRP suppresses PNPase-mediated mRNA decay and promotes polyadenylation in human mitochondria. Nucleic Acids Res 40: 8033–8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvillion MT, Soto IC, Shipkovenska G, Churchman LS. 2016. Synchronized mitochondrial and cytosolic translation programs. Nature 533: 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciat CM, Brunner S, Baumann F, Neupert W, Stuart RA. 2000. The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J Biol Chem 275: 18093–18098. [DOI] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. 2011. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet 12: 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Ferlito M, Kent OA, Fox-Talbot K, Wang R, Liu D, Raghavachari N, Yang Y, Wheelan SJ, Murphy E, et al. 2012. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ Res 110: 1596–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decoster E, Simon M, Hatat D, Faye G. 1990. The MSS51 gene product is required for the translation of the COX1 mRNA in yeast mitochondria. Mol Gen Genet 224: 111–118. [DOI] [PubMed] [Google Scholar]
- Dennerlein S, Wang C, Rehling P. 2017. Plasticity of mitochondrial translation. Trends Cell Biol 27: 712–721. [DOI] [PubMed] [Google Scholar]
- Desai N, Brown A, Amunts A, Ramakrishnan V. 2017. The structure of the yeast mitochondrial ribosome. Science 355: 528–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukonovic J, Rapaport D. 2011. Multiple pathways in the integration of proteins into the mitochondrial outer membrane. Biochim Biophys Acta 1808: 971–980. [DOI] [PubMed] [Google Scholar]
- Ellenrieder L, Mårtensson CU, Becker T. 2015. Biogenesis of mitochondrial outer membrane proteins, problems and diseases. Biol Chem 396: 1199–1213. [DOI] [PubMed] [Google Scholar]
- Endo T, Yamano K, Kawano S. 2011. Structural insight into the mitochondrial protein import system. Biochim Biophys Acta 1808: 955–970. [DOI] [PubMed] [Google Scholar]
- Fiorese CJ, Schulz AM, Lin YF, Rosin N, Pllegrina MW, Haynes CM. 2016. The transcription factor ATF5 mediates a mammalian mitochondrial UPR. Curr Biol 26: 2037–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanesi F, Soto IC, Horn D, Barrientos A. 2010. Mss51 and Ssc1 facilitate translational regulation of cytochrome c oxidase biogenesis. Mol Cell Biol 30: 245–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanesi F, Clemente P, Barrientos A. 2011. Cox25 teams up with Mss51, Ssc1, and Cox14 to regulate mitochondrial cytochrome c oxidase subunit 1 expression and assembly in Saccharomyces cerevisiae. J Biol Chem 286: 555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox TD. 2012. Mitochondrial protein synthesis, import and assembly. Genetics 192: 1203–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier AE, Taylor RD, Mick DU, Warscheid B, Stoepel N, Meyer HE, Ryan MT, Guiard B, Rehling P. 2006. Mdm38 interacts with ribosomes and is a component of the mitochondrial protein export machinery. J Cell Biol 172: 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Guerrero AE, Camacho-Villasana Y, Zamudio-Ochoa A, Pérez-Martínez X. 2018. Cbp3 and Cbp6 are dispensable for synthesis regulation of cytochrome b in yeast mitochondria. J Biol Chem 293: 5585–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbeth C, Schmidt O, Rao S, Harbauer AB, Mikropoulou D, Opalińska M, Guiard B, Pfanner N, Meisinger C. 2013. Glucose-induced regulation of protein import receptor Tom22 by cytosolic and mitochondria-bound kinases. Cell Metab 18: 578–587. [DOI] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Leitner A, Bieri P, Voigs-Hoffmann F, Erzberger JP, Leibundgut M, Aebersold R, Ban N. 2014. Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature 505: 515–519. [DOI] [PubMed] [Google Scholar]
- Greber BJ, Bieri P, Leibundgut M, Leitner A, Aebersold R, Boehringer D, Ban N. 2015. The complete structure of the 55S mammalian mitochondrial ribosome. Science 348: 303–308. [DOI] [PubMed] [Google Scholar]
- Gruschke S, Kehrein K, Römpler K, Gröne K, Israel L, Imhof A, Herrmann JM, Ott M. 2011. Cbp3–Cbp6 interacts with the yeast mitochondrial ribosomal tunnel exit and promotes cytochrome b synthesis and assembly. J Cell Biol 193: 1101–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruschke S, Römpler K, Hildenbeutel M, Kehrein K, Kühl I, Bonnefoy N, Ott M. 2012. The Cbp3–Cbp6 complex coordinates cytochrome b synthesis with bc1 complex assembly in yeast mitochondria. J Cell Biol 199: 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Wu M, Guo R, Yan K, Lei J, Gao N, Yang M. 2016. The architecture of the mammalian respirasome. Nature 537: 639–643. [DOI] [PubMed] [Google Scholar]
- Guo H, Bueler SA, Rubinstein JL. 2017. Atomic model for the dimeric Fo region of mitochondrial ATP synthase. Science 358: 936–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn A, Parey K, Bublitz M, Mills DJ, Zickermann V, Vonck J, Kühlbrandt W, Meier T. 2016. Structure of a complete ATP synthase dimer reveals the molecular basis of inner mitochondrial membrane morphology. Mol Cell 63: 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hällberg BM, Larsson NG. 2014. Making proteins in the powerhouse. Cell Metab 20: 226–240. [DOI] [PubMed] [Google Scholar]
- Harbauer AB, Opalińska M, Gerbeth C, Herman JS, Rao S, Schönfisch B, Guiard B, Schmidt O, Pfanner N, Meisinger C. 2014. Cell cycle-dependent regulation of mitochondrial preprotein translocase. Science 346: 1109–1113. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D. 2007. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev Cell 13: 467–480. [DOI] [PubMed] [Google Scholar]
- He J, Ford HC, Carroll J, Douglas C, Gonzales E, Ding S, Fearnley IM, Walker JE. 2018. Assembly of the membrane domain of ATP synthase in human mitochondria. Proc Natl Acad Sci 115: 2988–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfenbein KG, Ellis TP, Dieckmann CL, Tzagoloff A. 2003. ATP22, a nuclear gene required for expression of the Fo sector of mitochondrial ATPase in Saccharomyces cerevisiae. J Biol Chem 278: 19751–19756. [DOI] [PubMed] [Google Scholar]
- Hell K, Neupert W, Stuart RA. 2001. Oxa1p acts as general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J 20: 1281–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann JM, Woellhaf MW, Bonnefoy N. 2013. Control of protein synthesis in yeast mitochondria: the concept of translational activators. Biochim Biophys Acta 1833: 286–294. [DOI] [PubMed] [Google Scholar]
- Hewitt V, Alcock F, Lithgow T. 2011. Minor modifications and major adaptations: the evolution of molecular machines driving mitochondrial protein import. Biochim Biophys Acta 1808: 947–954. [DOI] [PubMed] [Google Scholar]
- Higuchi-Sanabria R, Frankino PA, Paul JW III, Tronnes SU, Dillin A. 2018. A futile battle? Protein quality control and the stress of aging. Dev Cell 44: 139–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildenbeutel M, Hegg EL, Stephan K, Gruschke S, Meunier B, Ott M. 2014. Assembly factors monitor sequential hemylation of cytochrome b to regulate mitochondrial translation. J Cell Biol 205: 511–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoseini H, Pandey S, Jores T, Schmitt A, Franz-Wachtel M, Macek B, Buchner J, Dimmer KS, Rapaport D. 2016. The cytosolic cochaperone Sti1 is relevant for mitochondrial biogenesis and morphology. FEBS J 283: 3338–3352. [DOI] [PubMed] [Google Scholar]
- Iwata S, Lee SW, Okada K, Lee JK, Iwata M, Rasmussen B, Link TA, Ramaswamy S, Jap BK. 1998. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science 281: 64–71. [DOI] [PubMed] [Google Scholar]
- Jia L, Dienhart M, Schramp M, McCauley M, Hell K, Stuart RA. 2003. Yeast Oxa1 interacts with mitochondrial ribosomes: the importance of the C-terminal region of Oxa1. EMBO J 22: 6438–6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jores T, Lawatscheck J, Beke V, Franz-Wachtel M, Yunoki K, Fitzgerald JC, Macek B, Endo T, Buchner J, Rapaport D. 2018. Cytosolic Hsp70 and Hsp40 chaperones enable the biogenesis of mitochondrial β-barrel proteins. J Cell Biol 217: 3091–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrein K, Schilling R, Möller-Hergt BV, Wurm CA, Jakobs S, Lamkemeyer T, Langer T, Ott M. 2015. Organization of mitochondrial gene expression in two distinct ribosome-containing assemblies. Cell Rep 10: 843–853. [DOI] [PubMed] [Google Scholar]
- Khalimonchuk O, Bird A, Winge DR. 2007. Evidence for a pro-oxidant intermediate in the assembly of the cytochrome oxidase. J Biol Chem 282: 17442–17449. [DOI] [PubMed] [Google Scholar]
- Khalimonchuk O, Bestwick M, Meunier B, Watts TC, Winge DR. 2010. Formation of the redox cofactor centers during Cox1 maturation in yeast cytochrome oxidase. Mol Cell Biol 30: 1004–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C, Hunte C. 2002. Crystal structure of the yeast cytochrome bc1 complex with its bound substrate cytochrome c. Proc Natl Acad Sci 99: 2800–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau WC, Baker LA, Rubinstein JL. 2008. Cryo-EM structure of the yeast ATP synthase. J Mol Biol 382: 1256–1264. [DOI] [PubMed] [Google Scholar]
- Letts JA, Fiedorczuk K, Sazanov LA. 2016. The architecture of respiratory supercomplexes. Nature 537: 644–648. [DOI] [PubMed] [Google Scholar]
- Luttik MA, Overkamp KM, Kötter P, de Vries S, van Dijken JP, Pronk JT. 1998. The Saccharomyces cerevisiae NDE1 and NDE2 genes encode separate mitochondrial NADH dehydrogenases catalyzeing the oxidation of cytosolic NADH. J Biol Chem 273: 24529–24534. [DOI] [PubMed] [Google Scholar]
- Marres CA, de Vries S, Grivell LA. 1991. Isolation and inactivation of the nuclear gene encoding the rotenone-insensitive internal NADH: ubiquinone oxidoreductase of mitochondria from Saccharomyces cerevisiae. Eur J Biochem 195: 857–862. [DOI] [PubMed] [Google Scholar]
- Mayorga JP, Camacho-Villasana Y, Shingú-Vázquez M, Garcia-Villegas R, Ramudio-Ochoa A, García-Guerrero AE, Hernández G, Pérez-Martínez X. 2016. A novel function of Pet54 in regulation of Cox1 synthesis in Saccharomyces cerevisiae. J Biol Chem 291: 9343–9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick DU, Vukotic M, Piechura H, Meyer HE, Warscheid B, Deckers M, Rehling P. 2010. Coa3 and Cox14 are essential for negative feedback regulation of COX1 translation in mitochondria. J Cell Biol 191: 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick DU, Dennerlein S, Wiese H, Reinhold R, Pacheu-Grau D, Lorenzi I, Sasaran F, Weraarpachai W, Shoubridge EA, Warscheid B, et al. 2012. MITRAC links mitochondrial protein translocation to respiratory-chain assembly and translational regulation. Cell 151: 1528–1541. [DOI] [PubMed] [Google Scholar]
- Morgenstern M, Stiller SB, Lübbert P, Peikert CD, Dannenmaier S, Drepper F, Weil U, Höß P, Feuerstein R, Gebert M, et al. 2017. Definition of a high-confidence mitochondrial proteome at quantitative scale. Cell Rep 19: 2836–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. 2012. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 337: 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Fiorese CJ, Pellegrino MW, Deng P, Haynes CM. 2015. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during UPRmt. Mol Cell 58: 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumenko N, Morgenstern M, Rucktäschel R, Warscheid B, Rehling P. 2017. INA complex liaises the F1Fo-ATP synthase membrane motor modules. Nat Commun 8: 1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W. 2015. A perspective on transport of proteins into mitochondria: a myriad of open questions. J Mol Biol 427: 1135–1158. [DOI] [PubMed] [Google Scholar]
- Okreglak V, Walter P. 2014. The conserved AAA–ATPase Msp1 confers organelle specificity to tail-anchored proteins. Proc Natl Acad Sci 111: 8019–8024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M, Prestele M, Bauerschmitt H, Funes S, Bonnefoy N, Herrmann JM. 2006. Mba1, a membrane-associated ribosome receptor in mitochondria. EMBO J 25: 1603–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M, Amunts A, Brown A. 2016. Organization and regulation of mitochondrial protein synthesis. Annu Rev Biochem 85: 77–101. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. 2008. A mitochondrial protein compendium elucidates complex I disease biology. Cell 134: 112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce SF, Rebelo-Guiomar P, D'Souza AR, Powell CA, van Haute L, Minczuk M. 2017. Regulation of mammalian mitochondrial gene expression: recent advances. Trends Biochem Sci 42: 625–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Martínez X, Broadley SA, Fox TD. 2003. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J 22: 5951–5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Martínez X, Butler CA, Shingu-Vazquez M, Fox TD. 2009. Dual functions of Mss51 couple synthesis of Cox1 to assembly of cytochrome c oxidase in Saccharomyces cerevisiae mitochondria. Mol Biol Cell 20: 4371–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Woellhaf MW, Herrmann JM, Förster F. 2015. Organization of the mitochondrial translation machinery studied in situ by cryoelectron tomography. Nat Commun 6: 6019. [DOI] [PubMed] [Google Scholar]
- Quirós PM, Mottis A, Auwerx J. 2016. Mitonuclear communication in homeostasis and stress. Nat Rev Mol Cell Biol 17: 213–226. [DOI] [PubMed] [Google Scholar]
- Rainbolt TK, Atanassova N, Genereux JC, Wiseman RL. 2013. Stress-regulated translational attenuation adapts mitochondrial protein import through TIM17A. Cell Metab 18: 908–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja SJ, Charapitsa I, Conrad T, Vaquerizas JM, Gebhardt P, Holz H, Kadlec J, Fraterman S, Luscombe NM, Akhtar A. 2010. The nonspecific lethal complex is a transcriptional regulator in Drosophila. Mol Cell 38: 827–841. [DOI] [PubMed] [Google Scholar]
- Rak M, Tzagoloff A. 2009. F1-dependent translation of mitochondrially encoded Atp6p and Atp8p subunits of yeast ATP synthase. Proc Natl Acad Sci 106: 18509–18514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak M, Zeng X, Brière JJ, Tzagoloff A. 2009. Assembly of Fo in Saccharomyces cerevisiae. Biochim Biophys Acta 1793: 108–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak M, Gokova S, Tzagoloff A. 2011. Modular assembly of yeast mitochondrial ATP synthase. EMBO J 30: 920–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak M, Su CH, Xu JT, Azpiroz R, Singh AM, Tzagoloff A. 2016. Regulation of mitochondrial translation of the ATP8/ATP6 mRNA by Smt1p. Mol Biol Cell 27: 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Schmidt O, Harbauer AB, Schönfisch B, Guiard B, Pfanner N, Meisinger C. 2012. Biogenesis of the preprotein translocase of the outer mitochondrial membrane: protein kinase A phosphorylates the precursor of Tom40 and impairs its import. Mol Biol Cell 23: 1618–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter-Dennerlein R, Oeljeklaus S, Lorenzi I, Ronsör C, Bareth B, Schendzielorz AB, Wang C, Warscheid B, Rehling P, Dennerlein S. 2016. Mitochondrial protein synthesis adapts to influx of nuclear-encoded protein. Cell 167: 471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein JL, Walker JE, Henderson R. 2003. Structure of the mitochondrial ATP synthase by electron cryomicroscopy. EMBO J 22: 6182–6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rühle T, Leister D. 2015. Assembly of F1Fo-ATP synthases. Biochim Biophys Acta 1847: 849–860. [DOI] [PubMed] [Google Scholar]
- Ryan MT, Hoogenraad NJ. 2007. Mitochondrial-nuclear communications. Annu Rev Biochem 76: 4.1–4.22. [DOI] [PubMed] [Google Scholar]
- Samluk L, Chroscicki P, Chacinska A. 2018. Mitochondrial protein import stress and signaling. Curr Opin Physiol 3: 41–48. [Google Scholar]
- Sasarman F, Bruned-Guitton C, Antonicka H, Wai T, Shoubridge EA, LSFC Consortium. 2010. LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Mol Biol Cell 21: 1315–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, Pfeiffer K. 2000. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J 19: 1777–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schendzielorz AB, Schulz C, Lytovshenko O, Clancy A, Guiard B, Ieva R, van der Laan M, Rehling P. 2017. Two distinct membrane potential – dependent steps drive mitochondrial matrix protein translocation. J Cell Biol 216: 83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff E, Becker T. 2011. Common ground for protein translocation: access control for mitochondria and chloroplasts. Nat Rev Mol Cell Biol 12: 48–59. [DOI] [PubMed] [Google Scholar]
- Schmidt O, Harbauer AB, Rao S, Eyrich B, Zahedi RP, Stojanovski D, Schönfisch B, Guiard B, Sickmann A, Pfanner N, et al. 2011. Regulation of mitochondrial protein import by cytosolic kinases. Cell 144: 227–239. [DOI] [PubMed] [Google Scholar]
- Shoubridge EA. 2001. Nuclear defects in respiratory chain disorders. Semin Neurol 21: 261–267. [DOI] [PubMed] [Google Scholar]
- Shpilka T, Haynes CM. 2018. The mitochondrial UPR: mechanisms, physiological functions and implications in aging. Nat Rev Mol Cell Biol 19: 109–120. [DOI] [PubMed] [Google Scholar]
- Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi RP, Meyer HE, Schönfisch B, Perschill I, Chacinska A, Guiard B, et al. 2003. The proteome of Saccharomyces cerevisae mitochondria. Proc NatI Acad Sci 100: 13207–13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siira SJ, Spähr H, Shearwood AJ, Ruzzenente B, Larsson NG, Rackham O, Filipovska A. 2017. LRPPRC-mediated folding of the mitochondrial transcriptome. Nat Commun 8: 1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small WC, McAlister-Henn L. 1998. Identification of a cytosolically directed NADH dehydrogenase in mitochondria of Saccharomyces cerevisiae. J Bacteriol 180: 4051–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeitink JA, Zeviani M, Turnbull DM, Jacobs HT. 2006. Mitochondrial medicine: a metabolic perspective on the pathology of oxidative phosphorylation. Cell Metab 3: 9–13. [DOI] [PubMed] [Google Scholar]
- Smith PM, Fox JL, Winge DR. 2012. Biogenesis of the cytochrome bc1 complex and role of assembly factors. Biochim Biophys Acta 1817: 276–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Pfanner N, Becker T. 2018. Assembling the mitochondrial ATP synthase. Proc Natl Acad Sci 20: 2850–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto IC, Fontanesi F, Myers RS, Hamel O, Barrientos A. 2012. A heme-sensing mechanism in the translational regulation of mitochondrial cytochrome c oxidase biogenesis. Cell Metab 16: 801–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripada L, Tomar D, Prajapati P, Singh R, Singh AK, Sing R. 2012. Systematic analysis of small RNAs associated with human mitochondria by deep sequencing: detailed analysis of mitochondrial associated miRNA. PLoS One 7: e44873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock D, Leslie AGW, Walker JE. 1999. Molecular architecture of the rotary motor in ATP synthase. Science 286: 1700–1705. [DOI] [PubMed] [Google Scholar]
- Stoldt S, Wenzel D, Kehrein K, Riedel D, Ott M, Jakobs S. 2018. Spatial orchestration of mitochondrial translation and OXPHOS complex assembly. Nat Cell Biol 20: 528–534. [DOI] [PubMed] [Google Scholar]
- Szyrach G, Ott M, Bonnefoy N, Neupert W, Herrmann JM. 2003. Ribosome binding of the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J 22: 6448–6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Garcia G, Bian Q, Steffen KK, Joe L, Wolff S, Meyer BJ, Dillin A. 2016. Mitochondrial stress induces chromatin reorganization to promote longevity and UPRmt. Cell 165: 1197–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timón-Gómez A, Nývltova E, Abriata LA, Hosler J, Barrientos A. 2018. Mitochondrial cytochrome c oxidase biogenesis. Recent developments. Semin Cell Dev Biol 76: 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topf U, Suppanz I, Samluk L, Wrobel L, Böser A, Sakowska P, Knapp B, Pietrzyk MK, Chacinska A, Warscheid B. 2018. Quantitative proteomics identifies redox switches for global translation modulation by mitochondrially produced reactive oxygen species. Nat Commun 9: 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. 1996. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science 272: 1136–1144. [DOI] [PubMed] [Google Scholar]
- van der Laan M, Wiedemann N, Mick DU, Guiard B, Rehling P, Pfanner N. 2006. A role for Tim21 in membrane potential-dependent preprotein sorting in mitochondria. Curr Biol 16: 2271–2276. [DOI] [PubMed] [Google Scholar]
- Vogel F, Bornhövd C, Neupert W, Reichert AS. 2006. Dynamic subcompartimentalization of the mitochondrial inner membrane. J Cell Biol 175: 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vögtle FN, Wortelkamp S, Zahedi RP, Becker D, Leidhold C, Gevaert K, Kellermann J, Voos W, Sickmann A, Pfanner N, et al. 2009. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 139: 428–439. [DOI] [PubMed] [Google Scholar]
- Wang X, Chen XJ. 2015. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature 524: 481–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Amon A. 2018. MitoCPR—a surveillance pathway that protects mitochondria in response to protein import stress. Science 360: eaan4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir NR, Kamber RA, Martenson JS, Denic V. 2017. The AAA protein Msp1 mediates clearance of excess tail-anchored proteins from the peroxisomal membrane. Elife 6: e28507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weraarpachai W, Antonicka H, Sasarman F, Seeger J, Schrank B, Kolesar JE, Lochmüller H, Chevrette M, Kaufman BA, Horvath R, et al. 2009. Mutation in TACO1, encoding a translational activator of COX I, results in cytochrome c oxidase deficiency and late-onset Leigh syndrome. Nat Genet 41: 833–837. [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Pfanner N. 2017. Mitochondrial machineries for protein import and assembly. Annu Rev Biochem 86: 685–714. [DOI] [PubMed] [Google Scholar]
- Wiedemann N, van der Laan M, Hutu DP, Rehling P, Pfanner N. 2007. Sorting switch of mitochondrial presequence translocase involves coupling of motor module to respiratory chain. J Cell Biol 179: 1115–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel L, Topf U, Bragoszewski P, Wiese S, Sztolsztener ME, Oeljeklaus S, Varabyova M, Lirski M, Chroscicki P, Mroszek S, et al. 2015. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature 524: 485–488. [DOI] [PubMed] [Google Scholar]
- Xia D, Yu CA, Kim H, Xia JZ, Kachurin AM, Zhang L, Yu L, Deisenhofer J. 1997. Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science 277: 60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Hoogenraad NJ, Hartl FU. 2003. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112: 41–50. [DOI] [PubMed] [Google Scholar]
- Zara V, Conte L, Trumpower BL. 2007. Identification and characterization of cytochrome bc1 subcomplexes in mitochondria from yeast with single and double deletions of genes encoding cytochrome bc1 subunits. FEBS J 274: 4526–4539. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zua X, Yang B, Li Z, Xue Y, Zhou Y, Huang J, Zhao X, Zhou J, Yan Y, et al. 2014. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell 158: 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimorski V, Ku C, Martin WF, Gould SB. 2014. Endosymbiotic theory for organelle origins. Curr Opin Microbiol 22: 38–48. [DOI] [PubMed] [Google Scholar]